Home
Query Results
"ENSGT00920000149015_4"
Found:
ENSGT00920000149015_4
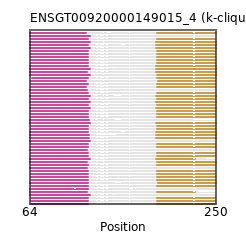

ENSMEUP00000004761 (view gene)
-----MNYSL---DDS-----PSQNHHESVLPLVARNTTITQVPPAKTITFFKKGDPRFG
GVR
PF03607 // ENSGT00920000149015_4_Domain_0 (64, 124)
MAVNQRMFKSFNALMDELSHHIPLSFGVRTITTPQGVHCVSSLEQLEDGAWYVCSDK
NPPENSGT00920000149015_4_Linker_0_1 (124, 190)
KALDG--SYRRPQ-ES--GFPTQN-------RQLEGWQE-TPSWQGHKAPKKLTLVK
NGDNGFRRSPF03607 // ENSGT00920000149015_4_Domain_1 (190, 250)
LILSPKNTQNLKAFLNDASEILHFPVNHLYSPDGSKV--ESLQSLLQGPSI
VVCAGQEPFRPMAVENSRRRLPERLPGLTSKKR-----NANWELKAKKSVIHSRSESSN-
RSRQLSLTSEKSYLNGSRASIV--GPVPS--QHPPENSAD--LIPSGANDDIEKKVHVNK
DGSLSVEMKVRFRLLSEEMLQWSTHVRKTSHLRAANRDDLGFSGVPLFHPSRENNVQRSS
DPWS-GD------------QVSRTGEASYQGCQQPQPKY-EIWK-NPMC-ISQGHG--AA
PGR-SSWITQSPC-RGLCSDRPSTKQRIDSLC---TTFGGEPL-ECSAPGPSCPSGTLEG
RVW---------HSDKHPASSDDEGEEEYTS------TGKNMVEEPDSIGRAPRKLKKSR
QMGSGHQMRRAWDTAEDVSASS-------------ICSRCREA-----------------
-CGGLWQSQTGRGFLNMRE-------------------------AQISWHSLWPYASDGE
GSGSSLVGP--------------------SSLKNKVLEDEEVEH----------------
-NKVIQDTSSGSVWRMPR------AMGNGKFSDLE-GV-YSTSSCPSAQW-KQRMGSLRS
DESLHNVSHSPRVYEGSKSSQPQMKNSGDRETSCILEDSPAKEHVQ-----FPPEKPRAD
NQTPMFSGSPSEAKQQGARNLGSSSSSPH--------------------------HLECL
SCLSDASIAQDD---------------------NPNQKKLENISFQPFNP-N--TL-EET
RAATKASISSLEPKSSNISDTFDFQG--I---VSLGEDPGGPSSFLELKIKESQMEEPNA
SRSTCS--------SQSDKLQVLS--VKGKNDHKGPGSQVDSVASGCLPTPPRGMPHIRK
ST-------SGSSKWSANLGEDGSWEEPVSMCL----APGSRSESS--NSKRARKGVWS-
------------WKMKENLRGSLKKPESSSGIIPSDLPSTSPEEVVYEWLSKIPEESILM
SYEMQDDSPRAA---VKAPEEDCAAKCSPEVPGEMAQAQE-HAADKEDSEEAPL---DEN
PRFDAASPEARGSY-----------KATLAT--VAKGSPGECPDLEA-VG-----WRLPS
TLYSSVQIMKTLLSSKQSPELGRFNSLPEVSQTMGRKLSHSAHMLITCLASLHFFDE-EL
VTVTNQLTYTNSPKYQELLSIFQALWTECSPN-----------TEESNSKALPSSKSQTP
MTGDFTPTSSSGVDVSSGSGGSGEGRVGGVVDX-X-------------------------
-------------------------------XXXXXXXXXXXXXXXXXXX----------
------------------------------------------------------------
--X---------------XXXXXXXXXXXXXXXXXXXXXXXXXXXX--------XXXXXX
XXXXXXXXXXXXX----XXXXX--XXXXXX--------------XXXXXX------XERT
DGKQGL--MKEIKVVEAEEEKMLGKEMQEAETQDDGMEVEKI--QEEKQ-----------
--AE---ETKDEK------------------------VFVERV------------QEEEG
IQKG-------------------------SIECDFTDVCPPVKAEE--PTEQVCNPSKDD
SNS-DNVHDSQELEPHLKEV-----TEGVAVTSEQSVVNTTAGTGGKRASIIQNTSPD--
PDPLWVLRLLKKIEKEFMTHYVNAMSEFKVRWNLQNNEMLDQMIIELKNEVNQRIQKSIE
KELRKIQSRAGKKLP--KPPKEAFKWETSLQTEQRRRRLKGM----RNHSAFTEQNKMPG
----KRNLSLAFTD-ARASSGSLADDLRELEGEEEYCPCDTCIRKKMTTLSLKNTAP---
ASDAPLRKAFDLQQILLK---KKEEKLNKGAAELATG------KVQRDYTS---------
----NEAVDKTEPGMASE--L----------------KQGRKT----------TDPAK--
------------------------------------------------------------
-----------------------------MEGRNCLK-EK---SRRL-------------
--IG---NDL--------E---E--E---E---A--D--LH--K----------T-----
-------E---E---R------HNEGK---A--E-------------NDSEEDEDK--GE
S-V--GDG-NE--R---K------------------------------------------
----------------ETESA---PQYGANSEGETSED-----------------GERCS
EQR---------DG---A---E-----------E---L---Q-------A--------T-
--E----S-----------------------------FSPEAEVEKQKKSGSGKPREPE-
SNCQESPGETW-------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
ENSRNOP00000064085 (view gene)
-----MNSTPGDTRDAPAPSHPAPSHRQCLLPSVAHTPSVTEVTPAKKITFLKRGDPQFA
GVRL
PF03607 // ENSGT00920000149015_4_Domain_0 (65, 122)
AVHQRTFKTFSALMDELSQRMPLSFGVRSVTTPRGLHGLSALEQLQDGGCYLCSDR
KENSGT00920000149015_4_Linker_0_1 (122, 191)
PPKTPRES--GR----LQ--RKSPPAGRSQVSQGGHEAPETSSYSWKNPMAPRRLTLVK
NGYPGCQQTVPF03607 // ENSGT00920000149015_4_Domain_1 (191, 249)
VLSHKNTRNLAGFLSKASELLRFPVKQVYTTHGKKV--DSLQTLLDGPSL
LVCAGNEAFRRLEMENDRGNRTRMVSGVTARSER-----GCWGPNAKQSVIHSRGRSGG-
KLRQFSLMSERSGLSDHPASGHQAWAGPALDRCPQDTPAP--PGSLVAADDVEKKVCMNE
DGSLSVEMKVRFQLLGEDTLRWSQRVGHASVFTAATGEDRDLREADRLCCRQEGYPWGVL
EPGAQGLGPHDG----------GCQGAFDVGQ-QSRPSY-DIWR-NPLT-APEGTG--PT
PRRRWGLAKLSRC----WREGTNDRKG-HDKANASPVS-TPRHPGSVQPG-SCCLWTPEV
ETGSDTLRPVSSASS-----RSETDLESGED------------PCLEDTGP---------
--------------------------------------CGLRLQTQSIDRALSDTSASAE
SR------EESCEGGGQLHRGSSQARVMA--SQGQATR-GDNPCIS--------------
-------------------------TQRHLSLNHTGLQTKKDRQSA--------------
---SCQEVRGDPVLRLPL------VPR-HSGSRDTQRDALPAPACAPAQRRQRKQKRPAG
VVCLPSVSIPHQVAQKGHARQCHCC----RDTQSSGDT--ALQMT----MPMEREQACPG
SPAPQFPSNSPRAGNQAPECLRPPFSSSLDFQEPQ-ATSI----AMSDS--------DCM
SSFCH-------------------PSTRSAEPAEDSKCQAPSSTPTPIHRGE--------
----------------------------VGYLWDKAGSTPEPFSSSVLLNRWPEA-DLRT
YQNFGCLQVAPTSTLATPSSQT-----------QASISEACLGDNSSCPTPPQDQICSRR
EPASSNSTH--SDHGRADGYT---------------------------------------
--GP---RRTLPGMP-LGIRGSLEEQEGDGGVTPSALPYTSPDAVVREWLGNIPEKPALM
TYEVADETIEVPSEGPEGPKEDLGDGCSLKVFGELTQARQ-QMLEGVTDEHPEP--AGVP
AGPGSVCCKLGGDMHSD--ATSVERLKAPAEAGTGEGTT---------VERGVSLCALPT
RVSASTQIMKALLGSKP----GRPSSLPEVSSTVAQRLSCSAGALIACLAKLHFFDE-DL
GPLDGKRRLEESPKYQEMLNISQTLWP-GSELGQGQL--DFSLRRLTSHQALLG------
-TEDFTPTSSSGVDVSSGSGGSGEGSVPCVMDS-TLASKKIDLPLK-----IPSQRPDSR
NQGYPEI----------VSPSTV--------SSGSQMRACTTSGE---------------
--------------------------------------------------------EAGK
G-GRKQTWGSAPEQSVENK-------------------VPEGDAQLEETEGRAR------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------ERLQENSVLG-EGLPEEGTR
VCSQGMLAANSRDGA-GSPEDTRVPNDEVGTD---GGLWPLDGRED--PTESPCHFEESN
SRV-SERQSAHVPELGL------DVRG--GKQACVKAS---SETRETNTSIAHRGTLD--
PDPIWVSKLLKKIEKAFMAHLTDATAELRARWNLHDNNLLDQMVTELEQDVGQRLQASTV
REVRKIQSRAGRMFP--EPPREALRGQSSLQTEQRRRRLQGL----RNLSAFP----GQG
------PLSFTLEDGA-TLGTALGTRSGAGLVEDEFCPCEICLKKKRTPRFPKDSTT---
VSGAPVRKAFDLQQILQS---KKDGSSNREAMEVSPQ-KTGMMLSQEDP-----------
-----RTVQGADEKQELRLAQGLGVAEGEE-------GEGKQRMRAEEDT----------
------------------------------------------------------------
------------------------------E----FS------K---A---E---G---D
---G---C---HA--P--K------E---D---A-TT---K--E-------D--------
-------G---KICIG------T---A---Q--D---G---Q--Q-LE----------G-
--T-EIG------K---E---E---T--L---H---Q------SF-R-----------D-
--G----------DTSE---A------------------------------PARQG----
-------------G-------H---S---V---E---S---Q---E---A---S----R-
--E----G--------------------------------------QPEVEGR-------
-NPDEKEESSWVSSEESQG-RVGSENNSLDQEGR--LQDHHQ-RPGPQSH----------
---HAARSSRAMSLDNCSQVSQKGSDRDFPSGDLKYNKAKN-------------------
----------SGVLPAERKVPMMYPESSSSE-----QEA---------PSSPRPPKQEKG
E-D---------------------------------------------------------
---GRSACTQVGG---RVDGFGQDDLDF--------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
ENSMUSP00000055449 (view gene)
-----MNSTPGDTRDAPAPSHPAPSHRQCLLPSVAHTPSVTEVTPAKKITFLKRGDPQFA
GVRL
PF03607 // ENSGT00920000149015_4_Domain_0 (65, 122)
AVHQRTFKTFSSLMDELSQRMPLSFGVRSVTTPRGLHGLSALEQLQDGGCYLCSDR
KENSGT00920000149015_4_Linker_0_1 (122, 191)
PPKTSREP--GR----LQ--RKSPSAGQAQVFQGGHEAPET-SYSWKGPVAPRRLTLVK
NGDPRRQQTVPF03607 // ENSGT00920000149015_4_Domain_1 (191, 249)
VLSHKNTRSLAAFLGKASELLRFPVKQVYTTRGKKV--DSLQTLLDGPSV
LVCAGNEAFRCLEMENDRGNRTRKLSSVTARSER-----GCWGPNAKQSVIHSRGRSGG-
KLRQVSLTSERSGLSDHPASGHRAWAGPALDRCPQDMPVP--PGSLVAADDVEKKVCMNE
DGSLSVEMKVRFQLLGEDTLRWSQRVGQASVFTAASGKGQDPREADRFCCRQEGYPWGIL
KPGAQGLGSYDG----------GCQEAFDVGQ-KSQPSY-DIWR-NPLA-TPEGTG--PT
PRRRWGLAKLSGC-KSHWRQEANHRKG-HDKDNLSRVS-TPRHPRSVQPG-SCCPWTPDG
DTGSDTLHPVSSASS-----HNETDLESGEG------------LCLEDTGP---------
--------------------------------------HGSRPETQSTERALSDTSVSAK
SR------EESSEGGGQLHRSSSQARVMA--SREQVTK-GDNPCIS--------------
-------------------------TQSHLPLNHMGLQTEKYRQGT--------------
---RGWEVSGEPELRLAL------VPG-HSGSQDTQRDALPAPACAPAQWRQRKQKRPAS
VECLPSVSVPYQVAQKGHARQDHYY----RDTQSSLDT--ALQMP----MPQEREQACPG
SPAPQSPSNSPSAGNQASEDLRSPFSSSLDLQEPQ-ATSKATTIAVSGS--------DCV
----C-------------------HSTRSVEPAGDTKCQAHSSTPTPAHRGE--------
----------------------------LGCLWDKAGTTPEPFSFSVLLDRCPEADDPRT
YHDCCCLQAVPSSPLAAPSGQT-----------QTSISEACLGGSSFCPTPPKEQTCFGR
ESASNGSTS--SGHSRADGFA---------------------------------------
--GP---RRTLLVKS-PGVRGSLEEREADGGVTPSALPYASPDAVVREWLGNIPEKPVLM
TYEMADENTEVPSDGPEGPKED-----SLKVLGEPSQAKQ-QPPEGATNEHPEP--AGVL
SGPGSVCCRLGGDLHPD--ATSGERLKAPAEAGIGEGAR---------VDHGVSLCALPT
KVAASTQIMKALLGSKP----GRPSSLPEVSSTVAQRLSSSAGAFIACLARLHFFDE-SL
GPLDGKVRLEESPKYQEMLRLFQTLWP-GSELWQGQL--DFSLRKLTSHQALLG------
-TEDFTPTSSSGVDVSSGSGGSGESSVPCVMDN-TLAPEKRDLPLK-----IPSQRPDSR
NQGYPEL----------VGHSTV--------SSVSQVRACATGGE---------------
--------------------------------------------------------ETGK
G-GRKQTWGNAPEQSVHST-------------------MLEGDALSEETEGRVR------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------ERLQENSVHG-KGLPEEGVR
VCSQEMLAAGSQDGA-GSPEDTRVPTDEAGADAASGGLWPLDGREE--PTESPQHFSESN
SRV-REHQSAHKLELGLEEVSRLDARG--CKQACIKAS---SGT------MAHKGSLD--
PDPIWVSKLLKKIEKAFMAHLADATAELRARWDLHDNHLLDQMVTELEQDVGRRLQASTV
MEVRKIQSRAGRMVP--EPPREALRGQASLQTEQRRRRLQGL----RNFSAVP----GQG
------PLSLTLEDGP-TLKTALGTKSGAEPAEDEFCPCEICLKKKRTPRFPKDAAT---
VSGAPVRKAFDLQQILQS---KKGGSSNREAMEVAPQ-RTGRMLSQEDL-----------
-----GTVQGADEKQ------GLGVAEGEE-------GEGKQRLRAEEDP----------
------------------------------------------------------------
------------------------------E----IL------K---T---E---G---S
---G---C---CA--P--E------E---D---E-AT---E--E-------D--------
-------G---EICIG------T---A---Q--E---S---Q--Q-LE----------G-
--T-EMG------K---E---G---T--L---P---Q------SF-R-----------D-
--G----------GTLE---A------------------------------PARQG----
-------------T-------H---S---V---E---I---Q---E---A---S----R-
--E----R--------------------------------------QQEVEGR-------
-HQDVKEDSPWVSSGESQG-RVGSENTSLDQEGR--LLNHHQ-RPGPQSH----------
---HTACSSRALSLDNSSQVSQKGSDGDLTSGDLKCTKAKN-------------------
----------SRVLHAEKKVPVMYPERSSSE-----QEV---------PSSPRLPKQGKG
E-DEGS------------------------------------------------------
--AGSLACTQVGG---KVDGFGQDDLDF--------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
ENSMAUP00000004680 (view gene)
-----MNNTPGDARDAPA-----PSHRQCLLPAVAHTPSVAEVTPAKKITFLKRGDPQFA
GVRL
PF03607 // ENSGT00920000149015_4_Domain_0 (65, 122)
AVHQRTFKTFSALMDELSQRMPLSFGVRSVTTPRGLHGLSDLEQLQDGGCYLCSDK
KENSGT00920000149015_4_Linker_0_1 (122, 191)
PPKTPREP--GR----LQ--RKSPSAGQSRGFEGGHEAPET-SSSWKGLKAPRQLTLVK
NGDPRCQQTVPF03607 // ENSGT00920000149015_4_Domain_1 (191, 249)
VVSHRNTRSLKAFFSKASELLRFPVKQVYTISGKKV--DSLRTLLDGPSL
LVCAGNEAFRHLEMENVGGTRTRTLSGVTARSGR-----GCWGPNAQQSVMHSRARSGS-
RPQQFSLMPERSGLSDRPAPGHQAWAGPALHRCPQDTPAP--PGSLVAADDVEKKVCMNE
DGSLSVEMKVRFQLLGEDTLRWPQRVGHASVFSPASGEGQVLREADPFCCRQEGHPWGFL
AHGAQGLGPYDG----------EYQGVFDLDQ-ESQPNY-DIWR-NPLT-TPGGAG--PT
RRRRWGLSKLSGC-RSHCSQGTNDRKG-RGNYSTSPVS-SPRHPRR---C-SCCPWSPEG
ETDSDTLHPVSSASS-----PSGAELEAGKG------------PCLEDTGP---------
--------------------------------------CGLGPETQGIERALSDTSVSAE
SH------EVSSECGDQHHRGSSQARVMV--SQGQTTQ-GDR------------------
-------------------------PG----ASTLSIQTEKYRQGT--------------
---RAQEARGEPELRLPL------TLS-SSDSQDTQGSALPAPATAPAQQGQKKQKRPAG
IVCLPSVSVPYQDAQKGHASRCHYC----RDTQPSLDT--ALQMT----ASQEGEQACPG
GPAPRFPPNSPSAGNQASGHLRSPLSRSLDFQDPQ-ATSKATIIPT-NS--------ACA
SSLYH-------------------PSTCSAEPAGDTECQAHPPDPTPAHREE--------
----------------------------VGCLWEEAGTTAEPSSSSVLLDRWPEAEDPGT
HQDCCCLQVGASSVLTARSGQT-----------QAPISEACWGASSFCPTPPQEQTCSRK
DPISSSGPS--SDHSQADGHA---------------------------------------
--EP---RRSLLGKS-PGVRGSLEEQEGDGGMVPSALPYTSPDATVREWLSNIPEKPVLV
TCEMEGETTEVASDGPESPEEDLG-------PRELAQARQ-QLPEGVTDKQPEP--E---
----------GGDPHHA--AASGEGVKAPAEAGVGEETT---------VDHGVSLYALPS
RVSASTQIMKALLGSKL----DRPSSLPELSTTVAQRLSYSAGALIACLARLHFFDG---
-----NK---ESPKYKEMLSISQTLWP-GNELGQGQL--DIGLRKLSSHQALLG------
-TEDFTPTSSSGVDVSSGSGGSGEGNVPCAMDN-TLAPHGVDLPSK-----IPSQRPDSG
NQGYPEA----------LSHSTA--------SSGSQVRACATSGE---------------
--------------------------------------------------------ESGK
S-GRKQTRGNTPEQSIESA-------------------TLEEEAQSEETGGRVR------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------ERLQEDGVRG-KGLPEERAE
VCSQEMFPA-----E-LSPEDTRVSKGEAGRDAASGGLWPLAGREE--PTEFPRRFRESH
SNA-SEGQSLHALELGLEGKSGEAATD--CEQACVKTS---SGTREMSTPVAHGRSLD--
PDPFWVSKLLKKVEKDFMAHLAHATTELRARWNLQDNSLLDQMVTELEQDVGRRLQASTV
REVRKIQSRAGRMVP--EPPREALRGQTSLQTALRRRRLQGL----RNFSAFP----GQG
------ALSLTLEDGP-TLSTALGTRPGVGAVGDEFCPCEVCLKKKMTPRFPKGATA---
VSSAPVRKAFDLQQILQS---KKGG-RNGEAMEEAPQ-KTGMMLPQEDP-----------
-----RAVQGTLRKQGLRL--------EEE-------GEGRQSQRGEKDP----------
------------------------------------------------------------
------------------------------E----FL------K---V---E---G---A
---V---C---HV--P--E------E---G---A-VT---A--G-------E--------
-------G---EIHVS------A---A---Q--E---N---Q--Q-LE----------E-
--S-VP-------E---E---E---I--P---H---Q------SF-R-----------D-
--G----------DPRE---A------------------------------SGRQR----
-----D---D---G-------H---S---V---E---T---L---E---A---S----R-
--E----R--------------------------------------QPEAEGG-------
-NQEEKENGPWGSSGESQS-GAVSENISLDRERR--LHNHYR-RPGPQHH----------
---HAAHSSRASSLGNCSQVSQKGSEEEFPSRELKCTKAKN-------------------
----------SGVKHAEREVTRMYPESSSSE-----QEA---------PSSPRPPKQG-E
EVYEGT------------------------------------------------------
--AGSLACTQAEG---RVDGFGQDDLDF--------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
ENSPPYP00000020575 (view gene)
-----MNSTP---RDAQA-----PSHRECLLPSVARTPSVTKVTPAKKITFLKRGDPQFA
GVR
PF03607 // ENSGT00920000149015_4_Domain_0 (64, 124)
LAVHQRAFKTFSALMDELSQRVPLSFGVRSVTTPRGLHSLSALEQLEDGGCYLCSDK
KPPENSGT00920000149015_4_Linker_0_1 (124, 190)
KTPSGP--GR----PQ--ERNPTAQQLRDIEGQREAPGT-SSSRKSLKTPRRILLIK
NTDPRLQQTPF03607 // ENSGT00920000149015_4_Domain_1 (190, 229)
VVLSHRNTRNLAAFLSKASDLLHFPVKQLYTTSGKKV--LQNMNFHQRWA-
-------EWRM---EVD-------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
MGP_CAROLIEiJ_P0037115 (view gene)
-----MNSTPGDMRDAPAPSHPAPSHRQCLLPSVAHTPSVTEVTPAKKITFLKRGDPQFA
GVRL
PF03607 // ENSGT00920000149015_4_Domain_0 (65, 122)
AVHQRTFKTFSSLMDELSQRMPLSFGVRSVTTPQGLHDLSALEQLQDGGCYLCSDR
KENSGT00920000149015_4_Linker_0_1 (122, 191)
PPKTHREP--GR----LQ--RKSPSAGQSQVFQGGHEAPET-SYSWKGPVAPRRMTLVK
NGDPRCQQTVPF03607 // ENSGT00920000149015_4_Domain_1 (191, 249)
VLSHKNTRSLAAFLGKASELLRFPVKQVYTSRGKKV--DSLQTLLDGPSV
LVCAGNEAFRCLEMENDRGNRTRKLSGVTARSER-----GCWGPNAKQSVIHSRGRSGG-
KLRQVSLTSERSGLSDHPASGHQAWAGPALDRCPQDMPVP--PGSLVAADDVEKKVCMNE
DGSLSVEMKVRFQLLGEDTLRWSQRVGQASVFTAASGKDQDPREADRFCCRQEGYPWGIL
KPGAQGLGSYDG----------GCQEAFDVGQ-QSQPSY-DIWK-NPLT-TPEGTG--PT
PRRRWGLAKLSGC-KSHWRQKANHRKG-HDKDNASRVS-TPRHPRSVQPG-SCCPWTPDS
DTGSDTLHPVSSASS-----HSETDLESGEG------------LCLEDTGP---------
--------------------------------------HGSRPETQSTERALSDTS-SAK
SR------EESSEGGGQLHRGSSQARVMA--SREQVTK-GDNPCIS--------------
-------------------------TQSHLPLNHTGLQTEKYRQGT--------------
---RGQEIRGEPELRLPL------VPS-HSGSQDTQRDALPAPACAPAQWRQRKQKRPAS
VVCLPSVSVPYQVAQKGHARQDHYC----RDTQSSLDT--ALQMP----MPQEREQACPG
SPAPQSPSNSPSAGNQASEDLRSPFSSSLDLQEPQ-ATSKATTIAVSGS--------DCV
----C-------------------HSTHSVEPAGDTKCQAHSSTPTPAHRGE--------
----------------------------LGCLWDKAGTTPEPFSSSVLLDRCSEAEDPRT
YHDCCCLQAAPSSPLAAPSGQT-----------QASISEACLGGSSFCPTPPKEQTCFGR
ESASNGSTS--SGHSRADGFA---------------------------------------
--RP---RRTLLVKS-PGVRSSLEEREADGGVTPSALPYASPDAVVREWLGNIPEKPVLM
TYEMADENTEVPSDGSEGPKEDPGDSCSLRVLGEPSQVKQ-QPPEGATDEHPEP--AGVL
SGPGSVCCRLGSDLHPD--TTSGERLKAPAEAGLGEGTR---------VDHGVSLCALPT
RVAASTQIMKALLGSKP----GRPSSLPEVSSTVAQRLSSSAGAFIACLARLHFFDE-GL
GPLDGKVRLEESPKYQEMLRLSQTLWP-GSELWQGQL--DFSLRKLTSHQALLG------
-TEDITPTSSSGVDVSSGSGGSGESSVPCVMDN-TLAPEKRDLPLK-----IPSQRPDSR
SQGYPEP----------MSHSTV--------SSVSQVRACATGGE---------------
--------------------------------------------------------ETGT
G-GRKQTWGNAPEQSVHST-------------------MLEGDALSEETEGRVR------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------ERLQENSVHG-KELPEEGAR
VCSQEMLAAGSQDGT-GSPEDTRVPTDEAGADAASGGLWPLDGREE--STESPQHFSKSN
SRV-REHQSAHKLELGLEEISRLDARD--CKQACIKAS---SGTREMTTSIAHKGSLD--
PDPIWVSKLLKKIEKAFMAHLADATAELRARWDLHDNHLLDQMVTELEQDVGRRLQASTV
MEVRKIQSRAGRMVP--EPPREALRGQASLQTEQRRRRLQGL----RNFSAIP----GQG
------PLSLTLEDGP-TLKTALGTKSGAGPAEDEFCPCEICLKKKRTPRFPKDAAT---
VSGAPVRKAFDLQQILQS---KKGGSSNREAMEVAPQ-RTGRMLSQEDL-----------
-----GTVQGADEKQGLKLAQGLRVAEGEE-------GEGKQRLRAEEDP----------
------------------------------------------------------------
------------------------------E----IL------K---T---E---G---S
---G---C---CA--P--E------E---D---E-AT---E--E-------D--------
-------G---EICIG------T---A---Q--E---S---Q--Q-LE----------G-
--T-EIG------K---E---D---T--L---H---Q------SF-R-----------D-
--G----------GTLE---A------------------------------PARQG----
-------------T-------H---S---V---E---I---Q---G---S---S----R-
--E----R--------------------------------------QQEVEGR-------
-NQDVKEDSPWVSSGESQG-RVGSENTSLDQEGR--LQNHHQ-RPGPQSH----------
---HTACSSRASSLDNCSQVSQKGSDGDLTSGDLKCTKAKN-------------------
----------SRVLHAERKVPMMYPESFSSE-----QEV---------PSSPRLPKQGKG
E-DEGS------------------------------------------------------
--AGSLACTQVGG---KVDGFGQDDLDF--------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
ENSCGRP00000017687 (view gene)
-----MNSTPGDTKDVPA-----PSRRQCLLPSVAHTPSVTEVTPAKKITFLKRGDPQFA
GVRL
PF03607 // ENSGT00920000149015_4_Domain_0 (65, 122)
AVHQRTFKTFSALMDELSQRMPLSFGVRSVTTPRGLHGLSALEQLQDGGCYLCSDK
KENSGT00920000149015_4_Linker_0_1 (122, 191)
PPRTPREP--GR----RQ--RKSPSAGQSRGFEGGHEAPET-SSSWKGLMPPRRLTLVK
NGDPRCQQTVPF03607 // ENSGT00920000149015_4_Domain_1 (191, 249)
VLSHRNTRSLKAFLSKASELLHFPVKQVYTISGKKV--DSLLTLLDGPSL
LVCAGNEAFRRLEMENVGGARTRTLSGVTARSGR-----GGWGPNAKQSVMHSRARSGS-
RPQQVSLMSERSGLSNHTALGHQDWGGPALDRRPQDTPAP--HGSLVAADDVEKKVCMNE
DGSLSVEMKVRFQLLGEDTLRWSQRVGHTSVFTPASGEGQVLREADPFCCRQEGYPWGFL
AHGAQGLGPYDG----------GYQGVFNLDQ-ESQPNY-DIWR-NPLT-TPGGTG--QT
RRRRWGLSKPSGC-RSHCSQVANDRKG-HGNNNASPVS-SPRHPRSAQSG-PCCPWTTEG
ETDSDTLHPASSAS------SSGVELEAEKG------------PCLA-TGP---------
--------------------------------------CGLGPETQGIERALSDTSVSAE
SH------EGSSECGGQCHRGSSQVRVMV--SQGQATQ-GIS------------------
-------------------------TQSLLSLSRMDFQTEQCNRST--------------
---RDQEARGEPDLRLPL------VPG-CSGSSDGERGALSAPASAPAQQRQKKQKRPVS
IVCLPDVSIPCQGAQKGHARQCHYC----RDTQPSLDT--ALQMP----MPQEGEQACPG
GPTPRFPPNSPSAENQASGHLRSPFSRSLDFQDPQ-ATSKANIIPTSDS--------DCA
SSFYH-------------------PSTCSAEPAGDTECQARPPAPTPVHREE--------
----------------------------VGCLWEDAGTTPEPFSPSVLLDPWPEVEDPGT
HQDCCCLQVGASSVLIVPTGQA-----------QAPISEACWGTSSFCPTPPQEQTCSRN
DPTSSSGPR--SDHNQADGHD---------------------------------------
--EP---RKTLLGKS-PGTRGSLEEQ-ADGGVTPTALPHTSPDAVVREWLGNIPEKPVLM
TWEMEGESTEVASDGPESPEGDLA-------LRELAQARQ-QPPEGVTDEQPEP--TGDL
PGSGPEYYKPGGDPHHD--AASGEGVKAPAEAGIGEETT---------VDHGVSLYALPS
RVSASTQIMKALLGSKP----DRPSSLPEISSTVAQRLSYSAGALIACLARLHFFDG---
-----NTRLEESPKYKEMLNISQALWP-GSELGQAQL--AIDLRKLSSHQALLG------
-TEDFTPTSSSGVDVSSGSGGSGEGSVPCAMDN-TLAPDRVDLPLK-----IPSQRPDSG
NQGYPGV----------LSHSTA--------SSGSQMWPCATSGQ---------------
--------------------------------------------------------ESGK
G-GRNQKWSNTPEQSIQST-------------------TLEEEAQSEEKKGRVK------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------ERLQGDGVHG-KGLPEERSG
VCSQEKFPAGSWDRE-SSPEDTRMSKDEAGRDVASGGLWPLDGREE--PTESPCRFRESN
SNT-NEGQSLHVLELGLEGKSRVAATG--SEQAGIK----------TNTPIAHRRALD--
PDPIWVSKLLKKVEKDFMAHLAGAITELRARWNLQDNSQLDQIVTELEQDVGRRLQASTV
REVRKIQSRAGRMVP--EPPREALRGQTSLQTALRRRRLQGL----RNFSAFP----GQG
------APSLALEDGS-TLSTALGTRPGVGGMGDEFCPCEVCLKKKMTPRFPKGATA---
VFSAPVRKAFDLQQILQS---KKEG-RNGEAMEVPPQ-KAGMMLPQEDP-----------
-----QTVQGTHQKQELRL--------EEE-------GKGRQRLRGEEDS----------
------------------------------------------------------------
------------------------------E----FL------K---A-------G---A
---G---C---HV--P--E------E---D---A-AT---A--R-------E--------
-------G---EIHIS------A---A---Q--E---N---Q--Q-LE----------E-
--C-MT-------E---E---E---T--Q---H---Q------SF-R-----------E-
--G----------DSWE---A------------------------------SGRQG----
-----D---D---G-------H---S---V---E---T---R---D---A---S----R-
--E----R--------------------------------------QSEVEGG-------
-NQDEKENGPWGSSGESQR-RAVSKNISLDQEGR--LYNYHR-RPGPQCH----------
---HTAHSSRASSLGICSQVSQKSSEEKFSSGELKFTKAKN-------------------
----------SRVLHAEREGTTMYPESSSSE-----QEA---------PSSPRSPKQREG
EVYEET------------------------------------------------------
--AGSLVCTQVKG---RVDGIGQDDLDF--------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
ENSPANP00000006324 (view gene)
-----MNSTP---RDAQA-----PRHRECLLPSVARTPSVTKVTPAKKITFLKRGDPQFA
GVRL
PF03607 // ENSGT00920000149015_4_Domain_0 (65, 122)
AVHQRAFKTFSALMDELSQRVPLSFGVRSVTTPRGLHSLSALEQLEDGGCYFCSDK
KENSGT00920000149015_4_Linker_0_1 (122, 191)
PPKTPSGP--GR----PQ--GRSPTAQQLRDIQGRREAPGT-SSSRKSLKTPRRILLVK
NMDPHLQQTVPF03607 // ENSGT00920000149015_4_Domain_1 (191, 249)
VLSHRNTRNLAAFLSKASDLLRFPVKQLYTTSGKKV--DSLQALLHSPSV
LVCAGHEAFRPPAMENARRSEAETLSGLTSRNKN-----RSWGPKTKPSVIHSRSRPGS-
RPQL----PERPVPSN-------PPVGPAPDRHPQDTPAQ--PGPLVAGDDMKKKVRMNE
DGSLSVEMKVRFHLVGEDTLLWSRRMGRASALTADSGEDPILGEVDPLCCVWEGYPWGFS
DPGVWGPRPCRV----------GCGEAFGRGG-QPGPKY-EIWT-NPLH-ASQGER--VA
TRKRWGLAQHVRC-SGLWGHGAAGRQR-CSLDSASPAS-STGHPEGSEPDSSCCPRTPED
RVDSG------SPSA-----QRGAEREAGGS------LGEDPSLCTDGAGLGSPE-----
------------------------------------QGGRLTPRAQSEEGASSDSLASTG
SH------EGSSEWGGQPRSCPGKARAET--SQQEATK-GGNPGSP--------------
-------------------------APSLSSLRSEDLQAEMQGRGT--------------
---EQ--ARGASVKREPL------VPG-LSGSWDSEGASSTPSTSASSQQERRRHRSRAS
ATSSPSSPGLGRVAPRGHPRQCHYC----KDTHSPLDSSVTKQVP----RPPERRRACRD
GSVPRSSGSSLSTRTQDSGNLRPPSLGSLPSQDLL-GISSATVTPAGHS--------DCA
SGVSP-------------------CNAPFVGWAGDTGSRTCSPDPTPPHTSD--------
--------------------SCSKSG--AASLEEQARDTPQPSSPLILQVGRPEQGAAGP
HQSCCHSQAGTQPAE---------------EAQRGPSPEARWLCGRYCPTPPRGRPCPQR
RSSSCGSTG--SSHRSAARRPGGSQQKEGTHQAGPTPPLGPGSG-------ASRRSSASQ
DVGS---GRLSQEET-SRSWGGLQGQEEASGVSPSSLPRSSPEAVVREWLDHIPEEPVLM
TYEMADDTTGAAGGGLRGPEVDPGDDHSLEGLGEPALARQ-QPLEGDPSQDLEP--EGAL
PGSSDAGPQSGEGVPQG--AAPEGVSEAPAEAGADREDP---------ASCGVSLRALPG
RVSASTQIMRALMGSKQ----GRPSSMPEVSRPMARRLSRSAGALITCLASLQFFDE-DL
GSPASKVRFKDSPRYQELLNISKDLWP-GCDLGEDQL--DSGLWELTWSQALPDLGSQ-A
VTENFTPTSSSGVDISSGSGGSGESSVPCAMDS-TLVPGGTELSLK-----TSNQRPDSR
TSENPGD-----LENQQQCCSPA--------FLNSRACACATNED---------------
--------------------------------------------------------EAER
D-GEEQRASLNLEQLAENT-------------------MQE-EVQLEESKE-ETEGGGL-
QEEGLQLEGTKETE----G--------------LQE------------------------
--------------------------------------------EGGLQLEGTKGTEG-E
GHQEEGAQLEEIKEETGGEGLQEEGAQLEEVKEGPEGGLQEEALEEGLKE-EGLPE-EGS
VGRQELSDAGSPDGE-GSWEDDPIQEEEAGRAS--AEPCPPEGTEE--PTEPASHLSETD
PSA-SESQSGSQLEPGLEKPPGAAMLG--QEHTQAQPP---LGAAERSSSVACRAALD--
CDPIWVSVLLKKMEKAFLAHLSGAVAELRARWGLQDSDLLDQMAAELQQNVAQRLQDSTE
RELQKLQGRVGRMVL--GPPREALTGELLQQTQQRRHRLRGL----RNLSAFSERTLGLG
------PFSFTLEDEP-ALSTALGSQLGEEGEGEEFCPCEACVRKKVSPASPKATMG---
ATRGPIKEAFDLQQILQK---KREE-HTHGEA----A-EVAPGETCTDP----TST----
-----RTVQGVEGGPGLGLGHGPGVDQGED-------GEGSQRLDRDKDP----------
------------------------------------------------------------
------------------------------K----L---G---E---A---E---G---D
---T---M---A---Q--E---R--E---G---K--I---H--N---S---K--T-----
-------S---V---G--S---K---L---G--E---A---E--Q--E---G---E--G-
--I---S---E--R---G---E---T--G---G---Q--G---S--G---H---E---D-
--N--L---Q---G--E---A---V---A---G---G---D---Q--D---P---G----
-Q---S---D---G---A---E---G---I---E---A---P---E---A---E----G-
--E----A--------------------------------------QPES--E-------
-GVEAPEVEGEMQE--------AEGEAQPESESV--EAQEAQ-PE---SEGVEAPEAEGE
VQEPESEGVEAPEAEKEAQPETESVEAE-------EAEGEAP----------------PE
SEDA-EAREAGE---------------EAQE-----AEGE--------------------
-----DKPESEV-------I--EGEMQEAEGEAQPESEGVEAPEAEGEAQEAEGEA-QPE
SEVVGAPEVEGEM---QEAEGETQPESEGVEAQEEAEGEAQSESEGVEAPEAEGEAQPES
EGVEAPEAEGEAQ-----EA--EGEAQEAEGEAQPESEGVEAQEAEGEAQPESEGVKAPE
AEGEAQEA--EGEAQEAEGEA-----------------------KPESEVVGAPEAE---
----GETQTESEGV------------EAQEAEGEAQPESEGVEAQEAEGEAQPESEGVEA
PEAEGEAQPETEGVEAPEAEG---------E--------E--AQPESEGVEALEAD----
----------EEAQPESEVVE--ALEAEGEAQPESEGE--------TQGEKKGSPQVSLG
DGQSVGASESNSPVPKDRPTPPPSPGGDTPHQKPA-SQTGPSSSRASSQG--DSWRKDSE
DGHVCGDTR-SPDAKSVGAPHAERKATRMYPESS-TSEQEEAPSDTRSPEQGAS------
---EG-CDPQEDQTLGNPAPTKAVGRADGFGQDDLDF
