Found:
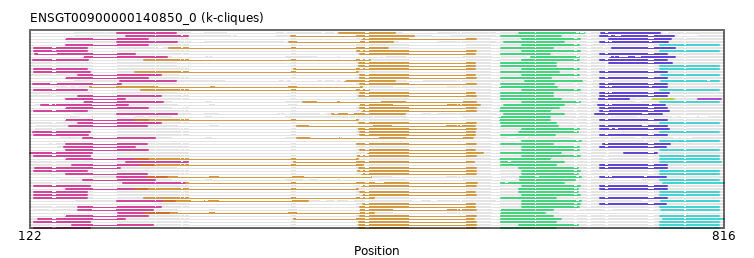
{"ENSMOCP00000023055": [["Domain_0", 170, 253, "PF01391"], ["Domain_1", 451, 568, "PF01391"], ["Linker_1_2", 568, 593], ["Domain_2", 593, 648, "PF01391"], ["Domain_5", 752, 811, "PF01391"]], "ENSACAP00000016639": [["Domain_0", 209, 268, "PF01391"], ["Linker_0_1", 268, 431], ["Domain_1", 431, 487, "PF01391"], ["Linker_1_2", 487, 611], ["Domain_2", 611, 676, "PF01391"], ["Linker_2_3", 676, 693], ["Domain_3", 693, 752, "PF01391"]], "ENSTSYP00000026073": [["Domain_0", 158, 240, "PF01391"], ["Domain_1", 452, 568, "PF01391"], ["Linker_1_2", 568, 593], ["Domain_2", 593, 645, "PF01391"], ["Domain_5", 752, 811, "PF01391"]], "ENSSBOP00000017733": [["Domain_0", 126, 179, "PF01391"], ["Linker_0_1", 179, 236], ["Domain_1", 236, 466, "PF01391"], ["Linker_1_2", 466, 593], ["Domain_2", 593, 648, "PF01391"], ["Linker_2_3", 648, 692], ["Domain_3", 692, 759, "PF01391"]], "ENSPANP00000011005": [["Domain_0", 158, 240, "PF01391"], ["Domain_1", 227, 450, "PF01391"], ["Domain_2", 592, 642, "PF01391"], ["Domain_5", 752, 811, "PF01391"]], "ENSANAP00000039724": [["Domain_0", 158, 240, "PF01391"], ["Domain_1", 454, 568, "PF01391"], ["Linker_1_2", 568, 614], ["Domain_2", 614, 672, "PF01391"], ["Linker_2_3", 672, 701], ["Domain_3", 701, 763, "PF01391"]], "ENSJJAP00000019986": [["Domain_0", 170, 253, "PF01391"], ["Domain_1", 451, 568, "PF01391"], ["Linker_1_2", 568, 593], ["Domain_2", 593, 645, "PF01391"], ["Linker_2_3", 645, 692], ["Domain_3", 692, 759, "PF01391"]], "ENSGALP00000010837": [["Domain_1", 182, 454, "PF01391"], ["Domain_2", 592, 649, "PF01391"], ["Linker_2_3", 649, 692], ["Domain_3", 692, 759, "PF01391"]], "ENSDORP00000010363": [["Domain_0", 170, 253, "PF01391"], ["Domain_1", 452, 567, "PF01391"], ["Linker_1_2", 567, 614], ["Domain_2", 614, 671, "PF01391"], ["Domain_5", 753, 811, "PF01391"]], "MGP_SPRETEiJ_P0027891": [["Domain_0", 126, 182, "PF01391"], ["Domain_1", 451, 568, "PF01391"], ["Linker_1_2", 568, 593], ["Domain_2", 593, 648, "PF01391"], ["Domain_5", 752, 811, "PF01391"]], "ENSCAFP00000041451": [["Domain_0", 126, 179, "PF01391"], ["Domain_1", 261, 480, "PF01391"], ["Linker_1_2", 480, 593], ["Domain_2", 593, 645, "PF01391"], ["Linker_2_3", 645, 704], ["Domain_3", 704, 767, "PF01391"]], "ENSCGRP00000019253": [["Domain_1", 451, 567, "PF01391"], ["Linker_1_2", 567, 593], ["Domain_2", 593, 648, "PF01391"], ["Linker_2_3", 648, 701], ["Domain_3", 701, 764, "PF01391"]], "ENSAMEP00000006758": [["Domain_0", 218, 280, "PF01391"], ["Linker_0_1", 280, 383], ["Domain_1", 383, 506, "PF01391"], ["Linker_1_2", 506, 