Home
Query Results
"ENSGT00900000140813_0"
Found:
ENSGT00900000140813_0
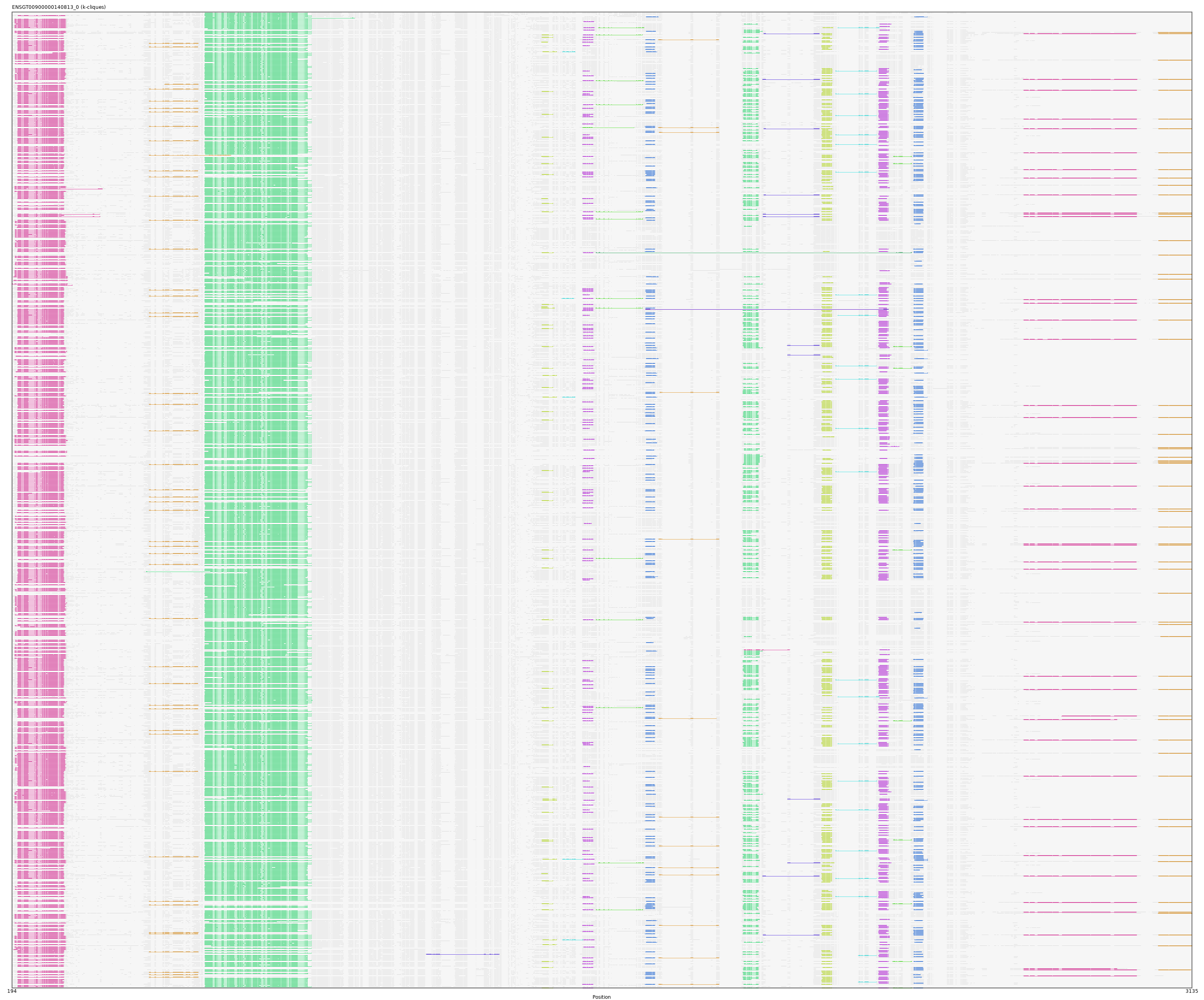
ENSOGAP00000021901 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------PRLE-VEEAN-GKELADVLIDRSPQILD
RG-VVNS----SE-KMNLTDLSKMAK-GQIPGSAQ-GQ------GKMAEL---EDTDT--
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------EDDLDVEMSGKKEECRNI-
---------TEI----------------LPLL--RKNKFQLVGSDNFLASVI--------
-----------------------------------------------QRSQKFIPFLQPR
IA--PQPIA----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
HTVVLYGPAGVGKSTLAKQV-----FLDWA--E---------NNLNQ
---------MFNY-VFYLRCKELNHVG-------ACSFAE-LIFK-DW-----PE-WLGC
AAEVFG-QA----QKILFIIDGFEE-LRVPAG------ALIH-D-----ICGNWERE-MP
-VPVLLGSLLKRKMLPKASLLVT-TRPEALRELRLLV-E-----QPL--FIEMEGLLEPD
RRAYF----------------L--RQ-FAD---------QDQALRALDLMKG-NV-----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------VQ-KLFSKEERV-RNPSLTQVGYFL----
----FGLSNEKRVQELESTFGCR---------------M--SLQ-IRE-ELLKYRANLDE
NK------------------------------------PFSRTDMKELLYCLYESQD-EG
LMRDAMVP---------------------F-------------------TEVSLHWMNTF
EV---MYS--S----FCLK-HCQN-LQK-LSLQ----------------IARG----IFL
-END-PVS----RE--------------------DAQLE-----------SWSRCD-HRS
LCVWMDFC-SVVSSNKNLRVLDMTQSFLSDS---------SVRILC--DHITR-F-----
SC--H-LQKVV--I-RNVTPD--SA----YR-DFCLAF----------------VGKKTL
THLT-LEGH-VQWDDMLLLLLGEVLKHVKCS----LQYL--------RLG---------F
H------S----------------------------------------------------
---------VT---TE-RWT---NLTLALKV-NQSLMCLDLSDNELLD-ESAKLLYTS--
LKG-PKCLLQRL------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSSARP00000012840 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------HWRCYRER-
--------MKAD----------------ILRM--WARAPWPEDHIHLLTP----------
-----------------------------------------------YEHQELERVLCP-
RG--ADMRP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
LTVVLEGSAGVGKTTVAAKL-----ALHWA--E---------GMLLH
R--------CFSF-VSYISCHQVNEMG-------DACLAD-LLSL-HW-----SPDPRFL
MQKFLS-HP----QRLLFIVDGIEE-LRCPVEAE--S------P-----HLADWFQR-LP
-ATQVLLSLLRRKALPTATLLVT-SRAQRRPALYRLL-R-----RPS--FVTLRGFSEDD
QEEYF----------------L--RF-FGD---------QDTGKAIMQWLRA-TGTWLPT
CCAPLMCRVVGTYLKRQRA-TN-SF-Q--LG-AQSPTALYAGFFSSLVSA-E-AGLQGVG
------------AHEQWGALCRLA---ARGM-WQ-ST----GAFKAD-------------
--DGSRA------LAAPFIQALLRLNILQAVAS-CE-DCVAFIHPSFQAFFGALFYVLR-
GTPGC--------------LGGL--TK-PQELH-M-LQLSLAD-RATHWEQTAIFF----
----FGLLHRDVARELEGAP------------------P--APATATD-QVLEWAEG-LD
GA-----------------------------------A-SL--WGMLLFQCLHEAQE-AS
MVKQVLSH---------------------L-------------------REAHVDVCGNR
QL---QDA--A----FCL-RDCRN-LQK-LRLS----------------LS---------
----GPS---------------------------------RG----SSLKTPGKMA-SSK
IRQWQDIC-SVFSYG-NVHELDLSNSTLNTL---------TMKKLC--RELRN-P-----
RC--K-LRQLT--C-RSVEP---RR---VLQ-ELPVVL----------------HGNS
PF00560 // ENSGT00900000140813_0_Domain_6 (1619, 1645)
RL
THLD-LSAN-DL-GV-TVPLIFKTLRHPACQ----LRSL--------CLE---------K
C------N----------------------------------------------------
---------LS---SD-TYR---GLASLLIS-AQKTTHLCLGFNPLQD-NGVQLLCAS--
LVL-PDCALQRL-E----------------------------------------------
------------------------LWFCQLG-----------------------------
----------------------------APSGQYLSR-----------------------
--------------------------------ALLQNRALTC---LN-LRRNR-LGD---
-------EGVEDLCVAL----RGPHC----------------------------------
--------------------------RLQ-RLD---------------------------
-------------------------------LSACSFSVPGCGALAAA-LRDNRSLQVLD
V-GENEFGDDGME-ALSQALGPA--P-A----L---------------------------
-----------------------TTLGL--QKC-----QLTAASNEHL------------
-------GHLL-RGSKSLVSLNLLGNDLQLQGLSM-LWKAVK-SS-QC-PLRVLG-----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCCAP00000028558 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----MSVD--M----QCVDIQCEE-LSD-ARWA----------------EL---------
----LPLL-------------------------------QRCQAV------RLDDC-GLT
EAQCKDVS-SALQA
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1542)
NPALTELNLRNNELGDA---------GVHRVL--QALQS-S-----
SC--K-IQKLS--F-QSCYLT--VA---GCG-VLSGTL----------------RS
PF13516 // ENSGT00900000140813_0_Domain_6 (1617, 1642)
LPTL
QELH-LSDN-PMGDA-GLQLLCEGLLDPQCR----LEKL--------QLE---------Y
C------S----------------------------------------------------
---------LS---SA-GCE---HLASVLRA-KPDFKELTVSNNDIDE-AGVRTLCQG--
LKD-SPCQLETL-K----------------------------------------------
------------------------LEKCGAT-----------------------------
----------------------------SDNCRDLCS-----------------------
--------------------------------ILASK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
ASLRE---LA-LGNNK-LGD---
-------VGIAELCPGL----LHPSS----------------------------------
--------------------------RLR-TLW---------------------------
-------------------------------IWECGLTAKSCVDLCRV-LRAKD
PF13516 // ENSGT00900000140813_0_Domain_13 (2215, 2237)
SLKELS
L-AGNNLGDEGAR-LLCESLLES--GCQ----L---------------------------
-----------------------ESLWV--KSC-----SLTAACCSHF------------
-------SSVL-AQ
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NKFLLELQISENRLGDAGVQE-LCQGLG-QP-GS-VLRSLW--LG-
--D-CDVSDSSCS--------------------SLAATLLA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2465)
NHSL-RELDLSNNCLGDEG
ILALMG--------SVRQPACL--LE--Q-------------------------------
---LVLYDI-YWSLEMDNQ------LQ----------ALE-KEKPSLRIIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSSBOP00000005889 (view gene)
ENSCGRP00000019004 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MSLDIQCEQ-LSD-ARWT----------------EL---------
----LPLI-------------------------------QQYQVV------RLDDC-GLT
EVRCKDIS-SAIQ
PF13516 // ENSGT00900000140813_0_Domain_4 (1514, 1542)
SNPTLTELSLCTNELGDP---------GVCLVL--QGLQN-P-----
TC--K-IQKLS--L-QNCSLT--EA---GCR-VLPDVL----------------RS
PF13516 // ENSGT00900000140813_0_Domain_6 (1617, 1642)
LPTL
RELH-LSDN-PLGDA-GLKLLCEGLLDPQCR----LEKL--------QLE---------Y
C------N----------------------------------------------------
---------LT---AT-SCE---PLASVFRV-KPNFKEIVLSNNDLHE-AGVQMLCQG--
LKD-SACQLESL-K----------------------------------------------
------------------------LENCGIT-----------------------------
----------------------------SANCKDLCD-----------------------
--------------------------------VVASK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
ASLQQ---LD-LGSNK-LGN---
-------AGIAALCSGL----LLPSC----------------------------------
--------------------------RIR-TLW---------------------------
-------------------------------LWECDITAEGCKDLCRV-LRT
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
KQSLKELS
L-AGNELRDEGAQ-LLCESLLEP--GCQ----L---------------------------
-----------------------ESLWI--KTC-----SLTAACCSHI------------
-------CSVL-T
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
KNRSLLELQMSSNPLGDLGVLE-LENSGT00900000140813_0_Linker_15_16 (2378, 2391)
CKALG-QP-DT-VPF13516 // ENSGT00900000140813_0_Domain_16 (2391, 2436)
LRVLW--LG-
--D-CDVTDNGCS--------------------SLENSGT00900000140813_0_Linker_16_17 (2436, 2441)
ATVLLPF13516 // ENSGT00900000140813_0_Domain_17 (2441, 2464)
HNRSL-RELDLSNNCMGDSG
VLRLVE--------SVRQPSCA--LQ--Q-------------------------------
---LVLYDI-YWTEEVEDQ------LR----------ALE-EERPSLRVIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCCAP00000023046 (view gene)
ENSFCAP00000013894 (view gene)
ENSCSAP00000016257 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------MQK----------------QSLV--WKNTFWQGDTDSFHDDVI--------
-----------------------------------------------RRNQRFIPFLNAR
TP--GKLTP----Y
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVLHGPAGVGKTTLAKKC-----MLDWS--D---------CNLSP
---------TVRY-AFYLSCKELSRMR-------PCSFAE-LISK-DW-----PE-LQDD
IPSILA-QA----QRILFVVDGLDE-LKVPPG------ALIQ-D-----ICGDWKQQ-KP
-VPVLLGSLLKRKMLPKATLLVT-TRPRALRDLQLLV-Q-----QPI--YIRVEGFLEED
RRAYF----------------L--RH-FGE---------EDQAMRAFELMRS-NAALFQL
GSAPAVCWIVCTTLKLQMENGE-DP-A--PT-CLTSTELFLRFLCSQFPQG-------AQ
------------LRGALRALSLLA---AQSL-WA-QM----SVLHRE-------------
--DLESAGVQES-DLRPFLDR----DILHQDRV-SK-SCYSFIHLSFQQFLTALFYALE-
KE--E---EE--------DRDGH--AWDIGDVQ-KLLSEEERL-KNPDLIQVGHFL----
----FGLANEKRVKELEATFGCR---------------M--SPE-IKQ-ELLRCKANLHA
NK-----------------------------------P-LSMTDLKEVLCCLYESQE-EE
LAKVVVAP---------------------L-------------------KKISIKLANTS
EI---MHC--S----FSLK-QCPD-LEK-VSLQ----------------VAKG----VFL
-ENY-VDI----EP--------------------DIEFERC-TYV--TLPSWAQQD-LRS
LRLWTDFC-SLFSSNSNLKFLEVKQSFLSDS---------SVRILC--DHITR-S-----
TC--H-LQKVE--I-KNVTPD--TA----YR-DFCLAF----------------IGKKTL
THLT-LEGH-IEWERTMLLLLCDLVRNHKCN----LQYL--------RLG---------G
H------C----------------------------------------------------
---------AT---SE-QWA---DFSYVLKA-NQSLRHLHLSASVLLD-EGAKLLYKT--
MTH-PKHFLQML-S----------------------------------------------
------------------------LENCRLT-----------------------------
----------------------------EASCKDLAA-----------------------
--------------------------------VLVVSQQLTH---LC-LAKNP-IGD---
-------TGVKFLCEGL----SYPEC----------------------------------
--------------------------KLQ-ALV---------------------------
-------------------------------LQQCSITKRGCRYLSEA-LQEACSLTNLD
L-SLNQIA-RGLW-ILCQALENP--NCN----L---------------------------
-----------------------KHLRL--WSC-----SLMPFYCQHL------------
-------GSAL-ISNQKLETLDLGQNHLWKSGIIQ-LFRVLR-QR-T-------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSBTAP00000054745 (view gene)
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------MA------------------
----------SPAGISWHDGA
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 326)
RDQ---LLSHLRGLD--P---WQLEDFKLALQCPELL--
--------------PEG--------A--RRI-PWAD-LRAAG-PADLLCLLEERFPGRRT
WEAALRV----FE-DLRLSSLCERMR-AELHEEVQ-SQNTQDTYLT------EMSLFICC
----IQRVPVPGFL-----------------------L----------PQEDW-------
---------LCYVESF-------SCNKMS----------QVCRLQDPNQ-----------
------EEPE-------------------------------TLEET----AHRRRYRER-
--------LRTR----------------ILAL--WDRTPWPEDHIYLRHVTQ--------
----------------------------------------------R-EHAELRGLLRPR
G---AGAPP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 929)
TMLLEGAAGVGKTTLATKL-----VLHWA--E---------GVLFR
G--------RFSY-VFYISGHWLAKLG-------DISFAG-LLAL-DW-----PD-SQVP
VEEFRA-HP----ERLLFVIDGAEE-VTLGSK------GSAS-R-----PGADWYQE-LP
-AASILVRLLKKELVPEATLLVT-AGPPGGHALRGLL-L-----RPC--RVGIPGFTEGD
RREYL----------------R--RF-LGD---------PDVAEEAWRRMQG-SETLVRL
CAAPLACWAVCAGLKRQLAGSP-A---PAPG-APTPTGLYAQLFCSLLAGAE-PGLRTGS
------------SAGQWRAFCSLA---AEGL-WL-AA----FTFAGD-------------
--ALERWRLEAP-FIDGLLRL----QILRRVSD-CE-HCVTFAHRSFQEFFGALFYVLW-
GAQGS--------------L-GG--VPRHQEMR-RWLNHAFAD-ANPYWRQMVRFF----
----FGLLGTDLARQLEEAVGCR---------------M--SPG-VAD-EVLDWAEELER
CG-----------------------------------AVSGRFDFLWLFQCLHETQD-EN
LARRVLNR---------------------L-------------------PVADLDIQGCE
HL---RVS--S----FCLK-HCQK-LRK-LRLS----------------VSH-----RVL
-EKKRT----------------------------------------SGL--GTPDA-DFR
MHQWEDIC-S-VFCSGNLSQLDLSNSKLSTA---------SMMRLC--LKLGN-P-----
QC--R-LQKLA--W-KSMAPV--EG---L-R-KLGLLL----------------RGDRHL
THLD-LSSN-SLDAA-VSRGVFRMLGHSACG----LKYL--------WLE---------K
G------N----------------------------------------------------
---------LS---AA-ACE---DL-----------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------AS-----------------------
--------------------------------LLTSTPRLTR---LC-LGLNP-LGD---
-------EGVQLLCGSL----TRPEC----------------------------------
--------------------------VLQ-RLE---------------------------
-------------------------------LWCCRLSTPSCRHLSDA-LLRSRSLTHLN
L-RRNSLGDGGVK-LLSSALGRA--DCA----L---------------------------
-----------------------QSLNL--SHC-----SLTVAGCREL------------
-------AHAL-KHNGHLKILDVGNNDIQDEGVKE-LCSVLK-SP-SC-VLQTLG--LE-
--K-CSLTAACCR--------------------PLSSVLGSSKSL-ENLNLLGNNLGPDG
VSRLWM--------PLRKPTCK--LR--K-------------------------------
---LG-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMOCP00000010558 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------------------FF
---HAEEQ------------------------------------DSEADKNGLHSYKAH-
--------VIAK----------------FDTH--VEVH-Y--------------------
--------------------------------------------DSPEM-QVLLKAFKSY
Q---KAFWP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
HTVILHGKPGVGKSSLARSI-----ILGWA--Q---------GKLYQ
D--------M-SY-AFFFSVREIKWTE-------KSSLAE-LISE-EW-----PY-LQAS
VAKIMS-KP----ERLLFVLDGFDD-LDLALLQD------DM-R-----PSGDWEDE-QP
-IYILMYSLLMKVLLPQSCLIIT-TRDTDLEKLKSLA-V-----SPL--YILVGGLSATR
RTQLL----------------L--EN-ISD---------DQRRKHVVRTVIE-NDQIFDQ
CQVPSICSLVCEALKLQEKLGE-RC-A--PV-CWTLTSLYATLVFHQLTPRE-PFQSCLS
----------HREQVALMGLCGMA---AEGV-WN-MR----SVFYDD-------------
--DLQNYGLEEA-AISALFHM----NVLLRVDD-STEQYYIFFHLSLQDFCAALYYLLK-
GLK-K--------------WNQY--CLFVENPKRNIMTLKRTN-FNTHLLEMKRFL----
----FGLMNKDTMRTLEALLGCP---------------V--SP-AVKQ-RLQHWVFEVGQ
QVS----------------------------------ATG-PEDVLDAFYYLFESQD-KE
FVCWALNS---------------------F-------------------QEVWLQVNQKM
DL---MVS--S----YCLQ-HCQN-LKT-LRVD----------------VRD-----IFL
-VDKNAEL----CP--------------------VA-----------LP--QAQGK-SLI
MELWEDFC-SVLSMHPKLNKLDLGSSVLNEW---------AMKILC--LKLRN-P-----
AC--N-IQNLM--L-KDAEI---AA---GLQ-YLWMTL----------------V
PF13516 // ENSGT00900000140813_0_Domain_6 (1616, 1641)
SNRNL
KYLN-LGNN-NLKDD-DIKLVCEALRHPNCS----LETL--------RLD---------S
C------E----------------------------------------------------
---------LT---SD-SYS---MISELLLS-STSLKSLSLARNGVTE-KSMGQLCGA--
LSS-SRCILQKL-I----------------------------------------------
------------------------LDSCELS-----------------------------
----------------------------LVSCHGLVS-----------------------
--------------------------------ALSGNMTLTH---LC-LSNNS-LRT---
-------EEVRLLCQFM----KGPRC----------------------------------
--------------------------ALQ-RLL---------------------------
-------------------------------LNQCDLKRDACGFLALM-LI
PF13516 // ENSGT00900000140813_0_Domain_13 (2212, 2237)
NNRTLTHLS
L-TKNPVVDDGVK-FLCEAIKEP--TLD----RGCG------------------------
---------------------FPVSRRL--VEC-----WLTGDCCENL------------
-------ASMI-TM
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NQHLKSLDLGYNNLGDNGVIA-LCKGLK-QN-NS-SLRRLG--LE-
--A-CGLTFESCE--------------------ALSSTLA
PF13516 // ENSGT00900000140813_0_Domain_17 (2441, 2465)
SNQCL-TSLNLVKNSFSTPG
MLKLCY--------AFRHFTSN--LR--I-------------------------------
---IGLWKQ-QHHAEVRRQ------LE----------EIQ-LTRPHTVITGDWYSLE---
--E---------------------------------------------------------
--------------------------------------------------DDRNWWKN--
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCSAP00000007255 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MSLDIQCEQ-LSD-ARWA----------------EL---------
----LPLL-------------------------------QQCQVV------RLDDC-GLT
EARCSDIS-SALRVNPALTELSLCGNELGDA---------GMHCVV--QGLQS-P-----
SC--K-IQKLS--L-QNCCLT--GA---GCG-VLSSTL----------------RYLPTL
QELH-LSDN-LLGDA-GLQLLCEGLLDPQCR----LEKL--------QLE---------Y
C------N----------------------------------------------------
---------LS---AA-SCE---SLASVLRA-KPDFKELTVSNNDINE-AGVRVLCQG--
LKD-SPCQLEAL-K----------------------------------------------
------------------------LESCGIT-----------------------------
----------------------------SNNCRDLCS-----------------------
--------------------------------IVASKASLRE---LA-LGSNK-LGD---
-------VGIAELCPGL----LHPSS----------------------------------
--------------------------SLR-TLW---------------------------
-------------------------------IWECGITAKGCADLCGV-LRTKESLKELS
L-AGNELGDEGAR-LLCETLLEP--GCQ----L---------------------------
-----------------------ESLWV--KSC-----SFTAACCSHF------------
-------SSVL-TQNKFLLELQISNNRLGDAGVQE-LCKGLG-QP-GS-VLRVLW--LG-
--D-CDMSDRSCS--------------------SLAATLLANQ
PF00560 // ENSGT00900000140813_0_Domain_17 (2444, 2458)
SL-RELDLSNNCLGDAG
VLQLVE--------SVRQPSCL--LE--Q-------------------------------
---LVLYDI-YWSEATEDR------LQ----------ALE-EEKPSLRVIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSP00000309767 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-MDQPEAPCSSTGPR--LAVAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLAALEELS--Q---EQLKRFRHKLRD------
------------VGPDG-----------RSI-PWGR-LERAD-AVDLAEQLAQFYGPEPA
LEVARKT----LK-RADARDVAAQLQ-ERRLQR---------L---------GLG-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------SG-TLLSVSEYKKKYREH-
--------VLQL----------------HARV--KERNARS------------VKITKRF
TKLLIAPESAAPE-E--AMGPAE-------EPEP--------GRARRSDTHTFNRLFRRD
E---EGRRP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYQ
G--------QVDF-AFFMPCGELLERPG------TRSLAD-LILD-QC-----PD-RGAP
VPQMLA-QP----QRLLFILDGADE-LPALGGP--------EAA-----PCTDPFEA-AS
-GARVLGGLLSKALLPTALLLVT-TRAAAPGRLQGRL-C-----SPQ--CAEVRGFSDKD
KKKYF----------------Y--KY-FRD---------ERRAERAYRFVKE-NETLFAL
CFVPFVCWIVCTVLRQQLELGR-DL-S--RT-SKTTTSVYLLFITSVLSSAPVADGP---
-----------RLQGDLRNLCRLA---REGVLGR--R----AQFAEK-------------
--ELEQLELRGSKVQTLFLSKKELPGVLET-----E-VTYQFIDQSFQEFLAALSYLLE-
DGGVP-------------------R-TAAGGVG-TLLRGDAQP--HSHLVLTTRFL----
----FGLLSAERMRDIERHFGC-----------------MVSER-VKQ-EALRWVQGQGQ
GCPGVAPEVTEGAKGLEDTEEPEEE-----------EEGEEPNYPLELLYCLYETQE-DA
FVRQALCR---------------------F-------------------PELALQRVRFC
RM-DVAVL--S----YCVR-CCPA-GQA-LRLI------------------SC----RLV
------AA----------------------------------------QEK--K---KKS
----------------------------------------LGKR----------------
------LQAS---L--------------GGG------S----------------SSQGTT
KQ---LPAS-------LLHPLFQAMTDPLCH----LSSL--------TLS---------H
C-----------------------------------------------------------
--------KLP---DA-VCR---DLSEALRA-APALTELGLLHNRLS-EAGLRMLSEG--
-LAWPQCRVQTV------------------------------------------------
----------------------------RVQ-----------------------------
---------------LPDP---------QRGLQYL-------------------------
------------------------------VGMLRQSPALTT---LD-LSGC-QLPAPMV
--T--------YLCAVL----QHQGCGLQTLSL--ASVELSEQ-----------------
----------SLQELQAVKRAKP-------DLV----------------------I--TH
PALD-------------------GHPQPP-KELISTF-----------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCLAP00000011529 (view gene)
ENSCSAP00000016258 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------MVSST----QV
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 326)
NFN---LQALLEQLS--Q---DELSKFKSLLRT------
---------V--SLGNE--------V--QKI-PQKE-VDEAD-GKQLAEILISHCHSYWT
ELATIQV----FE-KMHRVDLSERAK-DELREAAL-KSFN----KNK------PLSLG--
------ITRKERQP---------------------------------IDVEEMLERFK-A
GTLECT---------------------------------ET--EEDATV-----------
-------LTE-------------------------------VFKVKGEKSDNENRYRLI-
--------LKTK----------------FREM--WQ--SWPGDSKEVHV-----------
---------------------------------------------MAERYRMLIPFSNPR
VL--PGPFS----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
HTVVLHSPAGLGKTTLANKL-----MLDWT--E---------DNLIQ
---------KFKY-AFYLSCRELSRLG-------PCSFAE-MVFR-DW-----PE-LQDD
IPHILA-QA----QKILFVIDGFDE-LGAPPG------ALIQ-D-----ICGDWEQQ-KP
-VPVLLGSLLKRKMLPKANLLVT-TRPRALRDLRFLA-E-----QPI--YIRVEGFLESD
RRAYF----------------L--RH-FGE---------EDQAMRAFELMRS-NAALFQL
GSAPAVCWIVCTTLKLQMEKGE-DP-A--PT-CLTSTELFLRFLCSQFPQ-------GAQ
------------LWGALRALSLLA---AQSL-WA-QM----SVLHGE-------------
--DLESAGVQES-DLRLFLDR----DILRQDRV-SK-GCYSFIHLSFQQFLTALFYALE-
KEEEE-------------DRDGH--AWDIGDVQ-KLLSREERL-KNPDLIQAGRFL----
----FGLANEKRVKELEATFGCR---------------M--SPE-IKQ-ELLQCDISRKN
G------------------------------------H-STVADLKELLCCLYESQE-DE
LVKEVMAQ---------------------F-------------------KEISLHLN-AV
DI---APS--S----FCFK-HCPN-LQK-MSLQ----------------VIKK----ETL
-PENVAAS----ES--------------------NTEAER------------SQDD-QDM
LPFWTDLC-SIFGSNKDLMGLEINNSFLSAS---------LVRILC--EQIAS-D-----
SC--H-LQRVV--F-KNISPA--DA----PR-NLCLAL----------------RGHKTV
THLT-LQGT-DQKD--MLPALCEVLRHPECN----LRYL--------GLV---------S
C------S----------------------------------------------------
---------AT---TQ-QWA---DLSLALEA-NRSLMCVNLSDNELLD-EGAKLLYTT--
LRH-PKCFLQRL-S----------------------------------------------
------------------------LENCHLT-----------------------------
----------------------------EANCKDLAA-----------------------
--------------------------------VLVVSRELTH---LC-LAKNS-LKD---
-------TGVKFLCEGL----SYPEC----------------------------------
--------------------------KLQ-ALV---------------------------
-------------------------------LWNCDITSDGCCSLAKL-LQEKSSLSCLD
L-GLNHIGVTGVK-VLCEALSKP--LCN----L---------------------------
-----------------------RCLWL--WGC-----SIPPFSCEDL------------
-------CSAL-SCNQSLITLDLGQNPLGSSGVKM-LFKTLT-RP-G--TLQTLR--LK-
--I-DDFNDELHK--------------------LLEEIEENNPQLIIDTEKH----DPWK
---------------KRPSSHD--FM----------------------------------
----I-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSNGAP00000025650 (view gene)
ENSBTAP00000055476 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------DPQETYRDY-
--------VRRK----------------FRLM--EDRNARLGEFV---------NLSHRY
TRLLLAKEHSSPRWAQQKLL------DTGRGHAGTAG--------HPASLIQIEALFEPD
EE--RPEPP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
RTVVLQGAAGMGKSMLAHKV-----MLDWA--D---------GKLFQ
N--------RFDY-VCYINCREMNRGTA------ELSVRD-LLAS-CW-----PE-PCAP
LQELVR-VP----ERLLVIMDGFDE-LRPSFHEP------QG-P-----WCLCWEKK-AP
-AEVLLGSLIRKKLLPELSLLIT-TRPTALAKLQRSL-E-----HPR--HVAILGFSEAE
RKEYF----------------H--RY-FHD---------REQASQVFSFVRD-DEPLFTL
CFVPMVCWVVCTCLKQQLEGGG-LL-R--RR-SRTTTAVYLLYLLSLMQPKPGT-PTL--
-----------PSPPNRRGLCALA---AHGL-WE-QK----ILFAER-------------
--DLREHGLDAA-DVSSFLNM----NIFQKDII-CE-TLYSFIHLSFQEFFAAMHYVLD-
D--GA--------------S--G--SGPERNLS-RLLTEYAFS-DRSFLALTVRFL----
----FGLLNEETRSYLEKTLGWK---------------V--SPG-VKT-ELLEWIRRKAR
SE-----------------------------------GSTLKQGALEVFGCLYELQE-EE
FIQQALSH---------------------F-------------------QVVVVNNIT-T
KM-EHMVS--S----FCVK-SCRN-AEV-LQLF----------------GA------AYQ
-AHGEDRL--R-WP---------------------------------------LAT---S
H-------------------------SV------------------------I-------
VQ--L--------PERHVLPD-------IYSEQLAAAL----------------STNPSL
VELA-LHSN-ALGSQ-GVKLLCQGLRHPNCR----LQNL--------RLK---------R
C-----------------------------------------------------------
--------QVS---SS-VCQ---DLTTALIA-NKHLLRMDLSGNALGL-LGAQLLCQG--
LRH-PKCRLQVVQ-----------------------------------------------
------------------------LRKCHLE-----------------------------
----------------------------AGACQELAS-----------------------
--------------------------------VLSTSRHLLE---LD-LTGNA-LED---
-------SGLRLLCQGL----RHPVC----------------------------------
--------------------------RLR-ILW---------------------------
-------------------------------LKICLLTGAACEDLAST-LHMNQSLVELD
L-SLNDLGDPGVL-LLCEGLRHP--QCK----L---------------------------
-----------------------QTLRL--CIC-----RLSSVACEGL------------
-------SAVL-GVSSHLRELDLSFNDLGDRGMSL-LCEGLR-HP-TC-RLQKLW--LD-
--S-CSLTGKACE--------------------DISSALGINQTL-TDLYLTNNALGNTG
VRLLCE--------RLSHPGCK--LR--V-------------------------------
---LWLFGM-DLNKMTHRS------LE----------ALR-MTKPYLDVGC---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCGRP00001007377 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------M---ASFFSDFG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 323)
-LMWYLKELN--K---KEFMKFKEVLRQ------
---------E--IAESG--------L--QHI-PWNE-VKKAS-REHLANLLVKHYE-QKA
WDVTFRI----FQ-NINRKDLSERAA-EEM------------AG--------------HS
K---------------------------------------------------MYQA----
------------------------------------------------------------
----------------------------------------------------------Y-
--------LKNM----------------LSHD--WSRKFKIAIECF--------------
---------------------------------G-------NEDLTQDNCSRLWNLFVPK
AN---EK-P----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
HTVLLIGMSGVGKTVFLAQL-----MRAWS--E---------GLMFQ
N--------EFSY-VFYLCCHEIKQLK-------TASLAD-LLSR-EW-----PRP-SAP
IAEIVS-QP----AKLLFIIDSLEG-LNCDWSEP------ES-E-----LCDNWMQK-CP
-VSILLSSLLRRKLLPESSLLIT-ATPEAFEKLGDRI-E-----YTV--KNFVTGFNENS
RRSCF----------------Q--RV-FQD---------EDTAQEVFTLVRE-NEQLYNI
CHVPSLCWMVATCLNDEVKRGG-HL-S--AV-CRCTTSVFAAYVLNLFIPQS-AHYPSK-
-----------KSQALLRGLCSVA---AEGM-WT-DN----FAFNQK-------------
--DLEKNGVFHS-DISTLLNM----KILLKSRD-SK-NLYTFFHPSIQEFCAALFYLVN-
SHVDH-----------------P--SKDVLCVE-TLLFTFLKK-VRVQWIFLGCFI----
----FGLLHKSEQAKLEAFFGYQ---------------L--SQE-VKQ-TLCQSLEKISA
DKE-----------------------------------LRKQIDDMKLFYCLFELED-DG
FVTQVMTF---------------------L-------------------PQIKFVAKEYS
DL---IVA--A----YCLK-NCST-LQN-LSIS----------------TQD-----ILW
-KEN--DP-----------I-----------------------------------YLKKL
LLRWQDLC-SVLGSSEQIQVLQVKDTYLDEL---------AFLELY--NHLKNP------
RC--A-PQTFQVNN---------VTL-W-YNQLFFELL----------------TQNHNL
HHLD-LSLT-CLSHS-DVKLLYDALNQEECH----IEEL--------LL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------SGCQLS-----------------------------
----------------------------HDDCIIFAS-----------------------
--------------------------------ILISSKKLRH---LN-VSYNN-LD----
-------KGMYLLSKAL----CHPDCVLKDL-----------------------------
------------------------------------------------------------
------------------------------VLASCSLSKQCWNYLSDV-LKR
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
NKTLSHLD
V-SSNDLQDEGLK-VLCQALSLP--DSA----L---------------------------
-----------------------KSLFL--RRC-----LITSSGCQYL------------
-------AEVL-S
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
SNQNLISLQVSNNKLEDAGVKL-LCEAIK-EP-NC-RLENLG--LE-
--A-CELTGASCE--------------------YLVSALSQNETL-WAINLLKNALDLRG
LAMLFE--------ALNVHGCS--PN--V-------------------------------
---LGLQIT-IPDKETQEL------LI----------AEK-KKNPYLAILSR-V------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPANP00000000631 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MAMTKA-RNPREA---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWALSDLE--E---NDFKKLKFYLRD------
---------M--TLSEG--------Q--PPL-ARGE-LEGLI-PVDLAELLISKYGEKEA
VKVVLKG----LR-VMNLLELVDQLS-HVC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LH--DYREIYREH-
--------VRC-----------------LE------EW---QEA----------GVNGRY
NQVLLVA-KSSSESPESPAH------PI-------PE-----Q-ELDSV--MVEALFDSG
EK--PSLAP----SLVVLQG-------------------------------------L
PF05729 // ENSGT00900000140813_0_Domain_2 (719, 940)
YP
G--------RFDY-VFYVSCKEVVLLLE-------SKLEQ-LLFW-CC-GD-----NQAP
VTEILR-QP----ERLLFILDGFDE-LQRPFEEK------LK-K-----RG-----L-SP
-KESLLHLLIRRHTLPTCSLLIT-TRPLALRNLEPLL-K-----QAR--HVHILGFSEEE
RARYF----------------S--SY-FTD---------EKKADSAFDIVQK-NDILYKA
CQVPGICWVVCSWLKGQMERGK-AV-L--ET-PRNSTDIFMAYVSTFLPPVD-DGGCSEF
----------S-RHRVLRSLCSLA---AEGI-QN-QR----FLFEEA-------------
--ELRKHNLDGPRLA-AFLSS----NDYQLGLA-IK-KFYSFRHISFQDFFHAMSYLVK-
EDQSQL----------------G--KESRREVQ-RLLEAKEQE-GNDEMTLTMQFL----
----LDILKKDSFLNLELKFCFR-----------------ISPC-LAQ-DL-KHFKEQM-
E-------------------------------------SMKHNRTWDLEFSLYEAKI-KN
LVKGIQMN---------------------D-------------------VSFKIKHSN-E
KK-SQSQN--S----FSVK-SSLS-HGP-KEKQ------------------KC----PSV
---HGQKEGK---DNI--A-------------------GTHKE-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------AST----------------GKG---------R
-GTEE-TDEEVRS-PKN--TYI----YT--------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSBTAP00000048061 (view gene)
------------------------------------------------------------
------------------------------------------------------------
-------------M-GDMNLSS--------------DPSSCPSSP-----------LS-P
S---GVSPPSLESSSSSTLPFR
PF02758 // ENSGT00900000140813_0_Domain_0 (203, 326)
NG---VMPFMLSVS--A---EHLQRFKQLLVE------
---------E--NPRPG--------C--SPL-TWDQ-LKSAR-CGELVHLLTEYFPGQRA
WEMARDI----FA-KMNQTELCLQTQ-RELNEILP-NLEPEALSLR------K-------
------------------------------------------------------------
------------------------------------------------------------
-------REL-------------------------------TLEEDES--DKIREYKAR-
--------VMSE----------------HSTL--WDRTSWPGNNVDFFYQEP--------
----------------------------------------------CREDTLLQCLLLPR
KP--QGRQP----K
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVLQGDAGVGKTTVAKKV-----MQQWA--E---------NKFYA
H--------KGWL-AFYLHCQEMDRPE-------EQSFSE-LIAC-KL-----SG-SPAL
ASRILS-HP----EHLLLLLDGFEE-LTLTLI------DRRE-D-----LSEDWSQK-MP
-GSVLLTSLLSKRMLPDATIIVF-LRFSLWKTVGPFL-N-----SPS--LITLMGFSAPE
RSRYF----------------R--SY-FKN---------RRDADAALRFAMG-NAVLFSM
CRVPVICWLVCSCLKQQMERGS-SL-P--RA-FPNATAVFIHYLSSLLPTRV-RHVVGET
------------PQEQLKRLCSLA---VEGM-WK-NQ----WVFSEM-------------
--DLKLARLDDR-DVEVFLGV----RVLRRAVA-GG-DLIAFAHPSFQEFFAALLFVLC-
FPQRL--------------RNFQ--VLDRFRIF-QLLAHPG-R-RKNHLAGMALFL----
----FGLLNDTCALAVEQLFGCK---------------V--SLG-NKR-KLLKVATMPPD
GD-----------------------------------PLTPQHGLPQLFYCLHEIRE-EV
FVGQILHN---------------------H-------------------RKALLVISKIR
DL---QVS--A----FCLK-FCRQ-LRE-LEVT----------------ISW-----AVA
-KATSLSP----GP-------------------------------LPSP--QPEGS-DLR
SLWWQDFC-SVFKTHESLEVLTVRDSVMDTE---------AVETLA--AALRH-P-----
HC--N-LRKLI--F-KRVGSL--VV---S-E-GIIRVL----------------VENQYL
RHLQ-IQDT-EVGCQ-VIDALCNTLKHPWCF----LQFL--------RLE---------G
C------P----------------------------------------------------
---------FD---PS-NGA---DLTRSLKR-NIHLKTLMLRRGSLER-GEECPP-----
--V-LAPQLERL-S----------------------------------------------
------------------------LENCDLT-----------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------PLSCESLAFS-LVSNQSLTHLS
L-AENALQDDGVK-QLWNILQHF--PCP----L---------------------------
-----------------------QRLVL--RNC-----ALTSECCQDI------------
-------ASAL-DKNKTLRSLDLGANRLRDSGVVL-LCQPLL-NP-DS-GLQVLE--LE-
--E-CQFTSICCP--------------------ALTSVLLHNRTL-RYLDLSGNGVGLQG
AKLLQD--------AALKRVLR--PE--V-------------------------------
---V------LYV-P---------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSETEP00000006872 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------VEHR--
PF02758 // ENSGT00900000140813_0_Domain_0 (198, 332)
AAVARDL---LLAALEDLS--Q---EQLKRFRHKLRD------
------------ASGGS-----------RSI-PWGR-LERAD-ALDLVERLVEFYGPEPA
LEVARKA----LK-RSDVRDVATRLK-EQRLQC---------L---------GPG-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------PE-VELSVSEYKRTYRAH-
--------VLLL----------------HAKV--KERNARS------------VKITKRF
TKLLIAPESAAPR-P--TMRPPE-------EPVP--------RRARRSNTDTFNRLFGRD
E---EGERP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
LTVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYH
G--------QVDF-AFFLCCHELLEQAC------ARSLGD-LVLE-QC-----PD-RRAP
VRQILA-QP----ERLLFILDGADE-LLAAGSA--------EAA-----PCSDPFEA-VS
-GARLLGALLSKALLPSARLLVT-TRAALPAQLQGPL-R-----SPQ--CAEVRGFSDKD
KKKYF----------------Y--KF-FLD---------ERTAERAYRFVKE-NETLFAL
CFVPFVSWIVCSVLRRQLERGQ-DL-S--RT-SKTTTSVYLLFLISLLSSAPPAKEA---
-----------GLQGELRKLCRLA---REGVLGR--R----AQFTEQ-------------
--DLARQELRGSNVQGLFLSQKEMPGLLEA-----E-VAYQFTDHSFQEFFAALSYLLE-
EDASP-------------------E-VPAGSVE-ALLRGDTEL--RGHLTLTTRFL----
----FGLLSAERMGDIERYFGC-----------------VVSKR-VKQ-DALQWVLGQGR
PP-----ATPEGAEGLQGPEGAEEQEE-------EVEEEEEANYPLELLYCLYETQE-AE
FVRQALHG---------------------L-------------------PELTLERLRFS
RT-DVAVL--S----YCVQ-HCPR-GQA-LQLR------------------GC----GLN
------VV----------------------------------------REK--K---RKS
----------------------------------------LVKR----------------
------LQSS---L--------------GGR------N----------------SQ-SGQ
KQ---RQVS-------PLRPLCEAMVDPHCG----LSSL--------TLS---------H
C-----------------------------------------------------------
--------KLP---DS-VCR---DLAEALRL-APALTELDLQHNSLS-DMGLKVLCEG--
-LAWPRCQVQTL------------------------------------------------
----------------------------RVQ-----------------------------
---------------QPSL---------QKGLQYV-------------------------
------------------------------VSLLQQRPALAT---LD-LHGC-QLPEDTV
--K--------YLCATL----QHPACSLQSL-----------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
MGP_SPRETEiJ_P0086559 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MSLDIQCEQ-LSD-ARWT----------------EL---------
----LPLI-------------------------------QQYEVV------RLEDC-GLT
EVRCKDIS-SAVQA
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1542)
NPALTELSLRTNELGDG---------GVGMVL--QGLQN-P-----
TC--K-IQKLS--L-QNCGLT--EA---GCG-ILPGML----------------RS
PF13516 // ENSGT00900000140813_0_Domain_6 (1617, 1642)
LSTL
RELH-LNDN-PMGDA-GLKLLCEGLQDPQCR----LEKL--------QLE---------Y
C------N----------------------------------------------------
---------LT---AT-SCE---PLASVLRV-KADFKELVLSNNDLHE-PGVRILCQG--
LKD-SACQLESL-K----------------------------------------------
------------------------LENCGIT-----------------------------
----------------------------AANCKDLCD-----------------------
--------------------------------VVASK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
ASLQE---LD-LSSNK-LGN---
-------AGIAALCPGL----LLPSC----------------------------------
--------------------------KIR-TLW---------------------------
-------------------------------LWECDITAEGCKDLCRV-LRA
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
KQSLKELS
L-ASNELKDEGAR-LLCESLLEP--GCQ----L---------------------------
-----------------------ESLWI--KTC-----SLTAASCPYF------------
-------CSVL-T
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
KNRSLLELQMSSNPLGDEGVQQ-LENSGT00900000140813_0_Linker_15_16 (2378, 2390)
CKALS-QP-DT-PF13516 // ENSGT00900000140813_0_Domain_16 (2390, 2436)
VLRELW--LG-
--D-CNVTNSGCS--------------------SLENSGT00900000140813_0_Linker_16_17 (2436, 2442)
ANVLLAPF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2464)
NRSL-RELDLSNNCMGGPG
VLQLLE--------SLKQPSCT--LQ--Q-------------------------------
---LVLYDI-YWTNEVEDQ------LR----------ALE-EERPSLRIIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMLUP00000015241 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------DKEAHKML-
--------IRKK----------------L--Q--EERCLHNRIHDIFCQDMI--------
----------------------------------------------LELVNKFDFVFHHV
SP--EAAKA----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
LNVFLIGQEASGKTMVLRGA-----LFEWA--N---------GNLWA
D--------VFSY-VIHLSSHDINQMT-------SGSLVA-LISK-DW-----PE-GRAP
VADILS-EP----QKILFILEDLDN-VEHSFRIS------PQ-A-----LCSDGRQQ-VS
-TPVLVASLLTRKLAPGCRFLIT-SRPQGQAVTWTIM-K-----KTD--MLMTLSFAREK
RHKYF----------------T--IF-FRD---------GQRAEAAFQLVQR-NELFLHL
CQVPILCWVTCVALQRQMNQG-ADL-K--RS-CQTLTDIYTHFLAETLTAQA----AD--
----------QRPVVPLQSLCSLA---LEGL-LH-NT----LHFTEE-------------
--DLRGVGFTQA-DVSTLQTL----EILWPSSH-MK-GHYRFIHSKVQEFCAAVAFMLPL
VQHGI-----------------P----SAWG-R--C--KGKRE-LYDDFSPVIASM----
----FGLLNKKRRKTFETALGCR---------------LL-TDY-VSQ-YLLMETRCFGD
NL-------------------------------------KAIGHHTPLFHGLFENQE-EE
YVKRIMDS---------------------F-------------------SEANIYVRDHR
DL---MVS--S----YCLG-YCQH-LQK-LKLS----------------IQN-----IFE
--KRS-NE-------------------------------------------QSTPRTIRS
LVHWRDVC-TLLHTRETLRELELCNTDLDDT---------SDRVLC--KALSHP------
DC--Q-LRTLKLTY---------FSV-DTRFEEMFKAI----------------VHNRNL
TCLC-LSCM---------------------------------------------------
------------------------------------------------------------
--------------------------------------------PISS-KVFSLLHEV--
LAN-PRCSIQHL-S----------------------------------------------
------------------------LMKCDLE-----------------------------
----------------------------TSAYQELAS-----------------------
--------------------------------LLLSSKNLKK---LT-LSKNA-LSR---
-------DGMEVFCDAL----FHPNCVLESL-----------------------------
------------------------------------------------------------
------------------------------VLLFCCLTLRSLSLISRS-LVLTETLKHLD
V-SVNNIRDNGVL-VLMMSLVFP--TCS----V---------------------------
-----------------------QELEM--SGC-----FFTAKACEYM------------
-------VCVLQ-NNP
PF00560 // ENSGT00900000140813_0_Domain_15 (2357, 2405)
NLKSLELGSNNVGDAGLAP-T-------P-LC--ISPHR--LE-
--E-CMLTSACCA--------------------SLASVLISSKTL-RKINLLGNNLGDEG
IVQLLE--------GLGHPDCV--LQ--S-------------------------------
---VGLDIK-NMNAETMTL------FM----------TVR-NKNTHLHVTDQSWAKKEGR
EV----------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMLUP00000014098 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------SVLTANENLGELRVSDSNLPGP---------ALVTLS--NHLKHP------
RC--R-LQKLKMNN---------VSF-SAESRFFFEVF----------------TQSPVL
KHLD-LSDT-RISHD-DFKLLCNALNNPACN----IENL--------LL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------VECGLL-----------------------------
----------------------------ADDCEGLKE-----------------------
--------------------------------VLMRNT
PF00560 // ENSGT00900000140813_0_Domain_11 (2019, 2057)
KLKV---LD-LSYNC-LG----
-------NGLSLLCEAL----CLPACALRGL-----------------------------
------------------------------------------------------------
------------------------------GLVDCCLREPCWERLRDV-VLINRNLTHLD
L-STNGLEDKDLN-LLGEALKQP--SCY----L---------------------------
-----------------------KSLSL--FNC-----FITANGCQDL------------
-------ASIL-TGNP
PF00560 // ENSGT00900000140813_0_Domain_15 (2357, 2381)
NLRNLQIGHNNIEDDGVRL-LCEALL-HP-NC-HLENLG--LD-
--A-CQLTSACCE--------------------DLASVLRGSRCL-KELNLAGNPLGHHG
VQLLCA--------ALRHPECG--LQ--T-------------------------------
---LELEKA-EFDEETQEL------LV----------AEE-ERNPCLTITH---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
MGP_CAROLIEiJ_P0090776 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------M---ASFISDFG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 323)
-LMWYLKELN--K---KEFIKFKEFLIQ------
---------E--ILKLK--------L--KQI-SWTK-VKKAS-REDLANLLLKCYEENQA
WDMTFNI----LQ-KINRKDLTERAT-EEIADSSPLGLTD--SG--------------NP
K---------------------------------------------------LYRD----
------------------------------------------------------------
----------------------------------------------------------H-
--------LKKK----------------LTHD--CSKKFNVRIQDF--------------
---------------------------------I-------KETFIQNDYDTFENLLISK
GT--EKK-P----H
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
MVFLQGMAGIGKTMMLKNL-----MLAWS--K---------GLVFQ
N--------KFSY-TFYFCCQDMKQLK-------TASLAE-LISR-EW-----PSP-SAP
IEEILS-QP----EKLLFIIDSLEG-MECDLTKQ------ES-E-----LCDNCMKK-QP
-VSILLSSLLTRKMLPESSFLLS-TTPETFEKMEDRI-L-----CTD--VKTATAFDERS
IKIYF----------------H--RL-FQD---------KIRAQEAFSLVRE-NEQLFSI
CQVPLLCWMVATCLKKEIEKGR-DP-V--SV-CRHITSLYTTHILNLFIPQS-AQYPSK-
-----------KIQEQLQGLCSLA---AEGM-WT-DT----FVFGEE-------------
--ALRRNGILDS-DIPTLLDI----GMLGKIRE-FE-NSYIFLHPSVQEVCAAIFYLLK-
SHVDH-----------------P--SQDVKSIE-TVLFMFLKK-VKTQWIFLGCFI----
----FGLLQKSEQEKLVVFFGRR---------------L--SQK-IQH-KLYQCLETISG
NAE-----------------------------------LQEQIDGMKLFSCLSEIED-EA
FLVKVMSY---------------------M-------------------QQINFVAKNYS
DL---ILA--A----YCLK-HCST-LKK-LSFS----------------TQN-----VLN
-EKG--GQ-----------R-----------------------------------CMKKL
LICWNDMC-SVFVRSKDIQVLQIKDTSFSEP---------AIRVLY--ESLKYP------
SF--T-LNKLVANN---------VHF-FGDNHMFFEL-----------------IQNCSL
QYLD-LGCS-FLTHS-EVKLLCDILNQAECN----IEKL--------V------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSP00000478516 (view gene)
ENSBTAP00000017623 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MREAKIAPLS
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 326)
NYG---LQWCFEQLG--K---EEFQTFKALLKE------
---------H--ASESA--------A--CSF-PLVQ-VDRAD-AESLASLLHEHCRASLA
WKTSTDI----FE-KMSLSALSEMAR-DEMKKYLL-AEISEDSAPTKT--DQGPSMKEVP
G-----------------------------------------------------------
------------------------------------------------------------
---------------------------------------------PREDPQDSRDYRIH-
--------VMTT----------------FSTR--LDTP-QRFEEF---------------
-----------------------------------AS-------ECPDA-HALSGAFNPD
SS--GGFRP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 905)
TVVLHGPPGVGKSSLARRL-----LLFWA--Q---------GDLYK
G--------LFSY-VFLLRARDLQGSR-------ETSFAE-LISK-EW-----PD-APVP
VEKVLS-QP----ERLLIVVDGLEE-LELTFRDQ------DS-S-----LLADWAER-QP
-APVLAHSLLKKVLLPECALLLT-VQDAGLQRLQALL-R-----SPR--YLWVGGLSVEN
RMQLL----------------L--GG-GKH---------CRRKTCAWHAGAD-HQEVLDK
CQVPVVCALVREALELQGEPGK-GL-P--VP-GHTLTGLYATFVFQRLAPKD-AGWRALS
----------GEERCALKGLCRLA---ADGV-WN-AK----FVFDGD-------------
--DLGVHGLQGP-ELSALQQA----SILLPDGH-CGRG-HAFSHLSLQEFFAALFYVLR-
GVEGD--------------GEGY--PLFPQSTK-SLTELRHID-LNVQLVQMKRFL----
----FGLVSKEVMRALETLLGCP---------------V--AP-VAKQ-QLLHWICLLGQ
HPA----------------------------------A-AASPDLLEAFYCLFETQD-DE
FVRLALNG---------------------F-------------------QEVWLQLNRPM
DL---TVS--S----FCLR-RCQH-LRK-VRLD----------------VRG-----T-P
-KDEFAEA----WS--------------------GA-----------PQ--GLKIK-TL-
DEHWEDLC-SVLSTHPNLRQLDLSGSVLSKE---------AMKTLC--VKLRQ-P-----
AC--K-IQNLI--F-KGARV---TP---GLR-HLWMTL----------------IINRNI
TRLD-LTGC-RLREE-DVQTACEALRHPQCA----LESL--------RLD---------R
C------G----------------------------------------------------
---------LT---PA-SCR---EISQVLAT-SGSLKSLSLTGNKVVD-QGVKSLCDA--
LKV-TPCTLQKL-I----------------------------------------------
------------------------LGSCGLT-----------------------------
----------------------------AATCQDLAS-----------------------
--------------------------------ALIENQGLTH---LS-LSGDE-LGS---
-------KGMSLLCRAV----KLSSC----------------------------------
--------------------------GLQ-KLA---------------------------
-------------------------------LNACSLDVAGCGFLAFA-LMGNRHLTHLS
L-SMNPLEDPGMN-LLCEVMMEP--SCP----L---------------------------
-----------------------RDLDL--VNC-----RLTASCCKSL------------
-------SNVI-TRSPRLRSLDLAANALGDEGIAA-LCEGLK-QK--N-TLTRLG--LE-
--A-CGLTSEGCK--------------------ALSAALTCSRHL-ASLNLMRNDLGPRG
MTTLCS--------AFMHPTSN--LQ--T-------------------------------
---IGLWKE-QYPARVRRL------LE----------QVQ-RLKPHVVISDAWYTEE---
--E---------------------------------------------------------
--------------------------------------------------EDGPCWRI--
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMNEP00000029873 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MSLDIQCEQ-LSD-ARWA----------------EL---------
----LPLL-------------------------------QQCQVV------RLDDC-GIT
EARCGDIS-SALRV
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1542)
NPALTELSLRSNELGDA---------GVHCVL--QGLQS-P-----
SC--K-IQKLS--L-QNCYLT--GA---GCG-VLSGTL----------------RS
PF13516 // ENSGT00900000140813_0_Domain_6 (1617, 1642)
LPTL
QELH-LSDN-LLGDA-GLQLLCEGLLDPQCR----LEKL--------QLE---------Y
C------N----------------------------------------------------
---------LS---AA-SCE---PLASVLRA-KPDFKELTVSNNDINE-AGVRVLCQG--
LKD-SPCQLEAL-K----------------------------------------------
------------------------LESCGIT-----------------------------
----------------------------SNNCRDLCS-----------------------
--------------------------------IVASK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
ASLRE---LA-LGSNK-LGD---
-------VGIAELCPGL----LHPSS----------------------------------
--------------------------SLR-TLW---------------------------
-------------------------------IWECGITAKGCADLCRV-LRTKE
PF13516 // ENSGT00900000140813_0_Domain_13 (2215, 2237)
SLKELS
L-AGNELGDEGAR-LLCETLLEP--GCQ----L---------------------------
-----------------------ESLWV--KSC-----SFTAACCSHF------------
-------SSVL-AQS
PF13516 // ENSGT00900000140813_0_Domain_15 (2356, 2378)
KFLLELQISNNRLADVGVQE-LCKGLG-QP-GS-VLRVLW--LG-
--D-CDMSDSSCS--------------------SLAATLLA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2464)
NHSL-RELDLSNNCLGDAG
VLQLVE--------SVRQPGCL--LE--Q-------------------------------
---LVLYDI-YWSEAMEDR------LQ----------ALE-EEKPSLRVIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMFAP00000021717 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MSLDIQCEQ-LSD-ARWA----------------EL---------
----LPLL-------------------------------QQCQVV------RLDDC-GLT
EARCGDIS-SALRV
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1542)
NPALTELSLRSNELGDA---------GVHCVL--QGLQS-P-----
SC--K-IQKLS--L-QNCYLT--GA---GCG-VLSGTL----------------RS
PF13516 // ENSGT00900000140813_0_Domain_6 (1617, 1642)
LPTL
QELH-LSDN-LLGDA-GLQLLCEGLLDPQCR----LEKL--------QLE---------Y
C------N----------------------------------------------------
---------LS---AA-SCE---PLASVLRA-KPDFKELTVSNNDINE-AGVRVLCQG--
LKD-SPCQLEAL-K----------------------------------------------
------------------------LESCGIT-----------------------------
----------------------------SNNCRDLCS-----------------------
--------------------------------IVASK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
ASLRE---LA-LGSNK-LGD---
-------VGIAELCPGL----LHPSS----------------------------------
--------------------------SLR-TLW---------------------------
-------------------------------IWECGITAKGCADLCRV-LRTKE
PF13516 // ENSGT00900000140813_0_Domain_13 (2215, 2237)
SLKELS
L-AGNELGDEGAR-LLCETLLEP--GCQ----L---------------------------
-----------------------ESLWV--KSC-----SFTAACCSHF------------
-------SSVL-TQ
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NKFLLELQISNNRLGDAGVQE-LCKGLG-QP-GS-VLRVLW--LG-
--D-CDMSDSSCS--------------------SLAATLLA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2464)
NHSL-RELDLSNNCLGDAG
VLQLVE--------SVRQPGCL--LE--Q-------------------------------
---LVLYDI-YWSEAMEDR------LQ----------ALE-EEKPSLRVIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPCAP00000002371 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------------KINPYRKH
------------M-KEKFHLIWEK------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------ETCLQVPDS--------------
---------------------------------F-------YRETIKKEYE----ELN-N
VF--ETSEA----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
RQVVLKGAEGSGKTTLLRKA-----MLDWT--E---------GNFWK
G--------RFRF-VFFLSARELNCVT-------ETSLVE-LISR-DW-----PES-SEP
IEDIFS-QP----QDILFIMDGFEE-LKFALL-L------KN-N-----TSDDWRQQ-QP
-IEIILSSLLQKKMLPESSLLIA-LGLVGMQKNYSLL-Q-----HPK--YIVLSPFSEHE
RKLYC----------------C--HF-FQE---------KNKAMKAFSYVKD-NTFLFAL
CQNPLLCSLTCCCIKKQLEKGE-DL-E--IA-CQSTTSLYVYFFIHIVKAGC-E------
------------NRAQLKGLCSLA---AEGI-WT-HT----YLFCLG-------------
--DFRRKGVSES-SVLRWLGM----SLLRRSG-----DCITFIHKHVQEFCAAMFYLIK-
EPKDN-----------------S--HPVVGSIT-RFIATAQP---KASLPQMGLFL----
----FGLLTEKVTKILETSIGFQ---------------L--SQG-IKQ-EIIQGLKNLSK
CA-----------------------------------SSETELVFQDLFFCIYETRE-KE
FTAQVMNF---------------------F-------------------KEVIVYISSTE
EL----TS--S----YCLK-HCRI-CSG-XXXX----------------XXXXX-XXXXX
-XXXXXXX-----------X-------------------N------------------WL
LNFWQDLC-SVFSTNKNLQVLELDNCRFSED---------SLVILC--EALAHP------
TC--K-LHRLLLHC---------VST-TGERADFFWAV----------------LQKPHL
KYLN-LHDT-GLSYA-AVIQLSEMLKLPTCS----VEKL--------ML-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------GNCDIT-----------------------------
----------------------------DNKCEDIAS-----------------------
--------------------------------VLMCNNKLKL---LS-FVENP-LDD---
-------EGTKVLCYAL----KHPXXXXXXX-----------------------------
------------------------------------------------------------
------------------------------XLNHCCLSSVSCDYIAQV-LLCNQFLTLLD
L-GSNILKDDGVN-SLCKALKSP--NCH---
PF00560 // ENSGT00900000140813_0_Domain_14 (2252, 2355)
-L---------------------------
-----------------------EELWL--SNC-----FLTSDCCKDI------------
-------ASVL-TSNQKLRTLKLGKNYILDLGVKL-LCVALK-HP-NC-KLENLG--LE-
--M-CRLTSACCE--------------------DLASALTVCKTL-RCLNLQCNTFDHEA
VMVLC---------ALSHLHCD--LQ--T-------------------------------
---LGLNKC-AFAEGTQLL------LT----------AVE-EKKPHLAIIRQPWADN---
--EAE-----VT------------------------------------------------
--------------------------------------------------GVLFR-----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSJJAP00000023655 (view gene)
ENSMPUP00000006858 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------------------------YRER-
--------MKAK----------------ILMM--WDNMPWPEGHIYLRNVTE--------
----------------------------------------------K-EHKELKSLLYPN
R---MGAQP----Q
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TIVLEGVAGVGKTTLAMKA-----MLHWA--E---------GFLFQ
Q--------RFSY-VFFISCHKVKEVK-------DTTFAG-LLSW-DW-----PD-SQPP
FEELMS-HP----ERLLFVIDGFEE-MDMPSN------LDAS-P-----PCTDWFQR-LP
-VHRILFYLLRKELVPGATLLIT-TKEYRTKDLKKLL-L-----DPW--FVQISGFTEGD
REEYF----------------I--RY-FGD---------QNKAKKILRWVRK-NENLFYF
CSAPMVCWAVCSCLKWQMARSP-CF-Q--LS-TQTTTSLYVYFFSSLFAKAE-VSLSAQS
------------WPEQWILLCSLA---AEGM-WF-RN----FTFAKE-------------
--DLKHRHLEAS-LIDTLFSL----NILRKVSD-CD-GCVTFTHQSFQVFLGAMFYALG-
GTKGS--------------M-GS--PAKHEEMK-VLLTDALAD-TNFYWNQMALFF----
----FGLLKRNLAGELEATLRCK---------------V--SRR-ITD-ELLEWAEELGS
YD-----------------------------------IVSNCFEFLHFFECLYETQE-EN
FVRQILSP---------------------L-------------------LEADLDIFGNL
QL---QVA--S----FCLK-HCQR-LNK-LRLS----------------VTV-----PIP
-QTELTF-------------------------------------TTETL--ETSSH--SK
IHQWQDVC-S-VFCHGNVRELDMSNSKLNAS---------SMKKLC--YELRS-P-----
RC--K-LQRLT--C-KSVSPV--RV---L-K-ELVLVL----------------HGNQRL
THLN-LSSN-NLGIP-VSTMIFKTLRHSACN----LQYL--------WLE---------S
C------G----------------------------------------------------
---------LT---PL-TCC---HLFAELRK-NSSVRFLSLGDNDLSG-TEEKGLQGP--
STV-STSSLQEL-S----------------------------------------------
------------------------LEKCQLS-----------------------------
----------------------------AAVYRGLAS-----------------------
--------------------------------CLTSTQRTTR---LC-LGFNP-LQD---
-------DGVRLLCSSL----THPEC----------------------------------
--------------------------ALQ-RLV---------------------------
-------------------------------LWFCQLGAPSCKYLSDA-LLENKSLTHLN
L-RRNSLGDEGVK-LLCEALSRP--DCR----L---------------------------
-----------------------QNLDL--SGC-----SLTAGGCQEL------------
-------ANAL-RHNHNVKILDLGSNDVRDDGVIH-LCEALK-DP-SC-ALNTLG--LE-
--K-CNLMPACCQ--------------------HLSSVLGSSKSL-VHLNLLQNDLEPSG
VGILWK--------ALKKSTCK--LQ--K-------------------------------
---LGLKKE-LYD-IVKRE------LE----------ELQ-ERGSFLRVKCKWDFND---
--LE--------------------------------------------------------
---------------------------------------------------DKWWW----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCANP00000022391 (view gene)
ENSCLAP00000000175 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---SSFFSDFG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 323)
-LMWYLEELK--K---KEFMKFKELLKE------
---------E--TSHLG--------L--KQI-PWAE-VKKAS-REELACLLLKHYEEEQA
WDVTFRV----FQ-RIDRRDLCERAI-RES------------TG--------------HS
K---------------------------------------------------LYQA----
------------------------------------------------------------
----------------------------------------------------------H-
--------VREK----------------YSNI--LAKKSIATLHQN--------------
---------------------------------L-------DETITQEEIKSLELLFTPQ
KP--GKQ-A----H
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
AVLLKGNQGVGKTTFLIKF-----MLAWS--K---------GLIYK
D--------RFSY-VFYFCCQELKQLP-------VTSLAE-LITS-TW-----PST-SAP
VSEILA-HP----EKVLFMVDSFEM-LGSDLGEP------EG-E-----LCRDWVEV-RP
-PQLLLSSLLRRRMVPECSLLIT-VDIKARQSLEDRL-R-----DLD--VRVLGGFRESD
IKLYI----------------Y--GM-FQD---------LHRAAEAFSLIQS-KEWFLAL
CRFPVLCCLAGTALKHEMDEGK-DV-A--RT-FQRTTSLYTSFLFNLFTPRT-TDSPSW-
-----------QGQAQLQTLCSLA---AQGM-WT-NC----FVFWEK-------------
--DLRRNGIEGS-DVPALLEL----GILLRCRG-SE-PSYKFLHPSIQEFCAALFYLLK-
SPGHH-----------------P--HPAVQGTQ-DLLVTFLEK-SRAYWVFLGCFL----
----FGLLHERERQKLDAFFGFQ---------------L--SSE-VKQ-QVYQYLQSLST
VR-----------------------------------NLHERIVFLALFYCLFEMQE-ET
FVSQAMNL---------------------M-------------------QELTFTMSGKL
DL---EVA--A----YCLK-HCSK-LRQ-LSIS----------------IQD-----LPF
-F----FP-----------H-----------------------------------RSNAD
LVCWHQVC-SVLTTNKHLRVLQIKDSVLSDC---------ATGTLL--NQLRQP------
NC--L-VENLMINN---------ARL-HCDSWLLFEVF----------------IHNPNV
KYLS-LNNT-DLCRD-AVRNLCSVLSHPTCV----SCP----------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------RPRG-----------------------------
----------------------------HPDCKAFAS-----------------------
--------------------------------ILMEGRKLKQ---LN-LASNF-LK----
-------KGVRTLCKAL----CHPDCTLQFL-----------------------------
------------------------------------------------------------
------------------------------VLAFCYLSNWCWDYLAEV-LLT
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
SKSLTHLD
L-SLNTLRDEGLK-VLCEALRYP--TCM----L---------------------------
-----------------------KTLSL--MQC-----SITAGGCRDL------------
-------AAVL-T
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
SNQNLTSLLLAKNHLGDTGVKL-LCGAVA-QP-GC-RLENLG--LD-
--D-CGLTGACCE--------------------DLADMLTSSKTL-TTLSLIQNPLDHCG
VAVLCE--------GLRHPECT--LR--V-------------------------------
---LGLNKA-AFDKKTQAL------LI----------DEE-KRNPNLAVLES--------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSSSCP00000023003 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------LGDCGLT-----------------------------
----------------------------AADSQDLAS-----------------------
--------------------------------ALT
PF13516 // ENSGT00900000140813_0_Domain_11 (2016, 2050)
KNQSLTH---LY-LASNS-LGS---
-------EGVNLLGRAM----KLPNC----------------------------------
--------------------------SLQ-RL----------------------------
------------------------------ILNACNLDVAGCGFLAFA-LMG
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
NRRLTHLS
L-SVNPLEDDGMK-LLCEVMMEP--SCH----L---------------------------
-----------------------QDLEL--VKC-----QLTATCCKSL------------
-------SHVI-TR
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NKHLQSLDLAANALGDNGIEV-LCEGLK-QK--N-SLRRLG--LE-
--A-CELTSGCCE--------------------ALSSALSGS
PF13516 // ENSGT00900000140813_0_Domain_17 (2443, 2465)
EHL-TSLNLMQNNFSLGG
MTKLCS--------AFAQPTCD--LQ--M-------------------------------
---LG-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSRNOP00000046459 (view gene)
ENSPPAP00000034190 (view gene)
ENSPCAP00000003709 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------KKDKFRMLL-
---------KKT----------------YWHI--WQI-----------------------
-----------------------------------------------WRYKMQIPFCDPR
AL--SGMLP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
YTVVLQGPSGVGKTTLAKAV-----LLDWI--Q---------DSLAM
---------KFEY-AFYLSCEELNRMG-------PCTFAD-LITK-DF-----PE-LRDA
MPDDLD-RS----HNTLIIVDGFNE-LRA-PG------VLIS-G-----ICANWKVK-KP
-VPILLSSLLSRNLLPRATLLIT-MQTWAWNRLRYLV-E-----RPF--FVDMEGLSEVE
RKEYF----------------L--QH-FQD---------EDQALQAFNLMRS-NEALFRM
GSSPVVCWVVSTCLKLQMEKGE-DP-T--PT-CETITSLFLRFICSQFTSA-------WQ
------------LQAPVKSLCLLA---AEGV-WT-QI----TVFDEE-------------
--DLKRLEINDC-ELSPFLDK----NILQKSKD-RA-GYYSFIHLSVQQFFAAMFYVLR-
TKEEE-------------VQGIP--KGDIGDIQ-QLFSKEETL-KNPSLIEVGYFL----
----FGLANEKRVRELETTFGCP---------------VTRSVE-VKQ-EIVRWKVNTSG
RK-----------------------------------RPLLPMAMKEVVDCLCESQE-ED
FVKDAVVH---------------------F-------------------KEVSLKLKNEK
DL---VQV--S----FCLK-HCQN-LQK-LSLK----------------IDKG----IFL
-ENN-A-M----EV--------------------D------------TQDERSQKD-QHI
LPFWMDLC-SLFGSNKNPIFLEINRSFLSDS---------SVRILC--DQIAS-T-----
TH--S-LQNVV--L-DNIVPA--SA----YQ-DLCLTF----------------SGHESL
THLT-LQSI-GQND--VFPPLCEVLRHPKCN----LQYL--------RLG---------S
C------S----------------------------------------------------
---------AT---TQ-QWD---GLFSVLTI-NQSLTCLNLSASELLD-EGLKLLCRA--
LKH-PQCSLQRL-S----------------------------------------------
------------------------LENCHLT-----------------------------
----------------------------EASCKDLSS-----------------------
--------------------------------ALIINQ
PF00560 // ENSGT00900000140813_0_Domain_11 (2019, 2057)
KLTH---LS-LAKNK-LGD---
-------GGLQLLCEGL----SYPDC----------------------------------
--------------------------KLQ-TLV---------------------------
-------------------------------LWECDITDNGCSLLSKL-LQQKSSLTHLD
L-ELNYIGVTGLH-FLCEVLKDP--TCN----L---------------------------
-----------------------KHLVL--CV----------------------------
IPFSCMDFSVL-RCNQNLSTLDLDQ---NPL-------------------GYSRM--LK-
--V-DKSDTEMQK--------------------LVQEIKGSNPQVAIERHHL------KL
---------------NRSSSYD--FL----------------------------------
----P-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMPUP00000008374 (view gene)
------------------------------------------------------------
--------------------------------------------------RR--------
-------QSLGAKPRSPLHASPPRECSLEQRGLVGAADRRAPRGRVGWEAGQGRGPRNHM
RVDRHRPPAPSTEAR--AAKA
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 328)
RDL---LQAALEDLS--Q---EQLKRFRHKLRD------
------------AALDG-----------RSI-PWGR-LERAD-AVDLAEQMIQFYGPEPA
LDVARKT----LK-RADVRDVAARLK-EQGLQW---------F---------SPG-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------SS-ALLSVSEYKKKYREH-
--------VLQQ----------------HAKV--KERNARS------------VKINKRF
TKLLIARESAALVDE--APGLAE-------EPEP--------ERARRSDTHTFNRLFGRD
E---EGQRP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLQGPAGIGKTMAAKKI-----LYDWA--V---------GKLYH
D--------QVDF-AFFLPCRELLERPG------PCSLAR-LVLD-QC-----PH-RSAP
VRQMLA-RS----ERLLFILDGADE-LP-PGPA--------DAA-----PCTDPFEE-AD
-GARVLDGLMSKTLLPGARLLVT-ARATAPGRLQGRL-C-----SPQ--CAEVRGFSDKD
KKKYF----------------H--KF-FRD---------ERMAERAYRFVKE-NETLFAL
CFVPFVCWIVCTVLRQQLQLHQ-DL-S--RT-SKTTTSVYLLFITSVLSSAPATDGP---
-----------QMQEELRKLGRLA---REGTLRH--R----AQFPEK-------------
--DLERLELRGSKVQTLFLSKKELPGVLET-----E-VTYQFMDQSFQEFFAALSYLLE-
QEKAL-------------------E-APADSVG-ALLQGGTEL--RGHLTLTTRFL----
----FGLLNGERLRDIERHFGCG---------------GVVSKR-VKQ-DVLRWVQEQGQ
GHPRAAPEGTEETEAPEGSQEPEE------------EEEGELSYPLELLYCLYETQE-ST
FVHQALRG---------------------W-------------------PELLLERVHFS
RL-DLLVL--S----YCVR-SCPA-GQA-LRLV------------------SC----VLV
------PP----------------------------------------QEKK-----KKS
----------------------------------------LIKR----------------
------LHGS---L--------------SGS------S-----------------SQATA
RK---SQAS-------PLRTLFEAMTDQHCG----LHSL--------TLC---------R
C-----------------------------------------------------------
--------RLP---DS-ICR---DLSEALRE-APALRELGLLHNRLS-EAGLRVLSEG--
-LAWPQCRVQKL------------------------------------------------
----------------------------RVW-----------------------------
---------------QPRL---------QEAFQYL-------------------------
------------------------------TSVLRQSPALTI---LE-LGDC-QLSEPTV
--T--------DLCAVL----QHPGCRLQTLRL--TSVQLSEQ-----------------
----------SLKELQVVKTAKP-------DLV----------------------I--TH
PALD-------------------GHPRPPAGHSSAL------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPPAP00000011890 (view gene)
ENSCANP00000024527 (view gene)
ENSPTRP00000019830 (view gene)
ENSMLEP00000015777 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MTSP----QLEWT---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 323)
LQTLLEQLN--E---DELKSFKSLLRA------
---------L--PLEDV--------L--QKA-PWSE-VEEAD-GKKLAEILINTSSENWI
RSATVTV----LE-EMNLTELCK-MA-KSEM-----------------------------
------------------------------------------------------------
--L---------------------------------------------------------
--------------------------------------------------GEKEGRRNS-
---------MQK----------------QSLV--WKNTFWQGDIDSFHDDVI--------
-----------------------------------------------RRHQRFIPFLNPR
TP--GKLTP----Y
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVLHGPAGVGKTTLAKKC-----MLDWT--D---------CNLSP
---------TVRY-AFYLSCKELSRMG-------PCSFAE-LISK-DW-----PE-LQDD
IPSILA-QA----QRILFVVDGLDE-LKVPPG------ALIQ-D-----ICGDWTQQ-KP
-VPVLLGSLLKRKMLPKATLLVT-TRPRALRDLQLLV-Q-----QPI--YIRVEGFLESD
RRAYF----------------L--RH-FGD---------EDQAMRAFELMRS-NAALFQL
GSAPAVCWIVCTTLKLQMEKGE-DP-A--PT-CLTSTGLFLRFLCSQFPQG-------AQ
------------LWGALRALSLLA---AQGL-WA-QM----SVLHRE-------------
--DLESAGVQES-DLRLFLDR----DILRQDRV-SK-GCYSFIHLSFQQFLTALFYALE-
KE--E---EE--------DRDGH--AWDIGDVQ-KLLSREERL-KNPDLIQVGHFL----
----FGLANEKRVKELEATFGCR---------------M--SPE-IKQ-ELLRCKANLHA
NK-----------------------------------P-LSMTDLKEVLCCLYESQE-EE
LAKVVVAP---------------------L-------------------KKISINLTNTS
EM---MHC--S----FSLK-QCRD-LEK-VSLQ----------------VAKG----VFL
-ENY-VDF----EP--------------------DIEFERC-SYV--TLPSWAQQD-LRS
LRLWTDFC-SLFSSNSNLKFLEVKQSFLSDS---------SVRILC--DHITR-S-----
TC--H-LQKVE--I-KNVSPD--TA----YR-DFCLAF----------------IGKRTL
THLT-LEGH-IEWERTMLLLLCDLLRNHKCN----LQYL--------RLG---------G
H------C----------------------------------------------------
---------AT---SE-QWV---DFFYVLKA-NQSLRHLHLSASVLLD-EGAKLLYKT--
MTH-PKHFLQML-S----------------------------------------------
------------------------L-----------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------QQCSITKRGCRYLSEA-LQEACSLTNLD
L-SLNQIA-RGLW-ILCQALENP--NCN----L---------------------------
-----------------------KHLRL--WSC-----SLMPFYCQHL------------
-------GSAL-ISNQKLETLDLGQNHLWKSGIIQ-LFRVLR-QR-TG-SLKTLR--LK-
--T-YETNLEIKK--------------------LLEEVKEKNPKLTIDCNAS------GA
---------------TAPACCD--FF----------------------------------
----C-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMOCP00000012017 (view gene)
------------------------------------------------------------
------------------------------------------------------------
--------------YCP-----------------GAPSGECARR-EGCFGGLGTGLLQ--
VTSVI-TLFIPS--R--EALAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
ILAALEDLS--Q---EQLKRFRHKLRD------
------------APLDG-----------RSI-PWGR-LENSD-AVDLVDKLTQFYGSEPA
VDVTRKI----LK-KADARDVAVRLK-AQQLQQ---------L---------GPS-----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------S-VAVSLSEYKKKYREH-
--------VLLQ----------------HAKV--KERNARS------------VKINKRF
TKLLIAPGSNAVEDE--LLGPSE-------EPEP--------ERARRSDTHTFNKLFRGD
D---EGLRP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYH
G--------QVDF-AFFMPCGELLESPG------TRSLAD-LILD-QC-----PD-RKAP
VRQILA-LP----QRLLFILDGADE-LPAVGAP--------EAA-----PCKDPFEA-TS
-GLRVLRGLLSQELLPLARLLVT-TRNVAPGTLRGRL-G-----SPQ--CAEVRGFSDKD
KKKYF----------------F--KF-FRD---------ERKAERAYRFVKE-NETLFAL
CFVPFVCWIVCTVLQQQLELGQ-DL-S--RT-SKTTTSVYLLFIASMLKSA-GTNGP---
-----------RVQGELRTLCRLA---REGILKH--Q----AQFSEN-------------
--DLAQLKLQGSQVQTLFLSKKEVLGVLEK-----E-VSYQFIDQSFQEFLAALSYLLE-
AEGAP-------------------G-APAGGVK-MLLNSDAEL--RGHLALTTRFL----
----FGLLSAERVRDVERYFGC-----------------VVPER-VKQ-DTLRWVQGQGH
PK--VAPVGPEKNDELEDTEEPEE-------------DEEDLNFGLELLYCLYETQE-DA
FVRQSLTN---------------------L-------------------PEIVLERVRFT
RM-DLEVL--G----YCAQ-CCPA-GQA-LKLV------------------NC----GLV
------EV----------------------------------------KEK--K---KKS
----------------------------------------LMKR----------------
------LKGS----------------------------------------------QNTT
KQ---PPVS-------LLRPLCEAMTSQQCG----LRIL--------IFS---------R
C-----------------------------------------------------------
--------KLP---DA-VCR---DLSEALKV-APALRELGLLQCRLT-EAGLRLLSDG--
-LVWPTCQVQTL------------------------------------------------
----------------------------RMQ-----------------------------
---------------MPNL---------QEAIQYL-------------------------
------------------------------AILLQQSRVLTT---LD-LSGC-QLPNYMV
--S--------CLCSAL----KHPNCCLKTLSLT--SVELSDM-----------------
----------SLRDLQAVKTSKP-------DLT----------------------I--LH
PKL---------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMLEP00000025857 (view gene)
MGP_SPRETEiJ_P0084839 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MALARA-NSPQEA---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWALNDLE--E---NSFKTLKFHLRD------
---------V--T------------Q--FHL-ARGE-LESLS-QVDLASKLISMYGAQEA
VRVVSRS----LL-AMNLMELVDYLN-QVC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LN--DYREIYREH-
--------VGC-----------------LE------ER---QDW----------GVNSSH
NKLLLMA-TSSSGGRRSPSC------SD-------LQ-----Q-ELDPV--DVETLFAPE
AE--SYSTP----PI
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 930)
VVMQGSAGTGKTTLVKKL-----VQDWS--K---------GKLYP
G--------QFDY-VFYVSCREVVLLPK-------CDLPN-LICW-CC-GD-----DQAP
VTEILR-QP----GRLLFILDGYDE-LQK---------------------------S-SR
-AERVLHILMRRREVP-CSLLIT-TRPPALQSLEPML-G-----ERR--HVLVLGFSEEE
RETYF----------------S--SC-FTD---------KEQLKNALEFVQN-NAVLYKA
CQVPGICWVVCSWLKKKMARGQ-EV-S--ET-PSNSTDIFTAYVSTFLPTDG-NGDSSEL
----------T-RHKILKSLCSLA---AEGM-RH-QR----LLFEEE-------------
--VLRKHGLDGPSLT-AFLNC----IDYREGLG-IK-KFYSFRHISFQEFFYAMSFLVK-
EDQSQQ----------------G--EATHKEVA-KLVDP---E-NHEEVTLSLQFL----
----FDMLKTESTLSLGLKFCFR-----------------IAPS-VRQ-DL-KHFKEQI-
E-------------------------------------AIKYKRSWDLEFSLYDSKI-KK
LTQGIQMK---------------------D-------------------VILNVQHLD-E
KK-SDKKK--S----VLVT-SSFS-----SGKV------------------QS----PFL
--------GN---GKS--T-------------------RKQKK-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------ASN----------------GKS---------R
-GAEE-PAP-GVR-NRR-LASR----EK-GHMEMGD---KEDGGV---E-EQED-EEG-Q
T-LKK-DGEMI---DK-MNG----------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSSTOP00000015184 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MSLDIHCEQ-LSD-ARWT----------------EL---------
----LPLI-------------------------------QQYQVV------RLDDC-GLT
EGRCQDIS-SALRV
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1542)
NPSLTELSLRTNELGDA---------GVHLVL--QGLQS-P-----
VC--K-IQKLS--L-QNCCLT--EA---GCG-VLPGVL----------------RS
PF13516 // ENSGT00900000140813_0_Domain_6 (1617, 1642)
LSTL
RELH-LNDN-PLGDA-GLRLLENSGT00900000140813_0_Linker_6_7 (1642, 1650)
CEGLLDPQPF13516 // ENSGT00900000140813_0_Domain_7 (1650, 1766)
CH----LEKL--------QLE---------Y
C------N----------------------------------------------------
---------LT---AA-SCE---PLATVLRA-KPDFKELTVSNNDLQE-AGIQTLCQG--
LKD-SACQLEML-K----------------------------------------------
------------------------LENCGVT-----------------------------
----------------------------AANCKALSA-----------------------
--------------------------------VVASK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
ASLQE---LD-LGCNK-LGD---
-------EGIAELCPAL----LRASS----------------------------------
--------------------------RLR-TLW---------------------------
-------------------------------LWECDITAEGCKDLCRV-LKA
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
KQSLKELS
L-AENQLGDEGAR-LLCESLLEP--GCQ----L---------------------------
-----------------------ESLWV--KTC-----GLTAACCPYF------------
-------RSVL-A
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
QNKSLLELQISSNQLGDAGVQE-LCQGLS-QP-GA-VLRVLW--LG-
--E-CNVTDSGCD--------------------TLATALLV
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2464)
NRSL-RELDLSNNCVGDPG
IQRLVE--------SLRQPGCV--LE--Q-------------------------------
---LVLYDI-YWTEEMEEQ------LR----------TLE-EHKPSLRVIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCCAP00000024896 (view gene)
ENSCGRP00000011757 (view gene)
ENSMUSP00000139357 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--MDA-AGASC-SSV--DAVAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LMATLEELS--Q---EQLKRFRHKLRD------
------------APLDG-----------RSI-PWGR-LERSD-AVDLVDKLIEFYEPVPA
VEMTRQV----LK-RSDIRDVASRLK-QQQLQK---------L---------GPT-----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------S-VLLSVSAFKKKYREH-
--------VLRQ----------------HAKV--KERNARS------------VKINKRF
TKLLIAPGTGAVEDE--LLGPLG-------EPEP--------ERARRSDTHTFNRLFRGN
DE--ESSQP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYH
S--------QVDF-AFFMPCGELLERPG------KRSLAD-LVLD-QC-----PD-RAWP
VKRILA-QP----NRLLFILDGADE-LPTLPSS--------EAT-----PCKDPLEA-TS
-GLRVLSGLLSQELLPGARLLVT-TRHAATGRLQGRL-C-----SPQ--CAEIRGFSDKD
KKKYF----------------F--KF-FRD---------ERKAERAYRFVKE-NETLFAL
CFVPFVCWIVCTVLQQQLELGR-DL-S--RT-SKTTTSVYLLFITSMLKSA-GTNGP---
-----------RVQGELRTLCRLA---REGILDH-HK----AQFSEE-------------
--DLEKLKLRGSQVQTIFLNKKEIPGVLKT-----E-VTYQFIDQSFQEFLAALSYLLE-
AERTP-------------------G-TPAGGVQ-KLLNSDAEL--RGHLALTTRFL----
----FGLLNTEGLRDIGNHFGC-----------------VVPDH-VKK-DTLRWVQGQSH
PK--GPPVGAKKTAELEDIEDAEEE----------EEEEEDLNFGLELLYCLYETQE-ED
FVRQALSS---------------------L-------------------PEIVLERVRLT
RM-DLEVL--N----YCVQ-CCPD-GQA-LRLV------------------SC----GLV
------AA----------------------------------------KEKKKK---KKS
----------------------------------------LVKR----------------
------LKGS----------------------------------------------QSTK
KQ---PPVS-------LLRPLCETMTTPKCH----LSVL--------ILS---------H
C-----------------------------------------------------------
--------RLP---DA-VCR---DLSEALKV-APALRELGLLQSRLT-NTGLRLLCEG--
-LAWPKCQVKTL------------------------------------------------
----------------------------RMQ-----------------------------
---------------LPDL---------QEVINYL-------------------------
------------------------------VIVLQQSPVLTT---LD-LSGC-QLPGVIV
--E--------PLCAAL----KHPKCSLKTLSLT--SVELSEN-----------------
----------SLRDLQAVKTSKP-------DLS----------------------I--IY
SK----------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMLEP00000032802 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MAVTKA-RNPREA---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWALSDLE--E---NDFKKLKFYLRD------
---------M--TLSEG--------Q--PPL-ARGE-LEGLI-PVDLAELLISKYGEKEA
VKVVLKG----LR-VMNLLELVDQLS-HVC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LH--DYREIYREH-
--------VRC-----------------LE------EW---QEA----------GVNGRY
NQVLLVA-KSSSENPESPAH------PI-------PE-----Q-ELDSV--MVEALFDSG
EK--SSLAP----S
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
LVVLQGSAGTGKTTLARKM-----VLDWA--T---------GSLYP
G--------RFDY-VFYVSCKEVVLLLE-------SKLEQ-LLFW-CC-GD-----NQAP
VTEILR-QP----ERLLFILDGFDE-LQRPFEEK------LK-K-----RG-----L-SP
-KESLLHLLIRRHTLPTCSLLIT-TRPLALRNLEPLL-K-----QAR--HVHILGFSEEE
RARYF----------------S--SY-FTD---------EKKADSAFDIVQK-NDILYKA
CQVPGICWVVCSWLNGQMERGK-AV-L--ET-PRNSTDIFMAYVSTFLPPVD-DGGCSEF
----------S-RHRVLRSLCSLA---AEGI-QN-QR----FLFEEA-------------
--ELRKHNLDGPRLA-AFLSS----NDYQLGLA-IK-KFYSFHHISFQDFFHAMSYLVK-
EDQSQL----------------G--KESRRGVQ-RLLEAKEQE-GNDEMTLTMQFL----
----LDILKKDSFLNLELKFCFR-----------------ISPC-LAQ-DL-KHFKEQM-
E-------------------------------------SMKHNRTWDLEFSLYEAKI-KN
LVKGIQMN---------------------D-------------------VSFKIKHSN-E
KK-SQSQN--S----FSVK-SSLS-HGP-KEKQ------------------KR----PSV
---HGQKEGK---DNI--A-------------------GTHKE-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------AST----------------GKG---------R
-GTEE-TPKNTYI-Y---------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPTRP00000005809 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MAMAKA-RNPREA---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWALSDLE--E---NDFKKLKFYLRD------
---------M--TLSEG--------Q--PPL-ARGE-LEGLI-PVDLAELLISKYGEKEA
VKVVLKG----LK-VMNLLELVDQLS-HIC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LH--DYREVYREH-
--------VRC-----------------LE------EW---QEA----------GVNGRY
NQVLLVA-KPSSESPESLAC------PI-------PE-----Q-ELDSV--TVEALFDSG
EK--PSLAP----S
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
LVVLQGSAGTGKTTLARKM-----VLDWA--T---------GTLYP
G--------RFDY-VFYVSCKEVVLLLE-------SKLEQ-LLFW-CC-GD-----NQAP
VTEILR-QP----ERLLFILDGFDE-LQRPFEEK------LK-K-----RG-----L-SP
-KESLLHLLIRRHTLPTCSLLIT-TRPLALRNLEPLL-K-----QAR--HVHILGFSEEE
RARYF----------------S--SY-FTD---------EKQADRAFDIVQK-NDTLYKA
CQVPGICWVVCSWLQGQMERGK-VV-L--ET-PRNSTDIFMAYVSTFLPPDD-DGGCSEL
----------S-RHRVLRSLCSLA---AEGI-QH-QR----FLFEEA-------------
--ELRKHNLDGPRLA-AFLSS----NDYQLGLA-IK-KFYSFRHISFQDFFHAMSYLVK-
EDQSRL----------------G--KESRREVQ-RLLEVKEQE-GNDEMTLTMQFL----
----LDISKKDSFSNLELKFCFR-----------------ISPC-LAQ-DL-KHFKEQM-
E-------------------------------------SMKHNRTWDLEFSLYEAKI-KN
LVKGIQMN---------------------N-------------------VSFKIKHSN-E
KK-SQSQN--L----FSVK-SSLS-HGP-KEEQ------------------KC----PSV
---HGQKEGK---DNI--A-------------------GTPKE-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------AST----------------GKG---------R
-GTEE-TDEEVR--SPK-NTYI--------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSOGAP00000016874 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------IKIRDLFCPC
PE--TQEEP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
RIVILYGVDGIGKSTLARQV-----QKAWE--K---------GQIYQ
D--------HFQH-IFYFSCRELAQSN-------VISLAE-LITK-AQ-----AA-PSAP
LEQVLS-RP----EQLLFILDGIDE-PRWVLQEQ------ST-E-----VCLHWSQP-QE
-VAALLGSLLGKTILPEASFLIT-TRTVDLQKLIPSL-K-----QPR--WVEVLGFSESG
RKEYF----------------Y--NY-FTD---------ESQALRALFLVES-NPAVWTM
CLVPWLSWLVCTCLKQQMESGE-DL-T--LT-TTTATGLCLHYLSQVIP--------AQ-
-----------CLRHQLRGICSLA---AEGI-WQ-GK----SLFSLD-------------
--DLRKHRLDKP-IISAFLKM----RILQKH---PTSLTYSFIHLCFQEFFVAMSCALG-
D------EEE--------RSEHP--NSI-GGVR-RLLDAYERH--VLVRAPTRCFL----
----FGLLSEQVGREMESIFTCQ---------------R--APE-LKQ-ELLQWAKWEAQ
-----------------------------------EMRFPRQPGFLELFHCLYETQD-EE
FVTEVMGH---------------------F-------------------QGSSVYVQTDM
EL---LVF--T----FCIK-FCHH-VES-LQLH----------------EGGQ-------
------PRS-TWRPARVIL-----------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------HWESRI-S----------------------------------------------
-------VPNDDRCFCS-----QKVMQVPVT-----------------------------
----------------------------DACWQLLFS-----------------------
--------------------------------ILKDTGNLKE---LD-LSGNP-LSC---
-------SAVQSLSETL----RCPCCHLETLRLR--K-MT---------VVSVTHN----
--------------------------NLHSDFW---LILPE-------------------
N-----------------DMKAVLTNLIKLGLANCGLTAEGCKDLAFG-LSTSQTLTELE
L-SFNMVTDAGAK-HLCQRLRQQ--SCR----L---------------------------
-----------------------QRLQL--VSC-----GLTF------------------
------------------------------------------------------------
---------SCCQ--------------------DLASMLSTSRSL-KELDLQQNDLGDLG
VRLLCE--------GLRHPACS--LT--L-------------------------------
---L--------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSGGOP00000023539 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----MSLD--I----QSLDIQCEE-LSD-ARWA----------------EL---------
----LPLL-------------------------------QQCQVV------RLDDC-GLT
EARCKDIS-SALRV
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1542)
NPALAELNLRSNELGDV---------GVHCVL--QGLQS-P-----
SC--K-IQKLS--L-QNCCLT--GA---GCG-VLSSTL----------------RS
PF13516 // ENSGT00900000140813_0_Domain_6 (1617, 1642)
LPTL
QELH-LSDN-LLGDA-GLQLLCEGLLDPQCR----LEKL--------QLE---------Y
C------N----------------------------------------------------
---------LS---AA-SCE---PLASVLRA-KPDFKELTVSNNDINE-AGVRVLCQG--
LKD-SPCQLEAL-K----------------------------------------------
------------------------LESCGVT-----------------------------
----------------------------SDNCRDLCG-----------------------
--------------------------------IVASK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
ASLRE---LA-LGSNR-LGD---
-------VGIAELCPGL----LHPSS----------------------------------
--------------------------RLR-TLW---------------------------
-------------------------------IWECGITAKGCGDLCRV-LRAKE
PF13516 // ENSGT00900000140813_0_Domain_13 (2215, 2237)
SLKELS
L-AGNELGDEGAR-LLCETLLEP--GCQ----L---------------------------
-----------------------ESLWV--KSC-----SFTAACCSHF------------
-------SSVL-AQ
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NKFLLELQISNNRLEDAGVRE-LCQGLG-QP-GS-VLRVLW--LA-
--D-CDVSDSSCS--------------------SLAATLLA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2464)
NHSL-RELDLSNNCLGDAG
ILQLVE--------SVRQPGCL--LE--Q-------------------------------
---LVLYDI-YWSEEMEDR------LQ----------ALE-KDKPSLRVIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCJAP00000064897 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----XXXXX---------------------------------XXXXXXX-----------
------XXXX-------------------------------XXXXX----XHRRKYREK-
--------MRAT----------------VLEK--WDNNAWPEDHVYIHSTSE--------
----------------------------------------------D-EHQDLQRLLDPD
G---TGAQA----Q
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TIVLTGRAGVGKTTLAMKA-----LLHWA--D---------GVLFQ
Q--------RFSY-VFYLSCCRMRHMK-------ETTFAK-LISR-DW-----PG-SDAP
VEEFLS-QP----EKLLFIVDGCEN-MIAAEPCS--DGPDGG-P-----PCTDWYQE-LP
-VTRILLGLLKKELVPQATLLIT-IETFYLTDLRGLV-V-----NPR--FVHMTGFTDED
LWVYF----------------M--RH-FED---------PSEVRKILQQIRK-NETLFES
CSAPMVCWIVCACLKQPRVRYY-AL-R--SV-TETATALYACFVTNLFSTAE-VDLADDS
------------WPGQWRALCSLA---IEGL-WS-VA----FMFNTQ-------------
--ATQRQGLEAP-FLDYLHQL----NILQKVGD-GG-GCFTFTHPSLQEFFAAMSFMLE-
EPEEF--------------L-QH--SPKYEEMK-LLLQDALID-KEPCWTPVVLFL----
----FGLLNRSIARELESTLRCK---------------V--SPR-KTE-ELLEWWEGLDK
AE-----------------------------------SASVQFHVLRFFLCLHESQE-EG
FVEKMLDR---------------------T-------------------FEVDLHILREE
EL---KAA--S----FCLK-QCKG-LTK-LRLS----------------VGS-----HIL
-ERDVDTL----------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
MGP_CAROLIEiJ_P0078152 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------MMDSSGYG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 322)
-LLQYLQKLS--D---EEFQKFKERLRK------
---------E--PEKFN--------L--KPI-SWTK-IKNTS-KEDLAMQLYTHYPGK-A
WDMVLSL----FL-QVNRKDLSTMAQ-IERRD----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------KQTKYKEF-
--------MKNA----------------FQHI--WTTETNTYIPDR--------------
---------------------------------S-------YHEFIEVQYRALQDIFDCE
-----SE-P----V
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVVSGSRGGGKTTFLRKA-----MLDWA--S---------RNLLQ
N--------RFQY-VFYFSVFSLNNIT-------ELSLAE-LISS-TL-----PES-SET
VDDILS-DP----KRILFILDGFDY-LKFDLE-L------RT-N-----LCNDWRKK-LP
-TQIVLSSLLQKIMLPECSLLLE-LGNASLSNIIPLL-Q-----HPR--EIIMSGFSEQT
IEIYC----------------V--SF--FN---------TQTGVEIFENLKS-IKPLFNL
CCCPYLCWMICSTLKWQYERKE-VA-N--HF-GRTLALLYTIFMVSAFKSTY-ARNPSK-
-----------QNRARLRTLCTLA---VEGM-WK-QV----YVFDSD-------------
--DLRRNGISES-DKKVWLRM----KFLQNQG-----SNIVFYHSTLQWYFAVLFYFLQ-
QYKDA-----------------H--HPVIGNIA-QLLGEIYAH-KQNQWFHTRILL----
----FGMATEQVTSLLEPCFGCI---------------S--TKE-VRQ-EIIRYIKSLSQ
QEC----------------------------------NEKLVVHPQNLFFCILDNQE-ER
FVRQLMDL---------------------F-------------------EEMTVDISNVD
DM---SAT--Q----YCLH-RASK-VKN-LHLH----------------IQKR----VFL
-EIHDPEY-----------A-------------------D-L-------ELFK-LNQKLL
VNHWTRLC-TFLC---NLHVLDLDSCHFNEN---------AIEVLC--NCLSPTSSVPLT
GF--K-LHRLLCSF---------MTN-FGD-GLLFSTF----------------LHLPHL
KYMN-LYGT-NLSND-AVERLCSALKFSTCG----VEEL--------LL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------GKCDIS-----------------------------
----------------------------SEACGIIAT-----------------------
--------------------------------SLINS-EVKH---LS-LVENP-LKN---
-------KGVMSLCEML----KDPSCALESL-----------------------------
------------------------------------------------------------
------------------------------MLSYCCLTFIACGHLYEA-LLSNERLSLLD
L-GSNFLEDTGVN-LLCEALKVP--NCT----L---------------------------
-----------------------KELWL--PGC-----YLTSECCEEI------------
-------SAVL-TC
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NRNLKTLKLGNNNIQDTGVKR-LCEALC-HP-NC-EMQCLG--LD-
--M-CNFTSDCCE--------------------DLALVLTT
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2465)
CNTL-KSLNLDWNAFDHSG
LEMLCK--------ALNHKACN--LE--V-------------------------------
---LGLDKS-AFSEESQTL------LQ----------AVE-KKNNNLNVLHFPWVEE---
--ERE-----KR------------------------------------------------
--------------------------------------------------GVRPVWNSKN
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMLUP00000000748 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MALALT-SN
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
PQEA---LLLALSDLD--E---NDFKILKFHLRD------
---------R--TLLGG-----------QGL-ARGE-LEGLS-RVDLASRLVLRYGAQEA
VKMVCKV----LQ-VMNLLELVDQLS-HIC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LN--DYRETYREY-
--------VRC-----------------LE------ER---QEE----------GVSRTY
NQLLLVA-IASPGSPE-------------------PE-----Q-EQGSV--TVEALFDPG
EM--PTPGP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
ATVVLQGPAGTGKTTLARKI-----VLDWA--T---------GTLYK
D--------RFDY-VFYVGCREVVLLPE-------GTLDQ-LLLW-CC-GD-----SKAP
VTEIRR-QP----ERLLFILDGYDE-LQRPFAKR------LR-R-----PG-----P-SP
-AQDTLRRLIRREVLPTCSLLIT-TRPLALRNLQSLL-R-----QPR--HVQVRGFSEEE
REKYV----------------Q--AY-FTD---------GEQAQKAFDFIRG-NDVLYQA
CLVPGICWVVCSWLKGRMERGG-EV-S--ET-PSSGTDIFMAYVSTFLPPND-NEDPSEL
----------T-RDRVLRGLCSLA---AEGI-QH-QR----FLFEEA-------------
--DLRKHSLDGPSLA-AFLSS----HDYQEGLD-IK-KFYSFRHISFQEFFHAMSYLVK-
EDQSQL----------------G--EESLREVN-RLLGDREQA-AGEEMTLSMQFL----
----LDISKKESSSNLELTFCFK-----------------ISPS-IKQ-DL-KNFKEQM-
S-------------------------------------SLKHNRTWEWDFFLNEAKM-KS
LANCVQMS---------------------N-------------------VAFKVGHSN-E
KK-SHSRS--S----FSVT-TSLS-DAK-KEER------------------KC----LVV
--------NE---RNV--A-------------------GTQKQ-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------AAN----------------GKS---------R
-G----------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCGRP00000002480 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------M---ASFFSDFV---F
PF02758 // ENSGT00900000140813_0_Domain_0 (209, 323)
MWYLEELN--K---KEFMKFKEVLRK------
---------E--MAESG--------L--QHI-PWTE-VKKAS-REDLANLLVKHYE-KKA
WDVTFRI----FQ-NINRKDLSERAA-GEM------------AG--------------HS
K---------------------------------------------------IYQA----
------------------------------------------------------------
----------------------------------------------------------H-
--------LKKV----------------LSQD--CSRKFNIPIQHF--------------
---------------------------------L-------KEEFTPDKCSRLQQLFVPK
TT--EKK-P----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
HTVLLKGVSGVGKTVLLAEL-----MMAWS--E---------GLMFQ
N--------EFSY-VFYLCCHEIKQLK-------TASLAD-LLSR-EW-----PRP-SAP
IAEIVS-QP----AKLLFIIDSLEG-LNCDLSEP------ES-E-----LCDNWMQK-CP
-VSILLSSLLRRKLLPESSLLIT-ATPEAFEKLGDRI-E-----YTV--KNFVTGFNENS
RRSCF----------------Q--RV-FQD---------EDTAQEVFTLVRE-NEQLYNI
CHVPSLCWMVATCLNDEVKRGG-HL-S--AV-CRCTTSVFAAYVLNLFIPQS-AHYPSK-
-----------KSQALLRGLCSVA---AEGM-WT-DN----FAFNQK-------------
--DLEKNGVFHS-DISTLLNM----KILLKSRD-SK-NLYTFLHPSIQEFCAALFYLVN-
SHVDH-----------------P--SKDVLCVE-TLLFTFLKK-VRVQWIFLGCFI----
----FGLLHKSEQAKLEAFFGYQ---------------L--SQE-VKQ-TLCQSLEKISA
DEE-----------------------------------LRKQIDDMKLFYCLFELED-DG
FVTQVMTF---------------------L-------------------PQIKFVAKEYS
DL---IVA--A----YCLK-NCST-LQK-LSIS----------------TQD-----ILQ
-EEK--DP-----------R-------------------------------------KKL
LLCWQDLC-SVLGSSQQIQVLQVKDTYLNEL---------AFLELH--NHLRNP------
SC--A-PRKFLVNN---------VSF-LCNNQLFFELL----------------TQNHNL
HHLD-LSLT-SLSRS-DVKLLCEVLNQKGCH----IEEL--------LL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------ASCSLSKQCWNYLSDV-LKR
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
NKTLSHLD
V-SSNDLQDEGLK-VLCQALSLP--DSA----L---------------------------
-----------------------KSLLL--QIT-----EFDKETQELL------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------I----------AEK-EKNPYLTILSS-V------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMEUP00000000871 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------LPV----SGRH
PF02758 // ENSGT00900000140813_0_Domain_0 (204, 328)
ILCCLAAYLEELE--S---VELTKFKMYLGL------
---------E--AKREG--------E--GRI-PWGR-MEKAG-PLDMAQILVAHKGPAEA
WELALRV----FF-WINRKDLWERGC-RELLVRXX----X--XX--------XXXXXX--
------------------------------------------------------XX----
------------------------------------------------------------
X-----XXXX-------------------------------XXXXXXXXXXLQETYREH-
--------IRRK----------------YRFI--EDRNARLGELV---------NLSQRY
TRLLLVKEHATLTRAQHELL------ATGKQHALCLD--------QRASPIQIETLFEPD
EE--RPDAP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
HTVVLQGAAGIGKSILAQKV-----MLDWA--E---------GHLYS
G--------RFDY-LFYVNCREMNQV-R------ERSVED-LISV-CW-----PE-RNVP
ISEIVQ-VP----KRLLFIIDGFDE-LRTSFHES------RD-C-----CCIQWNER-KP
-VEDLLSCLLRKKLLPELSLLIT-TRPTTLETLHHLL-E-----QPR--HVKILGFSKVQ
RQEYF----------------Y--KF-FRH---------EEQAQQGFSLVRD-NEQLFTM
CFVPMICWIVCTCLKQQLEAGW-PF-S--QT-SKTTTAVYTFYLLSLLQPKQER-HRT--
-----------QLQPNLRGLCSLA---AAGL-WS-QR----ILFEED-------------
--DLHRHGLKGD-DVSAFLNM----NVFHKDIQ-CE-RYYSFIHLSFQEFFAAMFYVLE-
G---E--------------P--G--A-SPADLT-RLLNHYAKS-ERSYLALTVRFL----
----FGLLNEERRKSLEKQLGWK---------------T--SRR-TKW-DLLQWIQAKAQ
SE-----------------------------------GSTLQRGTLEIFECLYEIQE-ED
FIQRALDH---------------------F-------------------QVVVVSDMA-T
KT-EHVVS--S----FCVR-NCRS-ALV-VHLF----------------SA------MFS
-SDVMEDR--T-------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------KN-DLEDL-GIQELCRGLRHPNCK----VQEL--------KLK---------R
C-----------------------------------------------------------
--------QIS---SA-ACQ---DIALALMT-NQNLIRLDLSRNA-GC-PGVKLLCEG--
LQH-PKCHLQMLQ-----------------------------------------------
------------------------LRRCQVD-----------------------------
----------------------------EAACQELSK-----------------------
--------------------------------VLSTSQHLTE---LD-LTGNS-LXX---
-------XXXXXXXXXX----XXXXX----------------------------------
--------------------------XXX-XXX---------------------------
-------------------------------LKICHLTPSACHNLSTV-LSTNQ
PF00560 // ENSGT00900000140813_0_Domain_13 (2215, 2236)
NLTELD
L-SLNDLGDLGVK-SLCEGLCHS--KSQ----L---------------------------
-----------------------QTLRL--GIC-----RLTFVSCEAL------------
-------STTL-QSNI
PF00560 // ENSGT00900000140813_0_Domain_15 (2357, 2375)
HLKELDVSFNDLEDMGVHL-LCEGLQ-HP-KC-KLHKL------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMAUP00000020881 (view gene)
ENSCAFP00000003831 (view gene)
ENSMOCP00000000825 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------M---ASFFSDYG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 323)
-LMWYLEELN--K---KEFMKFKEFLKE------
---------D--IVQYG--------L--KQI-PWTE-VKKAS-REDLANLLVKYYEEKKA
WDVTFRI----FQ-KINRKDLSERAA-REI------------AG--------------YT
K---------------------------------------------------LYQA----
------------------------------------------------------------
----------------------------------------------------------H-
--------LKKK----------------LTRD--WSRKFDFPIQDF--------------
---------------------------------M-------KPKVSQDEYSRFEHLFTSK
TA--ETK-P----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
HTVLLKGMAGIGKTLTLARL-----MLAWS--D---------GLVFQ
N--------KFSY-IFYFCCREVTQLE-------TASLAE-LMSR-EW-----PSA-SAP
IMEIIS-QP----EKLLFIVDSLER-LNCDFTEP------EL-E-----LCHNWIEK-RP
-VPILLSSLLRRKILPESSLLIT-ATPETFDKLEDRI-D-----YTE--MRAMLGFDESS
RRMIF----------------R--DL-FPD---------KNRAQEAFRLVRE-NEQVFTL
CQVPLLCLAVATCLRNEIDKGR-DP-A--PV-CRRTTSVYTTYILNLLIPKS-AHGPNK-
-----------KSQDLLQGLCSLA---AEGM-WT-DR----FAFSEE-------------
--DLGKNGIVSS-DIPMLLNI----KIFLRSRD-FK-NFYTFLHPSVQEFCAAVFYLVK-
NHTDH-----------------P--SKDVRCIE-TLLFTFLKK-VNVHWIFGGCFI----
----FGLLHKCEQEKLEAFFGYQ---------------L--SQE-VKQ-KLYRCLETVSV
NED-----------------------------------LQEQVDSMRLFYCLFEMDD-EA
FVVQVVNF---------------------L-------------------QQVKFVAKDYS
DL---MVA--A----YCLK-HCST-LKK-LSFS----------------MQA-----VLQ
-LEQ--EH-----------S-----------------------------------YMEKL
LICWQDVC-SALVSSEQVQELQVKDTNLGEL---------AFSVLY--NHLKHP------
SC--T-PQILEVNN---------VSF-LCNNYMFFEML----------------AQNHNL
RYLN-LSFT-VLSHS-DVKLLCEVLNREECI----IEKL--------LV-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------AGCQLS-----------------------------
----------------------------PEDCKIFAS-----------------------
--------------------------------VLTNSKTLDH---LN-IASNS-LE----
-------KGVSSLCKAL----CQPDCVLKCL-----------------------------
------------------------------------------------------------
------------------------------MLANCSLSEQCWDYLSDV-LK
PF13516 // ENSGT00900000140813_0_Domain_13 (2212, 2233)
QNRTLEHLD
V-SSNDLQDEGLK-ILCKPLSLP--DSV----L---------------------------
-----------------------QTLHL--RHC-----LITASSCQDL------------
-------SEVL-R
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
NNQNLKSLQISNNKIEDAGVKF-LCDAIK-QP-SC-HLEILG--LQ-
--A-CELSDACCE--------------------DLASAFIQSQTL-RAVILAENALSRSG
LAVLCE--------ALQQHRNI--QR--I-------------------------------
---LGLQIT-DFDKETQAF------LL----------AEE-EKNPYMLIVS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSNLEP00000012957 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------MVSPA----PMGFN---LP
PF02758 // ENSGT00900000140813_0_Domain_0 (210, 323)
ALLEQLS--Q---DELSKFKSLITT------
---------V--SLANE--------L--QKI-PHKE-VDKAD-RKQLAEILTSHCHSYWV
EMATIQV----FE-KMHRMDLSESAK-DELREAAL-KSFN----KNK------PLSLG--
------VTRKERPP---------------------------------LDLEEMLERFK-A
EALAFT---------------------------------ET---EEDVT-----------
-------CLT-------------------------------KEVFKGEKPD--NRYRYI-
--------LKTK----------------FREM--WK--SWPGDSKEVQV-----------
---------------------------------------------MAERYKMLIPFSNPR
VL--PGPFS----YTVVLHGPAALGKPR--------------------------------
---------------------------------------------------------WDD
PF05729 // ENSGT00900000140813_0_Domain_2 (781, 930)
IPHILA-QA----QKILFVIDGFDE-LGAPPG------ALIQ-D-----ICGDWEKK-KP
-VPVLLGSLLKRSMLPKAGLLVT-TRSRALRDLRLLA-E-----EPI--YIRVEGFLEED
RRAYF----------------L--RH-FGD---------EDQAMRAFELMRS-NAALFQL
GSAPAVCWIVCTTLKLQMEKGE-DL-A--PT-CLTRTELFLRFLCSRFPQ-------GAQ
------------LRGALRKLSLLA---AQGL-WA-QM----SVLHRE-------------
--DLERLGVQES-DLRPFLDG----HILRQDTV-SK-GCYSFIHLSFQQFLAALFYALE-
KEEEE-------------DRDGH--AWDIGDVQ-KLLSGGERL-RNPDLIQAGYYS----
----FGLANERRAKELEATFGCR---------------I--SPD-IKQ-ELLRCDVSCKG
G------------------------------------H-STMTDLKELLGCLYESQE-EE
LVKEVMAQ---------------------F-------------------KEISLHLN-AV
DV---VPS--S----FCVK-HCRN-LQK-MSLQ----------------VIK-----AEL
-PENVTVS----ES--------------------DAEAER------------SQDD-EHV
LPFWTDLC-SIFGSNKDLMGLTINNSFLSAS---------LVRILC--EQIAS-D-----
TC--H-LQRVV--F-KNISPA--DA----LR-NLCLAL----------------RAHKTV
TYLT-LQGN-DQND--MLPTLCEVLRHPECN----LRYL--------GYS---------L
A-----------------------------------------------------------
------------------------------L-SPRLQCHNLGS---------LQPLWD--
YRH-PAVFFRR-------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------C-ITPLR-------
-------------THRF----GFSFF----------------------------------
--------------------------LWL-ISR---------------------------
-------------------------------LWNCDITSDGCCDLAKL-LQEKSSLSCLD
L-GLNHIGVIGVK-LLCEALRKP--LCN----L---------------------------
-----------------------KCLWL--WGC-----SIPPFSCEDL------------
-------CSAL-SCNQSLVTLDLGQNPLGSSGVKM-LFETLT-RP-NG-TLQTLR--LK-
--I-DNFNDELDK--------------------LLEEIEENNPQLIIDTEKH----DPWK
---------------KRPSSHD--FM----------------------------------
----I-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCATP00000031257 (view gene)
MDQPEDPSSRLRSLRTASPQRAFVKSVPHQRLRLRPWCKAPKLGLTK-RQPGHVTWPGVW
KNRSREGGAGEEGAPQSSGRRWPVSTSPSEGEEHQD------APTRVRGRWR--------
-------AKSGVTPRCPLPPPHG-----SS--LARGSLRPCPRWVLKRQSRGWAGVTRGP
LPDDPRPPVSSTGLR--LAVAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLAALEELS--Q---EQLKRFRHKLRD------
------------VGPDR-----------RSI-PWGR-LERAD-AVDLAEQLAQFYGPEPA
LEVARKT----LK-RADARDVAAQLK-EQRLRR---------L---------GLG-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------SA-ALLSVSEYKKKYREH-
--------VLQL----------------HARV--KERNARS------------VKITKRF
TKLLIAPESAAPEEE--TLGPTE-------EPEP--------GRARRSDTHTFNRLFRGD
E---DGRRP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYQ
G--------QVDF-AFFMPCGELLERPG------TRSLAD-LILD-QC-----PD-RGAP
VPQMLA-QP----QRLLFILDGADE-LPALEGP--------EAA-----PCTDPFEA-AS
-GARVLGGLLSKALLPTALLLVT-TRAAAPGRLQGRL-C-----SPQ--CAEVRGFSDKD
KKKYF----------------Y--KF-FQD---------ERRAERAYRFVKE-NETLFAL
CFVPFVCWIVCTVLRQQLELGR-DL-S--RT-SKTTTSVYLLFIASVLSSAPVADGP---
-----------RVQGDLRNLCRLA---REGVLGR--R----AQFAEK-------------
--QLEQLELRGSKVQTLFLNKKELPGVLET-----E-VTYQFIDQSFQEFLAALSYLLE-
DEGVP-------------------R-TAAGGVG-TLLRGDSQP--HSHLVLTTRFL----
----FGLLSAERMRDIERHFGC-----------------VVSER-VKQ-EALRWVQGQGQ
GCPGVAPEVTEETKGLEDTEEPEE-----------EEEGEEPNYPLELLYCLYETQE-DA
FVRQALCQ---------------------L-------------------PELALQGVRFC
RM-DVAVL--S----YCVR-CCPA-GQA-LRLI------------------SC----RLV
------AA----------------------------------------QEK--K---KKS
----------------------------------------LGKR----------------
------LQAS---L--------------GGS------S-----------------SRGTT
KQ---LPAS-------LLHPLFQTMTDPLCR----LTSL--------TLS---------H
C-----------------------------------------------------------
--------KVP---DA-VCR---DLSEALRA-APALTELGLLHNRLS-EAGLRMLSEG--
-LAWPQCRVQTV------------------------------------------------
----------------------------RVQ-----------------------------
---------------LPSS---------QQGLQYL-------------------------
------------------------------VGMLRQSPALTT---LD-LSGC-QLPAPMV
--T--------YLCAIL----QHQGCGLQTLRL--TSVELSEQ-----------------
----------SLQELQAVKRAKP-------DLV----------------------I--AH
PVLD-------------------GHPESP-KELVSTF-----------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSANAP00000003104 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----MSVD--M----PCLDIQCAE-LSD-ARWA----------------EL---------
----LPLL----QR---------------------------CQAV------RLNDC-GLT
EAQCTDIS-SALQA
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1542)
NPALTELNLRSNELGDA---------GVHRVL--QALQS-S-----
SC--K-ILKLS--F-QNCYLT--AA---GCG-VLSGTL----------------RSL
PF13516 // ENSGT00900000140813_0_Domain_6 (1618, 1642)
PTL
QELH-LSDN-PLGDA-GLQLLENSGT00900000140813_0_Linker_6_7 (1642, 1650)
CEGFLDPQPF13516 // ENSGT00900000140813_0_Domain_7 (1650, 1765)
CH----LEKL--------QLE---------Y
C------R----------------------------------------------------
---------LS---SA-GCE---PLGSAFRA-KPDFKELTVSNNDIDE-AGFCALCQG--
LKD-SPCQLETL-K----------------------------------------------
------------------------LENCGAT-----------------------------
----------------------------SDSCRDLCS-----------------------
--------------------------------ILASKASLWE---PA-LGNNK-LGD---
-------VGVAELCPEL----LHPSS----------------------------------
--------------------------SLK-TLW---------------------------
-------------------------------IWECGLTAKRCRDLCRV-LRAKD
PF13516 // ENSGT00900000140813_0_Domain_13 (2215, 2237)
SLKELS
L-AGNKLGDEGAR-LLCESLLEP--GCQ----L---------------------------
-----------------------ESLWL--KSC-----SLTGACCSHF------------
-------SSVL-AQ
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NKFLLEVQIRDNRLGDAGVQE-LCQGLG-QP-GS-VLRSLW--LG-
--D-CDVSDRICG--------------------SLPATRLA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2465)
NHSL-RELDLSNNCLGDAG
VLQLME--------ILRQPACL--LEQ-Q-------------------------------
---LVLYDI-YWSLEMEME------ME----------MAA-GPGEGEVIPEGHLLSR---
--FLP-----ARLLRPTLPGQ-------PGLEAPLG------------------------
--------------------------------------------------SHPAYPRFEE
KR----------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCSAP00000008835 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MAGG----A-WGR---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 328)
LACYLELLK--K---EELKEFQLLFTS------
---------K--VHSS--------GS--SGE-TPAR-PEKTS-GMEVASYLVAQYGEQRA
WDLALRT----WE-QMGLWSLCTQAQ-EGAG-------YS-PSFPY---SPIEPHLGSPS
QPTSTAVLRPWNRELP-----AECTQGSERRVLRQLPDTSGRRW-REISSSFLYQ--A--
---------LPSSPD-LESPSQESPNAPTSTAVLASWRSPPQPSLAPREQEAPGTQWPLD
ETSGNYYTGIREKEREESEKGR------PPWEAAVGTPPQVH---ASLQPPRHPWEPS--
--------ARES----------------LCSTWSW-------NNE---------DLNQNF
TQLLLLQRPHPRSQKP---L------VKGSWPHDVEE----DVEEDRGRLIEIRDLFGPG
LD--PQ-EP----H
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
IVILQGAAGIGKSTLARQV-----REAWG--R---------GQLYG
D--------RFQH-VFYFSCRDLAQCK-------VVSLAE-LIGK-DW-----TA-AQAP
IRQILS-RP----ERLLFILDGVDE-PRWVLQEP------SS-E-----LCLHWSQP-QP
-ADALLGSLLGKTILPEASFLIT-ARTTALQNLIPSL-E-----QAR--WVEVLGFSESS
RREYF----------------Y--KY-FTD---------ERQAIRAFGLIKS-NKELWAL
CLVPWVSWLACTCLMEQMKRKE-EL-T--LT-SKTTTTLCLHYLSQALQ--------AQ-
-----------PLGPQLGDVCSLA---AEGI-WQ-KK----TLFSRD-------------
--DLRKHELDGA-IISTLLKM----GILQEH---PIPLSYSFIHLCFQEFFAAMSYALE-
D------KKG--------RGKHA--NCI-IDLE-KLLEAYGTH--GLFGAPTTRFL----
----LGLLSDEGERAMENIFNCR---------------R--SQG-RN---LMQWVPC---
------------------------------------LRPLLQPHSLDFLHCLYETQN-KM
FLTHMMAD---------------------F-------------------QEMGMCVETDM
EL---LVC--T----FCIQ-FCRH-VKK-LQLI----------------EGRQ-------
------H-R---------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------P------------
------AWSP--------------------------------------------------
-------------------TGIVLFRWVPVT-----------------------------
----------------------------DAYWEILFS-----------------------
--------------------------------ILKVTR
PF00560 // ENSGT00900000140813_0_Domain_11 (2019, 2037)
NLKE---LD-LSGNS-LSH---
-------SAVKSLCKTL----RRPRCLLETLR----------------------------
------------------------------------------------------------
-------------------------------LASCGLTAQDCKDLASG-LRGNQNLTVLD
L-SFNVLTDAGAR-HLCQRLSWT--CCT----L---------------------------
-----------------------QRLRM--GFC-----H---------------------
------------------------------------------------------------
------VTSGCCQ--------------------DLASVLSASPFL-MELDLQQNNLGDTG
VRLLCE--------GLRHPACQ--LT--R-------------------------------
---LGLDQT-TLSDEMRQE------LR----------ALE-QKKPQLLIFS-RWKPTGMI
PNEGLGTGETTNS-----------------TSSRKRQRLGSETESSHVAQADFKLL-DWS
NPPAKA--SQS-AGITEGSSPEVEQ-LEPLCLPSPASLGD-QD-TPLG-TDDDFWGPMGP
VATEVVDK-ERSLYR---------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCCAP00000005912 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MAGG----A-WGR---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LACYLELLK--K---EELKKFQLLLTN------
---------K--AHSR--------GS--SGE-TPAQ-PEKMS-GMQVASHLVAQYGEQQA
WDLAIHT----WE-QMGLRSLCTQAQ-EEAG-------HY-PSFPS---SLSEPSLESPN
QPTSTAVLRSWIPEL------QGCTQGSERSILRHLPDTSGHHW-REITSSLLYQ--A--
---------PLSSPD-YGSPSHDSPNVPTSTAELGSWESPLHPSPVPREQEAPGTQWPLD
ETLGNYYTGETLLGSW-----W------GGRVGEANAGTKNP---LGTS-QCSVWLGSL-
--------WRGS----------------PCSTWSW-------KNE---------DLNQKF
TQLLLLQRPHPRSHEC---L------VRGRLLHNV--------EEDQGHLIEIGDLFGPG
LD--TQ-EP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
RVVILQGAAGIGKSTLARWV-----REAWG--R---------GQLYR
D--------CFQH-VFYFSCRELAQSK-------VLSLAE-LIGK-DW-----TA-TQAP
IRQILS-RP----ERLLFILDGVDE-PKWVLEEQ------SP-E-----LCLHWSQP-QP
-ADALLGSLLGKSILPKASFLIT-ARTTALQSLIPSL-E-----QAR--WVEVLGFSESS
RKEYF----------------Y--KY-FTN---------ERQASRAVRLVKS-NKELWAL
CLVPWVSWLACTCLMQQMKQKE-EL-T--LT-SKTTTTLCLHYFSQTLQ--------AQ-
-----------PLGSQLRGFCSLA---AEGI-WQ-RK----TLFSPD-------------
--DLKKHGLDGA-IISSLLKM----GILQEH---PIPLSYSFIHLCFQEFFAGMSCALE-
D------KKE--------GSEHS--NHI-MGLG-KLLETYGTH--DLFRAPTTRFL----
----LGLLSEQGAREMENIFDCR---------------L--SQE-RKR-RLLQCTQF---
------------------------------------LQPPLQSYPLEFLHCLYETQG-KR
FLTQMMAH---------------------F-------------------QEVCMCVETDM
EL---LVC--T----FCIK-FCHN-VKK-LQLI----------------EGRQ-------
------H-G---------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------P------------
------AWSL--------------------------------------------------
-------------------DRVVLFRWVPAT-----------------------------
----------------------------DAYWQILFS-----------------------
--------------------------------VLKVTGSLKE---LD-LSGNL-LSL---
-------SAVQSLCETL----RYPRCL
PF13516 // ENSGT00900000140813_0_Domain_12 (2068, 2206)
LETLR----------------------------
------------------------------------------------------------
-------------------------------LAGCGLTAEGCKDLENSGT00900000140813_0_Linker_12_13 (2206, 2212)
ASG-LRPF13516 // ENSGT00900000140813_0_Domain_13 (2212, 2237)
TNQTLTEVE
L-SFNVLTDAGAK-HLCQGLRRP--SCK----L---------------------------
-----------------------QRLQL--VSC-----GLTS------------------
------------------------------------------------------------
---------GCCR--------------------ALASVLSA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2464)
SPSL-KELDLQQNDLDDVG
VQLENSGT00900000140813_0_Linker_17_18 (2464, 2716)
LFK--------GLRHPTCK--LT--R-------------------------------
---LGLDQT-TLSDETRQE------LR----------ALV-QEKPRIQFFCMRRKPSVMI
PNEGPDTGEMSNS-----------------TSSLKRQRLRSETESSYVAQADLKPL-DWS
NPPAK------------GSVPRIAQ-QELACLPSPASQED-LHTKPLG-TDDDFWGPTGP
VATEVVDK-ERSLYRPF13553 // ENSGT00900000140813_0_Domain_18 (2716, 2997)
VHFPMAGTYRWP---NMGLCFVVREAV-------TTEIELCVWDK
LLGEINQQHSWMVAGPLLDIKAEPGA-IKAVHLPHFVALQGG--------HIDTSLFQVA
HFKEEGMLLEKPAKVELHHIVLEDPSFSPMGVLLRVIHNALRIFPVTSMVLLYHRIHPEE
V-TFHLYLIPSDGSIRKAVDDEETKFQFVRLHKPPPLTPLYMGSRYAVSGSEE-------
--LEIIPEELELCYRSPGEPQLFSEFYVGHLGSGIRLQMKNKTDETLVWEALVKPGENSGT00900000140813_0_Linker_18_19 (2997, 3052)
DLTP
ATTLVPP-----------------------------APKALPSPLDALGLLPF00619 // ENSGT00900000140813_0_Domain_19 (3052, 3134)
HFVDQYREQ
LVARVTSVEPVLDNLL-GHVLSQEQYEGVLAEDTRPSKMRKLFSFSQSWDRSCKDRLYQA
LKETHPYLIMELWEKGSKKGLLPLSI----------
ENSSSCP00000024653 (view gene)
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------MS------------------
----------SSAGISMDDTFLDK---F
PF02758 // ENSGT00900000140813_0_Domain_0 (209, 323)
MSYLKGLD--Q---YQLEEFKLCLQSPQLM--
--------------PEN--------F--PKI-PWTN-LKAVG-PSNLFCLLSDYLSVPQI
RDVTLCI----FE-NMKLMSRCEEIR-AEMNGEWD-AAWN--------------------
---------------------------------------------------SW-------
---------DCFHPGHLTSHHRCLVSEST----------QALGLQDPGQ-----------
------EEPE-------------------------------VPEEET---AHRRKYRER-
--------MKDK----------------ILLM--WDSIPWPEGHIYLRHMTE--------
----------------------------------------------Q-EHEELRSLLYPN
R---TGAQP----E
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TIILGGRAGVGKTILAMKV-----MLHWA--E---------GSLFQ
H--------RFSY-VFYISCHKLQRMR-------NTTFVG-LLSQ-DW-----PD-SQIP
LTELIN-HS----ERLLFIVDGFEE-SAVSST------LERS-P-----PCSDWYTP-LP
-GDRLILHLLKKELVPQATLLIT-ARDTQIAYLKKLL-P-----NPH--FIFISGFTEED
RGEYF----------------I--RF-FGD---------QDKANAILRWLRQ-NETLFDW
CSAPLVCWTVCSSLMWQMTRNP-SF-E--MS-TQTVTSIYAYFFSQLFTTAE-VNLSDQS
------------WPEQWRALCSLA---AEGM-WC-LN----SVFAKE-------------
--VLEHCHLEAS-FIDYLLRL----NILQKVID-CE-GCIAFTHQSFQMFFGAMFYLLR-
GTEGS--------------V-GG--LPKFDEIK-VFINGVFTD-TNLYWHQIALFL----
----FGLFKRDLARELEVSLHCE---------------M--SPR-VIY-ALLDWTEALAV
DD-----------------------------------PAYIHFEFLQFFQCLCEMQD-EN
LVQKVLNR---------------------L-------------------LEADIDIDKNE
GL---HAS--S----LCLK-HCPK-LTR-LRLS----------------ISS-----PIP
-QV------------------------------------------------QTRVV-DFK
MHQWQDIC-S-VFCNGHMRELDLSNSKLNTL---------YMKKLC--HELGS-S-----
RC--K-LQKLT--C-KSMSPV--RI---L-K-ELVLVL----------------HC
PF13516 // ENSGT00900000140813_0_Domain_6 (1617, 1633)
NPNL
THLD-LSSN-NLGIT-VSMMIFRTLRHPSCS----L------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------K----------------------------------------------
------------------------LEKCSLP-----------------------------
----------------------------VASCRDLAL-----------------------
--------------------------------LLISTH
PF13516 // ENSGT00900000140813_0_Domain_11 (2019, 2054)
RMTR---LC-LGFNQ-IQD---
-------AGVKLLCASL----AHPKC----------------------------------
--------------------------LLE-RLV---------------------------
-------------------------------LWFCQLSAASCRYLSNA-LL
PF13516 // ENSGT00900000140813_0_Domain_13 (2212, 2237)
QNKRLTHLN
L-RKNNLGDEGVK-FLENSGT00900000140813_0_Linker_13_14 (2237, 2247)
CEPLRCP--DPF13516 // ENSGT00900000140813_0_Domain_14 (2247, 2348)
CN----L---------------------------
-----------------------QDLNL--SDC-----SFSVEGCREL------------
-------ENSGT00900000140813_0_Linker_14_15 (2348, 2354)
ADAL-RPF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
HNQSLKILDLGENDVRDNGAKE-LCEVLK-HV-NY-ALKTLG--LE-
--K-CNLTHNCCQ--------------------HLCSVLR
PF13516 // ENSGT00900000140813_0_Domain_17 (2441, 2461)
NNRSL-VHLNLLGNDLGPDGVNRLWT--------SLKKSTCQ--LQ--K-------------------------------
---LG-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMLUP00000019706 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------LSLGIKEKYHNQ-
--------RRES----------------SCPTQSW-------RNE---------YLHQKF
TQLLLLHRSQPRGYDF---L------HWRKRYHEV--------VDEQGQLMEIGDLFGPG
LS--TQEEP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
PTVVVHGVAGIGKSTLARQV-----RRAWE--E---------GWLFK
D--------RFKH-VFYFNCRELAQSE-------TMSLAE-LITK-DW-----AA-PDAP
IGQILS-QP----EQLLFILDNLDE-PKWDLKEQ------SS-E-----LCPHWSQQ-QQ
-VHTLLGSLLKKTLLPTASLLIT-ARITALGKLIPSL-K-----HPH--WVEILGFSESA
RKEYF----------------Y--KY-FTD---------ESQAIRAFTLVES-NPALFTM
CLMPLVSWLVCTCLKQQMEQEQ-PL-S--LT-SQTTTALCLHYFFHVLQ--------AQ-
-----------SLGTKLRGFCSLA---AEGT-CQ-GK----TLFSPE-------------
--DFKKHGFYGA-NISTLLNM----GVLQEH---PTPQNYSFIHLCFQEFFAAMSYALG-
D------RVL--------KSGCF--NNI-TIIT-DLTDVYDRY--DLFGEPTICFL----
----FGLLSDQGMREMESIFKCS---------------L--SQE-THS-YLLQLAQEEVN
-----------------------------------YKHPNLEPYCWHLLHCFYEYQD-KR
FLTETMSN---------------------F-------------------YGMRIYVQTHM
EL---LVS--T----FCFK-FCHL-MDT-LQLN----------------DGGK-------
------K-K---------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------P------------
------AQRP--------------------------------------------------
-------------------SGVVLYSWGPFT-----------------------------
----------------------------DSWWQMFFS-----------------------
--------------------------------IFKVPGSLRE---LN-LSGNS-LSC---
-------SAVQSLCDAL----ESPHCRLETL-----------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSOGAP00000019290 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------MAESFSAA
PF02758 // ENSGT00900000140813_0_Domain_0 (203, 325)
FD---LLWYLEKLS--D---EEFHSFKSHLEQ------
---------E--IQQFG--------L--PQI-PLTD-LEV-S-KEVLVNTLRSSYEEQHT
WNMTFSI----LQ-KLGHKDLCEKI---TIRQN---------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------RNKEMHKCL-
--------MRTK----------------FLSQ--WETCRFSEIHYSFFSEVT--------
----------------------------------------------CDIFYILESSYDSS
T---SSKEN----LNVFLVGDRATGKSMITKMA-----VTKWT--V---------GEMWK
----------------------------------------------------------AP
IA
PF05729 // ENSGT00900000140813_0_Domain_2 (783, 930)
DILS-DP----RKVLFILEDLDN-MNVDLNVD------ES-A-----LCSDSRRK-VP
-VSVLLVSLLKRKMVPGCWVLVS-SRPSCEPSIKALM-N-----ERD--CYVTLQLSNEK
KQHYF----------------H--LF-FNN---------QERAVLTFKFVLM-SEILVDL
CEVPLLSWILCTVLNQQINQG-EDI-G--FS-CETPTDIYTHYLASVLASDPEVT-AR--
----------QHHLILLNRLCLLA---LEGL-SH-NT----LRFSRN-------------
--DLVSVGFTSD-DVSVLQAM----NILLRSSN-HQ-VRFEFIHLNIQEFCAALASLMVL
PRRWI-----------------P----SARR-R--K--KEKRE-LYDNFSPVVTSI----
----FGLLNEKRRKILETSLGCQ---------------LPMVES-LRQ-NLLMQMKYLGN
NP-------------------------------------NLMEHHMPLFYCFLENLE-EE
FVKEIMSF---------------------F-------------------SEATIFIQDNK
DL---MVS--L----YCLQ-HCQP-LQK-LKLS----------------VQQ-----VFR
--CETPIS-------------------------------------------MLTSSQIRG
LTFWMHIC-SLFYTKENFRELEICTSDFNRI---------SELLLC--KALRHP------
NC--Q-LQTLRLSY---------VSV-SSNFNDLFMSM----------------ACNQNL
TFLN-VNCI---------------------------------------------------
------------------------------------------------------------
--------------------------------------------PISP-KMFSLLCEA--
LSS-PTCNIQHL-S----------------------------------------------
------------------------LMRCDLQ-----------------------------
----------------------------ASACPEIAS-----------------------
--------------------------------ILISSKKLKK---LT-LSNNP-LKN---
-------DGVKILCDAL----FHPECVLESL-----------------------------
------------------------------------------------------------
------------------------------GLYFCCLTERCVAYIARV-LMFSTTLKHLD
L-SVNCLQNRAVL-ALTFPLSLP--ECH----L---------------------------
-----------------------QELEL--TGC-----FFNNEVCHYI------------
-------ASSLI-SNVHLRRLELGYNNLGDAGVES-LCNALQ-HQ-DC-KLETVG--LE-
--G-CMLTSACCT--------------------TLASVFSSSKTL-KKLNLIENNLGDEG
IMKLIQ--------GLGHPSCV--LE--L-------------------------------
---LGIGVS-DLSAETQSL------LM----------AVR-EKNTKLEFSSSFSTVKEGR
QFNDL-----LG------------------------------------------------
-----------------------------------------------------SGNPPEP
AST-----------------TPNYPWH---SI--FFKLPRKFSSMTEETTVSEQSL----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMLEP00000028379 (view gene)
ENSRNOP00000066377 (view gene)
ENSMUSP00000066841 (view gene)
ENSETEP00000012779 (view gene)
ENSHGLP00100023958 (view gene)
ENSRROP00000033846 (view gene)
ENSPTRP00000039083 (view gene)
ENSCATP00000028404 (view gene)
MGP_CAROLIEiJ_P0077856 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------MAGSSGYG---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLKPLQKLS--D---EEFQRFKELLRK------
---------E--PEKFK--------L--KPI-SWTK-IENSS-KEDLVTLLNTHYPGQ-A
WDMVLSL----FL-QVNREDLSIMAQ-KKKRH----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------KQTKYKKF-
--------MKTT----------------FQRI--WTMETNTHIPDR--------------
---------------------------------N-------YHLIVETQYKALQEIFDSE
-----SE-P----V
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TAIVTGATGEGKTTFLRKA-----MLDWA--S---------GVLLQ
N--------RFQY-VFFFSVFLLNKTT-------ELSLAE-LISS-TL-----PES-SET
VDDILS-DP----KRILFILDGFDY-LKFDLE-L------RT-N-----LCNDWRKR-LP
-TQIVLSSLLQKIMLPGCSLLLE-LGQISVPKIRHLL-K-----YPR--VITMQGFSERS
VEFYC----------------M--SF--FD---------NQRGIEVAENLRN-NE-VLHL
CSNPYLCWMFCSCLKWQFDREE-EG-Y--FK-AKTDAAFFTNFMVSAFKSTY-AHSPSK-
-----------QNRAQLKTLCTLA---VEGM-WK-EL----FVFDSE-------------
--DLRRNGISES-DKAVWLRM----QFLQNHG-----NHTVFYHPTLQSYFAAMFYFLK-
QDKDI-----------------C--VPVIGSIP-QLLGNMYAR-GQTQWLQLGTFL----
----FGLINEQVIALLQPCFGFI---------------Q--PIY-VRQ-EIISYFKCLGQ
QEC----------------------------------NEKL-ERAQNLFSCLRDSQE-ER
FVRQVVDL---------------------F-------------------EEITVDISSSD
VL---SVT--A----YALQ-KSSK-LKK-LHLH----------------IQKR----VFS
-EIYCPDH-----------C-------------------K-T-------RTSI-GKRRNT
AEYWKTLC-GIFC---NLHVLDLDSCQFNEK---------AIQDLC--HSMSPSPTVPLT
AF--K-LQSLLCSF---------MTD-FGD-GSLFHTL----------------LYLPHL
KYLN-LYGT-YLSSD-VIEKLCSALRCSACR----VEEL--------LL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------GKCGIS-----------------------------
----------------------------SKACGIIAT-----------------------
--------------------------------SLINS-KVKH---LS-LVENP-LKN---
-------KGVMSLCEML----KDPSCVLQSL-----------------------------
------------------------------------------------------------
------------------------------MLSYCCLTFIACGHLYEA-LLSNKHLSLLD
L-GSNFLEDTGVN-LLCEALKDP--NCT----L---------------------------
-----------------------KELWL--PGC-----FLTSECCEEI------------
-------SAVL-TCNRNLKTLKLGNNNIQDTGVRQ-LCEALS-HP-NC-NLECLG--LD-
--L-CEFTSDCCK--------------------DLALALTTCKTL-NSLNLDWKTLDHSG
LVVLCE--------ALNHKRCN--LK--M-------------------------------
---LG-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPPYP00000011653 (view gene)
------------------------------------------------------------
------------------------------------------------------------
----------------------R------------LEARP----DLREF------FSL-N
L---RSHTWSTMTSP----QL
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 326)
EWT---LQTLLEQLN--E---DELKSFKSLLWA------
---------L--PLEDV--------L--QKT-PWSE-VEEAD-GKKLAEILVNTSSENWI
RNATVNI----LE-EMNLMELCKMAK--AEM-----------------------------
------------------------------------------------------------
--MEYG---------------------------------QVQETDNPEL-----------
-------EDA-------------------------------EEDSELAKPGVKEGRRNS-
---------MEK----------------QSLV--WKNTFWQGDIDSFHDDVT--------
-----------------------------------------------LRNLRFIPFLNPI
TP--RKLTP----Y
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVLHGPAGVGKTTLAKKC-----MLDWT--D---------CNLSP
---------TVRY-AFYLSCKELSHMG-------PCSFAE-LISK-DW-----PE-LQDD
IPSILA-QA----QRILFVVDGLEE-LKVPPG------ALIQ-D-----ICGDWEKK-KP
-VPVLLGSLLNRYMLPKAALLVT-TRPRALRDLQLLA-Q-----QPI--YIRVEGFLEED
RRAYF----------------L--RH-FGD---------EDQAMRAFELMRS-NAALFQL
GSAPAVCWIVCTTLKLQMEKGE-DP-V--PT-CLTRTGLFLRFLCSRFPQG-------AQ
------------LQGALRTLSFLA---AQGL-WA-QM----SVFHQE-------------
--DLERLGVQES-DLRLFLDG----DILRQDRV-SK-GCYSFIHLSVQQFFAGLFYALE-
KE------EE--------DRDGH--AWDIGDIQ-KLLSGEERV-RNPDLIQVGHFL----
----FGLANEKRANELEAAFGCR---------------I--SPD-IKQ-ELLRCKAHLHA
NK-----------------------------------P-LSMTDLKEVLVCLYESQE-EE
LAKVVVAP---------------------F-------------------KEISIHLTNTS
EV---MHC--S----FSLK-HCQD-LQK-LSLQ----------------VAKG----VFL
-ENY-MDS----EL--------------------DIEFERC-TYL--TIPNWARQD-LRS
LCLWTDFC-SLFSSNSNLKFLEVKQSFLSDS---------SVRILC--DHITR-S-----
TC--H-LQKVV--I-KNVTPD--TA----YR-DFCLAF----------------IGKKTL
THLT-LEGH-IEWERTMMLMLCDLLRNHKCN----LQYL--------RLG---------G
H------C----------------------------------------------------
---------AT---PE-QWA---EFFYVLKA-NQSLKHLHISANVLLD-EGAKLLYKT--
MTH-PEHFLQML-S----------------------------------------------
------------------------LENCRLT-----------------------------
----------------------------EASCKDLAA-----------------------
--------------------------------VLVVSKKLTH---LC-LAKNL-IGD---
-------TGVKFLCEGL----SYPEC----------------------------------
--------------------------KLQ-TLV---------------------------
-------------------------------LQQCSITKLGCRYISEA-LQEACSLTNLD
L-SLNQIA-RGLW-ILCQALENP--NCN----L---------------------------
-----------------------KHLR---------------------------------
---------------------------------------------------------LK-