ENSRROP00000021794 (view gene)
-----MNSTP---RDAQA-----PRHRECLLPSVARTPSVTKVTPAKKITFLKRGDPQFA
GVRL
PF03607 // ENSGT00920000149015_4_Domain_0 (65, 122)
AVHQRAFKTFSALMDELSQRVPLSFGVRSVTTPRGLHSLSALEQLEDGGCYFCSDK
KENSGT00920000149015_4_Linker_0_1 (122, 191)
PPKTPSGP--GR----PQ--GRSPTAQQLRDIEGRREAPGT-SSSRKSLKTPRRILLVK
NMDPRLQQTVPF03607 // ENSGT00920000149015_4_Domain_1 (191, 249)
VLSHRNTRNLAAFLSKASDLLRFPVKQLYTTSGKKV--DSLQALLHSPSV
LVCAGHEAFRPPAMENARRSEAETLSGLTSRNKN-----RSWGPKTKPSVIHSRSRPGS-
RPQL----PERPVPSN-------PPVGPAPDRHPQDTPAQ--PGPLVAGDDMKKKVRMNE
DGSLSVEMKVRFHLVGEDTLLWSRRMGRASALTAASGEDPILGEVDPLCCVWEGYPWGFS
DPGVWGPRPCRV----------GCGEAFGRGG-QPGPKY-EIWT-NPLH-ASQGER--VA
TRKRWGLAQHVRC-SGLWGHRAAGRER-CSQDSASPAS-STGHPEGSEPDSSCCPRTPED
GVDSG------SPSA-----QRRAEREAGGS------LGEDPSLCTDGAGLGSPE-----
------------------------------------QGGCLTPRARSEEGASSDSLASTG
SH------EGSSEWGGRPRGCPGKARAET--SQQEAIK-GGNPGSS--------------
-------------------------APSLSSLRSEDLQAEMQGRST--------------
---EQ--AGGASVKRGHL------VPG-LSGSWDSEGASSTPSTSASSQQEQRRHRSRAS
ATSSPSSPGLGRVAPRGHPSQCRYC----KDTHNPLDSSVTKQVP----RPPERRRACRD
GSVKRSSGSSLSTRTQASGNLRPPSLGSLPSQDLL-GISSATVTHAGHS--------DCA
SGVSP-------------------YNAPSVGWAGDAGSRTCSLDPTPPHTSD--------
--------------------SCSKSG--AASLGEQARDTPQPSSPLVLQVGWPEQGAAGP
HQSCCHSQAGTQPAE---------------EAQRGPSPEAHWLCGRYCPTPPRGRPCPQR
RSSSCGSTG--SSHRSAARGPGGSQQKEGTHQAVPTLSLGPGSG-------ASRRSSASQ
DVGS---GGLSQEET-SRSWGGPQGQEEASGVSPSSLPRSSPEAVVREWLDNIPEEPVLM
TYEMADETTGAARGGLRGPEVDPGDDHSLEGLGEPALARQ-QRLEGDPGQDPEP--EGAL
PGSSDAGPQSGEGVPQG--AAPEGVSEAPAEARADREDP---------ASCRVSLRALPG
QVSASTQIMRALMGSKQ----GRPSSMPEVSRPMARRLSRSAGALITCLASLQFFDE-DL
GSPASKVRFKDSPRYQELLSISKDLWP-GCDLGEDQL--DSGLWELTWSQALPDPGSH-A
VTENFTPTSSSGVDISSGSGGSGESSVPCAMDG-TLVSEGTELPLK-----TSNQRPDPR
TSENPGD-----LENQQQCCSPA--------FLNSQACACATNED---------------
--------------------------------------------------------EAER
D-SEEQRASSNLEQLAENT-------------------MQEEEVQLEESKE-ETEGGGL-
QEEGLQLEGTKTE--G-LQEE---------------------------------------
---------------------------------------------GGAPLEGTKETEGER
HQQEEEAQLEEIKEETGGEGLQEEGAPLEEVKEGPEGGLREEALEEGLKE-EGLPE-EGS
VGRQELSAAGSPDGE-GSREDDPMQEEEAGRAS--AEPRPPEGTEE--PTEPASHLRETH
PSA-SESPSGSQLEPGLEKPPGAAVMG--PEHTQAQAP---PGAAERSSSVACGAALD--
CDPIWVSVLLKKTEKAFLAHLSGAVAELRARWGLQDSDLLDQMAAELQQDVARRLQDSTE
RELQKLQGRAGRMGL--GPPREALTGELLLQTQQRRHRLRGL----RNLSAFSERTLGLG
------PFSFTLEDQP-ALSAALGSQLGEDGEGEEFCPCEACVRKKVSPASPKATMG---
ATRGPIKEAFDLQQILQG---KREE-HTDGEA----A-EVAPGETCTDP----TST----
-----RTVQGAGGGPGPGLGCGPGVDQGED-------GEGSQRLDRDKDP----------
------------------------------------------------------------
------------------------------K----L---G---E---A---E---G---D
---A---K---A---Q--K---R--E---G---K--I---H--N---S---E--T-----
-------S---V---G--S---E---L---G--E---A---E--Q--E---G---E--G-
--I---S---E--R---G---E---T--G---G---Q--G---S--G---H---E---D-
--H--L---Q---G--E---A---V---A---G---G---D---Q--D---P---G----
-Q---S---D---G---A---E---S---I---E---A---L---E---A---E----G-
--E----A--------------------------------------QPES----------
-------------------------------EGV--EAPEAE-----------------G
EAQPESEGVEAQEAEKEAQPETESVEVPETEGEDQPESEVVEGEA-----------QEPE
SEVVEAPEEAEE---------------EAQE-----AEGE--------A-QSESEGVEAL
EAEGE-------------------------------------------------------
------------------------------------------------------------
--------------------------------AQPESEGVEAQEAEGEAQPESEGVEAPE
AEGEAQPEAEGVEAQEAEEEAQPESEGVEALEAEGEAQEAEGEAQPESEGGEALEAEGEA
QEAEGEAQPESEGV------------EAQEAEGEAQPESEGVEAPEAEGEAQPEAEGVEA
QEAEEEAQPEAEGVEAQEAEEEAQPEAEGVE--AQEAEGE--AQPEAEGVEAPEAE----
----------GEAQPEAEGVE--ALEAEGEAQPESEGE--------TQGEKKGSPQVSLG
DGQSGGASESSSPAPEDRPTPPPSPGGDTPHQKPT-FQTGPSSSRACSQG--SSWRKDSE
DSHVCGDTR-SPDAKSMGARHAERKATRMYPESS-TSEQEEAPSDTRTPEQGAR------
---EG-CDPQEDQTLGNPAPTKAVGRADGFGQDDLDF
ENSCANP00000004213 (view gene)
-----MNSTP---RDAQA-----PRHRECLLPSVARTPSVTKVTPAKKITFLKRGDPQFA
GVRL
PF03607 // ENSGT00920000149015_4_Domain_0 (65, 122)
AVHQRAFKTFSALMDELSHRVPLSFGVRSVTTPRGLHSLSALEQLEDGGCYFCSDK
KENSGT00920000149015_4_Linker_0_1 (122, 191)
PPKTPSGP--GR----PQ--GRSPTAPQLRDIEGRREAPGT-SSSRKSLKTPRRILLVK
NMDPRLQQTVPF03607 // ENSGT00920000149015_4_Domain_1 (191, 249)
VLSHRNTRNLAAFLSKASDLLRFPVKQLYTTSGKKV--DSLQALLHSPSV
LVCAGHEAFRPPAMENAR-SEAETLSGLTSRNKN-----RSWGPKTKPSVIHSRSRPGS-
RPQL----PERPVPSN-------PPVGPAPDRHPQDTPAQ--PGPLVAGDDMKKKVRMNE
DGSLSVEMKVRFHLVGEDTLLWSRRMGRASALTAASGEDPILGEVDPLCCVWEGYPWGFS
DPGVWGPRPCRV----------GCGEAFGRDG-QPGPKY-EIWT-NPLH-ASQGER--VA
TRKRWGLAQHVRC-SGLWGHRAAGRER-CSQDSASPAS-STGHPEGSEPDSSCCPRTLED
GVDSG------SPSA-----QRRAEREAGGS------LGEDPSLCTDGAGLGSPE-----
------------------------------------QGGRLTPRARSEEGASSDSLASTG
SH------EGSSEWGGRPRGCPGKARAET--SQQEATK-GGNPGSP--------------
-------------------------APSLSSLRSEDLQAEMQGWST--------------
---EQ--ARGASVKREPL------VPG-LSGSWDSEGASSTPSTSASSQQERRRHRSRAS
ATSSPSSPGLGRVAPRGHPRQCRYC----KDTHNPLDSSVTKQVP----RPPERRRACRD
GSVPRSSGSSLSTRTQASGNLRPPSLGSLPSQDLL-GISSATVTHAGHS--------DCA
SGVSP-------------------PNAPSVRCAGDVGSRTSSLNPTPPHTSD--------
--------------------SCSKSG--AASLGEQARDIPQPSSPLVLQVGWPEQGAVGP
HQSCCHSQAGTQPAE---------------DAQRGPSPEARWLCGRYCPTPPRGRPCPQR
RSSSCGSTG--SSHRSAARGPGGSQQNEGTHQAGPTLSLGPGSG-------ASRRSSASQ
DVGS---EGLSQEET-SRSWGGPQGQEEASSVSPSSLPRSSPEAVVREWLDNIPEEPVLM
TYEMVDETTGAAGGGLRGPEVDPGDDHSLEGLGEPALAGQ-QPLEGDPGQDPEP--EGAL
PGSSNAGPQSGEGVPQG--AAPEGVSEAPAEAGADRGDP---------ASCGVSLRALPG
RVSASTQIMRALMGSKQ----GRPSSMPEVSRPMARRLSHSAGALITCLASLQFFDE-DL
GSPASKVRFKDSPRYQELLSISKDLWP-GCDLGEDQL--DSGLWELTWSQALPDPGSH-A
VTENFTPTSSSGVDISSGSGGSGESSVPCAMDG-TPVSEGTELPLK-----TSNQRPDSR
TSENPGD-----LENQQQCCSPA--------FLKSQACACATNED---------------
--------------------------------------------------------EAER
D-SEEQRASSNLEQLAENT-------------------MQEEEVQLEEK---ETGGEGL-
QEEGVQLEGTKETE-G-LQEEGV-------------------------------------
------------------------------------------------QLEGTKGTEGE-
GQQEEEAQLEEIKEETGGEGLQEEGAQLEEVKEGPEGGLQEEGLEEGLKE-EGLPE-EGS
VGRQELSEAGSPDGE-GSREDDPIQEEEAGRAT--AEPRPPEGTEE--PTEPASHLSETD
PSA-SESQSGSQLEPGLEKPPRAAMMG--PEHTQAQPA---LGAAERSSPVACRAGLD--
CDPIWVSLLLKKTEKAFLAHLSGAVAELRARWGLQDSDLLDQMAAELQQDVARRLQDSTQ
RELQKLQ--AGRMVL--GPPREALTGELLLQTQQRRHRLRGL----RNLSAFSERTLGLG
------PFSFTLEDQP-ALSAALGSQLGEDGEGEEFCPCEACVRKKVSPASPKATMG---
ATRGPIKEAFDLQQILQR---KREE-HTDGEA----A-EVAPGETCTDP----TST----
-----RTVQGAGGGPGPGLGCGPGVDQGED-------GQGSQRLDRDKDP----------
------------------------------------------------------------
------------------------------K----L---G---E---A---E---G---D
---A---K---A---Q--E---R--E---G---K--I---H--N---S---E--T-----
-------S---V---G--S---E---L---G--E---A---E--Q--E---G---E--G-
--I---S---E--R---G---E---T--G---G---Q--G---S--G---H---E---D-
--H--L---Q---G--E---A---V---A---G---G---D---Q--D---P---G----
-Q---S---D---G---V---E---S---I---E---A---L---E---A---E----G-
--E----A--------------------------------------QPESE-G-------
-VEAPEPA----------------------SEGV--EASEAE-GEAQPESEGVGAPEAEG
EAQPESEGVEAQEAEKEAQPETESVEVPETEGEAPPESEGAEAQEAGEEAQEAEGEDQPE
SEVVQAPE-IEG---------------EMQE-----AEGE--------A-QPESEGIEAP
EFEGEAQEAEGX--XXXXXX--XXXXXXXXXXXXXXXXXX----------X-X-X-XX-X
-X-X-XX-X-X-X----XX-X-XX-X-X-XX-X-XX-X-XX-X-XX-X-XX-X-XX-X-X
X-XX-XXXXXXXXXXXXXXXXXXXXXXXXXXXX---------------------------
-----------XXXXX---------XXXXXXXXX--X---------------XXXXX---
------XXXXXXXX----------------------------------------------
--------------XXXXXXXXXXXXXXXXX--XPEAEGE--GQPESEGVEALEAD----
----------EEAQPESEGVE--ALEAEGEAQPESEGE--------TQGEIKGSPQVSLG
DGQSGRASESSSPVPEDRPTPPPSPGGDTPHQKPT-FQTGPSSSRASSQG--NSWRKDSE
DGQVCGDTR-SPDAKSMGAPHAERKATRMYPESS-TSEQEVAPSDTRTPEQGTS------
---EG-CDPQEDQTLGNPAPTKAVGRADGFGQDDLDF
ENSMLUP00000015234 (view gene)
-----MNSTP---RDAQA-----PSYRECLLPSVARTPSVTQVTPAKKITFLKRGDPRFS
GVR
PF03607 // ENSGT00920000149015_4_Domain_0 (64, 124)
LAVHQRTFKSLGTLMDELSQRVPLSFGVRSVTTPRGLHGLSALEQLEDGGCYLCSDR
KPPENSGT00920000149015_4_Linker_0_1 (124, 190)
KTSSGP--GR----PR--ERSPPAQQSQDVEDQCEGPGT-SSSRKNPKTPRRIMLVK
NGDPQFQQTPF03607 // ENSGT00920000149015_4_Domain_1 (190, 250)
MVLSHRDTRSLSAFLSRATGLLHFPVKQVYTPSGEKV--GSLKALLRSPSV
LVCAGHEPFRPP--EGAKRSGTGTLSGQTSRNKN-----GSWGPKAKPSMTHSRSRPGS-
RPQRSSLLSARAGLGDPLVSPHHTRVGPAPDRHPQDMPAQ--LGPLVARDGVEKKVHLNE
DGSLSVEMKVRFYLLGNDMLLWPRRAGRA------SGQGPVPREADPLHCVWEGHPGGSL
EPGTQGLGTREA----------GCTEACERGQWQPGSGY-EIWT-NPLY-TAQREG--TA
FWGSSRLAQHSPC-RGPWSRGVAGRTR-SSQDSGSPAS-SDRAPEGSEPN-SCCSRSPEG
SMGSCALHLASGAAP-----QRGSGWEAGGT------PQAAEGPGPEGAGPGSRG-----
-------------------------------------HRCLEPRAQGAAGAFSDVSASAG
AQ------GESSERGERHQGHASRTRLVT--SPRKATP-GAGPCSS--------------
-------------------------TADPSSLRNEDPRAEGSEQDT--------------
---GPPQPRGGPGRRLPL------ASG-RLGSGDTAEGCSPASTCTSATGRKREQESRAS
AVSSPSISGLGRGAQRGRPRQHHSQ----RHTHCPLDSPVSRPML----GPPSTGRLCPA
HPAPRFSGSSSSTRNPASRDPGPPSLASPNSQDAR-GVSSVPVTPVSSS--------DCA
SNFYP-------------------PFSPSAETEFRSHSL--------STTSD--------
--------------------PLSSPG---GGLGGKAGGGPKASRPLSLPVGQHEGDKPGA
HRGGCGSQMGTSRACRAHGGETQALLAPQPPGSQGPLSEPCLVCSGYCPTPPRARPPVRS
LSG-----------------------------------------GRGTAIRTVRRGSPSP
GPRP---GRKFQAQV-AGGGEGLEEQEDSGRVTPGALPRASPEAVVRRWLSNIPEEPVPL
SYEMVDDSMEVAGDGPEGPTEDPVDTHPPEGLEEPIQARQ-PSLEGAASEQADP--EGAL
PVTGDAGPQPGEV----------NLAEAPAEAGAGEGAA---------GDCGGSQCVLPR
SVSVSIQIMKALMGSKQ----GRPSSLPEVSGPEGRRLGHSARALITCLARLHFFNE-DR
GSPTGKARFTDSSRCQELLSTLQASWP-GCGLRRGEP--DSGPRGCGRSQALSGLGPH-A
VTEAFTPTSSSGVDVGSGSGGSGEGSGPCAVDC-ALVSERGELPSD-----IPYQRPDSR
TSENPEE-----PGNHESIGSMA--------SSSSQAWAGATRRD---------------
--------------------------------------------------------EAGG
S-CGEQARGRDLDQVVENR-------------------MRGEAVQLEKTQE-EKGRAEP-
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------HGEGV-RGFLE-EER
TTGQDSTGAGSQDGS-DAREDESVQEEEAGE-PAPTVLHPAGRRER--T---LGGLTERD
SNA-SGSQSSPSAEPGLEMLPRAADSG--HEQTQAKFT---QGAAEKGTSVAHRVSQD--
PDPLWVSRLLRKMEKAFLAHLASAMAELRARWSLQNNDLLDQMVAELQQDVSQRLQGSTA
NELRKIQSRAGRKAP--GPPRVALRWETSLQTEQRRSRLQGL----RNLSAFSEQTRALG
------APSLSLDDVP-NFSGALGTQLSGEAGPEEFCPCETCVRKTVTPVSPKDMMG---
IARAPIKKAFDLQLILQK---KQVGCANGETAGMAPE-KTGMEVLQRDP----SGT----
-----GTAQGADGGLELGLGRGPGAEEGDE-------DVG-----RGEGA----------
------------------------------------------------------------
------------------------------G----G---G---E---E---E---E---A
---T---Q---K---------------------S--G---G--N---T---D--P-----
-------Y---G---G--H---P---S---E--A---V---G--W--E---E---Q--G-
--A---G---D--Q---G---E---I----------------------------------
-----R---E---S--E---A---W---G---G---A---D---R--S---P---G-G--
-Q---N---D---T---S---E---V---------------Q---D---V---E----G-
--E----G--------------------------------------QPKSGRG-------
-NLGEKAGSPQAGPRQGQS-GEASGHSSPDQDGG--PTPPSA-SGGDTP-----GHRSGR
KTGL--SSSRTSSLGNCSQLSQKGSEEGSSNGDMSTFGDE------------PKGVPGSD
-----------------RNGTGVSLESSTSA-----REGP--------SS----------
-----------------------------------------------------GSGTPER
GTDEGLFGNLAMD---GTHGFGHDDLDF--------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
ENSRBIP00000035685 (view gene)
-----MNSTP---RDAQA-----PRHRECLLPSVARTPSVTKVTPAKKITFLKRGDPQFA
GVRL
PF03607 // ENSGT00920000149015_4_Domain_0 (65, 122)
AVHQRAFKTFSALMDELSQRVPLSFGVRSVTTPRGLHSLSALEQLEDGGCYFCSDK
KENSGT00920000149015_4_Linker_0_1 (122, 191)
PPKTPSGP--GR----PQ--GRSPTAQQLRDIEGRREAPGT-SSSRKSLKTPRRILLVK
NMDPRLQQTVPF03607 // ENSGT00920000149015_4_Domain_1 (191, 249)
VLSHRNTRNLTAFLSKASDLLRFPVKQLYTTSGKKV--DSLQALLHSPSV
LVCAGHEAFRPPAMENARRSEAETLSGLTSRNKT----VSCWGPKTKPSVIHSRSRPGS-
RPQL----PERPVPSN-------PPVGPAPDRHPQDTPAQ--PGPLVAGDDMKKKVRMNE
DGSLSVEMKVRFHLVGEDTLLWSRRMGRASALTAASGEDPILGEVDPLCCVWEGYPWGFS
DPGVWGPRPCRV----------GCGEAFGRGG-QPGPKY-EIWT-NPLH-ASQGER--VA
TRKRWGLAQHVRC-SGLWGHRAAGRER-CSQDSASPAS-STGHPEGSEPDSSCCPRTPED
GVDSG------SPSA-----QRRAEGEAGGS------LGEDPSLCTDGAGLGSPE-----
------------------------------------QGGCLTPRARSEEGASSDSLASTG
SH------EGSSEWGGRPRGCPGKARAET--SQQEAIK-GGNPGSS--------------
-------------------------APSLSSLRSEDLQAEMQGRST--------------
---EQ--AGGASVKRGRL------VPG-LSGSWDSEGASSTPSTSASSQQEQRRHRSRAS
ATSSPSSPGLGRVAPRGHPRQCRYC----KDTHNPLDSSVTKQVP----RPPERRRACRD
GSVKRSSGSSLSTRTQASGNLRPPSLGSLPSQDLL-GISSATVTHAGHS--------DCA
SGVSP-------------------YNAPSVGWAGDAGSRTCSLDPTPPHTSD--------
--------------------SCSKSG--AASLGEQARDTPQPSSPLVLQVGWPEQGAAGP
HQSCCHSQAGTEPAE---------------EAQWGPSPEAHWLCGRYCPTPPRGRPCPQR
RSSSCGSTG--SSHRSAARGPGGSQQKEGTHQAVPTLSLGPGSG-------ASRRSSASQ
DVGS---GGLSQEET-SRSWGGPQGQEEASSVSPSSLPRSSPEAVVREWLDNIPEEPVLM
TYEMADETTGAARGGLRGPEVDPGDDHSLEGLGEPALAGQ-QRLEGDPGQDPEP--EGAL
PGTSDAGPQSGEGVPQG--AAPEGVSEAPAEARADREDP---------ASCRVSLRALPG
QVSASTQIMRALMGSKQ----GRPSSMPEVSRPMARRLSRSAGALITCLASLQFFDE-DL
GSPASKVRFKDSPRYQELLSISKDLWP-GCDLGEDQL--DSGLWELTWSQALPDPGSH-A
VTENFTPTSSSGVDISSGSGGSGESSVPCAMDG-TLVSEGTELPLK-----TSNQRPDPR
TSENPGD-----LENQQQCCSPA--------FLNSQACACATNED---------------
--------------------------------------------------------EAER
D-SEEQRASSNLEQLAENT-------------------MQEEEVQLEE---------RL-
QEEGLQLEGTLKQK-G-PQEEGVSVGRQGSSPGKMT------------------------
------------------------------------------------QCRRRK------
------------------------------------------------------------
-RERSLCRAAAPEGS-R---------------------------GT--APGAASHLRETQ
PRAPRESKWRPQLEPGLEKPPGAANDGP-PEHTQAKPR---RGRAERELFVALRSGSGLA
TRILVFRCWLKRPEKPLGP------ADKMTCWT--------RWRPRLQAGVARRLQEAPR
-ESCRSPGPGGRMGP--GPAQGGPSNLGSCSCKTPAAQTAGLRALPQPSSAFSERTLGLG
------PFSFYAGESA-SAQVQPWGASGRGREGEEFCPSLRP---------PKPTNG---
GNRVPQKRALLNCSRFCR---GRGKNSTDGEA----S-GVAACT----------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------------------G-
--L---V---R--V---S---R---R--R---A---Q--G---Q--G---W---V---C-
--G--W---S-------------------------------------G---P---G----
-E---D----------------------------------------------------G-
--G---------------------------------------------------------
---------------------------------------------GKPRDIDRDKDPKPW
GRQ---------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------GGDGEAQKREGEEPHSENPVWARAGGRE-----------E
T-----------------------------------------------------------
-----------------------------------GGQGSGHGTELAEGEAQPESEGVEA
QEAEGEAQPESEGVEALEAEGEAQPESEGVE--APEAEGE--AQPEAEGVEAQEAE----
----------GEAQPEAEGVE--ALEAEGEAQPESEGE--------TQGEKKGSPQVSLG
DGQSGGASESSSPAPEDRPTPPPSPGGDTPHQKPT-FQTGPSSSRACSQG--SSWRKDSE
DSHVCGDTR-SPDAKSMGAPHADRKATRMYPESS-TSEQEEAPSDTRTPEQGAR------
---EG-CDPQEDQTLGNPAPTKAVGRADGFGQDDLDF
ENSCAFP00000011953 (view gene)
-----MNSTL---RDAQA-----PSHRECLLPSVARTPSVTQITPAKKITFLKRGDPRFA
GVR
PF03607 // ENSGT00920000149015_4_Domain_0 (64, 124)
LAVDQRAFKSFGALMDELSQRVPLSFGVRSVTTPRGLHGLSALEQLEDGGYYLCSDK
KPPENSGT00920000149015_4_Linker_0_1 (124, 190)
KTPSRP--GW----PQ--GRSSSAQQSRDFESRCEAPGT-SSSCKGPKAPRRIVLVK
NGDPRFQQTPF03607 // ENSGT00920000149015_4_Domain_1 (190, 250)
VVLSHRNTRNLMAFLSKASDLLHFPVKQVYTTSGKKV--DSLKGLLHSPSV
LVCAGYESFKPLAVEDARRYGTETLSGQTSRNKT-----GGWGPKAKQSVIHSRSRSGS-
RPRQCSLLSERSGLSDAPGSLHCAQTGPAPDGHPQDVPAH--PGPLVASDDVEKKVRMNE
DGSLSVEMKVRFHLLGKDAVLWSRRVGRASALPAASEGVPVLGEVDSLHCVWEGHLGGSS
EPGAQGLGPCGA---------GGCEEALGRSRWQPGSRY-EIWM-NPLY-STQGDG--TD
SRRRSRLTQHSHS-RRPCSQRFTSRKR-SSKDSVSPAS-SDRCPRDSEPNSSCCSRSPGS
SMGSCDLQSASGAIS-----QRGAGWEGGDP------SGTSESPGPQGAGQGSRE-----
------------------------------------CHSRLKAQTQGTAGALSDSSGSAG
SH------EESSERGEQPQGCPSKTRAVS--TSRPKITQREGLSPP--------------
-------------------------TVSPLFLRDEDSQEEESGQGT--------------
---RHSQAKDRSGMRWRL------ALR-HFGSRDIAGS------CSPVPGRRRKQKGRAS
AMSLPSQ-----GAQTGHPR-----------HHRPLDSPVPKKMP----GPPSRARACPD
GPAPHLSESSPSARNRASQDTGPSSSASLHSQYA-------LITPVSNS--------ECA
SNFYP-------------------PYSLSAETEGDPKLRASSPAPTPSNTSD--------
--------------------SLGNQA--DGLDKKPGGDIPKPSWPLDPPGGQPEGGMPGA
HRGCSCSPISTSMFHGTPSDETHAPQTSQPLGSQGVFSEVCLECSRYCPTPPRTWPFVKK
HPGAMGKAV--KA---------------------------------------SRRGSSSC
GPRS---GRVFQKKV-AGGGKSLEEEEKDGRKMLRALPRASPDTVVREWLSNIPEEPIPM
KCEMVGDSTDVAGDDPEGPKEDPVGRHSPECLGEPTQVRQ-LLHEGAASEKAEP--DGAL
PVPGDAHSKSGEGLPC------SGISQDPREAGSGKGMA---------VDRGVGQCLLPH
KLSASIQVMKALIGSKQ----GRPSSLPEVSSAVGRRLSRSAQALITCLAGLHFFDE-DL
GSPTGQVRFIDSPRYQELLSIFQALWP-ACGLRRGEV--DSGLWELGRCQALPGLRSH-V
MTEDLTPTSSSGVDVGSGSGGSGEGSGPCAMDC-AMVPERITLPLK-----IPSPRQDSR
TSENRED-----PGNQEPSGSTA--------SSNSQAWACATRKD---------------
--------------------------------------------------------EAER
N-SGQQVLGSNLDPVVDNT-------------------MQEEEVQVEKKVREEKEIAEL-
QG----------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------EGV-PGFPE-EET
VMGQELSEANSQAGE-GAPEDKSVQEEEAGD-PASPILCPPGRKEK--PTEPPRSLSEGH
PNA-SETESDPKVEPGLEKLPKAAETC--HEQSQVKCT---QGPGEKSSSAVRRVSLD--
PDPLWVSRLLKKMEKAFMAHLASATAELRARWSLQNNILLDQMVAELHQDLGRRLEDSIE
KELRKVQSRAGGKMQ--GPAREGLRWEMSLQTEQRRRRLQDL----RRLSAFSRQARSQG
------PLALPTEDMP-TLSGALGAWQVGEAEEEEFCPCEACMRKKVIPTSPKDTME---
ASRAPIKKAFDLQHILQK---KKGGCISGEAKEAAPE-KRGMEPLQGDP----FGT----
-----GTVQESDRGLELGLDQDSGTEEGGE-------GESSQTLGREGDP----------
------------------------------------------------------------
------------------------------R----V---G---E---G---E---A---S
---V---Q---K----------R--E---G---S--T-----------------G-----
-------G---P---G--S---I---P---E--G---A---G--G--E---E---L--S-
--S---G---G--K---E---E---N--T---E---E--G---S--G---A---E---G-
--S--L---E---G--E---D---S---G---G---G---N---R--S---P---G-D--
-Q---N---D---G---D---K---S---E---E---I---Q---E---A---E----G-
--E----E-------------------------------------GQPESGGGRG-----
EGEEEKEGSPQVDWGQGQP-GETSGNSSPDPEGK--PALPPT-TGAD-----SPCPRAGW
KAGLASS--SVASLGNCSQLSQKGSEEKPLNGDMRSIEEESEE------------IPGPE
R-----------------TGTGMYPESSTSE-----KEGA--------PSGPRTPEQEAG
KASGLEAEKVVK----------------------------------------------TL
AFTEMGFQNLPVD---RTDGFGQDDLDF--------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
ENSACAP00000018459 (view gene)
---------S---SSV-------PYNYDQPLPPLARTNAVTQVPPAKKITFYKSGDPQFG
GVK
PF03607 // ENSGT00920000149015_4_Domain_0 (64, 120)
MAINQRSFKSFNALMDDLSHRVPLPFGVRTITTPRGIHCISTLDQLEDGGCYLCSDENSGT00920000149015_4_Linker_0_1 (120, 191)
K
KYVTPISIGAAHRRTG-SQK----TSALRKAAQDAKQDDHSM-GFTQQSPRIPKKVTLVK
NGDITAQRPIPF03607 // ENSGT00920000149015_4_Domain_1 (191, 250)
ILNRRNARSLKALLDEISDIMQFTVKKLFTLDGKKI--DSMQALLHCPNV
LVCVGREPFKPMVKENPRKHSSEKLPGLTSRSSSNN--NVNFGLEAKKSVMHPRSSLSN-
RSQRFSLSSEKSSTNGLTTSPENGAPFPRNCPHS--KAGE--PTHSLINDDIEKKVHVNK
DGSLSVEMKVRFRLLNDETLHWSTQLKKS--MNKIPGEEEASSE---------------L
DDAS-LLYPCDIDSYMSNVDESELDEVRCRNCGKLCKDY-DIWK-NPMH-ISQKEE---P
GVK-STWYTHSSC-SSTSSHRRIVRKKVASVDSIRTSSSGEEYSKHVVRESSHYSETVEN
MVEYRTQNEEDAAEG--------QENIN--------------------------------
------------------------------------------------TGGRPGSNRSKS
SQGSAESQIRICDKS------SSLARTISSGSIKERGELEDVVACSSPPNSIY----SKS
GKGSVLDGAQENVDEVSENDHPVSSVSMDSCARKDAAEDEAEKEDEEINEIYSVSENPGS
NPSVKDDASECGRSSTARSIRSESSAGNHRRKNSNEVHLCSAASASSRT-----------
-------------SKKSKPSQNHLEVASNRGS--TSSKK--KKKP---NSSFQAEDARSN
SRASHYSRSSHEVEKRDSGHASQDSSRPHSHGS---SLSEAAAPPAYN-EGSTSSLSRCY
SKSSRNSQKENQIEGSIKVPSHCSSFSDCGGENGDGIPKIGSPEGSSPKGDVSESTCSKC
GHITKPCRDDLRSTSSKVS------------------------SH---------------
------------------------------------SEASESMYSLRYPSPPKGRPSSRK
SRTLLLKKSSKGTSACTESELIADKEEKEATASQPPEALNAE------------------
-------EGSCQVKLQEEAEMETSASCGGDQIIPSSLPNASLEEVVHEWLRKIPSETLLM
KCEMQDDGEGAE---L--DAEITGCSTSQELLEGDSEEKK-EDT-------KEV--ESTA
DAVEPMAEGAEEEAVNE------------------EPKEAMCNLSAL-TSQKNHKKDLPS
NVQTSVQIMKALLASKQEAKMDRAHSLPEVSPTMGRKLSNSANILITCLASLQFLDE-EL
DPSNKSSKGLNRPRYTELLNIFQALWCTGHTSEKGGP--SNGIGEESPAKVSAGYKGHCS
KDGDFTPMSSSGVDVSSGDGGSGEGSVAGGQDH-ALSQEKTEEAKPA------EED-VEN
TELEVRNEDDKEGGDEEVECVGGEE-------ATEATEDCEIEQAGEKEI----------
------------------------------------------------------------
--AEEGAMEESDQQENENVECGEVNADGDTIQVEAEEKAVEEPGQQEDVEAEEETVEEIS
QQEDENVEGVEAA----ADEETVQVEAKEE--------------TVEEISQ---QEDENV
EGVEAAADE-ETVQVEAEEETVEEISQQED-ENVEGVEAAAD--EETVQVEAEE------
ETVEETSQQKDENVVDGEADADGETAQVES----EEEAMEENV-----------QQEDEN
VEGGEVDA----DGE-----TVQVETQEDSLVKEAEDSPSQENVDQ--VTEEPSNHELIE
EPG-HN-DP-STKEGNEQEEPEIGGEKQSEADAQPSSQIIPLQTPVKVSPIVQQRSID--
PDPIWVLKLLKKIEKEFMTHYVNAMNEFKVKWNLPNTDEVNVMISELKEEVNRRIQKSIE
KELKKIRSRAGRMVP--RPPDE-LQRESTLQVENRRRRLQSI----RKMSLYNDQNANQS
NPQETFS--YEMGE-DFIFST-IRDDDSELPSEEEYCPCDACIRKKIEARALRGPVVLPV
AANAPVVKAFDLHQILKM---KKG------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
ENSCCAP00000013401 (view gene)
-----MNITP---RDTQA-----LSHRECLLPSVARTPSVTKVTPAKKITFLKRGDPQFA
GVRL
PF03607 // ENSGT00920000149015_4_Domain_0 (65, 122)
AVHQRAFKTFSALMDELSQRMPLSFGVRSVTTPRGLHSLSALEQLEDGGCYLCSDK
KENSGT00920000149015_4_Linker_0_1 (122, 191)
PPKTPSGP--GW----PQ--GRSPTAPHLRDIKGRREAPGT-SSSRKSLKTPRKILLVK
NTDPRLQQTVPF03607 // ENSGT00920000149015_4_Domain_1 (191, 249)
VLSHRNMRSLAAFLSKASDLLCFPVKQVYTTSGKKV--DSLQALLHSPSV
LVCAGYEAFRPPAMENARRSEAGTLSGLTSRNKN-----GSWGPKTKPSVIHSRSQSGS-
RPQL----PERPGPSD-------LLAGPAPDRHPQDTPAQ--PGPLVAGDDMKKKVRMNE
DGSLSVEMKVRFHLVGEDTLLWSRRMGRATALSTAGGEDPVLGEVDPLCYVWEGYPWGFS
QPGVWGPQPCRV----------GCGEAFGRGG-HPGPKY-EIWT-NPLH-ASQGER--VA
ARKRWGLAQHVRC-SGLWDCRAPGRER-CSQDSASPAS-STGHPEGSEPDSSCCPRIPED
RMDSG------SSSA-----RRGAEWEAGGR------LGEEPSMCTDGAGLGSPE-----
------------------------------------QGGRLTSRAQSEERTSSDSSAS--
SR------EGSSEWGGRTRGCLGKVRAET--SQQEATE-GGVLGSP--------------
-------------------------ALSLSSLRSEDLQAEMPGHGT--------------
---EHQQASGASVMREPL------ASG-LSSSWDSEGASSTPPTCTSAQQRRRRHKSRAS
AVSSPSSPGLGRVAPRDSPRQCRYC----RGTHSPLDSPATKQVP----RPPQQRRACRD
GSVPRSSGSSSSSRTQASRDLRPPSSGSFPSQDLL-GTSSATVTPAGHP--------DCA
SGISP-------------------NNVPSAGWAGEAGSRTCSPAPMPPHTSN--------
--------------------SCPKSG--AASLGDQAKDTPQPSLPLVLQVERPEQGALDP
HQGHSCSQTGTLSAQ---------------EAQWGPSLEARWLCGRYCPTPPRGRPCPER
RPSSCASTS--SSHRSAACGPGGSQQEEETHQAELTASSGPSSG-------ASRSSGASQ
GVRC---GVLSQEKT--ASRGDPQGQEEASGMSPGTLPRSTPEAVVREWLDNIPEEPVFM
TYEMVDETTGAAGDGLRGPEVDPGDDQALGGLVEPADAR--QPLEGDAGQDLEP--EGVL
LVSGDVGPPSGEGIPQG--ASPEGVSEAPAEAGVDRGAP---------VSCGVGLRALPG
RVSASTQIMRALMGSKH----NRPSSVPEVSRPLARRLSHSAGALITCLASLQFFDE-DS
GSPASKVRFKDSPRYQELLRISKDLWP-GCDLGQDQL--DSGLWELTWSQALPDLGSH-A
VTENFTPTSSSGVDISSGSGGSGESSIPCAMDG-TLVPEGTELSLK-----TSNQRSDSR
TSENPGD-----LEKQQHCCSP--------------ACACATKEE---------------
--------------------------------------------------------EAER
D-REE--------QMAENT-------------------MQEEEAQLEEMKE-ETG--EL-
QGEEAQLEEMKEET-GELQGEE--------------------------------------
------------VQLEEVEEAAEGERLQEEGTQLEETKEESELQEEM-QLE---------
-------ELEETKEE------------------TGGGELQEEVLEEVLKE-EELPE-EGH
VCGQELSEAGSLDGE-GSQEEDAVQEEEVGRASASAEPCPPEGTEE--PTEPLSHLNEMD
PSA-SESQSGSQLEPGLEKPPGAAWMG--QEHTRAPPT---QGAAEKSSSVACRVALD--
CDPILVSVLLKKTEKAFLAHLASSVAELRARWGLQDNDLLDQMVAELQQDVARRLQSSTE
RELQKLQSRAERMVL--GPPRETRTGELLLQTQQRRHRLWGL----RNLSAFSERTLGLE
------PLSFTLEDEP-ALSTALGTQLGAEAEGEEFCPCEACVRKKVSPTSPKDTRG---
ATRGPIREAFDLRQILQR---KKGE-RTDGEA----A-EVVPGRTPMDP----IST----
-----RTVQGADGRPGLGLGPGPGVDEEED-------SEGSQRLNRGKDP----------
------------------------------------------------------------
------------------------------K----L---G---E---A---E---R---A
---A---V---A---Q--E---R--E---G---K--T---H--S---S---K--T-----
-------S---A---S--S---E---L---E--E---A---E--L--E---G---E--G-
--V---G---E--R---G---E---T--G---D---Q--D---S--G---H---E---D-
--N--L---Q---G--E---A---A---A---G---G---D---Q--D---P---R----
-Q---S---D---G---A---E---G---I---E---A---Q---E---A---E----G-
--E----A--------------------------------------QPESEG--------
-------VEAQEAEGE--V-QEAEGEAQPETEGV--EAQEAE-GEAQPESEGVEA-----
---QESEGVEAQEAEGEAQPESEGVEAEEAEGEAQPESEGVEAQEAEEEAQEAEGEAQPE
SEGVEEA----------------------QE-----AEGE--------A-QPESEDVEAQ
AAEGEAQPESEGIEAQPESE--GVEAQEAEGEAQPESEGVEAQPESEG------------
----------------------------------------------VEAPEAEGEAQRKS
KGIE-------VLEAEQEAQEVEGETQEAEREAQPDSEGVEAPEVEGEDQPESEIVEALE
AEW----------------------------------------EAKEAEGVEAPEAEGEI
QEAEGEAQPESEGV------------EALEAEGETQEAE--GETQEAEEEAQPESESVEA
LEAKGEAQPESEGVEALEAEGEAQPKSEGVE--VSEAEGE--AQPESEGVEAPEAE--GE
A-----QEAEEEAQPESEGVE--APEAEGEAQPKSEGE--------TQ----GSPQVSLG
DGH---------------------------------------------------------
---------------------AERKTMSMYPESS-TSEQEEVPSGPRTPEQGAS------
---EG-CDLRDDQALGSLAPTEAVGRADGFGQNDLDF
ENSSBOP00000014364 (view gene)
