Found:
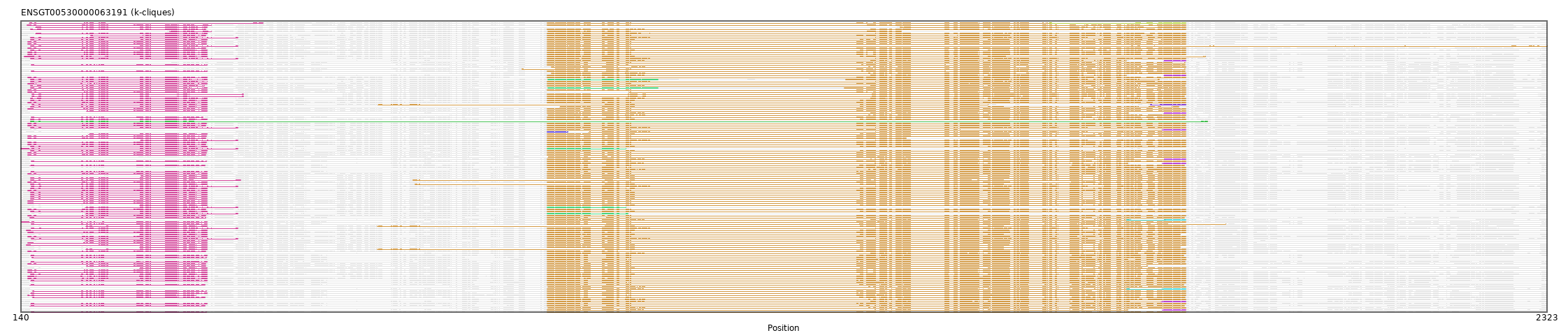
{"ENSRNOP00000058412": [["Domain_0", 156, 405, "PF15371"], ["Linker_0_1", 405, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSRNOP00000062200": [["Domain_1", 893, 1806, "PF14650"]], "ENSCLAP00000025044": [["Domain_0", 156, 458, "PF15371"], ["Linker_0_1", 458, 893], ["Domain_1", 893, 1804, "PF14650"]], "ENSMUSP00000097025": [["Domain_1", 893, 1723, "PF14650"], ["Linker_1_6", 1723, 1773], ["Domain_6", 1773, 1806, "PF14650"]], "MGP_PahariEiJ_P0041279": [["Domain_0", 157, 403, "PF15371"], ["Linker_0_1", 403, 893], ["Domain_1", 893, 1805, "PF14650"]], "MGP_PahariEiJ_P0041278": [["Domain_0", 150, 390, "PF15371"], ["Linker_0_1", 390, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSPEMP00000014222": [["Domain_1", 893, 1777, "PF14650"], ["Domain_5", 1722, 1806, "PF14650"]], "ENSP00000341988": [["Domain_0", 150, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "MGP_PahariEiJ_P0041277": [["Domain_0", 154, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "MGP_SPRETEiJ_P0036407": [["Domain_1", 893, 1723, "PF14650"], ["Linker_1_6", 1723, 1773], ["Domain_6", 1773, 1806, "PF14650"]], "ENSPTRP00000036038": [["Domain_0", 155, 450, "PF15371"], ["Linker_0_1", 450, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSCANP00000025219": [["Domain_0", 154, 450, "PF15371"], ["Linker_0_1", 450, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSCGRP00001009803": [["Domain_0", 155, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1805, "PF14650"]], "ENSCGRP00000017086": [["Domain_1", 893, 1805, "PF14650"]], "ENSCJAP00000006407": [["Domain_0", 150, 406, "PF15371"], ["Domain_1", 1010, 1806, "PF14650"]], "ENSMMUP00000019910": [["Domain_0", 150, 405, "PF15371"], ["Linker_0_1", 405, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSGGOP00000050518": [["Domain_0", 234, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSGGOP00000024081": [["Domain_0", 154, 406, "PF15371"], ["Linker_0_1", 406, 911], ["Domain_1", 911, 1796, "PF14650"]], "ENSCATP00000021179": [["Domain_0", 154, 450, "PF15371"], ["Linker_0_1", 450, 893], ["Domain_1", 893, 2323, "PF14650"]], "ENSCLAP00000001370": [["Domain_0", 156, 458, "PF15371"], ["Linker_0_1", 458, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSPTRP00000073385": [["Domain_0", 148, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSCHIP00000019878": [["Domain_0", 150, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSCCAP00000021405": [["Domain_0", 150, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "MGP_CAROLIEiJ_P0034740": [["Domain_1", 893, 1774, "PF14650"], ["Domain_5", 1722, 1806, "PF14650"]], "MGP_CAROLIEiJ_P0034741": [["Domain_0", 157, 405, "PF15371"], ["Linker_0_1", 405, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSCGRP00000024281": [["Domain_0", 155, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1805, "PF14650"]], "ENSCAFP00000002108": [["Domain_0", 149, 412, "PF15371"], ["Linker_0_1", 412, 893], ["Domain_1", 893, 1806, "PF14650"]], "MGP_PahariEiJ_P0041363": [["Domain_1", 893, 1772, "PF14650"], ["Domain_6", 1771, 1806, "PF14650"]], "ENSNLEP00000025394": [["Domain_0", 154, 454, "PF15371"], ["Linker_0_1", 454, 701], ["Domain_1", 701, 1806, "PF14650"]], "ENSCAFP00000040234": [["Domain_0", 353, 412, "PF15371"], ["Linker_0_1", 412, 894], ["Domain_1", 894, 1399, "PF14650"]], "MGP_SPRETEiJ_P0036408": [["Domain_0", 155, 405, "PF15371"], ["Linker_0_1", 405, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSP00000322640": [["Domain_0", 155, 450, "PF15371"], ["Linker_0_1", 450, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSP00000484807": [["Domain_0", 154, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSRROP00000016915": [["Domain_0", 