593], ["Domain_2", 593, 653, "PF01391"], ["Linker_2_3", 653, 692], ["Domain_3", 692, 766, "PF01391"]], "MGP_CAROLIEiJ_P0026519": [["Domain_0", 126, 182, "PF01391"], ["Domain_1", 451, 568, "PF01391"], ["Linker_1_2", 568, 593], ["Domain_2", 593, 648, "PF01391"], ["Domain_5", 752, 811, "PF01391"]], "ENSTGUP00000015550": [["Domain_0", 210, 261, "PF01391"], ["Domain_1", 236, 466, "PF01391"], ["Linker_1_2", 466, 593], ["Domain_2", 593, 637, "PF01391"]], "ENSOPRP00000006615": [["Domain_0", 122, 184, "PF01391"], ["Domain_1", 460, 575, "PF01391"], ["Linker_1_2", 575, 593], ["Domain_2", 593, 653, "PF01391"], ["Linker_2_3", 653, 716], ["Domain_3", 716, 749, "PF01391"]], "ENSPEMP00000027771": [["Domain_0", 127, 212, "PF01391"], ["Domain_1", 451, 568, "PF01391"], ["Linker_1_2", 568, 611], ["Domain_2", 611, 670, "PF01391"], ["Linker_2_3", 670, 692], ["Domain_3", 692, 759, "PF01391"]], "ENSMFAP00000002084": [["Domain_0", 176, 255, "PF01391"], ["Linker_0_1", 255, 395], ["Domain_1", 395, 568, "PF01391"], ["Linker_1_2", 568, 614], ["Domain_2", 614, 672, "PF01391"], ["Domain_5", 752, 811, "PF01391"]], "ENSCATP00000002267": [["Domain_0", 176, 259, "PF01391"], ["Domain_2", 623, 671, "PF01391"], ["Linker_2_3", 671, 702], ["Domain_3", 702, 767, "PF01391"]], "ENSOANP00000007926": [["Domain_2", 614, 673, "PF01391"], ["Domain_5", 752, 811, "PF01391"]], "ENSMUSP00000099826": [["Domain_0", 125, 182, "PF01391"], ["Domain_1", 452, 567, "PF01391"], ["Linker_1_2", 567, 611], ["Domain_2", 611, 670, "PF01391"], ["Domain_5", 752, 811, "PF01391"]], "ENSPCAP00000006490": [["Domain_0", 218, 280, "PF01391"], ["Linker_0_1", 280, 386], ["Domain_1", 386, 566, "PF01391"], ["Linker_1_2", 566, 593], ["Domain_2", 593, 656, "PF01391"], ["Domain_5", 752, 813, "PF01391"]], "ENSMICP00000004945": [["Domain_0", 158, 240, "PF01391"], ["Domain_1", 452, 568, "PF01391"], ["Linker_1_2", 568, 593], ["Domain_2", 593, 651, "PF01391"], ["Linker_2_3", 651, 692], ["Domain_3", 692, 759, "PF01391"]], "ENSTGUP00000001193": [["Domain_0", 170, 245, "PF01391"], ["Domain_1", 452, 568, "PF01391"], ["Linker_1_2", 568, 593], ["Domain_2", 593, 645, "PF01391"], ["Domain_5", 753, 811, "PF01391"]], "ENSTTRP00000010067": [["Domain_0", 130, 217, "PF01391"], ["Domain_1", 386, 566, "PF01391"], ["Linker_1_2", 566, 593], ["Domain_2", 593, 653, "PF01391"], ["Domain_5", 755, 816, "PF01391"]], "ENSCCAP00000013807": [["Domain_0", 125, 180, "PF01391"], ["Linker_0_1", 180, 236], ["Domain_1", 236, 462, "PF01391"], ["Linker_1_2", 462, 614], ["Domain_2", 614, 671, "PF01391"], ["Linker_2_3", 671, 692], ["Domain_3", 692, 