----MNSSTP---RDAQA-----PSHRECLLPSVARTPSVTKVTPAKKITFLKRGDPQFA
GVRL
PF03607 // ENSGT00920000149015_4_Domain_0 (65, 122)
AVHQRAFKTFSALMDELSQRMPLSFGVRSVTTPRGLHSLSALEQLEDGGCYLCSDK
KENSGT00920000149015_4_Linker_0_1 (122, 191)
PPKTPSGP--GW----PQ--GRSPAAPHLRDIKGQREAPGT-SSSWKSLKTPRRILLVK
NTDPRLQQTVPF03607 // ENSGT00920000149015_4_Domain_1 (191, 249)
VLSHRNVRSLAAFLSKASDLLCFPVKQVYTTSGKKV--DSLQALLHSPSV
LVCAGHEAFRPPAMENARRSEAGTLSGLTSRNKN-----GSWGPKTKPSVIHSRSQSGS-
RPRL----PERPGPSD-------PLAGPAPDRHPQDTPAQ--PGPLVAGDDMKKKVRMNE
DGSLSVEMKVRFHLVGEDTLLWSRRMGRASALTTASGEDPVLGEVDPLCCVWEGYPWGFS
QPGVWGPRPCRV----------GCGEAFGRGG-QPGPKY-EIWT-NPLH-ASQGER--VA
ARKRWGLAQHVRC-SGLWDRGAPGRER-CSQDSASPAS-STGHPEGSEPDSSCCPRIPED
RVDSG------SPST-----QRGAEWEAGGS------LGEEPGMCTDGAGPGGPE-----
------------------------------------QGGRLTSRAQSEEGASSDSSAS--
SR------EGSSEWGGRTRGCLGKTRAET--SQQEATE-GGVLGSP--------------
-------------------------ALSLSSLRSEDLQPEMPGHGT--------------
---EHQQARGASVMREPL------ASG-LSGSWDSEGASSTTPTCTSAQQGRRRHKSRAS
AVSSPSSPGLGRVAPRDSPRQCRYC----RGTHSPLDSPATKQVP----RPSERRRACRD
GLVPQSSGSSSSSRTQASGDLRPPSLGSLPSQDLL-GTSSATITPAGRS--------DCA
SGISP-------------------DNVPSAGWAGEVGSRTCSPAPTPPHTSN--------
--------------------SCSKSG--AASLGEQARDTPLPSMPLVLQVERPEQGASGP
HQGRSCSQIRTLLAQ---------------EAQWGPSLEARWPCGRYCPTPPRGRPCPQR
RPSSCASTS--SSHRSAACGPGGSQQEEKTHQAELTASSGPSSG-------ASRSSGASY
GVGS---GLLSQEKT--ASQGGPQGQEEASCMSPGTLPRSTPEAMVREWLDNIPEEPVLM
TYEMVDETTGAAGGVLRGPEVDPGDDHALEGLGEPADARQ-QPLEGDAEQDLEP--GGVL
QVSGDVGPQSGEGIPQG--ASPEGVSEAPAEARVDRGAP---------ASCGVGLRTLPG
RVSASTQIMRALMGSKH----DRPNSVPEVSRPVARRLSRSAGALITCLAGLQFFDE-DS
GTPASKVRFKDSPRYQELLRISKDLWP-GCDLGQDQL--DSGLWELTWSQALPDLGSH-A
VTENFTPTSSSGVDISSGSGGSGESSIPCAMDG-TLVPEGTELSLK-----TSNQRSDSR
TSENPGD-----LKKQQHCCSPA--------FLNSPACACATKEE---------------
--------------------------------------------------------ETER
D-REE--------QMAENT-------------------MQEEEAQLEETKE-GAEGE-L-
QEEGVQLEEMKEETDGEL------------------------------------------
------------------------------------------------------------
--QGEEAQFEETKKE---TAQLQEAAQLEEVKDGPEGGLQEEALEEGLQE-EELPE-EGH
VCGQELSEAGSLDGE-GSQEEDPFQEEEAGRASASAEPCPLEGTEE--PTEPPSHLSEMD
PSA-SESQSGSQLEPGLEKPPRATWMG--REHTQAPPT---QGAAEKSSSVACRVALD--
CDPILVSVLLKKTEKAFLAHLASSVAELRARWGLQDNDLLDQMVAELQQDVARRLQNSTE
RELQKLQSRAGRMVL--GPPRETRTGELLLQTQQRRHRLWGL----RNLSAFSERTLGLE
------PLSFTLEDEP-ALSTALGTQLGAEAEGEEFCPCEACVRKKVSPTSPKDTMG---
ATRGPIKEAFDLQQILQR---KKGE-RTDGEA----A-EVVPGRTPMDP----IST----
-----RTVQGAEGRPGLGLGPGPGVDEEED-------GEGSQRLDRDKEP----------
------------------------------------------------------------
------------------------------K----P---G---E---A---E---G---A
---A---V---A---Q--E---S--E---G---K--T---H--S---S---K--T-----
-------S---A---G--S---E---L---E--E---A---E--Q--E---E---E--G-
--L---G---E--R---G---G---T--G---G---Q--G---S--G---H---E---G-
--N--L---Q---G--E---A---A---A---G---G---D---Q--D---P---G----
-Q---S---D---G---A---E---G---I---E---A---P---E---A---K----E-
--E----A--------------------------------------QEAEEE--------
---------------------------------------------GQPESEDIEALEAEG
VAQPESEGVEAQEVEEEAQPESESIEAQETEEEAQP------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------ESED-------------------------------------------------
-----------------------------------------VEALEAEGVAQPESEGVEA
QEAEGEAQ--------EEAEGEAQPESEGVE--ALEAEGE--AQPESEGVEAPEAE----
----------GEAQPESEGVE--APEAEGEAQPESEGE--------TQGEKKGSPQVSLG
DGH---------------------------------------------------------
---------------------AERKTIRMYPESS-TSEQEEVPSIPRTPEQGAS------
---EG-RDLQDDQALGSLAHTEAVGRADGFGQDDLDF
ENSLAFP00000014874 (view gene)
-----MNSTP---QDAQA-----PRYHEFLLPSVARTSSITQVTPAKKITFFKRGDPRFS
GVR
PF03607 // ENSGT00920000149015_4_Domain_0 (64, 121)
LAVHQRVFKNFSALMDELSQRVPLSFGVRSVTTPQGLHCLSTLEQLEDGGCYLCSDKKPPQARSGP--GG----QQ--GTDPSAQPSGDLEGWYEVPGT-PSSWKGPKAPRRIMLVK
NTDPRFQRTVVLSHRNTRSLKTFISKASDLLQFPVKQVFTISGKKV--DSLQSLLHSPLV
LVCAGHESFRPPAMEESRRNGTETLPGLTSRNKN-----GSWGLKAKQSVIHSRSKSDG-
RLRQFSLLSERSGISDPPVSPHCTWMGPALHRHPQNTPVH--LGPLVADDDFEKKVHMNE
DGSLSVEMKVRFHLLSKDMLLWSRGARKASTLTTPSEECPVLGEVDPLHCVLEGSPGGFS
EPGAPGLRHCES----------GGEEAFNQGQ-QAGPGY-QIWT-NPLY-RTQGEE--TD
SWRRALLTKHCHS-TASWSQGVASGKR-TSKIRVSLAS-NDRPTAGSEANSSYCPRTPEG
SVGSCSGHQAARAAS-----KREVEQEAGCTHPAGGDPNL-----------G--------
----------------------------------------PKPEPCSLEVTLPDLSASAS
FL------KRSGETGEPCWGCASSEGTVI--SQQEHTQ-EGSTNCP--------------
-------------------------PLSPSSLKNGDLQAKECGQGT--------------
---GGHKARDGSVTRLPV------ALD-HSSSWYAGGCSSPLSSCTSAQRRSRKQKSQAS
AMSSPNTSDFGSISQRSHHRQHHHQ----RVVPYSLDLPTTKQVL----SPANTGGSCSG
GPELGFPESSSSVRKQASKDPGSPSSGSLCSQSTP-GVSSAAIILVSNSD---PQKTNCA
SHFYH-------------------SHFTSAETKGDSELRVSSP-PTPSNTSD--------
--------------------SFSSHVKVDHLDEKAGGNNPKLSSPLVWVVKWPDGVKSGD
HQGSCCSQIDTSLAS-------------QPQGGPRSISGACLVCSRYCPTPPRGQPCIKK
HSSSSSSNS--NSNQSANRGSGGREQ----------------------------------
--------GMSQGKV-AGEGESLEEQEEDGGMMPSALPHISPEAVIREWLSHIPEEPILM
KYEMMDESPNVAGDGPEGTQEDPVDKHSLEGLGELARVRE-QPLEGETSEKPEL--DRS-
SVTDDAIPKSKEAFPQR--GASRGVSDACEKDRASDRTT---------VGRGVSEGILPS
RIPASIQIMKELISSKQ----GRPSSLPEVSGVVGRRLSHSACALVTCLTRLHFFDE-HL
GSSASKMRFIDSPKFQELLVTLQSLWP-QCDLGKDKL--DLGLQELGSCQGLPGFTSH-A
VTEDFTPTSSSGVDVSSGSRGSREGSGPCPVDC-ALSPERMELPLKMVTSSISDQRPDSR
ASENPED-----PRNQHLSCSKV--------SSNSQAWACATNQD---------------
--------------------------------------------------------EAGE
N-SREQMLGNSLEQLVENT-------------------MF--------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------PE-TER
VNKQELSVDSTEDGE-GTQEDTRMQKEETGRDPDSVGLCPPGMSDK--PKELFSNPSECD
SHA-SEHQSGPTFDYGLKKLPGNAEMA--GGQTQVNST---QG------PGAHRGFLD--
LDPIWVFKLLKKMEKAFMTHFVSATAMVRAHWGLQNNDLLDQMVAELEQNVGWRLQDSIK
RELRKIQIRARRKAA------RPLRWEMSMQTEQRRRSLQGL----RNLSAFTEQTRAHG
------LFSLTLEDEP-TVSGAQRNELGGKAKGEEFCPCKACMRKKVSPIPSRDTMG---
AGSAPIKKAFDLQQILQK---RKGGPNNGEAVEEAPE-EGGMGLLQVAS----LRT----
-----RSVQGVYGELEPGLGLSSGADEGDE-------IEGKQKLSGDEDP----------
------------------------------------------------------------
------------------------------E----L---G---E---G---E---G---A
---S---Y---Q---E--G---E--E---D---T--D---T--Q---K---R--DR---K
---T--GI---S-V-G--N---N---L---E--K---E---E--W--E---E---Q--H-
--V---G---E--Q---E---E---N----------------------------------
-------------D--K---A---S---G---R---G---N---R--G---P---G-G--
-D---V---D---G---T---D---A---I---K---A---Q---K---A---E----G-
--E----G--------------------------------------QT------------
ELRDEKEGSPPVSQGEVQL-GKASENSCPDQEGK--LASPAA-PSGDTPYQRSAS-----
--KTVNSSSSMSSLGNCSQVSQKGSEEEHPNGDTRSIADEP------------K------
-----------GITHQERKVTRMYFKSSTSE-----GEGA--------PSGPSTPDQGAD
DDPDLEDRKIVK------------------------------------------------
----SLTFTKVVD---RADEFGQDDLDF--------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
ENSMPUP00000001748 (view gene)
-----MNSTL---RDAQA-----LSHRECLLPSMARTPSVTQVTPAKKITFLKRGDPRFA
GVR
PF03607 // ENSGT00920000149015_4_Domain_0 (64, 124)
LAVDQRAFKSFGALMDELSQRVPLSFGVRSVTTPRGLHGLSALEQLEDGGCYLCSDK
KPPENSGT00920000149015_4_Linker_0_1 (124, 190)
KTPSRP--GW----SQ--WRSPSTQQSRDFESRCEAPGT-SSSCKGPKAPRKIMLVK
NGDPRFQQTPF03607 // ENSGT00920000149015_4_Domain_1 (190, 250)
LVLSHRNTRNMTAFLSKASDLLHFPVKQVYTTSGKKV--DSLKGLIHSPSV
LVCAGYESFKPLAMEDSRRHGIETLSGQTPRNKT-----GNWGPKAKQSVIHSRSRSGS-
RPQQFSLLSERSGLSNPPVSLHHAQMDPAHDGHPQNIPAQ--LGPLVASDDVEKKIRMNE
DGSLSVEMKVRFHLLGEDALFWSRRVGRASALTAASEEVPVLGEVGSLHCVWEDHPGA-S
EHKAQGQGPCEA----------GCEEACGQGLWQPGSRY-EIWM-NPLY-STQGEG--MA
SRKRSRLTQHSHS-RRPCSKGFTRRKR-SSKDNVSPAS-SDRPPKDSESNPSCCSRSPEG
SVGSCDLHSASGAAS-----QRGAGWETDGP------SRTREAPGQQGAGHGSRE-----
------------------------------------HHSCLKPQTRGTVGALSDSSGSAG
SH------EQSSERSEQHQGFQSQTRAMT--TSRTKATQGEGPSLP--------------
-------------------------TVSPLSLRNEDSQAEENGQGT--------------
---RSTQAEGRSGMRLPL------APG-HSGSGDTAEGSSPPSACTSV-PGKRRKKGRAS
ALSSPSISGLSRGAQIGCPSQHHTR----RHTHCPPDSPMPRQMP----GPPSRGRACPN
GPAPPLSASSPSTRNQASQDTGSTFSASLHSQYAQ-GVSSALITPVSNL--------DCA
SNFYS-------------------PYPPSAEAEVDPEFRACSSAPTPSNTSD--------
--------------------SLSNQA--DGLDKKAGGSIPKSSWPLVLPVGQPDTGMPGA
HQGCSCSSISTTS---------------QPLRSLGPFSEVSLACSRYCPTPPRTWPFVKK
HPSCS-------SSPRADCRPGGGELREGMLDAWHPRPPGSQSGAMGKAVKALRRGSSSC
GPRS---GRIYQGKKVTGRGENLEGQEKDGRMVLGALPQASPDAVVREWLSNIPEEPIPM
KCEMVDESTDVAGNDPEGPKEDPVDQHSPECLREPTQVRQ-LSHEGANSEKAEP--GEAF
PITGDTDPKSGEGLPC------NGVSEDPREAGAGKGMA---------ADCRVGQCVLPH
KVSASIQIMKVLMGSKQ----GRPSSLPEVSNTVGRRLSHSAQALITCLAGLHFFEE-DL
GPPTGKVRFIDSSRYQELLSTIQALWP-GCDLRRGEL--DLGLWQLGWRQALPGLRSD-I
VTEDFTPTSSSGVDVHSGSGGSGEGSGPCVMEC-ALVPEKIALPLK-----IPSQRSDSR
NSESQED-----QGNQHPSGSTA--------SSNSQAWTCASRKD---------------
--------------------------------------------------------ETER
N-SGGQVLGHNLDQEVENT-------------------VQDEEALLEKVGE-EKEIAAL-
QG----------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------EGV-HGFPE-EER
VMGQEMSEANFQDGE-GAQEVKSGQEEEAGEDPASTLLCPPGEREK--PTELPRNLSERD
PNT-----SGPQMEHGLEKLPKAAEPC--HVQIQAKHT---QGPGDRSSSQAHRVSLD--
PDALWVSRLLRKMEKAFMAHLASATAELRARWSLQNDRLLDQMVAELQQDLDWRLRESTE
NELRKIQSRAGGKT----PAREALRWELSLQTEQRRRRLQGL----RHLSAFSGQAGSQG
------PGSFPMEEDASTLSGTLGTWLVGEPEGEEFCPCEACIRKKVIPMSPKDTTG---
APRAPIKKAFDLQHILQK---KKGGCASGEAAKTSSK-EERMEPLQVDP----LGT----
-----GTIQESNGGLELGLGQDPGPEEQDE-------GENSQTLGRDGDC----------
------------------------------------------------------------
------------------------------R----G---E---E---E---E---A---T
---A---Q---K----------R--E---G---I--T---G-------------A-----
-------D---R---G--S---V---P---E--G---A---G--E--E---E---Q--G-
--S---D---G--K---E---E---N--T---E---E--G---S--G---A---D---T-
--S--L---E---G--E---A---S---G---R---E---D---P--S---L---G-G--
-Q---D---D---G---V---K---T---P---E---T---W---E---A---E----G-
--E----G--------------------------------------QSELGGG-------
-NQGEKEGSPEAGSGQGQL-AETSGNSSPDQEGE--------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
ENSPVAP00000009834 (view gene)
-----MNSTP---RDAQA-----PTYRECLLPSVARTPSVTQVTPAKKITFLKRGDPQFA
GVR
PF03607 // ENSGT00920000149015_4_Domain_0 (64, 121)
LPVHQRTFKSFSALMDELSQRVPLSFGVRSVTTPRGLHGLSALEQLEDGGCYLCSDKKLPKTSSGP--GL----PQ--GRSPSAQQSRDVEGQCEAPGT-SASRRGPRAPRKIMLVK
NGEPGSQQTVTLSHRNTRSLTAFLSKASDLLHFPVKRVYTTSGEKV--DSLKALLRSPSV
LVCAGHESFRPLAMEGARRKGTETLLGQTSRNKN----XXXXXXXXXXXXXXXXXXXXX-
XXXXXXXXXXXXXXXX-------XXXXXXXXXXXXXXXXX--XXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXX------------XXXXXXXX-XXXXXX-XXXX-XXXX-XXXXXX--XX
XXXXXXXXXXXXX-XXXXXXXXXXXXX-X-XXXXX---------XXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXX-----XXXXGWEAGGN------PAGE-VPGPE--GARGSR-----
------------------------------------EQRCLKPRTQDVAGA-SDSSVSTG
SH------EASSERGEPHQGL-SKTRTVT--SLGKAT-EGGGPSSP--------------
-------------------------TLSASSLRNKDPQGEESGQGT--------------
---MRPQDRGGSGTRLPL------AAG-HCGSVDTAGGCSPPSACVSAMGGRRKQGSRAS
AFSSPSISVLSRGAQKGCPRQHHNR----RDAHCPLHSPVSRQRP----GPPSVSRARPV
-------PRSPSTRNRASRGAGPPSSASLHSQEAR-GVSSAPFTSVSDS--------DCA
SNFYP-------------------PYSPSAETEGDPEFRAHSPTPTSSNFF---------
-----------------------RSQ--ADGLGKKAWGMPQPPWSLVLPDGQDNGEKPES
RQGCCCSQAHPPQ-----------------DAWREPLSEASSVCSRYCPHPPRAQPSVKK
HPSGSSSHS--GGDHSADWGPRGAQQGRKSWTA--PTPPGSQSR-------ASRRGSPSL
GPRP---SRMFQGQV-TGAEEGLQEQEDGGSVSPGALPRVSPEAVVREWLNNIPEEPVPM
KYEMVDDSTDVAGDGQEGPTEDPIEKHSPEGLGEPSQPRG-PSLEGTANEKAEP--DGTL
PVMSDAGPQSAESLSA--------GSEAPKEAGAGGGTT---------EDRGPGQXXXXX
XXXXXXXXXXXXXXXXX----XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX-XX
XXXXXXXXXXXXXXXXXXXXXXXXXXX-XXXXXXXXX--XXXXXXXXXXXXXXXXXXX-X
XXXXXXXXXXXXXXXXX--------XXXXXXXX-XXXXXXXXXXXX-----XXXXXXX-X
X--XXXX-----XXXXXXXXXXX--------XXXXXXXXXXXXXX---------------
--------------------------------------------------------XXXX
X-XXXXXXXXXXXXXXXXX-------------------XXXXXXXXXXXXX-XXXXXXX-
XXXXXXXXXXXXXX-X-XXXXXXXXX-X-----XXX----XX-------XXXXXXXXXX-
------------XXXXXXXX------X----XXXXXXXXXXXXXXXXXXXXXXX------
--X----XXXXXX-X----XXX---XXXXXX-------X---XXXXXXXX-XXXXX-XXX
XXXXXXXXXXXXXXX-XXXXXXXXXX---XX-X--XXXXXXXXXXX--XXXXXXXXXXXX
XXX---XXXXXXXXXXXXXXXXXXXXXX-XXXXXXXXX---XXXXXXXXXXAHRMSLD--
PDPLWVSMLLKKMEKAFMAHLASATAMLRARWSLPSDDLLDQMVAELQQDMRQRLQDSTE
KELQKIRSRAREKAR--GPPRGALRWETSLQTEQRRRRLQGL----RNLSAFSEQTA-LG
------HPSLSQEDVP-AFSEALETPLGREAEGEEFCPCEACMKKNVIPASQKDTMG---
VARTPIKEAFDLKQILQK---K-GGCANRDTAEVVPE-RTGMELPQRVP----SGT----
-----GTVQGTDGGL----------EEEDE-------GKGSQTLGRGEDP----------
------------------------------------------------------------
------------------------------E----R---E---E---E---D---V---A
---T---Q---N---V---------E---G---S--------------------T-----
-------P---A---S--C---H---P---E--A---A---E--W--E---E---Q--G-
--V---G---E--G---G---E---H--G----------G---S--G---A---E---V-
--G--V---E---G--E---A---S---G---G---G---E---R--S---P---G----
-Q---N---D---G---A---D---T---I---E---A---Q---E---T---E----G-
--E----G--------------------------------------QPELRGG-------
-DQDEKERTPQVGSRQGQS-GEASGHSSPDQQGR--PMPPPA-PGGDAP-----GQRSGL
KTAP--SSSRTSSLGNCSRLSQKGSEEEPSKGDMRTTAD------------EPKGVPGP-
----------------ERTVIGMYPEGSTSE-----QEGA--------TSGSRTPERGTE
EGDS-----------------------------------------------REGKVIKSL
AWTE------MKN---TTDGFGQDDLDF--------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
ENSSTOP00000020260 (view gene)
-----MSSSP---RDTQG-----PSHRQGLLPSV------THVPPAKKITFLKRGDARFA
GVRL
PF03607 // ENSGT00920000149015_4_Domain_0 (65, 122)
AVHQRSFKTFSALMDELSQRVPLSFGVRSVTTPRGLHGLSALEQLQDGGCYLCSDR
KENSGT00920000149015_4_Linker_0_1 (122, 191)
PPKTPSGP--GQ----LR--GRSPSALQSRDLDGGREVSGT-SSSWKGPRAPRRITLVK
NRDPRFQETVPF03607 // ENSGT00920000149015_4_Domain_1 (191, 249)
VLSHRNTRSLAAFLSKASDLLRFPVRQIYTPSGKKV--DSLQALLRGPST
LVCAGNEAFRPPALESAQKNKTETLSGMTSRNKHSEFALGCWGPKAKQSVIHLRSTASS-
RPPQFSLS----GLRDPSASLH-----GAPHRHPQDT-----SGPLLAGDNVEKKVCMNE
DGSLSVEMKVRLHLLGEETLLWSHRAGRAGTLTAASGEGPVLGEGSPLCCRWGGHPWGFS
ESGAQRPGPCEA----------GPQGAFDGGQ-QPRPSF-EIWR-NPLY-VPQGEG--AA
PWRRSGLAQLSQC-RGRWSRGARNRER-RSRDSGSPSS-SAGHGEGSEPD-SCSPRTPEG
GVGSDSPCPASTAAS-----QCGDEREAGEG--------------------SCPG-----
------------------------------------EHGCPGPGIPGAKGALSDSLASTG
PR------EESSKWDGQHEGCLSPVRVRT--SQEKGSQ-GSRTDDP--------------
-------------------------TRSLSPLSDGDSQAEQCEQGT--------------
---THQQATEGPVAGPPL------ALG-RSHSWDAEGGCPGLLAHAPAQLGRRKRQSPSS
AVSSPGTA---GVVQRGRARRCHHC----RDTHCLPDSPTALQMP----R------TGPG
GPGPQLSQNSPSTRKQASRDLGPSVPGSLDSQDLP-EASSASTTLLSNS--------DCA
-----------------------------AELAGDTEGKVCSPEPLQ-------------
-------------------------G--GGSLGDQAEDTLEPSLPLALPVGCPEGEEPGA
HKSCCCPQIGSSPAA------------------WGPFSEACWVCRGYCPTPPRGRPCSRK
HPTSSSSTS--SDHQRADDGGPGS------------------------------------
--GP---KRVLLGTS-SGNGGGLEEPREDGTMTPHDLPHSSPDAMVREWLDNIPEEPLHV
KHEMVAEATSVVSNIPEGPREDPEGDHSLSSTGEQAQAGR-QALEGGT---HEP--DRTP
PGTGDAGSKPGDHSHQG--TAPGQASQAASEARVGEGAT---------VGPGMSPCVLPG
RDSASMQIMKALMGSKH----GRPSSLPEVSSAEARRLRRLAGDLIACLARLHFFDE-DL
GSPAGKVRPTHSARYQELLNISQTLWP-GCGLGRDML--DLSLGDLTSLQAPP-------
VTDNFTPTSSSGVDVSSGSGGSGEGSVPCATDG-ALVSERTELPLE------ISQRPDSR
TSGHLED-----LGTRQP----T--------APSSQVCACAASGE---------------
--------------------------------------------------------GGGK
----------GPEGFVGNT-------------------LQEAEAQSEETEETESP-----
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------------PE------EGLEE-VGL
LGEAGVSGEESPGGE-GTLGDPGVPGEEAGGDSASAGLCPPGSAEE--PAECPRS--ERD
SHA-SESQTGPTSKSSLEELSRADALG--CE------------PAPASASAAHRVFLE--
PDPAWVSKLLRKIEKAFMKHLATATAELRARWDLQDDPLLDQMVAELEQDMGLRLQESTN
RELQKIESRTGRMAL--GPPREVIRGQASLQTEQRRRRLQGL----HNLLAFSEQTYARA
------LHSLTLEDRP-TLHEALGDLPCGETEEDEFCPCEACIRKKVAPVPPKDTPG---
ASSAPIKKAFDLQQILQNLQKKKEGGTDGEAEERVPE-GTGTELPRDIP----ARA----
-----GSV----------QDLGPTVGQERE-------GEGSQRPSRDEDA----------
------------------------------------------------------------
------------------------------Q----L---E---E---A---E---E---A
---G---Y---Q---E--T---A--G---D---A--E---A--G---R---A--H-----
-------G---H-H-G--S---T---A---E--E---A---Q--Q--L---E---R--A-
--E---T---E--K---E---G---A--L---G---E--H---S--G---E---------
----------------------------G---D---A---S---E--A---A---G-G--
-P---G---D---L---V---D---A---I---E---A---Q---E---A---G----G-
--G----R--------------------------------------RLESTGG-------
-SQGEEEDGPLASPGQGQR-REASENSVTDQEDT--PHPEPD-PQT--------------
----HDRSSRTFTLGNCSLVSQKGSEEVQAGGEPGDTGAES-------------------
------------EPPAQEKVTSMYPESSTSE-----REEP--------PSDPETLEQGSG
RDAGEGNEKA--------------------------------------------------
--------PQSSP---RADGFHQEDLDF--------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
ENSMLEP00000032547 (view gene)
-----MNSTP---RDAQA-----PRHRECLLPSVARTPSVTKVTPAKKITFLKRGDPQFA
GVRL
PF03607 // ENSGT00920000149015_4_Domain_0 (65, 122)
AVHQRAFKTFSALMDELSQRVPLSFGVRSVTTPRGLHSLSALEQLEDGGCYFCSDK
KENSGT00920000149015_4_Linker_0_1 (122, 191)
PPKTPSGP--GR----PQ--GRSPTAQQLRDIQGRREAPGT-SSSRKSLKTPRRILLVK
NMDPHLQQTVPF03607 // ENSGT00920000149015_4_Domain_1 (191, 249)
VLSHRNTRNLAAFLSKASDLLRFPVKQLYTTSGKKV--DSLQALLHSPSV
LVCAGHEAFRPPAMENARRSEAETLSGLTSRNKN-----RSWGPKTKPSVIHSRSRPGS-
RPQL----PERPVPSN-------PPVGPAPDRHPQDTPAQ--PGPLVAGDDMKKKVRMNE
DGSLSVEMKVRFHLVGEDTLLWSRRMGRASALTAASGEDPILGEVDPLCCVWEGYPWGFS
DPGVWGPRPCRV----------GCGEAFGRGG-QPGPKY-EIWT-NPLH-ASQGER--VA
TRKRWGLAQHVRC-SGLWGHGAAGRQR-CSLDSASPAS-STGHPEGSEPDSSCCPRTPED
RVDSG------SPSA-----QRGAEQEAGGS------LGEDPSLCTDGAGLGSPE-----
------------------------------------QGGHLTPRAQSEEGASSDSLASTG
SH------EGSSEWGGQPRGCPGKARAET--SQQEATK-GGNPGSP--------------
-------------------------APSLSSLRSEDLQAEMQGRGT--------------
---KQ--ARGASVKREPL------VPG-LSGSWDSEGASSTPSTSASSQEERRRHRSRAS
ATSSPSSPGLGRVAPRGHPRQCHYC----KDTHSPLDSSVTKQVP----RPPERRRACRD
GSVPRSSGSSLSTRTQDSGNLRPPSLGSLPSQDLL-GISSATVTPAGRS--------DCA
SGVSP-------------------CNAPSVGWAGDTGSRTCSPDPTPPHTSD--------
--------------------SCSKSG--AASLEEQARDTPQPSSPLILQVGRPEQGAAGP
HQSCCHSQAGTQPAE---------------EAQRGPSPEAHWLCGRYCPTPPRGRPCPQR
RSSSCGSTG--SSHRSAARRPGGSQQKEGTHQAGPTPSLGPGSG-------ASRRSSASQ
DVGS---GGLSQEET-SRSWGGPQGQEEASGVSPSSLPRSSPEAVVREWLDHIPEEPVLM
TYEMADDTTGAAGGGLRGPEVDPGDDHSLEGLGEPALARQ-QPLEGDPGQDLEP--EGAL
PGSSDAGPQSGEGVPQG--AAPEGVSEAPAEAGADREDP---------ASCGVSLRALPG
RVSASTQIMRALMGSKQ----GRPSSMPEVSRPMARRLSRSAGALITCLASLQFFDE-DL
GSPASKVRFKDSPRYQELLNISKDLWP-GCDLGEDQL--DSGLWELTWSQALPDLGSQ-A
VTENFTPTSSSGVDISSGSGGSGESSVPCAMDS-TLVPGGTELSLK-----TSNQRPDSR
TSENPGD-----LENQQQCCSPA--------FLNSRACACATNED---------------
--------------------------------------------------------EAER
D-GEEQRASLNLEQLAQNT-------------------MQE-EVQLEEKET-ETE-EGL-
QEEGVQLEETKTEG-------------------RLQ------------------------
--------------------------------------------EEGVQLEETNGTEG-E
GQQE-EAQLEEIKEETGGEGLQEEGVQLEEVKEGPEGGLQEEALEEGLKE-EGLPE-EGS
VGRQELSDAGSPDGE-GSWEDDPIQEEEAGRAS--AEPCPPEGTEE--PTEPASHLSETD
PSA-SDSQSGSQLEPGLEKPPGAAMMG--QEHTQAQPP---LGAAERSSSVACRAALD--
CDPIWVSVLLKKMEKAFLAHLSGAVAELRARWGLQDSDLLDQMAAELQQDVARRLQDSTE
RELQKLQGRAGRMVL--GPPREALTGELLLQTQQRRHRLRGL----RNLSAFSERTLGLG
------PFSFTLEDEP-ALSTALGSQLGEEGEGEEFCPCEACVRKKVSPASPKATMG---
ATRGPIKEAFDLQQILQR---KREE-HTGGEA----A-EVAPGETCTDP----TST----
-----RTVQGAEGGPGLGLGHGPGVDQGED-------GEGSQRLDRDKDP----------
------------------------------------------------------------
------------------------------K----L---G---E---A---E---G---D
---T---M---A---Q--E---R--E---G---K--I---H--N---S---E--T-----
-------S---V---G--S---E---L---G--E---A---E--Q--E---G---E--G-
--I---N---E--R---G---E---T--G---G---Q--G---S--G---H---E---D-
--N--L---Q---G--E---A---V---A---G---G---D---Q--D---P---G----
-Q---S---D---G---A---E---G---I---E---A---L---E---A---E----G-
--E----A--------------------------------------QPES--R-------
-GIEAQEAEEEAPE--------AE------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------GEAQPESEGVE-------------------
-----APEVEGEM---QEAEGEAQPESE--------------------------------
-GIEAPEAEGE-------------------------------------------------
----AQEA--EGE-----------------------------------------------
----------------------------------AQPES---------------------
------------------------------------------------------------
----------EGAQPESEGVE--ALEAEGEAQPESEGE--------TQGETKGSPQVSLG
DGQSVGASESNSPVPEDGPTPPPSPGGDTPHQKPA-SQTGPASSRASSQG--HSWQKDSE
DGHVCGDTR-SPDAKSVGAPHAERKATRMYPESS-TSEQEEAPSDTRSPEQGAS------
---EG-CDPQEDQTLGNPAPTKAVGRADGFGQDDLDF
ENSFCAP00000028643 (view gene)
-----MNSTL---RDAQA-----PSHRECLLPSVARTPSVTQITPAKKITFLKRGDPRFA
GVRLA
PF03607 // ENSGT00920000149015_4_Domain_0 (66, 122)
VDQRAFKSFGALMDELSQRMPLSFGVRSVTTPRGLHGLSALEQLEDGGCYLCSDK
KENSGT00920000149015_4_Linker_0_1 (122, 191)
PPKTPSGP--GW----PQ--GGTSSAQQSREFESRGEAPGT-SSSCKGPKVPRRIMLIK
NGDPRFQKTVPF03607 // ENSGT00920000149015_4_Domain_1 (191, 249)
VLSHRNTRNLTAFLSKASDLLHFPVKQVYTTSGEKV--DSWNGLLHSPSV
LVCAGYESFKPLAMEDSRRYGIETLSGQTSRNKT-----GSWGPKAKQSVIHSRSRSGS-
RPRRFSLLSERSGLSDPPVSLHHAQMGPPSERYPQDTPTQ--LGPLVASDDVEKKVHMNE
DGSLSVEMKVRFHLLGEDALLWSRRVGRASVLTGANGEVPVLGEVDPLHCVCESHPGGSS
EPGAQGLGTCEA----------GCEKALGRGRWKPGSRY-EIWM-NPLY-SAQEEG--TA
SQRRSRLTQHSYS-RRPWSQGVTGRKR-SRKDSVSPDS-SDRPPKDSEPNSSCCSRSLEG
SVDSCDLHAASGAAS-----QREAGREAGNM------CRTSKDPGPQGTGQD-RE-----
------------------------------------HHRCLKQRSGGTASATCESSGSAG
SH------EESSEMGEQHQGCLSKTSVMT--TSRPKATQRKGPSLP--------------
-------------------------TVSPSSLRNKDSQAEESRQGS--------------
---RCPQTRGRSGMGSPL------VSD-QSGSGDTAESCSLPSACASV-PGRRKQKGRSR
AVSSPSISSHSRGARIGHPRQHPNR----RDIHCPLDSAVPRQVP----GPSSGGRACPD
SPAPHLSGSSPSARNQASWDAGPPSSASLHSQDVQ-GISSALMTPVSNS--------DCT
SNFYP-------------------PYSPSAETEGHSELRARSPAPSPSNTSD--------
--------------------PLSIQA--DGLDKKAGNDLPKPSWPLVPPVGEPEGGMPGA
HRDCCCSPISTPLFHETPSDNTKTLQTSQPSGSQGPSSEVSLVCSRYCPTPPRTWPFVKK
HPSGS-------SSLGADWGPEGRKLEEEKLDARPPRPPGSQSGAMGKAVKALRKGSSSC
GPRS---GRMLQGKV-AGGGESLEEQEEDGRMMLGALPRASPDAVVREWLSNIPEEPIPM
KCETVDESTVVVGGDPEGPKEDNADKHSQEGLGEPVLARQ-LSHEGATSEKAES--DGAL
PVTSDAHSKSGEGHPC------SRVSEVPRETGSGKGTA---------VDCGVGQCVLPH
KVSASIQIMKVLMGSKQ----GRPSSLPEVSSAVGRRLSHSARALITCLAGLHFFDE-DL
RSPTDKVRFTESPRYQELLSTFQALWP-GCGYRQGEL--DSGLWELGWCKALPGLRSH-V
MTEDFTPTSSSGVDVGSGSGGSGEGSGPCVMDS-ALVPERIELPLK-----IPSQRPDSR
TSKNQED-----PEKQQSSVSTA--------SSNSQEWACATRKD---------------
--------------------------------------------------------KAER
N-SGEQVLGSNLDQVVENA-------------------MQEEGVQLEKVEE-EKEIAEL-
PR----------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------EGV-QGSPE-EGR
VMGQELSEADSDDGE-GAQEDKSVQEEEAGGDPVSTTLCPPGTREK--PPEPPRSPSRRD
PDA-SESHSGPNNEPGLEKPPQTTETS--HEQTQTKFL---QGPGEKSSSTACRVSLD--
PDILWVTKLLRRMEKAFMADLARATAEIRARWSLQSNDLLDQMVAELQQDVGQRLQDSTE
KELHKIQSRAAGRMP--GPAREALRWEMSLQTEQRRRRLRDL----RNLSAFSEQARSQG
------LLSFPVEDVP-TLHGALGTWLVGEPEGEEFCPCEACMRKKVIPTSPKDTKG---
VTSAPIKKAFDLQHILQK---KKGGCANGEAAETVPE-KREMELLQGDA----SGI----
-----GTID--NGGLELGLGQDPGTEEGGE-------DESSQSLGRDGDP----------
------------------------------------------------------------
------------------------------Q----V---G---E---E---G---E---V
---A---Q---N---G---------E---E-------------K---T---D--P-----
-------G---Q---G--S---V---F---E--G---V---G--R--E---E---Q--G-
--F---G---E--K---D---E---N--T---E---K--G---S--R---T---E---G-
--S--L---E---G--E---A---W---G---E---G---D---Q--S---P---E-G--
-Q---N---D---G---A---V---T---T---E---G---D---R--------------
----------------------------------------------KPESGGG-------
-DQGEKEGSSQVVWVQGQQ-GETSGNSSPDQEGK--PAMLPT-PDG-----DTPCQRSGW
KAGLAAS--STFSLGNCSQLSQKGSEEKPSNGDMRSIEDESKV------------IPSP-
----------------ERKVTGMYPESSTSE-----QEVA--------PLGPRTPEQEVG
EASGLEAENVVE----------------------------------------------SL
SFTEMRSKNLTKD---RTDGFGRDDLDF--------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
ENSMFAP00000033163 (view gene)
-----MNSTP---RDAQA-----PRHRECLLPSVARTPSVTKVTPAKKITFLKRGDPQFA
GVRL
PF03607 // ENSGT00920000149015_4_Domain_0 (65, 122)
AVHQRAFKTFSALMDELSQRVPLSFGVRSVTTPRGLHSLSALEQLEDGGCYFCSDK
KENSGT00920000149015_4_Linker_0_1 (122, 191)
PPKTPSGP--GR----PQ--GRSPTAQQLRDIQGRREAPGS-SSSRKSLKTPRRMLLVK
NMDPRLQQTVPF03607 // ENSGT00920000149015_4_Domain_1 (191, 249)
VLSHRNTRNLAAFLSKASDLLRFPVKQLYTTSGKKV--DSLQALLHSPSV
LVCAGHEAFRPPAMENARRSEAETLSGLTSRNKN-----RSWGPKTKPSVIHSRSRPGS-
RPQP----PERPVPSN-------PPVGPAPDRHPQDTPAQ--PGPLVAGDDMKKKVRMNE
DGSLSVEMKVRFHLVGEDTLLWSRRMGRASALTAASGEDPILGEVDPLCCVWEGYPWGFS