--S-YGTSLEIKK--------------------LLEEVKEKNPKLTIDCNAS------GA
---------------MAPRCCD--FF----------------------------------
----C-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCJAP00000034052 (view gene)
MGP_CAROLIEiJ_P0077858 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------------------M
------------SPSENDS-----------------R----A------------------
--ILKDN-----GS-EEVEQT----SGFQAGSE-QE------------------------
---------------------Q---TLDNGGYL----QDYKTH-MIAKFDTSVDLY----
----YDSPEVKLLSDAFKPYQKTFQP
PF05729 // ENSGT00900000140813_0_Domain_3 (1227, 1408)
HTIILHGRPGVGK--SVL-ARS-IVLDWAQDKLF
------------------------------------------------------------
-------------------------------------------------QKMSY------
---VFSLD--L----FSVD-NTHE-LCP-VVAL----------------Q----------
------------------------------------------------------------
-------------------------ETQCKP---------LL------------------
----------------MEWWG--N----FCS-VLGSHWN---------LKELDLGLRQSE
LYLN-LGNT-PMKDD-DMKLACEALKHPKCS----LETL--------RLD---------S
CELTLIGY----------------------------------------------------
---------EM-------------ISTLLIS-SSSLKCLSLAKNTVGV-KSMISLGNA--
LSS-STCPLQKL-I----------------------------------------------
------------------------LDSYGLT-----------------------------
----------------------------HASCHLLVS-----------------------
--------------------------------SLF
PF13516 // ENSGT00900000140813_0_Domain_11 (2016, 2054)
SNQNLTH---LC-LSNNS-LGT---
-------EGVQLLCQFL----RNPEC----------------------------------
--------------------------ALQ-RL----------------------------
------------------------------ILNHCNIVDDAYSFLALR-LA
PF13516 // ENSGT00900000140813_0_Domain_13 (2212, 2237)
NNTKLTHLS
L-TMNPVGDDAMK-LLCEALKEP--TCY----L---------------------------
-----------------------XELEL--VDC-----QLTQNCCEDL------------
-------ACMI-TTT
PF13516 // ENSGT00900000140813_0_Domain_15 (2356, 2378)
KHLKSLDLGNNTLGDKGVIT-LCEGLK-QS-IS-SLRRLG--LG-
--A-CRLTSKCCE--------------------ALSLALSC
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2464)
NPHL-NSLNLVKNDFSTSG
KLKLCS--------AFQSPVSN--L-----------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSSBOP00000006129 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---ESFFSDFG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 322)
-LLWYLKELR--K---EEFWKFKELLKQ------
---------E--PLKFE--------L--KPI-PWTE-LKKAS-KEDVAKLLDTHYPGKRA
WELTLNL----FL-QISRKDLWTKAQ-EEM-S----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------NLNPYRKQ-
--------IKEK----------------FQLI--WEKETCLHVPER--------------
---------------------------------F-------YKESMRHECTVLNKAYTVA
------AAP----V
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVVEGPDGIGKTTLLRKV-----MLDWA--E---------GNLWK
D--------RFTF-VFFLTVYEMNSIT-------ETSLPE-ILSR-DW-----PES-SEK
IEDIFS-QP----ERILLIMDGFEQ-LKLDLD-H------QA-D-----VCDDWRQR-QP
-LPIILSSLLQKKMLPECSLLLA-LGQMGMRKNYFVL-E-----HPK--LIKLSGFTEPE
RKMYF----------------S--CF-FGE---------KNKALKVFNFVRE-IGPLFIL
CHNPFICWLVCTCMKYRLERGE-DL-E--IN-SQNTTSLHASFITNVFQAGS-QSSPPK-
-----------LNRARLRNLSALA---AEGL-WT-HT----FVFGRE-------------
--DLRRNGLSES-EGLMWVSM----KLLQRSG-----DCFTFIHVCIQEFCAAMFYLLR-
GPKDS-----------------P--NPAIGSIT-QLVRASVAQ-PQNFLTQMGVFV----
----FGISTEEIASMLEMSFGFP---------------L--SKD-LQR-EIAQCLKSFSQ
CE-----------------------------------ADREAIGFQELFSCLFETQE-KE
FVTQVMNF---------------------F-------------------EEVYIYIANIE
HL---VIA--S----FCVK-HCRN-LTT-LRLC----------------MEN-----IFP
-DDSGCVS-----------E-------------------R-N-------EK---------
LMYWRELC-SMFSNNKNFQTLDMENTSFDDA---------SLAILC--KALAQP------
VC--K-LQKLMLTF---------MSD-FGHDTELFRAV----------------LHNPHL
KILS-LYGT-SLCHT-KVKHLCETLRHPMCR----IEEL--------ML-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------MYCCLT-----------------------------
----------------------------CVSCEAISE-----------------------
--------------------------------G---------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------------------LARSKSLSLLD
L-GSNHLEDSAVA-LLCEALKHP--DCS----I---------------------------
-----------------------RELWL--MAC-----FLTSDSCKDI------------
-------ADVL-ICNEKLKILKLGHNEIGDAGVRQ-LCEALK-HP-NC-KLECLG--LQ-
--T-CRITHACCE--------------------DLAAALVASKTL-RSLNLDWITLDPEG
VVLLCA--------ALSHPDCA--LQ--M-------------------------------
---LGLQKS-AFDEESQKM------LM----------ALE-EKCPHLSISHEPWIEE---
--EYR-----IR------------------------------------------------
--------------------------------------------------GVIP------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPANP00000023613 (view gene)
ENSRBIP00000006660 (view gene)
ENSPTRP00000005475 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-MDQPEAPCSSTGPR--LAVAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLAALEELS--Q---EQLKRFRHKLRD------
------------VGPDG-----------RSI-PWGR-LERAD-AVDLAEQLAQFYGPEPA
LEVARKT----LK-RADARDVAAQLQ-EQRLQR---------L---------GLG-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------SG-TLLSVSEYKKKYREH-
--------VLQL----------------HARV--KERNARS------------VKITKRF
TKLLIAPESAAPEEE--ALGPAE-------EPEP--------GRARRSDTHTFNRLFRRD
E---EGRRP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYQ
G--------QVDF-AFFLPCGELLERPG------TRSLAD-LILD-QC-----PD-RGAP
VPQMLA-QP----QRLLFILDGADE-LPALGGP--------EAA-----PCTDPFEA-AS
-GARVLGGLLSKALLPTALLLVT-TRAAAPGRLQGRL-C-----SPQ--CAEVRGFSDKD
KKKYF----------------Y--KF-FRD---------ERRAERAYRFVKE-NETLFAL
CFVPFVCWIVCTVLRQQLELGR-DL-S--RT-SKTTTSVYLLFITSVLSSAPVADGP---
-----------RLQGDLRNLCRLA---REGVLGR--R----AQFAEK-------------
--ELEQLELRGSKVQTLFLSKKELPGVLET-----E-VTYQFIDQSFQEFLAALSYLLE-
DGGVP-------------------R-TAAGGVG-TLLRGDTQP--HSHLVLTTRFL----
----FGLLSAERMRDIQRHFGC-----------------MVSER-VKQ-EALRWVQGQGQ
GCPGVAPEVTEGAKGLEDTEEPEEE-----------EEGEEPNYPLELLYCLYETQE-DA
FVRQALCG---------------------L-------------------PELALQRVRFC
RM-DVAVL--S----YCVR-CCPA-GQA-LRLI------------------SC----RLV
------AA----------------------------------------QEK--K---KKS
----------------------------------------LGKR----------------
------LQAS---L--------------GGS------S-----------------SRGTT
KQ---LPAS-------LLHPLFQAMTDPLCH----LSSL--------TLS---------H
C-----------------------------------------------------------
--------KLP---DA-VCR---DLSEALRA-APALTELGLLHNRLS-EAGLRMLTEG--
-LAWPQCRVQTV------------------------------------------------
----------------------------RVQ-----------------------------
---------------LPDP---------QRGLQYL-------------------------
------------------------------VSVLRQSPALTT---LD-LSGC-QLPAPMV
--T--------YLCAVL----QHQGCGLQTLSL--ASVELSEQ-----------------
----------SLQELQAVKRAKP-------DLV----------------------I--TH
PALD-------------------GHPQPP-KELISTF-----------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCHOP00000001173 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------------INPYKKH-
--------MKEK----------------FRLI-------WKNEAC---------------
----------------LQVP------E-----CFYKE-------VTNNEFEELHDAYNA-
EE--FRQRS----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
LTVAMQGPRGVGKTTFLRRV-----MLEWA--E---------GNFCK
D--------RFTF-VFFLDARGLNHVK-------EASLVE-LISR-DW-----PE-CSEP
IEDIFS-QP----QKILFIIDGFEE-LKFDLELY-------T-N-----LCHDWTQQ-QP
-MPIVLSSLLQKKMLPESSLLIA-LGTVGMQKNHFLL-K-----HPK--YIRLVGLSEHE
RELYF----------------S--HF-FCE---------KKKVLQALSFVRD-NSPLFSI
CQSPLLCWLICTCMN-QLERGE-DF-E--IS-WQTTTSLYASFFTNVFNRGE-XXXXXXX
XXXXXXXXXXX----XXX-XXXXX---XXXX-XX-XXXXXXXXXXXX-------------
--XXXXXXXX-XXXXXXXXXX----XXXXXX-X-XX-X-XX-XXXXXXXXXXXXXXXXX-
XXXXX----------------XX--XXXXX-------XX-XXX--XXXXXXXXXXX----
----XXXXXXXXXXXXXXXXXXX---------------X--XXX----------------
XXX-----X-----XXXXXX-X---------------XXXXXXXXXXXXXXXXXXXX-XX
XX-XXXXX---------------------X-------------------XXXXX-XXXXX
XX-X-XXX--X--------XXXXX-XXX-XXXX----------------XX---------
----XXXX----XX---------------------------XXXXXX----XXXXH-NER
LGLWRDLC-SVFSTSKDLQVLELDNCSLDGA---------SLAILC--KALAQ-P-----
TC--T-LQRL--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPTRP00000093283 (view gene)
ENSMEUP00000011516 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------S
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
LRCR---LVRYLEELT--N---EEFKKFKLYLED------
---------T--SPREG--------A--LRI-PRGQ-VKKAD-STDMAELMVHHYGEWES
WEVALMI----WE-KMGLRELWEQAK-RESPQIXX-----XXXXXX------XXXXXXX-
------------------------------------------------------------
------------------------------------------XXXXXXX-----------
XXXXXX-XXX-------------------------------XXXXXXXXXXYKEKYRDQ-
--------VRKK----------------FQVL--VERNARLGEWV---------YLSHHY
TRLLLVSGHASPGSREQELL------ASGM---GISG-----R-SVEPI--ELQALFDAD
KG----EAV----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
PTVVLLGIAGIGKTTLARKV-----LLDWA--L---------GTFYP
G--------QFDY-AFYVNCRTVDVAAQ-------RTVAELLLSW-LE-GTGPAS-GAAP
VSSMLA-QP----ERLLFVLDGFDE-SGHWGPAW------VP-----------REEK-QR
-VDVHLRNLLGKRLLPESSLLIT-TRPMALEKLQQLL-E-----QPR--VARVLGFRALE
RKAYF----------------Y--RY-FPD---------PGQAGRAWATVQA-NEVLFAL
CFVPLVCWTVCTGLRQQMERGE-AL-A--QT-GRTTTAVYTAFLSSLLHPDRPT------
-----------QESANLWGLCSLA---ATGQ-QE-QR----LWFREQ-------------
--DLVRHGLDMATVS-TFLHM----DQFQDEAK-CE-AIYSFFHSTFQEFFAALFLVLK-
QLGGPS----------------------------VVLAENCRL-AEADLPLTIRFL----
----FGLSSRERAGTLEKMVGGP-----------------VPQR-SQR-ILLKWIRAQAA
Q-------------------------------------RPRQTKTLELLYCLFETQE-PE
FVRSAMAC---------------------F-------------------REVKVRVCT-R
-M-DGLVV--A----FCXX-XXXX-XXX-XXXXXXXXXXXX--XXXXXXXXXX----XXX
XXXXXXXXXX---XXX--XX------------------XXXXX-----------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------XXXX-------X--------XXX---------X
XXXXXXXXXXXXXXXXXXXXXX----XXXXXXXXXXX--XXXXXXX--X-XXXXXXXXXX
XXXXXXXXXXX--XXXXXXX---XXXXXXXX--------------------------X--
XXX-XXXXXXXX-X----------------------------------------------
------------------------XXXXXXX-----------------------------
----------------------------XXXXXXXXXXXXX-------------------
------------------------------XXXXXXXXXXXX---XXXXXXXX-XXXX--
-------XXXXXXXXXX----XXXXX----------------------------------
--------------------------XXX-XXX---------------------------
-------------------------------XXXXXXXXXXXXX------------XXXX
X-XXXXXXXXXXX-----XX----------------------------------------
---------------------------X--XXX-----XXXXXXXXXX------------
-------XXXX-XXXXXXXXXXWWQHLSTFGSLSL-QTLGEF-NN-DLGVLR-VL--GG-
----EV---LEQQSWWHL------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSRBIP00000014522 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MAMTKA-RNPREA---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWALSDLE--E---NDFKKLKFYLRD------
---------M--TLSE--------------------------------ELLISKYGEKEA
VTVVLKG----LR-VMNLLELVDQLS-HIC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LH--DYREIYREH-
--------VHC-----------------IE------EW---QEA----------GVNGRY
SQVLLVA-KSSSESPES-AH------PI-------PE-----Q-ELDSV--MVEALFDSG
EK--PSLAP----S
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
LVVLQGSAGTGKTTLARKM-----VLDWA--T---------GSLYP
G--------RFDY-VFYVSCKEVVLLLE-------SKLEQ-LLFW-CC-GD-----NQAP
VTEILR-QP----EWLLFILDGFDE-LQRPFEEK------LK-K-----RG-----L-SP
-KESLLHLLIRRHTLPTCSLLIT-TRPLALRNLEPLL-I-----QAR--HVHILGFSEEE
RARYF----------------S--SY-FTD---------EKKADSAFDIVQK-NDILYKA
CQVPGICWVVCSWLKGQMERGK-AV-------------------STFLPPVD-DGGCSEF
----------S-RHRVLRSLCSLA---AEGI-QN-QR----FLFEEA-------------
--ELRKHNLDGPRLA-AFLSS----NDYQLGLA-IK-KFYSFRHISFQDFFHAMSYLVK-
EDQSRL----------------G--KESRRELQ-RLLEAKEQE-GNDEMTVTMQFL----
----LDISKKDSFLNLELKFCCR-----------------MSPC-LAQ-DL-KHFKEQM-
E-------------------------------------SMKHNRTWDLEFSLYEAKI-KN
LVKGIQMK---------------------D-------------------VSFKIKHSN-E
KK-SQSQN--S----FSVK-SSLS-HGP-KEEQ------------------KR----PSV
---RGQKEGK---DNI--A-------------------GTHKE-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------AST----------------GKG---------R
-GTGE-TDEEVRS-PKN--TYI--------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSAMEP00000003008 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 940)
VVLQGCAGIGKTAVVHKF-----MFDWA--A---------AMVSR
G--------RFDY-LIYVTCRERSHIA-------NLSAAD-LIIN-TF-----QD-IDGP
ILDIILVYP----EKLLFILDGFPE-LQYLVGNQ------EE-D-----LSANPQEK-KP
-VETLLCSFVRKKLFPESSLLIT-ARPTTMKQLHSLL-K-----QPI--QAEIVWFTEAE
KRAYF----------------L--SQ-FSG---------ADAAMGVFYELRQ-NESLDIM
SFLPIISWMIC----PQGDGDR-TL-M--RS-LQTMTDVYLFYFSECLKTLTGIS-VW--
-----------KGQSCLWGLCSLA---AEGL-RN-HQ----VLFEVS-------------
--DLRRHGIGVYDINCTFLNH-----FLKKTEK-SV-NVYTFLHFSFQEFLTAVFYTLK-
NDSSW---------------------MFFDQVGKTWQEIFQQY-GKGFSSLTIQFL----
----FGLLHKGKGKAVETTFGRR---------------V--SP------ELLKWTEKEIK
DK-----------------------------------SSRLQIEPTDLFHCLYEIQE-EM
FLKSVLSA---------------------C-------------------QSTSGVH----
------------------------------------------------------------
--RH----------------------------KP--------------------------
---------PCFQ-----------------------------ALHC--TAQI--------
CS--K-IKK---------------------------------------------------
KDIN-EPRD-ISENA-GHKYT-----NSVCV----------------------------C
VH------------YI----SKTLSTSQWLTDLEFSETKLEASALKLLCEDLKDPNCK-L
QKLNDN------NIEY-ICGFIYFFLTVLIC-NP
PF00560 // ENSGT00900000140813_0_Domain_8 (1775, 1805)
NLTELDLSENPLGD-TGVKYLCEG--
LRH-SNCKVEKL-D----------------------------------------------
------------------------LSTCYLT-----------------------------
----------------------------DASCMELSS-----------------------
--------------------------------FLQVSQTLKE---LF-VFASA-LGD---
-------TGVQHLCEGL----RHAKGILENL-----------------------------
------------------------------------------------------------
------------------------------VLSECSLSAACCESLAQV-LSSTRSLTRLL
L-INNKIEDLGLK-LLCEGLKQP--DCQ----L---------------------------
-----------------------KDLAL--WNC-----HLTGECCQDL------------
-------CNAL-YTNEYLRDLDLSDNALGNGGMQV-LCEGLK-HP-SC-KLHTLW--LA-
--E-CHLTDACCG--------------------ALASVLNRNE
PF00560 // ENSGT00900000140813_0_Domain_17 (2444, 2475)
NL-TLLDLSGNDLKDFG
VQMLCD--------SLIQPICK--L-----------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMICP00000005760 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-MDGPEASCPSMGPR--AAVAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLAALEDLS--Q---EQLKRFRHKLRD------
------------SLADG-----------RSI-PWGR-LELAD-AVDLAEQLTNFYGPEPA
LDVARKT----LK-RADVRDVAARLK-EQRLQR---------L---------GRS-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------SS-ALLSLSEYKKKYREH-
--------VLRQ----------------HAKV--KERNARS------------VKISKRF
TKLLIAAESGALEEE--ALGPAE-------APEP--------QRARRSDTHTFSRLYRRD
K---EGQRP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYQ
S--------QVDF-AFFMPCGELLERPG------ARSLAD-LILD-QC-----PD-RGAP
VQQMLG-QP----QRLLFILDGADE-LPAPDAP--------EAA-----PCTDPFEA-TS
-GARVLCGLLNKALLPTARLLVT-ARASAPGRLQGRL-C-----SPQ--CAEVCGFSDKD
KKKYF----------------Y--KF-FQD---------EWRAERAYRFVKE-NETLFAL
CFVPFVCWIVCTVLRQQLELGR-DL-S--RT-SKTTTAMYLLFIASVLSSAPAADRP---
-----------RVQGELRKLCRLA---REGVLGH--R----AQFAEK-------------
--DLERLELRGSKVQTLFLSKKELPGVLET-----E-VTYQFTDQSFQEFFAALSYLLE-
DEGSP-------------------G-GEAGGLG-TLLRGDAQL--RHYLALTTRFL----
----FGLLSAERMRDIEHHFGC-----------------VVSER-VKQ-HALRWVQGQGC
PR-----ATPEGTEVLEDKEEPEE-E---------E-EEEELNYPLELLYCLYESQE-DA
FVRQALGS---------------------F-------------------PELALERVSFS
RM-DIAVL--S----YCVR-CCPT-GQA-LRLV------------------SC----RLA
------AA----------------------------------------QK-R-K---KKS
----------------------------------------LVKR----------------
------LQGH---L--------------GSS------S----------------SSRATT
KQ---LPPS-------SLCPLFEAMTDQQCG----LTSL--------TFS---------H
C-----------------------------------------------------------
--------KLP---DT-VCR---DLSEALRA-APALTELSLLHCRLS-EVGLRLLSEG--
-LAWPQCRVQTL------------------------------------------------
----------------------------RLQ-----------------------------
---------------LPGP---------QGALQYL-------------------------
------------------------------VAMLRQSPALTT---LD-LSGC-QLPGSMV
--T--------YLCAVL----QHPACSLQTLSL--TSVELSEG-----------------
----------SVQELQAVKTEKP-------DLV----------------------I--MH
PALD-------------------SRPEPPAGLSRAL------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSRROP00000035661 (view gene)
ENSCANP00000035667 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MAMTKA-RNPREA---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWALSDLE--E---NDFKKLKFYLRD------
---------M--TLSEG--------Q--PPL-ARGE-LEGLI-PVDLAELLISKYGEKEA
VTVVLKG----LR-VRNLLELVDQLS-HVC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LH--DYREIYREH-
--------VRC-----------------IE------EW---QEA----------GVNGRY
NQVLLVA-KSSSESPESSAH------PI-------PE-----Q-ELDSV--MVEALFDSG
EK--PSLAP----SL----
PF05729 // ENSGT00900000140813_0_Domain_2 (680, 940)
GSAGTGKTTLTRKM-----VLDWA--T---------GSLYP
G--------RFDY-VFYVSCKEGVLLLE-------SKLEP-LLFW-CC-GD-----NQAP
VTEILR-QP----ERLLFILDGFGE-LQRPFEEK------LK-K-----RG-----L-SS
-KESLLHLLIRRHTLPTCSLLIT-TRPLALRNLEPLL-K-----QAG--HVHILGFSEEE
RARYF----------------S--SY-FTD---------EKKADSAFDIVQK-NDILYKA
CQVPGICWVVCSWLKGQMERGK-AV-L--ET-PRNSTDIFMAYVSTFLPPVD-DGGCSEF
----------S-RHRVLRSLCSLA---AEGI-QN-QR----FLFEEA-------------
--ELRKHNLDGPRLA-AFLSS----NDYQLGLA-IK-KFYSFRHISFQDFFHAMSYLVK-
EDQSRL----------------G--KESHREVQ-RLLE----------MTLTMQFL----
----LDISKKDSFLNLELKFCCR-----------------ISPC-LAQ-DL-KHFKEQM-
E-------------------------------------SMKHNRTWDLEFSLYEAKI-KN
LVKGIQMK---------------------D-------------------VSFKIKHSN-E
KK-SQSQN--S----FSVK-SSLS-HGP-KDEQ------------------KR----PSV
---RGQKEGK---DNI--A-------------------GTHKE-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------AST----------------GKG---------R
-GTEE-A-----------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPCOP00000021743 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------LVTLPTGPVHSTLPKNP------------L-------------------FHQ-N
L---SFQSCIRMAEDKLPAFSIDG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 323)
-LQWCLKELD--K---EEFQTFKELLKK------
---------K--SSELT--------K--SSI-PLVE-VVNAD-VEHLAFLLHEYYTGPHA
WEMSINI----FE-EMNLPTLSEKAK-DEMKKHSV-AEIPGDFEPIKT--DQGPSKEEGS
EIS-QAMEIDG----A------------------------TEAE---TEKQEISQAIE--
QG-GATAA---------ETKEQV-------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------RK-----SIS
-KDE--EA----WS--------------------VI-----------PR--RVWGK-GLI
DQQWETFC-RVLCNHRNLQHLDLGTSILTEW---------AMKKLC--AELKS-P-----
TC--T-VQNLV--F-RDTQI---MA---GLQ-HLWLTL----------------I
PF13516 // ENSGT00900000140813_0_Domain_6 (1616, 1640)
SNHHI
KCLN-LGST-LLKDE-DVKAACEALKHPNCF----LESL--------RLD---------L
C------A----------------------------------------------------
---------IS---QA-GYL---MISQVLTTRS
PF13516 // ENSGT00900000140813_0_Domain_8 (1774, 1795)
ISLNYLGLAGNKVTD-EGVKPLFEA--
LKA-PQCTLQKL-T----------------------------------------------
------------------------LESCSLT-----------------------------
----------------------------AVGCHALAS-----------------------
--------------------------------ALD
PF13516 // ENSGT00900000140813_0_Domain_11 (2016, 2054)
SNRSLTH---LC-LSSNH-LGI---
-------EGMYMLCQSI----RLPSC----------------------------------
--------------------------SLR-RLI---------------------------
-------------------------------LNQCGVNVDTCDFLVIA-MMG
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
NTLLTHLS
L-NWNPLQDEGVN-LLCKVMLKT--WCQ----L---------------------------
-----------------------QDLEL--VGC-----HLTAACCTNL------------
-------SSVI-TR
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
SKHLKSLDLASNAVGDSGVAV-LCEGLK-HE-KG-SLRRLG--LQ-
--A-CGLTSNCCE--------------------VLSSTLSC
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2465)
NQSL-TSLNLVQNSFSPEG
MVKLCS--------AFAHPLCS--LQ--V-------------------------------
---IGLWKC-QYPVQVQKL-----------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMLUP00000000546 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------------EKYQNQ-
--------KRES----------------SCPTQSW-------RNE---------YLHQKF
TQLLLIRRSQPRGYDF---L------LRRRRNHEV--------VDEQRHLMEIGHLFGPG
LR--TRKEP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
PTVVVHGVAGIGKSTLARQV-----RRAWE--E---------DWQFR
D--------RFKH-VFYFNCRELAQSK-------TMSLAE-LITK-DW-----AA-PDAP
IGQILS-QP----EQLLFILDNLDE-PKWDLKKQ------SS-A-----LCPHWSQQ-QQ
-VHTLLGSLLKKTLLPTASLLIT-ARITALRKLIPSL-K-----HPR--WVEVLGFSESA
RKEYF----------------F--QY-FID---------ESQAIRAFTSVES-NPALFTM
CLMPLVSWLVCTCLKQQMEQEQ-PL-S--LT-SQTTTALFLHYFFHVLQ--------AQ-
-----------SLGAKLRGLCSLA---AKGT-CQ-RK----TLFSPE-------------
--DFRKYRIDEA-NIPILLNM----GVLQEH---PTLRSYSFLHRCFQEFFAAMSYASG-
D------RKL--------KRGGF--INI-KTIT-DLTDVYDRR--DLFGEPTTCFL----
----FGLLSDQGMRELESIFKRS---------------L--SRK-RHS-YLLQLAQREVD
-----------------------------------CKHPNLEPYCWHLLHCFYENQD-TR
FLTEMMSN---------------------F-------------------YGMRICVQTHM
EL---LVF--T----FCFK-FCHR-MMT-LQLN----------------DGGE-------
------K-K---------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------L------------
------AQSP--------------------------------------------------
-------------------SDVVLYSWGPFT-----------------------------
----------------------------DSWWQMFFS-----------------------
--------------------------------ILKVPG
PF00560 // ENSGT00900000140813_0_Domain_11 (2019, 2057)
SLRE---LD-LSGNY-LSC---
-------SAVKSLCDAL----KSPHCRLETLT----------------------------
------------------------------------------------------------
-------------------------------LVSCGL-----------------------
------------------------------------------------------------
----------------------------------------TS------------------
------------------------------------------------------------
---------GCCQ--------------------DLVSMLSNNIML-SELDLRQNDLGNLG
VRLLSE--------GLRQSNCR--LK--Y-------------------------------
---LWLDGT-QLSAEVTEM------LR----------LLQ-QDKSHLVISY-EWALRRIA
QMDEMDEETTPLQ-----------------R-----------------------------
----------------------------QEPESEPLGD---IPVSSIG-TDDDFWGPTGP
VTPEVGDE-DRGLYRVHFPVAGSYHWP---NTGIYFVVRGPV-------TIEIEFCAWDQ
FLDQIVPQHSWMVAGPLFDIKAEPGA-VEAVYLPHFIALQEE--------HVDISWFQVA
HFKEDGILLEKPTMVEPHYIVLENPSFSPMGVLLRKIHSFLN-IPVISNVLLYHRLHSEE
V-NFHLYLLPSDCTIEKAVDIEKKKFQFVRIHKPPPLTPLHIGSRYIVSGS---------
QKMEIIPEELELCYRSPGKAQLFSEFYVDHLGSGIRLQIKSKNDETVVWEALVKSGDLRP
AAPLVS-----------------------------QALTASPSSPNAPNL
PF00619 // ENSGT00900000140813_0_Domain_19 (3051, 3135)
LHFVDQHRAQ
LVARVTSVDAVLDKLH-GPVLSDEQYERVRAEPTNPDRMRKLFSIIKSCDWACKDQLYRA
LKEIHPHLIEELLEK---------------------
ENSOPRP00000007037 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------------SSQTAG--
HDGGAEGT---------Q--EQXXXXXXXXXXXXX------XXXXXXXX----------X
-----X---X-------------------------------XXXXXXXXXXXXXXXXXX-
--------XXXX----------------XXXX--XXXX-X--------------------
--------------------------------------------XXXXX-XXXXXXXXXX
X---XXXXX----XXXXXXXXXXXXXXXXXXXX-----X-XXX--X---------XXXXX
X--------X-XX-XXXXXXXX---XX-------XXXXXX-XXXX-XX-----XX-XXXX
XXXXXX-XX----XXXXXXXXXXXX-XXXXXXXX------XX-X-----XXXXXXXX-XX
-XXXXXXXXXXQVLLPGCFLVVT-IRNADLAKLQP-L-T-----APL--YLAMSGLSVQR
RIQLL----------------Q--EH-VPN---------EQGKMQVLHSVMD-NHQLFDQ
CQVSTVCLLVCRALELQEATGN-SAGA--PG-CQTLTALYAAFVFHQLAPEG-VLQHCLG
----------RPERAMLLGLCRLA---AESV-WA-KR----SVFHAE-------------
--ELRALGLSDE-ELSAPPLM----SVLLPANRNHAEERYAFLHLSLRDFFAALYYVLE-
GLGSL--------------PAPC--SLQSPGTG-----RSSAD-RRPHPLGMKRFL----
----FGLMSQEVLRALEVLLGCA---------------V--PQGTARQ-ELTSWISLLGL
QAG----------------------------------TL-APESAMESFYCLFETQD-KE
FVLLALDS---------------------F-------------------QDLWLRVSQKM
DL---IVS--S----FCLQ-CCRH-LRK-IRVD----------------VRD-----LFL
-KEEIP-T----WP--------------------RI-----------PQ--WMQCK-TIV
DEWWENFC-SVLATHPQLQQLDLGHSILNEW---------AMKMLC--AKLRH-P-----
TC--K-IQRLX--X-XXXXXX--XX---XXX-XXXXXX----------------X-XXXX
XXXX-XXXX-XXXXX-XXXXXXXXXXXXXXX----XXXX--------XLD---------F
C------G----------------------------------------------------
---------LN---QS-CCL---MISQVLHK-STSLKILGMAGNKVTD-WGLKVLCDA--
LRP-SWCALRRL-T----------------------------------------------
------------------------LEDCGLM-----------------------------
----------------------------ATSCHILAL-----------------------
--------------------------------ALTNNR
PF00560 // ENSGT00900000140813_0_Domain_9 (2019, 2132)
SLTH---LC-LSRNP-LDP---
-------EGLSPLCRAI------PDS----------------------------------
--------------------------SLQ-RLV---------------------------
-------------------------------LNHCGLDIAGCGYLVFA-LRDNRQLTHLS
L-SQNPLGDSGIK-LLYEAMREL--SCH----L---------------------------
-----------------------QDLEV--AGC-----QLTAACCESL------------
-------SHMI-MRSK
PF00560 // ENSGT00900000140813_0_Domain_15 (2357, 2381)
HLKSLDLGSNALGDSGVAA-LCEGLK-QR-SS-ALQRLG--XX-
--X-XXXXXXXXX--------------------XXXXXXXXXXXX-XXXXXXXXXXXXXX
XXXXXX--------XXXXXXXX--XX--X-------------------------------
---XXLCKE-QYPEETRKL------LE----------GVQ-HAKPHVVIDGDWYSHD---
--E---------------------------------------------------------
--------------------------------------------------EDQHWWRS--
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCATP00000001010 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MAESNSDFD---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWYLENLS--D---KEFQSFKKYLAR------
---------K--ILDFK--------L--PQV-PLIN-LRA-T-KEELANLLPISYEGQHI
WNMLYSI----FL-MMRKEDHCMKI---IGRRN---------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------RNQEACKAL-
--------MRRK----------------LMLR--WENHTFGEFHYKFFCNVA--------
----------------------------------------------SDVFYVLQLAYDST
SC--YSAND----LN
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 930)
VFLMGDRASGKTMTINLA-----VMKWI--N---------GEMWQ
N--------MISY-VVHLTSHEINQMT-------DSSLAE-LIAK-DW-----PD-GQAP
IADILS-DP----KKLLFILEDVDN-LRFELNVN------EQ-A-----LCSNSTQK-VP
-IPILLVSLLKRQMAPGCWFLIS-SRPTREENIKMFL-K-----TTD--CYVTLQFSNEK
RELYF----------------H--SF-FND---------RQRASTALMLVHD-DEILTGL
CRVAILCWITCTVLNRQMDKG-HDP-Q--HC-CETPTDLHAHFLADALTSKAGLT-AN--
----------QYHLGLLRRLCLLA---AGGL-FL-ST----LNFSGE-------------
--DLRCVGFTEV-DVSVLQAV----NILLPSST-HK-DRYKFIHLNVQEFCAAIAFLMAV
PNCLI-----------------P----SGSR-G--Y--KEKRE-QYCNFNQVFTFI----
----FGLLNENRRKILETTFGYQ---------------LPMVDS-FRR-YSVEYMKHLGR
DQ-------------------------------------EKLTHHGSLFYCLFENQE-EE
FVKKVVNT---------------------L-------------------KEVTVYLQSNK
DM---MVS--L----YCLE-YCCH-LRT-FKLS----------------VQR-----IFE
--YKEPLV-------------------------------------------RPTASQMKS
LVYWREIC-SLFCTMDTLWELHLFDSDLNDI---------SERILS--KALEHS------
SC--K-LRTLKLSY---------IST-ASGFEDLLKAL----------------ARNRSL
IHLS-LNCT---------------------------------------------------
------------------------------------------------------------
--------------------------------------------SISL-NMFSLLREI--
LDK-STCQIRHL-S----------------------------------------------
------------------------LMKCDLR-----------------------------
----------------------------ASECGEIAS-----------------------
--------------------------------LLVNGGSLRK---LT-LTNNP-LRN---
-------DGVNILCNAL----LHPNCTLTSL-----------------------------
------------------------------------------------------------
------------------------------ALVFCCLTEKCCSSLGRV-LLLGRTLKQLD
L-CVNHLQNYGVL-HVTFPLIFP--TCQ----L---------------------------
-----------------------EELYL--SGC-----FFTNDICQYI------------
-------AIVIA-T
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NETLRSLEIGSNSIEDAGMQL-LCGGLR-HP-DC-MLVNIG--LE-
--E-CMLTGACCE--------------------SLASVLT
PF13516 // ENSGT00900000140813_0_Domain_17 (2441, 2464)
TNKTL-ERLNILQNQLGDEG
VVKLLE--------SLILPECV--LQ--I-------------------------------
---IGLPLN-DVSAETRRM------VM----------TVK-ERKPNLIFLSETWSVKEGR
EIGVI-------------------------------------------------------
-------------------------------------------------------PASQP
GSV-----------------IPNSNLD---YM--FFKFPRMSTAMRMSSTASRQPL----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMICP00000046163 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MNN---------------------------QKKRKDMEPA-----
--------CTWS------------------------------------------LLSE--
-----------------------------------------KR-----------------
----VASSSL-------------------------------ASFSWRELTDYREVFRKH-
--------LKEK----------------YLKI-GVDKCYHHGK--------------HIE
SRLLLVKEHGISEKTPCGIP-------------------------QQDHFIEMEHIFDPD
EE--GSSSS----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 929)
RTVVLQGCAGSGKTAVVHKF-----MFDWA--A---------GMVTP
G--------RFDY-LIYVNCREISHIA-------NLSAAD-LITN-TF-----QD-INGP
ILDIILIYP----EKLLFILDGFPE-LQYPVGDR------EE-D-----LSAHPHER-KP
-VESLLCSFVRKKLFPESSLLIT-ARPTAMKKLHSLL-K-----QPI--QAEILWFTDAE
KKAYF----------------L--SQ-FSG---------ANAAMKVFYGLRE-NEGLDIM
SSLPIVSWMICSVLQSQGDGDR-TL-L--RS-LQTMTDVYLFYFSKCLKTLTGIS-VW--
-----------KGQSCLWGLCSLA---AEGL-QN-HQ----VLFAVS-------------
--DLRRHGIGVYDSNCAFLNH-----FLKKAEG-DI-NVYTFLHFSFQEFLTAVFYALK-
NDNSW---------------------MFFDQVGKTWQEIFQQY-GKGFSSLMIQFL----
----FGLLHKGKGKAVETTFGRK---------------I--SPG-LRE-ELLKWTEREIK
DK-----------------------------------SSRLQIEPVDLFHCLYEIQE-EE
YSKKIIDD---------------------L-------------------QSVILLQPTYT
KM-DILAM--S----FCVK-SSHSHLS----------------V-----SLKCQHLPGF-
-EEEEPAS----------------------AFVP--------------------------
---------PALTLNESCQ-----LHNLPVS---------RLHLLC--QALRNP------
YC--K-IKDLKL---IFCHLT------ASCGRD---------------LSAVFET
PF13516 // ENSGT00900000140813_0_Domain_6 (1616, 1642)
-NQYL
TDLE-FVKN-TLEDS-GMKFLCEGLKQPNCI----LQTLR--------LYRCLI-SPA-S
CG------------AL----AAVLSTNQWLTELEFSETKLEASALKLLCEGLKDPNCK-L
QKLKLCASLLPESSEA-VCK---YLASVLIC-
PF13516 // ENSGT00900000140813_0_Domain_8 (1773, 1796)
NPHLTELDLSENPLGD-RGVKYLCEG--
LRH-SNCKVEKL-D----------------------------------------------
------------------------LSTCYLT-----------------------------
----------------------------DASCVELSS-----------------------
--------------------------------FLQMSQTLKE---LF-VFANA-LGD---
-------TGVQHLCEGL----KHAKGIIENL-----------------------------
------------------------------------------------------------
------------------------------VLSECSLSAACCEPLAQV-LSSTRSLTRLL
L-INNKIEDLGLK-LLCEGLKQP--DCQ----L---------------------------
-----------------------KDLAL--WTC-----HLTGACCQDL------------
-------CSAL-Y
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
TNEHLRVLDLSDNALGDEGMQV-LCEGLK-HP-CC-KLQTLW--LA-
--E-CHLTDACCG--------------------ALASVLNR
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2465)
NENL-ILLDLSGNDLRDFG
VQTLCD--------ALIHPICK--LQTFY-------------------------------
------IDTDHLHEDTFRK------ME----------ALK-MSKPGITW-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
MGP_PahariEiJ_P0019911 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MSLDIQCEQ-LSD-ARWT----------------EL---------
----LPLI-------------------------------QQYEVV------RLDDC-GLT
EVRCKDIS-SAVQA
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1542)
NPALTELSLRTNELGDA---------GVGLVF--QGLQS-P-----
TC--K-IQKLS--L-QNCGLT--EA---GCG-ILPGML----------------RS
PF13516 // ENSGT00900000140813_0_Domain_6 (1617, 1642)
LSTL
RELH-LNDN-PMGDA-GLKLLCEGLQDPQCR----LEKL--------QLE---------Y
C------N----------------------------------------------------
---------LT---AT-SCE---PLASVLRV-KADFKELVLSNNDLHE-AGVRILCQG--
LKD-SACQLESL-K----------------------------------------------
------------------------LENCGIT-----------------------------
----------------------------SANCKDLCD-----------------------
--------------------------------VVASK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
ASLQE---LD-LGSNK-LGN---
-------AGIAALCPGL----LLPSC----------------------------------
--------------------------KLR-TLW---------------------------
-------------------------------LWECDITAEGCKDLCRV-LRAKE
PF13516 // ENSGT00900000140813_0_Domain_13 (2215, 2237)
SLKELS
L-ASNELKDEGAR-LLCESLLEP--GCQ----L---------------------------
-----------------------ESLWV--KTC-----SLTAASCPYF------------
-------CSVL-T
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
KNRSLLELQLSSNPLGDAGVQE-LCKALG-QP-DT-VLRDLW--LG-
--D-CDVTNSGCS--------------------SLANVLLA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2464)
NRSL-RELDLSNNLMGGHG
VLQLLE--------SLKQPSCA--LQ--Q-------------------------------
---LVLYDI-YWTNEVENQ------LR----------ALE-EQRPSLRIIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMAUP00000007100 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------MEETHGYN---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 321)
LTLCLLKLS--D---EDFMEFKDSLRK------
---------V---LGFK--------P--KPI-PRTK-IATAS-RRGLAMLLNKHYPGQ-V
WDIVLSL----SQ-RAKWKKLFDRAE-RLKRD----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------ERRPYKET-
--------MEII----------------FKYI--WDFENYIHVPSH--------------
---------------------------------E-------YNIITEMQHRTLKDIFDLQ
-----LE-S----VT
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 929)
AVILGNKGEGKTTFLRKA-----MLDWA--A---------GDLWQ
N--------RFHY-VFFFSLMSLNNIT-------ELSLAQ-LILS-KL-----SES-SET
LEDIVS-DP----RRILFILEGFDY-LKFDLG-L------QT-N-----LCNDWRKK-AP
-TQIVLSSLLQKVMLQESSLLLE-LGNLSVPKIYPLL-Q-----KPR--EITIGGYTEAS
VKYYC----------------C--SF-MHS---------FKKGSEIFNYLQA-IQLLLTL
CKNPYKRWMVCRTLKCQFKRGE-KL-N--IA-FKPDSVLYTSFMVSSFRAKY-AGYPSR-
-----------QNRNKLMTLCTLA---VEGM-WK-SV----FVFKYE-------------
--DLLRNGISES-EQILWMKM----YFLNTRG-----DCFVFYHPALQLYFAALFYFLR-
QDKDK-----------------H--HPIIGSLP-QLLREIYAH-GQNQWLLTGKFV----
----FGIATEKIAAILEPHFGFI---------------P--CEK-MRQ-EIFKCFRSLSQ
GEC----------------------------------GEKL-ISPQSLFDSLVENQE-EQ
FITQVMDM---------------------F-------------------EEMTVDISSAD
VL---SVA--T----TCLL-KSQS-LKK-LHLH----------------IQQR----VFS
-EIYNPED-----------G-------------------D-L-------E-VFKHEKNNA
IKHWRMLC-RVFQ---NLEVLDLDSCKFNVT---------VIFLLY--LSMSPTHRGPLN
AF--K-LQSFSCSY---------LTN-FGC-GLLFHTL----------------LQLPHL
KSLN-LYGT-NLSND-VVENMCCVLKSPSCR----VEEL--------LL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------GKCDIS-----------------------------
----------------------------SEACGVIAT-----------------------
--------------------------------SLTKC-KVKH---LS-LVENP-LKN---
-------KGVMLVCEIL----KHPSCVLETL-----------------------------
------------------------------------------------------------
------------------------------MLSYCCFTFIACGHLYEA-LLHNKYLSLLD
L-SSNFLEDTGVN-ILCEALKDP--RCN----L---------------------------
-----------------------RELWL--SGC-----YLTSDCCKAI------------
-------SAVL-SCNQNLKTLKLGSNNIQDTGIKQ-LCEALR-NP-GC-KLQCLG--LD-
--M-CEFTTDSCA--------------------DLALALTTCKTL-TVLSLDWMTLGHNG
LELLCE--------ALNHKGCT--LK--V-------------------------------
---L--------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSACAP00000017210 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------MSPDYREKYREH-
--------VRKK----------------YCSP--EDGNPCLMVQI---------PPNEKY
TKMVVAS----GKEKEHEIL------TRPWRHTQLLF----EQ--MP--ATTIDVLFDTE
ND-----AP----Q
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVLFGVAGVGKTLMARKI-----MQDWA--T---------RSQSP
E--------RFDS-VFYLSCRQMNLIVGE------VNLAE-LILN-NH-SE--PR-LGEA
INKLLI-NP----GKCLFIIDGFEE-LNCSIKQE------GD-N-----LSISPYEK-KP
-VELILSSLFCKTLFPESCLMIT-TRPNTLESLEDCL-P-----SPR--YVRHLGFSEEG
KKEYF----------------C--KF-FGE---------EELAAQALSYIKR-NEALLAI
SFVPGVCWIICTMMKEQLDRAEKDL-V--PV-SDTLTGVYALYLSHLFKSFCTGS-----
-----------EQLDLMKTLCSLA---ADGL-WE-KK----PFLKEE-------------
--DIRKE---------LALHG----TLFRKDPD-CE-GSYSFFHPSIQEFLAALFYVSE-
DLSRG-----------------T--ETTKENMN-IFLDACH-E-VRGDLTLTVQFV----
----FGLLNKEIMKYLKEQFEWN-----------------ISPK-IKT-DLIEWFQLDT-
QIE----------------------------------FIRPNYGQLERFYSLYEVKEEEE
FVKHALNH---------------------L-------------------TELTLQDLTFT
QM-DQVVL--C----YCIN-NCLN-LET-LS------------------LYRC----LFL
LEECETEVLQRLPNYP--DQPLQKTEK-PSAI--DLLCQTLKDPDGKIKKVELDGC-QFT
DTFWEDLS-VVLSTSLVLVELDLQMTNLKDS---------GVRLLR--EGLIH-P-----
NC--K-LNKLG--L-WGCDLT--HA----CGDDLFTIL----------------SHSHTL
MELK-LGCNSSLGIQ-GVQLLCEGFKHPNCK----LQSL--------GLD---------E
CGLNAAGCQ-----HFIPVLSI----SQTLRKLTLESNKLGDVGVKLLCEGLKHPQCQ-L
ETLSLATCELS---PA-CCG---DLSSVLSI-SQTLRELELEYNKLED-TGLRLLCEG--
LKQ-PQCKLEKL-G----------------------------------------------
------------------------LAGCGIT-----------------------------
----------------------------AA----SCGDLAC-------------------
------------------------------ALAVKW--TLKH---LD-IEYNK-LGD---
-------SGVKMLCEGL----KHPHC----------------------------------
--------------------------RME-TLN---------------------------
-------------------------------LFTCDLTAACTKDLASI-LCCNQTLAQLF
L-GGNMLGDSGLK-QLCEGLKHS--QCK----I---------------------------
-----------------------KKLCL--CDT-----GITAAGCMEL------------
-------SSVL-STSQALEDIDLWQNTLEDSGLRL-LCEGLK-HP-------------D-
----CK---LEVLSMWGCKFTSASCG-------DLQCVLSTNQNL-KKLQLENNKLGDSG
VLLLCE--------GLKHPNCK--LK--S-------------------------------
---IGLDIE-ELSEDTVRE------LK----------YLK-RMKPRLTIDT---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMFAP00000020036 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------MAESNSTDFD---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWYLENLS--D---KEFQSFKKYLAR------
---------K--ILDFK--------L--PQV-PLIN-LGA-T-KEELANLLPISYEGQHI
WNMLYSI----FL-MMRKEDHCMKI---IGRRN---------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------RNQEACKAL-
--------MRRK----------------ITLR--WENHTFGEFHYKFFCNVA--------
----------------------------------------------SDVFYVLQLAYDST
SC--YSAND----LN
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 930)
VFLMGDRASGKTMTINLA-----VMKWI--N---------GEMWQ
N--------MISY-VVHLTSHEINQMT-------DSSLAE-LIAK-DW-----PD-GQAP
IADILS-DP----KKLLFILEDVDN-LRFELNVN------EH-A-----LCSNSTQK-VP
-IPILLVSLLKRQMAPGCWFLIS-SRPTREENIKMFL-K-----TTD--CYVTLQFSNEK
RELYF----------------H--SF-FND---------RQRASTALMLVHD-DEILTGL
CRVAILCWITCTVLNRQMDKG-HDP-Q--HC-CETPTDLHAHFLADALTSKAGLT-AN--
----------QYHLGLLRRLCLLA---AGGL-FL-ST----LNFSGE-------------
--DLRCVGFTEV-DFSVLQAV----NILLPSST-HK-DRYKFIHLNVQEFCAAIAFLMAV
PNCLI-----------------P----SGSR-G--Y--KEKRE-QYCNFNQVFTFI----
----FGLLNENRRKILETTFGYQ---------------LPMVES-FRR-YSVEYMKHLGR
DQ-------------------------------------QKLTHHGSLFYCLFENQE-EE
FVKKVANT---------------------L-------------------KEVTLYLQSNK
DM---MVS--L----YCLE-YCCH-LRT-FKLS----------------VQR-----IFE
--YKEPLV-------------------------------------------RPTASQMKS
LVYWREIC-SLFCTMDTLWELHLFDSDLNDI---------SERILS--KALEHS------
SC--K-LRTLKLSY---------IST-ASGFEDLLKAL----------------ARNRSL
IHLS-LNCT---------------------------------------------------
------------------------------------------------------------
--------------------------------------------SISL-NMFSLLREI--
LDK-STCQIRHL-S----------------------------------------------
------------------------LMKCDLR-----------------------------
----------------------------ASECGEIAS-----------------------
--------------------------------LLVNGGSLRK---LT-LTNNP-LRN---
-------DGVNILCNAL----LHPNCTLTSL-----------------------------
------------------------------------------------------------
------------------------------ALVFCCLTEQCCSSLGRV-LLLGRTLKQLD
L-CVNHLQNYGVL-HVTFPLIFP--TCQ----L---------------------------
-----------------------EELYL--SGC-----FFTNDICQYI------------
-------AIVIA-T
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NETLRSLEIGSNSIEDAGMQL-LCGGLR-HP-DC-MLVNIG--LE-
--E-CMLTGACCA--------------------SLASVLT
PF13516 // ENSGT00900000140813_0_Domain_17 (2441, 2464)
TNKTL-ERLNILQNQLGDEG
VVKLLE--------SLILPECV--LQ--I-------------------------------
---IGLPLN-DVSAETRRM------VM----------TVK-ERKPNLIFLSETWSVKEGR
EIGVI-------------------------------------------------------
-------------------------------------------------------PASQP
GSV-----------------IPNSNLD---YM--FFKFPRMSTAMRMSSTASRQPL----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCSAP00000016343 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------LVTLSTGPTCSMLPKNP------------L-------------------FPQ-N
L---SSQPCLKMEGDKSLTFS
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 326)
SYG---LQWCLHELD--K---EEFQTFKELLKK------
---------K--SSEST--------T--CSI-PQFE-IENAN-VECLALLLHEYYGASLA
WATSISI----FE-SMNLPTLSEKAR-DDMKRHS-----PEDLEPTTT--GQGPSKEEVP
G-----NSHVVGQESA--AAAE-------------------------TKEQEISQAME--
QE-SATAA---------ETEEQEISQAMEQEGA---------------------------
-----T---A-------------------------------AETEEQECGGDKWDYKSH-
--------VVTK----------------FAVK--EDVR-RGFEKT---------------
------------------------------------A-------DWPEM-QMLAGAFNSD
Q---WGFRP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 922)
HTVVLHGKSGIGKSALARRI-----VLCWA--Q---------GGLYQ
G--------MFSY-VFFLPVREMQQKK-------ESSVAE-FISR-EW-----PD-AQAP
VTEIMS-RP----ERLLFIIDGFDD-LG-SVLNS------DT-K-----LCKDWAEK-QP
-PFILIQSLLRKVLLPESFLMVT-VRDVGIEKLTSEV-V-----SPR--YLFVRGITAEQ
RIHLL----------------L--ER-GIG---------EHQKTQGLCAIMN-DRKLFDQ
CQVPAVGSLICVALQLQDVVGE-SI-A--PF-GQTLTGLYAAFVFHQLTPRG-VARRCLN
----------LEERVVLKRFCRMA---VEGV-WN-MK----SVFDGD-------------
--DLMVHGLGES-ELRTLFHM----NILLRDSH-CE-EFYTFFHLSLQDFCAALYYVLE-
GLEIE--------------PALC--PLFVEKTK-RSMELQQAG-FHIHLLWMKRFL----
----FGLVSEDVRRPLEVLLGCP---------------V--PL-GVKQ-KLLHWVSLLGQ
QPN----------------------------------AA-TPGDTLDAFRCLFETQD-KE
FVRSALNG---------------------F-------------------QEVWLPINQNL
DL---IAS--S----FCLQ-HGPY-LRK-IRVD----------------VKG-----IFP
-RDELAEA----WP--------------------VV-----------PQ--WMRGK-TLM
EDQWEDFC-SVLSTHPHLRQLDLGSSILTER---------AMKTLC--AKLRH-S-----
TC--K-IQTLM--F-RNAQI---AS---GLQ-HLWRTL----------------IANRNL
RSLN-LGGT-HLKEE-DVRMACEALKHPKCL----LESL--------RLD---------R
C------G----------------------------------------------------
---------LT---HA-CYL---KISQILTT-SPSLKSLSLAGNEVTD-QGVTPLSDA--
LRV-SQCALQKL-T----------------------------------------------
------------------------LEDCGIT-----------------------------
----------------------------ATGCQSLAS-----------------------
--------------------------------ALVSNRSLTH---LC-LSNND-LGN---
-------EGVNLLCRSM----RLPHC----------------------------------
--------------------------SLR-RLM---------------------------
-------------------------------LNRCNLDTAGCGFLALA-LMGNAWLTHLS
L-SMNPVEDNGMK-LLCEVMREP--SCH----L---------------------------
-----------------------QDLEL--VKC-----HLTAACCESL------------
-------SCVI-SRSRHLKSLDLTDNALGDGGVTA-LCEGLK-QK-KS-VLTRLG--LK-
--A-CGLTSDCCE--------------------ALSL-ALSCNRHLTSLNLVQNNFSPEG
MMKLCS--------AFACPTSN--LQ--I-------------------------------
---IGLWKW-QYPVQIKKL------LE----------EVQ-LLKPRIVIDGSWHSFD---
--E---------------------------------------------------------
--------------------------------------------------DDRYWWKN--
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCGRP00001015691 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------MEEISGYD---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 321)
LALCLQKLS--Q---ADFMTFKVLLRK------
---------A--PEEFE--------L--NPI-PWTE-IKMAS-RKGLVMLLNKNYPGK-V
WDIALSL----FQ-KVKRKDLFAMTE-ALRRD----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------KGTPYKET-
--------MKTL----------------FNFI--WNFESGIYVPYH--------------
---------------------------------E-------FEIITEMQYSSLQVVFEPQ
-----SQ-P----VT
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 905)
AVILGNKGEGKTTFLRRA-----MLDWA--S---------GDLWQ
N--------RFQY-VFFFSLMSLNDIT-------ELSLAQ-LLLS-KL-----SES-SES
LEDILS-DP----RRILFILEGFDY-LKFDLE-L------RT-N-----LCNDWRKT-VP
-TQIVLSSLLQKVMLPESSLLLE-LGNLSVPKIYPLL-Q-----KPR--DITLGGYTDEA
IKYYC----------------M--SF-MYS---------YTEGSAIFNYLQT-MQPLLTL
CKNPYKRWMVCSTLKCQRKRGE-NM-N--ID-YKPDSALYASFMVNSFRAEY-ANRPSR-
-----------QNRDQLKPLCTLA---VEGM-WK-LV----FVFKPE-------------
--DLYRNGISES-GQIVWLKM----NFLNTRG-----DHFVFYHPALQLYFAALFYFLR-
QDEDR-----------------P--HPVIGSLS-QLLREIYAH-DQNQWLLTGIFV----
----FGIATEKVAAILEPHFGSI---------------P--SEE-MRQ-EILKCFRNLSR
GEC----------------------------------GEKL-RSPQSLFASLVDNQE-EG
FIRQVMDL---------------------F-------------------EEMTIDISSAD
AL---SVA--T----TCLL-KSQR-LKN-LHLH----------------IQHR----VFS
-EKYKPED-----------A-------------------T-R-------NNGMLSHCSNA
IKHWRMLC-RVFQ---NLEVLDLDSCNFNET---------AISLLY--LSMYPSHRGALN
AF--K-LQSFSCSF---------VTN-FGH-GVLFHTL----------------LQLPHL
KSLN-LYGT-NLSND-VVENMCCVLKCSTCR----VEEL--------LL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------GKCNLS-----------------------------
----------------------------SEACGIIAT-----------------------
--------------------------------SLIFC-KVKH---LS-LVENP-LKN---
-------KGVMLLCETL----KRPSCDLETL-----------------------------
------------------------------------------------------------
------------------------------MLSYCALTFIACGHLYEA-LLHNKYLSLLD
L-SSNFLGDTGVN-ILCKALKDP--MCP----L---------------------------
-----------------------QELWL--SGC-----YLTSDCCEEI------------
-------SAVL-T
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
HNQKLKTLKLGNNNIQDTGVRQ-LCEALS-NP-EC-KLQCLG--LD-
--M-CEFTTGSCA--------------------DLALALTTCKTL-TMLNLDRMTFDRDG
LKLLCE--------ALNHKDCN--LK--V-------------------------------
---LGLDKS-AFCEESQKI------LQ----------EVE-KKN-TLDILHYPWIKE---
--ESK-----KR------------------------------------------------
--------------------------------------------------GVRLVWNSEN
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMUSP00000152151 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------M---ASFISDFG---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 323)
LIWYLRELN--K---KEFMKFKDFLIQ------
---------E--ILELK--------L--KQV-SSTK-VKKAS-REDLANLLLKCG-ENQA
WDMTFRI----LQ-KINRKDLTERAT-GAI------------VG--------------NP
N---------------------------------------------------LYRD----
------------------------------------------------------------
----------------------------------------------------------H-
--------LKKK----------------LTHD--CPKKFNVRIQDF--------------
---------------------------------I-------KETFIQNDYDAFENLLISK
GT--ERK-P----H
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
MVFLKGMAGVGKTLMLKNL-----MLAWS--K---------GLVFQ
N--------KFSY-AFYFCCQDVKQLK-------TASLAE-LISR-EW-----PSP-SAP
IEEILS-QP----EKLLFIIDSLEG-MEWDLTKQ------ES-E-----LCDDCMEK-QP
-VSTLLSSLLRRKMLPESSLLLS-TTPETFEKMEDRI-Q-----CTD--VKTATAFDERS
MKIYF----------------H--RL-FQD---------RKRAQEAFSLVRE-NKQLFTI
CQVPLLCWMVATCLKEEIEKGG-DP-V--SL-CRRTTSLYTTHIFSLFIPQS-AQYPSK-
-----------KSQDQLQGLCSLA---AEGM-WT-DT----FVFGKE-------------
--ALRRNGIFDS-DIPTLLDI----GMLGKIRE-FE-NSYIFLHPSVQEVCAAIFYMLK-
RHVEH-----------------P--SQDVKNIE-TVLFMFLKK-VKTQWIFLGCFI----
----FGLLQKSEQEKLGVFFGHR---------------L--SKN-IHH-KLYQCLETLSG
NAE-----------------------------------LQEQIDGMRLFSCLFEMED-EA
FLVKAMNC---------------------M-------------------QQINFVAKNYS
DF---IVA--A----YCLK-HCST-LKK-LSFS----------------TEN-----VLN
--EG--DQ-----------S-----------------------------------YMEEL
LICWNNMC-SVFVRSKDIQELRIKDTNFNEP---------AIRVLY--ESLKYP------
SF--T-LNKLVANN---------VS--FGDNHVLFEL-----------------IQNSSL
QYLD-LSCS-FLSHN-EVKLLCDILNQAECN----IEKL--------MI-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------AHCKLS-----------------------------
----------------------------PDDCKIFGS-----------------------
--------------------------------ILMSSKSLKV---LN-LASNN-LN----
-------QGISSLCKAL----CHPHCTLEYL-----------------------------
------------------------------------------------------------
------------------------------VLSNCSLSEQCWDYLSEV-LR
PF13516 // ENSGT00900000140813_0_Domain_13 (2212, 2233)
QNKTLSHLD
I-SSNDLKDEGLK-ILCRSLILP--YCV----L---------------------------
-----------------------ESLCL--SCC-----GITERGCQDL------------
-------AEVL-K
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
NNQNLKYLHVSYNKLKDTGVML-LCDAIK-HP-NC-HLKDLQ--LE-
--A-CEITDASNE--------------------ELCYAFMQCETL-QTLNLMGNAFEVSR
MVFFPR--------F---------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMNEP00000023610 (view gene)
ENSHGLP00000021761 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------RWCGAPRGSSGCSGPRRQ
LSEEPTARARSLEPR--AAAAREL---LLAALQYLS--Q---DQVKRFLLKLRD------
------------AP------------------------------------LTHFYGPEPA
LDVTRRT----LK-RADVHDVAARLK-AQRLQPP--------S---------CRA-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------PT-SASLPKEFQKKYRER-
--------VLQQ----------------HAEV--KE----------------------RF
TKLLIAPESAAPGEKEVALGPAE-------KPKP--------GRAWRSDTRTFSRLLSSD
A---EGPRP----P
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 890)
TVLLQGPAGIGKTMAAKKI-----LYDWA--A---------EKLYR
G--------QVDF-AFFLQCRELLEWPG------ARSLAD-LILD-QC-----PD-RGAP
LSRMLA-QP----QRLLFILDGA---------------------------------A-AS
-GARVLGGLLRKALLPSARLLVT-TRATASGKLHFGL-C-----APK--CAAFPPETRRN
T----------------------------------------STSSSGRSAGR-TETLFAL
CFVPAVCWIVCTVLRRQLELGQ-DL-S--RT-SKTTTSVYLVFLASALSSV-GAEGP---
-----------GVRAKLPKLRRLA---REGVLGH--K----AQFSEG-------------
--DLERLELRGSVVQH-FLIKKELPSVLET-----E-VIYQFINQSFQEFFAALSYLLE-
DVGAP-------------------G-VSTGGLE-TLLCLGSEG--RSHLMLTTRFF----
----FGLLNMERVQDVERHFGC-----------------VVPAH-VKQ-DALRWVQGQSH
LR--VASEGTDGTKGLKDTEEPEEK-----------EEEEEPNFPLELLYCLYETQD-EA
FMYQALSS---------------------L-------------------PELMLERVCFH
RM-DLDVL--S----YCMR-CCPA-GQVL-WLV------------------SC----SLM
------A-----------------------------------------GQEKKR---KRS
----------------------------------------LVKR----------------
------LK-G---------------------------------------------SQAAM
KQ---LRVS-------PLHPLFG-------------------------MS---------R
C-----------------------------------------------------------
--------KLP---DR-IYR---DLSMALRA-APALTELGLLHCRLR-EAGLRVLNEG--
-LAWPQCRVQTL------------------------------------------------
----------------------------RVL-----------------------------
---------------LLKP---------LEAFQCL-------------------------
------------------------------VALPWQSPTLTT---PD-LSGC-YLPDPMV
--TY--------LCAAR----QHPGCSLQTLR----------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCANP00000035810 (view gene)
ENSRROP00000004574 (view gene)
ENSRNOP00000040262 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------MKTEDEE-MED------------
------------------------------------------------------------
----------------------QL------------------------------------
-------SKE-------------------------------EIVSEDKEF----DYRTI-
--------IKEN----------------FFII--WDKTSLLGESATLNCVTT--------
----------------------------------------------HKDQKLLEHIFDVN
IQ--TSKSP----Q
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVLYGAAGIGKTTLLKKA-----VLEWA--D---------GNLYQ
---------QFTH-VFYLSGREISQEK-------EKNFAQ-LISK-HW-----PS-SEGP
IEQILC-KP----SSLLFILDSFDE-LDFSFEEP------EF-A-----LCRDWTQK-HP
-VSFLISSLLRKVMLPESYLLVT-TRTTAWKRLVPLL-Q-----KPH--RVKLPGLSRSS
RIDYI----------------H--HL-LKD---------KTWAMNAVCSIRM-NAKLFNM
CHVCHVCQMICTFLKEQVEKGG-NV-E--MT-CQTSTALFTYYICSLFPRI-AGSSVTLP
------------NETLLRSLCQAA---VEGI-WT-MK----HVLYQQ-------------
--NLRKHELTRE-DLSLFLEA----NVLQQDTE-CE-NCYTFTHLQVQEYFAALFYLLR-
ENPEE--------------KDHP--LEPFENVH-LLLESNHFH--DPHLEQMKCFL----
----FGLLNKDRVQQLEETFNLT---------------I--SRA-VRE-ELLVCLEALEK
DD-----------------------------------SPLSQLRIQDLLHCIYETQD-EE
FITQALTY---------------------F-------------------QKIMVKVDEES
KL---LIY--S----YCLK-HCHS-LQT-VRLT----------------ARA-----DLR
-----NML------DP-----TA--E------------------------ICLEDAAVRI
IHYWQDLF-SVLHTNESLLEMDLYESRLDES---------LMKILN--EEFSHP------
KC--N-LQKVMFRA---------VCF-LNSCYD-FSFL----------------AF
PF13516 // ENSGT00900000140813_0_Domain_6 (1617, 1642)
NNTV
THLD-LKDT-DLGDN-GVKTLCEALKYQGCK----LRVL--------RLE---------S
CD----------------------------------------------------------
---------LN---VT-HCQ---NLSKALQS
PF13516 // ENSGT00900000140813_0_Domain_8 (1772, 1796)
-NRSLVFLNLSTNSLSN-DGVKSLCEV--
LEN-PNCPLERL-A----------------------------------------------
------------------------LASCGLT-----------------------------
----------------------------KVGCEVLSS-----------------------
--------------------------------ALTKTK
PF13516 // ENSGT00900000140813_0_Domain_11 (2019, 2054)
RLTH---LC-LAHNV-LED---
-------EGIELLSYTL----RHPRC----------------------------------
--------------------------TLQ-SL----------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------VL--MGC-----VLTSEACGDL------------
-------ASVL-VNNSNLWNLDLGNNILDDAGLNI-LCEALR-NP-NC-QIRRLG--LE-
--N-CGLTPGCCQ--------------------DLLDLLSNNQSV-IQMNLMKNSLDYEA
IRNLCK--------VLRSPTCK--ME--F-------------------------------
---LALDKKDLFKKKIKKF------LV----------DVR-INNPHLVIRPQCPN-----
--T---------------------------------------------------------
--------------------------------------------------ESGCWWQYF-
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMMUP00000012994 (view gene)
ENSPVAP00000000490 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MPRT----VVKD
PF02758 // ENSGT00900000140813_0_Domain_0 (204, 328)
GLCRLSVYLEELE--A---VELKKFKLHLGT------
---------A---TDMG--------E--DGI-PWGR-METAG-PLEMAQLLVAHYGIGEA
WVLALSI----FD-RINRKDLWERGQ-REDLVMDT----S--PG--------GLSSL---
------------------------------------------------------GS----
------------------------------------------------------------
---QS-ACSL-------------------------------KIFTGTPRKDPRETYRDY-
--------VRRK----------------FRLM--EDRNARLGECV---------NLSHRY
TRLLLVKEHSNPMWAQQKLL------DTGWGHARTIG--------HQTSLIQMETLFEPD
EE--RPEPP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
RTVVLQGAAGMGKSMLAHKV-----MLDWA--D---------GRLFR
D--------RFDY-VFYIHCREMNQSAS------DQSVQD-LLSS-CW-----PE-PSAP
LWELVQ-VP----ERLLFIIDGFDE-LRPSFHDS------QG-P-----CCLCWEEK-QP
-TELLLSSLIQKKLLPELSLLIT-TRPAALEKLHRLL-E-----HPR--HVEVLGFSEAE
RKEYF----------------Y--KY-FHN---------TEQAGQVFNFVRD-NEPLFTL
CFVPLVCWVVCTCLKQQLEGGG-LL-R--QT-SRTTTAVYMLYLLSLMQPKPGS-PTL--
-----------QPPPNQRGLCSLA---ADGL-WN-QK----ILFEEQ-------------
--DLRKHGLDGV-DVSSFLNM----NIFQKDIN-CE-KFYSFIHLSFQEFFAAMYYIMD-
G--GE--------------S--G--ASPHQNLR-RLLAEYAFS-ERSFLALTVRFL----
----FGLLNEETRSYLEKCLCWK---------------V--SPN-IKV-ELLQYIRSKAQ
SE-----------------------------------GSTLQRGSLEFLSCLYELQE-KE
FIQQALGH---------------------F-------------------QVVVVSSIT-T
KM-EHMVS--S----FCMK-NCRS-ILV-LHLY----------------GA------AYS
-ADEEDGA--R-WT---------------------------------------SGP---R
M-------------------------PL------------------------T-------
PS--R--------PERNILPD-------AYSEQLAAAL----------------STNPNL
MELA-LYRN-ALGNR-GVKLLCQGLQHPNCK----LQNL--------RLK---------R
C-----------------------------------------------------------
--------LVS---SS-ACQ---DLTAALMT-NKNLMRMDLSSNDLGL-PGLKLLCEG--
LQH-PKCRLQMIQ-----------------------------------------------
------------------------LRKCHLE-----------------------------
----------------------------ARACQEMAS-----------------------
--------------------------------VLSTSR
PF00560 // ENSGT00900000140813_0_Domain_11 (2019, 2052)
HLVE---LD-LTGNA-LED---
-------LGLKLLCQGL----RNPVC----------------------------------
--------------------------RLQ-ILW---------------------------
-------------------------------LKICHLTAASCEDLAST-LSVNQSLMELD
L-SLNDLGDPGVL-LLCEGLRHP--QCK----L---------------------------
-----------------------QTLRL--GIC-----RLSSAACEGL------------
-------STVL-QLNR
PF00560 // ENSGT00900000140813_0_Domain_15 (2357, 2385)
HLRELDLSFNDLGDRGMWL-LCEGLR-HP-TC-RLQKLW--LD-
--S-CGLTAKASE--------------------DLSSILGVNQTL-IELYLTNNSLGNTG
VRLLCK--------RLSHPGCK--LR--V-------------------------------
---LWLFGM-DLNKMTHRR------LA----------ALR-VTKPYLDIGC---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCJAP00000021695 (view gene)
ENSMICP00000050110 (view gene)
ENSMPUP00000010362 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------MGPDDART-QE------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------EQDGQETVQGDGTEYRIQ-
--------IKEK----------------FCIM--WDKKSLFGEPEDFHLGVA--------
----------------------------------------------QEHQELLEHLFDVD
VK--TGEQP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
QTVVLQGAAGVGKTTLVRKA-----IVDWA--Q---------GNLYQ
Q--------RFSY-VFYFNAREINQLR-------EQSFVQ-MISK-DW-----PS-REGP
IERIMS-EP----SSLLFIIDSFDE-LNFAFEEP------EF-V-----LCEDWTQV-HP
-VSFLMSSLLRKVMLPESSLLVT-TRPTACKKLKPLL-K----N-QR--SVELLGMSESA
RKEYI----------------Y--QF-FED---------KKRAFQVFNSLRS-NEKLFSM
CKVPLVCCAICTYLDQQMEMGG-DV-S--LT-CKTTTTLFTCYISSFFAPV-DGSCTSLP
------------NQSQLRSLCHLA---ARGV-WT-LT----SVFYRE-------------
--DLRKHGLTKS-DVSIFLDR----NILQKDTE-YE-NCYVFTHLHVQEFLAAMFYVLK-
DNWET--------------RDRS--LQSFEDVK-LLLEANSGQ--DPHLTQMKCFL----
----FGLLNEDLVKQLETTLNCT---------------M--SLE-IKG-KILQWLEILGK
SE-----------------------------------HFPTQLEFLELFLCLYETQD-EA
FISQAMKP---------------------F-------------------QKAVIDVCEKV
HL---LVS--S----FCLK-HCQY-LRA-VKLS----------------VTV-----SCE
-----TML----NSSR-----PA--A------------------------LWQR-DDGHF
THCWQDLC-SVLHTNEHLRELELCHSNLGEL---------AMKTFY--QELRHP------
NC--K-LQKLLLRF---------LSF-PGGCQDIANSL----------------IDNQNL
MHLD-LKGS-DIGDN-GVKSLCEALKHPECK----LQNL--------SLE---------S
CN----------------------------------------------------------
---------LT---TV-CCL---NISKALLR-SQSLRFLNLSTNHLLD-DGIKLLCEA--
LGH-PKCHLERL-S----------------------------------------------
------------------------LESCGLT-----------------------------
----------------------------VAGCEDLSL-----------------------
--------------------------------AVINNKRLTH---LC-LADNI-LGD---
-------DGIKFVTDAL----KNPQC----------------------------------
--------------------------NLQ-------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------SLVL--LGC-----ALTSTCCLDL------------
-------ASAI-LNNTHLRSLDLGHNDLQDYGVKV-LSEALR-HP-NC-NIERLG--LE-
--Y-CGLTSLCCQ--------------------DLSSVLRSNQNL-IKINLRQNTLGHEG
VMKLCE--------VLRSPECK--LK--V-------------------------------
---LGVCKE-AFDEEARKL------LE----------AVQ-VSNPHLIIKHDYTDHN---
--D---------------------------------------------------------
--------------------------------------------------EDGSWWRCT-
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSAMEP00000017920 (view gene)
ENSCSAP00000000724 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------MADSSSSSFFP
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 328)
DFG---LLLYLEELN--K---EELNTFKLFLK-------
---------E--TMEPE--------H--GLI-PWNE-VKKAR-RDDLANLMNKYYPGEKA
WSVTLKI----FG-KMNLKDLCQRAK-EEINWSAQ-SIGPDDAKV-GE------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------TQ-DQETVLGDGTEYRIR-
--------IKEK----------------FCIT--WDKKSLAGKPEDFHHGIV--------
----------------------------------------------EKDRKLLEHLFDVD
VK--TGEQP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
QTVVLQGAAGVGKTTLVRKA-----MLDWA--E---------GNLYQ
Q--------RFMY-VFYLNGREINQLR-------ERSFAQ-LISK-DW-----PS-TEGP
IERIIS-QP----SSLLFIIDSFDE-LNFAFEEP------EF-A-----LCEDWTQE-HP
-VSFLMSSLLRKVMLPEASLLVT-TRLTTSKRLKQLL-K----N-HR--YVELLGMSEGA
REEYI----------------Y--QF-FED---------KRWATKAFSSLKS-NEMLFSM
CQVPLVCWATCTCLKQQMEKGG-DV-T--LT-CQTTTALFTSYISSLFTPV-GGCSPRLP
------------NQVQLRRLCQVA---AKGI-WT-MT----YVFYRE-------------
--NLRRFGLTKS-DVSSFLGN----NIIQKDTE-YE-NCYVFTHLHVQEFFAAMFYMLK-
GNWEA--------------GNPS--CQPFEDLK-SLLQSTSYK--DPQLRQMKCFL----
----FGLLNEDRVKQLERTFNCK---------------M--SLK-IKS-KLLQCMEVLGN
SD-----------------------------------YSPSQLEFLELFHYLYETQD-KA
FISQAMRY---------------------F-------------------SKVAINICEKI
HL---LVS--S----FCLK-HCRC-LRT-IRLS----------------VTT-----VFE
--K--KTL----KTSL-----PT--N------------------------TW---DGDRI
THWWQDLC-SVLHTNEHLRELDLCHSNLDKS---------AMNILH--QELRHP------
NC--K-LQKLLLKC---------ITF-PDGCQDISTSL----------------IHNQNL
MHLD-LKGS-DIGDN-GVKSLCEALKHPECK----LQTL--------RLE---------S
CN----------------------------------------------------------
---------LT---VF-CCL---SISNVLIR-SQSLIFLNLSTNNLLD-DGVQLLCEA--
LRH-PKCYLQRL-S----------------------------------------------
------------------------LESCGLT-----------------------------
----------------------------EAGCEDLSL-----------------------
--------------------------------ALISNKRLTH---LC-LADNV-LGD---
-------GGLKLMSDAL----QHAQC----------------------------------
--------------------------TLQ-SLV---------------------------
-------------------------------LRHCHFTSLSSEYLSTS-LLHNKSLTHLD
L-GSNWLQDNGMK-LLCDVFRHP--SCN----L---------------------------
-----------------------QDLEL--MGC-----VLTNACCLDL------------
-------ASVI-VYNPNLRSLDLGNNNLQDDGVKI-LCDALR-YP-NC-NIQRLG--LE-
--Y-CGLTSLCCQ--------------------DLSSALICNQRL-IKMNLTQNTLGYDG
ITKLYK--------VLKSPRCK--LQ--V-------------------------------
---LGLCKE-AFDEEAQKL------LE----------AVG-VSNPQLIIKPDYNYHN---
--E---------------------------------------------------------
--------------------------------------------------ENESWWRCF-
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSRNOP00000038641 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------M---ASLFSDFG---F
PF02758 // ENSGT00900000140813_0_Domain_0 (209, 323)
MWYWKELN--K---VEFMKFKELLIL------
---------E--ILQMG--------L--KQI-SWTE-VKNAS-REDLAILLVKYCEGKQA
WDTTFKV----LQ-KISRHDLTERAT-GEI------------AA--------------HA
N---------------------------------------------------IYRA----
------------------------------------------------------------
----------------------------------------------------------H-
--------LKKK----------------LTRD--CSRKFSISIQNF--------------
---------------------------------F------------QDEYDHLENLLIPK
VT--EEK-P----Q
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
LVFLKGIAGIGKTILLKNL-----MIVWS--E---------GLVFQ
N--------KFSY-IFYFCCQDVKQLK-------TTSLTE-LISR-EW-----PSS-SAP
IEEILS-QP----EKLLFIIDSLEG-MEWDFTKQ------ES-E-----LCDNCLEK-QP
-VNVLLSSLLKKKILPESSLLIS-ATFETFEELKDWI-E-----YTN--VRTITGFKESN
IKMYF----------------H--SL-FQD---------RNRALEAFSFVRE-NEQLFTL
CQAPVICCMVATCLKNEIEKGK-DP-V--SI-CRRTTSLYTTHIFNLFIPPN-VQYPSK-
-----------KSQDQLQGLCFLA---VEGM-WT-DI----SVFSEE-------------
--TLRRNGILDS-DIPTLLDI----GILEQSRE-SQ-NSYIFFHPSVQEFCAAMFYLLQ-
THMNH-----------------P--SPDVLHVE-KLLFSFLKE-VNTQWIFLGRFI----
----FGLLNELEHEKLEAFFGYQ---------------L--SQQ-LKQ-ELFEWLELLLD
P--------------------------------------EVKVNTMKFFYCLFEMEE-EV
FVQSAMNC---------------------R-------------------EEIDVVAKDYY
DF---IVA--A----YCLN-HGSA-LRD-LSIS----------------TQN-----VLN
-EKL--NQ-----------R-----------------------------------YMENL
LMLWHNIC-SVFARNKDIHILQMKDTIFNEP---------VFQILY--NYLKNS------
SC--I-LEVLVAND---------VSF-LCDKYLFFDL-----------------IQSYNL
ELLD-LSGT-FLSHS-DVVILCNILNKAEYK----IQEL--------ET-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------TAFNLW-----------------------------
----------------------------T--CVLFFY-YPC-------------------
--------------------------------IDFLTLKLDH---NG-LSTGP-SQ----
-------NAMPMRSLTLFHTECASLLILFAI-----------------------------
------------------------------------------------------------
------------------------------MLANCSLSEQSWKYISDV-LC
PF13516 // ENSGT00900000140813_0_Domain_13 (2212, 2237)
QNKTLRHLD
I-SSNDLKDEGLK-VLCKALTLP--DSV----L---------------------------
-----------------------LTLSV--KAC-----FITTVGCQYL------------
-------AEVL-R
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
NNHKLMFLTLSENKLEDSGVKL-LCDAIK-HP-RC-QLAQLD--LE-
--A-CELTGACCE--------------------DLASTFTQCKTL-GWINLVKNALDFNG
LVVLCK--------ALKQKTSN--LK--E-------------------------------
---LRLRIT-DFDNKSQTF------LL----------SEE-KGNKFLNIEND-E------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPSIP00000006673 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 328)
LLAILEELE--E---EQRKKFKFKLNT------
---------T--ELREG--------Y--KNI-PKGL-LQKAD-VTDMVDLLMKYYQEEYA
VEVVINV----LD-QINNKDLAERLR-RNV------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------NWKLILGGH-
PN----------------------------------------------------------
-----LKKHHQVEDKEHELL------SRGKRHMEIMR----KPSSFEDSHVKVEHLFDTL
SD--G-TIT----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
RKVILQGVAGIGKTATVGKI-----MYDWS--C---------GTLYQ
G--------RFDY-VFHWSCRELNRRTE------KLSLVQ-LILQ-NW-GH-----PKPA
IGEILS-CP----SKVLFIIDGFDE-LRFPEDYD------ESKI-----LSCDLDTP-HS
-AAAVVESLLSSRSLSEACFLVT-TRPLAVAKLETHM-K-----ADL--WAEILGFLEED
RKEFF----------------K--KF-FKD---------EKKAAIAYHYIQE-NDVVFTM
CFVPLICWIVCTTLNLQMKHGE-EF-V--QK-VSTTTQIFTYFVATLLQSHGRS---QTS
------------TQNLFHNLGSLA---YAGM-RD-QK----VLFDES-------------
--MLRKCNLEVCKGPSTFLTE-----LLQGDIF-VE-TIYSFVHLSVQELFAALFTYWN-
KKEAF-----------------------------ELLKEAFVE-KKSHLILTVRFL----
----CGLNNDESLKPLKRYYETE------------------TGI-SKN-ELLKWIKKAL-
LIC----------------Q----------------GQEKQSNLLLELLHCLFEIQD-EE
FVIRALEE---------------------V-------------------KGLGFFVCLFL
FL-FCFVL--K----FTNG-MKRI-KYT-LDMR-----QRMKSLRTSNGR--Q----LWF
FESSNRD--------A--VHRIRFLNV-QSHL--RALCLDENSIDD--------------
------------------------------------------------------------
------------------------------------------------------------
--------------E-LMKTLCNSLNSTEYR----LQSL--------SLR---------S
NNLTQESIA-NICS----LFHC----NM-AVSLNLQSNSVGDEGVKILATCLKSKGCC-L
EKLSL-PGAGL--TED-CVP---AITSLFSQ-KNTLRWLDLSFNNLGD-GGVKALCSA--
LKD-KESRLEKL-I----------------------------------------------
------------------------LENNGIT-----------------------------
----------------------------NSCEKELIQLIET-------------------
------------------------------TP------SLQY---LC-LDNNE-FSST--
-------SRAHI------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------YDIWEEYHEEEE-------GGG-GR-D-------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCJAP00000059395 (view gene)
ENSGGOP00000004737 (view gene)
ENSMPUP00000012314 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MNLLELVDWLS-HIC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LN--DYREMYREH-
--------VRC-----------------LE------ER---QEG----------GIRGSY
NQLLLVT-RSNPESADPSAC------PL-------PE-----Q-ELASV--SVETLFDPG
EE--SYQAP----P
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLQGSAGTGKTTLARKM-----VLDWA--T---------GTLYP
G--------QFDY-VFYVSCREVVLLSE-------GTPDQ-LLFW-CC-GD-----NQAP
VKEMQR-EE----ERLLFILDGFDE-LQRPFATG------LK-R-----LS-----L-NP
-MENVLHRLIRREVFSRCSLLIT-TRPVAMCSLEPLL-K-----QSR--YIHILGFSEDE
RRRYF----------------S--SY-FAD---------EEQARNAFDIVQG-NDVLYKS
CQIPGICWVICSWLKEWMERGR-EI-S--ET-PSNGTDIFMAYVSTFLPPSH-NEDCAEL
----------T-SHRVLRGLCSLA---AEGI-QH-QR----FIFEEA-------------
--DLRKHNLDGASLD-AFLSS----MPYQEGGH-AK-TFYTFRHISFQEFFHAVSYLVK-
EDQSDL----------------G--KESHRELR-RLLEEEKEE-ENGEITLSMQFL----
----LDILKKETSSNFELKFFLK-----------------VSPS-LKQ-EL-MYFKEQM-
K-------------------------------------SIKHKRAWDLEFSLNPFKI-RN
LVKGIQIS---------------------D-------------------VSLKVGQSN-K
-K-SHDRK--S----FSVK-TSLR-DEQ-KEKQ-------------------G----PVV
--------GK---DNI--V-------------------KTQKE-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------TSN----------------GKG---------R
-EMEK-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMAUP00000016758 (view gene)
ENSPPAP00000002657 (view gene)
ENSGGOP00000008087 (view gene)
ENSCGRP00001001791 (view gene)
------------------------------------------------------------
------------------------------------------------------------
--------------CCP-----------------GAPCGECERWDPGHFGGSGTGPEQ--
VVSVI-TLSIPSRDT--DALAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLVALEDLS--Q---EQLKRFRHKLRD------
------------APLDG-----------RSI-PWGR-LENSD-PVDLVDKLTQFYGSEPA
VDVTRKI----LK-KADARDVAVRLK-AQQLQR---------L---------GSS-----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------S-AVVSLSEYKKRYREH-
--------VLLQ----------------HAKV--KERNSRS------------VKINKRF
TKLLIAPGSGAVEDE--LLGPSE-------EPQP--------ERARRSDTHTFNKLFRGD
D---EGPR-----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYH
G--------QVDF-AFFMPCGELLERPG------TRSLAD-LILD-QC-----PD-RKAP
VRQILA-HP----QRLLFILDGADE-LPAAAAP--------EAT-----PCTDPFEA-TS
-GLRVLSGLLSQELLPLARLLVT-TRNAAAGRLQGRL-C-----SPQ--CAEVRGFSDKD
KKKYF----------------F--KF-FRD---------ERKAERAYRFVKE-NETLFAL
CFVPFVCWIVCTVLQQQLELGQ-DL-S--RT-SKTTTSVYLLFIASVLKSA-GTNGP---
-----------RVQGELRTLCRLA---REGILKH--Q----AQFSEN-------------
--DLEQLKLQGSQVQTLFLSKKELPGVLDK-----E-VTYQFIDQSFQEFLAALSYLLE-
AEG-----------------------APAGGVQ-MLLNSDSEL--RGYLALTTRFL----
----FGLLSAERIRDIERHFGC-----------------VVPER-LKQ-DTLRWVQGQGH
PK--VAPVEAEKNDELKDTKEPEEEEEE-----EEEEEEEDLNFGLELLYCLYETQE-DA
FVRQALSS---------------------L-------------------PEMVLERVRFT
RM-DLGVL--S----YCVQ-CCPA-GQS-LRLV------------------NC----GLV
------EA----------------------------------------KEKK-----KKS
----------------------------------------LVKR----------------
------LKGS----------------------------------------------GNTT
KQ---PPVS-------LLRPLCEAMTSQQCG----LRIL--------ILS---------R
C-----------------------------------------------------------
--------KLP---DS-VCR---DLSEALKL-APALRELGLLQSRIT-ETGLRLLSEG--
-LAWPKCRVQTL------------------------------------------------
----------------------------RIQ-----------------------------
---------------LPNL---------QEAIQYL-------------------------
------------------------------VIVLQQSRVLTT---LD-LSGC-QLVTYMV
--E--------NLCSAL----KYPMCCLKTLSLT--SVELSEV-----------------
----------SLRELQIVKASKP-------DLT----------------------I--LH
SKP---------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSTBEP00000005214 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---SSFF
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
SDFG---LMWYLEELR--K---EEFRKFKEHLKK------
---------E--TLELG--------L--KQI-PWTE-VKKAS-REDLANLLIEHYEEQQA
WDVTFRI----FH-QISRKDLCEKAE-RER------------TG--------------HM
K---------------------------------------------------MYRA----
------------------------------------------------------------
----------------------------------------------------------H-
--------VKKK----------------FSNI--WSRESVTGIHDH--------------
---------------------------------F-------NQELTPEECKYLELFFAHK
ET--GER-S----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
RTVLLKGVQGVGKTTLLIKL-----MLAWS--E---------NTLYQ
D--------TFQY-VFYLSCREVKQLT-------ATSLAE-LISR-EW-----PNS-SAP
IAEIMF-QP----ERLLFIIDSFEE-LKCDLDEP------ES-N-----LHNDWMEK-QP
-VRVLLSSLLRKKMLSESSLLIA-ATHEYPREVEGRL-E-----CPE--IKTLMGFTEAD
RKLYF----------------C--CL-FQD---------RNKAMEAFHFVRE-NDHLFSM
CQIPLLCWIVCTSLKQEMEKGR-DL-A--LA-CRRSTSLFASFIFNLFTSSG-ARRTRQ-
-----------QSQGLLISLCSLA---AEGM-WT-DA----SAFSED-------------
--DLRRNGIADP-DIPMLLGV----KVLK-RRE-DE-SSYTFIHMRVQEFCAAMFYLLK-
SHSDH-----------------P--HPAVGCAE-ALLFNYLKT-VKTHWILLGCFV----
----FGLLNEKEQQRLDAFFGFQ---------------L--SQK-IKQ-QYYQYLKSLAE
R-E----------------------------------DLKRXXXXXXXXXXXXXXXX-XX
XXXXXXXX---------------------X-------------------XXXXXXXXXXX
XX---XXX--X----XXXX-XXXX-XXX-XXXX----------------XXX-----XXX
-XXX-XXX-----------X-----------------------------------XSSRG
IIHWHRIC-SVLTTNENLRELKVKNSDLNTS---------AFVALC--DQLRHP------
RC--H-LEKLEMNN---------VSF-SGNMWHFFEVL----------------VHNPGI
KFLS-LSST-KLARD-EVTLLCDALNHPVCN----IEEL--------RX-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------XXXXXX-----------------------------
----------------------------XXXXXXXXX-----------------------
--------------------------------XXXXXXXXXX---XX-XXXXX-XX----
-------XXXXXXXXXX----XXXXXXXXXX-----------------------------
------------------------------------------------------------
------------------------------XFAYCYLTEHCWEXXXXX-XXXXXXXXXXX
X-XXXXXXXXXXX-----XXXXX--XXXXXXXX---------------------------
-----------------------XXXXL--VKC-----FITTEGCQDL------------
-------TLVL-TSNQNLRSLQIGYNDVEDVGVKL-LCEALI-HP-NC-CLENLG--LE-
--A-CGLTSACCE--------------------ALSSALTSCKTL-RRLNLVQNALDHSG
VAVLCE--------ALRHPECA--LE--V-------------------------------
---L--------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSANAP00000039831 (view gene)
ENSRROP00000007398 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---ESFFSDFG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 322)
-LLWYLKELR--K---EEFWKFKELLKQ------
---------P--LEKFE--------L--KPI-PWAE-LKKAS-KEDVAKLLDKHYPGKQA
WEVTLNL----FL-QISRKDLWTKAQ-EEMRN----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------KLNPYRKH-
--------MKEK----------------FQHI--WEKETCLQVPEH--------------
---------------------------------F-------YKETMKNEYKELNGAYTAA
------ARL----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
HTVVLEGSDGIGKTTLLRKV-----MLDWA--E---------GNLWK
D--------RFTF-VFFLNVYEMNGVT-------ETSLLE-LLSR-DW-----PES-SET
IEDIFS-QP----ERILFILDGFEQ-LKFNLE-F------KA-D-----FSDDWRRR-QP
-TPIILSSLLQKKMLPESSLLIA-LGKLAMQKHYFTL-Q-----HPK--LIKLSGFTESE
KKSYF----------------S--YY-FGE---------KNKALKVFNFVRD-SGPLFIL
CHNPFICWLVCTCVKQRLERGE-DL-E--IN-SQNTTSLYASFLTNVFKAGS-QSFLPK-
-----------VNRARLKNLCALA---AEGV-WT-YT----FVFCHE-------------
--DLRRNGLSES-EGLMWVGM----RLLQRRG-----DCFTFVHLCIQEFCAAMFYLLK-
RPKDD-----------------P--NPAIGSIT-QLIKASMTQ-PQTHLTQVGVFT----
----FGISAEEIVSMLETSFGFP---------------L--SKD-LQQ-EITQCLKSLSQ
CE-----------------------------------ADREAIGFQELFVSLFETQE-KE
FVTQVMNF---------------------F-------------------EEVFIYIGNIQ
HL---AIA--S----FCLK-HCRR-LTT-LRMC----------------MES-----IFP
-DDSGCAS-----------G-------------------Y-N-------EK---------
LVYWRELC-SVFITNKNFQILDMENSSLDDA---------SLAILC--KALTQP------
VC--K-LRKLIFTS---------MLN-FGHDSELFKAV----------------LHNPHL
KLLS-LYGT-SLSHT-DVRHLCETLKHPMCK----IEEL--------IL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------GKCDIS-----------------------------
----------------------------SEACEDIAS-----------------------
--------------------------------VLAC
PF13516 // ENSGT00900000140813_0_Domain_11 (2017, 2054)
NSKLKH---LS-LVENS-LRD---
-------EGMMLLCEAL----KHPHCALETL-----------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------ML--MGC-----FLTSDSCKDI------------
-------AAVL-IC
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NEKLKTLKLGHNEIGDAGIRQ-LCAALK-HP-NC-KLEGLG--LQ-
--T-CPITRACCG--------------------DLAAALVACKTL-RSLNLDWITLEPDA
VAVLCE--------ALSHPDCA--LQ--M-------------------------------
---L--HKS-GFDEETQKM------LM----------SVEEKM-PHLTISHEPWINE---
--EYK-----IR------------------------------------------------
--------------------------------------------------GVLL------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPPAP00000014381 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-MDQPEAPCSSTGPR--LAVAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLAALEELS--Q---EQLKRFRHKLRD------
------------VGPDG-----------RSI-PWGR-LERAD-AVDLAEQLAQFYGPEPA
LEVARKT----LK-RADARDVAAQLQ-EQRLQR---------L---------GLG-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------SG-TLLSVSEYKKKYREH-
--------VLQL----------------HARV--KERNARS------------VKITKRF
TKLLIAPESAAPEEE--ALGPAE-------EPEP--------GRARRSDTHTFNRLFRRD
E---EGRRP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYQ
G--------QVDF-AFFLPCGELLERPG------TRSLAD-LILD-QC-----PD-RGAP
VPQMLA-QP----QRLLFILDGADE-LPALGGP--------EAA-----PCTDPFEA-AS
-GARVLGGLLSKALLPTALLLVT-TRAAAPGRLQGRL-C-----SPQ--CAEVRGFSDKD
KKKYF----------------Y--KF-FRD---------ERRAERAYRFVKE-NETLFAL
CFVPFVCWIVCTVLRQQLELGR-DL-S--RT-SKTTTSVYLLFITSVLSSAPVADGP---
-----------RLQGDLRNLCRLA---REGVLGR--R----AQFAEK-------------
--ELEQLELRGSKVQTLFLSKKELPGVLET-----E-VTYQFIDQSFQEFLAALSYLLE-
DGGVP-------------------R-TAAGGVG-TLLRGDAQP--HSHLVLTTRFL----
----FGLLSAERMRDIERHFGC-----------------MVSER-VKQ-EALRWVQGQGQ
GCPGVAPEVTEGAKGLEDTEEPEE------------EEGEEPNYPLELLYCLYETQE-DA
FVRQALCG---------------------L-------------------PELALQRVRFC
RM-DVAVL--S----YCVR-CCPA-GQA-LRLI------------------SC----RLV
------AA----------------------------------------QEK--K---KKS
----------------------------------------LGKR----------------
------LQAS---L--------------GGS------S----------------SSRGTT
KQ---LPAS-------LLHPLFQAMTDPLCH----LSSL--------TLS---------H
C-----------------------------------------------------------
--------KLP---DA-VCR---DLSEALRA-APALTELGLLHNRLS-EAGLRMLSEG--
-LAWPQCRVQTV------------------------------------------------
----------------------------RVQ-----------------------------
---------------LPDP---------QRGLQYL-------------------------
------------------------------VSVLRQSPALTT---LD-LSGC-QLPAPMV
--T--------YLCAVL----QHQGCGLQTLSL--ASVELSEQ-----------------
----------SLQELQAVKRAKP-------DLV----------------------I--TH
PALD-------------------GHPQPP-KELISTF-----------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMNEP00000040976 (view gene)
ENSOPRP00000003739 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---ETYF
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
SDFG---LLWYLEELR--R---EEFWKFKEHLRQ------
---------E--PLKFD--------L--KPI-PWAE-LKRAS-KGELASLLEKHYPGTQA
WELTLGL----FL-QLNRKDLWRKAK-EEMRS----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------KLNPYRNL-
--------MKDK----------------FQLV--WEKESCLPVPES--------------
---------------------------------F-------YRETVSNEWIS-------A
RA--VEDGP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
HTLVLHGPEGRGKTTFLRRV-----MLEWA--L---------GNFLR
D--------RFTY-VFFLNVLELNGLQ-------ETSLEE-LLSG-DW-----PAS-PGA
LGEVLS-EP----DRVLFLLDGFDE-LRFDLE-L------QA-D-----LCGDWRQR-RP
-PAVVLGSLLQKAMLPECSLLLA-LGTVGIRRYYPLL-R-----SPR--HMQLPGFSEQH
RKQYI----------------S--HF-FRE---------RDDALRVSSFVRD-IPSLFAM
CESPLVCWLVCNCLRWQLERGE-SL-H--VG-PSNTTSLQLFFLVSALQVGS-RASPPLP
----------APARVRLQSLCRLA---AEAT-WT-QA----SVFREE-------------
--DLRRHGLAVP-DTRVWVGT----HCLRPSG-----DAYVFSHASLRDFCAALFYLLR-
QPEDP-----------------P--QPGVGSVS-QVIRASLGY-DHSHLSRLGVFL----
----FGVSAGES-RVLETYFGMP---------------P--TSE-LKA-ES---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------------------EE---------
LSCWKEFC-SALGTRTSLRVLDLDNCKLNEA---------CLTVLW--NALAQP------
TC--K-LHSV--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMNEP00000015393 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------MVSST----QVNFN--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 323)
-LQALLEQLS--Q---DELSKFKSLLRT------
---------V--SLGNE--------V--QKI-PQKE-VDEAD-GKQLAEILISHCHSYWT
ELATIQV----FE-KMHRVDLSERAK-DELREAAL-KSFN----KNK------PLSLG--
------ITRKERQP---------------------------------VDVEEMLERFK-A
GTLECT---------------------------------ET--EEDVTV-----------
-------LTD-------------------------------IFKVKGEK---SGLYRLI-
--------LKTK----------------FREM--WQ--SWPGDSKEVHV-----------
---------------------------------------------MAERYRMLIPFSNPR
VL--PGPFS----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
HTVVLHSPAGLGKTTLANKL-----MLDWT--E---------DNLIQ
---------KFKY-AFYLSCRELSRLG-------PCSFAQ-MVFR-DW-----PE-LQDD
IPHILA-QA----QKILFVIDGFDE-LGAPPG------ALIQ-D-----ICGDWEQQ-KP
-VPVLLGSLLKRKMLPKATLLVT-TRPRALRDLRFLA-E-----QPI--YIRVEGFLEED
RRAYF----------------L--RH-FGD---------EDQAMRAFELMRS-NAALFQL
GSAPAVCWIVCTTLKLQMEKGE-DP-A--PT-CLTSTGLFLRFLCSQFPQ-------GAQ
------------LRGALRALSLLA---AQGL-WA-QM----SVLHGE-------------
--DLESAGVQES-DLRLFLDR----NILRQDGV-AK-GCYSFIHLSFQQFLTALFYALE-
KEEEE-------------DRDGH--AWDIGDVQ-KLLSREERL-KNPDLIQAGRFL----
----FGLANEERVKELEATFGWR---------------M--SPE-IKQ-ELLRCDVSRKN
G------------------------------------H-FTAADLRELLCCLYESQE-DE
LVKEVMAQ---------------------F-------------------KEISLHLN-AV
DI---APS--S----FCFK-HCQN-LQK-MSLQ----------------VIKK----ETL
-QENVAAS----ES--------------------NAEAER------------SQDD-QDM
LPFWTDLC-SIFGSNKDLMGLEINNSFLSAS---------LVKILC--EQIAS-D-----
NC--H-LQRLV--F-KNIFPA--DA----YR-NLCLAL----------------RGHKTV
THLT-LQGI-DQKD--MLPALCEVLRHPECN----LRYL--------GLV---------S
C------S----------------------------------------------------
---------AT---TQ-QWA---DLSLALEA-NRSLMCVNLSDNELLD-EGAKLLYTT--
LRH-PKCFLQRL-S----------------------------------------------
------------------------LENCHLT-----------------------------
----------------------------EANCKDLAA-----------------------
--------------------------------VLVVSRELTH---LC-LAKNS-LKD---
-------TGVKFLCEGL----SYPEC----------------------------------
--------------------------KLQ-ALV---------------------------
-------------------------------LWNCDITSDGCCSLAKL-LQEKSSLSCLD
L-GLNHIGVTGVK-VLCEALSKP--LCN----L---------------------------
-----------------------KY----------------------L------------
-------CSAL-SLQPEPHPLDLGQKSLGVSGLKI-LCKSR----------DIWT--LK-
--I-DDFNDELHK--------------------LLEEIEENNPQLIIDTEKH----DPWK
---------------KRPSSHD--FM----------------------------------
----I-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMPUP00000003790 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------E
PF05729 // ENSGT00900000140813_0_Domain_2 (687, 929)
TAVVYKF-----MFDWA--A---------GMVTP
G--------RCNY-LIYINCREINHIG-------NLT----VLIN-TL-----LN-KDGP
ILDTILVYP----EKVLFILNGFPE-LQYPVGNQ------EE-D-----LSANLQKK-KT
-VETLLCSFVKKKLFPESSLLIT-AQPTTMRRFHTLL-Q-----QPI--QAEIVWFTDAE
KRVYF----------------L--SQ-LSG---------ACAALRLFYEL----------
SSLPIISWMICSVLQSQGDGNR-TL-M--RS-LQTVTAVYLFYFSKCLKTLAGIS-VW--
-----------KGLSCLWGLCSLA---AEGL-QN-QQ----VLFEVS-------------
--DLRRHGIGVHDIKSLFEK-------TKQNKG-TV-NVYIFLHFSFQEFLNAVFYDLK-
NDSSW---------------------VFFDQVGETWKEIFQQY-GHGFSSLIIQFL----
----FSLLHKGKGKAMETTFGRK---------------V--SLG-LQE-ELLKWTEKEIK
DK-----------------------------------SSRRQIDPMDLFHCMRFRKK-NM
YKSIQAHH------------------------------------------HFL--ALSIN
KV-NHLHF--P----YFML-ASSQRVQR-FAHL-----QRVQRG-----LDNCISSLGSS
-METEGSM----------------------GYMH--------------------------
---------IVNFWKRSFQ-----LCSLPVS---------QLHLLC--QALYNP------
YC--K-IKDLNVLS-VHISST------QAQAPLFPNSLPHRQHRRVANLCCVVRPGLTTA
NTTL-TMES-ASQDS-GTKLLCEGLKQPNCI----LWTLR--------LYRCLI-SPA-S
CG------------VL----AAILSTSQWLTDLEFSETRLEASALKLLCEGLKDPNCK-L
RK----------------------------------------------------------
-----------F-N----------------------------------------------
------------------------LSTCYLP-----------------------------
----------------------------DASCMELSS-----------------------
--------------------------------FLHVSQTLKE---LF-AFANA-VGD---
-------TGVQLLCEGL----QHAKTILQNL-----------------------------
------------------------------------------------------------
------------------------------VLSECSLSAASCESLAQV-LSSTWRLTRLL
L-INNKIEDLGLK-LLCKGLKQP--DCQ----L---------------------------
-----------------------KDLAL--WTC-----HLTGEFCQDL------------
-------CNVL-YTNEYLRDLDLSDNASGDEGMQV-RCEGLK-DP-SC-KLQTLW-----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSSBOP00000008549 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------CLLLPR
RP--RGRQP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 928)
RTVVIQGAPGIGKTTLARRV-----MCEWA--R---------NKFYA
H--------KPWC-AFYFRCQEMNRMA-------EQSFSE-LIQH-KW-----PG-SQDL
VSKIMS-KP----DRLLLLLDGFED-FTPALI------DRSE-D-----LSEDWRQK-LP
-GSVLLSSLLSKRMLPEATLLIM-IRSTSWQTCEPLL-Q-----SPS--LVTLKGFNATE
KIKYF----------------R--AY-FGY---------TKEGDDVLSFAME-NTIIFSM
CQVPVVCWMVCSCLKQQMERGH-KL-T--QA-CPNATSVFVRYLSSLLPIRA-ENFSRKT
------------HRQQLEGLCRLA---ADGV-WH-RK----CVFGKK-------------
--DLERAKMDQM-GVATFLGV----NILRRIAA-EK-DRYAFTLLIFQDFFAALFYVLC-
FPQRL--------------KNFH--VLNHANIQ-RLVAGSC-G-SKHSLSHMGLFL----
----FGLLNRACAPIVEHSFRCR---------------V--SFG-NKR-KLLKVIPQLQK
SS-----------------------------------LRPLCIRSF---WSQG-------
-SCS--------------------------------------------------PVELNM
ML---EIS--C----VCLV-QCGN-HWL-HATT----------------E-------LLE
-CGWRCL-------------------------------------FSLPC--RAPES-NKL
LRWWQDLC-SVFAVNDKLEVLTLTTSVLQPP---------FLKALA--AALKH-P-----
QC--K-LQKLL--L-RRVNRT--GL---N-K-DLIGVL----------------TGNKHL
RYLE-IQHV-EVEWK-AMKLLCTALRSPRCR----LQCL--------RLE---------D
C------L----------------------------------------------------
---------AT---PR-VWT---YLGNNLRG-NRYLKTLMLRKNSLEN-RGLYY------
--L-SGGQLERL-L----------------------------------------------
------------------------IENCNLT-----------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------ELTCESLASC-LRQS
PF13516 // ENSGT00900000140813_0_Domain_13 (2214, 2236)
KMLTHLS
L-AENALKDEGAK-HIWKALQHL--RCP----L---------------------------
-----------------------QRLVL--RKC-----DLTFDCCQDM------------
-------ISAL-R
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
KNKTLKSLDLSFNSLKDDGVTL-LCEALK-NP-DC-TVQILE--LE-
--N-CLFTSSCYQ--------------------AMASMLCE
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2456)
NQHL-RHLDLRKNAIRVHS
ILTLCD--------AFPSQKKR--EI--I-------------------------------
---F------SWD-PQPHP------SN----------PTA-RYVPQRN------------
------------------------------------------------------------
----------------------------------------------------Q-------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSVPAP00000002974 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------PSAPNLQSPS
WPTSTEVLKFCNSEPP-----MPLHNSEKRDLRHLPSGTSGS-IRKENPSSLLYQ--D--
---------LLNSLD-YESPSKELPNAPVSTAVLSGWEPPVQPSPEPRKQETPGTVWPLA
ETPETPENSCTXXXXXXXXXXXXXXXXX-----X-XXXXX-----XXXXXXXXXXXXXX-
--------XXXX----------------XXXXXXXX------XXX---------XXXXXX
XXXXXXXXXXXXXXXX---X------XXXXXXXXX------XXXXXXXXXXXXXXXXXXX
XX--XXXXX----XXXXXXXXXXXXXXXXX
PF05729 // ENSGT00900000140813_0_Domain_2 (691, 940)
RQV-----RGAWE--E---------AQLYR
D--------HFQH-V-YFSCRELAQSK-------VMSLAE-LLTK-DW-----AG-PVAP
IGQILS-EP----EQLLFILNDLDE-PNWVLEEQ------RA-E-----LCLHWSQR-QP
-VHTLLGSLLEKTVLPEASLLIT-ARTTALQKFIPSL-K-----QPR--WVEVLGFSESS
RKEYF----------------Y--KY-FTD---------ESQAVRAFSLVAS-SQALSTM
CLVPSVSWLVCTCLQQQMESGE-EL-S--LT-SQTTTALYLRYLSQALP--------DQ-
-----------PLGPQLRGLCSLA---AEGT-WQ-GK----TLFSPG-------------
--DLRKHGLDGA-IISTLLKM----RVLQKH---PTSLSYSFMHLCFQEFFAAVSCALG-
D------KEE--------KGEYP--DSP-GSVE-KLPEVYGRR--DLFGAPIVRFL----
----FGLLSEPGVRAMESVFSCT---------------L--PPE-RRG-DLLSWAKSEVR
-----------------------------------RERSSLQPDSLQLLHCLYEIQD-EE
FLMRAMAH---------------------F-------------------QGSRMCVQTGV
EL---LVF--T----FCIK-FCCH-VKR-LQLN----------------EGG--------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------LSWVPVT-----------------------------
----------------------------DVCWQVLFS-----------------------
--------------------------------VLQVTGSLKE---LD-LSGNL-LSH---
-------SAVQGLCEAL----RCPSCHLETLR----------------------------
------------------------------------------------------------
-------------------------------XXXXXXXXXXXXXXXXX-XXXXXXXXXXX
X-XXXXXXXXXXX-XX--X--XX--XXX----X---------------------------
-----------------------XXXXX--XXX-----XXXX------------------
----------------------XXXXXXXXXXXXX----X--------------------
---------XXXX--------------------XXXXXXXXXXXX-XXXXXXXXXXXXXX
XXXXXX--------XXXXXXXX--XX--X-------------------------------
---XXXXXX-XXXXXXXXX------XX----------XXX-XXXXXXXXXX-XXXXXXXX
XXXXXXXXXXXXX-XX---------XXXXXXXXXXXXXXXXXX---X-XX----------
-XXXXX----XX------XXXXXXX--XXXXXXXXXXXXX-XXXXXXX-XXXXXXXXXXX
XXXXXXXX-XXXXXXXXXXXXXXXXXX---XXXXXXXXXXXX-------XXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXX-XXXXXXXXXXXXXXX--------XXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXAIDDEEQKFQFVRIHKPPPLTPLYMGSRYIVSGS---------
ERMEIMPEXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXEQEDGTVVWETLVKPGDLKH
SATLVRP-----------------------------GPKASPSPPDAPGL
PF00619 // ENSGT00900000140813_0_Domain_19 (3051, 3135)
LHFVDRYREQ
LVARVTSVDPILDKLH-GQVLSEEQYEGVRAEATKPGQMRKLFSFSRSWDWACKDCLYQA
LKETHPHLILELWEMWSTGGDQRGFLKLGS------
ENSMMUP00000031438 (view gene)
ENSETEP00000004560 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MLRTLV
PF02758 // ENSGT00900000140813_0_Domain_0 (198, 332)
-KNDLCC---LATYLEELA--A---MDLKKFKLYLGT------
---------T--V-ELG--------E--GKI-PWGR-METAG-TLEMAQLLVAHYGSGKA
WLLALSI----FE-RINRKDLWERGQ-REDLVR-D-----APPGA---------------
------------------------------------------------------------
----------------------------------------------PSS-----------
LGTQSR-CLV-------------------------------EVSASSPRKXXXXXXXXX-
--------XXXX----------------XXXX--XXXXXXXXXXX---------XXXXXX
XXXXXXXXX--XXXXXXXXX----XXXXXXXXXX-----------XXXXXXXXXXX----
-----------------------XXXXXXXXXX-----XXXXX--XXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXX------------------X-XXXX-XX-XX-----XXXX
XXXXXX-XXXXXXXXXXXXXXXXXXXXXXXXXXX------XX-X-----XXXXXXXX-XX
-XX--XXXX-XXXXXXXXXXXXXXXXXXXXXXXXXXX-X-----XXX--XXXXX-XXXXX
XXXXXXXXXXXXXXXXXXXXXX--XX-XXX---------XXXXXXXXXXXXX-XXXXXXX
XXXXXXXXXXXXXLKQQLEGGG-LI-T--QT-SRTTTAVYMLYLLSLMSPKPGTP----T
----------RQSPPNQKGLCSLA---AEGL-WN-QK----VLFEEQ-------------
--DLRKHGVDGVDAS-AFLNT----NIFQRDIN-CE-KFYSFIHLSFQEFFAAMYYILD-
-RRES-----------------G--PGPDPSVS-RLLAEYGCS-ARSFLAPAVRFL----
----FGLLNGETRSYVEKSLGWR-----------------VSPR-IKA-DVLEWIRSKAQ
S-----------------------------------EGSTLQQGSLELLSCLYEIQE-EP
FIQQALSG---------------------F-------------------QVVVVGDVT-T
KM-EHVVA--S----FGVK-NCQS-AVA--VR-----------------LYGA----SFG
------AGGHDGEG-----GGPRSAEG--------------------IQASSAEPX-XXX
XXXXXXXX-XXX------------------------------------------------
------------------------------------------------------------
-------------------------XXX--------------------------------
--------------------XX----XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX-X
XXXXLKKCRVS---AA-ACD---DLAAAVAA-NQ
PF00560 // ENSGT00900000140813_0_Domain_8 (1775, 1805)
SLRRLDLSNNSLGL-PGMKLLSAG--
LKH-PGCRLQKI-Q----------------------------------------------
------------------------LRKCQLE-----------------------------
----------------------------AGACEEISS-----------------------
--------------------------------ALRSNQ
PF00560 // ENSGT00900000140813_0_Domain_11 (2019, 2057)
HLVE---LD-LTGNA-LED---
-------MGLRLLCQGL----KHPMC----------------------------------
--------------------------RLQ-TLW---------------------------
-------------------------------LKICRLTVASCGDLASA-LQQRL
PF00560 // ENSGT00900000140813_0_Domain_13 (2215, 2237)
SLRELD
L-SLNDLGDPGVL-LLCEGLQHP--NCK----L---------------------------
-----------------------QTLRV--SIC-----RLGSAACEGL------------
-------SAVL-QANHHLWELDLSFNDLGDVGMKL-LCEGL-RHP-AC-RLQKLW--VD-
--S-CGLTAQAGE--------------------DLSSALAANQTL-TELYLTNNALRDAG
VQLLCH--------RLSHPGCK--LR--V-------------------------------
---L-LFGM-DLNRKTHRA------L-----------PLR-VKKPDLDIGC---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSETEP00000004566 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------MAASSSSSFFSDFG---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 332)
LLLYLQELN--K---DEFKRFKLLLKE------
---------D--LEL-E--------H--HHV-PWTE-VKKAN-REDLANLMKEHYPGEKT
WSMTLKL----FG-KMNLKDLCKKAK-AEISXXXX-XXXXXXXXXX------XXXXXXXX
----XXXXXXXXXX-----------------------X-X-------------XXXX---
------------------------------------------------------------
-----------------------------------------XXXXXXXXXXXXXXXXXX-
--------XXXX----------------XX-----------XXXX---------------
----------------XXXX--------------X-X-------XX-XXX---XXX-XX-
XX--XX-XX----XXXXXXXXX--------XXX-----XXXXX--X---------XXXXX
X--------XXXX-XXXXXXXXXXXXR-------EMSFVQ-LIS--KW-----
PF05729 // ENSGT00900000140813_0_Domain_2 (774, 940)
PS-E-GA
IE-RIM-QP----RNLLFIVDSFDE-LNFAFEEP------EI-V-----LSNDRTLE-QP
-VSFLMSSLLRRVMLPEASLLVT-TRSTAYKKLRSLL-K-----THV--SIHLLGMPENE
MHEYI----------------S--QF-CAS---------KDWNINALTAIRN-NEILLNL
CQVPLVCRIICTALKQKMEKGV-NV-T--LT-CQTTTSLFTCFLGSLLTPAD-GSCPS--
------LPGQI----SLKNMCHLA---AMG-IWN-MT----YVFYKD-------------
--NLKKHGLK-LSDISIFLDM----NIFQKDSA-YS-DCYVFAHLHVQEFFAAMFYMLP-
GNWET--------------MKYS--SQSFENLK-KLLKS-KAY-EDTHLMQMKCFL----
----FGLLNEETVKQLENIFNCT---------------M--SLE-LI-------------
--------------KWDFLH-WVETL-------GNNLCPPPQSRFLELFHCLFETQD-KE
FIIQAMSH---------------------F-------------------QNIII---NIC
GKTDLLLS--S----FCL-KHCNS-LLK-IKIT----------------VT---------
----VAFE----KM---------------------------LDLKPR----DSTDH-DHI
THCWQDLF-SMLHRNRYLKELDLFQSLLDDL---------AVMSLY--RELQR-P-----
DC--R-LQKLL--L-RSVSYP--D----GCK-GLFRSL----------------TENQNL
IHLD-LKKS-DINYT-GVQSLCTALKHPNCK----LQIL--------XXXXXX-XXXXXX
X------X----------------------------------------------------
---------XX---XX-XXX---XXXXX------------------XX-XXXXXX-XX--
XXX-XXXXXXXX-X----------------------------------------------
------------------------XXXXXXX-----------------------------
-------XX---X--XX-----------XXXXXXXXX-----------------------
--------------------------------XXXXXXXXXX---XX-XXXXX-XXX---
-------XXXXX---XX----XXXXX----------------------------------
--------------------------XXX-XXX---------------------------
-----------------------------XXXXXXXFTSLSIE--YLL-TSINWSLAHLN
L-ASNYLK-GDEK-LLCDAFWHP--NCS----L---------------------------
-----------------------D---LELADC-----SLTSACCPDL------------
-------ASVL-SNNSNLSLLDLGKNDLQDNGVKI-LCEALK-HP-GC-HIQSL------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPTRP00000080469 (view gene)
ENSMOCP00000002645 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------------MLRG-AR
LALRVF----------------LKSPPGPL-------------------NTWQSV---WT
RR-LHCTP--T----MSLDIQCEQ-LSD-ARWT----------------EV---------
----LPLI-------------------------------QQYQVV------RLDDC-GVT
EVRCKDLS-SAIQA
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1542)
NPTLTELSLCTNELGDA---------GVCLVL--QGLRN-P-----
TC--K-IQKLS--L-QNCSLT--EA---GCR-ALPDVL----------------RSL
PF13516 // ENSGT00900000140813_0_Domain_6 (1618, 1642)
PTL
RELH-LSDN-PLGDA-GLKLLCEGLLDPQCR----LEKL--------QLE---------Y
C------N----------------------------------------------------
---------LT---AA-SCE---PLASVLRA-KANFKELVLSNNDLHE-AGVQTLCQG--
LKD-SACQLESL-K----------------------------------------------
------------------------LENCGLT-----------------------------
----------------------------SANCRDLCD-----------------------
--------------------------------VLASK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
ASLQH---LD-LGSNK-LGN---
-------AGIAALCSGL----LLPSC----------------------------------
--------------------------RLK-TLW---------------------------
-------------------------------LWECDITAEGCKDLCRV-LRA
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
KQSLKELS
L-AGNELRDEGAQ-LLCESLLEP--GCQ----L---------------------------
-----------------------ESLWI--KTC-----SLTAACCSHV------------
-------CSVL-T
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
KNRSLVELQMSSNPLGDSGVLE-LCKALG-QP-DT-VLRVLW--LG-
--D-CDVTDNACS--------------------SLAAVLLA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2465)
NRSL-QELDLSNNCMGDLG
VLQLME--------SLKKPSCT--LQ--Q-------------------------------
---LVLYDI-YWTEEVEDQ------LR----------ALE-EERPSLRIIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSGGOP00000016043 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------MAESDSTDFD---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWYLENLS--D---KEFQSFKKYLAR------
---------K--ILDFK--------L--PQF-PLIQ-M---T-KEELANVLPISYEGQYI
WNMLFSI----FS-MMRKEDLCRKI---IGRRN---------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------RNQEACKAV-
--------MRRK----------------FMLQ--WESHTFGKFHYKFFRDVS--------
----------------------------------------------SDVFYILQLAYDST
SC--YSANN----LN
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 930)
VFLMGERASGKTMVINLA-----VLRWI--K---------GEMWQ
N--------MISY-VVHLTSHEINQMT-------NSSLAE-LIAK-DW-----PD-GQAP
IADILS-DP----KKLLVILEDLDN-IRFELNVN------ES-A-----LCSNSTQK-VP
-IPVLLVSLLKRKMAPGCWFLIS-SRPTRGNNVKTFL-K-----EVD--CCTTLQLSNEK
REIYF----------------N--SF-FKD---------RQRASAALQLVHE-DEILVGL
CRVAILCWITCTVLKRQMDKG-RDF-Q--LC-CQTPTDLHAHFLADALTSEAGLT-AN--
----------QYHLGLLKRLCLLA---AGGL-FL-ST----LNFSGE-------------
--DLRCVGFTEA-DVSVLQAA----NILLPSST-HK-DRYKFIHLNVQEFCTAIAFLMAV
PNYLI-----------------P----SGSR-E--Y--KEKRE-QYSDFNQVFTFI----
----FGLLNANRRRILETSFGYQ---------------LPMVDS-FKW-YSVGYMKHLDR
DP-------------------------------------EKLTHHMPLFYCLYENRE-EE
FVKKIVDA---------------------L-------------------MEVTVYLQSDK
DM---MVS--L----YCLD-YCCH-LRT-LKLS----------------VQR-----IFQ
--NKEPLI-------------------------------------------RPTASQMKS
LVYWREIC-SLFYTMESLQELHIFDNDLNGI---------SERILS--KALEHS------
SC--K-LRTLKLSY---------VST-ASGFEDLLKAL----------------ARNRSL
TYLS-INCT---------------------------------------------------
------------------------------------------------------------
--------------------------------------------SISL-NMFSLLHDI--
LDE-PTCQISHL-S----------------------------------------------
------------------------LMKCDLR-----------------------------
----------------------------ASECEEIAS-----------------------
--------------------------------LLISGGSLRK---LT-LSSNP-LRS---
-------DGMNILCDAL----LHPNCTLISL-----------------------------
------------------------------------------------------------
------------------------------VLVFCCLTENCCSALGRV-LLFSPTLRQLD
L-CVNRLKNYGVL-HVTFPLLFP--TCQ----L---------------------------
-----------------------EELHL--SGC-----FFSSDICQYI------------
-------AIVIA-T
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NEKLRSLEIGSNKIEDAGMQL-LCGGLR-HP-NC-MLVNIG--LE-
--E-CMLTSACCR--------------------SLASVLT
PF13516 // ENSGT00900000140813_0_Domain_17 (2441, 2464)
TNKTL-ERLNLLQNHLGNEG
VAKLLE--------SLISPDCV--LK--V-------------------------------
---VG-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCHOP00000007146 (view gene)
ENSCPOP00000007980 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MSLEIRCEQ-LSD-TRWT----------------EL---------
----LPLI-------------------------------QQHQVV------RLQDC-GLT
EERCRDIS-SALQANPTLIELCLSTNELGDA---------GVHLVL--QGLQS-P-----
TC--K-IQSLS--L-PYCGLT--ET---GCG-ILPSVL----------------PSM
PF13516 // ENSGT00900000140813_0_Domain_6 (1618, 1642)
PTL
RELR-LSGN-SLGDG-GLRLLENSGT00900000140813_0_Linker_6_7 (1642, 1650)
SRGLLDAQPF13516 // ENSGT00900000140813_0_Domain_7 (1650, 1766)
CH----LEKL--------HLD---------Y
C------S----------------------------------------------------
---------LS---DA-SCE---PLASMLRA-KTNIKKLVMSNNDIHE-AGIRMLLSG--
LKD-SACPLETL-W----------------------------------------------
------------------------LENCGVT-----------------------------
----------------------------AATCKDLCD-----------------------
--------------------------------VVAA
PF13516 // ENSGT00900000140813_0_Domain_11 (2017, 2054)
KPSLQD---LD-LGNNR-LGD---
-------AGLAVLCSQL----LHPSC----------------------------------
--------------------------RLR-KLW---------------------------
-------------------------------LWECDITTEGCKNLCQV-LMA
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
KQSLKALS
L-MLNRLGDEGAR-LLCEALREP--TCQ----L---------------------------
-----------------------ECLWV--KEC-----GFTAACCPYF------------
-------REVL-A
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
QNKFLTELLLSENNLGNTGVQE-LCQALC-QP-GS-VLQVLE--LV-
--D-CNLTNSSCS--------------------NLALVLLA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2464)
CHSL-RELDLSNNGLGDPG
VLQLVE--------SLRQPSCT--LE--C-------------------------------
---LLLFDI-YVSDEVRDQ------LE----------ALE-KEKPPLRIIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSRNOP00000029443 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------MVDTSGYG---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLKHLRKLS--D---GEFWSFKELLRK------
---------E--PEKFK--------L--KPI-SWMK-IENAS-KEELVMLLNTHYPKQ-A
WDMALSL----FL-QVNREDLSIMAQ-KKRRY----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------KQTKYKKF-
--------MKTT----------------FQSI--WTLESNICIPDR--------------
---------------------------------S-------YHLIVEHQYRKLQNIFDSE
-----SE-P----VT
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 930)
AVVTGPTGEGKTVFLRKA-----MLDWA--S---------GILWQ
N--------RFQY-VFFFSVLSLNNTT-------ELSLAE-LISS-KL-----PES-SET
LNDILS-DP----KKILFILDGFDY-LKFDLE-L------RT-N-----LCNDWRKI-LP
-TQIILSSLLQKIMLPESSLLLE-LGHISLPKIFPLL-Q-----YPR--DITIQGFSERC
LKTYF----------------I--SF--FN---------TEKGIEVFENLKS-NQ-MLKL
CSNPYLCWMFCSCLKWQFDREE-EA-Y--FQ-AKTDSVFFTSFMVSAFKSAY-ASNPPK-
-----------QNRAQLKSLCTLA---VEGM-WK-QL----FVFDSE-------------
--DLRRNGISES-DKAVWLRM----KFLQNHD-----NHIVFYHPTLQLYFASMFYFLK-
QDKDT-----------------H--VPVIGSIP-QLLRKIYAR-DHTQWLQIGIFL----
----FGLATEQVASLLKPYFGFI---------------Q--HRD-VRQ-EVMRYLKSLSQ
REC----------------------------------CEKL-ERPQNLFACLRDNKE-EK
FVREVVDL---------------------F-------------------EEITVDITNSH
VL---SII--A----NHLQ-KSSK-LKK-LHLH----------------IQKR----VFL
-EIHDPEY-----------S-------------------D-S-------ETFT-QDKKNA
AEYWKKLC-HIFV---NLHVLDLDSCNFNKQ---------VIEELC--NVLSPSPKIPLM
AF--K-LESLLCSF---------MTN-FGD-GSLFHTL----------------LQLPHL
KYLN-LYGT-NLSNS-RIENLCSALRRSTCK----VEEL--------LL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------GKCDIS-----------------------------
----------------------------SEACGIIAT-----------------------
--------------------------------SLMNS-KVKH---LS-LVENP-LKN---
-------KGVMSLCEML----KDPSCVLETL-----------------------------
------------------------------------------------------------
------------------------------MLSYCCLTFIACGHLYEA-LVSNKHLSLLD
L-GSNFLEDTGVN-LLCEALKDP--NCI----L---------------------------
-----------------------KELWL--SGC-----FLTSECCEEI------------
-------SAVL-TCNNNLKTLKLGNNDIEDTGVKH-LCEALS-HP-NC-KLECLG--LD-
--L-CKFTSDCCE--------------------DLASALTTCKTL-NSLNLDWKTLEHSG
VVALCE--------ALNHKKCN--LK--M-------------------------------
---LGLDKS-AFSVESQTL------LQ----------AVE-KKNNNLSILHYPWVEE---
--ERK-----KR------------------------------------------------
--------------------------------------------------GVRLVWNSKN
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSAMEP00000004746 (view gene)
------------------------------------------------------------
------------------------------------------------------------
----------------SILLKNP------------L-------------------FRQ-N
L---S-SDCTKMKEAKLPSF
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
SNYG---LQWCFKQLS--K---EEFQTFKKLLME------
---------N--ASELA--------M--CSF-PWAE-VNNAG-VEHLATLLHEHYKESLA
WKISIHI----FD-KMYLPTLSEKAR-EEMKKYSL-TEIPENSTRMKN--DQEPIMKEVP
GQC-RHQSSEA-KTKRERCRLG------------------------------IPQALE--
QG-GATAA---------ETKE-QVSFPRALE-----------------------------
----SSPRTE-------------------------------SPLSSLGHGGNMGDYKGY-
--------VMTK----------------FATK--LDEY-PGFQKL---------------
-----------------------------------AS-------HCPEM-QTLLAAFDPD
Q---RGFQP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
HTVVLHGKSGIGKSTLARRV-----LLYWA--R---------GELYR
G--------MFSY-VFFLRARDMQCMR-------EASFAE-LISR-EW-----PD-SQVP
MMEIMS-QP----ERLLLLIDGFDD-LDFAFPDD------DT-N-----LCEHWADK-QP
-TSVVMCSLLKKVLLPESYLMIT-VRDAGIEKLKSVL-T-----SPR--YLFVGGISVEK
RIQLL----------------L--HH-MKN---------EHQRTQILHSVVD-NHPLFDE
CQVPVVGSIICEALNLQEASGK-SL-P--PT-CQTLTGLYATFVFHQLTPRD-ASQRCLN
----------QEERAVLKSLCLMA---VQGV-WN-RR----FVFYGD-------------
--DLSVHRLQES-ELSALFHM----NILLQDGG-HGRC-CTFPHVSLQEFCAALYYVLE-
GLEME--------------GNPY--PVLMESLR-TLVELKEIN-SNAHLLQMKRFL----
----FGLMSKEVTGALEVLLGRR---------------V--RL-AVKQ-RLLSWISLLGQ
QAS----------------------------------AP-APPDLLDAFHCLFETQD-DE
FVCSALNG---------------------F-------------------REVWLPINRRM
DL---MVS--S----FCLQ-RCRY-LQK-IRVD----------------VHE-----SFS
-KDEFTDA----QT--------------------LI-----------PQ--GMPTR-TLA
HECWDSLC-FVLGTHPSLRQLDLSGSVLNER---------AMKTLC--ISLRQ-P-----
TC--K-VESLI--F-KGAQV---TR---GLR-HLWITL----------------ITNRNI
KYLN-LEST-HLKAA-DLTMACEALRHPNCL----LESL--------RLD---------H
C------G----------------------------------------------------
---------LT---PA-CCL---VISQILVT-SISLKFLSLVGNEVAN-QGIKSLCDT--
LTA-SQGTLQKL-V----------------------------------------------
------------------------LGNCGLT-----------------------------
----------------------------AAGCQDLAS-----------------------
--------------------------------ALISNQSLTH---LC-LSSNS-LGS---
-------EGVNLLCRAM----KFPNC----------------------------------
--------------------------GLQ-RLM---------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSANAP00000025829 (view gene)
ENSCLAP00000001546 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MTS---------------------------KQERKDMEAA-----
--------CSWS------------------------------------------LLPR--
-----------------------------------------E------------------
-------------------------------------------------KDYRELYKKH-
--------LREK----------------YLTI-GVDKCYHHCK--------------HIK
SRMLLVKEHAVSEKTPYGVP-------------------------QQDHFLEIEHVFDPN
EE--GSSSS----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 928)
RTVVLQGYPGTGKTALVHKF-----MFDWA--S---------GMVTP
G--------RFDF-LIYVNCREISHTV-------MLSAAD-LISS-AF-----QD-INGP
KLDIILTCP----EKLLFILDGFPE-LQYPVCDQ------QG-G-----LSANPWER-KP
-VETLLCSFVRKKLFPESSLLIT-ARFTAMRKLHSLL-K-----QPI--QAEILWFTDAE
KRAYF----------------L--SQ-FLD---------PNAAMEAFYGLRE-SEGSDIL
SSLPLVSWMLCSVLQSQGDGDS-RL-L--RS-LQTLTDMYLFYFSKCLKSLKGIS-VW--
-----------KGQSCLWGLCCLA---AEGL-QK-HQ----VLFTVS-------------
--DLRRHGVGVYDINGPFLTH-----FLKKAEG-CD-NIYTFLHFSFQEFLTAVFYALK-
NDNNC---------------------LFCDHMGKIWREIFQQY-GKGFSSLMIQFL----
----FGLLHKGKEKTLETTFGRK---------------V--SQG-LRG-ELLKWTEGEIK
DE-----------------------------------SSRLQIESVDLFHCLYEMQE-EE
YAEKIIDN---------------------L-------------------QSIMFLQPTYT
KM-DLLAM--S----FCVK-SSNSHLS----------------V-----SLKCQHLLGF-
-EEED-DS----------------------VFMP--------------------------
---------PTLTISQSFQ-----LCSLPVS---------RLHLLR--QALCNP------
YC--K-IKDLKL---IFCHLT------ASCGRD---------------LSSVFET-NHSL
TDLE-FVKS-TLEDS-GVKQLCEGLKQPNCI----LQALR--------LNRCLI-SPA-S
CG------------AL----AAVLSTSQWLTDLEFTETKLEASALKLLCEGLKDPGCK-L
QRLKLCDSLLPESSEA-VCN---YLASVLIC-
PF13516 // ENSGT00900000140813_0_Domain_8 (1773, 1796)
NPYLTELDLSENPLGD-KGVKYLCEG--
LRH-SNCHVEKL-E----------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------LSECSLSATCCESLAQI-LSSSRTLTRLL
L-INNKIGDLGLK-LLCEGLKQP--DCQ----L---------------------------
-----------------------RDLAL--WTC-----HLTGACCQDL------------
-------CNAL-Y
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
TNEHLRDLDLSDNALGDVGMQV-LCEGLK-YP-CC-KLQTLW--LA-
--E-CDLTDVCCR--------------------ALASVLYR
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2465)
NENL-MLLDLSGNDVKDFG
VQMLCD--------ALIYPTCK--LQTFY-------------------------------
------IDVDHLHEDTFRK------LE----------TLK-MIKPGINW-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSP00000301295 (view gene)
ENSCGRP00000007759 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------MGNPS----QQQFN---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 323)
LKDILKSLN--R---DDLDNFKCVLKN------
---------M--SLPRT--------L--KQI-PQII-LDMAD-GVHLAEILHEYCPSGWV
ETVTMEI----FQ-MINREDLADLII-FHNNQ-----LCS----I---------------
------------------------------------------------------------
------------------------------------------------------------
---------Y-------------------------------PSLPI--TDGKEHKHKKE-
--------WNKV----------------FRNK--WKFNFWPKCKENLYV-----------
---------------------------------------------ETESYRTLVTLCNPK
A---TMPFA----H
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 931)
AIVLHGPPGSGKTTVANRL-----MLQWS--E---------SKQAQ
---------MFPC-AFSISCREVNVTN-------SCTFAQ-LLSL-KN-----DH-WRDY
VK-SLD-LA----ENFLFVVDGFDE-LRVPAG------ALLQ-D-----ICSDWDTE-KP
-AAVLLSSLLKRKMAPHATLLVT-TQTQALRQIYLMI-D-----QPF--LIENQGFSEQD
IQEYF----------------K--RF-FKDEEE----D-EEKALRALNALRC-NAALFHM
ASIPAACMIFCLCLEKVMEKGE-DL-A--LT-CQSHTSMFLNFLCRMFSPETYERHLKKE
------------FQISFKKTCFLA---ADSL-LE-QL----PTFSKE-------------
--DILKLQPDMK-GLHPFVCR----YIINNDS--YN-NSCSFIYIRIQQLLAAIVFVQE-
LE--------------------K--SVSKYSMQ-NMLSKEARL-KNPNLSGVLLFV----
----FGLLNETRIQELATIFGCQ---------------I--SIS-VKR-KFLECE--SGK
NK-----------------------------------PFLLLMDVQDILSCLYESQE-EG
LVKEAMAL---------------------L-------------------EEISLHLKTNM
DL---MHA--S----FCLK-NSQN-LRT-MSLQ----------------VDKG----IFP
-END-AVL----ES--------------------TAWHQG------------SQND-QCL
LTFWTDFC-DMFNSNEKLVFLDISESFFSSS---------SLQILC--DKLSS-S-----
AC--H-LQKVI--L-KNISPA--DA----YK-KLCLTF----------------YGYETL
SHLT-LKGD-NLST--MLPSLSMVVKHPSCK----LKFL--------SLS---------S
C------S----------------------------------------------------
---------TT---IH-KWN---EFFSALKV-SQSLTCLDLTDNELLN-KGIWLLCKT--
WKQ-PNSILQRV-S----------------------------------------------
------------------------LENCHLT-----------------------------
----------------------------EACCKGLAS-----------------------
--------------------------------VLMV
PF13516 // ENSGT00900000140813_0_Domain_11 (2017, 2051)
SQTLTH---LN-LAKNT-LGD---
-------NGVKILCESL----SHPEC----------------------------------
--------------------------KLQ-TLV---------------------------
-------------------------------LWSCDITSDGCCHLSKM-LRQASSLEHLD
L-GLNYIRSVGAR-FLCEALKEP--QSN----L---------------------------
-----------------------KSLWL--YGC-----SMTPVNCKDF------------
-------SKIL-R
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
NNKSLNTLDLAQNSLGTDAVKT-FCEDLK-LR-IC-PLQTLR--LK-
--F-DDANPTIQK--------------------LIQNMKESHPQLTISSDQP----DPRK
---------------KEP-LPH--FI----------------------------------
----F-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCHIP00000029121 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------MF--------
--------------------------------------------QLGII-----------
-------AFT-------------------------------SMLSPCTSIQVKRRYRNT-
---------MEK----------------F-SV--WKDILWSGDDGGFHYNTT--------
-----------------------------------------------QRSPKLITYLNPQ
AP--TQSTP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TMVLHGPAGVGKTTLAKEL-----MLDWT--Q---------DDLAE
---------TSTS-AFYLSCKGLNHRG-------TCTFAE-LISA-NW-----PH-VQDD
IPAILA-RA----QKVLFILDGFDE-LKVPSG------ALIH-D-----LCGDWKKQ-KP
-VPVLLGSLLKRKMLPKATLLIT-TRPGALQELRLLT-E-----QPL--FIEIEGFLEED
RKAYF----------------L--KH-FEE---------ESQALRAFDLMKS-NAALFQL
GSAPSVCWLVCTCLRQQMERGE-DP-A--AT-CRTTTALFLRFLCSRFPPP-RGGGPRRG
------------LQAPLKPLCLLA---AEGV-WT-RS----SVFDGE-------------
--DLRRLGVHPS-ALSPFLDG----NILQKSED-GE-ACYSFIHLSVQHFLAAMFYVLE-
PEEQEE---E--------GLGGC--WWHVEDVG-KLLSKAERL-KNPSLTHVGYFL----
----FGLCNERRAMELEATFGCL---------------V--STE-IKR-ELLKYTLMPHG
KK------------------------------------SFSVMDTKEVLSCLYESQE-EQ
LVKDAMAH---------------------L-------------------KEMSIHLTNTS
EI---MQS--S----FCLK-HCAN-LQK-ISLQ----------------VGKK----IFL
-END-PAL----KS--------------------D------------PQNEISQHD-HRS
LQLWADLC-SVLSSNENLNFLDVKESFLSHS---------SVRILC--EQITH-V-----
TC--H-LQKVV--I-KNVSPA--SA----YR-DFCLAF----------------IGKKTL
THLA-LEGS-VHGDKMLLLLLCEILKHPRCN----LQYL--------RLG---------S
C------S----------------------------------------------------
---------DT---TW-QWA---DFSSALKV-NQSLKCLDLTASEFLD-AGVKLLCLT--
LRH-PKCVLQKL-S----------------------------------------------
------------------------LENCQLT-----------------------------
----------------------------EACCKELSS-----------------------
--------------------------------ALIV
PF13516 // ENSGT00900000140813_0_Domain_11 (2017, 2054)
NQRLTH---LC-LAKND-LGD---
-------GGVKTLCEGL----SYPEC----------------------------------
--------------------------QLQ-TLV---------------------------
-------------------------------LSHCNINRHGCKYISKL-LQGDCSLTSLD
L-GFNPIA-TGLC-FLYEALKKP--NCN----L---------------------------
-----------------------KCLGL--WGC-----PITPFSCQHL------------
-------ASAL-VSSRSLETLDLGQDAWGQSGIVV-LLKALK-QN-HG-SLKMLR--LK-
--M-DKSTVEIQR--------------------LLKDVKENNPRLTIECNDA------RT
---------------TRSSCCD--FF----------------------------------
----C-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPCAP00000003633 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------LVTLPTGS-TSILPKYL----------------LFP-----------------H
N---LSCQPIMLEEVKSSSF
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
SIYG---LNWCLMELD--K---DEFEMFKEFLKE------
---------E--SSKAM--------M--DLL-PWSE-VKNAD-VECLASLLHEYYMVPLA
WEISIHI----FE-RMKLSEVSEIAK-NALKAEDY-EQIEIYPQPS------LGEVPXXX
----XXXXXXXXXX-----------------------X-XXXXX---XT----PQTME--
QDDTS----------AAEIKGQX-XXXXX-------------------X-----------
------XXXX-------------------------------XXXXXXXXVDDKWGYRAH-
--------VMTR----------------FT-----------KELD---------------
----------------AYHD------F-----QEF-A-------TDWSQIQSLADAFNS-
DR--RG-QP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
LTVVLHGKAGTGKSTLVKRI-----MLHWA--Q---------GQIYQ
G--------MFSY-VFFLHAREVQHMR-------EGSFTE-LISK-GW-----PD-SCAP
MGEIMS-EP----ERLLFVIDDFDD-LDFVLQDC------PE-K-----LCDNCAEN-QP
-ISVLMRSLLNKVLLPESSLLIT-VRDVGIGKLTSMV-V-----SPR--YLFIGGIPVER
RMQLI----------------L--QR-IAD---------EQQQTQ-SQRVVV-NSQLFDE
CQVYKMCWLTSVAMQLXXXXXX-XX-X--XX-XXXXXXXXXXXXXXXXXXXX-XXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX-XXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXX-XXXXXXXXXX----X----------------XXXXXXX------XXXX-
XXX------------------------------------------XXXXXXXXXXX----
----XXXXXXXXXXXXXXXXXXX---------------X--XXX-XXX-XXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXX-XXXXX-------XXXXXXXXXXXXXXXXXXXXXXXX-XX
XX-XXXXX---------------------X-------------------XXXXXXXXXXX
XX-X--XX--X----XXX-XXXXX-XXX--------------------------------
------XX----XX---------------------------XXXX------XTKPK-TLV
TVLWQDLC-TVLSTHETLRVLDLSGSVLNEW---------AMRILC--SKLKH-P-----
TC--K-IQTLX--X-XXXXXX--X----XXX-XXXXXX----------------XXXXXX
XXXX-XXXX-XXXXX-XXXXXXXXXXXXXXX----XXXX--------XXXXXX-XXXXXX
X------X----------------------------------------------------
---------XX---XX-XXX---XXXXXXXX---------XXXXXXXX-XXXXXXXXX--
XXX-XXXXXXXX-X----------------------------------------------
------------------------LGNCGLT-----------------------------
----------------------------AAGCGDLAS-----------------------