759, "PF01391"]], "MGP_PahariEiJ_P0033290": [["Domain_0", 164, 245, "PF01391"], ["Domain_1", 451, 568, "PF01391"], ["Linker_1_2", 568, 611], ["Domain_2", 611, 670, "PF01391"], ["Domain_5", 752, 811, "PF01391"]], "ENSHGLP00100023903": [["Domain_1", 452, 567, "PF01391"], ["Linker_1_2", 567, 593], ["Domain_2", 593, 645, "PF01391"], ["Linker_2_3", 645, 692], ["Domain_3", 692, 761, "PF01391"]], "ENSOGAP00000015182": [["Domain_0", 158, 240, "PF01391"], ["Domain_1", 452, 567, "PF01391"], ["Linker_1_2", 567, 614], ["Domain_2", 614, 671, "PF01391"], ["Domain_5", 752, 811, "PF01391"]], "ENSSSCP00000052271": [["Domain_1", 455, 567, "PF01391"], ["Linker_1_2", 567, 614], ["Domain_2", 614, 672, "PF01391"], ["Linker_2_3", 672, 692], ["Domain_3", 692, 758, "PF01391"]], "ENSECAP00000006550": [["Domain_0", 158, 240, "PF01391"], ["Domain_1", 452, 568, "PF01391"], ["Linker_1_2", 568, 593], ["Domain_2", 593, 643, "PF01391"], ["Linker_2_3", 643, 692], ["Domain_3", 692, 759, "PF01391"]], "ENSRBIP00000028116": [["Domain_0", 126, 182, "PF01391"], ["Domain_1", 454, 568, "PF01391"], ["Linker_1_2", 568, 614], ["Domain_2", 614, 672, "PF01391"], ["Linker_2_3", 672, 692], ["Domain_3", 692, 759, "PF01391"]], "ENSMLUP00000003080": [["Domain_0", 209, 269, "PF01391"], ["Linker_0_1", 269, 424], ["Domain_1", 424, 497, "PF01391"], ["Linker_1_2", 497, 596], ["Domain_2", 596, 656, "PF01391"], ["Linker_2_3", 656, 687], ["Domain_3", 687, 754, "PF01391"]], "ENSPPAP00000037430": [["Domain_0", 126, 179, "PF01391"], ["Domain_1", 450, 568, "PF01391"], ["Linker_1_2", 568, 614], ["Domain_2", 614, 672, "PF01391"], ["Domain_5", 752, 811, "PF01391"]], "ENSBTAP00000017548": [["Domain_0", 158, 240, "PF01391"], ["Domain_2", 614, 671, "PF01391"], ["Domain_5", 752, 811, "PF01391"]], "ENSCGRP00001020731": [["Domain_1", 451, 567, "PF01391"], ["Linker_1_2", 567, 593], ["Domain_2", 593, 648, "PF01391"], ["Domain_5", 752, 811, "PF01391"]], "ENSRNOP00000004510": [["Domain_0", 158, 239, "PF01391"], ["Domain_1", 227, 450, "PF01391"], ["Domain_2", 611, 670, "PF01391"], ["Linker_2_3", 670, 692], ["Domain_3", 692, 759, "PF01391"]], "ENSNGAP00000023927": [["Domain_0", 126, 180, "PF01391"], ["Domain_1", 452, 567, "PF01391"], ["Linker_1_2", 567, 593], ["Domain_2", 593, 647, "PF01391"], ["Linker_2_3", 647, 692], ["Domain_3", 692, 759, "PF01391"]], "ENSOCUP00000002797": [["Domain_0", 156, 227, "PF01391"], ["Domain_2", 614, 671, "PF01391"], ["Linker_2_3", 671, 692], ["Domain_3", 692, 758, "PF01391"]], "ENSAPLP00000014398": [["Domain_0", 209, 269, "PF01391"], ["Linker_0_1", 269, 381], ["Domain_1", 381, 