DPGVWGPRPCRV----------GCGEAFGRGG-QPGPKY-EIWT-NPLH-ASQGER--VA
TRKRWGLAQHVRC-SGLWGHGAAGRQR-CSLDSASPAS-STGHPEGSEPDSSCCPRTPED
RVDSG------SPSA-----QRGAEREAGGS------LGEDPSLCTDGAGLGSPE-----
------------------------------------QGGRLTPRARSEEGASSDSLASTG
SH------EGSSEWGGQPRGCPGKARAET--SQQEATK-GGNPGSP--------------
-------------------------APSLSSLRSEDLQAEMQGRGT--------------
---EQ--ARGASVKREPL------VPG-LSGSWDLEGASSTPSTSASSQQERRRHRSRAS
ATSSPSSPGLGRVAPRGHPRQCHYC----KDTHSPLDSSVTKQVP----RPPERRRACRD
GSVPRSSGSSLSTRTQASGNLRPPSLGSLPSQDLL-GISSATVTPAGHS--------DCA
SGVSP-------------------CNAPSVGWAGDTGSRTCSPDPTPPHTSD--------
--------------------SCSKSG--AASLEEQARDTPQPSSPLILQVGRPEQGAAGP
HQSCCHSQAGTQPAE---------------EAQRGPSPEARWLCGRYCPTPPRGRPCPQR
RSSSCGSTG--SSHRSAAQRPGGSQQKEGTHQAGPTPSLGPGSG-------ASRRSSASQ
DVGS---GGLSQEET-SRSWGGPQGQEEASSVSPSSLPRSSPEAVVREWLDNIPEEPVLM
TYEMADDTTGAAGGGLRGPEVDPGDDHSLEGLGEPALARQ-QPLEGDPGQDLEP--EGAL
PGSSDAGPQSGEGVPQG--AAPEGVSEAPAEAGADREDP---------ASCGVSLRALPG
RVSASTQIMRALMGSKQ----GRPSSMPEVSRPMARRLSRSAGALITCLASLQFFDE-DL
GSPASKVRFKDSPRYQELLSISKDLWP-GCDLGEDQL--DSGLWELTWSQALPDLGSQ-A
VTENFTPTSSSGVDISSGSGGSGESSVPCATDS-ALVPEGTELSLK-----TSNQRPDSR
TSENPGD-----LENQQQCCSPA--------FLNSRACACATNED---------------
--------------------------------------------------------EAER
D-GEEQRASLNLEQLAENT-------------------MQE-EVQLEESKE-ETEGGGL-
QEEGLQLEGTKETE----G--------------LQE------------------------
---------------------------------------------EGVQLEETKT--E-E
GLQEEGVQLEEIKEETGEGLQEEGGAQLEEVKEGPEGGLQEEALEEGLKE-EGLPE-GGS
VGRQELSDAGSPGGE-GSWEDDPIQEEEGGRAS--AEPCPPEGTEE--PTEPASHLSETD
PSV-SESQSGSQLEPGLEKPPGAAMMG--QEHTQAHPP---LGAAERSSSVACRAALD--
CDPIWVSVLLKKMEKAFLAHLSGAVAELRARWGLQDSDLLDQMAAELQQDVARRLQDSTE
RELQKLQGRAGRMVL--GPPREALTGELLLQTQQRRHRLRGL----RNLSAFSERTLGLG
------PFSFTLEDEP-ALSTALGSQLGEEGEGEEFCPCEACVRKKVSPASPKATMG---
ATRGPIKEAFDLQQILQR---KREE-HTDGEA----A-EVAPGGTCTDP----TST----
-----RTVQGAEGGPGLGLGHGPGMDQGED-------GEGSQRLDRDKDP----------
------------------------------------------------------------
------------------------------K----L---G---E---A---E---G---D
---T---M---A---Q--E---R--E---G---K--I---H--N---S---E--T-----
-------S---V---G--S---E---L---G--E---A---E--Q--E---G---E--G-
--I---S---E--R---G---E---T--G---G---Q--G---S--G---H---E---D-
--N--L---Q---G--E---A---V---A---G---G---D---Q--D---P---G----
-Q---S---D---G---A---E---G---I---E---A---P---E---A---E----G-
--E----A--------------------------------------QPES--E-------
-GVEAREVEGEMLE--------AEGEAQPESESV--EAQEAQ-PE---SEGIEAPEAEGE
A-PPESEGVEAPEAEKEAQPETESVEAP-------ETEGEAP----------------PE
SEDA-EAREAGE---------------EAQE-----AEGE--------------------
-----DQPESEV-------I--EGEMQEAEGEAQPDSEGVEAPEAEGEAQEAEGEA-QPE
SEVVGAPEVEGEM---QEAEGETQPESEGVEAQEAEEK---AQEAGEEAQESEGEAQPES
ESVEALEAEGEAQ-----PESESVEAQEAEGEAQPESESVEAQEAEGEAQPESEGVETPE
AEEEGQPESEGVEALEADEEA-----------------------QPESEGVEAPEAE---
----GEAQPESEGV------------KAPEAEGEAQPESEG-EAQEAEGEAQPESEGVEA
PEAEGEAQPESEGVEAPEAEG---------E-----------AQPESEGVEALEAD----
----------EEAQPESEGVE--ALEAEGEAQPESEGE--------TQGEKKGSPQVSLG
DGQSVGASESNSPVPEDRPTPPPSPGGDTPHQKPA-SQTGPSSSRASSQG--NSWWKDSE
DGHVCGDTR-SPDAKSVGAPHAERKATRMYPESS-TSEQEEAPSDTRSPEQGAS------
---EG-CDPQEDQTLGNPAPTKAVGRADGFGQDDLDF
ENSNLEP00000018242 (view gene)
-----MNSTP---RDAQA-----PSHRECHLPSVARTPSVTKVAPAKKITFLKRGDPRFA
GVRL
PF03607 // ENSGT00920000149015_4_Domain_0 (65, 122)
AVHQRAFKTFSALMDELSQRVPLSFGVRSVTTPRGLHSLSALEQLEDGGCYLCSDK
KENSGT00920000149015_4_Linker_0_1 (122, 191)
PPKTPSGL--GP----PQ--ERNPTAQQLRDIEGQREAPGT-SSSRKSLKTPRRILLIK
NMDPRLQQTVPF03607 // ENSGT00920000149015_4_Domain_1 (191, 249)
VLSHRNTRNLAAFLSKASDLLRFPVKQLYTTSGKKV--DSLQALLHSPSV
LVCAGHEAFRPPAMKNARRSEAETLSGLTSRNKN-----GSWGPKTKPSVIHSRSPPVS-
RLRL----TERPSPSN-------PPVGPARDRHPQDTPVQ--PGPLVAGDDMKKKVRMNE
DGSLSVEMKVRFHLVGEDTLLWSRRMGRASALTAASGEDPVLGEVDPLCCVWEGYPWGFS
EPGVWGPRACRV----------GCREAFGRGG-QPGPKY-EIWT-NPLH-ASQGER--VA
ARKRWGLAQHVRC-SGLWGRGAAGRER-CSQDSASPAS-STGLPEGSEPDSSWCPRTPED
GVDSG------SPSA-----QIEAERKAGGS------LGDNPSLCTDGAGLGSPD-----
------------------------------------E-----------EGLSSDSSASTG
SH------EGSSEWGGRPQGCPGKARAET--SQQEATE-GGDPGSP--------------
-------------------------ALSLSSLRSEGLQAEMEGQGT--------------
---EQ--ARGASVTREPL------VPG-LSGSWDSEGTSSTPSTCASSQQGRRRHRSRTS
AMSSPSSPGLGRVAPRGHPRHSHCC----KDTHSPLDSSVTKQVP----RPPERRRACRD
GSVPRYSGSSSSTRIQASGNLRPPSSGSLPSQDFL-GTSSATVTPAGHS--------DFA
SGVSP-------------------HNAPCAELAGDTGSRTCSPAPIPPHTSD--------
--------------------SCSKSG--AASLGEQARDTPQPSSPSVLQVGQPEQGAVGP
HQSRCCSQAGTQPVQ---------------EAQWGPSPEARWLCGRYCPTPPRGRPCPKR
RSSSCGSTS--SSHRSAARGPGGSQQEEGTRQAAPRRP----------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------R--------
------------------------------------------------------------
------AP------FQ--------------------------------------------
------------------------------------------------------------
---GIEE-----SSASQAACFPT--------FLNSRACACAANED---------------
--------------------------------------------------------EAER
D-SEEQRASSNLEQLAENT-------------------MKEEEVQLEETKE-ETEGEGL-
QEEGVQLEESKETE-G-LQE----------------EGGVQLEESKTEGLQEE-------
-----------------------------------GGA----------QLEETKG-----
--------------TEGEGQQEEEGAQLEEVKEGPEGGLQEEALEEGLEE-EGLPE-EGS
VHRQELSEASSPDGK-GSLEDDPVPEEEAGRGS--AEACPPEGTEE--LTEHPSHLSETD
PSA-SESQSGSQLDPGLEKPPGATMMV--QEHTRAQPT---QGAAERSSSVACRAALD--
CDPIWVSVLLKKTEKAFLAHLASAVAELRARWGLQDNDLLDQMAAELQQDVAQRLQDSTK
RELQKLQGRAGRVVL--EPPREALSGELLLQTQQRRHRLRGL----RNLSAFSERTLGLG
------PLSFTLEDEP-ALSTALGSQLGEEAEGEEFCPCEACVRKKVSPTSPKTTMG---
ATRGPIKEAFDLQQILQR---KRGE-HTDGEA----A-EVAPGKTHTDP----KST----
-----RTVQGAEGGLG--LGQGPGVDEGED-------GERSQRLNSDKDP----------
------------------------------------------------------------
------------------------------K----L---G---E---A---E---G---D
---A---M---A---Q--E---R--E---G---K--T---H--N---S---E--T-----
-------S---A---G--S---K---P---G--E---A---E--Q--E---G---E--G-
--I---S---E--R---G---E---T--G---G---Q--G---S--G---H---E---D-
--N--L---Q---G--E---V---V---A---G---G---D---Q--D---P---G----
-Q---S---D---E---A---E---G---I---E---A---L---E---A---E----G-
--E----A--------------------------------------QPESE-G-------
-VEAQE----------------AKGEAQPESEGM--DAQEAE-KEAQPESEGVEAPEAEE
EAQPESEGVEAPEAEKEAQPETESLEAQEAEGEAQPESEGVGAQEAEK-------EAQPE
AESVEAQEAEKEAQPETEKSEGVEAQ----E-----AEKE--------A-QPETENAGGR
R------------------G--GP------------------------------------
---------------------------A-R--V-R----R----Y----R----------
------------------GPGGEGETQKTEGDAQPKSDSIEAQEAEEEA-----------
------------------------------------------------------------
--------------------------------------------------QEPESEGIEA
LAAGEEAQPEPEGVEALEAEGEAQPESEVVE--APEVEGE--M-----------------
------------------------QGAQGEAQPELEGE--------TQGEIKGSPQVSLG
DGQSEEASESSSPVPEDRPTPPPSPGGDTPHQRPG-SQTGPSSSRASSRG--TCWQTDSE
NDHVRGDIR-SPDAKSMGAPHAERKAPRMYPESS-TSEQEEAPSGPRTPELGAS------
---EG-CDLQEDQALGSLAPTKPVGRADGFGQDDLDF
ENSOGAP00000020959 (view gene)
-----MHSAQ---REAQA-----PGHRECLLPSVARTPSVTQVTPAKKITFLKRGDPRFA
GVR
PF03607 // ENSGT00920000149015_4_Domain_0 (64, 123)
LAVHQRAFKTFSALMDELSQRVPLSFGVRSVTTPRGLHGLSALEQLEDGGCYLCSDK
KAENSGT00920000149015_4_Linker_0_1 (123, 190)
PKTPSGP--GR----PQ--GRSPSAQRSWDCEGQREAPGT-SSSWKVPRAPRKILLVK
NRDPRVQQMPF03607 // ENSGT00920000149015_4_Domain_1 (190, 250)
VTLSQRDTKTLAAFLSKASDVLHFPVKQLYTMSGRKV--DSMQTLPRGTSV
LVCASNEAFRPLAVEDARKNGTETLAGLTSKNKT-----GSWGPKAKRSVIHARSRSGS-
RPRQSSLLSEQSRFSDPPASLDPAWVGPVPNRHPQDTPAQ--LSPLVASDDVKKKVRMNE
DGSLSVEMKVRFHLCGQDTLLWSRRVGKA------SGEGPDLGEADPLCCVWEGRPVGCS
VPGVWGPGPCGE----------GCEEVCGHGR-PSRPGY-EIWT-NPLH-TAQGKG--LA
SRRKLGLTQHPSC-RGPSSQGAIFRER-P-QDSISSPS-SSSHSEGSEPDSSCCPRTPEG
AVGSSGLSAQS-----------R--------------ARREDDPCLEAAGLGSRG-----
-----------------------------------------------AKGALSDSSAGAR
SL------QGASKWCRQHHSHSGEAGAVT--SQRKF-TQGSGPGPP--------------
-------------------------TQSPSSLKNEDLQAETCGQGT--------------
---GCRHAKWESVTRLPL------APG-HSGSWDTEGSSPTLPTCASAQRGTTNQ--RTQ
ALSLLNTPGLGRVAQRGHTRQCH---------HCLLNSPAAQPAS----RPPNQGRACPE
RPESQVSESSSSTQIRSSSYLRALCSGSLHSRDFS-GASSATISPASES--------DYT
PGSCH-------------------CDVPSAGWTGEAGSRAGSPPPSPPHTPQ--------
--------------------SCSQAR--AGSQREEAEDTPGPSSTLALQIGQPERGE---
-QGCCCSQIGAPPVQGVPRGNT--------QALWAPSPEAHWAGSRYCPTPPREK-----
RPPSCSS----GSFHSAARGPAGSQQGEESPVVGCMLTPGPQGP----------------
----------SQEKT-AVSGGGPEEREVCGGVTPGALPQASPDAVVREWLDNIPEEPVLM
KYEMVDEAACVAGDGPESPKEDPGGGRSLEGPGELVQVRP-QPPEGDAGENLEA--VGAL
LGAGSTGLPSGEGTPQG--AALGKDPKAPVEAREGGGVA---------VGCREGQRGLPP
RVSASTQILKVLMGFKQ----GRPSSLPEVSSPVAKRLSYSAGALVTCLARLHFFDE-DL
GSPTCKVKFTDSPQYQELLSISRTLWP-GCDLGRGQA--DLGLQDLTWSQALQGLGSH-A
VTENLTPTSSSGVDLSSGSGGSGESNVPCAMDC-TLGPERGELPQE-----TSHQRPDSR
ASEEAER-----LGHPHVSCSPA--------SPNSPALSE----G---------------
--------------------------------------------------------RAER
D---GGELGSDPGQVVGNT-------------------TQQEGAHLEKIGG-EIERQE--
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------LQEEGTEE-EGLPG-EGR
VGSQAWAGASCLGGE-GAPGEEAGKEEEDGRAS-----APVGRREQ--PTEPPGQPSERD
PDV-TGGQSGPRFEPSLEKLPEAARTG--REQTQALLP---LGARERSTRMGGRALLD--
PDRIWVSAMLKKMEKAFLAHLTSAVAELRARWSLQDHPLLDQMAAELQQDVGRRLQGSTQ
RELQKIQSRAGRGAL--GPPREALRGELSLKTEQRRRCFRDL----HKFSVFSQRTQGLG
------TLSLED-------GAALETQLGREVEGEEFCPCEACMKKKVSPVSPKGMVG---
TTRAPIKEAFDLQQILQK---RGGH-SGGGAAEVTPE-KTGMEQLQEDP----SSP----
-----QPSQGADDGL----ALGPGVQEEEE-------GQESQRCCRDDCP----------
------------------------------------------------------------
------------------------------E----P---E---G---A---E---G---P
---G---L---K---E--G---G--E---D---A--------------------------
----------------------A---A---Q--R---G---E--G--N---G---L--G-
--A---A---E--R---E---Q---A--R---N---Q--G---P--G---G---K---D-
--G--L---E---D--E---A---P---T---G---G---D---Q--V---L---V-G--
-Q---G---H---G---A---E---G---E---G---E---G---R--------------
-----------------------------------------------PGS----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
MGP_PahariEiJ_P0085219 (view gene)
-----MNITPGDRRDAPAPSHPAPSHRQCLLPSVAHTPSVTEVTPAKKITFLKRGDPQFA
GVRL
PF03607 // ENSGT00920000149015_4_Domain_0 (65, 122)
AVHQRTFKTFSALMDELSQRMPLSFGVRSVTTPRGLHGLSALEQLQDGGCYLCSDR
KENSGT00920000149015_4_Linker_0_1 (122, 191)
PPKIPREQ--GR----LP--RKSPSAGHSQVFQGGHEAPET-SYSWKSPVAPRKLTLVK
NGDPRSQQTVPF03607 // ENSGT00920000149015_4_Domain_1 (191, 249)
VLSHKNTRSLAAFLSQASELLRFPVKQVYTIRGKKV--DSLQTLLDGPSM
LVCAGNEAFRRLEMENDRGNRTRMLSGVTVRSER-----GCWGPNAKQSVIHSRGRSGG-
KLRQVSLTSERSGLSDHPASGRQAWAGPALDSCPQVTPVP--PGSLVAADDVEKKVCMNE
DGSLSVEMKVRFQLLGEDTLRWSQRVGQASVFTAASGEGQDPREADRFCCRQEGYPWGVL
KPGAQGLGPHDG----------GCQGAFDVDQ-QSRPSY-DIWR-NPLT-TPEGTG--PT
PRRRWGLAKLSGC-QSHWRQEANHRKG-RDKDNASPVS-TPRHPRSVQPG-SCCPWTPDG
DTGSDTLHPVSSASS-----HSETDLESGEG------------LCLEDTGP---------
--------------------------------------HGLRPE-----RALSDTSVSAE
SR------EESSEGGGQLHRGSSQARVMA--SQEQVAK-SDNPCIS--------------
-------------------------TQSHLPLNHTSLHTKKYRQGT--------------
---RGREVRGEPVLRLPL------VPG-HSGSRDTHRDALPAPARAPARRRQRKQKRPTG
VVCLPSVSVPYQVAQKDHAKKGHYC----RDTQSSLDT--ALQMS----MPREREQACLG
SPAPQSPSNSPSAGSQTPEDLRSPFSSSLDFQEPQ-ATSKATTISVSGP--------DCV
----R-------------------HSTCSAESAGDTKCQAHSSTPTPAHGGE--------
----------------------------LGCLWDKAGTTPEPFSSSVLLDRCPEAEDPRT
YHDCCCLQAAPSSPLAAPSGQT-----------QASVSEACLGGGSFCPTPPKEQTRSRR
ESA----SS--SDHSRADGFA---------------------------------------
--GP---RRTLLGKS-PGIRGSLEEREADGGVTPSALPYASPDAVVREWLGNIPEKPVLM
TYEMADENTEVPSGGLEGPKEDPGGSCSLKFLGELTQARQ-QPPEGATDEHPEP--VGVL
SGPGSVWCKLEGDLHLD--ATSGERLKAPAEAGIGEGTR---------VDHGVSLCALPT
RVAASTQIMKALLGSKP----GRPSSLPEVSSTVAQRLSCSAGALIACLARLQFFDE-GL
GPLDDKVRLEESPKYQEMLRLSQTLWP-GSELWQGQL--DFSFRKLTSHQALLG------
-TGDFTPTSSSGVDVSSGSGGSGESNVPCVMDN-TLAPEKRDLPLK-----TPSQRPDSR
NQGYPEL----------VSHPTV--------SSVSQVKTCATKGE---------------
--------------------------------------------------------EAGK
G-GRKQTWGSSPEPSVHST-------------------MLEGDALSEETEGRVR------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------ERLQENSVHG-KGLPEEGAR
VCSQEMLAAGSQDGA-SSPEDTRVPTDEAGTDVASGELWPLHGRQE--PTESPQHFSESN
SRV-HEHQSTHTLDLGLEEMSRLDARG--CKQACIKAS---SETREMTTSMAYKGSLD--
PDPVWVSKLLKKIEKAFMAHLADATAELRARWDLYDNNLLDQMVTELEQDVGRRLQASTV
MEVKKIQSRAGRMVP--EPPREALRGQTSLQTEQRRRRLQGL----RNFSAVP----GQA
------PLSLTLEDGP-TLKTALGTRSGAGPAEDEFCPCEICLKKTRTPGFPKDAAT---
VSGAPVRKAFDLQQILQS---KKGGSSNREAMEVATQ-RTGKMLSQEDL-----------
-----GTVQGADERQGLRLAQGLGVAEGEE-------EEGKQSLRAEEDP----------
------------------------------------------------------------
------------------------------K----IL------K---T---E---R---S
---G---C---HA--P--E------E---D---A-AT---E--E-------D--------
-------G---EICTG------T---A---Q--E---S---Q--Q-LE----------G-
--T-EIG------R---E---E---T--L---H---Q------SF-R-----------D-
--G----------GTLE---A------------------------------PARQG----
-------------T-------H---S---V---V---I---Q---E---A---S----R-
--E----K--------------------------------------EREVEGR-------
-NQDVKEDSLCVSSGESQE-RVGSENTSLEQEGR--LPSHHQ-RPGPQGH----------
---HTTCSSRASSLDNCSQVSQKGSDGDLTSGDLKCTKAKS-------------------
----------SRVLHAERKVPMMYPESSSSE-----QEV---------PSSPRPRKQGKG
E-DEGS------------------------------------------------------
--AGSLACTQVGG---KVDGFGQDDLDF--------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
ENSCSAP00000012938 (view gene)
-----MNSTP---RDAQA-----PRHRECLLPSVARTPSVTKVTPAKKITFLKRGDPQFA
GVR
PF03607 // ENSGT00920000149015_4_Domain_0 (64, 122)
LAVHQRAFKTFSALMDELSQRVPLSFGVRSVTTPRGLHSLSALEQLEDGGCYFCSDK
KENSGT00920000149015_4_Linker_0_1 (122, 190)
HPKTPSGP--GR----PQ--GRSPTAQQLRDIQGRREAPGT-SSSRKSLKTPQRILLVK
NMDPRLQQTPF03607 // ENSGT00920000149015_4_Domain_1 (190, 250)
VVLSHRNTRNLAAFLSKASDLLRFPVKQLYTTSGKKV--DSLQALLHSPSV
LVCAGHEAFRPPAMENARRSEAETLSGLTSRNKN-----RSWGPKTKPSVIHSRSRPGS-
RPQL----PERPVPSN-------PPVGPVPDRHPQDTPAQ--PGPLVAGDDMKKKVRMNE
DGSLSVEMKVRFHLVGEDTLLWSRRMGRASALTAASGEDPILGEVDPLCCVWEGYPWGFS
EPGVWGPRPCRV----------RCGEAFGRGG-QPGPKY-EIWT-NPLH-ASQEER--VA
TRKRWGLAQHVRC-SGLWGHGAAGRER-CSLDSASPAS-STGHPEGSEPDSSCCPRTPQD
RVDSG------SPSA-----QRGAEQEAGGS------PGEDPSLCTDGAGLGSPE-----
------------------------------------QGGRLTPRARSEEGASSDSSASTG
SH------EGSSEWGGRPRGCLGKARAET--SQQEATK-GGNPGSP--------------
-------------------------APSLSSLRSEDLQAEMQGRGT--------------
---EQ--ARGASVKREPL------VPG-LSGSWDSEGASSTPSTSASSQQERRSHRSRAS
ATSSPSSPGLGRVAPRGHPRQCRYC----KDTHSPLDSSVTKQVP----RPPERRRACRD
GSVPRSSGSSLSTRTQASGNLRPPSLGSLPSQDLL-GISSATVTPAGHS--------DCA
SGVSP-------------------NNAPSVGWAGDTGSRTCSPDPTPPHTSD--------
--------------------SCSKSG--AASLGEQARDTPQPSSPLVVQVGRPEQGAVGP
HQSCCHSQA-----E---------------EAQRGPSPEAHWLCGRYCPTPPRGRPCPQR
RSSSCGSTG--SSHRSAARRPGGSQQREGTYQAGPTLSLGPSSG-------ASRRSSASQ
DVGS---GGLSQEET-SRSWGGPQGQEEASGVSPSSLPRSSPEAVVREWLDNIPEEPVLM
TYEMVDETTGAAGGGLRGPEVDPGDDHSLEGLGEPALARQ-QPLEGDPSQDPEP--EGAL
PGSSDAGPQSGEGVPQG--AAPEGVSEAPAEAGADREDP---------ASCGVSLRALPG
RVSASTQMMRALMGSKQ----GRPSSMPEVSRPMARRLSRSAGALITCLASLQFFDE-DL
GSPASKVRFKDSPRYQELLSISKDLWP-GCDLGEDQL--DSGLWELTWNEALPDLGSH-A
VTENFTPTSSSGVDISSGSGGSGESSVPCAMDG-ALVPEGTELPLK-----TSNQRSDSR
TSENPGD-----LENQQQCCSPA--------FLNSRACACATNED---------------
--------------------------------------------------------EAER
D-SEEQRASSNLEQLAENT-------------------MQEEEVQLEESKE-ETEGGGL-
QEEGLQLEGTKETE-G-LQEEGVQLEETKTEEGLQEEGVPLEETKTEEGLQEEGVQLEET
NGTEGEGQQEEEVQLEEIKEETGGEGLQEEGVQLEETKTEEGLQEEGVQLEETKGTEG-E
GQQEEEAQLEEIKEETGGEGLQEEGAQLEEVKEGPEGGLQEEALEEGLKE-EGLPE-EGS
VGRQELSEAGSPDGE-GSWEGDPIQEEEAGRAS--AEPCPPEGTEE--PTEPASHLSETD
PSA-SESQSGSQLEPGLEKPPGAAMMG--QEHTQAQPL---LGAAERSSSVTCRAALD--
CDPIWVSVLLKKMEKAFLAHLSGAVAELRARWGLQDSDLLDQMTAELQQDVAQRLQDSTE
RELQKLQGRAGRMVL--GPPREALTGELLLQTQQRRHRLRGL----RNLSAFSERTLGLG
------PFSFTLEDEP-ALSTALGSQLGEEGEGEEFCPCEACVRKKVSPASPKATMG---
ATRGPIKEAFDLQQILQR---KREE-HTVGEA----A-EVAPGETCTDP----TSI----
-----RTVQGAEGGPGPGLGHGPGVDQGED-------GEGSQRLDRDKDP----------
------------------------------------------------------------
------------------------------K----L---E---E---A---E---G---D
---A---M---A---Q--E---R--E---G---K--I---H--N---S---E--T-----
-------S---V---G--S---E---L---G--E---A---E--Q--E---G---E--G-
--I---S---E--R---G---E---T--R---G---Q--G---S--G---H---E---D-
--S--L---Q---G--E---A---V---A---G---G---D---Q--D---P---G----
-Q---S---D---G---A---E---G---R---E---A---P---E---A---E----G-
--E----A--------------------------------------QPESE-E-------
-GVEAQEVEGEMQE--------AEGEAQPESEGV--EAPEAE-GE---AQEAE------G
EAQSESEGVEAQEAEEEAQPETESVEAPETEGETQEAEGETP----------------PE
SEGA-EAQEAGE---------------EAQE-----AEGE--------D-QPESEGEEAQ
EAEGEAQPESEV--VQAPEI--EGEMQEAEGEAQPESEGVEAPEVEGEAQEAEGEAQPEA
GV----------------------------EAQ---EAEEEAQEAEEEAQEAEGEAQPES
ESVEAQEAEEEAQEAEKEAQEAEGEAQEAEGEAQPESEGVEAQEAEGEAQPESEGVEALE
AEGEAQQESEGIEAQEAEEET-----------------------QPESEGVEAPEAEGEA
QEAEGEVQ---------------------EAEGEAQPESEGVEAQEAEGEAQPESEGVEA
PEAEGEAHPESEGVEAPEAEG---------E--AQEAEGE--GQPESEGVETPKVE----
----------GEGQPESEGVE--ALEAEGEAQPESEGE--------TQGEKKGSPQVSLG
DGQSGGASESSSPVPEDRPTPPPSPGGDTPHQKPA-SQTGPSSSRASSRG--DSWRKDSE
DGRVCGDTR-SPDAKSVGAPHAERKATRMYPESS-TSEPEEAPSDTRTPEQGAS------
---EG-CDPQEDQTLGNPAPTKAVGRADGFGQDDLDF
ENSGGOP00000000667 (view gene)
-----MNSTP---RDAQA-----PSHRECFLPSVARTPSVTKVTPAKKITFLKRGDPRFA
GVRL
PF03607 // ENSGT00920000149015_4_Domain_0 (65, 122)
AVHQRAFKTFSALMDELSQRMPLSFGVRSVTTPRGLHSLSALEQLEDGGCYLCSDK
KENSGT00920000149015_4_Linker_0_1 (122, 191)
PPKTPSGP--GW----PQ--ERNPTAQQLRDIEGQREAPGT-SSSRKSLKTPRRILLIK
NMDPRLQQTVPF03607 // ENSGT00920000149015_4_Domain_1 (191, 249)
VLSHRNTRNLAAFLSKASDLLRFPVKQLYTTSGKKV--DSLQALLHSPSV
LVCAGHEAFRTPAMKNARKSEAESLSGLTSRNKN-----GSWGPKTKPSVIHSRSPPGS-
RPRL----PERPGPSN-------PSVGPAPERHPQDTPAQ--SGPLVAGDDMKKKVRMNE
DGSLSVEMKVRFHLVGEDTLLWSRRMGRASALTAASGEDPILGEVDPVCCVWEGYPWGFS
EPGVWGPRPCRV----------GCREAFGRGG-QPGPKY-EIWT-NPLH-ASQGER--VA
ARKRWGLAQHIRC-SGLWGHGTAGRER-CSQDSASPAS-STGLPEGSEPESSCCPRTPED
GVDSA------SPSA-----QIGAERKAGRS------LGEDPGLCTDGAGLGGPE-----
------------------------------------QGGHLTPRARSEEGASSDSSASTG
SH------EGSSEWGGRPQGCPGKARAET--SQQEASE-GGDPGSP--------------
-------------------------ALSLSSLRSDDPQAETQGQGT--------------
---EQ--ATGAAVTREPL------VLG-LSGFWDSEGASSTPSTCTSSQQGRRRHRSRAS
AMSSPSSPGLGRVAPRGHPRHSHYC----KDTHSPLDSSVTKQVP----RPPERRRACQD
GSVPRYSGSSSSTRTQVSGNLRPPSSGSLPSQDLL-GTSSATVTPAGHS--------DFA
SGVSP-------------------HNAPSAGWAGDAGSRTCSPAPIPPHTSN--------
--------------------SCSKSG--AASLGEEARDTPPPSSPLVLQVGRPEQGAVGP
HQSRCCSQPGMQPAQ---------------EAQREPSPEASWLCGRYCPTPPRGRPCPQR
RSSSCGSTG--SSHQSTARGPGGSPQEGT-RQPGPTPPPGPNSG-------ASRRSSASQ
GAGS---GGLSQENT-LRSGGGPQGQEEASGVSPSSLPRSSPEAVVREWLDNIPEEPILM
TYEMADETTAAAGGGLRGPEVDPGDDHSLEGLGEPAEAGQ-QSLEGDPGQDPEP--EGAL
LGSSDTGPQSGEGVPQG--AAPEGVSEVPAEAGADREAP---------AGCTVSLRALPG
RVSASTQIMRALMGSKQ----GRPSSVPEVSRPMARRLSCSAGALITCLASLQLFDE-DL
GSPASKVRFKDSPRYQELLSISKDLWP-GCDIGEDQL--DSGLWELTWSQALPDLGSH-A
MTENFTPTSSSGVDISSGSGGSGESSVPCAMDG-TLVPQGTELPLK-----TSNQRPDSR
TSESPGD-----LENQQQCCFPT--------FLNARACACATNED---------------
--------------------------------------------------------EAER
D-SEEQRASSNLEQLAENT-------------------VQEEEVQL-EETK-GTEGEGL-
QEEGVQLEETKTEE-G-LQEEG--------------------------------------
----------------------------------------------GVQLEGTKET----
EGMQEEGGVQLEGTKEMEEGLQEEGVQLEEVKEGPEGGLQGEALEEGLKE-EGLPE-EGS
IHGQELSEASSPGGK-GSQEDDPVQEEEAGRASASAEPCPPEGTEE--PTEPPSQLSETD
PSA-SESQSGSQLEPGLEKLPGATMMG--QEHTQAQPT---QGAAERSSSVACRAALD--
CDPIWVSVLLKKTEKAFLAHLASAVAELRVRWGLQDNDLLDQMAAELQQDVAQCLQDSTK
RELQKLQGRAGWMVL--EPPREALTGELLLQTQQRRHRLRGL----RNLSAFSERTLGLG
------PLSFTLEDEP-ALSTALGSQLGEEAEGEEFCPCEACVRKKVSPMSPKATMG---
ATRGPIKEAFDLQQILQR---KRGE-HPDGEA----A-EVAPGKTHTDP----TST----
-----RTVQGAEGGLGLGLSQGPGVDEGED-------GKGSQRLNRDKDP----------
------------------------------------------------------------
------------------------------K----L---R---E---A---E---G---D
---A---M---A---Q--K---R--E---G---K--T---H--N---S---E--T-----
-------S---V---G--S---E---L---G--E---A---E--Q--E---G---E--G-
--I---S---E--R---G---E---T--G---G---Q--G---S--G---H---E---D-
--N--L---Q---G--E---A---V---A---G---G---D---Q--D---P---G----
-Q---S---D---G---A---E---G---I---E---A---P---E---A---E----G-
--E----A--------------------------------------QPDVE-A-------
-LETEGEAQPESEDVE--A-PEAEGEAQPESEGV--EAPEAE-GEAQPESEGVEAPEAEG
EAQPESEGVEAPEAEGEAQPESEGVEAPEAEGEAQPESEGIEAPEAE-------GEAQPE
SEGVEAPEAEGEAQPESEGIEAPEVE------------GE--------A-QPE-------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------SEGVEAPEAEGEAQPESEGIEAPEAEGEAQP---------------------
------------------------------------------------------------
------------------------------------------------------------
---------------ESEGVE--APEAEG------------------------EAQVSLG
DGQSEEASESSSPVPEDRPTPPPSPGGDTPHQRPG-SQTGPPSSRASSRGN--CWQKDSE
NDHVRGDKR-SPDAKSTGAPHAERKATRMYPE-SSTSEQEEDPLGSRTPGQGAS------
---EG-YDLREDQALGSLAPTEAVGRADGFGQDDLDF
ENSCATP00000038772 (view gene)
-----MNSTP---RDAQA-----PRHRECLLPSVARTPSVTKVTPAKKITFLKRGDPQFA
GVRL
PF03607 // ENSGT00920000149015_4_Domain_0 (65, 122)
AVHQRAFKTFSALMDELSQRVPLSFGVRSVTTPRGLHSLSALEQLEDGGCYFCSDK
KENSGT00920000149015_4_Linker_0_1 (122, 191)
PPKTPSGP--GR----PE--GRSPTAQQLRDIQGRREAPGT-SSSRKSLKTPRRILLVK
NMDPRLQQTVPF03607 // ENSGT00920000149015_4_Domain_1 (191, 249)
VLSHRNTRNLAAFLSKASDLLRFPVKQLYTTSGKKV--DSLQALLHSPSV
LVCAGHEAFRPPAMENARRSEAETLSGLTSRNKN-----RSWGPKTKPSVIHSRSRPGS-
RPQL----PERPVPSN-------PPVGPAPDRHPQDTPAQ--PGPLVAGDDMKKKVRMNE
DGSLSVEMKVRFHLVGEDTLLWSRRMGRASTLTAASGEDPILGEVDPLCCVWEGYPWGFS
DPGVWGPRPCRV----------GCGEAFGRGG-QPGPKY-EIWT-NPLH-ASQGER--VA
TRKRWGLAQHVRC-SGLWGHGAAGRQR-CSLDSASPAS-STGHPEGSEPDSSCCPRTPED
RVDSG------SPSA-----QRGAEREAGGS------LGEDPSLCTDGAGLGSPE-----
------------------------------------QGGRLTPRAQSEEGASSDSLASTG
SH------EGSSEWGGQPRGCPGKARAET--SQQEATK-GGNPGSP--------------
-------------------------APSLSSLRSEDLQAEMQGWGT--------------
---EQ--ARGASVKREPL------VPG-LSGSWDSEGASSTPSTSASSQQERRRHRSRAS
ATSSPSSPGLGRVAPRGHPRQCHYC----KDTHSPLDSSVTKQVP----RPPERRRACRD
GSVPRSSGSSLSTRTQASGNLRPPSLGSLPSQDLL-GISSATVTPAGHS--------DCA
SGVSP-------------------CNAPSVGWAGDTGSRTCSPDPTPPHTSD--------
--------------------SCSKSG--AASLEEQARDTPQPSSPLILQVGQPEQGAAGP
HQSCCHSQAGTQPAE---------------EAQRGPSPEARWLCGRYCPTPPRGRPCPQR
RSSSCGSTG--SSHRSAARRPGGSQQKEGTHQAGPTPSLGPGSG-------ASRRSSASQ
DVGS---GGLSQEET-SRSWGGPQGQEEASGVSPSSLPRSSPEAVVREWLDHIPEEPVLM
TYEMADDTTGAAGGGLRGPEVDPGDDHSLEGLGEPALARQ-QPLEGDPGQDLEP--EGAL
PGSSDAGPQSGEGVPQG--AAPEGVSEAPAEAGADREDP---------ASCGVNLRALPG
RVSASTQIMRALMGSKQ----GRPSSMPEVSRPMARRLSRSAGALITCLASLQFFDE-DL
GSPASKVRFKDSPRYQELLNISKDLWP-GCDLGEDQL--DSGLWELTWSQALPDLGSQ-A
VTENFTPTSSSGVDISSGSGGSGESSVPCAMDS-TLVPGGTELSLK-----TSNQRPDSR
TSENPGD-----LENQQQCCSPA--------FLNSRACACATNED---------------
--------------------------------------------------------EAGR
D-GEEERASLNLEQLAENT-------------------MQE-EGQLEEK---ETGGEGL-
QEEDVQLEGTKGTE-G-EG--------------HQE------------------------
--------------------------------------------EGDVQLEGTKGTEG-E
GPQEEGLQLEGTKGTEEEGHQEEGGAQLEEVKEGPEGGLQEEALEEGLKE-EGLPE-EGS
VGRRELSDAGSPDGE-GSWEDDPIQEEEAGRAS--AEPCPPEGTEE--PTEPASHLSETD
PSA-SESQSGSQLEPGLEKPPGAAMMG--QEHTQAQPP---LGAADRSSSVACRAALD--
CDPIWVSVLLKKMEKAFLAHLSGAVAELRARWGLQDSDLLDQMAAELQQDVARRLQDSTE
RELQKLQGRAGRMVL--GPPREALTGELLLQTQQRRHRLRGL----RNLSAFSERTLGLG
------PFSFSLEDEP-ALSTALGSQLGEEGEREEFCPCEACVRKKVSPASPKATMG---
ATRGPIKEAFDLQQILQR---KREE-HTGGEA----A-EVAPGETCTDP----TST----
-----RTVQGAEGGPGLGLGHGPGVDPGED-------GEGSQRLDRDKDP----------
------------------------------------------------------------
------------------------------K----L---G---E---A---E---G---D
---T---T---A---Q--E---R--E---G---K--I---H--N---S---E--T-----
-------S---V---G--S---E---L---G--E---A---E--Q--E---G---E--G-
--I---S---E--R---G---E---T--G---G---Q--G---S--G---H---E---D-
--N--L---Q---G--E---A---V---A---G---G---D---Q--D---P---G----
-Q---S---D---G---A---E---G---M---E---A---L---E---A---E----G-
--E----A--------------------------------------QPES--K-------
-GIEAQEAEEEAPE--------AEGEAQPESEST--EG----------------------
-EAPESEGVEAPEAEKEAQPETESVEAP-------ETEGEAP----------------PE
SEDA-EAREAGE---------------EAQE-----AEGE--------------------
-----DQPESEV-------I--EGEMQEAEGEAQPESEGVEAPEAEGEAQEAEGEA-QPE