--------------------------------VLITNQNLTH---LC-LSDNS-LGS---
-------EGMKLLCGAI----KNPNC----------------------------------
--------------------------GLK-RLI---------------------------
-------------------------------LSQCNLDVTSCGFLAFA-LMKNRRLTHLT
L-SMNPLEDNGIQ-VLCEVMKES--SCY----L---------------------------
-----------------------QDLEL--VRC-----HLTSACCEDL------------
-------SCVI-SRNS
PF00560 // ENSGT00900000140813_0_Domain_15 (2357, 2381)
RLKSLDLAYNSLGDSGAAM-LCKGLK-HK-HC-SLRRLG--LE-
--A-CGLTLDCCE--------------------ELSLAIYHNQ
PF00560 // ENSGT00900000140813_0_Domain_17 (2444, 2475)
SL-TSLNLMQNHLGPLG
ISKLYS--------ASACPKSN--LQ--I-------------------------------
---IGLWRG-CYPEQTKNL------LE----------ELK-RVKPQIVIDDN--------
------------------------------WYSFDQ------------------------
--------------------------------------------------DDRYWWQH--
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSRBIP00000003237 (view gene)
ENSMICP00000044792 (view gene)
ENSMLUP00000020634 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LS-VGIKEKYQNQ-
--------RRES----------------SCPTQSW-------RNE---------YLHQKI
TQLLFLHRYQPRGYDF---L------LRRR-NHEV--------VDEQENWMEIGDLIGPG
LS--TQEEP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
PTVVVHGVAGIGKSTLARQV-----RRAWE--E---------GWLFK
D--------RFKH-VFYFNCRELAQSE-------TMSLAE-LITK-DW-----AA-PDAP
IGQILS-QP----EQLLFILDNLDE-PKWDLKGK------SS-E-----LCPHWSQQ-QQ
-VHTLLGSLLKKTLLPTASLLIT-ARITALRKLIPSL-K-----HPR--WVEVLGFSESA
RKEYF----------------F--QY-FTD---------ESQAIRAFTSVES-NPALFTM
CLMPLVSWLVCTCLKQQMEQEQ-PL-S--LT-SQTTTALCLHYFFHVLQ--------AQ-
-----------SLGTKLRGFCSLA---VEGT-CQ-GK----TLFSPE-------------
--DFREHGLDGA-NISTLLNM----GVLQEH---PTPQSYSFIHLCFQEFFAAMSYALG-
D------GVL--------KGDCF--NNI-IITI-DLTGVYDRH--DLFGEPTTCFL----
----FGLLSDQGMREMESIFKCS---------------L--SRK-RLP-SLLHLAKREVL
-----------------------------------LKRLNLDPYSWHLLHCFYEIQD-ED
FMKNIF-I---------------------P-------------------FTLRICVQTDM
EL---LLF--T----FFFT-FCHR-IER-LQLN----------------ESGQ-------
------H-R---------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------Q------------
------AGRT--------------------------------------------------
-------------------SGAFLYSWGPFT-----------------------------
----------------------------DSWWKMFFS-----------------------
--------------------------------ILKVPR
PF00560 // ENSGT00900000140813_0_Domain_11 (2019, 2057)
SLQE---LD-LSGNS-LSC---
-------SAVQSLCDAL----KSPQCRLATLR----------------------------
------------------------------------------------------------
-------------------------------LVSCGL-----------------------
------------------------------------------------------------
----------------------------------------TY------------------
------------------------------------------------------------
---------NCCQ--------------------DLASMLGASP
PF00560 // ENSGT00900000140813_0_Domain_17 (2444, 2463)
SL-TELDLRHNDLGDLG
VRLLCE--------GLRQPACQ--LR--K-------------------------------
---LR-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPPAP00000019584 (view gene)
ENSCAFP00000015730 (view gene)
ENSOCUP00000002291 (view gene)
------------------------------------------------------------
------------------------------------------------------------
----------MASP----------------------SSRPCPPGSS-----------G-A
G---LSETNTLEKQMAMLDLTD
PF02758 // ENSGT00900000140813_0_Domain_0 (203, 330)
LD---LLWFLQTLS--D---KEFQSFKDCLEH------
---------D--PLALG--------L--TQL-PLLP-LRRAG-REELVDILTATYEAQDT
WMMAGNA----FD-KINRIDLCERI---RRRRR---------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------RHEEIDKVV-
--------VGRK----------------VLYQ--WEQCPFAEVHNYFYGEIA--------
----------------------------------------------INTLHVLQNSYTSN
SD--YSAKN----LN
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 930)
VFLFGEGAIGKTMVIRMA-----VLGWI--K---------EEIWK
D--------TISY-VVYLTSHEINQIT-------CCSLVE-LISK-DW-----PG-GYAP
VADILA-YS----KHLLFILEDLDN-INCSLNVD------EL-A-----LCSDSRQQ-VP
-MSVLLASLLQRKMAPGSWFLIS-SRFGGETAMRTLM-K-----HSD--YCLTLQFSDER
RKLYF----------------T--LF-FRN---------EQKVMTACRLVHD-NEMLISL
CQVPALCWVTCTALSRHLGKG-DDI-K--LS-YQTHTDVFTHFLANTLTPEAGAT-GG--
----------QHRLVLLERLCTLA---VEGL-LH-DT----QNFSDE-------------
--DLRSVGLTED-DVSLVQAR----KILVRCSN-IQ-DRHTFIHLKVQEFCAAIAYMMIL
NSCQI-----------------P----SASR-K--L--KEKRE-TYPDFSPVTVSI----
----FGLLNEKRRKILETSFGCQ---------------LL-VEP-LRQ-YFRQQMECLSN
NP-------------------------------------KAMEYHMPVFYCLFENQE-EE
FVKQIMGF---------------------F-------------------LQATVNIRTSR
DL---MVS--S----YCLK-HSQT-LQR-LKLG----------------VLF-----VFK
--HKDPNV-------------------------------------------KLTSRQVRN
LAYWRDIC-SLLHTNENLRELELCNSDLHST---------SERILC--KSLTHA------
SC--R-LQTLKLSN---------LAV-GSEFDDLFRAI----------------AQIKSL
TSLS-LNRM---------------------------------------------------
------------------------------------------------------------
--------------------------------------------DMSP-KMFSLLHEI--
LDS-PACNLQHL-C----------------------------------------------
------------------------LMKCDLK-----------------------------
----------------------------ASACRDVAS-----------------------
--------------------------------LLVDSKKLKK---LT-LSNNP-LKN---
-------DGVKILCDAL----LHPNCILESL-----------------------------
------------------------------------------------------------
------------------------------VLFFCCLTEACCSSIGRV-LLLSKTLKHLD
L-SVNFLQNHGVL-VLTLPLMFP--TCK----L---------------------------
-----------------------QELEL--SGC-----FFNGDVCRDI------------
-------ASAIV-NNQNLRSLELGSNYLGDAGVTL-LCEALR-HP-SC-KLENIG--LE-
--E-CMLTSASCE--------------------SLASVLRSSKTL-KRLNLLGNKLGDEG
IVQLVQ--------GLGEPSCV--LQ--T-------------------------------
---LGLEMS-NVGEETQKL------LQ----------AVK-EKNCSLEFVSQSWAKREGR
GAGGR-----TD------------------------------------------------
-----------------------------------------------------CPSPSEP
SFA-----------------RQKEAWH---LI--TSRMPRRFRPLEGSAAGSGQPP----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMICP00000045945 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---SSFFSDFG---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 323)
LIWYLEELK--K---DEFRKFKEHLKQ------
---------E--TLQLG--------L--QQI-PWAE-VKKAS-REDLANLLIKHYEEQQA
WDITLKI----FH-KISRQDLCDKVL-RET------------TG--------------NT
K---------------------------------------------------IYQD----
------------------------------------------------------------
----------------------------------------------------------H-
--------IKKK----------------FDDL--WSSKSITGVYAY--------------
---------------------------------I-------CSEARQEECDYMELIFAPR
ET--GKQ-P----Y
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TLVFEGLQGVGKTTFLMKA-----MLTWS--E---------SSVYQ
S--------RFLY-VFYFCCRELKQLT-------ETSLAE-LISR-DW-----PQP-SAP
ITEIMS-QP----ERLLFIIDSFEQ-LKCGLSQP------ES-E-----LCSDWTKT-QP
-VRVLMSSLLRRKMLPESSLVIA-VAPRCTVELQDRL-V-----CPE--FLIFPGFREID
MQAYF----------------C--CL-FQD---------RNRAMEAFSFVRE-NEQLFIM
CRVPLLCWIVGTCVKREMEKGK-DL-T--WT-CRRVTSLLASFVFHMFTPKG-ADCPSQ-
-----------QSQFQLKGLCSLA---AEGM-WT-DL----FEFSEQ-------------
--DLRRNGIVDS-HIPALLDT----NILVKC---TE-RSYMFLHMSIQEFCAAMFYLLN-
SDGVH-----------------P--NPAVRCRE-LLLFTSIKKARKLHWISLGSFT----
----FGLLNEREQQKLDAVFGFQ---------------L--SRE-TKQ-RFLQCLAYLGD
H-E----------------------------------DLQEQTDFLALFYCLFEMQD-EA
FVRQAMDF---------------------L-------------------QQVNFSIFENT
DL---VVC--A----YCLK-YCSG-LRK-LRFA----------------VQN-----IFK
-EEN-GQH-----------S-----------------------------------MPSCS
LINWHHVC-SVLATNEHLEKFKLVNSSLNES---------AMVTLC--NQLRLP------
SC--R-LQKLELNT---------VSF-SGKSLAFFEVL----------------SYNAHI
KNLK-LIKM-KLSRE-EVKLLCYGLACSVCN----VETL--------ML-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------GSCSLL-----------------------------
----------------------------PEDCAMFAV-----------------------
--------------------------------VLDVNKKLKH---LN-LTCNH-LD----
-------QGLGILCEAL----CQPDCVLEFL-----------------------------
------------------------------------------------------------
------------------------------VLAYCYLSEPCWEYLHEA-LRD
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
NKSLRHLD
L-SVNTVKDEGVT-LLCEALRRP--DCC----L---------------------------
-----------------------QSLCL--VKC-----SFTAAGCQDL------------
-------ASVL-I
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
SNQNLRNLQIGFNDIGDVGVKV-LCEALT-HP-NC-RLEKLG--LQ-
--A-CELTSACCK--------------------DLSSVLTSSKTL-WRLNLAWNALDHSG
VVVLCE--------ALRHPDCA--LR--T-------------------------------
---LVLNKA-EFDEETQMF------LM----------TEE-ERNPHLSIIVE-QSRDGTM
GVDT--------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPCOP00000013110 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MSLDIQCEQ-LSD-TRWK----------------EL---------
----LPRI-------------------------------QQSQVV------RLDDC-GLT
EVQCRDIS-SALRA
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1543)
NPALTELNLRTNELGDA---------GVRLVL--QGLQS-P-----
TC--K-IQKLS--L-QNCCLT--EA---GCG-VLPGVL----------------RSV
PF13516 // ENSGT00900000140813_0_Domain_6 (1618, 1642)
PTL
RELH-LSDN-PLGDA-GLRLLENSGT00900000140813_0_Linker_6_7 (1642, 1650)
CEGLLDPRPF13516 // ENSGT00900000140813_0_Domain_7 (1650, 1766)
CH----LERL--------QLE---------Y
C------N----------------------------------------------------
---------LT---AA-SCE---PLASVLRA-KPDFKELMLSNNDLGE-AGVRVLCQG--
LQD-STCQLESL-K----------------------------------------------
------------------------LESCGVT-----------------------------
----------------------------AANCRDLCG-----------------------
--------------------------------VVASK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
ASLRE---LD-LGSNQ-LGD---
-------AGLAELCSGL----LRPSS----------------------------------
--------------------------RLR-TLW---------------------------
-------------------------------LWECGITAEGCRELCRV-LRAKE
PF13516 // ENSGT00900000140813_0_Domain_13 (2215, 2237)
SLKELS
L-SGNELGDAGAR-LLCESLLEP--GCQ----L---------------------------
-----------------------ESLWV--KTC-----GLTAACCPHF------------
-------SSVL-A
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2377)
QNKSLLELQMSSNKLGDAGVRE-LGQGLR-QP-SS-VLRVLW--IG-
--D-CEVTDGGCG--------------------SLASTLLA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2464)
NRSL-RELDLSNNCMGDPG
VLQLIE--------SLRQPACA--LE--Q-------------------------------
---LVLYDI-YWSEEMDNQ------LR----------ALE-EDKPSLRIIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSLAFP00000022456 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----SSKP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
HTVVLQGPAGVGKTTLAKKL-----MLDWT--R---------DNFGK
---------AFKY-AFYLSCKELNHMG-------PRTCAE-LISK-DC-----PE-LQDA
ILEVLD-RS----QRILIIVDGFDE-LRFPPG------ALIS-D-----MCGDWKKQ-KP
-VAVLLSSLLTRRMFPKATLLIT-TRPWAMRELWFLV-E-----RPF--FIDIEGFLEVD
KKEYF----------------L--KH-FQD---------EDQALRAFNLMRG-NTALFSL
GSAPMVCWVVCTCLKLQMDKGE-DP-T--PT-CQTTTSLFLRFICSQFMPV-PDSCPDQC
------------FQAPVKALCLLA---AEGV-WT-QT----SVFDGE-------------
--DLERLGVKEC-ELSPFLDK----NILQKVKG-CE-GCYSFIHLSVQQFFAAIFYVLG-
SEEEK----E--------DRESH--REDIGNIQ-KLLSKEERL-KNPSLPQVGYFL----
----FGLSNEKRTRELETTFGCQ---------------L--SID-VKQ-ELLRWKVKSN-
EN-----------------------------------KPFSVLDMKDVFYCFYESQK-EQ
FVKDAMAH---------------------F-------------------REVSLKLKNET
DI---VHA--S----FCLK-HCQN-LEK-ISLK----------------YSS--------
-----------------------------------------------SLPHRSQRD-YRI
LPLWVDLC-SVVNLNKNLSVLDVSDSFLSDT---------SVRILC--ERMAR-V-----
TC--N-LQKVV--I-KNVSPV--DA----YR-DFCLAL----------------VGKTTL
THLI-LEGS-VHSEK-MLLLLYEALRHSKCN----LQFL--------RIE---------M
C------SSTCSVTTYM-ILSCFLTHKTKLVRIINRRSFQLSLSLQYLNSTNFGPDCLDI
SNFLLGSCAAI---AE-EWE---CLFSGLEL-NQSLTCLNLTDSELSV-EDVKLLCAA--
LKH-PNCSLRML-S----------------------------------------------
------------------------LENCHLT-----------------------------
----------------------------EASCKDLSS-----------------------
--------------------------------ALIINQKLTH---LS-LAKNE-LGD---
-------GGVQLLCEGL----SYPDC----------------------------------
--------------------------KLQ-TLV---------------------------
-------------------------------LWGCSITTLCCQDLASA-LINNE
PF00560 // ENSGT00900000140813_0_Domain_13 (2215, 2240)
SLETLD
L-GENILGQSGVT-VLLEALKQK--KGF----L---------------------------
-----------------------QKL----------------------------------
------------------------------------------------------R--LT-
--I-DESNVETQK--------------------LLEELKERNPQLTIECSD---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSGGOP00000011186 (view gene)
ENSEEUP00000014333 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------AAME----S
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
VRCK---LTRYLEDLG--D---IDFKKFKMHLED------
---------Y--PPEKG--------C--CPL-PRAK-TEKAN-HMDIATLMIDFNGEQNA
WDMALCI----FH-AINRRDLHEKAM-SDRPDWVPGNAQ--VS---------ESSVE---
--------ER-----------------------------------EESL-----------
-----------------------------------------------------EEEWKG-
------------------L-----------------LGYLSRISSCKKKKYYCKKYRKH-
--------IRSR----------------FQCI--KDRNARLGESV---------NLNKRY
TRLRLIQEHRSQQEREHELL------TIGRDCAKTLD------SPEFLSSTNVELLFE--
PDDQHTEPV----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
HTVVFQGTAGIGKTMLARKI-----MLDWA--S----------YMYQ
D--------RFDY-LFYIHCREVSLGA-------RRSLGD-LIVS-CC-----PD-PHPP
VGKIVS-KP----SRLLFLMDGFDE-LQGAFDER------TE-A-----LCTDWQKV-ER
-GDILLSSLIRKRLLPSASLLIT-TRPVALEKLQHLL-D-----RPR--HVEILGFSEAK
RKEYF----------------F--KY-FSD---------EHQAREAFTLTQE-NEVLFTM
CFIPLVCWIVCTGLKQQMDSGK-SL-EQ--T-SKTTTAVYTFFLSSLLQTRRPSQ-----
------------VSATIRGLCSLA---ADGI-WN-QK----ILFEEC-------------
--DLRHHGLQRA-DVSSFLRM----NLFQKEVD-CE-KLYSFIHMTFQEFFAAMYYLLE-
EEEE-------------VLWSCP--ELPSRDVT-VLLENYGKF-EKGYLIFVVRFL----
----FGLLNQERACYLEKKLSCR---------------L--SQQ-IRA-ELLQWIQVKAK
AK-----------------------------------KLQTQPSQLELFYCLYEMQE-ED
FVQEAMSY---------------------F-------------------PRVEVHLATRM
---DHVVS--S----FCIE-NCRS-VGS-LSLR----------------LLHS-----SP
KEEEEEEE-----D------------------------------------TR---H----
---------------AGV---NLHACCPRXX-----------------------------
------------------------------------------------------------
-------------------------XXXXXX----X-----------XXX---------X
X-----------------------------------------------------------
----------X---XX-XXX---XXXXXXXX-XXXXXXXXXXXXXXXX-XXXXXXXXX--
XXX-XXXXXXXX-X----------------------------------------------
------------------------XXX-XXX-----------------------------
----------------------------X---XXXXX-----------------------
--------------------------------XXXXXXXXXX---XX-XXXXX-XXX---
-------XXXXXXXXXX----X-XXXXXXXX-----------------------------
--------------------------------X---------------------------
-------------------------------XX---XXXXXXXXXXXX-XXXXXXXXXXX
X-XXXXXXXXXXX-XXXXXXXXX--XXX----X---------------------------
-----------------------XXXXL--VNS-----RLTAGCCPAL------------
-------TSVL-SSNQNLTHL-LRGNALQDRGVEL-LCEGLL-HP-SC-RLQLLE--LD-
--G-CSLTGHCCQ--------------------DLSKLLTAHHSL-RTLSLGANNLGDQG
IKLLSE--------VLRQQGSH--LR--N-------------------------------
---L--------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSNGAP00000019592 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------MKSEDVEAIKE------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------DHGDEEAMLGDRADYRTQ-
--------IKEK----------------FCIT--WNKTSLLGESADFNYGIT--------
----------------------------------------------QKNRKLLEHAFDVN
VK--THKMP----K
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 929)
IVLLLGAAGIGKTTLLRKV-----MVDWA--E---------GNLYH
---------QFTY-VFYLNGREISQLK-------ERSFAQ-LISK-DW-----PS-TEIP
IEKILS-QP----SSILFIIDSFDE-LNFAFEEP------EF-A-----LCGDWTQE-HP
-VSFLMSSLLRKVMLPESFLLVT-TRITTLNRLHPLL-K----K-PL--SVRLKELCKNA
RMDYI----------------H--QF-LKG---------KTWATNAARSIRK-NARLFSM
CRIPLVCWLVCTSLKQQVEKGG-SV-A--ET-CQTTTALFTCYICSLFPRV-DGHSANLP
------------NETVLRSLCQAA---ANGI-WT-MT----HVLYRD-------------
--SLRKHELTRS-DISVFLEA----NVLQKDTE-CE-NCYVFTHLHVQEFFAAMFYLLR-
DNLEG--------------VDYP--FQLFENLN-VLLQSEHYH--HPHLAQMKCFL----
----FGLLNKDRIKQLEETFNIT---------------I--SLE-IKK-ELLQCLELLGN
DD-----------------------------------SFPSQLRLEDLFHCLYEIQD-EV
FTTQAMSY---------------------F-------------------PKVTIKISEEI
NL---LVF--S----FCLK-LSEC-LQT-IKLT----------------VTA-----LLD
--T---IF----HS-Y-----PT--------------------------------YVDRV
THGWQDLF-SALHNNEDLRELCLYDSTFDEL---------MMNILS--QELRHP------
NC--K-LQKLVLRA---------VNF-LDYSQD-FSFL----------------P
PF13516 // ENSGT00900000140813_0_Domain_6 (1616, 1642)
HNQNL
IHLD-LKDM-DMQDK-GLKSLCDALKCLTCE----LQKL--------RLT---------C
CD----------------------------------------------------------
---------LK---PI-HCL---HLSEALIR-
PF13516 // ENSGT00900000140813_0_Domain_8 (1773, 1796)
NKRLVFLNLSTNNLSD-DGVKLLCKA--
LEH-PDCCLERL-S----------------------------------------------
------------------------LANSGLT-----------------------------
----------------------------KVGCEDLSL-----------------------
--------------------------------ALISNKRLTH---LC-LADNF-LGD---
-------DGITLISDAL----KYPQC----------------------------------
--------------------------TLK-SLV---------------------------
-------------------------------LRSCHFTPLGSEYLSTS-LL
PF13516 // ENSGT00900000140813_0_Domain_13 (2212, 2236)
HNKSLTHLD
L-GSNSLQDSGAK-LLFHVLQQR--TCN----L---------------------------
-----------------------QELEL--MGC-----ILTSECCVDL------------
-------VSAL-VNNPNLCSLQLGNNNLKDTGVHI-LCDALR-NP-NC-NIQRLG--LE-
--H-CGLTSACCQ--------------------DLSTSLHG
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2465)
NQKL-LRINLTKNTLGYEG
IRNLCE--------VLGSPECK--LE--V-------------------------------
---LGLDKE-KVDEKAKKL------LE----------AVR-VNNPLLVIKSHYD--N---
--E---------------------------------------------------------
--------------------------------------------------GSGCWWQYF-
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCHOP00000007242 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------IPF-DSPPEDLSPR------K-------
------------------------------------------------------------
------------------------------------------------------------
-------IQM-------------------------------DMKKEES--DKIQEYKLD-
--------VMSK----------------YSTR--WNRAAWPESHMDFIYQDV--------
----------------------------------------------HRDEKCIPLLFLPQ
KP--QCRQP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 941)
KTVVLQGYAGIGKTTLAKKV-----MLAWA--E---------NKFYS
H--------KFWY-AFHFHCQEVIQGA-------EHSFCE-LIVQ-KW-----PG-SQAL
LSKIMS-KP----DHILLLFDGFEE-LTSTLE------ERQA-E-----LSGNWSQ--LP
-GSILLGSLLSKRMLPEAALLIV-MRFASLEKLKPLL-K-----CPF--SVTLKGFSTDE
KIEYF----------------N--KY-FGA----------TEVDKALRVVMG-DRMFFSM
CRTPVVCWIVCSVLKQLMERGA-DL-R--ET-CPNSTAVFIWYLSSLFPTRA-GDPHKQT
------------FQEQLNAVCHLA---AKGM-WD-MR----WVFDKK-------------
--DLEQSKLDET-GVTTLISV----NILQRAGD-NE-DRYTFAHLSFQEFFAALLYVLC-
YPQRF--------------QTFQ--ALDHGDIK-RLIANPE-G-SKNYLSQMGLFL----
----FGLLNDMRALAVEQALGCK---------------L--TTG-NKK-KLLNSITQVHE
WD-----------------------------------PLPPCCAVPQLFYYLHEIQE-EA
FVRQALKD---------------------Y-------------------YKATLKIKKKI
DM---QVS--A----FCLR-HCQH-LQE-MKLS----------------IEA-----RFN
-TESFSSP----RX--------------------XXXXXXXXXXXXXXX-----------
----------XXXXXXXXXXXXXXXXXXXXX---------XXXXXX--XXXXX-X-----
XX--X-XXXXX--F-RSVNPP--ML---S-E-DLIRVL----------------TDNQYL
RYLE-ISHT-KVESE-TMRLLCSALKHPQCH----LQCL--------RLE---------D
F------P----------------------------------------------------
---------VT---PE-HCC---KIAWALKD-NVHLKTLLLRRSSPQD-LGAYY------
--L-FMARLERL-S----------------------------------------------
------------------------LENCGLT-----------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------ESSCKSLAPS-LRSSKMLTHLS
L-AENSLKDEGVK-MLWEALKHS--ECP----L---------------------------
-----------------------QRLVL--WTC-----HLTGACCQDL------------
-------CNAL-YTSEHLRDLDLGDDALRDKGMQL-LCEGLK-HP-CC-KLQL-------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSSBOP00000010225 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-MDHSDAPCSSAGLR--FAVAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LQAALEDLS--Q---EQLKRFRHKLRD------
------------AGSDG-----------RSI-PWGR-LERAD-AMDLAEQLAQFYGPEPA
LEVTCKT----LK-RADARDVAAQLQ-EQRLRX---------L---------GLG-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------PA-AQLSVSEYKKKYREH-
--------VLQR----------------HARV--KERNARS------------VKITKRF
TKLLIAPESAAPLEE--ALGPAE-------EPEP--------QRARRSDTHTFNRLFRGD
E---EGRRP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYQ
A--------QVDF-AFFLPCGELLERPG------ARSLAD-LILD-QC-----PD-RGAP
VQRMLA-QP----ERLLFILDGADE-LPALGGP--------EAA-----PCTDPFEA-AS
-GARVLGGLLNKALLPSALLLVT-TRAADPGRLQGRL-C-----SSQ--CAEVRGFSDKD
KKKYF----------------Y--KF-FGN---------ERRAESAYRFVKE-NETLFAL
CFVPFVCWIVCTVLHQQLELGR-DL-S--RT-SKTTTSVYLLFVASVLSSAPAADRP---
-----------RVQGDLRKLCRLA---REGVLQR--R----AQFAEE-------------
--ELKQLELRDSKVQTQFFSKKERPGVLET-----E-VTYQFLDHSFQEFLAALSYLLE-
DEGVP-------------------G-TAAGGVG-TLLRGDAQP--HSHLVLTTRFL----
----FGLLSAERMGDIQRHFGC-----------------RASER-VKQ-EALRWVQE--Q
GCPGAAPEVTKEAKGFEDTEEPEEEEE-------EGEEGEEPNYSLELLYCLYETQE-DA
FVRQALSR---------------------L-------------------PELALQRVRFS
RM-DVAVL--S----YCVR-CCPP-GQA-LRLI------------------SC----RLV
------AA----------------------------------------QEK--K---KKS
----------------------------------------LGKR----------------
------LQAS---L--------------GGS------S----------------SSRATA
KQ---LPAS-------LLHPLFQAMTDPLCR----LSSL--------TLS---------H
C-----------------------------------------------------------
--------KLP---DA-VCR---DLSEALRA-APTLTELSLLHSRLS-EAGLRMLSEG--
-LAWPQCQVQTF------------------------------------------------
----------------------------RVQ-----------------------------
---------------LSGP---------QRGLQYL-------------------------
------------------------------VPMLRESPALTT---LD-LSGC-QLPAPMV
--T--------YLCAVL----QHPGCSLQTLSL--ASVELSEQ-----------------
----------SLQELRAVKRAKP-------DLV----------------------I--AH
PALD-------------------GHPEPP-EDLSSIF-----------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMLUP00000021000 (view gene)
ENSTSYP00000003797 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------SEKMHKVL-
--------MKEK----------------FMLQ--WEKSPLEEFHHQFFCEVA--------
----------------------------------------------MDVFYVLQLAIDPH
DS--YPKEN----LN
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 930)
ILLTGHAAAGKSMIVKMA-----VFQWASDS---------GNMWK
G--------RISY-VVHLTAHEMNQMT-------HSSLVE-LISK-NW-----SE-GPAP
IADILS-DS----RRLLFILEDLDN-MSFTVSTE------KS-F-----LCSDSREQ-VP
-VPVLLASLLQRKMAPGSWFLLT-SRPIGEEALKAFM-K-----PTD--CNVFLQLSDEM
RRIYF----------------N--LF-FKD---------SQRAAAAFKFVNE-NEVLVGL
CRVPILCWITCSVLNQQMDMG-DNI-P--LS-YQMPTDIYAHFLARALTSGAGVP-SG--
----------QSPLVLLKRLGVLA---VEGL-FQ-NT----LNFSVG-------------
--DLSTVGLTEA-DVGMLQAL----NILLPSNS-CE-NHYQFAHVNIQEFCGAIGYLMTV
ADCPL-----------------P----SGNK-V--N--KDSRD-QYNDFEPIINFI----
----FGLLNERRRKVLETSLGCQ---------------LPMAIC-FKE-YLLAQMQQLGD
EL-------------------------------------KAMEHHTPLFYCLFENQE-EE
FVSQMMGF---------------------F-------------------SEATIYIQANR
DL---MVS--L----YCLK-HCCS-LRK-LKLS----------------V-H-----IFG
--DKEFAG-------------------------------------------KLTPSQMKS
LLYWRDTC-SLLHTMETLRELEICNSNLDDT---------SERVLS--KALRHP------
DC--K-LEMLSLSY---------LSA-GSDFDDVFKAV----------------VHNQNL
HCLS-LNCM---------------------------------------------------
------------------------------------------------------------
--------------------------------------------TISL-RMFSLLHEV--
LSH-PLCSIRHL-S----------------------------------------------
------------------------LMKCGLQ-----------------------------
----------------------------PIVCGEIAS-----------------------
--------------------------------FLVNSQNLRK---LT-LSNNP-LSH---
-------DGVKILCNAL----LQPECALNSL-----------------------------
------------------------------------------------------------
------------------------------VLLFCCLTADCCYPIGRV-LLFS
PF13516 // ENSGT00900000140813_0_Domain_13 (2214, 2237)
KTLVHLD
L-SVNCLRDRGML-ALSLPLFLP--TCP----L---------------------------
-----------------------RELEL--CGC-----FFTGEICQYI------------
-------ASIII-
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
SNENLKSLELGSNNIGDAGVEQ-LCHALR-HP-NC-KLENIG--LE-
--E-NMLTGACCE--------------------SLAYVLI
PF13516 // ENSGT00900000140813_0_Domain_17 (2441, 2464)
SNKTL-KRLNLLNNKLGDEG
IAKLLE--------SLGHPDCM--LE--E-------------------------------
---IGLPES-NVSAETQRL------LK----------AVR-EKNTNLLFVSQSWAGKEGR
EIGDR-------------------------------------------------------
-------------------------------------------------------P----
GSM-----------------KPNLHWY---SI--FFKFPTRVMTVI-STTVSGQVL----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCHOP00000003625 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------LITFPVGPTCSISPKYL------------L-------------------FYQ-N
V---SSQPFIKMEEIELPSF
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
SIYG---LQWCFKELD--K---EEFQRFKELLME------
---------E--TSELA--------K--GHF-PWFE-VMNAN-TERLASLLHEYYIAPLA
WKVSINI----FE-KMNLSELAEKAK-DEMEXXXX-XXXXXXXXXXXX------------
--------------------------------------------------------XX--
XX----------------XXX-X-------X-X---------------------------
-------------------------------------------------X----------
-----------------------------------XXX-X--------------------
--------------------------------------------XXXXX-X----XXXXX
X-----------------------------------------------------------
--------------------------X-----------X---XXX-X------XX-----
-----------------------------XXXX-------XX-X----------------
------------------------------------------------------------
X--------------------X--------------------------------------
---------XXXXXXXXXXXXX-XX-X--XX-XQTSTSLYATLCSHQPTPQN-EAQRCLN
----------QKERAVLKGLCVMA---LEGV-WH-MK----FVFYGD-------------
--DLGFHGLEES-ELSTLFRM----KVLPQDIL-DKR-CYTFFHLSLQEFCAALCYVLE-
GIERD--------------RDPY--PLSIGNVR-SLMELTQID-WNPHLLQMKRFL----
----FGLMNRKSMSKLETLLGCR---------------V--PL-EIKQ-ELLQWVSLLGK
QAK----------------------------------AP-SPLDILDSFHCLFETQD-EE
FVRLALEP---------------------F-------------------QEVWLPINQQM
DL---LVS--S----FCLQ-YCQH-LQK-IRLS----------------VKE-----IFS
-KDEVTEA----QP--------------------IN-----------PS--XXXXX-XXX
XXXXXXXX-XXXXXXXXXXXXXXXXXXXXXX---------XXXXXX--XXXXX-X-----
XX--X-XXXXX--X-XXXXX---XX---XXX-XXXXXX----------------XXXXXX
XXXX-XXXX-XXXXX-XXXXXXXXXXXXXXX----XXXX--------XLN---------S
C------G----------------------------------------------------
---------LT---HT-CCL---MISKALVV-CT
PF00560 // ENSGT00900000140813_0_Domain_8 (1775, 1801)
SLKSLSLTGNNVTD-KGLRPLCEA--LKY-PTCNLQKL-A----------------------------------------------
------------------------LGHCGLT-----------------------------
----------------------------ESSCQDLAI-----------------------
--------------------------------VFIINQ
PF00560 // ENSGT00900000140813_0_Domain_11 (2019, 2056)
NLTH---LY-LAGNN-LGN---
-------DGVKALCRFI----KHPSC----------------------------------
--------------------------YLQ-RLI---------------------------
-------------------------------LNQCNLQVASCGFLALA-LISNRSLTHLS
L-SLNTLQDDGMN-LLCEVMREP--YCH----L---------------------------
-----------------------QDLEV--VGC-----CLSAACCEGL------------
-------SSVV-TRSS
PF00560 // ENSGT00900000140813_0_Domain_15 (2357, 2381)
RLTSLDLSSNDLGDRGVIA-LCEGLK-QE-TS-SLRRLG--XX-
--X-XXXXXXXXX--------------------XXXXXXXXXXXX-XXXXXXXXXXXXXX
XXXXXX--------XXXXXXXX--XX--X-------------------------------
---XXMWKV-VFTKQIKDL------PE----------EVE-RIKPNFVTDENWYVFD---
--E---------------------------------------------------------
--------------------------------------------------DDQYWQN---
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSOCUP00000012880 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---ASYFA
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 328)
EFG---LLWYLEELR--R---EEFWKFKELLNQ------
---------E--PAKLG--------L--KPI-PWAE-IKRAS-KEDLANMLAERYSRKQA
WEVTLSI----FL-QINRQDLWTKAK-EEMES----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------KPNPYKNR-
--------MKER----------------FRLV--WIQESNLPVPES--------------
---------------------------------F-------YKDSVGVEYSTLTRAAAEA
------PGP----A
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVAMNGPEGCGKTTILRKV-----MLEWA--E---------GTLWR
D--------RYTY-VFFLSVDELNRVV-------ETSLAE-LLSK-DW-----PES-AES
LEDALA-HP----EKILFIMDGFEE-LKFDLN-V------KT-D-----ACSDWRRR-WP
-TPVLLSSLLQKSMLPGCSLVLG-FGTRSMQSNRFLL-K-----HPV--LVGLQPFSEHH
RKLYF----------------F--HF-FDD---------TDEAMRAFSFVSN-IPPLFVL
CESPLVCWLVCTCVKWQLDRGE-SL-C--ID-PETGTSLYVSFAATAFKAGR-SDS-LL-
-----------QNRARMKSLCSLA---AEGM-WT-QA----FFFVQE-------------
--DIGRNGISES-DLLMWAGM----RLLRRRG-----DGFTFNNRSLQEFCAALYYYLR-
QPREQ-----------------Q--NTAIGTVA-QMVAASMGH-VHTHLSQVGGFL----
----LGISTHRILNLLETSFGFV---------------L--SKA-MKP-EIIQSLKSLGQ
HE-----------------------------------PDKGEVNFQRLFSALFETQD-ED
FVIKVMNF---------------------F-------------------EEIFINIEDTD
QM---VIA--S----YCLK-HCRS-LKK-LHLC----------------INN-----VFP
-YDLSSPI-----------S-------------------E-I-------QT---------
IFFWRDLC-SVFTNSRKFQVLKLDSCTLDDV---------SLGIIC--KALARP------
GC--R-IESLWCNF---------IYN-FGKGLDFSKAI----------------LHSFHL
KYLN-LYGT-SLSTG-AVTYLCETLRYPACN----VEEL--------VL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------GKCDLP-----------------------------
----------------------------SEVSKDLAC-----------------------
--------------------------------VLICN-KLKS---LS-LIENA-LGD---
-------EGANVLCEGL----KHSKCALQTL-----------------------------
------------------------------------------------------------
------------------------------IVNFCCLTRVSCDYFSQT-LPNNISLSTLD
L-GSNILGDEGVA-TLCEALKHQ--NCT----L---------------------------
-----------------------AELCL--TSC-----YLTPACCKDI------------
-------ATAL-TCNESLKTLKLGNNEIKDAGVKQ-LCEALK-HP-KI-QLQRLG--LE-
--M-CQLTMACVE--------------------DLASALITCKSL-KGLNLDWIVLDHDG
VVKLCE--------ALTHADCV--LE--L-------------------------------
---LGLDKS-VYEEESQML------LS----------STE-EKVSHLTIVHEPWLQE---
--EYR-----MR------------------------------------------------
--------------------------------------------------GVVPLGSNLK
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSOPRP00000011804 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------HKEINKAL-
--------IETR----------------VLCQ--WEKDYFGSSHDNFASGIP--------
----------------------------------------------AF---VHPGSNFHS
DG--YYEEN----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
LNTFMLGDNAVGKTMVVRAV-----LASWV--S---------EELWQ
D---------ISH-VVYLTSQEVNQTP-------HCSLVD-LISK-DW-----PG-GTAP
IADILS-EP----NRLVFILEDLDN-INWPLDVE------ES-A-----LCSDIREQ-VS
-MSVLLASLLQRTLLPGCWFLIT-LRCSGRGTMRALM-K-----DSD--CFLTMQFGDDK
RERYF----------------T--LF-YKD---------QQRASTACKLVND-NEVLRSL
CRAPVLCWILCIALGSHIDRA-DDL-S--SS-YQTRADVFTRFLAAVLSSNVSRA-DR--
----------PRRLALLERLCALA---VEGL-LH-NT----LNFQDE-------------
--DLRSVGLSQE-DVSTLETF----RLLLPSNS-LP-GHYTFIHPKMQEFCAAMAYMMVL
VHHQL-----------------P----RLRN-K--P--EDKRE-VYSDFSPVVAWI----
----FGLLNEKRRRILEACVGCR---------------LL--------------------
--------------------------------------------HMPLFYCLFENQE-EA
FVRQLMDF---------------------F-------------------PTVNAYVQSSR
DL---MVS--A----YCLQ-RSQA-LHK-LKLS----------------LLD-----LFK
--PQDPHL-------------------------------------------RLSTCQVKS
LAYWRDIC-SLLHTNENFRDLEISNSEFDTT---------SGRILC--KALMHR------
HC--R-LQTVRLSH---------VTV-GSDLDDLLKVI----------------VGLRNL
TFLS-LNGA---------------------------------------------------
------------------------------------------------------------
--------------------------------------------ADFP-DICRLREAL--
--E-STWHVRHL-R----------------------------------------------
------------------------LMRCDIK-----------------------------
----------------------------ANVCGEVAS-----------------------
--------------------------------LLVNSR
PF00560 // ENSGT00900000140813_0_Domain_11 (2019, 2057)
NLKK---LT-LSNNP-LKD---
-------DGMKTLCDVL----RQPGCSLESL-----------------------------
------------------------------------------------------------
------------------------------VXXXCCLTEACCNSLATV-MLLTKTLKHLD
L-SVNFLQNSGVS-ILLMPLVFP--ICT----I---------------------------
-----------------------QELEL--SGC-----FFSD-VC-YI------------
-------STAIL-RNPNLRSLELGSNNLGNEGVVL-LCQALE-LP-SC-NLENLG--LE-
--E-CKITGACCE--------------------FLASMLVTNKKL-KKLNLLGNKLGEAG
ITQLLV------VLMLGPPNCV--LE--T-------------------------------
---L--------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPVAP00000011856 (view gene)
ENSSARP00000006107 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------------LDDC-GLT
EGRCKDIS-SALQANP
PF00560 // ENSGT00900000140813_0_Domain_4 (1517, 1551)
SLTELSLRTNELGDA---------GVHLVL--QGENSGT00900000140813_0_Linker_4_5 (1551, 1566)
LQA-P-----
TC--KPF00560 // ENSGT00900000140813_0_Domain_5 (1566, 1617)
-IRKLS--L-QNCSLT--EA---GCG-ILPSAL----------------RSENSGT00900000140813_0_Linker_5_6 (1617, 1619)
LPPF00560 // ENSGT00900000140813_0_Domain_6 (1619, 1645)
TL
RELQ-LNDN-PLGDA-GLRLLCEGFLDTQCH----LERL--------QLE---------Y
C------N----------------------------------------------------
---------LT---AT-GCE---ALASVLRV-KPNFKELVVSNNTIGE-AGARELCQG--
LTK-STCQLETL-X----------------------------------------------
----------------------XXXXXXXXXXXXXXXXXXXXXXXXX-------------
----------------------------XXX-XX-XX-----------------------
--------------------------------XXXXXXXXXX---XX-XXXXX-XXX---
-------XXXXX---XX----XXXXX----------------------------------
--------------------------XXX-XXX---------------------------
-------------------------------------XXX-------X-XXXXX--XXXX
X-XXXXXXXXXXX-XXXXXXXXX--XXX----X---------------------------
-----------------------XXXXXXVKDC-----SFTGACCQHF------------
-------SAVL-AQNK
PF00560 // ENSGT00900000140813_0_Domain_15 (2357, 2381)
HLVELQLSNNKLEDAGVQQ-LCQALS-QP-NT-RLQMLC--LG-
--D-CEVTDSSCR--------------------ALAALLCANQ
PF00560 // ENSGT00900000140813_0_Domain_17 (2444, 2476)
SL-KELDLSNNGLGDPG
IQLLVT--------SLEHPDSH--LE--Q-------------------------------
---LVLYDI-YWSEAMEDR------LR----------ALE-ESKPNLKVIF---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMLEP00000009094 (view gene)
ENSMLUP00000017003 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------IKEKYQNQ-
--------RRES----------------CCPTQSW-------RNE---------YLHQKF
TQLLLLHRSQHRGYDF---L------LRRRRNHEV--------VDEQGQLMEIGDLFGPG
LR--TQEEP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
PTVVVHGVAGIGKSTLARQV-----RRAWE--E---------GWLFR
N--------RFKH-VFYFNCRELAQSE-------TMSLAE-LITK-DW-----AA-PDAP
IGQILS-QP----EQLLFILDNIDE-PKWDLKEQ------SS-E-----LCPHWSQQ-QQ
-VHTLLGSLLKKTLLPTTSLLIT-ARITALRKLIPSL-K-----HPR--WVEVLGFSESA
RKDYF----------------F--QY-FTD---------ESQAIRAFTSVES-NPALFTM
CLMPLSYSLVCTLPE-AADVPG-AT-S--LT-SQTTTALLSALLFPRSP--------AS-
-----------VPRTKLRDFCSLA---AEGT-CQ-GK----TLFSPE-------------
--DFRKHGLDET-NISTLLNM----GVLQEH---PTTQSYSFIHLCFQEFFAAMSYALG-
N------GVL--------KGDCF--NNI-KIIT-DLTDVYDRH--DLFEEPTTCFL----
----FGLLSDQGMREMESIFKCS---------------L--SRE-RHC-SLLHLAKSEVL
-----------------------------------AKYLNLDTYSWHLLHCFYEIQD-EN
FMKNIF-I---------------------P-------------------FTLRICVQTDM
EL---LLF--T----FFFT-FCHR-IER-LQLN----------------EDEQ-------
------H-R---------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------Q------------
------AGRT--------------------------------------------------
-------------------SGAFLYNWGLFT-----------------------------
----------------------------DSWWQMFFS-----------------------
--------------------------------ILKVPG
PF00560 // ENSGT00900000140813_0_Domain_11 (2019, 2057)
SLRE---LD-LSGNS-LSC---
-------SAVQSLCDAL----KSPHCHLVTLR----------------------------
------------------------------------------------------------
-------------------------------LVSCGL-----------------------
------------------------------------------------------------
----------------------------------------TY------------------
------------------------------------------------------------
---------NCCK--------------------DLVSMLQDNFML-SDLDLWQNDLGDLG
VKLLCE--------GLRQSNCQ--LK--H-------------------------------
---LWLDQT-QLSEEVKKM------LS----------LLQ-QEKPQLVISS-RQKSLYYI
PTGNLG--KVSVI-----------------S-----------------------------
----------------------------LLRRSHSSGNKH-S--PQTQ-MDDDFWGPTGP
VATEMVDK-HRSLYRVHFPVAGSYHCP---NVGLYFVVREPV-------TIEFEFCVWDQ
FLDQIVPQHSWMVAGPLFDIKAEPGA-VEAVYLPHFIALQGQ--------HMDISWFQVA
HIKEDGILMEKPASVEPHYIVLENPSFSPMGVLLRMIHTALC-IPVISNVLLYYRLQSEE
V-NFHLYLIPSDCTIQKAIDDEEKKFQFFQIRKPPPLTPLYMGSRYTVSGS---------
DKLKIIPEELELCYRSPGKPQLFSEFYVGHLGSGIWLQIKSKDDETVVWEALVKSVNLPP
EVPQQ-------------------------------AFQASLSSPDAPTL
PF00619 // ENSGT00900000140813_0_Domain_19 (3051, 3135)
LHFVDQHRED
LVTRVTSVDAVLDKLH-GQVLSEEQYERVRAEPTNTEKMRTLFSFSKSWDWACKDQLYRA
LKKTHPHLIVDLWENMSSVSIN--I-----------
ENSPPYP00000008863 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MAGG----A-WGR---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 328)
LACYLEFLK--K---EELKEFQLLLAN------
---------K--AHSR--------SS--SGE-TPAQ-PEKTS-GMEVASYLVAQYGEQRA
WDLALHT----WE-QMGLRSLCAQAQ-EGAG-------HS-PSFPY---SPSEPHLGSPS
QPTSTAVLMPWIHELP-----AGCTQGSERRVLRQLPDTSGRRW-REISSSLLYQ--A--
---------LPSSPD-YESPSQESPNAPTSTAVLGSWGSPPQPSLAPREQEAPGTQWPLD
EMSGIYYAEIREREREKSEKGR------PPWAAAVGTPPQAH---TSLQPHRHPWEPS--
--------VRES----------------LCSTWPW-------KNE---------DLNQKF
TQLLLLQRPHPRSQVP---L------FKGSWPHYV--------EENRGHLIEIRDLFGPG
LD--TQ-EP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
RIVILQGAAGIGKSTLARQV-----KEAWG--R---------GQLYG
E--------RFQH-VFYFSCRELAQSK-------VVSLAE-LIGK-DW-----TV-TQAP
IRQILS-MP----ERLLFILDGVDE-PGWVLQEP------SS-E-----LCLHWSQP-QQ
-ADALLGSLLGKTILPEASFLIT-ARTTALQNLIPSL-E-----QAR--WVEVLGFSESS
RKEYF----------------Y--KY-FTD---------ERQAIRAFRLVKS-NKELWAL
CLVPWVSWLACACLMQQMKRKE-KL-T--LT-SKTTTTLCLHYLSQALQ--------AQ-
-----------PLGPQLRDLCSLA---AEGI-WQ-KK----TLFSPD-------------
--DLRKHGLDGA-VISTLLKM----GILQEH---PIPLSYSFIHLCFQEFFAAMSYALE-
N------KKG--------KGKHS--NCI-IDLE-KLLEAYGTH--GLFGASTTRFL----
----LGLLSDEGEREMENIFHCQ---------------L--SQG-RN---LMRWVPS---
------------------------------------LQLLLQPHSLESLHCLYETRN-KT
FLTRVMAH---------------------F-------------------EEMGMCVETDM
EL---LVS--T----FCIK-FSRH-VKK-LQLI----------------EGRQ-------
------H-R---------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------S------------
------TWSP--------------------------------------------------
-------------------TMVVLFRWAPVT-----------------------------
----------------------------DAYWQILFS-----------------------
--------------------------------VLKVTRNLKE---LD-LSGNS-LSR---
-------SAVKSLCKTL----RCPRCLLETLR----------------------------
------------------------------------------------------------
-------------------------------LAGCGLIAEDCKDLASG-LRANWTLTKLD
L-SFNVLTDAGAK-HLCQRLRQP--SCK----L---------------------------
-----------------------QRLQL--VSC-----GVTS------------------
------------------------------------------------------------
---------DCCQ--------------------DLASVLSASPIL-KELDLQQNNLDDAG
VRLLCE--------GLRHPACK--LI--R-------------------------------
---LGLDQT-TLSDEMRQE------LR----------ALE-QEKPQLLIFS-RRKPSVMT
PIEGLDTGETSNS-----------------TSSLKRQRLRSETEPSHIAQADLKRL-DWS
NPPAKA--SQS-AGITEGSSPEVAQ-VEPLCLPSPASQGD-LHTKALG-TDDDFWGPTGP
VAAEIVDK-EKSLYRVHFPVAGSYRWP---NMGLCFVVREAV-------TVEIEFCVWDQ
FLGEINPQHSWMVAGPLLDIKAEPGA-VEAVHLPHFVALQGG--------HVDTSLFQVA
HFKEEGMLLEKPARVELHHIVLENPSFSPLGVLLKMIHNALRFLPVTSVVLLYHRLHPEE
V-TFHLYLIPSDCSIRKAIDDLEMKFQFVRIHKPPPLTPLYMGCRYTVSGS-------GS
GMLEILPKELELCYRSPGEDQLFSEFYVGHLGSGIRLQVKDKKDETLVWEALVKPGDLMP
ATALVPP-----------------------------ACIAVPSPLDAPRLL
PF00619 // ENSGT00900000140813_0_Domain_19 (3052, 3133)
HFVDQYREQ
LIARVTSVEVVLDKLH-GQVLSQEQYERVLAEDTRPSQMRKLFSLSQSWDRRCKDRLYQA
LKETHPHLIMELWEKGSKKGLLPLSS----------
ENSEEUP00000008107 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MTNMFSSPL
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
SIFE---LLLYLEKLN--I---EELNRFKLLLKD------
---------E--MMGTG--------D--YQI-PWTQ-VKTK--ASVLAKLMDEYYSVEQV
WNVTLKI----FD-QMNLKFLCGRLK-AQRTXXXX-XXXXXXXXXX------X-------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------XXXXXXXXXXEGEEYRIR-
--------IKEK----------------FGIL-------YSKLLP---------------
----------------GESD------F-----CDG-I-------LR-EDEKLLVYLFEV-
VT--TGKEP----ERVVXXXXXXXX
PF05729 // ENSGT00900000140813_0_Domain_2 (686, 940)
KTTLMIKA-----MLAWE------------GRLF-
Q--------KFDY-VFYLNAKEINHLK-------ENSFLQ-MISK-YW-----PS-TEGP
IEKIMM-KP----SRLLFIIDGIDE-MNFAFEEP------VD-A-----LCKEWNQL-HP
-ASFLMSSLLRNRMLPESSLLLT-ARLTDSKKLK-LL-N-----KAR--FIEMHGMTEDA
IKEFI----------------F--QL-FED---------KNWALEMFNSLQD-NETLFSM
CKAPQVCWDICNSLKQQVEKGG-NV-T--LA-CQTITEVLTCCISDLFQP-------TKD
-C-PR-VP-SE-IQ-LKR-LC-YL-AA-KGI-WN--MT-Y-V-FYRE-------------
--TLRKHGLL-NSDISVFLGM----NIIQQDVE-YE-SCYVFTHIHVQEYLAALSYMLE-
GDWGM--------------KSDS--FQSFEHLK-LLLES-KMY-NSPHLINMKCFL----
----FGLLNENRIRRLER------------------------------------------
----------------------------------------------------------CL
NC-TMLLD---------------------V-------------------KDKILQWVKRL
GS-RKYFP--SRPEFLDLLHYLYE-TQD-EAWV----------------KQ---------
----A-------------------------------------------------------
--------------MKSFQKVEIDICERR-------------------------------
----H-MTVSSYCL-KNCQSL--QHI-KPCK-KVVSQX----------------XXXXXX
XXXX-XXXX-XX------------------------XXX--------XXX----------
------------------------------------------------------------
----------------------------------------XXXXX-XX-XXXXXXXXX--
XXX-XX-XXXXX-X----------------------------------------------
----------------------------XXX-----------------------------
-------XXXXXXXXXXXXXXXLKYVSFPDGCQDIFS-----------------------
--------------------------------SLTQSCNLNH---LY-VKGYN-MKD---
-------QDVKSLCEGL----KHPKC----------------------------------
--------------------------KLQ-DLG---------------------------
-------------------------------LESCSLTTDSCVNISEA-LIRYQNLINLS
L-STNSLSDKGVK-LLCEALRHP--KCQ----L---------------------------
-----------------------KRLSL--MDC-----NLTRACCLEL------------
-------ASAI-SNSPNLYRLNLGENNLEDDGVKI-LNEALR-NP-TC-NIQRLG--LE-
--Y-CGLTSHCCQ--------------------DLASTLRTSPKL-IEINLIKNKLDLEG
IKKLCD--------VLKSPQCK--LQ--V-------------------------------
---LGLCKD-DFDEVVKKM------LE----------EVE-LSNSQLTIKSNYK------
------------------------------------------------------------
--------------------------------------------------EDR-SWCWFL
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMMUP00000049609 (view gene)
ENSSTOP00000012288 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
MHLD-LKGS-KIENN-GIKSLCEALKHPDCK----LQNL--------RLE---------F
CE----------------------------------------------------------
---------LT---AV-CCL---NISKALIK-
PF13516 // ENSGT00900000140813_0_Domain_8 (1773, 1796)
SQSLIFLNLSANNLLD-DGVKLLCEA--
LRH-PKCCLERL-S----------------------------------------------
------------------------LESCGLT-----------------------------
----------------------------EVGCEDLSL-----------------------
--------------------------------ALL
PF13516 // ENSGT00900000140813_0_Domain_11 (2016, 2052)
SNKTLTH---LC-LADNV-LGD---
-------DGVKLVSDAL----KDPQC----------------------------------
--------------------------TLQ-SLV---------------------------
-------------------------------LRCCHFTSLSSEYLSTS-LV
PF13516 // ENSGT00900000140813_0_Domain_13 (2212, 2237)
HNKSLKHLD
L-GSNYLQDDGVK-LLCDVFRQP--SCN----L---------------------------
-----------------------QDLEL--MGC-----VLTDACCLDL------------
-------ASVI-L
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2374)
NNPNLHSLDLGNNELKDDGMKI-LCDALR-HP-NS-NIQKLG--LE-
--Y-CSLTSLCCQ--------------------DLSSTLLSNQRL-INMNLTQNALGCEG
IGKLCE--------ALKSQECK--LQ--I-------------------------------
---LGLCKE-AFDEEAQKL------LE----------EVE-VSNPHLVIKPYCNDYG---
--E---------------------------------------------------------
--------------------------------------------------EDGSWWWCS-
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSOARP00000003372 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----GAPP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 929)
TVLLEGAAGVGKTTLATKL-----VLHWA--E---------GVLFR
G--------RFSY-VFYISGHRLAELG-------DTSFAG-LLAL-DW-----PD-SQGP
VEELRA-HP----ERLLFVIDGAEE-VILGSD------GSAS-Q-----PSADWYQE-LP
-AASILVRLLKRELVPEATLLVT-AGPPGGRALRRLL-L-----SPC--CVDIPGFTEGD
RREYL----------------R--RF-LGD---------PDVAEEAWRRMRG-SETLVRS
CAAPLACWAVCAGLKRQLAASP-AS-APAPG-APTPTSLYARLFCSLLEEAE-PGLRAGS
------------SAGQWRAFCSLA---AEGL-WL-AA----FTFAGD-------------
--ALERRRLEAP-FIDGLLRL----QILRRVSD-CE-HCVTFAHRSFQEFFGALFYVLR-
GSRGS--------------L-GG--VPRHQEMR-RWLNHAFAD-ANPYWRQTVRFF----
----FGLLGTDLAGQLEEALGCQ---------------M--YPG-AAD-ELLDWAEELER
CG-----------------------------------AVSAHFDFLWLFQCLHETQD-EN
LVRRVLNH---------------------L-------------------PEADLDIQGCE
HL---RVS--S----FCLK-HCQK-LRK-LRLS----------------VRG-----DFP
-KGPSESL----RS-----------------------------LLFPPR--RMRAA-DFQ
MHQWEDIC-S-VFCSGNLSELDLSNSKLSTA---------SMMRLC--LNLGN-P-----
RC--R-LKKLA--W-KSVSPV--EG---L-R-KLGLLL----------------HGNRHL
THLD-LSSN-ILRAA-VSPGVFRMLGHSACS----LKYL--------WLE---------S
C------G----------------------------------------------------
---------LS---AE-VCH---QLFMELGR-NSSLRFLSLADNNLSH-VELERLREP--
PGT-ATCALKEL-S----------------------------------------------
------------------------LLQTAVI-----------------------------
---------------------------RASGALERVG-----------------------
--------------------------------LLTSTPRLTR---LC-LGLNP-LGN---
-------EGVRLLCGSL----TWPAC----------------------------------
--------------------------VLQ-RLE---------------------------
-------------------------------LWWCLLGAPSCRHLSDA-LCRSRSLTHLN
L-RKNHLGDGGVK-LLCAALGRA--YCA----L---------------------------
-----------------------QSLNL--SHC-----SLTVAGCCEL------------
-------AYAL-KRNGHLKILDVGNNAVQDEGVRE-LCSVLK-SP-SC-VLQTLG--LE-
--K-CSLTAVCCR--------------------PLSCVLGSSKSL-ENLNLLGNNLGPDG
PLWLCF--------SLECPVLG--LF--H-------------------------------
---HLSWG--FPV-CVSAE------LE----------KLH-EKGCSLRVTSQWDFND---
--PE--------------------------------------------------------
---------------------------------------------------ERWWW----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMUSP00000045077 (view gene)
ENSECAP00000014445 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------FHYSIT--------
-----------------------------------------------QRSQKFITFLNPQ
IS--TQLMP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLHGPAGVGKTTLAKKW-----MVDWT--Q---------DNHAK
---------TFNS-TFYLSCKELNHKG-------ECSFAD-LLSK-NW-----PD-LQDA
VPEILT-QA----QKILFIIDGFEE-LRVPSE------ALIH-D-----MCCDWQKQ-KP
-VPVLLGSLLKRKMLPKATLLIT-TRPGALRELGLLV-E-----QPF--FIEIEGFLELD
RKAYF----------------L--KH-FEE---------EAQALRAFDLMKS-NAALFHL
GSAPAVCWIVCTCMKLQMEKGE-DP-A--PT-CQTITSLFLHFLCSQFTSP-PGSCPGPF
------------LQVPVTALCLLA---AEGI-WT-QT----SMFDGE-------------
--DLERLGVKES-DFHPFLDK----NILQKGKD-CE-GCYSFIHLSVQQFLAALFYVLE-
REEEE-------------DGDSC--RWDIGGVQ-KLLSKEERL-KNPSLTHVGYFL----
----FGLLNEKRAGELETTFGCQ---------------V--SRK-VKQ-ELLEFRVKSNK
NK-----------------------------------RLSSVTDMKEVLYCLYESQE-EQ
LVKNAMSY---------------------V-------------------KEISIHLTNTF
EM---MQS--S----FCLK-HCEN-LQK-ISLQ----------------VEKG----IFL
-END-TES----EL--------------------D-----------------AQCD-HRS
LPLWTDLC-SVFSSNKNLSFLDVSQSFLSHS---------SVRILC--EQITR-V-----
TC--H-LQKVV--I-KNISPS--DA----YR-DLCLAF----------------IGKKTL
THLI-LEGD-VHTDKMLLLLLCEMLKHSRCN----LQYL--------RLG---------S
C------C----------------------------------------------------
---------DA---AQ-KWD---DSSLALGI-NQSLRCLDLTASELLD-EGVKLLCTT--
LRH-PKCFLQKL-S----------------------------------------------
------------------------LENCHLT-----------------------------
----------------------------EACCKELSS-----------------------
--------------------------------TLIVNQRLTY---LC-LANNK-LGD---
-------GGVKLLCEGL----SYPEC----------------------------------
--------------------------QLQ-TLV---------------------------
-------------------------------LWHCNITRRGCKLISKL-LQGDSSLTNLD
L-GLNPIA-TGLW-FLCEALKKP--NCN----L---------------------------
-----------------------KYLGL--WGC-----SITPSCCQGL------------
-------ASAL-ISNQRLETLDLGQNVLGKNGIMM-LFEALK-QN-DG-PLKTLR--LK-
--V-DESSVEVQK--------------------LLKDVKDSNPKLTIECNNA------RT
---------------TRSSCCD--FF----------------------------------
----S-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPSIP00000012294 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MGNQSSR--I
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 326)
SDL---LVQALDDLS--Q---EDVKRFKDKLSH------
---------S--AFE-G--------K--RTI-PRGR-LENAD-RIDTKNLLMEFYGGAAA
VEVTINV----LT-QINHRDSAAKLR-GESE-KEI------------------P-LSQ--
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------KSSELSAKAYRKKYKED-
--------IKKK----------------YKLI--KDMNSRFGDKM---------LLNRRY
TNLTIVDRPRQAKEREHEIM------ATGWRHAEVMT-------ERASSAVTLATLYKPD
ED--GQTPQ-----
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 929)
VVVLLGAAGIGKTMTARKI-----MYDWA--A---------GELYK
E--------KFDY-VFYINCREVNFLN------KQGSVAD-LMFK-NW-----LH-TNPP
INHILM-NP----EKFLFIIDGLDE-LRFSFDQP------ED-D-----LCFDPWEK-KS
-MEIILSSLFRKTVLPESYLMIT-TRPAALEKLEQCL-E-----CSR--YAEILGFSEAE
REEYF----------------H--KF-FEM---------ETQARQALQFVKA-NEILFTM
CFVPFMCWIICTVVKQQLEKGE-NL-T--QT-SKTVTGVYVLYLSSLVKPLS-NDL-KQN
------------LHGNLRGLCSLA---ADGI-WN-QK----ILFEEE-------------
--EIKKFGLDQENSLPLFLNE----NIFQKETD-CE-CLYSFIHLSFQEFFAALLYVLE-
EDEA---------------AIDS--GTLGRNVK-ALLENYGNS-KN-YLMLTVRFL----
----FGLLNEERMKDMQEKFRCK---------------F--SPA-IKA-DLLEWVQANQE
TDLKTLPF---------------------------AHEGIHQRHQLELFHCVYEMYE-ED
FAITALDH---------------------F-------------------TELKLNQNKFT
QM-DQMAL--S----FCVK-HCRS-LES-LCIY----------------DCT-----FSL
-EDHEVGL----PR----------------------------LQK------WLHQK-H--
-----------------------HEDKPKHS---------PIYLLC--QALKD-P-----
YC--K-LKKLE--L-CDCRLT--AA---CGE-DLASLV-----------------TKQTL
VELN-LAGN-SLGDS-GVKLLCEGLKRANCK----LQKL--------DLK---------A
C------K----------------------------------------------------
---------LS---AA-GCG---ELSSVLST-SQWLSHLNLSGNQLGA-LGIQLLCEG--
LKQ-PNCKLQRL-V----------------------------------------------
------------------------LRFCNLS-----------------------------
----------------------------GECFGDLST-----------------------
--------------------------------VLSTSQHLTE---LD-LSYNFSLGD---
-------VGVQLLCEGL----KHANC----------------------------------
--------------------------HLQ-RLE---------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMLEP00000022240 (view gene)
ENSOARP00000003407 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------SSP-----------LS-R
S---GVSPPSLEPS-SSTLPFRNG---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 326)
VMLFMLSVS--A---EQLQRFKELLVE------
---------E--DPSPG--------C--TPL-TWGQ-LKSAR-CGEVVHLLTEYFPGRRA
WEVARDI----FT-KMNQTELCLQTQ-RELNEMLP-NLEPEASSLR------R-------
------------------------------------------------------------
------------------------------------------------------------
-------RVL-------------------------------TLEEDES--DKIREYKAR-
--------VMSE----------------HSTL--CDRTCWPGNNVDFFYQEP--------
----------------------------------------------WREDTLLQCLLLPR
KP--QGRQP----G
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVLQGDAGVGKTTVANTL-----MLRWA--Q---------DTFYA
H--------KGWL-AFYLHCRELDRPE-------EQSFSE-LIAH-KL-----SG-SLAL
ASRILS-HP----EHLLLLLDGFEE-LTLTLI------DRPE-D-----LSEDWGQK-MP
-GSVLLTSLLSKRMLPDATLIVF-LRSSLWKTVSPFL-K-----GPS--LIPLTGFSAPE
RSMYF----------------R--SY-FKN---------RHDADAALSFATG-NAVLFSM
CRVPVTCWLVCSCLKRQMERGS-GL-P--QA-FPNATAVFAHYLSSLLPTRV-RHAVGET
------------HQEQLKRLCSLA---AEGM-WK-NQ----WVFGET-------------
--DLSLARLDER-DVEAFLGV----RVLRRAVA-GE-D----------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSTSYP00000021583 (view gene)
ENSMICP00000049318 (view gene)
ENSRNOP00000031864 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------MKLFYCLFEMED-EA
FLIQAMNC---------------------I-------------------EQINFVAKDYS
DI---IVA--A----YCLK-HCST-LKK-VSFS----------------TQN-----ILN
-EE----Q-----------E-----------------------------------YREKL
LTYWNQMC-SIFISNKDIQELQIKQTCLSDP---------AFSVFY--NHWKHR------
TC--T-IKILKANM---------VSF-LCEKHLFFEF-----------------IQNHNL
QYLD-FSFT-CLSHI-NVELLCDVLNQAECN----VEKL--------VV-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------EHCNLS-----------------------------
----------------------------PDDCQNFAF-----------------------
--------------------------------VLMN
PF13516 // ENSGT00900000140813_0_Domain_11 (2017, 2036)
SKTLKI---LN-LACNN-LD----
-------KGIYSLCKAL----CHPNCILENL-----------------------------
------------------------------------------------------------
------------------------------VLDNCSLSEQCWDYLSDA-LRQNKILC
PF13516 // ENSGT00900000140813_0_Domain_13 (2218, 2233)
HLD
I-SNNELKDKGLK-ILCKALTLP--NCV----L---------------------------
-----------------------KSLCM--THC-----LITSSGCQDL------------
-------AEVL-R
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
NNQNLKGLQISNNKLEDAGVKL-LCDAIK-HP-NC-QLMDLG--LE-
--A-CEITGASSE--------------------DLSSAFLQSNTL-GRVNLCGNAFEISG
LALFPR--------F---------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCJAP00000010235 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-MDQSDAPCSSTGAR--FAVAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LQAALEDLS--Q---EQLKRFRHKLRD------
------------VGSDG-----------RSI-PWGR-LERAD-PMDLAEQLAQFYGPEPA
LEVARKT----LK-RADARDVAAQLQ-EQQLRR---------L---------GFD-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------PA-ALLSVSEYKKKYREH-
--------VLQR----------------HARV--TERNARS------------VKITKRF
TKLLIAHESAAPLGE--ALGTAE-------EPEP--------QRARRSDTHTFNRLFRGD
E---EGRRP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYQ
G--------QVDF-AFFLPCGELLERPG------ARSLAD-LILD-QC-----PD-RGAP
VQRMLA-QP----ERLLFILDGADE-LPALEGP--------EAA-----PCTDPFEA-AS
-GARVLGGLLNKALLPSALLLVT-TRATDPGRLQGRL-C-----SPQ--CAEVRGFSDKD
KKKYF----------------Y--KF-FGN---------EKRAERAYRFVKE-NETLFAL
CFVPFVCWIVCTVLHQQLELGR-DL-S--RT-SKTTTSVYLLFVASVLSSAPAADRP---
-----------RVQGDLRKLCRLA---RKGVLER--R----AQFAEE-------------
--ELEQLELRDSKVQTQFFSKKERPGVLET-----E-VTYQFLDLSFQEFLAALSYLLE-
DEGVP-------------------G-TAAGGVR-TLLRGDAQP--HSHLVLTTRFL----
----FGLLSAERMGDIQRHFGC-----------------RVSDH-VKQ-EALRWVQG--Q
GCRGAAPEVTEGAKGLEDTEEPEEEEE-------EEEEGEEPNYSLELLYCLYETQE-DA
FVRQALSG---------------------L-------------------PELVLQRMRFS
RM-DVAVL--S----YCVR-CCPP-GQA-LRLI------------------SC----RLV
------AA----------------------------------------QEK--K---KKS
----------------------------------------LGKR----------------
------LQAS---L--------------GGS------S----------------SSRATA
KQ---LPAS-------LLHPLFQAMTDPLCC----LNSL--------TLS---------H
C-----------------------------------------------------------
--------KLP---DA-VCR---DLSEALRA-APTLTELSLLHNRLS-EAGLRMLSEG--
-LAWPRCQVQTF------------------------------------------------
----------------------------RVQ-----------------------------
---------------LSGP---------QRGLQYL-------------------------
------------------------------MPMLRESPALTT---LD-LSGC-QLPAPMV
--T--------YLCAVL----QHPGCSLQTLSL--ASVELSEQ-----------------
----------SLQELRDVKRAKP-------DLV----------------------I--AH
PALD-------------------GHPEPP-KELSSTF-----------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCGRP00000000203 (view gene)
ENSOCUP00000021941 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------GAC--ATVA
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 328)
REL---LLATLEDLS--Q---EQLKRFRHKLRD------
------------ARTGS-----------RSI-PWGR-LELAD-ALELAEQLVHFYGPEPA
LDVARAT----LK-RADVRDVAARLK-EQRLQRK--------L---------GAR-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------AG-VLGGAAEYAKKYREH-
--------VLRQ----------------HAKV--RERNARS------------VRLDRRF
TKLLIAPEA--RGD--PAPEAAQ-------ESQK--------GAARRSDTRTFNRLFGRD
D---EGQRP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 783)
TVVLQGPAGIGKTMAARKI-----LYDWA--A---------GKLYQ
G--------QVDF-AFFVPCRELLGRPD------ERSLAD-LILD-QC-----PD-PGAP
LPQML-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMGAP00000003606 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MA-GE
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
ESAI---LLEALEGLT--L---EDFQEFKKKLSH------
---------I---DIKE--------G--WNI-HRDE-LEKFTHPSTLISYMGDSYGEAAA
MDIAIGL----FE-KMNQRHLAEKIL-DEK------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------VK--EYKQKYREH-
--------VARE----------------FLQY--KEVNSWLGENL---------SVSDRY
TNLTIARRSWNQ-HGGE------------------PG----DV-SSDTV--TIQTLFEPS
KD--G-QLQ----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
RVTVLVGATGMGKTMTIRKV-----MMDWV--E---------GTLC-
T--------QFDY-AFCIDCKELSFSKE-------VSMVD-LISK-CC-PQ-----QRTP
AGRIFD-NP----EKILFIFDSFEV-LGLPLVQH------KG-E-----LSTDPTEA-KP
-LETTLLSLLKRKVLPESSVLIA-MRPTALQSLGQCL-E-----GEH--YVEILGFAPAA
REEYF----------------H--RY-FGN---------DNKADVAFRFTRG-NGVLYSL
CVIPIMSWTVCTILEQELHKRN-QL-L--AC-SKTTTQMILFYLSWLTKHRVSNA--WQ-
-----------NLQQFLHKLCSLA---ADGI-WK-HK----VLFEEK-------------
--EIEDQGLNQPELLSLFLNE----KGLEKGTH-HG-NVYSFSHLHLQEFFAAMFYVLE-
DQEEML--------------SDS--QIFAKDVN-MLLKSYHTS--RMDLNVTVRLL----
----FGLVNPKSVEYADEGIGCR-----------------ISLR-PRE-DLLRWLQTRHR
DLS----------------H----------------PSKIMTVKDLDTFHFLFETNE-ES
FVQSVLGS---------------------F-------------------TGIVLQDAKLM
LY-DQVAL--C----FCIK-QWAG-LLS-VT------------------LRSC----SFH
QQHRRQEPGK---GLP--WQSWRQEEL-HSPL--YPLCQALRH-----------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------PGSL----LQSL--------RLQ---------W
CGLTE-GDSKTLGTLL---ATL----PS-LVYFEVGDGALGDDGVRMLCTGLRQPDCH-L
RVLR--------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSRNOP00000043888 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------M-KVLSDAFKPH
Q---KTFRP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
HTIILHGRPGVGKSALARSI-----ILGWA--Q---------GKLYQ
D--------M-SY-VFLFSVREMKWTE-------KSSLAQ-LMAN-EW-----PH-SQAP
VTKIMS-QP----ERLLFVIDGLDD-MDSALQHD------DM-T-----LSRDWKDK-QP
-IYILVYSLLRKALLSQSFLIIT-TRNTGLENLRSMV-V-----SPL--YILVEGLSASR
RSQLV----------------L--EN-IPD---------DHQKIQVFHSVIE-NHQLFDQ
SQAPSFCSLVCEALQLQEKLGK-RC-T--LP-CQTLTDLYATLVFHQLTSRG-PSQRALS
----------KEEQITLVGLCRMA---LEGV-WT-MR----SVFYDD-------------
--DLKSYSLKQS-EISALFHV----NILLQVGH-SSEQCYVFFDLSLQDFFAALYYVLE-
GLG-E--------------WNQY--FSYITNQR-NIMEVKRSS--DSHLLEMKRFL----
----FGLMNKDTLKTVEVLFRCP---------------V--TP-PIKQ-KLQHCLSLIGQ
QVN----------------------------------GSS-PMDTLDAFYYLFESQD-EE
FVHMALKS---------------------F-------------------QEVWLLINQKM
DL---MVS--S----YCLQ-QCQN-LKA-IRVD----------------ISD-----LFS
-VDNTPKL----CP--------------------VA-----------LQ--KTQCK-PLI
IEWWENFC-TVLSTHPNLKTLDLGHSILNEW---------SMKILC--LKLRN-S-----
SC--S-IQNLT--F-KNSEV---VS---GLQ-YLWMLL----------------VSNRNL
KYLN-LGNT-PMKDD-DIKLACEALKHPSCS----LETL--------RLD---------S
C------E----------------------------------------------------
---------LT---LT-GYE---LISKLLLS-ASSLKCLSLARNKVGV-KSMTSLGEA--
LSS-STC-----------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------TLQ-KLI---------------------------
-------------------------------LNHCNIIKDAYGFLALI-LA
PF13516 // ENSGT00900000140813_0_Domain_13 (2212, 2237)
NNRKLTHLS
L-TMNPVGNSAMK-LLCEALKEP--TCY----L---------------------------
-----------------------QDLEL--VDC-----QLTENCCEDL------------
-------ACMI-TTT
PF13516 // ENSGT00900000140813_0_Domain_15 (2356, 2378)
KHLQSLDLGNNALGDKGVIT-LCKGLK-QS-SS-SLRRLG--LA-
--A-CGLTSKCCE--------------------VLSSALSC
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2465)
NPYL-NSLNLVRNDFNTSG
MLKLCS--------AFQNPASN--LW--I-------------------------------
---IGLWKQ-QFYAQVRRQ------LE----------EVQ-FIKPHVVIDGDWYSID---
--E---------------------------------------------------------
--------------------------------------------------DDRNWWKN--
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCAFP00000022816 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MACR----V-
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 324)
QWQ---LAWYLEMMG--K---EELKEFQLRLLE------
---------Q--QF-WG-------DP--PHA-LRAQ-LGKAR-GLEVASRLVAQYGEQQA
WVLALRT----WE-EMGLSRLCAQSR-AEAGLIP--------LSPG---SPMAPSMQSPN
SPISTEVLRFIQDPRT-----RNSG----QPLPPLLPNTSGHMW-NEDPSSCKYQ-EG--
---------LPDSPEFQGSPHQESPSAPMPTAVLGGWEAPPQPSPVPGKQEAPKAGWPVA
GTSGNHNPEKEGREGGRKGRRRE----EARKGGSPTQPRA--GRQPQVQPEGHPWPGPR-
--------GRKK----------------PYPTWSW-------EDK---------DLHQIF
IQLLLLHQPYPRGHES---L------TKGRWHHGA---------EKQGHLIEVEDLFGPG
LG--AQEEP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
QTVILHGVAGIGKSTLARQI-----RTAWK--E---------GQLYK
D--------RFRH-VFYFNCRELARSE-------TMSLTE-LITK-AQ-----AG-PVLP
AEQILS-QP----GRLLFILDDLDE-PRWVLGEQ------ES-E-----LHSSWSKQ-RP
-VHALLGSLLGKTVLPEVSLLVT-VRTTALRRLVPSV-G-----QLC--WVEVLGFSESG
RREYF----------------Y--KY-FKN---------KSQATQAFSLVEL-NPTLLTM
CLVPLVSWLLCTCLKKQMQRGE-EL-S--LGSSQTTTALWLYYLSQALP--------AQ-
-----------LLGAQLRGFCSLA---AEGF-WQ-GT----NLLSRR-------------
--DLRKHGFDEA-TISTFLKT----GILRKH---PSPLNYHLAHRCLQEFFAAVSCVLG-
G------EDG--------GSPHP--DSI-RGAE-KLLEVYEGH--DLFGAPTTHFL----
----FGLLSEQGEREMEAVFQCK---------------L--PRE-RKR-DLLRWAEAGLW
-----------------------------------PRLSAPQPCSLHLLHCLYEIQD-ED
LLTRAMAH---------------------F-------------------HGRSMCVQTGV
EF---LVC--T----FCFK-FCSH-VKR-LQLN----------------ESRW-------
------D-G---------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------Q------------
------AWRP--------------------------------------------------
-------------------SSVALFSWVPVT-----------------------------
----------------------------DACWQALFS-----------------------
--------------------------------TLRASG
PF13516 // ENSGT00900000140813_0_Domain_11 (2019, 2055)
SLKE---LD-LSGNH-LSH---
-------PAVQSLCENSGT00900000140813_0_Linker_11_12 (2055, 2065)
KVL----KFPPF13516 // ENSGT00900000140813_0_Domain_12 (2065, 2207)
CCHLETVR----------------------------
------------------------------------------------------------
-------------------------------LAGCGLTAEGCSDLASA-LSASPALAELE
L-SFNLLLDAGAQ-HLSRGLRQP--ACR----L---------------------------
-----------------------RRLLL--AGC-----SLTS------------------
------------------------------------------------------------
---------RCCG--------------------ALASALSTSPLL
PF13516 // ENSGT00900000140813_0_Domain_17 (2446, 2465)
-TELDLQQNELGDVG
VRLLENSGT00900000140813_0_Linker_17_18 (2465, 2715)
CG--------GLGHPTCQ--LT--L-------------------------------
---LWLDQT-QLSEEMTQM------LR----------ALQ-EEKPQLFVCS-KWKPRATV
PTEGLSRGETSGT------------------SSLKQQRQGSGDQKLPDRE---SGADS--
----CSAPFPER------SCPQVGQ-VQLGRPPSPAAPWD-LNTEPLR-IGDGFWGPLGP
VAPEWLAE-DGSLYPF13553 // ENSGT00900000140813_0_Domain_18 (2715, 2998)
RVHLPVAGSYHWP---DTGLRFEVRGPA-------TIDIEFCVWDQ
HLPRAGLWHSWMVAGPLFDIKAEPGA-VATVCLPHFVDLQGS--------SVDTSLFYAA
HLKDEGMLVETPARVEADHVVLENPSFSPIGVLLRTIHAALRFLPVTSVVLLYHHLRPGE
V-AFHLYLIPSDYSIRKAIDNEEKKFQFVQIHKPPPLASLYMGSRYTVSGS---------
EKLEIIPKELELCYRSPREPQLFSEFYVSHLGSGIRLQVRDKKDDTVVWEALVKPGDENSGT00900000140813_0_Linker_18_19 (2998, 3052)
LRP
VASLAPP-----------------------------APAARPFPTDAPAWLPF00619 // ENSGT00900000140813_0_Domain_19 (3052, 3133)
HFVDRHREQ
LVARVTSVDPVLDMLH-GQVLSEEQYERVRAEATTPSQMRKLFSFSRSWDRACKDQVYQA
LKEAHPHLIMELWEKWGGNSGKQAPPG---------
ENSCATP00000010211 (view gene)
ENSSBOP00000005801 (view gene)
ENSMLUP00000007905 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------------HT-
---------MEK----------------FCSI--WKHTCWDGVGDDLHYKII--------
-----------------------------------------------GGSQ-FTWLSSSR
TS--SQRRL----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
LTVVLQGAAGVGKTTLAKKW-----MLDWA--Q---------DHLPG
---------TFHS-AFYLSCKELGRQG-------PCTFAE-LLAK-KC-----PD-AQEA
GPQVLA-QG----QKILVVIDGFNE-LRVP-G------SLIH-D-----ICGNWEKP-KP
-VPVLLGSLLKRKLLPKATLLVT-TRPEALGELRLLV-D-----QPL--FIEVEGFLEAD
RRGYF----------------L--KH-FEQ---------QDQALRAFEGMKG-SPALFHM
G----ACWGFCTCLKGQLEKGE-DP-A--PT-CRTPTSLFLRFLCNQCSPA-A-----PH
------------LPTALHALSLLA---ARGI-WA-QT----SLFDGE-------------
--DLGQLGLSES-DLRPFLDG----KVLQADAD-CE-GCYAFLHLSVQQFLAALFYLLD-
GAEPG-------------DAGGW--AADIGNMR-ALLSKEERL-RNPSLTRVGYFL----
----FGLANQERARELEATFGCR---------------V--SLD-VRQ-ELLRVTRSRD-
-W-----------------------------------SFPSRADTRELMRCLYESQE-EA
LVREALVP---------------------I-------------------TELALHLQSER
DL---VHA--S----FCLR-HGQA-LRR-LWLQ----------------VEKG----IFL
-END-GAS----EP--------------------GCAVNRCC-SP--SRPARSQQD-HHA
LCRWTELC-SVLSSNENLTFLEVSQSFLSHS---------SVRILC--EQITR-V-----
TC--N-LQKVV--I-KNVSPV--HA----YR-DFCLAL----------------VGKKTL
THLI-LEGS-VPCDNMLLLLLCEALKHRRCN----LQYL--------RLG---------S
C------S----------------------------------------------------
---------DF---TQ-HLN---DFSLALKV-NQSLQSLDLTASELGD-EGVKLLCGS--
LRH-PSCFLQWL-S----------------------------------------------
------------------------LENCHLT-----------------------------
----------------------------EVCCKELAS-----------------------
--------------------------------TLIVNQRLTH---LC-LAKNN-LGD---
-------GGVRLLSEGL----SYPEC----------------------------------
--------------------------QLQ-TLV---------------------------
-------------------------------LCQCSITRRGCKHISDF-LQGDSCLTLLD
L-SLNPIA-TGLW-FLCEALKKP--SCN----L---------------------------
-----------------------KCLGL--CGC-----SLPSFCWQDL------------
-------ASAL-ASNQ
PF00560 // ENSGT00900000140813_0_Domain_15 (2357, 2381)
RLEVLDLGQNSLGQSGVTV-LFEALK-QR-SS-PLKRLR--LR-
--T-DESSAQTQK--------------------LLKEVRDSNPKLNIDCKAA------RA
---------------ASSSYCE--FP----------------------------------
----F-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPANP00000002923 (view gene)
ENSOANP00000014644 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------MTCSI
PF02758 // ENSGT00900000140813_0_Domain_0 (205, 326)
RSRLAEYLEELR--D---PELRKFKFHLED------
---------L--APAAG--------W--API-PWGR-TEKAD-SLDLAHLLVAHVGERGA
WELAVHV----FE-RIHRKDLWERAR-AEAEVRGK----S--RQ--------IIPGVLIV
--------CR--------------------------------AL---SCLKRWRGL----
------------------------------------------------------------
-----------------------------------------RLLYVPVYPDPRDVYREH-
--------IRRK----------------FRFF--EDRNARPGECV---------NLSQRY
TQLLLMERHPTPRDAAPVSP------TAQREDAGAPE--------RQPGPLQLETLLEPD
AS--WPEPP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
RTVVLRGAAGIGKSMLARKI-----MLDWA--D---------GLLYR
A--------RFDY-LFYVSCREMSRV-G------RGSLAG-LISG-RW-----PR-REAP
LADILR-RP----ERLLFVIDGFDE-LGRT----------------------PWKAK-MP
-VAGLLGGLLGKDLLPEASLLIT-TRPAALARLQPLL-E-----QPR--HAEILGFSRAG
RRDYF----------------R--KF-FGD---------GRQASRALGLVRD-VEALSAV
CFVPLVCWIVCTCLKQEMERGE-PP-G--QP-SKTTTAVYVFYLLSLLRPDPGG-SGP--
-----------RGRPVLAPLCSLA---AHGI-WA-RK----VLFDEG-------------
--DLSRHGLAGS-DVSAFLNL----SVFQKDIH-CQ-RLYSFVHRSFQEFFAALFYLTD-
EGGRE--------------G--G--RGSRHSVT-RLLEHYGRS-ETGYLALTVRFL----
----FGLLNGENKSYLERQLGCP---------------L--SPT-VKG-ELLAWIEAGAR
IG-----------------------------------GRTLERGALDLFSCLYETQE-EG
FI------------------------------------------------VIVACDLS-T
KM-DHVIS--A----FCVG-NCRN-ASV-VHLG----------------SE------EFS
-SEEAAEG--Q-GP---------------------------------------VGM---E
GTHLGDIL-VIISS---------SSTSI------------------------T-------
PC--S--------VEKCWLPD-------TYCEHLSSAL----------------RTNQNL
AELV-LDHN-ALGNR-GVKLLCQGLGHANCK----LQNL--------GLK---------K
C-----------------------------------------------------------
--------RFS---SA-ACQ---DISSALSA-NQNLVMMDLSNNALED-VGVKLNPP---
------LPFPLL------------------------------------------------
----------------PSLSHGHRLKKCYFS-----------------------------
----------------------------WAACEDLSS-----------------------
--------------------------------VLSTNPHLME---LD-LTGNA-LGD---
-------AGVQLLCVGM----KQTSC----------------------------------
--------------------------RLK-TLW---------------------------
-------------------------------LKICHLTRASCMELASV-LSVDCSLTELD
L-SLNDLEDAGVR-LLCEGLGQP--KCK----L---------------------------
-----------------------QKLRY--TRC-----FL--------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMLUP00000022275 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------HK----H----SPKEKGES--HSKYSEGKRIKEKYQNQ-
--------RRES----------------SCPTQSW-------RNE---------YLHQKF
TELLLLHRSQPRGYDF---L------LRRRWNHEV--------VDEQGNLMEIGDLFGPG
LS--TQEEP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
PTVVVHGVAGIGKSTLARQV-----RRAWE--E---------GWLFR
D--------RFKH-VFYFNCRELAQSE-------TMSLAE-LIKK-DW-----AA-PDAP
IGQILS-QP----EQLLFILDNLDE-PKWDLKEQ------SS-E-----FCPHWSQQ-QQ
-VHTLLGSLLKKTLLPTASLLIT-ARITALRKLIPSL-K-----HPR--WVEVMGFSESA
RKDHL----------------F--QY-FTD---------ESQAIRAFISVES-NPALFTM
CLMPLVSWLVCTCLKQQMEQEQ-PL-S--LT-SQTTTALCLHYFFHVLQ--------AQ-
-----------SLGTKLRGFCSLA---AEGT-WQ-GK----TLFSPE-------------
--DFREHGLDGA-NISTLLNM----GVLREH---PTAQSYSFIHLCFQEFFAAMSYALG-
N------RVL--------KSGCF--NNI-KIIT-DLTDVYDRH--NLFGEPTTCFL----
----FGLLSDQGMREMESIFKCS---------------L--SRE-GHC-SLLQLAKSEVL
-----------------------------------EKHLNLHPYSWHLLHCFYEIQD-EN
FMKNIF-T---------------------F-------------------NEMRICVQTDM
EL---LLF--T----FFFT-SCHH-IKR-LQLN----------------ESEQ-------
------H-R---------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------K------------
------AGRT--------------------------------------------------
-------------------FGAFLYSWGPFT-----------------------------
----------------------------DSWWQMFFS-----------------------
--------------------------------ILKVPG
PF00560 // ENSGT00900000140813_0_Domain_11 (2019, 2057)
SLRE---LD-LSGNS-LSC---
-------SAVQSLCDAL----KSPHCHLVTLR----------------------------
------------------------------------------------------------
-------------------------------LVSCGL-----------------------
------------------------------------------------------------
----------------------------------------TY------------------
------------------------------------------------------------
---------NCCQ--------------------DLASMLGACP
PF00560 // ENSGT00900000140813_0_Domain_17 (2444, 2463)
SL-RELDLWQNDLGDLG
VRLLCE--------GLRHPACQ--LR--N-------------------------------
---LWIERL-NILNTIIFI------II----------ILE-TVNNLEFLLK-TMTQWQKI
PTERG-VGKS-AL-----------------T-----------------------------
----------------------------EQGRQRPRGGDK-DKERELG-GSEQHWGPTGP
ATSRMVDK-HRSLYRFGLYLNRPHHGP---NTGLYFVVRGPV-------TIEIEFCAWDQ
FLDQIVPQHSWMVAGPLFDIKAEPGA-VEAVYLPHFIALQGQ--------HVDISWFQVA
HIKEDGILMEKPASVEPHYIVLENPSFSPIGVLLRKIHTALN-IAVSANVLLYYRRQPEE
V-NFHLYLIPSDCTVRKAIDDEEKKFQFFQIRKSAVQPHLHMGFRYTVSGS---------
DKLEIIPEELELCYRSPAKPQVFSEFYVGHFGSGIRLQIKSKIDETVVWEALVKSVNLPP
EVPQQ-------------------------------AFQASPSPPDAPTS
PF00619 // ENSGT00900000140813_0_Domain_19 (3051, 3135)
LHFVDQHRED
LVTRVTSVDAVLDKLH-GQVLSEEQYERVRAEPTNPDKMRMLFSFSKSWDWACKDQLYRA
LKKTHPHLIVDLWENMSSVSIN--I-----------
ENSSBOP00000016702 (view gene)
ENSPANP00000033792 (view gene)
ENSMICP00000037714 (view gene)
ENSOARP00000008143 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MIMA----SV
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 328)
RCK---LARYLEDLE--D---IDFKKFKMHLED------
---------Y--PSQKG--------C--TSI-PRGQ-TEKAD-HVDIATLMIDFNGEEKA
WAMAKWI----FA-AINRRDLYEKAK-RDEPEWE--N-----R---------NVSVL---
--------SQ-----------------------------------EESL-----------
-----------------------------------------------------EEEWMG-
------------------L-----------------LGYLSRISICRKKKDYCKKYRKY-
--------VRSK----------------FQCI--EDRNARLGESV---------NLNKRF
TRLRLIKEHRSQQEREHELL------AIGRTWAKIQD------SP--VSSVNLELLFD--
PEDQHSEPV----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
HTVVFQGAAGIGKTILARKI-----MLDWA--S---------EKLYQ
D--------RFDY-LFYIHCREVSLGT-------QRSLGD-LIAS-CC-----PG-PNPP
IGKIVS-KS----SRILFLMDGFDE-LQGAFDEH------TE-A-----LCTNWQKV-ER
-GDILLSSLIRKRLLPEASLLIT-TRPVALEKLQHLL-G-----QAR--HVEILGFSEAR
RKEYF----------------L--KY-FSD---------EQQAREAFRLIQE-NEILFTM
CFIPLVCWIVCTGLKQQMDSGK-SL-AR--T-SKTTTAVYIFFLSSLLQSQGGSQENH--
------------NSATLWGLCSLA---ADGI-WN-QK----ILFEEC-------------
--DLRNHGLQKA-DVSAFLRM----NLFQKEVD-CE-KFYSFIHMTFQEFFAAMYYLLE-
EDNH--------GEMRNMPQACS--KLPNRDVK-VLLENYGKF-EKGYLIFVVRFL----
----FGLINQERTSYLEKKLSCK---------------I--SQK-IRL-ELLKWIEAKAK
AK-----------------------------------TLQIEPSQLELFYCLYEMQE-ED
FVQRAMGH---------------------F-------------------PKIEIKLSTRM
---DHVVS--S----FCIE-NCRH-VES-LSLR----------------LLHN-----SP
KEEEEE------------------------------------------------------
---------------EEVRHSHMDHSVLSDS-----------------------------
------------------------------------------------------------
--------------------------EVAYS----Q-----------GLV---------N
Y-----------------------------------------------------------
---------LT---SS-ICR---GIFSVLSN-NWNLTELNLSGNTLGD-PGMKVLCET--
LQQ-PGCNIRRL-W----------------------------------------------
------------------------LGQCCLS-----------------------------
----------------------------HQCCFSISS-----------------------
--------------------------------VLSSNQKLVE---LD-LSHNS-LGD---
-------FGIRLLCVGL----RHLFCNLKKL-----------------------------
--------------------------------W---------------------------
-------------------------------LVSCCLTSACCEDLASV-LSTNHSLTRLY
L-GENALGDSGVG-ILCEKVKNP--HCN----L---------------------------
-----------------------QKLGL--VNS-----GLTSGCCPAL------------
-------SSVL-STNQNLTHLYLQGNALGDMGVKL-LCEGLL-HH-NC-KLQVLE--LD-
--N-CSLTSHCCW--------------------DLSTLLTSNQSL-RKLSLGNNDLGDLG
VMLLCE--------VLKQQGCL--LK--S-------------------------------
---LRLCEM-YFNYDTKRA------LE----------TLQ-EEKPELTIVFEPSR-----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSAMEP00000003461 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------MEPSA----Q
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
LGVS---LQRLLEQLG--Q---HEPSQFKSLLRP------
---------L--SPQGE--------L--QHM-PATG-VKEAD-GKQLAEILSNRCPSHWV
EMVTIQV----FD-KMNRTDLSERAK-DELGGHGG-------------------------
---------IARTPEPK--------------------------C---ANLEETQQRAE-E
K-----------------------------------------------------------
------------------------------------------------------EPAHN-
--------KEEK----------------VQQI--WKENFWPAASENIHT-----------
---------------------------------------------VTQKYKTLIPSCNPK
ML--AGPFR----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
HVVVLHGLVGVGKTTLAKKC-----LRDWT--Q---------DNLAR
---------PRPT-A-FLGCRTLSRRG-------TRSLAE-LLAH-SA-----LA-SQHA
LPQVLA-QA----QELLLVFDSFKE-LRVPSGLG----ALVR-D-----LRGDRRTQ-RP
-APVLLGSLLKGSLHP--MLVVT-TRPEVLSKLQLLL-E-----QPL--LV---GLSERD
RSAYF----------------R--RH-FAG---------EAQARHAFHLMQS-NAALFRL
GAAPAVCWTVCPCLRLQMHRGG-DS-R--PT-CQTARAL---------------------
------------------------------------------------------------
-------------------------------RL-CQ-GCYCFVRPSVQ-LLATAFYVLR-
GEDGQ-------------DAGSP--TWTS-GVQ-RLLSKQEGL-KNPHLAPVGRFL----
----FGLANEKRARELGTTFGCP---------------V--STE-VKQ-ELLECRVRSAQ
NR-----------------------------------HFSSVPDTKETLYCLYKSQE-EP
LVKEAMAQ---------------------V-------------------TEVSLHLQDAS
DI---VHA--S----FCLK-HCEN-LQK-MWLR----------------VERG----CSW
-RAM-LHGNQRLRL--------------------TAIFGKF-PPL--PLPSRFRND-HSV
LPFWMDPF-STFDSNKNLVFLDISQSFLSSS---------SVKLLC--EQIAS-A-----
TH--N-LPKVV--L-KNISPA--DA----YR-NLCIAF----------------GGHET-
THLT-LQGN-DQNN--MLPIWREILRHPRCN----LQYL--------RLE---------T
C------S----------------------------------------------------
---------AT---TQ-PWA---HLSSSLKI-SESLLCLDPTASERPD-KSARLLCTT--
PRH-QGA---GA-E----------------------------------------------
------------------------LESCHLT-----------------------------
----------------------------EDCCGDLSS-----------------------
--------------------------------ALIINQRLTH---PC-LAKNT--GD---
-------GGVELLCEGL----RSPDC----------------------------------
--------------------------QLL-MLV---------------------------
-------------------------------LWYCSITSSGCNDPSTL-LQPNSSL-TLD
L-GLNHIGIPGLK-FLGEALKKP--TCN----L---------------------------
-----------------------KCLWL--WGC-----AITPLSSEVP------------
-------SCAL-GSKKSLVTLDLGQNSL--------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPTRP00000069850 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----MSLD--I----QSLDIQCEE-LSD-ARWA----------------EL---------
----LPLL-------------------------------QQCQVV------RLDDC-GLT
EARCKDIS-SALRV
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1542)
NPALAELNLRSNELGDV---------GVHCVL--QGLQS-P-----
SC--K-IQKLS--L-QNCCLT--GA---GCG-VLSSTL----------------RTL
PF13516 // ENSGT00900000140813_0_Domain_6 (1618, 1642)
PTL
QELH-LSDN-LLGDA-GLQLLCEGLLDPQCR----LEKL--------QLE---------Y
C------N----------------------------------------------------
---------LS---AA-SCK---PLASVLRA-KPDFKELTVSNNDINE-AGVRVLCQG--
LKD-SPCQLEAL-K----------------------------------------------
------------------------LESCGVT-----------------------------
----------------------------SDNCRDLCG-----------------------
--------------------------------IVASK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
ASLRE---LA-LGSNK-LGD---
-------VGMAELCPGL----LHPSS----------------------------------
--------------------------RLR-TLW---------------------------
-------------------------------IWECGITAKGCGDLCRV-LRAKE
PF13516 // ENSGT00900000140813_0_Domain_13 (2215, 2237)
SLKELS
L-AGNELGDEGAR-LLCETLLEP--GCQ----L---------------------------
-----------------------ESLWV--KSC-----SFTAACCSHF------------
-------SSVL-AQ
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NKFLLELQISNNRLEDAGVQE-LCQGLG-QP-GS-VLRVLW--LA-
--D-CDVSDSSCS--------------------SLAATLLA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2464)
NHSL-RELDLSNNCLGDAG
ILQLVE--------SVRQPGCL--LE--Q-------------------------------
---LVLYDI-YWSEEMEDR------LQ----------ALE-KDKPSLRVIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMLUP00000022171 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------------------
PF05729 // ENSGT00900000140813_0_Domain_2 (779, 940)
AP
IGQILS-QP----EQLLFILDNLDE-PKWDLKKQ------SS-A-----LCPHWSQQ-QQ
-VHTLLGSLLKKTLLPTASLLIT-ARITALRKLIPSL-K-----HPR--WVEVLGFSESA
RKEYF----------------F--QY-FTD---------EGQAIRAFTSVES-NPALFTM
CLMPLVSWLVCTCLKQQMEQEQ-PL-S--LT-SQTTTALFLHYFFHVLQ--------AQ-
-----------SLGAKLRGLCSLA---AKGT-CQ-RK----TLFSPE-------------
--DFRKYGLDEA-NIPILLNM----GVLQEH---PTPRSYSFLHRCFQEFFAAMSYASG-
D------RKL--------KRGGF--INI-KTIT-DLTDVYDRR--DLFGEPTTCFL----
----FGLLSDQGMRELESIFKHS---------------L--SRK-RHS-YLLQLAQREVD
-----------------------------------CKHPNLEPYCWHLLHCFYENQD-TR
FLTEMMSN---------------------F-------------------YGMRICVQTHM
EL---LVF--T----FCFK-FCHR-MKT-LQLN----------------LLCW-------
------YHRPGPWGTPTKPGPSGFMT----------------------------EITSNF
FSHMVCLRAV---KDMKGKMNGVGD---------------------------I-L-----
GA--L-FW-QEN-----R-DG-------SLQL----------------------EIEF--
-----ESL---SHG--KED------CL---------------------------------
------------------------------------------------------------
----HPQALCW---A--LQ----IKKMNIYA-LSQKNFPHSGRNRPTK------------
------GVQQ--------------------------------------------------
-------------------CRMARYSWGPFT-----------------------------
----------------------------DSWWQMFFS-----------------------
--------------------------------IFKVPG
PF00560 // ENSGT00900000140813_0_Domain_11 (2019, 2057)
SLRE---LD-LSGNS-LSC---
-------SAVQSLCDAL----KSPHCHLETLT----------------------------
------------------------------------------------------------
-------------------------------LANCGLTAEDCRVLAHV-LSDNW
PF00560 // ENSGT00900000140813_0_Domain_13 (2215, 2240)
HLIELD
L-SFNKLQDTGAR-HLFQELTHS--HCK----L---------------------------
-----------------------WWLLL--VSC-----GLTS------------------
------------------------------------------------------------
---------GCCQ--------------------DLVSMLSNNVML-IVLDLQQNDLGDLG
VRLLSE--------GLRQSNCR--LK--Y-------------------------------
---LG-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSGGOP00000002851 (view gene)
ENSAMEP00000003422 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------PGDGD------E-------FHY-------NVTQRSQRFLSFLNPQ
TP--TQLTS----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 1048)
HAVVLHGPTGVGKTTLAKKCLKKKCLRDWT--Q---------DNLTQ
P--------S-ST-AFSLGGRALSRRG-------TRSLAE-LLLA-HS-----APAPQHA
LPQVLA-QA----QELPLVFDAFED-LRVPSGLG--A--LVR-D-----LCCDWRTP-RP
-APVLLGSLLKGRLLPM--LVVT-TWPEALSELRLLL-E-----QPL--LVEMEGLSERD
RSAYF----------------W--------------------------------------
------------------------------------------------------------
-----------------G--GAGA---ECAY-WR-PQS---ALCDAH-------------
--DLRRLRVH-ESALRPFLDR----GILQKGRD-CQ-GCYCFVRLSVQL-LATAFYVLR-
GEDGQ--------------DAGS--PTWTSGVQ-RLLSKQEGL-KNPHLAPVGRFL----
----FGLANEKRARELGTTFGAR---------------S--MDVK--Q-ELLECRVRSAQ
NR-----------------------------------HFSSVPDTKETLYCLYKSQE-EP
LVKEAMAQ---------------------V-------------------TEVALHLQDPS
DI---VHA--S----FCL-KHCEN-LQK-MWLR-----------------V---------
----KGMF----LE-------------------------SDAAWEPEAQADGSQHD-LVS
LHPWIDLC-SVFSSNKNLCFLDVSRSFLSPS---------SVRILC--EQIAR-V-----
TC--H-LQKVA--S-KRL----------AYR-DLCFAL----------------VGKMPL
THLV-LEGG-VR-SD-ELLLLRETLSRPRSN----VQHL--------RLG---------S
C------S----------------------------------------------------
---------AT---TR-QWA---DFSLTLEV-NQPLKCLDLTASELPG-GCAELLCTA--
LRH-PKCSLQRL-S----------------------------------------------
------------------------SGATQFR-----------------------------
----------------------------SQGD----------------------------
-----------------------------------------D---AC-ILLAL-FSC---
-------LVLQALITST----RYRGC----------------------------------
--------------------------RIS-KKG---------------------------
-------------------------------LSQCSISRHGCRCISKL-LHEDTGLSDLD
L-GLSPIA-AGPW-FLWEALEKP--RCN----L---------------------------
-----------------------RCLRL--YGT-----SITPSCCRGL------------
-------APAL-LSARRLETLDLGQNILGHSSVTA-LFEALK-QN-NS-PLKILR--LE-
--T-DESSVEIQK--------------------LLKDVKVHNLNL-TIERGDTR------
--------------ASRSPPCD--FL--S-------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMODP00000022770 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------PL-------NQ-----Y-DSSS--ISLETLFNSA
GN--TSNVP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 929)
QTVVLQGVAGIGKSTLARKV-----LQGWA--A---------GTLYP
D--------RFNY-VFYIGCQEAHFLRE-------SSMEQ-LITI-LC-GD-----NNAP
ITDILK-QP----QKLLFVLDGLDE-LQFLSDEQ---------------FYPSVNEK-DS
-VNALWGSLIQKKLLPQSTLLIT-IRIPVLEKLEPWL-K-----HSR--HVKVLGFSQTQ
KKEYF----------------D--LY-FTN---------GDLGKKALDFVLK-NKELSNE
CCAPLMCWIICSWMKQRQDQGK-SP-T--KA-LKNRTDVYMAYISTFLTTEK----SLSK
----------STQHVALRGLCFLA---AEGI-QS-QK----VLFTEG-------------
--DLRRHSLDGMDIS-SILNV----SGAQAGLS-GT-MLYSFRHLSFQEFFNAMFYLLE-
EKSLLG---------------------KSQNVK-ELIQ-------KQEASGTMQFL----
----YGLLEKENEKNLELKFDLR-----------------ISMP-LRK-EV-ETLKKKM-
D-------------------------------------KVKNTIDTLQDF-LYE-KK-DF
LVKVLFIN---------------------N-------------------VSYKKAETQ-S
FK-SQS-K--R----YPVK-MNTS-RLP-REES------------------KV----KSI
---YSKTEGK---GLK--K-------------------KTQRR-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------TTQ----------------GKG---------R
-GFDQ-GPK---------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMPUP00000008675 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------MG----------------------------------GA
AQ-EPSLP--Q----VELLVH-E--------QA----------------RT---------
----CRVV-------------------------------TWCPVH------RLDDC-GLT
EERCRDIR-SALQANSSLTELNLCSNMLGDG---------GVRLVL--QGLQS-P-----
AC--K-IRKLS--L-ENCCLT--KA---GCG-VLPDVL----------------RSVPT
PF00560 // ENSGT00900000140813_0_Domain_6 (1620, 1638)
L
RELD-LSNN-PLEDA-GLQLLCEGLLDPQCH----LENL--------HLE---------Y
C------N----------------------------------------------------
---------LT---GA-SCA---SLASALKA-KQHFKALVVSNNELEE-AGVRVLCQG--
LVD-SACQLETL-R----------------------------------------------
------------------------LENCGLT-----------------------------
----------------------------AASCEDLCG-----------------------
--------------------------------VVAAKPSLQV---LD-LGDNK-LGD---
-------QGIATLCSSL----LHSSS----------------------------------
--------------------------QIR-VLW---------------------------
-------------------------------LWHCDITTTGCRDLCRV-IRAKQSLKEMN
L-ADNVLGDEGAR-LLCESVLEP--GCQ----L---------------------------
-----------------------QALWM--KTC-----SFTTASCASF------------
-------GEML-AQNKHLMELHLSCNKLGDAGVRE-LCEGLR-QP-GT-VLRELW--IG-
--D-CDVGDEGCA--------------------SLASLLLVNC
PF00560 // ENSGT00900000140813_0_Domain_17 (2444, 2458)
SL-RELDLSNNCMSEEG
IRWLME--------SVEQPGCM--LE--Q-------------------------------
---LV-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSLAFP00000011752 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------LVSCD-----------K-HLN--CDQK-EE------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------SDLGKDEAIRDGTEYRIQ-
--------IKEK----------------FCIM--RDKNPLLRESEDFHHEIT--------
----------------------------------------------PEDRELLEHLFDGD
VE--TGEQP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
RTVVLQGMAGVGKTTLVRKA-----MLDWA--E---------GNLYQ
Q--------KFEY-VFYLSAREINQLK-------EKSFVQ-LVSK-NW-----PS-REGP
I-----------------------------------------------------------
------SSLLRKVMLPESSLLVT-TRLTAYKRLKPLL-K----S-HH--CVELVGMSEDE
RKEYI----------------Y--QF-FED---------KEWAIQAFSSLRN-NETLFNM
CQVPLMCRVICTCLKQQMQKRD-DV-T--LT-CQTTTALFTC----LFTLV-GRSCPSLP
------------DQIQLKNLCHLA---AKGI-WT-MT----YVFYRE-------------
--SLRKHGLSKS-DVSVFLDM----NILQKDSE-YE-NCYVFTHLHVQEYFAAMFYTLK-
GNRGT--------------RDHS--LQPFEDLK-LLLESKAYE--DTHLMQMKCFL----
----FGLLNEDRVMQLEKTFYCT---------------M--SLG-LKW-ELIQWIETLGN
SE-----------------------------------YSPSQLRFLELFHCLYETQD-GA
FISQAMSC---------------------F-------------------QKVDIDICGKI
HL---LVS--S----FCLK-HCQC-LRT-VKLT----------------VTI-----ALE
-----KML----DSRP-----PD--E------------------------TWQI-DHDHI
THCWQDLC-SVLHTNEDLRELDLFHNHLDEL---------VIKILC--QELRHP------
NC--K-LQKLLLRF---------VSF-PDGCKDIFSVL----------------TQNQNL
IHLD-LKGS-DVGDN-GVKSLCEALKCPDCK----LQNL--------RLE---------S
CN----------------------------------------------------------
---------LS---TV-CCL---NISNALIR-NQSLIFLNLSTNNLLD-DGVELLCEA--
LRH-PMCYLQRL-S----------------------------------------------
------------------------IERCGLT-----------------------------
----------------------------VAGCEDLSS-----------------------
--------------------------------SLISSKRLTH---LS-LADNF-LGD---
-------DGVKLISDAL----KHPRC----------------------------------
--------------------------
PF00560 // ENSGT00900000140813_0_Domain_12 (2127, 2208)
TLR-SLV---------------------------
-------------------------------LRRCNFTSLSTEYLSAS-LLLNKSLTHLD
L-GSNCLKDDGVK-LLCDAVRHP--SCN----L---------------------------
-----------------------QDLGL--MGC-----ALTSACCLDL------------
-------ASAI-LNNP
PF00560 // ENSGT00900000140813_0_Domain_15 (2357, 2385)
NLRILDLGNNNLQDDGAKI-LCEALR-HP-SC-NIERLE--LE-
--Y-CGLTALCCQ--------------------ELSATLRSNQRL-TKINLTRNILGREG
IKTLCD--------ALQSPRCK--LQ--I-------------------------------
---LGLCKE-AFDKEGQKL------LK----------AVE-VNNPHLTIKSTCNDQN---
--E---------------------------------------------------------
--------------------------------------------------EDGSWWQHF-
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCATP00000024116 (view gene)
ENSCAFP00000010008 (view gene)
ENSMUSP00000081819 (view gene)
ENSCATP00000004222 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------T----P--PG--------GPSSL---
------------------------------------------------------GN----
------------------------------------------------------------
---QS-TCLL-------------------------------EVSPVTPRKDPQEA
PF14484 // ENSGT00900000140813_0_Domain_1 (536, 658)
YRDY-
--------VRRK----------------FRLM--EDRNARLGECV---------NLSHRY
TRLLLVKEHSNPMQAQQQLL------DIGRGHTRTEG--------HQASPIKIETLFEPD
EE--RPEPP----RTVVMQGVAGIGKSMLAHKV-----MLDWA--D---------GKLFQ
G--------RFDY-LFYINCREMNQSDT------ECSMQD-LISS-CW-----PE-PSAP
LQELVR-VP----ERLLFIIDGFDE-LKPSFHDP------QG-P-----WCLCWAEK-RP
-MELLLNSLIRKKLLPELSLLVT-TRPTALEKLHRLL-E-----HPR--HVEILGFSEAE
RKEYF----------------Y--KY-FHD---------TEQAGQVFNYVRD-NEPLFTM
CFVPLVCWVVCTCLQQQLEGGG-LL-R--QT-SRTTTAVYMLYLLSLMQPKLRT-PRL--
-----------QPPPNQRGLCSLA---ADGL-WN-QK----ILFEEQ-------------
--DLRKHGLDGA-DVSAFLNM----NIFQKDIN-CE-RYYSFIHLSFQEFFAAMYYILD-
D--GE--------------R--G--AGPEQDVI-RLLTEYGFS-ERSFLALTIRFL----
----FGLLNEETRSYLENSLCWK---------------V--SPH-IKR-ELLQWIRSKAQ
SD-----------------------------------GSTLQQGSLEFFSCLYEIQE-EE
FIQQALSH---------------------F-------------------QVIVVSNIA-S
KM-EHMVC--S----FCVK-NCRS-AQV-LHLY----------------GA------TYS
-ADGEDWA--R-CS---------------------------------------AGA---H
TL------------------------L-------------------------V-------
QL--R--------PERTVLLD-------AYSEHLAAAL----------------C
PF13516 // ENSGT00900000140813_0_Domain_6 (1616, 1642)
TNPNL
VELS-LYRN-ALGSR-GVKLLCQGLRHPSCK----LQNL--------RLK---------R
C-----------------------------------------------------------
--------RIS---SS-ACE---DLSAALIA-
PF13516 // ENSGT00900000140813_0_Domain_8 (1773, 1796)
NKNLTRMDLSGNGVGF-PGVTLLCEG--
LRH-PQCRLQMIH-----------------------------------------------
---------------------------GQLE-----------------------------
----------------------------SGACIPRV------------------------
---------------------------------------GFC---LD-LTGNA-LED---
-------LGLRLLCQGL----RHPVC----------------------------------
--------------------------RLR-TLC---------------------------
-------------------------------LSPRQPIV----------------VFSVE
T-GLHHVGQTGLE-LLTSGDSPTSTSQT----S---------------------------
-----------------------CLCRL--GIC-----RLGSAACEGL------------
-------SAVL-QV
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2373)
NHHLRELDLSFNDLGDWGLWL-LAEGLQ-HP-TC-RLQKLW--LD-
--S-CGLTAKACK--------------------NLYFTLGI
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2465)
NQTL-TELYLTNNALGDTG
VRLLCK--------RLNHPGCK--LR--V-------------------------------
---LWLFGM-DLNKMTHSR------LA----------ALR-VTKPYLDIGC---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSNGAP00000012000 (view gene)
ENSCJAP00000015134 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---ASFFSDFG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 322)
-LMWYLEELK--K---EEFRRFKEYLKQ------
---------L--TSHLE--------L--KPI-PWAE-VKKAS-REELANLLLKHYEEQQA
WNVTLRI----FQ-KMDRKDLCVKVT-RER------------TG--------------YT
K---------------------------------------------------TYQD----
------------------------------------------------------------
----------------------------------------------------------H-
--------AKKK----------------FSRL--WSSNSVTEIHQY--------------
---------------------------------F-------HQEVKQEECDHLDRLFASK
EP--GKH-P----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
RTVVIQGAQGIGKTTLLMKL-----MWAWS--D---------SRVYQ
D--------RFLY-AFYFCCRELRELP-------AVSLAD-LICR-EW-----PDP-AAP
VTEIVS-QP----ERLLFVIDSFEQ-LEADVNQP------DS-D-----LCGDWKEK-RP
-VQVVLSSLLRRKMLPEASLLVA-VKPTCPKELRERV-A-----ISE--IYEPRGFDESD
RLVYF----------------C--CF-FQD---------PKRAMEAFTLVRE-SQQLFAT
CQVPLLCWLLCTALKQEMQKGK-DL-A--LT-CQNTTSVYASFIFNLFTPDG-AAGPTE-
-----------DSRRQLKGLCALA---AEGM-WT-DT----FEFGDE-------------
--ALRRHGIADA-DIPALLGT----KVLLKFGE-RV-SSYAFIHVCIQEFCAAMFYLLK-
SHLDH-----------------P--HPAVRSVQ-ELLVNNFQKARRPHWIFLGCFL----
----TGLLNKKEQEKLNAFFGFQ---------------L--SQE-IKQ-QFEQCLKSLGE
H-E----------------------------------DLQEQVDALAIFYCLFEMQE-PA
FVRQAVNL---------------------L-------------------QEGSFRIVDSS
DL---VVS--A----YCLK-YCSS-LKK-LSFS----------------IQN-----VFK
-KED-THS-----------P-----------------------------------VSDYS
LTCWHHIC-SVFTTSGCLREVQVQDSTLSES---------AFATWC--NQLRHP------
SC--R-LQKLGINN---------VSF-SGQSSQLFEVL----------------FHQPDL
KYLS-LTLT-KLSRD-DVKSLCDALNAPAGS----VEEL---------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPEMP00000026823 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------MDESAVK---MI
PF02758 // ENSGT00900000140813_0_Domain_0 (210, 322)
QCLQKLS--A---AEFLMFKELFRR------
---------E--MEEYE--------H--MQV-SWDI-IKMAS-ERDLILLLNRNYLRH-L
WGVLSAL----FV-QVNRTDLWTMAQ-TLKID----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------DQNSYKEI-
--------MKII----------------FQHI--WSKETFVQMDEE--------------
---------------------------------N-------YQITVEEEYNALQEVFDCP
-----LE-P----V
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TTVVLGSKGKGKTTFLRKA-----MLDWA--S---------GNLWQ
N--------RFQY-VFFISLISLNNVT-------ELSLAE-LMLA-KL-----SQS-SET
LEKVIS-DP----KRVLFILEGLEY-LKFDLE-L------RT-N-----LCNDWGKT-LP
-TQVVFSSLLQKVMLPGSSLLIE-LGNPSVPKIYPLL-Q-----YPK--KITIQGFTDEI
TRFYC----------------M--CF-FSD---------YQKGLHVFDYLKK-SEQLLTL
CRNPYMCWVSCSTLMWQWDREE-EM-N--LV-GVTDSIIYTSFMVSAFRSVY-DNCPPN-
-----------QNRARLKTLCTLA---AEAM-WK-QV----FVFTSE-------------
--DLRRNGISES-EKAVWLGM----NFLCTQG-----EDFMFYHPTLQSYFTALFYFLR-
QDEDT-----------------P--HPVIGSLP-QLLREVYDH-GQTIWLLTGIFV----
----FGIATEKVTNILKPHFDFI---------------P--SKD-IKQ-EILKCFRSLSQ
AEC----------------------------------SEKL-MNTQRLFESLLDNQE-ES
FETEVMDL---------------------F-------------------EEMTVDISNVD
AL---LVA--T----HGLL-KSQK-LKK-LHLH----------------IQHK----VFS
-EIYNPED-----------G-------------------D-L-------EEFE-YNKKKA
IKYWSMLC-KIFQ---DLQVLDLDCCNFNET---------AIRRLC--DSMYPSSKDPLT
AF--K-LQSLSCPF---------MTN-FGDDGALFHTI----------------LLLPHL
KSVN-LYGT-NLSND-AVENMCSVLKCPACR----VEEL--------LL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------GKCDIS-----------------------------
----------------------------SEACGMIAT-----------------------
--------------------------------CLIYR-KVKH---LS-LVENP-LKN---
-------KGVRLLCEIL----KHPSCVMETL-----------------------------
------------------------------------------------------------
------------------------------MLSCCCLNFNACCHLYEA-LLCNKYLSLLD
L-SSNVLEDFGVN-ILCEALKDP--KCT----L---------------------------
-----------------------QELWL--SGC-----FLTSNCCGGI------------
-------SAVL-TSKKNLKTLKLGKNNIEDAGIKQ-LCEALR-NP-NC-KLQCLG--LD-
--M-NDFKTDCCA--------------------DLASALTTCRTL-TSLNLDWITFDHDG
LQLLCE--------ALSHKSCN--LK--V-------------------------------
---LGLDKS-ALTEESQML------LQ----------AVE-MIG-KLDILHCPWVKE---
--ERE-----RR------------------------------------------------
--------------------------------------------------GVRLVWNSTN
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSRROP00000045869 (view gene)
ENSAMEP00000004163 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MS---ESFF
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
TDFG---LLWYLEELR--K---EEFWKFKELLRQ------
---------E--PLQLD--------L--KPI-PWTE-LKKAS-REDLAKLLDKHYPGKWA
WEVTLNL----FL-QIDRRDLWTKAQ-EEIRN----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------KPNPYRNH-
--------MKEK----------------FRLI--WEKETCLPVPSD--------------
---------------------------------F-------YKEITKREYEKLCAAYH-A
SQ--AGEPS----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
PTVVLQGPEGIGKTTLLRKV-----MLEWA--E---------GNLWK
D--------RFTF-IFFLNGCEMNAIT-------ETSLVE-LISR-DW-----PPS-SDH
IEGIFS-QP----ERILFIMDSLEE-LRFDLE-L------TT-Q-----LCDDQRQR-QP
-TQIILSSLLRKKMLPQSSLLIA-LGTVGMRKNYFLL-Q-----CPK--YITLSGLSEHE
RKLYF----------------Y--HF-FRE---------RSKALKAFSFVRG-NVPLFLF
CHNPLICWLICTCVKWQLEKGE-DL-R--IV-SESTTSLYASFFISVFKSRN-ENCPPK-
-----------QSRARLKGLCTLA---AEGM-WT-RV----FVFGRG-------------
--DLRRNGVSES-DALMWVST----RLLQRNG-----ECFTFTHVCLQQFCASMFYLLK-
EPKDG-----------------A--HPAIGSVT-QLVTAGVSQ-VQGRLSQMVTFL----
----FAFSTEKITNMLETSFGFL---------------L--SKE-IKQ-EIIQCLKSLSG
CN-----------------------------------PDQKAVNFQELFSALFETHE-KE
FVKQVMDF---------------------F-------------------EEVSIYIGNTE
DL---VIS--A----FCLK-HCQN-LQK-LHLC----------------IEN-----VFS
-DDSESST-----------N-------------------L-S-------KK---------
LSFWQDLC-SAFTTSKNFQMLDLDNCTFSEA---------SLAILC--KALAQP------
VC--K-LEKFMYNF---------ASH-LGSGPYLHKAV----------------LHNP
PF00560 // ENSGT00900000140813_0_Domain_6 (1619, 1644)
HL
KHLN-LHGT-SLSHA-DVGQLCEMLRHPRCS----VEEL--------ML-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------GMCDVT-----------------------------
----------------------------TEACEDIAS-----------------------
--------------------------------VLVCNKKLKL---LS-LVENP-LRN---
-------EGMLLLCDAL----KHPDCVLERL-----------------------------
------------------------------------------------------------
------------------------------LLMCCCLTSVACDYLSEA-LLCNTSLSFLD
L-GSNFLEDKGVA-SLCETLKHP--HCN----I---------------------------
-----------------------EQLGL--EDC-----YLTSVCCKDL------------
-------SAVL-VGNEKLKTLKLGSNDIQDVGVEQ-LCEALK-HP-DC-KVENLG--LE-
--M-CQLTSACCG--------------------ALASALSVCRSL-KSLNLDWIGLDRAG
ALVLCE--------ALSHPDCA--LR--Q-------------------------------
---LGLRKS-PFDKDIQML------LT----------AVE-ENNPRLTISHLLWVSE---
--EYR-----IR------------------------------------------------
--------------------------------------------------GVPT------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSTBEP00000004281 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------MSLSLCLCED
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
PRDG---LLSHLVALD--Q---YQLEEFKLGLQTQQLCS-
----------------N--------S--RPL-PWAD-VKAAD-PLNLYLLLGEHFSERQA
WEVALSI----FK-SMNLTSFWEKVR-AEMAGTVQ-AQGPQDPKQG------DPEVSEEA
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------------AGHRER-
--------MKAK----------------ILAM--WDNTSWPEDHIYVRNMTE--------
----------------------------------------------Q-EHEELQNLLDPN
R---TGTQA----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
QTIVLEGRAGV-KTTLAVKA-----MLHWA--C---------GLLFE
H--------KFSY-VFYLSCHAMRGVT-------ETSFAE-LLSW-DW-----PN-SQAP
IEEFMS-RP----ERLLFIVDGFEE-MSIPEA-------SDS-P-----PCTDWYQK-LP
-VNKILFSLLKKHLVPFATLLIT-TRILSTNHLKDLL-V-----NPC--CVQIVGFTDKD
QEEYF----------------I--RY-FSD---------QKEARKILHWIRK-NETLFNA
CCAPMMCWAVCSSLKQQKAR-R-DL-Q--LV-ARTTTGLYACFVSSLFSVAE-VSWAERS
------------WPRQWRAFCALA---AEGM-WN-AA----FTFGKE-------------
--DTGI-CLEAP-FLDSLFKF----SILQKVGD-CE-DCVTFTHASFQEFFAAMFYVLE-
GTEGS--------------L-GH--STKYQEMK-TVLSSVLVN-RSIYWTPVVRLL----
----FGLLNQNLARKLEGTLHCK---------------V--SLQ-IME-DLLEWEEELHE
PE-----------------------------------MVSVPFDVLQFFHCLYETQE-ED
FVKKMLSH---------------------I-------------------FEADLDLSGSM
EL---RVS--A----FCLK-HCGR-LNK-LRLS----------------VSG-----PIF
-QRVSDVE----TA--------------------------------------XXXX-XXX
XXXXXXXXXXXXXXXXXXXXXXXXNSKLP-A---------SEEVFC--HALRS-Q-----
RC--R-LQKLT--X-XXXXXX--XX---X-X-XXXXXX----------------XXXXXX
XXXX-XXXX-XXXXX-X-XXXXXXXXXXXXX----XXXX--------XXX---------X
X------X----------------------------------------------------
---------XX---XX------------XXX-XXX----XXXXXXXXX-X--X-------
------------------------------------------------------------
---------------------------XXXX-----------------------------
----------------------------XXXXXXXXX-----------------------
--------------------------------XXXXXXXXX--------XXXX-XXX---
-------XXXXX---XX----XX-------------------------------------
------------------------------------------------------------
--------------------------------------XXXXXXXXXX-XXXXXXXXXXX
X-XXXXXXXXXXX-XXXXXXXXX--XXX----X---------------------------
-----------------------XXXXL--SDC-----AITRQGCQEL------------
-------ANAL-KHNHNVQVLDLGNNDLQDEGVKP-LCEALR-PQ--G-ALTTLG--LE-
--K-CSLTPACCQ--------------------PLSSVLSSSQSL-VSLNLLGNDLEVDG
VKVLCR--------ALRKPTCK--LR--K-------------------------------
---LG-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
MGP_CAROLIEiJ_P0034749 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------M---ASFISDFG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 323)
-LLWYLRELN--K---KEFMKFKDFLIQ------
---------E--ILELK--------L--KQV-SSTK-VKKAS-REDLANLLLKCG-ESQA
WDMTFKI----LQ-KINRKDLTERAT-GAI------------AG--------------NP
T---------------------------------------------------LYRD----
------------------------------------------------------------
----------------------------------------------------------H-
--------LKKK----------------LTHD--CPKKFNVRIQDF--------------
---------------------------------I-------KETFIQNDYDTFENLLISK
GT--EGK-P----H
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
MVFLKGMAGVGKTLMLKNL-----MLAWS--K---------GLVFQ
N--------KFSY-VFYFCCQDVKKMK-------RASLAE-LISK-EW-----PNA-SAP
IEEILS-QP----EKLLFIIDSLEG-MEWDLTKQ------ES-E-----LCDNCMEK-QP
-VSTLLSSLLTRKMLPESSFLLS-TTPETFEKMEDRI-Q-----CTD--VKTATAFDEKS
IKIYF----------------H--KL-FQD---------RKRAQEAFSLVRE-NKQLFTI
CQAPLLCWMVATCLKEEIEKGG-DP-V--SL-CRRTTSLYTTHIFNLFIPQS-AQYPSK-
-----------KSQDQLQGLCSLA---AEGM-WT-DT----FVFGKE-------------
--ALRRNGIFDS-DIPTLLDI----GMLGKIRE-FE-NSFIFLHPSVQEVCAAIFYMLK-
RHVDH-----------------P--SQDVKNIE-TVLFMFLKK-VKTQWIFLGCFI----
----FGLLQKSEQEKLGEFFGHR---------------L--SKN-IHH-KLYQCLKTLSG
NAE-----------------------------------LQEQIDGMKLFSCLFEMED-EA
FLVKAMNC---------------------M-------------------QQINFVAKNYS
DF---IVA--A----YCLK-HCST-LKK-LSFS----------------TEN-----VLN
--EG--DQ-----------S-----------------------------------YMDEL
LICWNNMC-SVFVRSKDIQELRIKDTNFNER---------AFLVLY--SHLKYS------
SC--I-LKVLEANN---------VS--FGDNHVLFEL-----------------IQNSSL
QYLD-LSCS-FLSHN-EVKLLCDVLXQAECN----IEKL--------ML-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------ANCSLSEQCWGYLSDV-LM
PF13516 // ENSGT00900000140813_0_Domain_13 (2212, 2233)
QNKTLSHLD
I-SSNDLKDEGLK-ILCRSLTLP--HCV----L---------------------------
-----------------------NSLCL--SCC-----GITEKGCQDL------------
-------AEVL-K
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
NNQNLKYLHVSYNKLKDTGVML-LCDAIK-HP-NC-HLKDLR--LE-
--A-CEITDASNE--------------------ELCYAFTQCETL-QTLNLMGNAFEVSR
MVFFPR--------F---------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMICP00000046552 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MPRP----PVRDGLCL
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 323)
LSTYLEELE--A---VELKKFKLYLGT------
---------V--TERGE--------D---KI-PWGR-METAG-PLDMAQLLTGHFGTEAA
WLLALGL----FE-RINRRDLWEKGQ-REDLVRDT----P--PG--------GPSSL---
------------------------------------------------------GS----
------------------------------------------------------------
---QS-ACLL-------------------------------EVSPGTPRKDPGEAYRDS-
--------VRRR----------------FRLM--QDRNARPGECV---------NLSRRY
TRLLLVREHSHPTRARQKLL------DTGRGRARAAA--------QQASPIRLETLFEPD
AE--RPEPP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
RTVVLQGAAGIGKSMLARKV-----MLDWA--D---------GKLFR
G--------RFDY-VFYVNCRDMNQG-A------ERSARD-LVAG-CW-----PE-PRAP
LHELLR-VP----ERLLLIIDGFDE-LEPSFHDA------QG-P-----WCACWEEK-RP
-AELLLSSLIGRKLLPELSLLIT-TRPAALEQLHGLL-E-----QPR--CVEILGFSEAE
RKEYF----------------H--KY-FPD---------AQQASQAFHFVRD-NEPLFTM
CFVPMVCWVVCTCLRQQLEGGG-LL-R--QA-CRTTTAVYLLYLLSLTPPAPAT-R----
---------------SLRGLCSLA---CDGL-WH-QK----VLFEER-------------
--DLGRHGLRGA-EVSAFLNM----DIFQKDIN-CE-KYYSFIHLSFQEFFAAMHYVLD-
G--AE--------------R--G--SGPGGTVT-RLLAEYGFA-ERSFLALTVRFL----
----FGLLNEETRGYLEKSLSWK---------------V--SPG-IKA-ELLGWIESKAQ
SE-----------------------------------GSTLQRGSLELFSCLHEIQD-EE
FIQRALSP---------------------F-------------------RVVVVSDIA-S
RM-EHMVS--S----FCVK-SCRN-VQV-LHLR----------------GA------AHR
-AHEDDRA--G-GT---------------------------------------LGA---Q
TP------------------------L-------------------------A-------
------------------------------------------------------------
--------------------------------------Q--------SLK---------K
C-----------------------------------------------------------
--------HIS---SS-ACQ---DLAATLIA-NKHLIRMDLSGNSLGL-PGIELLCEG--
LQH-PKCRLQMIR-----------------------------------------------
------------------------LK----------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------ICHLTAAACEDLAPT-LRE
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2236)
NRSLVELD
L-GLNELGDPGTL-LLCQGLRHP--TCT----L---------------------------
-----------------------QTLRL--GIC-----RLGAVACAAL------------
-------CDVL-QA
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2377)
SRHLLELDLSFNDLGDGGLRL-LGAGLR-HP-AC-RLQKLW-----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSDNOP00000034243 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MSN---------------------------QKKRKDMEPA-----
--------CTWS------------------------------------------LLCE--
-----------------------------------------EK-----------------
----AASSSL-------------------------------ASYSQKELADYREIFRKH-
--------LKEK----------------YLKI-GVDKCYHHGK--------------HIE
SRLLLVKEHGISEKTPCGIP-------------------------QQDHFIEMEHVFDPD
EE--GFSSS----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
QTVVIEGCAGIGKTAVVHKF-----MFDWA--A---------GMVTP
G--------RFDC-LIYLNCREISHIS-------NLSAAD-LITN-TF-----QD-INVP
IWDIILAYP----EKLLFILDGFPE-LQYPVSVQ------EE-N-----LSGNPQEK-KP
-VETILFSFVRKKLFPESSLLIT-ARPTSMKKLHTLL-K-----QPI--QAEILWFSDAE
KRAYF----------------L--SQ-FSG---------ANVAMRVLCSLQE-SEGLDIM
SSLPIVSWMLCSVLQLQGDGDR-SL-M--RS-LQTMTDMYLFYFSKCLKTLKGIS-VW--
-----------KGQSCLWGLCSLA---AEGL-QN-QR----VLFEVS-------------
--DLRKHGIGVYDTHCVFLNY-----FLKKAEG-HV-NVYTFLHFSFQEFLTAVFYALK-
NDSSW---------------------MFFDQVGKTWQELFQQY-GKGFSSLTIQFL----
----FGLLHKAKGKAVETTFGRK---------------V--SPG-LRE-ELLKWTEREIK
DK-----------------------------------SSRLQIEPMDLFHCLYEIQE-EE
YAKRIIDD---------------------L-------------------QSIILLQPTYT
KM-DILAM--S----FCLK-SSHSHLS----------------V-----SLKCQHLLGF-
-EEEERAT----------------------AFMP--------------------------
---------PALTPNQSFQ-----LCTLPVS---------RLHLFC--QALCNP------
CC--K-IKDLKL---IFCHLT------ASCGRD---------------LSFVFET-NQYL
TDLE-FVKN-TLEDS-GMKFLCEGLKQPKCV----IHTLR--------FYRCLI-SPA-S
CG------------EL----AAVLTTSRWLTELEFSETKLEASALKLLCEGLKDPNCK-L
QKLKLCASLLPESSEI-VCK---YLASVLIC-NPNLTELDLSENPLGD-TGVKYLCEG--
LRH-SNCKLEKL-D----------------------------------------------
------------------------LSTCYLT-----------------------------
----------------------------DVSCLELSS-----------------------
--------------------------------FLQVSQTLKE---LF-VFANS-LGD---
-------MGVQHLCEGL----RHTKGIIENL-----------------------------
------------------------------------------------------------
------------------------------VLSECSLSATCCEPLAQV-LRSTRSLTRLL
L-INNRIEDLGLK-LLCEGLKQP--DCQ----L---------------------------
-----------------------KDLAL--WTC-----HLTGACCQDL------------
-------CNAL-YTNEHLRDLDLSDNALGDKGMQV-LCEGLK-HP-CC-KLQTLW--LA-
--E-CHLTDACCG--------------------ALASVLNRNETL-TLLDLSGNDLKDFG
VQMLCD--------ALIHPICK--LQTFY-------------------------------
------IDTDHLHEETFRK------ME----------ALK-TSKPGITW-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSOGAP00000006687 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MKMA----ST
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 326)
RCR---LARYLEDLE--D---VDFKKFKMHLED------
---------Y--PPQKG--------C--ISL-PRGQ-TEKAD-HVDLATLMIDFNGEERA
WAMAVWI----FA-AINRRDLYEKAK-MDEPEWGN--TQ--VS---------NPHVI---
--------CQ-----------------------------------QESI-----------
-----------------------------------------------------EEEWMG-
------------------L-----------------LGCLAKLSICKKKKDYCKKYRKY-
--------VRSR----------------FQCI--EDRNARLGESV---------SLSRRY
TRLQLVKEHRSQQEREQELV------AIGRT--KIWD------KP--MSPIKMELLFE--
PECEFSEPI----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
HTVVFQGAAGIGKTILARKI-----MLDWA--S---------GKLYQ
D--------KFDY-LFYIHCREVSLVT-------ERSLRD-LMVS-CC-----PD-PNPP
ICKIVS-RP----SRILFLMDGFDE-LQGAFDEH------TG-A-----LCTDWQKA-ER
-GDILLSSLIRKKLLPEASLLIT-TRPVALEKLQHLL-Y-----HPR--HIEVLGFSEAK
RKEYF----------------F--KY-FSD---------ETQAREAFRLIQE-NEVLFTM
CFIPLVCWIVCTGLKQQMESGK-SL-AQ--T-SKTTTAVYTFFLSSLLQSRGGSHEHY--
------------LAAHLRGLCSLA---ADGI-WN-QK----ILFDES-------------
--DLRKHGLQKV-DVSSFLRM----NLFQKEVD-CE-KFYSFIHMTFQEFFAAMYYLLE-
EEEVEEE-----ERMRNVLGRSS--KLPNRDVT-VLLENYGKF-EKGYLIFVVRFL----
----FGLVNQERTSYLEKKLSCK---------------I--SQQ-IRL-ELLKWIEVKAK
AK-----------------------------------KLQIQPSQLELFYCLYEMQE-ED
FVQRAMNH---------------------F-------------------PKIEINLSTRM
---DHVVS--S----FCIA-NCHS-VES-LSLG----------------FLHN-----TP
KEEEE--------E------------------------------------E---------
---------------EEGRHLDMVQRVLPGP-----------------------------
------------------------------------------------------------
--------------------------HAACF----H-----------RLV---------N
C-----------------------------------------------------------
--------YLT---SS-FCR---GLFSVLST-NQSLTELDLSDNTLGD-SGMKVLCEA--
LQH-PSCNIQRL-W----------------------------------------------
------------------------LGRCGLS-----------------------------
----------------------------HQCCFDISL-----------------------
--------------------------------VLSSNQKLAE---LD-LSDNA-LGD---
-------FGIRLLCVGL----KHLFCNLKKL-----------------------------
--------------------------------W---------------------------
-------------------------------LVSCCLTSACCQDLASV-LSPSRSLTRLY
V-GENTLGDSGVG-ILCEKAKHP--QCN----L---------------------------
-----------------------QKLGL--VNS-----GLSSVCCTAL------------
-------SSVL-SSNPNFTHLYLRGNTLGDMGVKL-LCEGLL-HP-NC-KLQVLE--LE-
--N-CSLTSHCCW--------------------DLSTLLTSNQRL-RKLSLGNNDLGDLG
VMMLCE--------VLRQQSCL--LQ--S-------------------------------
---LQLCEM-YFNYETKRA------LE----------TLR-EEKPELTVVFEPSW-----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSNLEP00000028046 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MAMAKA-RNPREA---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWALSDLE--E---NDFKKLKFYLRD------
---------M--TLSEG--------Q--PPL-ARGE-LGGLI-PVDLAELLISKYGEKEA
VKVVLKG----LK-VMNLLELVDQLS-HIC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LH--DYREIYREH-
--------VCC-----------------LE------EW---QEA----------GVNGRY
NQVLLVA-KPSSESPESLAC------PI-------PE-----Q-ELDSV--MVEALFDSG
EK--PSLAP----S
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
LVVLQGSAGTGKTTLARKM-----VLDWA--T---------GTLYP
G--------RFDY-VFYVSCKEVVLLLE-------SKLEQ-LLFW-CC-GD-----NQAP
VTEILR-QP----ERLLFILDGFDE-LQRPFEEK------LK-K-----RG-----L-SP
-KESLLHLLIRRHTLPTCSLLIT-TRPLALRNLEPLL-K-----QAR--HVHILGFSEEE
RARYF----------------S--SY-FTD---------EKQAARAFDIVQK-NDILYKA
CQIPGICWVVCSWLKGQMERGK-VV-L--ET-PRNSTDIFMAYVSTFLPPDD-DGGCSEL
----------S-RHRVLRSLCSLA---AEGI-QH-QR----FLFEEA-------------
--ELRKHNLDGPRLA-AFLSS----NDYQLGLA-IK-KFYSFRHISFQDFFHAMSYLVK-
EDQSRL----------------G--KGSRREVQ-RLLEVKEQE-RNDEMTLTMQFL----
----LDISKKDSFSNLELKFCFR-----------------ISPC-LAQ-DL-KHFKEQM-
E-------------------------------------SMKHNRTWDLEFSLYEAKI-KN
LVKGIQMN---------------------N-------------------VSFKIKHSN-E
KK-SQSQN--L----FSIK-SSLS-HRP-KKEQ------------------KC----PSV
---HGQKEGK---DNI--A-------------------GTQKE-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------AST----------------GKG---------R
-GTEE-ADEEVRY-TPK-NTYI--------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPEMP00000002485 (view gene)
ENSCGRP00000011966 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------MEEISGYD---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 325)
LALCLQKLS--Q---ADFMTFKVLLRK------
---------A--PEEFE--------L--NPK-PWTE-IKMAS-RKGLVMLLNKNYPGK-V
WDIALRQ----ES-MN--------QT-ARWKW----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------LGTPYKET-
--------MKTL----------------FNFI--WNFESGIYVPYH--------------
---------------------------------E-------FEIITEMQYSSLQVVFEPK
-----SQPP----VT
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 905)
AVILGNKGEGKTTFLRRA-----MLDWA--S---------GDLWQ
N--------RFQY-VFFFSLMSLNDIT-------ELSLAQ-LLLS-KL-----SES-SES
LEDILS-DP----RRILFILEGFDY-LKFDLE-L------RT-N-----LCNDWRKT-VP
-TQIVLSSLLQKVMLPESSLLLE-LGNLSVPKIYPLL-Q-----KPR--DITLGGYTDEA
IKYYC----------------M--SF-MYS---------YTEGSAIFNYLQT-MQPLLTL
CKNPYKRWMVCSTLKCQRKRGE-NM-N--ID-YKPDSALYASFMVNSFRAEY-ANRPSR-
-----------QNRDQLKPLCTLA---VEGM-WK-LV----FVFKPE-------------
--DLYRNGISES-GQIVWLKM----NFLNTRG-----DHFVFYHPALQLYFAALFYFLR-
QDEDR-----------------P--HPVIGSLS-QLLREIYAH-DQNQWLLTGIFV----
----FGIATEKVAAILEPHFGE--------------------------------------
--G----------------------------------GEGQ-RSPQSLFASLVDNQE-EG
FIRQVMDL---------------------F-------------------EEMTIDISSAD
AL---SVA--T----TCLL-KSQR-LKN-LHLH----------------IQHR----VFS
-EKYKPED-----------G-------------------D-L-------E-AFKHEKINA
IKHWRMLC-RVFQ---NLEVLDLDSCNFNET---------AISLLY--LSMYPSHRGALN
AF--K-LQSFSCSF---------VTN-FGH-GVLFHTL----------------LQLPHL
KSLN-LYGT-NLSND-VVENMCCVLKCSTCR----VEEL--------LL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------GKCNLS-----------------------------
----------------------------SEACGIIAT-----------------------
--------------------------------SLIFC-KVKH---LS-LVENP-LKN---
-------KGVMLLCETL----KRPSCDLETL-----------------------------
------------------------------------------------------------
------------------------------MLSYCALTFIACGHLYEA-LLHNKYLSLLD
L-SSNFLGDTGVN-ILCKALKDP--MCP----L---------------------------
-----------------------QELWL--SGC-----YLTSDCCEEI------------
-------SAVL-T