485, "PF01391"], ["Linker_1_2", 485, 596], ["Domain_2", 596, 656, "PF01391"], ["Linker_2_3", 656, 692], ["Domain_3", 692, 760, "PF01391"]], "ENSCAPP00000012912": [["Domain_0", 127, 227, "PF01391"], ["Domain_1", 452, 567, "PF01391"], ["Linker_1_2", 567, 593], ["Domain_2", 593, 642, "PF01391"], ["Linker_2_3", 642, 692], ["Domain_3", 692, 759, "PF01391"]], "ENSMGAP00000007865": [["Domain_0", 133, 220, "PF01391"], ["Domain_1", 452, 572, "PF01391"], ["Linker_1_2", 572, 596], ["Domain_2", 596, 656, "PF01391"], ["Linker_2_3", 656, 690], ["Domain_3", 690, 757, "PF01391"]], "ENSMMUP00000005337": [["Domain_2", 592, 642, "PF01391"]], "ENSSHAP00000018791": [["Domain_0", 209, 267, "PF01391"], ["Domain_1", 255, 400, "PF01391"], ["Domain_2", 614, 673, "PF01391"], ["Domain_5", 753, 811, "PF01391"]], "ENSPVAP00000015801": [["Domain_0", 215, 277, "PF01391"], ["Linker_0_1", 277, 278], ["Domain_1", 278, 569, "PF01391"], ["Linker_1_2", 569, 593], ["Domain_2", 593, 656, "PF01391"], ["Domain_5", 755, 816, "PF01391"]], "ENSRROP00000019272": [["Domain_0", 126, 179, "PF01391"], ["Domain_1", 454, 568, "PF01391"], ["Linker_1_2", 568, 614], ["Domain_2", 614, 672, "PF01391"], ["Domain_5", 752, 811, "PF01391"]], "ENSPCOP00000011629": [["Domain_0", 164, 246, "PF01391"], ["Domain_1", 233, 455, "PF01391"], ["Domain_2", 593, 648, "PF01391"], ["Linker_2_3", 648, 692], ["Domain_3", 692, 759, "PF01391"]], "ENSPPYP00000018024": [["Domain_0", 158, 239, "PF01391"], ["Domain_1", 454, 568, "PF01391"], ["Linker_1_2", 568, 614], ["Domain_2", 614, 670, "PF01391"], ["Domain_5", 752, 811, "PF01391"]], "ENSMNEP00000025487": [["Domain_0", 158, 240, "PF01391"], ["Domain_1", 227, 450, "PF01391"], ["Domain_2", 592, 642, "PF01391"], ["Linker_2_3", 642, 692], ["Domain_3", 692, 759, "PF01391"]], "ENSNLEP00000019127": [["Domain_0", 175, 247, "PF01391"], ["Linker_0_1", 247, 389], ["Domain_1", 389, 565, "PF01391"]], "ENSCLAP00000016620": [["Domain_0", 126, 182, "PF01391"], ["Linker_0_1", 182, 227], ["Domain_1", 227, 450, "PF01391"], ["Domain_2", 593, 651, "PF01391"], ["Linker_2_3", 651, 692], ["Domain_3", 692, 759, "PF01391"]], "ENSPSIP00000019151": [["Domain_0", 209, 259, "PF01391"], ["Domain_1", 239, 463, "PF01391"], ["Linker_1_2", 463, 612], ["Domain_2", 612, 673, "PF01391"], ["Linker_2_3", 673, 692], ["Domain_3", 692, 758, "PF01391"]], "ENSLAFP00000021248": [["Domain_0", 158, 217, "PF01391"], ["Domain_2", 593, 653, "PF01391"], ["Linker_2_3", 653, 691], ["Domain_3", 691, 721, "PF01391"], ["Linker_3_4", 721, 744], ["Domain_4", 744, 766, "PF01391"], ["Linker_4_6", 766, 790], ["Domain_6", 790, 