SEVVGAPEVEGEM---QEAQ----PESEGVEAQ-----------------EAEEEAQPES
EGVEAPEAEG---------------------EAQPESEGVEALEAEGEAQPESEGVEAPK
AEGEAQEA--EGET---------------------------------------PEAE---
----EEAQPESEGV------------EALEADEEAQPESEGVEALEADEEAQPESEGVEA
LEAEGEAQPESEGVEAPEAEG---------E-----------------------------
-----------------------ALEAEGEAQPESEGE--------TQGEKKGSPQVSLG
DGQSVGASESNSPVPEDRPTPPPSPGGGTPHQKPA-SQTGPSSSRASSQG--DSWRKDSE
DGHVCGDTR-SPDAKSVGAPHAERKATRMYPESS-TSEQEEAPSDTRSPEQGAS------
---EG-CDPQEDQTLGNPAPTKAVGRADGFGQDDLDF
ENSTBEP00000001026 (view gene)
-----MNSTL---KDAQA-----PNHRECLLPSVARTPSVIQVTPAKKITFLKRGDPRFA
GVR
PF03607 // ENSGT00920000149015_4_Domain_0 (64, 122)
LAVNQRTFKTFSALMDELSQRMPLSFGVRSVITLRGLHGLSTLQ--LDWVCYLCSDR
LKTPNXX----XX----XX--XXXXXXXXXXXXXXXXXXXXX----XX-XXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX---XXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXWGPKAQQSVIHSRSTSGS-
RPRWFSLLSEKPGVSD-------APVGPALDRHPQDTSAQPQPGALVAGDDMEKKVHVNE
DGSLSVEMRVRFHLLGDDMLLWSRRLGRASGLTAASGADPGLGEADDVRCMWEGHPWGFV
GPGARGRRPCEA----------GCDTAFDRSC-QPGPRC-EIWT-NPLY-ATQDED--PA
SRSGAGRAQHSCC-RGPWSPGAAGGDR-SSQDSGSLAS-SARCFEGSEPDSSCCPRSLEG
SVGSVSLHQASAAAE-----QAA-----GE------GPGVGPSSEPEGAEPGSQD-----
------------------------------------QCSCLTPGNQGVEGAPSDPSARAR
SC------AEPRERGEQHQGCLGKSRARA--PQQKATQ-GDGSGSP--------------
-------------------------TPSPSSLRNEGLQ--KCGQVS--------------
---EHHQARSGSITRLSL------ALS-HPSSWDTEGGSLPLSAYASAQQ-----RSPDR
AMSSPNVSDLDQVGLRGHPGKCCHC----RDTCCPLDSPATSRVP----RPPDRGGACAH
SPALRS-ERSPSNRRQGSRALRPSSSSSLHSQDLP-GVSSATINPVSNS--------DCA
SSFSR-------------------PDATSAXXXXXXXXXXXXXXXXXXXXXX--------
--------------------XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXX--------XXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXX--XXXXXXXXXXXXX-X--XXXXXXXXXX-----X-------XXX------
-XXX---XXXXXXXX-XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX-XXXXXXXXXXXXX---XXX
XXXXXXXXXXXXXXXXX-----XXXXXXXXXXXXXXXXX---------XXXXXXXXXXXX
XXXXXXXXXXXXXXXXX----XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX-XX
XXXXXXXXXXXXXXXXXXXXXXXXXXX-XXXXXXXXXXXXXXXXXXXXXXXXXXXXXX-X
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX-XXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXX-----XXXXXXXXXXX--------XXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXEQSVENT-----------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------MHEEGLTLEEA---KEEGENEESQEEDSKE-EGPPE-EGG
VSGEELSGTGSLDGE-TTRDGQGVQEEEAGRDSTCSGLCPPEKSEEPEPTEAPSQLSDGD
TNA-SESQSGPQLEPGLEKLPMAAVMG--QEQGQAKSA---QDAGEEGTSLACRGSPD--
ADLIWVSKLLKKMEKAFMAHLTSAVVELRVRWSLQDNRLLDQMVAELEQAVGRRLQASTE
KELRKVQSWVERKAL--GSPRGVLRGQASLQTEQRRRRLQGL----RNLSAFSEQTRGRE
------LLSLSLEDAP-SLSVALGSQLSWEDEGEEFCPCETCVRKKVVMVSPKDTMG---
AASAPIKKAFDLQQILQK---KKEEFAN---EEVAPE-GSRTKLLQEDP----SCT----
-----RADQGAEGGLASVSGPGPGVSGEDE-------EEGCQRLSREEDP----------
------------------------------------------------------------
------------------------------D----G---G---E---A---E--GG---A
---A---S---Q---Q--E------E---E---K--S---H-------------T-----
-------S---A---V--S---C---L---E--E---A---E--V--E---A---Q--G-
--K---S--QE--E---G-------A--V---D---E--G---S--G---D---E---D-
--N--L-------GSVA---A-----------G---E--DD---Q--A---P---E----
-E---S---D---G---A---E---A---V---E---T---Q---E---A---G----G-
--E------------------------------------------ERPESEGG-------
-NESEKEDSPPDSRGHSQS-GEAPGNSSLDLEGK--STPHPG-PGGDTSHQSPDPDT---
-----GPSSRASSLGHCWQKDSEDHSSVN-----RSTEDE--------------------
---------PKEDPPAESKAPRMYPESSTSE-----QEGT--------PSGLQTPEQGAD
EGSDLQDKKAAA------------------------------------------------
----NLTLTKAVG---KAEGFDQDDFDF--------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
ENSOCUP00000009111 (view gene)
-----------------A-----PSHRECLLPAVARALSATQVTPAKKITFLKRGDPQFA
GVR
PF03607 // ENSGT00920000149015_4_Domain_0 (64, 123)
LAVHQRTFKTFSALMDELSQRVPLAFGVRSVTTPRGLHGLSALEQLQDGGCYLCSDK
QPENSGT00920000149015_4_Linker_0_1 (123, 190)
PKTPGWP--G-----------------SLDSEGQGEAPGT-TSSWKGPKAPRRMTLVK
NGDPRLRQTPF03607 // ENSGT00920000149015_4_Domain_1 (190, 250)
VVLSPRNTRNLAAFLSQASAALRFPVKQVYTVSGKKV--DSLQCLLRCPST
LVCAGDEAFRPPVMEDARRNGTEMLPGLSSRSKT-----GSWGPKAKQSVMHSRSGSGSG
RPRRLSPLSETSAPHRPPASLHRAQGAPAPDRHLRDTPAA--PGPLVAGDGVEKKVHLNE
DGSLSVEMRVRFQLLGQDALLWSQRVGRAGALTAASREDLSLGEADPPCCTWQGHPWGFS
EPGAGASGPCEA----------GFKCALGHGQ-QPGPSY-EIWR-NPPV-RLPGRA--A-
---------ACSR-QARWSRGTAGKHG-RGQDSASPAS-SAGHSEGSEQDTCSSPRTMGG
GVSSGSLHPASASPA-----SPR-----------------SPSPRSEGGRLAGGG-----
------------------------------------PRSCLRLETGDTDEADSDSSASAG
PL------EGPSEQGEQLRGCQRQTGAMP--SQRLLAQ-GGSLGAP--------------
-------------------------TLSPSSLGTEDLQAEKPGQDT--------------
---GHDQARGGSVMGLLL------APG-HPGPGDTEGGCSPLPARASARQGRRRQRSPAS
AVCSPSSPG----------RVCCHH----RDAPCLLHAPAALQAP----RPPDRGGASPG
GPVPPFSENVLMSRKQVSGDLRPPSPGSLYSQEPS-GARSVTLTPVSNS--------DCA
SSVHP-------------------TSTPSAEPAGDIKYREHPPAPALAHTPH--------
--------------------PCPQAG--AGRQRGKTGGTPRPSPPLVLLAGQPEAGEPGA
QQGHSCSPIGT----------------------PASFPEGAWACSGYCPTPPEGRPSAKK
PPLSPRNTS--SGDQSADDWDA--------------------------------------
--GP---GRALEGKA-ASAGQGLEGLEDDGSVTPSALPHASPDAVVREWLGNIPEEPVLM
RYEM-DEPTGAARDGPGGPEEAPADERPPESLGELAQGGQ-LLLEGDAGEHPEPEPEGAL
PGPR-------QAPPLGHAAAPAGVPDSPMGAGTREGAA---------GAQGRAPCALPG
RVSASTQIMKALLGAKQ----SRPSSLPELSDVVAKRLSHSAGALVACLARLHFFDE-DP
GSPADKVRFIDSPRYRELLSISQALWP-GYSPGRDQL--DPSLGELAPHKALL-------
VTEDFTPTSSSGVDVSSGSGGSGDGSVPCVLDG-TLVPEKTEPPLE-----IPSQGPDSS
ISEHPEG-----LGNLRLGCPTA--------STSSQTWACATSKP---------------
--------------------------------------------------------EAAE
GGGGEQPLDNASEHEVEKV-------------------VRGDGGQVEEIEGTRRE-----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------EPQEGTQE-GGLPG-EAS
GHGPGVPVAGAADGDLCTLEDQREHKEEAASRPVSAGLCPPDGSEN--PQETPCPPSEGA
SNA-SDSQGGPRSEPGVEEPPAAAGGE--REQAQARWR---QGAGEAGRCTACRACLD--
PDPVWVSKLLKKLQASFLAHLADATAQLRARWNLQDHRLLDQMVAELEQDVSERLQASTD
KELQKIQSRAGRKAL--GPPREALRGQVSLQTEQRRRRLHGL----RNLSAFPEPTCGRG
------LFSCSLEDAS-------------SAPGEEFCPCEACVRKKGTRKFPKDTPG---
AAGAPIKKAFDLQQILQK---KKEGCPEGEAA----E-GGGVTPGQEDP----AMA----
-----GDTPAASGGQGLGLGPGPGADEGDE-------GEGNQVLSRDKDP----------
------------------------------------------------------------
------------------------------P----G---G---E---A---E---G---T
---G---D---L---E--S---G--D---S---V--A---R--S----------------
--------------------------------------------Q--A---E---Q--G-
--V----------Q---E---G---P--V---Q---E--D---S--G---D---G---Q-
--D--C---E---A--A---A---S---E---G---G---D---L--N---P---G-G--
-Q---G---D---G---A---K---A---A---E---V---Q---E---A---G----G-
--E----G--------------------------------------WPGSEGR-------
-NGTEMEGSPLGHPGESQI-GEASETSTPAQEDR--PASPSA-PRGDSPNQRLGPPTPPP
ACASCS-------SSAAAVSSQEDPAAECPGGDRRGSD------------AEPQGLPYP-
----------------DRKVPRMYPGSSTSE-----QDGP--------PSCPGTPEQGSG
GGCDVQDEKVAG------------------------------------------------
----GLTSTQVGG---RADGFGGDDFDF--------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
ENSMGAP00000015014 (view gene)
------------------------------LPSVGRTSTVTQVPPAKKITFFKSGDPQFG
GVK
PF03607 // ENSGT00920000149015_4_Domain_0 (64, 124)
MAINQRSFKSFNALMDDLSHRVPLPFGVRTITTPRGIHCISELDQLEDGGCYLCSDK
KYVENSGT00920000149015_4_Linker_0_1 (124, 190)
KPINIISAGHRPG-PPRNGRPSSTLRRAAQESRLDDYST-SFTQHGPRIPKKITLVK
NGETSFRRSPF03607 // ENSGT00920000149015_4_Domain_1 (190, 250)
IILNRRNARSFKTLLDEISEILQFPVKKLFTVDGKKI--DSMQALLHCPNV
LVCVGREPFKPVSMENLRKHSVEKLPDLAPRSN-GNNTAVNFGLKAKKSIIHPRSASSN-
RSMRFSLSSEKSYPNGLATSPDNGAPFSNNCSHP--KPGD--LVHSLVNDDIEKRVHVNK
DGSLSVEMKVRFRLLNDETLQWSTQIKKSNLMNRMPCEESGLEEDSGVDPLQKMNPEASS
EADD-SLYPCDVDSYMSKLEESECDEAHCHSCGKKHQDY-DIWK-NPMH-TSQREE---P
SVR-STWHTRSSC-SSTSSRRRVIHKKKASVDSLRTTS-SEEFSEHIVQESSSYSETIEN
RVAYRSIKN---------------------------------------------------
---------------------KTNSQKEDENCFEISSVKCLKDDAEEVESSRVGSVISRS
SV-QSRQSKNICEET------SSVGRTMSSSSLQKA-NEGDNTMCSTPANSVH----SRS
SNCSTPRA-KSEHGDHSENNAVVSSFSIKD------------------------------
------EGSEVEQCSAPGTPPSIASVRNSRRSDSDEIVPCNA-SCSSRG-----------
-------------SKKSKHSHLHLDAQSEMSVSSLESSQ--NKKK---NSL-HADD----
-----------------------------SNIS---STSEAEAIAVIAEEKSSKSTTNGY
SASSKGSKRGSHREENSKPSSLCSSVSSPNRKGGEGHSKMNSEVHSSRHGSVSESVCSKG
DHL---------------------------------------------------------
TETGIR-NGNPESSSKSRQIKSRNLHVAMSNSSRASPSETRSICSKHCPAPPKDSSK---
-----------------------------------PTSFGSKSVASEANDQK--------
---NTKLNGSCEGKVEEVLE-DTGVQEEWSNLMPSSLPNTSPEEVVQEWLSKIPSETLLM
KYEMEDNAEEEC---IEATTEMSCCTNAEETPEDEKAEKK-SAEEAENETVDEV--EKET
VEVTAVNEEAEEYANQE------QIPEVLSE--AAKADQAECSQSVT-SSQNNNKKDLPT
SVQTSVQIMKALLSSKHEAKFDRSNSLPEVSPTMGRKLSNSATVLITCLASLQLLDE-ES
EDPSDKSKCLKKTRYMELLNIFQALWF-GQTAEKSGP--SSGQEASGQEKSASGFKVPKS
VNGDFTPMSSSGVDVSSGSGGSGEEITAGAKNC-NLLAQKTVEFKSD------GQV-EDG
-ETATE--LTEAEE----QSKQEEH-------SSRPSTACSKSKGEEKAN----------
------------------------------------------------------------
--S---------------------------------------------------------
AREESLI-------------QD-------------------------QCSN---CENDQE
EGEENENTE-----------------------NSRLVEN--G--EQE-------------
----------------------------------ESEAVN--------------------
------------------------------------EALLKENLEE--ATEQLSATIET-
-NV-SDADCGTELKADIQEQ--L--EKGSTKDPESKVDM----ATSVGTSLVQQNSVD--
PDPVWVLRLLKKIEKEFMAHYVNAMNELKVRWNLQNNQQMDEMILELKEEVSKRIQRSIE
KELKKIKSRAGTKMP--RPPDGPLKRESTLQTEQRRRRLQTM----HKNDKTGT----HS
RLADTSDLSLDIDE-DTAFSTAFEASISEQPSEEEYCPCDTCVRKKMASKPIRNPVL---
ATNAPVAKAFDLQQILKS---QKEVKENEEKKSVLDE-----------------------
-------MDGSVLK-------------------------------------EDEEGPI--
------------------------------------------------------------
-----------------------------LESQNCAI-EE---EVTDE---ENKEE---E
KEET---KEV--------E---E--E---E---E--K--VE--D--IT------------
-----------------------LKAE---E--E---E---E--T-KEE-----------
---------------------------------------V---E--GEI-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
ENSGALP00000040738 (view gene)
MTQVPADYLS---TTS-------SYNYDQPLPSVGRTSTVTQVPPAKKITFFKSGDPQFG
GVKM
PF03607 // ENSGT00920000149015_4_Domain_0 (65, 120)
AINQRSFKSFNALMDDLSHRVPLPFGVRTITTPRGIHCISELDQLEDGGCYLCSDENSGT00920000149015_4_Linker_0_1 (120, 192)
K
KYVKPINITTAGHRPG-PPRNGRPSSTLRRAAQEGRLDDYST-PFTQHGPRIPKKITLVK
NGETSFRRSIIPF03607 // ENSGT00920000149015_4_Domain_1 (192, 249)
LNRRNARSFKTLLDEISEILQFPVKKLFTVDGKKI--DSMQALLHCPNV
LVCVGREPFKPVSMENLRKHSVEKLPDLAPRSN------VNFGLKAKKSVIHPRSASSN-
RSMRFSLSSEKSYPNGLAASPDNGAPF---CSHP--KPGD--LVHSLVNDDIEKRVHVNK
DGSLSVEMKVRFRLLNDETLQWSTQIKKSNLMNRMPCEESGLEEDSGVDPIQKMNPEASS
EADD-SLYPCDVDSYMSKLEESECDEAHCHSCGKKHQDY-DIWK-NPMH-TSQREE---P
SVR-STWHTRSSC-SSTSSRRRVIHKKKASVDSLRTTS-SEEFSEHIVHESSSYSETIEN
RVAYRSIKKCTCRSDLSTGASNGEDQQEYTR------TSTSNSQRPSSLGLMSHSSCENN
-----LDTQEA---PEDYEASKTNSQKEDENCFEISSVKCLKDDAGEAESSRVGSVISRS
SV-QSRQSKNICEET------SSVGRTMSSSSLQKE-NEGDNTMCSTPANSVH----SRS
SNCSTPRA-KSEQDDNSENNAVVSSFSSESCPKKEAEEIEEEQEENEVNEISSVSTNTKP
NQSVKDESSEGEQCSNKKKNSLHADDLRSCSRVSNY------------------------
----------------SKSSSILK----NSSISSIGTES--VF-----------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------------TTGRQQKDVVDSSK---
-----------------------------------PTSFGSKSVATEANDQK--------
---NTKLNGSCSGKVEEVLE-DTGVQEEWSGLMPSSLPNTSPEEVVQEWLSKIPSETLLM
KYEMEDNAEEEC---IEATTEMSCCTNAEEIPEDEKGEEK-NAAEAENEAVDEV--EKET
VEVIAVNEEAEEYANQE------QIPEVMSE--AAKADQAECSQSVT-SSQNNNKKDLPT
SVQTSVQIMKALLSSKHEAKFDRSNSLPEVSPTMGRKLSNSATVLITCLASLQLLDE-ES
EDPSDKSKCLNKPRYMELLNIFQALWF-GQTAEKSGP--SSGQEASGQEKSTSGFKVPKS
VNGDFTPMSSSGVDVSSGSGGSGEEITAGAKDC-NLLAQKTVEFKSD------GQA-EDG
-ETATE--LTETEE----QSKQEEH-------SSRPSTACSKSKAEEKEN----------
------------------------------------------------------------
--S---------------------------------------------------------
TREESLL-------------QD-------------------------QCSN---CENDQE
KGEENENTE-----------------------NSRLVEN--G--EKE-------------
----------------------------------EAEAVN--------------------
------------------------------------EALLKENLEE--AADQLSTTIET-
-NV-SDADCGTELKADIQEQ--L--EKGSTKDPEPKVDT----ATSMGTSLVQQNSVD--
PDPVWVLRLLKKIEKEFMAHYVNAMNELKVRWNLQNNQQMDEMILELKEEVSKRIQRSIE
KELKKIKSRAGTKMP--RPPDGPLKRESTLQTEQRRRRLQTM----HKKS----------
-FFNTSDLSLDVDE-DTAFSAAFEASISEQPSEEEYCPCDACVRKKMASKPARNPVV---
ATNAPVVKAFDLQQILRR---KK-------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
ENSPCAP00000010849 (view gene)
-----MNNTP---RDIQV-----QSYRECLLPSVARTSSITQVVPARD--FLKR-APRFS
GVR
PF03607 // ENSGT00920000149015_4_Domain_0 (64, 121)
LAVHQRNFKSFSALMDELSQRMPFSFGVRSVTTPRGLHCLSTLEQLEDGGCYLCSDKKLPQVSSGP--DR----QQ--GTDVSAQPLGDVNGWYEAPGT-PSSGKSPKAPRRVMLVK
NADPRLQRTVVLSHRNTRSLTAFLSKASDLLHFPVKQVFTISGKKXXXXXXXXXXXXXXX
XXXXX--XXXXXXXX-----XXXXXXXXXXXXXXXXXXXXSWGLKAKQSIIHSRPKSSG-
R-------SQRSGISD-------FPVGSTPHSHPEDTSDH--LCPLVANDNFEKKVHMNE
DGSLSVEMKVRFHLLSEDTPLWSMGVRRASALVTASRECPVPGEVDSLRCVWEGSPGNFS
EPGALRLRCYKS----------EAEEAFKQGQ-QADPSC-EIWT-NPLC-WTQRKE--VD
S----QVTQHCRS-TAFWSQGVAGGKR-TSKASVSSAS-SDRPTAGLKNKSSNYPRTLEG
NTGSA------KPAS-----KRVGEQEAGCIP-----QGGVPNLCQKGAGXXXXX-----
------------------------------------XXXXXXXXXXXXXXXXXXXXXXXX
XX------XXXXXXXXXXXXXXXXXXXXX--XXXXXXX-XXXXXX---------------
-------------------------XXXXXXXXXXXXXXXXXXXXX--------------
---XXXXXXXXXXXXXXX------XXX-XX-XXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXHHHHH----RDVPCSLDST-TKHTL----SPTNRGGTCSC
GLEPSFPESSSSVRKQAFKDPKFPSSGSLCSQYTQ-GIGSAAIILVIHT--------NCA
SHFYH-------------------PHFPSAKPEGDPEIRVSSALPTPSLTSD--------
--------------------SFSSHVKIDHLNEKTRSNIPKLSSPLVW-VKWPDGMKSGD
HQGSCCSQSGTH------GGKMQALKA----SQREPSSWACLVCRRYRPTPPREQPCIKK
RFSGNSNKS--NINQSADRGSGGGEQERKTMDHTLLLG--SESV-------VPGRSESIQ
EASP---REMSQRKI--LMVEGLEQQEEDGSMTLSSLPHISPEAVICDWLSHIPEKPVLM
KYEIMDKSPNVAGHGPEGTQEDPADKHSLEGLQESPQARE-QSLEGETSERPEP---DRA
SVTYDASPKSTEAFPQR--GASRGISDTCEKARDSDRAP---------MGQSVNENVLHR
RIPASIQIMKELISSKR----GRTSSLPEVSGMVGRRLSHSACALITCLARLRFFDE-DL
RSPATKGRFIDSPKFQELLLTFQSLWP-QGDLGKDEA--DLDLQELRLCQVLPAFTSH-T
VTEECTPTSSSGVDVSSGSRGSGEGSGPCPVDC-ALTRERMELPLKMVTSIISYQKPDSI
SSENPED-----PRDQHLSCCKV--------FCNVPACSCATKWN---------------
--------------------------------------------------------KSGE
N-SREQILDNSLEQLVENT-------------------IPEESMQVEKTQE-EIEKKEM-
Q-----------------------------------------------------------
------------------------------------------------------------
--------------------------------------------EKHTSE-EEFPE-EEG
VSKQKLPVASTQDGE-DTQGDVTVQKEEVRRDNDPIELCPPAMSDK--PTEPSSSPGETE
SHS--SEQSGLMSEPSLKNLPGNAEMA--GEQTQVNSTHSTQVTGEKSSSMVHRGFLD--
PDPIWVFRLLKKMEKAFMTHFASAVAAVRAHWGLQNNDMLDQMVAELEQNVARWLQDSTE
KELRKIQSSAREKVP--RPPWEALQQETSLQTEQRRYHLQGL----RNLSAFTEQPQAYR
------LFSPTLENEL-SLSSALGSDLGEKGKGEEFCPCEACTRKKVSPTSLRDTKG---
EGSAPIKKAFDLQQILQK---RKGGPTNNAEVERASA-EGGMMLLHETS----SKT----
-----RSIQAAGREVXXXXXXXXXXXXXXX-------XXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX-----
XXXGRKEC---SLQSQGES-HGASENSNPNQEGR--LTSPAP-G-GNTHYQKA-------
-SKTVIFSSSMSSLGNCSQVSQKDSEEEHSNRDTRNMADEP------------KG-----
------------LTYQERKVTKIYLESSTSE-----QEGT--------PSGPSTLDQGTD
EDPDLEDRKILE------------------------------------------------
----SLILTRVVD---WTDEFGQDDLNF--------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
ENSETEP00000014171 (view gene)
-----MSGH---QRDGQV-----PTYRECLLPFVARTSSVAQITPAKKITFLKRGDARFS
GVQ
PF03607 // ENSGT00920000149015_4_Domain_0 (64, 121)
LAVHQRTFKSFGALMDELSQRVPLSFGVRSVTTPRGLHCLSTLEQLEDGGCYFCSDRENSGT00920000149015_4_Linker_0_1 (121, 190)
KAPQASGGL--GR----QQ--GRGPAAQLSGDFEGQRDASGT-PSSWKGPKAPRRILLVK
NTEPRFQRTPF03607 // ENSGT00920000149015_4_Domain_1 (190, 250)
VVLSPRDTSSFAAFLRKASDLLHVPVKRVFTASGKKV--DSLHGLLHSPLV
LVCAGQEPFRPPAMEHFRRSGTETISDLTSKNKN-----GDWGPKAKPSIIHSRSKSGG-
RSRPLSGLSERSGLSN-------PPVGSALHRHPQDTPAH--LGPLVAGDDFEKQVHMNE
DGSLSVEMKIRLHLLSEDTLLWSTGVRRASDLTTANEEHSIPGDVAPVHCVWEGSPGCFS
EPGALRRRPCES----------GGQEDFKQEQ-QAGPGF-EIWT-NPLF-RSQEEE--ID
AWRRGRLTQNRHS-RPLWSQGGASKKT--SKANASSVS-NQRPAAGSEPDSSYRPRTTAG
SMGSCSGHQASRVAS-----ESRAEQGS----WLY-TLEGESNPCPKGAGKGNQE-----
------------------------------------QDGCWKPEPCGQEDALPDLSAGAS
VL------EKSSEWGEPGWDCANRSGTTT--SKRELSL-EGGTHCS--------------
-------------------------ASSPLSLKNRGLQAEVSGRAM--------------
---ERHQTRGRSVSRLPM------ALG-HSGSWDAEECLSSLSSCAPAQRRGRKLKSQAP
GVSLPSTPGSGTVSQKS------CQ----RAFHCPLESPTTKQAL----SPANSGGPCLG
GPAPCFPESPCGVRKRAAEDSGSPFSGSLHSQYPS-GLSTTTYTPGVNT--------DCT
SHLYH-------------------PHFASAGTEGAPGLGVSSAAPTPSDTSD--------
--------------------SFSSHVRADRMDKKAGGSIPKLSSPAARPLQWPHRLKSGG
HQRSCCSPPGLQGPP-------------RQKAQQEPISGACFVCTRYCPTPPRGQPCVKK
HSSSSSSNSSSRSNQRADWRPDGSKHQENSTLPLS------------TDLGASRRS----
SPRP---RGTSQGKM-SGDGADLEEPEEDGGMVPSALPHFSPEAVVCEWLTHIPEDPILM
KYEMADESPTVAGVGPEGPQEGPVDRYSLEGLGELARAREPQPLEGAASERPKL---DTG
PGTDDAGPQSEEALSQR--GASEKGSDVYEEARASKRRA---------VGHSISKGALPI
RMPTSVQIMRAFISSKP----GRPSSMPEVSTVVGRRLGHFAHSLLTCLAGLHFFDE-DH
GSPARRVRFTDSPRFQELLATLQSLWP-QCDEG------ELGHQEFRLGHALSILASH-P
VTEDITLTSSSGVDVSSGSAGSGEGSGPCPGKG-TFPPGRMELPLQ-----TLVRSPGTR
TSEEQEG-----LGNQHPICPKD--------SSEAPVWACTPSPD---------------
--------------------------------------------------------EAGE
K-GRDHVLGSSLEQLVEST-------------------TQKEAYQPENPEEIEKEERQ--
-E----------------------------------------------------------
------------------------------------------------------------
---------------------------------------------ASDQE-EEFLD-EEG
INRQEY-----------------QQGRGAGSDRDSTDVCPAGMNQE--PTELWNRARESS
SPA-RELQSGLAFEPDLKKLAEM---MA-WEQSQVTPS---QGTGANRSPIAPRVPLD--
PGAIWVHKLLKKMEKAFMTHFASAATSVRAHWGLQHHHVLDQMVAELEQDLGRRLQDSIT
KELSKIPRRTGRKAP--RPPREALHWEMSLQTGQRRRRLQGL----HNLSAFTEPRS---
-------QWFSLEDEP-FSSGTLGAALGAKVEEEEFCPCETCLRKNMSPTSPRAKME---
AGGAPIEKAFDLQQILQK---RKGGRPDGEAA----E-DGGVGWF-----WASLRT----
-----RTVQGADEELEPGFVPGSGVDEGAK-------KEGHPKPGRTEAL----------
------------------------------------------------------------
------------------------------Q----L---G---E---A---E---G---L
---V---P---R---R--D---R--K---T---Y--N---D-------------------
-------T---G---V--G---N---D---L--E---S---E--W--E---E---Q--G-
--V---N---E--K---E---E---S--M---N---E--D---T--R---D---E---G-
--I--L---E---G--E---A---S---R---R---G---D---P--V---Q---G----
-E---D---D---S---A---D---A---I---K---A---Q---G---K---Q----P-
--G----R-------------------------------------DQPME----------
VPRGEKEGSLPVSPGQSQV-EEKRLPPSPA-------------PDADPHQR-S-------
APQTSLSSSSRSSLGNCSRVSQKGSEEARSNGDTRSVAGES-------------------
--------------HSERKVPRMHSESSTSE-----QDGT--------LWGPSAPEQGAD
EEPDLEDRKSMQ------------------------------------------------
----NLTFTKMAD---RTDGFGQDDFDF--------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
ENSPSIP00000006505 (view gene)
MMQAPANYLP---TSS-------PYNYEQALPSVTRINSIPQVPPTKKITFFKSGDPQFG
GVK
PF03607 // ENSGT00920000149015_4_Domain_0 (64, 121)
MAINPRSFKSFNALMDDLSHRVPLPFGVRTITTPRGVHCISALDQLEDGGCYLCSDKENSGT00920000149015_4_Linker_0_1 (121, 191)
KYTKPFSIG-MGRRGG-PQWNGRPVSALRRGGPEGRHEDQSM-PFSQQAPKIPKKITLVK
NGDVGVRRSIPF03607 // ENSGT00920000149015_4_Domain_1 (191, 250)
ILNRRNARSFKTLLDEISELLQFTVKKLYTVDGKKI--DSMQALLHCPSV
LVCVGREPFKPLVLENSRRQLSEKLPGLATQPS-GNPVSENDGSKKDGGRV----ARDT-
PHM--PLPSAPPLVWGYGAVEHPGE-----------RIKS--APGVLVNDDIEKRVHMNK
DGSLSVEMKVRFRLLNDETLHWSTRIKKSNLMSNTPCEESGID----------------S
EAEE-SLYPCDADSYISKLDESESEETHCHSCGKQHLDY-DIWK-NPMH-ASRKEE---P
NVR-NTWHTRSSR-SSTSSRRRIVRKKITSMDSLHTTS-SEEYSEHIVQESSCYSETTDD
RVECCNVKKHNYRSTLSTAASRGEESQDCSR------RNTSSSQKASSVGPGSQSSCENN
-----LDSEEA---VDEAEASQAHSQNEDENDMEVSSVKCSKEEMDEIEGSRVGSRISRS
SL-HSRQKAAS-------------------------------------------------
------------------------------------------------------------
--------------------KARAWDGNIRRSDSDEILVCNA-SCSSKA-----------
-------------SRRTKHSQIHLDAHSEMSVSSRGSTA--KKKK---KNSLHAEVSRSN
SKASNYSKSSY---------------SPLSSTS---SVNEAAPPPIHNEDQCTSSTSKCY
SKSSKGTKQGSQSEASSKMSSPSSSVSNSNRNDGEGHAQVSSETPSSRHGSMSESLCSKC
EQPKAAGNDYHPSSSSRVS------------------------SYSDLQGKRAETEVSRT
SNDGCS-QSSVTRSSKPRRKKMKSLHAGVENSSRASASEVASMGSLHCPTPPKGKPSRKK
LRPAVFKN-SSSISACTESVLTEGKEQKEILDSSKPTSSGSKSAEMNAMNGA--------
---E---NKSHDEQKEDVVS-CKEKMDDPEDLAPSALPNTSPEEVVQEWLSKIPPETLLM
KYEMEDDGAEQC---TEATTEISHCARNQETSEEETAERQ-DAGEGENGAEEEL--EEEA
AEAEATEEGAEKCTQEE------KTSEGMLE--GAEAKQAECSQSIETSSQNNHRKDLPS
TIQTSVQIMKALLSSKQEAKFDRSNSLPEVSPTMGRKLSNSANILITCLASLQLLDE-EP
LGPSDKPKSLNKPRYTELLNIFQALWF-GCTVEKSGP--SSGQPGSDQIKMSSGFKGQK-
----STPMSSSGVDVGSGSGESREGSMASARDC-ILLPQKTVTSKLA------DQG-ASV
-ENEPE--LSETGG----QSKAGEQ-------CSRPSTSCSKSKEGREAA----------
------------------------------------------------------------
--S---------------------------------------------------------
IEEDSPEMGAEAD----HDEDD-------------------------QGSN---CENRE-
-GGENEVEE-----------------------NSKAEAG--D------------------
------------------------------------------------------------
------------------------------------EIPPEANTDE--VTEQPSNLAETD
AIA-DDANGEKEAKSNVEEA--V--GEGSPENQDSNVQAIPE-TSVKTSSIAQQKSVD--
PDPVWVLRFLKKIEKEFMTHYISAMNEFKIRWNLENNELLDKMISELKDEVSKRIQKSIE
KELRKIKSRAGRRHP--KPPNETLRRESTIQTEQRRQRLQTM----LNRSLFNDRTVTQS
KPQGTTDLSFDVGEEDFGISVALVDEVSEQPSEEEYCPCDACIRKKMASKPTRNPIV---
AANAPIVKAFDLKQILRG---KEENETENNMVDCASE------VVQDSEAIPSEHTQEAG
EETNSEEADGEYEPEPPE--QKPDAEEEKEPEQSETDEQGEQEVNEDETAAPAEGDEI--
------------------------------------------------------------
-----------------------------SDNQSCEV-EE---EAKEQ---EEEQE---E
EEEE---RAGKE---E--E---E--E---E---A--K--EE--E--EE--QE--Q-----
-------E---E---E------EQAGK---E--E---E---E--V-KEEADETKEE--EE
E-VKEEGG-NE--E---E---E---V------K---QEEG---N--GEVEE--EEKGEDD
TPN----------ENQECEQS---PKSEEDTEGKVEGAEGEAD-R--DN--NEENASECE
ANAGVS---DAAED---A---E-----------E---A---P------LA--------R-
--N----E-----------------------------EEPQADVGDEAVEETG--PEVEC
EACSEAPAAEEEHENEGDP--------ECAQNGD--------EAAGDEPQEDNEEAREAR
NGEVPEMASTVVEDQDCSKQSQKGSEDGSEGEEGEDGN--------------PNDDPSRE
-----------ANADGGSKSAKMYPDIEEED-----EEEEEDRA----------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
ENSOARP00000016221 (view gene)
-----MNSTP---RDAPA-----PGHREDLLPSVARTPSVTQVTPAKKITFLKRGDPRFA
GVR
PF03607 // ENSGT00920000149015_4_Domain_0 (64, 122)
LAVHQRAFRSFGALMDELSQRVPLSFGVRSVTTPRGLHGLNTLEQLQDGGCYLCSDK
KENSGT00920000149015_4_Linker_0_1 (122, 190)
PPRTPGGL--GQ----PQ--GRSPSAQQLREFE----APGT-ASTCKGLKASRRITLVK
NGDPRLQQTPF03607 // ENSGT00920000149015_4_Domain_1 (190, 250)
VVLSHRNTRNLTAFLSKASDLLRFPVKHVYTTSGKRV--DSLKALLRSPSV
LVCAGLEPFRPLVMEDVRKKGSELSSGQTSRNKS-----GSWGPKAKQSVIHTRSRLEN-
RPRQFSLMSERSGHSDPSVCPHRACMCPAPNRHPLNTPAR--LGPLVAGEDIEKNVSMNE
DGSLSVEMKVRFHLLGEEPLLWSRRLGRASTLTATSREGPDDGEADLHGCVWKGLSRDPS
EPGAQGLGPCEA----------GGVGAFDQGQQQQGSRY-EIWM-NPLY-TSQGE-WSAS
QWR-SGLTQNSHC-RAPRSQRVTSRKR-GSKDRASPAS-SGTPPGGSEPSSSCCSGSLED
AVASCGPHLASGAGE-----GRG-----------------------NGAALGCRD-----
------------------------------------PDGCLKPRTRGLTDALPDSCASAR
SH------EECGERDKLSQGRPSKTPR-------RATQ-QGVPGSP--------------
-------------------------TVSPSSIGNKDPQAEEFGQGA--------------
---GHPQARNRSGVRPAS------TRG-PAASGDDAGGYTPPSACASAMSRSRKQESRAS
TLTSASISVCSRGAQRARPREHHSP----KDINCPLDSPVSQPMP----GPSSGDRAHPH
GPQPSLSESPQGTWNRASWDPGPPFSASLYSQDTR-GLSSIPITPGSNS--------DCV
SNFHL-------------------PDSPSAETEGGPEFRPCSPAPTPSNTSG--------
--------------------PFSAPA--DGLGEKTGGDKSQPSWPLVLPAGWPEGGRLGA
HWGSFCSHLGTSLAHGSSCGTIQTMPAPQPQGSQGPSSKACWVCSRYCPTPPRTRPSGKR
HPSG--SRR--EGDRSADGEPSGEQVGEEKLDAWPPRPPGSQGAASGRAVRAVRRGNPRS
GPQL---GRLVQGKL-SGRGGGLEEPEEGGGELLGALPRASPEAVVRAWLSNIPEEP--M
KHELEDKGEDMAGHGLEGLQEDPVDKHSPDGLERSVWARQ-VPLEQAASEEAEP--DKAF
AAASSAGPKSGEGLPCH------GVSETPTEAGTGKGVA---------VDGGRGQRVLPS
RVSTSLQIMKALMGSRP----GRPSSLPEVSHSAGRRLSRSAQALITCLARLHFFDDDDL
GSPASKVRFTDSPRYQELLSTFQALWP-GSGLRNGEL--ESDLRELGWRQALPALGPH-V
ATEDFRPTSSSGVDVGSGSGGSGEGSGPCTVDC-SLVSEKMELPLS-----ISCQGPDSR
TSGNQEV-----PGNQQLSSYE---------------AEGAASED---------------
--------------------------------------------------------GAEK
N-GREQMLGGDPDQGDKNT-------------------MQEEGVQSREIKE-EKERAEL-
QAEG--------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------VRGFPE-EEG
IMGLELSGPVSQDGA-GVWENKSLHEEEPGDPTAVDTQSSSGRRGS--PPEPPRSLSERC
LDA-SGSQSGPKAEPPLEKLLGAAETG--CGQTLDKLT---QGAGERATCKGRRGSLI--
SDPVWVSKLLKKMEKAFMDHLAAATAELCTRWGLQGDHLLDQMVAELQRDVGQRLQDSVE