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
HNQKLKTLKLGNNNIQDTGVRQ-LCEALS-NP-EC-KLQCLG--LD-
--M-CEFTTGSCA--------------------DLALALTTCKTL-TMLNLDRMTFDRDG
LKLLCE--------ALNHKDCN--LK--V-------------------------------
---LGLDKS-AFCEESQKI------LQ----------EVE-KKN-TLDILHYPWIKE---
--ESK-----KR------------------------------------------------
--------------------------------------------------GVRLVWNSEN
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSGGOP00000008682 (view gene)
ENSCATP00000029059 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MAMTKA-RKPREA---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWALSDLE--E---NDFKKLKFYLRD------
---------M--TLSEG--------Q--PPL-ARGE-LEGLI-PVDLAELLISKYGEKEA
VKVVLKG----LR-VMNLLELVDQLS-HVC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LH--DYREIYREH-
--------VRC-----------------LE------EW---QEA----------GVNGRY
NQVLLVA-KSSSESPESPAH------PI-------PE-----Q-ELDSV--MVEALFDSG
EK--PSLAP----S
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
LVVLQGSAGTGKTTLARKM-----MLDWA--T---------GSLYP
G--------RFDY-VFYVSCKEVVLLLE-------SKLEQ-LLFW-CC-GD-----NQAP
VTEILR-QP----ERLLFILDGFDE-LQRPFEEK------LK-K-----RG-----L-SP
-KESLLHLLIRRHTLPTCSLLIT-TRPLALRNLEPLL-K-----QAR--HVHILGFSEEE
RARYF----------------S--SY-FTD---------EKKADSAFDIVQK-NDILYKA
CQVPGICWVVCSWLNGQMERGK-AV-L--ET-PRNSTDIFMAYVSTFLPPVD-DGGCSKF
----------S-RHRVLRSLCSLA---AEGI-QN-QR----FLFEEA-------------
--ELRKHNLDGPRLA-AFLSS----NDYQLGLA-IK-KFYSFRHISFQDFFHAMSYLVK-
EDQSQL----------------G--KDSRREVQ-RLLEAKEQE-GNDEMTLTMQFL----
----LDILKKDSFLNLELKFCFR-----------------ISPC-LAQ-DL-KHFKEQM-
E-------------------------------------SMKHNRTWDLEFSLYEAKI-KN
LVKGIQMN---------------------D-------------------VSFKIKHSN-E
KK-SQSQN--S----FSVK-SSLS-HGP-KEKQ------------------KR----PSV
---HGQKEGK---DNI--A-------------------GTHKE-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------AST----------------GKG---------R
-GTEE-TDEEVRS-PKN--TYI----YT--------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSOANP00000018896 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------RELNLER-------
PF05729 // ENSGT00900000140813_0_Domain_2 (755, 930)
KRSAVQ-LIQQ-CC-----DD-DGVP
LSEICR-RS----DRLLFLVDGFDE-LGWSSGGWPGAADDDD-D-----LFVDWKDK-RP
-VGSLLAHLIRKRLFPKASLLIT-TRPAAAESLRPLL-R-----WPR--RAEILGFSEAE
RAEYF----------------H--HY-FLD---------SGRANRALAFIQE-NDVLFTM
CFVPLICWIVCTGLRQQMDRRE-DL-A--QA-SKTTTAVYLSFLSSLLRPAR-HLP-SRV
------------PPAHLRGLCSLA---ADGI-LS-QR----ILFRKA-------------
--DLRKHGLPEDGL-SAFLHV----DVFEKDVD-CE-TLFSFVHLTFQEFFAALFYLLG-
PDETG--------------PGP---LPDARTLL----QHYRLC-DTGFLTLTVRFF----
----FGLLNRERVADLKATMDCV---------VRP--------E-ARR-ALVEWIDTSAR
KEA--------------------------------L-----PGPTLQWLYCFYEIQE-VD
FVERAMGS---------------------F-------------------RKIDID--VRT
RM-DQIVV--A----FCLR-NSRN-LCS-IELM----------------RFF-----NVA
-EADAA------------------------------------------------------
-AAAAERT----------------PEVAADR---------RP------------------
----------------QDWLQ--DT---LCE-SLSEAS------------ANNRG----L
THLA-LRNK-NLGRR-GAELLSKGLSHPNCK----LKSL--------RLP---------N
CTRRPQDW----------------------------------------------------
---------LQ---DT-LCE---SLSEASAN-NRGLTHLALRNKNLGR-RGAELLSKG--
LSH-PNCKLKSL-R----------------------------------------------
------------------------LVNCLLT-----------------------------
----------------------------PDSCRDFSS-----------------------
--------------------------------ILSTDRNLSE---LD-LSSNA-LKD---
-------SGLSLLCEGL----RHPSC----------------------------------
--------------------------KLQ-TL----------------------------
------------------------------WLQSCALTSACGPALSSL-LTTSANLTKLD
L-FNNALGDTGVS-LICEGLKHP--NCK----L---------------------------
-----------------------QALLL--RHC-----ALTLNCCPDL------------
-------SSVL-SINRDLSELDLSYNSLEDAGVCL-LCEGLR-HP-NC-KLQTLR--LQ-
--K-CKLTSRSCS--------------------DLSQALGINREL-RELILHDNVLGDAG
ISVIWK--------GIQQPSCG--LK--I-------------------------------
---LRLGKT-HFSEKMEEE------MR----------TVQ-KMKPELKIRY---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSTTRP00000014925 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------YGEMYREH-
--------VHG-----------------LE------ER---QEG----------GIG-GY
NQLLLVA-TPSSGSPASPVC------PSP----------------LDSV--TVEALLDPG
GK--SSQVP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
PIVVLQGSAGMGKTTLARK------VLDWT--T---------GTLYP
G--------QLDD-VFYVSCREVVLLPE-------GRLDQ-LLFW-RC-GE-----NQAP
ITETLR-QL----EWLLFILDDCDE-LQRPFAER------VT-T-----PR-----S-SP
-TEDVLHFLLGRNACSQVRLLVT-TRVLAL-NLESLL-R-----EPPHVHI-----EKPE
RRRYF----------------N--SY-FTD---------KEQARNALDIVQR-NDVLYKA
CQVPGICCVVCSWLKGQMEQGR-EV-L--EM-PGNCTDIFTAYVSTFLPHSA-PGVCSEP
----------T-RDKVLR-LCPLA---AEGI-----------VFEEG-------------
--DVRKHSLDGLSLA-AFVSS----NSYQEGLD-IK-KY-SFYRIS-LEFFHAMSYLVE-
EDRSQL-------------AGGG--GECCREVN-RLLDEKKQT-GGEAMALSMQFL----
----LDISKKEGSSNWELKFCLK-----------------ISPS-IKQ-DL-KCLKERV-
D-------------------------------------SIKHFRTWDLEFSLYGSKV-KG
LIKGVRMS---------------------D-------------------VSFKMEHFS-E
KK-FHRRN--S----FSVK-ISLS-NGQ-E--K------------------KC----PIV
--------GK---SNR--A-------------------GIQKS-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------ASS----------------GKG---------R
-GTEE-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPANP00000019578 (view gene)
ENSMLEP00000031413 (view gene)
ENSCGRP00001016160 (view gene)
ENSTBEP00000005770 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------MALA-RS
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
PREA---LLWALNDLD--E---NSFKTLKFYLLD------
---------HARSMSGD--------Q--RLL-TRGE-LENMG-QVDLASRLILKYGAQEA
VKVVCQV----LE-AMNLLELVNQLS-HIC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LN--DYRDIYREH-
--------VLN-----------------SE------ER---REG----------DVGSSY
SQRLLVA-KLHSGSPGSSAC------SIP------EQ-----E-ELDLV--MVDALFDP-
-------VP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
SLVVLQGVAGTGKTTLARKI-----LLDWA--S---------GTLYP
G--------RFDY-VFHVSCRDVVLLPW-------NQLDK-LLCW-CC-GD-----DQAP
VAEIQR-EA----ERLLFILDGFDE-LQRPFANQ------SV-R-----QS-----L-SC
-MEDVLRLLIKRQKLPKCSLLIT-TRPLALQNLEPLL-K-----EPL--HIHILGFSEEE
RESYF----------------S--SY-FAD---------KEQARKAFEFVQG-NSVLFKA
CQVPSICWMVCSWLKEQKERDK-AT-S--ET-PSNSTDIFMAYVTTFLPPNG-SGDCSEL
----------S-RHKILRGLCSLA---VEGV-QR-QR----LVFEEA-------------
--DLKKHNLDGPSLT-AFLSS----YDNPAGLD-IK-KFYSFRHITFQEFFYAMSYLVK-
EDQSQL----------------G--EASRKEVE-RLLKVEKEN-KSEMMTLTVQFL----
----LDLLQKESSLNFQLKFCFN-----------------ISPS-LKQ-DL-RRFKEQI-
E-------------------------------------AIKHKRPWDMEFSLYKPKM-KS
MSGGIEMS---------------------D-------------------VLFKVGHSN-E
IK-PHGKS--S----VSVK-ASLG-TGQ-EKEQ------------------WC----PLV
---SKK--------ES--K----------------------GI-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------KWN----------------GKD---------R
-QVEE-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSODEP00000019883 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------L---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLIALEDLS--Q---DQLKRFRHKLRD------
------------ASVNG-----------RSI-PWGR-LECAD-AVDLADQLTHFYGPEPA
LDVACKT----LK-RADVRDVAARLK-GQRLQR---------L---------GPS-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------SV-ALLSVSEFQKKYRQH-
--------VLLQ----------------HAKV--RERNARS------------VKISQRF
TKLLIEPESAAPREEAAALGPAE-------EPEP--------GRAGRSDTHTFNRLFGRD
N---EGRRP----LTVVLQGPAGIGKTMAAKKI-----LYDWA--A---------ARHC-
------------------------------------------------------------
------------------------------------------------------------
--------------CPPARLLVT-KRTTATWKLHCSL-Y-----APQ--RAEVRGFSDKD
KKKYF----------------Y--KF-FQE---------EHRAKRAYRFVKE-NETLFAL
CFVPVVCWIVCTVLRQQLELDR-DL-S--RT-SKTTTSVYLLFLGSALSSM-GAEEA---
-----------RVRGELRKLCHLA---RDGVLGC--K----AQFSER-------------
--DLKCLELQDSDVQH-FLTKKELPGVLET-----E-VIYQFIDQSFQEFFAALSFLLE-
DEGPP-------------------G-VSVGRLG-MLLHKGMEQ--HSHLTLTMRFF----
----FGLLSMEHVQDVQRHFSC-----------------VVPAQ-VKQ-DALKWVQNQGH
SW--VAPQGTDRAEGLKDTEKPEEE-----------EEEEDPNFLLELLYCLYETQD-DV
FVYQALRS---------------------L-------------------PELTLEHVCFS
RM-DLNVL--S----YCIR-CCPA-GQV-LQLV------------------SC----SLV
------TG----------------------------------------QEKK-K---KGN
----------------------------------------LVRW----------------
------LKGN---------------------------------------------SQAAM
KQ---LQVS-------PLHPLCEAMTDMWCG----LSSL--------RLS---------H
C-----------------------------------------------------------
--------KLS---DR-VCR---DLSVALRK-APALTELSLLHCRLS-EAGLRVISEG--
-LAWPQCQVQTL------------------------------------------------
----------------------------RP------------------------------
----------------------------LEAFQCL-------------------------
------------------------------IALLQQSHTLTT---LD-LSGC-HLPGSMV
--T--------YLCTVL----QHPACSLQTLSL--ASVELNET-----------------
----------SVQELRDVKTAKP-------SLA----------------------I--IH
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSECAP00000016749 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------MALT-RSPQ
PF02758 // ENSGT00900000140813_0_Domain_0 (203, 325)
EA---LLWALSDLE--E---NEFKMLKFRLRD------
---------R--ALLEG--------Q--CQL-ARGE-LEGLS-RVDLASRLTLMYGAQEA
VKVLLKV----LR-DMNLLELVEQLG-HIC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LN--DYREIYRER-
--------VRR-----------------LE------ER---QDV----------EINWRY
NQLLLVT-KPSSGSPESPAC------SI-------LE-----Q-ELDSI--MVEALFDPG
EK--PYQVP----PI
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 940)
VVLQGSAGIGKTTLSKKM-----VLDWA--T---------DTLYP
G--------RFDY-VFYVSCREVVLLQE-------GKLDR-LLFW-CC-ED-----NQAP
VTEILR-QP----ERLLFILDGYDE-LR----------------------------S-SP
-MEDMLHLLIRRKVLSTSSLLIT-TRPLALQNLESLL-E-----QPC--HVHILGFSEDE
RRRYF----------------S--SY-FTD---------EEQARNAFDIVQG-NDFLYKA
CQVPGICWVVCSWLKGQMERGR-AV-S--ET-PSNGTDIFMAYVSTFLPPSD-N-GSSEL
----------T-RDRVLRSLCSLA---AEGI-QH-RR----FLFEEA-------------
--DLRKHNLDGPSLA-AFLSS----NDYQEGLE-VK-KFYSFHHISFQEFFHAMSYLVK-
EDQ--------------------------REVK-RLLDEKKQE-GNEEMTLSMQFL----
----LDIAKKERFSNFELKFCLK-----------------ISPT-MKK-DL-KHFKEQM-
E-------------------------------------SIKHNRPWDLEFSLYESKI-KN
LVKGVQIS---------------------D-------------------ISFKMEHSK-E
KK-SYSKN--A----FSVK-TSLS-DGQ-KEKQ------------------KS----PSV
--------DK---GNI--A-------------------GSLKE-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------ASN----------------GKG---------R
-GTEK-LETGRFG-SSR-MESR----GS-ETAELKG---QEDGGV---D-GQED-E----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPEMP00000011414 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MA-NSPQEA---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWALNDLE--E---NSFKTLKFHLRD------
---------M--T------------Q--FHL-ARGE-LEGLS-RVEVASKLISMYGAQEA
VRVVSRS----LR-AMNLMELVDYLS-RVC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LN--DYREIYREH-
--------VRC-----------------LE------ER---QDW----------GVNSSH
NELLLVA-MSSPASPGSPSC------LD-------LE-----Q-EL--V--SVESLFAPE
AE--SYSTP----PI
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 930)
VVMQGSAGTGKTTLVKKL-----VQDWA--K---------GTLYP
G--------RFDY-VFYVSCREVVLLPK-------CDLPN-LICW-CC-GD-----DQAP
VTEILR-QP----ERLLFILDGFDE-LQK---------------------------S-SR
-AECVLHILIRRREVP-CSLLIT-TRPPALQSLEPML-G-----ERR--HVHVLGFSEEE
RKKYF----------------N--SY-FTD---------KEQSRNALEFVQN-NDVLYKA
CQVPGICWVVCSWLKRKMARGQ-EL-S--ET-PNNSTDIFTAYVSTFLPTDG-NGDSSEL
----------T-RHRVLRSLCSLA---AEGI-QH-QR----LLFEED-------------
--VLRKYSLDGPSLT-AFLNS----IDYREGLG-IK-KLYSFRHISFQEFFYAMSYLVK-
EDQSQL----------------G--EATRREVA-KLVEQ---E-KDEEMTLSLQFL----
----FDMLKTDSTLGLGLKFDFT-----------------ITPS-LRR-DL-NYFKEKI-
E-------------------------------------TMKHNRSWDLEFSLYNSKI-KK
IAQGIQIK---------------------D-------------------VRFNMQHSV-K
KE-PGKGK--P----ISVK-SSLG-----KGQV------------------QS----PLL
--------KT---NDS--T-------------------EEPKG-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------ASN----------------GKS---------R
-GAEE-PAR-RVR-NSR-LAST----EK-GHMEMKA---QEGGGV---E-RQED-GEG-Q
R-VKK-DGEMR---DQ-MNG---C-QRE--------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCGRP00001023035 (view gene)
ENSPTRP00000014740 (view gene)
ENSMPUP00000006824 (view gene)
------------------------------------------------------------
------------------------------------------------------------
-------------M-SDVNLNSS------------SQFSPCSS-SSSSS------TPS-P
C---YLISS-LMSSASSNSPFE
PF02758 // ENSGT00900000140813_0_Domain_0 (203, 326)
NG---VMLYMAYLS--K---EGLQTFKQLLVD------
---------E--NPRPG--------S--VQI-TWDQ-VKTAR-WGEMVHLLVEFFPGRLA
WDVTHDI----FA-KMNQTELCLRVQ-MELNYILP-NLELKDLNPR------E-------
------------------------------------------------------------
------------------------------------------------------------
-------MPV-------------------------------NLEEGES--DKIQEYKVH-
--------VTNE----------------YSMM--RTMTTWSGSHADFFYQDI--------
----------------------------------------------ERHERFLPCLFLPK
RP--QAKQP----K
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVLQGVAGVGKTSLAKKV-----MLEWA--E---------NKFYC
H--------KFRC-AFYFHCQEMAQVD-------EQSFAE-LIAH-KW-----PG-SRSL
MSKIMS-KP----DQLLLLFDGFEE-VSLTLT------DRPA-D-----LSDDWIQK-LP
-GPVLLASLLSKRMLPEATLLIT-FRFTSWRKLKPFL-K-----HPS--FITLTGFSMAE
RAKYF----------------R--TY-FGN---------KRDADEALRFVTG-NTVLFSM
CQVPAVCWMVCSCLRQQMQRGA-DL-A--QA-YPNATAVFVQYLSGFFATKP-GSPPRNT
------------HQTRLAGLCYLA---AEGM-WN-MR----WVFDTK-------------
--DLEQAKLDET-AVATFLHM----NIFQRVAG-DG-DRYAFAILSFQEFFAAFLYVLC-
FPQRL--------------RNFR--VLDRVQII-RLIAYPG-R-KKNYLAQMGLFL----
----FGLLNEKCASAAEKSFGCK---------------L--SLG-NKK-KLLKVAALLHE
CS-----------------------------------PPTLHHGVPQLFYCLHEIQE-EA
FVRQTLSD---------------------C-------------------QNATLIISKKK
DM---QVS--A----FCLK-YCQR-LRA-LELT----------------MTL-----TIP
-HVGTLSV----D--------------------------P-----LHSA--EPESS-DQC
FLWWQEFC-SVFRTHENLEVLTLTNTSLEAE---------SVKVLS--AALRH-P-----
GC--K-LQKLI--F-RRVHPS--ML---N-E-DLIQVL----------------TENQYL
SYLE-IQGT-EVRRE-AMEFLCTALKCPQCY----LQCL--------RLE---------D
C------S----------------------------------------------------
---------VT---PK-SWI---DLAKDLGS-NKSCKSLSLTDSSVFS-QEGCY------
--P-LLQDLLAL-R----------------------------------------------
------------------------LENCELA-----------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------LLTCKTLTSS-LKSNEMLTHLS
L-AENTLKDEGTK-ELCNALQCS--TCP----L---------------------------
-----------------------QRLVL--RNC-----ALTSDCCPDL------------
-------VSAL-DKNKNLKSLDLGFNSLKDDGVIL-LFEALM-KP-QS-GLQILE--LE-
--K-CLLTAVCCQ--------------------AMASMVLHNQSL-RYLDVSKNDIGLRG
RELLRE--------AFQQRKGE--KK--V-------------------------------
---V------L-------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCANP00000026970 (view gene)
ENSBTAP00000002954 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------MTHLSSSSFFS
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 326)
DFG---LLLYLEELN--K---EELIRFKSFLKN------
---------E--TLEPR--------S--CRI-PWSE-VKKAK-RKDLADLMSKYYPGEQA
WEVALSI----FG-KMNLKDLCQRAK-AEINGTAQ-TMRTEDTEA-RE------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------VQGDQEAVLGDGAEYRIQ-
--------IREK----------------FRNM--LDKNHLLGKSGDFCHEIA--------
----------------------------------------------QEGRELLERLFGEE
VG--TEEQP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
QTVVLQGAAGIGKTTLVRMM-----MLDWA--Q---------GNLYQ
Q--------KFTY-VFYLNAREINQLR-------ERSFVQ-LLSK-DW-----PS-TEGP
IERIMS-QP----SRLLFIIDSFDE-LNFAFEEP------EF-V-----LCADWTQV-HP
-VSFLMSSLLRKVMLPESSLLVT-TKLTAWRKLKPLL-K----N-RH--SIQLLGMSEDA
RQEYI----------------Y--QF-FED---------QSWALQVFSCLRN-SVMLFNM
CKVPAVCWVVCNCLEQQMEKGA-DI-T--LT-CKTTTSLFACCISSLLTQV-DRSFPGLP
------------SQTQLRSLCHLA---AKGV-WT-MT----YVFYKE-------------
--NLRRHGLVKS-DVSIFLDT----NILQKDTE-YE-NCYVFTHLHIQEFFAAMFYVLE-
GNWDA--------------RDDS--LQSFENLE-LVLESSSYK--DPHLSQMKCFL----
----FGLLNEDRMKQLEETFNCT---------------M--SLE-VKW-TILQWMETLGS
SE-----------------------------------QLPSHLVFLELFFHLYETQD-EV
FVSQAMRY---------------------F-------------------QKVVINICEEI
HL---LLS--S----FCLK-HCQC-LRT-MKLM----------------VTG-----VFE
--K--KML----NSSF-----PP--E------------------------TWQR-AGSHI
FHWWQDLC-SVLHTNKHLREVILCHSNLDEL---------AMKIFN--LELRHP------
NC--K-LQKLRLRF---------TSF-PDLCQGISGSL----------------THNQNL
IHLD-LKGS-DIGDD-GVKSLCEALKHPDCK----LQSL--------SLE---------S
CD----------------------------------------------------------
---------LT---TV-CCL---NISKALVR-SQSLLSLNLSTNNLLD-DGVKLLCEG--
LMH-PKCNLEKL-S----------------------------------------------
------------------------LESCGLT-----------------------------
----------------------------VACCEDLSL-----------------------
--------------------------------VLISNKRLTH---LC-LADNM-LGD---
-------GGVKFMSEAL----KHPQC----------------------------------
--------------------------ILQ-SLV---------------------------
-------------------------------LRRCHFTSLSSESLSAS-LLHNKSLMHLD
L-GSNCLQDDGVK-LLCDAFRHP--SCS----L---------------------------
-----------------------QDLEL--MGC-----VLTSACCLDL------------
-------ASAI-LNNPNLQSLDLGNNDLRDDGVKF-LFEALR-HP-NC-NIQRLG--LE-
--H-CGLTSLCCQ--------------------DLSSTLSSNQGL-IKISLTLNTLQCEE
IMKL-K--------VLRSTECK--LQ--V-------------------------------
---LGLCKE-ALDEEAQKL------LE----------AVA-SSNLRLAVKQDCNDHE---
--E---------------------------------------------------------
--------------------------------------------------EDGSWWRCF-
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPTRP00000003728 (view gene)
ENSHGLP00000014368 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---SPFFSDFG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 323)
-LMWYLEELK--K---KEFMKFKELLKQ------
---------E--TPLLG--------L--KQI-SWAE-VKKAS-REELANLLLRHYEEQQA
WDVTFRV----FQ-KLGRRDLCEKAM-RES------------TG--------------NA
K---------------------------------------------------IYQT----
------------------------------------------------------------
----------------------------------------------------------H-
--------MREK----------------FTDF--LSRESIPTMHEQ--------------
---------------------------------L-------DMNITQEECKFLELLFEP-
----EKQ-T----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
RILCLLGAQGVGKTTFLAKM-----ILAWS--K---------GFVYQ
D--------RFSY-IFYFCCKELKQLP-------ATSLAG-LITR-AW-----PGT-SAP
VSEIMA-HP----EKLLFMADSFEE-LGSDLAEP------ED-E-----LCSDWAEV-QP
-PKLLLSSLLRKKMAPECCLLVT-VNPRAGQDLEDRL-E-----DPD--IRELTGLKEFD
VKFYI----------------C--HM-FPD---------RRRALEAFNVILN-NEQLLSM
CRLPVLCWLVCTTLKQEMDKGI-DV-A--LT-CQRPTALYSSFLLDLFTPEG-ADGPSW-
-----------QGQAQLQSLCSLA---VQGM-WT-NT----MVFREK-------------
--DLRRNGIGSS-DLSALLEL----RVLLPCWE-TE-GLYKFLHSSVQEFCAALFYLLK-
SPGHH-----------------S--HPAVRGTQ-DLLLTFLEK-SRAHWVFVGCFL----
----FGLLHERECQKLDAFFGFQ---------------L--SSE-VKR-QLYQCLQQLAK
VAD----------------------------------KLQGQVSFLALFYALFEMQE-EA
FLNQVMDL---------------------M-------------------QDLAFSVFIET
DL---RVA--G----YCLK-HCSN-LRK-LSLS----------------AQG-----VFK
-EEGREQS-----------S-----------------------------------VSSSE
LLCWHQVW-SVLRTCSQLQMLQVKDGTLNES---------AMLSLC--QQLRQP------
NC--L-LENLFVNN---------VTF-HCESWTLFEAF----------------PHNPNL
KYVN-LSNT-QLGRD-DVKVLHGALSHPTSS----VEKL--------LL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------VNCNLS-----------------------------
----------------------------ADDCKALAA-----------------------
--------------------------------TLMESKKLDS---LN-LSCNN-LK----
-------KGVRALCKAL----CHPNCTLKFL-----------------------------
------------------------------------------------------------
------------------------------ALSFCTLREWCWDYLAEV-LLIS
PF13516 // ENSGT00900000140813_0_Domain_13 (2214, 2237)
KSLSHLD
L-SINSLRDEGLK-VLCDTLRYP--VCA----L---------------------------
-----------------------QILCL--MKC-----SITAEGCRDL------------
-------ASVL-TS
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
SRNLRSLQIGGNNIEDAGVKL-LCQALA-QP-SC-LLETLG--VE-
--D-CGLTSACCE--------------------DLVTVLTTSKTL-FMLNLLQNPLDHGG
VALLCQ--------GLRHPECA--LR--V-------------------------------
---LGLNSA-EFGEETQAL------LT----------AEE-ERNPDLVILDT-W------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSANAP00000003731 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---ESFFSDFG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 322)
-LLWYLKELR--K---EEFWKFKELLKQ------
---------E--ALKFE--------L--KPI-PWTE-LKKAS-KEDVAKLLDTHYPGKRA
WEVTRSL----FL-QISRKDLWTKAQ-EEM-S----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------KLNPYRKQ-
--------IKEK----------------FQLI--WEKETCLHVPEH--------------
---------------------------------F-------YRESMKHERTVLSKAFTAA
------ATP----V
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVVEGPDGIGKTTLLRKV-----MLDWA--E---------GNFWK
D--------RFTF-VFFLTVYEMNSIT-------ETSLLE-LLSR-DW-----PES-SEK
IEDIFA-QP----ERILLIMDGFEQ-LKLDLD-H------KA-D-----LCDDWRQR-QP
-LPVILSSLLQKKMLPECSLLLA-LGQVGMRKNYFVL-E-----HPK--LIKLSGFTEPE
RKMYF----------------S--CF-FGE---------KNKALKVFNFVRD-IGPLFIL
CHNPFICWLVCTCMKYRLERGE-DL-E--IN-SQNTTSLHASFMTSVFKAGS-QSSPPK-
-----------LNRARLKNLSALA---AEGL-WT-HT----FVFCRE-------------
--DLRRNGLSES-EGLMWVGM----KLLQRSG-----DCFTFIHVCIQEFCAAMFYLLK-
GPGDS-----------------P--NPAIGSIT-QLVRASVAQ-PQNFLTQMGVFV----
----FGISTEEIASMLETSFGFP---------------L--SKD-LQQ-EIAQCLQSFSQ
CE-----------------------------------ADREAIGFQELFSCLFETQE-KG
FVTQVMNF---------------------F-------------------EEVYIYIANIE
HL---VIA--S----FCVK-HCHS-LTT-LRLC----------------MEN-----IFP
-DDSGCVS-----------E-------------------H-N-------EK---------
LMYWRELC-SMFSNNKNFQTLDMENTSFDDA---------SLAILC--KALAQP------
VC--K-LQKLMLTF---------MSN-FGHDSELFRAV----------------LHNPHL
KILS-LYGS-SLCRT-EVKYLCETLKHPMCK----IEEL--------IL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------GKCDLS-----------------------------
----------------------------HEACEDIAS-----------------------
--------------------------------V---------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------------------LACNSKLKLLS
L-VENPLRDEGVA-FLCEALKHP--NCA----L---------------------------
-----------------------EKLML--MYS-----CLTCVSCEAI------------
-------SQGL-ARSKSLSLLDLGSNHLEDSAVAL-LCEALK-HP-DC-SIRELW--LQ-
--T-CRITHACCE--------------------DLAAALVTSKTL-RSLNLDWITLDPEG
VVLLCG--------ALSHPDCA--LQ--M-------------------------------
---LGLQKS-AFDEESQKM------LM----------ALE-EKCPHLSISHEPWVEE---
--EYR-----IR------------------------------------------------
--------------------------------------------------GVIP------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSJJAP00000024566 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------------MLRC-AR
LVLRAV----------------LGSPPGPV-------------------NTWRSV---RT
GR-QHPAS--T----MSLDIQCEQ-LSD-ARWM----------------EL---------
----LPQI-------------------------------QQYQVV------RLDDC-GLT
EGRCKDIS-SAVQA
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1542)
NPALTELSLRTNELGDG---------GVHLVL--QGLQN-S-----
TC--K-IQKLS--L-QNCCLT--EA---GCG-VLPGVL----------------RS
PF13516 // ENSGT00900000140813_0_Domain_6 (1617, 1642)
LPTL
RELH-LNDN-PLGDA-GLQLLCEGLLDPQCH----LEKL--------QLE---------Y
C------N----------------------------------------------------
---------LT---VT-SCE---PLASVLKA-KPDFKELVLSNNDFHE-AGVQALCRG--
LKN-SACQLETL-K----------------------------------------------
------------------------LENCGIT-----------------------------
----------------------------SANCRDLCD-----------------------
--------------------------------VIASK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
ASLQE---LD-LGSNK-LGN---
-------AGIAALCPGL----LGPSC----------------------------------
--------------------------KLK-TLW---------------------------
-------------------------------LWECNITAEGCRDLCRV-LRS
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
KSNLKELS
L-ANNELGDEGAR-LLCESLLEP--GCQ----L---------------------------
-----------------------ELLWV--KTC-----SLTPACCSHF------------
-------RSML-A
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
QNRSLLELQMSSNPLGDSGVQE-LENSGT00900000140813_0_Linker_15_16 (2378, 2391)
CEALS-QP-GT-VPF13516 // ENSGT00900000140813_0_Domain_16 (2391, 2436)
LRVLC--LG-
--D-CDVTDAGCS--------------------SLENSGT00900000140813_0_Linker_16_17 (2436, 2442)
ARVLLAPF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2465)
NHTL-QELDLSNNCMGDLG
ILQLME--------NLRKPSCV--LQ--Q-------------------------------
---LVLYDI-YWSEEVEDQ------LR----------ALE-EEKPSLKIIF---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMFAP00000000969 (view gene)
MDQPEDPGSRLRSLRTASPQRAFVESVPHQRLRLRPWCKAPKLGLTK-RQPGHVTWPGVW
KNRSRKGGAGEEGAPQSSGRRWPVPTSPSEGEEHQD------APTRVRGRRR--------
-------AKSGVTPRCPLPPPHG-----SS--LARGSLRPCPRWVLKRQSRGWAGVTRGP
LPDDPRPPVSSTGLR--LAVAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLAALEELS--Q---EQLKRFRHKLRD------
------------VGPDR-----------RSI-PWGR-LERAD-AVDLAEQLAQFYGPEPA
LEVARKT----LK-RADARDVAAQLK-EQRLRR---------L---------GPG-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------SA-ALLSVSEYKKKYREH-
--------VLQL----------------HARV--KERNARS------------VKITKRF
TKLLIAPESAAPEEE--ALGPAE-------EPEP--------GRARRSDTHTFNRLFRGD
E---DGRRP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYQ
G--------QVDF-AFFMPCGELLERPG------TRSLAD-LILD-QC-----PD-RGAP
VPQMLA-QP----QRLLFILDGADE-LPALEGP--------EAA-----PCTDPFEA-AS
-GARVLGGLLSKALLPTALLLVT-TRAASPGRLQGRL-C-----SPQ--CAEVRGFSDKD
KKKYF----------------Y--KF-FQD---------ERRAERAYRFVKE-NETLFAL
CFVPFVCWIVCTVLRQQLELGR-DL-S--RT-SKTTTSVYLLFIASVLSSAPVADGP---
-----------RVQGDLRNLCRLA---REGVLGR--R----AQFAEK-------------
--QLEQLELRGSKVQTLFLSKKELPGVLET-----E-VTYQFIDQSFQEFLAALSYLLE-
DEGVP-------------------R-TAAGDVG-TLLRGDAQP--HSHLVLTTRFL----
----FGLLSAERMRDIERHFGC-----------------VVSER-VKQ-EALRWVQGQGQ
GCPGVAPEVTEETKGLEDTEEPEE-----------EEEGEEPNYPLELLYCLYETQE-DA
FVRQALCR---------------------L-------------------PELALQGVRFC
RM-DVAVL--S----YCVR-CCPA-GQA-LRLI------------------SC----RLV
------AA----------------------------------------QEK--K---KKS
----------------------------------------LGKR----------------
------LQAS---L--------------GGS------S-----------------SRGTT
KQ---LPAS-------LLHPLFQTMTDPLCH----LTSL--------TLS---------H
C-----------------------------------------------------------
--------KVP---DA-VCR---DLSEALRA-APALTELGLLHNRLS-EAGLRMLSEG--
-LAWPQCRVQTV------------------------------------------------
----------------------------RVQ-----------------------------
---------------LPSS---------QQGLQYL-------------------------
------------------------------VGMLRQSPALTT---LD-LSGC-QLPAPMV
--T--------YLCAVL----QHQGCGLQTLRL--TSVELSEQ-----------------
----------SLQELQAVKRAKP-------DLV----------------------I--AH
PVLD-------------------GHPESP-KELVSTF-----------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSVPAP00000002585 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------MTDVSSSSYF
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
SDFG---LLLYLEELN--K---EELNIFKSFLKN------
---------E--TVRPG--------S--RRI-PWSD-VKKAK-REDLANLMNQYYPGGQA
WDVALKI----FG-KMNLKGLCQRAK-AEISWIAK-TVRPEDTGA-RE------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------VQGHQEAVLGDGTKYRIQ-
--------IEEK----------------FRFM--SNKNCLLAESGNFCHEIT--------
----------------------------------------------QESRELLERLFGED
VR--TGVQP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
QTVVLQGPAGVGKTTLVRKM-----MLDWA--Q---------GNLYQ
Q--------KFTY-VFYLNAREINPLR-------EKSFAE-MLSD-DW-----PS-TEGP
IERIMS-QP----SGLLFIIDSFDE-LHFAFEEP------EF-V-----LCGDWTQV-HP
-VSFLVSSLLRKVMLPQSSLLVT-TKLTAFKKLKPLL-K----S-PR--SIQLLGMSENA
REEYI----------------C--QF-FED---------ERWALQVSSCLRN-NEMLFSM
CKVPLVCWVICNCLEQQMEKGG-DI-T--LT-CRTTTALFTCCISSLFTRV-DGRFPSLP
------------SETQLRSLCHLA---ARGV-WT-MT----YVFYKE-------------
--NLRKHGLVKS-DVSVFLDM----NILQRDAE-YE-NCYMFPHLHIQEFFAAMFYMLQ-
DSWDT--------------GDNS--FQSFEDPE-LLLESSSCN--DPHLLQMKCFL----
----FGLLNEDLVKQLEETFNCK---------------M--SLE-MKW-KTLQWMETLGN
SD-----------------------------------PFPSHLVFLDLFHILYETQD-EA
FINQAMRY---------------------F-------------------PKVVINICEEI
HL---LVS--S----FCLK-HCQY-LRT-IKLS----------------VTV-----VFE
--K--KEL----NSSP-----QA--E------------------------TWD---GGRI
TRWWQDLC-SVLHTNE
PF00560 // ENSGT00900000140813_0_Domain_4 (1517, 1550)
HLRELDLCHSNLDKL---------AMKTFN--QELRHP------
DC--K-LRKLLLRF---------ISF-PDACQDISSSL----------------THNQNL
MHLD-LKGS-DIGDN-GVKSLCDALKHPECK----LQNL--------SLE---------S
CS----------------------------------------------------------
---------LT---TV-CCL---NISKALIR-SQSLRSLNLSTNNLLD-DGVMLLCEA--
LMH-PKCYLERL-S----------------------------------------------
------------------------LEHCGLT-----------------------------
----------------------------AAGCEDLSL-----------------------
--------------------------------ALISNERLTH---LC-LADNV-LGD---
-------GGVKLISHAL----KHPQC----------------------------------
--------------------------TLQ-SLV---------------------------
-------------------------------XXXXXXXXXXXXXXXXX-XXXXXXXXXXX
X-XXXXXXXXXXX-XXXXXXXXX--XXX----X---------------------------
-----------------------XXXXL--MGC-----VLTSACCLDL------------
-------ASAI-LNNP
PF00560 // ENSGT00900000140813_0_Domain_15 (2357, 2381)
NLRSLDLGNNDLKDDGVKI-LFEALR-HP-DC-NIQRLG--LE-
--Y-CGLTALCCQ--------------------DLSSTLSSNQRL-IKINLTLNTLGHEG
IKKLCE--------VLRSPECK--LQ--V-------------------------------
---LGLCKE-AFDEEAQEL------LE----------AVC-SSNPHLAIKQDCDDHD---
--E---------------------------------------------------------
--------------------------------------------------EDGSWWRCL-
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMMUP00000045983 (view gene)
ENSNLEP00000021218 (view gene)
ENSTTRP00000012845 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--GDQPEAPSGTEAR--AAA
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
AREV---LLAALEDLS--Q---ERLKRFRHKLRD------
------------APLHG-----------RRK-PWGR-LEGTD-AVDLAEHLIHFDGPERA
LDAARKT----LK-RAGVRDVATLLK-EQRLRR---------L---------GLS-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------S--ALLSASEYKKKYREH-
--------VLRQ----------------HAKV--KERDARS------------VKMSERF
TKLLIAPDSAALEHE--APGPAE-------EPEL--------ERARRSDTHTFNRLLGHD
G---EGQRP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
PTVVLQGPAGIGKTMAARKI-----LYDWA--A---------GKVYH
G--------QVDF-AFFVSCREVLERPR------TCSLAD-LTLD-QY-----PD-RNAP
VRQMLA-QS----TRLLSILDGVNE-LPAPGPA--------EAA-----PCTDPLEA-AS
-GARMLGGLPSKALLPTARVLVT-ARAAAPGRLQSRL-C-----SPQ--RAEVRGPSDND
KKKYF----------------Y--RF-FQD---------EWRAERACRCAKE-NETLFSL
CFA--VCWTVCPVLRQL--PGP-DL-S--RT-SKTTTSVYLLFIGSVLSSAPAADRP---
-----------RLQEELHKLCRLA---CEGVLRG--R----AQVSEK-------------
--DLERLELCGSKVQAQFLSKK-MTGVLET-----E-VTYRFLDHTFQEFLAVLSYLLE-
DEGLP---------------------TPAGGVG-ALLRGHPEL--RGHLALTTRFL----
----FGLLNTERMRDIEHHFSC-----------------AVSER-GKQ-DVLRWVQDQGQ
GCPRVAPEGTEGTQALRGAGESEE------------EEGEELSDLLELLYCRYETQE-GA
LVHQALRG---------------------L-------------------PELALE-VHFS
RM-DLAVP--S----YCVL-CCPA-GQA-LQLV------------------SC----RLA
------TA----------------------------------------QEKK-----KKS
----------------------------------------LMKW----------------
------LQGS---L--------------GGS------S----------------SSRTTM
RK---LLGS-------PLHPLCE-WTDRQCG----LNTL--------MLS---------R
C-----------------------------------------------------------
--------KLP---AL-VCR---DLSEA-------LTELGLFHSWLS-EAGLHALSEG--
-LA-PQCRLQTL------------------------------------------------
----------------------------RVQ-----------------------------
---------------QLGL---------QG-SQYV-------------------------
------------------------------FGLLQQSPALTT---LD-LSGC-QLSGTMA
-----------TLCAVL----QLPGCRLQSLSL--TSVELSER-----------------
----------SLQERRAVGTAKA-------GLA----------------------V--TH
PALD-------------------SDPQPHKGLGSDP------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSP00000291971 (view gene)
ENSCGRP00000010646 (view gene)
------------------------------------------------------------
------------------------------------------------------------
--------------CCP-----------------GAPCGECERWDPGHFGGSGTGPEQ--
VVSVI-TLSIPSRDT--DALAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLVALEDLS--Q---EQLKRFRHKLRD------
------------APLDG-----------RSI-PWGR-LENSD-PVDLVDKLTQFYGSEPA
VDVTRKI----LK-KADARDVAVRLK-AQQLQR---------L---------GSS-----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------S-AVVSLSEYKKR-----
----------------------------HAKV--KERNSRS------------VKINKRF
TKLLIAPGSGAVEDE--LLGPSE-------EPQP--------ERARRSDTHTFNKLFRGD
D---EGPR-----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYH
G--------QVDF-AFFMPCGELLERPG------TRSLAD-LILD-QC-----PD-RKAP
VRQILA-HP----QRLLFILDGADE-LPAAAAP--------EAT-----PCTDPFEA-TS
-GLRVLSGLLSQELLPLARLLVT-TRNAAAGRLQGRL-C-----SPQ--CAEVRGFSDKD
KKKYF----------------F--KF-FRD---------ERKAERAYRFVKE-NETLFAL
CFVPFVCWIVCTVLQQQLELGQ-DL-S--RT-SKTTTSVYLLFIASVLKSA-GTNGP---
-----------RVQGELRTLCRLA---REGILKH--Q----AQFSEN-------------
--DLEQLKLQGSQVQTLFLSKKELPGVLDK-----E-VTYQFIDQSFQEFLAALSYLLE-
AEG-----------------------APAGGVQ-MLLNSDSEL--RGYLALTTRFL----
----FGLLSAERIRDIERHFGC-----------------VVPER-LKQ-DTLRWVQGQGH
PK--VAPVEAEKNDELKDTKEPEEEEEE-----EEEEEEEDLNFGLELLYCLYETQE-DA
FVRQALSS---------------------L-------------------PEMVLERVRFT
RM-DLGVL--S----YCVQ-CCPA-GQS-LRLV------------------NC----GLV
------EA----------------------------------------KEKK-----KKS
----------------------------------------LVKR----------------
------LKGS----------------------------------------------GNTT
KQ---PPVS-------LLRPLCEAMTSQQCG----LRIL--------ILS---------R
C-----------------------------------------------------------
--------KLP---DS-VCR---DLSEALKL-APALRELGLLQSRIT-ETGLRLLSEG--
-LAWPKCRVQTL------------------------------------------------
----------------------------RIQ-----------------------------
---------------LPNL---------QEAIQYL-------------------------
------------------------------VIVLQQSRVLTT---LD-LSGC-QLVTYMV
--E--------NLCSAL----KYPMCCLKTLSLT--SVELSEV-----------------
----------SLRELQIVKASKP-------DLT----------------------I--LH
SKP---------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPEMP00000002722 (view gene)
ENSRROP00000025388 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------FT-EAGQ-VQEID-SPQ--------------
-LGDAED----DS-ELANANCCVRLT-SKYF-----------------------------
------------------------------------------------------------
--TEAG---------------------------------QVQEIDSPQL-----------
-------GDA-------------------------------EDDSELAKPGEKEGRRNS-
---------VEK----------------QSLV--WKNTFWQGDTDSFHDDVI--------
-----------------------------------------------RRNQRFIPFLNPR
TP--RKLTP----Y
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVLHGPAGVGKTTLAKKC-----MLDWT--D---------CNLSP
---------TARY-AFYLSCKELSHMG-------PCSFAE-LISK-DW-----PE-WQDD
IPHILA-QA----QRILFVVDGLDE-LKVPPG------ALIQ-D-----LCGDWEQQ-KP
-VPVLLGSLLKRKMLPKAALLVT-TRPRALRDLQLLV-Q-----QPI--YIRVEGFLEED
RRAYF----------------L--RH-FGD---------EDQAMRAFELMRS-NAALFQL
GSAPAVCWIVCTTLKLQMEKGE-DP-A--PT-CLTSTGLFLRFLCSQFPQG-------AQ
------------LRGALRALSLLA---AQSL-WA-QM----SVLHGE-------------
--DLERVGVQES-DLRPFLDR----DILRQDRV-SK-GCYSFIHLSFQQFLTALFYVLE-
EEEEE---EE--------DRDGH--AWDIGDVQ-KLLSEEERL-KNPDLIQVGHFL----
----FGLANEKRAKELEATFGCQ---------------M--SPE-IKQ-GLLRCKANLHA
NK-----------------------------------P-LSMTELKEVLCCLYESQE-EE
LAKVVVAP---------------------F-------------------KEISINLTNTS
EM---MHC--S----FSLK-HCRD-LEK-VSLQ----------------VAKG----VFL
-ENY-VDF----EP--------------------DIEFERC-TYV--SLPSWAQQD-LRS
LRLWTDFC-SLFSSKSNLKFLEVKQSFLSDS---------SVRILC--DHITR-S-----
TC--H-LQKVE--I-KNVTPD--TA----YR-DFCLAF----------------IGKKTL
THLT-LEGH-VEWERTMLLLLCDLLRNHKCN----LQYL--------RLG---------G
H------C----------------------------------------------------
---------AT---SE-QWA---DFSYVLKA-NQSLNHLHLSASVLLD-EGAKLLYKT--
MTH-PNHFLQML-S----------------------------------------------
------------------------LENCRLT-----------------------------
----------------------------EASCKDLAA-----------------------
--------------------------------VLVVSQQLTH---LC-LAKNP-IGD---
-------TGVKFLCEGL----SYPEC----------------------------------
--------------------------KLQ-ALV---------------------------
-------------------------------LQQCSITKRGCRYLSEA-LQEACSLTNLD
L-SLNQIA-RGMW-ILCQALENP--NCN----L---------------------------
-----------------------KHLRL--WSC-----SLMPFYCQHL------------
-------GSAL-ISNQKLETLDLGQNHLWKSGIIK-LFRVLR-QR-TG-SLKTLR--LK-
--T-YETNSEIKK--------------------LLEEVKEKNPKLTIDCNAS------GA
---------------TAPACCD--FF----------------------------------
----C-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCAFP00000003989 (view gene)
ENSMLUP00000016674 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------VHFPVAGSYHCP---NMGLYFVLRGPV-------TIEIEFCAWDQ
FLDQIVPQHSWMVAGPLFDIKAEPGA-VAAVYLPHFIALQEE--------HVDISWFHIA
HFKEDGILLEKPTRVEPRYTVLENPSFSPMGVLLRKIHSFLS-IPVISNVLLYHRLHSEE
V-NFHLYLLPSDCTIEKAVDNEKQKFQFVRIHKPPPLTPLHIGSRYIVSGS---------
QKMEIIPEELELCYRSPGKPQLFSEFYIDHLGSGIRLQIKSKNDETVVWEALVKSVF---
-----------------------------------MSLSASPSPPDAPDL
PF00619 // ENSGT00900000140813_0_Domain_19 (3051, 3135)
LHFMDQHRAQ
LVTRVTSVDAVLDKLH-GQMLSEEQYERVRAEPTNPDKMRKLFSFSKSWDWACKDQLYRA
LKETHPHLIIDLWEK---------------------
ENSRBIP00000031628 (view gene)
MGP_PahariEiJ_P0034731 (view gene)
MGP_PahariEiJ_P0034732 (view gene)
ENSSBOP00000014848 (view gene)
ENSSSCP00000057427 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------------SRKPSLTI
SNEGPDGEKMSGD------------------GSSPKQQ----------------------
------------RQESEGSSPQVAQ-EKPLCLSSPAAPDD-LHAESLR-PEDDFWGPTGP
VAIKVVDQ-ERSLYR
PF13553 // ENSGT00900000140813_0_Domain_18 (2716, 2997)
VHFPMAGSYHWP---HTGLHFVVRGPV-------TIEIEFCAWDQ
FLNKTIPRPNWRVAGPLFDIKAEPGV-VAAVYLPYFVALQES--------CVDISWFQVA
HFKEEGMVLEKPAMVAPSYAVLENPSFSPMGVLLRMVHTALRFIPVISTVMLYHHPHPEE
V-TFHLYLVPSDCSIQKAIDDEERKFQFVRIHKPPPLNPLYLGSRYIVSGS---------
ENLEIIPEELELCYRSPREPQLFSEFYVGHWRSGIKLQVTNKKDGIVVWEALVKPGENSGT00900000140813_0_Linker_18_19 (2997, 3052)
DLRR
AAAQVPT-----------------------------GPIASPSPSKAPGSLPF00619 // ENSGT00900000140813_0_Domain_19 (3052, 3133)
HFVDRYREQ
LVARVTSVDPILDKLH-GQVLSEEQYEGVRAEVTKPDQMRKLFGFSRSWDWACKDRLYQA
LKETHPHLIVELWERWGLEKTTGASCQTQKLSEHLL
ENSMMUP00000014119 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MSLDIQCEQ-LSD-ARWA----------------EL---------
----LPLL-------------------------------QQCQVV------RLDDC-GLT
EARCGDIS-SALRV
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1542)
NPALTELSLRSNELGDA---------GVHCVL--QGLQS-P-----
SC--K-IQKLS--L-QNCYLT--GA---GCG-VLSGTL----------------RSL
PF13516 // ENSGT00900000140813_0_Domain_6 (1618, 1642)
PTL
QELH-LSDN-LLGDA-GLQLLCEGLLDPQCR----LEKL--------QLE---------Y
C------N----------------------------------------------------
---------LS---AA-SCE---PLASVLRA-KPDFKELTVSNNDINE-AGVRVLCQG--
LKD-SPCQLEAL-K----------------------------------------------
------------------------LESCGIT-----------------------------
----------------------------SNNCRDLCS-----------------------
--------------------------------IVASK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
ASLRE---LA-LGSNK-LGD---
-------VGIAELCPGL----LHPSS----------------------------------
--------------------------SLR-TLW---------------------------
-------------------------------IWECGITAKGCADLCRV-LRTKE
PF13516 // ENSGT00900000140813_0_Domain_13 (2215, 2237)
SLKELS
L-AGNELGDEGAR-LLCETLLEP--GCQ----L---------------------------
-----------------------ESLWV--KSC-----SFTAACCSHF------------
-------SSVL-TQ
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NKFLLELQISNNRLGDAGVQE-LCKGLG-QP-GS-VLRVLW--LG-
--D-CDMSDSSCS--------------------SLAATLLA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2464)
NHSL-RELDLSNNCLGDAG
VLQLVE--------SVRQPGCL--LE--Q-------------------------------
---LVLYDI-YWSEAMEDR------LQ----------ALE-EEKPSLRVIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPPAP00000037748 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----MSLD--I----QSLDIQCEE-LSD-ARWA----------------EL---------
----LPLL-------------------------------QQCQVV------RLDDC-GLT
EARCKDIS-SALRV
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1542)
NPALAELNLRSNELGDV---------GVHCVL--QGLQS-P-----
SC--K-IQKLS--L-QNCCLT--GA---GCG-VLSSTL----------------RTL
PF13516 // ENSGT00900000140813_0_Domain_6 (1618, 1642)
PTL
QELH-LSDN-LLGDA-GLQLLCEGLLDPQCR----LEKL--------QLE---------Y
C------N----------------------------------------------------
---------LS---AA-SCK---PLASVLRA-KPDFKELTVSNNDINE-AGVRVLCQG--
LKD-SPCQLEAL-K----------------------------------------------
------------------------LESCGVT-----------------------------
----------------------------SDNCRDLCG-----------------------
--------------------------------IVASK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
ASLRE---LA-LGSNK-LGD---
-------VGMAELCPGL----LHPSS----------------------------------
--------------------------RLR-TLW---------------------------
-------------------------------IWECGITAKGCGDLCRV-LRAKE
PF13516 // ENSGT00900000140813_0_Domain_13 (2215, 2237)
SLKELS
L-AGNELGDEGAR-LLCETLLEP--GCQ----L---------------------------
-----------------------ESLWV--KSC-----SFTAACCSHF------------
-------SSVL-AQ
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NKFLLELQISNNRLEDAGVQE-LCQGLG-QP-GS-VLRVLW--LA-
--D-CDVSDSSCS--------------------SLAATLLA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2464)
NHSL-RELDLSNNCLGDAG
ILQLVE--------SVRQPGCL--LE--Q-------------------------------
---LVLYDI-YWSEEMEDR------LQ----------ALE-KDKPSLRVIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSSTOP00000009174 (view gene)
ENSECAP00000013016 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------MA----SV
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 326)
RCK---LARYLEDLE--D---ADLKKFKMHLED------
---------Y--PPQKG--------C--TRP-PRGQ-TEKAD-PVDLATLMIDFNGEEKA
WAMAVWI----FA-AINRRDLHEKAK-LEEPEWA--HSC--CY---------RSHLI---
--------CQ-----------------------------------EESI-----------
-----------------------------------------------------EEEWIG-
------------------L-----------------LEYLSRISICKKKKDYCKKYRKH-
--------VRSR----------------FQCI--KDRNARLGESV---------NLNKRY
TRLCLLKEHRSQQEREQELL------AIGRTSPKTQD------CP--MSSMNLELLFE--
PDDQHSEPV----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
HTVVFQGAAGIGKTILARKI-----MLDWA--S---------EKLYK
N--------RFDY-LFYIHCREVSLGT-------RRSLGD-LIVS-CC-----PD-PKPP
IGKILS-KP----SRILFLMDGFDE-LQGAFDEH------TE-A-----LCTDWQKV-ER
-GDILLSSLIRKRLLPEASLLIT-TRPVALEKLQHLL-D-----RPR--HVEILGFSEAK
RKEYF----------------F--KY-FSD---------EQQAHEAFRLIQE-NEILFTM
CFIPLVCWIVCTGLKQQMDSGK-SL-AQ--T-SKTTTAVYVFFLSSLLQSRGESQKQH--
------------LSANVRGLCSLA---ADGI-WN-QK----ILFEES-------------
--DLRKHGLQRA-DASAFLRM----NLFQKEVD-CE-KFYSFIHMTFQEFFAAMYYLLE-
DEEQ--------GSVRNMPWGRS--NLPNRDVS-VLLENYGKF-EKGYLIFVVRFL----
----FGLVNQERTSYLEKKLSCK---------------I--SQQ-IRL-ELLKWISAKAE
VK-----------------------------------KLQTQPSQLELFYCLYEMQE-ED
FVRRAMGY---------------------F-------------------PKIEISLSTRM
---DHVVS--S----FCIE-NCRS-VES-LSLR----------------LLYN-----SP
KEEEEEEE-----E------------------------------------E---------
---------------EDGIDF------LFSP-----------------------------
------------------------------------------------------------
-------------------------SSFLPI----S-----------RLV---------N
C-----------------------------------------------------------
--------SLT---SS-FCQ---SLFSALST-NWSLTELNLGDNTLGD-SGMKVLCDM--
LQH-PGCNIQRL-C----------------------------------------------
------------------------ATHWGLK-----------------------------
----------------------------QQK-FTFSH-----------------------
--------------------------------LITSLKALSP---NT-VTFSG-CGE---
-------TGTTLHCG--------WKCKMVQL-----------------------------
--------------------------------L---------------------------
-------------------------------LVGCCLTSACCEDLASV-LSTSHSLIRLY
L-GENALGDSGVG-ILCEKAKHP--RCN----L---------------------------
-----------------------QVLGL--VNS-----GLTSGCCPAL------------
-------SSVL-SANHNLKQLYLRGNALGDTGVKL-LCEGLS-HP-NC-KLQMLE--LD-
--S-CSLTSHCCW--------------------DLSTLLTSNQSL-RELSLGGNDLGDLG
VMLLCE--------VLKQQGCL--LR--S-------------------------------
---LGLCEM-YFNYETKCA------LE----------TLQ-EEKPELSVVFEPSW-----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSSSCP00000058159 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MNLDIHCEQ-LSD-ARWT----------------EL---------
----LPLL-------------------------------QQYEVV------RLDDC-GLT
EEHCKDIG-SALRA
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1542)
NPSLTELCLRTNELGDA---------GVHLVL--QGLQS-P-----
TC--K-IQKLS--L-QNCSLT--EA---GCG-VLPSTL----------------RS
PF13516 // ENSGT00900000140813_0_Domain_6 (1617, 1642)
LPTL
RELH-LSDN-PLGDA-GLRLLCEGLLDPQCH----LEKL--------QLE---------Y
C------R----------------------------------------------------
---------LT---AA-SCE---PLASVLRA-TRA
PF13516 // ENSGT00900000140813_0_Domain_8 (1776, 1795)
LKELTVSNNDIGE-AGARVLGQG--
LAD-SACQLETL-R----------------------------------------------
------------------------LENCGLT-----------------------------
----------------------------PANCKDLCG-----------------------
--------------------------------IVASQA
PF13516 // ENSGT00900000140813_0_Domain_11 (2019, 2054)
SLRE---LD-LGSNG-LGD---
-------AGIAELCPGL----LSPAS----------------------------------
--------------------------RLK-TLW---------------------------
-------------------------------LWECDITASGCRDLCRV-LQAK
PF13516 // ENSGT00900000140813_0_Domain_13 (2214, 2237)
ETLKELS
L-AGNKLGDEGAR-LLCESLLQP--GCQ----L---------------------------
-----------------------ESLWV--KSC-----SLTAACCQHV------------
-------SLML-T
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
QNKHLLELQLSSNKLGDSGIQE-LENSGT00900000140813_0_Linker_15_16 (2378, 2389)
CQALS-QP-GTPF13516 // ENSGT00900000140813_0_Domain_16 (2389, 2436)
-TLRVLC--LG-
--D-CEVTNSGCS--------------------SLENSGT00900000140813_0_Linker_16_17 (2436, 2442)
ASLLLAPF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2464)
NRSL-RELDLSNNCVGDPG
VLQLLG--------SLEQPGCA--LE--Q-------------------------------
---LVLYDT-YWTEEVEDR------LQ----------ALE-GSKPGLRVIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSNLEP00000013011 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---ASFFSDFG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 322)
-LMWYLEELK--K---EEFRKFKEHLKQ------
---------M--TLHLE--------L--KQI-PWTE-VKKAS-REELANLLIKHYEEQQA
WNITLRI----FQ-KLDRKDLCMKVM-RER------------TG--------------YT
K---------------------------------------------------AYQA----
------------------------------------------------------------
----------------------------------------------------------H-
--------AKQK----------------FSHL--WSSKSVTEIHLY--------------
---------------------------------F-------EEEVKQEECDHLDRLFAPK
ET--GKQ-P----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
RTVIIQGPQGIGKTTFLMKL-----MMAWS--D---------NKIFR
D--------RFLY-TFYFCCRELRELP-------AMSLAD-LISR-EW-----PDP-AAP
ITEIVA-QP----ERLLFIIDSFEE-LKGGLNEP------DS-D-----LCGDLMEK-QP
-VQVLLSSLLRKKMLPDASLLIA-VKPVCPKELRDQV-T-----ISE--IYQPRGFNESD
RLVYF----------------C--CF-FKD---------PKRAMEAFNLVRE-SEQLFSI
CQIPLLCWILCTSLKQEMQKGK-DL-A--LT-CQSTTSVYSSFVFNLFTPEG-AEGPTP-
-----------QSQRQLKALCSLA---AEGM-WT-DA----FEFRED-------------
--DLRRNGVVDA-DIPALLGT----KILLKYGE-RE-SSYVFLHVCVQEFCAALFYLLK-
GHFDH-----------------P--HPAVRCVQ-ELLVANFKKARRAHWIFLGCFL----
----TGLLNKKEQEKLDAFFGFQ---------------L--SQE-IKQ-QFHQCLKSLGE
C-G----------------------------------DPQGQVDSLAIFYCLFEMQD-PA
FVKQAVNL---------------------L-------------------QEANFHIIDNA
DL---VVS--A----YCLK-YCSS-LRK-LCFS----------------VQN-----VFE
-TED-EHS-----------S-----------------------------------TSDYS
LVCWHHIC-SVLTTSGHLRELQVEDSTLSES---------TFVTWC--NQLRHP------
SC--R-LQKLGINN---------VSF-SGQSAQLFEVL----------------FYQPDL
KYLS-LTLT-KLSRD-DVRSLCDALNYPAGN----VKEL--------AL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------VNCRLS-----------------------------
----------------------------PIDCEVLAG-----------------------
--------------------------------LLTNNKKLAY---LN-VSCNQ-LD----
-------AGLPLLCAAL----CSPDAVLVYL-----------------------------
------------------------------------------------------------
------------------------------MLAFCHLSEQCCEYISEM-LLR
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
NKSVRYLD
L-SANVLKDEGLK-TLCEALKRP--DCCLDCCL---------------------------
-----------------------DSLCL--VKC-----FITADGCEDL------------
-------ASAL-I
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
SNQNLKILQIGCNEIGDVGVQL-LCRALT-HP-DC-RLEILG--LE-
--E-CGLTSACCK--------------------DLASVLTSSKML-QQLNLTLNALDHTG
VVVLCE--------ALRHPACA--LQ--V-------------------------------
---LGLRKT-DFDEETQAL------LT----------AEE-ERNPDLTITDE-FDTITRV
EI----------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPCOP00000023924 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------MEPSG----QLDFD---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 323)
LQALLEQLD--Q---DELSKFKSLVRT------
---------L--RQERE--------L--PQV-IQTE-VDKAD-GKQLAEIFSNHYPSYWV
QWAAIMV----FN-KMNRTDLSERVR-NELREAAM-KSLE----ENK------PRSLD--
------IK--RTEA-PN--------------------------R---EDEEEIIRR-LEE
RDPELT---------------------------------KIQIKDITNL-----------
-------GRP-------------------------------RRFLEGQKPGKEDTYRGI-
--------IMKK----------------FWKA--WNNN-GPGAGEHISD-----------
---------------------------------------------VTQRDKMLIPFCDPR
TL--PGVFP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
HTLVLHGPAGFGKTTLTRKV-----ILDWT--E---------NHLNQ
---------TFRY-AFYFTCKELNRMG-------PCTFAE-LISK-DW-----PE-WRND
LPEVLA-RA----YRILVIIDGFEE-LKVPEG------ALIR-D-----ICGNWEEQ-KP
-VPVLLGSLLIGTMLPNATLLVT-TRSEALGDLRLLL-Q-----QPL--FLEIEGFLEQD
KKAYF----------------L--KH-FED---------EEEALRAFDLMRS-NSALFRM
CAAPAVCWVVCTCLKLQMERGQ-DP-A--PT-CLSATSLFLRFLCGQFMPE---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
NK-----------------------------------P-FSETDTKEFLYCLYESQD-EG
LVKDAMAP---------------------F-------------------KEISLHLRNKM
DM---VHS--S----FCLK-HCQN-LQK-IWLH----------------VAKG----VFL
-ESG-PAS----EP--------------------H------------AQLERSQDD-QHI
IPFWRDLC-SMFGSNMNLKFLKISESFLSTS---------SVQLLC--QEIVS-T-----
TC--Y-LQKVV--L-KDISPA--DF----CQ-NFCLAL----------------RRHKTL
THLT-LQGN-NQND--TLPSLCEILRHPKCN----LQYL--------RLV---------S
C------S----------------------------------------------------
---------AT---PQ-QWA---GLSSALEA-NESLTCLILSANKLMD-EGVKLLHTT--
LRH-PKSVLQKL-S----------------------------------------------
------------------------LENCHLT-----------------------------
----------------------------EACCKNLAS-----------------------
--------------------------------ALIISQRLTH---LC-LAENT-LGD---
-------RGVEILCEGL----RYPEC----------------------------------
--------------------------KLQ-ILV---------------------------
-------------------------------LWHCNITDNGCKHLSKV-IEQQSSLTHLD
L-GLNHVGITGLR-SLCEALKEP--LCN----L---------------------------
-----------------------RCLWL--WGC-----SITRFSCEDL------------
-------SAAL-GSNRSLITLDLGQNSLGFNGVKM-LCEALK-RQ-NC-PLQTLR--LK-
--I-ERYDAPMQK--------------------LLEEMKESNPQLTIEDDRY----DPRK
---------------KRPSSRY--FV----------------------------------
----L-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSAPLP00000009811 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------
PF02758 // ENSGT00900000140813_0_Domain_0 (295, 328)
YGEDAA
MDIAIGL----FE-EMNQRDLAEKIL-DEKVKGKELGSCSPEDLGC-------QEVWL--
------------------------------------------------------------
------------------------------------------------------------
--------TL-------------------------------ALTLLLSGVEYKQKYREH-
--------VARE----------------FLQY--KEVNSCLGENL---------SVSSRY
TALTITKKPWSQRGGEPEGA------DVSWGCAD------------TTSAVTAQTLFKPD
ED--GQTPQ----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
-TVVLVGAPGMGKTMMVRKM-----MVEWV--E---------GALYM
---------QFDY-VFCIDCKELSL-S------KQVSVLD-LVSK-CC-----PH-QRIP
AGSILD-NQ----EKVLFIFDGFEA-LGFPLAQP------KD-E-----LSSDPREA-KP
-LETTLMSLLKRTVLPASSLLIT-TRPMALQNLGRCL-E-----GEC--YVEILGFSAAA
REEYF----------------H--RY-FKN---------DNKADVAFRFARG-NESLYSL
CVIPIMSWTICTILEQELYKKN-NI-L--EC-SKATTWMGMFYLSWLMKCRG-SNA-QQD
------------LQQFLHKLCSLA---ADGI-WK-HK----VLFEEK-------------
--EVKDCGLDWPDLLSLFLNE----KSLKKGID-QD-NVYSFTHLHLQEFFAAMFYVLD-
DDEET--------------VSDP--EALKKNVN-TLLESYSKS-RK-DLNLTIRFL----
----FGLVNPKSIEYAGERIGCR---------------I--SPR-AQE-DLLRWLQTRHR
GLS--------------------------------HPSEALMIKDLDTFHFLFEMNE-KS
FAQNVLGC---------------------F-------------------TGIDLHDIKLT
LY-DQMAL--C----FCIK-QWDG-VDS-VTLR----------------SCS-----FHQ
-QQCREE-----------------------------------PAT------VLPGFYLFL
GR-----------------------------------------I-------LR-DY--GH
TC--P--------------LV--P------------RQ----------------------
-----HPRQ-EELRS-PLHPLCQALGHTGSS----LQNL--------RLQ---------W
C------G----------------------------------------------------
---------LT---EG-GCE---ALGTLLAT-HPSLACLELGDGALGD-SGVRLLCTG--
LRQ-PGCHLRIL-R----------------------------------------------
------------------------LRYTRLT-----------------------------
----------------------------SACCQDLAA-----------------------
--------------------------------VLGTSP
PF00560 // ENSGT00900000140813_0_Domain_11 (2019, 2057)
HLEE---LD-LSFNTGLRD---
-------DGVQLLCEGL----RHRAC----------------------------------
--------------------------QLR-VLR---------------------------
-------------------------------LGSCHLTGSCCQALATH-IGESCS-----
------------------------------------------------------------
------------------------------------------------------------
-----------------LSCLDLSDTELGAGAV-L-LLRQLR-HP-AC-PLQAL------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSSSCP00000003594 (view gene)
ENSSBOP00000019275 (view gene)
ENSOGAP00000015737 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-SPLTTLACSSGPGA--AAVA
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 328)
REL---LLAALEDLS--Q---EQLKRFRHKLRD------
------------APANQ-----------RSI-PWGR-LEHAD-AVDLAEHLTHFYGPEPA
LDVARKT----LK-RADVRDVAARLK-EQRLQR---------L---------GRS-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------SS-ALLSVSEYKKRYREH-
--------VLRE----------------HAKV--KERNARS------------VKISKRF
TKLLIAPESAAA-EQ--VLGLAE-------AAER--------ERARRSDTHTFNRLFRRD
R---EGQRP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYQ
G--------QVDF-AFFLPCGELLERPG------ARSLAD-LILD-QC-----PD-RGAP
MQLILG-QP----QRLLFILDGADE-LSLLDPR--------EAR-----PCTDPFEA-TS
-GERVLGGLLNKALLPSARLLVT-RRAAAPRRLQGCL-C-----SPQ--CAEVCGFSDKD
KKKYF----------------Y--KF-FQD---------EWRAERAYRFVKE-NETLFAL
CFVPFVCWIVCTVLRQQLELCQ-DL-S--RT-SKTTTSVYLFYIASELNSALTVNGP---
-----------QVREQLLKLCRLA---REGVLGR--R----AQFVEK-------------
--DLERLQLRGSKVQRLFLIEKELPGVLET-----E-VTYQFMDQSFQEFFAALSYLLE-
EEESP-------------------R-AAADGVG-TLLRGDPQL--RRHLVLTRRFL----
----FGLLSAERMGDIERHFGC-----------------AVSER-VKQ-EALQWVQGQGC
PP-----AALEETLGLQDVEEPEE-E-----------EGEELSYPLELLYCLYETQE-DT
FVRQALSG---------------------F-------------------RELVLERVRFS
RM-DIAVL--S----YCVR-CCPT-GQA-LRLV------------------SC----SLA
------AA----------------------------------------QEK--K---KKS
----------------------------------------LVKR----------------
------LQGS---L--------------GGS------S----------------SSRTTM
KQ---LPAS-------PLCPLFEAMTDQQCG----LSSL--------TLS---------H
C-----------------------------------------------------------
--------KLS---DA-VCR---DLSEALRA-APALTELGLRHSRLS-EAALRLLSEG--
-LAWPQCQVQTL------------------------------------------------
----------------------------RLQ-----------------------------
---------------VPGL---------QGAFQYL-------------------------
------------------------------VGMLQQSPALTT---LD-LSGC-QLPDSMV
--T--------YLCAVL----QHPGCKLQTLSL--ASMELNEG-----------------
----------CMQELRAVKTAKP-------DLV----------------------I--IH
PALD-------------------SHSAPPTGFSSAL------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSFCAP00000018243 (view gene)
ENSCAFP00000040841 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MTS---------------------------QKKRKDMEPA-----
--------CTWS------------------------------------------LLSE--
-----------------------------------------QK-----------------
----AASNSQ-------------------------------SPSSWKEFRDYQEMFRKH-
--------LKEK----------------YLNI-GAHKCYHHGK--------------HIE
SRLLVVKEHGISEKTPCGIP-------------------------QQDNFIDMEHVFDPD
EE--GSSSS----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
QTVVLQGCAGIGKTAVVHKF-----MYDWA--A---------GMVTP
G--------RFDY-LIYVNCREINSIA-------NLSAAD-LITN-TF-----QD-IDGP
ILDVIPMYP----EKLLFILDGFPE-LQCPVDNQ------EE-D-----LSANPQEK-KP
-VETLLCSFVRKKLFPESSLLIT-ARPTTMKKLHSLL-K-----QHI--QAEIVWFTDAE
KRAYF----------------L--SQ-FSG---------ATVAMKVFYELQQ-NESLDIM
SSLPIISWMICRVLQSQGDGDR-TL-M--RS-LQTVTDVYLFYFSKCLKTLTGIS-VW--
-----------KGQSCLWGLCSLA---ADGL-QN-QQ----VLFEVS-------------
--DLRRHGIGIYDMNCTFLNH-----FLKKVKG-SA-SVYTFLHFSFQEFLTAVFYVLK-
NDSNW---------------------MFFGQVGKTWQEIFQQY-GKGFSSLTIQFL----
----FGLLHKGKGKAVETIFGRK---------------I--SPG-LRE-ELLKWTEKEIK
DK-----------------------------------SSRLQIEPMDLFHCLYEIQE-EE
YTKGIIDD---------------------L-------------------QSIILLQPTYT
KM-DILAM--S----FCVK-SSHNHLS----------------V-----SLKCQHLLGF-
-EEEERAS----------------------AFIP--------------------------
---------PASALNQSSQ-----LCNLPAS---------RLQLLC--QALCNP------
YC--K-IKDLKL---IFCHLT------ASCGKD---------------LSLVFET-NQYL
TDLE-FVKN-TLEDS-GMKLLCEGLRKPNCV----LQTLR--------LYRCLL-SPA-S
CG------------AL----AAVLSTSQWLTDLEFSETKLDALALKLLCEGLKDPNCK-L
QKLKLCASFLPESSEV-VCK---YLASVLIC-
PF13516 // ENSGT00900000140813_0_Domain_8 (1773, 1797)
NPNLTELDLSENPLGD-TGVKYLCEG--
LRH-SNCKVEKL-D----------------------------------------------
------------------------LSTCCLT-----------------------------
----------------------------DASCMELSS-----------------------
--------------------------------FLQVSQTLKE---LF-VFANT-LGD---
-------TGVQHLCEGL----QHTKGILENL-----------------------------
------------------------------------------------------------
------------------------------VLSECSLSAACCESLSQV-LSSSRSLTRLL
L-INNKIEDLGLK-LLCEGLKQP--DCP----L---------------------------
-----------------------KDLAL--WTC-----HLTGECCQDL------------
-------CNAL-YTNEY
PF13516 // ENSGT00900000140813_0_Domain_15 (2358, 2379)
LRDLDLSDNALGDEGMQV-LCEGLK-QP-SC-KLQTLW--LA-
--E-CHLTDACCG--------------------ALASVLSRN
PF13516 // ENSGT00900000140813_0_Domain_17 (2443, 2465)
ENL-TLLDLSGNDLKDFG
VQMLCD--------ALLQPICK--LQTFY-------------------------------
------IDTDHLREETFRK------ME----------ALK-MSKPGITW-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSSSCP00000003593 (view gene)
ENSRBIP00000018340 (view gene)
MDQPEAPCSRLRSLRTASSQRAFVESVPHQRLRLRPWCKAPKLGLTK-RQPGHMTWPGVW
KNRSRKGGAGEEGAPQSSGRRWPVPTSPSEGEEHQD------APTRVRGRRR--------
-------AKSGVTPHCPLPPPHG-----SS--LARGSLRPCARWVLERQSRGGR----GS
PEDDPRPPVSSTGLR--LAVAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLAALEELS--Q---EQLKRFRHKLRD------
------------VGPDG-----------RSI-PWGR-LERAD-AVDLAEQLTQFYGPEPA
LEVARKT----LK-RADARDVAAQLQ-EQRLRR---------L---------GLG-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------SV-ALLSVSEYKKKYREH-
--------VLQL----------------HARV--KERNARS------------VKITKRF
TKLLIAPESAAPEEE--ALGPAE-------EPEP--------GRARRSDTHTFNRLFRGD
E---EGRQP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYQ
G--------QVDF-AFFMPCGELLERPG------TRSLAD-LILD-QC-----PD-RGAP
VPQMLA-QP----QRLLFILDGADE-LPALEGP--------EAA-----PCTDPFEA-AS
-GARVLGGLLSKALLPTALLLVT-TRAAAPGRLQGRL-G-----SPQ--CAEVRGFSDKD
KKKYF----------------Y--KF-FQD---------ERRAERAYRFVKE-NETLFAL
CFVPFVCWIVCTVLRQQLELGR-DL-S--RT-SKTTTSVYLLFIASVLSSAPVADGP---
-----------RVQGDLRNLCRLA---REGVLGR--R----AQFAEK-------------
--QLEQLELRGSKVQTLFLSKKELPGVLET-----E-VTYQFIDQSFQEFLAALSYLLE-
DEGVP-------------------R-TAAGGVG-TLLRGDSQP--HSHLVLTTRFL----
----FGLLSAERMRDIERHFGC-----------------VVSER-VKQ-EALRWVQGQGQ
GCPGVAPEVTEETKGLEDTEEPEEE----------EEEGEEPNYPLELLYCLYETQE-DA
FVRQALCR---------------------L-------------------PELALQGVRFC
RM-DVAVL--S----YCVR-CCPA-GQA-LRLI------------------SC----RLV
------TA----------------------------------------QEK--K---KKS
----------------------------------------LGKR----------------
------LQAS---L--------------GGS------S-----------------SRGTT
KQ---LPAS-------LLHPLFQAMTDPLCR----LTSL--------TLS---------H
C-----------------------------------------------------------
--------KVP---DA-VCR---DLSEALRA-APALTELGLLHNRLS-EAGLRMLSEG--
-LAWPQCRVQTV------------------------------------------------
----------------------------RVQ-----------------------------
---------------LPSS---------QQGLQYL-------------------------
------------------------------VGMLRQSPALTT---LD-LSGC-QLPAPMV
--T--------YLCAIL----QHQGCGLQTLS--LTSVELSEQ-----------------
----------SLQELQAVKRVKP-------DLV----------------------I--AH
PALD-------------------GHPEPP-KEVISTF-----------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMPUP00000006685 (view gene)
------------------------------------------------------------
------------------------------------------------------------
----------------SILLRNP------------L-------------------FQQ-N
L---RSQPCTKMREAKLPSFS
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 326)
NYA---LQWCFMQLS--K---EEFQMFKGLLME------
---------K--ASEVA--------V--CSF-PWVE-VNNAD-MEHLASLLHEHYEESLL
WKISNHI----FE-TMGLSTLSEKAR-QEMKKYSL-AEIQENSTQMKK---EEPSMKEVP
---------------------E------------------------------ISQAIE--
QG-GAISA---------EIKE-QETVEEARSMEVKFFHSLGARQQDASK-------LPFR
ES--AGFLFS-------------------------------LPLSNLGRGDKTRDYKGY-
--------VMTK----------------FAAK--SDEF-HGFEHL---------------
-----------------------------------AS-------DCPEI-QMLLGAFNPD
Q---TGFQP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
RTVVLHGKSGIGKSALAQRV-----LQCWA--Q---------GELYQ
V--------RFSY-IFFLRARDMQCMR-------EGSFAE-LISG-EW-----PD-SPVP
MMEIMS-QP----ERLLFIVDGFDD-LHFALQDD------NA-S-----LCEDWAVK-RP
-PSVLMRSLLKKVLLPESSLMIT-VRDTGIEKLKSML-T-----SPC--YLLVGGISVEK
RVQLL----------------L--RH-MKN---------EHQKTQALRAVAD-SRSLLDA
CQVPVLGSLICEALNLQEACGE-HL-S--PP-CHTLTGLYTTFVFHQLGPQD-ASRRRLS
----------QDETAVLRSLCLLA---VQGV-WH-RK----FVFYAD-------------
--DLRAHGLRES-ELSALLHT----DLLLQDGR-QERCCYSFLHISLQEFCAALHYVLE-
GLETG--------------SHPC--LVLMQGL----AALRPAG-SDSHLHQMKRFL----
----FGLMSKEGTGALEVLLGCC---------------V--PR-ALKR-GLLSWISLLGR
QPG----------------------------------TC-TPPDLLDAFHCLFETQD-EE
FVRLALDS---------------------F-------------------QEVWLPIGHRT
DV---LVS--A----FCLQ-RCRY-LQR-IHIE----------------VPQ-----SLS
-KDEFSDA----QP--------------------LI-----------PQ--GMPAK-TLA
AECWESLC-IVLSTHPSLRELDLSGSVLNEQ---------AMKTLC--IKLRQ-P-----
TC--K-IQNLI--F-KDAQV---PL---GLR-HLWMVL----------------ITNHNM
KYLN-LEST-RLKEA-DLMMACQALRHPSCL----LESL--------RLD---------H
C------G----------------------------------------------------
---------LT---PA-CCL---VISQILIT-SISLKSLSLVGNKLAT-QGVKALCES--
LTA-SQCALQKL-I----------------------------------------------
------------------------LGNCGLT-----------------------------
----------------------------AADCRDLAS-----------------------
--------------------------------ALISNQSLTH---LC-LSSNS-LGR---
-------EGVNLLCRAI----KLPSC----------------------------------
--------------------------GLQ-RLI---------------------------
-------------------------------LNECNLDVAGCGFLAFA-LMGNKRLTHLS
L-SMNPVEDDGMN-LLCEVLMEP--SCH----L---------------------------
-----------------------QDLEM--VKC-----HLTATCCKKL------------
-------SCVI-TRNKHLKSLDLAVNALGDDGIMA-LCEGLR-HR-RA-SVWRLG--LE-
--A-CGLTSGCCE--------------------ALATAVLGSQCL-TSLNLMRNDLSPEG
MTTLCL--------ALAQPTCN--LQ--I-------------------------------
---IGLWKW-QYPTQTRKL------LE----------EVQ-RQKPQMIIGDSWYSTD---
--E---------------------------------------------------------
--------------------------------------------------DDRYWWKN--
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCAPP00000001320 (view gene)
ENSPANP00000007375 (view gene)
ENSRBIP00000027142 (view gene)
------------------------------------------------------------
------------------------------------------------------------
-------------M-SDVNPPSD------------T-PVPFSSS-------------C-T
H---SSCNPPWTFSCYPGSPCENG---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
VMLYMKYVS--N---EELRWFKQLLLT------
---------E--F-RTG--------T--MPI-TWDQ-VKTAS-WAEMVHLLIERFPGRRA
WDVTSNI----FA-MMNRENMCAVVC-REINGILH-TLEPEDLNVG------E-------
------------------------------------------------------------
------------------------------------------------------------
-------TQV-------------------------------NLEEGES--GKIRQYKLN-
--------VIDK----------------CFPT--WDFMTWPGNQRDFFYQDV--------
----------------------------------------------HRHTEYLPCLLLPK
RP--HGRQP----K
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 929)
TVVIQGVPGIGKTTLAKRV-----MLEWA--S---------NRFYT
H--------KLWC-AFYFHCQEMNQTT-------EHSFSE-LIEH-KW-----PG-SQDL
VAKIMS-KP----DQLLLLLDGFEE-LTSTLI------NRPE-D-----LSEDWRQK-LP
-GSVLLSSLLSKRMLPEATLLIM-IRCTSWPTCKPLL-K-----CPS--LVTLPGFNTME
KIKYF----------------Q--MY-FGH---------RE-RDRVLSFAIE-NTILFSM
CQVPVVCWMVCSGLKQQMERGN-NL-T--QA-CPNATSVFVRYISSLFPTRA-ENFSRKI
------------HQAQLEGLCHLA---ADSM-WH-RK----WVFGKE-------------
--DLEEAKLDQT-GVAAFLGM----SVLRRIAG-EE-DRYAFTLLIFQEFFAALFYVLC-
FPQRL--------------KNFH--VLSHVNIQ-HLIASPR-G-SKSYLSHMGLFL----
----FGFLNGTCASAVEQSFGCK---------------V--SFN-NKR-KLLKVISRLQK
CE-----------------------------------PPSPCYGVPQLFYCLHEIQE-EA
FVSQALNG---------------------Y-------------------RKVVLKIGNNK
DV---QVS--A----FCLK-RCEH-LQE-VELT----------------VTL-----NFK
-NMWKLSS----S-------------------------------SHPGS--EAPES-KAL
HGWWQDLC-SVFATNDKLEVLTVTNSVLEAP---------FWKALA--AALKH-P-----
QC--K-LQKLL--L-RRVNST--VL---N-Q-DLIGVL----------------TGNQHL
RYLE-IQRV-EVDSN-AMKLLCRALRSPLCC----LQCL--------RLE---------D
C------L----------------------------------------------------
---------AT---PR-IWM---DLGSNLQV-NRHLKTLILTKISLEN-CGAYY------
--P-SMAQMERL-S----------------------------------------------
------------------------IENCNIT-----------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------QLSCESLAFC-VGE
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2236)
SKTLTHLS
L-AENALKDEGAK-HIWNALSYL--RCP----L---------------------------
-----------------------QRLVL--RKC-----DLTLNCCQNM------------
-------ISAL-C
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
KNKTLKSLDLSFNSLKDDGVIL-LCEALK-NP-DC-TLQILE-----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSOCUP00000014512 (view gene)
------------------------------------------------------------
------------------------------------------------------------
-------------M-SDAI---P------------G-PSALSSS-------------V-V
Y---ASFLPFWITSCCSASPYE
PF02758 // ENSGT00900000140813_0_Domain_0 (203, 323)
KG---LMLYLLYLS--K---EELRQFKQLLVR------
---------Q--F-SIS--------A--QPA--GEQ-LEAAS-WAELAHFLTQYFPGRQA
WDVTHNL----FA-KMNQQNMCFLVQ-KELNALLP-TLVSSE----------E-------
------------------------------------------------------------
------------------------------------------------------------
-------VLL-------------------------------KLE-KEEV-DQLREYKLR-
--------VLRK----------------FFPE--WDRIAWPGNRVDFFYQEV--------
----------------------------------------------HKHKTFLPCLFLPR
KP--QGRPA----K
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 929)
TIIIDGAPGVGKTTLARKL-----MVAWA--R---------NQFYV
H--------KGWF-VFYFQCQEVTGVG-------EQSCSE-LIAR-QW-----PG-CQAL
VSKILS-KP----DQLLLLFDGFEK-LTRPLF------VRQR-E-----LSADWSRK-LP
-GAVLLSSLLNKTMLPEASVLLT-MRMNSWWVVRPLL-K-----SPF--FVTLRGFTMME
KIRYL----------------R--TY-FGN---------TEEGERAVDFARR-NTILFSL
CRVPALCWMVGSCLRQQMQAGV-DL-A--EA-CPNATAVVALYVSSLFPFRA-EPLPGRT
------------HLEHLEGLCHLA---ALGM-WS-MK----WVFGKE-------------
--DLEEAKVDEL-DVSSFLGA----DILQEVEG-EE-PRYTFAFAVFQEFLAALFHVLY-
FPGRP--------------TPFH--LLGHGDIQ-YLIASPG-G-SRSHLAHMGMFL----
----FGLFNVSCAAMVRRVFHCE---------------L--SWG-NKR-KVLQTVSLLLE
GD-----------------------------------LPALCCGVPQLFYCLTEVRE-ED
YVSQALSG---------------------Y-------------------RKAVLKMQSNK
DV---QLS--A----FCLK-RCRR-LQE-VELT----------------LMV-----NFS
-QVWWPGP----GD------------------------------SLHAA--EIQES-DMQ
YQWWQDIC-SVLTTNEGLALLTLTKSILEAP---------LVKVLA--AALRH-P-----
QC--K-LQKLS--L-RHLGPS--VL---H-E-DLFCVL----------------VENRHL
SCLE-IQYT-EVGTE-AMRDLCAALRHPQCA----LQCL--------RLE---------Y
C------M----------------------------------------------------
---------AA---PE-DWI---DLAKGLQN-DIHLGTLLLKRNPLES-FMANY------
--V-SASRLERL-S----------------------------------------------
------------------------LETCNLT-----------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------GLACERLASS-LQQSKMLSHLS
L-ADNPLGDEGVK-PIWKALEQL--ACP----L---------------------------
-----------------------QRLVL--RKC-----ALTSACCQDV------------
-------ASVL-CQSQTLRSLDLSFNSLMDKGVTL-LCEALK-NP-DC-SLQVLD--FK-
--Y-CLFTLVCCS--------------------AMSSMLLSNPSL-RYLDLSKNNIGLLG
ILTLVG--------AFPSQRQR--DK--V-------------------------------
---L------L-------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSNLEP00000012949 (view gene)
ENSMFAP00000035233 (view gene)
ENSPPAP00000033490 (view gene)
ENSMLEP00000013154 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MSLDIQCEQ-LSD-ARWA----------------EL---------
----LPLL-------------------------------QQCQVV------RLDDC-GLT
EARCGDIS-SALRV
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1542)
NPALTELSLRSNELGDA---------GVHCVL--QGLQS-P-----
SC--K-IQKLS--L-QNCYLT--GA---GCG-VLSGTL----------------RS
PF13516 // ENSGT00900000140813_0_Domain_6 (1617, 1642)
LPTL
QELH-LSDN-LLGDA-GLQLLCEGLLDPQCR----LEKL--------QLE---------Y
C------N----------------------------------------------------
---------LS---AA-SCE---PLASVLRA-KPDFKELTVSNNDINE-AGVRVLCQG--
LKD-SPCQLEAL-R----------------------------------------------
------------------------LESCGIT-----------------------------
----------------------------SNNCRDLCS-----------------------
--------------------------------IVASK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
ASLRE---LA-LGSNK-LGD---
-------VGIAELCPGL----LHPSS----------------------------------
--------------------------SLR-TLW---------------------------
-------------------------------IWECGITAKGCADLCRV-LRTKE
PF13516 // ENSGT00900000140813_0_Domain_13 (2215, 2237)
SLKELS
L-AGNELGDEGAR-LLCETLLEP--GCQ----L---------------------------
-----------------------ESLWV--KSC-----SFTAACCSHF------------
-------SSVL-TQ
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NKFLLELQISNNRLGDAGVQE-LCKGLG-QP-GS-VLRVLW--LG-
--D-CDMSDSSCS--------------------SLAATLLA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2464)
NHSL-RELDLSNNCLGDAG
VLQLVE--------SVRQPGCL--LE--Q-------------------------------
---LVLYDI-YWSEAMEDR------LQ----------ALE-EEKPSLRVIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
MGP_CAROLIEiJ_P0083139 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MSLDIQCEQ-LSD-ARWT----------------EL---------
----LPLI-------------------------------QQYEVV------RLDDC-GLT
EVRCKDIS-SAVQA
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1542)
NPALTELSLRTNELGDG---------GVGLVL--QGLQN-P-----
TC--K-IQKLS--L-QNCGLT--EA---GCG-ILPGML----------------RS
PF13516 // ENSGT00900000140813_0_Domain_6 (1617, 1642)
LSTL
RELH-LNDN-PMGDA-GLKLLCEGLQDPQCR----LEKL--------QLE---------Y
C------N----------------------------------------------------
---------LT---AT-SCE---PLASVLRV-KADFKELVLSNNDLHE-PGVRILCQG--
LKD-SACQLESL-K----------------------------------------------
------------------------LENCGIT-----------------------------
----------------------------AANCKDLCN-----------------------
--------------------------------VVASK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
ASLQE---LD-LSSNK-LGN---
-------AGIAALCPGL----LLSSC----------------------------------
--------------------------KIR-TLW---------------------------
-------------------------------LWECDITAEGCKDLCRV-LSA
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
KQSLKELS
L-ASNELKDEGAR-LLCESLLEP--GCQ----L---------------------------
-----------------------ESLWI--KTC-----SLTAASCPYF------------
-------CSVL-T
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
KNRSLLELQMSSNPLGDEGVRE-LENSGT00900000140813_0_Linker_15_16 (2378, 2389)
CKALS-QP-DTPF13516 // ENSGT00900000140813_0_Domain_16 (2389, 2436)
-VLRELW--LG-
--D-CDVTNNGCS--------------------SLENSGT00900000140813_0_Linker_16_17 (2436, 2442)
ANVLLVPF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2464)
NRSL-RELDLSNNCMGGPG
VLQLLE--------SLKQPSCT--LQ--Q-------------------------------
---LVLYDI-YWTNEVENQ------LR----------ALE-EERPSLRIIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMICP00000048054 (view gene)
ENSPEMP00000002348 (view gene)
ENSAPLP00000008960 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----SSTS--K----MDLDIQCEE-MDP-SRWA----------------EL---------
----LSTM-------------------------------KSCRSI------RLDDC-NLS
SSNCKDLS-SVINTNP
PF00560 // ENSGT00900000140813_0_Domain_4 (1517, 1551)
SLTELKLNNNELGDA---------GIEYLC--KGENSGT00900000140813_0_Linker_4_5 (1551, 1566)
LLT-P-----
SC--SPF00560 // ENSGT00900000140813_0_Domain_5 (1566, 1597)
-LQKLW--L-QNCNLT--SA---SCE-TLRSVL----------------SAQPSL
TELH-VGDN-KLGTA-GVKVLCQGLMNPNCK----LQKL--------QLE---------Y
C------E----------------------------------------------------
---------LT---AD-IVE---ALNAALQT-KP
PF00560 // ENSGT00900000140813_0_Domain_8 (1775, 1801)
TLKELSLSNNTLGD-TAVKQLCRG--LVE-ASCNLELL-H----------------------------------------------
------------------------LENCGIT-----------------------------
----------------------------SDSCRDISA-----------------------
--------------------------------VLSNKSSLID---LS-VGDNK-IGD---
-------SGLALLCHGL----LHPHC----------------------------------
--------------------------RIQ-KLW---------------------------
-------------------------------LWDCDLTSASCKDLSRV-LSTKETLLEVS
L-IDNNLRDSGME-MLCQALKDP--KAN----L---------------------------
-----------------------QELWV--REC-----GLTTACCKAV------------
-------SSVL-SVNKHLKVLHIGENKLGDAGVEL-LCEGLL-HP-NC-NIHSLW--LG-
--N-CDLTAACCA--------------------TLATLMVAKPCL-TELDLSYNTLEDEG
VRKLCE--------AVRNPNCK--LQ--Q-------------------------------
---LILYDI-FWSSEVDDE------LR----------ALE-ESKPEVKIIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSECAP00000018270 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---SSFFS
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 328)
DFG---LLWYLGELK--K---DEFRKFKDFLKQ------
---------E--PLQLG--------L--KPI-PWAE-VRKAT-REGLANLLVKHYEEQEA
WNVTFSI----FH-KIHRKDLCEKAK-KEI------------TG--------------HT
K---------------------------------------------------TYQA----
------------------------------------------------------------
----------------------------------------------------------H-
--------IQEK----------------FNNM--WFRESVTRIHDY--------------
---------------------------------F-------DQKTTQKERESLERLFAPK
ET--GKQ-P----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
RTVVFKGIQGIGKTTFLIKL-----MLAWA--E---------GSLYQ
E--------RFFY-VFYFCCREMKQLT-------QTSLAA-LISR-DW-----PNS-HVP
LTEIMS-QP----ERLLFVIDSFEE-LKHNLDEP------ES-D-----LCSDWMEQ-RP
-VRVLLSSLLRKKMLPECSLLIA-AAPVYPGEIEDRL-V-----CPE--IKTFLGFSESE
RKQYF----------------C--CI-FQD---------RNRAMEAFSFVRE-NEQLFCM
CQIPTLCWIVCTCLKHEMERGK-DL-A--LT-CRRTTSLYTSFIFNLFTAKG-TSCPDQ-
-----------QSQSQLKGLCSLA---AEGM-WT-DT----FVFSEE-------------
--DLRRNGLVDS-DIPTLLDM----KALQKYRE-CE-NSYLFIHVCVQEFCAAMFYFLK-
SHIDH-----------------P--NPAVGCTE-TLLFTYLKK-VKVHWIYLGCFM----
----FGLLNEKEQQKLDAFFGFH---------------L--SQE-IQQ-KFRQCVQRIGE
S-E----------------------------------HLQGEVDFLALCYCLFEMQN-EA
FVKWAMAL---------------------F-------------------QDVTFSIVDSA
DL---VTS--A----YCLK-HCSR-LRK-LWLS----------------IQN-----VFK
-EEN-ANN-----------P-----------------------------------TSNYS
FICWHHVC-SVLTTNENLTELQVSDSSLNES---------AFVTLC--NQLRHP------
SC--H-LQKLWINN---------VSF-SAESWFFFEVL----------------TLSPDL
KYLN-LNGT-ALSRK-DVKLLCNALSNPKCN----VEEL--------LL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------TNCCLS-----------------------------
----------------------------ADDCEAFTW-----------------------
--------------------------------VLNSNNKLKL---LN-VSYNY-LD----
-------KGVPLLCKAL----RHPDCALEVL-----------------------------
------------------------------------------------------------
------------------------------VLGYCYLSGHCWKSLSDA-LLCNKSLVHLD
L-SANVLKYEELK-LLCEALKQP--SCQ----L---------------------------
-----------------------QSLCL--LKC-----CITARGCQDL------------
-------ASVL-TSNQNLTGLQIGYNDIEDVGAKL-LCEALT-HP-QC-RLENLG--LG-
--A-CKLTSACCE--------------------DLSSALTSSKTL-RRLNLSGNALDHGG
VVVLCE--------ALRCPECP--LQ--V-------------------------------
---LGLKKD-EFDEETQRL------LM----------AEE-ERNPHLTIFS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSDNOP00000026801 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------MGSST----KL
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 324)
DFN---LQLLLEQLD--Q---DELVMFKYFLRT------
---------F--PPPSG--------L--QQI-PPTE-VEEAD-AKQLAEILINTYPSSWV
ETVTIQA----LI-KMSRVDLCTREK-EEAGGKQT-PVYS----GFRKRK---PRTSS--
------VGIRYA-D-KF--------------------------N---INLDD---KLNL-
LQVELS---------------------------------RVQEEDVKSL-----------
-------GGA-------------------------------KESLEREKPGKGDQYSSI-
---------LRS----------------FWQT--QKKNFYPGAGESFHPSYF--------
---------------------------------------------VTQRYKTLIPLYNPK
AL--AGPFP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
HTVVLQGAAGVGKTTLAKKL-----MLDWT--E---------DELGQ
---------TFRY-AFYLCCRRLNRVG-------PCTFAE-LVLQ-GW-----PA-LPEA
IADILA-QP----QKVLFVVDGFDE-LRVPPG------ALIH-D-----LCGDWRTQ-KP
-VPVLLGSLLKRKMCPKATLLVT-TRPWALRELRLLV-E-----QPV--VLEVEGFSESD
REEYF----------------L--KH-FED---------EAGALRALAAARS-NAALFGL
GAAPPVCWIICSCLKLQMLKGQ-DP-T--LT-CQTTTSLFLRFLCGQFTPT-PGAHPTER
------------LQAPMKALCLLA---VQGI-WA-RT----STFDGE-------------
--DLRRLGLEEG-ELGLFLDK----SVLLRDRD-CE-GAYCFAHLSVQQFLAAVFYLLG-
GEE-E-------------DRDRS--RWDIGDVE-ELLSKEERL-KNPSLTQVGYFL----
----FGLFNEERVKELETTFGCQ---------------I--SME-VKR-QLLRCKAKYIE
NK-----------------------------------PFPSMMGMKELFYCLYESQE-EE
FVKDFVGH---------------------L-------------------KEMSLHLKNKT
DI---IHS--S----FCLK-HCPN-LQK-ISLQ----------------VEKG----LFL
-END-AAL----ES--------------------D------------SQ-VRSQID-RHI
LPFWTDLC-SMFATNNNLIFLDISESFLSNS---------SVRILC--ENIVS-A-----
TC--N-LQKVV--L-KNIFPV--DA----YQ-NLCLTF----------------SGQGYL
THLD-LQGN-DHID--MLPLVCEVLQQPKCN----LQYL--------RLE---------S
C------L----------------------------------------------------
---------VT---TQ-QWA---DLFSTLRI-NQSLTCLNLTSSELSE-EGVRLLCAT--
LTH-PKCSLQRL-S----------------------------------------------
------------------------LESCRLT-----------------------------
----------------------------EACCEDLSS-----------------------
--------------------------------ALVANQRLTH---LC-LAKND-LGD---
-------EGVKLLCKGL----SHPNC----------------------------------
--------------------------KLQ-RLV---------------------------
-------------------------------LWCCNITEVGCDHLSKL-LQWKTSLTHLD
L-GLNRIGTIGLK-SLCKALKEP--LCS----L---------------------------
-----------------------KCLWL--WGC-----ALPPFSCMDL------------
-------SSAL-SINQSLISLDLGQNTLGHSGIMI-LCEALK-LQ-TC-PLQTLR--LK-
--I-DAPGPETQK--------------------LLNEVRESNPQLTIESETP----NPRE
---------------HRPSSCD--FL----------------------------------
----F-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSECAP00000018272 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MNLDIQCEQ-LSD-ARWT----------------EL---------
----LPLI-------------------------------QQYAVV------RLDDC-GLT
EVRCRDIS-SALQANPSLTELSLRTNELGDA---------GVHLVL--QGLQS-P-----
TC--K-IQKLS--L-QNCCLT--EA---GCG-VLPSVL----------------RSVHSL
RELH-LSDN-PLGDV-GLQLLCEGLLHPQCH----LEKL--------QLE---------Y
C------N----------------------------------------------------
---------LT---AA-SCG---PLAAVLRA-KQDFKELTVSNNDMGE-AGVRVLCQG--
LAE-SACQLETL-K----------------------------------------------
------------------------LENCGLT-----------------------------
----------------------------PANCKDLCG-----------------------
--------------------------------IVASKASLRE---LD-LGSNK-LGD---
-------VGIAELCPGL----LSPSS----------------------------------
--------------------------QLK-TLW---------------------------
-------------------------------LWECDITAGGCRDLCRV-LRAKENLKELS
L-AGNALGDEGAQ-LLCESLLEP--RCQ----L---------------------------
-----------------------ESLWV--KSC-----SLTAACCHHF------------
-------STML-TQNRHLLELQMSSNKLGDSGVQE-LCQGLG-QP-GS-TLRVLW--LG-
--D-CDVSNSGCS--------------------SLASLLLANR
PF00560 // ENSGT00900000140813_0_Domain_17 (2444, 2462)
SL-RELDLSNNALGDPG
LLELLQ--------SLEQPGCA--LE--Q-------------------------------
---LVLYDI-YWTQELEDR------LQ----------ALE-ESKPGLRLIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMNEP00000038446 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MTSP----QLEWT---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 323)
LQTLLEQLN--E---DELKSFKSLLRA------
---------L--PLEDV--------L--QKA-PWSE-VEEAD-GKKLAEILIHTSSENWI
RSATVTV----LE-EMNLTELCKMAK--SEM-----------------------------
------------------------------------------------------------
--LEAG---------------------------------QVQEIDNPQL-----------
-------GDA-------------------------------EEDSELAKPGEKEGRRNS-
---------VQK----------------QSLV--WKNTFWQGDIDSFHDDVI--------
-----------------------------------------------QRNQRFIPFLNPR
TP--RKLTPYTTPY
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVLHGPAGVGKTTLAKKC-----MLDWT--D---------CNLSP
---------TVRY-AFYLSCKELSRMG-------PCSFAE-LISK-DW-----PE-LQDD
IPSILA-QA----QRILFVVDGLDE-LKVPPG------ALIQ-D-----ICGDWEQQ-KP
-VPVLLGSLLKRKMLPKATLLVT-TRPRALRDLQLLV-Q-----QPI--YIRVEGFLEED
RRAYF----------------L--RH-FGD---------EDQAMRAFELMRS-NAALFQL
GSAPAVCWIVCTTLKLQMEKGE-DP-A--PT-CLTSTGLFLRFLCSQFPQG-------AQ
------------LRGALRALSLLA---AQGL-WA-QM----SVLHGE-------------
--DLESAGVQES-DLRLFLDR----NILRQDGV-AK-GCYSFIHLSFQQFLTALFYALE-
KE--E---EE--------DRDGH--AWDIGDVQ-KLLSEEERL-KNPDLIQVGHFL----
----FGLANEERVKELEATFGWR---------------M--SPE-IKQ-ELLRCKASLHA
NK-----------------------------------P-LSMTDLREVLCCLYESQE-EE
LAKVVVAP---------------------L-------------------KKISMNLTNTS
EM---MHC--S----FSLK-HCRD-LEK-VSLQ----------------IAKG----MFL
-ENY-VDF----EP--------------------DIEFERC-TYV--TLPSWAQQD-LRS
LRLWTDFC-SLFSSNSNLNFSFIHVSTQCE--------------DC--S--LK-V-----
ES--L-LLFPR--I-KNVTPD--TA----YR-DFCLAF----------------IGKRTL
THLT-LEGH-IEWERTMLLLLCDLLRNHKCN----LQYL--------RLG---------G
H------C----------------------------------------------------
---------AT---SE-QWV---DFFYVLKA-NQSLRHLHLSASVLLD-EGAKLLYKT--
MTH-PKHFLQML-S----------------------------------------------
------------------------C-----------------------------------
--------------------------------KDAFA-----------------------
--------------------------------AVWLQPAADT---PC-AAKNP-HRD---
----------------I----TGPPS----------------------------------
--------------------------SLD-ILA---------------------------
-------------------------------VQQCTNQQRG-EYPTEQ-LQETAGLTKPD
AESERQIA-RDLW-ISLSASELL--PS---------------------------------
----------------------------------------LLSYCQHW------------
-------DL-----------------------LSH-NQKVLR-TK-TG-SLKTLR--LK-
--T-YETNLEIKK--------------------LLEEVKEKNPKLTIDCNAS------GA
---------------TAPACCD--FF----------------------------------
----C-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSOARP00000001012 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---DSFFS
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 328)
DFG---LLWYLEELK--K---EEFWKFKELLKQ------
---------E--SLKFK--------L--KPI-PWTE-LKKAS-RENVSKLLSKHYPGKLA
WDVTLSL----FL-QISRDDLWRKAR-NEIRQ----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------TINPYRSH-
--------MKQK----------------FQVL--WEKEACLLVPED--------------
---------------------------------F-------YRETTKSEYEYLNAVFLAA
LQ--PGESS----P
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVILHGPEGIGKTTLLRKV-----MLEWA--K---------GNLWR
D--------RFSF-VFLLTGREMNGVT-------DMSLVE-LLSR-DW-----PES-SEP
IEDIFS-QP----ERILFILDGVEE-LKFDLD-C------DT-D-----LCEDWEEP-HS
-MQVVLRSLLQKQMLPECSLLLA-LSKTGMRKNHSLL-K-----HIK--CIFLLGFSEHQ
RKLYF----------------S--HY-FQE---------NDASSRAFSFVRE-KRSLFFL
CQSPFLCWLVCTGLKCQLDKGE-DL-E--LD-SETITGLYVSFFTEVFKSGS-ETCPLK-
-----------QRRACLKSLCTLA---AEGM-WT-RT----FLFCPE-------------
--DLRRNGVSES-DTLMWLDM----KLLHRSG-----DCLAFIHACIQEFCAAMFYMFK-
RPKDP-----------------P--HSVIGNVT-QLITRAVSE-HYSHLSWTAVFL----
----FVFSTERMTNRLETSFGFP---------------L--SKE-IKQ-EITQSLDTLSQ
YD-----------------------------------PNNVTVSFQALFNCLFETQD-PE
FVAQVVNF---------------------F-------------------KDIDIYIGTKE
EL---IIC--A----ACLR-HCHS-LQK-FHLC----------------MER-----VFP
-DESGCIS-----------N-------------------T-I-------EK---------
LTLWRDVC-SAFTASEDFEMLNLENCQFDEA---------SLAVLC--RMLSQP------
VC--K-LRKFVCNF---------ASN-LANSLELFKAI----------------LHNPHL
KHLN-LYGS-SLSHM-DAKQLCEALKHPMCN----IEEL--------ML-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------GKCDIT-----------------------------
----------------------------GEACEDIAS-----------------------
--------------------------------VIIHNKKLKL---LS-MCENA-LKD---
-------DGVQVLCEAL----KSPDCALEAL-----------------------------
------------------------------------------------------------
------------------------------LLSHCCFTSAACDYLSQV-LLGNRSLTVLD
L-GSNVLKDEGVT-TLCESLRHP--SCN----L---------------------------
-----------------------QELWL--MNC-----YFTSVCCVDI------------
-------ATVL-VHSEKLTTLKLGNNKIYDAGAKQ-LCKALK-HP-KC-KLEHLG--GVA
LAP-RGWSPASCE--------------------DLASALTTCKSL-TCVNLEWITLDYDG
AAVLCE--------ALVSLECS--LQ--V-------------------------------
---LGLNKS-SYDEEIKMM------LT----------QVE-EMNPNLIISHHLWTHN---
--EGR-----LR------------------------------------------------
--------------------------------------------------GVLV------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSDNOP00000020663 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---ESFFS
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 328)
DFG---LLWYLEELR--K---EEFWKFKELLKQ------
---------Q--PPKLE--------F--KSI-PWSE-LKKAS-REDLAKLLDKHYPGKMA
WEVTLSI----FL-QMDRRDLWLKAQ-EEMSA----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------DINTYRKH-
--------MKEK----------------FHHI--WKKETCLPVPES--------------
---------------------------------F-------YKEVTNNEYEELNGAYT-A
KE--FRECS----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 891)
HTVVLQGPQGIGKTTFLRKV-----MLEWA--E---------GNFCK
D--------RFKS-VFFLNAHEMNHIK-------EANLLE-LIPR-DW-----PE-----
------------------------------------------------------------
-------------------------------------------------XICLPGLSEYE
RELYF----------------S--HF-FSE---------KKKVLKAFSFVRD-NGPLFSM
CQNPLLCWLICTCIKIQLERGE-DL-E--VA-WQTNTFLYASFFTNVFKTGG-ENRSPK-
-----------QNRTRLKGLCSLA---AEGI-WT-QK----FVFSHR-------------
--DLRRNGVSES-DLLMWVNM----SLLLRSG-----DCFAFIHVCVQEFCAAMFYLLK-
QPKEN-----------------P--NAAIGSVT-QLVEAGMTG-VQDRLSQTGLFI----
----FGLLNEKITKILEISFGFQ---------------L--PQE-IKQ-EIIQCLKSLSH
YG-----------------------------------PNKAEVNFKELFKGIFETQS-KI
FVEQVLDF---------------------F-------------------EEVAIHISNRE
EL---AIS--S----FCLQ-ECHN-LQT-FRLC----------------IEN-----VFS
-DAFEFIT-----------S-------------------D-H-------KI---------
LTFWRDLC-SVFSTSKNLQVLELDNCILDGI---------SWTILC--KALAQP------
GC--K-LQSLKVHF---------ASN-LGNDSDFFKAI----------------LHNPHL
NFLS-LHGT-SLSLI-AVRQLCEALKHPMCN----IEQL--------LL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------GKCDIT-----------------------------
----------------------------KDACEAIAS-----------------------
--------------------------------ALICNNKVKR---LS-LRENP-LMD---
-------EGVELLCDAL----KHPSHALETL-----------------------------
------------------------------------------------------------
------------------------------LLMHCGLTSISCDYISQV-LLCTSSLTFLN
L-GSNFLGDKGMK-SLCEGLKHP--NCN----I---------------------------
-----------------------QELWL--VGC-----YLTSDCCTDL------------
-------SSVL-ISNEKLKTLKLGNNEIQDDGVKQ-LCEALK-HP-KC-KLQNLG--LE-
--R-CHLTSACCE--------------------DLASALTSSRTL-RSLSLHCIPLDHDG
ALVLCE--------ALVHIDCP--LQ--T-------------------------------
---LGLNKS-SFDEETQML------LS----------AVE-EKNPQLIISHDPW------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMMUP00000033280 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---ESFFSDFG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 322)
-LLWYLKELR--K---EEFWKFKELLKQ------
---------P--LEKFE--------L--KPI-PWAE-LKKAS-KEDVAKLLDKHYPGKQA
WEVTLHL----FL-QIDRKDLWTKAQ-EEMRN----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------KLNPYRKH-
--------MKEK----------------FQLI--WEKETCLPVPEH--------------
---------------------------------F-------YKETMKNKYKELNGAYTAA
------ARL----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
HTVVLEGSDGIGKTTLLRKV-----MLDWA--E---------GNLWK
D--------RFTF-VFFLNVYEMNGVT-------ETSLLE-LLSR-DW-----PES-SET
IEDIFS-QP----EGILFIMDGFEH-LKFNLK-F------KA-D-----LSDDWRRQ-QP
-TSIILSSLLQKKMLPECSLLIA-LGKLAMQKHYFML-Q-----HPK--LIKLSGFTESE
KKSYI----------------S--YY-FGE---------KNKALKVFNFVRD-SGPLFIL
CHNPFICWLVCTCMKRRLERGE-DL-E--IS-SQNTTSLYASFLTNVFKAGS-QSFVPK-
-----------VNRARLKNLCALA---AEGV-WT-YT----FVFCHE-------------
--DLRRNGLSES-EGLMWVDM----RLLQRRG-----DCFTFIHLCIQEFCAAMFYLLK-
RPKDN-----------------P--NSAIGSIT-QLVKATVIQ-PQTHLTQVGIFM----
----FGISAEEIVSMLETSFGFP---------------L--SKD-LQQ-EITQSLKSLSQ
CE-----------------------------------ADREAIGFQELFVSLFETQE-KE
FVTQVMNF---------------------F-------------------EEVFIYIGNIE
HL---VIA--S----FCLK-HCRG-LTT-LRMC----------------MEN-----IFP
-DDSGCAS-----------G-------------------Y-N-------EK---------
LVYWRELC-SVFITNKDFQILDMENSSLDNA---------SLAILC--KALTQP------
AC--K-LRKLIFTS---------MLN-FGHDSELFKAV----------------LHNPHL
KLLS-LYGT-SLSHT-DVRHLCETLKHPMCK----IEEL--------IL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------GKCDIS-----------------------------
----------------------------SEDCEDVAS-----------------------
--------------------------------VLAC
PF13516 // ENSGT00900000140813_0_Domain_11 (2017, 2054)
NSTLKH---LS-LVENS-LRD---
-------EGMMLLCEAL----KHPHYGVFSL-----------------------------
------------------------------------------------------------
------------------------------QVMYCCLTCVSCTYHLPSSLLGSKSL--WS
L-VTGLISTHVCV-CVCVCVCLC---VC----V---------------------------
-----------------------SSPRL--MGC-----FLTSDSCKDI------------
-------AAVL-IC
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NEKLKTLKLGHNEIGDAGVRQ-LCAALK-HP-NC-KLEQLG--LQ-
--T-CPITRACCG--------------------DLAAALVACKTL-RSLNLDWITLEPDA
VAVLCE--------ALSHPDCA--LQ--M-------------------------------
---LGAPEI-VSPGDTVAPAENHCSIK----------TCSTDVGHKYATPAQTWE-----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMNEP00000026032 (view gene)
ENSPEMP00000014889 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-MDVA-GASCSS--S--EALAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLAALEDLS--Q---EQLKRFRHKLRD------
------------APLEG-----------RSI-PWGR-LENSD-PVDLVDKLTQFYGSEPA
VDVTRKI----LK-KADARDVAMRLK-EQQLQR---------L---------GPS-----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------S-AVLSVSEYKKKYREH-
--------VLLQ----------------HAKV--KERNARS------------VKINKRF
TKLLIAPGSGAVEDE--LLGRSE-------EPEP--------ERARRSDTHTFNRLFRGD
D---QGLRP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLQGPAGIGKTMAAKKI-----LYDWA--S---------GKLYH
G--------QVDF-AFFMPCGELLERPG------TRSLAD-LILD-QC-----PD-RKAP
VRQMLA-HP----QRLLFILDGADE-LPAVVAP--------EAA-----PCTDPFEA-TS
-GLRVLSGLLSQELLPLARLLVT-TRNVASGRLQGRL-C-----SPQ--CAEVRGFSDKD
KKKYF----------------F--KF-FQD---------ERKAERAYRFVKE-NETLFAL
CFVPFVCWIVCTVLQQQLELGR-DL-S--RT-SKTTTSVYLLFIASMLKSA-GANGP---
-----------RVQGELRTLCRLA---REGILKH--Q----AQFSEN-------------
--EIERLKLQGSQVQALFLSKKELLGVLEK-----E-VTYQFVDQSFQEFLAALSYLLE-
AEGAP-------------------G-APGGGVQ-MLLNSDLEL--RGYLALTTRFL----
----FGLLSAERIRDIERHFGC-----------------VVPPR-VKQ-DTLRWVQGQGH
PK--VAPVGADKKDELEDTEEAEE-------------EEEDLDFGLELLYCLYETQE-DA
FVRQALGS---------------------L-------------------PEMVLERIRFT
RM-DLEVL--S----YCVQ-CCPP-GQA-LRLV------------------NC----GLV
------EA----------------------------------------KQK--K---KKS
----------------------------------------LVKR----------------
------LTGS----------------------------------------------QNTT
KQ---TPVS-------LLRPLCEAMTAQQCG----LSIL--------ILS---------H
C-----------------------------------------------------------
--------KLP---DT-VCR---DLSEALKV-APALKELGLLQNRLT-EAGLRLLCEG--
-LAWPKCQVQTL------------------------------------------------
----------------------------RVQ-----------------------------
---------------LPSL---------QEAIQYL-------------------------
------------------------------VIVLQQSRALTT---LD-LSGC-QLLGYMV
--S--------CLCSAL----KYPTCCLKTLSLT--SVELSDV-----------------
----------SLRELQAVKTSKP-------DLT----------------------I--LH
SKLD-------------------TKPL--KG-----------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMMUP00000046788 (view gene)
MDQPEDPGSRLRSLRTASPQRAFVESVPHQRLRLRPWCKAPKLGLTK-RQPGHVTWPGVW
KNRSRKGGAGEEGAPQSSGRRWPVPTSPSEGEEHQD------APTRVRGRRR--------
-------AKSGVTPRCPLPPPHG-----SS--LARGSLRPCPRWVLKRQSRGWAGVTRGP
LPDDPRPPVSSTGLR--LAVAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLAALEELS--Q---EQLKRFRHKLRD------
------------VGPDR-----------RSI-PWGR-LERAD-AVDLAEQLAQFYGPEPA
LEVARKT----LK-KADARDVAAQLK-EQRLRR---------E---------FRA-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------GG-PSCLDREYKKKYREH-
--------VLQL----------------HARV--KERNARS------------VKITKRF
TKLLIAPESAAPEEE--ALGPAE-------EPEP--------GRARRSDTHTFNRLFRGD
E---DGRRP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYQ
G--------QVDF-AFFMPCGELLERPG------TRSLAD-LILD-QC-----PD-RGAP
VPQMLA-QP----QRLLFILDGADE-LPALEGP--------EAA-----PCTDPFEA-AS
-GARVLGGLLSKALLPTALLLVT-TRAASPGRLQGRL-C-----SPQ--CAEVRGFSDKD
KKKYF----------------Y--KF-FQD---------ERRAERAYRFVKE-NETLFAL
CFVPFVCWIVCTVLRQQLELGR-DL-S--RT-SKTTTSVYLLFIASVLSSAPVADGP---
-----------RVQGDLRNLCRLA---REGVLGR--R----AQFAEK-------------
--QLEQLELRGSKVQTLFLSKKELPGVLET-----E-VTYQFIDQSFQEFLAALSYLLE-
DEGVP-------------------R-TAAGGVG-TLLRGDAQP--HSHLVLTTRFL----
----FGLLSAERMRDIERHFGC-----------------VVSER-VKQ-EALRWVQGQGQ
GCPGVAPEVTEETKGLEDTEEPEE-----------EEEGEEPNYPLELLYCLYETQE-DA
FVRQALCR---------------------L-------------------PELALQGVRFC
RM-DVAVL--S----YCVR-CCPA-GQA-LRLI------------------SC----RLV
------AA----------------------------------------QEK--K---KKS
----------------------------------------LGKR----------------
------LQAS---L--------------GGS------S-----------------SRGTT
KQ---LPAS-------LLHPLFQTMTDPLCH----LTSL--------TLS---------H
C-----------------------------------------------------------
--------KVP---DA-VCR---DLSEALRA-APALTELGLLHNRLS-EAGLRMLSEG--
-LAWPQCRVQTV------------------------------------------------
----------------------------RVQ-----------------------------
---------------LPSS---------QQGLQYL-------------------------
------------------------------VGMLRQSPALTT---LD-LSGC-QLPAPMV
--T--------YLCAVL----QHQGCGLQTLRL--TSVELSEQ-----------------
----------SLQELQAVKRAKP-------DLV----------------------I--AH
PVLD-------------------GHPESP-KELVSTF-----------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPSIP00000017030 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------MENR-SGI
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 326)
SDL---LLSILEDLP--Q---EAFKRFKDKLSH------
---------T---DFEG--------K--GPI-RRGR-LENAD-WTDTKNLLMESYGGAAA
VDVTIDV----LT-QINLQESAAQLR-AGSEKVLS-----KEKKKT------AQVKEETR
G-----------------------------------------------N-----------
---------------------------------------NRQWNQGPYA-----------
ETHHAK-DLR-------------------------------IDNPSLKGSAYQRKYREH-
--------IQEE----------------YQVI--ENRKALLGEHV---------SLTKRY
TELLIVKEHRGREEKKHEII------TRGRRHAELMA----QQ----TSLTQINLLFDSD
GN--L-QNP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
RTVVLQGAAGIGKTVTARKI-----MLDWA--A---------GKLYW
K--------KFDY-VFYINCREINHILKNQSGAKNRSAAD-LVLN-NC-PD-----REGP
IKEIFE-KP----EKLLFIIDGFDE-LNSLLDID------DV-S-----LYMDPFEK-RP
-VETTLCSLLRKKVLQKSFLLIT-TRPTALEKLKENL-K-----LPR--FAEIMGFSADE
RKEYF----------------H--KF-FCD---------EKKATEAFNFVKE-NEMLFTM
CFVPIVCWIICTVLKQQMEKKE-DL-T--QT-SKTTTSVYLLFLSTLITDHITDT--IEQ
----------S--KTNLWALCSLA---ADGI-LE-RK----ILFEED-------------
--DLKKHNLSLSNIHSLFLSK----NIFQKDTE-YE-NVYSFIHLSFQEFFAALFYVLE-
EDEEIM--------------KNS--LSSKKDVK-SLLKNYGIS-TNNYLMLTVRFL----
----FGLSNRERLNDTEKKLHCK-----------------TSSG-IRD-DLLQWFEEETK
KIS-------------SLSY----------------KTYEELHDQLELFHCL--------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMUSP00000015866 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----MGP----PE-KESKAILKARGLEEEQKSERK-MTSPENDS-KSIQKDQGPEQEQTS
EST-MGPPEKDSKAILKARGLEEE------------QK-SESTM---SPSENVSRAIL--
KDSGSEEV---------EQASERKMTSPENDSK------SIQKDQGPEQ-------EQTS
ETLQSKEEDE-------------------------------VTEADKDNGGDLQDYKAH-
--------VIAK----------------FDTS--VDLH-Y--------------------
--------------------------------------------DSPEM-KLLSDAFKPY
Q---KTFQP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
HTIILHGRPGVGKSALARSI-----VLGWA--Q---------GKLFQ
K--------M-SF-VIFFSVREIKWTE-------KSSLAQ-LIAK-EC-----PD-SWDL
VTKIMS-QP----ERLLFVIDGLDD-MDSVLQHD------DM-T-----LSRDWKDE-QP
-IYILMYSLLRKALLPQSFLIIT-TRNTGLEKLKSMV-V-----SPL--YILVEGLSASR
RSQLV----------------L--EN-ISN---------ESDRIQVFHSLIE-NHQLFDQ
CQAPSVCSLVCEALQLQKKLGK-RC-T--LP-CQTLTGLYATLVFHQLTLKR-PSQSALS
----------QEEQITLVGLCMMA---AEGV-WT-MR----SVFYDD-------------
--DLKNYSLKES-EILALFHM----NILLQVGH-NSEQCYVFSHLSLQDFFAALYYVLE-
GLE-E--------------WNQH--FCFIENQR-SIMEVKRTD--DTRLLGMKRFL----
----FGLMNKDILKTLEVLFEYP---------------V--IP-TVEQ-KLQHWVSLIAQ
QVN----------------------------------GTS-PMDTLDAFYCLFESQD-EE
FVGGALKR---------------------F-------------------QEVWLLINQKM
DL---KVS--S----YCLK-HCQN-LKA-IRVD----------------IRD-----LLS
-VDNTLEL----CP--------------------VV----------TVQ--ETQCK-PLL
MEWWGNFC-SVLGSLRNLKELDLGDSILSQR---------AMKILC--LELRN-Q-----
SC--R-IQKLT--F-KSAEV---VS---GLK-HLWKLL----------------FSNQNL
KYLN-LGNT-PMKDD-DMKLACEALKHPKCS----VETL--------RLD---------S
C------E----------------------------------------------------
---------LT---II-GYE---MISTLLIS-TTRLKCLSLAKNRVGV-KSMISLGNA--
LSS-SMCLLQKL-I----------------------------------------------
------------------------LDNCGLT-----------------------------
----------------------------PASCHLLVS-----------------------
--------------------------------ALF
PF13516 // ENSGT00900000140813_0_Domain_11 (2016, 2054)
SNQNLTH---LC-LSNNS-LGT---
-------EGVQQLCQFL----RNPEC----------------------------------
--------------------------ALQ-RLI---------------------------
-------------------------------LNHCNIVDDAYGFLAMR-LA
PF13516 // ENSGT00900000140813_0_Domain_13 (2212, 2237)
NNTKLTHLS
L-TMNPVGDGAMK-LLCEALKEP--TCY----L---------------------------
-----------------------QELEL--VDC-----QLTQNCCEDL------------
-------ACMI-TTT
PF13516 // ENSGT00900000140813_0_Domain_15 (2356, 2378)
KHLKSLDLGNNALGDKGVIT-LCEGLK-QS-SS-SLRRLG--LG-
--A-CKLTSNCCE--------------------ALSLAISC
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2465)
NPHL-NSLNLVKNDFSTSG
MLKLCS--------AFQCPVSN--LG--I-------------------------------
---IGLWKQ-EYYARVRRQ------LE----------EVE-FVKPHVVIDGDWYASD---
--E---------------------------------------------------------
--------------------------------------------------DDRNWWKN--
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMUSP00000113095 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------M---ASLFSDFG---F
PF02758 // ENSGT00900000140813_0_Domain_0 (209, 319)
IWYWKELN--K---IEFMYFKELLIH------
---------E--ILQMG--------L--KQI-SWTE-VKEAS-REDLAILLVKHCDGNQA
WDTTFRV----FQ-MIGRNVITNRAT-GEI------------AA--------------HS
T---------------------------------------------------IYRA----
------------------------------------------------------------
----------------------------------------------------------H-
--------LKEK----------------LTHD--CSRKFNISIQNF--------------
---------------------------------F------------QDEYDHLENLLVPN
GT--ENN-P----K
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
MVVLQGVAGIGKTILLKNL-----MIVWS--E---------GLVFQ
N--------KFSY-IFYFCCHDVKQLQ-------TASLAD-LISR-EW-----PSP-SAP
MEEILS-QP----EKLLFIIDSLEG-MEWNVTQQ------DS-Q-----LCYNCMEK-QP
-VNVLLSSLLRKKILPESSLLIS-TSCETFKDLKDWI-E-----YTN--VRTITGFKENN
INMCF----------------H--SL-FQD---------RNIAQEAFSLIRE-NEQLFTV
CQAPVVCYMVATCLKNEIESGK-DP-V--SI-CRRTTSLYTTHILNLFIPHN-AQNPSN-
-----------NSEDLLDNLCFLA---VEGM-WT-DI----SVFNEE-------------
--ALRRNGIMDS-DIPTLLDI----GILEQSRE-SE-NSYIFLHPSVQEFCAAMFYLLH-
SEMDH-----------------S--CQGVYFIE-TFLFTFLNK-IKKQWVFLGCFF----
----FGLLHETEQEKLEAFFGYH---------------L--SKE-LRR-QLFLWLELLLD
TLH----------------------------------PDVKKINTMKFFYCLFEMEE-EV
FVQSAMNC---------------------R-------------------EQIDVVVKGYS
DF---IVA--A----YCLS-HGSA-LTD-FSIS----------------AQN-----VLN
-EEL--GQ-----------R-------------------------------------GKL
LILWHQIC-SVFLRNKDIKTLRIEDTIFNEP---------VFKIFY--SYLKNS------
SC--I-LKTLVAYN---------VSF-LCDKRLFLEL-----------------IQSYNL
EELY-LRGT-FLSHS-DVEMLCDILNQAECN----IRI----------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------L-----------------------------
------------------------------------------------------------
------------------------------DLANCSLCEHSWDYLSDV-LR
PF13516 // ENSGT00900000140813_0_Domain_13 (2212, 2237)
QNKSLRYLN
I-SYNNLKDEGLK-ALCRALTLP--NSA----L---------------------------
-----------------------HS-----------------------------------
-----------------------------------------------------LS--LE-
--A-CQLTGACCK--------------------DLASTFTRYKCL-RRINLAKNSLGFSG
LFVLCK--------AMKDQTCT--LY--E-------------------------------
---LKLRMA-DFDSDSQEF------LL----------SEM-ERNKILSIENG-V------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMPUP00000011174 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MASG----V-
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 326)
QWR---LARYLDMLG--K---EELKEFQLRLPE------
---------K--PL-GG-------PR--PHL-TSAC-GQQPG-ALEAASQLVAQHGEQQA
WDLALHT----WE-QMGLSQLCAQAR-AEAALMSGEG----SSLPC---SPGAPSMQSPS
GPTSTVVLSFVQDPPT-----TS---------SKQPPGTSRCFL-QNRYPSFSFN-RV--
---------LPCSSN-QKSLLQESPSAPTSTTVLEDWVPPPQFSRKPRKQEAPRAEWLLV
EMSGEHSK------------------------------------------GIRERRQHQ-
--------KRKR----------------PRPTKSR-------GKE---------GLHEEF
IELLLLHQPRGGLLES---Q------SKGSWHHSG--------AEEQGRLIEVKDLFGPG
LG--AQEEG----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
RAVVLHGVAGVGKSVMARQV-----RRAWE--E---------GRLYR
D--------RFQH-VFYFSCRQLAQSK-------TMSLKE-LITK-DW-----AA-PTAP
SEQILS-QP----GQLLLILDGLDE-LKWVFVKE------KS-E-----VCPYWDQP-QP
-VKTLLLGLLRKTILPGASLLIT-ARTTALHLFILSV-E-----QPR--WVKVLGFSESS
RKEYF----------------Y--KY-FKS---------KNKANRAFHSVEL-NQPLLTM
CLVPLVSWLVCTCMKKQMERGE-SL-G--LP-SQTATALCLHYLAQALP--------AQ-
-----------PLGTQLRGLCSLA---AKGI-RQ-RE----TLLIWE-------------
--DLRKHELED--TVSALLEM----GILRKH---PSSPDYDFAHQYLQEFFAAVSCVLG-
S------EDK--------ECHPP--DSI-QGLE-DLLKAYGGL--DPFGVPPTCFV----
----LGLLSEPVQREMEAVFQGK---------------V--PAE-RKR-KLLCWAKEAVR
-----------------------------------NQLPVLQQSSLPMLHCLYEIQD-KD
FLTHAMAP---------------------F-------------------HGARVRVHTSL
EL---LVF--M----FCIK-FCT--VKE-LRLN----------------ERGQ-------
------H-G---------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------H------------
------TGSP--------------------------------------------------
-------------------LHVALLRGFPVT-----------------------------
----------------------------DAWWEVLFF-----------------------
--------------------------------TLRTTGKLKE---LD-LSGNC-LSN---
-------TVVQSLCKAL----RFPSCCLETLR----------------------------
------------------------------------------------------------
-------------------------------LTHCSLTAEGCIGLAWV-LDTPETLTELE
L-SFNLLLDLGVE-RLCNGLKEP--GCR----L---------------------------
-----------------------HRLRL--VGC-----GLTS------------------
------------------------------------------------------------
---------RCCR--------------------ALASALSASPRL-TELDLQQNELEDAG
VRLLCE--------GLRRPTCQ--LR--L-------------------------------
---LWLDQT-GLSEEVTRM------LR----------ALE-DEKPPLLIPS-RWKPSETM
PNEGPRETVPNEDPRETMPNEGPHEGETSGTSSLQQDRRESEG-----------------
------------------LSPPVVQ-EGLVGLLSPAPPEDLQHKGPLG-TEDDFWGPLGP
VTIEMVDK-DRSLHRVHLPMAGSYRWP---STGLRFVVREPA-------TIDIEFCASDK
FLHQMAWEQSWMVVGPLFDIKVKPGA-VATVYLPHFVDLRGG--------CVDTSLFQVA
HFKEPGMLLEKPTRVELYHVVLEDPSFSPLGVFLRTIHAALRFLPITSMVLLYHHTRPDE
T-VFHLYLIPSDCCVRKAIDDEEKAFQFVQIRKPPPLGSLYLGSRYTVSSS---------
ETLEAIPKELELCYRSPGQPQLYSEFYVGDLGSGLRLQLKDG-SGAVVWEALVKPGDLQL
TSSLVPP-----------------------------APAASPAPPGAPASL
PF00619 // ENSGT00900000140813_0_Domain_19 (3052, 3133)
HFLDRHREQ
LVARVTTVDPVLDKLL-GQVLSEEQYERVRAKATAANQMRELFRFSRSWDPACKDQVYQA
LKETHPYLIMELWETWGGDRERGTSPPTQPLP----
ENSRBIP00000037431 (view gene)
ENSNLEP00000012932 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---ESFFSDFG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 322)
-LLWYLKELR--K---EEFWKFKELLKQ------
---------P--LEKFE--------L--KPI-PWAE-LKKAS-KEDVAKLLDKHYPGKQA
WEVTLNL----FL-QINRKDLWTKAQ-EEMRN----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------KLNPYRKH-
--------MKEK----------------FQLI--WEKETCLHVPVH--------------
---------------------------------F-------YKETMKNEYKELNDAYTAA
------ARR----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 929)
RTVVLEGPDGIGKTTLLRKV-----MLDWA--E---------GNLWK
D--------RFTF-VFFLNVYEMNGIA-------ETSLLE-LLSR-DW-----PEP-SEK
IEDIFS-QP----ERILFIMDGFEQ-LKFNLQ-L------KA-D-----LSNDWRQR-QP
-MPIILSSLLQKKMLPESSLLIA-LGKLAMQKHYFLL-Q-----HPK--LIKLLGFTESE
KKSYF----------------S--YF-FGE---------KSKALKVINFVRD-SGPLFIL
CHNPFICWLVCTCVKQRLERGE-DL-E--IN-SQNTTSLYASFLTTVFKAGS-QSFPPK-
-----------VNRARLRSLCALA---AEGI-WT-YT----FVFSHG-------------
--DLRRNGLSES-EGLMWVGM----RLLQRRG-----DCFAFTHLCLQEFCAAVFYLLK-
RPKDD-----------------P--NPAIGSVT-QLVRASVVQ-PQTLLTQVGIFM----
----FGISAEDIVNMLETSFGFP---------------L--SKD-LKR-EITQCLEGVSQ
CE-----------------------------------ADREAIGFQELFTGLFETQE-KE
FVTKVMNF---------------------F-------------------EEVFIYIGSIE
HL---VIA--S----FCLK-HCQH-LTT-LRMC----------------MEN-----IFP
-DDSGCIS-----------D-------------------Y-N-------EK---------
LVYWRELC-LVFITNKNFQMLDMENTSLDDA---------SLAILC--KALAQP------
VC--K-LQKLIFTS---------VSN-FGHDSELFKAV----------------LHNPHL
KLLS-LYGT-SLSQS-DVRHLCETLKHPMY------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------NGVA-SLCGALKHP--GCS----I---------------------------
-----------------------RELWL--MGC-----FLTSDSCKDI------------
-------AAVL-ICNEKLKTLKLGHNEIGDTGVRQ-LCAALK-HP-NC-KLECLG--LQ-
--T-CPITHVCCG--------------------DLATALIACKTL-RSLNLDWITLDADA
VVVLCE--------ALSHPDCA--LQ--M-------------------------------
---LGLHKS-VFDEEAQKI------LM----------SVEEK-IPHLTISHGPWIDE---
--EYK-----IR------------------------------------------------
--------------------------------------------------GVLL------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
MGP_SPRETEiJ_P0086520 (view gene)
------------------------------------------------------------
------------------------------------------------------------
-------------MYCP-----------------GAPCGDCAMCDPGCFGGSGTGPIQ--
VASVM-ITLLI-PSV--DAVAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LMATLEELS--Q---EQLKRFRHKLRD------
------------APLDG-----------RSI-PWGR-LERSD-AVDLVDKLIEFYEPVPA
VEMTRQV----LK-RSDIRDVASRLK-QQQLQK---------L---------GPT-----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------S-VLLSVSAFKKKYREH-
--------VLRQ----------------HAKV--KERNARS------------VKINKRF
TKLLIAPGTGAVEDE--LLGPLG-------EPEP--------ERARRSDTHTFNRLFRGN
DE--ESSQP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYH
S--------QVDF-AFFMPCGELLERPG------KRSLAD-LVLD-QC-----PD-RAWP
VKRILA-QP----NRLLFILDGADE-LPTLPSS--------EAT-----PCKDPLEA-TS
-GLRVLSGLLSQELLPGARLLVT-TRNAATGRLQGRL-C-----SPQ--CAEIRGFSDKD
KKKYF----------------F--KF-FRD---------ERKAERAYRFVKE-NETLFAL
CFVPFVCWIVCTVLQQQLELGR-DL-S--RT-SKTTTSVYLLFITSMLKSA-GTNGP---
-----------RVQGELRTLCRLA---REGILDH-HK----AQFSEE-------------
--DLEKLKLRGSQVQTIFLNKKEIPGVLKT-----E-VTYQFIDQSFQEFLAALSYLLE-
AERTP-------------------G-TPAGGVQ-KLLNSNAEL--RGHLALTTRFL----
----FGLLNTEGLRDIGNHFGC-----------------VVPDH-VKK-DTLQWVQGQNH
PK--GPPVGAKKNAELEDIEEAEEEEEE-----EEEEEEEDLNFGLELLYCLYETQE-ED
FVRQALSS---------------------L-------------------PEIVLERVRLT
RM-DLEVV--N----YCVQ-CCPD-GQA-LRLV------------------SC----GLV
------AA----------------------------------------KEKKKK---KKS
----------------------------------------LVKR----------------
------LKGS----------------------------------------------QSTK
KQ---PPVS-------LLRPLCETMTSPKCH----LSVL--------ILS---------H
C-----------------------------------------------------------
--------KLP---DA-VCR---DLSEALKV-APALRELGLLQSRLT-NTGLRLLCEG--
-LAWPKCQVKTL------------------------------------------------
----------------------------RMQ-----------------------------
---------------LPDL---------QEVINYL-------------------------
------------------------------VIVLQQSPVLTT---LD-LSGC-QLPGVIV
--E--------PLCAAL----KHPKCCLKTLSLT--SVELSEN-----------------
----------SLRDLQAVKTSKP-------DLS----------------------I--IY
SK----------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSTSYP00000030146 (view gene)
ENSMFAP00000014158 (view gene)
ENSGGOP00000026379 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---ESFFSDFG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 322)
-LLWYLKELR--K---EEFWKLKELLKQ------
---------P--LEKFE--------L--KPI-PWAE-LKKAS-KEDVAKLLDKHYPGKQA
WEVTLNL----FL-QINRKDLWTKAQ-EEMRN----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------KLNPYRKH-
--------MKEK----------------FQLI--WKKETCLHVPEH--------------
---------------------------------F-------YKETMKNEYKELNDAYTAA
------ARR----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 929)
HTVVLEGPDGIGKTTLLRKV-----MLDWA--E---------GNLWK
D--------RFTF-VFFLNVYEMNGIA-------ETSLLE-LLSR-DW-----PEP-SEK
IEDIFS-QP----ERILFIMDGFEQ-LKFNLQ-L------KA-D-----LSDDWRQR-QP
-MPIILSSLLQKKMLPESSLLVA-LGKLAMQKHYFML-R-----HPK--LIKLLGFSESE
KKSYF----------------S--YF-FGE---------KSKALKVFNFVRD-NGPLFIL
CHNPFICWLVCTCVKPRLERGE-DL-E--IN-SQNTTYLYASFLTTVFKAGS-QSFPPK-
-----------VNRARLKSLCALA---AEGI-WT-YT----YVFSHG-------------
--DLRRNGLSES-EGLMWVGM----RLLQRRG-----DCFAFMHLCIQEFCAAMFYLLK-
RPKDD-----------------P--NPAIGSIT-QLVRASVVQ-PQTLLTQVGIFM----
----FGISTEEIVSMLETSFGFP---------------L--SKD-LKQ-EITQCLESLSQ
CE-----------------------------------ADREAIAFQELFIGLFETQE-KE
FVTKVMNF---------------------F-------------------EEVFIYIGNIE
HL---VIA--S----FCLK-HCQH-LTT-LRMC----------------VEN-----IFP
-DDSGCIS-----------D-------------------Y-N-------EK---------
LVYWRELC-SMFITNKNFQILDMENTSLDDP---------SLAILC--KALAQP------
VC--K-LRKLIFTS---------V-Y-FGHDSELFKAV----------------LHNPHL
KLLS-LYGT-SLSQS-DIRHLCETLKHPMCK----IEEL--------IL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------GKCDIS-----------------------------
----------------------------SEACEDIAS-----------------------
--------------------------------VLAC
PF13516 // ENSGT00900000140813_0_Domain_11 (2017, 2054)
NSKLKH---LS-LVENP-LRD---
-------EGMTLLCEAL----KHPHCALERL-----------------------------
------------------------------------------------------------
------------------------------M-----------------------------
------------------------------------------------------------
---------------------------L--MGC-----FITSDSCKDI------------
-------AAVL-IC
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NEKLKTLKLGHNEIGDTGVRQ-LCAALQ-HP-HC-KLECLG--LQ-
--T-CPITRACCG--------------------DLAAALITCKTL-RSLNLDWIALDADA
VVVLCE--------ALSHPDCA--LQ--M-------------------------------
---LGLHKS-GFDEETQKI------LM----------SVEEK-IPHLTISHGPWIDE---
--EYK-----IR------------------------------------------------
--------------------------------------------------GVLL------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSTSYP00000012813 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------MAPT-QNPREA---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWALSDLE--E---KDFKILKFHLRN------
---------M--TPPDG--------Q--PHL-ARGE-LEGLS-QVDLAHLLILKYGEQDA
VRVVLKG----LK-IMNLLELVDQLS-HIS------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LH--DYREIYREH-
--------MRC-----------------LE------EL---QEG----------GVNGCY
NQLLLVA-RPSSEDPESPAC------PI-------PE-----Q-ELDSV--MVETLFGSR
EK--PCPAP----S
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
LVVLQGSAGTGKTTLVRKM-----VLDWA--T---------GTLYP
G--------HFDY-VFYVSCKEVVLLPE-------STLDQ-LLFW-CC-GD-----SQAP
VREIVR-QP----ERLLFILDGFDE-LQRPYEEN------WK-K-----RG-----S-NP
-KDDLLYLLIRRHILSTCSLLIT-TRPLALQNLKPLL-D-----KAC--HIHVLGFSEEE
RWRYI----------------N--AY-FTN---------EEQARDAFAIVQK-NAFLYKA
CQVPGICWVVCSWLKGQMERGK-EV-L--ET-PRTSTDIFMAYVSTFLPPND-DAGCSQL
----------S-RHRVLQSLCSLA---AEGI-QH-QR----FLFEEA-------------