150, 404, "PF15371"], ["Linker_0_1", 404, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSMLEP00000012431": [["Domain_0", 237, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSP00000488251": [["Domain_0", 150, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSPPAP00000013004": [["Domain_0", 150, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSGGOP00000015790": [["Domain_0", 150, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSMLEP00000016173": [["Domain_0", 150, 405, "PF15371"], ["Linker_0_1", 405, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSGGOP00000033589": [["Domain_0", 150, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSPEMP00000026167": [["Domain_0", 155, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1863, "PF14650"]], "ENSMOCP00000003603": [["Domain_1", 893, 1775, "PF14650"], ["Linker_1_6", 1775, 1776], ["Domain_6", 1776, 1806, "PF14650"]], "ENSMNEP00000001359": [["Domain_0", 140, 450, "PF15371"], ["Linker_0_2", 450, 893], ["Domain_2", 893, 1005, "PF14650"]], "ENSCCAP00000019788": [["Domain_0", 150, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSPPAP00000004293": [["Domain_0", 233, 406, "PF15371"], ["Linker_0_1", 406, 650], ["Domain_1", 650, 1806, "PF14650"]], "ENSPTRP00000062919": [["Domain_0", 232, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSMNEP00000027722": [["Domain_0", 150, 405, "PF15371"], ["Linker_0_1", 405, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSRNOP00000058061": [["Domain_0", 154, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1803, "PF14650"]], "ENSPPAP00000007724": [["Domain_0", 148, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSRROP00000027566": [["Domain_0", 310, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSCGRP00001024169": [["Domain_1", 899, 1806, "PF14650"]], "ENSCHIP00000030624": [["Domain_0", 154, 402, "PF15371"], ["Linker_0_1", 402, 893], ["Domain_1", 893, 1805, "PF14650"]], "ENSCHIP00000001790": [["Domain_0", 154, 406, "PF15371"], ["Linker_0_2", 406, 894], ["Domain_2", 894, 1051, "PF14650"], ["Linker_2_1", 1051, 1320], ["Domain_1", 1320, 1768, "PF14650"]], "ENSMMUP00000048861": [["Domain_0", 154, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSMOCP00000001139": [["Domain_1", 894, 1806, "PF14650"]], "ENSRNOP00000024504": [["Domain_1", 893, 1806, "PF14650"]], "ENSMFAP00000021357": [["Domain_0", 150, 405, "PF15371"], ["Linker_0_1", 405, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSMUSP00000128200": [["Domain_0", 153, 403, "PF15371"], ["Linker_0_1", 403, 893], ["Domain_1", 893, 1804, "PF14650"]], "ENSCCAP00000014615": [["Domain_0", 154, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSNLEP00000026503": [["Domain_0", 154, 406, "PF15371"], ["Linker_0_1", 406, 704], ["Domain_1", 704, 1806, "PF14650"]], "ENSCGRP00001028170": [["Domain_1", 893, 1721, "PF14650"], ["Linker_1_6", 1721, 1775], ["Domain_6", 1775, 1806, "PF14650"]], "ENSCANP00000039217": [["Domain_0", 150, 405, "PF15371"], ["Linker_0_1", 405, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSP00000485628": [["Domain_0", 154, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSSBOP00000038899": [["Domain_0", 154, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSP00000488117": [["Domain_0", 150, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSCATP00000031773": [["Domain_0", 150, 404, "PF15371"], ["Linker_0_1", 404, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSMUSP00000136424": [["Domain_1", 894, 1806, "PF14650"]], "ENSSSCP00000056301": [["Domain_0", 150, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSCCAP00000035621": [["Domain_0", 154, 450, "PF15371"], ["Linker_0_1", 450, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSMNEP00000013565": [["Domain_0", 154, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSPANP00000022503": [["Domain_0", 232, 450, "PF15371"], ["Linker_0_2", 450, 893], ["Domain_2", 893, 1008, "PF14650"]], "ENSMNEP00000034680": [["Domain_0", 150, 404, "PF15371"], ["Linker_0_1", 404, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSPANP00000019326": [["Domain_0", 150, 404, "PF15371"], ["Linker_0_1", 404, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSPPAP00000007248": [["Domain_0", 155, 450, "PF15371"], ["Linker_0_1", 450, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSCGRP00001000422": [["Domain_1", 857, 1805, "PF14650"]], "ENSPANP00000005054": [["Domain_0", 310, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSNLEP00000003918": [["Domain_0", 150, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSPPAP00000020711": [["Domain_0", 310, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1798, "PF14650"]], "ENSNLEP00000026440": [["Domain_0", 150, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSMMUP00000048831": [["Domain_0", 150, 404, "PF15371"], ["Linker_0_1", 404, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSSBOP00000020639": [["Domain_0", 150, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSP00000366875": [["Domain_0", 154, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSCANP00000028918": [["Domain_0", 154, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "MGP_CAROLIEiJ_P0034660": [["Domain_1", 894, 1806, "PF14650"]], "ENSCHIP00000000750": [["Domain_0", 150, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSCAFP00000003154": [["Domain_0", 161, 406, "PF15371"], ["Domain_1", 1398, 1806, "PF14650"]], "ENSPANP00000016170": [["Domain_0", 232, 450, "PF15371"], ["Linker_0_2", 450, 893], ["Domain_2", 893, 1005, "PF14650"]], "ENSCANP00000023985": [["Domain_0", 240, 404, "PF15371"], ["Linker_0_1", 404, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSMFAP00000005735": [["Domain_0", 154, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSMFAP00000025842": [["Domain_1", 893, 1772, "PF14650"], ["Linker_1_6", 1772, 1773], ["Domain_6", 1773, 1806, "PF14650"]], "ENSMLEP00000037542": [["Domain_0", 150, 404, "PF15371"], ["Linker_0_1", 404, 893], ["Domain_1", 893, 1406, "PF14650"]], "ENSMLEP00000011912": [["Domain_3", 893, 922, "PF14650"], ["Linker_3_1", 922, 952], ["Domain_1", 952, 1806, "PF14650"]], "ENSCAFP00000042043": [["Domain_0", 161, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSPTRP00000091227": [["Domain_0", 233, 406, "PF15371"], ["Linker_0_1", 406, 650], ["Domain_1", 650, 1806, "PF14650"]], "ENSMFAP00000014937": [["Domain_1", 154, 1837, "PF15371"], ["Domain_2", 154, 1837, "PF15371"]], "ENSCHIP00000032764": [["Domain_0", 154, 406, "PF15371"], ["Linker_0_2", 406, 894], ["Domain_2", 894, 1051, "PF14650"], ["Linker_2_1", 1051, 1318], ["Domain_1", 1318, 1806, "PF14650"]], "ENSNLEP00000025117": [["Domain_0", 154, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSCCAP00000029575": [["Domain_0", 145, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1834, "PF14650"]], "ENSGGOP00000021229": [["Domain_0", 156, 406, "PF15371"], ["Linker_0_1", 406, 651], ["Domain_1", 651, 1523, "PF14650"], ["Linker_1_5", 1523, 1756], ["Domain_5", 1756, 1806, "PF14650"], ["Domain_6", 1756, 1806, "PF14650"]], "ENSPEMP00000007144": [["Domain_0", 153, 404, "PF15371"], ["Linker_0_1", 404, 893], ["Domain_1", 893, 1804, "PF14650"]], "MGP_CAROLIEiJ_P0034658": [["Domain_1", 893, 1806, "PF14650"]], "MGP_CAROLIEiJ_P0034659": [["Domain_0", 153, 403, "PF15371"], ["Linker_0_1", 403, 893], ["Domain_1", 893, 1803, "PF14650"]], "ENSRBIP00000033419": [["Domain_0", 150, 404, "PF15371"], ["Linker_0_1", 404, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSANAP00000025214": [["Domain_0", 152, 486, "PF15371"], ["Linker_0_1", 486, 893], ["Domain_1", 893, 1613, "PF14650"], ["Domain_4", 1611, 1806, "PF14650"]], "MGP_CAROLIEiJ_P0034657": [["Domain_0", 154, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSCHIP00000018596": [["Domain_0", 154, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSCGRP00000024397": [["Domain_1", 899, 1806, "PF14650"]], "ENSCAFP00000019990": [["Domain_0", 154, 404, "PF15371"], ["Linker_0_1", 404, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSMAUP00000022680": [["Domain_0", 155, 400, "PF15371"], ["Linker_0_1", 400, 893], ["Domain_1", 893, 1805, "PF14650"]], "MGP_PahariEiJ_P0041280": [["Domain_1", 894, 1806, "PF14650"]], "ENSMFAP00000021787": [["Domain_0", 150, 404, "PF15371"], ["Linker_0_1", 404, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSCHIP00000032922": [["Domain_0", 150, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSMUSP00000097024": [["Domain_0", 155, 405, "PF15371"], ["Linker_0_1", 405, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSCATP00000007146": [["Domain_0", 150, 405, "PF15371"], ["Linker_0_1", 405, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSMAUP00000020557": [["Domain_1", 893, 1724, "PF14650"], ["Linker_1_6", 1724, 1775], ["Domain_6", 1775, 1806, "PF14650"]], "ENSRNOP00000069160": [["Domain_0", 154, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1805, "PF14650"]], "ENSSBOP00000006577": [["Domain_0", 150, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSCGRP00000016066": [["Domain_1", 893, 1721, "PF14650"], ["Linker_1_6", 1721, 1775], ["Domain_6", 1775, 1806, "PF14650"]], "MGP_SPRETEiJ_P0036314": [["Domain_0", 154, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "MGP_SPRETEiJ_P0036315": [["Domain_0", 153, 403, "PF15371"], ["Linker_0_1", 403, 893], ["Domain_1", 893, 1804, "PF14650"]], "MGP_SPRETEiJ_P0036316": [["Domain_1", 894, 1806, "PF14650"]], "ENSRROP00000020609": [["Domain_0", 234, 404, "PF15371"], ["Linker_0_1", 404, 893], ["Domain_1", 893, 1751, "PF14650"]], "ENSP00000329825": [["Domain_0", 154, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSMAUP00000022662": [["Domain_1", 894, 1806, "PF14650"]], "ENSMFAP00000024653": [["Domain_0", 154, 450, "PF15371"], ["Linker_0_1", 450, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSSSCP00000052107": [["Domain_0", 150, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSPANP00000019452": [["Domain_0", 154, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSNLEP00000005339": [["Domain_0", 312, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSPTRP00000071382": [["Domain_0", 150, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSMMUP00000012616": [["Domain_0", 311, 450, "PF15371"], ["Linker_0_1", 450, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSMUSP00000152919": [["Domain_0", 154, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSPTRP00000082829": [["Domain_0", 310, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1798, "PF14650"]], "ENSMUSP00000130813": [["Domain_1", 893, 1806, "PF14650"]], "ENSPANP00000041496": [["Domain_0", 150, 404, "PF15371"], ["Linker_0_1", 404, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSGGOP00000000162": [["Domain_0", 154, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSFCAP00000007858": [["Domain_0", 154, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSP00000485118": [["Domain_0", 154, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSPEMP00000018614": [["Domain_0", 141, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]], "ENSPPAP00000026962": [["Domain_0", 150, 406, "PF15371"], ["Linker_0_1", 406, 893], ["Domain_1", 893, 1806, "PF14650"]]}