KELLKVQSWAGGMGT--GPPREALRWEASLQTEERRRRLQAL----RNLSPFSEQTRGRE
------APSFSLEDLP-IFR--------VEAKGEEFCPCEACVRKKISPASPVEAAG---
SASAPIRKAFDLQHILQK---KQGGCADGEAAEVGPAVEREVEPLHRDP----SGT----
-----GTARGTDGDPEPGSGQGPGAEEG------------DQTLGRDEEPWGTEE-----
------------------------------------------------------------
-------------------------------------------E---A-------G---A
-------Q---E---K--E---G--E----------T-----------------------
-----------EPCAG--SDPWE--GL---GRGDVASP---E--G--E---E------G-
--P---G---D--R---G---G---P-------------G---SRSG---D---R---G-
--R----------S--G---P---R---AGD-R---G----------G---PGSRS----
-G---D---RGRSG---P--RE---T---E---E---D---L---A---P----------
--E------------------------------------------QETEEG---------
-PAPDQ-----ETEEDLAPEQETEEGPAPDQETEEDLAPEQETEEGPAPDQETEEDL--A
PEQETEEGPAPDQETEEDLAPEQETEEGPAPDQETEEDLAP-E-------QETEEGPAPD
----------------QETEEDLAPEQETEEGPAPDQETEEDLAPEQETEEGPAPDQETE
EDLAPEQETEEGPAPDQETEEDLAPEQETEEGPAPDQETEEDLAPEQ---ETEEGPAPDQ
ETEEDLAPEQETEEGPAPDQETEEDLAP----------------------EQETE-----
EGPAPDQETEEDLAPEQETEEGPAPDQETEEDLAPEQETEEGPAPEQE----AEEGPAPE
QEGEEGPA----PDQETEEDLAPEQETEEGPAPEQEA---EEGPAPEQEGEEGPAPDQ--
-ETEEDLAPEQETEEGPAPEQETEEGLAPEQETEEGPAP----EQETEEGLAPEQETE--
----EGPAPEQETEEDL------APEQETEEGLAPEQETEEGPAPEQEGGEGPAPEQETE
EDLAPEQETEEGLAPEQETEEGPAPEQEGGEGPAPEQETEEDLAPEQEGEEGLAPEQET-
---------EEGPAPEQEGEEGPA------------------------------------
---------------------PEQE-----------------TEEDLAPEQETEESSDLE
AKKVGKCLASIE----TRFRNLRMDRTDGFDQDDLDF
ENSMODP00000028679 (view gene)
---------------------------------------ITQVPPAKTITFFKKGDPRFG
GVR
PF03607 // ENSGT00920000149015_4_Domain_0 (64, 123)
MVVNQRMFKSFNALMDELSHHIPLSFGVRTITTPQGVHCVSSLEQLEDGGCYVCSDR
NPSKAFGG--PSRRPT-GSSGGLPTQRI------SGSGGWQE-TPSWQGPKVPKKLILIK
NGDIGFRRSMILGPKNTQNFKTFLDDASDILHFPIKHLYSTDGRKK--------------
------------------------------------------------------------
-----------SYLNGSRASIRSGSPSPT--ANAPEKSTG--LIAIGANDDIEKKVHVNK
DGSLSVEMKVRFRLLGEEMLQWSTHVRKTDDLRMTLEDHPEVS-----------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------LELKIKEPQLEESNA
CRSRCS--------SQSSKTQPFR--T---------------------------------
------------------------------------------------------------
------------YRKPQEGKKNLGMPEGNTDMIPSDFPSTSPEEIIYEWLIKIPEESILM
SYEMQDDSPGAN---VGAPEEDLR----------------------------------VN
PRVDAASPEVIGSLTEE------KSSSTLAT--VAEGSPGECADLEV-KSNLSKMENLPH
SLFSSVQIMKTLLSSKQNHKLGRFNSLPEVSQTMGRKLSHSAHVLITCLASLHFFDE-EL
VTVANQQTYMNSPKYQELLSIFQSMWTECSPN-----------IGELSSKVLPGPKSQTP
VTGDFTPMSSSGVDVSSGSAGSGEGSVAGMEDS-K-------------------------
-------------------------------LKRQDSIASHIDMDL--------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------TT------TEKV
EDKGSF--SKGQ-------VKM---ETDSNEEDEEGRSTHNV------------------
------------------------------------------------------------
-------------------------------------------------------TSKDE
AN---------------------------ETTKE----QILENSMDIRASIIQKASPD--
PDPLWVLRLLKKIEKEFMAHYVNAMSEFKIRWNLQNNEMLDQMIIELKNEVNQRIQKSIE
KELRKIQSRAGKKLP--KPPREAFRWETSLQTEQRRRRLKVM----HNHSQY--------
-------FSLVFVD-GMASSETLGDDLREQGGEEEYCPCDTCIRKKMTTMSLKNTTP---
ATDAPRKKDFDLQQILLK---KKEEKLNEGATELATE------ERGSE------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
ENSTTRP00000014723 (view gene)
-----MNGTP---RDAQA-----PGQRERLLPSVARTPSVTQVTPAKKITFLKRGDPQFA
GVR
PF03607 // ENSGT00920000149015_4_Domain_0 (64, 124)
LAVHRRAFRSFGALMDELSQRVPLSFGVRSVTTPRGLHGLSTLEQLQDGGCYLCSDK
KPPKTPSGP--GR----PR--GRSPS-QQSRGFEGCCEAPGT-ASSCKSLRAPRRITLVK
NGDPRYQQTAVLSHRNTRSLTAFLSKASDLLHFPVKQVYTTGGKKV--DSLKGLLHSPSV
LVCAGHEPFRPLAVEDVRRNGSEPLSGQTSRRKS-----GSWGPKAKQSVIHARSRPGS-
GPRRFSLLSERSGLGD-------PSVCPAPDGNPQGTPGQ--LSPSVAGEDIEKNVRVNE
DGSLSVEMKVRFHLQGEDLLLWSRRVGRA------SGEGPVLGEADPLGCVWSGPPRDPS
EPGAQGLGPCEA----------GGVGAFDRGRWQPGSRY-EIWM-NPLY-APQGEQ--TA
SQRRSGLPQKSHS-RTPRSQGVASRKR-RGKDSASPSS-SDRAPEGSETNSSFCSRSLED
GVGSCGPS------G-----GAG-------------------EGTGEGAMPGRRE-----
------------------------------------HRGCLKPGTRGPADAFPDSSASGG
SH------EESSERGELHQGRPSKKSWAV--TSWGA-TQGGGPSSP--------------
-------------------------TVSPSSVRKEGSRAEESGQGA--------------
---GHPGQP--------L------D-----------AGPPSTSACASARQ-EQEQQSRAG
TLSLPSISVLSRGSQRGRPGQHHHP----KDTHCPLGLPVSRPVP----GPPSRGRARPD
SPAPGPSESPHSRGSWASRDPGPCFSASLCSWDTR-GFSSIPITPVSHS--------DCA
SNFHP-------------------PTSPAAETEGDPEFRACSPAPTPSNTSD--------
--------------------SFNSQA--DGLGEKAGGDSPKPSWPLALPVGWPEGGRPGV
HGASCCSQLGTSPAQDA-----------QSWGGQGPFSEACLVCSGYCPVPRKPRPSGKK
HPSRTLSRS--EGTHSADGEPGGTERGRKSWTPA--PPPGSPGG-------AVGRSRAHR
GPKL---GRTFQEKV--SGGGDPEEHKEGGCVTLGTLPRASPGAVVREWLSNISEEPV--
KYETADEGEDLAGRSPEGPEEDPVDQYPPEGLEGSVQARQ-VPLEGAASEKAEP--DAAL
PGTGSAGPRSGAGLPA-------GVSEASTEAGKGKGAA---------VDRAMGQCVLPS
RVSSSIHIMKALMGSRP----GRPSSVPEVSSSVGRRLGRSAQALVTCLARLRFFDD-DL
GSPAGKVRFTDSPRYQELLSTLQALWP-GSGLGHREL--DSGLQELGWGQALPALGSH-A
AAEDFTPTSSSG-DVGSGSGGSGEGHGPCTVXX-XXXXXXXXXXXX-----XXXXXXXXX
XXXXXXX-----XXXXXXXXXXX--------XXX--XXXXX---X---------------
--------------------------------------------------------XXXX
X---X-XX-XXXXXXXXXX-------------------XXXXXXXXXXXXX-XXXXXXX-
XXXXXXXXXXXXXX-X-XX--------------XXX----XX-------XXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXXMQEEGVHLKE------------------------
--------------------------------KGKERGLQ-------EEG-VGFPE-EEG
ITGRESSGAGSQDGA-GAREDKSLQEA--G-RPTSATLSPPGKRES--PTERPKSLRESD
PDA-SGSQGGPNAEPSLEKLSTAAETG--CGQTQAKLT---QGAEERGTSTARRGPLD--
PDPLWVSKLLKNMEKAFMAHLATATAELQARWGLESNDLLDQMVAELQQDMGRRLQDHVE
KELCKVQSRAGGTAP--GTPREAPHWETPLQTKQYRQHLQGL----SDLSASSEHMRGRG
------PPSFSLEDMP-TFRRVLGTRLVEQVEEEGFCPCEACMRKKVALASPRDAVR---
AASAPVREAFDLQQILQK---KKGRYVDGETAEAAPA-KRGMEPLERDP----SGM----
-----DPVRGADGGPELGSGQGPGVGG-------------HETLSRDEAP----------
------------------------------------------------------------
------------------------------W----G---E---E---E---G---A---T
---T---E---K---R--E---R--K---T---D--------------------P-----
-------F---V---G---------------------S---T--G--A---G---E--G-
--V---G------R---E---E---G--P---G---G--G---S--G---A---E---D-
--T--G---A---G--E---T---P---G---G---G---D---P--S---P---G-G--
-Q---N---A---G---A---E---S---A---E---A---Q---G---A---D----G-
--E----Q--------------------------------------QAESGGG-------
-NQGKREARPQLGLGHRQS-REASRNSSPDQEGR--PTPSLA-PGADSPRQ-----RS--
----GLSSSSASSLGNCSRLSQKSSEDEPSDRHTRSLEDD------------AKGVPDPE
T-----------------KVTGTCPES-SSE-----QDGA--------SSGSRMPEQGTD
EGSDLEAEVV--------------------------------------------------
----KSLASTEME---RTDGFGD-DLDF--------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
ENSBTAP00000054813 (view gene)
-----MNSTP---RDAPA-----PGHRECLLPSAARTPSVTQVTPAKKITFLKRGDPQFA
GVR
PF03607 // ENSGT00920000149015_4_Domain_0 (64, 122)
VAVHQRAFRSFGALMDELSQRVPLSFGVRSVTTPRGLHGLSTLEQLQDGGCYLCSDK
KENSGT00920000149015_4_Linker_0_1 (122, 190)
PPRTPGGL--GQ----PQ--GRSPSAQQLREFE----APGT-ASTCKGLKASRRITLVK
NGDPQLQQTPF03607 // ENSGT00920000149015_4_Domain_1 (190, 250)
VVLSHRNTRNLTAFLSKASDLLRFPVKHVYTTGGKRV--DSLKALLRSPSV
LVCAGLEPFRPLVTEDIRRKGSEPSSGQTSRNKS-----GSWGPKAKQSVIHARSRLEN-
RPRQFSLMSERSGHSDPSVFPHRACMCPAPDRHRLNTPAQ--LGPLVTGEDIEKNVSVNE
DGSLSVEMKVRFHLLGEDPLLWSRRLGRASTLTATSGEGPDAGEVDLLGCVWKGHSGDPS
EPGARGLGPCEV----------GGVGAFDQGQRQQGSRY-EIWM-NPLY-ISQGE-WSAS
RWR-SALTQNCHS-RAPRSQRVASRKR-GSKDRASPAS-SDTPPGGSELNSSCCSGSLED
AVASCGPHLASGAGE-----GKG-----------------------EGAALGCRD-----
------------------------------------HDGCLKPRTRGLTDALPDSSASAR
SH------EECSKRDKLSQGRPSETPQ-------GATQ-QGVPGSP--------------
-------------------------TVSPLSIANKDPQAEEFGQGA--------------
---GHPQARNRSGVRPPS------TRG-PAASGDDAGGYTPPSACASALGRSRKQESRAS
TLYSASSSVPSQGAQRACPRQHHSP----KDIHCPVHSSVSRPMP----GLSSRDRAHPH
GPPPSLSESPQGTWNQASWDPRPPFSGSLYSQDTR-RLSSIPITPGSNS--------DCV
SNFYL-------------------PDSPSPETEGGPEFRPCSPALTPSNTSG--------
--------------------PFSAPA--DGLGEKTGGDRFQPSWPLVLPAGWPEGGRLGA
QWGSCCSHLGTSLARGAS---------------QGPSSKACWVCSRYCPTPPRTRPLGKR
HPSG--SRS--EGDRSADGKPSGEQVGEEKLVAWHPRPPGSQGAALGRAVRAVRRGRPRS
GPQL---RRMVQGKL-SGRGGGLEEPEEGGSELLGALPRASPEAVVRAWLSNIPEEP--I
KHELEDKGEDVAGHGLEGPQEDPVDKHSPDSLERSVWARQ-LPLEQAASEEAEP--DKAF
PVASSASPKSGEGMPCR------GVSETPTEAGTGCGR--------------WEGRVLPS
RVSTSLQIMKALMGSRP----GRPSSLPEVSHSAGRRLSRSAQALITCLARLHFFDDDDL
GSPASKVRFTDSPRYQELLSTFQALWP-GRGLRNTEL--ELDLRELGWRQALPALGPH-I
ATEDFRPTSSSGVDVGSGSGGSGEGSGPCTVDC-SLVSEKMELPLS-----ISCQGPDSR
TLGNPEV-----PGNQQLSSYE---------------AEGAASDN---------------
--------------------------------------------------------GAEK
N-GREQTLGGDPDQGDKNT-------------------MQEEGVQLEEIKE-EKERAEL-
QVEG--------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------VRGFPE-EEG
VMELGLSGAGSQDGA-GVWENKSLNEEEPGDPTAVATQSSSERRGS--PPEPPRSLSERY
LDA-SGSQSGPKAEPPLEKLLGAAETG--CGQTLDKIT---QGAGERGTCKGRRGSLI--
LDPVWVSKLLKKMEKAFMDHLATATAELCTRWGLQGDHLLDQMVAELQRDVGQRLQDSVE
KELLKVQSWAGGAGT--GLPREALHWEASLQTEQRRRRLQAL----RNLSPFSEQMRGRG
------PPSFSLEDLP-IFRGALGTQLAVEAKGEEFCPCEACVRKKMAPASPVDAAG---
SASAPIRKAFDLQHILQK---KKGGCADGETVEVAPAVEREVEPLHRDP----SRT----
-----STVQGADGDPELGSGQGPRAEEG------------DQTLGRDEDPWGTEE-----
------------------------------------------------------------
-------------------------------------------E---A-------G---A
-------Q---E---E--E---R--K----------T-----------------------
-----------EPCVG--SDPWE--GL---GRGEQGMG---G--E--E---G------D-
--L---E------------------------------------ADCG---A---E---D-
--T----------G--E---A---H---GLG-G---G----------G---PGPCG----
-Q---D---AGA---------E---T---T---E---A---L---E---A---A----R-
--E------------------------------------------ERSETGGG-------
-HGGDEEGGLQSKEEGSAPEQETEEG--PAPEQET--------EEGPAPEQ---------
-----------------------ETEEGPA----------P-E-------QETEEGPAPE
----------------QETEEGPAPEQETEEGPAPEQEG----------EEGPAPEQEGE
EGPAPEQEGEEGPAPEQE----------TEEGPAPEQETEEGPAPEQ---ETEEGPAPEQ
ETEEGPAPEQEGEEGPAPEQEGEEGPAP----------------------EQETE-----
EGPAPEQETEEGPAPEQETEEGPAPEQEGEEGPAPEQEGEEGPAPEQE----TEEGPAPE
QEGEEGPA----PEQEGEEGPAPEQEGEEGPAPEQEG---EEGPAPEQEGEE--------
-------------------------GPAPEQEGEEGPAP----EQETEEGPAPEQETE--
----EGPAPEQEGEEGP------APEQEGEEGPAPEQETEEGPAPEQEGEEGPAPEQEGE
----------EGLAPEQEGEEGPAPEQETEEGPAPEQEGEEGLASEQEGEEGLAPEHEG-
---------E----------EGS-------------------------------------
-----------------------------------------------APEQETEEGSNLE
AKK----------------------------------
ENSCGRP00001001143 (view gene)
-----MNSTPGDTKDVPA-----PSRRQCLLPSVAHTPSVTEVTPAKKITFLKRGDPQFA
GVRL
PF03607 // ENSGT00920000149015_4_Domain_0 (65, 122)
AVHQRTFKTFSALMDELSQRMPLSFGVRSVTTPRGLHGLSALEQLQDGGCYLCSDK
KENSGT00920000149015_4_Linker_0_1 (122, 191)
PPRTPREP--GR----RQ--RKSPSAGQSRGFEGGHEAPET-SSSWKGLMPPRRLTLVK
NGDPRCQQTVPF03607 // ENSGT00920000149015_4_Domain_1 (191, 249)
VLSHRNTRSLKAFLSKASELLHFPVKQVYTISGKKV--DSLLTLLDGPSL
LVCAGNEAFRRLEMENVGGARTRTLSGVTARSGR-----GGWGPNAKQSVMHSRARSGS-
RPQQVSLMSERSGLSNHTALGHQDWGGPALDRRPQDTPAP--HGSLVAADDVEKKVCMNE
DGSLSVEMKVRFQLLGEDTLRWSQRVGHTSVFTPASGEGQVLREADPFCCRQEGYPWGFL
AHGAQGLGPYDG----------GYQGVFNLDQ-ESQPNY-DIWR-NPLT-TPGGTG--QT
RRRRWGLSKPSGC-RSHCSQVANDRKG-HGNNNASPVS-SPRHPRSAQSG-PCCPWTTEG
ETDSDTLHPASSAS------SSGVELEAEKG------------PCLA-TGP---------
--------------------------------------CGLGPETQGIERALSDTSVSAE
SH------EGSSECGGQCHRGSSQVRVMV--SQGQATQ-GIS------------------
-------------------------TQSLLSLSRMDFQTEQCNRST--------------
---RDQEARGEPDLRLPL------VPG-CSGSSDGERGALSAPASAPAQQRQKKQKRPVS
IVCLPDVSIPCQGAQKGHARQCHYC----RDTQPSLDT--ALQMP----MPQEGEQACPG
GPTPRFPPNSPSAENQASGHLRSPFSRSLDFQDPQ-ATSKANIIPTSDS--------DCA
SSFYH-------------------PSTCSAEPAGDTECQARPPAPTPVHREE--------
----------------------------VGCLWEDAGTTPEPFSPSVLLDPWPEVEDPGT
HQDCCCLQVGASSVLIVPTGQA-----------QAPISEACWGTSSFCPTPPQEQTCSRN
DPTSSSGPR--SDHNQADGHD---------------------------------------
--EP---RKTLLGKS-PGTRGSLEEQ-ADGGVTPTALPHTSPDAVVREWLGNIPEKPVLM
TWEMEGESTEVASDGPESPEGDLA-------LRELAQARQ-QPPEGVTDEQPEP--TGDL
PGSGPEYYKPGGDPHHD--AASGEGVKAPAEAGIGEETT---------VDHGVSLYALPS
RVSASTQIMKALLGSKP----DRPSSLPEISSTVAQRLSYSAGALIACLARLHFFDG---
-----NTRLEESPKYKEMLNISQALWP-GSELGQAQL--AIDLRKLSSHQALLG------
-TEDFTPTSSSGVDVSSGSGGSGEGSVPCAMDN-TLAPDRVDLPLK-----IPSQRPDSG
NQGYPGV----------LSHSTA--------SSGSQMWPCATSGQ---------------
--------------------------------------------------------ESGK
G-GRNQKWSNTPEQSIQST-------------------TLEEEAQSEEKKGRVK------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------ERLQGDGVHG-KGLPEERSG
VCSQEKFPAGSWDRE-SSPEDTRMSKDEAGRDVASGGLWPLDGREE--PTESPCRFRESN
SNT-NEGQSLHVLELGLEGKSRVAATG--SEQAGIK----------TNTPIAHRRALD--
PDPIWVSKLLKKVEKDFMAHLAGAITELRARWNLQDNSQLDQIVTELEQDVGRRLQASTV
REVRKIQSRAGRMVP--EPPREALRGQTSLQTALRRRRLQGL----RNFSAFP----GQG
------APSLALEDGS-TLSTALGTRPGVGGMGDEFCPCEVCLKKKMTPRFPKGATA---
VFSAPVRKAFDLQQILQS---KKEG-RNGEAMEVPPQ-KAGMMLPQEDP-----------
-----QTVQGTHQKQELRL--------EEE-------GKGRQRLRGEEDS----------
------------------------------------------------------------
------------------------------E----FL------K---A-------G---A
---G---C---HV--P--E------E---D---A-AT---A--R-------E--------
-------G---EIHIS------A---A---Q--E---N---Q--Q-LE----------E-
--C-MT-------E---E---E---T--Q---H---Q------SF-R-----------E-
--G----------DSWE---A------------------------------SGRQG----
-----D---D---G-------H---S---V---E---T---R---D---A---S----R-
--E----R--------------------------------------QSEVEGG-------
-NQDEKENGPWGSSGESQR-RAVSKNISLDQEGR--LYNYHR-RPGPQCH----------
---HTAHSSRASSLGICSQVSQKSSEEKFSSGELKFTKAKN-------------------
----------SRVLHAEREGTTMYPESSSSE-----QEA---------PSSPRSPKQREG
EVYEET------------------------------------------------------
--AGSLVCTQVKG---RVDGIGQDDLDF--------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
ENSAMEP00000017019 (view gene)
-----MNSTL---RDAQA-----PSHRECLLPSMARTPSVTQVTPAKKITFLKRGDPRFA
GVR
PF03607 // ENSGT00920000149015_4_Domain_0 (64, 124)
LAVDQRAFKSFGALMDELSQRVPLSFGVRSVTTPRGLHGLNALEQLEDGGCYLCSDK
KPPKTPSRP--GW----PQ--GRNPSAQQLRDFESQHEAPGT-SSSCNGPKAPRRIMLVK
NGDPRIQQTVVLSHRNTRNLTAFLSKASDLLHFPVKQVYTTSGKKV--DSLKGLLHSPSV
LVCAGYESFKPLAIEDSRRYGTETLSGQTPRNKT-----GSWGPKAKQSVIHSRSGSGS-
RPRRFSLLSERSGLSNFPVSLHCAQMGPAPDGQPQDMPAQ--LGPLVASDDVEKKVRMNE
DGSLSVEMKVRFHLLGEDALLWSRRVGRASALTAASEEVPVLGEVDSFHCVWEDHPGGSL
EPGAQGRGPCEA----------GCEEALSQGRWQPGSRY-EIWM-NPLY-STQGEG--TA
SQRRSRLTQHSHS-RRPCSQGFTSRKR-NSKDSVSPAS-SDRASKDSVPNSSCCSRSSEG
SVGSCDLHSASGAAS-----QRGAGWEVGSP------CGTSESPGPQGAGQGSRE-----
------------------------------------RHCCLKPQTQDSAGALSDSSGSAG
SH------EESSERGEEHQGCLNKTRAVT--TSRTKAT-QG-------------------
--------------------------EGPLSLRHKDSQTEESGQGT--------------
---THSQAKGRSGIRPPL------PLG-HSGSGDTAEGHSLPSACVSD-PGRRRKKGRAS
AVSSPSISGLSRGAQLGCPKQHHTR----RHTHCPCDSPVPRQIP----GPPSGGRAHPD
GPAPPLSASSPSARNQASQDREPSSPASLHSQYAQ-EVSSALITPVRNS--------CSA
SNFYP-------------------PYSPSAETEGDPEFRACSPAPTPSNISD--------
--------------------SVSNQA--DGLDKKSGGGIPKPSWPL---VGQPEAGRPGT
HRGCCCSTISTSLFPGTPSDKTHVLQTSQPLDSQGPFSEVCLVCSRYCPTPPRTWPFVKK
HLL-CS------SSPGADWGPGGGQLRSGRIFQGKVA-----------------------
-----------------GGGESLEWQEKDGRMALGALPRASPDAVVREWLSNIPEEPISM
KCEMVGESTDVTRDDPEGPKEDPVDKHSPACLGEPMQVRQ-LSHEGAANEKAE-------
PVTGEACPRSGQGLPC------SGVSEDSREAGSGKGMA---------MDCGVGPCVLLH
KVSASVQIMKALMGSKR----GRPSSLPEVSSTAGRRLSRSAQALITCLSGLRFFDE-DL
GSTTGQVRFIDSPRYQELLSTFQALWP-GCGPRRGEP--DSGLWELGWCQALPGLRSN-I
MTEDFTPTSSSGVDVGSGSGGSGEGSGPCAMEC-ALVPESITLPLK-----ISCQRPDSR
TSKNQED-----PGNQQPSGSKA--------FSNSQAWACASRED---------------
--------------------------------------------------------EAER
N-SRGQVLGSNLDQGVENT-------------------MQEEEVQLEKVRE-EKEITAL-
QGEG--------------------------------------------------------
------------------------------------------------------------
-------------------------------------------------V-QGFPE-KGR
VMGQELSEVNSQEGE-GAQEGKSIQKEEAGRDPASTILCPPGKREK--PTELPKSFSEKD
E-----NASGPKLEPGLEKLPKAADTH--HVQTQAKCT---QGPGETSSSANRRVSLD--
PDPLWVSKLLRKMEKAFMAHLSSATAELRARWSLQSNPLLDQMVVELQQDLGRRLRESTE
NELWKIQSRAGGRTP--GLAREALRWEMSLQTELRRRRLQGL----RHLSAFSGQAGSLG
------PLSFPSEEEASTLSGALGTWLVGEAEGEEFCPCEACMRKKAIPTSPKDTMG---
ASRAPIRKAFDLQHILQK---KKGGCASGEAA----E-KRGMEPLQGDP----LGT----
-----GTVHGSDGGLELGLDQDPGTEEGGE-------GESSQTLGRDGDP----------
------------------------------------------------------------
------------------------------P----G---G---E---E---E---A---A
---V---Q---E---R---------E---E---S--T---G-------------A-----
-------G---Q---G--N---G---P---E--G---A---G--G--E---E---Q--G-
--S---G---G--K---G---E---N--M---E---E--G---S--G---A---D---A-
--S--L---E---G--E---A---S---G---G---G---D---P--S---P---R-G--
-Q---N---N---G---V---K---T---A---E---T---R---E---A---E----G-
--E----G--------------------------------------QPESGGG-------
-NQREKEGSLQVGWGDGQP-GETSGNSSPDQEGK--TALPP----------------TPG
GHSP-CQSISSL--GNCSQLSQKGSEEKPLNGDMRNIK---------E---KPKGVPGP-
----------------KRKITGMNLESSTSE-----QEGA--------PLGPRSSEQEAG
EASG-----------------------------------LE-------------------
---ETRFKNLT-------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------LDRTDGFGQDDLDF
ENSMNEP00000027443 (view gene)
-----MNSTP---RDAQA-----PRHRECLLPSVARTPSVTKVTPAKKITFLKRGDPQFA
GVRL
PF03607 // ENSGT00920000149015_4_Domain_0 (65, 122)
AVHQRAFKTFSALMDELSQRVPLSFGVRSVTTPRGLHSLSALEQLEDGGCYFCSDK
KENSGT00920000149015_4_Linker_0_1 (122, 191)
PPKTPSGP--GW----PQ--GRSPTAQQLRDIQGRHEAPGS-SSSRKSLKTPRRMLLVK
NMDPRLQQTVPF03607 // ENSGT00920000149015_4_Domain_1 (191, 249)
VLSHRNTRNLAAFLSKASDLLRFPVKQLYTTSGKKV--DSLQALLHSPSV
LVCAGHEAFRPPAMENARRSEAETLSGLTSRNKN-----RSWGPKTKPSVIHSRSRPGS-
RPQP----PERPVPSN-------PPVGPAPDRHPQDAPAQ--PGPLVAGDDMKKKVRMNE
DGSLSVEMKVRFHLVGEDTLLWSRRMGRASALTAASGEDPILGEVDPLCCVWEGYPWGFS
DPGVWGPRPCRV----------GCGEAFGRGG-QPGPKY-EIWT-NPLH-ASQGER--VA
TRKRWGLAQHVRC-SGLWGHGAAGRQR-CSLDSASPAS-STGHPEGSEPDSSCCPRTPED
RVDSG------SPSA-----QRGAEREAGGS------LGEDPSLCTDGAGLGSPE-----
------------------------------------QGSRLTPRARSEEGASSDSLASTG
SH------EESSEWGGQPRGCPGKARAET--SQQEATK-GGNPGSP--------------
-------------------------APSLSSLRSEDLQAEMQGQGT--------------
---EQ--ARGASVKREPL------VPG-LSGSWDSEGASSTPSTSASSQQERRRHRSRAS
ATSSPSSPGLGRVAPRGHPRQRHYC----KDTHSPLDSSVTKQVP----RPPDRRRACRD
GSVPRSSGSSLSTRTQASGNLRPPSLGSLPSQDLL-GISSATVTPAGHS--------DCA
SGVSP-------------------CNAPSVGWAGDTGSRTCSPDPTPPHTSD--------
--------------------SCSKSG--AASLEEQARDTPQPSSPLILQVGRPEQGAAGP
HQSCCHSQAGTQPTE---------------EAQRGPSPEARWLCGRYCPTPPRERPYPQR
RSSSCGSTG--SSHRSAARRPGGSQQKEGTHQAGPTPSLGPGSG-------ASRRSSASQ
DVGS---GGLSQEET-SRSWGGPQGQEEASGVSPSSLPRSSPEAVVREWLDNIPEEPVLM
TYEMADDTTGAAGGGLRGPEVDPGDDHSLEGLGEPALARQ-QPLEGDPGQDLEP--EGAL
PGSSDAGPQSGEGVPRG--VAPEGVSEAPAEAGADREDP---------ASCGVSLRALPG
RVSASTQIMRALMGSKQ----GRPSSMPEVSRPMARRLSRSAGALITCLASLQFFDE-DL
GSPASKVRFKDSPRYQELLNISKDLWP-GCDLGEDQL--DSGLWELTWSQALPDLGSQ-A
VTENFTPTSSSGVDISSGSGGSGESSVPYAMDS-ALVPEGTELSLK-----ASHQRPDSR
TSENPGD-----LENQQQCCSPA--------FLNSQACACATNED---------------
--------------------------------------------------------EAER
D-GEEQRASLNLEQLAENT-------------------MQE-EVQLEESKE-ETEGGGL-
QEEGLQLEGTKETE----G--------------LQE------------------------
--------------------------------------------EGS---EETKGTEG--
GQQEEEAQLEEIKEET-EEGPQEEGAQLEEVKEGPEGGLQEEALEEGLKE-EGLPE-EGS
VGRQELSDAGSPDGE-GSWEDDPIQEEEAGRAS--AEPCPPEGTEE--PTEPASHLSETD
PSA-SESQSGSQLEPGLEKPPGAAMMG--QEHTQAQPP---LGAAERSSSVACRAALD--
CDPIWVSLLLKKMEKAFLAHLSGAVAEVRVRWGLQDSDLLDQMAAELQQDVARRLQDSTE
RELQKLQGRAGRMVL--GPPREALTGELLLQTQQRRHRLRGL----RNLSAFSERTLGLG
------PFSFTLEDEP-ALSTALGSQLGEEGEGEEFCPCEACVRKKVSPASPKATMG---
ATRGPIKEAFDLQQILQR---KREE-HTDGEA----A-EVAPGETCTDP----RST----
-----RTVQGAEGGPGLGLSHRPGVDQGED-------GEGSQRLDRDKDP----------
------------------------------------------------------------
------------------------------K----L---G---E---A---E---G---D
---T---M---A---Q--E---R--E---G---K--I---H--N---S---E--T-----
-------S---V---G--S---E---L---G--E---A---E--Q--E---G---E--G-
--I---S---E--K---G---E---T--G---G---Q--G---S--G---H---E---D-
--N--L---Q---G--E---A---V---A---G---G---D---Q--D---P---G----
-Q---S---D---G---A---E---G---I---E---A---L---E---A---E----G-
--E----A--------------------------------------QPES--K-------
-GIEAQEVEGEMQE--------AEGEAQPESEGI--EAPEAE-GE---AQEAE------G
EAQPESEGVEALEAEKEAQPETESVEAPETE-------GEAP----------------PE
SEDA-EAREAGE---------------EAQE-----AEGE--------------------
-----DQPESEV-------I--EGEMQEAEGEAQPESEGVEAPEAEGEAQEAEGEAQEPE
SEVVGAPEIEGEM---QEAEGEAQPESKGEAQ----------------------------
----------------------------------------------------------PE
SEREAQPESEGVEAQEAEGEA-----------------------QPESEVVGAPEAEGEA
QEAEGEVQPESEGV------------ETPEAEEEGQPESEGVEALEADEEAQPESEGVEA
LEAKGEAQPESEGVEAPEAE------------------GE--TQPESEGVETPEAE----
----------EEGQPESEGVE--ALEAEGEAQPESEGE--------TQGEKKGSPQVSLG
DGQSGGASESNSPVPEDRPTPPPSPGGDTPHQKPA-SQTGPSSSRASSQG--NSWRKDSE
DGHVCGDTR-SPDAKSVGAPHAERKATRMYPESS-TSEQEEAPSDTRSPEQGAS------
---EG-CDPQEDQTLGNPAPTKAVGRADGFGQDDLDF
ENSMMUP00000044936 (view gene)
-----MNSTP---RDAQA-----PRHRECLLPSVARTPSVTKVTPAKKITFLKRGDPQFA
GVRL
PF03607 // ENSGT00920000149015_4_Domain_0 (65, 122)
AVHQRAFKTFSALMDELSQRVPLSFGVRSVTTPRGLHSLSALEQLEDGGCYFCSDK
KENSGT00920000149015_4_Linker_0_1 (122, 191)
PPKTPSGP--GR----PQ--GRSPTAQQLRDIQGRREAPGS-SSSRKSLKTPRRMLLVK
NMDPRLQQTVPF03607 // ENSGT00920000149015_4_Domain_1 (191, 249)
VLSHRNTRNLAAFLSKASDLLRFPVKQLYTTSGKKV--DSLQALLHSPSV
LVCAGHEAFRPPAMENARRSEAETLSGLTSRNKN-----RSWGPKTKPSVIHSRSRPGS-
RPQP----PERPVPSN-------PPVGPAPDRHPQDTPAQ--PGPLVAGDDMKKKVRMNE
DGSLSVEMKVRFHLVGEDTLLWSRRMGRASALTAASGEDPILGEVDPLCCVWEGYPWGFS
DPGVWGPRPCRV----------GCGEAFGRGG-QPGPKY-EIWT-NPLH-ASQGER--VA
TRKRWGLAQHVRC-SGLWGHGAAGRQR-CSLDSASPAS-STGHPEGSEPDSSCCPRTPED
REDSG------SPSA-----QRGAEREAGGS------LGEDPSLCTDGAGLGSPE-----
------------------------------------QGGRLTPRARSEEGASSDSLASTG
SH------EGSSEWGGQPRGCPGKARAET--SQQEATK-GGNPGSP--------------
-------------------------APSLSSLRSEDLQAEMQGRGT--------------
---EQ--ARGASVKREPL------VPG-LSGSWDLEGASSTPSTSASSQQERRRHRSRAS
ATSSPSSPGLGRVAPRGHPRQCHYC----KDTHSPLDSSVTKQVP----RPPERRRACRD
GSVPRSSGSSLSTRTQASGNLRPPSLGSLPSQDLL-GISSATVTPAGHS--------DCA
SGVSP-------------------CNAPSVGWAGDTGSRTCSPDPTPPHTSD--------
--------------------SCSKSG--AASLEEQARDTPQPSSPLILQVGRPEQGAAGP
HQSCCHSQAGTQPAE---------------EAQRGPSPEARWLCGRYCPTPPRGRPCPQR
HSSSCGSTG--SSHRSAAQRPGGSQQKEGTHQAGPTPSLGPGSG-------ASRRSSASQ
DVGS---GGLSQEET-SRSWGGPQGQEEASGVSPSSLPRSSPEAVVREWLDNILEEPVLM
TYEMADDTTGAAGGGLRGPEVDPGDDHSLEGLGEPALARQ-QPLEGDPGQDLEP--EGAL
LGSSDASPQSGEGVPRG--AAPEGVSEAPAEAGADREDP---------ASCGVSLRALPG
RVSASTQIMRALMGSKQ----GRPSSMPEVSRPMARRLSRSAGALITCLASLQFFDE-DL
GSPASKVRFKDSPRYQELLNISKDLWP-GCDLGEDQL--DSGLWELTWSQALPDLGSQ-A
VTENFTPTSSSGVDISSGSGGSGESSVPYAMDS-ALVPERTELSLK-----ASNQRPDSR
TSENPGD-----LENQQQCCSPA--------FLNSQACACATNED---------------
--------------------------------------------------------EAER
D-GEEQRASLNLEQLAENT-------------------MQE-EVQLEESKE-ETEGGGL-
QEEGLQLEGTKETE----G--------------LQE------------------------
--------------------------------------------EGGAQSEETKGTEG--
GQQEEEAQLEEIKEETGGEGLQEEGAQLEEVKEGPEGGLQEEALEEGLKE-EGLPE-GGS
VGLQELSDAGSPDGE-GSWEDDPIQEEERGRAS--AEPCPPEGTEE--PTEPASHLSETD
PSV-SESQSGSQLEPGLEKPPGAAMMG--QEHTQAHPP---LGAAERSSSVACRAALD--
CDPIWVSVLLKKMEKAFLAHLSGAVAEVRVRWGLQDSDLLDQMVAELQQDVARRLQDSTE
RELQKLQGRAGRMVL--GPPREALTGELLLQTQQRRHRLRGL----RNLSAFSERTLGLG
------PFSFTLEDEP-ALSTALGSQLGEEGEGEEFCPCEACVRKKVSPASPKATMG---
ATRGPIKEAFDLQQILQR---KREE-HTDGEA----A-EVAPGGTCTDP----TST----
-----RTVQGAEGGPGLGLGRGPGMDQGED-------GEGSQRLDRDKDP----------
------------------------------------------------------------
------------------------------K----L---G---E---A---E---G---D
---T---M---A---Q--E---R--E---E---K--I---H--N---S---E--T-----
-------S---V---G--S---E---L---G--E---A---E--Q--E---G---E--G-
--I---S---E--K---G---E---T--G---G---Q--G---S--G---H---E---D-
--N--L---Q---G--E---A---V---A---G---G---D---Q--D---P---G----
-Q---S---D---G---A---K---G---I---E---A---L---E---A---E----G-
--E----A--------------------------------------QPES--E-------
-GIEAPEAEGEAQE--------AEGEAQPESEGV--E-----------------------
----------ALEAEKEAQPETESVEAPETEGEAQEAEGEAP----------------PE
SEDA-EAREAGE---------------EAQE-----AEGE--------------------
-----DQPESEV--VQAPEI--EGEMQEAEGEAQPDSEGVEAQ-------EAEGEA-QPE