--ELRKHNLDGSKLA-AFLCS----NDYQKGLD-IK-KFYSFRHISFQEFFHAMFYLVK-
EDQRQL----------------R--EESCREVK-RLLEVTEQK-EDEEMTLSLHFL----
----LDVLKKESSTNLELQFCFK-----------------ISPC-IMQ-DL-KHLKAQV-
E-------------------------------------SMKHNRTWDLEFSLYESKK-KN
MVRSVQMS---------------------S-------------------ALFKVKHSN-E
RK-SHSRN--G----FHVK-TSLS-HGQ-KEEQ------------------KC----LFV
--------CK---RNI--A-------------------GTPKE-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------ASN----------------GKG---------G
-GTEE-LEDDVGS-SGM-GS--------------RK---WDNSTD---G-SKE-------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSP00000327763 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MAMAKA-RKPREA---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWALSDLE--E---NDFKKLKFYLRD------
---------M--TLSEG--------Q--PPL-ARGE-LEGLI-PVDLAELLISKYGEKEA
VKVVLKG----LK-VMNLLELVDQLS-HIC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LH--DYREVYREH-
--------VRC-----------------LE------EW---QEA----------GVNGRY
NQVLLVA-KPSSESPESLAC------PF-------PE-----Q-ELESV--TVEALFDSG
EK--PSLAP----S
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
LVVLQGSAGTGKTTLARKM-----VLDWA--T---------GTLYP
G--------RFDY-VFYVSCKEVVLLLE-------SKLEQ-LLFW-CC-GD-----NQAP
VTEILR-QP----ERLLFILDGFDE-LQRPFEEK------LK-K-----RG-----L-SP
-KESLLHLLIRRHTLPTCSLLIT-TRPLALRNLEPLL-K-----QAR--HVHILGFSEEE
RARYF----------------S--SY-FTD---------EKQADRAFDIVQK-NDILYKA
CQVPGICWVVCSWLQGQMERGK-VV-L--ET-PRNSTDIFMAYVSTFLPPDD-DGGCSEL
----------S-RHRVLRSLCSLA---AEGI-QH-QR----FLFEEA-------------
--ELRKHNLDGPRLA-AFLSS----NDYQLGLA-IK-KFYSFRHISFQDFFHAMSYLVK-
EDQSRL----------------G--KESRREVQ-RLLEVKEQE-GNDEMTLTMQFL----
----LDISKKDSFSNLELKFCFR-----------------ISPC-LAQ-DL-KHFKEQM-
E-------------------------------------SMKHNRTWDLEFSLYEAKI-KN
LVKGIQMN---------------------N-------------------VSFKIKHSN-E
KK-SQSQN--L----FSVK-SSLS-HGP-KEEQ------------------KC----PSV
---HGQKEGK---DNI--A-------------------GTQKE-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------AST----------------GKG---------R
-GTEE-T--------PK-NTYI--------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSAMEP00000003541 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------PPAPSTEFR--AAA
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
AREL---LRAALEDLS--Q---EQLKRFRHKLRD------
------------APLDG-----------RSI-PWGR-LERAD-AMDLAEQLTQFYGPEPA
LDVTRKT----LK-RADVRDVAARLK-EQRLQR---------L---------GPS-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------SS-ALLSVSEYKKKYREH-
--------VLQQ----------------HAKV--KERNARS------------VKINKRF
TKLLIARESATLVDE--VPGLAE-------EQEP--------ERARRSDTHTFNRLFGRD
G---EGQRP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
LTVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYH
D--------QVDF-AFFLPCRELLERPG------TCSLAG-LILD-QC-----PH-RSAP
VRQMLA-RS----ERLLFILDGADE-LPPPGPA--------EAA-----PCTDPFEA-AD
-GARVLGGLMSKTLLPHARLLVT-ARATAPGRLQSRL-C-----SPQ--CAEVRGFSDKD
KKKYF----------------H--KF-FRD---------ERMAERAYRFVKE-NETLFAL
CFVPFVCWIVCTVLRQQLELGQ-DL-S--RT-SKTTTSVYLLFIASVLSSAPAADGP---
-----------QMQGELRKLCRLA---REGILRH--R----AQFGEK-------------
--DLERLALRGSKVQTLFLSKKELPGVLET-----E-VTYQFMDQSFQEFFAALSYLLE-
DERDL-------------------R-APADSVG-ALLRGDTEL--RGHLTLTTRFL----
----FGLLNTERMRDVERHFG-----------------GVASER-VKQ-EVLRWVQEQGQ
GRPRAAAEGTEETEEP-------E------------EEEGELNYPLELLYCLYETQE-GA
FVRQALRG---------------------L-------------------PELELERVRFS
RM-DLVVL--S----YCVR-CCPA-GQA-LRLV------------------SC----GLV
------PA----------------------------------------QEKK-----KKG
----------------------------------------LLKR----------------
------LHGS---L--------------SGS------S----------------SSQATM
RK---PQAS-------PLRSLFEAMTDQHCG----LNSL--------TLS---------R
C-----------------------------------------------------------
--------KLP---DS-VCQ---DLSEALRE-APALRELSLLHNRLS-EAGLRVLSEG--
-LAWPQCRVQKL------------------------------------------------
----------------------------WVQ-----------------------------
---------------QPGL---------QEALRYL-------------------------
------------------------------ISVLRQSPALTT---LD-LSDC-HLSESMV
--T--------YLCAVL----QQPGCRLQTLSL--TSMALSEQ-----------------
----------SLKELQTVRTAKP-------GLV----------------------I--SH
PALD-------------------SHPSPPTGFSSAL------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCGRP00001022670 (view gene)
ENSPVAP00000008200 (view gene)
------------------------------------------------------------
------------------------------------------------------------
-------VVLPTGPICSLLPKNL------------L-------------------IHP-H
L---SLQPCIKMEEANLPSF
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
SSYG---LWWCFKQLD--E---EEFQAFKDLLKE------
---------N--ASELA--------M--CSF-PWVE-VDKAS-VEHLAYLLHEHCRIPLV
WKISTTI----FE-QMNLPMLSQKAR-DEMEKCFM-VKIPEDSTPMKI--DHGTSMEEVP
EM---------------------------------------------------SQAME--
QD-GTTAA---------ETKDQXXXXXXXXXXXXXXXXXXXXXXXXXXX-----------
---------------------------------------------XXXXXXXXXXXXXX-
--------XXXX----------------XXXX--XXXX-XX-------------------
--------------------------------------------XXXXX-XXXXXXXXXX
X---XXXXX----XXXXXXXXXXXXXXXXXXXX-----XXXXX--X---------XXXXX
X--------X-XX-XXXXXXXXXX-XX-------XX--XX--X-X-X------XX-XXXX
XXXXX--XX----XX-XXXXXXXXX-XXXXXXX--------X-X-----XXXXXXXX-XX
-XXXXX----XXXX-XX-XXXXX-XXXXXXX---XXX-X-----XXX--XXXXXXX----
XXXXX----------------X--XX-XXX---------XXXXXXXXXXXXX-XXXXXXX
XXXXXXXXXXXXXLQIQEASGK-SL-P--CT-CQVLTGLYVTFVVHQLTPQD-AFGRCLS
----------PEERVVLKGLCQMA---TEGV-WG-TK----FVFYND-------------
--DLGVHGLTDS-KLSTLFHM----NILHQDGH-NDSC-YTFLHVSLQEFCAALYYILE-
GLESD--------------WDPH--PLLTENIR-TLMELKQIG-INVHLLQMKRFL----
----FGLMNKEVTRALGDLLGCP---------------V--PL-VVKR-VLLHWVCLLGQ
RAN----------------------------------TA-APQDFLDSFYCLFETQD-EE
FVRLALNG---------------------F-------------------QEVWLSVNRQM
DL---LVS--S----YCLQ-HCHC-LRR-IRMD----------------VQE-----IFS
-EDEPTEA----WP--------------------VI-----------PQ--GMQFK-VLV
DNWWENLC-FMLGTHPSLQELDLSGSILNEW---------AMKTLC--IKLRQ-P-----
TC--K-IQNLI--F-KSVQI---TF---GLH-YLWNTI----------------VTNCNV
KYLS-LEGT-HLKDE-DIGIAYEALRHPRCL----LQSL--------XXX---------X
X------X----------------------------------------------------
---------XX---XX-XXX---XXXXXXXX-XXXXXXXXXXXXXXXX-XXXXXXXXX--
XXX-XXXXXXXX-X----------------------------------------------
------------------------LGNCDLS-----------------------------
----------------------------AASCQDLAM-----------------------
--------------------------------ALIGNQSLTH---LY-LSTNS-LGS---
-------EGVNLLCRAL----KLFSC----------------------------------
--------------------------ALQ-RLI---------------------------
-------------------------------LNECNVDVAGCGFLALA-LTSNQHLTHLS
L-SMNPLGDNGMN-LLCEVMAEP--SCH----L---------------------------
-----------------------QDLEL--VQC-----HLTACCN--V------------
-------SYMI-TS-KH-----SDLLNLGDSGMAE-LCEGLK-QR-KS-SLRRLG--LE-
--A-CDLTSDCCE--------------------GLSLALSCNQCL-ISLNLMRNNFSPEG
MMKLCP--------AFAHPRSN--LQ--I-------------------------------
---VGLWKW-QYPVQIRKL------LE----------HVQ-LIKPHIVIDDGWYSLD---
--E---------------------------------------------------------
--------------------------------------------------DDRYWWKN--
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMOCP00000012739 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------MA------ESSGLG---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
VALCLKQLS--D---ATFLHFKNLLIK------
---------E--PE-LQ--------V--NPV-LCAN-IKLAS-RKGLITLLHAYYP-GQV
WDIMSRT----FL-QVNRRDLWTKVQ-EL-RRDK--------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------------RTYKEI-
--------MKTA----------------FHQI-------WTFKDN---------------
----------------LYVL------D-----AEYEV-------VTHMHFKVLQEVFDP-
-----QLKP----VT
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 928)
AVILGAEAIGKSTFLRKA-----VLDWA--S---------GNLWK
N--------RFQY-VFFFSLISLNSTT-------ELSLAQ-LLLN-QL-----SA-SSET
PDDVLS-DP----RKILFILDGFDH-LKFDLDIR-------T-N-----LCDDWRKM-LP
-TQIVLSSLIQKIMLPESSLLLE-LGNPSIPKIYPLL-Q-----YPR--EITMGGFSEQS
IKFYC----------------V--YF-FND---------FEKGLEIFGYLQS-MQALLDL
CISPYMCWMFCRTLKGQRDRGE-KM-N--LV-YEPDSALYTSFMVSSFRSVY-ESRRSRQ
------------NRARLKTLCTLA---AEGM-WK-QV----FVFKSE-------------
--DLRRNGIT-EPEQTSWLRI----NFLNYR----G-DCFMFYHPTLQSYFAALFYFLR-
EDKDT-----------------P--HPIIGSLL-QFLREVYAH-GQTQWLRTGMFV----
----FGIATEKVAAMLNPHFGFI---------------P--SRD-VRG-VIFNCFRN---
-----------------MNQ----------------EEFSKTLRAQSLFKSLFGNKE-EE
FVTQVMGL---------------------F-------------------EEMTVDISNVD
VL---TVA--T----YSL-LRSQK-LKK-LHLH----------------IQ---------
----ERVF----TE--------------------------TYNPRDGDIETFRRHK-KIA
NKYWRSLC-GIFW---HLQVLDLDSCNFNET---------AIQHLY--ELLSPAP-----
PI--T-LNM---------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------FKLQSL-S----------------------------------------------
------------------------LGKCGIS-----------------------------
----------------------------SNACVEIVT-----------------------
--------------------------------SLN-LGKLKH---LS-LVENP-LNN---
-------KGVVALCEIL----KDPSC----------------------------------
--------------------------VLE-KL----------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSRBIP00000003387 (view gene)
ENSNGAP00000001161 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---SSFFSDFG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 323)
-LMWYLEELN--K---KEFMKFKELLKQ------
---------E--ILHLG--------L--KHI-PRME-VKKAS-REDLANLLMKYYEEKQA
WNVTFEI----FQ-KINRLDLCERAA-REI------------AG--------------HP
K---------------------------------------------------IYQA----
------------------------------------------------------------
----------------------------------------------------------H-
--------IKTK----------------FIGI--WSRKFTPMVHDF--------------
---------------------------------F-------DQEITQEEHDYFEYFFPSK
TT--GNR-S----H
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
ILLLKGVPGIGKTMMLTKL-----MLAWA--E---------GKIYQ
N--------KFTY-VFYFCCQEVKPLT--------TSLSE-LISR-EW-----PNP-SAP
IAEIMS-QP----EKLLFIIDSFEW-LRCDLTEP------ES-E-----LCTDWIEE-RP
-MKTLLSSLFRRKILPESSLLIA-GTPESLQTLENKF-E-----NPE--IRTIIGFDESN
RKLFF----------------R--CL-FPD---------KYRAQEAYGFVRE-NEQLFTI
CQIPLLCWVVAMCLKQEMEKGN-DL-T--PI-CRSTTSLYVAYILNLFLPRG-TCYPSK-
-----------KSQEQLQGLCSLA---AEGL-WT-DT----FVFSEE-------------
--ALRRNGLVDS-DVPVLLNI----KILVKSRE-SE-NSYTFLHPSVQEFCAAVFYLLK-
RHVDH-----------------P--STDVKCID-ILLIMFLKK-AQARWIFVGCFI----
----FGLLHEIEQEKLRVFFGCQ---------------I--SKE-RKQ-QLHQCLETLSK
SEE-----------------------------------LWEQVDCLKLFYCLFELRN-EA
FVIQAMDL---------------------I-------------------QEVKFEIKDNS
DL---IVS--A----YCLK-HCTA-LKK-LSIS----------------VRD-----ILR
-GDQ--AP-----------K-----------------------------------SMANR
LICWHQIC-SVLTTNENLQVLQVKNSGLNES---------AFMILY--NQLRHP------
NC--P-LQVLEINN---------VSF-FCDNHLFFEVF----------------LQNPNL
KHLN-LSLT-VLSHS-DVKLLCDVLNHPESN----IEKL--------LL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------TDCNLS-----------------------------
----------------------------SSDFEIFAP-----------------------
--------------------------------VLISNKTLKH---LN-IGSNH-LD----
-------KGIQSLCQSL----CSQNCILKFL-----------------------------
------------------------------------------------------------
------------------------------VLANCHLSDNSWGYIREV-LLTNKTLSHLD
L-SSNSLKDEGLK-ILCEALSLP--ESG----L---------------------------
-----------------------KSLCL--RNC-----LITASGCQDL------------
-------AAVL-RSNGNLRSLQISENKVKDAGVKL-LCDAVK-HP-NC-HLENLG--LE-
--A-CELTGACCE--------------------DLASAFTQSETL-WGLNLLENTLDSSG
VIVLCE--------ALKQPKCN--LH--V-------------------------------
---LGLRVT-DFDEETQAF------LI----------AEE-ERNPYLTIISN-V------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSP00000466285 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------MAESDSTDFD---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWYLENLS--D---KEFQSFKKYLAR------
---------K--ILDFK--------L--PQF-PLIQ-M---T-KEELANVLPISYEGQYI
WNMLFSI----FS-MMRKEDLCRKI---IGRRN---------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------RNQEACKAV-
--------MRRK----------------FMLQ--WESHTFGKFHYKFFRDVS--------
----------------------------------------------SDVFYILQLAYDST
SY--YSANN----LN
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 930)
VFLMGERASGKTIVINLA-----VLRWI--K---------GEMWQ
N--------MISY-VVHLTAHEINQMT-------NSSLAE-LIAK-DW-----PD-GQAP
IADILS-DP----KKLLFILEDLDN-IRFELNVN------ES-A-----LCSNSTQK-VP
-IPVLLVSLLKRKMAPGCWFLIS-SRPTRGNNVKTFL-K-----EVD--CCTTLQLSNGK
REIYF----------------N--SF-FKD---------RQRASAALQLVHE-DEILVGL
CRVAILCWITCTVLKRQMDKG-RDF-Q--LC-CQTPTDLHAHFLADALTSEAGLT-AN--
----------QYHLGLLKRLCLLA---AGGL-FL-ST----LNFSGE-------------
--DLRCVGFTEA-DVSVLQAA----NILLPSNT-HK-DRYKFIHLNVQEFCTAIAFLMAV
PNYLI-----------------P----SGSR-E--Y--KEKRE-QYSDFNQVFTFI----
----FGLLNANRRKILETSFGYQ---------------LPMVDS-FKW-YSVGYMKHLDR
DP-------------------------------------EKLTHHMPLFYCLYENRE-EE
FVKTIVDA---------------------L-------------------MEVTVYLQSDK
DM---MVS--L----YCLD-YCCH-LRT-LKLS----------------VQR-----IFQ
--NKEPLI-------------------------------------------RPTASQMKS
LVYWREIC-SLFYTMESLRELHIFDNDLNGI---------SERILS--KALEHS------
SC--K-LRTLKLSY---------VST-ASGFEDLLKAL----------------ARNRSL
TYLS-INCT---------------------------------------------------
------------------------------------------------------------
--------------------------------------------SISL-NMFSLLHDI--
LHE-PTCQISHL-S----------------------------------------------
------------------------LMKCDLR-----------------------------
----------------------------ASECEEIAS-----------------------
--------------------------------LLISGGSLRK---LT-LSSNP-LRS---
-------DGMNILCDAL----LHPNCTLISL-----------------------------
------------------------------------------------------------
------------------------------VLVFCCLTENCCSALGRV-LLFSPTLRQLD
L-CVNRLKNYGVL-HVTFPLLFP--TCQ----L---------------------------
-----------------------EELHL--SGC-----FFSSDICQYI------------
-------AIVIA-T
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NEKLRSLEIGSNKIEDAGMQL-LCGGLR-HP-NC-MLVNIG--LE-
--E-CMLTSACCR--------------------SLASVLT
PF13516 // ENSGT00900000140813_0_Domain_17 (2441, 2464)
TNKTL-ERLNLLQNHLGNDG
VAKLLE--------SLISPDCV--LK--V-------------------------------
---VGLPLT-GLNTQTQQL------LM----------TVK-ERKPSLIFLSETWSLKEGR
EIGVT-------------------------------------------------------
-------------------------------------------------------PASQP
GSI-----------------IPNSNLD---YM--FFKFPRMSAAMRTSNTASRQPL----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSTSYP00000028812 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MNS---------------------------QKKRKDMEPA-----
--------CTWS------------------------------------------LLSK--
-----------------------------------------EK-----------------
----VAPSSL-------------------------------ASFSWKELTDYREVFRKH-
--------LKEK----------------YLKI-GVDKCYYHGK--------------HIE
SRLLLIEEHGISEKTPCGIP-------------------------QQDHFIEMEHVFDPD
EE--GSSSS----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 929)
RTVVLQGCAGIGKTAVVHKF-----MFDWA--A---------GMVTP
G--------RFDY-LIYINCREISHIA-------NLSAAD-LITN-TF-----QD-INGP
VLDIILLYP----EKLLFILDGFPE-LQYPVGDH------EE-D-----LRAHPKER-KP
-VDTLLCSFVRKELFPESSLLIT-ARPTSMKKLHSLL-K-----QPI--QVEILWFTDAE
KRAYF----------------L--SQ-FSG---------VNAAMKVFYGLQE-NEGLSIM
SSLPIVSWMICSVLQSQGDGDR-TL-L--RS-LQTMTDVYLFYFSKCLKTLTGIS-IW--
-----------KGQSCLWGLCSLA---AEGL-QN-HQ----VLFAVS-------------
--DLRRHGIGVYDTNCAFLNH-----FLKKADG-GL--VYTFLHFSFQEFLTAVFYVLK-
NDNSW---------------------MFFDQGGKMWQEIFQQY-GKGFSSLMIQFL----
----FGLLHRGKGKAVETTFGRK---------------V--SPG-LRE-ELLKWTEKEIK
DK-----------------------------------SSRLQIEPMDLFHCLYEIQE-EE
YAKRIIDD---------------------L-------------------QSIILLQPTYT
KM-DILAM--S----FCVK-SSHNYLS----------------M-----SLKCQHLLGF-
-EEEERAS----------------------AFMP--------------------------
---------PALTLNQSFQ-----LHDLSMS---------RLYLLC--QALRNP------
YC--K-IKDLKL---IFCHLT------ASCGRD---------------LSSVFET
PF13516 // ENSGT00900000140813_0_Domain_6 (1616, 1642)
-NQYL
TDLE-FVKN-TLEDS-GVKLLCEGLKQPNCI----LQTLR--------LYRCLI-SPA-S
CG------------AL----ATVLSTNQWLTELEFSETKLEASALKLLCEGLKDPNCK-L
QKLKLCASLLPESSEA-VCK---YLASVLIC-
PF13516 // ENSGT00900000140813_0_Domain_8 (1773, 1796)
NPHLTELDLSENPLGD-IGVKYLCEG--
LRH-SNCKVEKL-D----------------------------------------------
------------------------LSTCYLT-----------------------------
----------------------------DASCVELSS-----------------------
--------------------------------FLQVSQTLKE---LF-VFANA-LGD---
-------TGVQYLCKGL----QYAKGIIENL-----------------------------
------------------------------------------------------------
------------------------------VLSECSLSAACCKPLAQV-LSSTQSLTRLL
L-INNKIEDLGLK-LLCEGLKRP--DL---------------------------------
---------------------------L--WT------VATGACCQDL------------
-------CNAL-Y
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
TNIHLRDLDLSDNALGDEGLLV-LCDGLK-HP-CC-KLQTLW--LA-
--D-CHLTDACCG--------------------VLASVLNR
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2465)
NENL-TLLDLSGNDLKDFG
VQMLCD--------ALIHPICK--LQTFY-------------------------------
------IDTDHLREDTFRK------ME----------ALK-ISKPGITW-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSRROP00000041080 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------MAESNSTDFD---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWYLENLS--D---KEFQSFKKYLAR------
---------K--ILDFK--------L--PQV-PLIN-LRA-T-KEELANLLPISYEGQYI
WNMLYSI----FL-MMRKEDHCMKI---IGN-----------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------------QEACKAL-
--------MRRK----------------VTLQ--WENHTFGEFHYKFFCNVA--------
----------------------------------------------SDVFYVLQLAYDST
SC--YSAND----LN
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 930)
VFLMGDRASGKTMTINLA-----VMKWI--N---------GEMWQ
N--------MISY-VVHLTSHEINQMT-------DSSLAE-LIAK-DW-----PD-GQAP
IADILS-DP----KKLLFILEDVDN-LRFELNVN------EH-A-----LCSNSTQK-VP
-IPILLVSLLKRQMAPGCWFLIS-SRPTREENIKVFL-K-----TTD--CYVTLQLSNEK
RELYF----------------H--SF-FND---------RQRASTALMLVHD-NEILTSL
CRVAILCWITCTVLNRQMDKG-HDP-Q--HC-CETPTDLHAHFLADALTSEARLT-AN--
----------QYHLGLLKRLCLLA---AGGL-FL-ST----LNFSGE-------------
--DFRCVGFTEA-DVSVLQAV----NILLPSST-HK-DRYKFIHLNVQEFCAAIAFLMAV
PNCLI-----------------P----SGSR-E--Y--KEKRE-QYCDFNQVFTFI----
----FGLLNENRRKILETTFGYQ---------------LPMVES-FRR-YSVEYMKHLGR
DQ-------------------------------------EKLTHHGSLFYCLFENQE-EE
FVKKVVNP---------------------L-------------------KEVTVYLQSNK
DM---MVS--L----YCLE-YCCH-LRT-FKLS----------------VQR-----IFE
--YKEPLV-------------------------------------------RPTASQMKS
LVYWREIC-SLFCTMDTLWELHLFDSDLNDI---------SERILS--KALEHS------
SC--K-LRTLKLSY---------IST-ASGFEELLKAL----------------ARNRSV
IYLG-LNCT---------------------------------------------------
------------------------------------------------------------
--------------------------------------------SISL-NMFSLLREI--
LDK-PTCQISHL-S----------------------------------------------
------------------------LMKCDLR-----------------------------
----------------------------ASECREIAS-----------------------
--------------------------------LLVNGGSLRK---LT-LTNNP-LRS---
-------DGVNILCNAL----LHPNCTLTSL-----------------------------
------------------------------------------------------------
------------------------------ALVFCCLTEKCCSSLGRV-LLLSPTLKQLD
L-CVNHLRNYGVL-HVTFPLIFP--TCQ----L---------------------------
-----------------------EELYL--SGC-----FFTNDICQYI------------
-------AIVIA-T
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NETLRSLEIGSNSIEDAGMQL-LCGGLR-HP-DC-MLVNIG--LE-
--E-CMLTGACCE--------------------SLASVLT
PF13516 // ENSGT00900000140813_0_Domain_17 (2441, 2464)
TNKTL-ERLNILQNQLGDEG
IVKLLE--------SLILPDCV--LQ--I-------------------------------
---IGLPLN-NVSAETQRM------VM----------TVK-ERKPDLIFLSETWSVKEGR
EIGVI-------------------------------------------------------
-------------------------------------------------------PASQP
GSV-----------------IPNSNLD---YM--FFKFPRMSTAMRMSNTASRQRL----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMODP00000015678 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------MD----MADTILC
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 324)
CLVTYLEELE--C---GELSKFKMYLGL------
---------E--AEREG--------E--ERV-PRGR-MEKAG-PLDLAKILVAQWGPAEA
WELALRV----FN-RINRKDLWERGC-QEPLVRDA----P--RS--------LELIAL--
------------------------------------------------------DS----
------------------------------------------------------------
CRRVS-GSLA-------------------------------GACPGAPSKDPQETYREH-
--------VRRK----------------YRFI--EDRNARLGELV---------NLSQRY
TRLLLVKEHATLIRAQRELL------ALGRQHALCLD--------QRASPIHIETLFEPD
EE--RPEAP----HS
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 930)
VVLQGAAGIGKSILAQKV-----MLDWA--E---------GHLYS
D--------RFDY-LFYVNCREMNQV-R------ERSLED-LISL-CW-----PE-RNVP
ISEIVR-VP----ERLLFIIDGFDE-LQPFFHEP------TD-C-----CCLRQGER-RP
-VEALLSCLLTRKLLPERSVLIT-TRPTTLETLRHLL-E-----HPR--HVEILGFSEAQ
RQEYF----------------Y--KF-FGN---------EEQARQGLAVVRD-NEPLFTL
CFVPMMCWIVCTCLKQQLDVGR-PF-P--HT-SKTTTAVYMFYLLSLLQPKAGG-SRA--
-----------QPPPSLRGLCSLA---AAGL-WR-QR----ILFDED-------------
--ELRRHGLEAA-HVSAFLNM----NVFHKDIQ-RE-RYYSFIHLSFQEFFAAMFYALE-
G---E--------------L--G--A-SPAGLT-GLLDHYAKC-ERSSLALAVRFL----
----FGLLNEETRGYLDRRLGWR---------------I--SPR-TKR-DLLRWIRAKAR
SE-----------------------------------GSTLQRGALELFTCLYEIQE-ED
FIQRALDY---------------------F-------------------QVVVVSEMA-T
KM-EHVAS--S----FCVR-NCWS-ALV-VHLC----------------SA------PFS
-ADGEQEP--E-GA---------------------------------------EG-----
PQ----------------------N--Q------------------------P-------
LK--P--------SEKNILPE-------TYCEHLAAAL----------------STNRNL
VELV-LYRN-ALGDR-GVRLLCQGLRHPNCQ----LQNL--------RLK---------R
C-----------------------------------------------------------
--------RIS---AA-ACQ---DIAAALVT-NPNLIRLDLSRNVLGA-AGVRLLAGG--
LLH-PRCRLQMLQ-----------------------------------------------
------------------------LRRCHVD-----------------------------
----------------------------GEACQELSK-----------------------
--------------------------------VLSTSQHLAE---LD-LTGNA-LGE---
-------SGLQLLCPGL----GHSAC----------------------------------
--------------------------KLQ-TLW---------------------------
-------------------------------LKICHLTPAACQNLSSV-LSTNQSLTELD
L-SLNDLGDAGVT-SLCEGLCHS--KSQ----L---------------------------
-----------------------HTLRL--GIC-----RLTFVSCEAL------------
-------STTL-QSNVHLKALDVSFNDLEDMGVHL-LCEGLQ-HP-TC-KLQKLW--LD-
--S-CCLSGAACR--------------------DLASALGANQTL-RELYLTNNALGDAG
VQLLCQ--------RLSQASCR--LR--T-------------------------------
---LWLFGT-ELTDGTQQA------LA----------ALR-GSKPHLDIGS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMMUP00000009423 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MNN---------------------------QKKRKYMELA-----
--------CTWS------------------------------------------LLSG--
-----------------------------------------EN-----------------
----VASSSL-------------------------------ASFSCKELTDYWEIFRRH-
--------LKEK----------------YLKI-GVDKCYHHGK--------------HIE
SQLLLVKDHGISEETPCGIP-------------------------QQDHFIDMEHVFDPS
EE--GSRMC----W---------DWETAVVHKF-----MFDWA--A---------GTVTP
G--------REIC-HIA-NLSAA----------------D-LITN-TF-----QD-INGP
ILDTILIYP----EKILFILDGFPE-LQDPVGDQ------EE-D-----LNSIF------
------------------------------------------------------------
------------------------------------------------------------
------TEPVFSVLQSQGDGDR-TL-L--RS-LQTMTDVYLFYFSKCLKTLTGIS-VW--
-----------ERQSCLWGLCYLA---AEGL-QN-HQ----VLFAVS-------------
--DLRRHGIGVCDTNCIFLSR-----FLKKAEG-AV-SVYTFLHFSFQEFLTAVFHALK-
NDNSW---------------------IF-------------FF-LSSFSSLMMQFL----
----FGLLHKGKGKAVETTFGIK---------------L--SPG-LRE-GLLKWTEREIK
DK-----------------------------------ASRLQIEPVDLFHCL--------
------------------------------------------------------------
-------------------------YE----------------I-----QEEENHLLGF-
-EEEERAS----------------------TFVP--------------------------
---------LALTLNQSFQ-----LCSLPVS---------QLHLLC--QALRNP------
YC--K-IKDL---------------------RD---------------LSLVFET
PF13516 // ENSGT00900000140813_0_Domain_6 (1616, 1642)
-NQHL
TDLE-FVKN-TLKDS-GMKLLCEGLKQPNCV----LQT-----------LRFFRRLISSS
CG------------AL----AAVLSTNQWLTELEFSETKLEASALKLLCEGLKDTNCK-L
QKLKLCASLLPEGSEA-VCR---YLSCVLIC-NPDLTELDPSENPLGD-IGVKYLCVG--
LRH-SNCKVEKL-D----------------------------------------------
------------------------LSTCHST-----------------------------
----------------------------DASCVDESG-----------------------
--------------------------------F-------KR---AV-LFANV-LGD---
-------TGVQHLCEGL----QHTKGIIENL-----------------------------
------------------------------------------------------------
-------------------------------LSEYSLSAACCEPLAQV-LSSTRSLTRLI
L-INNKIEDMGLK-LLCEGLKQP--DCH----L---------------------------
-----------------------KDLAL--WTC-----HLSGACCQDL------------
-------CNAL-Y
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
TNEHLRDLDFSDNALGDEGMRV-LCEGLK-CP-CC-KLQTLW--LA-
--E-CHLTDACCG--------------------ALASVLNRDE
PF13516 // ENSGT00900000140813_0_Domain_17 (2444, 2465)
NL-TLLDLSGNDLKDFG
VQMLCD--------ALIHPICK--LQTFY-------------------------------
------LDTDPLREDAFRK------ME----------VLK-MSKPGI-CCLRECLI----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCANP00000020630 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---ESFFSDFG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 322)
-LLWYLKELR--K---EEFWKFKELLKQ------
---------P--LEKFE--------L--KPI-PWAE-LKKAS-KEDVAKLLGKHYPGKQA
WEVTLNL----FL-QISRKDLWTKAR-EEMRN----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------KLNPYRKH-
--------MKEK----------------FQHI--WEKETCLQVPEH--------------
---------------------------------F-------YKETMKNEYKELNGAYTAA
------ARL----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
HTVVLEGSDGIGKTTLLRKV-----MLDWA--E---------GNLWK
D--------RFTF-VFFLNVYEMNGIT-------ETSLLE-LLSR-DW-----PES-SET
IEDIFS-QP----ERILFILDGFEQ-LKFNLE-F------KA-D-----FSDDWRRR-QP
-TPIILSSLLQKKMLPESSLLIA-LGQLAMQKHYFTL-Q-----HPK--LIKLSGFTESE
KKSYF----------------S--YY-FGE---------KNKALKVFNFVRD-SGPLFIL
CHNPFICWLVCTCVKQRLERGE-DL-E--IN-SQNTTSLYASFLTNVFKAGS-QSFPPK-
-----------VNRAGLKNLCALA---AEGV-WT-YT----FVFCHE-------------
--DLRRNGLSES-EGLMWVGM----RLLQRRG-----DCFTFVHLCIQEFCAAMFYLLK-
RPKDD-----------------P--NPAIGSIS-QLVKASMTQ-PQTHLTQVGIFT----
----FGISAEEIVSMLETSFGFP---------------L--SKD-LQQ-EITQCLKSLSQ
CE-----------------------------------ADREAMGFQELFVSLFETQE-KE
FVTQVMNF---------------------F-------------------EEVFIYIGNIQ
HL---AVA--S----FCLK-HCRR-LTT-LRMC----------------MEN-----IFP
-DDSGCAS-----------G-------------------Y-N-------EK---------
LVYWRELC-SVFITNKNFQILDMENSSLDDA---------SLAILC--KALTQP------
VC--K-LRKLIFTS---------MLN-FGHDSELFKAV----------------LHNPHL
KLLS-LYGT-SLSHT-DVRHLGEALRHPMCK----IEEL--------IL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------GKCDIS-----------------------------
----------------------------SEACEDIAS-----------------------
--------------------------------VLACNSKLKH---LS-LVENS-LRD---
-------EGMMLLCEAL----KHPHCALETL-----------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------M---------------------------------
---------------------------------------------------------LQ-
--T-CPITRACCG--------------------DLAAALVACKTL-RSLNLDWITLEPDA
VAVLCE--------ALSHPDCA--LQ--M-------------------------------
---L--HKS-GFDEETQKM------LM----------SVEEKM-PHLTISHEPWIDE---
--EYK-----LR------------------------------------------------
--------------------------------------------------GVLL------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCATP00000004673 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MS-DLHSANCCVSLT-SKYF-----------------------------
------------------------------------------------------------
--TEAG---------------------------------QVQEIDNPQL-----------
-------GDA-------------------------------EEDSELAKPGEKEGRRNS-
---------MQK----------------QSLV--WKNTFWQGDIDSFHDDVI--------
-----------------------------------------------RRHQQFIPFLNPR
TP--GKLTP----Y
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVLHGPAGVGKTTLAKKC-----MLDWT--D---------CNLSP
---------TVRY-AFYLSCKELSRMG-------PCSFAE-LISK-DW-----PE-LQDD
IPSILA-QA----QRILFVVDGLDE-LKVPPG------ALIQ-D-----ICGDWEQQ-KP
-VPVLLGSLLKRKMLPKATLLVT-TRPRALRDLQLLV-Q-----QPI--YIRVEGFLESD
RRAYF----------------L--RH-FGD---------EDQAMRAFELMRS-NAALFQL
GSAPAVCGIVCTTLKLQMEKGE-DP-A--PT-CLTSTGLFLRFLCSQFPQG-------AQ
------------LWGALRALSLLA---AQGL-WA-QM----SVLHGE-------------
--DLESAGVQES-DLRLFLDR----DILRQDRV-SK-GCYSFIHLSFQQFLTALFYALE-
KE------EE--------DRDGH--AWDIGDVQ-KLLSREERL-KNPDLIQVGRFL----
----FGLANEKRVTELEATFGCR---------------M--SPE-IKQ-ELLRCKANLHA
NK-----------------------------------P-LSMTDLKEVLCCLYESQE-EE
LAKVVVAP---------------------L-------------------KKISINLTNTS
EM---MHC--S----FSLK-QCRD-LEK-VSLQ----------------VAKG----VFL
-ENY-VDF----EP--------------------DIEFE---------------------
------------SSNSNLKFLEVKQSFLSDS---------SVRILC--DHITR-S-----
TC--H-LQKVE--I-KNVSPD--TA----YR-DFCLAF----------------IGKRTL
THLT-LEGH-IEWERTMLLLLCDLLRNHKCN----LQYL--------RLG---------G
H------C----------------------------------------------------
---------AT---SE-QWV---DFFYVLKA-NQSLRHLHLSASVLLD-EGAKLLYKT--
MTH-PKHFLQML-S----------------------------------------------
------------------------LENCRLT-----------------------------
----------------------------EASCKDLAA-----------------------
--------------------------------VLVV
PF13516 // ENSGT00900000140813_0_Domain_11 (2017, 2054)
SQQLTH---LC-LAKNP-IGD---
-------TGVKFLCEGL----SYPEC----------------------------------
--------------------------KLQ-ALV---------------------------
-------------------------------LQQCSITKRGCRYLSEA-LQEACSLTNLD
L-SLNQIA-RGLW-ILCQALENP--NCN----L---------------------------
-----------------------KHLRL--WSC-----SLMPFYCQHL------------
-------GSAL-ISNQKLETLDLGQNHLWKSGIIQ-LFRVLR-QR-TG-SLKTLR--LK-
--T-YETNLEIKK--------------------LLEEVKEKNPKLTIDCNAS------GA
---------------TAPPCCD--FF----------------------------------
----C-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSRNOP00000047770 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------M---ASLFSDFG---F
PF02758 // ENSGT00900000140813_0_Domain_0 (209, 323)
MWYWKELN--K---VEFMKFKELLIL------
---------E--ILQMG--------L--KQI-SWTE-VKNAS-REDLAILLVKYCEGKQA
WDTTFKV----LQ-KISRHDLTERAT-GEI------------AA--------------HA
N---------------------------------------------------IYRA----
------------------------------------------------------------
----------------------------------------------------------H-
--------LKKK----------------LTRD--CSRKFSISIQNF--------------
---------------------------------F------------QDEYDHLENLLIPK
VT--EEK-P----Q
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
LVFLKGIAGIGKTILLKNL-----MIVWS--E---------GLVFQ
N--------KFSY-IFYFCCQDVKQLK-------TTSLTE-LISR-EW-----PSS-SAP
IEEILS-QP----EKLLFIIDSLEG-MEWDFTKQ------ES-E-----LCDNCLEK-QP
-VNVLLSSLLKKKILPESSLLIS-ATFETFEELKDWI-E-----YTN--VRTITGFKESN
IKMYF----------------H--SL-FQD---------RNRALEAFSFVRE-NEQLFTL
CQAPVICCMVATCLKNEIEKGK-DP-V--SI-CRRTTSLYTTHIFNLFIPPN-VQYPSK-
-----------KSQDQLQGLCFLA---VEGM-WT-DI----SVFSEE-------------
--TLRRNGILDS-DIPTLLDI----GILEQSRE-SQ-NSYIFFHPSVQEFCAAMFYLLQ-
THMNH-----------------P--SPDVLHVE-KLLFSFLKE-VNTQWIFLGRFI----
----FGLLNELEHEKLEAFFGYQ---------------L--SQQ-LKQ-ELFEWLELLLD
P--------------------------------------EVKVNTMKFFYCLFEMEE-EV
FVQSAMNC---------------------R-------------------EEIDVVAKDYY
DF---IVA--A----YCLN-HGSA-LRD-LSIS----------------TQN-----VLN
-EKL--NQ-----------R-------------------------------------ENL
LMLWHNIC-SVFARNKDIHILQMKDTIFNEP---------VFQILY--NYLKNS------
SC--I-LEVLVAND---------VSF-LCDKYLFFDL-----------------IQSYNL
ELLD-LSGT-FLSHS-DVVILCNILNKAEYK----IQEL--------E------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------LANCSLSEQSWKYISDV-LC
PF13516 // ENSGT00900000140813_0_Domain_13 (2212, 2237)
QNKTLRHLD
I-SSNDLKDEGLK-VLCKALTLP--DSV----L---------------------------
-----------------------LTLS---------------------------------
---------------------------------------------------------LE-
--A-CELTGACCE--------------------DLASTFTQCKTL-GWINLVKNALDFNG
LVVLCK--------ALKQKTSN--LK--E-------------------------------
---LR-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSRROP00000041840 (view gene)
ENSRROP00000044374 (view gene)
ENSHGLP00100021899 (view gene)
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------EWGSGPRRQ
LSEEPTARARSLEPR--AAAAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLAALQYLS--Q---DQVKRFLLKLRD------
------------APHAV-----------AV---------------DLADQLTHFYGPEPA
LDVTRRT----LK-RADVHDVAARLK-AQRLQPP--------S---------CRA-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------PT-SASLPKEFQKKYRER-
--------VLQQ----------------HAEV--KE----------------------RF
TKLLIAPESAAPGEKEVALGPAE-------KPKP--------GRAWRSDTRTFSRLLSSD
A---EGPRP----P
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 839)
TVLLQGPAGIGKTMAAKKI-----LYDWA--A---------EKLYR
G--------QVDF-AFFLQCRELLEWPG------ARSLAD-LILD-QC-----PD-RGAP
LSRMLA-QP----QRLLFILDGA---------------------------------A-RG
-R----------------------------------------------------------
-------------------------------------------------ALR-TETLFAL
CFVPAVCWIVCTVLRRQLELGQ-DL-S--RT-SKTTTSVYLVFLASALSSV-GAEGP---
-----------GVRAKLPKLRRLA---REGVLGH--K----AQFSEG-------------
--DLERLELRGSVVQH-FLIKKELPSVLET-----E-VIYQFINQSFQEFFAALSYLLE-
DVGAP-------------------G-VSTGGLE-TLLCLGSEG--RSHLMLTTRFF----
----FGLLNMERVQDVECHFGC-----------------VVPAH-VKQ-DALRWVQGQSH
LR--VASEGTDGTKGLKDTEEPEEK-----------EEEEEPNFPLELLYCLYETQD-EA
FMYQALSS---------------------L-------------------PELMLERVCFH
RM-DLDVL--S----YCMR-CCPA-GQVL-WLV------------------SC----SLM
------A-----------------------------------------GQEKKR---KRS
----------------------------------------LVKR----------------
------LK-G---------------------------------------------SQAAM
KQ---LRVS-------PLHPLFG-------------------------MS---------R
C-----------------------------------------------------------
--------KLP---DR-IYR---DLSMALRA-APALTELGLLHCRLR-EAGLRVLNEG--
-LAWPQCRVQTL------------------------------------------------
----------------------------RVL-----------------------------
---------------LLKP---------LEAFQCL-------------------------
------------------------------VALPWQSPTLTT---PD-LSGC-YLPDPMV
--TY--------LCAAR----QHPGCSLQTLR----------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPTRP00000019737 (view gene)
ENSJJAP00000003663 (view gene)
ENSMNEP00000042530 (view gene)
ENSOCUP00000008333 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MLPA----KLGGGLC
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 328)
RLSAYLEELE--A---VDLKKFKLYLGT------
---------A---AELG--------E--DRI-PWGR-METAG-PLDVAQLLVAHCGPERA
WQLALRV----FE-RMHRRDLWERGQ-REHPAGDA----A--PG--------GPSSL---
------------------------------------------------------GS----
------------------------------------------------------------
---HP-ARLL-------------------------------EVSPDAPKPDPREAYRDY-
--------VRRR----------------FRLM--EDRNARLGECV---------NLSHRY
TRLLLVKEHSSPLRAQQMLL------DMGRGHARSGG--------SQASPIRVEMLFEPD
EE--RPEPP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
RTVVLQGAAGMGKSMLAHKV-----MLDWA--D---------GRLFQ
H--------RFDF-LFYVNCREMNQAAP------EQSAQD-LLAS-CW-----PE-AGAP
LRELAH-LR----GRLLLILDGFDE-LKPSFHEA------RG-P-----WCRCWEEK-RP
-TELVLSSLIQKKLLPELCLLIT-TRPSALEKLQRLL-E-----HPR--HVEILGFPEAE
RKEYF----------------L--RY-FQD---------AAQAAQAFGFVRD-SEPLFTM
CFVPMVCWVLCTCLKQQLEGGG-LL-G--QT-SRTTTAVYTLYLLSLLQP----------
---------------NQRGLCSLA---ADGL-WA-QK----VLFEER-------------
--DLQRHGLDGA-DVSAFLNM----NIFQRDIN-CE-KFYSFIHLSFQEFFAAMHYALE-
G--TE--------------G--G--AGPDQNLA-RLLAEYGFS-ERSFLALTVHFL----
----FGLLSEETRSYLERTLCWK---------------V--SPQ-VKE-ALLRAIQSKAR
SE-----------------------------------GSTLQAGSLEVFSCLYEIQE-EE
FVQQALSH---------------------F-------------------QVVVVGHIA-T
KM-EHMVS--A----FCAK-HCRS-AVV-LHLH----------------GA------AYG
-MDADDGT--G-WT---------------------------------------GAQ---T
P-------------------------LA--------------------------------
-E--P--------AERSILPD-------AYSQHLAAAL----------------CCCPDL
VELA-LYRN-ALGSR-GLRLLCQGLRHPGCR----LQNL--------RLK---------R
C-----------------------------------------------------------
--------RIP---SS-ACG---DLATALTA-NRHLLRLDLSGNSLGL-PGMELLCEG--
LRQ-PRCRLQMVQ-----------------------------------------------
------------------------LRKCLLD-----------------------------
----------------------------EGACRELAS-----------------------
--------------------------------VLCTNPHLAE---LD-LTGNA-LED---
-------AGVGRLCQGL----RHPGC----------------------------------
--------------------------RLR-TLW---------------------------
-------------------------------LKICRLTGAACGELAAT-LPVNQSLVELD
L-SLNDLGDPGAL-LLCEGLRHP--SCR----L---------------------------
-----------------------HTLRL--GIC-----RLGSPACGGL------------
-------STVL-QANPHLRELDLSFNDLGDPGVWL-LGEGLR-HP-TC-RLQKLW--LD-
--S-CSLTAKACE--------------------DLSCTLATSQTL-TELYLTNNALGNTG
VLLLCR--------RLRHSSCK--LR--V-------------------------------
---LWLFGM-DLNKLTHKR------LA----------ALR-ATKPYLDIGC---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPVAP00000004083 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---EFLF
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
SDIG---LFWYLKELQ--K---EEFWKFKELLKQ------
---------E--LLKFE--------L--KPI-PWPM-LKKAS-REDLAKLLD-HYPGK-A
WEVTLSL----FL-QINRCDLWTKAQ-EEIRN----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------KLNPYKSH-
--------MKDK----------------FRLI--WEKETCLQVPDD--------------
---------------------------------F-------YTEATKNEYEELSAAYT-A
GQ--TGQHS----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
LTMVLQGSQGIGKTTLLRKV-----MLEWA--E---------GNLWR
E--------KFTF-VFFLDGCDMNSIT-------ETSLVG-LLSR-DW-----PGS-AEP
IEDIFS-QP----EKVLFIIDGFEE-LKFDLD-L------NT-N-----WCSDWKQR-QP
-MQVILNSLLQKKLLPESSLLVA-LGMTGMKKIFYLL-Q-----HPK--YIALPGLSEYN
RKLYF----------------S--YF-FRE---------KNKASRAFSFVRD-NVSLFVL
CHYPLACWLVCMCLKWQLERGE-DL-G--IA-SQTTTSLYVSFFTSVFKSGL-GNCPPK-
-----------QSRAQLKSLCTLA---ADGI-WT-DT----FVFHSG-------------
--DLRRNGISEP-DAFMWVDL----RLLQRNS-----DRFTFVHVCIQEFCAAMLYLFR-
RPQDN-----------------A--NPAIGSVA-QLVKATVTQ-APTYLLQTGIFL----
----FGFSTEKIMNMLETSFGFP---------------L--SKE-IKQ-EITECLQSLSQ
DD-----------------------------------PNQVVVSFHELFNSLFETQD-KE
FITQVMDF---------------------F-------------------EDVNIYINNTD
DL---VVS--A----FCLK-HSQN-LQK-LHLC----------------IQN-----VFS
-DDCGAIL-----------N-------------------D-T-------QK---------
LCFWRDLC-SVFTTSRNFQVLDLDNCTFDEA---------SQTILC--KALAQP------
IC--R-LQKLVYNF---------ASS-LGNGLDFFKAV----------------LHNP
PF00560 // ENSGT00900000140813_0_Domain_6 (1619, 1644)
HL
KYLN-LHGT-SLSPV-EVRQLCEMLKHPMCN----VEQL--------ML-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------GKCNIT-----------------------------
----------------------------VEACREITS-----------------------
--------------------------------VLVCNQRLKL---LS-LIENP-VMN---
-------DGVMLLCNAL----KHPHCALETL-----------------------------
------------------------------------------------------------
------------------------------LLTYCCLNSVACDYISQA-LLCNKSLSLLD
L-GSNFLEDDGVM-SLCETLKHP--TCN----L---------------------------
-----------------------QQLWL--AGC-----YLTPVCCKDL------------
-------SVVL-SSNEKLKTLKLGDNKIQDSGVKQ-LCEALK-HP-NC-KLENLG--LE-
--I-CQLTSACCK--------------------DLASALTVCKSL-RGLNLDWNSLDHDG
LVVLCE--------ALSHPDCA--LQ--L-------------------------------
---LGLDKS-AFDEETQLL------LT----------AME-DRSPHLTILHQPWGKD---
--EYM-----IK------------------------------------------------
--------------------------------------------------GVLL------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSLAFP00000005807 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------MAASSS
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 330)
TDSN---LLWYLEKLN--N---KEFQSVKEHLIQ------
---------A--PLEFE--------L--PEI-PWLQ-LRKAQ-REDLDKILTTNYEAHIT
WNMMFSI----FQ-KINRQDLCEKI---DARRN---------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------RNKEAHKVL-
--------VSKK----------------PLLQ--WKEGIYPKANDEFYNSIA--------
----------------------------------------------PGLLHVFDMIFDPR
NN--LSSDN----FN
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 940)
MFLMGERATGKTVLIKTA-----MILWL--R---------GDMWK
D--------TFSY-VVHLTSHEINQMA-------RSSFVE-LLSK-DW-----PD-SQAP
VADILS-DS----RKLLFVLEDLDN-IKFTLNMD------ES-A-----LCSDSRNQ-VP
-VLVLLVSLLKRKMAPGSSFLIS-ARPGHQDAMDVLL-K-----ETD--FSIILNFSDEI
RQAYF----------------S--SF-FKD---------SRRATIAFKFIHE-NEILVDL
CSVPILCWVVCTTLNQQMHEA-DDV-K--LE-FQTSTDIYSHFLTNALTSEAGVV-AG--
----------QHRLVLLERLCWLA---LEGL-MQ-DV----LDFSDQ-------------
--DLRSVGLTEA-DISVLQTT----NILMQSSN-CE-DHCTFVHLHVQEFCAAIAIMRPL
VQTVL-----------------P----STFK----E--KERRA-EYNDLSPIMACV----
----FGLLNKKTRDILETSFDCP---------------LL-SGM-HRR-YFLEQMEYLGA
NP-------------------------------------RVMEHHMPLFYCLFENQE-EE
FVKQIMGF---------------------F-------------------VEATVLIRENK
DL---MVS--S----YCLK-RCHP-LQK-LRLC----------------VQD-----IFE
--KKVIVV-------------------------------------------KTSSSQMRS
LDHWRDFC-SLLYTKKNLRELEICNSKLDST---------SERILC--KALRHP------
SC--Q-LRTLK-------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMOCP00000021229 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------MESEDAEVMED------------
------------------------------------------------------------
----------------------QI------------------------------------
-------SQE-------------------------------KMLFEDKDFGDGTDYRIV-
--------IKEK----------------FCVA--WDKNSLTGESATLYQRII--------
----------------------------------------------KKDQKLLRNIFDTA
IN--TYKMR----Q
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 928)
TVLLYGDAGVGKTTLLRRA-----MLEWA--D---------GNLFE
---------KFAY-VFYLSGREISQAK-------EKSFAQ-LISK-NW-----PR-SEVP
IEQILS-ES----RNLLFLIDSFDE-LDFSFEEP------EF-A-----LCNDWTQE-YP
-VSILISSLLRKVMLPESFLLVT-ARSTARKKLMPLL-H-----KPH--CVELPGLSENA
RMDYI----------------Q--HFFLND---------KKWAKGVIDSIRK-NPRLFNM
CHVCQMCWVVCTCLKQQVEKGG-DI-A--MT-CQTTTALFTCYVCSLFSRT-DGSSVTLP
------------NETLLRSLCQAA---AEGI-WT-MK----HILYKK-------------
--NLRKHELTRN-DISVFLDA----KVLHEDTE-YE-NCYTFTHLHVQEFFGALFYLLR-
ENPEK--------------KDYP--LKPFESLY-LLLESNHSE--DPHLEQMKCFL----
----FGLLNKEQVRQLEETFNVT---------------I--SMD-VRG-DLLMCLEAIEK
DD-----------------------------------SYPSQMRLLDLFHCLYETQD-ND
FITQALSS---------------------F-------------------QKIAVQVVEEK
QL---LIY--S----FCFK-HCYS-LQS-VKLT----------------IKS-----ELR
-----EIL----DIDP-----TD--E------------------------TCLKDTVARI
THCWEDLF-SVLHTNESLREMDLYESILDEA---------LMKIIN--EELSHP------
KC--K-LQKLQFGS---------VCF-LNSCQD-FSFL----------------AA
PF13516 // ENSGT00900000140813_0_Domain_6 (1617, 1642)
CHTL
THLD-LKHT-DLEDN-GLRSLCTGLKCQQCK----LRVL--------RLE---------A
CD----------------------------------------------------------
---------VN---VN-RCQ---QLSEALER-
PF13516 // ENSGT00900000140813_0_Domain_8 (1773, 1796)
NSSLVFLNLSTNNLTN-YGVKSLCKF--
FGN-PNCCLERL-S----------------------------------------------
------------------------LASCGFT-----------------------------
----------------------------KDVCGILSF-----------------------
--------------------------------ALTE
PF13516 // ENSGT00900000140813_0_Domain_11 (2017, 2054)
SRRLTH---LC-LADNL-LGD---
-------EGIELLSDAL----KHPQC----------------------------------
--------------------------TLQ-SLV---------------------------
-------------------------------LRCGFFTPVGSEFLSTA-LGI
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
NRSLVHLD
L-GGNRLEDSGIK-LLCHVLQQP--TCT----L---------------------------
-----------------------EELEL--TGC-----LLTSESCLDL------------
-------ASVL-VNSPNLWSLDLGNNYLLDAGLHI-LCDALK-NP-NC-QIQRLG--LE-
--N-CGLTSACCQ--------------------DLLSLFCTNQSL-VQMNLMKNALGYDT
IKNLCK--------VLRCPTCK--MK--F-------------------------------
---LALDKREIDRKKIKKF------LN----------DVK-INNPELVIRPHCSA-----
--T---------------------------------------------------------
--------------------------------------------------ESGCWWQYF-
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMICP00000008087 (view gene)
ENSPANP00000034802 (view gene)
MDQPEDPGSRLRSLRTASPQRVFVASVPHQRLRLRPRCKAPKLGLTK-RQPGHVTWPGVW
KNRGRKGGAGEEGAPQKSGRRWPVPTSPSEGEEHQD------APTRVRGRWR--------
-------AKSGVTPRCPLPPPHG-----SS--LARGSLRPCPRWVLKRQSRGWAGVTQGP
LPDDPRPPVSSTGLR--LAVAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLAALEELS--Q---EQLKRFRHKLRD------
------------VGPDR-----------RSI-PWGR-LERAD-AVDLAEQLAQFYGPEPA
LEVARKT----LK-RADARDVAAQLK-EQRLRR---------L---------GLG-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------SA-ALLSVSDYKKKYREH-
--------VLQL----------------HARV--KERNARS------------VKITKRF
TKLLIAPESAAPEEE--ALGPAE-------EPEP--------GRARRSDTQTFNRLFRGD
E---DGRRP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYQ
G--------QVDF-AFFMPCGELLERPG------TRSLAD-LILD-QC-----PD-RGAP
VPQMLA-QP----QRLLFILDGADE-LPALEGP--------EAA-----PCTDPFEA-AS
-GARVLGGLLSKALLPTALLLVT-TRAAAPGRLQGRL-C-----SPQ--CAEVRGFSDKD
KKKYF----------------Y--KF-FQD---------ERRAERAYRFVKE-NETLFAL
CFVPFVCWIVCTVLRQQLELGR-DL-S--RT-SKTTTSVYLLFIASVLSSAPVADGP---
-----------RVQGDLRNLCRLA---REGVLGR--R----AQFAEK-------------
--QLEQLELRGSKVQTLFLSKKELPGVLET-----E-VTYQFIDQSFQEFLAALSYLLE-
DEGVP-------------------R-TAAGGVG-TLLRGDAQP--HSHLVLTTRFL----
----FGLLSAERMRDIERHFGC-----------------VVSER-VKQ-EALRWVQGQGQ
GCPGVAPEVTEGTKGLEDPEEPEEE----------DEEGEEPNYPLELLYCLYETQE-DA
FVRQALCR---------------------L-------------------PELALQGVRFC
RM-DVAVL--S----YCVR-CCPA-GQA-LRLI------------------SC----RLV
------AA----------------------------------------QEK--K---KRS
----------------------------------------LGKR----------------
------LQAS---L--------------GGS------S-----------------SRGTT
KQ---LPAS-------LLHPLFQTMTDPLCG----LTSL--------TLS---------H
C-----------------------------------------------------------
--------KVP---DA-VCR---DLSEALRA-APALTELGLLHNRFS-EAGLRMLSEG--
-LAWPQCRVQTV------------------------------------------------
----------------------------RVQ-----------------------------
---------------LPSS---------QQGLQYL-------------------------
------------------------------VGMLRQSPALTT---LD-LSGC-QLPAPMV
--T--------YLCAVL----QHQGCGLQTLRL--TSVELSEQ-----------------
----------SLQELQAVKRAKP-------DLV----------------------I--AH
PVLD-------------------GHPESP-KELVSTF-----------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCCAP00000018679 (view gene)
------------------------------------------------------------
----------------------------------------------------Q-TP----
---LLSLDTLSTDP----------------------------------------------
----TCPPRINMEENKLLTFSSYG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 323)
-LQWCLDELD--K---EEFQTFKKLLKK------
---------I--SSEST--------T--CTI-PQSE-IENAN-VECLALLLHEYYGRSLA
WSTSISI----FE-TMNLRTLSEKAR-DDMKIHS-----PEDLQPRTT--YQGPNKKEMP
EIS-QAIKQDSATASE--AEEQGE------------CN-ICLLP---SWKASLFSKSL--
LS-TNTER---------MPVVAEISQPTEQEGA---------------------------
-----M---A-------------------------------AETEEQGPGGDTWDYKRH-
--------VMTK----------------FTAE--EDVH-RGFGKP---------------
-----------------------------------AA-------DWLEM-QMLVGAFGAD
Q---WGFRP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 926)
HTVVLHGKSGIGKSALARRI-----VLSWA--Q---------GRLFQ
N--------MFSY-VFFIPIREVQRRN-------EASLAE-LISR-EW-----PD-SQVP
VTEIMS-QP----ERLLFILDGFDD-LG-SNVHS------DT-K-----LCKAWAER-QP
-MGTLIHSLLRKALLPESFLIVT-VRDVGVERLKSQV-V-----SPR--YLLVRGISAER
RIHLL----------------L--EH-GTR---------EHQKAQVVRAMMS-NRGLFDQ
CEVPAVGMLVCVALQLQDVEEE-GV-------GLTLTGLYASFVFRQLTPRD-VAQRCLD
----------LEEKAVLKRLCRMA---VEGV-WN-MK----SVFDSD-------------
--DLLMHNLGES-ELRALLGM----DMLLRDGR-CEEEHFTFFHLSLQDFCAALYYVLE-
GLDVE--------------PALC--PLFMEKGK-RSVELKQAG-FHSHLLWMKRFL----
----FGLMNENVRRPLEVLLGCP---------------V--PL-GVKQ-KLLHWVSLLGQ
QTN----------------------------------AT-APADTLDAFHCLFETQD-KE
FVRSALNS---------------------F-------------------QEVWLPICQNM
DL---TAS--S----FCLQ-HCPN-LRK-VRVD----------------VKG-----IFL
-RDESAEA----WP--------------------VV-----------PQ--WLRSK-TLV
DEQWENFC-SVLGTHPHLQQLDLGNSVLTER---------AMKTLC--AKLRP-P-----
TC--K-IQNLR--L-RNAQM---TP---GLQ-HLWRTL----------------ITNRNL
RSLN-LGST-HLKEE-DVRMACEALKHPRCM----LECL--------RLD---------R
C------G----------------------------------------------------
---------LT---HT-CYL---KISQILTT-
PF13516 // ENSGT00900000140813_0_Domain_8 (1773, 1795)
SPSLSSLSLAGNEVTD-EGVKAENSGT00900000140813_0_Linker_8_10 (1795, 1807)
LGEV--
LKV-SQPF13516 // ENSGT00900000140813_0_Domain_10 (1807, 1956)
CTLQKL-T----------------------------------------------
------------------------LQDCSIT-----------------------------
----------------------------ATGCQSLENSGT00900000140813_0_Linker_10_11 (1956, 2016)
AS-----------------------
--------------------------------ALVPF13516 // ENSGT00900000140813_0_Domain_11 (2016, 2054)
TNHSLTH---LC-LSNNN-LGN---
-------EGVSLLCRSL----RLPHC----------------------------------
--------------------------GLQ-RLM---------------------------
-------------------------------LNHCNLDTAGCGFLALT-LLG
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
NARLMHLS
L-SMNPLEDSGVK-LLCEVMREP--SCH----L---------------------------
-----------------------QDLEL--VSC-----RLTAACCESL------------
-------SYVI-AR
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
SRYLKSLDLAENAVGDVGVTV-LCEGLK-QR-RS-VLRRLG--LK-
--A-CGLTSGCCE--------------------ELSLALSC
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2465)
NRHL-ASLNLVQNNFSPTG
MRKLCS--------AFACPTSN--LQ--I-------------------------------
---IGLWKW-QYPVQIRKL------LE----------EVQ-LLKPQMVIDGSWYSFD---
--E---------------------------------------------------------
--------------------------------------------------DDRYWWKN--
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSOANP00000022985 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------P----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVLQGAAGIGKSYTAHKI-----MLAWA--S---------QRLYH
D--------RFDW-VFLFNCRELGVEPR------PRSLVD-LVLS-DC-PA-----LGPH
VGQIFS-SP----QRLLFLLDGFDE-LQLPRSVR--------------------------
-------LLLRQRLLPGCLVVVT-TRPSALEQVEGCI-R-----ADV--HLEVLGFLEPE
RQAFF----------------T--RF-FGD---------AKRGREAYEAVQG-NEALSTM
CFVPLVCWIVCTVLHKQLEKGQ-GL-D---G-LQTTTQVFLHFLSILLRFHRRRG-ARAP
------------ADSLLEQLGTLA---VHGL-MA-RK----VIFDQE-------------
--DLEAHGLPAAAPPTIFLSA-----VLRQGVT-VE-TVYSFGHVMLQEVFAAIFCFLP-
GRGAR----------------------PELGLA-HLLEAGLQA-ENGHLLQTIRFL----
----FGLSHPQCQAMLRQLLPHR---------------AALTPS-PEG-ALLSWVQRSA-
ES----------------------------------PRAEDPRFLLELLHCLYEWHC-DG
LVGRVASE---------------------L-------------------N-IRFLLFPLK
RS-DCLAL--A----YCLG-C---------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------CASVSC---------------------------------
------------LH----LYSC----GL-DQ-------------VPPDRSI--FWVCG-F
CPFRI-SGNLL--TKG-CIP---HLELLVRA-NTGLTHLDLSGNSLGD-EAVVLLCAH--
LGA-AAGQLRNL------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSRBIP00000010509 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------MAESNSTDFD---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWYLENLS--D---KEFQSFKKYLAR------
---------K--ILDFK--------L--PQV-PLIN-LRA-T-KEELANLLPISYEGQYI
WNMLYSI----FL-MMRKEDHCMKI---IGRRN---------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------RNQEACKAL-
--------MRRK----------------VTLQ--WENHTFGEFHYKFFCNVA--------
----------------------------------------------SDVFYVLQLAYDST
SC--YSAND----LN
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 930)
VFLMGDRASGKTMTINLA-----VMKWI--N---------GEMWQ
N--------MISY-VVHLTSHEINQMT-------DSSLAE-LIAK-DW-----PD-GQAP
IADILS-DP----KKLLFILEDVDN-LRFELNVN------EH-A-----LCSNSTQK-VP
-IPILLVSLLKRQMAPGCWFLIS-SRPTREENIKVFL-K-----TTD--CYVTLQLSNEK
RELYF----------------H--SF-FND---------RQRASTALMLVHD-NEILTSL
CRVAILCWITCTVLNRQMDKG-HDP-Q--HC-CETPTDLHAHFLADALTSEARLT-AN--
----------QYHLGLLKRLCLLA---AGGL-FL-ST----LNFSGE-------------
--DFRCVGFTEA-DVSVLQAV----NILLPSST-HK-DRYKFIHLNVQEFCAAIAFLMAV
PNCLI-----------------P----SGSR-E--Y--KEKRE-QYCDFNQVFTFI----
----FGLLNENRRKILETTFGYQ---------------LPMVES-FRR-YSVEYMKHLGR
DQ-------------------------------------EKLTHHGSLFYCLFENQE-EE
FVKKVVNP---------------------L-------------------KEVTVYLQSNK
DM---MVS--L----YCLE-YCCH-LRT-FKLS----------------VQR-----IFE
--YKEPLV-------------------------------------------RPTASQMKS
LVYWREIC-SLFCTMDTLWELHLFDSDLNDI---------SERILS--KALEHS------
SC--K-LRTLKLSY---------IST-ASGFEELLKAL----------------ARNRSV
IYLG-LNCT---------------------------------------------------
------------------------------------------------------------
--------------------------------------------SISL-NMFSLLREI--
LDK-PTCQISHL-S----------------------------------------------
------------------------LMKCDLR-----------------------------
----------------------------ASECGEIAS-----------------------
--------------------------------LLVNGGSLRK---LT-LTNNP-LRS---
-------DGVNILCNAL----LHPNCTLTSL-----------------------------
------------------------------------------------------------
------------------------------ALVFCCLTEKCCSSLGRV-LLLSPTLKQLD
L-CVNHLRNYGVL-HVTFPLIFP--TCQ----L---------------------------
-----------------------EELYL--SGC-----FFTNDICQYI------------
-------AIVIA-T
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NETLRSLEIGSNSIEDAGMQL-LCGGLR-HP-DC-MLVNIG--LE-
--E-CMLTGACCE--------------------SLASVLT
PF13516 // ENSGT00900000140813_0_Domain_17 (2441, 2464)
TNKTL-ERLNILQNQLGDEG
IVKLLE--------SLILPDCV--LQ--I-------------------------------
---IGLPLN-NVSAETQRM------VM----------TVK-ERKPDLIFLSETWSVKEGR
EIGVI-------------------------------------------------------
-------------------------------------------------------PASQP
GSV-----------------IPNSNLD---YM--FFKFPRMSTAMRMSNTASRQRL----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSOCUP00000004733 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------MA----GT
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 328)
RCK---LAQYLEDLE--E---LDLKKFKMHLED------
---------Y--PPDQG--------C--IPL-PRGQ-AARAD-PVDLATLMIDFNGEEKA
WAMALWI----FT-AINRRDLYERAK-RDEPVWE--PAH--VS---------NATVI---
--------CH-----------------------------------EEII-----------
-----------------------------------------------------EEERMG-
------------------I-----------------LGHLSRISICTKRKDYCKKYRKH-
--------VRSR----------------FQCI--EDRNARLGESV---------NLNKRY
TRLRLAKEHLSRQEREQELV------AIGRT--HVWD------SP--ASPVRVELLFD--
ADEECLEPV----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 929)
RTVVLQGAAGIGKTILARKI-----MLDWA--S---------GQLFQ
D--------RFDY-LFYIHCREMSLGM-------HRSLVD-LIAG-CC-----SD-PSPP
VSKMLH-RP----SRVLFLMDGFDE-LQGAFDES------SG-A-----LCSDWRVA-VR
-GDVLLGSLIRKKLLPEASLLIT-TRPVALEKLQHLL-D-----RPR--HVEVLGFSEAR
RKEYF----------------F--KY-FLE---------EAQAREAFSLLQE-NEVLLSM
CCIPLVCWIVCTGLKQQMQGGK-HR-AKTTT-SKTTTAVYTFFLSSLLQSRGGSQEQR--
------------LSGHLRGLCSLA---ADGI-WN-QK----ILFEER-------------
--DLRRHGLQEA-EVSAFLRM----SLFQKEVD-CE-AFYSFVHLTFQEFFAAMYYVLE-
EEEEEKEEEEGAGTGRSPPARGL--DLPHRDVR-VLLENYGKF-EKGYLIFVVRFL----
----FGLVNQERASYLEKKLSCK---------------I--SQQ-VRP-ELLRWIETKAN
AK-----------------------------------KLQIQPSQLELFYCLYEMQE-EA
FVREAMGH---------------------F-------------------PRIEINLSTRM
---DHVVS--S----FCIG-NCHS-VES-LSLG----------------FLHN-----SP
KEEEEEEE-----E------------------------------------E---------
---------------EVESPLFDVGCPNRQG-----------------------------
------------------------------------------------------------
-------------------------PHAACS----W-----------RLV---------N
S-----------------------------------------------------------
--------CLT---SS-FCR---GLFSVLST-NPSLTELDLSDNSLGD-PGMRVLCEA--
LQH-PGCNIRRL-W----------------------------------------------
------------------------LGRCGLS-----------------------------
----------------------------HQCCFDISL-----------------------
--------------------------------VLSGSQKLVE---LD-LSDNA-LGD---
-------FGVRLLCVGL----RHLSCQLQKL-----------------------------
--------------------------------W---------------------------
-------------------------------LVSCCLTSMCCQDLASV-LSTNHCLTRLY
L-GENTLGDYGVG-LLCEKVKHP--QCN----L---------------------------
-----------------------QKLGL--VNS-----GLTSACCAAL------------
-------SAAL-STSRSLTHLYLRGNALGDAGVKL-LCEGLL-HP-SC-RLQMLE--LD-
--S-CSLTSHCCW--------------------NLCTILTCSRSL-RKLSLGNNDLGDLG
VMMLCE--------VLRQPDCP--LQ--R-------------------------------
---LQLCEM-YFNPETIHA------LE----------TLR-EEKPALTVVFEPSW-----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSSHAP00000002191 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MG-ITP
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 326)
RNI---LMNILEDLI--E---EELKKFKFQLED------
---------P---PLKD--------F--GPL-PRSQ-LHSAQ-PVDLAELLIRHYGVNYA
VEVTKAV----LE-IINQRDLVEKLE-QAI------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------EKGK---NSYQKR-
--------IKVQ----------------YE------EK---S-V----------GFILRI
SELKSVK-SPKTEDSDNCIC------SF-------GQ-----Y-NSNS--ISLETLFDFD
IN--TPHDL----QT
PF00004 // ENSGT00900000140813_0_Domain_2 (676, 862)
VVLQGPAGIGKSTLARKV-----VQDWA--A---------GILYT
G--------RFDY-VFYISCQEVSLLRE-------NSVDH-LITV-LC-GN-----NNAP
ITDILR-QP----QKLLFVLDDLDE-LQFLSAEQ------SS-D-----FHSKVKEK-DP
-VHVLLESLIQKKLLPQSTLLIT-IRPPTLEKLEPWL-V-----CSH--HVQVLGFFQEQ
KKEYF----------------S--LY-FMD---------DDLAKKAFDFVQT-NKAISDE
CHAPLICWIICCWMKQRLKLGK-SP-T--KD-LKNRTDIYMAYISTFLPTGK----NLSR
----------PTQHAVLRGLCSLA---TEGM-QR-QK----ILFTEG-------------
--DLKRHSLDGVDVS-SVLNV----NGSQARLS-GT-LMYSFRHLSFQEFFNAMFYLLE-
-EISDR---------------------ERQNLK-GLIE-------KQETSGTVQFL----
----HGLLEKENEKNLELKFNIR-----------------ISLP-LKK-EV-MILTKRM-
N-------------------------------------EMK-------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCANP00000022505 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MSLDIQCEQ-LSD-ARWA----------------EL---------
----LPLL-------------------------------QQCQVV------RLDDC-GLT
EARCSDIS-SALRV
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1542)
NPALTELSLRGNELGDA---------GVHCVL--QGLQS-P-----
SC--K-IQKLS--L-QDCYLT--GA---GCG-VLSSTL----------------RS
PF13516 // ENSGT00900000140813_0_Domain_6 (1617, 1642)
LPTL
QELH-LSDN-LLGDA-GLQLLCEGLLDPQCR----LEKL--------QLE---------Y
C------N----------------------------------------------------
---------LS---AA-SCE---PLASVLRA-KPDFKELTVSNNNIGE-AGVRVLCQG--
LKD-SPCQLEAL-K----------------------------------------------
------------------------LEGCGIT-----------------------------
----------------------------SDNCRDLCS-----------------------
--------------------------------IVASK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
ASLRE---LA-LGSNK-LGD---
-------VGLAELCPGL----LHPSS----------------------------------
--------------------------SLR-TLW---------------------------
-------------------------------IWECGITAKGCGDLCRV-LRTKE
PF13516 // ENSGT00900000140813_0_Domain_13 (2215, 2237)
SLKELS
L-AGNELGDEGAR-LLCETLLEP--SCQ----L---------------------------
-----------------------ESLWV--KSC-----SLTAACCSHF------------
-------SSVL-T
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
QNKCLLELQISNNRLGDAGVQE-LCQGLG-QP-GS-VLRVLW--LG-
--N-CDVSDRSCS--------------------SLAATLLA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2464)
NHSL-RELDLSNNCLGDAG
VLQLVE--------SVRQPGCL--LE--Q-------------------------------
---LVLYDI-YWSEAMEDR------LQ----------ALE-EEKPSLRIIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSP00000375063 (view gene)
ENSCGRP00001014679 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------MEETTGYR---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LTLCLQKLS--D---AEFLKFKELLRN------
---------A--PEKHK--------L--KPI-PWTT-IKRTS-RKDMEMLLNTHYPER-V
WGISLRL----FL-DINREDLWIMTQ-ELKRD----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------KQRPYKET-
--------MKTI----------------FKYI--WTQENYIHFQNE--------------
---------------------------------E-------YQVIIKKQYSALQETFDHQ
-----LK-P----VT
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 898)
AVVLGNIGEGKSTFLRKA-----MLDWA--S---------GNLWQ
N--------RFQY-VFFFSLISLNNIT-------ELSLAQ-LLLS-KL-----SES-SEA
LDDILS-NP----KRILFILEGFDY-LNFDLE-L------RT-N-----LCSDWRKH-LP
-TQIVLGSLLQKVMLPESSLLLE-LGPQSEPKIYPLL-Q-----YPK--NIIIGGFNSA-
MQLYC----------------M--LF--FN---------IDKGLEVFKYIQT-SKPLFSL
CSNPYACWMVCSTLKQQCDRGD-RI-I--LP-LGTDTILYTMFMLSSFRSVY-ANCSSK-
-----------ENRARLKTLCTLA---VEGM-WK-RV----FVFNSE-------------
--DLRRNRICES-EQTVWLQV----NFLNTRG-----DCFVFYNPALQWYFAALFYFLR-
EDKDK-----------------P--HPIIGSLP-QLLKEIYAC-GETQWSMTGIFV----
----FGIATEIVAAMLKPHFGFI---------------P--SKE-VRQ-VILKCFRSLSQ
GDC----------------------------------SGKL--SPQRLFDGLLDSQE-EP
FETQVLDL---------------------F-------------------EEMTIDISSTD
EL---SLA--K----IYLL-KSQK-LKK-LHLH----------------IQHR----IFS
-EIYNPEE-----------G-------------------D-L-------EEFTQ------
--EKTELC-RIFQ---NLQVLDLDSCNFSET---------AIQDLC--SSMSPPLMMPLN
AF--K-LQRMSCSF---------MTN-FGN-GALFPTL----------------LRLPHL
KSLN-LYGT-NLSSD-VVENMCSVLKCPTCR----VEEL--------LL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------GNCDIS-----------------------------
----------------------------SEACGVIAT-----------------------
--------------------------------SLT
PF13516 // ENSGT00900000140813_0_Domain_11 (2016, 2054)
KC-KVKH---LS-LVENP-LKN---
-------EGVMLLCQIL----KHPSCV-ELL-----------------------------
------------------------------------------------------------
------------------------------MLSYCCLTFIACGHLYEA-LLCNRSLSLLD
L-GSNFLEDTGVN-ILCEALKDP--TCC----L---------------------------
-----------------------QELWL--SGC-----YLTTACCEGI------------
-------SAVL-LR
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NKNLKTLKLGNNSIQDTGVQQ-LCEALR-NP-EC-KLQCLG--LD-
--M-CEFTTGSCA--------------------DLALALTKCKTL-TSLNLDWMTFDPDG
LELLCE--------ALNHKACN--LK--V-------------------------------
---LGLDKS-ASSKESQML------LE----------GVE-KKN--LIILHYPWINE---
--ERK-----KR------------------------------------------------
--------------------------------------------------GVCLVWNSKN
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSFALP00000011314 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 930)
VVLVGAPGMGKTTMVRKV-----MVEWG--E---------GTLHT
---------QFDF-VFCIDCKAMVL-P------MEASVAE-LVSQ-CC-----PG-LRVP
AGKILD-GQ----KKILFIFDGFEA-LGFSLVQP------EA-E-----LSSDPREV-KP
-LELTLMSLLKKTVLPEASLLIT-VRPAALGRLRQCL-K-----GEY--YVEILGFSAAR
RKEYF----------------H--RY-FEK---------SNKADMAFRFARG-NEILYNL
CVIPVMSWTTCTVLEQELCGKK-NL-L--DC-SKTTTGMIMFYLSQILKHRE-RDN-PQV
------------LQQFLLQLCSLA---AEGI-WK-HK----VLFEEK-------------
--EIKDCGLDQPSLLPLFLNE----RIPREGED-HE-SVYAFTHLHLQEFFAAMFYVLE-
DDGEM--------------ASSL--GKLEKDVN-KLLDSYNKS-RK-DLNVTVRFL----
----FGLVSQKSIEYMDKTIGCR---------------I--SAQ-ARE-ELLEWLQGRHR
GLS--------------------------------LPGQALKVSETDTFHFLFEMNE-KS
FAQSALNH---------------------F-------------------TDLDLQDTKLT
LY-DQMAL--S----FCIQ-HWAG-LGC-VTLQ----------------VNK-----SST
-EDLLIVG----EN-------------------------PLLPQR------EIPGAGMLS
APCDSPS----------ISPGGCSSNARGPSPLQSHRGLSGGALPC--AAGSA-RG--GG
LC--AAVAAGS--R-RHCSAV--SA---AGA-HPGALV----------------------
-----VRQR-EELPC-PIHLLCRALRQRDSA----LQVL--------RLQ---------W
C------Q----------------------------------------------------
---------LS---ES-CCA---ELAALLAE-QPGLTRLELADSSLGD-SGVRLLCEG--
LRA-PGCCLRIL-R----------------------------------------------
------------------------LRYSRIT-----------------------------
----------------------------SACCEDLAA-----------------------
--------------------------------VLSTNTRLEE---LD-LSFSEGLRD---
-------AGVELLCEGL----QHCAC----------------------------------
--------------------------PLQ-TLR---------------------------
-------------------------------LGSCRLSGTCCQSLAVQ-LLQGPRLTCL-
------------------------------------------------------------
------------------------------------------------------------
-------------DGPRLSCPHLSDNELGAHGVLQ-LCRQLR-HP-AC-PLK--------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSECAP00000021583 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MEEAEVPSLS
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 326)
NYG---LQWCFKQLS--K---EEFQTFKELLNE------
---------K--ASELE--------A--GSF-PWVE-VNNAN-IEHLASLLHEHYRGSLA
WKISINI----FE-KMNLSALSEKAR-DEMKKYSL-AEIPKASTPK-T--DQEPSMEEVP
GPR-S-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------DEDDKWYYKSY-
--------VMTK----------------FAMK--LDAC-YGFKNF---------------
-----------------------------------TS-------EHPEM-QTLLGAFKPD
Q---NGFQP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
HTVVLHGEPGIGKSTLARRI-----LLSWA--Q---------GEVYQ
D--------LFSY-AFFLCAREIQQIR-------ESTLAE-LISR-EW-----PD-SPVP
VMEIMS-QP----ERLLFVVDGFDD-LDFVFKDG------DI-N-----LCEDWAEK-QP
-MSILMYSLLKKVLLPESSLIIT-VRDAGIEKLKSIV-M-----SPR--YLLLEGISVEK
RIQLL----------------L--QH-IKN---------DHQKMQVLQSVVD-NHLLFDK
CQVSVVCSLICEALKLQEMSGK-SP-P--PT-CQTLTGLYATFVFHQLTPQD-AFPRALS
----------QEERVVLKGVCRLA---VEGM-WN-MK----SVFYGD-------------
--DLGVHGLRKS-ELSALFHM----NILLQDSH-CERC-YSFLHLSLQEFCAALYYVLV-
GLERE--------------GDLY--PLFIENIG-SLTELKQFG-ISVHLLQMKRFL----
----FGLMSKEVIKTLEALLGCP---------------V--PV-VVKQ-KLLHWISLLGQ
QAN----------------------------------ST-APLDFLDAFYCLFETQD-EE
FIRLALNS---------------------F-------------------QEVRLPMNRRM
DL---IVS--S----FCLP-HCRY-LRK-IRMD----------------VRE-----ILS
-RDELTEA----WP--------------------MI-----------PE--GMQTK-TFI
DKSWENFC-FVLSTHPNLQELDLSGSVLDEW---------AIKTFC--VKLRQ-P-----
TC--K-IQNLI--F-KGAQI---TL---GLR-HLWMTL----------------LVNRNI
KYLN-LERT-RLKNE-DVGMACEALRHPNCL----LESL--------RLD---------H
C------G----------------------------------------------------
---------LT---HA-CCV---VISQILMT-SINLKSLSLAGNKVTD-QGMKPLCDA--
LKV-SQCTLQRL-I----------------------------------------------
------------------------LGNCGLT-----------------------------
----------------------------AADCQDLAS-----------------------
--------------------------------SLISNLSLTH---LD-LSSNS-LGS---
-------EGMNLLYRTI----KLPNC----------------------------------
--------------------------GLQ-RLI---------------------------
-------------------------------LNECNLDIASCGFLGFA-LVGNRRLTHLS
L-SMNPLEDAGVN-LLCEVMMEP--SCH----L---------------------------
-----------------------QDLEL--VKC-----NLTAACCKNL------------
-------SLMI-STSKRLRSLDLAANALRDEGIVA-LCQGLK-QR-KS-SLRRLG--LE-
--A-CELTSGCCE--------------------VLSSALSCNQHL-TSLNLIRNNFSPGG
MMKLCS--------AFAHPTSN--LQ--I-------------------------------
---IGLWKL-QFPAQVRKM------LE----------EVQ-LLKPHIIIDDGWYS-----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMLUP00000021929 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------IL----------------KVLG
PF00560 // ENSGT00900000140813_0_Domain_6 (1619, 1645)
SL
RELD-LSGN-SLSCS-AVQSLCDALKSPCCH----LDTL--------RLV---------R
C-----------------------------------------------------------
--------GLT---NN-CCK---DLASMLGA-SP
PF00560 // ENSGT00900000140813_0_Domain_8 (1775, 1794)
SLTELDLRQNDLGD-LGLKLLCEG--
LRQ-PACQLRNLR-----------------------------------------------
------------------------LVRCGLT-----------------------------
----------------------------NNCCKDLAS-----------------------
--------------------------------MLGASP
PF00560 // ENSGT00900000140813_0_Domain_11 (2019, 2057)
SLTE---LD-LGQND-LGD---
-------LGLKRLCEGL----RQPAC----------------------------------
--------------------------QLR-KLR---------------------------
-------------------------------LVSCSLTNNCCKDLASM-LGASP
PF00560 // ENSGT00900000140813_0_Domain_13 (2215, 2235)
SLTELD
L-GQNDLGDLGVK-LLCEGLRQP--ACQ----F---------------------------
-----------------------RKMRL--VRC-----GLTNNCCKDL------------
-------ASML-GASP
PF00560 // ENSGT00900000140813_0_Domain_15 (2357, 2375)
SLTELDLGQNDLGDLGVKL-LCEGLR-QP-AC-QLRKLR--LV-
--R-CGLTNNCCQ--------------------DLASMLGASP
PF00560 // ENSGT00900000140813_0_Domain_17 (2444, 2463)
NL-TELDLGQNDLGDLG
VKLLCE--------GLRQPACQ--LR--KLRLDQ--------------------------
---TW------MSKKVKTM------LT----------LLQ-QEKPQLVISS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSECAP00000006832 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MAGG----A-ELQ---WPWYLESRE--K---KEPKEFQLRLPD------
---------A--H--SR-------HS--PGE-TVAQ--VEAS-GPEVASRLVALYGQRQA
WDLALLT----QR-RMDLSRVSIQAH-REAASIPIS--SL-HASLG---KPN---LEFSS
APTSTMVLGHWVPEAP-----GLLQDS-QRKNLRHLPGTSGHTW-KENPPSFPFQ--D--
---------FPYSPV-CESPSQESPNAPTSTAVLGGWEFPPQSMLACRKQEPPRTAWPLA
ETSGKHSTVVEGRDRRGHLTGVP----L----MAAEPGSE--PQAHTEHPGLWERYQKQ-
--------WRET----------------PCPTQSR-------QNE---------DLHQKF
VQLLLLHTSDPGSHES---L------VRRCCPHGV--------VEEEGHLIEIGDLFGPG
LG--TQRGP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
HTVVLHGVTGIGKSTLARQV-----RGAWE--E---------GRLYR
D--------RFQH-VFYFNCTELAQSK-------PVSLAK-LMGE-DC-----AA-PAPP
I---RS-QQ----EKMLFILDGLDE-PELVLEEQ-------R-P-----LCEDWSLR-QP
-VPRVLHSLLRKVLLPEASLLVT-ARTPAPQKFILSL-E-----HPH--WVEVLGFSESS
RKEYF----------------Y--RY-FTE---------ESQAVQAFSLVES-NPALLTL
CLVPFVSWMVCTCLKQQMERGE-EL-S--LT-SQTTTALCLHYLSQALP--------AQ-
-----------PLRTQLRCLCSLA---AEGV-GQ-GE----TLFSLN-------------
--DLRKHRLDGA-CISTLLKM----GLLQKH---PTSQRYSFIHPFFQGFFAAMSYALG-
D------KE---------KSDHL--NST-RRIK-KLLEKCERH--DPFGTPTTPFL----
----FGLLSEQGVRKMEHIFRCK---------------L--SQE-RKG-DLLQK----VR
-----------------------------------RGHPSLPPYSLQVLRCLYEIQD-ED
FLTQAMAH---------------------F-------------------QRTRMYVQTDM
EL---LVF--T----FCVK-FCHH-VER-LQLN----------------ESGP-------
------H-R---------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------Q------------
------AWRP--------------------------------------------------
-------------------SSVVLLSRAPVT-----------------------------
----------------------------DACWKVLFS-----------------------
--------------------------------VLQVTGSLKE---LD-LSGNF-LSL---
-------SAVQSLCEAL----RHPRCHLQTLR----------------------------
------------------------------------------------------------
-------------------------------LASCGLTAEGCKDLASG-LSTSQTLTELE
L-SCNVLEDAGVK-HLCQGLRQP--SCV----L---------------------------
-----------------------QRLLL--VSC-----GLTS------------------
------------------------------------------------------------
---------GFCQ--------------------DLASVLSAGSRL-TELDLQQNKLGDLG
VKLLCE--------GLRRPTCQ--LM--L-------------------------------
---LWLDQT-QLNEEVTEM------LR----------ALE-KNAQLL-ILS-RWKPSVTT
PNEGRDGGEMSEN-----------------RCSLKRQRAESARAQSPKAH---SGLLSLE
LTTALP--VPES------SSPQVVQ-VQPLFLPFPASPGD-PHVEPLE-TKDDFWGPMGP
VATEVVDK-ERSLYRVHLPMAGSYRWP---NTGLHFVVRGPV-------TIEIEFCAWGQ
FLDRTVPQHSWMVVGPLFDIKAEPGA-VAAVYLPHFVVLQGA--------HVDISLFQVA
HFKEEGMLLEKPARVEPCYAVLENPSFSPVGVLLRMIHAALPFIPVTSTVLLYYHFQPEE
V-TFHLYLIPNDCTIRKAIDDEEKKFQFVRIHKPPPLTPLYVGSRYTVSGS---------
KKLEIIPKELELCYRSPGESQLFSEFYIDHFGTGIKLQMRDKKDGTVVWEALVKRGKVET
KAVVKPS----------------------------CLLAVSPSHGLAPASL
PF00619 // ENSGT00900000140813_0_Domain_19 (3052, 3133)
HFVDRYREQ
LVARVTSVDPVLDKLH-GLVLSEEQYEKVRAEATTPSQMRKLFSFSKSWDWACKDKFYQA
LKETHPHLIVELWEK---------------------
ENSACAP00000020972 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------LFLAS-IVCRTS---E
PF02758 // ENSGT00900000140813_0_Domain_0 (209, 326)
LDALEDLG--E---EELKRFKFKLHT------
---------T---TAPG--------D--KNI-PLGR-LERAD-RENLVGLLVEFYEDKAA
VLM-VSV----FE-DMGLKYNASKLS-KAESKFSS-----EDAN----------IVSPSD
N-----------------------------------------------C-----------
---------------------------------------NVKAVKRRVP-----------
GLFNAY-HTM-------------------------------LLKK--ALKGYKKKYAET-
--------VMEE----------------YQLT--EDQNSLFGEST---------PLKDRY
IELLIIRKHRPQKHREHELS------ATGRLHMELLE----SN-RSEYSPTDIESLFEPD
NS--R-RTP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
RTVVLQGPAGIGKTMTVQKI-----MLDWS--S---------GELYP
D--------VFDY-VFCFHCRELRFRE------EPHSLAD-LLIE-QC-QD-----MYAP
VKKILA-QP----EKLLFIIDGFDE-LRSFQDPG------GD-N-----CCDDPYSK-VS
-VEVVLSSLLRKTLLPKSYLLIT-TRPGAAERLQECL-K-----FPR--FAEILGFSEKG
RRKYF----------------L--KF-FED---------ERKANIALRFIEN-TIAVSTI
CFIPMVCWVICSVIKVELETED-EI-A--DT-LDTTTKVFMQFSHGLLKHDPKT--RMIP
----------S--LKEFRKLCSLA---RDGI-LE-QK----ILFEEQ-------------
--DIKKHDLDASSLKDLFLDR----RTFHKGIG-RH-TLYSFLHLCFQEFFAAMWYILQ-
KESTE----------------EL--DNNKLDLK-RLLVECEKP-GNEHLTFTVHFI----
----FGLSSEKVCTFLEQMLECK-----------------TSSD-LVK-PLLLHRAEEVA
A------------------E----------------DPPRKGHRLLKFFHCLFEIQE-TE
FARRVMNH---------------------F-------------------QMINLSFKILS
VL-DCRVL--A----FCLQ-HSEI-RDH-SIDLTFCRNKELHHIRVSDSILLC----SMN
--------------NT--C-----------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------L--------GIS----------
-GFSL-PI--TVCFDLQNLKSI----GSNISQPLFGSNRLGNSGVKVLCAILKETGCN-M
TTLEL-QKSII--SRN-LLG---DLQVLLLL--------------------------E--
LVK-QLCNTKNV-W----------------------------------------------
------------------------WNSCYI------------------------------
-------------------------------SNACIQEL-C-------------------
------------------------------DVLST-SHTLET---LS-LQENS-FTEV--
-------S-VPFI-----------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSP00000433999 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----MSLD--I----QSLDIQCEE-LSD-ARWA----------------EL---------
----LPLL-------------------------------QQCQVV------RLDDC-GLT
EARCKDIS-SALRV
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1542)
NPALAELNLRSNELGDV---------GVHCVL--QGLQT-P-----
SC--K-IQKLS--L-QNCCLT--GA---GCG-VLSSTL----------------RTL
PF13516 // ENSGT00900000140813_0_Domain_6 (1618, 1642)
PTL
QELH-LSDN-LLGDA-GLQLLCEGLLDPQCR----LEKL--------QLE---------Y
C------S----------------------------------------------------
---------LS---AA-SCE---PLASVLRA-KPDFKELTVSNNDINE-AGVRVLCQG--
LKD-SPCQLEAL-K----------------------------------------------
------------------------LESCGVT-----------------------------
----------------------------SDNCRDLCG-----------------------
--------------------------------IVASK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
ASLRE---LA-LGSNK-LGD---
-------VGMAELCPGL----LHPSS----------------------------------
--------------------------RLR-TLW---------------------------
-------------------------------IWECGITAKGCGDLCRV-LRAKE
PF13516 // ENSGT00900000140813_0_Domain_13 (2215, 2237)
SLKELS
L-AGNELGDEGAR-LLCETLLEP--GCQ----L---------------------------
-----------------------ESLWV--KSC-----SFTAACCSHF------------
-------SSVL-A
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
QNRFLLELQISNNRLEDAGVRE-LCQGLG-QP-GS-VLRVLW--LA-
--D-CDVSDSSCS--------------------SLAATLLA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2464)
NHSL-RELDLSNNCLGDAG
ILQLVE--------SVRQPGCL--LE--Q-------------------------------
---LVLYDI-YWSEEMEDR------LQ----------ALE-KDKPSLRVIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMUSP00000104155 (view gene)
ENSMFAP00000034555 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MTSP----QLEWT---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 323)
LQTLLEQLN--E---DELKSFKSLLRA------
---------L--PLEDV--------L--QKA-PWSE-VEEAD-GKKLAEILIHTSSENWI
RSATVTV----LE-EMNLTELCK-MA-KSEM-----------------------------
------------------------------------------------------------
--LEAG---------------------------------QVQEIDNPQL-----------
-------GDA-------------------------------EEDSELAKPGEKEGQRNS-
---------VQK----------------QSLV--WKNTFWQGDIDSFHDDVI--------
-----------------------------------------------QRNQRFIPFLNPS
TP--RKLTP----Y
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVLHGPAGVGKTTLAKKC-----MLDWT--D---------CNLSP
---------TARY-AFYLSCKELSRMG-------PCSFAE-LISK-DW-----PE-LQDD
IPSILA-QA----QRILFVVDGLDE-LKVPPG------ALIQ-D-----ICGDWEQQ-KP
-VPVLLGSLLKRKMLPKATLLVT-TRPRALRDLQLLV-Q-----QPI--YIRVEGFLEED
RRAYF----------------L--RH-FGD---------EDQAMRAFELMRS-NAALFQL
GSAPAVCWIVCTTLKLQMEKGE-DP-A--PT-CLTSTGLFLRFLCSQFPQG-------AQ
------------LRGALRALSLLA---AQGL-WA-QM----SVLHGE-------------
--DLESAGVQES-DLRLFLDR----NILRQDGV-AK-GCYSFIHLSFQQFLTALFYALE-
EE--E---EE--------DRDGH--AWDIGDVQ-KLLSEEERL-KNPDLIQVGHFL----
----FGLANEKRVKELEATFGCQ---------------M--SPE-IKQ-ELLRCKASLHA
DK-----------------------------------P-LSMTDLKEVLCCLYESQE-EE
LAKVVVAP---------------------L-------------------KKISINLTNTS
EM---MHC--S----FSLK-HCRD-LEK-VSLQ----------------IAKG----VFL
-ENY-VDF----EA--------------------DIEFERC-TYV--TLPSWAQQD-LRS
LRLWTDFC-SLFSSNSNLKFLEVKQSFLSDS---------SVRILC--DHITR-S-----
TC--H-LQKVE--I-KNVTPD--TA----YR-DFCLAF----------------IGKRTL
THLT-LEGH-IEWERTMLLLLCDLLRNHKCN----LQYL--------RLG---------G
H------C----------------------------------------------------
---------AT---SE-QWV---DFFYVLKA-NQSLRHLHLSASVLLD-EGAKLLYKT--
MTH-PKHFLQML-S----------------------------------------------
------------------------LENCRLT-----------------------------
----------------------------EASCKDLAA-----------------------
--------------------------------VLVV
PF13516 // ENSGT00900000140813_0_Domain_11 (2017, 2054)
SQQLTH---LC-LAKNP-IGD---
-------TGVKFLCEGL----SYPEC----------------------------------
--------------------------KLQ-ALV---------------------------
-------------------------------LQQCSITKRGCRYLSEA-LQEACSLTNLD
L-SLNQIA-RGLW-ILCQALENP--NCN----L---------------------------
-----------------------KHLRL--WSC-----SLMPFYCQHL------------
-------GSAL-ISNQKLETLDLGQNHLWKSGIIQ-LFRVLR-QR-TG-SLKTLR--LK-
--T-YETNLEIKK--------------------LLEEVKEKNPNLTIDCNAS------GA
---------------TAPACCD--FF----------------------------------
----C-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMUSP00000104158 (view gene)
ENSCATP00000003382 (view gene)
MGP_SPRETEiJ_P0028307 (view gene)
ENSBTAP00000006201 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---ESFFS
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 328)
DFG---LLWYLEELK--K---EEFWKFKELLKQ------
---------E--PLKLK--------L--KPI-PWTE-LKKAS-RENVSKLLSKHYPGKLA
WDVTLNL----FL-QISRDDLWRKAR-NEIRQ----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------KINPYRSH-
--------MKQK----------------FQVL--WEKEPCLLVPED--------------
---------------------------------F-------YEETTKIEYELLSTVYLDA
FK--PGESS----P
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVLHGPEGIGKTTFLRKV-----MLEWA--K---------GNLWR
D--------RFSF-VFFLTGREMNGVT-------DMSLVE-LLSR-DW-----PES-SEP
IEDIFS-QP----ERILFILDGMEE-LKFDLD-C------NA-D-----LCEDWEQP-QS
-MQVVLQSLLQKQMLPECSLLLA-LSKMGMRKNYSLL-K-----HMK--CIFLLGFSEHQ
RKLYF----------------S--HY-FQE---------KDASSRAFSFVRE-KSSLFVL
CQSPFLCWLVCTSLKCQLEKGE-DL-E--LD-SETITGLYVSFFTKVFRSGS-ETCPLK-
-----------QRRARLKSLCTLA---AEGM-WT-CT----FLFCPE-------------
--DLRRNGVSES-DTSMWLDM----KLLHRSG-----DCLAFIHTCIQEFCAAMFYMFT-
RPKDP-----------------P--HSVIGNVT-QLITRAVSE-HYSRLSWTAVFL----
----FVFSTERMTHRLETSFGFP---------------L--SKE-IKQ-EITQSLDTLSQ
CD-----------------------------------PNNVMMSFQALFNCLFETQD-PE
FVAQVVNF---------------------F-------------------KDIDIYIGTKE
EL---IIC--A----ACLR-HCHS-LQK-FHLC----------------MEH-----VFP
-DESGCIS-----------N-------------------T-I-------EK---------
LTLWRDVC-SAFTASEDFEILNLDNCRFDEP---------SLAVLC--RTLSQP------
VC--K-LRKFVCNF---------ASN-LANSLELFKVI----------------LHNPHL
KHLN-FYGS-SLSHM-DARQLCEALKHPMCN----IEEL--------ML-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------GKCDIT-----------------------------
----------------------------GEACEDIAS-----------------------
--------------------------------VLVHNKKLNL---LS-LCDNA-LKD---
-------DGVLVLCEAL----KNPDCALEAL-----------------------------
------------------------------------------------------------
------------------------------LLSHCCFSSAACDHLSQV-LLYNRSLTFLD
L-GSNVLKDEGVT-TLCESLKHP--SCN----L---------------------------
-----------------------QELWL--MNC-----YFTSVCCVDI------------
-------ATVL-IHSEKLKTLKLGNNKIYDAGAKQ-LCKALK-HP-KC-KLENLG--LE-
--A-CELSPASCE--------------------DLASALTTCKSL-TCVNLEWITLDYDG
AAVLCE--------ALVSLECS--LQ--L-------------------------------
---LGLNIS-SYDEEIKMM------LT----------QVE-EMNPNLIISHHLWTDD---
--EGR-----RR------------------------------------------------
--------------------------------------------------GILV------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSSSCP00000059045 (view gene)
ENSCHIP00000026281 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MREAKTAPFSSYG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 323)
-LQWCFEQLG--K---EELQTFKALLKE------
---------H--ASEPA--------A--CSF-PLVQ-VDRAD-AVSLASLLHEHCRASLA
WKTSIGI----FE-KMRLSALSEMAR-DEMKKCLL-AETSEDSAPAKT--DQGPSMKEVP
G-----------------------------------------------------------
------------------------------------------------------------
---------------------------------------------PREDPQDSRDYRIH-
--------VMTT----------------FSTR--LDTP-QHFEEF---------------
-----------------------------------AS-------ECPDA-HALSGAFNPD
PR--GGFRP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 905)
TVVLHGPPGVGKSMLARRL-----LLFWA--Q---------GDLYQ
G--------LFSY-VFLLRARDLQGSR-------ETSFAE-LISR-EW-----PD-APVP
VEKVLS-QS----KRLLIVVDGLEE-LELTLGDQ------DS-S-----LSADWVER-QP
-VAVLAHSLLKKVLLPECALLLT-VQDAGVQRLQALL-R-----SPR--YLWVGGLSVEN
RVQLL----------------L--GG-GKH---------GRQKTRAWHAGTD-HQEVLDK
CQVPVVCALVREALELHGEPEK-GL-P--VP-SHTLTGLYATFVFQRLAPKD-AGRRTLS
----------REERSALKGLCRLA---VDGV-WN-AK----FVFDGD-------------
--DLGVHGLKGP-ELSALQQA----SILLPDGR-CGRG-HAFSHLSLQEFFAALFYVLR-
GVEGD--------------GEGY--PLFPQSTK-SLTELRHVD-LNVHLVQMKRFL----
----FGLVSKEATRALETLLGCP---------------A--AP-VAKQ-RLLHWICLLGQ
HPA----------------------------------APASSPDLLEAFYCLFETQD-DE
FVRLALNG---------------------F-------------------QEVWLQLNRPM
DL---TVS--S----FCLR-RCQH-LRK-VRLD----------------VRE-----T-P
-KDESAEA----WP--------------------GA-----------PQ--GLKIK-TL-
HEHWEDLC-SVLGTHPNLRELDLSGSVLSKE---------AMKTLC--VKLRQ-P-----
AC--K-IQNLI--C-TGARV---TA---GLR-HLWMTL----------------VVNRSV
TRLD-LTGS-RLGEE-DVRAAGEALRHPHCA----LQTL--------RLD---------R
C------G----------------------------------------------------
---------LT---PA-SCR---VISQVLAT-SV
PF13516 // ENSGT00900000140813_0_Domain_8 (1775, 1796)
SLKSLSLMGNKVAD-QGVKSLCDA--
LKV-APCTLQKL-I----------------------------------------------
------------------------LGSCGLA-----------------------------
----------------------------AATCQDLAS-----------------------
--------------------------------ALIENQGLTH---LS-LSGDE-LGS---
-------EGMSLLCRAV----KLPSC----------------------------------
--------------------------GLQ-KLA---------------------------
-------------------------------LNACSLDVAGCGFLALA-LMG
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
NRHLTHLS
L-SMNPLEDPGMN-LLCEVMMEP--SLS----P---------------------------
-----------------------LVG----------------------------------
--------------------------------------------------DQIYH--TE-
--A-CGLTSEGCK--------------------ALSAALIC
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2465)
SRHL-ASLNLMRNDLGPGG
MATLCS--------AFMHPTSN--LQ--T-------------------------------
---IGLWKE-QYPARVRRL------LE----------QVQ-RLKPHVVISDAWYSEE---
--E---------------------------------------------------------
--------------------------------------------------DQ-RWWRI--
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCANP00000019338 (view gene)
MDQPEAPCSRLRSLRTASPHRAFVGSVPHQRLRLRPWCKASKLGLTK-RQPGHVTWPGV-
----RKGGAGEEGAPQSSGRRWPVPTSPSEGEEHQD------APTRVRGRRR--------
-------AKSRVTPRCPLPPPHG-----SS--LARGSLRPCPRWVLERQSRGWAGVTRGP
LPDDPRPPVSSTGLR--LAVAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLAALEELS--Q---EQLKRFRHKLRD------
------------VGPDG-----------RSI-PWGR-LERAD-AVDLAEQLAQFYGPEPA
LEVARKT----LK-RADARDVAAQLQ-EQRLRR---------L---------GPG-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------SV-ALLSVSEYKKKYREH-
--------VLQL----------------HARV--KERNARS------------VKITKRF
TKLLIAPESAAPEEE--ALGPAE-------EPEP--------GRARRSDTHTFNRLFRGD
E---EGRQP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYQ
G--------QVDF-AFFMPCGELLERPG------TRSLAD-LILD-QC-----PD-RGAP
VPQMLA-QP----QRLLFILDGADE-LPALEGP--------EAA-----PCTDPFEA-AS
-GARVLGGLLSKALLPTALLLVT-TRAAAPGRLQGRL-G-----SPQ--CAEVRGFSDKD
KKKYF----------------Y--KF-FQD---------ERRAERAYRFVKE-NETLFAL
CFVPFVCWIVCTVLRQQLELGR-DL-S--RT-SKTTTSVYLLFIASVLSSAPVADGP---
-----------RVQGDLRNLCRLA---REGVLGR--R----AQFAEK-------------
--QLEQLELRGSKVQTLFLSKKELPGVLET-----E-VTYQFIDQSFQEFLAALSYLLE-
DEGVP-------------------R-TAAGGVG-TLLRGDAQP--HSHLVLTTRFL----
----FGLLSAERMRDIERHFGC-----------------VVSER-VKQ-EALRWVQGQGQ
GCPGVAPEVTEGTKGLEDTEEPEEE-----------EEGEEPNYPLELLYCLYETQE-DA
FVRQALCR---------------------L-------------------PELALQGVRFC
RM-DVAVL--S----YCVR-CCPA-GQA-LRLI------------------SC----RLV
------AA----------------------------------------QEK--K---KKS
----------------------------------------LGKR----------------
------LQAS---L--------------GGS------S-----------------SRGTT
KQ---LPAS-------LLHPLFQAMTDPLCR----LTSL--------TLS---------H
C-----------------------------------------------------------
--------KVP---DA-VCR---DLSEALRA-APALTELGLLHNRLS-EAGLRMLSEG--
-LAWPQCRVQTV------------------------------------------------
----------------------------RVQ-----------------------------
---------------LPSS---------QQGLQYL-------------------------
------------------------------VGMLRQSPALTT---LD-LSGC-QLPAPTV
--T--------YLCAVL----QHQECGLQTLRWSLISVELSEQ-----------------
----------SLQELQAVKRAKP-------DLV----------------------I--AH
PALD-------------------GHPEPP-KELISTF-----------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPPAP00000039657 (view gene)
ENSOCUP00000000403 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MAASSSSYFS
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 326)
DFG---LLLYLEELN--K---EEFNKFKLLLR-------
---------E--TMEPG--------P--CLI-PWTA-VKKAK-REDLANLMNRYYPGEQA
WSVTLKI----FG-KMNLRGLCERAK-AEINRTAT-AMEQEDVEP-RE------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------MQGDQEAMLDSGTEYRIQ-
--------IKEK----------------FCII--WDESSWLGELETFHRAIS--------
----------------------------------------------QEDEDLVEHLFDMD
VK--TGEQP----Q
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
IVVLQGAAGVGKTKLARKA-----MLDWA--K---------GNFYQ
---------KFSY-VFYLNGREMNQLK-------ETSFAQ-LISK-DW-----PS-AESP
IEKILS-QP----SGLLFIIDSFDE-LNFAFEEP------EF-A-----LCEDWTQV-HP
-VSFLLSSLLRKVMLPESSLLLT-MRLTSSKRLKNVL-K----S-QR--YVEIPGMPEQI
RKEYI----------------Y--QF-FED---------EEWGKKAFSLLRS-NEMLFSM
CRVPQVCWIACTCLKREMEKGG-DV-T--VT-CQTTTALFTSYISSLFTSV-DECCLSLP
------------NQDQLGRLCHLA---AKGI-WT-MT----HVFYRD-------------
--NLRKHGLTKD-DVAIFLDM----NILQRDRE-YE-NCYVFTHLHVQEFFAAMFYMLK-
DNWEA--------------RDYP--VESFEDLK-LLLESKNDK--DSNLMQMRCFL----
----FGLLNEKKIKQLEEALNCK---------------T--SMD-IKQ-ESLQWMEELGN
SD-----------------------------------YSPSELGFLELFHHLYETQD-EA
FIGQAMSY---------------------F-------------------PKIDIKMCEKN
HL---LVT--S----FCLK-HCRC-LQT-IKLS----------------VSV-----IFE
--K--KIV----NSSC-----PA--K------------------------TWQN-DEDLI
IRCWKDLC-SVIHTNEHLKELNLCHSNLDEL---------AMRIFY--QELSHP------
NC--K-LQKLLLRF---------VSF-PDGCLGISNFL----------------TQNQHL
MHLD-LTGS-DIGDN-GVKALCEALKHPRCK----LQSL--------RLE---------S
CD----------------------------------------------------------
---------LT---TV-CCL---NISKALIR-NQSLGFLNLSTNNLLD-DGVKLLCEA--
LRH-PKCPLERL-S----------------------------------------------
------------------------LESCGLT-----------------------------
----------------------------EAGCEDLSL-----------------------
--------------------------------ALITNTRLTH---LC-LTDNV-LGD---
-------GGVKFMSDAL----QHPQC----------------------------------
--------------------------TLQ-SLV---------------------------
-------------------------------LRRCHLTSLSSRCLSTS-LLYNKSLRHLD
L-ALNFLQDEGAK-LLCDVFRHP--SCS----L---------------------------
-----------------------QDVEL--VGC-----AITSACCLDL------------
-------ASAI-LNNPNIWSMDLGNNNLQDGGVKI-LCDALR-HP-NC-NIQKLG--LA-
--Y-CGLTSLCCR--------------------DLSSTLISNQRL-RKINLTQNPLGSEG
IKKLCE--------VLRSPQCK--LQ--T-------------------------------
---LGLCKE-AFDEEAQKL------LE----------AVT-LKNPHLVIKPDYNDHN---
--E---------------------------------------------------------
--------------------------------------------------DDGSWWQFND
SSE-----------------EDG-SW----------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSOCUP00000019453 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------MEAVAP----SRSFN---LP
PF02758 // ENSGT00900000140813_0_Domain_0 (210, 326)
VLLEQLD--Q---DELTTFKNILKN------
---------L--CQQNE--------L--QRV-APAE-IKEAD-GRRLAEMLAAHCPRYWV
ETVTVQV----FE-KMNRMDLSEQAK-DELGAMAP-VD--------K------PAAAG--
------AEL---------------------------------------------------
-----------------------------------------GSKAGN-------------
-----------------------------------------------QKP-GPSRDWEP-
--------SLQK----------------FAKA--WEDDLRLGDAESIQA-----------
---------------------------------------------FARRYEALIPFCSPK
AH--EGPFP----HS
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 931)
VVLQGPAGVGKSVLARKV-----LRDWL--Q---------RGLGL
---------PFQF-AFYLSCPELRGEP-------ACTFAE-LLSR-DC-----PE-LRDA
VVRGAT-QA----RHVLLVLDGFDE-LRAPTG------ALVS-D-----ICGDWTQP-KP
-VPVLLSSLLKRKLLPRATVLVT-TRPQALRELRILV-E-----QPL--LIQVRGFSEQD
RKLYF----------------L--RH-FSNEGD------EAQALPSLDAARG-NAALFDL
GAAPAVCWLTCACLGAQLERGE-DP-A--AA-CATTTELFLRFLCGCCAP-------GPH
------------LRAALGALSLLA---ARGV-WA-QV----SGFCGP-------------
--ELRALGLAEP-ELRPFVAG----GVLREDGD-RA-GSYSFVHLSVQQCLGALFYLLE-
AAEGG---E---------AGGAD--DEDVGAVR-QLFSTKARA-DNPGLAWTGRFL----
----FGLLNRGRARELEAAFGCR---------------M--STR-VLQ-ELLDAE----E
GR-----------------------------------PVLADLDTAEALCCLHESQD-DA
LVRKAMVP---------------------F-------------------QELTVHLRDAT
EL---AQA--A----FCLQ-RSQN-LQS-LRLQ----------------VDKA----VFT
-PDA-ADE----EA--------------------AGAREHV-DHQ--KLPPASRDD-KHL
LLCWTNLC-SVLDSSSKLTFLDISQSVLSAP---------AVQALC--DHLAA-S-----
SC--C-LQKVA--L-RNTQPA--DA----YR-DLCLSV----------------HGHRTV
THLT-LQGC-DQVD--VLPALCEVLRHPYCR----LQYL--------QLA---------S
C------S----------------------------------------------------
---------AT---TQ-QWA---ALALALAE-NQSLMFLNLTAVELLD-TGAKLLCVA--
LRH-PRCLLQRL-S----------------------------------------------
------------------------LESCHLT-----------------------------
----------------------------EASCKELSS-----------------------
--------------------------------ALIVNQRLTH---LS-LAEND-LGD---
-------GGVKVLCEGL----SYPEC----------------------------------
--------------------------CLQ-TLV---------------------------
-------------------------------LWCCNVTSEGCAALSRL-LQQTSSLTHLD
L-GLNHIGLAGLR-FLCEALKDP--CCN----L---------------------------
-----------------------QCLWL--WGC-----SIPPFSCADL------------
-------SSAL-AINQTLVSLDLGQNSLGFSGVKM-LCEALK-LA-RC-PLRTLR--MK-
--I-DESDTHFQK--------------------VLREARESNPRLLIESDSR----DPRR
--------------PPRPSAQD--FV----------------------------------
----F-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSP00000299481 (view gene)
ENSMMUP00000031420 (view gene)
ENSCSAP00000007374 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-PDDPRPPVSSTGLR--LAVA
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 328)
REL---LLAALEELS--Q---EQLKRFRHKLRD------
------------VGPDR-----------RSI-PWGR-LERAD-AVDLAEQLAQFYGPEPA
LEVARKT----LK-RADARDVAAQLK-EQRLRR---------L---------GLG-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------ST-ALLSVSEYKKKYREH-
--------VLQL----------------HARV--KERNARS------------VKITKRF
TKLLIAPESAAPEEE--ALGPAE-------EPEP--------GRARRSDTHTFNRLFRGD
E---DGRRP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYQ
G--------QVDF-AFFMPCGELLERPG------TRSLAD-LILD-QC-----PD-RGAP
VPQMLA-QP----QRLLFILDGADE-LPALEGP--------EAA-----PCTDPFEA-AS
-GARVLGGLLSKALLPTALLLVT-TRAAAPGRLQGRL-C-----SPQ--CAEVRGFSDKD
KKKYF----------------Y--KF-FQD---------ERRAERAYRFVKE-NETLFAL
CFVPFVCWIVCTVLRQQLELGR-DL-S--RT-SKTTTSVYLLFIASVLSSAPVADGP---
-----------RVQGDLRNLCRLA---REGVLGR--R----AQFAEK-------------
--QLEQLELRGSKVQTLFLSKKELPGVLET-----E-VTYQFIDQSFQEFLAALSYLLE-
DEGVP-------------------R-AAAGGVG-TLLRGDAQP--HSHLVLTTRFL----
----FGLLSAERMRDIERHFGC-----------------VVSER-VKQ-EALRWVQGQGQ
GCPGVAPEATEGTKGLEDPEEPEEE-----------EEGEEPNYPLELLYCLYETQE-DA
FVRQALCR---------------------L-------------------PELALQGVRFC
RM-DMAVL--S----YCVR-CCPA-GQA-LRLI------------------SC----RLV
------AA----------------------------------------QEK--K---KKS
----------------------------------------LGKR----------------
------LQAS---L--------------GGS------S----------------SSRGTT
KQ---LPAS-------LLHPLFQTMTDPLCR----LTSL--------TLS---------H
C-----------------------------------------------------------
--------KVP---DA-VCR---DLSEALRA-APALTELGLLHNRLS-EAGLRMLSEG--
-LAWPQCRVQTV------------------------------------------------
----------------------------RVQ-----------------------------
---------------LPSS---------QQGLQYL-------------------------
------------------------------VGMLRQSPALTT---LD-LSGC-QLPAPMV
--T--------YLCAVL----QHQGCGLQTLRL--TSVELSEP-----------------
----------SLQELQAVKRAKP-------DLV----------------------I--AH
PVLD-------------------GHPESATKELVSTF-----------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSNLEP00000007510 (view gene)
ENSCJAP00000055581 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------------YPS
WYNPITLLFYATA-PGLNFYLCISFC-HVPADL---------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------GNVEACKAL-
--------MRRK----------------FTLQ--WENQTLGEFHYKFFCDTA--------
----------------------------------------------ADVFYVIHLAYDPT
SP--YSARD----LN
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 930)
VFLMGEAASGKTMIVRMA-----VTRWL--S---------GDMWP
N--------TISY-VVPLTAHEINQMS-------SLSLAE-LIAK-DW-----PH-RQAP
VADILS-DP----GQVLFILEDLDN-VSFQLNID------EH-A-----LCSDSTQR-VP
-IPILLVSLLRRKMAPGCWFLIS-SRFTNEGAVNMFW-K-----TAD--IFTSLHLSNGK
REIYF----------------N--SF-FND---------RQKASAALRLVRG-NDMLSGL
CRVPILCWITCSALSQQRDTGEQDL-E--LC-FQTPTDLHAHFLAGAFTSEAGAA-AS--
----------EYHLNLLKRLCSLA---AGGL-FK-NT----LHFSRE-------------
--DFSCVGLTEA-DVSLLQTL----NIVLPSST-HK-DCCTFIHLTLQEFCAAIAFLMPV
PPCQL-----------------P----SGAS-G--H--KEKRE-QYCNFSQVYSFI----
----YGLLNEKRRRILETSFGFQ---------------LPTVDA-FRQ-HSLAYLKHVGR
DA-------------------------------------EKLTHHVSLFYCLFENRE-EE
FVKMTVDP---------------------L-------------------QEVTVYLEANN
DM---MVS--L----YCLG-HCRH-VQT-LKLS----------------VQR-----IFE
--GREPTV-------------------------------------------RPTASQMKS
LVYWRDLC-SLSRTMENLQELHVFDSDLNNL---------SERILS--QALEHP------
SC--R-LRTLRFSY---------VST-GTGFEDILSAV----------------ASNRSL
TCLS-LNCM---------------------------------------------------
------------------------------------------------------------
--------------------------------------------SISL-NMFSLLHEI--
LTE-PTCQITHL-S----------------------------------------------
------------------------LMKCDLR-----------------------------
----------------------------ACDCRQIAL-----------------------
--------------------------------LITSG
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
CSLRK---LT-LSHNP-LHD---
-------DGVHTLCEAL----VHPDCTLRSL-----------------------------
------------------------------------------------------------
------------------------------VLVFCCLTAGCCNFLGKA-FLLSPSLREVD
L-SVNHLKNYGAL-ILLFPLVF---SCR----L---------------------------
-----------------------EELQL--FGC-----FLTPEICEQI------------
-------AVVLA-ANG
PF13516 // ENSGT00900000140813_0_Domain_15 (2357, 2378)
NLKSLELGSNHVGDTGMQQ-ICDALK-EP-SC-KLANIG--LE-
--E-CMLTTACCD--------------------ALASVFA
PF13516 // ENSGT00900000140813_0_Domain_17 (2441, 2464)
TNTTL-KRLNVLQNSFSNEG
IVKLLE--------SLMHPNCT--LE--I-------------------------------
---VGLPLS-GVSTETQQL------LR----------TVK-ERKPELIFLSETWSAKEGR
EIGVR-------------------------------------------------------
-------------------------------------------------------PSSRL
SSE-----------------IPHCNLQ---NM--FSEFSPTL------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSSHAP00000003923 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------------------------LFDPF
KE--TGVQP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 805)
RTVVLQGAAGIGKTTLASKV-----MLDWA--E---------GNLFQ
E--------RFDY-VFYLTCRELNPLGE-----REISFAD-LIAN-DW-----PG-PKAP
MAEIMS-QP----ERLLFIIDGLEE-EDL-------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----RRNTLE-AAVVSAFLDM----NIFQKDSG-CE-NCYSFIHLSFQEFFAAIFYVMN-
TEKEG--------------MKSS--DASIPGVR-ELLEECSGL-NSSFVVLVVRFL----
----FGFLNVETAKELERKFRCR---------------M--SLK-IKS-QLLQWAQG---
-------------------E-T---E-------RNIQYHQSMLYFREFYSHLYETQD-SE
FVTQVLDN---------------------I-------------------QEIKVQVYHQY
DA---LNA--A----FCI-KHCYK-IKT-VNTY----------------CR---------
----IHLT----GI-------------------------LLYYC-------TSYVI-LIM
ISIWQELF-SIVNKNPDLKELMLGSFEFDDS---------CMENLC--KELNN-P-----
NC--K-LEKLTRRN-DKYYTL--S-----AL-KDFMHL----------------FSAPSL
KSLH-LKGE-IVKND-DMKLPRETLRKTNCQ----LEIL--------SLV---------N
C------L----------------------------------------------------
---------LQ---PT-FCY---KLFSVLGS-SKSLKNLDLSLNFFGK-EEIKCLCKT--
MEN-QVWKLETL-R----------------------------------------------
------------------------MSRCKLT-----------------------------
----------------------------IDHLQNLLP-----------------------
--------------------------------FFCSIKNLKK---LD-LSHNF-FGE---
-------DGINLVCEAL----EKQDC----------------------------------
--------------------------NLQ-ILK---------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCATP00000010619 (view gene)
ENSCATP00000025317 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MAGG----A-WGR---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LACYLELLK--K---EELKEFQLLFTS------
---------K--VHSS--------GS--SGE-TPAR-PEKTS-GMEVASYLVAQYGEQRA
WDLALRT----WE-QMGLWSLCTQAW-EGAS-------YS-PSFPY---SPSEPHLGSPS
QPTSTAVLRPWNRELP-----AECTQGSERRVLRQLPDTSGHRW-REISSSLLYQ--A--
---------FPSSPD-FESPSQESPNAPTSTAVLASWGSPPQPSLAPREQEAPGTQWPLD
ETSGNYYTGIREREREESEKGR------PPWEAAVGTPPQVH---TSLQPHHHPWEPS--
--------ARES----------------LCSTWSW-------KNE---------DLNQNF
TQLLLLQRPHPRNHKP---L------VKGSWPHDAEE----DVEEDRGRLIEIRDLFGPG
LD--TQ-EP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
RIVILQGAAGIGKSTLARQV-----REAWG--R---------GQLYG
D--------RFQH-VFYLSCRELAQCE-------AVSLAE-LIGK-DW-----TA-AQAP
IRQILS-RP----ERLLFILDGVDE-PRWVLQES------SS-E-----LCLHWSQP-QP
-ADALLGSLLGKTILPEASFLIT-ARTTALQNLIPSL-E-----QAR--WVEVLGFSESS
RREYF----------------Y--KY-FTD---------ERQAIRAFGLIKT-NKELWAL
CLVPWVSWLACTCLMEQMKRKE-KL-T--LT-SKTTTTLCLHYLSQALQ--------AQ-
-----------PLGPQLGDVCSLA---AEGI-WQ-KK----TLFSRD-------------
--DLRKHELDGA-IISTLLKM----GILQEH---PIPLSYSFIHLCFQEFFAAMSYALE-
D------KKG--------RSKHA--NCI-IDLE-KLLEAYGTH--GLFGAPTTRFL----
----LGLLSDEGKRAMENIFNCR---------------L--SQG-RN---LMQWVPS---
------------------------------------LWPLLQPHSLDFLHCLYETQN-KM
FLTQMMAN---------------------F-------------------QEMGMCVETDM
EL---LVC--T----FCIQ-FCRH-VKK-LQLI----------------EGRQ-------
------H-R---------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------P------------
------AWSP--------------------------------------------------
-------------------TGIVLFRWVPVT-----------------------------
----------------------------DAYWEILFS-----------------------
--------------------------------VFKVT
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
RNLKE---LD-LSGNS-LSH---
-------SAVKSLCKTL----RRPRCLLETLR----------------------------
------------------------------------------------------------
-------------------------------LASCGLTAEDCKDLASG-LRV
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
NQNLTVLD
L-SFNVLTDAGAR-HLCQRLSWP--RCT----L---------------------------
-----------------------QRLQL--VSC-----GLTSGCCLDL------------
-------AYML-STSPI
PF13516 // ENSGT00900000140813_0_Domain_15 (2358, 2378)
LMELDLQQNNLGDTGVRL-LCEGFR-HP-AC-QLTRLG--LV-
--S-CGLTSGCCQ--------------------DLASVLSA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2465)
SPFL-MELDLQQNNLGDTG
VRLLENSGT00900000140813_0_Linker_17_18 (2465, 2716)
CE--------GLRHPACQ--LT--R-------------------------------
---LGLDQA-TLSDEMRQE------LR----------ALE-QKKPELLIFS-RWKPTGMI
PDEGLGTGETSNS-----------------TSSLKRQRLGSE------------------
-----------------GSSPEVEQ-VEPLCLPSPASLGD-QD-TPLG-TDDDFWGPTGP
VATEVVDK-ERSRYRPF13553 // ENSGT00900000140813_0_Domain_18 (2716, 2997)
VHFPVAGSYRWP---NTGLCFVVREAV-------TTEIEFCVWDQ
FLGEINPQHSWMVAGPLLDLKAEPGA-VEAVHLPHFVALQGG--------HVDTSLFQVA
HFKEEGMLLEKPARVELHHIVLENPSFSPLGVLLKVIHGTLRFFPVNSLVLLYHRLHPEE
V-TFHLYLIPSDCSIRKAIDDEETKFQFVRIHKPPPLTPLYMGCRYTVSGS-------GL
GMLEILPKELELCYRSPGEAQLFSEFYVGHLGSGIRLQMKDKKDETLVWEALVKPGENSGT00900000140813_0_Linker_18_19 (2997, 3052)
DLAP
AATLVPP-----------------------------APIALPSSLDAPGLLPF00619 // ENSGT00900000140813_0_Domain_19 (3052, 3134)
HFVDQYREQ
LIARVTSVEVVLDKLY-GQVLSQEQYERVLAEDTRPSQMRKLFSLSQSWDRRCKDQLYQA
LKETHPHLIMELWEKGSKKGLLPLSS----------
ENSOARP00000017438 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------MTHLSSSSFFS
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 326)
DFG---LLLYLEELN--K---QELIRFKSFLKN------
---------E--TLEPG--------S--CRI-PWSE-VKKAK-RKDLADLMSKYYPGEQA
WKVALSI----FG-KMNLKELCERAK-AEINWTVQ-TMRPEDTEA-GE------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------VQGDQEAVLGDGAEYRIQ-
--------IREK----------------FHIM--LDKNHLLGKSENFCHEIA--------
----------------------------------------------QGGRELLERLFEED
VG--TREQP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
QTVVLQGAAGIGKTTLVRMM-----MLDWA--Q---------GNLYQ
Q--------KFTY-VFYLNAREINQLR-------ERSFVQ-LLSK-DW-----PS-TEGP
IERIMS-QP----SRLLFIIDSFDE-LNFAFEEP------EF-V-----LCTDWTQV-HP
-VSFLMSSLLRKVMLPESSLLVT-TKLTAWGKLKPLL-K----N-QH--SLQLLGMSEDA
RQEYI----------------Y--QF-FED---------QSWALQVFSSLRN-NVMLFNM
CQVPAVCWIVCNCLEQQMEKGG-DI-T--LT-CKTTTSLFACCISSLLTQV-DRSFPGLP
------------SQTQLRSLCHLA---AKGV-WT-MT----YVFYKE-------------
--NLRRHGLVKS-DVSIFLDT----NILQKDTV-YE-NCYVFTHLHIQEFFAAMFYVLQ-
DSWDT--------------RDDS--LQSFENLE-LVLESSSYK--DPHLLQMKCFL----
----FGLLNEDRMKQLEETFNCT---------------M--SLE-VKW-MILQWMETLGN
SE-----------------------------------QLPSHLVFLELFAHLYETQD-EV
FVSQAMRY---------------------F-------------------QKVVINICEEI
HL---LLS--S----FCLK-HCQC-LRT-MKLM----------------VTE-----VFE
--K--KML----NSSF-----PP--E------------------------TWQR-ADSRI
FHWWQDLC-SVLHTNEHLREVILCHSNLDEF---------AMKILN--QELRHP------
NC--K-LQKLLLRF---------TSF-PDVCQGISGSL----------------THNQNL
IHLD-LKGT-DIGDD-GVKSLCEALKHPDCK----LQNL--------SLE---------S
CN----------------------------------------------------------
---------LT---TV-CCL---NISKALVR-SQSLLSLNLSTNNLLD-DGVKLLCEG--
LMH-PKCNLEKL-S----------------------------------------------
------------------------LESCGLT-----------------------------
----------------------------VACCEDLSL-----------------------
--------------------------------VLISNKRLTH---LC-LADNI-LGD---
-------GGVKLMSEAL----KHPQC----------------------------------
--------------------------ILQ-SLV---------------------------
-------------------------------LRHCHFTSLSSESLSAS-LLHNKSLKHLD
L-GSNCLQDDGVK-LLCDAFRHP--SCS----L---------------------------
-----------------------QDLEL--MGC-----VLTSACCLDL------------
-------ASAI-LNNPNLQSLDLGNNDLQDDGVKL-LFEALR-HP-NC-NIQRLG--LE-
--H-CGLTSLCCQ--------------------DLSSTLSSNQSL-IKISLTVNTIEHEE
ITKLSE--------ALRSTECK--LQ--V-------------------------------
---LGGKE---ALEETQKL------LE----------AEG-SSNLRLAVKQYCNDHE---
--E---------------------------------------------------------
--------------------------------------------------EDGS------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSSARP00000006776 (view gene)
ENSNLEP00000010087 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----MSLD--I----RSLDIQCEE-LSD-ARWA----------------EL---------
----LPLL-------------------------------QQCQVV------RLDDC-GLT
EARCKDIS-SALRV
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1542)
NPTLAELNLRSNELGDA---------GVHCVL--QGLQS-P-----
SR--K-IQKL--------------------------------------------------
----------------------EGLLDPQCR----LEKL--------QLE---------Y
C------N----------------------------------------------------
---------LS---AA-SCE---PLASVLRA-KPDFKELTVSNNDINE-AGIRVLCQG--
LKD-SPCQLEAL-K----------------------------------------------
------------------------LESCGVT-----------------------------
----------------------------SDNCRDLCS-----------------------
--------------------------------VVASK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2053)
ASLRE---LA-LGSNK-LGD---
-------GGIEELGPGL----LHPSS----------------------------------
--------------------------RLR-TLW---------------------------
-------------------------------IWECGITAKGCGDLCRV-LRAKE
PF13516 // ENSGT00900000140813_0_Domain_13 (2215, 2237)
SLKELS
L-AGNELGDEGAR-LLCETLLEP--GCR----L---------------------------
-----------------------ESLWV--KSC-----SFTAACCSHF------------
-------SSVL-AQ
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NKFLLELQISNNRLGDAGVRE-LCQGLG-QP-GS-VLRVLW--LA-
--D-CDVSDSSCS--------------------SLATTLLA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2464)
NHSL-RELDLSNNCLGDAG
VLQLVE--------SVRQPGCL--LE--Q-------------------------------
---LVLYDI-YWSEEMEDR------LQ----------ALE-KDKPSLRVIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCCAP00000038497 (view gene)
ENSNGAP00000014801 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-KDMAGTSSSSTETR--NALAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLAALEDLT--Q---EQLKRFCYKLRD------
------------AQLDG-----------RSI-PRGR-LEHVD-RVDLVERLTQFYGPEPA
LDVTRKV----LK-KADARDVAARLK-AQRLEG---------L---------GPS-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------CS-ALLSVSKYKKKYREH-
--------VLLQ----------------HAKV--KERNARS------------VKISKRF
TKLLIAPGM---EDE--VLGTEE-------EPEP--------ERARRSDSHTFNRLFRGD
E---EGQRP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYH
G--------LVDF-AFFLLCREVLERPG------ECSLAD-LILD-QC-----PD-RVAP
VRQMLA-QP----QRLLFILDGADE-LPGLGAP--------EAA-----PCTDPFQA-TS
-GLRILSGLLSQELLPSVRLLVT-TRTVAPGRLQGRL-S-----SPQ--CAEVCGFSDKD
KKKYF----------------F--KF-FKD---------EKKAERAYRFVKE-NETLFTL
CFVPFVCWIVCTVLQEQLQLGQ-NL-S--RT-SKTTTSVYLHFVISTLKSP-GADGP---
-----------QVPRELCKLCLLA---REGILKG--Q----AHFSEK-------------
--DLERLKLQGSQVQTLFLSKKEVPDVQEK-----K-VTYSFIDLSFQEFLAALSYLLE-
DKGAP-------------------G-APVGGVQ-MLLNSDAEL--RGHLVLTTRFL----
----FGLLNVEWIGDIERHFGC-----------------VVPER-VKQ-DTLRWVQGQGR
RR--VAPVGAQRNDKLEDTEKPEEEEEE------EEEKEEEINLGLELLYCLYETQD-DT
FVHQALSS---------------------L-------------------PELVLERVRFS
WM-DLEVL--G----YCVK-RCPE-GQA-LKLV------------------SC----GLV
------AA----------------------------------------REK--K---KKN
----------------------------------------LVKR----------------
------LKGS----------------------------------------------RATK
KQ---PLAS-------LLHPLCEVMIDQQCN----LSSL--------TLS---------C
C-----------------------------------------------------------
--------KLH---DA-VCQ---DLAKALRV-ASNLRELGILQCRLT-KIGLHLLSEG--
-LVWPKCQVQTL------------------------------------------------
----------------------------RMQ-----------------------------
---------------LLDL---------REAIQYL-------------------------
------------------------------VIVLQQSPVLTT---LD-LSGC-QLSGPMV
--T--------NLCSAL----KHPTCCLKTLSLA--SVELSEL-----------------
----------SLRELRAVKTAKP-------NLT----------------------I--MH
SAL---------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSFCAP00000001890 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---ESFFSDFG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 322)
-LLWYLEELR--K---EEFWKFKELLKQ------
---------E--PLQLE--------L--KPI-PWTE-LKKAS-REDVAKLLGKHYPGKQA
WELTLNL----FL-QIERRDLWTKAQ-EEIRN----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------KPNPYRNH-
--------MREK----------------FRLI--WEKETCLPVPSD--------------
---------------------------------F-------YKETTKNEYETLKAAYP-A
SQ--AGEPT----P
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVLQGPEGIGKTTLLRKA-----VLEWA--E---------GNLWK
D--------RFAF-IFFLNGCEMNAIA-------ETSLVE-LISR-DW-----PQS-SEP
VEDIFS-QP----ERILFIMDGFEE-LKFDLE-I------TT-Q-----LCDDQKQR-KP
-MQIIVRSLLQRKMLPKSSLLVA-LGTVGMRRYYFLL-Q-----HPK--YIVLPGFSHHE
RKLYF----------------Y--HF-FRE---------RSKAMKAFSFVRD-NVPLCMF
CHNPLVCWLVCTCMKWQLEKGK-DL-E--VA-SESTTSLYASFLINVFKSRN-ENCPPK-
-----------QSRARLKGLCTLA---AEGI-WT-RT----FVFRHG-------------
--DLRRSGVSES-DTLMWVGM----RLLLRNG-----HCFTFKHLCIQEFCAAMFYLLR-
QPEDN-----------------P--DPAIGSVT-QLVTAGVSR-VQSHLSQMITFL----
----FGVSTGKITNMLETSFGFL---------------L--SKE-IRQ-EITQCLKSLSQ
CD-----------------------------------TNQVVVNFQELFSGLFETHE-KE
FVRQVMDF---------------------F-------------------EEVDIYIGNIE
EL---VIS--A----FCMK-HCQN-LQK-LHLS----------------IEN-----VFS
-DDSGSIT-----------N-------------------D-E-------EK---------
LSFWKDLC-SVFTANKNFQMLDLDNCMFDEP---------SLEILW--KALAHP------
VC--K-LQKFGYNF---------ASY-VGNGPDFFEAI----------------LHNPYL
KCLN-LHGT-RLSLM-NVRQLCETLKHPVCI----VEQL--------ML-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------GRCDIT-----------------------------
----------------------------AKACDDIAS-----------------------
--------------------------------VLVCNKNLKL---LS-LVENP-LRN---
-------EGMLMLCDAL----KHPDCALETL-----------------------------
------------------------------------------------------------
------------------------------LLMCCGLTSVACDYLSQA-LV
PF13516 // ENSGT00900000140813_0_Domain_13 (2212, 2237)
YNTSLSVLD
L-ESNLLEDSGVA-SLCEALKHP--NCN----I---------------------------
-----------------------EQLWL--SDC-----YLTPLCCKHL------------
-------SAVL-VC
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NEKLKTLKLGSNDIQDAGVKE-LCEALK-HP-NC-KVETLG--LE-
--I-CHLTTACCE--------------------ALASALTGCKSL-RNLNLDWISLDHDG
AVVLCE--------AMCHLDCT--LQ--R-------------------------------
---LGLDKS-AFDEETQML------LT----------AVE-EKNPHLTISHQLWVNE---
--EYR-----VR------------------------------------------------
--------------------------------------------------GVLS------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCAPP00000012130 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------MIRGT-R-LT
QC-ACLSS---------------------F-------------------SGLNIWWSIQS
GR-QHCVP--A----MSLEIRCEQ-LSD-TRWT----------------EL---------
----LPLI----QQ---------------------------HQVV------RLQDC-GLT
EERCRDIS-SALQANPTLIELCLSTNELGDA---------GVHLVL--QGLQS-P-----
TC--K-IQSLS--L-PYCGLT--ET---GCG-ILPSVL----------------PSM
PF13516 // ENSGT00900000140813_0_Domain_6 (1618, 1642)
PTL
RELR-LSGN-SLGDG-GLRLLENSGT00900000140813_0_Linker_6_7 (1642, 1650)
SRGLLDAQPF13516 // ENSGT00900000140813_0_Domain_7 (1650, 1766)
CH----LEKL--------HLD---------Y
C------S----------------------------------------------------
---------LS---DA-SCE---PLASMLRA-KTNIKKLVMSNNDIHE-AGIRMLLSG--
LKD-SACPLETL-W----------------------------------------------
------------------------LENCGVT-----------------------------
----------------------------AATCKDLCD-----------------------
--------------------------------VVAA
PF13516 // ENSGT00900000140813_0_Domain_11 (2017, 2054)
KPSLQD---LD-LGNNR-LGD---
-------AGLAVLCSQL----LHPSC----------------------------------
--------------------------RLR-KLW---------------------------
-------------------------------LWECDITTEGCKNLCQV-LMA
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
KQSLKALS
L-MLNRLGDEGAR-LLCEALREP--TCQ----L---------------------------
-----------------------ECLWV--KEC-----GFTAACCPYF------------
-------REVL-A
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
QNKFLTELLLSENNLGNTGVQE-LCQALC-QP-GS-VLQVL------
-----------CS--------------------NLALVLLA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2464)
CHSL-RELDLSNNGLGDPG
VLQLVE--------SLRQPSCT--LE--C-------------------------------
---LLLFDI-YVSDEVRDQ------LE----------ALE-KEKPPLRIIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMLUP00000017257 (view gene)
ENSCANP00000036520 (view gene)
ENSMNEP00000020184 (view gene)
ENSGGOP00000007590 (view gene)
ENSTSYP00000002782 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------MTDSSLSYSFFSDFG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 323)
-LLLYLEELN--K---EELNKFKSFLK-------
---------E--TMEPG--------H--CLI-PWTE-VKKAR-REDLANLMKKYYPGQEA
WNVALKI----FG-KMNLKSLCERAK-AEINWSAQ-SVGPEDTKS-GE------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------TC-DRKAVLGDGIEYRIR-
--------IKEK----------------FCNG--RDKNSLLGEPEDFHHELS--------
----------------------------------------------QENRKLLEHLFDVD
VK--TGEQP----Q
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
IVVLQGAAGVGKTTLMRKA-----MLDWA--E---------GNLYQ
Q--------RFTY-IFYLNGREINQLR-------ESTFVQ-LISK-DW-----PS-TESP
IEGIMS-QP----SGLLFIIDSFDE-LSFAFEEP------EF-A-----LCEDWTQE-HP
-VSFLMSSLLRKVMLPESFLLVT-ARHTASKRLKPLL-K----S-YR--YVELLGMSVNA
REEYI----------------Y--RF-FED---------KKWAMKVFSSLRN-NEVLFSM
CQVRLVCWVFCTCLKQQMEKGG-DV-T--LT-CQTTTALFTCYISSLFAPE---RSPRLP
------------NQNQLRSLCHLA---AKGI-WT-MT----YVFYRE-------------
--NLRRHGLTKY-DVSIFLDM----NILQKDTE-YE-NCYVFTHLHVQEFFAAVYYLLK-
GSWEA--------------RDVS--FQPFEDLK-LLIESTSYK--NPQLTQMKCFL----