813, "PF01391"]], "ENSCJAP00000022389": [["Domain_0", 126, 179, "PF01391"], ["Domain_1", 386, 567, "PF01391"], ["Linker_1_2", 567, 614], ["Domain_2", 614, 671, "PF01391"], ["Domain_5", 752, 811, "PF01391"]], "ENSOARP00000006997": [["Domain_0", 158, 239, "PF01391"], ["Domain_1", 449, 568, "PF01391"], ["Linker_1_2", 568, 593], ["Domain_2", 593, 645, "PF01391"], ["Linker_2_3", 645, 695], ["Domain_3", 695, 762, "PF01391"]], "ENSMODP00000005144": [["Domain_0", 161, 239, "PF01391"], ["Linker_0_1", 239, 389], ["Domain_1", 389, 567, "PF01391"], ["Linker_1_2", 567, 593], ["Domain_2", 593, 650, "PF01391"], ["Linker_2_3", 650, 692], ["Domain_3", 692, 759, "PF01391"]], "ENSFALP00000008010": [["Domain_0", 212, 268, "PF01391"], ["Linker_0_1", 268, 437], ["Domain_1", 437, 487, "PF01391"], ["Linker_1_2", 487, 617], ["Domain_2", 617, 674, "PF01391"], ["Domain_5", 753, 811, "PF01391"]], "ENSCANP00000020721": [["Domain_0", 126, 179, "PF01391"], ["Domain_1", 449, 568, "PF01391"], ["Linker_1_2", 568, 614], ["Domain_2", 614, 672, "PF01391"], ["Domain_5", 752, 811, "PF01391"]], "ENSGGOP00000006764": [["Domain_0", 126, 179, "PF01391"], ["Linker_0_1", 179, 233], ["Domain_1", 233, 461, "PF01391"], ["Linker_1_2", 461, 614], ["Domain_2", 614, 672, "PF01391"], ["Domain_5", 752, 811, "PF01391"]], "ENSMPUP00000000735": [["Domain_1", 451, 568, "PF01391"], ["Linker_1_2", 568, 614], ["Domain_2", 614, 671, "PF01391"], ["Linker_2_3", 671, 692], ["Domain_3", 692, 761, "PF01391"]], "ENSRNOP00000062298": [["Domain_0", 126, 179, "PF01391"], ["Domain_1", 452, 567, "PF01391"], ["Linker_1_2", 567, 611], ["Domain_2", 611, 670, "PF01391"], ["Domain_5", 752, 811, "PF01391"]], "ENSSTOP00000004457": [["Domain_0", 170, 253, "PF01391"], ["Domain_1", 452, 567, "PF01391"], ["Linker_1_2", 567, 614], ["Domain_2", 614, 671, "PF01391"], ["Domain_5", 752, 811, "PF01391"]], "ENSFDAP00000004266": [["Domain_0", 125, 213, "PF01391"], ["Domain_1", 452, 568, "PF01391"], ["Linker_1_2", 568, 593], ["Domain_2", 593, 646, "PF01391"], ["Linker_2_3", 646, 692], ["Domain_3", 692, 759, "PF01391"]], "ENSMLEP00000035911": [["Domain_0", 126, 179, "PF01391"], ["Domain_1", 454, 568, "PF01391"], ["Linker_1_2", 568, 592], ["Domain_2", 592, 642, "PF01391"], ["Linker_2_3", 642, 692], ["Domain_3", 692, 761, "PF01391"]], "ENSPTRP00000030067": [["Domain_0", 175, 247, "PF01391"], ["Domain_2", 612, 667, "PF01391"], ["Domain_5", 752, 811, "PF01391"]], "ENSP00000375069": [["Domain_0", 126, 182, "PF01391"], ["Domain_1", 450, 568, "PF01391"], ["Linker_1_2", 568, 614], ["Domain_2", 614, 672, "PF01391"], ["Domain_5", 752, 811, "PF01391"]]}