SEVVGAPEVEGEM---QEAEGETQPESEGVEAQEAEEK---AQEAGEEAQEAEGEAQPES
ESVEALEAEGE--------------AQEAEGEAQPESESIEAQEAEGEAQPESEGVETPE
AEEEGQPESEGVEALEADEEA-----------------------QPESEGVEAPEAEGEA
QEAEGEAQPESEGV------------KAPEAEGEAQPESEGVKAQEAEGEAQPESEGVEA
PEAEGEAQPESEGVEAPEAEG---------E--AQEAEGE--AQPESEGVEALEAD----
----------EEAQPESEGVE--ALEAEGEAQPESEGE--------TQGEKKGSPQVSLG
DGQSVGASESNSPVPEDRPTPPPSPGGDTPHQKPA-SQTGPSSSRASSQG--NSWWKDSE
DGHVCGDTR-SPDAKSVGAPHAERKATRMYPESS-TSEQEEAPSDTRSPEQGAS------
---EG-CDPQEDQTLGNPAPTKAVGRADGFGQDDLDF
ENSANAP00000018750 (view gene)
-----MNSTP---RDAQA-----PSHRECLLPSVARTPSVTKVTPAKKITFLKRGDPQFA
GVRL
PF03607 // ENSGT00920000149015_4_Domain_0 (65, 122)
AVHQRTFKTFSALMDELSQRMPLSFGVRSVTTPRGLHSLSALEQLEDGGCYLCSDK
KENSGT00920000149015_4_Linker_0_1 (122, 191)
PPKTPSGP--GW----PQ--GRSPAAPHLRDIEGRREAPGT-SSSRKSLKTPRRILLVK
NTDPRLQQTVPF03607 // ENSGT00920000149015_4_Domain_1 (191, 249)
VLSHRNMRSLAAFLSKASDLLCFPVKQVYTTSGKKV--DSLQALLYSPSV
LVCAGHEAFRPPAMENARRSEAGTLSGLTSRNKN-----GSWGPKTKPSMIHSRSQSGS-
RPRL----LERPGPSD-------PLVGPAPDRHPQDTLAQ--PGPLVASDDMKKKVCMNE
DGSLSVEMKVRFHLVGEDTLLWSRRMGRASALTAASREDPVLGEVDPLSYVWEGYPWGFS
QPGVWGPRPCRV----------GCGEAFGRGG-QPGPKY-EIWR-NPLH-ASQGEI--VA
ARKRWGLAQHVRC-SGLWDRGAPRRER-CSQDSASPAS-STSHPEGSEPDSSCCPRIPED
RVDSG------SPSA-----QRGAEWEAGRS------LGEDPGMYTDGAQLGGPE-----
------------------------------------QGDRLTSRARSEEGASSDSSASTG
--------EGSSEWDGWTRGCLGKARAET--SQQEATK-GGVLGSS--------------
-------------------------GLSLSSLRSEDLQAEMPGHGT--------------
---EH--QQAR------G------TAV-LSSSWDSEGASSTPPTCTSAQQGQKRHKSRAS
AVSLPSS--LGRVAPRDSPRQCHYC----RGTHSPLDSPATKQVP----RPPEWRRACRD
GSVPRSSGSSSSFRTQASRDLRPPSWGSLPSQDLL-GTSSATVTPAGHS--------DCA
SGLSP-------------------DNAPSAGWVGEAGSRTCSPAPTPPHTSN--------
--------------------SCSKFG--AASLQEPARDTPQPSLPLVLQVEWPEQGASGP
HQGRSCSQTRTLPAQ---------------EAQWGPSLEARWLCGRYCPTPPRGRPCPER
RPSSCASTS--SIHRSAAHGPGGSQQEEGTHQAELTASSGPSSG-------ASRRNGTNR
GVRS---GVLSQEKT--ANLGGPQGQEEASGMSPGTLPHLSPEAVVREWLDNIPEEPVLM
TYEMVDETTGPAGGGLRGPEVDPGDDHALEGLGEPADAR--QQLEGDAGQDLEP--EGVL
PVSGDVGPQSGEGIPQE--ASPEGVSEALTEAGVDRGAL---------ASCRVGLLALPG
RVSASTQIMRALMGSKH----DRPSSVPEVSRPMARRLSCSAGALITCLAGLQFFDE-DS
GSPASKVRFKDSPRYQELLRISKDLWP-GCDLGQDQL--DSGLWELTWSQALPDLGSH-A
VTNNFTPTSSSGVDISSGSGGSGESSMPCAMDG-TLVPEGTELSLK-----TSNQRSDSR
TSENPGD-----LEKQQHCCSPA--------FLNSRACACATQEE---------------
--------------------------------------------------------EAER
D-REE--------QMAENT-------------------MQEEEAQLEETKE-GTEGELP-
-EEGGQLEEMKEEM-GELQGEEVPLEKMEEETGEDL--------------QEEAPFEEVE
E-----------------------------------AAAAEVLQEEGARLEETKEESG-G
ELQEEEAQFEETKEET--EERLQEEAQLEEVKEGPEGGLQEETLEEGLTE-EECPE-EGR
VRGQELSEAGSLDGE-GSQEEDPVQEEEAGRASASAEPCPPEGTEE--ATEPPSHLSEMD
PSA-SESQSGSQLEPGLEKPPGAVWMG--QEHTQAPPT---QGAAEKSSSVACRVALD--
CDPILVSVLLKKTEKAFLAHLASAVAELRARWGLQDDDLLDQMAAELQQDVARRLQNSTE
SELQKLQSRAGRMVL--GPPRETLTGELLLQTQQRRHRLWGL----RNLSAFSERILGLE
------PLSFTLEDEP-ALSTALGTQLGAEAEGEEFCPCEACMRKKVSPTSPKDTMG---
ATRGPIREAFDLRQILQR---KKGE-RTDGEA----T-EVAPGRTPMDP----IST----
-----RTVQGAEGRPGLGLGPGPGVDEEED-------GEGSQRLDRDKDP----------
------------------------------------------------------------
------------------------------K----P---G---E---A---E---G---A
---A---L---A---Q--E---R--E---G---K--T---H--N---S---K--T-----
-------S---T---S--S---E---L---E--E---A---G--Q--E---G---E--G-
--L---G---E--R---G---E---T--G---G---Q--G---S--G---H---E---D-
--N--L---Q---G--G---A---A---A---G---G---D---Q--D---P---G----
-H---S---D---G---A---E---G---I---E---T---P---E---A---K----E-
--E----A--------------------------------------QEAEEEG-------
-QEAEQ--EAQEAEEE--A-QEA------------------E-EEVQPESEGVEAQEAEE
EVQPESEGIEAQEAEEEVQLESEGVEAQEAEGEAQPESESVEAQEAEEEAQEAEEEA---
------QEAEEEAQQKSEGIEALEAEKEAPE-----AEGE--------A-QRKSEGIEAL
EAEKEAQEAEGE--VQPESE--CAEALEIEE-----------------------------
------------------------------------------------------------
-------------------------------ETQPESEIIEVP---------EVEGQTQE
AEREAQPDSEDAEAPEVEGEAQPESEIVEALEAEWE------------------------
-------------------------------------------AKEAEEEAQPEAEGIE-
-------APEAEGIEAPEAEGEAQPEAEGVE--APEAEGE--T---------QEAE----
----------EEAQPESEGAE--APEAEGEAQPESEGE--------TQGEKKGSPQVSLG
DGH---------------------------------------------------------
---------------------AERKTIRMYPESS-TSEQEEVPSGPRTPEQGAS------
---EG-CNLQDDQALGSLAPTEAVGRADGFDQDDLDF
ENSTGUP00000013707 (view gene)
MTQVPADYLS---TTS-------SYNYEPPLPSVARASTLTKVPPAKKITFFKSGDPQFA
GVK
PF03607 // ENSGT00920000149015_4_Domain_0 (64, 121)
MAINQRSFKSFNALMDDLSHRVPLPFGVRTITTPRGIHCISELDQLEDGGCYLCSDKENSGT00920000149015_4_Linker_0_1 (121, 191)
KYVKPISITSGGHRPA-PPRNGRPSSTMRRLAQEGKLEDYSP-HFTHHGPRIPKKITLVK
NGESGFRRSIPF03607 // ENSGT00920000149015_4_Domain_1 (191, 229)
ILNRRNARSFKTLLDEITEILQFPVKKLYTIDGKKV--NIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
ENSFALP00000011381 (view gene)
------------------------------LPSVARASTFTKVPPAKKITFFKSGDPQFA
GVK
PF03607 // ENSGT00920000149015_4_Domain_0 (64, 124)
MAINQRSFKSFNALMDDLSHRVPLPFGVRTITTPRGIHCISELDQLEDGGCYLCSDK
KFVENSGT00920000149015_4_Linker_0_1 (124, 191)
KPISIASAGHRPA-PPRSSRPSSTVRRAAQEGKLEDYPT-PFTHQGPRIPKKITLVK
NGESGFRRSIPF03607 // ENSGT00920000149015_4_Domain_1 (191, 250)
ILNRRNARSFKTLLDEITEILQFPVKKLYTVDGKKI--DSMQALLHCPNV
LVCVGREPFKPISTENLRKHSLEKLPNLPPRSNNGNNTPVNFGLKAKKSVIHPRSASSN-
RSMRFSLSSEKSYPNGLAASPDNGAPFSNSCSHP--KAGD--LVQSLVNDDIEKRVHVNK
DGSLSVEMKVRFRLLNDETLQWSTQIKKSNLMNQLPCEESGVE-DSEVDPLQKMNPEASS
EADD-SLYPCDIDSYMSKLEESECDEAHCHSCGKKHQDY-DIWK-NPMH-MSQREE---P
SVR-STWHTRSSC-SSTSSRRRVVHKKMTSVDSIHTT--------------------SKN
RVAYRSVKKCMCRSDLSTGASNGEEQQEHTR------TSTGNSQRPSSLGLMSHSRQ---
------------------------------------------------------------
------SRKNVCEET------SSVGRSMSSSSLQKA-NQGDNSQCSTPANSVH----SKS
SNCTTPKE-KTEQGDLSDDNPVVSSFSSESCPKKEAEEVEAEQEESEVNEVSSVSEKTKP
SHSIRADSSEGEQCSMQSTHRSKASDRNSRESGSDENVPCSV-SCSSRG-----------
-------------TKKSKRSHIHLDARSEMSVSSLGSTQ---------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------NKKKNSLHADN---
-----------------------------------G-RSGSR---------A--------
---SIYSKSSCETKKKSVGD-AM---SHKSGSLHSNFSSISEAEA---------------
------------------------------TAGPAEGNSS-KST----------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
MGP_SPRETEiJ_P0038846 (view gene)
-----MNSTPGDRRDAPAPSHPAPSHRQCLLPSVAHTPSVTEVTPAKKITFLKRGDPQFA
GVRL
PF03607 // ENSGT00920000149015_4_Domain_0 (65, 122)
AVHQRTFKTFSSLMDELSQRMPLSFGVRSVTTPRGLHGLSALEQLQDGGCYLCSDR
KENSGT00920000149015_4_Linker_0_1 (122, 191)
PPKTSREP--GR----LQ--RKSPSAGQSQVFQGGHEAPET-SYSWKGPVAPRRLTLVK
NGDLRRQQTVPF03607 // ENSGT00920000149015_4_Domain_1 (191, 249)
VLSHKNTRSLAAFLGKASELLRFPVKQVYTPRGKKV--DSLQTLLDGPSV
LVCAGDEAFRRLEMENDGGNRTRMLSGVTARSER-----GCWGPNAKQSVIHSRGRSGG-
KLRQVSLTSERSGLSDHPASGHRAWAGPALDRCPQDMPVP--PGSLVAADDVEKKVCMNE
DGSLSVEMKVRFQLLGEDTLRWSQRVGQASVFTAASGKGQDPREADRFCCRQEGYPWGIL
KPGAQGLGSYDG----------GCQEAFDVGQ-QSQPSY-DIWR-NPLA-TPEGTG--PT
PRRRWGLAKLSGC-KSHWRQEANHRKV-HDKDNLSRVS-TPRHPRSVQPG-SCCPWTPDS
DTGSDTLHPVSSASS-----HSETDLESGEG------------LCLEDTGP---------
--------------------------------------HGSRPETQSTERALSDTSVSAK
SH------EESSEGGGQLHRSSSQARVMA--SREQVTK-GDNPCIS--------------
-------------------------TQSHLPLNHMGLQTEKYRQGT--------------
---RGREVRGEPELRLAL------VPG-HSGSQDTQRDALPAPACAPAQWRQRKQKRPAS
VECLPSVSVPYQVAQKGHARQDHYC----RDTQSSLDT--ALQMP----MPHEREQACPG
SPAPQSPSNSPSAGNQDSEDLRSPFSSSLDLQEPQ-ATSKATTIAVSGS--------DCV
----C-------------------HSTHSVEPAGDTKCQTHSSTPKPVHRGE--------
----------------------------LGCLWDKAGTTPEPFSSSVLLDRCPEAEDPRT
YHNCCCLQAVPSSPLAASSGQT-----------QTSISEACLGGSSFCPTPPKEQTCFGR
ESASNGSTS--SGHSRADGFA---------------------------------------
--GP---RRTLLVKS-PGVRGSLEEREADGGVTPSALPYASPDAVVREWLGNIPEKPVLM
TYEMSDENTEVPSDGPEGPKEDPGDSCSLKVLGEPSQAKQ-QPPEGATDEHPEP--AGVL
SGPGSVCCRLGGDLHPD--ATSGERLKAPAEAGIGEGAR---------VDHGVSLCALPT
KVAASTQIMKALLGSKP----GRPSSLPEVSSTVAQRLSSSAGAFIACLARLHFFDE-SL
GPLDGKVRLEESPKYQEMLRLSQTLWP-GSELWQGQL--DFSLRKLTSHQALLG------
-TEDFTPTSSSGVDVSSGSGGSGESSVPCVMDN-TLAPEKRDLPLK-----IPSQRPDSR
NQGYPEL----------VGHSTV--------SSISQVRACATGGE---------------
--------------------------------------------------------ETGK
G-GRKQTWGNAPEQSVHST-------------------MLEGDALSEETEGRVR------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------ERLQENSVHG-KGLPEEGAR
VCSQEMLAAGSQDGA-GSPEDTRVPTDEAGADAASGGLWPLDGREE--PTESPQHFSESN
SRV-REHQSAHKLELGLEEVSRLDARG--CKQACIKAS---SAT------MAHKGSLD--
PDPIWVSKLLKKIEKAFMAHLADATAELRARWDLHDNHLLDQMVTELEQDVGRRLQASTV
MEVRKIQSRAGRMVP--EPPREALRGQASLQTEQRRRRLQGL----RNFSAVP----GQG
------LLSLTLEDGP-TLKTALGTKSGAGPAEDEFCPCEICLKKKRTPRFPKDAAT---
VSGAPVRKAFDLQQILQS---KKGGSSNREAMEV----RTGRMLSQEDL-----------
-----GTVQGADEKQGLKLAQGLGVAEGEE-------GEGKQRLRAEEDP----------
------------------------------------------------------------
------------------------------E----IL------K---T---E---G---S
---G---C---CA--P--E------E---D---E-AT---E--E-------D--------
-------G---EICIG------T---A---Q--E---S---Q--Q-LE----------G-
--T-EMG------K---E---G---T--L---P---Q------SL-R-----------D-
--G----------GTLE---A------------------------------PARRG----
-------------T-------H---S---V---E---I---Q---E---A---S----R-
--E----R--------------------------------------QQEVEGR-------
-HQDVKEDSPWVSSGESQG-RVGSENTSLDQEGR--LLNRHQ-RPGPQSH----------
---HTACSSRASSLDNCSQVSQKGSDGDLTSGDLKCTKAKN-------------------
----------SRVLHAEKKVPMMYPESSSSE-----QEV---------PSSPRLPKQEKG
E-DEGS------------------------------------------------------
--AGSLACTQVGG---KVDGFGQDDLDF--------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
ENSECAP00000010663 (view gene)
-----------------A-----PSHRGCLLPSVARTPSVTQITPAKKITFLKRGDPRFA
GVR
PF03607 // ENSGT00920000149015_4_Domain_0 (64, 124)
LAVHQRNFKSFGSLMDELSQRVPLSFGVRSVTTPRGLHGLSALEQLEDGGCYLCSDK
KPPENSGT00920000149015_4_Linker_0_1 (124, 190)
KTPNGP--GQ----PQ--GRSPS-QQSQDFEGRCEVPGT-SSSCRGLKAPRRIILVK
NGDPRVQQTPF03607 // ENSGT00920000149015_4_Domain_1 (190, 250)
VVLSHRNTRNLMAFLSKASSLLHFPVKQVYTTNGKKVGWDSLKGLLCSSSV
LVCAGHESFRPLAMEDARRNGTETLPGQTSRNKN-----GDWGPKAKQSVIHSRARSGS-
R--RFSLLSEKSGLSDPPVSLHHAWMGLAPDRHPQDTPAQ--LGPLVAGDDVEKKVRMNE
DGSLSVEMKVRFHLLGEDMLQWSRRVGRASPLTAAGGEGPVLGEVDSLHRMWEGHPGGCS
EPGARPLGPCKT----------GCKETSDRGRWQPGSRY-EIWT-NPLY-ASQGER--TA
SRRRTRLTPHSHS-RSPWSQGVTGRKR-SSKDSFSPAS-SDRHPEDSEPNSSCCSSSLEG
SKGSCGLHPASGAAS-----PRGAGQGAGSRPWAAGGPGQGASPGPEGAGPGSR------
--------------------------------------------ARGAAGALSDSSASSG
PH------EESSEKDEWPQGHPSKMKAVT--SLQKATP-GGGPHSP--------------
-------------------------TVSPSSLRNEDPQAEECGQST--------------
---GHRQARAGSGMRLSP------ALG-LPGSGDAVGGCSPPSSCISAPGGRRKQESRAR
AVSSPSISGLSEGAQRGRPRQRGNQ----RATHCPLDSPVSRQVL----GPPSGGRACPA
GPTPHLSESASSARNQASWDPGPPPSASFHSQDAR-GISSAPITPVSNS--------DCT
SNFYP-------------------PYSPSAETEGDPEFRAHSPAPIPSNISD--------
--------------------SLSSRA--DGL----GGDVPKPSWPLVLPVQQPEGRKPGA
HRGGCCSHIGTSPVPGTPGGKTQALQGSQPRG-------HCLVCSRYCPTPPRAQPSVKK
HPPHSSSHS--DGDRSADWAPGGS------------------------------------
--RP---GRMFHGKA-AGGGESLEEHEEGSGTMPGALPRVSPEAVVREWLSNIPEEPLPM
KYEMVEESTDVAGDCLEGSMEDPVDKHSPEGLEGSTQAGQ-ALLEEAASEKAEP--DGSP
PATGDAGPESGEVEF----------QKSPKRLEQAKGVA---------VDCGAGQCMLP-
RVSASIQIMKALMGS-Q----GRPSSLPEVSSTVGRRRSHSAQALITCLASLHFFDD-DL
GSPAGKVRFIDSPRYQELLSTFRALCP-GCGLRRGEL--DSGLQELGWSQA---------
VTEDFTPTSSSGVDVGSGSGGSGEGSGPGAVDC-TLVPERMELPLK-----IPSE--DSR
TPENPED-----LGNQQLSGSEA--------SSNSQALACAARKE---------------
--------------------------------------------------------GAER
S-SVEQTVGSDLDQAVEST-------------------MQEEGVPSEQVKE-EEERAEL-
Q-----------------------------------------------------------
------------------------------------------------------------
----------------------------------------------QEGV-EGFPE-EAR
VTGQELSGAGSQDGE-RTQEDKSLQEEEAGRDPPSAVLHPPERREK--PTGPPRSLRERD
PNA-SGSPSGPMAEPGWEKLPRAAEAG--PEQSQAKST---QGTGEKGTSTARRASLD--
PDALWVSTLLRKMQKAFMAHLASATAELRARWSLQSNDLLDQMVAELQQDIGQRLQNSTE
KELLKLHSRAGGKAP--GPHREAVRRETSLQTEQRRHRLQGL----RNLSAFTEQTRSWG
------PPSLSLEDMS-TLSGAPGIRLGGEATGEEFCPCEACMRKKVVPTSPKDVTG---
VASAPIKEAFDLQQILQK---KKGGCASGNTAETAPE-KRG-------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
ENSPPAP00000002626 (view gene)
-----MNRTP---RDVQA-----PSHRECFLPSVARTPSVTKVTPAKKITFLKRGDPQFA
GVRL
PF03607 // ENSGT00920000149015_4_Domain_0 (65, 122)
AVHQRAFKTFSALMDELSQRMPLSFGVRSVTTPRGLHSLSALEQLEDGGCYLCSDK
KENSGT00920000149015_4_Linker_0_1 (122, 191)
PPKTPSGP--GR----PQ--ERNPTAQQLRDVEGQREAPGT-SSSRNSLKTPRRILLIK
NTDPHLQQTVPF03607 // ENSGT00920000149015_4_Domain_1 (191, 249)
VLSHRNTRNLAAFLSKASDLLRFPVKQLYTTSGKKV--DSLQALLHSPSV
LVCAGHEAFRTPAMKNARRSEAETLSGLTSRNKN-----GSWGPKTKPSVIHSRSPPGS-
RQRL----PERPGPSN-------PPVGPAPDRHPQDTPAQ--SGPLVAGDDMKKKVRMNE
DGSLSVEMKVRFHLVGEDTLLWSRRMGRASALTAASGEDPVLGEVDPLCCVWEGYPWGFS
EPGVWGPRPCRV----------GCREAFGRGG-QPGPKY-EIWT-NPLH-ASQGER--VA
ARKRWGLAQHVRC-SGLWGHGTAGRER-CSQDSASPAS-STGLPEGSEPESSCCPRTPED
GVDSA------SPSA-----QIGAERKAGGS------LGEDPGLCIDGAGLGGPE-----
------------------------------------QGGRLTPRARSEEGASSDSSASTG
SH------EGSSEWGGRPQGCPGKARAET--SQQEASE-GGDPGSP--------------
-------------------------ALSLSSLRSDDLQAETQGQGT--------------
---EQ--ATGAAVTREPL------VLG-LSGSWDSEGASSTPSTCTSSQQGRRRHRSRAS
AMSSPSSPGLGRVAPRGHPRHSHYR----KDTHSPLDSSVTKQVP----RPPEQRRACQD
GSVPRYSGSSSSTRTQASGNLRPPSSGSLPSQDLL-GTSSATVTPAGHS--------DFA
SGVSP-------------------HNAPSAGWAGDAGSRTCSPAPIPPHTSD--------
--------------------SCSKSG--AASLGEEARDTPQPSSALVLQVGRPEQGAVGP
HQSRCCSQPGTQPAQ---------------EAQRGPSPEASWLCGRYCPTPPRGRPCPQR
RSSSCGSTG--SSHQSTARGPGGSPQEGT-RQPGPTPPPGRNSG-------ASRRSSASQ
GAGS---GGLSQENT-LRSGGGPQGQEEASGVSPSSLPRSSPEAVVREWLDNIPEEPILM
TYEMADETTAVAGGGLRGPEVDPGDDHSLEGLGEPAEARQ-QSLEGDPGQDPEP--EGAL
LGSSDTGPQSGEGVPQG--AAPEGVSEAPAEARTAREAP---------ASCRVSLRALPG
RVSASTQIMRALMGSKQ----GRPSSVPEVSRPMARRLSCSAGALITCLASLQLFDE-DL
GSPASKVRFKDSPRYQELLSISKDLWP-GCDVGEDQL--DSGLWELTWSQALPDLGSH-A
MTENFTPTSSSGVDISSGSGGSGESSVPCAMDS-TLVPQGTELPLK-----TSNQRPDSR
TSESPGD-----LENQQQCCFPT--------FLNARACACATNED---------------
--------------------------------------------------------EAER
D-SEEQRASSNLEQLAENT-------------------VQEEEVQL-EETK-GTEGEGL-
QEEGVQLEETKTEE-G-LQEEG----------------GVQLEETKTEGLQ---------
------------------------------------------------------------
----------------------EEGVQLEEVKEGPEGGLQGEALEEGLKE-EGLPE-EGS
VHGQELSEASSPDGK-GSQEDDPVQEEEAERASASAEPCPPEGTEE--PTEPPSHPSETD
PSA-SESQSGSQLEPGLEKLPGATVMG--QEHTQAQPT---QGAAERSSSVACRAALD--
CDPIWVSVLLKKTEKAFLAHLASAVAELRARWGLQDNDLLDQMAAELQQDVAQRLQDSTK
RELQKLQGRAGRMVL--EPPREALTGELLLQTQQRRHRLRGL----RNLSAFSERTLGLG
------PLSFTLEDEP-ALGTALGSQLGEEAEGEEFCPCEACVRKKVSPMSPKATMG---
ATRGPIKEAFDLQQILQR---KRGE-HTDGEA----A-EVAPGKTHMDP----TST----
-----RTVKGAEGGLGPGLSQGPGVDKGED-------GEGSQRLNRDKDS----------
------------------------------------------------------------
------------------------------K----L---R---E---A---E---G---D
---A---M---A---Q--G---R--E---G---K--T---H--N---S---E--T-----
-------S---A---G--S---E---L---G--E---A---E--Q--E---G---E--G-
--I---S---E--R---G---E---T--G---G---Q--G---S--G---H---E---D-
--N--L---Q---G--E---A---A---A---G---G---D---Q--D---P---G----
-Q---S---D---G---A---E---G---I---E---A---P---E---A---E----G-
--E----A--------------------------------------QPESE-G-------
-VEAQE------AEE----------EAQPESEGV--EAPEAE-GEAQPESEDVESPEAEW
EAPARVRRCKAPEGRRGGPARGRRFRGP----------------------TRQKGRPTPE
SEV---------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
ENSJJAP00000006578 (view gene)
-----MTSTPGDANDTQS-----PSHRQCLLPSVAPTPSVTEVTPAKKITFLKRGDPQFA
GVRL
PF03607 // ENSGT00920000149015_4_Domain_0 (65, 120)
AVHQRTFKTFSALMDELSRRVPLSFGVRSVTTPRGLHSLSALEQLQDGGCYFCSDENSGT00920000149015_4_Linker_0_1 (120, 191)
K
RLSKTPRGP--ER----PL--RRNPSTVPSRGFEGGREEPGT-ASSWKAPTAQRRMTLLK
NGDPTCQQTVPF03607 // ENSGT00920000149015_4_Domain_1 (191, 249)
VLSHRNTRNLAAFLRKASDLLHFPVKQAYTTSGKKV--DTLQTLLQGPSV
LVCAGNEAFRHPAVENNQGNRMDALSGVTARSKT-----GLGGPRARQSVIHSRSRTGG-
RLQL----SERSGLGDCLASGHHDWVGPATDSCPQDTPAA--PGALMVGDDVEKKVCMNE
DGSLSVEMKVRFQLLGEDALRWSQRAGHNSVLTAASRAGRVLEEAHPLCYRQEGHPWGFL
EPRAQGLGPCEV----------GCQRVFDRGQQQPGSSY-DIWR-NPLA-TSQGER--PA
PRRRWGLAQLAHC-RGRWIQGFDVRKR-YDKDSSNPAS-NPGHPENIQA-SSHCPQSPEG
GVGSDSPCPTSSAAS-----RSEAELETG------------KGLCPKEPGPGAQ------
----------------------------------------------DMERTRSDTSASAG
SH------EESSEWGDGHQSCLSQARAAA--SQGKATH-IDSPHVP--------------
-------------------------LLRPSSVSHMVVPLNKHGQGT--------------
---RSCQDRGGPGQSLPV------VPG-HCTSCDAVGTAYPTLTATPSQRGKRRQKRPAS
AVCIP---VHCCVAQRGHDGQCHQC----RDTHCSLASPEALQVP----RPPEKGRVCSR
GPDPQCPKNSSRARNQASRDPGSPSSSSFDSQDLP-TASRATIPHMSNT--------DHA
SGSNC-----------------------STESAGATEHRAHSSAPTLAHT----------
------------------------DG--AGGLGDKAGGNPESPSSSALLGGCPEAGEPGV
HQDGCRSQLAASSILGTPSGQT-----------QAPFSEACWVGSSYCPTPPGERPCAKK
HPSSSGSTS--SGQQRVDDNA---------------------------------------
--RP---RRVLLGKSPGARESFEEQEEDDGGMTPYALPSASPDAVVREWLDNIPEEPVLM
TYETAGETTEVAGDETKDSEEDPRND------GELAQARQ-QLLEGDTGLHPDP--AEVL
PVTGDALPKSGDGPHQD--ADPGDVSQDPAEAKGREGTS---------VVGDVSPRVLPN
KVSVSTQIMKALLGSKQ----GRPSSLPEVSGTVAQRLSHSTRALITCLARLHFFDE-DL
GLLAGKMRLEDSPRYQELLSISQSMWP-GNGLGQGRL--DLGLGRLTSHQALLG------
-SENFTPTSSSGVDVSSGSGGSGEGSVPCAVDT-ILVPERMEPPLT-----TSSQRPTFK
TQGHPE----------VSSCSTE--------SSNSQVRARAPSRE---------------
--------------------------------------------------------ETER
G-GTEQ------EQMLDNVLEH----SGM---------MQEEEEQLEEREEI--------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------GRERPQEEGLGE-ESFPE-KVE
VSSRELLGAGSQDGE-GSPEDKEVLKEET----EPSELCPPGQGEE--PTKPPYHLSERD
SNA-TGCQSGPRLAPGLEELPRAAGMG--CEQTQT------------STFTVHKVSLD--
PDAIWVSKLLRKMEKAFMAHLASATAELRARWSLQNNGLLDQMVVELVQDVGLRLQASTD
KELRKIQSRAGRTVP--GPPREALRGQTSLQTEQRRRRLQGL----RNFSAFP----GLG
------PLSLTLEDGP-AYGAALGTRPGGGTTGDDFCPCDVCMKKKLASMSSKDTAV---
ATSAPIRKAFDLRQILQS---KKAGCGAGVEAGLSPE-KTGILLQ---------------
---------GADRAQELQ--PAPQAGEELE-------ETGGQRSSRAEDT----------
------------------------------------------------------------
------------------------------G----F---Q---E---A---E---R---G
---G---Y---H---E--P---E--N---E---K--E---G--G---T---H--P-----
-------C---L---A--H---A---T---Q--D---A---W--Q--L---E---E--G-
--V---T---K--T---E---G---P--L---S---Q--G--------------------
----------------S---G---D---G---G---A---S---E--S---I---G-Q--
-E---D---D---N---S---R---S---V---E---S---Q---E---A---A----G-
--E----G--------------------------------------QPEAEGG-------
-IQCEQENAPQVSLGERPR-GDVLANSSLDQEGR--LRTPHR-RPGSQAQ----------
--AAARDFSNASSLSNCSLVSQKGSEKDCPSRDLKDIGVK--------------------
---------DSGVLHTERKVTTMYPESSTSE-----QEGT--------LSGPRTPEQETG
EGCDVRGERFAC------------------------------------------------
--------VQTRV---RADGFDQHDLDF--------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
ENSPTRP00000092368 (view gene)
-----MNRTP---RDAQA-----PSHRECFLPSVARTPSVTKVTPAKKITFLKRGDPQFA
GVRL
PF03607 // ENSGT00920000149015_4_Domain_0 (65, 122)
AVHQRAFKTFSALMDELSQRMPLSFGVRSVTTPRGLHSLSALEQLEDGGCYLCSDK
KENSGT00920000149015_4_Linker_0_1 (122, 191)
PPKTPSGP--GR----PQ--ERNPTAQQLRDVEGQREAPGT-SSSRKSLKTPRRILLMK
NTDPHLQQTVPF03607 // ENSGT00920000149015_4_Domain_1 (191, 249)
VLSHRNTRNLAAFLSKASDLLRFPVKQLYTTSGKKV--DSLQALLHSPSV
LVCAGHEAFRTPAMKNARRSEAETLSGLTSRNKN-----GSWGPKTKPSVIHSRSPPGN-
RQRL----PERPSPSN-------PPVGPAPDRHPQDTPAQ--SGPLVAGDDMKKKVRMNE
DGSLSVEMKVRFHLVGEDTLLWSRRMGRASALTAASGEDPVLREVDPLCCVWEGYPWGFS
EPGVWGPRPCRV----------GCREAFGRGG-QPGPKY-EIWT-NPLH-ASQGER--VA
ARKRWGLAQHVRC-SGLWGHGTAGRER-CSQDSASPAS-STGLPEGSEPESSCCPRTPED
GVDSA------SPSA-----QIGAERKAGGS------LGEDPGLCIDGAGLGGPE-----
------------------------------------QGGRLTPRARSEEGASSDSSASTG
SH------EGSSEWGGRPQGCPGKARAET--SQQEASE-GGDPGSP--------------
-------------------------ALSLSSLRSDDLQAETQGQGT--------------
---EQ--ATGAAVTREPL------VLG-LSGSWDSEGASSTPSTCTSSQQGRRRHRSRAS
AMSSPSSPGLGRVAPRGHPRHSHYR----KDTHSPLDSSVTKQVP----RPPEQRRACQD
GSVPRYSGSSSSTRTQASGNLRPPSSGSLPSQDLL-GTSSATVTPAGHS--------DFA
SGVSP-------------------HNAPSAGWAGDAGSRTCSPAPIPPHTSD--------
--------------------SCSKSG--AASLGEEARDTPQPSSALVLQVGRPEQGAVGP
HQSRCCSQPGTQPAQ---------------EAQRGPSPEASWLCGRYCPTPPRGRPCPQR
RSSSCGSTG--SSHQSTARGPGGSPQEGT-RQPGPTPPPGRNSG-------ASRRSSASQ
GAGS---GGLSQENT-LRSGGGPQGQEEASGVSPSSLPRSSPEAVVREWLDNIPEEPILM
TYEMADETTAAAGGGLRGPEVDPGDDHSLGGLGEPAEARQ-QSLEGDPGQDPEP--EGAL
LGSSDTGPQSGEGVPQG--AAPEGVSEAPAEARADREAP---------AGCRVSLRALPG
RVSASTQIMRALMGSKQ----GRPSSVPEVSRPMARRLSCSAGALITCLASLQLFDE-DL
GSPASKVRFKDSPRYQELLSISKDLWP-GCDVGEDQL--DSGLWELTWSQALPDLGSH-A
MTENFTPTSSSGVDISSGSGGSGESSVPCAMDS-TLVPQGTELPLK-----TSNQRPDSR
TSESPGD-----LENQQQCCFPT--------FLNARACACATNED---------------
--------------------------------------------------------EAER
D-SEEQRASSNLEQLAENT-------------------VQEEEVQL-EETK-GTEGEGL-
QEEGVQLEETKTEE-G-LQEEGVQLEGTKETEEWLQEEGVQLEETKTEGLQEEGVQLEET
---------------------------KTEGLQEKGGTKTKGLQGEGVQLEGTKET----
EGLQEEGGVQLEGTKETEGLQEEEGVQLEEVKEGPEGGLQGEALEEGLKE-EGLPE-EGS
VHGQELSEASSPDGK-GSQEDDPVQEEEAGRASASAEPCPPEGTEE--PTEPPSHPSETD
PSA-SESQSGSQLEPGLEKLPGATVMG--QEHTQAQPT---QGAAERSSSVACRAALD--
CDPIWVSVLLKKTEKAFLAHLASAVAELRARWGLQDNDLLDQMAAELQQDVAQRLQDSTK
RELQKLQGRAGRMVL--EPPREALTGELLLQTQQRRHRLRGL----RNLSAFSERTLGLG
------PLSFTLEDEP-ALGTALGSQLGEEAEGEEFCPCEACVRKKVSPMSPKATMG---
ATRGPIKEAFDLQQILQR---KRGE-HTDGEA----A-EVAPGKTHMDP----TST----
-----RTVKGAEGGLGPGLSQGPGVDKGED-------GEGSQRLNRDKDP----------
------------------------------------------------------------
------------------------------K----L---R---E---A---E---G---D
---A---M---A---Q--G---R--E---G---K--T---H--N---S---E--T-----
-------S---A---G--S---E---L---G--E---A---E--Q--E---G---E--G-
--I---S---E--R---G---E---T--G---G---Q--G---S--G---H---E---D-
--N--L---Q---G--E---A---A---A---G---G---D---Q--D---P---G----
-Q---S---D---G---A---E---G---I---E---A---P---E---A---E----G-
--E----A--------------------------------------QPESE-G-------
-VEAQE------AEEE--A-PEAEGEAQPESEDV--EAPEAE-GEAQPESEDVESPEAEW
EAQPESEGVEAPEAEKEAQPETESVEALETEGEDQPESEGAEAQEAEEEAQEAEGEAQPE
SEVIESQEAEEEVQPESEDVEALEVE----------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------VETQEAEGEAQPESEGVEAQEAEREASQCQSVEAQ--------------
-RERRQKR----------------------------------------------------
------RRP--QE------------------------------------RVAMQRE----
----------REAQRQSQRHR--GPGAEGEAQTRE--N--------SKVRKRASPQVSLG
DGQLREA------------TPTPSPG-ETSHQRPRLAKTGPSSSRAFSISGQLLAQKDSE
NDACPGCTPWGPSCREEGPPQP-----GCYRRSSNLLRQEEARFGLKRPEQGAM------
---MKAMTYKRTQAPKSAPPPWQ----AGRADDDLVF
ENSP00000371923 (view gene)
-----MNSTP---RNAQA-----PSHRECFLPSVARTPSVTKVTPAKKITFLKRGDPRFA
GVRL
PF03607 // ENSGT00920000149015_4_Domain_0 (65, 122)
AVHQRAFKTFSALMDELSQRVPLSFGVRSVTTPRGLHSLSALEQLEDGGCYLCSDK
KENSGT00920000149015_4_Linker_0_1 (122, 191)
PPKTPSGP--GR----PQ--ERNPTAQQLRDVEGQREAPGT-SSSRKSLKTPRRILLIK
NMDPRLQQTVPF03607 // ENSGT00920000149015_4_Domain_1 (191, 249)
VLSHRNTRNLAAFLGKASDLLRFPVKQLYTTSGKKV--DSLQALLHSPSV
LVCAGHEAFRTPAMKNARRSEAETLSGLTSRNKN-----GSWGPKTKPSVIHSRSPPGS-
TPRL----PERPGPSN-------PPVGPAPGRHPQDTPAQ--SGPLVAGDDMKKKVRMNE
DGSLSVEMKVRFHLVGEDTLLWSRRMGRASALTAASGEDPVLGEVDPLCCVWEGYPWGFS
EPGVWGPRPCRV----------GCREVFGRGG-QPGPKY-EIWT-NPLH-ASQGER--VA
ARKRWGLAQHVRC-SGLWGHGTAGRER-CSQDSASPAS-STGLPEGSEPESSCCPRTPED
GVDSA------SPSA-----QIGAERKAGGS------LGEDPGLCIDGAGLGGPE-----
------------------------------------QGGRLTPRARSEEGASSDSSASTG
SH------EGSSEWGGRPQGCPGKARAET--SQQEASE-GGDPASP--------------
-------------------------ALSLSSLRSDDLQAETQGQGT--------------
---EQ--ATGAAVTREPL------VLG-LSCSWDSEGASSTPSTCTSSQQGQRRHRSRAS
AMSSPSSPGLGRVAPRGHPRHSHYR----KDTHSPLDSSVTKQVP----RPPERRRACQD
GSVPRYSGSSSSTRTQASGNLRPPSSGSLPSQDLL-GTSSATVTPAVHS--------DFV
SGVSP-------------------HNAPSAGWAGDAGSRTCSPAPIPPHTSD--------
--------------------SCSKSG--AASLGEEARDTPQPSSPLVLQVGRPEQGAVGP
HRSHCCSQPGTQPAQ---------------EAQRGPSPEASWLCGRYCPTPPRGRPCPQR
RSSSCGSTG--SSHQSTARGPGGSPQEGT-RQPGPTPSPGPNSG-------ASRRSSASQ
GAGS---RGLSEEKT-LRSGGGPQGQEEASGVSPSSLPRSSPEAVVREWLDNIPEEPILM
TYELADETTGAAGGGLRGPEVDPGDDHSLEGLGEPAQAGQ-QSLEGDPGQDPEP--EGAL
LGSSDTGPQSGEGVPQG--AAPEGVSEAPAEAGADREAP---------AGCRVSLRALPG
RVSASTQIMRALMGSKQ----GRPSSVPEVSRPMARRLSCSAGALITCLASLQLFEE-DL
GSPASKVRFKDSPRYQELLSISKDLWP-GCDVGEDQL--DSGLWELTWSQALPDLGSH-A
MTENFTPTSSSGVDISSGSGGSGESSVPCAMDG-TLVTQGTELPLK-----TSNQRPDSR