----FGLLNEDRVKQLEKTFNCK---------------M--SLE-IKS-KLLQQMEVLGN
SE-----------------------------------YSPSQLGFLELFHCLYETQD-EA
FISQTMRY---------------------F-------------------SKIVINICGKL
HL---LVS--S----FCLK-HCQC-LQT-IKLS----------------ITM-----VFE
--K--KML----NTNP-----PA--E------------------------TW---DGDHI
IHYWQDLC-SVLHTNEHLRELDLYQSNLDKS---------AMKIFH--QELRHP------
NC--K-LQKLLLRC---------VTF-PDGCQDISQSL----------------I
PF13516 // ENSGT00900000140813_0_Domain_6 (1616, 1642)
HNQNL
IHLD-LKGS-DIGDN-GVESLCETLKRPECK----LQKL--------RLE---------S
CN----------------------------------------------------------
---------LT---VV-CCL---NISKALIR-
PF13516 // ENSGT00900000140813_0_Domain_8 (1773, 1796)
SQNLISLNLSTNNLLD-DGVELLENSGT00900000140813_0_Linker_8_10 (1796, 1807)
CEA--
LGH-PKPF13516 // ENSGT00900000140813_0_Domain_10 (1807, 1955)
CYLERL-S----------------------------------------------
------------------------LESCGLT-----------------------------
----------------------------DAGCEDENSGT00900000140813_0_Linker_10_11 (1955, 2016)
LSL-----------------------
--------------------------------ALIPF13516 // ENSGT00900000140813_0_Domain_11 (2016, 2054)
SNKKLTH---LC-LADNV-LGD---
-------GGVKLLSNVL----QHPQC----------------------------------
--------------------------TLQ-SLV---------------------------
-------------------------------LRCCHFTSLSGEYLSIS-LL
PF13516 // ENSGT00900000140813_0_Domain_13 (2212, 2237)
HNKSLTHLD
L-GSNWLQDGGAK-LLCDVLRHP--SCN----L---------------------------
-----------------------QDLEL--MGC-----VLTSACCLDL------------
-------ASVI-L
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2374)
SNPNLWSLDLSNNDLQNEGVKI-LCDALR-HP-SC-NIQRLG--LE-
--Y-CGLTSLCCQ--------------------DLSSVLRSNQQL-IKMSLTQNTFGCEG
IMELCK--------VLRSPKCK--LQ--V-------------------------------
---LGLCKE-AFDEKSQKL------LE----------AVG-VSNPHLIIKPDCNDQN---
------------------------------------------------------------
--------------------------------------------------EDMSWWRCF-
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSVPAP00000004578 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---ESFF
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
PDFG---LLWYLEELK--K---EEFWKFKELLKQ------
---------E--PLKFE--------L--KPI-PWTE-LKKAS-RESLSELLSKHYPGKLA
WEVTLSL----FL-QVNRRDLWAKAQ-EEMRXXXXXXXXX--XX--------------XX
X-----------------------------------------------------------
------------------------------------------------------------
--------XX-------------------------------XXXXXXXXXXXXXXXXXX-
--------XX--------------------------XXXXXXXXXX--------------
---------------------------------X-------XXXXXXXXXXXXXXXXX-X
X-----XXX----XXXXXXXXXXXXXXXXXXXX-----XXXXX--X---------XXXXX
X--------XXXX-XXXXXXXXXXXXX-------XXXXXX-XXXX-XX-----XXX-XXX
XXXXXX-XX----XXX--XXXXXXX------X-X------XX-X--------XXXXX-XX
-XXXXX----XXXXXXX---XXX-XXXXXXXXXXXXX-X-----XXX--XXXXXXXXXXX
XXXXX----------------X--XX-XXX---------XXXXXXXXXXXXX-XXXXXXX
XXXXXVCWLVCTCLKCQLEKGE-DV-Q--MD-SESITSLYASFLMSVFKSVS-ENCAPK-
-----------QNRARLKGLCALA---AEGM-WT-HT----FLFCPE-------------
--DLRRSGVSEA-DTLVWVDV----RLLQRSG-----GCFSFIHTCIQEFCAAMFYLFQ-
RPKDG-----------------P--HPAIGSMA-QLVAAAVND-VQTYWSWVGIFL----
----FALSTKKITTVLETSFGFP---------------L--SKE-LRQ-EITQCLKTLSR
FD-----------------------------------PDQAVVSFQALFNCLFETQE-EE
FVAQVMNV---------------------F-------------------EDVSVYIGSRD
EL---TMA--A----FCLK-HCQN-LQV-FRLC----------------IEN-----IFS
-DASEPIS-----------S-------------------K-I-------QV---------
LLLWRDLC-SLFTTNKNFQMLDLDNV-SDEA---------SLAILC--KTLAQP------
AC--K-LQKFVCNF---------ASN-LINGLDFFKAV----------------LHNP
PF00560 // ENSGT00900000140813_0_Domain_6 (1619, 1645)
HL
KYLN-LSGT-GLSHP-DVRRLCETLRHPMCS----IEKL--------XXXXXXXXXXXXX
XXXXX-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------XXX-XXXXX-----------------------
--------------------------------XX----X---------XXXXX-XXX---
-------XX---------------------------------------------------
---------------------------------------------------XXXXXXXXX
---------------------XXXXXXXXXXLMQCCLTSAACGAISQA-LLCNRSLTLLD
L-GSNFLEDAGVT-SLCAALKHP--DCP----L---------------------------
-----------------------RELRL--EGC-----GLTSACCGDI------------
-------AAAL-TSNERLQTLKLGNNGLRDAGARR-LCAALR-HP-SC-KLANLG--LG-
--I-CELTSACCE--------------------DLASALTVCS-L-RGLDLNLI-INYEG
ALVLRE--------ALTHPECE--LQ--T-------------------------------
---LGLQTS-KLSEDIKLL------LT----------EVE-ERKPRLTISPSLWVED---
--MSR-----VR------------------------------------------------
--------------------------------------------------GTAI------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMLUP00000008763 (view gene)
ENSOGAP00000010273 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------S--T----MSLDIQCEQ-LSD-TRWT----------------EL---------
----LPLI-------------------------------QQCQVV------RLDDC-GLT
EVRCQDMS-SALRANPALTELDLRTNELGDA---------GVRLVL--QALQG-P-----
TC--T-IQKLS--L-QNCCLT--EA---GCG-VLPSML----------------RSVPSL
RELH-LSDN-PLGDA-GLKLLCEGLLNPQCH----LEKL--------QLE---------Y
C------N----------------------------------------------------
---------LT---AA-SCE---FLASVLRA-KPDFKELIVSNNDLSE-AGVRTLCQG--
LKD-SACPLEAL-K----------------------------------------------
------------------------LENCGVT-----------------------------
----------------------------AANCRDLCS-----------------------
--------------------------------AVASKTSLQA---LD-LGSNR-LGD---
-------AGLTELCQGL----LRPSF----------------------------------
--------------------------RLR-TLW---------------------------
-------------------------------LWECGITAEGCHELCRM-LCSLESLRELS
L-AGNELGDAGAQ-LLCESLLAP--GCR----L---------------------------
-----------------------ESLWV--KTC-----SLTAACCPLF------------
-------SSVL-AQNKCLLELQMSSNPLGDVGVQT-LCQGLG-QP-GT-VLRVLW--LG-
--D-CDVSDVGCS--------------------SLASTLLANH
PF00560 // ENSGT00900000140813_0_Domain_17 (2444, 2457)
SL-RELDLSNNRMGDEG
VRQLMG--------SLRQPGCA--LE--Q-------------------------------
---LVLYDI-YWSEEMDNQ------LR----------ALE-EDKPSLRIIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSECAP00000015250 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MPQI----PVRDGLC
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 328)
RLSAYLEELE--A---VELKKFKLYLGT------
---------A---TEPG--------E--AKI-PRGR-MEQAG-PLEMAQLLVAHCGMQEA
WLLALSI----FE-RINRKDLWERGR-REDLVRDT----P--PG--------GLSSL---
------------------------------------------------------GS----
------------------------------------------------------------
---QS-ACSL-------------------------------EVFPGTPRKDPRESYRNH-
--------VRRK----------------FRLM--EDRNARLGECV---------NLSQRY
TRLLLVKEHSNPTWAQQKLL------DTGRGHTRTVG--------QQASLIQMETLFEPD
EE--RPEPP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
RTVVLQGAAGMGKSMLAHKV-----MLDWA--D---------GKLFQ
D--------RFDY-VFYIHCREMNQNAS------ERSAQD-LISS-CW-----PE-PGAP
LQELLR-AP----ERLLFILDGFDE-LKPSFHDP------QG-P-----WCLCWEEK-RP
-TELLLSSLIRKKLLPELSLLIT-TRPTALEKLHRLL-E-----HPR--HVEILGFSEAE
RKEYF----------------Y--KY-FHN---------AEQAGQVFHFVRD-NEPLFTL
CFVPMVCWVICTCLKQQLEGGR-LL-R--QT-SRTTTAVYTLYLLSLMQPKPGT-PTL--
-----------QIPPNQRGLCSLA---ADGL-WN-QK----ILFEEQ-------------
--DLRKHGLDGV-DVSSFLNM----NIFQKDIN-CE-RFYSFIHLSFQEFFAAMHYILE-
D--GE--------------S--G--SGPDQNVA-RLLAECGFS-ERSFLALTVRFL----
----FGLLNEQTRSYLEKSLCWK---------------V--SPR-VKA-ELLEWIESKAQ
SE-----------------------------------GSMLQQGCLELFSCLYEIQE-EE
FIQQALSH---------------------F-------------------QVVVVSNIA-T
RM-EHMVS--C----FCVK-NCRN-VVV-LHLC----------------GA------AYS
-ADEEDGL--R-GA---------------------------------------EGP---Q
A-------------------------PL------------------------A-------
Q---P--------PERNILPD-------AYSEQLASAL----------------STNPNL
IELV-LYRN-ALGSR-GVKLLCQGLRHPNCK----LQNL--------RLK---------R
C-----------------------------------------------------------
--------QLS---GP-ACW---DLAAALIA-NKNLIRMDLSGNGLGL-PGIRLLCEG--
LRH-PRCRLQMIQ-----------------------------------------------
------------------------LRKCQLE-----------------------------
----------------------------AGACREIAS-----------------------
--------------------------------VLSTNRHLVE---LD-LTGNA-LED---
-------LGGRLLCEGL----SNPVC----------------------------------
--------------------------RLQ-ILW---------------------------
-------------------------------LKICRLTAAACEDLAST-LSVNQSLVELD
L-SLNDLGDAGVL-LLCEGLRHP--QCK----L---------------------------
-----------------------QTLRL--GIC-----QLSSAACEGL------------
-------SAVL-QVNPHLRELDLSFNDLGDRGVWL-LCEGLR-HP-TC-RLQKLW--LD-
--S-CGLTAKACE--------------------ALSCSLGVNRTL-RELYLTHNALGDTG
VRLLCK--------RLSHAGCP--LR--V-------------------------------
---LWLFGM-ALNPMTHRR------LA----------ALR-VAKPSLDLGC---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSFDAP00000022072 (view gene)
ENSCCAP00000002303 (view gene)
ENSLAFP00000001775 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----PSSV--T----MNLDIQCEH-LSD-TQWK----------------EL---------
----LPRI-------------------------------QQVEVI------RLDDC-GLT
EARCKDIS-SALQANPSLTELSLHNSELGDA---------GVCQVL--QALQG-P-----
AC--K-ICKLS--L-QNCSLT--AA---CCT-VLPGTL----------------RWLP
PF00560 // ENSGT00900000140813_0_Domain_6 (1619, 1645)
TL
LELR-LSDN-PLGDE-GLQLLCEGLRDPQCH----LERV--------QLE---------Y
C------N----------------------------------------------------
---------LT---AA-SCE---PLAAVLPA-KL
PF00560 // ENSGT00900000140813_0_Domain_8 (1775, 1801)
GLRELVLSNNDLGE-AGVQVLCRG--LLG-ATCPLETL-K----------------------------------------------
------------------------LASCGVT-----------------------------
----------------------------AANCKDLCS-----------------------
--------------------------------IVATKDSLQE---LD-LGESK-LGD---
-------AGMAALCPGL----LSPSS----------------------------------
--------------------------KLK-TLW---------------------------
-------------------------------LWECDISSEGCREISQV-LRAKESLKELS
L-MGNELGDQGAR-LLCEALREP--GCR----L---------------------------
-----------------------ESLWV--KAC-----GFTDACCPDF------------
-------STML-AQNKFLLELQLSNNKLGDTGVQQ-LCQGLS-QP-GA-TLQMLC--LG-
--D-CDVTDSGCT--------------------ALASLLLSSR
PF00560 // ENSGT00900000140813_0_Domain_17 (2444, 2476)
SL-RELDLSNNCMGDAG
VLQLVE--------SLRQPGCA--LQ--Q-------------------------------
---LVLYDT-YLSEAVDDS------LQ----------ALE-ESKPSLRIIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPTRP00000005793 (view gene)
ENSMMUP00000007829 (view gene)
ENSCANP00000036805 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------MVSST----QVNFN--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 323)
-LQALLEQLS--Q---DELSKFKSLLRT------
---------V--SLGNE--------A--QKI-PQKE-VDEAD-GKQLAEILISHCHSYWT
ELATIQV----FE-KMHRMDLSERAK-DELREAAL-KSLN----KNK------PLSLG--
------ITRKEREP---------------------------------LDVEEMLERFK-A
GTLECT---------------------------------ET--EEDVPV-----------
-------LTE-------------------------------VFKVKGEKSDNENRYRLI-
--------LKTK----------------FREM--WQ--SWPGDSKEVRV-----------
---------------------------------------------MAERYRMLIPFSNPR
ML--PGPFS----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
HTVVLHSPAGLGKTTLANKL-----MLDWT--E---------DNLIQ
---------KFKY-AFYLSCRELSRLG-------PCSFAE-MVFR-DW-----PE-WQED
IPHILA-QA----QRILFVIDGFDE-LGAPPG------ALIQ-D-----LCGDWEQQ-KP
-VPVLLGSLLKRKMLPKAALLVT-TRPRALRDLRFLA-E-----QPI--YIRVEGFLEED
RRAYF----------------L--RR-FGD---------EDQAMRAFELMRS-NAALFQL
GSAPAVCWIVCTTLKLQMEKGE-DP-A--PT-CLTSTGLFLRFLCSQFPQ-------GAQ
------------LRGALRALSLLA---AQGL-WA-QM----SVFHGE-------------
--DLERLGVQES-DLRPFLDR----DILCQDRA-SK-GCYSFIHLSFQQFLTALFYALE-
KE-EE-------------DRDGH--AWDIGDVQ-KMLTREERL-KNPDLIQAGRFF----
----FGLANEKRAKELEATFGCQ---------------M--SPE-IKQ-ELLRCDVSRKN
G------------------------------------T---VADLKELLCCLYESQE-DE
LVKEVMAQ---------------------F-------------------KEISLHLN-AV
DI---ASS--S----FCVK-HCRN-LQK-MALQ----------------VIK-----ETL
-PENVAAS----ES--------------------NAEPER------------SQDD-QDM
LPFWTDLC-SIFGSNKDLMGLEINNSFLSAS---------LVKILC--EQIAS-D-----
NC--H-LQRVV--F-KNLSPA--DA----HR-NLCLAL----------------RGHKTV
THLT-LQGN-DQKD--MLPALCEVLRHPECN----LRYL--------GLV---------S
C------S----------------------------------------------------
---------AT---TQ-QWA---DLSLALEA-NQSLMCVNLSDNELLD-EGAKLLYTT--
LRH-PKCFLQRL-S----------------------------------------------
------------------------LENCHLT-----------------------------
----------------------------EANCKDLAA-----------------------
--------------------------------VLVVSRELTH---LC-LAKNS-LKD---
-------TGVKFLCEGL----SYPEC----------------------------------
--------------------------KLQ-ALV---------------------------
-------------------------------LWNCDITSDGCCSLAKL-LQEKSSLSCLD
L-GLNHIGVTGVK-VLCEALSKP--LCN----L---------------------------
-----------------------RCL----------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSOCUP00000023322 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------MKTK----------------FHLI--WEEETCLQVPEH--------------
---------------------------------F-------YRETVFNEYISLYVAYK--
------VGP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 929)
TVVLSGPKGSGKTTLLRKL-----MLEWA--N---------GNLWR
D--------RFAF-VFFFSVYELNGVA-------ETSLAE-LVSR-DW-----PAS-EEV
LEDVFA-QP----VKILFIMDGFEE-LKFDLE-Q------QT-D-----LCSDWRQR-KP
-TQVILSSLLQKAMLPECSLLLG-YGKTSIGKYCSFL-E-----NPT--YLRLKGFSEQQ
RKLYF----------------S--YF-FGE---------KKDALRALSFVRA-IPSLFAL
CENPLISWLLCTCMKWQQERGE-QL-H--VI-FENTTSLHAFFLTGAFRVGR-ENCPPL-
-----------QNRAQMKSLCTLA---AEGI-WT-QT----YVFSQA-------------
--DLRRHGVSES-DMVMWLDV----RFLRPRG-----DGFVFNHTPLQEFCAALFYFLG-
QPGDQ-----------------L--NPAIGSIA-QLVTATMGQ-LQSRLYRMGIFL----
----FGICSGRITGMLGKWFGML---------------L--SEE-IKP-KISQCLQRLSK
GE-----------------------------------PGE-VVNFQNLSSALFEIQE-RE
FVAQVMDF---------------------F-------------------EEIFIYIDSLE
NL---AMS--S----FCLK-SSRN-VKK-LHLC----------------VDD-----DFP
-GDLGAIP-----------D-------------------H-S-------KK---------
LAYWRDLC-SVFSTSKKFELLDMDNCKLDDA---------SLTILW--KALAHP------
TC--K-IQALAFNF---------MSN-FGNGVDFSTKM----------------LLHPHL
KYLN-LYRT-NVSSI-GVRYLCEMLKKPKCN----LEVL--------ML-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------GKCDIK-----------------------------
----------------------------EDHCDDIAS-----------------------
--------------------------------VLVCNSKLKC---LS-LVENP-LNN---
-------TGVMILCKGL----KRPECVLESL-----------------------------
------------------------------------------------------------
------------------------------ILNCCCLTSVSCDYFSKA-LLCNRALSLLD
L-GSNMLEDTGVA-TLCEVLKHQ--NCT----L---------------------------
-----------------------KELWL--VGC-----YLTADCCKDI------------
-------VATL-ICNESLKTLKLGSNEIQDAGVRQ-LCEALR-HP-NF-RLQRLG--LE-
--M-CQLTTACVE--------------------DLASALITCKSL-KGLNLDGIMLDHDG
VVKLCE--------ALTHVDCV--LE--L-------------------------------
---LGLDKT-AYGKESWQL------LS----------AAE-ERKPDLTIQHEPWAAE---
--ENK-----MK------------------------------------------------
--------------------------------------------------GVAL------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMEUP00000004119 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------ERNKYRER-
--------MRKK----------------FQLM--RDRNHRPGENE---------LFQHRF
TQK------------------------------------------D-G--DMLEHLFDPD
QK--AGIQP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
HTVVLQGAAGIGKTTLAIKV-----MLDWA--E---------GKLYQ
E--------RFDY-VFYLTCRELNTIGV-----NVLSFAH-LIAR-HW-----PG-LQAP
MTEIMS-QP----ERLLFIIDGLDE-LKFPCNEN------RC-D-----LCQNWNQK-RP
-VSILLSSLLRKMLLPEASLLLT-TRLTALGKFSPLL-E-----NPR--HVEILGFSVSQ
RTEYF----------------C--KF-FRN---------EKLGKKAFDLVEG-NAPLFTM
CSVPLMNWIICTCLKQQEERGQ-DP-L--QA-LKTTTALYMCYFNSLT-PDD-----KNF
--------------AHLKGLCGLA---AEGI-WQ-RK----VLFEEE-------------
--DLRRHNLEAGDV-SAFLDM----NIFQKDSG-CE-NSYSFIHLSFQEFFAAMFYLMK-
NGRRA--------------L------TSPSQIS-ENLREYSEN-DASFLALVVQFL----
----FGFLNAEIASELQRKFRCR---------------T--SLE-MKS-QLLEWVERKTK
GD---------------------------------------NGYLFLVYLFSYETQD-AE
IVTQASRQ---------------------R-------------------NEVNV----FV
-P-MVLMN--T----FCIK-HCXX-XXX-XXXX----------------XXX-----XXX
-XXXXXXX-------------------------------XXXXXX------YI------F
TCYWQDLS-TVINKNQNLKELQLFCTRFADS---------YMENLS--AALRN-P-----
KC--E-LEKLT--L-KVVSLT--DA---CCQ-NLSTCI----------------K-NK
PF00560 // ENSGT00900000140813_0_Domain_6 (1619, 1645)
NL
TYLD-LSGN-SLHDC-GMRLLCEGLRHPACN----LQKL--------S------------
------------------------------------------------------------
------------------CE---YLSCALIS-NQ
PF00560 // ENSGT00900000140813_0_Domain_8 (1775, 1801)
SLTLLDLSGNNLGD-YGIKLLCEA--LGN-QYCSVNSL-M----------------------------------------------
------------------------LRSCGLT-----------------------------
----------------------------ASCCQDLST-----------------------
--------------------------------ALKRNRSVTH---LN-LADNA-LGN---
-------HGVTLLCDAL----GYPDC----------------------------------
--------------------------HLQ-EL----------------------------
------------------------------XXXXXXXXXXXXXXXXXX-XXXXXXXXXXX
X-XXXXXXXXXXX-XXXXXXXXX--XXX----X---------------------------
-----------------------XXXXL--DDC-----HLTAACCQDL------------
-------SYAL-GSNHQLKNLNLSVNSLGDDGMKL-LCEALG-NQ-DC-SIQELR--LK-
--N-CSLTT-CSQ--------------------NLFSALICNK
PF00560 // ENSGT00900000140813_0_Domain_17 (2444, 2476)
SL-KNINLSGNSLEQSG
IKLLCK--------ALKPKQQ-------V-------------------------------
---LRLWRE-AFSEDAQQM------LK----------AAQ-ESKPELIITGDWYSHD---
--A---------------------------------------------------------
------------------------------------------------EEYGFSWW----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMLUP00000019239 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------MAESDS
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 330)
SDSD---LLWYLEKLS--R---KKCHSLRNQLGQ------
---------E--CMEIG--------L--PPI-PRLE-LSE-E-NPHLANQLTTFYEAQHL
WNLMYNI----FQ-KIPRKDLCEKI---NARRN---------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------LLSVLRGDC-
--------KHDA----------------L--G--YNCKIWPKKSVGLCANIA--------
---------------------------------FL-------KESESESSHFLLFLFR--
NK--EAHKM----LIKKNLQEEA
PF05729 // ENSGT00900000140813_0_Domain_2 (684, 940)
SGKTMVLRGA-----LFEWA--N---------GNIWA
D--------MFSS-VIHLSSHDINQMT-------SGSLVP-LISK-DW-----PS-GQAP
VADILS-EP----QKILFILEDLDD-VELSFRIS------PQ-A-----LCTNSRQQ-VS
-MPILMASLLTRKLAPGCRFLIT-SRPQGQAVTWTIM-K-----KTD--MLMTLSFSHEK
RHKYF----------------T--LF-FKD---------GQSAESAFQLVQR-NELFLHL
CQAPILCWVTCVALLRQMNQG-ADL-K--RS-CQTLTDIYTNFLADTLTAQA----AH--
----------QQPIVPLQSLCSLA---LEGL-LH-NT----LHFTEE-------------
--DLRSVGFTQA-DVSTLQTL----EILWPSSH-LK-GHYRFIHSKVQEFCAAVAFILPL
AEHGI-----------------P----SAWG-R--C--KGKRE------CPVIPSM----
----FDL-NKKRRKTFEKALGCH---------------LL-TDY-ISQ-YLLMERSHWAP
---------------------------------------------HTLFHGLFENQE-EE
YMKWILDS---------------------F-------------------SEANIYVRDHQ
DL---MVS--S----YCL--YCQH-LQK-LKLS----------------IQN-----IFE
--KRS-NE-------------------------------------------QSTPRYQLL
LMGHKVSI----------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
MGP_PahariEiJ_P0018223 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------MKTEDDE-MEY------------
------------------------------------------------------------
----------------------EA------------------------------------
-------SKQ-------------------------------EISSEDKDFDDGIDYRTV-
--------IKEK----------------IFTM--WYKTSSPGEFATLNCIIT--------
----------------------------------------------HKDQNLLQHIFDED
IQ--ASKAP----Q
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TIVLQGAAGIGKTTLLKKA-----ILEWA--D---------GNLYQ
---------QFTH-VFYLSGREISQVK-------EKSFAQ-LISK-YW-----PS-SEGP
IEQVLS-KP----SSLLFIIDSFEE-LDFSFEEP------EF-A-----LCKDWTQN-SP
-VSFLISSLLRKVMLPESYLLVA-TRSTAWKRLVPLL-Q-----KPH--RVKLSGLSKHA
RMDYI----------------H--HL-LKD---------KTWAMSAIYSIRM-NWRLFYM
CHVCHVCQMICTVLKEQVEKGG-SV-E--KI-CQASTALFTYYICSLFPRI-PVGCVTLP
------------NETLLRSLCKAA---VEGI-WS-MK----HVLYQQ-------------
--NLRKHELTRE-DILLFLDA----KVLQQDAE-YE-NCYMFTHLHVQEFFAALFYLLR-
ENLEE--------------KDYP--SEPFENLY-LLLESNHIH--DPHLEQMKCFL----
----FGLLNKDRVRQLEETFNLT---------------I--SME-VRE-ELLACLEALEK
DD-----------------------------------SSLSQLRFQDLLHCIYETQD-EE
FITQALMY---------------------F-------------------QKIIVRVDEEP
QL---RIY--S----FCLK-HCHT-LET-MRLT----------------ARA-----DLK
-----NML------D------TA--E------------------------MCLEGAAARI
IHYWQDLF-SVLHTNESLIEMDLYESRIDKS---------LMKILN--EELSHP------
RC--N-LKKLMFRA---------VDF-LNSCQD-FTFL----------------A
PF13516 // ENSGT00900000140813_0_Domain_6 (1616, 1642)
SNKKV
THLD-LKET-DLGDN-GLKTLCEALKCQGCK----LRVL--------RLA---------S
CD----------------------------------------------------------
---------LN---VA-RCQ---KLSNALQT
PF13516 // ENSGT00900000140813_0_Domain_8 (1772, 1796)
-NRSLVFLNLSLNNLSN-DGVKSLCEV--
LEN-PNCPLERL-A----------------------------------------------
------------------------LASCGLT-----------------------------
----------------------------KAGCKVLSS-----------------------
--------------------------------ALTKTK
PF13516 // ENSGT00900000140813_0_Domain_11 (2019, 2054)
RLTH---LC-LSDNV-LED---
-------EGIELLSHTL----KHPQC----------------------------------
--------------------------TLQ-SLV---------------------------
-------------------------------LRSCSFTPIGSEHLSTA-LL
PF13516 // ENSGT00900000140813_0_Domain_13 (2212, 2237)
HNRSLVHLD
L-GQNKLADNGVK-LLCHSLQQP--HCN----L---------------------------
-----------------------QELEL--MSC-----VLTSKACGDL------------
-------ASVL-VNNSNLWNLDLGHNILDDAGLNI-LCEALR-NP-NC-HVQRLG--LE-
--N-CGLTPGCCQ--------------------DLLGILSNNKSV-IQMNLMKNALDHEA
IKNLCK--------VLRSPTCK--ME--F-------------------------------
---LALDKKEILKKRIKKF------LV----------DVK-INNPHLVIEPQCPN-----
--T---------------------------------------------------------
--------------------------------------------------ESGGWWNYF-
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSOPRP00000010415 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MLPGAA-CR
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
DLSL---LATYLEELE--A---MELKKFKLYLGV------
---------V--AAKLG--------E--DRI-PWGR-MEGAG-PLELAQLLVAHCGPQGA
WRLALSV----FQ-RMHRKDLWERGH-GENLAG-X-----XXXXX---------------
------------------------------------------------------------
----------------------------------------------XXX-----------
XXXXXX-XXX-------------------------------XXXXXXXXXXPQKAYRDY-
--------VRRK----------------FRLM--EDRNARLGECV---------NLSHRY
TRLLLVQEH--SSLRGAQQK----LLDRGWGQAR---T-----AAQQASPIRVEMLFEPD
EE--RPEPP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 879)
RTVVLQGAAGMGKSMLARKV-----MVDWA--D---------GRLFQ
G--------RFDF-VCYINCRDMNQAGP------EQSALD-LLAS-CW-PE-----PGAQ
VRELLQ-RP----ERLLLIIDGFH-ELQPAFHEA------RG-P-----WCFSWEER-WP
-TALLLGSLIGKKLXXXXXXXXX-XXXXXXXXXXXXX-X-----XXX--XXXXXXXXXXX
XXXX-X---------------X--XX-XXX---------XXXXXXXXXXXXX-XXXXXXX
XXXXXXXXXXXXXXXXXXXXXX-XX-X--XX-XXXXXXXXXXXXXXXXXXXXXXX----X
----------XXXXXXXXXXXXXX---XXXX-XX-XX----XXXXXX-------------
--XXXXXXXXXXXXX-XXXXX----XXXXXXXX-XX-XXXXXXXXXXXXXXXXXXXXXX-
XXXXX-----------------X--XXXXXXXX-XXXXXXXXX-XXXXXXXXXXXX----
----XXXXXXXXXXXXXXXXXXX-----------------XXXX-XXX-XXXXXXXXXXX
X-----------------------------------XXXXXXXXXXXXX--XXXXXX-XX
XXXXXXXX---------------------X-------------------XXXXXXXXX-X
XX-XXXXX--X----XXXX-X-XX-XXX--XX-----------------XXXX----XXX
------XXXXXXXXX----XXXXXXXX--------------------XXXXXXXXX-XXX
XXXXXX---XXX------------------------------------------------
------------------------------------------------------------
-------------------------XXX--------------------------------
--------------------XX----XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX-X
XXXXXXXXXXP---SS-ACQ---DLAAALTA-NDHLLRLDLSGNKLGL-PGLRLLCEG--
LQH-PRCGLQML-Q----------------------------------------------
------------------------LRKCLLD-----------------------------
----------------------------AGACQELAS-----------------------
--------------------------------VLGTNP
PF00560 // ENSGT00900000140813_0_Domain_11 (2019, 2057)
HLAE---LD-LTGNA-LQD---
-------VGVAWLCQGL----RLPGC----------------------------------
-------------------------R-LR-TLW---------------------------
-------------------------------LKICHLSGTACRELAST-LSVNR
PF00560 // ENSGT00900000140813_0_Domain_13 (2215, 2237)
SLTELD
L-SLNDLGDPGVL-LLCDGLRHP--ACQ----L---------------------------
-----------------------QTLRL--GIC-----RLGPAACEGL------------
-------SAVL-RVNLRLRELDLSFNDGDPAV--GLLGEGL-RPP-TC-RLQKLW--LD-
--N-CSLTAKACE--------------------TLSSALATNQTL-TELYLTNNALGDAG
VRLLCR--------RLHHRTCK--LR--V-------------------------------
---LWLFGM-DLNKLTHKR------LA----------ALR-TTKPYLDIGC---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSHGLP00100012129 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---MLEC----LT-LTCCGLFC--SC-APD------------LG--------------NA
K---------------------------------------------------IYQT----
------------------------------------------------------------
----------------------------------------------------------H-
--------MREK----------------FTDF--LSRESIPTMHEQ--------------
---------------------------------L-------DMNITQEECKFLELLFEP-
----EKQ-T----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
RILCLLGAQGVGKTTFLAKM-----ILAWS--K---------GFVYQ
D--------RFSY-IFYFCCKELKQLP-------ATSLAG-LITR-AW-----PGT-SAP
VSEIMA-HP----EKLLFMADSFEE-LGSDLAEP------ED-E-----LCSDWAEV-QP
-PKLLLSSLLRKKMAPECCLLVT-VNPRAGQDLEDRL-E-----DPD--IRELTGLKEFD
VKFYI----------------C--HM-FPD---------RRRALEAFNVILN-NEQLLSM
CRLPVLCWLVCTTLKQEMDKGI-DV-A--LT-CQRPTALYSSFLLDLFTPEG-ADGPSW-
-----------QGQAQLQSLCSLA---VQGM-WT-NT----MVFREK-------------
--DLRRNGIGSS-DLSALLEL----RVLLPCWE-TE-GLYKFLHSSVQEFCAALFYLLK-
SPGHH-----------------S--HPAVRGTQ-DLLLTFLEK-SRAHWVFVGCFL----
----FGLLHERECQKLDAFFGFQ---------------L--SSE-VKR-QLYQCLQQLAK
VAD----------------------------------KLQGQVSFLALFYALFEMQE-EA
FLNQVMDL---------------------M-------------------QDLAFSVFIET
DL---RVA--G----YCLK-HCSN-LRK-LSLS----------------AQG-----VFK
-EEGREQS-----------S-----------------------------------VSSSE
LLCWHQVW-SVLRTCSQLQMLQVKDGTLNES---------AMLSLC--QQLRQP------
NC--L-LENLFVNN---------VTF-HCESWTLFEAF----------------PHNPNL
KYVN-LSNT-QLGRD-DVKVLHGALSHPTSS----VEKL--------LL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------VNCNLS-----------------------------
----------------------------ADDCKALAA-----------------------
--------------------------------TLMESKKLDS---LN-LSCNN-LK----
-------KGVRALCKAL----CHPNCTLKFL-----------------------------
------------------------------------------------------------
------------------------------ALSFCTLREWCWDYLAEV-LLIS
PF13516 // ENSGT00900000140813_0_Domain_13 (2214, 2237)
KSLSHLD
L-SINSLRDEGLK-VLCDTLRYP--VCA----L---------------------------
-----------------------QILCL--MKC-----SITAEGCRDL------------
-------ASVL-TS
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
SRNLRSLQIGGNNIEDAGVKL-LCQALA-QP-SC-LLETLG--VE-
--D-CGLTSACCE--------------------DLVTVLTTSKTL-FMLNLLQNPLDHGG
VALLCQ--------GLRHPECA--LR--V-------------------------------
---LGLNSA-EFGEETQAL------LT----------AEE-ERNPDLVILDT-W------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMUSP00000072895 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------MAGSSGYG---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLKLLQKLS--D---EEFQRFKELLRE------
---------E--PEKFK--------L--KPI-SWTK-IENSS-KESLVTLLNTHYPGQ-A
WNMMLSL----FL-QVNREDLSIMAQ-KKKRH----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------KQTKYKKF-
--------MKTT----------------FERI--WTLETNTHIPDR--------------
---------------------------------N-------YHLIVEVQYKALQEIFDSE
-----SE-P----VT
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 940)
AIVAGTTGEGKTTFLRKA-----MLDWA--S---------GVLLQ
N--------RFQY-VFFFSVFSLNNTT-------ELSLAE-LISS-TL-----PES-SET
VDDILS-DP----KRILFILDGFDY-LKFDLE-L------RT-N-----LCNDWRKR-LP
-TQIVLSSLLQKIMLPGCSLLLE-LGQISVPKIRHLL-K-----YPR--VITMQGFSERS
VEFYC----------------M--SF--FD---------NQRGIEVAENLRN-NE-VLHL
CSNPYLCWMFCSCLKWQFDREE-EG-Y--FK-AKTDAAFFTNFMVSAFKSTY-AHSPSK-
-----------QNRARLKTLCTLA---VEGM-WK-EL----FVFDSE-------------
--DLRRNGISES-DKAVWLKM----QFLQTHG-----NHTVFYHPTLQSYFAAMFYFLK-
QDKDI-----------------C--VPVIGSIP-QLLGNMYAR-GQTQWLQLGTFL----
----FGLINEQVAALLQPCFGFI---------------Q--PIY-VRQ-EIICYFKCLGQ
QEC----------------------------------NEKL-ERSQTLFSCLRDSQE-ER
FVRQVVDL---------------------L-------------------EEITVDISSSD
VL---SVT--A----YALQ-KSSK-LKK-LHLH----------------IQKT----VFS
-EIYCPDH-----------C-------------------K-T-------RTSI-GKRRNT
AEYWKTLC-GIFC---NLYVLDLDSCQFNKR---------AIQDLC--NSMSPTPTVPLT
AF--K-LQSLSCSF---------MAD-FGD-GSLFHTL----------------LQLPHL
KYLN-LYGT-YLSMD-VTEKLCAALRCSACR----VEEL--------LL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------GKCGIS-----------------------------
----------------------------SKACGIIAI-----------------------
--------------------------------SLINS-KVKH---LS-LVENP-LKN---
-------KGVMSLCEML----KDPSCVLQSL-----------------------------
------------------------------------------------------------
------------------------------MLSYCCLTFIACGHLYEA-LLSNKHLSLLD
L-GSNFLEDTGVN-LLCEALKDP--NCT----L---------------------------
-----------------------KELWL--PGC-----FLTSQCCEEI------------
-------SAVL-ICNRNLKTLKLGNNNIQDTGVRQ-LCEALS-HP-NC-NLECLG--LD-
--L-CEFTSDCCK--------------------DLALALTTCKTL-NSLNLDWKTLDHSG
LVVLCE--------ALNHKRCN--LK--M-------------------------------
---LGLDKS-AFSEESQTL------LQ----------DVE-KKNNNLNILHHPWFEA---
--ERN-----KR------------------------------------------------
--------------------------------------------------GTRLVWNSRN
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSGALP00000011167 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MDLDIQCEE-MNP-SRWA----------------EL---------
----LSTM-------------------------------KSCSTI------RLDDC-NLS
SSNCKDLS-SIIH
PF13516 // ENSGT00900000140813_0_Domain_4 (1514, 1546)
TNPSLKELKLNNNELGDA---------GIEYLC--KGLLT-P-----
SC--S-LQKLW--L-QNCNLT--SA---SCE-TLRSVL----------------SAQ
PF13516 // ENSGT00900000140813_0_Domain_6 (1618, 1642)
PSL
TELH-VGDN-KLGTA-GVKVLENSGT00900000140813_0_Linker_6_7 (1642, 1649)
CQGLMNPPF13516 // ENSGT00900000140813_0_Domain_7 (1649, 1765)
NCK----LQKL--------QLE---------Y
C------E----------------------------------------------------
---------LT---AD-IVE---AENSGT00900000140813_0_Linker_7_8 (1765, 1773)
LNAALQA-PF13516 // ENSGT00900000140813_0_Domain_8 (1773, 1796)
KPTLKELSLSNNTLGD-TAVKQLCRG--
LVE-ASCDLELL-H----------------------------------------------
------------------------LENCGIT-----------------------------
----------------------------SDSCRDISA-----------------------
--------------------------------VLSS
PF13516 // ENSGT00900000140813_0_Domain_11 (2017, 2054)
KPSLLD---LA-VGDNK-IGD---
-------TGLALLCQGL----LHPNC----------------------------------
--------------------------KIQ-KLW---------------------------
-------------------------------LWDCDLTSASCKDLSRV-FSTK
PF13516 // ENSGT00900000140813_0_Domain_13 (2214, 2237)
ETLLEVS
L-IDNNLRDSGME-MLCQALKDP--KAH----L---------------------------
-----------------------QELWV--REC-----GLTAACCKAV------------
-------SSVL-SV
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2374)
NKHLQVLHIGENKLGNAGVEI-LCEGLL-HP-NC-NIHSLW--LG-
--N-CDITAACCA--------------------TLANVMVT
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2465)
KQNL-TELDLSYNTLEDEG
VMKLCE--------AVRNPNCK--MQ--Q-------------------------------
---LILYDI-FWGPEVDDE------LK----------ALE-EARPDVKIIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSOPRP00000011835 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------MAASFF
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
TDFG---LLWYLEELK--R---KEFVQFKEHLKQ------
---------E--TLQLG--------L--EPI-PWAK-VKKAS-RADLADLMMKSYE-RRT
WEVTFAV----FH-RLGRKDLCDRAR-SESTGHA--------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------------TLYQDH-
--------IRKK----------------FSPL-------LQ-------------------
-------------------P------V-----KSA-T-------CSQAEREFMSPLFAP-
GD--AGEQG----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
HTVVFQGSPGMGKTALMWKL-----LLAWA--E---------GSLYQ
D--------RFTY-IFYLCCHKLKPLA-------PMSLAE-LLAR-DW-----PD-AAAP
AAEILA-LP----ERLLFMVDSLEE-LGCDLLEP------ES-P-----LCSSWAER-QP
-IHVLLNSLLGKTLLPEASLLIA-TTPVCRETCYQ---------VQN--PKIPWGFSENH
RKLYF----------------C--NW-FQS---------NSRAMEAFRCVRE-NQYIFSM
CQNPQLCWLTCASMKEEMDRGK-DP-A--LS-CRCVTSLFVSFILNSFTPKG-ARGPPQQ
------------SWGQLKGLCSLA---AEGV-WT-NMST----FSRE-------------
--ALRRNGIM-DSDVATLLDA----RVLRRR-A-AE-DSYALLHVVVQEFCAVLFHLLQ-
RGD------------------HP--HPAVQDVE-TLAEV-PEK--ASPWIFLGCFV----
----FGLLNEREQERLDALFGCH---------------R--SQE-SKQ-QL---------
---------------LGLLQ-S---------------LDEQQVNFALLFSCLAEIQD-EA
FVQQTMES---------------------F-------------------QEVEVAVVHPF
QL---AAS--A----YCL-GHCSG-LRK-LCLS----------------IQ---------
----NVFP----EE-------------------------------DG----GGQRS-QPN
LVLWRQIC-TVLTTNRHLYAFQVKDSSLPMS---------AVVALC--HQLKQ-P-----
KC--R-LQKLQ--M-NNVSLH-------CES-GFFEVL----------------IHNPHL
TCLD-LSCT-GLSQH-SVKLLSKALKHPACN----VREL--------LXXXXX-XXXXXX
X------X----------------------------------------------------
---------XX---XX-XXX---XXX---------XX---XXXXXXXX-XXXXXXXXX--
XXX-XXXXXXXX-X----------------------------------------------
----------------------XXLASCHLG-----------------------------
----------------------------EPCWEVLSE-----------------------
--------------------------------ALQCSR
PF00560 // ENSGT00900000140813_0_Domain_11 (2019, 2056)
KLTH---LD-LSANM-LQD---
-------EGLRALSAAL----GCPSC----------------------------------
--------------------------PLQ-SLX---------------------------
-----------------------------XXXXXXXXXXXXXXXXXXX-XXXXXXXXX--
---XXXXXXXXXX-XXXXXXXXX--XXX----X---------------------------
-----------------------XXXXXXLQLC-----GLTSACCLAL------------
-------SSAL-TSSK
PF00560 // ENSGT00900000140813_0_Domain_15 (2357, 2381)
TLRSLDLSGNALDLSGLAV-LASALR-QP-TC-ALEVLG--LK-
--G-MELGEDARA--------------------L-----LMEDRE-SRLTIVDCC-----
-----R----------RAQAC---------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSRNOP00000051298 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------M---ASFFSDFG---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LMWYLEELN--K---KEFMKFKEFLKQ------
---------E--ILQLG--------L--KQV-SWTE-VKKAS-REDLASLLLKHYEEKQA
WDMTFNF----FQ-KINRKDLIKRAK-REI------------DG--------------YP
K---------------------------------------------------LYRA----
------------------------------------------------------------
----------------------------------------------------------H-
--------LKTK----------------LTHD--SSRIFNINIQDF--------------
---------------------------------L-------KEKFTKDDHDRFETLLQSK
GT--ESK-P----Q
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
VVVLSGVAGIGKTLMLKRL-----MLAWI--E---------GLVFP
Y--------KFSY-IFYFCCQEVKQLK-------TASLAE-LISR-EW-----PDP-SAP
IEEILS-EP----EKLLFIIDSLEG-IECDLFKC------DS-E-----LCDNCMEK-QP
-VNILLSSLLRKKMLLEASLLIS-TTPETFEKMEDRI-E-----YTH--VKIITGLNESD
IKMCF----------------H--RL-FQD---------RNRAQEAFSLVRE-NEQMLTI
CQVPVLCWMVATCLKTEIEKGR-NL-V--ST-CRRTTSLYTTYIFNLFIPQS-AHSPSK-
-----------KSQDQLQGLCSLA---AEGI-WT-DT----FVFTEE-------------
--ALRRNGILDS-DIPTLLDI----GMLGKMGK-FE-NCYIFLHPSLQEACAAIFYLLK-
SHGDH-----------------P--IEDVKSVE-ALLFTFLKK-AKVQWILGGRFI----
----FGLLHESEQKKLEAFFGHQ---------------L--SQE-IKH-QLYQCLETISV
NEE-----------------------------------LQEQIDGMKLFYCLFEMED-ED
FLMEAMNY---------------------M-------------------EQINFVAKDYS
DV---IVA--A----YCLK-HCYT-LKK-LSFS----------------TQN-----ILN
-EEH--EH-----------S-----------------------------------CMENL
LTCWRHIC-SVLINSKDIQELQIKDTNLNEP---------AFSVLY--NSLKYC------
ND--T-LKVLVENN---------VFF-LCEKYLFFEL-----------------IQNCNL
QHLN-LNLT-FLSHS-DVKLLCDVLNQAECN----IEKL--------EV-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------AACNLS-----------------------------
----------------------------PDDCKMFAS-----------------------
--------------------------------ILMSSKTLKQ---LN-LSSNI-LG----
-------KGISSLCKSL----CHPDCILEHL-----------------------------
------------------------------------------------------------
------------------------------VLAKCSLSDQCWDYLSDG-IR
PF13516 // ENSGT00900000140813_0_Domain_13 (2212, 2237)
QNKTLNHLD
I-SSNDLKDEGLK-VLCRALALP--NSV----L---------------------------
-----------------------KSLCL--RHC-----LITTSGCQDL------------
-------AEVL-R
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
NNQNLTSLQISYNKIEDAGVKL-LCDAIK-QP-NC-HLEDLG--LE-
--A-CELTGACCE--------------------DLASTFTQCRTL-WGINLLNNTLDYTG
LVVLCE--------ALRQQKCT--PH--V-------------------------------
---LGLRIT-DFDNETQAF------LV----------AEQ-EKNPDLSILSG-V------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
MGP_CAROLIEiJ_P0083101 (view gene)
------------------------------------------------------------
------------------------------------------------------------
-------------MYCP-----------------GAPCGECAMCDLGCFGGSGTGPIQ--
VASVM-ITLLI-PSV--DAVAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LMATLEELS--Q---EQLKRFRHKLRD------
------------APLDG-----------RSI-PWGR-LERSD-AVDLVDKLIEFYEPVPA
VEMTRQV----LK-RSDIRDVASRLK-QQQLQK---------L---------GPT-----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------S-VLLSVSAFKKKYREH-
--------VLRQ----------------HAKV--KERNARS------------VKINKRF
TKLLIAPGTGAVEDE--LLGPLG-------EPEP--------ERARRSDTHTFNRLFRGN
EE--ESSQP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYH
S--------QVDF-AFFMPCGELLERPG------TRSLAD-LVLD-QC-----PD-RAWP
VKRILA-QP----NRLLFILDGADE-LPTLASS--------EAT-----PCKDPMEA-TS
-GLRVLSGLLSQELLPGARLLVT-TRNAATGRLQGRL-C-----SPQ--CAEIRGFSDKD
KKKYF----------------F--KF-FRD---------ERKAERAYRFVKE-NETLFAL
CFVPFVCWIVCTVLQQQLELGR-DL-S--RT-SKTTTSVYLLFITSMLKSA-GTNGP---
-----------RVQGELRTLCRLA---REGILDH-HK----AQFSEE-------------
--DLEKLKLRGSQVQTIFLNKKEIPGVLKT-----E-VTYQFIDQSFQEFLAALSYLLE-
AERTP-------------------G-TPAGGVQ-KLLNSDAEL--RGHLALTTRFL----
----FGLLNTEGIRDIGNHFGC-----------------VVPDH-VKK-DTLQWVQGQSH
PK--GPPAGAKKNAELEDIEEAEEEE--------EEEEEEDLNFGLELLYCLYETQD-ED
FVRQALSS---------------------L-------------------PEIVLERVRLT
RM-DLEVL--N----YCVQ-CCPD-GQA-LRLV------------------SC----GLV
------AA----------------------------------------KEKKKK---KKS
----------------------------------------LVKR----------------
------LKGS----------------------------------------------QSTK
KQ---PPVS-------LLRPLCETMTTPKCH----LSVL--------ILS---------H
C-----------------------------------------------------------
--------KLP---DA-VCR---DLSEALKV-APALRELGLLQSRLT-NTGLRLLCEG--
-LAWPKCQVKTL------------------------------------------------
----------------------------RIQ-----------------------------
---------------LPDL---------QEVVNYL-------------------------
------------------------------VIVLQQSPVLTT---LD-LSGC-QLPGVIV
--E--------PLCAAL----KHPKCCLKTLSLT--SVELSEN-----------------
----------SLRDLQAVKTSKP-------DLS----------------------I--IY
SK----------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCHIP00000027263 (view gene)
ENSPCOP00000009760 (view gene)
ENSRROP00000044645 (view gene)
ENSCHIP00000016479 (view gene)
ENSCGRP00001004006 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------MGNPS----QQQFN---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 323)
LKDILKSLN--R---DDLDNFKCVLKN------
---------M--SLPRT--------L--KQI-PQII-LDMAD-GVHLAEILHEYCPSGWV
ETVTMEI----FQ-MINREDLADLII-FHNNQ-----LCS----I---------------
------------------------------------------------------------
------------------------------------------------------------
---------Y-------------------------------PSLPI--TDGKEHKHKKE-
--------WNKV----------------FRNK--WKFNFWPKCKENLYV-----------
---------------------------------------------ETESYRTLVTLCNPK
A---TMPFA----H
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 931)
AIVLHGPPGSGKTTVANRL-----MLQWS--E---------SKQAQ
---------MFPC-AFSISCREVNVTN-------SCTFAQ-LLSL-KN-----DH-WRDY
VK-SLD-LA----ENFLFVVDGFDE-LRVPAG------ALLQ-D-----ICSDWDTE-KP
-AAVLLSSLLKRKMAPHATLLVT-TQTQALRQIYLMI-D-----QPF--LIENQGFSEQD
IQEYF----------------K--RF-FKDEEE----D-EEKALRALNALRC-NAALFHM
ASIPAACMIFCLCLEKVMEKGE-DL-A--LT-CQSHTSMFLNFLCRMFSPETYERHLKKE
------------FQISFKKTCFLA---ADSL-LE-QL----PTFSKE-------------
--DILKLQPDMK-GLHPFVCR----YIINNDS--YN-NSCSFIYIRIQQLLAAIVFVQE-
LE--------------------K--SVSKYSMQ-NMLSKEARL-KNPNLSGVLLFV----
----FGLLNETRIQELATIFGCQ---------------I--SIS-VKR-KFLECE--SGK
NK-----------------------------------PFLLLMDVQDILSCLYESQE-EG
LVKEAMAL---------------------L-------------------EEISLHLKTNM
DL---MHA--S----FCLK-NSQN-LRT-MSLQ----------------VDKG----IFP
-END-AVL----ES--------------------TAWHQG------------SQND-QCL
LTFWTDFC-DMFNSNEKLVFLDISESFFSSS---------SLQILC--DKLSS-S-----
AC--H-LQKVI--L-KNISPA--DA----YK-KLCLTF----------------YGYETL
SHLT-LKGD-NLST--MLPSLSMVVKHPSCK----LKFL--------SLS---------S
C------S----------------------------------------------------
---------TT---IH-KWN---EFFSALKV-SQSLTCLDLTDNELLN-KGIWLLCKT--
WKQ-PNSILQRV-S----------------------------------------------
------------------------LENCHLT-----------------------------
----------------------------EACCKGLAS-----------------------
--------------------------------VLMV
PF13516 // ENSGT00900000140813_0_Domain_11 (2017, 2051)
SQTLTH---LN-LAKNT-LGD---
-------NGVKILCESL----SHPEC----------------------------------
--------------------------KLQ-TLV---------------------------
-------------------------------LWSCDITSDGCCHLSKM-LRQASSLEHLD
L-GLNYIRSVGAR-FLCEALKEP--QSN----L---------------------------
-----------------------KSLWL--YGC-----SMTPVNCKDF------------
-------SKIL-R
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
NNKSLNTLDLAQNSLGTDAVKT-FCEDLK-LR-IC-PLQTLR--LK-
--F-DDANPTIQK--------------------LIQNMKESHPQLTISSDQP----DPRK
---------------KEP-LPH--FI----------------------------------
----F-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPPYP00000011692 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------MAESDST
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 325)
DFD---LLWYLENLS--D---KEFQSFKKYLAR------
---------K--ILDFK--------L--PQV-PLIQ-L---T-KEELANVLPISYEGQYI
WNMLFSI----FL-MMRKEDLCRKI---IGRRN---------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------RNQEACKAV-
--------LRRK----------------FILQ--WERHTFGKFHYKFFCDIT--------
----------------------------------------------SDVFYILQLAYDST
SC--YSAND----LN
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 930)
VFLMGDRASGKTMVVNLA-----VLRWI--N---------GEIWQ
N--------MISY-VVHLTSQEINQMT-------DSSLAE-LIAK-DW-----PD-RQAP
IADILS-DP----KKLLFILEDLDN-IRFELNVN------DS-A-----LCSNSTQK-VP
-IPILLVSLLKRKMAPGCWFLIS-SRPTRGNNIKMFL-K-----EAD--CYTTLQLSNEK
REIYF----------------N--SF-FKD---------RQRASAALRHVHE-DEILVGL
CRVTILCWITCTVLNRQMDKG-RDF-Q--LC-CQTPTDLHAHFLADALTSEAGLT-AN--
----------QYHLGLLKRLCLLA---AGGL-FL-ST----LNFSGE-------------
--DLRCVGFTEA-DVSVLQAA----NILLPSST-HK-DRYKFIHLNVQEFCAAIAFLMAV
PNCLI-----------------P----SGSR-A--Y--KEKRE-QYSDFNQVFTFI----
----FGLLNANRRKILETSFGCQ---------------LPMVDS-FKW-YSVGYMKLSGP
DR-------------------------------------EKLRHHMPLFYCLYENRE-EE
FVKKIVDA---------------------L-------------------EEVTVYLQSDK
DM---MVS--L----YCLE-YCCH-LRT-LKLS----------------VQR-----IFQ
--NKEPLV-------------------------------------------RPTASQMKS
LVYWREIC-SLFYTMETLRELHIFDNDLNDI---------SERILS--KALKHS------
SC--K-LRTLKLSY---------VST-ASGFEDLLKAL----------------ARNRSL
TYLS-LNCT---------------------------------------------------
------------------------------------------------------------
--------------------------------------------SISL-NMFSLLHDI--
LDE-PTCQISHL-G----------------------------------------------
------------------------LMKCDLQ-----------------------------
----------------------------ASECEEIAS-----------------------
--------------------------------LLINGGSLRK---LT-LSNNP-LRS---
-------DGVNILCDAL----LHPNCTLISL-----------------------------
------------------------------------------------------------
------------------------------VLVLCCLTENCCSALGRV-LLFSPNLKQLD
L-CVNHLKNYGVL-HVTFPLIFP--TCQ----L---------------------------
-----------------------EELHL--SGC-----FFSSDICQYI------------
-------AIVIA-TNEKLRSLEIGSNSIEDAGMQL-LCGGLR-HP-NC-MLVNIG--LE-
--E-CRLTSACCR--------------------SLASVLTTNKTL-ERLNLLQNHLGNEG
VAKLLE--------SLIRPDCA--LK--V-------------------------------
---VGLPLT-GLSTETQRL------LM----------TVK-ERKPSLIFLSETWSLKEGR
EIGVT-------------------------------------------------------
-------------------------------------------------------PASQP
GSI-----------------IPNSNLD---YM--FFKFPRMSAAMRTSNTASRQPL----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPPYP00000011693 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---ASFFS
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 326)
DFG---LMWYLEELK--K---EEFRKFKEHLKQ------
---------M--TLHLE--------L--KQI-PWTE-VKKAS-REELANLLIKHYEEQQA
WNITLRI----FQ-KMDRKDLCMKVM-RER------------TG--------------YT
K---------------------------------------------------TYQA----
------------------------------------------------------------
----------------------------------------------------------H-
--------AKQK----------------FSHL--WSSKSVTEIHLY--------------
---------------------------------F-------EEEVKQEECDHLDRLFAPK
ET--GKQ-P----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
RTVVIQGPQGIGKTTLLMKL-----MMAWS--D---------NKIFR
D--------KFLY-TFYFCCRELRELA-------PMSLAD-LISR-EW-----PDP-AAP
ITEIVS-QP----ERLLFVIDSFEE-LKGGLNEP------DS-D-----LCGDLMEK-RP
-VQVLLSSLLRRKMLPEASLLIA-VKPVCPKELRDQV-T-----ISE--IYQPRGFNESD
RLVYF----------------C--CF-FKD---------PKRAMEAFNLVRE-SEQLFSI
CQIPLLCWILCTSLKQEMQKGK-DL-A--LT-CQSTTSVYSSFVFNLFTHEG-AEGPTP-
-----------QSQHQLKALCSLA---AEGM-WT-DT----FEFCED-------------
--DLRRNGVIDA-DIPVLLGT----KILLKYGE-RE-SSYVFLHVCIQEFCAALFYLLK-
SHLDH-----------------P--HPAVRCVQ-ELLVANFEKARRAHWIFLGCFL----
----TGLLNKKEQEKLDAFFGFQ---------------L--SQE-IKQ-QFHQCLKSLGE
R-G----------------------------------QPQGQVDSLAIFYCLFEMQD-PA
FVKQAVNL---------------------L-------------------QEAKFHIIDNA
DL---VVS--A----YCLK-YCSS-LRK-LCFS----------------VQN-----VFE
-KED-EHS-----------S-----------------------------------TSDYS
LICWHHIC-SVLTTSGHLRELQVQDSTLSES---------TFVTWC--NQLRHP------
SC--R-LQKLGINN---------VSF-SGQSALLFEVL----------------FYQPDL
KYLS-FTLT-KLSRD-DIRCLCDALNYPAGN----VKEL--------GL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------VNCHLS-----------------------------
----------------------------PIDCEVLAG-----------------------
--------------------------------LLTNNKKLTY---LN-VSCNQ-LD----
-------TGVPLLCEAL----CSPDTVLVYL-----------------------------
------------------------------------------------------------
------------------------------MLAFCHLSEQCCEYISEM-LLRNKSVRYLD
L-GANVLKDEGLK-TLCEALKRP--DCC----L---------------------------
-----------------------DSLCL--VKC-----FITAAGCEDL------------
-------ASAL-INNQNLKILQIGCNEIGDVGVQL-LCRALT-HP-DC-RLEILG--LE-
--E-CGLTSACCK--------------------DLASVLTCSKNL-QQLNLTLNALDHTG
VVALCE--------ALRHPECA--LQ--V-------------------------------
---LGLRKT-DFDEETQAL------LT----------AEE-ERNPNLTITDD-CDTITRV
EI----------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPPYP00000011694 (view gene)
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------MN------------------
----------FSVITCPNGGT
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 326)
NQG---LLPYLIALD--Q---YQLEEFKLCLEPQQLMDF
WSA----------PQGH--------F--PRI-PWAN-LRAAD-PLNLSFLLDEYFPKGQA
WKVILGI----FQ-TMNLTSLCEKVR-TKMRENVQ-TQELQDPTQE------DVEMLEAA
----AGNMQTQGCQ-----------------------D----------PNQEELEELE--
E-----------------------ETGNV----------QAQRCQDPNQ-----------
------EEPE-------------------------------MLEEA----DHRRKYREK-
--------MKAE----------------LLET--WDNINWPKDHVYIRNTSK--------
----------------------------------------------D-EHEELQRLLDPN
R---TRAQA----Q
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TIVLVGKAGVGKTTLAMQA-----MLHWA--N---------GVLFQ
Q--------RFSY-VFYLNCHKIRYMK-------ETTFAE-LISL-DW-----PD-FDAP
IEEFMS-QP----EKLLFIIDGFEE-IIISESRS--ESLDDG-S-----PCTDWYQE-LP
-VTKILHSLLKKELVPLATLLIT-IKTWFVRDLKASL-V-----NPC--FVQITGFTGDD
LRVYF----------------M--RN-FDD---------LSEVEKILQQIRK-NETLFDS
CSVPMVCWTVCSCLKQPKVRYY-DL-Q--SI-TQTTTSVYAYFFSNLFSRAE-VDLADDS
------------WPGQWRALCSLA---IEGL-WS-MN----FTFNKE-------------
--DTEIEGLEVP-FIDSLYEF----NILQKIND-CG-GCTTFTHLSFQEFFAAMFFVLE-
EPREF--------------P-PH--STKPQEMK-MLLQHVLLD-KEAYWTPVVLFF----
----FGLLNKNIARELEDTLHCK---------------I--SPR-VME-ELLKWGEELGK
AE-----------------------------------SASLQFHILRLFHCLHESQE-ED
FTKKMLGH---------------------I-------------------FEVDLNILEDE
EL---QAS--S----FCLK-HCKR-LNK-LRLS----------------VSS-----HIL
-ERDLEIL----ET--------------------------------------S-KF-DSR
MHAWNSIC-STLVTNENLHELDLSNSKLHGS---------SVKGLC--LALKN-P-----
RC--K-VQKLT--C-KSVTPE--WV---L-Q-DLIIVL----------------QGNSKL
THLN-FSSN-KLGMT-V-PLILKALRHSACN----LKYL--------CL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------EKCNLS-----------------------------
----------------------------AASCQDLAL-----------------------
--------------------------------FLTSIQHVTR---LC-LGFNR-LQD---
-------DGIKLLCAAL----THPKC----------------------------------
--------------------------ALE-RLE---------------------------
-------------------------------LWFCQLGAPACEHLSDA-LLQNRSLMHLN
L-SKNSLRDEGVK-FLCEALGHP--DCN----L---------------------------
-----------------------QSLNL--SGC-----SFTREGCREL------------
-------ANAL-RHNHNVKILDLGENDLQDDGVKL-LCEALK-PS-HR-ALHTLG--LA-
--K-CNLTTACCQ--------------------HLFSVLSSSKSL-VNLNLLGNELDPDG
VKMLCK--------ALRKSTCR--LQ--K-------------------------------
---LG-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPPYP00000011695 (view gene)
------------------------------------------------------------
--------------------------------------------MKIAGGLEL-GAVALL
A-SPRDLVTLSTGPTCSILPKNP------------L-------------------FPK-N
L---SSQPCIKMEGDKSLTFS
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 326)
SYG---LQWCLYELD--K---EEFQTFKELLKK------
---------K--SSEST--------T--CSI-PQFE-IENAN-VECLALLLHEYYGASLA
WAMSISI----FE-NMNLRTLSEKAR-DDMKRHS-----PEDLEATTT--DQGPSKEKVP
GIS-Q----AVQQHSA--TA-A-------------------------ETEQEISQAME--
QE-GATA----------ETEEQEISQAMEQEDA---------------------------
-----T---A-------------------------------AETEEQGPGGDKWDYKSH-
--------VMTK----------------FTEK--EVVR-RSFEKT---------------
-----------------------------------AA-------DLPEM-QMLAGAFNSD
Q---WGFRP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 925)
RTVVLHGKSGIGKTALARRI-----VLCWA--Q---------GGLYQ
G--------MFSY-VFFLPVREMQQKK-------ESSVTE-FISR-EW-----PD-SQAP
VTEIMS-RP----ERLLFIIDGFDD-LG-SVLNN------DT-K-----LCKDWAEK-QP
-PFILISSLLRKVLLPESFLIIT-VRDVGIEKLKSEV-V-----SPR--YLLVRGISAEQ
RIHLL----------------L--ER-GIG---------EHQKTQGLRVIMN-NRKLFDQ
CQVPAVGSLICVALQLQDVVGE-SV-A--PF-NQTLTGLYAAFVFNQLTPRG-VVRRCLN
----------REERVVLKRFCRMA---VEGV-WN-RK----SVFDGD-------------
--DLMVQGLGES-ELRALFHM----NILVPDSH-CE-EYYTFFHLSLQDFCAALYYVLE-
GLEIE--------------PALC--PLYVEKTK-RSMEFKQAG-FHIHSLWMKRFL----
----FGLVSEDVRRPLEVLLGCP---------------V--PL-GVKQ-KLLHWVSLLGQ
QPN----------------------------------AT-TPGDTLDAFHCLFETQD-KE
FVRLALNS---------------------F-------------------QEVWLPINQNL
DL---IAS--S----FCLQ-NCPY-LQK-IRVD----------------VKG-----IFP
-RDESAEA----CP--------------------VV-----------PL--WMRGK-TLI
EEQWEDFC-SVLGTHPHLRQLDLGSSILTER---------AMKTLC--AKLRH-H-----
TC--K-IQTLM--F-RSAQI---TP---GVQ-HLWRTL----------------IANRNL
RSLN-LGGT-HLKEE-DVRMACEALKHPKCL----LESL--------RLD---------C
C------G----------------------------------------------------
---------LP---HA-CYL---KISQTLTT-PPSLKSLSLAGNKVTD-QGVTPLSDA--
LRVSSQCTLQKL-I----------------------------------------------
------------------------LQDCGIT-----------------------------
----------------------------ATGCQSLAS-----------------------
--------------------------------ALVSNRSLTH---LC-LSNNG-LGN---
-------EGVNLLCRSM----RLPHC----------------------------------
--------------------------SLQ-RLM---------------------------
-------------------------------LNQCHLDTAGCGFLALA-LMGNSWLTHLS
L-SMNPVENNGVK-LLCEVMREP--SCH----L---------------------------
-----------------------QDLEL--IKC-----HLTAACCESL------------
-------SCVI-SRSRHLKSLDLTDNALGDGGVAA-LCEGLK-QK-KS-VLTRLG--LK-
--A-CGLTSDCCE--------------------ALSLALSCNQHL-TSLNLVQNNFSPKG
MMKLCS--------AFACPTSH--LQ--I-------------------------------
---IGLWKW-QYPVQIK-L------LE----------EVQ-LLKPRVVIDGSWHSFD---
--E---------------------------------------------------------
--------------------------------------------------DDRYWWKN--
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSLAFP00000017981 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------MAEC
PF02758 // ENSGT00900000140813_0_Domain_0 (204, 328)
VLHRLVTYLEELE--A---VDLKKFKLHLGT------
---------A---AQLG--------E--DKI-PWGR-METAG-PLEMAQLLVAHHGSGEA
WLLALSV----FE-RINRKDLWERGQ-RGPEYTQQ----N--IH--------QFNSFYVL
--------Q---------------------------------EK---VSHEEWLHP----
------------------------------------------------------------
---QSYLSLL-------------------------------NIIDFKKTSDPWETYRNY-
--------VRRK----------------FLLM--EDRNARLGECV---------SLSHRY
TRLLLVKEHSNPMRTQEKLL------ATGRRHSRTVD--------HQASPIWVETLFEPD
EE--RPEPP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
QTVVLQGAAGMGKSMLAHKM-----MLDWA--E---------GSLFQ
G--------RFDF-LFYINCREMNQK-A------EWILQD-LISS-CW-----PE-QNVP
LLELAR-IP----ERLLFIIDGFDE-LKPSFHNP------QS-P-----RCFCWQEK-QP
-TELLLSSLIRKKLLPELSLLIT-TRPMALEKLHRLL-E-----HPR--HVEILGFSEAE
RKEYV----------------Y--KY-FHS---------VEQAGQVFNFVRD-NEPLFTM
CFVPMVCWVVCTCLKQQLEGGR-LV-R--QM-SRTTTAVYVLYLLSLMQPKPGT-PTH--
-----------QTPPNQKGLCSLA---ADGL-WN-QK----VLFEEQ-------------
--DLRKHGLDGA-DVSAFLNM----NIFQKDIN-CE-KFYSFIHLSFQEFFAAMYYILG-
G--GE--------------T--G--LGPDNNLS-RLLAEYGSS-DRSFLAPTVRFV----
----FGLLNKEMRSYLEKNLCWK---------------V--SPH-IRM-ELLKRI----Q
SE-----------------------------------GSTLQQGSLEFLSCLYEIQE-EE
FIQQALSH---------------------F-------------------QVVTVSHMA-T
KM-EHMIS--S----FCVK-NCRS-VLV-VHLY----------------GA------AFS
-ADDDDGA--K-WA---------------------------------------LGS---Q
A-------------------------PL------------------------A-------
QS--------------SVLPD-------HDEV----------------------------
-----------------------------------TCLH--------ELK---------R
C-----------------------------------------------------------
--------RVS---SS-ACQ---DLAAALTV-NRNLLRVDLSSNRLGL-TGVKLLCEG--
LRH-PKCRLQMIQ-----------------------------------------------
------------------------LRKCQLE-----------------------------
----------------------------AGACQEISS-----------------------
--------------------------------VLSNNQHLME---LD-LTGNA-LED---
-------LGLRLLCQGL----KHPTC----------------------------------
--------------------------HLQ-TLW---------------------------
-------------------------------LKICHLTAVACEDMAST-LSVNQ
PF00560 // ENSGT00900000140813_0_Domain_13 (2215, 2237)
SLRELD
L-SLNDLGDPGVL-LLCEGLSHP--KCK----L---------------------------
-----------------------QTLRL--GIC-----RLGSAACEGL------------
-------SAVL-QTNH
PF00560 // ENSGT00900000140813_0_Domain_15 (2357, 2381)
HLRELDLSFNDLGDWGMRL-LCEGLR-HP-TC-RLQKLW--LD-
--S-CGLTAKACE--------------------DLSSMLAVNQTL-TELYLTNNTLKDTG
VRLLCK--------RLGHPSCK--L-----------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSOGAP00000011492 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------DLGDRTEYRIQ-
--------IKEK----------------FGIV--WDRNSLLGKPEDFHQGIS--------
-----------------------------------------------QGQKLLEHLFDVD
VK--TGEQP----Q
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
VVVLQGAAGVGKTTLVRKA-----MLDWA--E---------GNLYQ
Q--------KFAY-VFYLSGREINLMK-------ERSFAQ-LISQ-DW-----PS-IEGS
IDRIMS-QP----SSLLFIIDSFDE-LNFAFEEP------EF-A-----LCKDWTQE-HP
-VSFLISSLLRKVMLPESSLLVT-LRLTASKRLKPLL-K----SHYH--FVELPGMSEDA
RKECI----------------Y--QF-FED---------EKSAMNAFSSLRN-NEMIFSI
CQVPLVCRLICTCLKQQIEKGS-NV-S--LT-CQTTTALFTCYICSLFTVI-EGHFPRLP
------------SQDQLSSLCHLA---AKGV-WT-MT----YVFYRE-------------
--DLRRHGLTKL-DVSIFLDM----NILQRDPE-YE-NCFVFIHLHVQEIFAAMFYVLE-
GSQKA--------------RGHS--FQPFEDIT-LLLESKSDE--DSHLTQMKCFL----
----FGLLNEDLAKQLEKTLNCK---------------M--SLE-IKS-KVLQWLEALGN
SE-----------------------------------SSPPQLECLEWFHYLYETQD-EA
FVSQAMKY---------------------F-------------------PKVVINICGKT
HL---FVS--S----FCLK-HCQC-LQT-IKLS----------------ITV-----VFE
--K--KML----NTSL-----PA--E------------------------TCRQSDVDRI
THYWQELC-SVLHTNEDLRELELCHSNLDKL---------AMKIFN--QELRHP------
KC--K-LQKLRLSF---------VTFPVTACRGISSSL----------------THNKNL
MHLD-LKGN-DIGDN-GVKLLCEALKHPECK----LQKL--------SLE---------S
CN----------------------------------------------------------
---------LT---EV-CCL---DIFNALIR-SQSLIFLNLSTNNLLD-DGVKLLCEA--
LRY-PKCYLERL-S----------------------------------------------
------------------------LESCGLT-----------------------------
----------------------------EDVCEDLSL-----------------------
--------------------------------ALISNKRLTH---LC-LANNA-LGD---
-------SGIKLMSDVL----QNPCC----------------------------------
--------------------------TLQLSLW---------------------------
-------------------------------LRHCHFTLLSSEYLSTS-LLHNKSLTHLD
L-GSNRLQDDGAK-LLCDAFRHP--SCN----L---------------------------
-----------------------RDLEL--MGC-----VLTSACCLDL------------
-------ASVV-LSNPNLWSLDLGKNDLQDEGVKI-LCDALR-HP-DC-SIQRLG--LE-
--C-CGLTSLSCE--------------------DLSSTLSSNQRL-KKMNLAQNDLRCEG
IRKLCQ--------ALKSPDCK--LQ--V-------------------------------
---LGLCKE-AFDEEAQKL------LE----------AVR-VSNPHLIIKPDCNDNN---
--E---------------------------------------------------------
--------------------------------------------------EDGSWWRCF-
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMODP00000025242 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------MTES--V
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 328)
RCI---LVRYLEELT--F---EELKKFKLFLED------
---------S--PPKEK--------F--LRI-PRSQ-MKKAD-NTDMAELMVHHYGEGEA
WDVALMI----WE-KMGLRELWEQAK-REAPQMEKLRWNSRFLKGKDK----KSE--RRK
----LGFWKH--------------------------------------------------
------------------------VDKLM-------------------K-----------
------KWSG-------------------------------QDHMDQWNEGCQTTYRNR-
--------IMEK----------------FRVM--RDRNSCPGEYE---------NLHHRF
TRLLLLQEYRHREQKEHELL------AIGWEHAEIME-------NQ-GQLIEVASLFDPD
KK--REAHP----Q
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
VVVLQGAAGIGKTTLARKV-----ILDWA--E---------GNLYQ
D--------KFDY-VFYLHCREMNHLRE-----KKIRFAD-LIAN-DW-----PF-LEAS
MKRILS-HP----EKLLFVIDGLDE-LKFSCDEH------SY-D-----LCKDWNLQ-QP
-VPILLSSLLRKTMLLESSLLIT-TRLTALEKCDFLF-E-----NPR--HVEILGFSEED
RWKYF----------------C--KF-FGD---------EDQAQQAFSLIVN-NETLFTM
CFVPLMCWIICTCMKQQMRRKK-DL-K--QA-SKTTTTLYMCYLSSLATPGA-----RSC
------------SVQHHRSLCRMA---AEGI-WE-GK----ILFDGD-------------
--DLRRHGLEASDV-SAFLDM----HIFQKDSD-SE-NCYSFIHLSFQEFFAAMFYALG-
EEKET--------------GGRA--VPPIPGVK-DLLAKCRDS-ATSFVALTVHFL----
----FGFLNADNRKRLEEIYDCK---------------I--SQE-IKL-ELLQWIQAVTE
QQH--------------------------------KDPSLRN-DLLEMFSHLYETQD-EE
FVTTAMAH---------------------F-------------------QEISLN--IYS
KL-HFLIS--V----FCIK-FCHN-LGK-IRLN----------------IAR-----FQM
-K-----S-------------------------------LQFKVF------QL------A
VVDWQHLF-SVLSKNQNLRTLDLSGTELANS---------AVNALC--SELSQ-S-----
SC--K-LQKLS--L-SKCQLT--AS---CCQ-HLSSLL----------------KNNKNL
TYLD-LSMN-FLGDE-GIKALCEVLRNQNCN----IQEL--------NLS---------R
C------R----------------------------------------------------
---------FS---DA-CCK---NLSSALMA-HGNINILDLSENTLAD-VGMNLLCEA--
LGS-SDCNLQEL-K----------------------------------------------
------------------------LSQCHFT-----------------------------
----------------------------DACCQDLST-----------------------
--------------------------------CIK-NQSLIH---LD-VSGNF-LQD---
-------SGIRLLCEAL----RHPTC----------------------------------
--------------------------NLQ-KL----------------------------
------------------------------LLSLCHITDSSCQDVSFA-LKNNRSLIHLD
L-GCNNLYNHGVK-LLCEALENQ--NCN----L---------------------------
-----------------------QTLEL--WNC-----QLSAACCHEF------------
-------SSVL-KKNQSLTHLNLGANPLRNNGVKV-LCEALG-NQ-NC-KLQKLK--LC-
--K-CLLSAVGCR--------------------YLSTALKSNQNL-THLKLTGNGLGDDG
VKLLFE--------ALKNSDCK--LQ--K-------------------------------
---LTLDKW-KLSADTQRI------QD----------ELR-QRKPHLIIENSSLC-----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSBTAP00000001428 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------APAPSMEPR--AAAA
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 328)
REL---LLGALEDLS--Q---EQLKRFRHKLRD------
------------ERVDG-----------RSI-PWGR-LERAD-TLDLVEQLVHFYGPERA
LDVAKKT----LK-RADVRDVAARLK-ERRLQS---------L---------GPS-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------SP-ALLSVSEYKKKYREH-
--------VLRQ----------------HAKV--KERDARS------------VKINKRF
TKLLIAE-----------------------EPEA--------ERARRSDTHTFNRLFGRD
E---EGERR----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVALQGPAGIGKTMAARKI-----LYDWA--A---------GKLYH
S--------QVDF-AFFLSCRELLERPG------TCSLAD-LILD-QC-----PD-REAP
VQQMLA-QS----ERLLFILDGVDE-MLAPEPA--------EAA-----PCRDPHEA-AS
-GARVLGSLLSKELLPAARLLVT-RRGATPGRLQARL-C-----SPQ--CAEVCGFSDKD
RKKYF----------------Y--KF-FQD---------EWKAEHAYRSVKE-NEMLFSL
CFVPGVCWIVCTVLRQQLQRGQ-DL-W--RT-SKTTTAVYLLFVASVLRSAPAADAA---
-----------QLQRELRKLCRLA---CDGVLGG--R----ARFSEE-------------
--DLERLQLCGSKVQTQFLSKKEMPGVLET-----E-VTYQFIDQSFQEFLAALSYLLE-
DEGAP-------------------R-APAGSIE-ALLQGDPEL--RGHLNLTTRFL----
----FGLLNTERMCEIKRHFGC-----------------TVSER-VKQ-RVLRWVQDQGQ
GRPRAAPEGTEEILAPQDTEESEE------------EEGEQLNYPLELLYCLHETQE-DA
FVHQALRG---------------------L-------------------PELVLERVHFS
RM-DLAVL--S----YCVR-CCPA-GQA-LRLV------------------SC----RLA
------TA----------------------------------------QEKK-----KKS
----------------------------------------LMKR----------------
------LQGS---------------------------------------------SRATT
RK---SPGS-------PLRPLCEAMTDRQCG----LNSL--------TLS---------R
C-----------------------------------------------------------
--------KLP---DS-VCR---DLSEALRE-APALAELGLFHNGLS-EAGLRVLSEG--
-LASPQCRVQTL------------------------------------------------
----------------------------RVQ-----------------------------
---------------QPGL---------QEALQYM-------------------------
------------------------------VGKLQQIPALTT---LD-LSGC-QLSGPTV
--T--------CLCTTL----KCPGCHLQTLRL--TSVELSEQ-----------------
----------ALQELQAVETANP-------RLV----------------------I--TH
PALD-------------------AHPKPPTVPISAL------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSJJAP00000002893 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MASALS-QSPQEA---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWALSDLE--E---NSFKTLKFHLRA------
---------M--T------------Q--PHL-ARGE-LERLS-RVDLASRLILMYGAQEA
VRVVLKV----LK-IMNLLELVDQLS-QVH------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LN--DYREIYREH-
--------VRSL----------------AE-----------EDQ----------GVKGSF
NRLLLVA-AEPPAGPG-------------------LE-----Q-ELHSV--GMETLFGPG
EE--P--HP----S
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
VVVLQGSAGTGKTTLVKNL-----VQEWA--R---------GALYP
D--------RFDY-VFYVSCREVVLLPR-------CDLHQ-LLFW-CC-GD-----SQAP
ATEILR-QP----ERLLFILDGYDE-LQRPFEE-------------------------HC
-MEKVLHLLIRRRLLPTCSLLIT-TRPPALRNLQSML-G-----DQHHVHVHILGFSEEE
RKRYF----------------D--FY-FAD---------EERARKAFGFVQK-SAILYKA
CQVPGICWVLCSWLKAHLDRGQ-EV-S--EP-PSNSTDIYTAYVSTFLPADG-DGDCSDL
----------T-RYRVLKGLCSLA---AEGI-QH-HR----LLFKED-------------
--DLKRHNLDGPRLT-AFLSS----SEYQEGLG-IK-KFYSFRHISFQEFFHAMSFLMK-
EDQNQL----------------G--EETRREVD-RLLEVKDQE-EYEEVSLSMQFL----
----FDILKTDSALNWKFKNLFK-----------------TAPS-LMQ-DL-KHFKEQM-
E-------------------------------------CAKHHWSWELEFSLYESKI-KK
LTKGFQMK---------------------D-------------------ILFKIDYL---
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCAFP00000041931 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------M-
PF14484 // ENSGT00900000140813_0_Domain_1 (574, 658)
-EDRNARLGECV---------NLSHRY
TRLLLVKEHSNPMWAQQKLL------DTGWGQARTVG--------HQASFIQMETLFEPD
EE--RPEPP----RTVVLQGAAGMGKSMLAHKV-----MLDWA--D---------GRLFQ
D--------RFDY-LFYINCRKMNQSTA------EQSAQD-LISS-CW-----PE-PSVP
LQELVR-VP----ERLLFIIDGFHE-LKPSFHDP------QG-P-----WCLCWEEK-RP
-TELLLSSLIRKKLLPELSVLIT-IRPTALEKLHRLL-E-----HPR--HVEILGFSEAE
RKEYF----------------Y--KY-FHN---------AEQAGQVFNFIRD-NEPLFTL
CFVPMVCWVVCTCLKQQLEDGG-LL-R--QT-SRTTTAVYMLYLLSLMQPKPGS-PIL--
-----------QSPPNQRGLCSLA---ADGL-WN-QK----ILFEEQ-------------
--DLRKHGLDGA-DVSSFLNM----NIFQKDIN-CE-KFYSFIHLSFQEFFAAMYYILD-
P--GE--------------S--R--SSPEHNVT-RLLAEYEFS-ERSFLALTVRFL----
----FGLLNEETRSYLEKSLCWK---------------V--SPH-VKV-ELLEWIQRKAQ
SE-----------------------------------GSTLQQGSLELFSCLYEIQE-ED
FIQQALSP---------------------F-------------------QVVVVNNIA-T
KM-EHMIS--S----FCVK-NCRS-ALV-LHLH----------------GA------AYS
-PDEDDGG--R-WA---------------------------------------SGP---Q
M-------------------------LP------------------------T-------
Q---I--------PEKNVLPD-------AYSKQLAAAL----------------STNPNL
TELV-LYRS-ALGSR-GVRLLCQGLRHPSCK----LQNL--------SLK---------R
C-----------------------------------------------------------
--------CVA---SS-ACQ---DLAAALMA-
PF13516 // ENSGT00900000140813_0_Domain_8 (1773, 1797)
NQNLRRMDLSSNRLGL-PGLRALCKG--
LRH-PRCKLQVIQ-----------------------------------------------
------------------------LRKCQLE-----------------------------
----------------------------AEACQEIAS-----------------------
--------------------------------VLSTS
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2055)
RHLEE---LD-LTGNA-LED---
-------LGLKLLCQGL----RHPVC----------------------------------
--------------------------RLQ-ILW---------------------------
-------------------------------LKICHLTAAACEDLAST-LGV
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
NQSLIELD
L-SLNDLGDPGVL-LLCEGLRHP--QCR----L---------------------------
-----------------------QALRL--GIC-----RLSSAACKGL------------
-------CTVL-QV
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2373)
NPCLRDLDLSFNDLGDAGVWP-LCEGLR-HP-TC-RLQKLW--LD-
--S-CGLTAKACE--------------------DLSSALGVS
PF13516 // ENSGT00900000140813_0_Domain_17 (2443, 2466)
QTL-RELYLTNNALGNAG
VRLLCK--------GLSHPGCK--LQ--V-------------------------------
---LWLFGM-ELNKMTHRR------LA----------ALR-VVKPQLDIGC---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSJJAP00000013786 (view gene)
ENSTBEP00000013080 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------MTDSSSSSFF
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
SDFG---LLLYLEELN--K---EELNKFKLLLKE------
---------T--MEP-G--------H--CPI-PWTE-MKKAK-REDLANLIKKYYPGELA
WNVTLKI----FG-KMNLKGLCERAK-AEINWTAQ-TVK-LE-DTN------TEEAQADQ
----EAVLXXXXXX-----------------------X-XXXXX---X-----XXXXX--
XXXXX-------------------------------------------------------
-----------------------------------------XXXXXXXXXXXXXXXXXX-
--------XXXX----------------XX-----------XXXX---------------
----------------XXXX------X-----XXX-X-------XX-XXXXXXXXXXXX-
XX--XXXXX----XXXXXXXXXXXXXX---XXX-----XXXXX------------XXXX-
X--------XXXX-XXXXXXXXXXXXX-------XX-XXX-XXX--XX-----XX-X--X
XX-XXX-XX----XXXXXXXXXXXX-XXXXXXXX------XX----------XXXXX-XX
-XXXX-----XXXXXXXX--XXX-XXXXXXXXXX-XX-X-----XXX--X---XXXXXXX
XXXXX----------------X--XX-XXX---------XXXXXXXXXXXXX-XXXXXXX
XXXXXXXWIICTCLKQQMEKGD-V--A--SI-CQTV-TAFTCYLSGLFTLAG-GSPS---
------LPNE----AQLKSLCHLA---AEGI------QTMTYVFYRE-------------
--NFRKHGLT-KSVVSIFLDM----NILQKNTE-YE-NCYGFIHLYIQEFFAAMFYMLK-
GNWEA--------------SDHS--FQSFEDLK-LLLGN-KIN-EEPHLMQVKCFL----
----FGLLSEDRIKQLEETFNCK---------------M--SLE----------------
-------------IKWELLQ-WMEIL-------GTSKCSLSQLGFLELFHYLYETQD-KA
FINQAMGY---------------------F-------------------PKVVI---NIS
GIVDLLVS--S----FCL-KHCQC-LQS-IELS----------------IT---------
----MVFE----KK-------------------------MLNSNPTA----ETWDG-GHI
ICCWQDLC-SVLHTNEHLRELVLCHSYLDEL---------AMKTLY--EELRH-P-----
NC--K-LQKLL--L-RFVSFP--D----SCQ-DISSSL----------------IHNQNL
MHLD-MKGS-NIGDN-GVKSLCEALTFPECK----LQNL--------RLE---------S
C------N----------------------------------------------------
---------LT---TV-CCQ---SISKVLTG-NQSLIFLNLSTNNLLD-NGVQQLCEG--
LRH-PTCHLERL-S----------------------------------------------
------------------------LSTCYLT-----------------------------
----------------------------EASCVELSS-----------------------
--------------------------------LASESDFKR-----L-LCLNA-LGD---
-------TGVQHLCEGL----QHAKG----------------------------------
--------------------------IIE-NLV---------------------------
-----------------------------XXXXXXXXXXXXXXXXXXX-XXXXXX--X--
X-XXXXXXXXXXX-XXXXXXXXX--XXX----X---------------------------
-----------------------XXXXXXLMGC-----VLTSACCLDL------------
-------ASVL-LNNPNL-SLDLGNNDLQDDGVKI-LCDALR-HP-NC-NIQRLG--LA-
--Y-CGLTSLCCQ--------------------DLASMLSSNQRL-TQMNLTQNTLGCGG
IMELYE--------TLRSPKCK--LR--V-------------------------------
---LGLCKE-AFDKEAQKL------LR----------AVE-INNPNLVINPYSDH-----
------------------------------------------------------------
-----------------------------------------------AEEDGAFWWRCF-
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSODEP00000019410 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------------MASS-TR
LAQHAF----------------LRSLSGSF-------------------NTWSIH---SS
-G-RHRIY--I----MSLKICCEQ-LSD-ARWT----------------EL---------
----LPLI-------------------------------QQHQVV------RLEDC-GLT
EGRCRDLS-AMLRA
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1542)
SPTLTELCLSNNELGDA---------GVHLVL--QGLQS-P-----
TC--K-IQSLS--L-PYCGLT--ET---GCG-VLPSVL----------------PSM
PF13516 // ENSGT00900000140813_0_Domain_6 (1618, 1642)
PTL
RELR-LSGN-QLGDG-GLRLLENSGT00900000140813_0_Linker_6_7 (1642, 1650)
AEGLMGTQPF13516 // ENSGT00900000140813_0_Domain_7 (1650, 1766)
CH----LEKL--------QLD---------Y
C------G----------------------------------------------------
---------LS---DA-SCE---PLASMLRA-KSSIKELVMSNNDIHE-AGIRTLLRG--
LKD-SACPLETL-W----------------------------------------------
------------------------LENCGVT-----------------------------
----------------------------AASCKELCD-----------------------
--------------------------------VLAA
PF13516 // ENSGT00900000140813_0_Domain_11 (2017, 2054)
KPSLQD---LD-LGNNR-LGD---
-------VGIAALCSCL----LHPSC----------------------------------
--------------------------RLK-KLW---------------------------
-------------------------------LWECDITAGGCRDLCQV-LRA
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
KQSLKALS
L-MLNGLGDEGAR-LLCEALQAP--TCQ----L---------------------------
-----------------------ECLWV--KEC-----SFTAACCPYF------------
-------RSVL-A
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
QNKFLTELQLSENNLGDSGVQE-LCQALG-QP-HS-VLQVLG--LG-
--D-CNLTSSSCS--------------------NLALVLLA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2464)
CHSL-RELDLSNNGLGDPG
ILQLVE--------GLRQPTCA--LE--C-------------------------------
---LFLFDV-YLSDDMRDQ------LE----------ALE-REKPSLKIIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSECAP00000021325 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
M---TGAQP----Q
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TIVLEGRAGVGKTTLAMKV-----MLHWA--E---------GFLFQ
H--------RFSY-VFYISCHKMKEME-------DTTFAD-LLSQ-DW-----PD-SQAP
IEEFIC-HP----ERLLFVIDGFEE-MAMPSK------LSDS-P-----PCTDWYQQ-LP
-VARILLHLLKKELIPTATLLIT-TRDYRNGALKKLL-L-----NPW--FVLISGFTEGD
RSEYF----------------F--RY-FGD---------RNIAKKILHWIRK-NETLFHS
CSAPLVCWTVCSSLKRQMARNP-HF-Q--LS-TQTTTSLYAHFFSSLFATAE-VSLSDQS
------------WPEQWRALCSLA---AEGM-WF-SN----FTFAKE-------------
--AIEHRRLEAS-LIDSLLSL----NILRKASD-CE-DCIAFTHQSFQVFFGAMFYVLR-
GTEGS--------------I-GG--PTRHQEMR-VLLNDALAD-TNFYWNQMALFF----
----FGLLKRDLARELEDTLHCK---------------M--SSR-IMD-ELLKWSEELDK
FD-----------------------------------TVSNHFEFLQFFHCLYETQE-EN
FVRQILNH---------------------L-------------------LEADLNIFGNL
QL---QVS--S----FCLK-HCQR-LSK-LRLS----------------VSR-----PLI
-PKIIPF-------------------------------------IV-RI--DTQLV-DSK
IRQWQDIC-S-VFCNGNVSELDLSNSELNTS---------SMKKLC--YKLRN-P-----
WC--K-LQKLT--C-KSITPV--RI---L-K-ELVLVL----------------YGNNKL
THLN-LSSN-NLGVT-VSTMIFRTLRHSACN----LKYL--------WLE---------S
C------G----------------------------------------------------
---------LS---SR-VCH---QLFLELSK-NSSLSFLSLGDNDLSD-IEVKSLQEP--
FEM-SKCSLKEL-S----------------------------------------------
------------------------LEKCNLS-----------------------------
----------------------------ASSYQNLAL-----------------------
--------------------------------LLPSTQRLTR---LC-LGFNQ-LQD---
-------DGVKLLCSSL----SHPEC----------------------------------
--------------------------ALE-RLV---------------------------
-------------------------------LWFCQLGTPSCRYLSEA-LLRNKSLTHLN
L-RKNNLGDEGVK-VLCKALSHL--DCN----L---------------------------
-----------------------QNLDL--SDC-----SFTTESCGEL------------
-------ANVL-KHNHNVKILDIGNNDVQDDGVKR-LCKVLK-HS-NC-ALNTLG-----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSOPRP00000002921 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MADSSSSYF
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
SDFG---LLLYFEELN--K---EEFNNFKLLLKQ------
---------R------T--------D--ALI-PWSA-LKNAK-REDLANLMNKYYPREQA
WNVTLQI----FG-KMNLKGLCERAK-AEINCSAK-TMEPEDVEDKEI------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------HSDQDLVLXXXXXXXXX-
--------XXXX----------------XXXX--XXXXXXXXXXXXXXXXXX--------
----------------------------------------------XXKKKLLEHLFDVD
IK--TGEQP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
KTVVLHGAAGVGKTQLARQA-----MLDWA--E---------GNLYQ
Q--------RFAY-VFYLNAREMNQLN-------ESSFAE-LISK-DW-----PS-TKYP
IEKILS-QP----NSLLFIIDSFDE-LNFAFEEP------EF-A-----LCNDWSQV-HP
-VSFLLSSLLRKVMLPESSLLLT-LRLTNAKKLKHLL-K-----NEH--YVELPGMPELA
RKEYI----------------C--RF-FED---------KNSGMKAFHLLRS-NEMLFSM
CQAPLVCWIACTCLKQIMKKGG-DI-T--MT-CRTPTDLFTCYISGLFTSV-DGCSFNLP
------------NQDQLGRLCYLA---AKGI-WT-MT----YVFYRE-------------
--NFRKYGLTKY-DVSMFLDM----NILQRDRE-LE-NCYVFTHLHVQEFFAAMFYMLK-
DSWET--------------RDS----CPFENLK-SVFESKNDK--DSHLMQMKCFL----
----FGLLNEERIKQLEKALNCK---------------L--SLD-VRQ-ELLQTMEVLGK
SD-----------------------------------YSPSELGFLELFHYLYETQD-EA
FVCQAMSY---------------------F-------------------PKIDVNICAKI
HL---FVT--S----FCLK-HCRD-LQT-IKFS----------------VSI-----FEK
------MS----SSSW-----LP--E---------------------------ISDGEII
THCWEDLC-SVLHTNEHLKELSLCKSKLDKL---------AMKTFY--QGLRHP------
NC--K-PQTLLXXX---------XXXXXXXXXXXXXXX----------------XXX-XX
XXXX-XXXX-XXXXX-XXXXXXXXXXXXXXX----XXXX--------XLE---------C
CD----------------------------------------------------------
---------LT---AV-CCL---NISKALIS-NQNLIILNLSTNNLLD-DGVKLLCEA--
LSH-PKCRLERL-S----------------------------------------------
------------------------XXXXXXX-----------------------------
----------------------------XXXXXXXXX-----------------------
--------------------------------XXXXXXXX--------------------
------------------------------------------------------------
----------------------------X-------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------X-----------X---------------
---------------------XXXXXXXXXXXXXX-XXXXXX-XX-XX-XXXXXX--LE-
--C-CSLTALSCD--------------------ALVSTLIGNQKL-KIMNLTQNALGYEG
IKKLSE--------VLRSAICK--LQ--V-------------------------------
---L--------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSSHAP00000001069 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------ALAMG----MADAILCC
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 324)
LAAYLEELE--S---VELIKFKMYLGL------
---------E--AEKVG--------E--GRV-PRGR-MEKAG-PLDMAQILVAHWGPAEA
WELALRV----FN-RINRKDLWARGC-QEVLVREC----S--SS--------PPESEG--
------------------------------------------------------SL----
------------------------------------------------------------
LPTIS-FPLI-------------------------------PTSSPTPQLDPQEMYREH-
--------VRRK----------------YRFI--EDRNARLGELV---------NLSQRY
TPLLLVKEHTTLTRAQHELL------AVGRQHALGLD--------QRASPIQIETLFEPD
EE--RPHAP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
HTVVLQGVAGIGKSILAQKV-----MLDWA--E---------GLLYS
D--------RFDY-VFYMNCREMNQF-Q------ERSMDD-LISV-CW-----PE-RNVP
ISEIVR-VP----ERLLFIIDGFNE-LRTSFHEP------RN-C-----GCIQWGER-KP
-VEALLSCLLSRKLLRELSLLIT-TRPTTLETLHHML-E-----HPR--HVAILGFSEAL
RQEYF----------------Y--KF-FRH---------EEQARQGFSLVRD-NEPLFTM
CFVPLMCWIVCPCLKQQLEAGW-PF-P--HT-SKTTTAVYMFYLMSLLQPTPGR-SRA--
-----------QPPPNLRGLCSLA---AWGL-WT-QR----ILFEEE-------------
--DLQKHGLGGD-DFSAFLNM----NIFHKDIQ-CE-RYYSFIHLSFQEFFGAMFYVLE-
E---E--------------P--G--A-PSADLA-KLLKHYSKS-ERSYLTLTVRFL----
----FGLLNDERRNSLEKQLGGK---------------T--SAR-TKR-ELLRWIQVKAL
SE-----------------------------------GSTLQRGSLELFQCLYEIQE-ED
FIQRALDH---------------------F-------------------QVVVVSDMA-T
KM-EHMVS--S----FCVR-NCCS-AVV-VHLG----------------ST------MFS
-ADCEEDT--Q-NQ---------------------------------------GGR---A
SM----------------------R--L------------------------D-------
LH--W--------SEKDVLPE-------AYCEHLATAL----------------STNRNL
IELV-LYRN-ILGNR-GVRLLCQGLRHPNCQ----LQNL--------RLK---------R
C-----------------------------------------------------------
--------RLS---SA-SCQ---DLSSALVS-NQNLTRLDLSRNSLGA-AGVKLLAEG--
LRH-PKCRLQMLQ-----------------------------------------------
------------------------VRRGEPD-----------------------------
----------------------------WEACRALSE-----------------------
--------------------------------VLSASRHLTE---LD-LTGNA-LGD---
-------WGLRPLCAGL----SHPAC----------------------------------
--------------------------RLQ-TLW---------------------------
-------------------------------LKICHLPPSACQDLASV-LSINQNLTELD
L-SLNELGDQGVK-LLCEGLCHS--KSQ----L---------------------------
-----------------------QTLRL--GIC-----RLTFVSCDAL------------
-------STTL-QSNIHLKALDVSFNDLEDLGVHL-LCQGLQ-HP-NC-KLHKLW--LD-
--S-CSLSGAACG--------------------DLAAALGINQTL-RELYLTNNALGDAG
VQLLCE--------RLSQPNCR--LR--T-------------------------------
---LWLFGT-ELTEGTQQA------LA----------ALR-GAKPHLDIGS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPANP00000004746 (view gene)
ENSPPYP00000011607 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MLRT----AGRDGLC
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 328)
RLSTYLEELE--A---VELKKFKLYLGT------
---------A--TELGE--------G---KI-PWGR-MEKAG-PLEMAQLLITHFGPEEA
WRLALST----FE-RMNRKDLWERGQ-REDLVRDT----P--PG--------GPSSL---
------------------------------------------------------GN----
------------------------------------------------------------
---QS-TCLL-------------------------------EVSLVTPRKDPQETYRDY-
--------VRRK----------------FRLM--EDRNARLGECV---------NLSHRY
TRLLLVKEHSNPMQAQQQLL------DTGRGHARTVG--------HQASPIKIETLFEPD
EE--RPEPP----C
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVMQGAAGIGKSMLAHKV-----MLDWA--D---------GKLFQ
G--------RFDY-LFYINCREMNQSAT------ECSMQD-LISS-CW-----PE-PSAP
LQELIR-VP----ERLLFIIDGFDE-LKPSFHDP------QG-P-----WCLCWEEK-PP
-TELLLNSLIRKKLLPELSLLIT-TRPTALEKLHRLL-E-----HPR--HVEILGFSEAE
RKEYF----------------Y--KY-FHN---------AEQAGQVFNYVRD-NEPLFTM
CFVPLVCWVVCTCLQQQLEGGG-LL-R--QM-SRTTTAVYMLYLLSLMQPKPGA-PCL--
-----------QPPPNQRGLCSLA---ADGL-WN-QK----ILFEEQ-------------
--DLRKHGLDGA-DVSAFLNM----NIFQKDIN-CE-RYYSFIHLSFQEFFAAMYYILD-
E--GE--------------R--G--AGPDQDVT-RLLTEYAFS-ERSFLALTSRFL----
----FGLLNEETRSHLEKSLCWK---------------V--SPH-IKM-DLLQWIQSKAQ
SD-----------------------------------GSTLQQGSLEFFSCLYEIQE-EE
FIQQALSH---------------------F-------------------QVIVVSNIA-S
KM-EHMVS--S----FCLK-HCRS-AQV-LHLY----------------GT------TYS
-ADGEDRA--R-CS---------------------------------------AGV---H
TL------------------------L-------------------------V-------
QL-----------PERTVLLD-------AYSEHLAAAL----------------CTNPNL
IELS-LYRN-ALGSR-GVKLLCQGLRHPNCK----LQNL--------RLF---------G
M-----------------------------------------------------------
--------DLN---KM-THS---RLAALRVT-K----------------PYLDIGC----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCSAP00000005252 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MKMA----ST
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 328)
RCK---LARYLEDLE--D---VDLKKFKMHLED------
---------Y--PPQKG--------C--ISL-PRGQ-TEKAD-HVDLATLMIDFNGEEKA
WAMAVWI----FA-AINRRDLYEKAK-RDEPKWGSDNAR--VS---------NPTMI---
--------CQ-----------------------------------EDSI-----------
-----------------------------------------------------EEEWMG-
------------------L-----------------LEYLSRISICKKKKDYCKKYRKY-
--------VRSR----------------FQCI--EDRNARLGESV---------SLNKRY
TRLRLIKEHRSQQEREHELL------AIGKT--KTWE------SP--VSPIKMELLFD--
PDDEHSEPV----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
HTVVFQGAAGIGKTILARKI-----MLDWA--S---------GTLYQ
D--------RFDY-LFYIHCREVSLVT-------QRSLGD-LIMS-CC-----PD-PNPP
IRKIVS-KP----SRILFLMDGFDE-LQGAFDEH------IG-P-----LCTDWQKA-ER
-GDILLSSLIRKKLLPEASLLIT-TRPVALEKLQHLL-D-----HPR--HVEILGFSEAK
RKEYF----------------F--KY-FSD---------EAQARAAFSLIQE-NEVLFTM
CFIPLVCWIVCTGLKQQMESGK-SL-AQ--T-SKTTTAVYTFFLSSLLQPRGGSQEHR--
------------LCAPLWGLCSLA---ADGI-WN-QK----ILFEES-------------
--DLRNHGLQKA-DVSAFLRM----NLFQKEVD-CE-KFYSFIHMTFQEFFAAMYYLLE-
EEKE--------GR-TNVPGSCL--KLPSRDVT-VLLENYGKF-EKGYLIFVVRFL----
----FGLVNQERTCYLEKKLSCK---------------I--SQQ-IRL-ELLKWIEEKAK
AK-----------------------------------KLQTQPSQLELFYCLYEMQE-ED
FVRRAMDY---------------------F-------------------PKIEINLSTRM
---DHVVS--S----FCIE-NCHR-VES-LSLG----------------FLHN-----MP
KEEEE--------E------------------------------------E---------
---------------KEGRHLDMVQCVLPGS-----------------------------
------------------------------------------------------------
--------------------------HAACS----H-----------RLV---------N
S-----------------------------------------------------------
--------DLT---SS-FCR---GLFSVLST-SQSLTELDLSDNSLGD-PGMRVLCET--
LQH-PDCNIRRL-W----------------------------------------------
------------------------LGRCGLS-----------------------------
----------------------------HECCFDISL-----------------------
--------------------------------VLSSNQKLVE---LD-LSDNA-LGD---
-------FGIRLLCVGL----KHLLCNLKKL-----------------------------
--------------------------------W---------------------------
-------------------------------LVSCCLTSACCQDLASV-LSASRSLTRLY
V-GENALGDAGVA-ILCEKAKNP--QCN----L---------------------------
-----------------------QKLGL--VNS-----GLTSVCCSAL------------
-------SSVL-STNQNLTHLYLRGNTLGDKGIKL-LCEGLL-HP-DC-KLQVLE--LD-
--N-CNLTSHCCW--------------------DLSTLLTSSQSL-QKLSLGNNDLGDLG
VMMFCE--------VLKQQSCL--LQ--N-------------------------------
---LGLSEM-YFNYETKSA------LE----------TLQ-EEKPGLTIIFEPSW-----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCGRP00000016706 (view gene)
ENSMPUP00000009935 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MPRA----PVSHGLC
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 325)
RLSAYLEELE--A---VELKKFKLYLGT------
---------E---AE----------A--GRI-PWGR-LETAG-PLDTAQLLVAHYGPDAA
WPLALGL----FQ-RISRRDLWERGL-REDLVRDT----A--PG--------GPSSL---
------------------------------------------------------GS----
------------------------------------------------------------
---QS-ACSL-------------------------------EVFPGAPTRDPQETYRDY-
--------VRRK----------------FRLM--EDRNARLGECV---------NLSHRY
TRLLLVKEHSNPMWAQQKLL------DTSWGQARTVG--------HQTSFIQLETLFEPD
EE--RPEPP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
RTVVLQGAAGMGKSMLAHKV-----MLDWA--D---------GRLFQ
D--------RFDY-VFYINCREMNQSTA------EQSAQD-LISS-CW-----PE-PSVP
LRELVR-VP----ERLLFIIDGFDE-LKPSFHDP------RG-P-----WCLCWEEK-RP
-TELLLCSLIRKKLLPELSVLIT-TRPTALEKLHRWL-E-----HPR--HVEILGFSEAE
RKEYF----------------Y--KY-FHN---------TEQAAQVFHFVRD-NEPLFTL
CFVPMVCWVVCTCLKQQLEDGG-LL-R--QT-SRTTTAVYMLYLLSLMQPKPGS-PTL--
-----------QPPLNQRGLCSLA---ADGL-WN-QK----ILFEER-------------
--DLRKHGLDGA-DVSSFLNM----NIFQKDIN-CE-KFYSFIHLSFQEFFAAMYYLLD-
P--GE--------------A--G--ASPELTVA-RLLVEYGFS-ERSFLALTVRFL----
----FGLLNEETRSYVEKSLCWT---------------V--SPH-VKA-ELLRWIQRKAQ
SE-----------------------------------GSTLQQGSLELFSCLYEIQE-ED
FIQQALSP---------------------F-------------------QVVVVSNMA-T
RM-EHLIA--S----FCVK-SCRN-ALV-LRLH----------------GA------AYS
-TDEDDGG--R-WA---------------------------------------SGP---Q
M-------------------------PP------------------------A-------
Q---S--------AEKNLLPE-------AYSKQLAAAL----------------STKPNL
TELV-LSRN-ALGSR-GIRLLCQGLRHPGCK----LQNL--------SLK---------R
C-----------------------------------------------------------
--------CVA---SS-ACQ---DLAATLMA-SRTLTRMDLSCNGLGL-PGVRLLCEG--
LRH-PRCRLQVIQ-----------------------------------------------
------------------------LRKCQLE-----------------------------
----------------------------AGACQEIAS-----------------------
--------------------------------VLSINQHLLE---LD-LTGNA-LED---
-------LGLRFLCQGL----KHPIC----------------------------------
--------------------------RLR-ILW---------------------------
-------------------------------LKICHLTAAACEDLAST-LRVNQSLMELD
L-SLNDLGDPGVL-LLCEGLRCH--QCR----L---------------------------
-----------------------QTLRL--GIC-----RLSSAACEGF------------
-------CTVL-QINHHLRELDLSFNDLGDTGMWP-LCKGLR-HP-TC-RLEKLW--LD-
--S-CGLTAKACE--------------------DLSSTLGVNQTL-RELYLTHNTLGNGG
VRLLCK--------GLSHPGCK--LQ--V-------------------------------
---LWLFGM-DLNKMTQRR------LA----------ALR-LTKPHLDIGC---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCCAP00000000787 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-MDQSDAPCSSAGPR--FAVAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LQAALEDLS--Q---EQLKRFRHKLRD------
------------VRSDG-----------RSI-PWGR-LERAD-AMDLAEQLAQFYGPEPA
LEVARKT----LK-RADARDVAAQLQ-EQRLRR---------L---------GLG-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------PA-AQVSVSEYKKKYREH-
--------VLRR----------------HARV--KERNARS------------VKITKRF
TKLLIAPESAAALEE--ALGPAE-------EPEP--------QRARRSDTHTFNRLFRGD
E---GGRRP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYQ
A--------QVDF-AFFLPCGELLERPG------ARSLAD-LILD-QC-----PD-RGAP
VQQMLA-QP----ERLLFILDGADE-LPALGGP--------EAA-----PCTDPFEA-AS
-GARVLGGLLNKVLLPSALLLVT-TRAADPGRLQGRL-C-----SPQ--CAEVRGFSDKD
KKKYF----------------Y--KF-FGN---------ERRAERAYRFVKE-NETLFAL
CFVPFVCWIVCTVLHQQLELGR-DL-S--RT-SKTTTSVYLLFVASVLSSAPAADRP---
-----------RVQGDLRKLCRLA---REGVLEC--R----AQFAEE-------------
--ELEQLELRDSKVQTQFFSKKEQPGVLET-----E-VTYQFLDHSFQEFLAALSYLLE-
DEGVS-------------------G-TAAGGVG-TLLRGDAQP--HSHLVLTTRFL----
----FGLLSAERMGDIQRHFGC-----------------RVSER-VKQ-EALRWVQG--Q
GCPGAAPEVTEGAKGLEDTEEPEEEE---------EEEGEEPNYSLELLYCLYETQE-DA
FVRQALSR---------------------L-------------------PVLALQRVRFS
RM-DVAVL--S----YCVR-CCPP-GQA-LWLI------------------SC----RLV
------AA----------------------------------------QEK--K---KKS
----------------------------------------LGKR----------------
------LQAS---L--------------GGS------S-----------------SRASA
KQ---LPPS-------LLHPLFQAMTDPLCG----LSSL--------TLS---------H
C-----------------------------------------------------------
--------KLP---DA-VCR---DLSEALRA-APTLMELSLLHNRLS-EAGLRMLSEG--
-LAWRQCQVQTF------------------------------------------------
----------------------------RVQ-----------------------------
---------------LSGP---------QRGLQYL-------------------------
------------------------------VPMLRESPTLTT---LD-LSGC-QLPAPIV
--T--------YLCAVL----QLPGCSLQTLSL--ASVELSEQ-----------------
----------SLQELRAVKRAKP-------DLV----------------------I--AH
PALD-------------------GHPEPP-KDLSSIF-----------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMODP00000014409 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------MFQGNRRKYINR-
--------MKEK----------------FSLTWDGSSSS-LGNLR---------DLNKEF
VPLMLLHKYKE--LKN---I------CRKNTPVSV--------MTKQAHIIDMSELFGSD
ER--KY-VP----Q
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVLCGGAGIGKTILTRKI-----MLDWA--E---------GDLFQ
D--------RFKN-VIYVNCKTINLTL-------EMTLVD-LISK-DW-----PD-LQAS
IEAIMS-NP----KQLLFIIDGFDE-VKWPFVEP------EP-E-----LCHHWKEK-CP
-VPVLLRSLLWKTIFPRSSLLIA-TRSSALMRFRCLI-D-----DPN--CVEVLGFTEAN
IEMYI----------------S--KF-FKS---------EAKVLKVLDLLKV-SDTLSEK
CSVPLECWIICTCLKQQMERGE-VP-V--LL-PQTTTDLYVSYLSFGLSHDGSS--SSQ-
-----------HHDLQLRRLCSLA---AEEV-WA-GK----FTFNED-------------
--DLKRHGLDVS--DDSFLNM----YILEKK---DKENCYSFIHGRFLEFFAAMFYVLE-
E------KRE--------ERDYP--AVAVKDVI-EL-LVQTRE--SDSEVSIMNFL----
----FGLLNEETMRQLDRNFVYP---------------T--IQR-GKA-ELLQWMILESN
IQR--------------------------------MINPLLHPDPLELFQCLYEIRQ-EE
LVRKTMKY---------------------F-------------------QSMRLNLHTSK
DI---LVS--I----FCIK-LCYN-LVN-IHLH----------------ISRT-------
------L-----------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------MYNSPHI-----------------------------
----------------------------LIQWKELIS-----------------------
--------------------------------VLLFKKSLIE---LE-LSGSF-PDD---
-------RLMKIVYKE--------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------IMHPNCK--LH--T-------------------------------
---LRFHQV-ELSEEMKKE------FE----------NLP-KMKNFL-------KNVEII
PDSEMKEEEYENSPQD-------------------HQRFEPEE-----------------
------------------VTPQTEAEVTSLL----ASFDP-LKEEQLDLEKCLFLGPNGP
VEPEWIDK-KRNIYKVRLSMAGFYHCP---YTGLGFDVRSEV-------TVDIEFGTWNQ
HLDWI-QQQSWMIAGPLFHITVDPGV-VTSVHLPHFVHLQGG--------KVDISLFKIA
HFKEEGMVLENPTRMESSHAILENPSFSPMGVLLRIVHSTFRFIPIYSTTLIYFRLRAED
I-TLHLYLIPSDCTIRKAIDEEEAKFHFLRLHKPSPVTPLYIGSRFLVSGS---------
KHLEIMPTELELCYQSAGECQFFSEIYTSNVKEIISLDLRDPK-GTVVWGTLVRSGDVGT
VCAPISP---------------------------------------ATTEL
PF00619 // ENSGT00900000140813_0_Domain_19 (3052, 3134)
HFVDQHRNQ
IISRVTSVDIILDRLH-SQFLSNEDYEHIRAEKTNPEKMRRLFSFSPSWSKDYKDHLYRV
LREIHPHLMNELEHMEKILVRK--------------
ENSTBEP00000012634 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-MVESEVPCSSMELV--TATA-KL---LLTTLKDPG--L---E--LKCILKLWD------
------------EWIGV-----------HRI-FWRQ-LEC-M-VVDFTKQLTHLHQLEPA
LEVTCKT----LQ-RADMRDVTSQLK-EQQQGQLL-------L---------GPR-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------SS-ALLFLSE-SKNYQTW-
--------LLKS----------------S-KV--------E------------RKINKCF
ILLKMMSWEEALHL---------------VEPES--------ECTQRWDMYNFD--FHHD
----RKFPS----LTMVLQAQEVSY
PF05729 // ENSGT00900000140813_0_Domain_2 (686, 930)
KTMAAKKI-----LCDWA--A---------GKLYQ
G--------EVDF-AFLIQWYELLGQPG------MCNLPD-LVLY-QC-----PD-HGTL
KQQMLG-QP----QWLLFILDCTEE-SQVLFAP--------NAV-----PCTGPFKT-TS
-STQILGGLLSKV-----------------------------------------------
-------------------------F-FRG---------EQRVKHAHSFMKK-TEMLFTL
CFEPFLCWIVCILL-HQLKLDQ-DL-L--CT-SKTTMSVYF-FIASVLRIPPAADP----
-----------C----------LP---YEGIFWC--K----VQF-TM-------------
--DLEELELLSSKVQTLLLSKE-KVLLLET-----Q-VTYEPIQQSFQEFCVALFYLLE-
KRGLK-------------------V---------------------LHLDFTMNFL----
----FVLVTMEQLHN-THHFGC-----------------LISVP-VKR-EALQ---VQGQ
VLLSVAPEGTKGSKVD--TKE------------TKEEEGEEFSLELELLHCLYETE--VV
FVGQGLCG---------------------L-------------------PESVLEPVCFS
CT-DLLVL--S----YCVD-CCQL-GKA-LKPV------------------RF----RQA
------SA----------------------------------------QGK--K---KKI
----------------------------------------LVKW----------------
------LQGS---L--------------GGX------X----------------XXXXXX
XXX----XX-------XXXXXXXXXXXXXXX----XXXX--------XLS---------R
C-----------------------------------------------------------
--------KLS---DT-MCR---DLSEALRA-APALTELCLLQVRLS-EAGLRLLIEG--
-LAWPQCRVHTL------------------------------------------------
----------------------------RMQ-----------------------------
---------------VPGL---------QRVLQYV-------------------------
------------------------------IGLLQQRPALIT---LD-LRGC-QLCGSLL
--T--------SLCAVL----QHPGCGLQTL-----------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSFCAP00000020543 (view gene)
ENSMPUP00000012189 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MNMT----SV
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 328)
RCK---LARYLEDLE--D---ADFKKFKMHLQD------
---------Y--PSQKG--------C--RPL-PRSQ-TEKAD-HMDLATLMIDFNGEEKA
WAMAVWI----FA-AINRRDLYEKAK-RDAPEWD------------------NVHVV---
--------CQ-----------------------------------EESL-----------
-----------------------------------------------------EEEWMG-
------------------L-----------------LGYLTRISICKKRKDYCKKYKKH-
--------VRSR----------------FQCI--KDRNARLGESV---------NLNKRY
TRLRLVKEHQSQQEREHELL------AIGRTSARMLD------SP--VSSLNMELLFE--
PDDQHSEPV----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
HTVVFQGAAGIGKTILARKI-----MLDWA--S---------EKIYQ
D--------RFDY-LFYIHCREVSLGT-------RRSLGD-LIVS-CC-----PD-PNPP
ICKIVS-KP----SRILFLMDGFDE-LQGAFDEH------TE-T-----LCTNWQKV-ER
-GDILLSSLIRKRLLPEASLLIT-TRPVALEKLQHLL-D-----RPR--HVEILGFSEAK
RKEYF----------------F--KY-FSD---------EQQATEAFRLIQE-NEVLFTM
CFIPLVCWIVCTGLKQQMDSGK-SL-AQ--T-SKTTTAVYVFFLSSLLQSHGENQKHQ--
------------VSANIRGLCSLA---ADGI-WN-QK----ILFEEC-------------
--DLRNHGLQRA-DVSAFLRM----NLFQKEVD-CE-KFYSFIHMTFQEFFAAMYYLLE-
EEEQ--------GELRNLPWRSS--KLPNRDVT-VLLENYGKF-EKGYLIFVVRFL----
----FGLVNQERTSYLEKKLSCK---------------I--SQH-IRL-ELLKWIKEKAK
AK-----------------------------------NLQIQPSQLELFYCLYEMQE-ED
FVQKAMGH---------------------F-------------------PKIEISLSTRM
---DHVVS--S----FCIE-NCHS-MES-LSLR----------------LLHN-----SS
KEEEEEEE-------------------------------------------E---E----
---------------GEMQLSEVDHCVRLDP-----------------------------
------------------------------------------------------------
--------------------------HAACT----H-----------RLV---------N
C-----------------------------------------------------------
--------YVT---LS-LWQ---GLFSVLSTNSLNLTELNLSDNALGD-PGVQVLCEM--
LQR-PGCNIRRL-W----------------------------------------------
------------------------LGRCCLS-----------------------------
----------------------------HHCCFNISS-----------------------
--------------------------------VLSSNQKLME---LD-LSHNT-LGD---
-------FGIRLLCVGL----RHLFCNLKKL-----------------------------
--------------------------------W---------------------------
-------------------------------LVSCCLTSACCEDLASV-LSTNQSLTRLY
L-GENALGDAGVG-ILCEKAKHP--QCK----L---------------------------
-----------------------QKLGL--MNS-----GLTSGCCPAL------------
-------SSVL-STNQKLTHLYLRGNSLGDTGVKL-LCEGLL-HP-NC-KLQILE--LD-
--G-CSLTSHCCW--------------------DLSTLLTSNRSL-RELSLGSNDLGDLG
VMLLCE--------VLKQQGCL--LK--S-------------------------------
---LKLCEM-YFNYDTKCA------LE----------TLQ-EEKPELTIVFEPFW-----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPPAP00000004041 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------MAESDSTDFD---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWYLENLS--D---KEFQSFKKYLAR------
---------K--ILDFK--------L--PQF-PLIQ-M---T-KEELANVLPISYEGQYI
WNMLFSI----FS-MMRKEDLCRKI---IGRRN---------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------RNQEACKAV-
--------MRRK----------------FMLQ--WESHTFGKFHYKFFRDVS--------
----------------------------------------------SDVFYILQLAYDST
SY--YSANN----LN
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 930)
VFLMGERASGKTMVINLA-----VLRWI--K---------GEMWQ
N--------MISY-VVHLTSHEINQMT-------NSSLAE-LIAK-DW-----PD-GQAP
IADILS-DP----KKLLFILEDLDN-IRFELNVN------ES-A-----LCSNSTQK-VP
-IPVLLVSLLKRKMAPGCWFLIS-SRPTRGNNVKSFL-K-----EVD--CCTTLQLSKEK
REIYF----------------N--SF-FKD---------RQRASAALQLVHE-DEILVGL
CRVAILCWITCTVLKRQMDKG-RDF-Q--LC-CQTPTDLHAHFLADALTSEAGLT-AN--
----------QYHLGLLKRLCLLA---AGGL-FL-ST----LNFSGE-------------
--DLRCVGFTEA-DVSVLQAA----NILLPSST-HK-DRYKFIHLNVQEFCTAIAFLMAV
PNYLI-----------------P----SGSR-E--Y--KEKRE-QYSDFNQVFTFI----
----FGLLNANRRKILETSFGYQ---------------LPMVDS-FKW-YSVGYMKYLDR
DP-------------------------------------EKLTHHMPLFYCLYENRE-EE
FVKTIVDA---------------------L-------------------MEVTVYLRSDK
DM---MVS--L----YCLD-YCCH-LRT-LKLS----------------VQH-----IFQ
--NKEPLI-------------------------------------------RPTASQMKS
LVYWREIC-SLFYTMESLWELHIFDNDLNGI---------SERILS--KALEHS------
SC--K-LRTLKLSY---------VST-ASGFEDLLKAL----------------ARNRSL
TYLS-INCT---------------------------------------------------
------------------------------------------------------------
--------------------------------------------SISL-NMFSLLHDI--
LHE-PTCQISHL-S----------------------------------------------
------------------------LMKCDLR-----------------------------
----------------------------ASECEEIAS-----------------------
--------------------------------LLISGGSLRK---LT-LSSNP-LRN---
-------DGMNILCDAL----LHPNCTLISL-----------------------------
------------------------------------------------------------
------------------------------VLVFCCLTENCCSALGRV-LLFSPTLRQLD
L-CVNRLKNYGVL-HVTFPLLFP--TCQ----L---------------------------
-----------------------EELHL--SGC-----FFSSDICQYI------------
-------AIVIA-TNEKLRSLEIGSNKIEDAGMQL-LCGGLR-HP-NC-MLVNIG--LE-
--E-CMLTSACCR--------------------SLASVLTTNKTL-ERLNLLQNHLGNDG
VAKLLE--------SLISPDCV--LK--V-------------------------------
---VGLPLT-GLNTQTQQL------LM----------TVK-ERKPSLTFLSETWSLKEGR
EIGVT-------------------------------------------------------
-------------------------------------------------------PASQP
GSI-----------------IPNSNLD---YM--FFKFPRMSAAMRTSNTASRQPL----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMICP00000009458 (view gene)
ENSPTRP00000072999 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------P-SPFSEAAL-KSLI----KEALSLGKLLIITF--
------ILLKERPP---------------------------------LDVDEMLERLK-T
EALAFP---------------------------------ET---KGNVI-----------
-------CLG-------------------------------KDVFKGKKPDKDNRCRYI-
--------LKTK----------------FREM--WK--SWPGDSKEVQV-----------
---------------------------------------------MAERYKMLIPFSNPR
VL--PGPFS----Y
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVLYGPAGLGKTTLAQKL-----MLDWA--E---------DNLIH
---------KFKY-AFYLSCRELSRLG-------PCSFAE-LVFR-DW-----PE-LQDD
IPHILA-QA----RKILFVIDGFDE-LGAAPG------ALIE-D-----ICGDWEKK-KP
-VPVLLGSLLNRVMLPKAALLVT-TRPRALRDLRILA-E-----EPI--YVRVEGFLEED
RRAYF----------------L--RH-FGD---------EDRAMRAFELMRS-NADLFQR
GSAPAVCWIVCTTLKLQMEKGE-DP-V--PT-CLTRTGLFLRFLCSRFPQ-------GAQ
------------LRGALRTLSLLA---AQGL-WA-QT----SVLHRE-------------
--DLERLGVQES-DLRLFLDG----DILRQDRV-SK-GCYSFIHLSFQQFLTALFYALE-
KEEEE-------------DRDGH--TSDIGDVQ-KLLSGVERL-RNPDLIQAGYYY----
----FGLANEKRAKELEATFGCR---------------M--SPD-IKQ-ELLRCDISCKG
G------------------------------------H-STVTDLQELLGCLYESQE-EE
LVKEVMAQ---------------------F-------------------KEISLHLN-AV
DV---VPS--S----FCVK-HCRN-LQK-MSLQ----------------VIK-----ENL
-PENVTAS----ES--------------------DAEVER------------SQDD-QHM
LPFWTDLC-SIFGSNKDLMGLAINDSFLSAS---------LVRILC--EQIAS-D-----
TC--H-LQRVV--F-KNISPA--DL----I------------------------------
--------G-TSDD--MFPALCERRVFENIRKMSNPAKL--------RLI---------S
A------G----------------------------------------------------
---------ES---PN-S-------------------------------GGGRLQLS---
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMLEP00000006325 (view gene)
MGP_SPRETEiJ_P0081462 (view gene)
ENSSHAP00000000660 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------------EKYRAQ-
--------VRKK----------------FQVL--EERNARLGEWV---------DLSQHY
TRLLLVRGHASPWRREQELL------ASGGLHGLLRE-------EH-RRPIELQTLFDAD
AEE-AGSPP----T
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVLQGVAGIGKTTLARKV-----LLDWA--A---------GTLYP
G--------RFDY-VFYVNCRTVEVVA-------PLSVAE-LLSG-WLEDTGPGS-GPAP
VPALLA-RP----ERLLFILDGFDE-LLWGPQAQEAGP------------RVPGEQR-QQ
-VDVHLRSLLGKRLLPEASLLIT-TRPTALEKLQPLL-E-----RPW--LAEVLGFRASE
RRAYF----------------D--KY-FPA---------PGQADRAWAAVQA-NKVLFTM
CFVPLVCWVLCTGLRQQMERGE-AP-A--QS-ARTTTAVYVAFLSSLVRPDR-HGP-PRI
------------PAAGLRGLCSLA---ATGQ-RE-RR----LWFRER-------------
--DLARHGLDAATV-STFLQM----DQFQHEAE-CR-ATYSFFHSTFQEFFAALFLVME-
RPRW---------------PFR---RPG-WGLL----LKGPSG-LQADLPLTIRFL----
----FGLSSRERADALETMVGE---------------PV--PRG-SQR-VLLRWIREQAA
QGP--------------------------------G-----RAKTLELLYCLFETQE-PE
FVRRAMGC---------------------F-------------------REVKVR--VRT
RM-DGLVA--T----FCLR-ASRS-TLA-VELD----------------GLG-----SGA
-A----------------------------------------------------------
--GLGFQE----------------PG-LADQ---------ET------------------
----------------GTYSL--KD---ICQ-DLSSGL----------------WVQRSL
SELG-LGED-ALGDS-GMRWLCEDLQRPRCP----LQSL--------RLL---------N
C------W----------------------------------------------------
---------LT---SD-FAE---SLSCALNV-SHTLTSLDLSYNPLED-QGAALLFRA--
LEH-PRCRLQKL-C----------------------------------------------
------------------------LKKCHLT-----------------------------
----------------------------EKSCENLGP-----------------------
--------------------------------VLGVSRTLQW---LD-LGFNV-LGD---
-------RGVGLLCGGL----RQATC----------------------------------
--------------------------QLQ-TL----------------------------
--------------------------------RTCGLTCQCCLDLASV-LSLGARLLCLF
L-RNNALGDSGVQ-QLCQGLRAP--RCR----L---------------------------
-----------------------RELSL--ANC-----RLTHACCASI------------
-------SSVL-SASNSIESLFLSGNDLEDVGIQE-LARGLR-HP-NC-KVQELK--VQ-
--M-CGLTRLGCE--------------------TLALVLSSCPSL-RQLDLSNNCVGEEG
TKLLGR--------GLLQPGCR--LQ--S-------------------------------
---LKLFWA-ALSEADKAE------LA----------AVA-RAKPGLEISY---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMGAP00000009467 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------FQKNIFCRQ-ASW-ASWG----------------AV---------
----FAQP-------------------------------VSWEAA------MLDDC-NLS
SNNCKDLS-SIIHTNP
PF00560 // ENSGT00900000140813_0_Domain_4 (1517, 1551)
SLKELKLNNNELGDA---------GIEYLS--KGENSGT00900000140813_0_Linker_4_5 (1551, 1566)
LLT-P-----
SC--SPF00560 // ENSGT00900000140813_0_Domain_5 (1566, 1597)
-LQKLW--L-QNCNLT--SA---SCE-TLRSVL----------------SAQPSL
TELH-VGDN-KLGTA-GVKVLCQGLMNPNCK----LQKL--------QLE---------Y
C------E----------------------------------------------------
---------LT---AD-IVE---ALNAALQA-KP
PF00560 // ENSGT00900000140813_0_Domain_8 (1775, 1801)
TLKELSLSNNTLGD-TAVKQLCRG--LVE-ASCDLELL-H----------------------------------------------
------------------------LENCGIT-----------------------------
----------------------------SDSCRDISA-----------------------
--------------------------------VLSNKPSLLD---LA-VGDNK-IGD---
-------AGLALLCQGL----LHPNC----------------------------------
--------------------------KIQ-KLW---------------------------
-------------------------------LWDCDLTSASCKDLSRV-FSTKETLLEVS
L-IDNNLRDSGME-MLCQALKDP--KAH----L---------------------------
-----------------------QELWV--REC-----GLTAACCKAV------------
-------SSVL-SVNKHLQVLHIGENKLGNAGVEI-LCEGLL-HP-NA-TSTLYG--LG-
--N-CDITAACCA--------------------TLANVMVTKQ
PF00560 // ENSGT00900000140813_0_Domain_17 (2444, 2475)
NL-TELDLSYNTLEDEG
VMKLCE--------AVRNPNCK--MQ--Q-------------------------------
---LILYDI-FWGPEVDDE------LK----------ALE-EARPDVKIVS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
MGP_SPRETEiJ_P0081466 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------MMDSSGYG---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLQYLQKLS--D---EEFQRFKEHLRK------
---------E--PEKFK--------L--KPI-SWTK-IKNTS-KEDLVMQLYTHYPGK-A
WDMVLSL----FL-QVNREDLSTMAQ-TERRD----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------KQTKYKEF-
--------MKNT----------------FQHI--WTMETNTYIPDR--------------
---------------------------------S-------YHEFIEVQYRALQDIFDCE
-----SE-P----V
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVVSGSRGGGKTTFLRKA-----MLDWA--S---------RNLLQ
N--------RFQY-VFYFSVFSLNNIT-------ELSLAE-LISS-TL-----PES-SET
VDDILS-DP----KRILFILDGFDY-LKFDLE-L------RT-N-----LCNDWRKK-LP
-IQIVLSSLLQKIMLPECSLLLE-LGNASLSNIIPLL-Q-----YPR--EIIMSGFSEQT
IEIYC----------------V--SF--FN---------TQTGIEIFKNLKS-IKPLFNL
CRCPHLCWMICSTLKWQYERRE-VA-S--RF-GRTLGLLYTIFMVSAFKSTY-ARNPSK-
-----------QNRARIRTLCTLA---VEGM-WK-QV----YVFDSD-------------
--DLRRNGISES-DKKVWLRM----AFLQNQG-----SNIVFYHSTLQWYFAVLFYFL--
QYKDT-----------------R--HPVIGNLA-QLLGEIYAH-KQNQWFHTRILL----
----FGMATEQVTSLLEPCFGCI---------------S--TKE-VRQ-EIIRYIKSLSQ
QEC----------------------------------NEKLVVHPQNLFFCILDNQE-ER
FVRQLMDR---------------------F-------------------EEMTVDISDVD
DM---SAT--P----YCLH-RAPK-VKN-LHLH----------------IQKR----VFL
-EIHDPEY-----------G-------------------D-L-------ELFK-LDQKLL
AKHWTTLC-TFLC---NLHVLDLDSCHFNEN---------AIEVLC--NCLPLTSLVPLT
GF--K-LHRLLCSF---------TTN-FGD-GLLFCTF----------------LHLPHL
KYMN-LYGT-NLSND-AVERMCSALKFSTCG----VEEL--------LL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------GKCDIS-----------------------------
----------------------------SEACGIIAT-----------------------
--------------------------------SLINS-EVKH---LS-LVENP-LKN---
-------KGVMSLCEML----KDPSCVLESL-----------------------------
------------------------------------------------------------
------------------------------MLSYCCLTFIACGHLYEA-LLSNEHLSLLD
L-GSNFLEDTGVN-LLCEALKDP--KCT----L---------------------------
-----------------------KELWL--PGC-----YLTSECCEEI------------
-------SAVL-TC
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NTNLKTLKLGNNNIQDAGVKR-LCEALC-HP-NC-EMQCLG--LD-
--M-CDFTSDCCE--------------------DLALVLTT
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2465)
CNTL-KSLNLDWNAFDHSG
LEMLCE--------ALNHKACN--LE--V-------------------------------
---LGLDKS-LFSEESQTL------LQ----------AVE-KKKKNLKVLHFPWVEE---
--ELE-----KR------------------------------------------------
--------------------------------------------------GVRLVWNSKN
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSFDAP00000016473 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MSLDIRCEQ-LND-ARWI----------------EL---------
----LPLI-------------------------------QQHQVV------RLEDC-GLT
EGRCKDIS-TALRA
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1542)
NSALTELSLRTNELGDA---------GVCLVL--QGLQS-S-----
TC--K-VQNLS--L-QNCGLS--ET---GCH-VLASVL----------------RCT
PF13516 // ENSGT00900000140813_0_Domain_6 (1618, 1642)
PTL
RELH-LSDN-PLGDT-GLRLLSEGLLDPQSC----LEKL--------ELE---------S
C------N----------------------------------------------------
---------LS---AA-SCE---PLAAILRA-KPNFKELVLSNNCIQE-AGIRVLCGG--
LKD-STCLLEML-R----------------------------------------------
------------------------LDNCGVT-----------------------------
----------------------------SANCKDLGD-----------------------
--------------------------------VVAA
PF13516 // ENSGT00900000140813_0_Domain_11 (2017, 2054)
KPSLQE---LD-LGGNR-LGD---
-------VGIVALCPGL----LHPNS----------------------------------
--------------------------RLR-KLW---------------------------
-------------------------------LWECDITAEGCRDLCQV-LRA
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
KQSLKALS
L-MLNELGNEGAR-LLCEALQEP--SCQ----L---------------------------
-----------------------ECLWV--RVC-----SLTDTCCPHF------------
-------RVML-A
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
QNKFLTELQLGENKLGDTGVQE-LCEGLG-QP-GS-VLRVLG--LG-
--D-CDLTDNSCS--------------------NLASVLLA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2464)
SCSL-RELDLSNNGLGDPG
VLQLVE--------SLRQPGCI--LE--C-------------------------------
---LLLFDV-YLSDGVNDQ------LQ----------ALE-EEKPSLRIIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMFAP00000026016 (view gene)
MGP_PahariEiJ_P0019874 (view gene)
------------------------------------------------------------
------------------------------------------------------------
-------------VYCP-----------------GSPCGECAMCDPGCFGVSGTGPIQ--
VASVM-ITLLITSSA--DAVAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LMAALQELT--Q---EQLKRFRHKLRD------
------------APLDG-----------RSI-PWGR-LERSD-ALDLVDKLIEFYEPVPA
VEMTRQV----LK-RSDIRDVALKLK-EQQLQK---------F---------GPS-----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------S-VLLFVSEFKKKYREH-
--------VLRQ----------------HAKV--KERNARS------------VKINKRF
TKLLIAPGTGAVENE--LLGPSG-------EPEP--------ERARRSDTHTFNRLFRGN
EE--EGSHP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYH
S--------QVDF-AFFIPCGELLERPG------TRSLAD-LVLD-QC-----PD-RSWP
VRRILA-QP----SRLLFILDGVDE-LPTLASS--------EAT-----PCKDPFEA-TS
-GLRVLSGLLSQELLPSARLLVT-TRNATTGRLQGRL-C-----SPQ--CAEIRGFSDKD
KKKYF----------------F--KF-FRD---------ERKAERAYRFVKE-NETLFAL
CFVPFVCWIVCTVLQQQLELGR-DL-S--RT-SKTTTSVYLLFITSMLKSA-GTNGP---
-----------RVQGELRTLCRLA---REGILDH-HK----AQFSEE-------------
--DLEKLKLRGSQVQTIFLNKKELPGVLKN-----E-VTYQFIDQSFQEFLAALSYLLE-
AERTP-------------------G-TPAGGVQ-KLLNSNAEL--HGPLALTTRFL----
----FGLLNTEGIRDIGNHFGC-----------------VVPDH-VKK-DTLQWVQGQSH
PK--GPPGGAKKNAELEDIKEAEEEEE-------EEEEEEDLNFGLELLYCLYETQE-ED
FVRQALSS---------------------L-------------------PEMVLERVRLT
RM-DLEVL--N----YCVQ-CCPD-GQT-LRLV------------------SC----GLV
------AA----------------------------------------KEKK-K---KKS
----------------------------------------LVKR----------------
------LKGS----------------------------------------------QSTK
KK---PPAS-------LLRPLCETMITPQCH----LSVL--------ILS---------H
C-----------------------------------------------------------
--------KLP---DA-VCR---DLSEALKV-APALRELGLLQSQLT-DTGLRLLCQG--
-LAWPKCQVKTL------------------------------------------------
----------------------------RMQ-----------------------------
---------------LPGL---------QEVINYL-------------------------
------------------------------VIVLQQSTALTT---LD-LSGC-QLPGVMV
--E--------PLCSAL----KHPKCCLKTLSLT--SVELSEN-----------------
----------SLRELQGVKTSKP-------DLS----------------------I--IY
SK----------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPPYP00000000034 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MKMA----ST
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 328)
RCK---LARYLEDLE--D---VDLKKFKMHLED------
---------Y--PPQKG--------C--IPL-PRGQ-TEKAD-HVDLATLMIDFNGEEKA
WAMAVWI----FA-AINRRDLYEKAK-RDEPKWGSDNAR--VS---------NPTVI---
--------CR-----------------------------------EDST-----------
-----------------------------------------------------EEEWMG-
------------------L-----------------LEYLSRISICKMKKDYRKKYRKY-
--------VRSR----------------FQCI--EDRNARLGESV---------SLNKRY
TRLRLIKEHRSQQEREQELL------AIGKT--KMCE------SP--VSPIKMELLFD--
PDDEHSEPV----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
HTVVFQGAAGIGKTILARKI-----MLDWA--S---------GTLYQ
D--------RFDY-LFYIHCREVSLVT-------QRSLGD-LIMS-CC-----PD-PNPP
IHKIVR-KP----SRILFLMDGFDE-LQGAFDEH------IG-P-----LCTDWQKA-ER
-GDILLSSLIRKKLLPEASLLIT-TRPVALEKLQHLL-D-----HPR--HVEILGFSEAK
RKEYF----------------F--KY-FSD---------EAQAKAAFSLIQE-NEVLFTM
CFIPLVCWIVCTGLKQQMESGK-SL-AQ--T-SKTTTAVYVFFLSSLLQPRGGSQEHC--
------------LCAHLWGLCSLA---ADGI-WN-QK----ILFEES-------------
--DLRNHGLQKA-DVSAFLRM----NLFQKEVD-CE-KFYSFIHMTFQEFFAAMYYLLE-
EEKE--------GR-TNVPGSLL--KLPSRDVT-VLLENYGKF-EKGYLIFVVRFL----
----FGLVNQERTSYLEKKLSCK---------------I--SQQ-IRL-ELLKWIEVKAK
AK-----------------------------------KLQIQPSQLELFYCLYEMQE-ED
FVQRAMDY---------------------F-------------------PKIEINLSTRM
---DHVVS--S----FCIE-NCHR-VES-LSLG----------------FLHN-----MP
KEEEE--------E------------------------------------E---------
---------------KEGRHLDMVQCVLPGS-----------------------------
------------------------------------------------------------
-------------------------SHAACS----H------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMLUP00000021206 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----RSLDHDSRGSQVGHLYHHK----H----SPKEKGQS--HSNILK-GRIKEKYQNQ-
--------RRES----------------SCPTQSW-------KNE---------YLHQKF
TQLLFLHRSQPRGYDF---L------LRRR-NHEV--------VDEQENWMEIGDLIGPG
LS--TQEEP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
PTVVVHGVAGIGKSTLARQV-----RRAWE--E---------GWLFR
D--------LFKH-VFYFNCRELAQSE-------TMSLAE-LITK-DW-----AA-PDAP
IGQILS-QP----EQLLFILDNLDE-PKWDLKEQ------SS-E-----LCPHWSQQ-QQ
-VHTLLGSLLKKTLLPTASLLIT-ARITALRKLIPSL-K-----HPH--WVEVLGFSESA
RKEYF----------------F--QY-FTD---------ESQAIRAFTLVES-NPALFTM
CLMPLVSWLVCTCLKQQMEQEQ-PL-S--LT-SQTTTALCLHYFFHVLQ--------AQ-
-----------SLGTKLRGFCSLA---VEGT-CQ-GK----TLFSPK-------------
--DFREHGLDGA-NISTLLNM----GVLKEH---PTPQSYSFIHLCFQEFFAAMSYALG-
D------GVL--------KGDCF--NNI-IITI-DLTGVYDRH--DLFGEPTTCFL----
----FGLLSDQGMREMETIFKCS---------------L--SRK-RLP-SLLHLAKREVL
-----------------------------------LKRLNLDPYSWHLLHCFYEIQD-ED
FMKNIF-I---------------------P-------------------FTLRICVQTDM
EL---LLF--T----FFFT-FCHR-IER-LQLN----------------EGGQ-------
------H-R---------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------Q------------
------AGRT--------------------------------------------------
-------------------SGAFLYSWGPFT-----------------------------
----------------------------DSWWQMFFS-----------------------
--------------------------------ILKVPR
PF00560 // ENSGT00900000140813_0_Domain_11 (2019, 2057)
SLRE---LD-LSGNS-LSC---
-------SAVQSLCDAL----KSPHCRLATLR----------------------------
------------------------------------------------------------
-------------------------------LVRCGL-----------------------
------------------------------------------------------------
----------------------------------------TY------------------
------------------------------------------------------------
---------NCCQ--------------------DLASMLGASP
PF00560 // ENSGT00900000140813_0_Domain_17 (2444, 2463)
NL-TELDLRQNDLGDLG
VRLLCE--------GLRQPACQ--LR--N-------------------------------
---LWLNLT-RLSEEVRQA------ER----------ALE-RIDHSHSIST-HINNHARC
PGMEGGQGPETEL-----------------S-----------------------------
----------------------------SLSSCEPLGN---QPVEPLT-MDNYFWGPTEP
VATEMVDK-HRNLYRAHFPVAGSYHWP---NTGLYFVVRGPV-------TIEIEFCAWDQ
FLDQTVPQHSWMVAGPLFDIKAKPGA-VAAVYLPHFIALQGQ--------HVDISLFQVA
HIKEDGILMEKPASVEPHYIVLENPSFSPMGVLQRMIHTALC-IPVISNVLLYYRLQSEE
V-NFHLYLIPSDCTVRKAIDDEEKKFQFFQIRKPPPLTPLYLGSRYTVSGS---------
DKLEIIPEELELCYRSPGKPQLFSEFYVGHFVSGIRLQIKSKDDETVVWKALVKSGDLRP
EAPLV-----------------------------------SQAMTDAPTL
PF00619 // ENSGT00900000140813_0_Domain_19 (3051, 3135)
LHFVDQHRED
LVTRVTSVDTVLDKLH-GQVLSEEQYERVRAEPTNPDKMRKLFSFSKSWDWACKDQLYQA
LKVTHPHLTEDLWEK---------------------
ENSCHOP00000002517 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------PS-PSFFG---SPNVASLESPS
WPTSTEVLRFCDPEQP------SPQDSDQRRVLRYLTNLSGSSW-RENPSSLYFS-----
-----------HSPS-HTSPNQESPTPPTSTAVLGGWELPSQSRTDAREQTAPRMIGLLV
ESRGNKERSS---------------------------------------PED-----RE-
--------ALHH----------------TGNSQSW-------KKG---------ILHQKF
TELLLLHRPHPQGHGP---L------VSRSWPQEV--------LEERGHLIEVQDLFGPA
LG--IQEEA----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
PTVILLGAAGIGKSTLARQV-----RGAWE--D---------GQLYR
D--------RFQH-VFYLDCRELAQFK-------VASLVE-LLEK-DW-----PT-PLVP
TGQILS-QP----EQLLFILDGLDE-PVWVFEEQ------CS-E-----LCLHWSQQ-QP
-VAPM----WWNTTLPEASLLIT-VRNTALEKLIPSL-K-----QPH--WVEVLGFSEAG
RKEYF----------------H--KY-FTG---------ESQAIRAFSLVES-NQALSTL
CLVPWVSWMVCTCLKQQMEKGE-EL-P--LA-SQTTTALCLHFLSQTLP--------SQ-
-----------PFSTHLRNLCSLA---AKGT-QK-GK----TLFSPA-------------
--EFRMQGLDRG-LILTFIKM----GVLQKH---PTSLSYSFIHLCFQEFLAAIFCVLG-
DLELG-DEEE--------TRDHP--SSI-QRVK-KLLRRYGRH--DLFGAPTTR------
----------HGARELENIFNCK---------------P--S---GGW-KLLRWTEAEVQ
-----------------------------------HRESSLRLCSLELFHCLYEIQD-EE
FLTQAMAH---------------------L-------------------QGTRVQVQTDP
DL---LVL--A----FCLK-FCHN-VKR-LQVN----------------DVGQ-------
------H-G---------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------P------------
------ASWD--------------------------------------------------
-------------------PRVALFIKGPIT-----------------------------
----------------------------DAQWKIFFS-----------------------
--------------------------------ILGVNGSLQE---LD-LSGNS-LSH---
-------SAVQALCEAL----GHPHCHLETLR----------------------------
------------------------------------------------------------
-------------------------------XXXXXXXXXXXXXXXXX-XXXXXXXXXXX
X-XXXXXXXXXXX-XXXXXXXXX--XXX----X---------------------------
-----------------------XXXXL--AGC-----VLTS------------------
------------------------------------------------------------
---------SCFQ--------------------DLASVLSASPSL-XXXXXXXXXXXXXX
XXXXXX--------XXXXXXXX--XX--X-------------------------------
---XXXXXX-XXXXXXXXX------XX----------XXX-XXXXXXXXXX-XXKQNVAI
PNEDSDGSQMDRS-----------------KSSLKRQRSES-------------------
-----H--VPQG------QSPQVAQ-VKPHWVPGSTLHKD-LHMEPLA-TEDDFWGPTGP
VATQVVDK-EKSLYRVHFPVAGSYYWP---NTGLCFVVRWAV-------TLEIEFCAWKQ
LLKGIGLQHSWMVAGALFDIKAEPGA-VAAVHLPHFVVLEGG--------HVDISLFQVA
HFKEEGMLLEKPARVEPSYIILESPSFSPMGVLLRVIHAALR-IPITSTVLLYHHPHPEE
V-SFHLYLIPSDCTIQKAIDDEEKKFQFVRILKPPPLTPLFMGSRYTVCGP---------
GKLEIIPKELELCY-SPGESQLFSEFYVGHLGSGIRLQVKDKKNETMVWEALVKAGDLRP
VVTLLPP-----------------------------TLTASSPFPE-APA
PF00619 // ENSGT00900000140813_0_Domain_19 (3051, 3135)
LHFVDRYREQ
LVARVTSVDPVLDKLY-GQVLSQEQYERVQAEATKPSQMRKLFSFSPSWDWACKDQLYRA
LKETHPHLIVELWEKWGSSRDG--VLMLS-------
ENSDNOP00000005182 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------E-VEEAK-GKQLAEILTN-SPDSWT
SLYTLRA----ED-FTVQLSLSSMFQ-WALN-------LK------HLKK---PNSTP--
------SENTWSSK-AL--------------------------D---AHLQEKKEHAEK-
GELECI---------------------------------QVKTMDDPKL-----------
-------GEA-------------------------------EEDLEVEKPGKKESDRNM-
---------VEV----------------ISSL--WKNKFCPADSENFHYNV---------
--------------------------------------------------IPFVPFFNPK
AP--VELTS----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
HTLVLQGAAGVGKTMLAKKL-----MLDWT--E---------DELGQ
---------TFRY-AFYLCCRRLNRVG-------PCTFAE-LVLQ-GW-----PA-LPEA
IVDVLA-QP----QKVLFVVDGFDE-LRVPPG------ALIH-D-----LCGDWRTQ-KP
-VPILLGSLLKRKMCPKATLLVT-TRPWALRELRLLV-E-----QPV--VLEVEGFSESD
REEYF----------------L--KH-FED---------EAGALRALAAARS-NAALFGL
GAAPPVCWIICSCLKLQMLKGQ-DP-T--LT-CQTTTSLFLRFLCSQFTPM-PGARPTER
------------LQAPMKALCLLA---VQGI-WA-RT----STFDSE-------------
--DLRRLGLEES-ELGLFLDK----SILLRDRD-CE-GAYCFVHLSIQQFLAAMFYLLG-
GEE-E-------------DRDSS--RWDIWDVE-ELLSKEERL-KNPSLTQVGYFL----
----FGLFNEERVKELETTFGCQ---------------I--SVE-VKR-QLLRCTAKYNE
NK-----------------------------------PFSSMMGMKELFYCLYESQE-EE
FVKDVVGH---------------------F-------------------KEMSFHLINTS
EM---LYS--S----FCLK-HCQN-LQK-ISLQ----------------VEKG----IFL
-END-STS----EL--------------------H------------SQAERSQND-HHS
LHLWMEFC-CVFSSNRNLSFLEVSQSFLSDS---------SVRILC--AQIVQ-G-----
TC--N-LQKVV--I-KHVSPA--DA----YR-DFCLAL----------------IGKKTL
THLI-LEGS-VQGE--PLLLLCESLKHSGCN----LQYL--------RLE---------S
C------S----------------------------------------------------
---------GT---AQ-QWA---DLFSTLRI-NQSLTCLNLTSSELSE-EGVRLLCAT--
LTH-PKCSLQRL-S----------------------------------------------
------------------------LENCCLT-----------------------------
----------------------------EACCKDLSS-----------------------
--------------------------------ALVVNQRLTH---LC-LAKNH-LGD---
-------EGVKLLCEGL----SYPDC----------------------------------
--------------------------KLQ-MLV---------------------------
-------------------------------LWSCNITRCSCKHLAKL-LQGNSSLTHLD
L-GLNPIA-AGLW-FLCEALKEP--NCN----L---------------------------
-----------------------KSLWL--WGC-----SITPYYCQDF------------
-------ASAL-ISNQSLETLDLGQNILGQSGVTV-LFEALK-QK-NG-PLKTLR--LR-
--I-DQSSVEIQK--------------------LLKEVKESNPELTIDCND------AKT
---------------NRPSCCD--FL----------------------------------
----S-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPCOP00000010092 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MNN---------------------------QKKRKDMEPA-----
--------CTWS------------------------------------------LLSE--
-----------------------------------------KR-----------------
----VASSSL-------------------------------ASFSWRELTDYREVFRKH-
--------LKEK----------------YLKI-GVDKCYHHGK--------------HIE
SRLLLVKEHGISEKTPCGIP-------------------------QQDHFIEMEHVFDPD
EE--GSSSS----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 929)
RTVVLQGCAGIGKTAVVHKF-----MFDWA--A---------GMVTP
G--------RFDY-LIYINCREISHIA-------NLSAAD-LITN-TF-----QD-INGP
ILDIILIYP----EKLLFILDGFPE-LQYPVGDR------EE-D-----LSAHPQER-KP
-VESLLCSFVRKKLFPESSLLIT-ARPTAMKKLHSLL-K-----QPI--QAEILWFTDAE
KKAYF----------------L--SR-FSG---------ANAAMKVFYGLRE-NEGLDIM
SSLPIVSWMICSVLQSQGDGDR-TL-L--RS-LQTMTDVYLFYFSKCLKTLTGIS-VW--
-----------KGQSCLWGLCSLA---AEGL-QN-HQ----VLFAVS-------------
--DLRRHGIGVYDSNCAFLNH-----FLKKAEG-DI-NVYTFLHFSFQEFLTAVFYALK-
NDNSW---------------------MFFDQVGKTWQEIFQQY-GKGFSSLMIQFL----
----FGLLHKGKGKAVETTFGRK---------------I--SPG-LRE-ELLKWTEREIK
DK-----------------------------------SSRLQIEPVDLFHCLYEIQE-EE
YSKKIIDD---------------------L-------------------QSVILLQPTYT
KM-DILAM--S----FCVK-SSHSHLS----------------V-----SLKCQHLPGF-
-EEEEPAS----------------------AFVP--------------------------
---------PALTLNESCQ-----LHDLPMS---------QLHLLC--QALRNP------
YC--K-IKDLKL---IFCHLT------ASCGRD---------------LSSVFET
PF13516 // ENSGT00900000140813_0_Domain_6 (1616, 1642)
-NQYL
TDLE-FVKN-TLEDS-GMKLLCEGLKQPNCI----LQTLR--------LYRCLI-SPA-S
CG------------AL----AAVLSTNQWLTELEFSETKLEASALKLLCEGLKDPNCK-L
QKLKLCASLLPESSEA-VCK---YLASVLIC-
PF13516 // ENSGT00900000140813_0_Domain_8 (1773, 1796)
NPHLTELDLSENPLGD-RGVKYLCEG--
LRH-SNCKVEKL-D----------------------------------------------
------------------------LSTCYLT-----------------------------
----------------------------DASCVELSS-----------------------
--------------------------------FLQMSQTLKE---LF-VFANA-LGD---
-------TGVQHLCEGL----QHAKGIIENL-----------------------------
------------------------------------------------------------
------------------------------VLSECSLSAACCEPLAQV-LSSTRSLTRLL
L-INNKIEDLGLK-LLCEGLKQP--DCQ----L---------------------------
-----------------------KDLAL--WTC-----HLTGACCQDL------------
-------CNAL-Y
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
TNEHLRVLDLSDNALGDEGMQV-LCEGLK-HP-CC-KLQTLW--LA-
--E-CHLTDACCG--------------------ALASVLKR
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2465)
NENL-TLLDLSGNDLRDFG
VQMLCD--------ALIHPICK--LQTFY-------------------------------
------IDTDHLREDTFRK------MD----------ALK-MSKPGITW-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
MGP_PahariEiJ_P0045529 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------M-HLGKHANFVYLF-FFFAEAVL-EAPK----EKVSSK----------
----------QREP----------------------------------------------
----------------------------------------------SET-----------
-------LPA-------------------------------PRNYTQRAPNPEDKQKVE-
--------WKTG----------------YTAK--WKKNFWPKCNKEIYV-----------
---------------------------------------------ATESYRTLLALCNPK
I---ETPFS----H
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 931)
AIVLHGPPGSGKTTIARRL-----MLEWS--E---------SKQAQ
---------IFSC-VFYISCRELNNTK-------PCTFAH-LLSM-DN-----PS-WRDY
VIRGLL-LG----QEFLFVVDGFDE-LTFPAG------AMIH-D-----LCGDWNTV-KP
-VEVLLGSLLKRKMAPHATLLVT-TRTQSLRQIFIMM-D-----QPL--LVETRGFLEQE
KQEYF----------------Q--KY-FEDEEGEEEDEGEGKALRALKEVRC-NADLYQM
ASLPAACGIFCLCLELRMKKGE-RL-A--LT-CQTYTSMFLNFLCEVFSPETCEGHLNKE
------------LQILFKKICILA---ANSL-LE-QV----PVLCEE-------------
--DFLKLKLNLN-QLHPIVSR----YILFKDSS-ST-HCLSFICLGIQQFLAAVIFVQE-
LGQES-----------------K--DVSKYSIQ-NMLSREARF-KNPDLSGMLPFV----
----FGLLNETRIQELETIFGCQ---------------I--SPG-VKR-KFLERE--SGE
NK-----------------------------------PLLLMMNMQEILSCLYESQE-EG
FVKEAMVL---------------------F-------------------EDISLHLKTNT
DL---IHA--S----FCLK-NSQN-LQT-MSLK----------------VEKA----IFP
-ENV-SAL----ES--------------------TAKHQR------------SPDE-QRM
LDFWTDFC-DTFKSNEKLVFLNIHESFLNSS---------ALEILC--EKLPS-A-----
SC--C-LQNVV--L-KNISPD--DA----YE-KVCLVF----------------NGYKTI
SHLT-LQGG-NLDS--MHHSLCEVLKNPACN----LKFL--------SLG---------S
C------S----------------------------------------------------
---------TA---DQ-KWD---GFFPVLKV-
PF13516 // ENSGT00900000140813_0_Domain_8 (1773, 1796)
NQSLIYLDLTDNNLLD-KGAKLLCSI--
WKE-PTCILQRV-S----------------------------------------------
------------------------LENCQLT-----------------------------
----------------------------EACCKDLSS-----------------------
--------------------------------VLMV
PF13516 // ENSGT00900000140813_0_Domain_11 (2017, 2054)
SQTLTH---LS-LANNK-LGD---
-------NGVKYLCESL----SHTAC----------------------------------
--------------------------KLQ-TLV---------------------------
-------------------------------LWSCNITNTGCHHVSKM-LGQTL
PF13516 // ENSGT00900000140813_0_Domain_13 (2215, 2237)
SLKHLD
L-GLNRIGTTGAK-FLCEALKNP--KSK----L---------------------------
-----------------------KSLWL--CGC-----SITPLSCQDF------------
-------SETL-R
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
SNKSLNTLDLSQNTLGTDAVKT-FCEALK-LQ-IC-PLQILR--VR-
--F-DEAKPTIKN--------------------LIQEMKVSHPQLSISSDRV----DLKN
-----------------PPLPH--FI----------------------------------
----F-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSOGAP00000013513 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------MAESFSAA
PF02758 // ENSGT00900000140813_0_Domain_0 (203, 325)
FD---LLWYLQKLS--D---EEFHSFKSHLIA------
---------E--IQKFG--------P--PEI-LWTD-LEV-S-KEVLVNMLTVSYEEQHT
WNMTFSI----LQ-KLGRKDLCQKI---TIRRN---------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------RDKEMYKSL-
--------MRTK----------------FLNQ--WETCPFSEIHYLYFCEVT--------
----------------------------------------------KDIFYTLELSYDSS
T---SSTEN----LN
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 930)
VFLVGDRAKGKSMIIRVA-----VIEWT--L---------GEMWN
D--------TISY-IIHITSHEINEMT-------NVSLVE-LISK-DW-----PE-DQAP
IADILS-EP----QKVLFILEDLDN-MNISLNVD------ES-A-----LCSDSRHK-VP
-VSVLFVSLLKRKVVPGCRVLVS-SRPSCEPCIKALM-N-----ERD--CYVTLQLSKEK
KQNYF----------------Y--LF-FKN---------KVRAERAFKRVLD-SEILVDL
CEIPLLSWILCTVLNQQMNR--LDF-E--YS-CQTPTDIYTYYLASLLTSDPDVT-AQ--
----------QYHLILLNRLCLLA---LEGL-SH-NT----ISFSGS-------------
--DLVSVGLTCY-DVSVLQAM----NILLRSSN-HE-VHYEFIHLNIQEYCAALACLMVL
PHCSI-----------------P----SARS-T--D--KEKRE-LYDNFSPVITFI----
----FGLLNERRRKILETSLGCQ---------------FPMVES-VTQ-NLVMHMKYLAA
NP-------------------------------------NLMEHHMPLFYCLLEYVD-EE
FVTQVMGF---------------------F-------------------SEATIFIQDHK
DL---MAS--L----HCLR-HCQP-LQK-LKLI----------------VQQ-----VFQ
--YGTPIE-------------------------------------------LLTYNQMRG
LAYWREIC-SIFHNRENFRELEIYNSDLNRV---------SEMLLS--KALRHP------
NC--Q-LQTLRLSY---------VSV-SADFDDLFMSV----------------AQNQNL
TFLN-MNCI---------------------------------------------------
------------------------------------------------------------
--------------------------------------------PFSL-EMFSLLREA--
LSS-PTCNIQHL-S----------------------------------------------
------------------------LMRCDLQ-----------------------------
----------------------------ASACREIAF-----------------------
--------------------------------ILISSRKLKK---LT-LSNNP-LKN---
-------DGAIILCEAL----LHPDCVLESL-----------------------------
------------------------------------------------------------
------------------------------GLYFCCLTEGCACYISRL-LICTRTLKHLD
L-SVNYLQSHAVM-TLTFPLSLS--DCQ----L---------------------------
-----------------------RELEL--TGC-----FFTGNACQCI------------
-------ADAIV-CNVNLRRLELGYNNLGDAGVEA-LCSGLQ-DP-DC-KLESVG--LE-
--C-CMLTSACCT--------------------TLATVLSISKTL-KKLNLKENNLGDEG
IIKLIQ--------GLGHPSCV--LE--L-------------------------------
---LGIRVS-DFSAETQSL------LI----------AVR-EKNNKLVFSSSFSTAQESR
QFTDL-----LG------------------------------------------------
-----------------------------------------------------SGSLPEP
VSK-----------------TPNYPWH---SI--FFKLPRKFSSTTEETIVSEQSL----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSFCAP00000029963 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MNLDIQCQQ-LSD-ARWT----------------EL---------
----LPLI-------------------------------QQYQVV------RLDDC-GLT
EVRCKDIC-SALQANPSLTELSLCTNELHDA---------GVRLVL--QGLQS-P-----
SC-
PF13516 // ENSGT00900000140813_0_Domain_5 (1564, 1594)
-R-IQKLS--L-RNCCLT--NT---GCE-VENSGT00900000140813_0_Linker_5_6 (1594, 1617)
LPDAL----------------RSPF13516 // ENSGT00900000140813_0_Domain_6 (1617, 1642)
LPTL
RELQ-LSDN-PLGDA-GLQLLENSGT00900000140813_0_Linker_6_7 (1642, 1650)
CKGLLDPQPF13516 // ENSGT00900000140813_0_Domain_7 (1650, 1766)
CH----LEKL--------QLE---------Y
C------N----------------------------------------------------
---------LT---AA-SCE---PLENSGT00900000140813_0_Linker_7_8 (1766, 1774)
AAVLRA-WPF13516 // ENSGT00900000140813_0_Domain_8 (1774, 1796)
RHVKELVVSNNDIGE-AGVQALCRG--
LVD-SACQLETL-K----------------------------------------------
------------------------LENCGLT-----------------------------
----------------------------PASCKDLCG-----------------------
--------------------------------VVASK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
ASLQE---LD-LGDNK-LGD---
-------QGIATLCPAL----LHPSS----------------------------------
--------------------------RLR-VLW---------------------------
-------------------------------LWDCDITTTGCRDLCQV-LRAKE
PF13516 // ENSGT00900000140813_0_Domain_13 (2215, 2237)
SLKELS
L-AGNALGDEGAR-LLCESLLEP--GCQ----L---------------------------
-----------------------QSLWV--KSC-----GLTAVCCPHV------------
-------SAML-T
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
HNKHLVELQMSDNKLGDSGVQE-LCQGLS-QP-SA-MLRVLW--LG-
--D-CDVANGGCN--------------------SLASLLVV
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2465)
NRSL-RELDLSNNCMDDRG
ILQLME--------SLERPDCA--LE--Q-------------------------------
---LVLYDI-YWTQQTEDL------LQ----------ALG-ERKPGLRIIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCSAP00000016337 (view gene)
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------MS------------------
----------FSVTTWADGGT
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 326)
NQG---LLPYLVALD--Q---YQLEEFKLCLEPQQLMDF
WSA----------PQGH--------F--PHI-PWAN-LRAAD-PFNLSFLLDEHFPKGQA
WKVVLGI----FQ-TMNLTSLCEEVR-AKMKGNVQ-TQELQGPTQE------DVEMLEAA
----AGNMQTQGCQ-----------------------D----------PNQEEPEELE--
E-----------------------ETGNV----------QVQGCQDPNQ-----------
------EEPE-------------------------------MLEEA----DHRRKYREK-
--------MKAK----------------VLET--WDNIDWPKDHVYVRNTSE--------
----------------------------------------------G-EHEELQRLLDPK
R---TRTQA----Q
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TIVLVGRAGVGKTTLAMQA-----MLHWA--N---------GVLFQ
Q--------RFSY-VFFLSCHKIRYMK-------ETSFAE-LISL-DW-----PD-SDAP
IEEFMS-QP----EKLLFIIDGFEE-IIISEPSS--ESLNDG-S-----PCIDWYQE-LP
-VTKILLSLVKKELVPLATLLIT-IKTCFVIDLKPSL-V-----NPC--FVQITGFTGED
LWVYF----------------M--RH-FED---------SREVEKILQQIRK-NENLFHS
CSVPMVCWTVCSCLKQSKVRYY-DL-Q--SI-TQTTTSLYAHFFSNLFSTAE-VDLADES
------------WPGQWRALCNLA---MEGL-WS-VN----LTFDRE-------------
--DTELEGLEVP-FIDCLYKF----NIIQKGND-CR-GCITFTHLSFQEFFAAMSFVLE-
EPREL--------------L-PH--STKPQEMK-ILLRDVLFN-KEAYWTPVVLFF----
----FGLLKKNLARELEDTLHCK---------------M--SPR-VME-ELLKWGEELGK
AE-----------------------------------SASLQFHVLQLFHCLHESQE-ED
LAKKMLGH---------------------I-------------------FEVDLNILGNE
EL---QAS--S----FCLK-HCKR-LDK-LRLS----------------VSS-----HIL
-ERDLEIL----ET--------------------------------------S-EV-DSR
MHAWNSIC-STLVTNENLHELDLSNSKLHAS---------SVKGLC--LALKN-P-----
RC--K-VQKLT--C-KSVTPE--CV---L-Q-DLILVL----------------QGNSKL
THLN-FSSN-KLGAT-V-PLILKALKHSACN----LKYL--------CL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------EKCNLS-----------------------------
----------------------------AASCQDLAL-----------------------
--------------------------------FLTSIQHVTR---LC-LGFNQ-LQD---
-------DGIKLLCVAL----THPKC----------------------------------
--------------------------ALE-RLE---------------------------
-------------------------------LWFCQLGAPSCRYMSDA-LLQNRSLAHLN
L-SKNHLGDEGVK-FLCEALGHP--DCN----L---------------------------
-----------------------QSLKM--KGH-----EANQQRERDL------------
-------GQSPELHQHSVMGLNLAVTRLECSGDLS-SLQP-L-PPGFK-QFSCLR--LA-
--K-CNVTTACCQ--------------------HLSSVLSSSKSL-VNLNLLGNELDHDG
VKMLCK--------ALRKSTCR--LQ--K-------------------------------
---LG-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCSAP00000016339 (view gene)
------------------------------------------------------------
------------------------------------------------------------
-------------M-SDVNPPSD------------T-PVPFSSS-------------S-T
H---SSCNPPWTFSCYPGSPC
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 325)
ENG---VMLYMRYVS--N---KELQWFKQFLLN------
---------E--L-RAG--------T--TPI-TWDQ-VKRAS-WAEVVHLLIERFPGRRA
WDVTSNI----FA-VMNRDKMCALVH-REINAILP-TLEPEDLNVG------E-------
------------------------------------------------------------
------------------------------------------------------------
-------TQM-------------------------------NLEEGDS--GKIQQYKLN-
--------VIVK----------------CFPT--WDFMIWPGNQRDFFYQDV--------
----------------------------------------------HRHMEYLPCLLLPK
RP--QGRQP----K
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 928)
TVVIQGVSGIGKTTLAKTV-----MLEWA--S---------NRFYA
H--------KPWC-AFYFHCQEMNQTT-------EHSFSE-LIEH-KW-----PG-SQDL
VAKIMS-KP----DQLLLLLDGFEE-LTSTLI------NRPE-D-----LSEDWRQK-LP
-GSVLLSSLLSKKMLPEATLLIM-IRCTSWPTCKPLL-K-----CPS--LITLPGFNTTE
KIKYF----------------Q--MY-FGY---------GE-TDRVLSFAME-NTILFSM
CQVPVVCWMVCSGLKQQMERGN-NL-T--QA-CPNATSVFVRYISSLFPTRA-ENFSRKL
------------HQAQLEGLCHLA---ADGM-WH-RQ----WVFGKE-------------
--DLEEAKLDQT-GVSAFLGM----SVLRRIAG-EE-DRYTFTLLIFQEFFAALFYVLC-
FPQRL--------------QNFH--VLSHVNIQ-RLIASPR-G-SKSYLSHMGLFL----
----FGFLNGTCASAVEQSFQCK---------------V--SFN-NKR-KLLKVIPLLQK
CE-----------------------------------PPSPCSGVPQLFYCLREIRE-EA
FVSQALNG---------------------Y-------------------RKVVLKIGNSK
DV---QVS--A----FCLK-RCAH-LQE-VELT----------------VTP-----SFK
-NVWKLSS----G-------------------------------SHPGS--EAPES-KEL
HCWWQDLC-SVFATNDKLEALTLTSSALGPP---------FLKALA--AALKH-P-----
QC--K-LQKLL--L-RRVNSA--ML---N-Q-DLIGVL----------------MGNQHL
RYLE-IQHV-EVESN-VVKLLCKALRSPRCR----LQCL--------RLE---------D
C------L----------------------------------------------------
---------AT---PR-IWT---DLGSNLQV-NRHLKTLILTKNSLEN-CGACY------
--L-SVAQLERL-S----------------------------------------------
------------------------IENCKLT-----------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------QLSCESLASC-LRKSKMLTHLS
L-AENALKDEGVK-HIWNALSHL--RCP----L---------------------------
-----------------------QRLVL--RKC-----DLTFNCCQDM------------
-------ISAL-CKNKTLKSLDLSFNSLKDDGVIL-LCEALK-NP-DC-TLQILE--LE-
--N-CLFTSICCQ--------------------AVASMLCKNQHL-RHLDLSKNVIEVDG
ILTLCE--------AFSSQKKR--EE--V-------------------------------
---I------L-------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSNLEP00000027320 (view gene)
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------MN------------------
----------FSVTTCAGGDTIQG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 323)
-LLPYLMALD--Q---YQLEEFKLCLEPRQLMDF
WSA----------PQGH--------F--PCI-PWAN-LRAAD-PLNLSFLLDEHFPKGQA
WKVVLGI----FQ-TMNLTSLCEKVT-AKMKENVQ-TQELQDPTQE------DVEMLEAA
----AGNMQTQGCQ-----------------------D----------PNQEELEELE--
D-----------------------ETGNV----------QAQECQDPNQ-----------
------EEPE-------------------------------MLEEA----DHRRKYREK-
--------MKAE----------------LLET--WDNISWPKDHIYIRNTSK--------
----------------------------------------------D-EHEELQRLLDPN
R---TRAQA----Q
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TIVLVGSAGVGKTTLAMQA-----MLHWA--S---------GVLFQ
Q--------RFSY-VFYLSCHKIRYMK-------ETTFAE-LISL-DW-----PD-FDAP
IEEFMS-QP----EKLLFIIDGFEE-ILISESHS--ESLDDG-S-----PCTDWYQE-LP
-VTKILHSLLKKELVPLATLLIT-IKTWFVTDLKASL-V-----NPC--FVQITGFTGED
LWVYF----------------M--RH-FDD---------SSEVEKILQQIRK-NETLFHS
CSAPMVCWTVCSCLKQPKVRYY-DL-Q--SI-TQTTTSLYACFFSNLFSTAE-VDLADDS
------------WPGQWKALCSLA---IEGL-WS-MN----FTFNKV-------------
--DTEIEGLEVP-FIDSLYEF----NILQKIND-RG-GCTTFTHLSFQEFFAAMSFVLE-
EPGEF--------------L-SH--STKPQEMK-MLLRDVLLN-KEDYWTSVVLFF----
----FGLLNKNIARELEDTLHCK---------------I--SPR-VTE-ELLKWGEELGK
AE-----------------------------------SASLQFHILQLFHCLRESQE-ED
FAKKMLGH---------------------I-------------------FEVDLNILGDE
EL---QAS--S----FCLK-HCKR-LNK-LRLS----------------VSS-----HIL
-ERDLEIL----ET--------------------------------------S-KV-DSR
MHAWNNIC-STLVTNENLHELDLSNSKLHAS---------SVKGLC--LALKN-P-----
RC--K-VQKLT--C-KSVTPE--WV---L-Q-DLIIVL----------------QG
PF13516 // ENSGT00900000140813_0_Domain_6 (1617, 1633)
NSKL
THLN-FSSN-KLGMT-V-PLILKALRHSDCN----LKYL--------CL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------SGC-----SFTREGCREL------------
-------ANAL-R
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
HNHNVKILDLGENDLKDDGVKL-LCEALK-PS-PR-ALHTLG--LA-
--K-CSLTTACCQ--------------------HLFSVLSSRKSL-VNLNLLGNELDPDG
VKMLCK--------ALRKSTCR--LQ--K-------------------------------
---LG-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCJAP00000044881 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----MSVD--T----QCLDIQCEE-LS--GRWA----------------KL---------
----LPLL-------------------------------QQCHTV------RLDDC-SLT
EAHCEGIS-SALQA
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1542)
NPVLTELNLRSNGLGDA---------GVHRIL--QALQS-S-----
SC--K-IQKLS--F-ECCKLT--VD---GCR-VLSGSL----------------RS
PF13516 // ENSGT00900000140813_0_Domain_6 (1617, 1642)
LPTL
QELN-LTCN-ALKDA-GLQLLCEGLLDPQCH----VEKL--------QLE---------F
C------R----------------------------------------------------
---------LS---SA-SCE---PLASVLRA-KPDFKELTVGNNDFDE-AGIRVLCQG--
LKE-SPCQLETL-K----------------------------------------------
------------------------LERCGVT-----------------------------
----------------------------SDNCRDLCS-----------------------
--------------------------------ILASK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
ASLRE---LA-LGHNN-LGD---
-------MGIAELCPGL----LHPSS----------------------------------
--------------------------RLR-TLW---------------------------
-------------------------------IWECGLTAKSCEDLGRV-LRAKD
PF13516 // ENSGT00900000140813_0_Domain_13 (2215, 2237)
SLKELS
L-AGNKLGDEGAQ-VLCESLLEP--GCQ----L---------------------------
-----------------------ESLWV--KYC-----GLTAACCSHF------------
-------SSVL-AQ
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NKFLLELQISENRLGDAGVQE-LCQGLG-QP-GS-VMRSL------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSOGAP00000016459 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------EP----H
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
IVILHGAAGIGKSTLARQV-----QRGWE--K---------GQLYQ
D--------RFQH-VFYFSCAELAQSK-------VMSLAD-LITK-AH-----GC-PPVP
LQQVLS-QP----EQLLFILDGVDE-PRWVLWQE------ST-E-----LCQHWSQP-QQ
-VATLLGSLLGKTILPEASFLIT-TRTVALQKLIPSL-E-----QPR--WVEVLGFSESS
RKEYF----------------Y--NY-FID---------ESQALRALSLVES-NPAVWTM
CLVPWLSWLVCTCLKQQMEMGE-DL-T--LT-TPTATGFCLRYLSQALL--------AQ-
-----------PLGPRLRGICYLA---AEGI-RQ-KR----SLFTPR-------------
--DLRKHGLEEA-IISTFVKI----GVLQKH---PNSRGYSFIHLCFQEFFAAVACALG-
D------EKE--------RREHP--DSI-KGVG-GLLQAYETH--NLLWTPTARFL----
----FGLLSEQAGREMESIFTCQ---------------R--APE-LRW-ELLQWAKREAQ
-----------------------------------GLWSPWKPGFLELFHCLYETQD-KE
FLTQVMCH---------------------F-------------------LASSVCVQTHM
EL---LVF--T----FCIK-FCHH-VKR-LQLH----------------DVGQ-------
------HRP-RWRPPGIVLHPIRTTE----------------------------CDPLLV
LKVWIRYYCS---PGKKVSTLETRK----------------EKILR------G-R-----
NV--R-LRSFIFRF---FSVA-------SGSGMMWRHK----------------MANAST
QFCR-FGGR-WIKD--GEKLECIKSKNRKMS-----------------------------
------------------------------------------------------------
LKLEIELKKVN---H--GC----SLCHLLLQ-TLSVVLLSLEGDRVPR------------
------HWESRI-S----------------------------------------------
-------MPKENRRVCIKEKKAVHFMQVPIT-----------------------------
----------------------------GACWELLFS-----------------------
--------------------------------ILEVTGSLKE---LD-LSGNL-LSH---
-------SAVQSLSETL----RRPRCHLETLR----------------------------
------------------------------------------------------------
-------------------------------LACCDLTLEDCKDLALG-LS--QTLTELE
L-SFNMLMDAGAK-YLCWELR-Q--NCT----L---------------------------
-----------------------QRLQL--VSC-----GLTS------------------
------------------------------------------------------------
---------SCCQ--------------------DLASVLSTSPSL-KELDLQQNDLGDHG
VRLLCE--------GLRHPVCS--LT--L-------------------------------
---L--------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSETEP00000006696 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------LVADSGRLRD
GVAPSLS----FL-QMGLWSPCPTLE-APS------------AVFG---FASSPRLGSPN
GPTSTEVLRFWDPDHPSRKRRE-------------LADSYEHRXXXXXXXXXXXX--X--
---------XXXXXX----------X-----XXXXXXXXX-X---XXXXXXXXXXX----
-XXXXXXXXX---X-------------------XXXXXXXXXXXXXXXXXXNRESSEPQ-
--------QRGL----------------EANTQSW-------KDE---------DSHQKF
TQLLLLHTPHPRGRES---W------GKASWHDGV--------LQEQGRLIEVQDLFGPD
LD--TQDAP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
HTVILQGAAGTGKSTLARQV-----RRAWE--Q---------GQLFK
D--------RFRY-VFYLNCRELAQSR-------TVSLPE-LLTQ-DW-----AA-PPAP
AGQILS-RP----EQLLFILDGLDE-PKWVFGEQ------TS-E-----LCLHWSQQ-QP
-VPTLLGSLLSKAILPGVSLLIT-ARTKALGELIPSS-E-----HPR--WVEVLGFSRAG
RKDYF----------------Y--NY-FPD---------ESQAAQAFSLVES-NSALLAM
CQVPWVSWLLCSCLQLQMEQGK-EL-S--LT-CQTTTTLSLHYLSLALP--------AQ-
-----------ALGTQLRALCSLA---AEGT-WQ-GK----TLFRSX-------------
--XXXXXXXXXX-XXXXXXXX----XXXXXX---XXXXXXXXXXXXXXXXXXXXXXXXX-
X------XXX--------XXXXX--XXX-X-XX-XXXXXXXXX--XXXXXXXXXXX----
----XXXXXXXXXXXXXXNMVHS---------------L--AQG-PKG-ELLRLVQ----
------------------------------------HQMPRPTCSLELLGCLYEIQD-EA
FLTRAWPH---------------------V-------------------QG-AGCAHTTP
EL---WVF--A----FCFG-FCCN-VRT-FRVS----------------EDSQ-------
------HD-QALRPPGVLLXXXXXXX----------------------------XXXXXX
XXXXXXXXXX--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------X------------
------XXXX--------------------------------------------------
-------------------XXXXXXXXXXXX-----------------------------
----------------------------XXXXXX--------------------------
-----------------------------------XX---XX---XX-XXXXX-XXX---
-------XXXXXXXXXX----XXXXXXXXXXX----------------------------
------------------------------------------------------------
-------------------------------XXXXXX-----------------------
----XXXXXXX-------------------------------------------------
------------------------------------------------------------
-------------------------------XXXX-XXXXXX-XX-XX-XXXXXX--LV-
--S-CGLTSSCCQ--------------------DLASVLSASPSL-MELDLQQNDLGRQG
VNLLCE--------GLRHSXXX--XX--X-------------------------------
---XXXXXX-XXXXXXXXX------XX----------XXX-XXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXX-XX---XXXXXXXXXXXXXXXXXXXXXXXX--XXXXX----XX-X--
-XXXXX----XX-----XXXXXXXX-XXXXXXXXXXXXXX-XXXXXXX-XXXXXXXXXXX
XXXXXXXXXXXXXXXVHLPTAGSYHWP---STGLHLVVKEAV-------TVGIGFCSWGQ
YLDGIASRHNWKVAGPLLDIKAEPGA-VAAVYLPHFVAFQXXXXXXXXXXXXXXXXXXXX
XXXXXXXX--------XXXXXXXXXXXXXXXXXXXXXXXXXX-XXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXAIDDEEKKFQFVQLHKPPPLTPLYIGSRYTVCSS---------
KNLEINPEXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXDLRP
AASLDLP-----------------------------EQVALPPIPDITDP
PF00619 // ENSGT00900000140813_0_Domain_19 (3051, 3135)
LHFVDRYREQ
LIARVTSVDPVLDKLH-GHMLSEEQYESVRAEATKQDQMRKLFSFSRSWNSACKDRLYQA
LKKIHPYLIVDLWEEWGTRKGLLTLGV---------
ENSEEUP00000003038 (view gene)
------------------------------------------------------------
------------------------------------------------------------
-----------------ILPQ-D----------------PFA-----------------S
Q---LSHPCLMMKEAVPSPF
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
SRCG---MLWCFQQLS--H---EEFQTFKHLLRE------
---------K--PSA-------------TDL-TWED-VQNAH-AEKLASILHQQCPASLV
WATASSI----LE-QMNLSLLTEVAR-EKMKXXXX-XXXXXXXXXX------XXXXXXXX
----XXXXXXXXXX-----------------------X-XXXXX---XXXXXXXXXXX--
XXXXX----------XXXXXXXX-XXXXX-------------------X-----------
------XXXX-------------------------------XXXXXXXXGGEASQYKIH-
--------MMSK----------------FA-----------TKLE---------------
----------------GQQD------L-----TQL-A-------CDILEKEVLAAAFSP-
EL--SGFRP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 894)
HTVVLHGMPSCGEWALARAV-----QLCWL--Q---------GELYQ
D--------LFSY-VLLLRLGALQPSE-------ESSLAE-LLCA-EW-----PG-A--P
LPDILA-QP----ELMLLVVDGLDG-LADALGEEAEA--RAG-C-----MCTDWTER-VP
-VWVLLRELLCKALLPKCSLLLT-VGDGALPALGPWL-Q-----APR--FLXXXXXXXXX
XXXXX----------------X--XX-XXX---------XXXXXX-XXXXXX-XXXXXXX
XXXXXX--XXXXXXXXXXXXXX-XX-X--XX-XXXXXXXXXXXXXXXXXXXX-XXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX-XXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXX-XXXXXXXXXX----XXXXXXXX-XX-XXXXXXXXXXXX------XXXX-
XXX------------------------------------------XXXLLLMVRFL----
----YGLMNEEATQTLETVLG---------------------------------------
RPP-----DPSVR--WDLQH-C-VSL-------LGH-RAAAHTDVLEAFYCLFETQ--NP
NF-VR-SA---------------------L-------------------SGFQELWLAAS
DL-H--VV--A----FCL-QHCPH-LSK--------------------------------
------LR----LE---------------------------VRVF------YPLQK-NSV
AESWEKLC-SMFGTNSELHQLDLSSSILTEG---------AMKVLC--AKLCQ-A-----
TC--K-IQKLT--F-KYAQIP--L----GLN-CLWMTL----------------ISNHNI
KHLD-LQGT-PL------------------------------------------------
------------------------------------------------------------
----------------------------------------------LY-EDVQMACEA--
LKH-PSCVLESL-X----------------------------------------------
------------------------XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX-----------------------
--------------------------------XXXXXXXXXX---XX-XXXXX-XXX---
-------XXXXXXXXXX----XXXXX----------------------------------
--------------------------XXX-XXX---------------------------
-------------------------------LNECDLDETTCGLLAIA-LQGKSHLTHLS
L-SVNPLGDGGVT-LLCASLKTP--CCH----L---------------------------
-----------------------QDLEL--VRC-----DLTEACCEKL------------
-------SCVI-TENK
PF00560 // ENSGT00900000140813_0_Domain_15 (2357, 2381)
RLKSLDLGGNALGDAGVLA-LCKGLR-QE-HS-SLRRLG--LE-
--A-CGLTAECCE--------------------ALA----DSQHL-TSLNLLHNDLGEAG
LKRLCE--------AFACSTCS--LQ--R-------------------------------
---LGLCVW-QNPAPMQKL------LD----------KLQ-QLRPQLEMGED--------
------------------------------WYTWGQ------------------------
--------------------------------------------------EDRFWWRY--
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMNEP00000036266 (view gene)
ENSCCAP00000031788 (view gene)
ENSGGOP00000000443 (view gene)
ENSMICP00000002549 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------MVLA-EDPQKA---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWALSDLE--E---DDFKTLKFHIRG------
---------M--ALPEG--------Q--PHL-GRGE-LEGLS-RVDLADKLILTYGAQEA
VRVVLKV----LK-VMNLLELVDQLS-LIC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LN--DYRETYREY-
--------VRS-----------------LE------G----REG----------GVGSSY
IQLLLVA-KPSSESPESSAC------TV-------PE-----Q-ELDSV--VVKDLFGSV
EK--PDSGP----SS
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 940)
VVLQGSAGTGKTTLAKKM-----VLDWA--T---------GTLYR
D--------RFDY-VFYVSCKEVVLLPK-------GQMEQ-LLFW-CC-GD-----SQAP
VTEILR-QP----ERLLFILDGFDE-LQRPFEEQ------LR-R-----MS-----L-NP
-KEDLLRRLIGRRILPTCSLLIT-TRPQALHNLKPLL-K-----EAC--HVHTLGFSEEE
RRSYF----------------R--SY-FKD---------EEQARKAFEVVEG-NAVLYKA
CQVPGICWVVSSWLKRQMERGQ-AV-S--ET-PRNSTDIFMAYVSTFLPSDD-NEGCCEP
----------S-RHRVLRSLCSLA---TEGI-QH-QR----FLFEEA-------------
--ELRKHNLDGPSLA-AFLSS----NDYQEGLD-IK-KFYSFRHISFQEFFHAMSYLVA-
EDQSRL----------------G--EASRREVE-RLLQEKDPE-GNQEMTLSVQFL----
----LDISKKESFSNLELKFHLK-----------------ISPC-IMQ-DL-KHIKEQV-
E-------------------------------------SLKLNRTWDLELSLHESKI-KS
LVNSIQMS---------------------D-------------------VSFKVEHSN-E
RK-SHSSK--S----FSVK-TSLN-NGQ-KEEP------------------KC----LPV
--------GA---Q---------------------------KE-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------VSH----------------GKG---------G
-GAEE-PDKDAGG-REM-GT-----------IALEG---QKNGGM---E-GQED-GER-P
--TAS-KTDWR--AER-SE-----------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSANAP00000020408 (view gene)
ENSMOCP00000005221 (view gene)
ENSCHIP00000021210 (view gene)
ENSNLEP00000005171 (view gene)
ENSP00000375653 (view gene)
ENSCLAP00000024372 (view gene)
ENSPVAP00000008198 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------MISATRLDGCSCDKLLS
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 328)
HLLTLDQYL--EEFMLGLKSLQLLIPW------
---------T--TPQEY--------F--QQI-PWAS-LKASD-PTNLLCLLS-EYFPAWM
WEVALRV----FE-IMNLTSLCEKVR-AKINEMAQ-AQRPQDPKQK------EPEMPEEE
----SXXXXXXXXX-----------------------X-XXXXX---XXX----------
--XXX----------XXXXXXX----XXX-------------------X-----------
------XXXX-------------------------------XXXXXXXXXXHRRIYRER-
--------MRAK----------------ILVM--WDSTPWPKDHIYLCSVME--------
-----------------------------------------------QEYEKLMCLVYPN
RA----GAW----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
LTKVLERMAGVRKTTLAMKV-----MVH-V--E---------GFLFQ
Y--------KFSY-VFYISYYAVREME-------DTT----------F-----APESQAP
IEELMT-HP----ERFLFVIDGFEE-LTVPFNLA--D----G-L-----PYTDQHQY-LP
-ETRILFHFLKTLLITTRD-----CRSKDLKKLD-----------PW--FVLILRFTEED
WEDY-----------------S--RY-FGD---------QSKAKKILHWIRS-FETPSQS
CP-----LLVCSCLKRQMVSHP-HF-Q--LS-TQTATSLYAYFFSRWFATAE-LSWSGQS
------------WPEQWRALCSLA---ADRV-WS-SN----FTFAKE-------------
--DIEHQGL-----QAPLIDFLFRLNILQKVSD-CE-DCITFTHCSFQMFWGAMIYVLG-
GTKGS--------------IGGV--SK-HQEMR-VLLSDAFAD-TNFYWHQISLFY----
----FSLLKRDLAREVKDTLHCR---------------M--SPR-VMD-ELLEWAED-LS
KY-----------------------------------DISFGFEFLLFFQCLCEMQE-ED
FVKQILSH---------------------L-------------------LEADL------
DI---LVT--S----FCL-KHCQR-LSK-LRLS----------------VS---------
----HLNR----HT-------------------------ES-------------------
------------------------------------------------------------
-T--F-NETLG--C-KSITP---VG---ILK-ELILVL----------------HGNDRL
THLN-LSSI-YL-GI-TVYMTFRTLRHSACN----LKYL--------CLE---------K
N------L----------------------------------------------------
---------LA---A--NSE---ELALLLT--STRRTALCLRFSLLQD-NGILWLCAS--
LSD-PECDL----E----------------------------------------------
------------------------LWFGQLG-----------------------------
----------------------------ALSCRSLSD-----------------------
--------------------------------TLLQNTTPIH---LD-LRKNP-LGD---
-------MGLRFLCEAL----SHPDC----------------------------------
--------------------------NLQ-NLN---------------------------
-------------------------------LFYCSFTADGCRELTDC-LKHNHNVKALV
I-GKNCIQDDRAK-QLCEILKHS--HCA----L---------------------------
-----------------------KL-----------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCLAP00000000911 (view gene)
ENSRNOP00000069711 (view gene)
ENSDNOP00000027235 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MKMA----SV
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 326)
RCK---LAYFLEDLE--D---IDFKKFKMHLED------
---------Y--PSEKG--------C--IPL-PRGQ-TEKAD-HLDLATLMIDFNGEKKA
WGMAVCI----FA-AINRRDLYEKAK-SDEPVWDN--AH--DP---------KAVVV---
--------CR-----------------------------------EESF-----------
-----------------------------------------------------EGDWMG-
------------------F-----------------LGHLSRLPFCKKKKDYYRKYTKY-
--------IRSR----------------FQCI--EDRNARLGESV---------NLNKRF
TRLLLVKEHQSQKEREHELL------AIAKTSAKTRE------SP--VSSIKVEFLFD--
PDDEKSEPV----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
HTVVFQGAAGIGKTILTRKI-----MLDWA--S---------GKVFK
D--------RFDY-LFYIHCREVSLVT-------RRSLGD-LIVS-CC-----PD-PNPP
ISKILR-KP----SKILFLMDGFDE-LQGAFDEH------TG-E-----LCTDWQKE-ER
-VDILLSSLIRKKLLPEASLLIT-TRPVALEKLQHLL-D-----HPR--HVEILGFSEMK
RKEYF----------------F--KY-FSD---------EDQAREAFRLIQE-NEILFTM
CFIPLVCWIVCTGLKQQMESGK-SL-AQ--T-SKTTTAVYIFFLSSLLQPRGGSQGYH--
------------LSANLRGLCSLA---ADGI-WN-QK----ILFEEC-------------
--DLRNHGLQKA-DVSAFLRM----NLFQKEVE-CE-KFYSFIHMTFQEFFAAMYYLLE-
EKEE--------GGKKNMPRRCS--KLPSRDVT-VLLENYGKF-EKGYLIFVVRFL----
----FGLINQERTSYLEKKLSCK---------------I--SQQ-IRL-ELLKWIEVKAK
AK-----------------------------------KLQIQPSQLELFYCLYEMQE-ED
FVQQAMDH---------------------F-------------------PKIETNLSTRM
---DHVVS--S----FCIK-NSRH-VES-LSLG----------------FLYN-----LK
VEE-E--------E------------------------------------E---------
---------------EEGQCLEVVDCFSPCP-----------------------------
------------------------------------------------------------
--------------------------HAACS----N-----------WLV---------K
C-----------------------------------------------------------
--------HLT---SQ-YCR---SLFSVLSM-NQSLTELDLSDNILGD-SGVRVLCET--
LQH-PSCNIQKL-W----------------------------------------------
------------------------LGRCGLT-----------------------------
----------------------------HHCCFHISS-----------------------
--------------------------------VLSSNQNLVE---LD-LSNNV-LGD---
-------TGVNFLCVGL----RHLFCNLQKL-----------------------------
--------------------------------W---------------------------
-------------------------------LVSCSLTSACCQDLASV-LSTNQTLSRLY
M-GENPLGDSGVG-ILCEKMKQS--QCK----L---------------------------
-----------------------KKLGL--VNS-----GLTSASCPAL------------
-------ASVL-STNQNLTHLYLRDNALGDKGIKL-LCEGLM-HP-SC-KLQMLE--LD-
--N-CSLTSHCCW--------------------NLSTLLTSNQSL-RELSLGNNDLGDLG
VMLVCE--------VLKQQSCH--LE--S-------------------------------
---LQLHEM-YFNHETKCA------LE----------TLK-EEKPELTIVFEPSW-----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPANP00000008010 (view gene)
ENSPANP00000008011 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---ESFFSDFG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 322)
-LLWYLKELR--K---EEFWKFKELLKQ------
---------P--LEKFE--------L--KPI-PWAE-LKKAS-KEDVAKLLDKHYPGKQA
WEVTLHL----FL-QINRKDLWTKAQ-EEMRN----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------KLNPYRKH-
--------MKEK----------------FQLI--WEKETCLPVPEH--------------
---------------------------------F-------YKETMRNKYKELTGAYTAA
------ARL----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
HTVVLEGSDGIGKTTLLRKV-----MLDWA--E---------GNLWK
D--------RFTF-VFFLNVYEMNGVT-------ETSLLE-LLSR-DW-----PES-SET
IEDIFS-QP----ERILFIMDGFEH-LKFNLK-F------KD-D-----LSDDWRRQ-QP
-TSIILSSLLQKKMLPECSLLIA-LGKLAMQKHYFML-Q-----HPK--LIKLSGFTESE
KKSYF----------------S--YY-FGE---------KSKALKVFNFVRD-SGPLFIL
CHNPFICWLVCTCVKQRLERGE-DL-E--IN-SQNTTSLYASFLTNVFKAGS-QSFVPK-
-----------VNRARLKNLCALA---AEGV-WT-YT----FVFCHE-------------
--DLRRNGLSES-EGLMWVDM----RLLQRRG-----DCFTFIHLCIQEFCAAMFYLLK-
RPKDD-----------------P--NPAIGSIT-QLVKATVIQ-PQTHLTQVGIFM----
----FGISAEEIVSMLETSFGFP---------------L--SKD-LQQ-EITQSLKSLSQ
CE-----------------------------------ADREAMGFQELFVSLFETQE-KE
FVTQVMNF---------------------F-------------------EEVFIYIGNIE
HL---VIA--S----FCLK-HCRG-LTT-LRMC----------------MEN-----IFP
-DDSGCAS-----------G-------------------Y-N-------EK---------
LVYWRELC-SVFITNKNFQILDMENSSLDDA---------SLAILC--KALTQP------
VC--K-LRKLIFTS---------MLN-LGHDSELFKAV----------------LHNPHL
KLLS-LYGT-SLSRT-DVRHLCETLNHPMCK----IEEL--------IEC----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------GNITIG-----------------------------
----------------------------S--FSGLGD-----------------------
--------------------------------GV-------S---LQ-VDERP-FG----
----------------------GLGDGVFLC-----------------------------
------------------------------------------------------------
------------------------------RLTYCCLTCVSCDSISQVL-LC
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
NKSLSLLD
L-SSNALEDNGVA-SLCGALKHP--ACS----I---------------------------
-----------------------RELWL--MGC-----FLTSDSCEDI------------
-------AAVL-IC
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NEKLKTLKLGHNEIGDAGVRQ-LCAALK-HP-NC-KLEQLG--LQ-
--T-SPITRACCG--------------------DLAAALVACKTL-RSLNLDWITLEPDA
VAVLCE--------ALSHPDL---------------------------------------
-----LHKS-GFDEETQKI------LM----------SVEEKM-PHLTISHEPWIDE---
--EYK-----IR------------------------------------------------
--------------------------------------------------GVLL------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPPYP00000011655 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------MVSSA----QMGFN--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 326)
-LQALLEQLS--Q---DELSKFKFLITT------
---------A--SLAHE--------L--QKI-PHKE-VDKAD-GKQLAEILTSHCDSYWV
EMASIQV----FE-KMHRMDLSQRAK-DELREAAL-KSLN----KRK------PLSLG--
------ITRKEQPP---------------------------------LDVEEMLEHFK-A
EALAFT---------------------------------ET--KEGNVT-----------
-------CLG-------------------------------KEVFKGEKPDKDNRYRYI-
--------LKTK----------------FREM--WK--SWPGDSKEVQV-----------
---------------------------------------------MAERYKMLIPFSNPR
VL--PGPFS----Y
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVLYGPAGLGKTTLAQKL-----MLDWA--E---------DNLIH
---------KFKY-AFYLSCRELGRLG-------PCSFAE-LISK-DW-----PE-LQDD
IPHILA-QA----QKILFVIDGFDE-LGAAPG------ALIQ-D-----MCRDWEKK-KP
-VPVLLGSLLNRYMLPKAALLVT-TRPRALRDLRFLV-E-----EPI--YIRVEGFLEED
RRAYF----------------L--RH-FGD---------EDQATRAFELMRS-NAALFQL
GSAPAVCWIVCTTLKLQMEKGE-DT-A--PT-CLTRTGLFLRFLCSRFPQ-------GAQ
------------LRGALWMLSFLA---AQGL-WA-QM----SVLHRE-------------
--DLERLRVQES-DLRLFLDG----DILRQDTV-SK-GCYSFIHLSFQQFLTALFYALE-
KEEEE-------------DRDGH--AWDIGDIQ-KLLSGEERL-RNPDLIQAGYYS----
----FGLANEKRAKELEATFGCR---------------I--SPD-IKQ-ELLQCDISCKG
G------------------------------------H-STVTDLKELLGCLYESQE-EE
LVKEVMAQ---------------------F-------------------KEISLHLN-AV
DV---VPS--S----FCLK-HCQN-LQK-MSLQ----------------VIQ-----ANL
-PENVTAS----ES--------------------DAEVER------------SQDD-QHV
LPFWMDLC-SIFGSNKDLMGLAINNSFLSAS---------LVRILC--EGIAS-D-----
TC--H-LQRVV--F-KNISPA--VF----NR-NLCLAL----------------RGHKTV
TYLT--QGN-DQND--MLPALCEVLRHPECN----LRYL--------GLV---------S
C------S----------------------------------------------------
---------TT---TQ-QWA---DLSLALEV-NQSLTCVNLSDNELLD-EGAKLLYTT--
LRH-PKCLLQRL-S----------------------------------------------
------------------------LENCHLT-----------------------------
----------------------------EANCKDLAA-----------------------
--------------------------------VLVVSRELTH---LC-LAKNP-IGD---
-------TGVELLCEGL----RYPEC----------------------------------
--------------------------KLQ-TLV---------------------------
-------------------------------LWNCNITSNGCCNLAKL-LQEKSSLSCLD
L-GLNYIGVSGMK-LLCEALRKP--LCN----L---------------------------
-----------------------RCLWL--WGC-----SIPLFSCEDL------------
-------CSAL-SCNQSLVTLDLGQNPLGSSGVKM-LFETLT-HP-NG-TLRTLR--LK-
--I-DDFNDELNK--------------------LLEEIEEKNPQLIIDTEKH----DPWK
---------------KRPSSHD--FV----------------------------------
----I-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSP00000467123 (view gene)
ENSTBEP00000006180 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------M----S----------------LDIQCEH-------LSD-
TRW------------------------------------------TE-LLPLIQQ-----
----YSVVXX-XXXXXXX-XXXX---------------X--X-X-XXX-XXX-XXXXXX-
XXXXXXX-XXXXXX-XXXXX-X-XXX-------XXX-XXXXXX-XXXXX-XXXXXX--XX
XX-X--XX---------------------X-------------------XXXXXXXXXXX
XX-X--XX--X----XXX-XXXXX-XXX--------------------------------
------XX----XX---------------------------XXXX------XLEYC-NLT
AASCEALA-AVLRAKPGFKELMVSNNGI--------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------GE-AGVRVLCQG--
LRD-SACQLETL-K----------------------------------------------
------------------------LENCGVT-----------------------------
----------------------------PANCKDLCG-----------------------
--------------------------------IVA-EA
PF00560 // ENSGT00900000140813_0_Domain_11 (2019, 2057)
SLQE---VD-LGSND-LGD---
-------AGIAELCPGL----LRPSS----------------------------------
--------------------------RVK-TLW---------------------------
-------------------------------LWECGITAEGCRHLCQV-LRAKGSLKELS
L-AGNELGDEGAR-LLCESLLDP--ACQ----L---------------------------
-----------------------QSLWV--KSC-----CLTATCCPHF------------
-------SSVL-AQNK
PF00560 // ENSGT00900000140813_0_Domain_15 (2357, 2381)
SLQELQISSNKLGDAGVQQ-LCQGLS-QP-SS-TLRVL------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSRBIP00000027983 (view gene)
ENSMEUP00000005340 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MSLEIQCED-LSS-ARWT----------------EL---------
----LPSM-------------------------------QQYGVI------RLDDC-GLT
NAMCSSIG-SVLQANS
PF00560 // ENSGT00900000140813_0_Domain_4 (1517, 1551)
SLTELSLANNELGDA---------GAQMIL--KGLQS-P-----
TC--K-IQKLX--X-XXXXXX--XX---XXX-XXXXXX----------------XXXXXX
XXXXXXXXX-XXXXX-XXXXXXXXXXXXXXX----XXXX--------RLE---------Y
C------E----------------------------------------------------
---------FT---TA-SCE---A-RSVLKA-KG
PF00560 // ENSGT00900000140813_0_Domain_8 (1775, 1801)
SLRELTLNNNELGE-AGVAVLCQG--LMD-PSCELQVL-K----------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------LWECDITAEGCRALAQV-LKNKPCLTKLS
L-ICNQLGDEGAE-LLCEALLDP--GCQ----L---------------------------
-----------------------EELWL--RTC-----GFTVASCASF------------
-------CTVL-EKNRTLKELQLSTNMLEDVGIEQ-MSKGLM-HP-DC-PVQSL------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMAUP00000023936 (view gene)
------------------------------------------------------------
------------------------------------------------------------
--------------YCP-----------------GAPCGECERWDSGSFGGSGTGPVQ--
VTSVI-TRPIPSLDT--DALAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLVSLEDLS--Q---EQLKRFRHKLRD------
------------APLDG-----------RSI-PWGR-LENSD-PVDLVDKLIQFYGSEPA
VDVTRKI----LK-KADARDVAMRLK-AQQLQR---------L---------GSS-----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------S-AVVSLSEYKKKYREH-
--------VLLQ----------------HAKV--KERNARS------------VKINKRF
TKLLIAPGGSAVEDE--LLGPSE-------EPQP--------ERARRSDTHTFNKLFRGD
D---QGLRP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYH
G--------QVDF-AFFMPCGELLERPG------TRSLAD-LILD-QC-----PD-RKAP
VRQILA-HP----QRLLFILDGADE-LPAVAAP--------EAA-----PCKDPFEA-TS
-GLRVLSGLLSQELLPLARLLVT-TRNVAAGRLQGKL-C-----SPQ--CAEVCGFSDKD
KKKYF----------------F--KF-FRD---------ERKAERAYRFVKE-NETLFAL
CFVPFVCWIVCTVLQQQLELGQ-DL-S--RT-SKTTTSVYLLFIASVLKSA-GTNGP---
-----------RVQGELRTLCRLA---REGILKH--Q----AQFSEN-------------
--DLEQLKLQGSQVQTLFLNKKELLGVLEK-----E-VTYQFIDQSFQEFLAALSYLLE-
AQG-----------------------TPTGGVQ-MLLNSDSEL--RDHLALTTRFL----
----FGLLSAERIRDIERHFGC-----------------VVPER-VKQ-DTLRWVQEQGH
PK--VAPVGAEKNGELEDTEEPEEEE---------EEEEEELNFGLELLYCLYETQE-DA
FVRQALSS---------------------L-------------------PEMVLERVRFT
QM-DLEVL--S----YCVQ-CCPA-GQS-LKLV------------------NC----GLV
------GA----------------------------------------KEKK-K---KKS
----------------------------------------LVKR----------------
------LKGS----------------------------------------------QNTT
KQ---PPVS-------LLRPLCEAMTSQQCG----LRIL--------ILS---------H
C-----------------------------------------------------------
--------KLP---ES-VCR---DLSEALKL-APALRELGLLQSRVT-ETGLRLLSEG--
-LAWPKCQVQTL------------------------------------------------
----------------------------RIQ-----------------------------
---------------LPNL---------QEAIQYL-------------------------
------------------------------VIVLQKNPVLTT---LE-LSGY-ELLLYMV
--E--------CLCSAL----KYPACCLKTLSLT-TSVELSDS-----------------
----------SKRELQTVKASKP-------DLT----------------------I--LY
SKL---------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
MGP_SPRETEiJ_P0080288 (view gene)
ENSMUSP00000104024 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------MMDSSGYG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 322)
-LLQYLQKLS--D---EEFQRFKEHLRK------
---------E--PEKFK--------L--KPI-SWTK-IKNTS-KEDLVMQLYTHYPGK-A
WDMVLSL----FL-QVNREDLSTMAQ-TERRD----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------KQTKYKEF-
--------MKNT----------------FQHI--WTMETNTYIPDR--------------
---------------------------------S-------YHEFIEVQYRALQDIFDCE
-----SE-P----V
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVVSGSRGGGKTTFLRKA-----MLDWA--S---------RNLLQ
N--------RFQY-VFYFSVFSLNNIT-------ELSLAE-LISS-TL-----PES-SET
VDDILS-DP----KRILFILDGFDY-LKFDLE-L------RT-N-----LCNDWRKK-LP
-IQIVLSSLLQKIMLPECSLLLE-LGNASLSNIIPLL-Q-----YPR--EIIMSGFSEQT
IEIYC----------------V--SF--FN---------TQTGVEIFKNLKS-IKPLFNL
CRCPHLCWMICSTIKWQYERRE-VA-S--RF-GRTLGLLYTIFMVSAFKSTY-ARNPSK-
-----------QNRARIRTLCTLA---VEGM-WK-QV----YVFDSD-------------
--DLRRNGISES-DKKVWLRM----KFLQNQG-----SNIVFYHSTLQWYFAVLFYFL--
QYKDT-----------------R--HPVIGNLA-QLLGEIYAH-KQNQWFHTRILL----
----FGMATEQVNSLLEPCFGCI---------------S--SKE-VRQ-EIIRYIKSLSQ
QEC----------------------------------NEKLVVHPQNLFFCILDNQE-ER
FVRQLMDR---------------------F-------------------EEMTVDISDVD
DM---SAT--P----YCLH-RAPK-VKN-LHLH----------------IQKR----VFL
-EIHDPEY-----------G-------------------D-L-------ELFK-LDQKLL
AKHWTTLC-TFLC---NLHVLDLDSCHFNEK---------AIEVLC--NCLPLTSLVPLT
GF--K-LHRLLCSF---------TTN-FGD-GLLFCTF----------------LHLPHL
KYMN-LYGT-NLSND-AVERLCSALKFSTCG----VEEL--------LL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------GKCDIS-----------------------------
----------------------------SEACGIIAA-----------------------
--------------------------------SLINS-EVKH---LS-LVENP-LKN---
-------KGVMSLCEML----KDPSCVLESL-----------------------------
------------------------------------------------------------
------------------------------MLSYCCLTFIACGHLYEA-LLSNEHLSLLD
L-GSNFLEDTGVN-LLCEALKDP--NCT----L---------------------------
-----------------------KELWL--PGC-----YLTSECCEEI------------
-------SAVL-TC
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NTNLKTLKLGNNNIQDTGVKR-LCEALC-HP-NC-EMQCLG--LD-
--M-CDFTSDCCE--------------------DLALVLTT
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2465)
CNTL-KSLNLDWNAFDHSG
LEMLCK--------ALNHKACN--LE--V-------------------------------
---LGLDKS-LFSEESQTL------LQ----------AVE-KKNKNLKVLHFPWLKE---
--ELE-----KR------------------------------------------------
--------------------------------------------------GVRLVWNSKN
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCATP00000008305 (view gene)
ENSPCAP00000005877 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------PREAYRSD-
--------VRRQ----------------F-LM--EDHSVRLGECV---------NLS-HY
TQLLLVKEH--SNLLRAQEK----LLAAD-WHMR---T-----VDHQASPIWVEAFFKPD
EE--QPEPP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 942)
HTVVLQEVAGMGKSMLAHNV-----MLDWA--G---------GKHFQ
V--------RFDV-VFYINLREMNKAQS------I----D-LISS-YR-PK-----QSVP
LLERVQ-VP----KHLLFIISGFD-ELKPSFHNP------QG-S-----CCFCWQEK-QS
-SNLI-----QKKLLPELFLLVT--TSMVFEKLYHLL-E-----IPQ--HVEVLGSSETE
KNHF-Y---------------K--YF-------------DMQAGRVFNSVKG-NDPFFTM
CFVPMVCWVICTSLKQQLEGGG-LV-R--QT-SRTTTAKYILYLLSLIHHKPGTH----H
----------HRTPSSQKGLCSLA---ADGL-WS-QK----VLSEKQ-------------
--DLRKHMLRGTDVS-AFLNM----NIFQKDIN-CE-KVYGFICFSLQEFFATMYYNLD-
-GGDS-----------------G--LGPDQNMS-RLP-EYRSS-DRSFLAPTXXXX----
----XXX-XXXXXXXXXXXXXXX-----------------XXXX--XX-XXXXXXXXXXX
X-----------------------------------XXXXX-XXXXXXX--XXXXXX-XX
XXXX-XXX---------------------X-------------------XXXXXXXXX-X
XX-XXX-X--X----XXXX-X-XX-XXX--XX-----------------XXXX----X-X
------XXXXXXXXX-----XXXXXXX--------------------XXX-XXXXX-XXX
XXXXXX---XXX------------------------------------------------
------------------------------------------------------------
-------------------------XX---------------------------------
--------------------XX----XXXXXXXXXXXXXXXXX-XXXXXXXXXXXXXX-X
XXXX-XXXXXX---XX-XXX---XXXXXXXX-XXXXXXXXXXXXXXXX-XXXXXXXXX--
XXX-XXXXXXXX-X----------------------------------------------
------------------------XXXXXXX-----------------------------
----------------------------XXXXXLKSP-----------------------
--------------------------------VLSAKQHLKE---LD-VTDSA-LED---
-------SGLR-LCQEL----KHPTC----------------------------------
-------------------------RHLQ-TLW---------------------------
-------------------------------LKICHLTAVASEDLASV-LTVNQSLRELD
L-SLNDLGDPVVL-LLGEGLSHP--KKF----Q---------------------------
-----------------------A-VRX--XXX-----XXXXXXXXXX------------
-------XXXX-XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX-XXXXXXXXX--XX-
--XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX-XXXXXXXXXLCETD
LNEMTQ--------GLA----A--LH--V-------------------------------
---M---------------------------------------KPEVAIGC---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPEMP00000021727 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------------LPFT
EALQTEEEDD-------------------------------A-SGEARQQGDIQGYKTH-
--------VITK----------------FNTH--VVVH-G--------------------
--------------------------------------------DNPEM-RVLLEALKSQ
Q---KTFSP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
HTVILHGRQGVGKSALARSI-----ILGWA--Q---------GKLYQ
D--------V-SY-AFFFSVREIKWRE-------KSSLAQ-LISR-EW-----PD-SRAP
MTEIVS-KP----ERLLFVMDGFDD-LGSALLLD------DM-R-----LPEGLEDE-QP
-TYILLYSLLTKALLPESFLVIT-TRDTGLEKLKSMV-V-----SPL--YIFVEGLSATR
RAELV----------------L--QN-ISD---------NHKSHQVVHSVID-NHQLFDQ
CQVPPICSLVCEALQLQEKLGE-KP-P--PV-CQTFTSLYATLVFYQLTPRD-PSRSCLN
----------PEERVTLIGLCRMA---AEGV-WS-MR----SVFYWT-------------
--DLHTHGLRES-EISALFHM----NILLRVDH-GAEWYYVFFHLSLQDFCAALYFVLE-
GLE-K--------------WEQP--SSITKNPR----SLRRTD-FNTHLLGMKRFL----
----FGLMNKDTMATLEVLLGCS---------------V--LP-TVRQ-TLQDWVLELGQ
RVS----------------------------------ATS-PMDVLDAFHYLFESRD-EE
FVCGALNS---------------------F-------------------QEVYLLVNQKM
DL---MVS--S----YCLQ-HCQN-LKT-LRLD----------------IRD-----IFS
-VNKNTDL----CP--------------------FV-----------PP--GTQNR-PLI
LEWWEKFC-CVLGTHKNLRQLDLGNSVLNEW---------AMKTLC--LKLRT-R-----
TC--N-VQTLT--F-KGAEV---TL---GLK-YLWVTL----------------ISNRKL
KYLN-LGNT-LLKDD-DIKLACEALKHWKCS----LETL--------RLD---------S
C------E----------------------------------------------------
---------LT---SA-CYL---EISKLLLS-TTSLKSLSLARNKVVD-KSMRSLCEA--
LSS-SQCSLQKL-I----------------------------------------------
------------------------LDGCDLT-----------------------------
----------------------------SASCQALAS-----------------------
--------------------------------ALLSNRTLTH---LC-LSNNS-LRA---
-------EEVRQLCQFI----RHPDC----------------------------------
--------------------------ALQ-RLI---------------------------
-------------------------------LNQCGLKGDAYGFLALM-LI
PF13516 // ENSGT00900000140813_0_Domain_13 (2212, 2237)
NNKKLTHLS
L-TMNPMEDDGVK-LLCEALREP--TCH----L---------------------------
-----------------------QELEL--VGC-----QLTEDCCEDL------------
-------ACVI-TA
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NKQLTSLDLGNNALGDVGVMA-LCKGLR-HE-DS-SLKRLG--LE-
--A-CGLTSECCE--------------------ALSLALS
PF13516 // ENSGT00900000140813_0_Domain_17 (2441, 2465)
SNQLL-TSLNLVKNDFSTSG
MLKLCS--------AFLHSTSK--LR--V-------------------------------
---IGLWKQ-QYYAQVRRQ------LE----------ELQ-FTKPQMVIDGDWYSFD---
--DD--------------------------------------------------------
--------------------------------------------------DDRNWWKN--
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMMUP00000031417 (view gene)
ENSSTOP00000030969 (view gene)
ENSBTAP00000051793 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------
PF05729 // ENSGT00900000140813_0_Domain_2 (699, 930)
MFDWA--A---------GVVTP
G--------RFDY-VIYINCREMR-FA-------NLSGAD-LINN-TF-----QD-TGGP
IMDIILVYP----EKLLFILDGFPE-LQYSVSDQ------EE-D-----LSTNPLER-KP
-VETLLHSFMKTKLFLESSLLIT-AQPMTMKKLHSLL-K-----QP-----EILWFTDVE
KRAYF----------------L--SQ-FSG---------ANVATRVLYDLRE-NEGLDIT
SSLPITSW---------EDGGR-TL-T--RS-RQTMTDTYLVLLFQLSQ-----------
-----------HPYKNLGGLCSLA---VEAL-QN-QQ----VLFEVS-------------
--DLRRHGIGVYDTNCTFLNH-----FLKKVE--VV-GVYTFLHFSFQEFLTAVFYTLK-
NDSSW---------------------MFFDQVGKTWQEIFQQY-EKGFSSLTIQFL----
----FGLLHKGKEKAVLTTFGKK---------------V--SPG-LQE-ELLKWTEKEIK
DK-----------------------------------PYRLQIEPMDLFHCLYDTQE-EE
YAKRIIND---------------------L-------------------RSALIVSHN-I
WL-RMGHM--S----HCCK-SSKNAQV----------------L-----CVRGSVRA---
-HAQERAR----------------------A-----------------------------
---------------------------------------------------CVG------
AC--V-HTYVSVIL-DKGVEA------SSYGRD---------------LALVFET-NQYL
TDLV-FVKN-TLEDS-GVKLLCEGLKQPNCV----VKVQP--------LEICLI-SPA-S
C-----------------------------------------------------------
-------------------G---YLASVLIC-NPNLTELDLSENPLGD-TGVKFLCEG--
LRH-LNCKVEKQ-D----------------------------------------------
------------------------LNTCYLT-----------------------------
----------------------------DASCVYLFS-----------------------
--------------------------------LLQVSQTLKE---LF-VFANS-LRD---
-------TGVQHLCEGL----CHVNSIIKNL-----------------------------
------------------------------------------------------------
------------------------------VLSECSLSPACCESLTQV-LSSTWSLTRLL
L-INNKIGDLGLK-LLCEGLKQP--DCQ----L---------------------------
-----------------------KDLVL--WTC-----HLIGECCQDS------------
-------CNAL-YTNEHLRVLELSDSALGDEGMQV-PCEGLK-HP-SC-KLQTLG--LA-
--E-CHFTGACCG--------------------ALTSVLSRNENL-TLLDLSGNHLKDFG
VQMLCD--------ALLHAICK--LQ----------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPVAP00000016707 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------VEPR--DAA
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
AREL---LLAALEDLS--Q---EQLKRFRHKLRD------
------------AQLDG-----------RSI-PWGR-LEGAD-AVDLAEQQTHFYGPEPA
LDVARKT----LK-RADVRDVAARLK-QQRLQR---------L---------GAS-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------SS-VLLSVSEYRKKYREH-
--------VLRQ----------------HAKV--KERNARS------------VKITKRF
TKLLIAPESAALESE--GPGPEE-------EPPP--------ERARRSDTHTFNRLFGRD
E---EGQRP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
LTVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYH
G--------QVDF-AFFLPCRELLERPG------ACSLAD-LILD-QC-----PD-RGAP
VRQMLA-QS----ERLLFILDGVDD-LPAP-----------EAA-----PCADPFEA-TT
-GARVLGGLLSKVLLPGARLLVT-TRTAAPGRLQDRL-C-----SPQ--CAEVRGFSDKD
KKKYF----------------F--KF-FQD---------EWQAERAYRFVKE-NETLFAL
CFVPFVCWIVCTVLRQQLELGQ-DL-S--RT-SKTTTSVYLLFIASVLSSAPAADGP---
-----------RLRGELRKLCRLA---REGVLRR--R----AQFAEA-------------
--DLEQLELRGSKVQMLFLSKKELAGVLET-----E-VTYQFIDQSFQEFFAALSYMLE-
GEGPP-------------------G-APAGGVG-ELLRGDAEL--RCHLALTTRFL----
----FGLLSAERMRDIERHFGC-----------------VVSEH-VKQ-DVLRWVQGQGW
GRARAELEGMQEAEALEGAEEPED-------------EESEFNSPLELLYCLYETQD-DA
FVRQALRG---------------------L-------------------PELALQRVRFS
RM-DLTVL--S----YCVR-CCPA-GQA-LRLD------------------SC----RLT
------PV----------------------------------------QDKK-----KKS
----------------------------------------SLMK----------------
------RLQS---S--------------GSS------S----------------SSQATA
RK---PQAS-------PLRPLCEAMADQQCG----LNSL--------TLS---------H
C-----------------------------------------------------------
--------KLP---DA-ACS---DLSEALRV-APALTELSLLHSGLS-DSGLRVLCAG--
-LAWPRCRVQTL------------------------------------------------
----------------------------RVQ-----------------------------
---------------QPGL---------QEALQYL-------------------------
------------------------------VSTLRQGPALST---LD-LSGC-QLSGPMV
--T--------YLCAVL----QHPGCRLQTL-----------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSP00000343891 (view gene)
ENSPTRP00000038834 (view gene)
ENSCANP00000031685 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MTSP----QLEWT---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 323)
LQTLLEQLN--E---DELKSFKSLLRA------
---------L--PLEDV--------L--QKT-PWSE-VEEAD-GKKLAEILINTSSENWI
RNATVTV----LE-EMNLTELCKMAK-SEM------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------------------LGEKEGRRNS-
---------VEK----------------QSLV--WKNTFWQGDIDSFHDDVI--------
-----------------------------------------------RRNQRFIPFLNPT
TP--RKLTP----Y
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVLHGLAGVGKTTLAKKC-----MRDWT--D---------CNLSP
---------TARY-AFYLSCKELSRMG-------PCSFAE-LISK-DW-----PE-LQDD
IPSLLA-QA----QRILFVVDGLDE-LKVPPG------ALIQ-D-----ICGDWEQQ-KP
-VPVLLGSLLKRKMLPKAALLVT-TRPRALRDLQLLV-Q-----QPI--YIRVEGFLEED
RRAYF----------------L--RR-FGD---------EDQAMRAFELMRS-NAALFQL
GSAPAVCWIVCTTLKLQMEKGE-GP-A--PT-CLTRTGLFLRFLCSQFPQG-------AQ
------------LRGKLRALSLLA---AQGL-WA-QM----SVFHGE-------------
--DLERAGVQES-DLPPFLDR----DILRQDRA-SK-GCYSFIHLSFQQFLTALFYALE-
EE--E---EE--------DRDGH--AWDIGDVQ-KLLSEEERL-KNPDLIQVGHFL----
----FGLANEKRAKELEATFGCQ---------------M--SPE-IKQ-ELLRCKANLHA
NK-----------------------------------P-LSMTELKEVLCCLYESQE-EE
LAKVMVAP---------------------F-------------------KEISINLTNTS
EM---MHC--S----FSLK-HCQD-LEK-VSLQ----------------VAKG----VFL
-ENY-VDF----EP--------------------DIEFER--------------------
------------------------------------------------------------
-------------I-KNVTPD--TA----YQ-DFCLAF----------------IGKKTL
THLT-LEGH-VEWERTMLLLLCDLLRNHKCN----LQYL--------R------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------LENCRLT-----------------------------
----------------------------EASCKDLAA-----------------------
--------------------------------VMVV
PF13516 // ENSGT00900000140813_0_Domain_11 (2017, 2054)
SQQLTH---LC-LAKNP-IGD---
-------TGVKFLCEGL----SYPEC----------------------------------
--------------------------KLQ-ALV---------------------------
-------------------------------LQQCSITKRGCRYLSEA-LQEACSLTNLD
L-SLNQIA-RGLW-ILCQALENP--NCN----L---------------------------
-----------------------KHLRL--WSC-----SLMPFYCQHL------------
-------GSAL-I
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2377)
SNQKLETLDLGQNHLWKSGIIK-LFRVLR-QR-TG-SLKTLR--LK-
--T-YGTNSEITK--------------------LLEEVKEKNPKLTIDCNAS------GA
---------------TAPACCD--FF----------------------------------
----C-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCCAP00000025568 (view gene)
ENSANAP00000013563 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---ASFFSDFG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 322)
-LMWYLEELK--K---EEFRKFKEHLKQ------
---------L--TSHLE--------L--KPI-PWGE-IKKAS-REELANLLIQHYEEQQA
WNVTLRI----FQ-KMDRKDLCVKVT-RER------------TG--------------YT
K---------------------------------------------------TYQD----
------------------------------------------------------------
----------------------------------------------------------H-
--------AKKK----------------FSHL--WASKSVTDIDRY--------------
---------------------------------F-------HQEVKREECDHLDRLFASK
ET--GKH-P----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
RTVVIQGAQGIGKTTLLMKL-----MWAWS--D---------SRIYQ
D--------RFLY-AFYFCCREMRELP-------AVSLAD-LICR-EW-----PYP-AAP
VTEIVS-QP----ERLLFVIDSFEQ-LEADVNQP------DS-D-----LCGDWKEK-RP
-VQVVLSSLLRRKMLAEASLLVA-VKPTCPKELRDRV-A-----ISE--IYEPRGFDESD
RLVYF----------------C--CF-FQD---------PQRAMEAFSLVRE-SQQLFAI
CQIPLLCWLLCTALKQEMQKGK-DL-A--LT-CQSTTSVYACFILNLFTPKG-ATGPTE-
-----------QSQRQLKGLCALA---AEGM-WT-DT----FEFGDE-------------
--DLRRHGIADA-DIPALLGT----KVLLKFGE-RA-SSYAFIHVCIQEFCAAMFYLLK-
SHLDH-----------------P--HPAVRSVE-ELLVNNFKKGRRPHWIFFGCFL----
----IGLLNKKEQEKLNGFFGFQ---------------L--SRE-IKQ-QFEQYLKRLGE
H-G----------------------------------DLQGQVDALAIFYCLFEMQE-PA
FVRQAVNL---------------------L-------------------QEGSFHIVDNS
DL---VVS--A----YCLK-YCSS-LRK-LSFS----------------IQN-----VFK
-KED-THS-----------S-----------------------------------VLDYS
LICWHHIC-SVFTASGYLREVQVQDSTLSEP---------AFVTWC--NQLRHP------
SC--R-LQKLRINN---------VSF-SGQSSHLFQVL----------------FHQPDL
KYLS-LTLT-KLSRD-DVKSLCDALNSPAGN----IEEL--------A------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------LAYCHLSEQCWEYFSEM-LLR
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
NSSLRHVD
L-GANVLKDEGLK-TLCKALKRP--DCH----L---------------------------
-----------------------DSLCL--VKC-----FITAAGCEDL------------
-------ASVL-I
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
SNQNLKNLEIGCNEIGDVGVKL-LCGALT-HP-DC-RVEVLG--LE-
--D-CALTSACCK--------------------DLASVLTGSKTL-QQLNLTLNALDHGG
VVVLCE--------ALRHPECA--LR--V-------------------------------
---LGLKKA-DFGEETQAL------LM----------AEE-ERNPKLTIVDH-LDTTTWA
EI----------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSGGOP00000047842 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------VLP-RLV-LNWA----------------QV---------
----ILLL----Q-----------------------------PPM------ALDDC-GLT
EARCKDIS-SALRV
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1542)
NPALAELNLRSNELGDV---------GVHCVL--QGLQS-P-----
SC--K-IQKLS--L-QNCCLT--GA---GCG-VLSSTL----------------RS
PF13516 // ENSGT00900000140813_0_Domain_6 (1617, 1642)
LPTL
QELH-LSDN-LLGDA-GLQLLCEGLLDPQCR----LEKL--------QLE---------Y
C------N----------------------------------------------------
---------LS---AA-SCE---PLASVLRA-KPDFKELTVSNNDINE-AGVRVLCQG--
LKD-SPCQLEAL-K----------------------------------------------
------------------------LESCGVT-----------------------------
----------------------------SDNCRDLCG-----------------------
--------------------------------IVASK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
ASLRE---LA-LGSNR-LGD---
-------VGIAELCPGL----LHPSS----------------------------------
--------------------------RLR-TLW---------------------------
-------------------------------IWECGITAKGCGDLCRV-LRAKE
PF13516 // ENSGT00900000140813_0_Domain_13 (2215, 2237)
SLKELS
L-AGNELGDEGAR-LLCETLLEP--GCQ----L---------------------------
-----------------------ESLWV--KSC-----SFTAACCSHF------------
-------SSVL-AQ
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NKFLLELQISNNRLEDAGVRE-LCQGLG-QP-GS-VLRVLC--TL-
-----------CP--------------------AHP-----------------ACRGRRG
AITLPLQAVLFCFSFETVSLCS--SSL-N-------------------------------
---IMQAGV-QWRNL---------------------------------------------
--------------GSLQPLP-------PGFKQFCL------------------------
--------------------------------------------------SHPSSWDYRH
TPP-----------------RPAN-----------------FCICIR-DRVSPCWRGWSR
T----------------------PGLK---------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSFDAP00000005705 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------MDKASV--FSTAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLAALEDLS--Q---DQLKRFRHKLRD------
------------APVDG-----------RSI-PWGR-LERAD-AVDLVDQLTHFYGPESA
LDVTRRT----LK-RADVRDVAARLK-AQRLQRE--------R---------PLP-----
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------PCLCPTEFQKKYREH-
--------VLQQ----------------HAKV--RERNARS------------VKISQRF
TKLLIAPESAAPREEEAALGPVE-------EREP--------GRAWRSDTHTFNRLFGSD
A---EGHRP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 795)
TVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYR
G--------QVDF-AFFLQCRELLDRPG------ARSLAD-LVLD-QC-----PD-RLAP
VAQMLA-QP----QRFKA------------------------------------------
-------------LLPSARLLVT-MRATASRKLHFDQ-C-----ALQ--CAEVRGFSDKD
KKKYF----------------Y--KF-FQE---------ERRAERAYRFVKE-NETLFAL
CFVPAVCWIVCTVLRRQLELGR-DL-S--RT-SKTTTSVYLLFLASALNSM-GAERA---
-----------RMRGELRKLCLLA---REGILGY--K----AQFSER-------------
--DMERLELRDSDIQH-FLIKKELPSVLET-----E-VIYQFIDQSFQEFFAALSYLLE-
DVGAP-------------------G-VPTGGLE-TLLHLGSEG--RSHLMLTTRFF----
----FGLLNMEHVQDVERHFGC-----------------VVPAH-VKQ-NALQWVQGQGH
LR--MAPKGADGSKGLKDKEEPKEEEEK------EEEEEEELSFPLELLYCLYETQD-EA
FVNQALRS---------------------L-------------------PELTLECVHFS
RM-DLDVL--S----YCMK-CCPA-GQV-LQLV------------------SC----SLV
------A-----------------------------------------GQEKKK---KRS
----------------------------------------LLKR----------------
-------LKG---N-----------------------------------------SQAAM
KQL---RIT-------PLHPLFE-------------------------A----------M
T-----------------------------------------------------------
--------DQQ---------------------CGLSK-LVLLHCRLS-EAGLRALSEG--
-LAWPQCRE-AEGH-DPT------------------------------------------
-------------------P-NQLFPRV-LL-----------------------------
---------------LNPP----------EAFQYL-------------------------
------------------------------VALLW-QSSTL-TTLD--LSSC-YLP-DSV
V-TYL-CA-AL----------QHPGCSLQTLSL--ASVELSEP-----------------
----------SVQELKAVKIAKP-------RLA----------------------I--VH
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMODP00000017539 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------MEQQQ-ESSAEQSLR-PYF----
-----------------------VFR-AHRPELGI-CWDPSEPLRG------SPEAAQTR
------------------------------------------------------------
--------------------GP----GGA-------------------G-----------
------PGED-------------------------------GDGEAEDDSDDQRRYRAW-
--------VAGR----------------FCSL--RAGEPWPGAPENFLYGPVTLQFHHNC
AKLRVVVGGEGPERPPGEGP------ERPPSEGPPAA-------GAPERAGHLAALFQPD
ER--TQRRP----N
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 929)
TVILQGAVGIGKTVLARKL-----VLDWA--R---------GRPRR
D--------AFRY-VFYLDAGDLDAAA-------DRSLAE-LMAA-QW-----PD-PPAP
VSEILS-RP----RRLLFVVDGLED-LKPFLDGP--E----A-E-----LCRDWGAP-RP
-AGRLLQALLRKALVPEARLLVV-VRPSGWARLAPWL-G-----CPR--LVEVLGLCAAD
RDDYF----------------H--RF-FGA---------PAGARAALAVVRD-NETLSAL
CRAPLVCWLVCACLKRRLDGGD-DL-R--RG-PLTASSLLASYVAGLFRG-R----PAPR
------------ELPLWG-LCRLA---AEGV-LA-RA----TRFGRG-------------
--DLERLGLA-EADLEPFVASRLL------LPR-PG-GAFAFLHLTLQEFLAALYSAME-
VTR-W--------------PGLA--PS-SQDLA-TELEKVSGE-DASFVGTAVHFL----
----FGLLNPECAGPVEEGLGCR---------------L--APQ-LRR-DLLLWAAV-VS
AA-----------------------------------P-PHSHSLHQFFLSLYEAQD-AA
FLTQVMDH---------------------F-------------------RDLRLVLRGKA
DL---LVS--S----FCL-RHCRR-LEK-LVLH----------------LD---------
----PDVF----SD-------------------------DDG----EPQAGPQASD-LAT
ASLWKALC-SVLTTSTSLRELDLRFFSLNDL---------CMKSLS--EQLRH-Q-----
DC--R-LQNLR--V-QQTAFV--PF---ACG-DFPLAL----------------SGHQHL
TWLD-LNLQ-LG-DD-GMKLLCEALRHPQCN----LQYL--------RLR---------S
C------A----------------------------------------------------
---------LT---EA-CCP---DLALALTC-NQSLSHLDMGGNQLLD-RGVQVLCEA--
LSQ-PKCRLQSL-L----------------------------------------------
------------------------LPACGLT-----------------------------
----------------------------AGVCQDLSA-----------------------
--------------------------------ALTSNQSLTR---LC-LASNS-LRD---
-------DGLKVLSSAL----KSPEC----------------------------------
--------------------------PLQ-RLA---------------------------
-------------------------------LWSCELTAEGCQALSAA-LHSNKNLTHLD
L-GENDLRDDGMK-LLCEALGQP--QCP----L---------------------------
-----------------------QALDM--LVC-----FLTEACCQDL------------
-------SAAL-VLNQNLRSLNLGHNALRDEGVKL-LCEALR-HP-SC-PLQRIG--LE-
--R-CQLNTACCQ--------------------DLSSVLLCNPRL-KSLNLAQNALWDEG
VRLLCE--------ALEKPECG--LE--I-------------------------------
---LSLWKE-AFSNSAQQM------LK----------AAE-ERKPHLIITGDWYS-----
---------------------------------QDP------------------------
------------------------------------------------EDYGSSWWLET-
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSSBOP00000017295 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------MASSV----RLDFN--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 323)
-LQALLEELS--Q---DELSQFKSLIRT------
---------I--TLGNG--------L--QKI-PQTE-VDEAD-GKQLAEILSSHCPSYWL
EMATLQV----LE-KMHRADLCERVK-EEL------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------------------RGKEDRHRSI-
--------LKMK----------------LKQM--WE--AWPGDSKEVQA-----------
---------------------------------------------VAQRYTTLIPFSNPR
ML--PGPFP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
HTVVLHGPAGLGKTTLAKKL-----MLDWT--E---------DNLSH
---------KFKY-AFYLSCKKLSRLG-------LCSFAE-LVFR-DW-----PE-LQDD
IPNILA-EA----QKILFVVDGLDE-LGAPPG------ALIH-D-----ICGDWLKP-KP
-VPVLLGSLLKRKMLPKAALLVT-TRSWALRDLRPLM-E-----QPI--FIRVEGFLEKD
RRAYF----------------L--RH-FGD---------EDQAV----------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------------------ELLCCDIICDE
DR-----------------------------------Y-STLTDLKEVLHCLYESQE-EE
LVKGAMAP---------------------L-------------------KEISLHLS-AA
DV---VPS--S----FCIK-HCRN-LEK-MSLQ----------------LIK-----AKL
-PENVTAS----ES--------------------DAEVER------------SQDD-QHM
LPFWTDLC-SIFGSNKDLIILEINNSFLSAS---------LLRILC--EQIAS-D-----
AC--H-LQRVV--I-TNISPA--DA----YR-NLCLAL----------------QGHKTV
THLR-LGGS-AQND--TLPALCEVLRHPKCN----LQYL--------GLV---------S
C------S----------------------------------------------------
---------AT---TQ-QWA---DLSLALEV-NRSLTCVNVSNSELLD-EGAKLLYTT--
LKH-PKCLLQRL-S----------------------------------------------
------------------------LENCHLT-----------------------------
----------------------------EANCKDLAA-----------------------
--------------------------------ILVVSRELMH---LC-LAENP-LGD---
-------TGVKFLCGGL----SYPDC----------------------------------
--------------------------KLQ-TLV---------------------------
-------------------------------LWNCDITSNGCCHLAKL-LHE
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
KSSLTNLD
L-GRNDIGVPGVR-GLCEALKKP--LCN----L---------------------------
-----------------------KRLWL--WGC-----SIPPASCENL------------
-------CCVL-SC
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2377)
NQTLITLDLGENSLGYSGVKL-LFETLT-RA-NG-TLQTLR--LK-
--I-DDVTDELDT--------------------LLKEIEGSNPQLTIATENQ----EPWK
---------------KRPSSGY--FI----------------------------------
----V-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSFCAP00000018506 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-PSCAGGSPSSLSFR--AATAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 323)
LQRALESLS--Q---EHLKRFRHKLRD------
------------AQQDG-----------RSI-PWGR-LQGAD-ALNLVEELIQFYGPEPA
LDVTRKT----LK-RADIRDVAERLK-EDRLKR---------E---------RSP-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------GSPRLRSLPPEAE------
------------------------------------SGP-------------------R-
------PPSFPLPQK--VP----------ARPPP--------PRPRAR------------
--------------HPHIQGPQGSDPG---------------------------------
------------------------------------------------------------
-----------------------PQ-LP-------------TXP-----PPPSPLHI-RQ
-G------LT--TALPSRARPQL-LLAALRVRLQSRL-C-----SPQ--CAEVCGFSDKD
KKKYF----------------H--KF-FRE---------EWLAEQAYRSVKE-NETLFAL
CFVPFVCWIVCTVLRQQLDLGQ-DL-S--RT-SKTTTSVYLFFVASFLSSAPAADGP---
-----------QVQGELRKLCRLA---REGLLGH--R----AQFAEK-------------
--DLERLQLRGSKIQRLFLSKKELPGVLET-----E-VTYQFMDQSFQEFFAALSYLLD-
DKGD--------------------R-APADSVG-ALLRGNAEL--RGHLALTTRFL----
----FGLLNTERMRDVERHFGC-----------------VVSER-VKQ-DVLRWVQDQG-
-HQRAAPEGTQETEAPEGREELEE-------------DEGELNYVLELLYCLYETQE-GA
FVSQALRG---------------------L-------------------PELALERVCLS
RM-DVVVL--S----YCIR-CCPE-GQA-LRLV------------------SC----GLA
------PA----------------------------------------HEKK-K---KKN
----------------------------------------LMKR----------------
------LHGS---L--------------GGS------S-----------------SQATM
KK---TQAS-------PLRSLFEAMTDQHCG----LSSL--------TLC---------R
C-----------------------------------------------------------
--------KLS---DS-VCR---DLSEALRA-APALTELCLLQNGLS-EAGLRVLSEG--
-LAWPQCRVRKL------------------------------------------------
----------------------------RVQ-----------------------------
---------------QPTL---------QEALHYL-------------------------
------------------------------IGALSQSSALTA---LD-LSDC-QLPEPVV
--T--------YLCRVL----QQSGCRLQTLSL--TSVKLSAQ-----------------
----------SLQELREVKKAKP-------GLV----------------------I--TH
PLLE-------------------SHP----------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCANP00000025648 (view gene)
ENSCCAP00000012304 (view gene)
ENSODEP00000001307 (view gene)
ENSTTRP00000012884 (view gene)
MGP_CAROLIEiJ_P0077022 (view gene)
ENSSSCP00000003586 (view gene)
ENSCGRP00001005030 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------------MLRG-AW
LALRVF----------------LESPPGPR-------------------NTWQSV---WT
RR-LHCAP--T----MSLDIQCEQ-LSD-ARWT----------------EL---------
----LPLI-------------------------------QQYQVV------RLDDC-GLT
EVRCKDIS-SAIQ
PF13516 // ENSGT00900000140813_0_Domain_4 (1514, 1542)
SNPTLTELSLCTNELGDP---------GVCLVL--QGLQN-P-----
TC--K-IQKLS--L-QNCSLT--EA---GCR-VLPDVL----------------RS
PF13516 // ENSGT00900000140813_0_Domain_6 (1617, 1642)
LPTL
RELH-LSDN-PLGDA-GLKLLCEGLLDPQCR----LEKL--------QLE---------Y
C------N----------------------------------------------------
---------LT---AT-SCE---PLASVFRV-KPNFKEIVLSNNDLHE-AGVQMLCQG--
LKD-SACQLESL-K----------------------------------------------
------------------------LENCGIT-----------------------------
----------------------------SANCKDLCD-----------------------
--------------------------------VVASK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
ASLQQ---LD-LGSNK-LGN---
-------AGIAALCSGL----LLPSC----------------------------------
--------------------------RIR-TLW---------------------------
-------------------------------LWECDITAEGCKDLCRV-LRT
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
KQSLKELS
L-AGNELRDEGAQ-LLCESLLEP--GCQ----L---------------------------
-----------------------ESLWI--KTC-----SLTAACCSHI------------
-------CSVL-T
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
KNRSLLELQMSSNPLGDLGVLE-LENSGT00900000140813_0_Linker_15_16 (2378, 2391)
CKALG-QP-DT-VPF13516 // ENSGT00900000140813_0_Domain_16 (2391, 2436)
LRVLW--LG-
--D-CDVTDNGCS--------------------SLENSGT00900000140813_0_Linker_16_17 (2436, 2441)
ATVLLPF13516 // ENSGT00900000140813_0_Domain_17 (2441, 2464)
HNRSL-RELDLSNNCMGDSG
VLRLVE--------SVRQPSCA--LQ--Q-------------------------------
---LVLYDI-YWTEEVEDQ------LR----------ALE-EERPSLRVIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
MGP_PahariEiJ_P0018264 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MALARA-NSPQEA---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWALNDLE--E---NSFKMLKFHLRD------
---------V--T------------Q--FHL-ARGE-LEGLS-QVDLASKLISMYGAQEA
VRVVSRS----LQ-AMNLMELVDYLN-QIC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LN--DYREIYRER-
--------VRC-----------------LE------ER---QDW----------GVNSSH
NKLLLVA-TSSSGSRRSLSR------SD-------LE-----Q-ELDPV--DVETLFAPE
AE--SYSTP----PI
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 930)
VVMQGSAGTGKTTLVKKL-----VQDWV--K---------GKLYP
G--------QFDY-VFYVSCREVVLLPK-------CDLPN-LICW-CC-GD-----DQAP
VTEILR-QP----ERLLFILDGYDE-LQK---------------------------S-SR
-AERVLHILMRRREVP-CSLLIT-TRPPALASLEPML-G-----ERR--HVHVLGFSEEE
RETYF----------------S--SC-FTD---------KEQLKNALEFVQN-NAVLYKA
CQVPGICWVVCSWLKKKMARGQ-EV-S--ET-PSNSTDIFTAYVSTFLPTDG-NGDSSEL
----------T-RHKVLKSLCSLA---AEGM-RH-QR----LLFEED-------------
--VLRKHGLDGPSLT-AFLNC----IDYREGLG-IK-KLYSFRHISFQEFFYAMSFLVK-
EDQSQL----------------G--EATHKEVA-KLVDP---E-NYEEVTLSLQFL----
----FDMLKTDNTLSLGLKFYFK-----------------IAPS-IRQ-DL-KHFKEQI-
E-------------------------------------AIKYNRPWDLEFSLYDSKI-KK
LTQGIQIK---------------------D-------------------VSINVQHLD-E
KK-SSKGK--S----VLVT-SSIS-R---RGEV------------------QS----SFL
--------GT---GKS--T-------------------RKQKK-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------ASN----------------GKS---------R
-GTEE-PAR-GIR-NGR-LASR----EK-GHMEMND---KEDGGV---E-GQED-EEG-Q
T-LKN-DGEMI---DK-MNG----------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSHGLP00000021086 (view gene)
ENSP00000331857 (view gene)
ENSLAFP00000028929 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---ESFF
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
SDFG---LLWYLEELR--K---EEFWKFKELLLR------
---------E--PLKYE--------L--KPI-PRTE-LKKAS-REDLAKLLDKYYPGKQA
WEVTLSI----FL-QINRRDLWKKAQ-EEIRLF---------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------------LRCISR--
-------------------------------H--LLQRTCPNQLSSFNLICK--------
----------------------------G----F-------SNGKVTPSHEERPTNLL-L
XE--TSEGS----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
PTVVLKGSEGTGKTTLLRKV-----MLEWA--E---------GNFWK
G--------RFTF-VFFLSACEMNCVT-------ETSLVE-LISR-DW-----PES-SEP
IKDIFS-QP----QHILFIMDGFEE-LKFNLR-L------KD-N-----TCNDWRQQ-QP
-IQIILSSLLQKKMLPESSLLIA-LGPMGMQKHYFLL-Q-----HPK--YIVVSGFSEHE
RKLYC----------------S--HF-FHE---------KNKAMKAFSFVRN-DGSLFSV
CQNPLLCWLVCCCMKWQLERGE-DL-N--IV-CQSSTFLYASFFINAVKAGG-E------
------------NRAQLKGLCSLA---AEGI-WT-RT----YLFCHG-------------
--DFRSNGLSES-DVWRWLGM----NLLRRNG-----DRITFIHRHVQEFYAAMFYLLK-
QPKDN-----------------S--NPIIGSIT-QFVATGLSQ-IELCLSHMGLFL----
----FGLSTEKITKMLEISTGFQ---------------L--SKE-IKQ-EIIQCLKSVSQ
YG-----------------------------------SNETELDFQELFNGIYETRE-KE
FAAQVMDF---------------------F-------------------EEVIVYISNTE
EL---IIT--S----YCLE-HCRK-LQR-LVLG----------------IEN-----VFS
-DDSEFIS-----------R-------------------D------------------WK
LISWQNLC-SVFSTNKNLKVFELDNCHLSYD---------SLVILC--KAMAQP------
TC--K-LQRLLFHC---------VSD-IGDSPDFFKAI----------------LHNP
PF00560 // ENSGT00900000140813_0_Domain_6 (1619, 1644)
HL
TYLN-LYDT-SLSYA-AVRQLSRMLKRPTCN----IEKL--------ML-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------GKCNIR-----------------------------
----------------------------LEACEDIAS-----------------------
--------------------------------VLICNNKLKH---LS-LAENP-LNE---
-------EGTMVLCDAL----KHPNCALETL-----------------------------
------------------------------------------------------------
------------------------------VIPNSLLEKQAWD--DLS-WNRNQVPDPLD
L-GSNNLEDSGVN-SLCEALKSS--YCH----I---------------------------
-----------------------EELWL--ANC-----FLTADCCKDI------------
-------SSVL-ISNEKLKTLKLGNNTILDVGVKQ-LCVALK-HP-NC-KLQNLG--LE-
--R-CSLTTACCE--------------------DLASALTACKTL-RSLNLDWNAFDHDG
VVLLCE--------ALSQLDCA--LQ--T-------------------------------
---LGLNKC-AFDEETQLL------LT----------AVE-ENNPHLAIIHQP-------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSRNOP00000074961 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------------MFWG-SR
LPLRVF----------------FGSLPGLL-------------------NTWQSV---WT
RR-LHCAP--T----MSLDIQCEQ-LSD-ARWT----------------EL---------
----LPLI-------------------------------QQYQVV------RLDDC-GLT
EVRCKDIR-SAIQA
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1542)
NPALTELSLRTNELGDA---------GVGLVL--QGLQN-P-----
TC--K-IQKLS--L-QNCSLT--EA---GCG-VLPDVL----------------RS
PF13516 // ENSGT00900000140813_0_Domain_6 (1617, 1642)
LSTL
RELH-LNDN-PLGDE-GLKLLCEGLRDPQCR----LEKL--------QLE---------Y
C------N----------------------------------------------------
---------LT---AT-SCE---PLASVLRV-KPDFKELVLSNNDFHE-AGIHTLCQG--
LKD-SACQLESL-K----------------------------------------------
------------------------LENCGIT-----------------------------
----------------------------SANCKDLCD-----------------------
--------------------------------VVASK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
ASLQE---LD-LGSNK-LGN---
-------TGIAALCSGL----LLPSC----------------------------------
--------------------------RLR-TLW---------------------------
-------------------------------LWDCDVTAEGCKDLCRV-LRA
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
KQSLKELS
L-AGNELKDEGAQ-LLCESLLEP--GCQ----L---------------------------
-----------------------ESLWV--KTC-----SLTAASCPHF------------
-------CSVL-T
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
KNRSLFELQMSSNPLGDSGVVE-LENSGT00900000140813_0_Linker_15_16 (2378, 2391)
CKALG-YP-DT-VPF13516 // ENSGT00900000140813_0_Domain_16 (2391, 2436)
LRVLW--LG-
--D-CDVTDSGCS--------------------SLENSGT00900000140813_0_Linker_16_17 (2436, 2442)
ATVLLAPF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2464)
NRSL-RELDLSNNCMGDTG
VLQLLE--------SLKQPSCA--LQ--Q-------------------------------
---LVLYDI-YWTDEVEDQ------LR----------ALE-EERPSLRIIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMLUP00000019501 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------MAESDS
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 330)
SDSD---LLWYLEKLS--R---KECHSLRNQLGQ------
---------E--CMEIG--------L--PPI-PRLE-LSE-E-NPHLANQLTTFYEAQHL
WNLMYNI----FQ-KIPRKDLCEKI---NARRN---------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------RNKEAHKML-
--------IKKK----------------L--Q--EERCLHNRIHDIFCQDMI--------
----------------------------------------------LELVNKFDFVFRHV
SS--EAVKA----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
LNVFLIGQEASGKTMVLRGA-----LFEWA--N---------GNIWA
D--------MFSY-VIHLSSHDINQMT-------SGSLVP-LIFK-DW-----PK-GQDP
IADILS-EP----QKILFILEDLDD-VELSFHIG------PQ-A-----LCTDSRQQ-VS
-MPILVASLLTRKLAPGCRFLIT-SRPQGQPVTWTIM-K-----KTD--MLMALSFSHEK
RHKYF----------------T--LF-FKD---------GQRAEAAFQLVQR-NELFLHL
CQAPILCWVTCVALQRQMNQG-ADL-K--RS-CQTLTDIYTNFLADTLTAQA----AH--
----------QQPIVPLQSLCSLA---LEGL-LH-NT----LHFTEE-------------
--DLRSVGFTQA-DVSTLQTL----EILWPSSH-LK-GHYRFIHSKVQEFCAAVAFMLPL
AEHGI-----------------P----SAWG-R--C--KGKWE------RPIIASM----
----FGLLNKKRRKTFETALGCH---------------LL-TDY-VSQ-YLLTETRCFGD
NL-------------------------------------KAIGHHTPLLHGLFENQE-EE
YMKWILDS---------------------F-------------------SEANIYVRDHQ
DL---MVS--S----YCLG-YCQH-LQK-LKLS----------------IQN-----IFE
--KR--------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSGGOP00000008815 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MAMAKA-RNPREA---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWALSDLE--E---NDFKKLKFYLRD------
---------M--TLSEG--------Q--PPL-ARGE-LEGLI-PVDLAELLISKYGEKEA
VKVVLKG----LK-VMNLLELVDQLS-HIC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LH--DYREVYREH-
--------VRC-----------------LE------EW---QEA----------GVNGRY
NQVLLVA-KPSSESPESLAC------PI-------PE-----Q-ELDSV--TVEALFDSG
EK--PSLAP----S
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
LVVLQGSAGTGKTTLARKM-----VLDWA--T---------GTLYP
G--------RFDY-VFYVSCKEVVLLLE-------SKLEQ-LLFW-CC-GD-----NQAP
VTEILR-QP----ERLLFILDGFDE-LQRPFEEK------LK-K-----RG-----L-SP
-KESLLHLLIRRHTLPTCSLLIT-TWPLALRNLEPLL-K-----QAR--HVHILGFSEEE
RARYF----------------S--SY-FTD---------EKQADRAFDIVQK-NDILYKA
CQVPGICWVVCSWLQGQMERGK-VV-L--ET-PRNSTDIFMAYVSTFLPPDD-DGGCSEL
----------S-RHRVLRSLCSLA---AEGI-QH-QR----FLFEEA-------------
--ELRKHHLDGPRLA-AFLSS----NDYQLGLA-IK-KFYSFRHVSFQDFFHAMSYLVK-
EDQSRL----------------G--KESRREVQ-RLLEVKEQE-GNDEMTLTMQFL----
----LDILKKDSFSNLELKFCFR-----------------ISPC-LAQ-DL-KHFKEQM-
E-------------------------------------SMKHNRTWDLEFSLYEAKI-KN
LVKGIQMN---------------------N-------------------VSFKIKHSN-E
KK-SQSQN--L----FSVK-SSLS-HGP-KEEQ------------------KC----PSV
---HGQKEGK---DNI--A-------------------GKPKE-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------AST----------------GKG---------R
-GTEE-TDEEVR--SPK-NTYI--------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCAFP00000003649 (view gene)
ENSVPAP00000007865 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MPAT----PLRN
PF02758 // ENSGT00900000140813_0_Domain_0 (204, 328)
GLCRLCAYLEELE--A---AELKKFKLYLGT------
---------A---AGIG--------E--GRI-PWGR-MEPAG-PLEMAQLLAAHCGVRQA
WLLTLSI----FE-RINRKDLWERGQ-REDLVRDP----P--SG--------GPSSL---
------------------------------------------------------GS----
------------------------------------------------------------
---QS-ACFL-------------------------------EVFPETPRKDPRETYRDY-
--------VRRK----------------FRLM--EDRNARLGECV---------NLSHRY
TRLLLVKEHSNPMWTQQKLL------DTGRGHARTAG--------HQASLIQMEALFEPD
EE--RPEPP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
RTVVLQGAPGMGKSMLAHKV-----MLDWA--D---------GKLFQ
D--------RFDY-LFYINCREMNQGTT------ERSAQD-LIAS-CW-----PE-PSVP
LQDLVQ-AP----ERLLFIIDGFDE-LKPSFHDP------QG-P-----WCLCWEKE-RP
-TEVLLSSLIRKKLLPELSLLIT-TRPTALEKLHRLL-E-----HPR--HVEILGFSEAE
RREYF----------------Y--KY-FHS---------EEQAGQAFSFVRD-NEPLFTL
CFVPMV------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----R--------PERNVLPD-------AYSEQLASAL----------------STNPNL
VELV-LYSN-ALGGR-GVKLLCQGLRHPNCK----LQNL--------XXX---------X
X-----------------------------------------------------------
--------XXX---XX-XXX---XXXXXXXX-XXXXXXXXXXXXXXXX-XXXXXXXXX--
XXX-XXXXXXXXX-----------------------------------------------
------------------------LRKCQLD-----------------------------
----------------------------AGACQEMAS-----------------------
--------------------------------VLSTSRHLAE---LD-LTGNA-LEG---
-------SGLRLLGRGL----RHPDC----------------------------------
--------------------------RLR-ILW---------------------------
-------------------------------LKICHLTAAACEDLAST-LSVNQSLMELD
L-SLNDLGDPGVL-LLCEGLRHP--QCK----L---------------------------
-----------------------QTLRL--GIC-----RLSSAACEGL------------
-------SAVL-QVNQ
PF00560 // ENSGT00900000140813_0_Domain_15 (2357, 2378)
HLRELDLSFNDLGDLGMWL-LCEGLR-HP-TC-RLQKLX--XX-
--X-XXXXXXXXX--------------------XXXXXXXXXXXX-XXXXXXXXXXXXXX
XXXXXX--------XXXXXXXX--XX--X-------------------------------
---XXLFGR-ELN--THRI------LA----------ALC-LTKPYLDISC---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSOARP00000002811 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------MKAWSQV------
---------S--KSQEKEDSEGHRTS--PEV-IHAQ-PEKPR-SQKI------AIKKVEL
RCKASHT----KGPHMNKQMACTKTL-EETDLNPA---LS-SSFSH---SPSAPNLKSPS
WPTSTEVLKVCKPKQP-----VLKQSK----NRSPSPLILASGYRK-DLSLGIH--QG--
---------FPSSPY-HESPSQELPNDPTSTAVLRVCELPR---PEPREKEGPGTVWPLV
ETSGNSCTG------------------------------------------SRKRYQYQ-
--------RRET----------------PYSTWSW-------KNE---------DLHPKF
VQLLLLHTPYHKDYES---L------NRESWDHGV--------VEEQGHLIEVRDLFGSD
LG--PQEGP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
HTVVLHGVAGIGKSTLARHV-----RRAWE--E---------GQLYR
N--------HFQR-VFYFNCRELATSK-------LTSLEE-LITK-DQ-----SA-TAAS
AGQVQS-QP----EQLLFILDGLDE-LEWVSEEQ------RA-E-----LCLNWSQL-QP
-VQTLLGSLLEKTVFPEASLLIT-ARTAALQKFIPSL-K-----QPC--WVEVLGFSESS
RKDYF----------------Y--EY-FTE---------ESQATRAFSVVES-NQALLAM
CLMPLVSWLACTCLKQQMEAGE-EL-S--LR-SQTMTALYLHYLAQAIQ--------AQ-
-----------PLGTQLRGFCTLA---AEGI-WQ-GK----TLFSPG-------------
--DLKKHGLGGV-INATPLKM----DVLQKH---PTSLNYSFTHLCFQEFFAAVSFALG-
D------MED--------KSDHP--NST-GSME-KLVEVYGIN--NLFRAPTVHFL----
----SGLLSEQGAREMENIFQCK---------------L--SWE-RKRESLLRRAELELW
-----------------------------------C-----EHYSLQLFHCLYEIQD-EE
FLTQAMAH---------------------F-------------------QGTRVYIRTSM
EL---LMF--T----FCIR-FCSH-VQR-LQLN----------------EGRQ-------
------HAG---------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------ACRP--------------------------------------------------
-------------------TDTVLLSWVPAT-----------------------------
----------------------------EARWQVLCS-----------------------
--------------------------------VLQVTGNLKE---LD-LSGNF-LSR---
-------SAVQSLGEAL----RCPGCQLETLR----------------------------
------------------------------------------------------------
-------------------------------LASCGLTAESCKDIASG-LCASQTLMQLE
L-SFNTLLDAGAE-HLCQGLRQP--TCK----L---------------------------
-----------------------QRLLL--AGC-----GLTS------------------
------------------------------------------------------------
---------GCCQ--------------------DLASVLRVSPRL-TELNLLQNDLDDLG
VRLLCE--------GLRHPSCQ--LM--F-------------------------------
---LWLDQT-QLSKEVTEM------LR----------ALE-REKPQLLVSR-MWKPSLRI
LHEGPGGAEMSNT------------------SSLKRQRQA--------------------
---------------SEGSSPQAAQ-GRPFCPSSPTPPGN-EHMESLG-TEDNLWGPTGP
VATEVVDE-ERSLYRVHLPVAGFYHWP---NTGLRFEVRGPA-------TVEIEFCAWDQ
FLSRNFPQHSWLAAGPLFDIKAEPGA-VAAVFLPHFVAFRGN--------DVDISQFQVA
HFKEEGMLLEKPVRVTPCYAVLENPSFSPMGVLLRMVHTALRFIPITSTVLLYHYLQPEE
V-TFHLYLIPSDCSIQKAIDDEEKKFQFVRIHKPPPLTPLYMGSRYIVSGS---------
EKLEILPEELELCYRSPRESQLFSEFYVRHLRSGIRLQIRSKKDGTVVWEALVKPGDLRR
AVTLVPP-----------------------------GLRASPDSPAALRL
PF00619 // ENSGT00900000140813_0_Domain_19 (3051, 3133)
RHFVDRYREQ
LVARVTSVEPVLDKLH-GLVLSEEQYEGVRAEATAPGQMRKLFGFSRSWDWACKDRLYQA
LKETHPHLIMELWEKWGNEETRSGS-----------
ENSBTAP00000001675 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MRMV----SV
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 328)
RCK---LARYLEDLE--D---IDFKKFKMHLED------
---------Y--PSQKG--------C--TSI-PRGQ-TEKAD-HVDLATLMIDFNGEEKA
WAMAKWI----FA-AINRRDLYEKAK-REEPEWE--N-----A---------NISVL---
--------SQ-----------------------------------EESL-----------
-----------------------------------------------------EEEWMG-
------------------L-----------------LGYLSRISICRKKKDYCKKYRKY-
--------VRSK----------------FQCI--KDRNARLGESV---------NLNKRF
TRLRLIKEHRSQQEREHELL------AIGRTWAKIQD------SP--VSSVNLELLFD--
PEDQHSEPV----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
HTVVFQGAAGIGKTILARKI-----MLDWA--S---------EKLYQ
D--------RFDY-LFYIHCREVSLGT-------QRSLGD-LIAS-CC-----PG-PNPP
IGKIVS-KP----SRILFLMDGFDE-LQGAFDEH------TE-A-----LCTNWRKV-ER
-GDILLSSLIRKRLLPEASLLIT-TRPVALEKLQHLL-G-----QAR--HVEILGFSEAR
RKEYF----------------L--KY-FSD---------EQQAREAFRLIQE-NEILFTM
CFIPLVCWIVCTGLKQQMDSGK-SL-AR--T-SKTTTAVYIFFLSSLLQSQGGSQENH--
------------NSATLWGLCSLA---ADGI-WN-QK----ILFQEC-------------
--DLRNHGLQKA-DVSAFLRM----NLFQKEVD-CE-KFYSFIHMTFQEFFAAMYYLLE-
EDNH--------GEMRNTPQACS--KLPNRDVK-VLLENYGKF-EKGYLIFVVRFL----
----FGLINQERTSYLEKKLSCK---------------I--SQK-IRL-ELLKWIEAKAN
AK-----------------------------------TLQIEPSQLELFYCLYEMQE-ED
FVQRAMSH---------------------F-------------------PKIEIKLSTRM
---DHVVS--S----FCIE-NCRH-VES-LSLR----------------LLHN-----SP
KEEEEE------------------------------------------------------
---------------EEVRHSHMDRSVLSDF-----------------------------
------------------------------------------------------------
--------------------------EVAYS----Q-----------GLV---------N
Y-----------------------------------------------------------
---------LT---SS-ICR---GIFSVLSN-NWNLTELNLSGNTLGD-PGMNVLCET--
LQQ-PGCNIRRL-W----------------------------------------------
------------------------LGQCCLS-----------------------------
----------------------------HQCCFNISS-----------------------
--------------------------------VLSNNQKLVE---LD-LSHNA-LGD---
-------FGIRLLCVGL----RHLFCNLKKL-----------------------------
--------------------------------W---------------------------
-------------------------------LVSCCLTSASCEDLASV-LSTNHSLTRLY
L-GENALGDSGVG-ILCEKVKNP--HCN----L---------------------------
-----------------------QKLGL--VNS-----GLTSGCCPAL------------
-------SSVL-STNQNLTHLYLQGNALGDMGVKL-LCEGLL-HR-NC-KLQVLE--LD-
--N-CSLTSHCCW--------------------DLSTLLTSNQSL-RKLCLGNNDLGDLG
VMLLCE--------VLKQQGCL--LK--S-------------------------------
---LRLCEM-YFNYDTKRA------LE----------TLQ-EEKPELTIVFEPSR-----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMUSP00000147928 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------------MFRG-AR
LPLRVF----------------FGSLPGPL-------------------NTWQSG---WT
RR-LHCAP--T----MSLDIQCEQ-LSD-ARWT----------------EL---------
----LPLI-------------------------------QQYEVV------RLDDC-GLT
EVRCKDIS-SAVQA
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1542)
NPALTELSLRTNELGDG---------GVGLVL--QGLQN-P-----
TC--K-IQKLS--L-QNCGLT--EA---GCG-ILPGML----------------RS
PF13516 // ENSGT00900000140813_0_Domain_6 (1617, 1642)
LSTL
RELH-LNDN-PMGDA-GLKLLCEGLQDPQCR----LEKL--------QLE---------Y
C------N----------------------------------------------------
---------LT---AT-SCE---PLASVLRV-KADFKELVLSNNDLHE-PGVRILCQG--
LKD-SACQLESL-K----------------------------------------------
------------------------LENCGIT-----------------------------
----------------------------AANCKDLCD-----------------------
--------------------------------VVASK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
ASLQE---LD-LSSNK-LGN---
-------AGIAALCPGL----LLPSC----------------------------------
--------------------------KLR-TLW---------------------------
-------------------------------LWECDITAEGCKDLCRV-LRA
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
KQSLKELS
L-ASNELKDEGAR-LLCESLLEP--GCQ----L---------------------------
-----------------------ESLWI--KTC-----SLTAASCPYF------------
-------CSVL-TK
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
SRSLLELQMSSNPLGDEGVQE-LENSGT00900000140813_0_Linker_15_16 (2378, 2389)
CKALS-QP-DTPF13516 // ENSGT00900000140813_0_Domain_16 (2389, 2436)
-VLRELW--LG-
--D-CDVTNSGCS--------------------SLENSGT00900000140813_0_Linker_16_17 (2436, 2442)
ANVLLAPF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2464)
NRSL-RELDLSNNCMGGPG
VLQLLE--------SLKQPSCT--LQ--Q-------------------------------
---LVLYDI-YWTNEVEEQ------LR----------ALE-EERPSLRIIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPPAP00000030933 (view gene)
ENSODEP00000023419 (view gene)
ENSCJAP00000020787 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MAGG----A-WGC---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LACYLELLE--K---EELKKFQFLLTN------
---------K--VHSR--------GS--SGE-TPAQ-PEKTS-GMQVASHLVAQYGEQQA
WDLAIHI----WE-QMGLRSLCAQAQ-EEAG-------HS-LSFHY---SLSEPSMGSPN
QPTSTTVLRSQIPEL------HGCTQRSERSVSRRLPDTSGHQW-REITSSLLYQ--A--
---------PLSSPD-YGSPSYESPSAPISTAELGSWISPLQPSPVPREQEAPGTQWPLD
ETLGNYYTGETLLRIR-------------------------------------ERDQSL-
--------WRES----------------PCSTWSW-------KNE---------DLNQKF
TQLLLLQRPHPRSHEC---L------VGGRLLHNV--------EENQGHLIEIGDLFGPG
LD--TQ-EP----H
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
IVILQGAAGIGKSTLARRV-----REAWG--R---------GQLYR
D--------CFQH-VFYFSCRELAQSK-------VLSLAE-LIEK-DW-----TA-TQAP
IRQILS-RP----ERLLFILDGVDE-PKWVLEEQ------SS-E-----LCLHWSQP-QP
-ADALLGSLLGKTILPKASFLIT-ARTTALQHLIPSL-E-----QAR--WVEVLGFSESS
RKEYF----------------Y--KY-FTD---------EMQASRAFRLVKS-NKELWAL
CLVPWVSWLACTCLMQQMKQKE-EL-T--LT-SKTTTTLCLHYFSQALQ--------AQ-
-----------PLGSQLQGFCSLA---AEGT-WQ-RK----TLFSPD-------------
--DLKKHGLDGA-IISSLLKM----DILQEH---PIPLSYSFIHLCFQEFFAAMSCALE-
D------KKG--------RGEHS--NHI-MGLG-KLLEAYGTH--DLLRAPTTRFL----
----LGLLSEQGAREMENIFDCR---------------L--SRE-RKK-RLLQCAQF---
------------------------------------LQPPLQSHSLEFLHCLYETQD-KS
FLTQMMAH---------------------F-------------------QEVCMCVETDM
EL---LVC--T----FCIK-FCHN-VKK-LQLI----------------EGRQ-------
------H-G---------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------P------------
------ARSL--------------------------------------------------
-------------------DRVVLFRWVPAT-----------------------------
----------------------------DAYWQILFS-----------------------
--------------------------------VLKVT
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
RSLKE---LD-LSGNL-LSL---
-------SAVQSLENSGT00900000140813_0_Linker_11_12 (2054, 2068)
CETL----KYPRCLPF13516 // ENSGT00900000140813_0_Domain_12 (2068, 2206)
LETLR----------------------------
------------------------------------------------------------
-------------------------------LAGCGLTAEGCKDLENSGT00900000140813_0_Linker_12_13 (2206, 2212)
ASG-LRPF13516 // ENSGT00900000140813_0_Domain_13 (2212, 2237)
TNQTLTEVE
L-SFNVLTDAGAK-HLCQGLRWP--TCK----L---------------------------
-----------------------QRLQL--VSC-----GLTS------------------
------------------------------------------------------------
---------GCCR--------------------ALASVLSA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2465)
SPSL-TDLDLQQNDLDDVG
VQLLENSGT00900000140813_0_Linker_17_18 (2465, 2716)
CK--------GLRRPTCK--LT--R-------------------------------
---LG-------------------------------------------------KPSAMI
PNEGLDIGDMSNS-----------------TSSLNWEHHDGLGEVEE-------------
----CG--ADHCLA-VSESVPRIAQ-QELAYLPSPASQED-LHTKPLG-NDDDFWGPTGP
VATEVVDK-ERSLYRPF13553 // ENSGT00900000140813_0_Domain_18 (2716, 2997)
VHFPVAGTYRWP---NMGLCFVVREAV-------TTEIEFCVWDK
LLGEINQQQSWMVAGPLLDIKAEPEA-IKAVHLPHFVSLQGG--------HMDTSLFQVA
HFKEEGMLLEKPAKVELHHIVLEDPSFSPIGVLLRVIHNALRIFPITSMVLLYHRIHPEE
V-TFHLYLIPSDCSIRKAIDDEEMKFQFVRLHKPPPLTPLYMGSRYAVSGSEE-------
--LEIIPEELELCYRSPGEPQLFSEFYVGHLGSGIRLQMKNKTDETLVWEALVKPGENSGT00900000140813_0_Linker_18_19 (2997, 3052)
DLTP
ATTLVPP-----------------------------APKALPSPLDAPGLLPF00619 // ENSGT00900000140813_0_Domain_19 (3052, 3134)
HFVDQYREQ
LVARVTSVEPVLDNLL-GHVLSQEQYERVLAEDTRPSKMRKLFSFSRSWDRSCKDRLYQA
LKDTHPYLIMELWEKGSKKGLLPFSI----------
ENSPPAP00000010743 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------EMNLTELCKMAK--AEM-----------------------------
------------------------------------------------------------
--MEDG---------------------------------QVQEIDNPEL-----------
-------GDA-------------------------------EEDSELAKPGEKEGWRNS-
---------MEK----------------QSSV--WKNTFWQGDIDNFHDAVT--------
-----------------------------------------------LRNQRFIPFLNPR
TP--GKLTP----Y
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVLHGPAGVGKTTLAKKC-----MLDWT--D---------CNLSP
---------TVRY-AFYLSCKELSRMG-------PCSFAE-LISK-DW-----PE-LQDD
IPSILA-QA----QRILFVVDGLDE-LKVPPG------ALIQ-D-----ICGDWEKK-KP
-VPVLLGSLLKRKMLPKAALLVT-TRPRALRDLQLLA-Q-----QPI--YVRVEGFLEED
RRAYF----------------L--RH-FGD---------EDQAMRAFELMRS-NAALFQL
GSAPAVCWIVCTTLKLQMEKGE-DP-V--PT-CLTRTGLFLRFLCSRFPQG-------AQ
------------LRGALRTLSLLA---AQGL-WA-QM----SVFHRE-------------
--DLERLGVQES-DLRLFLDG----DILRHDRV-SK-GCYSFIHLSFQQFFAALFYALE-
KE--E---EE--------DRDGH--AWDIGDVQ-KLLSGEERL-KNPDLIQVGHFL----
----FGLANEKRAKELEATFGCR---------------M--SPD-IKQ-ELLRCKAHLHA
NK-----------------------------------S-LSVTDLREVLGCLYESQE-EE
LAKVVVAP---------------------L-------------------KEISIHLTNTS
EV---RHC--S----FSLK-HCQD-LQK-LSLQ----------------VAKG----VFL
-ENY-MDF----EL--------------------DIEFERC-TYL--TIPNWARQD-LRS
LRLWTDFC-SLFSSNSNLKFLEVKQSFLSDS---------SVRILC--DHVTR-S-----
TC--H-LQKVE--I-KNVTPD--TA----YR-DFCLAF----------------IGKKTL
THLT-LAGH-IEWERTMMLMLCDLLRNHKCN----LQYL--------RLG---------G
H------C----------------------------------------------------
---------AT---PE-QWA---EFFCVLKA-
PF13516 // ENSGT00900000140813_0_Domain_8 (1773, 1795)
NQSLKHLRLSANVLLD-EGAMLLYKT--
MTR-PKHFLQML-S----------------------------------------------
------------------------LENCRLT-----------------------------
----------------------------EASCKDLAA-----------------------
--------------------------------VLVVS
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
KKLTH---LC-LAKNP-IGD---
-------TGVKFLCEGL----SYPDC----------------------------------
--------------------------KLQ-TLV---------------------------
-------------------------------LQQCSITKLGCRYLSEA-LQEACSLTNLD
L-SVNQIA-RGLW-ILCQALENP--NCN----L---------------------------
-----------------------KHLRL--WSC-----SLMPFYCQHL------------
-------GSAL-L
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2377)
SNQKLETLDLGQNHLWKSGIIK-LFGVLR-QR-TG-SLKTLR--LK-
--T-YETNLEVKK--------------------LLEEVKEKNPKLTIDCNAS------GA
---------------TAPPCCD--FF----------------------------------
----C-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSRNOP00000004280 (view gene)
ENSJJAP00000000928 (view gene)
ENSMLEP00000020588 (view gene)
ENSOANP00000018607 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------R----------------LQDV--EDENSLL-------------GANPEY
LRLLLVKEHPGQL-------------------GSAGH-------KQ-MSATQVDALFDPE
EE--GRKPF----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
RTVVLQGAAGIGKTTLATKI-----LEDWA--A---------GRLFH
G--------RFDY-VFYVSCREMNLVR-------ERSVAD-LISH-CC-----AD-KNGP
VTDIVW-LP----KRLLFIVDGFDE-LRCSNERS------RK-D-----LCSDCKEK-RS
-TDTLLASLMSKRLLPEASLLIT-TRPTTLEKMQALL-E-----QPR--WAEILGFAEAE
RKDYF----------------Y--RY-FKE---------KKLARQAFGFLQG-NDGLFTL
CMVPIVCRIVCASLKRQMERGE-AL-T--QT-SQTTTAGYLSYLFGLLREDG-TGP-KAP
------------RPLNLRRLCSLT---AERI-QN-PK----ILLEED-------------
--DLRRHGLGSSGS-TTFLKI----SILKKNHH-PQ-KFYSFPHLTFQEFFAAVSYVPT-
SSQFV--------------KK--------------LVGQYGQF-GSGYLTLTMRFL----
----FGLLNEERLEK---NLGCE---------------I--SHK-IKK-DLLKWIKAEAS
-----------------------------------------KSNPLDLFHCLYEIQE-EE
FVKRATDH---------------------F-------------------HSIEIGA-LAT
KL-DHTVA--S----FCVK-NCLG-VCS-VTLS----------------SVY-----SAG
-GKG--------------------------------------------------------
------------------------------------------------------------
-----------------------DD---VNA-VVEEGL----------------------
---S-LGSQ-H--------------------------------ESSHRLG---------K
C------F----------------------------------------------------
---------LP---DA-FCQ---NLSSALHT-DPNLLELNLSHIALGN-AGLRVPSEG--
LMD-QGCRLQKL------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCSAP00000016111 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MLRT----AGRDGLC
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 328)
RLSAYLEELE--A---VELKKFKLYLGT------
---------A--TELGE--------D---KI-PWGR-MEMAS-PLEMAQLLITHFGTEEA
WRLALST----FE-RMNRKDLWERGQ-REDLVRDT----P--PG--------GPSSL---
------------------------------------------------------GN----
------------------------------------------------------------
---QS-TCLL-------------------------------EVSPVTPRKDPQEAYRDY-
--------VRRK----------------FRLM--EDRNARLGECV---------NLSHRY
TRLLLVKEHSNPMQAQQQLL------DIGRGHARTVG--------HQASPIEIETLFEPD
EE--RPEPP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
RTVVMQGVAGIGKSMLAHKV-----MLDWA--D---------GKLFQ
G--------RFDY-LFYINCREMNQSDT------ECSMQD-LISS-CW-----PE-PSAP
LQELVR-VP----ERLLFIVDGFDE-LKPSFYDP------QG-P-----WCLCWAEK-RP
-TELLLNSLIRKKLLPELSLLVT-TRPTALEKLHPLL-E-----HPR--HVEILGFSEAE
RKEYF----------------Y--KY-FHN---------TEQAGQVFNYVRD-NEPLFTM
CFVPLVCWVVCTCLQQQLEGGG-LL-R--QT-SRTTTAVYTLYLLSLMQPKLRT-PHL--
-----------QPPPNQRGLCSLA---ADGL-WN-QK----ILFEEQ-------------
--DLRKHGLDGA-DVSAFLNM----NIFQKDIN-CE-RYYSFLHLSFQEFFAAMYYILD-
D--GE--------------R--G--AGPDQDVI-RLLTEYGFS-ERSFLALTIRFL----
----FGLLNEETRSYLENSLCWK---------------V--SPH-IKR-ELLQWIRSKAQ
SD-----------------------------------GSTLQQGSLEFFSCLYEIQE-EE
FIQQALSH---------------------F-------------------QVIVVSNIA-S
KM-EHMVC--S----FCVK-NCRS-AQV-LHLY----------------GA------TYS
-ADGEDRA--R-CS---------------------------------------AGA---H
TL------------------------L-------------------------V-------
QL-----------PERTVLLD-------AYSEYLAAAL----------------CTNPNL
VELS-LYRN-ALGSR-GVKLLCQGLRHPSCK----LQNL--------RLK---------R
C-----------------------------------------------------------
--------RIS---SS-ACE---DLSAALIA-NKNLTRMDLSGNGVGF-PGMTLLCEG--
LRH-PQCRLQMIQ-----------------------------------------------
------------------------LRKCQLE-----------------------------
----------------------------SGACQELAS-----------------------
--------------------------------VLSTNPHLVE---LD-LTGNA-LED---
-------LGLKLLCQGL----RHPVC----------------------------------
--------------------------RLR-TLW---------------------------
-------------------------------LKICHLTAAACEELAST-LSVNHSLRELD
L-SLNELGDPGVL-LLCDSLRHP--TCK----L---------------------------
-----------------------QTLRL--GIC-----RLGSAACEGL------------
-------SAVL-QANHHLRELDLSFNDLGDWGLWL-LAEGLQ-HP-TC-RLQKLW--LD-
--S-CGLTAKACK--------------------NLYFTLGINQTL-TELYLTNNALGDTG
VRLLCK--------RLNHPGCK--LR--V-------------------------------
---LWLFGM-DLNKMTHSR------MA----------ALR-VTKPYLDIGC---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSBTAP00000056628 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------KEDLEGEKPGKQDKYRNI-
--------LKKK----------------FCQP--W-KNFWPRLSEDVCI-----------
---------------------------------------------VTQRYETLIPFCNPK
MP--AGPFP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
HTVVLHGPAGVGKTTLAKKL-----MLDWT--Q---------DNLAE
---------TFNF-AFYLSCKELNHMG-------TCTFAE-LIST-NW-----PH-VQED
IPAILA-QA----QKVLFILDSFDE-LKVPSE------ALIH-D-----ICGDWKKQ-KP
-VPVLLGSLLKRKMLPKATLLIT-TRPGALRELRLLT-E-----QPL--FIEIEGFLEED
RKAYF----------------L--KH-FEE---------ESQALRAFDLMKN-NEALFQL
GSAPSVCWMVCTCLRQQMERGE-DP-A--AT-CRTTTALFLRFLCSRFPPPHG-GGPRRG
------------LRAPLKPLCLLA---AEGV-WM-RS----SVFDGE-------------
--DLRRLGVDPS-ALCPFLDG----NILQKSED-GE-ACYSFIHLSVQQLLAAMFYVLE-
LEEQE---EE--------GLGGR--QWHVGNVG-KLLSKEERL-KNPSLTHVGYFL----
----FGLCNERRAMELETTFGCL---------------V--STE-IKR-ELLKYTLMPHG
KK-----------------------------------SFS-VMDTKEVLSCLYESQE-EQ
LVKDAMAH---------------------V-------------------KEMSLHLKNET
DV---VHS--S----FCLK-HCGN-LQK-LSLQ----------------VEEG----IFL
-DND-TAL----ES--------------------GTQVER------------SQNE-QHM
LPLWMDLC-SVFDSSRSLLFLDISQSFLSTS---------SVRILC--EKIAS-A-----
AS--S-LQKVV--L-KNISPA--DT----YR-NFCMAF----------------GGHKTL
THLT-LQGN-DQND--MLPPLCEVLRNPKCN----LQYL--------RLV---------S
C------S----------------------------------------------------
---------AT---TQ-QWA---DLSCCLKT-NQSLTCLNLTANEFLD-EGAKLLYMT--
LRY-PTCFLQRL-S----------------------------------------------
------------------------LENCQLT-----------------------------
----------------------------EAYCKDLSS-----------------------
--------------------------------ALIVNQRLTH---LC-LAKNA-LGD---
-------RGVKLLCEGL----TYPEC----------------------------------
--------------------------QLQ-TLV---------------------------
-------------------------------LWCCNITSDGCIHLSTL-LQQNSSLTHLD
L-GLNHIGIIGLK-FLCEALKKP--LCK----L---------------------------
-----------------------RCLWL--WGC-----AITPFSCAEL------------
-------SSAL-RSNQNLITLDLGQNSLGSSGVNM-LCDALK-LQ-SC-PLQTLR--LK-
--I-DESDAQIQK--------------------LLREMKESNPQLTIESDHR----DPKD
---------------NRPSSHD--FI----------------------------------
----F-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCANP00000018943 (view gene)
ENSAMEP00000004586 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------NTMAQT-HN
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
PQEA---LLWALRDLE--E---KSFKLFKFHLWD------
---------G--SLLEG--------Q--PRL-ARGG-LEGLG-PVAPASRPVSL-GSTGA
VKVVLRA----LR-VMNLLESADQLS-HVC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LN--GSSADSLPLH
PALSRTWWIDAA----------------LE------NR---V---------------GME
------T-RSSV---PSR--------GV-------PG-----Y-THMSR--GLPLEVDPG
EK--PYQAP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
PTVVLQGSAGTGKTTLARKI-----VLDWA--T---------GTLYP
G--------RFDY-VF-VSCREVALLQE-------GNPEQ-LLFW-CC-GD-----NRAP
VEEMLR-EE----ERLLFILDGFDE-LQRPFAKG------WK-R-----LS-----P-NP
-VENVLHRLIRRKLFPTCSLLIT-TRPVALQNLKPSL-Q-----QSR--HIHVLGFSEDE
RRRYF----------------S--SY-FAG---------EEQARNAFDIVQG-KDVLYKS
CQIPGTCWVICSWLKEQMERGR-EI-S--ET-PSNGTDIFMAYVSTFLPPSD-NAGCSEL
----------T-GHRVLSGLCSLA---AEGI-QH-QR----FMFEEA-------------
--DLRKHDLDATNLD-AFLSS----MRYQEEHT-RK-TFCAFRHISFQEFFHAMSYLAK-
EDQSQL----------------G--KESHKELK-KLLDDEKEA-ENEEMTLSVQFL----
----LDMLKKETSSSFELKFSLK-----------------VSPL-VKQ-EW-MYFKEQM-
K-------------------------------------SRKHKGAWDLEFSLNPSKI-RN
LVKGVRIS---------------------D-------------------VSFKIGQSN-K
-K-SHDRN--S----FSVK-TSLS-NGW-EEKQ------------------KC----PVV
--------DK---DNT--A-------------------ETQKE-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------GPN----------------GKG---------R
-ETEK-RETRRVG-VVD-GKQ-----IN-GTVEMKG---QGG--G---E-GRDN-GGG-R