TYESPGD-----LENQQQCCFPT--------FLNARACACATNED---------------
--------------------------------------------------------EAER
D-SEEQRASSNLEQLAENT-------------------VQEEVQLE-ETKE-GTEGEGL-
QEEAVQLEETKTEE-G-LQEEGVQLEETKETEGEGQ--------------QEEE------
-----------------------------------------------AQLEE--------
--IE----------ETGGEGLQEEGVQLEEVKEGPEGGLQGEALEEGLKE-EGLPE-EGS
VHGQELSEASSPDGK-GSQEDDPVQEEEAGRASASAEPCPAEGTEE--PTEPPSHLSETD
PSA-SERQSGSQLEPGLEKPPGATMMG--QEHTQAQPT---QGAAERSSSVACSAALD--
CDPIWVSVLLKKTEKAFLAHLASAVAELRARWGLQDNDLLDQMAAELQQDVAQRLQDSTK
RELQKLQGRAGRMVL--EPPREALTGELLLQTQQRRHRLRGL----RNLSAFSERTLGLG
------PLSFTLEDEP-ALSTALGSQLGEEAEGEEFCPCEACVRKKVSPMSPKATMG---
ATRGPIKEAFDLQQILQR---KRGE-HTDGEA----A-EVAPGKTHTDP----TST----
-----RTVQGAEGGLGPGLSQGPGVDEGED-------GEGSQRLNRDKDP----------
------------------------------------------------------------
------------------------------K----L---G---E---A---E---G---D
---A---M---A---Q--E---R--E---G---K--T---H--N---S---E--T-----
-------S---A---G--S---E---L---G--E---A---E--Q--E---G---E--G-
--I---S---E--R---G---E---T--G---G---Q--G---S--G---H---E---D-
--N--L---Q---G--E---A---A---A---G---G---D---Q--D---P---G----
-Q---S---D---G---A---E---G---I---E---A---P---E---A---E----G-
--E----A--------------------------------------QPESE-G-------
-VEAPE------AEGD--A-QEAEGEAQPESEDV--EAPEAE-GEAQPESEDVETPEAEW
EVQPESEGAEAPEAEKEAQPETESVEALETEGEDEPESEGAEAQEAEEAAQEAEGQTQPE
SEVIESQEAEEEAQPESEDVEALEVEVETQE-----AEGE--------A-QPESEDVEAP
EAEGEMQEAEEE--AQPESD--GVEA----------------------------------
----------------------Q----P-K--S-E----G----E----E----------
------------------AQEVEGETQKTEGDAQPESDGVEAPEAEEEAQEA--EGEVQE
AEGEAHPESEDVDAQEAEGEAQPESEGVEAPEAEGEAQKAEGIEAP--------------
-ETEGEAQPESEGI------------EAPEAEGEAQPESEGVEAQDAEGEAQPESEGIEA
QEAEEEAQPELEGVEAPEAEGEAQPESEGIE--APEAEGE--AQPELEGVEAPEAE----
----------EEAQPEPEGVE--TPEAEGEAQPESEGE--------TQGEKKGSPQVSLG
DGQSEEASESSSPVPEDRPTPPPSPGGDTPHQRPG-SQTGPSSSRASSWGN--CWQKDSE
NDHVLGDTR-SPDAKSTGTPHAERKATRMYPESS-TSEQEEAPLGSRTPEQGAS------
---EG-YDLQEDQALGSLAPTEAVGRADGFGQDDLDF
ENSOPRP00000002130 (view gene)
-----MNGAP---RDGQG-----PSHRECLLPSVARTLSATQVTPAKKITFLKRGDPRFA
GVR
PF03607 // ENSGT00920000149015_4_Domain_0 (64, 124)
LAVHQRTFKTFNALMDELSQRVPLAFGVRSVTTPRGLHGLNALEQLQDGGCYLCSDK
QPPKTPS-----R----LQ--RRSSSALQSWDPEGQVEVPGT-TPSGKAPRAPRRMTLVR
NGDPRFRQTVVLSHRNTRNLAAFLSKASDLLHFPVKQVYTLSGKKV--DSLQSLLHCPSV
LVCAGDEAFRPLLMEDAARNGSEMLSGLSSRGKN-----GSWGPESKQSVIHARTGSGS-
GQARRLALLSESGPN-------HAQRGPAPIRHPQDTPAA--PGPLVAGDDVEKKVHLNE
DGSLSVEMRVRFQLLGEEALLWSQRVGRAASR----EDCSSLGEAEPFSCTWQGAPWGSS
ESGARASRLWKQ-----------DFTALGHSQ-QPEPSY-EIWR-NPLY-TCQGEG--PA
AERRPGVAQHTRC-RRRWSQGTTGRQR-SLQDSASPAS-TVGCLENSELEFRSSSRTVEG
DLNTGSPAGLSCP------------------ERANDGLKERPSPCPEAVVP---------
------------------------------------QSDCRRHESQGSEEALSDWSTSAG
SP------EVPIEGNGLHQGCQHQARAPS--SQASA--QGSSLCAP--------------
-------------------------TLSPLSLANENLQGEKPGQDI--------------
---WHGQARSGSGSMGPL------LADGHDDLQHPE------RGHSTSQQGGGRQMSPAN
AVSSPC------------SGMCAHH----RGTHHLLHSPAAQQVP----RPPDSVRASPC
GSMPQLSEHEPMIRKLASGELRSPSLGSVYSQDPQSGACGVTLTPGSNP--------YCA
C--SS-------------------PTTPSTEPAGGTEDREHLPAPTPASTPH--------
--------------------SYPQSG--AGSQWGKAMSTPGPSPPLILLAGQPEQGARSP
A-SCDCSPIGM----------------------PEHFPEAPWACSGYHPTPPEGRPCAKK
PLRNLSHTS--SGDPSADGWEADGGQQGGKLKEGPLQLSGPGSAAVGTTVKS----SLNP
GTGP---RWMLEVKA-ARIGPDLKELEDDGSMTPSALPHASPDAVVREWLGNIPEEPVLM
RYEM-DETSGMAREGPEGPQGSPADDQG--------QGGQ-LLLEVDISEHLEA--NGVL
PGPSDTGTESRQASPQGQGVAPAPASDSPVEVRAGRGAL------------GRASCALPG
RVSASTQIMKVLMGAKP----GRPSSLPELSDVVAKRLSHSAVALIACLARLHFFDK-DP
GSPVDKVSFTDSPQYQELLSISQTLWP-RCSSGQDLL--DPELQELPPHKPLP-------
MSEDLTPTSSSGVDVSSGSGGSGEGSVPCAMDS-TGVPERTEPLLK-----SSSPRPGSR
ASGHAEN-----VEEPQPGCSAA--------SASSQIGTYATSKQ---------------
--------------------------------------------------------AAEG
G-SREQLVGNTLEQEIENV-------------------VQEDGVQLEETKEAENEEPH--
------------------------------------------------------------
------------------------------------------------------------
------------------------------------AG---------IQE-QGLPM-GTC
LSRLGSSEAGSAGSE-GSQDDQRKHKEE-------ARPCPSDRSEN--PSEPSQPLSKGP
SKT-SDSQRGPKSDPGLEEPPTAAAAVG-VSHKQAQVR--PWAACGRGKQVACRACLD--
PDPTWVSKLLKKMQMSFLTHLADATAQLRARWNLQDDGLLDQMVAELEQDLGQRLQASTD
KELRKIQSRAGRRAQGSGPAWEVLRGQTSLQTEQRRHRLHGL----RNLSALTDQTCGRG
------LFSLSLEDPL---------------SGEEFCPCEVCLRKKVTPMVSPRE-----
-AGAPIKKAFDLQQILKK---KEG-CPKGEAEAVATK------RTQKDPGVAMDMS----
-----NSGQGASGRQELGSDIELGAKEGDG-------GEGSQRFSRDKEP----------
------------------------------------------------------------
------------------------------P----A---G---D---A---E---G---A
---V---Q---E---S--G---D--S---D---A--R---G-------------------
-----------------------------------------------Q---G---G--A-
--V---V---G--K---A---K---T--H---A---E--G---S--E---D---E---D-
--S--L---E---A--A---A---L---E---G---S---S---W--T---A-----G--
-Q---D---D---A---A---A---A---A---E---V---P---G---A---G----G-
--G----G--------------------------------------QSGSEGG-------
-SAGDTEGSSQGHSGASQL-AEASENSSPDQGRR--PASSPA-PQGDTPKQRPDP-----
--QPRSSSSLSAPGCVCPPAHQKGPASELPSGDRRNSR------------AEPQGTPPP-
----------------DTRVTARCPGSSPLE-----QEGC--------PSCPRTPEQGAG
YACYESQGEEVA------------------------------------------------
---RGIAHTQVGD---KADGFGQDDLDF--------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
ENSMOCP00000013009 (view gene)
-----MNSTPGDMRDAPA-----PSHRQCLLPSVAHTPSVTEVTPAKKITFLKRGDSRFA
GVRL
PF03607 // ENSGT00920000149015_4_Domain_0 (65, 122)
AVHQRTFKTFSALMDELSQRVPLSFGVRSVTTPRGLHGLSALEQLQDGGCYLCSDK
KENSGT00920000149015_4_Linker_0_1 (122, 191)
PLKTPREP--GQ----LQ--RKSPSAGQSRGFEGGHEAPEI-SSSWKGPVAPRRLTLVK
NGNPRCQQTVPF03607 // ENSGT00920000149015_4_Domain_1 (191, 249)
VVSHRNTRNLKAFLSKASELLRFPVKQVYTPSGKKV--DSLQTLLDGPSL
LVCAGNEAFRHLEMENDGRHRTRTLSGVTARSGR-----GCWGPNAKQSVMHSRARSGS-
RLQQFSLMSERSGPSGHPTSGHQTWAGPTLDKHSRDTPGP--PGSLVAADDVEKKVCMNE
DGSLSVEMKVRFQILGEDTLRWPQRVGHASVFAPASGEGQVPREADPFCCKQEGHPWGFL
THGAQGLEPCDG----------RYQGAFDMDQ-QSQPNY-DIWR-NPLT-TSGGAG--PT
RRRRWGLSKLSGC-RSHWNQTANDKKG---HSNNSPVS-GPRHPRSAQPG-SSCPWTSEG
ETDSDTLHPASSTSS-----LSGAELGAGKD------------RCLEDT-----------
------------------------------------ASCGLGSETQGIEKALSDTSASAE
SH------EMSRECREQHHRGSGQARVTV--SQGQATQGCDRPCVS--------------
-------------------------TQSPSSLSHMHLQTEKYTQGT--------------
---TYQEVRGEPELRLPL------VPG-HSASQDTQRHTLPAPGGAPAQRRQKKQKRPAG
IVCLPGASVPYQGAQKGHTRQCHCC----RDTQSSLDTVLQRQMP----E--EGEQAYPG
GLAPRFSPHSPSAENEVSGDLKSPLSSSLDFQDPQ-ATSVATILPTSDS--------DCA
SSFCH-------------------PNTRSAEPAGDTECPAHSPASIPVHSG---------
-------------------------E--VGYLWDKAGTNPEPFSASVLLDKWPEAEDPRT
HQDCCGLQVTASPVLTGPSGQT-----------QAPISEAYWGTPSFCPTPPQEQTCSRK
DPTSSNSPS--SDHGQTDGQA---------------------------------------
--EH---RKTLLGKS-PGNKGSLEEQEGDGSVTPSALPYTSPDAVVREWLGNIPEKPALM
TCDMEGEATKVASDGPESAEEDLV-------LRELAQARQ-QPPEGATDEQPET--TGDL
PGTGSVCCKPGGDACQD--AASGRGVEAATEAGIEEGTA---------VDHGVSLCALPS
RVSASTQIMKALLGSKP----GRPSSLPEISSTAAQRLSYSAGTLIACLARLHFFDE-DL
GPPDGNARFEESPKYKEILSISKALWP-ASVLGQGQLDMDSGLRKLTSHQALLG------
-TEDFTPTSSSGVDVSSGSGGSGEGSVPCAT-----APDRADLPLK-----IPSPRPDSR
KQGYPEV----------PSHSTA--------SPDPQVGACATSGE---------------
--------------------------------------------------------ESSK
S-GREQTWDKAPEQSMEST-------------------MLEEEAQSQETEGR--------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------VRERLQEDGVHGKEVPQE-RAG
VCSRGMLQAGSRDGE-HSPEDTRASKDEEGRDATSGALWPLDGREE--PTETPHSFSESN
SNV-SESQRVRVLEMRLEEMSGVAAMD--CKQAY--------------VKATHRESLD--
PDPIWVSKLLKKIEKDFMAHLAGATAELRARWNLPDDSLLDQMVTELEQDVGRRIQASTV
REVRKIQSRVGRMVP--EPPREALRGQTSLQTELRRRRLQGL----RNFSAFP----GQG
------ALSLSLEDGP-ILSTALGTSPGVGIMRDEFCPCEVCLKKKMSPQFPKDATA---
AFSAPVRKAFDLQQILQN---KKGG-RNGEAMELAPQ-KTGMLQL---------------
-------------------TAGLGIDEGEE-------GEGRPRMREEKDP----------
------------------------------------------------------------
------------------------------E----F---F---N---V---E---G---A
---G---C---H---G--A---G--E---D---A--A---T--V---T---E--R-----
-------E---L-H-V--S---V---A---R--E---S---Q--Q--L---E---E--G-
--G---T---K--K---G---E---A--P---Y---Q--S--------------------
----------------F---R---D---R---E---A---S---E--A---S---G-R--
-Q---S---D---G---G---H---S---V---D---T---Q---D---T---S----R-
--E----R--------------------------------------QPEVEGG-------
-SQDEKENGTWVSPGEGQR-RVVSENNSLDQEGG--LHKCHR-KPGPQC------HR---
----AALCSRASSSGSCSHVSQKSSEEDLSRGELKCTKDKH-------------------
----------SRVSQAERKVTTMCSESSSE------QE-V--------PSSPRPPKQGEG
EEV---------------------------------------------------------
----SLVCSQLGG---RSDGFGQDDLDF--------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
ENSPEMP00000027522 (view gene)
-----MNSTPGGMREAPA-----PSHRQCLLPSVAHTPSVTEVTPAKKITFLKRGDPRFA
GVRL
PF03607 // ENSGT00920000149015_4_Domain_0 (65, 122)
AVHQRTFKTFSALMDELSQRMPLSFGVRSVTTPRGLHGLSALEQLQDGACYLCSDK
KENSGT00920000149015_4_Linker_0_1 (122, 191)
PPKTPREP--GR----LQ--RKSPSAGQSRGHEGGREAPET-SSSWKGPMAPRRMTLVK
NGNPRCQQTVPF03607 // ENSGT00920000149015_4_Domain_1 (191, 249)
VLSHRDTRNLTAFLSKASELLRFPVKQVYTISGKKV--DSLQTLLDGPSL
LVCAGNEAFRRLEMENDGGNRTRTFSGVTARSER-----GCWGPNAKQSVMHSRARSGR-
RLRQFSLMSE----SDHPASGHQAWPGPALNRCPQDTPGP--PGSLVAADDVEKKVCMNE
DGSLSVEMKVRFHLLGEDTLRWSQRVGHASVCTPASGEGQVLREADPFCYRQEGHPWRFL
AQGTQGWGPCDG----------GYQRAFDVDQ-ESQPNY-DIWR-NPLT-TPGGAG--ST
QRRRWGLSKLSGC-RS---QGANDRKG-HSNNSVSPLS-SPRHPRSAQPG-SRCPWTPEG
ETDSDTLHPVSSASS-----QSGAELEAGKG------------SCLEDTGP---------
--------------------------------------CDLGPETRGIERALSDTSVSAE
SP------EGSSECGSQHHRGSSQVRVTV--SQRQATQ-GDGSCVP--------------
-------------------------THSLFSLSHMDPQTEKCRQST--------------
---RCQEARGEPELRLPL------VPS-RFSSLDTQRDALPAPASAPAQRRQKKQKRPAG
IMCLPSVPASCQVAQNGHARQCDYC----RNTQSSLDT--N---A----IPQEREQACPG
GPTPLSPPNSPRAGHQASSNVRSPFSSSLDFQGPQ-ATSKATTIPMSDS--------DCA
SSVYH-------------------PRTHSAEPAGDPECPAHSPTPTPVHTGD--------
----------------------------VGCLWDKAGTTPEPPSSSVSLDRQPEAEDPRP
HQDCCCSQVAASSVLTAPSGQT-----------QAPISEACWGASSFCPTAPQEQICSRK
DPTSCSSTS--SGQKQADDHA---------------------------------------
--EP---RKTLLGKS-PGIRGSLEEQEGDGGVTPSALPYTSPDAVVREWLGNIPEKPVPM
TYEIEGETTEVAHDGPESPEDDLA-------RRELAQARQ-QPPEGVTDEQPEP--AGAL
PRTGSVCCKLGDDPRHD--TASGEGAKAPAEAGIGEGTT---------VDHGVSLCALPS
RVSASTQIMKALLGSKP----GRPSSLPEVSSTVAQRLSFSAGALIACLARLHFFDE-DL
GPLDSNARLEESPRYKEMLSISRALWP-GSELGQSQL--DIGLRKLTSHQALLG------
-TEDCTPTSSSGVDVSSGSGGSGESSVPCALDNNTLAPERVDLPLK-----IPSQRPDSR
NQGYPEG----------LSHSAA--------SSGSQAWAGPASGE---------------
--------------------------------------------------------ESGK
G-GRKRKWGNAPEQSAEST-------------------RPEEEAQSEEAEGRVR------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------ETLQKDGVQG-EGLPEEKAG
VCSPEMPRAGSRDGE-SYPEDTKVSKDEAGRDAASGGLWPLDGREE--PTESPHRFSGSN
SND-RESPSVHILEQGLEEVSTVAAMG--CEQASIKT----------STSTAHRGCLD--
PDPVWVSKLLKKIEKDFMAHLAGATTELRARWNLHDNSLLDQMVTELEQDVGRRLQASTV
REVRKIQSRQGRMVP--EPPREPLRGQTSLQTEQRRRRLQGL----RNFSAFP----GQG
------PLSLTLEDGP-TLSTALGTRPGVGAVGDEFCPCEVCLKKKKIPKSPKDAAG---
VSSAPVKKAFDLQQILQS---KKGG-RNREAMEVAPQ-KAGMMLPQEDP-----------
-----RTVQATDGKQELNE--------GEE-------GEERQRLRGETDP----------
------------------------------------------------------------
------------------------------E----FL------K---E---E---G---A
---G---C---HV--P--E------E---D---A-AT---S--R-------E--------
-------R---EIHIS------A---A---Q--E---S---R--Q-LE----------E-
--S-VTK------K---E---E---T--P---H---P------RF-R-----------D-
--G----------DASE---V------------------------------PGRQG----
-----D---G---S-------H---S---V---E---T---Q---D---A---S----R-
--E----R--------------------------------------QPEVEGG-------
-NQDEKENGPWVSSGESQR-GVVSENISLDHEGR--LHNRHR-RPGPQRR----------
---PAAHCTRASSPSSCSQVSQKGSEEDFPSGALKCTKAKS-------------------
----------SRDLDAERKVTMMYPESSSSE-----RET---------PSSPRPPKQEEG
D----T------------------------------------------------------
--AGSLICTQVGG---RVDGFGQDDLDF--------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
ENSSHAP00000013165 (view gene)
-----MNYAL---DDV-----PSQNHHEYAFPLVARATTVTQVPPAKTITFFKKGDPRFG
GVR
PF03607 // ENSGT00920000149015_4_Domain_0 (64, 122)
MVVNQRMFKSFNALMDELSHHIPLSFGVRTITTPQGVHCINSLEQLEDGGCYICSDR
NENSGT00920000149015_4_Linker_0_1 (122, 192)
PSKALGG--SHKQSTASSASGYPTQRL------GNSESWQE-TPPWQGPNAPRKLTLVK
NGDIGFRRSLIPF03607 // ENSGT00920000149015_4_Domain_1 (192, 250)
LSLKNTQNLKAFLNDASEVLHFPVNQLYNTDGSKV--ESLQSLLQGPSI
VVCAGQEPFRPMAVESSRRRLPEKLPGLSSKKS-----NANWELKAKKSVIHSRSESSN-
RLRQLSLTSENS-----------GSPVPTTNTNASERSPC--AIPFGPNDDIEKKVHVNK
DGSLSVEMKVRFRLLSEEMLQWSTQVRKLSHLREANGDEPGLRGVA-FHPSRENNVQRSS
EMGT-FLCP----------NETNSQGLEEVSYLQPQPNY-EIWK-NPMF-VSQGHG--AA
PGR-SSWVTQSPC-SGASGEPDSPRKRSQKTQ---E--------------------PKAG
REW---------PPDEEEG---------------------------------------NK
ACSLDHLKDEAWVATED---SS-------------VCSRCREKRMPMVEG----------
-CGSPRQEGAFCPYVREAKGEATVAHYLP--SLD--------TACDLSRQVMPQVNDAQE
GSCASLVGT--------------------PSLKNKALEAEEDAH----------------
-SRVRQDTSSGSVQNMPR------SVINGEFSDPE-YNCIPYSSCPSPGSGSKGWKSPRS
DGSLHNISHTSTAHEGSKSSQLKIKDNGDRGTSSILEGSPERSLPRSVLSSPLLEKSRAD
NQTPISSRSSSETKQQTARDPGSSSSSPH--------------------------YLECL
SWLSDASNHN------------------------SGQKKLRNISNSSQSGSN--SLEKES
REATKA-----------------------------------SVSSLELKNKEPQVAESNT
RRSSCS--------SQSSRTQPYRTYTKGKNDSRGPHSWASSAASKSLPTPPRGTPHFRK
SKHAAIKHSSSNSKWNANLEGSGSLEEKANLSLSGSITPGSRSVTPSQTAKRARKGGPSP
GRERKMLRKNSREGKPHKEKRNVGKPEDSTGMKLSDLPSTSPEEVVYEWLSKIPEESILM
RCEMQDDSPVTA---MEASEENLDVNCSSEVPSEMAQAQE-NVLERKDSGGTSS---DEN
PRLDAASPKARSSLSKE------RSSSTLAT--ITEGSPGECADLGG-KNRLSKIEDLPS
PLYSSVQIMKALLSSKQSPKLGRFNSLPEVSQSTGRKLSHSAHVLITCLASLHFFDE-EL
VTVTNQLTYMNSPKYQELLSIFQDLWTESSPS-----------TDESH------PKLQTP
VTGDFSPTSSSGVDVSSGSAGSGEGSVPGVADC-VPLPGKIEISKPE------VGG-GSA
ASGEE----NVAG---VVDCVPLSEKMEDSKLKRQDSITCHDDMDIEKKK----------
------------------------------------------------------------
--S---------------FSNDQVKIEGQSAE----------------------------
--------GDEEG----SSTHD--VTNRDE--------------TNEDIK------EEML
ESSTGMVDEDEIKEVGMEEEKMLEKEMQEEETQDEVMEAEMI--EEEKQ-----------
--AE---ETKDEK------------------------VLMERM------------QEEEG
IQEEE-----------------------ASKKCDSIDSCPPVKTEE--VTEQVSNPSKDD
SNS-DHIHDSQELEPHLKEV-----AEGAVVVAEQSVVNTNSGTGGKRASIIQNTSPD--
PDPLWVLRLLKKIEKEFMTHYVNAMSEFKVRWNLQNNEILDQMIIELKNEVNQRIQKSIE
KELRKIQTRAGKKLP--KPPREAFRWDDSLQTEQRRRRLKGM----RNHSAFTEQNKIPE
----KRYLSLAFTG-ALASSETLADDLRELEGEGEYCPCDTCIRKKITSMSLKNPAP---
VTDAPLRKAFDLQQILLK---KKREHTGNEAVDEAE----------PEPTS---------
----NGCMDGTDAGEENH--R-----------------------------------SR--
------------------------------------------------------------
-----------------------------KDGESDLH-ER---EEQEA-----------N
HEET---MTG--------E---E--E---E---A--D--LQ--N----------S-----
-------E---E---G------EKEEK---A--E---S---D--L-KEDEDEKQDE--GE
S-A--ADG-SE--R---K------------------------------------------
----------------ETDSV---SEHGANPEGEATGD-----------------EDGCS
EVS---------GD---A---E-----------E---L---Q-------A--------I-
--E----P-----------------------------VSPEAEVEKQIESQEGSEKEVSN
GDCHEAENLEAEPNGE--------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
ENSAPLP00000012830 (view gene)
------------------------------LPSVARASTVTQVPPAKKITFFKSGDPQFG
GVK
PF03607 // ENSGT00920000149015_4_Domain_0 (64, 124)
MAINQRSFKSFNALMDDLSHRVPLPFGVRTITTPRGIHCISELDQLEDGGCYLCSDK
KYVENSGT00920000149015_4_Linker_0_1 (124, 190)
KPISITSAGHRPG-PPRSSRPSSSLRRAAQEGRFEDYST-PFTQHGPRIPKKITLVK
NGETSFRRSPF03607 // ENSGT00920000149015_4_Domain_1 (190, 250)
IILNRRNARSFKTLLDEISEILQFPVKKLYTADGKKI--DSMQALLHCPNM
LVCVGREPFKPVSMENLRKHSVEKLPDLAPRSN-SNNTPVNFGLKAKKSIIHPRSASSN-
RSMRFSLSSEKSYPNGLAASPDTGAPFSNNCSHP--KPGE--LVHSLVNDDIEKRVHVNK
DGSLSVEMKVRFRLLNDETLQWSTQIRKSNLMNQMPCEESGVE-DSGVDPIQKMNPEASS
EADD-SLYPCDVDSYMSKLEESECDEAHCHSCGKKHQDY-DIWK-NPMH-TSQREE---P
SVR-STWHTRSSC-SSTSSRRRVIHKKKASVDSLHTTS-SEEFSEHIVQESSSYSETIEN
RVAYRSIRKCTCRSDLSTGASNGEDQQEYTR------TSTSNSQRPSSVGLVSHSSCENN
-----LDTQEA---PEDYEASKTNSQKDDENCFEVSSVKCLKHDAEEVESSRAGSVI---
--------------------------------LQKA-NQEDNAMCSTPANSVH----SRS
TNCSTPRA-KMSPALRKKLRRLN---------KKKMKSMKSAQF----------------
----QQKQSAANQLEMRAVK-----VNN--ALHKEPPVPY--------------------
----------------------------AMPLA-LPEGP--DPMS---HNS-GSFH----
-----------------------------SNIS---STSEAEAIAVVTEDKSSKSTTKGY
SESSKGSKKKSHGEENSKPSSLCSSISSPNGKAGEGHYKMNSEAPSSRHGSISESESDD-
------------------------------------------------------------
---RCS-QSNISQSSKSRQIKTRNLHVKTSNSSRVSLSETVSICSMHCPAPPKDSSK---
-----------------------------------PTSFGSKSVATEANDQK--------
---NKELDDSCNGKVEEVLE-DTGMQEEWSNLMPSILPNTSPEEVVQEWLSKIPAETLLM
KYEMEDGAEEEC---VEAATEVSYCTNAEEISEDEKAEKK-NAEEAENEAADEV--EKET
AEVTAVDEGTEECVNQE------QIAETLSE--AAKADQAECSRSVT-SSQNNNRRDLPV
SVQTSVQIMKALLSSKHEAKFDRSNSLPEVSPTMGRKLSNSANILITCLASLQLLDE-ES
EDPSNKSKCLNKPRYMELLNIFQALWF-GRTAEKSGP--SSGLEASALEKTACGFKAPKS
VDDDFTPMSSSGVDVSSGSGGSGEESTAGARDC-NLLAQKTVELKSD------GQV-ENV
-ETVTE--QMEAEE----QSKQEEH-------SSRPSTACSKSKGEKKAR----------
------------------------------------------------------------
--S---------------------------------------------------------
TREDSVTQ--KAE----ENKED-------------------------QCSN---CENGQE
ERGENENEE-----------------------NIKLLEN--G--EQEVLVE---------
----------------------------------EVEAVN--------------------
------------------------------------KALPKENIEE--AADQLATNIGA-
-NV-SDADCGTNLKDDLDDH--L--GKESVKDPESKVNI----ATSVGASLAQQNSVD--
PDPIWVLRLLKKIEKEFMAHYVNAMNEFKVRWNLQDNQQMDEMILELKEEVSKRIQKSIE
KELKKIKSRAGAKMP--RPPDEPLRRESTLQTEQRRRRLQTM----HKKSL---------
-FTDTSDLSLDADD-DTVFSAAFEASISEQPSEEEYCPCDTCVRKKMASKPTKTPVV---
ASNAPVMKAFDLQQILRG---KK-------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------
{"ENSMEUP00000004761": [["Domain_0", 64, 124, "PF03607"], ["Linker_0_1", 124, 190], ["Domain_1", 190, 250, "PF03607"]], "ENSRNOP00000064085": [["Domain_0", 65, 122, "PF03607"], ["Linker_0_1", 122, 191], ["Domain_1", 191, 249, "PF03607"]], "ENSMUSP00000055449": [["Domain_0", 65, 122, "PF03607"], ["Linker_0_1", 122, 191], ["Domain_1", 191, 249, "PF03607"]], "ENSMAUP00000004680": [["Domain_0", 65, 122, "PF03607"], ["Linker_0_1", 122, 191], ["Domain_1", 191, 249, "PF03607"]], "ENSPPYP00000020575": [["Domain_0", 64, 124, "PF03607"], ["Linker_0_1", 124, 190], ["Domain_1", 190, 229, "PF03607"]], "MGP_CAROLIEiJ_P0037115": [["Domain_0", 65, 122, "PF03607"], ["Linker_0_1", 122, 191], ["Domain_1", 191, 249, "PF03607"]], "ENSCGRP00000017687": [["Domain_0", 65, 122, "PF03607"], ["Linker_0_1", 122, 191], ["Domain_1", 191, 249, "PF03607"]], "ENSPANP00000006324": [["Domain_0", 65, 122, "PF03607"], ["Linker_0_1", 122, 191], ["Domain_1", 191, 249, "PF03607"]], "ENSRROP00000021794": [["Domain_0", 65, 122, "PF03607"], ["Linker_0_1", 122, 191], ["Domain_1", 191, 249, "PF03607"]], "ENSCANP00000004213": [["Domain_0", 65, 122, "PF03607"], ["Linker_0_1", 122, 191], ["Domain_1", 191, 249, "PF03607"]], "ENSMLUP00000015234": [["Domain_0", 64, 124, "PF03607"], ["Linker_0_1", 124, 190], ["Domain_1", 190, 250, "PF03607"]], "ENSRBIP00000035685": [["Domain_0", 65, 122, "PF03607"], ["Linker_0_1", 122, 191], ["Domain_1", 191, 249, "PF03607"]], "ENSCAFP00000011953": [["Domain_0", 64, 124, "PF03607"], ["Linker_0_1", 124, 190], ["Domain_1", 190, 250, "PF03607"]], "ENSACAP00000018459": [["Domain_0", 64, 120, "PF03607"], ["Linker_0_1", 120, 191], ["Domain_1", 191, 250, "PF03607"]], "ENSCCAP00000013401": [["Domain_0", 65, 122, "PF03607"], ["Linker_0_1", 122, 191], ["Domain_1", 191, 249, "PF03607"]], "ENSSBOP00000014364": [["Domain_0", 65, 122, "PF03607"], ["Linker_0_1", 122, 191], ["Domain_1", 191, 249, "PF03607"]], "ENSLAFP00000014874": [["Domain_0", 64, 121, "PF03607"]], "ENSMPUP00000001748": [["Domain_0", 64, 124, "PF03607"], ["Linker_0_1", 124, 190], ["Domain_1", 190, 250, "PF03607"]], "ENSPVAP00000009834": [["Domain_0", 64, 121, "PF03607"]], "ENSSTOP00000020260": [["Domain_0", 65, 122, "PF03607"], ["Linker_0_1", 122, 191], ["Domain_1", 191, 249, "PF03607"]], "ENSMLEP00000032547": [["Domain_0", 65, 122, "PF03607"], ["Linker_0_1", 122, 191], ["Domain_1", 191, 249, "PF03607"]], "ENSFCAP00000028643": [["Domain_0", 66, 122, "PF03607"], ["Linker_0_1", 122, 191], ["Domain_1", 191, 249, "PF03607"]], "ENSMFAP00000033163": [["Domain_0", 65, 122, "PF03607"], ["Linker_0_1", 122, 191], ["Domain_1", 191, 249, "PF03607"]], "ENSNLEP00000018242": [["Domain_0", 65, 122, "PF03607"], ["Linker_0_1", 122, 191], ["Domain_1", 191, 249, "PF03607"]], "ENSOGAP00000020959": [["Domain_0", 64, 123, "PF03607"], ["Linker_0_1", 123, 190], ["Domain_1", 190, 250, "PF03607"]], "MGP_PahariEiJ_P0085219": [["Domain_0", 65, 122, "PF03607"], ["Linker_0_1", 122, 191], ["Domain_1", 191, 249, "PF03607"]], "ENSCSAP00000012938": [["Domain_0", 64, 122, "PF03607"], ["Linker_0_1", 122, 190], ["Domain_1", 190, 250, "PF03607"]], "ENSGGOP00000000667": [["Domain_0", 65, 122, "PF03607"], ["Linker_0_1", 122, 191], ["Domain_1", 191, 249, "PF03607"]], "ENSCATP00000038772": [["Domain_0", 65, 122, "PF03607"], ["Linker_0_1", 122, 191], ["Domain_1", 191, 249, "PF03607"]], "ENSTBEP00000001026": [["Domain_0", 64, 122, "PF03607"]], "ENSOCUP00000009111": [["Domain_0", 64, 123, "PF03607"], ["Linker_0_1", 123, 190], ["Domain_1", 190, 250, "PF03607"]], "ENSMGAP00000015014": [["Domain_0", 64, 124, "PF03607"], ["Linker_0_1", 124, 190], ["Domain_1", 190, 250, "PF03607"]], "ENSGALP00000040738": [["Domain_0", 65, 120, "PF03607"], ["Linker_0_1", 120, 192], ["Domain_1", 192, 249, "PF03607"]], "ENSPCAP00000010849": [["Domain_0", 64, 121, "PF03607"]], "ENSETEP00000014171": [["Domain_0", 64, 121, "PF03607"], ["Linker_0_1", 121, 190], ["Domain_1", 190, 250, "PF03607"]], "ENSPSIP00000006505": [["Domain_0", 64, 121, "PF03607"], ["Linker_0_1", 121, 191], ["Domain_1", 191, 250, "PF03607"]], "ENSOARP00000016221": [["Domain_0", 64, 122, "PF03607"], ["Linker_0_1", 122, 190], ["Domain_1", 190, 250, "PF03607"]], "ENSMODP00000028679": [["Domain_0", 64, 123, "PF03607"]], "ENSTTRP00000014723": [["Domain_0", 64, 124, "PF03607"]], "ENSBTAP00000054813": [["Domain_0", 64, 122, "PF03607"], ["Linker_0_1", 122, 190], ["Domain_1", 190, 250, "PF03607"]], "ENSCGRP00001001143": [["Domain_0", 65, 122, "PF03607"], ["Linker_0_1", 122, 191], ["Domain_1", 191, 249, "PF03607"]], "ENSAMEP00000017019": [["Domain_0", 64, 124, "PF03607"]], "ENSMNEP00000027443": [["Domain_0", 65, 122, "PF03607"], ["Linker_0_1", 122, 191], ["Domain_1", 191, 249, "PF03607"]], "ENSMMUP00000044936": [["Domain_0", 65, 122, "PF03607"], ["Linker_0_1", 122, 191], ["Domain_1", 191, 249, "PF03607"]], "ENSANAP00000018750": [["Domain_0", 65, 122, "PF03607"], ["Linker_0_1", 122, 191], ["Domain_1", 191, 249, "PF03607"]], "ENSTGUP00000013707": [["Domain_0", 64, 121, "PF03607"], ["Linker_0_1", 121, 191], ["Domain_1", 191, 229, "PF03607"]], "ENSFALP00000011381": [["Domain_0", 64, 124, "PF03607"], ["Linker_0_1", 124, 191], ["Domain_1", 191, 250, "PF03607"]], "MGP_SPRETEiJ_P0038846": [["Domain_0", 65, 122, "PF03607"], ["Linker_0_1", 122, 191], ["Domain_1", 191, 249, "PF03607"]], "ENSECAP00000010663": [["Domain_0", 64, 124, "PF03607"], ["Linker_0_1", 124, 190], ["Domain_1", 190, 250, "PF03607"]], "ENSPPAP00000002626": [["Domain_0", 65, 122, "PF03607"], ["Linker_0_1", 122, 191], ["Domain_1", 191, 249, "PF03607"]], "ENSJJAP00000006578": [["Domain_0", 65, 120, "PF03607"], ["Linker_0_1", 120, 191], ["Domain_1", 191, 249, "PF03607"]], "ENSPTRP00000092368": [["Domain_0", 65, 122, "PF03607"], ["Linker_0_1", 122, 191], ["Domain_1", 191, 249, "PF03607"]], "ENSP00000371923": [["Domain_0", 65, 122, "PF03607"], ["Linker_0_1", 122, 191], ["Domain_1", 191, 249, "PF03607"]], "ENSOPRP00000002130": [["Domain_0", 64, 124, "PF03607"]], "ENSMOCP00000013009": [["Domain_0", 65, 122, "PF03607"], ["Linker_0_1", 122, 191], ["Domain_1", 191, 249, "PF03607"]], "ENSPEMP00000027522": [["Domain_0", 65, 122, "PF03607"], ["Linker_0_1", 122, 191], ["Domain_1", 191, 249, "PF03607"]], "ENSSHAP00000013165": [["Domain_0", 64, 122, "PF03607"], ["Linker_0_1", 122, 192], ["Domain_1", 192, 250, "PF03607"]], "ENSAPLP00000012830": [["Domain_0", 64, 124, "PF03607"], ["Linker_0_1", 124, 190], ["Domain_1", 190, 250, "PF03607"]]}