QSIKR-NGEVL---EG-MEG----------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPSIP00000012133 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------MMENRSSR--IS
PF02758 // ENSGT00900000140813_0_Domain_0 (203, 328)
DL---LVYAFNNLS--Q---EEFKGFKDKLSH------
---------S--DMG-G--------K--GTI-PRGR-LENAD-RIDTKNLLMAFYGGDIA
VDVTVEI----FS-QIHLRESAAKLK-EQRN-WQK-RSIRPDQPAT-------S-SYH--
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------FSQEVAAGYYTVKYREH-
--------IQNK----------------YRVI--KDLNSRLGENV---------TLNSRY
TKLMIV---------------------------------------------NLSNLFKCD
ED--GQTPQ-----
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
IVVLLGAAGIGKTMTARKI-----MFDWA--G---------GELYR
G--------KFDY-AFYINCREVNFGS------EQGRIAD-LIIK-NC-----PN-KNAP
IEEILI-NP----EKLLFIIDGFDE-LRFSLDQP------ED-N-----LCSDPCEK-QP
-LEITLSSLVRKTVLLESYVIIT-TRPTALEKLRQCL-E-----CAR--YAEILGFSTAE
REEYF----------------H--KF-FGN---------EEKARKAFNFVKE-NEMLFTM
CFVPIACWIICTVLKQHMEAGE-DL-A--QT-SKTITGVYMLYLFSLLKGHS-SNA-KQP
------------TPTNLKGLCSLA---ADGI-WR-QK----ILFGEE-------------
--EIKKHGLDTSD--SLFLNE----NMFQKDID-CE-CVYSFIHLSFQEFFAALFYVLE-
KEETT--------------VVDT--GTAMNNVK-KLLENYGKS-RN-YLMLTVRFL----
----FGLLNEERMKDMEKKLSCK---------------I--SPK-IKP-DLLQWPDEELS
SF----------------------------------KRNAEMIYQLEEFHCLYEIQE-EN
FVQSALNR---------------------F-------------------TELYLAEYNFT
KM-DQVAL--S----FCLK-NCCR-LET-LHLN----------------LCM-----FGL
-EDSSEDS----LR-------------------------PPKQPR------LLTNG-HLT
TACYRDLA-SALSTKQTLSELDLSMNSLGDS---------GVRLLC--EGLTH-P-----
TC--K-LQKLG--L-SNCHLT--AT---CGG-DLASLL----------------STKPTL
TELD-IGEN-NLGDS-GVQLLCGGLKHPHCK----LQKL--------RLM---------N
C------G----------------------------------------------------
---------LT---AG-SCG---NLSSVLRA-KPTLNELDLALNNLGEAGGMKLLCEG--
LKH-PDCKLQKL-R----------------------------------------------
------------------------LMGCQLT-----------------------------
----------------------------AACCEDLSS-----------------------
--------------------------------VLSINQTLTE---LE-LRGNL-LRD---
-------SGVQLLCEGL----KHANC----------------------------------
--------------------------KLQ-KLG---------------------------
-------------------------------LKNFLLTDACCEDLAAV-LSISESLTELQ
L-GIDYLRDSGVQ-LLCEGLKHP--NCK----L---------------------------
-----------------------QKLGL--WGC-----DLTADCCGDL------------
-------SSAL-RTSQYLTELELGYNELGDSGVQL-LCEGVK-HP-NC-KLQKLG--LE-
--H-CSLTAACCG--------------------DLSSALSMNQNL-SELDIGRNKLGDSG
VQLLCE--------GLKHPNCK--LH--K-------------------------------
---ISLGYQ-SVTEETQAE------LA----------AVK-DTKPDLLIEIWVWCFY---
--T---------------------------------------------------------
------------------------------------------------TPQPLSWHVPY-
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPEMP00000020353 (view gene)
ENSPCOP00000019391 (view gene)
ENSCAFP00000034020 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MSLSIQCQQ-LSD-ARWT----------------EL---------
----LPLI-------------------------------QQYEVV------RLDDC-GLT
EVRCKDIS-SALQANPSLTELSLCTNELGDA---------GVHLVL--QGLQS-P-----
TC--K-IQKLS--L-QNCCLT--KT---GCG-VLPAVL----------------RS
PF13516 // ENSGT00900000140813_0_Domain_6 (1617, 1643)
MPTL
RELH-LSDN-PLEDA-GLQLLCENSGT00900000140813_0_Linker_6_7 (1643, 1650)
EGLLDPQPF13516 // ENSGT00900000140813_0_Domain_7 (1650, 1767)
CH----LEKL--------QLE---------Y
C------N----------------------------------------------------
---------LT---AA-SCE---SLASALRN-KQHFKELAVSNNEIGE-AGVRVLCQG--
LVE-SACQLETL-K----------------------------------------------
------------------------LENCGLT-----------------------------
----------------------------PASCEDLRA-----------------------
--------------------------------VVASK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
TSLRE---LD-LGDNK-LGD---
-------QGIAVLCPSL----LHPSC----------------------------------
--------------------------QIR-VLW---------------------------
-------------------------------LWECDVTATGCRDLCHV-VSTKE
PF13516 // ENSGT00900000140813_0_Domain_13 (2215, 2238)
SLEELS
L-ACNALGDEGAQ-LLCESLLEP--GSR----L---------------------------
-----------------------QSLWV--KSC-----NFTAACCHHF------------
-------GTML-PRRPALLRVSWPSGRVAAAPAGE-LRARLA-SG-GA-ACRVSF--LG-
--D-CDVANDGCA--------------------SLASLLLA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2462)
NRSL-RELDLSNNRMNDQG
IRRLME--------SVEQPGCA--LE--Q-------------------------------
---LVLYDI-YWSQDIEDS------LR----------ALE-ERKPSLKIIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSTBEP00000002286 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------HS-PSVLY---SPSAPNLESPS
WPTSTDVLSSLGSGSL-----GHPEDS-KRRLVRWLPDTSGPSL-KEIHPSFFYQ--A--
---------LPRSPE-HESPHQESPNAPTSTAVLRAWESISLPSPEPRKQETPWTRRHLA
AVSGKFCSEV------------------------------------------TERYQ-S-
--------EREK----------------SCSTPPW-------KNK---------FLHKKF
TQMLLLHRPHPRYHKP---L------VRGSWHHST--------AKERGHLIEITDLFGPG
LS--TQKGP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
HIVILQGAPGIGKSTLASQV-----RGAWE--E---------GQLYR
D--------RFQH-IFYFSCRELAQSK-------MMSLPE-LLTK-DT-----GA-LRAP
IEQILS-QP----GQLLLIFDDLDE-PKWVLEEQ------SP-E-----FCLPWSQP-QP
-VQALLRSLLRRTILPEVSLLIT-VRTTVLQKLIFSL-K-----QPR--WVEVLGFSESS
RKDYF----------------Y--KY-FTD---------ESQAIRALSLVES-NPALLTL
CLTPLVSWMVCTCLKEQMERGE-EP-T--VT-SQTTTALFLHYIFQALS--------AQ-
-----------SLRTELRGICSLA---SEGI-FQ-GK----TLFSSG-------------
--DFRKHGLDKA-IVFTFLKM----GVLQTN---TFSLSYSFTHL-FQEFFAAMFCALG-
D------KKK--------RSKEP--YNI-S-IE-MFLEVCRMC--DWFGAPTMRFL----
----FGLLSEQGMREMENIFHCQ---------------L--SQK-RKW-ELLRWAQEEAK
-----------------------------------VQHPFFQPYSLELLHCLYETQD-EV
FLTKVMPH---------------------F-------------------LGTKICVQTNV
EL---LVV--T----FCIK-FCQH-VKX-XXXX----------------XXXX-------
------XX----------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------X------------
------XXXX--------------------------------------------------
-------------------XXXXXLMWVPVT-----------------------------
----------------------------GACWQILFS-----------------------
--------------------------------NLGDAR
PF00560 // ENSGT00900000140813_0_Domain_11 (2019, 2057)
NLKE---LD-LSRNF-LTY---
-------SAVQSLCEAL----RHPCCHLETLR----------------------------
------------------------------------------------------------
-------------------------------LAGCGLTAECCKDLAFG-LNTSQTLTELE
V-SFNMLTDAGAK-HLCHGLRQP--SCK----L---------------------------
-----------------------QRIQL--VSC-----GLTS------------------
------------------------------------------------------------
---------SCCQ--------------------NLATVLSANPSL-TEMDLLHNCLGDLG
VGLLCE--------GLKHPACQ--LT--L-------------------------------
---LWLDQT-HLSDEVREA------LR----------TEK-EK--QLRISS-RXXXXXXX
XXXXXXXXXXXXX-----------------XX----------------------------
--------XXXXXXXXXGSSPQAAQ--EPFRLSSPASPRD-LHMEPLG-TEEDFWGPTGP
VTTEVVDK-EQSLYRVHFPMAGFYYWP---NTGLRFMVKGAV-------TIEIAFCAWNQ
FLE-TVPQHSWMVAGPLFDIKAEPGV-VAALYLPHFLSLQGK--------EMNTSLFQVA
HFKEEGMLLEKPSRVEPYYTVLENPSFSPMGVLLRMIHIALPFIPITSTVLVYYRLNAEE
V-IFHLYLIPTDCSIQKAIDDEEKKFQFVQIRKPPPLTPLYMGSRFTLSSS---------
RQLEIIPEELEFCYRSPGESQLFSEFYVDQLESGIRLQMRDKKDGTVVWKGLVKPGDLRP
KATLIPP-----------------------------TSIVSPSTRDIPDL
PF00619 // ENSGT00900000140813_0_Domain_19 (3051, 3135)
LHFVDQYREE
LIARVTSVDPVLDKLY-GQVLSEEQYERVRAEATMPDKMRKLFSFSQSWNQACKDQLYQA
LKETHPHLIVELWEKGWKTGLWILS--S--------
ENSPCAP00000008455 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---SSFF
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
SDFG---LLWYLEL----N---KEFRRFKELRQ-------
------------ELQHG--------L--EQI-PWAE-VKKAT-REVLANLLLKHYQEEQA
WDMTYRI----FH-KMGRKDLCVKAK-RES------------T---------------YT
K---------------------------------------------------MYQA----
------------------------------------------------------------
----------------------------------------------------------H-
--------VKKK----------------FSDI--WSRESLIKIQDC--------------
---------------------------------F-------EQEVMQKKXXXXXXXXXXX
XX--XXX-X----XXX--XXXXXXXXXXXXX
PF05729 // ENSGT00900000140813_0_Domain_2 (692, 940)
XI-----KLMWA--E---------GTVCQ
D--------RL---YFYFCCREVKQLT-------STSLAE-LISG-D------PSP-LPP
IAEILS-QP----ERLLFIIDSFEE-LTRALKEP------ES-E-----LCSDWMQQ-RT
-GEVVLGSLLRKKMLPESSLLIA-ATPSCQQELEAQL-E-----HPE--IKSIIGLNDHD
RKLYF----------------S--CV-FQD---------KKRAMEAYSSVRE-NQQLFHM
CQIPVLCWVVSTCLKQEMERGR-DL-A--LA-CRRTTSLYVSFIFNTFTPKG-ATCLHE-
-----------QSCVQLKGLCSLA---VEGM-WT-GT----FKFSEE-------------
--DLRRNGLVES-DIPALLDM----KLLQKCRG-CE--LYTFIHLSIQEFCAAMFYLLK-
SPGYH-----------------S--SPAIGSTK-ELLLSCLKK-ERVHWFFGGCFV----
----FGLLNAEEVKKLNAFFGFQ---------------L--SLE-VKQ-QFHQCLKSLGA
H-E----------------------------------HLEGEIDFLFLFYCLFEMRD-KV
FVRQALDC---------------------F-------------------QEVHFSVTEDV
DL---KVS--A----YCLR-YCSG-LKK-ISFS----------------IRN-----VFE
-GNR--DN-----------S-----------------------------------TSSCS
LLLWYHIC-SVLTTNVHLRELQVNNSVVSEP---------AFTVLC--KELKHP------
NC--H-LQKLAIND---------GS---------FEVF----------------IYNPYL
KYLD-LSSS-RLFHN-GVRFLCEVSNYPL-T----IEDL--------LL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------TNCSLS-----------------------------
----------------------------ANDCEAFAS-----------------------
--------------------------------VLEYNKKLKY---LX-XXXXX-XX----
-------XXXXXXXXXX----XXXXXXXXXX-----------------------------
------------------------------------------------------------
------------------------------XXXXXXXXXXXXXXXXX-----XXXXXXXX
X-XXXXXXXXXXX-XXXXXXXXX--XXXXXXXX---------------------------
-----------------------XXXXL--VKC-----FITREGCQNI------------
-------ASVL-TSNPNLRSLEIGNNKIQDEGAKL-LCEALT-HP-SC-HLENL------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCCAP00000039880 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------MAMA-RSPRKA---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWALSDLE--E---KDFKKLKFYLRD------
---------M--TLSEG--------Q--APL-ARGE-LEGLI-AVDLAELLISKYGEKEA
VKVVFES----LK-AMNVLELVDQLR-HIC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LH--DYREIYREH-
--------VRC-----------------LE------EW---QEV----------GVNGRY
NQVLLVA-KPSSESPESPAC------PIT------LE-----Q-ELDSV--MVEALFDSG
EK--PSLAP----S
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
LVVLEGSAGSGKTTLSRKI-----VLDWA--A---------GTMYP
G--------RFDY-VFYVSCKEVVLLLE-------SKLNE-LLFW-CC-GD-----NQAP
VTEILG-QP----ERLLFILDGFDE-LQRPFEEK------LK-K-----RG-----S-SP
-KEGILHLLIRRRTLPTCSLLIT-TRSLALRNLGPLL-K-----QAR--HVHILGFSEEE
RGRYF----------------S--SY-FMD---------EKQADRAFKIVQK-NDILYKA
CQVPGICWMVCSWLKQQMEKGE-AV-L--ET-PRNSTDIFMAYVSTFLLPDD-DGGCSES
----------S-RHRVLMSLCSLA---AEGI-QH-QR----FLFEEA-------------
--ELRKHNLDGPRLA-AFLSS----NDYQLGLA-IK-KFYSFRHISFQEFFYAMSFLVK-
KDQSWL----------------G--KESRREVQ-RLLEVKKQE-GNNEMTLTMQFL----
----LDISKKDSVSNLELKFCFR-----------------ISPC-IAQ-DL-KHFKKQL-
E-------------------------------------SIKQKRPWDLEFSMCEAKI-KN
LVKGIQMN---------------------D-------------------VSFKIAHSN-E
KK-SQSQN--S----FSVK-ARLS-HGQ-KEKQ------------------KC----PLV
--------GK---DNT--A-------------------GTQKE-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------SSI----------------GKG---------R
-GTEE-PDEEHQK-N----TYI--------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSFCAP00000016408 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MNS---------------------------QKKRKDMEPA-----
--------CTWS------------------------------------------LLSE--
-----------------------------------------QK-----------------
----AASNSQ-------------------------------ASLSWKELIDSREIFRKH-
--------LKEK----------------YLNI-GVHKCYH-GN--------------HIE
SRLLLVKEHGISEKTPCGIP-------------------------QQDNFIDMGHVFDPD
EE--GCSSS----Q
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 929)
TVVLQGCAGIGKTAVVHKF-----MFDWA--A---------GMVTP
G--------RFDS-LIYVNCREISHIA-------NLSAAD-LITN-TF-----QD-IDGP
ILDVILVYP----EKLLFILDGFPE-LQHPVGNL------EE-D-----LSANPQEQ-KP
-VETLLHSFVRKKLFPESSLLII-AQPATMKKLHSLL-K-----QSI--QAEIIWFTNAE
KRAYF----------------L--SQ-FSG---------ANAAMRVFYELQQ-NQSLDIM
SSLPIISWMICSVLQSQEDGDR-SL-M--RS-LQTMTDVYLFYFSKCLKTLTGIS-VW--
-----------KGQSCLWGLCSLA---AEGL-QN-QQ----VLFEIH-------------
--DLRIHGIGVYDINCTFLNH-----FLKKSEG-SV-NVYTFLHFSFQEFLTAVFYALK-
NDGNW---------------------MFFDQVGKTWQEIFQQY-GKGFSSLTIQFL----
----FGLLRKGKGKAVETTFGRK---------------V--SPG-LRE-ELLKWTEKEIK
DK-----------------------------------YSMLQIEPMDLFHCLYEIQE-EE
YAEKIIGD---------------------L-------------------QSIILLQPTYT
KM-DILAM--S----FCVK-SSHSHLS----------------V-----SLKCQHLIGF-
-EEEEQAS----------------------AFMT--------------------------
---------PVFTLNQSFK-----LHNLPVS---------RLHLLC--QALRNP------
CC--K-IKDLKL---IFCHLT------ASCGRD---------------LSLVFET
PF13516 // ENSGT00900000140813_0_Domain_6 (1616, 1642)
-NQYL
TDLE-FVKN-TLEDS-GMRLLCEGLKQPNCV----LQTLR--------LYRCLI-SPA-S
CG------------AL----AAVLSTSQWLTDLEFRETKLEASALKLLCEGLKDPNCK-L
QKLKLCASFLPESSEV-VCN---YLASVLIC-
PF13516 // ENSGT00900000140813_0_Domain_8 (1773, 1796)
NPNLTELDLSENPLGD-TGVKYLCEG--
LRH-SNCKV---------------------------------------------------
------------------------LSTCYLT-----------------------------
----------------------------DASCMELSS-----------------------
--------------------------------FLQ----------LF-VFANT-LGD---
-------IGVQHLCEGL----QHAQGIIESL-----------------------------
------------------------------------------------------------
------------------------------VLSQCSLSAACCESLAQV-LSSTWSLIRLL
L-INNKIEDLGLK-WLCEGLKQP--DCQ----L---------------------------
-----------------------KDLAL--WTC-----HLTGECCQDF------------
-------CNAL-YT
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NEYLRDLDLSDNALGDEGMQV-LCEGLK-HP-SC-KLQTLW--LA-
--E-CHLTDACCG--------------------ALASVLNR
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2465)
NENL-TLLDLSGNDLKDFG
VQMLCD--------ALIQPICK--LQTFY-------------------------------
------IDSDHLHEETFRK------ME----------ALK-MSKPGITW-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSOARP00000000498 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MSPA----PLRSELGR
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 326)
LCTYLEELE--A---VELKKFKLYLGT------
---------A---TEMG--------E--AKI-PWGR-MEPAG-PLEMAQLLVAHFGPREA
WLLALGV----FQ-RINRKDLWERGH-REDQGRDA----P--PG--------GPPSL---
------------------------------------------------------GS----
------------------------------------------------------------
---ES-ACSL-------------------------------DVLPAAPRKDSRETYRDY-
--------VRRK----------------FRLM--EDRNARLGEFV---------NLSHRY
TRLLLAKEHSSPRWAQQKLL------DTGQGNTGTAG--------QPASLIQIEALFEPD
EE--RPEPP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
RTVVLQGAAGMGKSMLAHKV-----MLDWA--D---------GKLFQ
D--------RFDY-VCYINCREMNRGTA------ERSARD-LLAG-CW-----PE-PCAP
LQELVR-VP----ERLLVIMDGFDE-LRPSFHDP------QG-P-----WCLCWEKK-AP
-VEVLLGSLIRKKLLPELSLLIT-TRPTALAKLQRSL-E-----HPR--HVAILGFSEAE
RKEYF----------------H--RY-FQD---------REQAGQVFSFVRD-DEPLFTL
CFVPMVCWVVCTCLKQQLEGGG-LL-R--RR-SRTTTAVYLLYLLSLMQPKPGT-PTL--
-----------PSPPNRRGLCALA---AHGL-WE-QK----ILFAER-------------
--DLREHGLDAA-DVSSFLNM----NIFQKDIN-CE-TLYSFIHLSFQEFFAAMHYVLD-
D--GA--------------S--G--SGPERDLS-RLLTEYAFS-ERSFLALTVRFL----
----FGLLNEETRSYLEKTLGWK---------------V--SPG-VKT-ELLEWIRRKAR
SE-----------------------------------GSTLKQGALEVFGCLYELQE-EE
FIQQALSH---------------------F-------------------QVVVVNNIT-T
KM-EYIVS--S----FCVK-SCRK-AEV-LQLF----------------GA------AYQ
-ADGEDGL--R-WP---------------------------------------LAT---S
------------------------------------------------------------
-------------PERHVLPD-------VYSEQLAAAL----------------STNPSL
VELA-LHSN-ALGSQ-GVKLLCQGLRHPNCR----LQNL--------RLK---------R
C-----------------------------------------------------------
--------QVS---SS-VCQ---DLTTALIA-NNHLLRMDLSGNALGL-LGVQLLCQG--
LRH-PKCRLQVVQ-----------------------------------------------
------------------------LRKCQLE-----------------------------
----------------------------AEACQELAS-----------------------
--------------------------------VLSASCHLLE---LD-LTGNA-LED---
-------SGLRLLCQGL----RHPVC----------------------------------
--------------------------RLR-ILW---------------------------
-------------------------------LKICLLTGAACEDLAST-LRVNQSLVELD
L-SLNDLGDPGVL-LLCEGLRHP--QCK----L---------------------------
-----------------------QTLRL--GIC-----RLSSAACEGL------------
-------SAVL-GVSSHLWELDLSFNDLGDRGMSL-LCEGLR-HP-TC-RLRKLW--LD-
--S-CSLTGKACE--------------------DISSALGVNQTL-TDLYLTNNALGNTG
VRLLCE--------RLSHPGCK--LR--V-------------------------------
---LWLFGM-DLNKMTHRS------LA----------ALR-VTKPYLDIGC---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSANAP00000026326 (view gene)
ENSMFAP00000041029 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MAMTKA-RNPREA---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWALSDLE--E---NDFKKLKFYLRD------
---------M--TLSEG--------Q--PRL-ARGE-LEGLI-PVDLAELLISKYGEKEA
VKVVLKG----LR-VMNLLELVDQLS-HVC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LH--DYREIYREH-
--------VRC-----------------LE------EW---QEA----------GVNGRF
NQVLLVA-KSSSESPESPAH------PI-------PE-----Q-ELDSV--MVEALFDSG
EK--PSLAP----S
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
LVVLQGSAGTGKTTLARKM-----VLDWA--T---------GSLYP
G--------RFDY-VFYVSCKEVVLLLE-------SKLEQ-LLFW-CC-GD-----NQAP
VTEILR-QP----ERLLFILDGFDE-LQRPFEEK------LK-K-----RG-----L-SP
-KESLLHLLIRRHTLPTCSLLIT-TRPLALRNLEPLL-K-----QAR--HVHILGFSEEE
RARYF----------------S--SY-FTD---------EKKADSAFDIVQK-NDILYKA
CQVPGICWVVCSWLKGQMERGK-AV-L--ET-PRNSTDIFMAYVSTFLPPVD-DGGCSEF
----------S-RHRVLRSLCSLA---AEGI-QN-QR----FLFEEA-------------
--ELRKHNLDGPRLA-AFLSS----NDYQLGLA-IK-KFYSFRHISFQDFFHAMSYLVK-
EDQSRL----------------G--KESRREVQ-RLLEAKEQE-GNDEMTLTMQFL----
----LDILKKDSFLNLELKFCFR-----------------ISPC-LAQ-DL-KHFKEQM-
E-------------------------------------SMKHNRTWDLEFSLYEAKI-KN
LVKGIQMN---------------------D-------------------VSFKIKHSN-E
KK-SQSQN--S----FSVK-SSLS-HGP-KEKQ------------------KH----PSV
---HGQKEGK---DNI--A-------------------GTHKE-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------AST----------------GKG---------R
-GTEE-TDEEVRC-SGM--GSR----EM-ATITLKD---WRNEGV---K-GQT-------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSANAP00000022958 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------MAMA-RKPRQA---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWALSDLE--E---KDFKKLKFYLRD------
---------M--TLSEG--------Q--APL-ARGE-LEGLI-AVDLAELLISKYGEKEA
VKVVFEG----LK-AMNLLELVDQLS-HIC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LH--DYREIYRKQ-
--------ARC-----------------LE------EG---QEV----------GVNGRY
NQVLLVA-KPSSESPESPAC------PIT------PE-----Q-ELDYV--MVEALFDSG
EK--SSLAP----S
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
LVVLEGSAGTGKTTLSRKM-----VLDWA--A---------GTLYP
G--------QFDY-VFYVSCKEVVLLLE-------SKLEE-LLFW-CC-KD-----NQAP
VTEILR-QP----ERLLFILDGFDE-LQRPFEEK------LK-K-----RG-----L-SP
-KEGLLHRLIKRRILPTCSLLIT-TRPLALRNLGPLL-K-----QAR--HVHILGFSEEE
RGRYF----------------S--SY-FTD---------EKQADRAFDIVQK-NDILYKA
CQVPGICWMVCSWLKGQMERGE-AV-L--ET-PRNNTDIFMAYVSTFLLPDD-DGGCSES
----------S-RHRVLMSLCSLA---AEGM-QH-QR----FLFEEA-------------
--ELRKHNLDGPRLA-AFLSS----KNYEFGLA-IK-KFYSFRHISFQEFFYAMSFLVK-
EDQSRL----------------G--KESHREVQ-RLLEVKKQE-ENDEMTLTMQFL----
----LDISKKDSVSKLELKFCFR-----------------ISPC-VVQ-DL-KHFKEQL-
A-------------------------------------SVKQKRTWDLEFSLYEAKM-KN
LVKGIRMN---------------------D-------------------ASFKIAHSN-K
KK-SQSQN--S----FSVK-TSLS-HGQ-KEKQ------------------KC----PLV
--------GK---DNI--A-------------------GTQKE-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------AST----------------GKG---------R
-GTEE-AP----K-N----TYI--------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPCOP00000019540 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------MVLA-NDPQNA---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWALSELE--E---NNFKILKFHLRG------
---------M--ALPEG--------Q--PHL-ARGE-LEGLS-RVDLASKLILTYGAREA
VRVVLKA----LK-VMNLLELVDQLS-LIC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LN--DYRETYREH-
--------VRS-----------------LE------ER---REG----------EVGSSY
IQLLLVA-KPSSGSPESSAC------LV-------PE-----Q-EVDSV--VVEALFGFG
EK--PHPAP----SS
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 940)
VVLQGSAGTGKTTLAKKM-----VLDWA--T---------GTLYP
G--------WFDY-VFYVSCKEVVLLPK-------GKIEQ-LLFW-CC-GD-----SQAP
VTEILR-QP----ERLLFILDGFDE-LQRPFEEQ------LK-K-----VS-----S-NP
-KEDVLHRLIRRRILPTCSLLIT-TRPQALQNLEPLL-N-----GAC--HVRILGFSEEE
RRSYF----------------S--SY-FKD---------EEQARKAFEVVEG-NAILYQA
CQVPGICWVVCSWLKRQMERGQ-AI-S--ET-PRNSTDIFMAYVSTFLPSDD-KEGRSEP
----------S-RHRVLRSLCSLA---AEGI-QH-QR----FLFGEA-------------
--ELRKHNLDGPSLA-AFLSS----NDYQEGLD-IK-KFYSFRHISFQEFFHAMSYLVT-
QDQSRL----------------G--EASRREMK-RLLQVQDPE-GNKEMTLSVQFL----
----LDMSRKESSSNLELKFRLK-----------------IPPC-IMQ-DL-KHIKEQM-
E-------------------------------------SLKLNRTWDLELSLRESKI-KN
LVNSIQMS---------------------D-------------------VSFKVEHSH-E
RK-SHSSK--S----FSVK-TSLN-NGQ-KEEP------------------KC----PLL
--------GA---Q---------------------------KE-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------ASH----------------GKE---------P
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSOGAP00000013522 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------EMA---SSFFS
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 328)
DFG---LIWYLEELN--K---KEFKKFKEHFKQ------
---------E--TLKLE--------L--KQI-SWTK-VKKSS-REDLANLLIQQYEAQQA
WNLTLRI----FH-RLGRQDLCDKVQ-RER------------TG--------------DC
H---------------------------------------------------HLLFLV--
--DG-----FCACPSQ----GQNVLWENI-----VLFLFQI-------------------
-------NLR-------------------------------VQSLDYTVFISKSKLRTC-
--------IQARNSRQCFSVFLFSHLSLIKEK--EKSKSITDFFEC--------------
---------------------------------F-------GENVRPEELKYMHLLFAAK
ET--GQR-S----H
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 929)
TTVLQGAQGVGKTTFLMKL-----VLGWA--E---------GSLFQ
D--------KFLY-VFYFCCRELRQVK-------ETSLAD-LMSR-NW-----PYPSPAP
ITEIVT-HP----EKLLFIIDSFEE-LKCDLSEP------ES-D-----LCGDWMKQ-QP
-VRALLSGLLRKKMLPEASLLVA-VTPVCPAELQARL-V-----CPE--LITLSGFSEGE
IELYF----------------R--SL-FPS---------RNRAAEAFRSVRE-NAQIYSL
CHAPRICWVIGTCMKQAVEKGR-EL-A--PI-CRCLTSLYASSILNLFTDKD-ASCPSP-
-----------QSRVRLRGLCSLA---VEGM-WT-DV----FEFHDE-------------
--DLRRNGIADL-DVPALLDT----NIFLKCT---E-SSYTFAHRSIQEFCAAMFYLLR-
--RDH-----------------P--NPAVRSRE-TLLAAHLKKVKKARWIHWGSFL----
----FGFLNEKEQEKVDKFFGFQ---------------P--SQE-IRQ-QFHQILKNLEE
CEG-----------------------------------LRGQTDFLALFYCLFEMED-AV
FVKEAVNF---------------------L-------------------QDINFSINNEL
DL---EVC--A----YCLK-YCSG-LRR-LSFS----------------VPK-----VIE
-EDGRGCR-----------RPWLTTGC----V-----VDM-KQHFSESMSFLFDHRSSCN
VVHWHHIF-CVFITNEHLKELEIINTDFCQS---------SLEILC--NQLRFP------
TC--H-LEKLQLNC---------VSF-SGGSSVFFEVL----------------SHSPDL
KYLN-LRNV-NLSCD-DMKILCKVLSSSMCN----IEQL--------VF-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------FCCCLA-----------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------CVICMDLL-HA-VLSKKSLCSLD
L-STSSLYNKGLK-VLCAALRHP--DCC----L---------------------------
-----------------------QSLGL--ANC-----SFTAAGCQDL------------
-------ASVL-TSNQKLTSLQIGLNKIGDVGVKV-LCDALT-HP-NC-HLEILG--LK-
--D-CGLTSACCR--------------------DLYSALTNSRTL-RVLDLTLNTLDHRG
VAVLCE--------ALKYTACP--LW--F-------------------------------
---LGLPKT-RVDEETQML------LT----------AVE-ERNPHLTVHRE-PGL----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMUSP00000046503 (view gene)
ENSOGAP00000013525 (view gene)
------------------------------------------------------------
--------------------------------------------IKRGGGLNLLTGGSLL
SNSTEHLIILPTGLIGCLLPKNP------------L-------------------FHQ-H
L---CFQRRIEMEEDDLSSFS
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 326)
SYG---LQWFLKELD--K---EELQAFKELLKE------
---------K--SSESA--------I--CSV-PQAE-VDSAD-VERLAFLLHEYYTGPQA
WAMSLNI----FE-EMNLPTLLKKAK-DAMKKHSL-AEIPEDAEQTKI--DQGPSQGGGS
EIS-QAEEQDD----A------------------------VPTE---TKEQEISQATK--
QD-GATAA---------ETKEQGEEKHTD-SFV---------------------------
-----ATLAG-------------------------------SQEEKPVHAGDKWTYKRH-
--------VMTK----------------FSTK--MSVY-HGSERS---------------
-----------------------------------AS-------EWPEV-QILASAFTPD
Q---GGFQP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVLHGKSGIGKSTLVRRI-----MLSWA--R---------GDLYQ
G--------TFSY-VFLLHAREIEWRR-------ESSFAE-LISR-EW-----PD-SQAP
VLEILS-QP----ERLLFVLDSFDD-MDSALKNG------DP-R-----LCEDWGTR-QP
-TFILIRSLLRKVLLPKSSLIIT-VRDVGLDKLKSVV-V-----SPR--YLLVGGISVEK
RLQLL----------------L--EH-IPD---------EHQKMQVL-C-LD-NCELLDQ
CQVLTVCSLIRKALELQKTVGG-SS-T--PV-CQTLTGLYATFMFHQLTPQE-MSKPCLS
----------REERAVLKSLCRMA---VEGV-WD-MK----SVFDSD-------------
--DLMVHGLRED-ELCALFHM----DILPRDSH-CE-ESYTFFHLSLQHFCAALYYVLE-
GLDRE---------------GIC--PLFIEKIK-NLMELKQTG-FNTHLLWMKRFL----
----FGLMSRDVLQSLERLLACP---------------I--PV-TVRQ-KLLHWVSLLGQ
QPG----------------------------------PT-APVDTLDSFHCLFETQD-EE
FVCLALNS---------------------F-------------------QEVWLPINQKM
DL---IVS--S----FCLQ-RCQH-LQK-IRVD----------------VKE-----IFS
-REQ--TM----WP--------------------GI-----------PH--WMQNK-VFI
NEQWKNFC-SVLCTHPNLQQLDLGSSILNEW---------AMKALC--AKLKH-P-----
TC--K-IQNLT--F-KNAQI---MS---GLQ-YLWMTL----------------ITNRNI
RHLN-LGST-YLKDE-DVKIACEALKHPNCL----LESL--------RLD---------H
C------G----------------------------------------------------
---------LT---HT-CYL---MISQILMST--SLKSLGLAGNKVTD-QGMKPLCDA--
LKA-PQCTLQKL-M----------------------------------------------
------------------------LENCSLS-----------------------------
----------------------------ALSCQVLAS-----------------------
--------------------------------ALVGNRSLTH---LC-LANNC-LGK---
-------EGVSLLSRSL----RLPGC----------------------------------
--------------------------NLR-RLI---------------------------
-------------------------------LNHCGLDITGCGFLAFA-LMGNARLTHLS
L-SLNPLEDDGMK-LLCEAMREP--SCQ----L---------------------------
-----------------------QDLEL--VRC-----QLTATCCTDL------------
-------SYVI-SRSKYLKSLDLAANTLGDSGVMV-LCEGLT-QK-KS-LLTRLG--LE-
--A-CGLTSDCCE--------------------VLSSALSSNQRL-TSINLVQNNFSPDG
MMKLCS--------AFACSMSN--LQ--I-------------------------------
---IGLWKW-QFPAQIQKL------LQ----------EVQ-LLKPHVIVDGNWYSFD---
--K---------------------------------------------------------
--------------------------------------------------EDRYWWKN--
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCGRP00000003281 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------MARA-NSPQEA---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWALKDLE--E---NSFKMLKFHLRD------
---------M--T------------Q--FHL-ARGE-LEGLS-RVDLASKLISMYGAPEA
VRVVSRS----LQ-AMNLMELVNYLS-QVC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LH--DYREIYREH-
--------VRC-----------------LE------ER---QDW----------GVNSSR
NELLMVA-TSSPGSPGSPPC------SD-------LQ-----Q-ELGPV--SVEALFTPE
AE--SYSTL----PI
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 930)
VVMQGSAGIGKTTLVKRL-----VQDWA--K---------GILYP
G--------QFDY-VFYVSCRDVVLLPK-------CDLPN-LICW-CC-GD-----NQAP
VTEILR-QP----ERLLFILDGYDE-LQK---------------------------S-SR
-AESVLHILIRRREVP-CSLIIT-TRPPALQSLEPML-G-----ERR--HVHVLGFSEQE
RENYF----------------N--SY-FMD---------KEQARNTLEFVQN-NDVLYKA
CQVPGICWVVCSWLERKMARGQ-EV-S--ET-PSNSTDIFTAYVSTFLPTDG-NEDSSGL
----------T-RHRVLRSLCSLA---AEGI-QH-QR----LLFEEN-------------
--ILRKHSLDGPSLT-AFLNS----IDYREGFS-IK-KLYSFRHISFQEFFYAMSFLVK-
EDQNQQ----------------G--EATRREVA-KLLEQ---E-KYEDMTLSLQFL----
----FDMLKIEGTLSLGLTFYFK-----------------IAPS-VRR-EL-NYFKERI-
Q-------------------------------------TIKCNRSWDLEFSLYDSKI-KK
LTQGIHMK---------------------D-------------------ITVNIQRSG-E
KK-PGKRK--T----VTVE-SSLG-----KGQV------------------QS----PLL
--------KT---NNS--T-------------------EKQKG-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------ASN----------------GKS---------R
-ETEE-PTG-KVR-NSR-LADT----ER-GNMEMKG---QEGGGV---E-RQED-GKG-Q
R-VTK-DGEMR---DQ-MNG---Y-QRE--------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCJAP00000031587 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------KLNPYRKQ-
--------IKEK----------------FQLI--WEKETCLQVPEH--------------
---------------------------------F-------YKESMKHECTVLNKAYAAA
------AAP----V
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVVEGPDGIGKTTLLRKV-----MLDWA--E---------GNLWK
D--------RFTF-VFFLTVYEMNSVT-------ETSLPE-LLSR-DW-----PES-SEK
MEDILS-QP----ERILLIMDGFEQ-LRLDLD-Q------EA-D-----LCGDWRQR-LP
-LPVILSSLLQRKMLPECSLLLA-LGQVGMRKNYFLL-Q-----RPK--LIKLSGFTEPE
RKMYF----------------S--CF-FGE---------KSKASKVFNFVRE-IGPLFIL
CHNPFICWLVCTCMKYRLERGE-DL-E--IN-SQNTTSLHASFITNVFKAGS-QSSPPK-
-----------LNRARLKNLSALA---AEGL-WT-HT----FVFCRE-------------
--DLRRNGLSES-EGWMWVGM----NLLRRSG-----DRFTFSHVCIQEFCAAMFYLLR-
GPKDS-----------------P--NPAVGSVT-QLVRASVAR-PQNVLTQMGVFV----
----FGVSSEEIAGMLEASFGFP---------------L--SKD-LQQ-EIAQCLRGFSQ
RE-----------------------------------ADREAIGFQELFSCLFETQE-EA
FVTRVMSF---------------------F-------------------EEVYVYIANIE
HL---VIA--S----FCVK-RSHN-LTT-LRLC----------------MEN-----IFP
-DDSGCAS-----------E-------------------H-N-------EK---------
LMYWRELC-SMFSNNKNFQVLDVENTSFDDA---------SLAVLC--KALARP------
VC--K-LQKLMLTF---------MSN-FGQDSGLFRAV----------------LHNPHL
KMLS-LYGT-SLCHS-EVKYLCETLKHPVCK----IQEL--------IL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------GKCDLS-----------------------------
----------------------------HEACEDIAS-----------------------
--------------------------------VLACNTKLKL---LS-LVENP-LRD---
-------EGMLFLCEGL----KHPNCALEKL-----------------------------
------------------------------------------------------------
------------------------------MLMYCCLTCVSCEAISEG-LARSKSLSLLD
L-GSNHLEDSAVA-LLCEALKHP--DCS----I---------------------------
-----------------------RELWL--MAC-----FLTSDSCKDI------------
-------ADTL-ICNEKLKILKLGHNEIGDAGVRQ-LCEALK-HP-NC-KLECLG--LQ-
--T-CRITHACCE--------------------DLAAALIASKTL-RSLNLDWITLDPEG
VVLLCE--------ALSHPDCA--LQ--M-------------------------------
---LGLQKS-AFDEESQKM------LT----------ALE-EKCPHLSIAHEPWVDE---
--EYR-----IR------------------------------------------------
--------------------------------------------------GVIP------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPPAP00000037285 (view gene)
ENSCANP00000021141 (view gene)
ENSNLEP00000011124 (view gene)
ENSETEP00000016431 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---ASFFSDFG---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 344)
LMWYLGELK--K---EEFKRFKLLLRK------
---------E--AVDLG--------L--EPI-PWTE-VRKAT-REDLANLLIKCYKEEHA
WEVTFRV----FH-QMDRKDLCEKAK-RES------------SG--------------YT
K---------------------------------------------------MYKD----
------------------------------------------------------------
----------------------------------------------------------H-
--------IKKN----------------LITR--WSQGLLTKIYEH--------------
---------------------------------F-------E----PKERDYLELLFAPT
AT--GER-P----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
QTVVILGSPKFGKTTFLLKL-----LMAWA--V---------DAVYR
D--------KFLY-VFYFCCREVKGLA-------ATSLAE-LISR-DW-----PNS-SVP
IAEILA-QP----ERLLFIVDSFEQ-MPRTLSEL------ES-D-----LCSDWAQREQP
-GEVVLGSLMKKKMLPESSVLIA-TTSARQEDLQAQL-E-----RPE--IIKLMGFDEGD
RKLFF----------------A--CF-FQD---------RKRSTEAFGLIQE-NKQLFSM
CKIPVLCWVVCTCLKQEMERGR-DP-A--LA-CHSVTSVYTSFLLNSFAPKG-ALAASA-
-----------Q-PCQMKSLCSLA---VDGM-WI-NA----AEFTDK-------------
--DLRRNGLVES-DVSALLDL----KLLRKCRG-CE-GLYTFIHFCVQEFCAALFYMIA-
DPRDH-----------------P--NPAVGSTE-ALVASR----ERRPWVFLGCFL----
----FGLLHAEERQKVEAFFGYQ---------------L--SQS-VQQ-QIYG-LKGFTE
H-E----------------------------------LLEAEVDFLALFYSLFEMQD-DA
FVRKAMGC---------------------F-------------------D-VQICITDTM
DL---MVS--A----YCXX-XXXX-XXX-XXXX----------------XXN-----VFD
-EKN-GSQ-----------F-----------------------------------TASCN
LVHWLCIC-SVLTTNKNLRTLQLMESTFDES---------ACVALC--KQLRDP------
KC--Q-VQNLXXXX---------XXX-XXXXXXXXXXX----------------XXXXXX
XXXX-XXXX-XXXXX-XXXXXXXXXXXXXXX----XXXX--------XL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------VNCNLS-----------------------------
----------------------------SVDGEIFAS-----------------------
--------------------------------VLKTNKKLKY---LN-LSYND-LD----
-------QSLSVLFKAL----CLPDCRLEAL-----------------------------
------------------------------------------------------------
------------------------------AVAGCNIREPFWENLCEV-LLCSKKLVHLD
V-SANIVKDHDLK-QLCEAMRHP--NCI----L---------------------------
-----------------------ESLC-----------------CQNI------------
-------TTVL-ISKPNMITLGIGENDIQDEGMKV-LCEAVA-HP-NC-RLQNLG--XX-
--X-XXLTSACCE--------------------DLASALAHNPAL-WRVNMVGNALDHNG
LEVLFE--------ALRHSQCP--LK--V-------------------------------
---L--------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSANAP00000015859 (view gene)
ENSECAP00000009788 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------MTDSLSSSFFP
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 326)
DFG---LLLYLEELN--K---EELNKFKLLLRN------
---------A--TMEPE--------H--CQI-PWAE-VKKAK-REDLANLMNRYYPGEQA
WDVALKI----FG-KMNLKDLCERAK-AEINWTTQ-SMGPEDARA-GE------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------AQGDQEAVLGDGTEYRSR-
--------IKEK----------------FCIM--WDKECLLGEPKDFCHEIA--------
----------------------------------------------QKGQELLEHLFDVD
VK--TGEQP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
QTVVLQGAAGVGKTTLVRKA-----MLDWA--Q---------GNLYQ
Q--------RFAY-VFYLNAREINQLR-------ERSFVQ-MISK-DW-----PS-TEGP
IERIMS-QP----SSLLFIIDSFDE-LNFAFEEP------EF-A-----LCEDWTQV-HP
-VSFLMSSLLRKVMLPESSLLVT-TRLTACKKLEPLL-K----S-QR--SVELLGMSEDA
REEYI----------------Y--QF-FED---------KKWALQVFGSLRN-SEMLFSL
CKVPLVCRVICTCLEQQMEKG---V-T--LT-CKTTTALFTCYISSLFTPA-DGSCPSLP
------------NQTQLRSLCHLA---AKGV-WT-MT----YVFYRE-------------
--NLRKHGLTKC-DVTIFLDV----NILQKDTE-YE-NCYVFTHLHIQEFFAAMFYILK-
DSMET--------------RNLF--FQSFEDVK-LLFESNSYK--DPHLMQMKCFL----
----FGLLNEGLVKQLEDTFNCK---------------M--SLG-VKW-KILQWMEILGN
SD-----------------------------------CFPSQLGFLELFHYLYETQD-EG
FISQAMRY---------------------F-------------------RKVVINIHGKI
HL---LVS--S----FCLK-HCQS-LQT-IKLS----------------VTV-----ESE
--R--KML----NSSH-----SA--E------------------------TWD---FGRI
THYWQDLC-SVLHTNEQLRELDLCHSNLDKL---------AMKTFY--QELRHP------
NC--K-LQKLRLRF---------LSF-PDGCQDISRSL----------------THNQNL
MQLN-LKGS-DIGDN-GVKSLCEALKRPECK----LQNL--------TLE---------S
CD----------------------------------------------------------
---------LT---TV-CCL---DISKALVR-SQSLIFLNLSTNNLLD-DGVKLLCEA--
LRH-PKCYLERL-S----------------------------------------------
------------------------LESCGLT-----------------------------
----------------------------VAGCEDLSS-----------------------
--------------------------------ALISNKRLTH---LC-LADNN-LGD---
-------GGVKLMSDAL----KHPQC----------------------------------
--------------------------TLQ-SLV---------------------------
-------------------------------LRRCHFTALSSEHLSSS-LLHNKSLTHLD
L-GSNQLQDDGVK-LLCDVFRHP--SCN----L---------------------------
-----------------------QDLEL--MGC-----VLTSACCLDL------------
-------SSAI-LNNPNLRSLYLGNNDLQDDGVKI-LFEALR-HP-NC-NIQRLR--LE-
--Y-CGLTSLCCQ--------------------DLSSTLSSNQRL-IKINLTQNTLGCEG
IMKLCE--------VLKSPECK--LQ--V-------------------------------
---LGLCKE-ALDEEAQKL------LE----------AVG-VSNPHLVIKHDCNDHN---
--E---------------------------------------------------------
--------------------------------------------------EDDSWWRCF-
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSEEUP00000001468 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------------SKEM
HRVLM------------KEK----LM-HKWEKHYSGYTENEFYKDLREEILKILDFIYSS
TR--RCSKD----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
SNLFVLGEQGVGKTLVITWA-----VLEWA--K---------GSLWK
D--------MFSY-IVFITSHEINQMGY------I-SLVD-LISR-DW-PN-----GQAP
VADILS-DS----KELLFIVEDLD-NVHGQNI---------N-S-----SNLCVDSR-QS
-VSFLLASLLRRKMTPDASFLIS-LRSESE---PIVV-Q-----ALG--HSMMMGLSSKK
REKY-F---------------T--DF-FEG---------GEKAEIAYQFVQD-NEVIMSL
CLSPALCWVTGMALQRQMDQRV-DV-K--HS-LQTPTDIYAHFLTNSLTSEAGNS----A
----------GHSH---LGLLGRL---XXXX-XX-XX----XXXXXX-------------
--XXXXXXXXXXXXX-XXXXX----XXXXXXX---X-XXXXX------XXXXXXXXXXX-
XXXXX-----------------X--XXXXXXXX---XXXXXXX-XXXXXXXXXXXX----
-------XXXXXXXXXXXXXXXX----------------------XXX-XXXXXXXXXXX
X-----------------------------------------XXXXXXX--XXXX-----
-----XXX---------------------X-------------------XXXXXXX----
-------X--X----XXXX-X-XX-X---------------------------------X
------XXXXXXXX------XXXXXXX--------------------XXX-XXXXX-XXX
XXX---------------------------------------------------------
------------------------------------------------------------
-------------------------X----------------------------------
--------------------XX----XXXXXXXXXX--------XXXXXXXXXXXXXX-X
XXXXXXXKVKS---LI-YWR---DLCSLLHT-KETLQELEISNAYFDD-TSERILCKA--
LQN-PSCRIQTF-K----------------------------------------------
------------------------LAYLPTS-----------------------------
----------------------------FKFESIFNTLAXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXLTKCELSLTVYDGIALLINSKKLKK---LS-LISIQ-MNN---
-------EEVKILSKAL----DSPDCPLESLTXX--XXXXXXXX-XXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXX-XXX---------------------------
-------------------------------LCGCMITSDITHNIASM-AITSPNLKSME
L-GSNHFGNRGLA-NLCEALEDD--SCK----L---------------------------
-----------------------ENLGR--EEC-----NFTSAASRVP------------
-------ILAL-MRNKTLKRLNLIGKQLGD------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCANP00000033003 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------MAESNSTDFD---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWYLENLS--D---KEFQSFKKYLAR------
---------K--ILDFK--------L--PQV-PLIN-LRA-T-KEELANLLPISYEGQYI
WNMLYSI----FL-MMRKEEHCMKI---IGRRR---------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------RNQEACKAL-
--------MRRK----------------VTLR--WENHTFGEFHYKFFCNVA--------
----------------------------------------------SDVFYVLQLAYDST
SC--YSAND----LN
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 930)
VFLIGDRASGKTMTINLA-----VMKWI--S---------GEMWQ
N--------MISY-VVHLTSHEINQMT-------NSSLAE-LIAK-DW-----PD-GQAP
IADILS-DP----KKLLFILEDVDN-LRFELNVN------EH-A-----LCSNSTQK-VP
-IPILLVSLLKRQMAPGCWFLIS-SRPTREENIKMFL-K-----TTD--CYVTLQFSNEK
RELYF----------------H--SF-FND---------HQRASTALMLVHD-NEILTSL
CRVANLCWITCTVLNRQMDKG-HDP-Q--HC-CETPTDLHAHFLADALTSEARLT-AN--
----------QYHLGLLKRLCLLA---AGGL-FL-ST----LNFSGE-------------
--DLRCVGFTEA-DVSVLQAV----NILLPSST-HK-DRYKFIHLNVQEFCAAIAFLMAV
PNCLI-----------------P----SGSR-E--Y--KEKRE-QYCDFNRVFTFI----
----FGLLNENRRKILETTFGCQ---------------LPMVES-FRR-YSVEYMKHLGR
DQ-------------------------------------EKLTHHGSLFYCLFENQE-EE
FVKKVVNP---------------------L-------------------KEVTVYLQSNK
DM---MVS--L----YCLE-YCCH-LRT-FKLS----------------VQR-----IFE
--YKEPLV-------------------------------------------RPTASQMKS
LVYWREIC-SLFCTMDTLWELHLFDSDLNDI---------SERILS--KALEHS------
SC--K-LRTLKLSY---------IST-ASGFEELLKAL----------------ARNQSV
IYLS-LNCT---------------------------------------------------
------------------------------------------------------------
--------------------------------------------SISL-NMFSLLREI--
LDK-PTCQISHL------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------RLVFCCLTEKCCSSLGRV-LLLSPTLKQLD
L-CVNHLQNYGVL-HVTFPLIFP--TCQ----L---------------------------
-----------------------EELYL--SGC-----FFTNDICQYI------------
-------AIVIA-T
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NETLRSLEIGSNSIEDAGMQL-LCGGLR-HP-DC-MLVNIG--LE-
--E-CMLTGACCE--------------------SLASVLT
PF13516 // ENSGT00900000140813_0_Domain_17 (2441, 2464)
TNKTL-ERLNILQNQLGDEG
IVKLLE--------SLILPDCV--LQ--I-------------------------------
---IGLPLN-NVSAETLRM------VM----------TVK-ERKPDLVFLSETWSVKEGR
EIGVI-------------------------------------------------------
-------------------------------------------------------PASQP
GSV-----------------IPNSNLD---YM--FFKFPRMSTAMRMSNTASRQRL----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSETEP00000010989 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------MALP-HDLQKA---LLFALNDLE--E---ESFKTLKFYLQT------
---------M--TLPDG--------S--GQL-GRGE-LKGLA-NS-------LDSGAHEV
VKVVLES----LN---KSLELVGQLS-FVR------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LD--DYRDRYREH-
--------VCC-----------------LE------ET---QKG----------GLNGSD
SQLLLMA-NPKSGSSEVF---------------------------LEEI--TVKDLFGSG
GK--S--PK----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
STVVLQGNAGTGKTTLARKM-----VQDWA--P---------GTS-A
G--------QFDS-VFYVSCRAVTLLPK-------CELTL-FLWW-CG--D---------
--------------ALLLAVDGFDE-LPTSFTGR------SM-R-----PT-----W-SP
-LENLLHRLIRREIP--RSLLIM-TRPQAVGNLRCSL-K-----ESY--RVHVLGFSEGE
RRRYF----------------S--SY-LTD---------EERARNT-DFVQR-SDIFNKL
CQVPG-CWMVCSWLKGLAERGR-DV-S--DI-PSNGTDIYT-IVSTVLPPND-NEDCLEL
----------SRRHRVLRGLCTLA---AEGT-QPQQR----ILFEEA-------------
--DLRKHNVDGLN-H-AFLRT----LDYQEGHV-IK-KFDCFRHIGFQEFF-AMIHLVK-
EDHGQL----------------G--KAAER--E-RLLE----T-KNEPMALSTQFL----
----LAVLKPEG-PDLEVKRCLK-----------------ISSR-TVQ-DV-RHFQKQL-
K-------------------------------------SIKPSSIWELESS--------N
LAKGVRMS---------------------G-------------------VSVDLRDNG--
-R-SHDSL--S----FSS----LS-REQ-EKEQ------------------N--------
----GQEESE---HHT--R-------------------GTQTG-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------PSE----------------GKE---------S
-GRG--------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSOGAP00000017957 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------FTMFLP-HDPQ
PF02758 // ENSGT00900000140813_0_Domain_0 (203, 325)
EA---LLWALHDLE--E---NDFKTLKFHLRG------
---------I--TVAEG--------H--LYL-ARGQ-LEGLS-RVDLASKLISVYGAQEA
VRVVLKG----LK-VMNLLELVDQLS-HIC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LNGSDYRETYREH-
--------VRK-----------------VE------ER---KEG----------GVDSSY
RQLLLVA-KPSLGSPESPAC------PI-------LE-----E-ELDSV--TVETLFDSG
GK--PYPDP----S
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
FVVLQGSAGTGKTTLAKKI-----LLDWA--S---------GTLYP
G--------RFDY-VFYVSCKEMLLLPK-------GQLDQ-LLYW-CC-ED-----NQAP
VTEIVR-QP----ERLLFILDGFDE-LQRPFAAQ------LK-K-----RS-----A-GS
-KEDVLHILIRRRKLPTCSLLIT-TRSLALQNLEPLP-E-----NAH--HVHILGFSEEE
RRGYF----------------S--SY-FME---------EEQAKSAFAFVQG-NDILYKA
CQVPGLCWVVCSWLKRQMERGQ-KV-S--ET-PRNSTDIFMAYVSTFLPSDD--EACSEL
----------S-RHRVLRSLCALA---AEGI-QH-QR----FLFEEA-------------
--ELRKHNLDGSSLA-AFLSS----SDYQEGLE-VK-KFYSFHHISFQEFFHAMSYLVK-
EDQSQL----------------G--KESRREVQ-RLLQVNELE-ESKEMTLSMQFL----
----LDIAKKGSFSSFGLNLSLE-----------------ISPC-LMQ-DL-RHFKEQM-
E-------------------------------------SWKCKRMWDLEFSLHDAKI-KN
LVRSIQVS---------------------N-------------------VSFNMQRLK-E
RS-SQSGT--S----FSVK-SSLN-NGQ-KEES------------------KC----PLV
--------GK---GDG--A-------------------ETQNE-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------ASN----------------GKG---------R
-GAEE-PDEQIKN-SRM-------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSHGLP00100018962 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MSLDIRCEQ-LSD-ARWT----------------EL---------
----LPLI-------------------------------QQHQVV------RLEDC-GLT
EGRCKDIG-SALRANSALAELSLRTNALGDA---------GVCLVL--QGLQS-P-----
TC--K-VQSLS--L-QNCGLT--ET---GCR-VLASVL----------------RCT
PF13516 // ENSGT00900000140813_0_Domain_6 (1618, 1642)
PTL
RELH-LSDN-PLGDP-GLRLLSEGLLDPQSC----LEKL--------ELE---------S
C------N----------------------------------------------------
---------LS---AA-SCE---PLAAMLRA-KPNFKELVVSNNCIQE-AGIRMLCGG--
LKE-STCLLETL-R----------------------------------------------
------------------------LDNCGVT-----------------------------
----------------------------SANCKDLCD-----------------------
--------------------------------VVAA
PF13516 // ENSGT00900000140813_0_Domain_11 (2017, 2054)
KPSLQE---LD-LGGNR-LGD---
-------AGIAALCPGL----LHTSC----------------------------------
--------------------------RLR-KLW---------------------------
-------------------------------LWECDITAEGCRDLGQV-LRA
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
KQSLKALS
L-MLNELGDEGAR-LLCEALQEP--SCQ----L---------------------------
-----------------------ECLWV--RVC-----SFTATCCPHF------------
-------RALL-A
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
QNKFLTELQLGENRLGDAGVQE-LCQGLS-QP-GS-VLRVLG--IG-
--D-CDLSDNCCS--------------------NLASVLLA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2464)
SCSL-RELDLSNNGLGDPG
VLRLVE--------SLRQPGCV--LE--C-------------------------------
---LLLFDV-YLSDGVNDQ------LQ----------ALE-EEKPTLRIIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSTBEP00000003854 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---ESFF
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
SDFG---LLWYLEELR--K---EEFWKFKELLKQ------
---------E--PLKFE--------L--RPM-PWTE-LKKAS-KGDLAQLLEKHYPAEQA
WAVTLSL----FL-QLGRRDLWTKAQ-DEIRN----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------RLNPYRKH-
--------VKEK----------------FRLV--WEKEIRLQVPEH--------------
---------------------------------F-------YRETTRSAREALADVFA--
------AGP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
ATVVLRGPEGSGKTTLLRKV-----MLEWA--E---------GNFWK
D--------RFTF-VFFLNVCEMSCRA-------ETSLVE-FISK-DW-----PEA-PAA
MEDTFS-QP----ERILFVMDSFEA-LKWDLA-L------KG-H-----SCSDWRQR-RP
-AQVILGSLLQKQLLAESCLLLA-LGKGGVQKYGLLL-R-----DAQ--HIVLSGFSERE
RKSYF----------------S--HF-FCE---------KNKALKAFGFVRD-TRPLLTL
CQNPLVCWLVCTCIKCQLETGP-DL-S--IS-SQNMSALYVSFLTSVLKAGA-ESRPPR-
-----------QSRARLTSLCALA---AEGM-WT-QT----FVFGRE-------------
--ELRRKGMSDS-DVSLWVAA----RLLQRSG-----EHFIFKHVCIQEFCAAVFYLLQ-
RPRDS-----------------P--HLAIGSVT-QLVTAWVAN-VQTHLSRLGVFV----
----FGISTEEVIGQLETSFGFL---------------L--SRD-IKQ-EITACVQSLSR
CK-----------------------------------TDQEVVSFNELFNGLFETQD-EE
FVAHVMNF---------------------F-------------------EEVSTYIGDVG
QL---LVS--S----YCLK-LCQN-LKK-LHLC----------------IDD-----VFS
-DDSGSVA-----------D-------------------D-S-------EK---------
LLYWQDLC-SVFRTCKGLEMLDLDNCSLDGA---------STAVLR--EALAQP------
DC--K-LQSLLXXX---------XXX-XXXXXXXXXXX----------------XXXXXX
XXXX-XXXX-XX-XX-XXXX---------------------------XX-----------
------------------------------------------------------------
--------------------------------------------------------XX--
XXX--XXXXXXXXX----------------------------------------------
------------------------LGKCDIT-----------------------------
----------------------------EDGCDDLAP-----------------------
--------------------------------VLACSDRLKH---LS-LAENP-VKD---
-------QGVTVLCRAL----R-PGCALQSLXXX--XXXXXXXXXXXXXX----------
------------------------------------------------------------
-----------------------------------------------X-XXXXXXXXXXX
X-XXXXXXXXXXX-XXXXXXKHP--ACS----L---------------------------
-----------------------QELWL--AGC-----FLGPDCCKDI------------
-------SAVL-THNGKLKTLKLGNNRVGDAGARQ-LCAALR-HP-GC-ALQTLG--XXX
XXX-XXXXXXXXX--------------------XXXXXXXX---X-XXXXXXXXXXXXXX
XXXXXX--------XXXXXXXX--XX--X-------------------------------
---XXLDKY-AFDEEIQML------LT----------SVE-EKNPHLTISHQPWIEE---
--EYR-----IK------------------------------------------------
--------------------------------------------------GMLV------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSSSCP00000038303 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------MLPSA----QLHFH---LQPLLEQLD--Q---QELSKFKSLPKA------
---------L--SLQEE--------L--QHV-PQMA-LDKAS-GRNAHQTLSQLLSGD--
--------------------------------------------GR---H----------
-------------P-AK--------------------------N---KNLYF--------
--AELT---------------------------------KIQEEDVAHP-----------
-------GEA-------------------------------KE-------GLEGEKNAL-
---------KKK----------------SWQI--WQNS---RPGASEHVHIF--------
----------------------------------------------TRRYEALVPFCNPQ
VP--TGPFP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
HTVVLHGPAGVGKTTVAKKL-----MLDWT--Q---------DGLAQ
---------AVHS-AFYLSCKDLSHRG-------TCTFAE-LIAA-AQ-----PG-AREA
VPQLPA-RA----QEILLVVDGFEE-LRVPSG------ALVR-D-----LCGDWKQP-KP
-VAVLLGSLLRRKMLPRATLVIT-TRPGALRELRLLA-E-----QPL--FVEIEGFLEPD
KKAYF----------------L--EH-FGE---------ERQALRAFALMKG-NAALFRL
GSAPAVCRIVCTCLKAQMEGGE-DP-A--ST-CQTTTSLFLRFLCSQFAPA---------
---------------ALRAACLLA---AEGA-WT-QT----SVFDGD-------------
--DLGRLGLTPS-ALCPFLDK----QILQKNGD-CG-GCYSFVHLSVQQLLAAM------
----EAAARG--------EAGGC--TWDIGDVQ-KLLSKEERR-KNPDLTRVGSFL----
----FGLLNEERARELETTFGCR---------------V--SGA-VRR-ELLKCRSESDG
NG-----------------------------------PFSSMMDTKEMLHCLHESQE-EQ
LVKDAMAH---------------------V-------------------PEIAVHLTSAF
EM---VEA--S----FCLK-HCQD-LWK-ISLQ----------------VGKR----IFL
-END-SMQ----KS--------------------S----------------TQAER-L--
-------------RTHFLLYELSHRPFLSFS---------AVRILC--EKVAS-A-----
TS--S-LQKVI--L-KNMSPV--DA----YW-NLCIAF----------------GGQQTL
THLT-LQGN-DQDD--MLPPLCEVLRHPKCN----LQYL--------RLA---------S
C------A----------------------------------------------------
---------AT---RR-QWA---DLSCCLHI-NQSLTRLNLTANEVLD-EVAKRRYVT--
LGH-PKCCLQRLSS----------------------------------------------
------------------------LENCQLT-----------------------------
----------------------------EASCKELSS-----------------------
--------------------------------ALTVK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
QRLTH---LC-LAKNA-LGD---
-------RGVKLLCEDL----SYPEC----------------------------------
--------------------------QLQ-TLV---------------------------
-------------------------------LWCFNITGDGCSHLSTL-VQ
PF13516 // ENSGT00900000140813_0_Domain_13 (2212, 2237)
QNSNLTHLD
L-GLNHIGIVGLK-FLCEAHFSH--THT----H---------------------------
-----------------------TNL-------------IPKFTVNCR------------
-------RHQI-VKS------TLTK-NNKAGGIML-PDFKLL-QN-SS-NQNSMA--LA-
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMUSP00000098707 (view gene)
ENSCGRP00000001860 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------HLGDLQTYKAH-
--------MITK----------------LSTS--LEIC-Y--------------------
--------------------------------------------DHPEM-RVLSEAFKPD
Q---NTFRP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
HTVVLHGKPGIGKSALARSV-----ILGWA--Q---------DKLYQ
D--------I-SC-DFFFSPRKMQWAE-------KSSLAQ-LISR-EW-----AD-SQAP
MTEIMS-QP----ERLLFVVDDFDD-LDSALPQE------DT-R-----LCEDWKDE-QP
-IHTLLYSLLKKTLLPQSFLLVT-TRDTGLQNLKSMV-V-----SPL--YILVEGLSATK
RTQLV----------------L--EN-VSD---------DHRRKQVIRSMLD-NQWLAEQ
CRVPSICSLVCKALKIQEKLGG-SC-T--PV-CQTLTSLYSTLVFHQLTPRE-PFRGCLS
----------PEERGTLMGLCRMA---AEGV-WS-KR----SVFYDN-------------
--DLKTYGLKES-EISALFGM----DVF-QASH-SAKRCYAFFHLSLQDFFAALHYVLE-
GLG-T--------------WNQS--CFVIKNLR-SLSELKGTH-FNAHLGGMKRFL----
----FGLVNKDTMRTLEVLLGYP---------------L--SP-VVNQ-VLQHWVFMLGQ
QAN----------------------------------DTS-PMDVLDAFRHVFESQD-KD
FVCSALKS---------------------F-------------------REVEIQVNQRT
DL---VVS--S----YCLQ-HCKN-LEA-LRVD----------------VSD-----IFS
-VEMNTEL----CP--------------------V-----------VPQ--QTQGK-PLI
MECWEDFC-SILGTHPKLKQLDLGSSILSEW---------AMKIMC--LKLKN-Q-----
AC--K-VQSLT--F-KNAEV---VS---GLR-HLWMTL----------------VSNRNL
KYLH-LGNT-LMEEN-VIKLACQALQHPKCS----LETL--------RLD---------S
C------A----------------------------------------------------
---------LT---PD-CYL---MLSNLLLS-TTSLRCLSLAGNKVTE-KSMRWLCDA--
LNS-SKCALQKL-I----------------------------------------------
------------------------LDSCELM-----------------------------
----------------------------PVSCHILAS-----------------------
--------------------------------ALCGNRTLTH---LC-LSDNS-LRT---
-------EEVRRLCRFM----SNPEC----------------------------------
--------------------------ALQ-RLI---------------------------
-------------------------------LKQCNLRGDAXXXXXXM-LSH
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
NGKLTHLN
L-TMNPVEDGGMK-LLCDAIKKP--TCH----L---------------------------
-----------------------QELEL--VSC-----QLTRDCCADL------------
-------ASVI-TR
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NQHLKSLDLSSNALGDSGVIA-LCKALK-QN-NS-SLRRLG--LE-
--A-CGLTSDCCT--------------------ELSLALSC
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2465)
NSRL-TSLNLMKNDFSTPG
MLKLCS--------AFLHPTSN--LW--I-------------------------------
---I--------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSSSCP00000003558 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------MEETSSLC-
----------------------------NHTF--WPSD------GGFHYNNI--------
----------------------------------------------AHRSQKLITYLNPT
IP--TQPMP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVLHGPAGVGKTTVAKKL-----MLDWT--Q---------DGLAQ
---------AVHS-AFYLSCKDLSHRG-------TCTFAE-LIAA-AQ-----PG-AREA
VPQLPA-RA----QEILLVVDGFEE-LRVPSG------ALVR-D-----LCGDWKQP-KP
-VAVLLGSLLRRKMLPRATLVIT-TRPGALRELRLLA-E-----QPL--FVEIEGFLEPD
KKAYF----------------L--EH-FGE---------ERQALRAFALMKG-NAALFRL
GSAPAVCRIVCTCLKAQMEGGE-DP-A--ST-CQTTTSLFLRFLCSQFAPA-PGACPHPR
------------LQAALRAACLLA---AEGA-WT-QT----SVFDGD-------------
--DLGRLGLTPS-ALCPFLDK----QILQKNGD-CG-GCYSFVHLSVQQLLAAMVYVLE-
TEEEEAAAGG--------EAGGC--TWDIGDVQ-KLLSKEERR-KNPDLTRVGSFL----
----FGLLNEERARELETTFGCR---------------V--SGA-VRR-ELLKCRSESDG
NG-----------------------------------PFSSMMDTKEMLHCLHESQE-EQ
LVKDAMAH---------------------V-------------------PEIAVHLTSAF
EM---VEA--S----FCLK-HCQD-LRK-ISLQ----------------VGKR----IFL
-END-ADA----EV--------------------D----------------RSQHD-QHS
LQVWADLC-SVFSSNENLLFLDVRDSVLSRS---------SVRILC--EQVTH-A-----
TC--H-LQKVV--I-KNISPA--DA----YR-DFCLAF----------------IGKKTL
THLV-LEGN-VHSDKMLLLLLCEILKHARCN----LQYL--------RLG---------S
C------S----------------------------------------------------
---------DT---TQ-RWA---DFSSALKI-NQSLKCLDLTASEFLD-EGVKLLCAT--
LRH-PKCSLQKL-S----------------------------------------------
------------------------LEDCQLT-----------------------------
----------------------------EACCKELSS-----------------------
--------------------------------ALIV
PF13516 // ENSGT00900000140813_0_Domain_11 (2017, 2054)
NQRLTH---LC-LAKNN-LGD---
-------HGVKLLCEGL----SYPEC----------------------------------
--------------------------QLQ-TLV---------------------------
-------------------------------LRHCSINRHGCKYISKL-LQG
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2228)
DCSLTSLD
V-GFNPIT-TGLY-FLCEALKKP--NC---------------------------------
-------------------------------------------KLKCL------------
-------GQGP-VPD-----GGLGDTGEEEQRITL-PRIQA----------ESVT--LQ-
--L---------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPCOP00000029322 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-MDEPEASCPSTGPS--AAVAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLAALEDLS--Q---EQLKRFRHKLRD------
------------APADG-----------RSI-PWGR-LEHAD-AVDLAEQLTHFYGPEPA
LDVARKT----LK-RADVRDVAARLK-EQRLQR---------L---------GRS-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------SS-ALLSVSEYRKEYREH-
--------VLRQ----------------HAKV--KERNAHS------------VKISKRF
TKLLIAPESGALEEE--ALGLAE-------APEP--------RRARRSDTHTFNRLYRRD
K---EGRRP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYQ
G--------QVDF-AFFVPCGELLERPG------ARSLAD-LILD-QC-----PD-RGAP
VRQMLR-QP----QRLLFILDGADE-LPALDAP--------EVA-----PCTDPFEA-AS
-GARVLGGLLNKALLPTARLLVT-ARASAPGRLQGRL-C-----SPQ--CAEVRGFSDKD
KKKYF----------------Y--KF-FQD---------EWRAERAYRFVKE-NETLFAL
CFVPFVCWIVCTVLRQQLELGR-DL-S--RT-SKTTTSMYLLFIASVLSSAPAADRP---
-----------RVQGELRKLCRLA---REGVLGH--R----ARFAEK-------------
--DLERLELRGSKVQTLFLNKKELPGVLET-----E-VTYQFTDQSFQEFFAALSYLLE-
DEGSP-------------------G-AEAGGIG-TLLRGDAQL--RHHLALTTRFL----
----FGLLSEERMRDVERHFGC-----------------AVSER-VKQ-HALRWVQGQGC
PR-----VAPEGTEVLEDTEEPEE-E---------EEEEEELNYPLELLYCLYESQE-DA
FVRQALGS---------------------F-------------------RELALERVCFS
RM-DVAVL--S----YCVR-CCSA-GQA-LRLV------------------SC----RLA
------AA----------------------------------------QEKK-K---KKS
----------------------------------------LVKR----------------
------LQGR---L--------------GSS------S----------------SSRATT
KQ---FPPS-------PLCPLFEAMTDQQCR----LSSL--------TLA---------R
C-----------------------------------------------------------
--------RLP---DT-VCR---DLSEALRA-APALTELSLLHSRLS-EVGLRLLSEG--
-LAWPQCRVQTL------------------------------------------------
----------------------------RLQ-----------------------------
---------------LPGP---------QGALQYL-------------------------
------------------------------IAMLRQSPTLTT---LD-LSGC-QLPSSML
--T--------YLCAVL----QHPACSLQTLSL--TSVELSEG-----------------
----------SVQELRAVKTAKP-------DLV----------------------I--LH
PALD-------------------SHPEPPTGLSRAL------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSANAP00000031681 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MTSA----QLECT---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LQALLEQLS--E---GELGTFKSILWA------
---------F--PLEDV--------L--QKT-PWSE-VEEAS-GKKLAEILVNTSSECWI
RNATVKV----FE-ELNLPELSKMAK--AEL-----------------------------
------------------------------------------------------------
--LEDG---------------------------------RVEDADSREL-----------
-------GDS-------------------------------EEDLELAKPGEKEGWRNV-
---------MDE----------------QSLV--WRNSFWRGDSDNFHDHVT--------
-----------------------------------------------QRNQRFIPFLNLR
RP--TQPSP----Y
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVLHGPAGVGKTTLAKKC-----MLDWK--D---------YNLSP
---------KARY-AFYLSCKELSRLG-------PCSFAE-LITK-DR-----PE-LQDD
IPNILA-QA----QRILFVVDGLDE-LRVPPG------ALIY-D-----ICGDWKKP-KP
-VPVLLGSLLKRKMLPKAALLVT-TRPRSLKDLQLLA-E-----KPI--YIRVEGFLEED
RRAYF----------------L--TH-FGE---------EDQAMRAFELMRS-NAALFQL
GSAPAVCWMVCTSLKLQMENGE-DP-A--ST-CHTRTGLFLHFLCSRFPKG-------AQ
------------LGGTLRALSLLA---AQGL-WA-QM----SVFHAE-------------
--DLESFGVQES-DLRPFLDR----GILRQDRD-SE-DCYSFLHLSVQQFFTALFYALE-
KE------EE--------DGDGH--AWDIKDVQ-KLLSGEERL-QNPDLIQAGHFL----
----FGLANEKRAKELETTFGCV---------------M--SRE-IKQ-ELLRCKANLHV
RK-----------------------------------P-SSMSGLKELLCCLYESQE-DE
LAKVVMAP---------------------F-------------------KEISIHLINTS
EM---MQC--Y----FSLK-HCPD-LEK-LSLQ----------------VAKG----VFL
-ENY-VDF----EL--------------------DFELERC-TYV--SLPNWARQD-LHS
LHVWKDFC-SLVSSNSNLKFLEVRQSFLSES---------SVRVLC--DHIAR-N-----
TC--H-LQKAE--I-KNVTPD--TA----YR-DFCLAF----------------IGKKTL
THLT-LEGH-IEWERTMLLLMCDLFRTPKGN----LQYL--------RFG---------G
H------S----------------------------------------------------
---------AT---PE-QWH---DFSFLLKA-NQSLRHLHLSASLLLD-EGAKLLYET--
VGH-PTNLLQKL-S----------------------------------------------
------------------------LENCRLT-----------------------------
----------------------------EASCKELAD-----------------------
--------------------------------VLIV
PF13516 // ENSGT00900000140813_0_Domain_11 (2017, 2054)
NQHLTH---LC-LAKNP-IGD---
-------KGVRLLCEGL----SYPDC----------------------------------
--------------------------KLQ-TLV---------------------------
-------------------------------LQQCNITKHGCRPISKV-LQGV
PF13516 // ENSGT00900000140813_0_Domain_13 (2214, 2228)
CSLTDLD
L-SFNHIG-CGLW-ILCKALENP--DCN----L---------------------------
-----------------------KHLRX--XXX-----XXXXXXXXXX------------
-------XXXX-XXXXXXXXXXXXXXXXXXXXXXX-XX----------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSOGAP00000020003 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------QQDHFINMEHVFDPD
EA--GFSLS----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 929)
RTVVLQGCAGIGKTALVHKF-----MFDWA--A---------GIVSP
G--------RFDY-LIYINCREISHIA-------SLSAAD-LITN-TF-----QD-VRGP
ILDIVLIYP----EKLLFILDGFPE-LQHPMGDQ------EE-A-----LSVQPQER-KP
-VESLLCSFVRKKLLPESSLLIT-ARPTAMKKLHSLL-R-----QPI--QAEILWFTSAE
KRAYF----------------L--SQ-FSG---------VKTAMKLFYGLRE-NEGLDVM
SSLPIVSWMICSVLKSQGDGDR-PL-L--RS-LQTMTDVYLFYFSKCLRTLTDVG-VW--
-----------KGQNCLWGLCCLA---AEAL-QS-RQ----VLFAAS-------------
--DLRRHGIGVCDSSYTFLSH-----FLKKAD--DI-NAYTFLHFSFQEFLTAVFYVLK-
NDNKW---------------------MYLSQVGKAWHEIFQRY-GKGFSSLTIRFL----
----FGLLRKGKGKIMETTFGRK---------------I--SPG-LRE-ELLKWTEREIK
DK-----------------------------------SSRLQIEPVDLFHCLYEIRE-EE
YSRRIMDD---------------------L-------------------HQVFSYSSIST
VD-----------------------NS----------------V-----EVECSR-----
------------------------------------------------------------
------------------------------------------------------------
HC--C-LAGL----------K------KLCLKD---------------KYIVFPV-----
-------------------------------------TSA--------HYKCFF-FFLMY
LE------------SA----CLFIITSEWLTELEFNETKLEVSALTLLCEGLKDPNCR-L
QKL----NFLPENSEA-VCK---CLASVVIC-NP
PF00560 // ENSGT00900000140813_0_Domain_8 (1775, 1792)
NLTELDLSENPLGD-IGVKYLCEG--
LKH-SDCKVEKL-D----------------------------------------------
------------------------LSTCCLT-----------------------------
----------------------------DASCVELSS-----------------------
--------------------------------CLQVSQTLKE---LF-VFANA-VGD---
-------TGVQHLCKGL----QHPKCAIENL-----------------------------
------------------------------------------------------------
------------------------------VLSECSLSAACCQPLGQV-LRASQSLTRLL
L-INNKIEDLGLK-LLCEGLKQP--DCQ----L---------------------------
-----------------------KDLAL--WTC-----HLTPACCKDL------------
-------CNAL-YTNENLRVLDLSDNALGDEGMRV-LCEGLK-HP-CC-RLQTLW--LA-
--E-CHLTDACCG--------------------ALASVLNRNENL-TLLDLTGNDLRDFG
VQMLCD--------ALIHPICK--LQ----------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPVAP00000010051 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------MIPSA----Q
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
LDFS---LQLLLQQLD--Q---AELSKFKSLLRM------
---------L--CPKDE--------L--EHI-PQME-VDEAD-GKQLAEILTNHCASYWV
EMVAIQI----FD-MMDRTDLSQRAK-DELRXXXX-XXXX----XXXXXX---XXXXX--
------XXXXXXXX-XX--------------------------X---XXXXX--------
---XXX--------------------------------------XXXXX-----------
-------XXX-------------------------------XXXXXXXXXXXXXXXXXX-
---------XXX----------------XXXX--XXXXXXXXXXXXXXXXXX--------
-----------------------------------------------XXXXXXXXXXXXX
XX--XXXXXXXXXXXXXXXXXXXXXXXXXXXXX-----XXXXX--X---------XXXXX
--------
PF05729 // ENSGT00900000140813_0_Domain_2 (729, 940)
-XXXX-XXXXXXXXXXXXX-------XXTFAE-LISK-N------TD-AQEA
RPEARA-PA----QKLLFVVDGFDE-LRVPSG------SLIH-D-----ICSDWNMR-KP
-APVLLGSLLKRQLSPQATLLVT-TRPEALPELRRLL-D-----QPL--LVEVEGLSEPD
RKASI----------------L--KH-FED---------EGQALRALALMQS-HPGLWRL
GAAPAVCGLLCACLKLQLEHGE-DP-A--TT-CGTATALLLHFLCGQFTPA-------SR
------------PPAPLQAACLLA---AGGM-WA-RT----SVFDAE-------------
--DLAALGLTEA-ALRPFLDA----EVLQKDVD-SV-GSYSFVHLSIQQFLTAVFYILE-
DDGEG-------------ARGRL--GGDIRDAR-ELLSKEERL-KNPGLAHVARFL----
----FGLANERMAGELETTFGCR---------------V--TTG-VAQ-ELLK-------
--------------------------------------SPSTMDLSEVMRCLHESQE-EL
LVRDAMDH---------------------V-------------------TEVSLRVKTRM
DL---VHS--S----FCLR-HCQN-LQK-ILLQ----------------VEKG----IFL
-ESD-TAL----ES--------------------D------------TPVERSKND-QHM
LPFWVDLC-SMFGSNKNLMFLDISQSFLSTS---------SVRILY--EKIAS-A-----
TH--N-LQKVV--L-KNISPA--DA----YQ-KFCIIF----------------RGHKTL
THLT-LQGN-DQNN--MLPSLCEVLRHPKCN----LKYL--------RLV---------S
C------S----------------------------------------------------
---------AT---TQ-QWA---DLSSSLEM-NQSLICLNLMANELLD-EGAKLLYMT--
LRH-PKCFLQRL-L----------------------------------------------
------------------------LEDCDLT-----------------------------
----------------------------GAYCKELSS-----------------------
--------------------------------ALIVNQRLTH---LC-LAKNA-LGN---
-------HGVKLLCEGL----SYPEC----------------------------------
--------------------------QLQ-TLV---------------------------
-------------------------------LYYCNITSDGCVNLSVL-LQQNSNLTHLD
L-GLNHIGITGLK-FLCESLKTP--LCN----L---------------------------
-----------------------RCLWL--WGC-----AITPLSCADL------------
-------SSAL-SSNQNLITLDLGQNSLGYSGVKM-LCDALK-RQ-SC-PLQTLR--PS-
--S-HDFIF---------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSP00000337383 (view gene)
ENSHGLP00000006766 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MSLDIRCEQ-LSD-ARWT----------------EL---------
----LPLI-------------------------------QQHQVV------RLEDC-GLT
EGRCKDIG-SALRANSALAELSLRTNALGDA---------GVCLVL--QGLQS-P-----
TC--K-VQSLS--L-QNCGLT--ET---GCR-VLASVL----------------RCT
PF13516 // ENSGT00900000140813_0_Domain_6 (1618, 1642)
PTL
RELY-LSDN-PLGDP-GLRLLSEGLLDPQSC----LEKL--------ELE---------S
C------N----------------------------------------------------
---------LS---AA-SCE---PLAAMLRA-KPNFKELVVSNNCIQE-AGIRMLCGG--
LKE-STCLLETL-R----------------------------------------------
------------------------LDNCGVT-----------------------------
----------------------------SANCKDLCD-----------------------
--------------------------------VVAA
PF13516 // ENSGT00900000140813_0_Domain_11 (2017, 2054)
KPSLQE---LD-LGGNR-LGD---
-------AGIAALCPGL----LHTSC----------------------------------
--------------------------RLR-KLW---------------------------
-------------------------------LWECDITAEGCRDLGQV-LRA
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
KQSLKALS
L-MLNELGDEGAR-LLCEALQEP--SCQ----L---------------------------
-----------------------ECLWV--RVC-----SFTATCCPHF------------
-------RALL-A
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
QNKFLTELQLGENRLGDAGVQE-LCQGLS-QP-GS-VLRVLG--IG-
--D-CDLSDNCCS--------------------NLASVLLA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2464)
SCSL-RELDLSNNGLGDPG
VLRLVE--------SLRQPGCV--LE--C-------------------------------
---LLLFDV-YLSDGVNDQ------LQ----------ALE-EEKPTLRIIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMICP00000014662 (view gene)
MGP_CAROLIEiJ_P0081408 (view gene)
ENSMNEP00000023257 (view gene)
ENSOGAP00000007576 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------
PF02758 // ENSGT00900000140813_0_Domain_0 (215, 328)
LD--Q---YQLEEFRLHLRPLE--RS
WQVPDVNITQ--LSQAR--------F--PEI-PWAD-LRAAD-PVNLFCLLKEHFSEWQL
WEVVLSI----LG-KMNLTSLCEQVR-EEMK-----------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------NHRREFRKR-
--------LKTN----------------ILAL--WDNIPCPEGHIYICNTSE--------
----------------------------------------------E-EHKEMQRLLDPN
G---TKAQA----Q
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 926)
TIVLQGRAGTGKTTLVLKA-----MLHWA--E---------GTLFQ
H--------RFSY-IFYLSCLKMKNMQ-------EITFAD-LLCWDDK-----PN-SEAL
IEEFVS-QP----EKLLFVVDGFEE-MTIPEPGP--HHLEDS-L-----PHIGWHEK-LP
-VTKILLSLLKKELVPMATLLIT-IRAPYVKNLTSLL-V-----NPC--FVCVTGFAEED
REVYF----------------S--SC-LGD---------QKEAKAMLEQVKR-NEALFNS
CSAPMVCWTICSCLKQLKARSS-DL--------QTTTSLYACFFSNLFSTAD-MGLPDES
------------LPGQWYTLCALA---AEGM-WS-LN----FTFGEE-------------
--DAQYQLLEVP-FIDSLFHL----DILQAVRD-HE-NCITFTHPSFQEFFAAMFYVLE-
EPRES--------------FHSS--STKHGEMK-IILRAVLVN-QESYWTPMVLFF----
----FGLLKKDLVRKLEDTLHCK---------------M--SPR-IME-ELLEWKEELDE
SE-----------------------------------TASVQFDTQQFFCCLHETQE-ED
FVKKMLGH---------------------V-------------------FEADLDILGNT
EL---QVS--S----FCLK-HCER-LSK-LRLS----------------VSR-----PIP
-EREGEVL----EY--------------------------------------MNTS-DSG
KQQWKDIC-TAFVTNRHLRELDLSNSKLSIS---------SMRSLC--HALRN-P-----
RC--K-LQKLT--C-KSITPV--SV---L-P-KLDLVL----------------HNNKKL
THLN-LSSN-RLE---V-GWIFKAVRYPACN----IKCL--------CLE---------R
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------CNLS-----------------------------
----------------------------ATSCPYLAS-----------------------
--------------------------------LLVNVQNMTR---LC-LGLNK-LQD---
-------EGAELLCDAL----TQTTC----------------------------------
--------------------------TLK-RLE---------------------------
-------------------------------LWLCQLGAPSCRHLSDA-LLKNKSLTHLN
L-SRNHLGDEGVK-FLSEALSCS--DCS----L---------------------------
-----------------------QCLNL--SGC-----SFTTEGCQEL------------
-------SDAL-RHNHNVHVLDIGRNDLGDDGAKL-LCEAMR-AS-SC-PLTTLG--LE-
--K-CNLTAACCQ--------------------HLSSLLSSHERL-AELNLIGNDLEPDG
VKTLCE--------ALRKSTCK--LQ--K-------------------------------
---LG-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPANP00000045277 (view gene)
ENSOGAP00000001153 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------QRSQKFIPFLQPR
IA--PQPIA----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 819)
HTVVLYGPAGVGKSTLAKQV-----FLDWA--E---------NNLNQ
---------MFNY-VFYLRCKELNHVG-------ACSFAE-LIFK-DW-----PE-WLGC
AAEVFG-QA----QKILFIIDGFEE-LRVPAG------ALIH-D-----ICGNWER----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------E-------------
--DLGKLEMEEE-DLCPFLDK----GVLQR--G-CD-GCYSLIHLSVQQFLAAMYYILD-
SEEQE-------------EVECH--RWDIREVQ-KLFSKEERV-RNPSLTQVGYFL----
----FGLSNEKRVQELESTFGCR---------------M--SLQ-IRE-ELLKYRANLDE
NK-----------------------------------P-FSRTDMKELLYCLYESQD-EG
LMRDAMVP---------------------F-------------------TEVSLHWMNTF
EV---MYS--S----FCLK-HCQN-LQK-LSLQ----------------IARG----IFL
-END-PVS----RE--------------------DAQLE-----------SWSRCD-HRS
LCVWMDFC-SVVSSNKNLKVLDMTQSFLSDS---------SVRILC--DHITR-F-----
SC--H-LQKVV--KFYQEIKT--SR----KW-RHMLGI----------------PGRETG
KYVS-TFSN-S-NDELMTSVTGMALRNESGQ-----------------GR---------V
K------Q----------------------------------------------------
---------MQ---TQ-MWT---GSILLVMF-SLDFTCHVMSDKENSE-QCLYLFNTV--
TGR-HRRGLIETRK----------------------------------------------
------------------------LENCHLT-----------------------------
----------------------------EACCKQLAS-----------------------
--------------------------------VLVVSQQLTY---LS-LAKND-LGD---
-------GGVKFLCEGL----GYPEC----------------------------------
--------------------------KLQ-TLV---------------------------
-----------------------------LVLRQCNISRLGCKHLSKL-FQGHSSLTSLD
L-GLNCIT-TGLW-FLCKALKNP--FCN----L---------------------------
-----------------------KYLGL--WGC-----SLTPFSCQDL------------
-------SSAL-LSNQKLESLDLGQNNLGQSGVTL-LFEALK-QK-NG-PLKTLR--LK-
--A-YESNLKIQV--------------------LLEYIKENNPQLTIDCDAA------RT
---------------TRSSSCY--YL----------------------------------
----S-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSTSYP00000003410 (view gene)
ENSCPOP00000004339 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------EYRRR-
--------IKEK----------------FQIL---NKNSLLGEPSDFHHGAS--------
----------------------------------------------KDEQKLLECLFDVK
VK--GGEPL----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
RTVVLRGAAGVGKTALMRKA-----MLHWA--D---------GGLYQ
H--------KFTY-VFYLGGKEISQVK-------EKSFAQ-LISK-DW-----PS-TEIP
IEKILS-QP----SNLLFIIDSFDE-LNFAFEEP------AF-A-----LCSDWTQE-HP
-VSFLISSLLRKVMLPESSLLVT-VRLTASNRLH-----PRLLKNQC--YVDLKGMCQDE
RKEYI----------------Y--HF-FED---------KTWATDVFNSIRS-NERLFNM
CHVPMVCRLICTCLKQQIEKND-DI-M--MV-CQTTTSVFVCCIFSLFSLG-EKYISNLP
------------NKAQLRSMCYLA---AKGV-WT-MT----HVLYKN-------------
--DFRKHGLTTC-DISVFLDM----NVLQKDTD-HE-NCYVFTHLHIQEFFAAMFYLLK-
SAWRD--------------REHS--SQSFEDLK-LLLEGNSDY--DPHLTQLKYFL----
----FGLLNEDLLKQLELILKCN---------------I--SLK-IKQ-EILQWVKGLEN
RD-----------------------------------TFPSQIVFLDLFHYLYETQD-EA
FVVQAMEN---------------------F-------------------SKLVIHIFMEV
HL---RVS--S----FCLK-HCRC-LET-FKLI----------------IA---------
------IV----NSNP-----AA--E------------------------TRQM-DD-QI
THWLQNLF-SVLNSNERLTELDLCNSNLDGL---------IMKTFC--QELRHP------
NC--K-LQKLLLKF---------VAF-PDDYQDIFSFL----------------TY
PF13516 // ENSGT00900000140813_0_Domain_6 (1617, 1642)
KQTL
MHLD-LKGS-YITDN-GVKSLCEALKHPECK----LQHL--------GLE---------S
CS----------------------------------------------------------
---------LT---AT-CCA---DIAKALVK-
PF13516 // ENSGT00900000140813_0_Domain_8 (1773, 1796)
SQSLVFLNLSSNNLLD-DGMMMLCEA--
LGH-PECYLQRL-V----------------------------------------------
------------------------LQNCGFT-----------------------------
----------------------------EVSCEDLSL-----------------------
--------------------------------VLLR
PF13516 // ENSGT00900000140813_0_Domain_11 (2017, 2054)
NKRLTH---LC-LADNS-VGD---
-------SGVKFLSTAL----AYSEC----------------------------------
--------------------------TLQ-SLV---------------------------
-------------------------------LRRCHFTTISIEYLSTS-LLCNESLIHLD
L-GSNSLHDDGLN-FVCNVLQKP--TCN----L---------------------------
-----------------------QELEL--MGC-----VLSTECCPDL------------
-------VSAI-L
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2374)
NSPNLLSLDLGYNDLQDEGVKI-LCEALR-DP-NC-NIQRLG--LE-
--Y-CSLTSVCCQ--------------------DLSSALVSNQKL-IKINLTQNTLGREG
IRTLCE--------VLTCPECK--LQ--V-------------------------------
---LGNFPV-VRDLGDHS-----------------------------FIGRSQNQ-----
--L---------------------------------------------------------
--------------------------------------------------DSGSWEY---
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSSBOP00000000646 (view gene)
ENSRNOP00000044415 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---MD-AAGASCSSS--EAVAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 321)
LLAALQDLS--Q---EQLKRFRHKLRD------
------------APLDG-----------RSI-PWGR-LEHSD-AVDLTDKLIEFYAPEPA
VDVTRKI----LK-KADIRDVSLRLK-EQQLQR---------L---------GSS-----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------S-ALLTVSEYKKKYREH-
--------VLRQ----------------HAKV--KERNARS------------VKINKRF
TKLLIAPGSGAGEDE--LLGTSG-------EPEP--------ERARRSDTHTFNRLFRGN
DD--EGPRP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLQGPAGIGKTMAAKKI-----LYDWA--G---------GKLYH
S--------QVDF-AFFMPCGELLERPG------TRSLAD-LILE-QC-----PD-RTAP
VRRILA-QP----HRLLFILDGADE-LPTLAAP--------EAT-----PCRDPFEA-TS
-GLRVLSGLLSQELLPSARLLVT-SRNATLGRLQGRL-C-----SPQ--CAEVRGFSDKD
KKKYF----------------F--KF-FRD---------ERKAERAYRFVKE-NETLYAL
CFVPFVCWIVCTVLLQQMELGR-DL-S--RT-SKTTTSVYLLFITSMLKSA-GTNGP---
-----------RVQGELRMLCRLA---REGILKH--Q----AQFSEK-------------
--DLERLKLQGSQVQTMFLSKKELPGVLET-----V-VTYQFIDQSFQEFLAALSYLLD-
AEGAP-------------------G-NSAGSVQ-MLVNSDAGL--RGHLALTTRFL----
----FGLLSTERIRDIGNHFGC-----------------VVPGR-VKQ-DTLRWVQGQSQ
PK--VATVGAEKKDELKDEEAE--EEEE-----EEEEEEEELNFGLELLYCLYETQE-DD
FVRQALSS---------------------L-------------------PEMVLERVRLT
RM-DLEVL--S----YCVQ-CCPD-GQA-LRLV------------------SC----GLV
------AA----------------------------------------KEKKKK---KKS
----------------------------------------FMNR----------------
------LKGS----------------------------------------------QSTG
KQ---PPAS-------LLRPLCEAMITQQCG----LSIL--------TLS---------H
C-----------------------------------------------------------
--------KLP---DA-VCR---DLSEALKV-APSLRELGLLQNRLT-EAGLRLLSQG--
-LAWPKCKVQTL------------------------------------------------
----------------------------RIQ-----------------------------
---------------MPGL---------QEVIHYL-------------------------
------------------------------VIVLQQSPVLTT---LD-LSGC-QLPGTVV
--E--------PLCSAL----KHPKCGLKTLSLT--SVELTEN-----------------
----------PLRELQAVKTLKP-------DLA----------------------I--IH
SKLG-------------------THPQPLKG-----------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSOPRP00000013263 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------MAVV-PN
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
PQQA---LIWALNELE--E---KNFKLLKYHLQN------
---------K--TPAKG--------H--RQL-TRGE-LEGLS-QPDLASQLILIYGAQEA
VKVVLEV----LK-EMHLMELVDQLN-LVC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LN--DYREIYREH-
--------VRC-----------------LE------EM---QEQ----------GINSTY
KQLLLVI-KPSAESPKSPDC------LS-------SE-----W-ELDSV--QVEALFGSG
EM--PDPAP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
HVVVLQGSAGTGKTTLTKKM-----LLNWA--N---------GTLYQ
D--------WFDY-VFYLSCREVGLLPG-------RKLDQ-LLFW-CC-GD-----QEAP
ITEILK-QP----ERLLFILDGFDE-LQRPFRQ-----------------------S-SP
-IENLLHLLIRRKVLPTCSLVIT-TRPSALRNLEPLL-H-----RPS--HVHVLGFSEEE
RREYF----------------S--SY-FTN---------EEQARNAFEVVQG-NDVLYKA
CQVPGICWMVCSWLRKKMERGX-XX-X--XX-XXXXXXXXXXXXXXXXXXXX-XXXXXXX
----------X-XXXXXXXXXXXX---XXXX-XX-XX----XXXXXA-------------
--DLRRHNLDGSRLV-AFLSS----NDYKNGRD-MK-KFYSFRHISFQEYFHAMSYLVK-
DNHNQL----------------G--KKSCREVE-KLLEEK-----DKEMNLCMQFL----
----WDISRKESFSNLELKFCFR-----------------IAPS-IAQ-EL-KYFKQQM-
E-------------------------------------SMKHKRTWDLEFSLYETTV-KN
LARSVQMT---------------------D-------------------VSFKMEHSN-T
MR-PRGKK--S----FCVK-TSLL-DGQ-REEQ------------------QC----ALM
--------GQ---SNM--V-------------------GAQKA-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------ISS----------------GKD---------R
-ETEE-V-----------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSSTOP00000011399 (view gene)
ENSPPAP00000017303 (view gene)
ENSMLEP00000024232 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---ESFFSDFG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 322)
-LLWYLKELR--K---EEFWKFKELLKQ------
---------P--LEKFE--------L--KPI-PWAE-LKKAS-KEDVAKLLDKHYPGKQA
WEVTLHL----FL-QINRKDLWTKAQ-EEMRN----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------KLNPYRKH-
--------MKEK----------------FQLI--WEKETCLPVPEH--------------
---------------------------------F-------YKETMKNKYKELNGAYTAA
------ARL----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
HTVVLEGSDGIGKTTLLRKV-----MLDWA--E---------GNLWK
D--------RFTF-VFFLNVYEINGVT-------ETSLLE-LLSR-DW-----PES-SET
IEDIFS-QP----ERILFIMDGFEQ-LKFNLK-F------KD-D-----LSDDWRRQ-QP
-TSIILSSLLQKKMLPECSLLIA-LGKLAMQKHYFML-Q-----HPK--LIKLSGFTESE
KKSYF----------------S--YY-FGE---------KNKALKVFNFVRD-SGPLFIL
CHNPFICWLVCACVKQRLERGE-DL-E--IN-SQNTTSLYASFLTNVFKAGS-QSFVPK-
-----------VNRARLKNLCALA---AEGV-WT-YT----FVFCHE-------------
--DLRRNGLSES-EGLMWVGM----RLLQRRG-----DYFTFIHLCIQEFCAAMFYLLK-
RPKDD-----------------P--NPAIGSIT-QLVKATVIQ-PQTHLTQVGIFM----
----FGISAEEIVSMLETSFGFP---------------L--SKD-LQQ-EITQSLKSLSQ
CE-----------------------------------ADREAIGFQELFVSLFETQE-KE
FVTQVMNF---------------------F-------------------EEVFIYIGNIE
HL---VIA--S----FCLK-HCRG-LTT-LRMC----------------IEN-----IFP
-DDSGCAS-----------G-------------------Y-N-------EK---------
LVYWRELC-SVFITNKNFQILDMENSSLDDA---------SLAILC--KALTQP------
VC--K-LRKLIFTS---------MLN-LGHDSELFKAV----------------LHNPHL
KLLS-LYGT-SLSRT-DVRHLCETLKHPMCK----IEE----------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------L-----------------------------
------------------------------------------------------------
------------------------------MLTYCCLTCVSCDSISQVL-LCNKSLSLLD
L-SSNALEDNGVA-SLCGALKHP--AC---------------------------------
------------------------------------------------------------
------------------------------------------------------G--LQ-
--T-CPITRACCG--------------------DLAAALVACKTL-RSLNLDWITLEPDA
VAVLCE--------ALSHPDCA--LQ--M-------------------------------
---LGLHKS-GFDEEAQKM------LM----------SVEEKM-PHLTISHEPWIDE---
--EYK-----IR------------------------------------------------
--------------------------------------------------GVLL------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSSARP00000010938 (view gene)
ENSANAP00000007054 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----MSVD--V----QRVDIQCEE-LSD-ARWA----------------EL---------
----LPLL-------------------------------QRCQVV------RLDDC-NLT
EAQCEDIS-SALQA
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1542)
NPALMELSLRSNELGDA---------GVRRVL--QALQS-S-----
SC--K-IQKLS--F-QNCYLT--AA---GCG-VLPGTL----------------RS
PF13516 // ENSGT00900000140813_0_Domain_6 (1617, 1642)
LPTL
QELH-LSDN-PLGDA-GLQLLCEVLLDPQCH----LEKL--------QLE---------Y
C------G----------------------------------------------------
---------LS---PA-SCE---PLAAVLRA-KPDFKELTVSNNDIEE-AGVHVLCQG--
LKD-SPCRLETL-K----------------------------------------------
------------------------LENCGAT-----------------------------
----------------------------SDSCRDLCG-----------------------
--------------------------------ILAS
PF13516 // ENSGT00900000140813_0_Domain_11 (2017, 2054)
KTSLRE---LA-LGNNK-LGD---
-------VGIAELCPGL----LHPGS----------------------------------
--------------------------RLK-TLW---------------------------
-------------------------------IWECGLTAKSCVDLCRV-LRAKD
PF13516 // ENSGT00900000140813_0_Domain_13 (2215, 2232)
SLKELS
L-AGNKLGDEGAW-LLCESLLEP--GCQ----L---------------------------
-----------------------ESLWV--KSC-----SLTAACCSHF------------
-------SSVL-AQ
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NKFLLELQISENRLGDAGVQE-LCQGLG-QP-GS-VLRSLW--LG-
--D-CDVSDSSCG--------------------SLAETLLA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2465)
NRSL-RELDLSNNCVGDAG
ILQLME--------SLRQPACL--LE--Q-------------------------------
---LVLYDI-YWSLDMDNR------LQ----------ALE-KEKSSLRVIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCHIP00000006950 (view gene)
ENSPEMP00000019983 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------M---ASFFSDYG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 323)
-LMWYLEELN--K---KEFMKFKEVLKQ------
---------E--IIQYG--------L--KQI-PWTE-VKKAS-RENLANLLVKYYDEKKA
WDVTFRI----FQ-NINRNDLSERAA-REI------------AG--------------HS
K---------------------------------------------------IYQA----
------------------------------------------------------------
----------------------------------------------------------H-
--------LKNK----------------LTED--WSRKFNIPTQDF--------------
---------------------------------L-------KQKFTQDECNRFELLFVSK
VT--ERK-T----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
HTVLLKGVAGIGKTLMLAKL-----MLAWS--E---------GLVFQ
N--------KFSY-IFYLCCQDVKQLK-------TTSLAE-LISR-EW-----PSP-SAP
IVEILS-QP----EKLLFIVDSLEG-LNCDLTEP------EW-K-----LCDNWVEK-RP
-VNILLSSLLRRKLLPEASLLLT-ATPETFDKLEDRI-D-----YTE--MKIMMGFDESS
RKMYI----------------R--GV-FQD---------KTRAQEVLRLVRA-NEQVFSI
CQVPLLCWVVATCLKNEVEKGG-DL-V--SV-CRRTTSVYTAYILNLFTPRS-AQYPSQ-
-----------KSRELLQSLCFLA---AEGV-WT-DK----FAFSEE-------------
--DLRKGGILSD-DISALLDI----RLLLKSRE-SK-LFYTFFHPSIQEFCAAVFYLVK-
SHGDH-----------------P--SKDVRSVK-ALLFTFLKK-VKVQWIFVGCFI----
----FGLLHKSEQEKLEVFFGHQ---------------L--SQE-VKQ-TLYQCLETISV
NED-----------------------------------LQEEVDGMKLFYCLYEMED-DV
FTMQAMSL---------------------M-------------------QHIKFVAKDYS
DL---IVA--A----YCLK-YCST-LRK-LSFS----------------TQD-----ILQ
-HEQ--EC-----------S-----------------------------------YMEKL
LVCWQEVC-SVLVRSNHINELQVKDTNLDES---------SFLVLY--NHLKHP------
SC--A-PQVLEVNN---------VSF-LCDNHLFFDLL----------------THNHNL
QYLN-LSLT-FLSHS-DVKLLCDIFSQTECN----IEKL--------LV-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------AACELS-----------------------------
----------------------------PDDCKIFAS-----------------------
--------------------------------VLISSKTLRH---LN-IASNN-LD----
-------KGIYSVCKAL----CHPDCVLKDL-----------------------------
------------------------------------------------------------
------------------------------VLANCSLSEPCWDYLSDV-LRRNKTLSHLD
I-GTNDLKDEGLK-ILCEALSLP--DSV----L---------------------------
-----------------------KSLCL--RCC-----LITSSGCQHL------------
-------AEVL-SSNQNLRCLQLAHNKIEDTGVKI-LCDAIK-QP-NC-RLENLG--LE-
--A-CKLTSACCE--------------------DLASTFTQNKAL-WGVNLLQNALDRSG
LALLCE--------ALKDHNCT--LH--V-------------------------------
---LGLRIT-DYDKETQEF------LI----------AEE-EKNPYLTILSS-I------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMLUP00000005806 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------MTDSSSSSFF
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
SDFG---LLLYLEELN--K---NELSKFKSFLKN------
---------E--TPGPG--------N--CLL-PWTE-VKKAN-REGLANLMRKHYPGEQA
WDVALKI----FG-KMNLKDLCERAK-AEMSWTSQ-TMEPEDTTA-GE------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------AQGEQEAVLGDGTKYRIQ-
--------IQEK----------------FSLE--PDEKCWLGEPKDFRHEIA--------
----------------------------------------------EDGRELLEHLFDVD
VN--TGEQP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
QTVVLQGAAGVGKTTLVRKA-----MLDWA--Q---------GTLYQ
E--------KFTY-IFYLNAREINQWK-------ERSFVE-MIST-DW-----PG-TEGP
IEKIMS-QP----SSLLFIIDSFDE-LNFAFEEP------EF-V-----LCEDWTQV-HP
-VSFLMSSLLRKVMLPESFLLVT-TRLTTCEKLKLLL-K----N-QR--SVELLGMSEDA
REEYV----------------Y--GF-FED---------KKWALLVLSLLRN-NEMLFSM
CNVPLVCWVICNCLEQQMEKGG-DI-T--LT-CKSTTALFTCYVSSLFTQV-DGRSPSLP
------------EQAQLRNLCQLA---ARGV-WT-MT----HVFYRA-------------
--NFRKHELMKS-DISSFLDM----NILQQDTD-YE-NCYEFTHLHIQEFFAAMFYMLE-
DGSKL--------------EDHA--SQSFADLK-LLLESTNCE--HPHLTQMKCFL----
----FGLLNEDRAKQLEETLNCN---------------L--SLD-LKG-KILQGMEILGK
SD-----------------------------------SFI-QLGCLELFHCLYETQD-EA
FIIQAMRY---------------------F-------------------QKVVINICEKF
HL---LVS--S----FCLK-HCQC-LRT-IKLS----------------VTV-----VFE
-----KML----KSSF-----PV--E------------------------TWDT------
YYCWQDLC-SVLHTNEHLRELELCHSNLNEL---------AEKIFY--KELRHP------
QC--K-LQKLLLRF---------FTF-ADGCQNFASSL----------------IHNQNL
LHLD-LKGS-DIGDG-GVKSLCEALRHPDCK----LQNL--------SLE---------S
CS----------------------------------------------------------
---------LT---TL-CCL---NISKILIR-SHSLIFLNLSTNNILD-DGVELLCEA--
LRH-PNCHLERL-S----------------------------------------------
------------------------LESCGLT-----------------------------
----------------------------VAGCEDLSL-----------------------
--------------------------------ALISNKGLTH---LC-LADNF-LGD---
-------GGVKLMSDVL----KHPQC----------------------------------
--------------------------TLQ-SLV---------------------------
-------------------------------LRRCHFTSGSCEHLSSS-LRSNK
PF00560 // ENSGT00900000140813_0_Domain_13 (2215, 2243)
SLTHLD
L-ASNQLQDDGVK-LLCDVFRHP--GCN----L---------------------------
-----------------------QDLEL--MGC-----VLTSACCLDL------------
-------ASAI-LNNP
PF00560 // ENSGT00900000140813_0_Domain_15 (2357, 2381)
NLRSLDLGNNDLQDDGVKI-LCEALK-HP-NC-NIQRLG--LE-
--Y-CGLTSLCCQ--------------------DLSSTLSSNQRL-IKMDLVQNTLGCEG
IKKLCE--------VLRSPECK--LQ--V-------------------------------
---LGLCTE-AFDEEAQEL------LE----------AVE-VSNPNLVIKQDCSDQN---
--E---------------------------------------------------------
--------------------------------------------------EDGSWWKCF-
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSAMEP00000007804 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------
PF05729 // ENSGT00900000140813_0_Domain_2 (682, 940)
AGVGKTTLGRKG-----TGDWA--Y---------KGL--
-----------AV-LLDFNAREIDQLR-------EGIFVQ-MIAK-DW-----PS-REGP
IERIMS-QQ----SSLLFLTDSFDE-LNFAFEEP------EF-V-----LCRDWTQL-HP
-VSFLMSRLLRKGMLPESSLLVT-TRLTAFKKLKPLL-K-----NQR--SVELLGMSKGA
RQEYI----------------Y--QF-FED---------KKRALKVFSSLRS-NEMLCSM
CKVPLVSWAICTCLEQQMEMGA-DV-S--LT-CKTATAVFTCSLSSL-----------LP
------------NQTQLRSLCHLA---ARGV-WT-MT----SVFYRE-------------
--DLRSMG-TKA-DVSIFLDM----KILQKDME-YE-NCC-FTHLHVQEFLAAMFYVLK-
---------------------------SLEGFK-LLLESSSGK--DAHLMQMKCVL----
----FGLLNEDLVKPLETTLNCK---------------M--SLD-VKG-RILQWLEILGN
---------------------------------------------------LYETQD-EA
FVSQAMRC---------------------F-------------------QKVVIDVYGKV
HL---LVS--S----FCLK-HCLC-LWT-IKLS----------------VTA-----IAY
--L--GLS----FVKF-----KA--SGQHARL------------LYILIMPNRKGMVVTL
LIAGKDVC-SVLHTNKHLREQDLCHSSLDEL---------AMKTFY--QDLRHP------
NC--K-LQKLLMRF---------LSF-PGGCQDIASSL----------------THNQNL
MHLD-LKGS-DIGDD-GVES-CKALKHLECK----LQNL--------RIR---------G
RC----------------------------------------------------------
---------LR---MT-TTA---LTLLLTRI-SQSLRFLNLSTNHLLD-DGITLLCEA--
LGH-PKCYLERL-S----------------------------------------------
------------------------LESCGLT-----------------------------
----------------------------VAGCEDLSL-----------------------
--------------------------------ALISNKRLTH---LC-LADNI-LGD---
-------DGVKLVSDAL----KNPQC----------------------------------
--------------------------NLQ-SLV---------------------------
-------------------------------LQITYLLLSYA--TRAG-VKLAARSQELD
L-GSNWLQDDGVK-LLCDAFWHP--SCH----L---------------------------
-----------------------QDLEL--MGC-----VLTGTCCLCL------------
-------ASAV-LNNAHLRSLDLGYSDLQDDG----------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSFCAP00000010670 (view gene)
ENSFCAP00000025751 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------MALA-HSPREA---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWVLSDLE--E---KSFKLFKFHLRD------
---------R--SLLER--------Q--PQL-ARGE-LEDLS-PVDLASLLISLYGAQEA
AKVVLRI----LK-VMNLLELVDQLS-HIC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LN--DYREIYREH-
--------VRG-----------------LE------ER---QEG----------GPCGSH
SQLLLVA-RPSPESPESSAL------PV-------LE-----Q-ELDSV--SLQALFDPG
ER--PYQAP----P
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVILQGSAGTGKTTLARKM-----VLDWA--T---------GTLYQ
G--------RFDY-VFYVSCREVVLLQE-------GRLRQ-LLFW-CC-GD-----DQAP
VDEMLR-EP----ERLLFILDGFDE-LQRPFAKG------LK-R-----LS-----P-SS
-MEETLHSLIKREALSTSSLLIT-TRPLALPNLKPLL-K-----QSH--HIHILGFSEDE
RRRYF----------------S--SY-FTD---------EEQARNAFDIVQG-NDALYKS
CQIPGICWVVCSWLKGQMEKGR-EI-S--ET-PSNDTDIFMAYVSTFLPPSD-NGGSSEL
----------T-RHRVLRGLCSLA---AEGI-QH-QR----FMFEEA-------------
--DLRKHNLDGTRLG-AFLSS----MDYQEGFD-IK-KFYTFRHISFQEFFYAMSYLVK-
EDQSQL----------------G--KESHRKVN-SLLDEEGQA-GNEEMTLSTRFL----
----LDISRKETSSNFELKFSLK-----------------ISPS-VQQ-DL-KHFKEKM-
E-------------------------------------SIKCNRAWDLELSLYKSKI-KN
LVKGVQMS---------------------D-------------------VSFKMEHSN-K
KK-SHGKN--S----FSVK-TSLS-NGW-KEKQ------------------KR----PCD
-------------NNI--A-------------------KAQEK-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------SSN----------------GKG---------R
-EAEE-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPSIP00000007091 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------MMENQ-SRIRGL--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 326)
-LQTALDDLP--Q---EVFRRFREKLSH------
---------S---AFEG--------K--SAI-PRGQ-LEKAD-RTDTKNLLMEFYGGAAA
VEVTINV----LT-QINLRESATKLR-AESEKVFS-----KAKKKT------AKI-EEGR
G-----------------------------------------------T-----------
---------------------------------------DRQWDQEPCA-----------
ETHHAK-EQG-------------------------------FNNPSLKGSDYQKKYREH-
--------VQDE----------------YQVV--EDRNVHLGEHV---------CLDKRY
TELLIVKERRGREEKEHEII------TRGRRHAELMA----QQ----TSLTQISLLFDPD
GN--R-QST----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
RTVVLQGVAGIGKTMTARKI-----MWDWA--A---------GKLYR
K--------KFDY-VFYINCREIKHIIKNQSGAKNRSAAD-LVLN-NC-PD-----GGGP
IKEIFE-KP----EKLLFIIDGFDE-LNSLLDID------DI-S-----LCTDPFEK-RP
-MEIILCSLLRKKVLKKSFLLIT-TRLTSLEKLHQIL-K-----FPR--YAEIMGFSADQ
RKEYF----------------Q--KY-FCD---------EKKATQAFNFVKE-NEVLFTV
CFVPIVCWIICTVIKQQMVKGE-DL-T--QT-SKTTTSVYLLFLTRLLGDHFTNATCVQS
----------F--KVNLWELCSLA---ADGI-LE-RK----ILFEED-------------
--DLKKHNLSVSNIHTLFLNQ----SIFQTDTE-CE-IFYSFIHLSFQEFFAALYFVLE-
EDEETI--------------KKS--LMSKKDVN-RLLKIYGAS-KNN-LMLTVRFL----
----FGLLNKERLNDIEEKLHCK-----------------TSSG-IKD-DLLKWFEAEAK
NIS-------------SLSY----------------KTYEVQHGQLELFHCLHESQE-EE
FAKCVMDS---------------------F-------------------QEIRVEHIRLT
TM-DEIAL--S----FCVK-NCCS-KQS---------------------LYLC----NC-
------------------T-----------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------L--------GIG----------
-EREE-EW------------------KAKLRKRLFPL-----------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSHGLP00100026435 (view gene)
ENSAMEP00000004644 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MPRA----PASD
PF02758 // ENSGT00900000140813_0_Domain_0 (204, 328)
GLCRLSAYLEELE--A---VELKKFKLYLGT------
---------E---AG----------A--SRI-PWGR-LEPAG-PLDTARLLVAHCGPDAA
WPLALGL----FQ-RINRRDLWERGQ-REEPLRGT----P--PG--------GPSSL---
------------------------------------------------------GS----
------------------------------------------------------------
---QS-ACSL-------------------------------EVFPGAPRRDPQETYRDY-
--------VRRK----------------FRLM--EDRNARLGECV---------NLSLRY
TRLLLVKEHSNPMRAQQKLL------DTSWGQARTLG--------HQASFIQMETLFEPD
EE--RPEPP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
RTVVLQGAAGMGKSMLAHKV-----MLDWA--D---------GRLFQ
D--------RFDY-VFYVNCREMNQGTA------EQSAQD-LISS-CW-----PE-PSVP
LQELVR-AP----ERLLFIIDGFDE-LKPSFHDP------QG-P-----WCLCWEEK-RP
-TELLLCSLIRKKLLPELSLLIT-SRPTALERLHRWL-E-----HPR--HVEILGFSEAE
RKEYF----------------Y--KY-FHD---------AEQARQVFHFVRD-NEPLFTL
CFVPMVCWVVCTCLKQQLEDGG-LL-R--QT-SRTTTAVYVLYLLSLMQPKPGS-PTL--
-----------QPLPDQRGLCSLA---ADGL-WN-RK----VLFEER-------------
--DLRKHGLDGA-DVSSFLNV----NIFQKDIH-RA-KFHSFIHLSFQELFAAMYYLLD-
P--GE--------------M--G--SGPEQNVT-RLLAEYGFS-DRSFVALTVRFL----
----FGLLNEDTRSYLEKSLGWK---------------V--SPH-VKA-ELLRCIQGKAQ
SE-----------------------------------GSTLRQGALELFSCLYEIQE-ER
FIRQALSH---------------------F-------------------QVVVVGNMA-T
KM-EHVIA--S----FCVQ-SCRH-ALV-LHLH----------------GA------AYS
-TDEDDGG--R-RA---------------------------------------PGP---Q
T-------------------------LP------------------------A-------
P---S--------PEKHLLPE-------AYSKQLAAAL----------------STNPNL
TELV-LYRN-ALGSR-GVRLLCQGLRHPGCK----LQNL--------SLK---------R
C-----------------------------------------------------------
--------CVP---SS-ACQ---DLAAALVA-NRSLTRVDLSCNGLGL-PGVRLLCEG--
LQH-PGCRLQAIQ-----------------------------------------------
------------------------LRKCQLE-----------------------------
----------------------------AGACREIAS-----------------------
--------------------------------VLSTNR
PF00560 // ENSGT00900000140813_0_Domain_11 (2019, 2052)
HLVE---LD-LTGNV-LED---
-------LGLRLLCQGL----RHPGC----------------------------------
--------------------------RLQ-ILW---------------------------
-------------------------------LKICHLTAAACEELAST-LAVNRSLMELD
L-SLNDLGDPGAL-LLCEGLRHP--QCS----L---------------------------
-----------------------QTVRL--GIC-----RLSSATCEGI------------
-------CTVL-RVNP
PF00560 // ENSGT00900000140813_0_Domain_15 (2357, 2385)
HLRELDLSFNDLGDTGMWP-LCEGLR-HP-TC-RLQKLW--LD-
--S-CGLTAKACE--------------------DLSSALGVSQTL-TELYLTNNALGNAG
VRLLCK--------GLSHPGCK--LQ--V-------------------------------
---LWLFGM-PLNKTTHRR------LA----------ALR-LMKPHLDIGC---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMUSP00000083106 (view gene)
ENSTTRP00000002365 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---ESFF
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
SDFG---LLWYLEELK--K---EEFWKFKELLKQ------
---------E--SLKFK--------L--KPI-PWTE-LKKAS-RENLSQLLSKHYPGKLA
WDVTLSL----FL-LINRGDLWTKAR-DEIRH----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------KLNPYRNH-
--------MKKK----------------FRDI--WEKETCLEVPED--------------
---------------------------------F-------YKETTKDEYEQLNAVYLTA
DK--PGNPS----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
PTVILQGPEGVGKTTLLRKV-----MLEWA--E---------GNLWR
D--------RFSF-VFFLKGSEMNSIT-------DTSLVE-LISR-DW-----PES-SEP
IEDIFS-QP----ERILFILDGLEE-LKFDLT-L------DA-D-----LCGDWEKQ-RP
-TQIVLSSLLHKQMLPESSLLIA-LGKMGMKKNYFLL-C-----QTK--CICLPGFSERQ
RKLYF----------------S--HY-FRE---------KNKSSRAFSFVRE-KRPLFVL
CKSPFVCWLVCTCLKWQLEKGE-DL-E--VA-SESITCLYVSFLTNVFKAAS-GNCPPK-
-----------QNRTRLQSLCTLA---AEGM-WT-RT----FLFCPG-------------
--DLRRNGVSES-DTSMWVDV----RLLQRSG-----NCLTFIHTCVQEFCAALFYLFK-
RPKDR-----------------P--HPVIGKVT-QFVTAAMSE-ACSHLSWMGLFL----
----FAFSTEKITNMLETSFGFP---------------L--STD-IKQ-EVTQSLETLSQ
CD-----------------------------------PGKMVVSFQELFNCLFETQD-QE
FVIQVMNF---------------------F-------------------KEIDIYIGTIE
EL---TVS--A----FCLK-HCQS-LQK-FHLC----------------IEN-----VFS
-DESGSIS-----------N-------------------T-I-------EK---------
LSLWRDLC-SAFTASENFQMLDLDNV-SDEA---------SLAILC--RTLAQP------
VC--K-LQKFVYNF---------ASN-LANSLYFFKAI----------------LHNPHL
KYLN-LYGT-GLSYM-DVRQLCETLKHPVCN----IEAL--------LX-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------XXXXXX-----------------------------
----------------------------XXACEDIAS-----------------------
--------------------------------VLVHNKKLKL---LS-LVDNP-LKD---
-------EGVRVLCDAL----KDPACALEAL-----------------------------
------------------------------------------------------------
------------------------------LLTYCCLTSVACDYISQV-LLCNKSLSFLD
L-GLNFLEDDGVI-TLCESLKHP--NCN----L---------------------------
-----------------------EELWL--VGC-----CFTSVCCVDL------------
-------AAVL-IHSEKLKTLKLGNNKIYDAGVKQ-LCEALK-HP-KC-KLETLG--LE-
--M-CELTSACCE--------------------DLALAFTLCKSL-RCMNLEWLSMDRDG
AVVLCE--------ALISLECG--LQ--T-------------------------------
---LGLNKS-SYDEEIQML------LT----------EVE-ETNPQLTISHSLWTDY---
--EHR-----IR------------------------------------------------
--------------------------------------------------GVIG------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSDNOP00000018711 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------MAECVI
PF02758 // ENSGT00900000140813_0_Domain_0 (206, 328)
CRLITYLEELE--A---VELKKFKLYLGC------
---------L---AELG--------E--GKI-PWGR-LEMAG-PLEMAQLLVAYRGSGDA
WLLALSI----FD-RINRKDLWEQGQ-REHLVRDA----P--PG--------GLSSL---
------------------------------------------------------RR----
------------------------------------------------------------
---QS-GCPM-------------------------------EVSSGTPRKGPQESYRDY-
--------VRRK----------------FRLM--EDRNARLGECV---------NISHRY
TRLVLVKEHLTPTRAQQKLL------AMGRGHARTVG--------YQASPIQVDTLFQPD
EE--RPEPP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
RTVVLQGAAGMGKSMLAHKV-----MLDWA--E---------GRLFQ
G--------RFDY-LFYINCREMNQR-A------EQSLLD-LILS-CW-----PE-LSTP
-QELLG-VP----ERLLFIIDGFDE-LKPSFHNP------QG-S-----GCLHWEEK-QP
-TELLLSSLIQKRLLPELSLLIT-TRPTALEKLHRLL-E-----HPR--HVEILGFSEAE
RKEYF----------------Y--KY-FRH---------AEQAAQVFSFVRN-NEPLFTM
CFVPLVCWVICTCLKQQLEDGE-LL-R--QT-SRTTTAVYTLYLLSLMQPKPGT-PTH--
-----------LPPPNQRGLCSLA---ADGL-WN-QK----ILFEEQ-------------
--DLRKHGLDGA-DVSAFLNM----NIFQKDIN-RE-KFYSFIHLSFQEFFAAMYYILD-
D--RD--------------G--G--PGPEHNVD-RLLAEYGLS-ERSFLALTVRFL----
----FGLLNEQTRSYLEKSLCWK---------------V--SPH-IKT-ELLKWIQSKAQ
SE-----------------------------------GSTLQQGSLEFFSCLYEIQE-EE
FIQQALSH---------------------F-------------------QVVVVSNIA-T
EM-EHMVS--S----FCVK-HCSG-VLV-LHLN----------------GA------AFG
-ADEDRGV--R-WA---------------------------------------SGT---Q
E-------------------------LL------------------------A-------
Q---S--------PERAILPD-------AYSEHLAAAL----------------RASPHL
TELV-LYRS-ALGSR-GVKLLCEGLGHPNCQ----LQNL--------RLK---------R
C-----------------------------------------------------------
--------QIS---SS-ACQ---DLAAALTA-NKNLIRMDLSSNSLGL-PGVRLLCEG--
LRH-PKCRLQMIQ-----------------------------------------------
------------------------LRKCHLE-----------------------------
----------------------------AGACQEMAS-----------------------
--------------------------------VLSTNRHLVE---MD-LTGNA-LGD---
-------WGLRLLCQGL----KHPVC----------------------------------
--------------------------RLR-TLW---------------------------
-------------------------------LKICRLTAVACEDLAST-LGVNQSLSELD
L-SLNELGDPGVL-LLCEGLRHP--KCK----I---------------------------
-----------------------QSLRL--GIC-----RLGPAACEGL------------
-------SAVL-QTNQHLRELDLSFNDLGDGGMHL-LSEGLQ-HP-TC-RLQKLW--LD-
--S-CGLTAKACE--------------------DLASTLEANQTL-MELYLTHNALRDRG
VHLLCK--------GLCHPGCK--LQ--V-------------------------------
---LWLFGM-DLNIMTHRE------LA----------KLR-VKKPYLDIGC---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
MGP_PahariEiJ_P0041372 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------M---ASFISDFG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 323)
-LMWYLRELS--K---EEFTKFKDFLIQ------
---------E--ILELK--------L--KQI-SLTE-VKKAS-REDLANLLLKCG-ENQA
WDVTFKI----LQ-KINRNDLTERVT-EET------------AG--------------NP
K---------------------------------------------------LYRD----
------------------------------------------------------------
----------------------------------------------------------H-
--------LKKK----------------LTHD--CSKKFNDRIQDF--------------
---------------------------------I-------KETFIQNDYDTFENLLISK
GT--EKK-P----H
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
MVFLKGMAGVGKTMMLKNL-----MLAWS--K---------GLVLQ
N--------KFSY-TFYFCCQDVKQLK-------TASLAE-LISR-EW-----PSP-SAP
VEEILS-QP----EKLLFIIDSLEG-MEWDLTKQ------ES-E-----LCDNCMEK-QP
-VSTLLSSLLRRKMLPESSFLLS-TTPETFEKMEDRI-Q-----CTD--VKTATAFDERS
XKIYF----------------Q--RL-FQD---------RIRAQEAFSLVRE-NEQLFTI
CQVPLLCWMVATCLKEEIEKGR-DP-V--SL-CRXITSLYTTHIFNLFIPQS-AQYPSK-
-----------KSQDQLQGLCSLA---AEGM-WT-DT----FVFGKE-------------
--ALRRNGILDS-DIPTLLDI----GMLGKISE-FE-NSYIFLHPSVQEVCAAIFYLLK-
RHVDH-----------------P--SQDVKNIE-TVLFMFLKK-VKTQWIFLGCFI----
----FGLLQKSEQEKLLVFFGHR---------------L--SKN-IQH-KLYQCLETLSG
NAE-----------------------------------LQEQIDGMKLFSCLFEMED-KA
FLVKAMNC---------------------M-------------------QQINFVAKNYS
DF---IVA--A----YCLK-HCST-LNK-LSFS----------------TEN-----VLN
-EEG--DQ-----------R-----------------------------------YMEEL
FICWKNMC-SVFVRSKDIQELRIKDTNFNEP---------AIRVLY--ESLKYP------
SF--T-LNKLVANN---------VS--FGDNYVLFEL-----------------IQNSSL
QYLD-LSCS-FLSNS-DVKLLCDVLNQAECN----IEKL--------ML-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------ANCSLSEQCWDYLSEV-LR
PF13516 // ENSGT00900000140813_0_Domain_13 (2212, 2237)
QNKTLSHLD
I-SSNDLKDEGLK-VLCRSLTLP--YCV----L---------------------------
-----------------------KSLCL--SCC-----GITSRGCQDL------------
-------AEVL-R
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
NNQNLKYLHVSYNKLKDTGVRL-LCDAIK-HP-NC-HLKDLR--LE-
--A-CEITDASNE--------------------ELCFAFIQCKTL-QTLNLMGNAFEVSR
MVFFPR--------F---------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSOARP00000009163 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------APAPSMEPR--AAAA
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 326)
REL---LLGALEDLS--Q---EHLKRFRHKLRD------
------------QRVDG-----------RNI-PWGR-LEGAD-TLDLVELLVHFYGPERA
LDVAQKT----LQ-RADVRDVAARLE-ERRLQS---------E---------CRA-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------SP-ARAGSFGFPQ------
----------------------------------------------------------RA
SACPGAPSPDAPFHT--PPGPRD-------------------------------------
--------P----PQLSL---PGLGPSSPAQ-----------------------------
-----------------------------------------L------------------
-----S-VS----GRSLSAESAGPE-LP-------------GHF-----GCPQS------
-----LGGS----------------AQLAPGEERGE------------------------
------------------------------------------------------------
-----------GVVRQQGKVRP-EP-A--GE-GTD------TAPPGRRSPARRGEGA---
-----------QLRRELRQLCRLA---RDGVLGG--R----AWFSEE-------------
--DLERLQLCGAKVQTQFLSKKEMPGVLET-----E-VTYQFIDQSFQEFLAALSYLLE-
DEGAP-------------------R-AAAGGIE-ALLQGDPEL--HSHLKLTTRFL----
----FGLLSTERMCEIERHFGC-----------------TASER-VKQ-QVLRWVQDQGQ
GRPRAAPEGTEEILAPQGTEESEE------------EEGEELDSPLELLYCLYETQE-DA
FVHQALRG---------------------L-------------------PELVLERVHFG
RM-DLAVL--S----YCVR-CCPA-GQA-LRLV------------------SC----RLA
------AA----------------------------------------QEKK-----KRS
----------------------------------------LMKR----------------
------LQGS---------------------------------------------SRATE
RK---PPGS-------PLRPLCEAMIDRQCG----LNSL--------TLS---------R
C-----------------------------------------------------------
--------KLS---DS-VCR---DLSEALRE-APALAELGLFHNGLS-EAGLRVLSEG--
-LASPQCRVQTL------------------------------------------------
----------------------------RVQ-----------------------------
---------------QPGL---------QEALQYM-------------------------
------------------------------VGKLQQIPALTT---LD-LSGC-QLSGPAL
--T--------CLCTTL----KGPGCRLQTLRL--TSVELSEQ-----------------
----------SLQELRAVETANP-------RLV----------------------I--TH
PALD-------------------VHPKPPKVSISAL------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSRBIP00000022982 (view gene)
ENSAMEP00000004761 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------SSNSP
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
FENG---VMLYMVYLS--K---EELQTFKQLLVD------
---------E--NPRPG--------S--VQI-TWDQ-VKTAR-WGEVVHLLVEFFPGRLA
WDVTHDI----FA-KMNQTELCLRVQ-MELNDTAY-VVSIDTLSVG------R-------
------------------------------------------------------------
------------------------------------------------------------
-------SST-------------------------------RSKLPPV--YQLEEYRLH-
--------MMDE----------------YSTI--RIMTTWFGSHVDFFYQDI--------
----------------------------------------------ERHERFLPCLFLPK
RP--QGRQP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
KTVVLQGVAGVGKTSLAKKV-----MLEWA--E---------NKFYP
H--------KFWC-AFYFHCREVVQVD-------EQSFEE-LIAC-KW-----PR-VQSI
VSKIMS-KP----DQLLLLFDGFEE-VTLTLT------DRPA-D-----LSDDWSQK-LP
-GSVLLTSLLSKRMLPEATLLIT-LRFTSWRKLKPFL-K-----RPS--FITLTGFSMAE
RAKYF----------------R--TY-FRN---------KREADEALSFVMG-NTILFSM
CQVPAVCWMVCSCLRQQVERGA-DL-A--QA-YPNATAVFVQYLSSLFPSKP-GSLPRNT
------------HQKQLAGLCYLA---AEGM-WN-MR----WVFDTK-------------
--DLEQAKLDET-TVANFLRV----NIFQRVAG-DR-DHYAFAIISFQEFFAALLYVLC-
FPQRL--------------RNFW--VLDRVQIT-RLIAYPG-R-KKNYLAQMGLFL----
----FGLLNEKCASATGKSFGCK---------------L--SLG-NKK-KLLKVAALSYE
CS-----------------------------------PPTLHHGVPQLFYCLHEIRE-EA
FVSQILSN---------------------C-------------------QKAALIISKKK
DM---QVS--A----FCLQ-HCQR-LRE-LELT----------------ITV-----TVT
-QPQYLVF----SS--------------------SVSDDELCSVFSLAH--RPESS-DQC
FLWWQDFC-SVFRTHENLEVLALTNTSMETE---------SVNVLS--AALRH-P-----
GC--K-LRKLT--F-RCLHPS--ML---N-E-DLIQVL----------------TENRYL
RYLE-IQGT-EVRCE-AMEFLCTALKCPQCS----LQCV--------RLE---------D
C------S----------------------------------------------------
---------VT---PK-SWV---DLARDLGS-NKHLKTLMLRSSYLET-FGDIACQWP--
NWR-GCQHIYIF-A----------------------------------------------
------------------------LENCDLT-----------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------SLMCKSLSSS-LKSNGMLTHLS
L-AENALKDEGAK-ELCSALWHS--TCP----L---------------------------
-----------------------QRLVL--RNC-----ALTAACCQDM------------
-------ASAL-DKNK
PF00560 // ENSGT00900000140813_0_Domain_15 (2357, 2381)
TLRSLDLGFNSLKDDGAIL-LFEALT-KP-KS-GLQILE--LE-
--Q-CLFTSDCCP--------------------AMASMLLHNQSL-RYLDVSKNDIGLRG
TVLLRE--------AFQQRKGE--KV--V-------------------------------
---L--------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSAMEP00000004763 (view gene)
ENSCGRP00000003634 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------M---ASFFSDFG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 323)
-LMWYLKELN--K---KEFMKFKEVLRQ------
---------E--IAESG--------L--QHI-PWNE-VKKAS-REHLANLLVKHYE-QKA
WDVTFRI----FQ-NINRKDLSERAA-EEM------------AG--------------HS
K---------------------------------------------------MYQA----
------------------------------------------------------------
----------------------------------------------------------Y-
--------LKNM----------------LSHV--WSRKFKIAIECF--------------
---------------------------------G-------NEDLTQDNCSRLWNLFVPK
AN---EK-P----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
HTVLLIGMSGVGKTVFLAQL-----MRAWS--E---------GLMFQ
N--------EFSY-VFYLCCHEIKQLK-------TASLAD-LLSR-EW-----PRP-SAP
IAEIVS-QP----AKLLFIIDSLEG-LNCDWSEP------ES-E-----LCDNWMQK-CP
-VSILLSSLLRRKLLPESSLLIT-ATPEAFEKLGDRI-E-----YTV--KNFVTGFNENS
RRSCF----------------Q--RV-FQD---------EDTAQEVFTLVRE-NEQLYNI
CHVPSLCWMVATCLNDEVKRGG-HL-S--AV-CRCTTSVFAAYVLNLFIPQS-AHYPSK-
-----------KSQALLRGLCSVA---AEGM-WT-DN----FAFNQK-------------
--DLEKNGVFHS-DISTLLNM----KILLKSRD-SK-NLYTFFHPSIQEFCAALFYLVN-
SHVDH-----------------P--SKDVLCVE-TLLFTFLKK-VRVQWIFLGCFI----
----FGLLHKSEQAKLEAFFGYQ---------------L--SQE-VKQ-TLCQSLEKISA
DEE-----------------------------------LRKQIDDMKLFYCLFELED-DG
FVTQVMTF---------------------L-------------------PQIKFVAKEYS
DL---IVA--A----YCLK-NCST-LQK-LSIS----------------TQD-----ILQ
-EEK--DP-----------S-----------------------------------YLKKL
LLRWQDLC-SVLGSSQQIQVLQVKDTYLNEL---------AFLELH--NHLRNP------
SC--A-PRKFLVNN---------VSF-LCNNQLFFELL----------------TQNHNL
HHLD-LSLT-SLSRS-DVKLLCEVLNQKGCH----IEEL--------LL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------SGCQLS-----------------------------
----------------------------HDDCIIFAS-----------------------
--------------------------------ILISSKKLRH---LN-VSYNN-LD----
-------KGMYLLSKAL----CHPDCVLKDL-----------------------------
------------------------------------------------------------
------------------------------VLASCSLSKQCWNYLSDV-LKR
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
NKTLSHLD
I-SSNDLQDEGLK-VLCQALSLP--DSA----L---------------------------
-----------------------KSLFL--RRC-----LITSSGCQYL------------
-------AEVL-S
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
SNQNLISLQVSNNKLEDAGVKL-LCEAIK-EP-NC-RLENLG--LE-
--A-CELTGACCE--------------------DLVSALSQNETL-WAINLLKNALDLRG
LAMLFE--------ALNVHGCS--PN--V-------------------------------
---LGLQIT-IPDKETQEL------LI----------AEK-KKNPYLAILSR-V------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSRNOP00000034130 (view gene)
ENSPPYP00000003320 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-MDQPEAPCSGTGPR--LAVA
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 328)
REL---LLAALEELS--Q---EQLKRFRHKLRD------
------------VGPDG-----------RSI-PWGR-LERAD-AVDLAEQLAQFYGPEPA
LEVARKT----LK-RADARDVAAQLQ-EQRLQR---------L---------GLG-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------SG-ALLSVSEYKKKYREH-
--------FLQL----------------HARV--KERNARS------------YSITKRF
TKLLIAPESA--------------------------------------------------
------------------------------------------------------------
--------------------GEL
PF05729 // ENSGT00900000140813_0_Domain_2 (744, 940)
LERPG------TRSLAD-LILD-QC-----PD-RGAP
VPQMLA-QP----QRLLFILDGADE-LPALGGP--------EAA-----PCTDPFEA-AS
-GARVLGGLLSKALLPTALLLVT-TRAAAPGRLQGRL-C-----SPQ--CAEVRGFSDRD
KKKYF----------------Y--KF-FRD---------ERRAERAYRFVKE-NETLFAL
CFVPFVCWIVCTVLRQQLELGQ-DL-S--RT-SKTTTSVYLLFIASVLSSAPVADAP---
-----------RVQGDLRNLCRLA---REGVLGR--R----AQFAEK-------------
--ELEQLELRGSKVQTLFLSKKELPGVLET-----E-VTYQFIDQSFQEFLAALSYLLE-
DGGVP-------------------R-TAAGGVG-TLLRGDAQP--HSHLVLTTRFL----
----FGLLSAERMHDIERHFGC-----------------MVSER-VKQ-EALRWVQGQGQ
GCPGATPEVTEGAKGLEDTEEPEEE-----------EEGEEPNYPLELLYCLYETQE-DA
FVRQALCR---------------------L-------------------PQLALQRVPFC
RM-DVAVL--S----YCVR-CCPA-GQA-LRLI------------------SC----RLV
------AA----------------------------------------QEK--K---KKS
----------------------------------------LGKR----------------
------LQAS---L--------------GGS------S----------------SSRGTT
KQ---LPAS-------LLHPLFQAMTDPLCH----LSSL--------TLS---------H
C-----------------------------------------------------------
--------KVP---DA-VCR---DLSEALRA-APALTELGLLHNRLS-EAGLRMLSEG--
-LAWPQCRVQTV------------------------------------------------
----------------------------RVQ-----------------------------
---------------LPNP---------QRGLQYL-------------------------
------------------------------VGMLRQSPALTT---LD-LSGC-QLPSPMV
--T--------YLCAAL----QHQGCGLQTLSL--ASVELSEQ-----------------
----------SLQELQAVKRAKP-------DLV----------------------I--TH
PALD-------------------SHPEPP-KELISTF-----------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSRROP00000000525 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------------------------M----
----FPELRHEE------LLTDG---------------N--RG-----------RVQSFQ
EGPAALQNLSSQPRPFRVSH-N---------------NSSSICSIFSSLVAKRGVSE-SP
AC-ETVTG---------------------F--------------------GQDVLFLLPP
TQ-PLFTS--T----MSLDIQCEQ-LSD-ARWA----------------EL---------
----LPLL----QQ---------------------------CQVV------RLDDC-GLT
EARCSDIS-SALRV
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1542)
NPALTELSLRGNELGDA---------GVHCVL--QGLQN-P-----
SC--K-IQKLS--L-QNCCLT--GA---GCG-VLSSTL----------------RSL
PF13516 // ENSGT00900000140813_0_Domain_6 (1618, 1642)
PTL
QELH-LSDN-LLGDA-GLQLLCEGLLDPQCR----LEKL--------QLE---------Y
C------N----------------------------------------------------
---------LS---AA-SCE---PLASVLRA-KPDFKELTVSNNDINE-AGVRVLCQG--
LKD-SPCQLEAL-K----------------------------------------------
------------------------LESCGIT-----------------------------
----------------------------SDNCRDLCS-----------------------
--------------------------------IVASK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
ASLRE---LA-LGSNK-LGD---
-------VGLAELCPGL----LHPSS----------------------------------
--------------------------SLR-TLW---------------------------
-------------------------------IWECGITAKGCGDLCRV-LRTKE
PF13516 // ENSGT00900000140813_0_Domain_13 (2215, 2237)
SLKELS
L-AGNELGDEGAR-LLCETLLEP--GCQ----L---------------------------
-----------------------ESLWV--KSC-----SFTAACCSHF------------
-------SSVL-TQ
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NKFLLELQISNNRLGDAGVQE-LCQGLG-QP-GS-VLRVLW--LG-
--D-CDVSDRSCS--------------------SLAATLLA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2464)
NHSL-RELDLSNNCLGDAG
VLQLVE--------SVRQPGCL--LE--Q-------------------------------
---LVLYDI-YWSEAMEDR------LQ----------ALE-EEKPS-----LRVIS----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCHIP00000015005 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------MYYMGPA-----
--------CTWS------------------------------------------LLCE--
-----------------------------------------QK-----------------
--------------------------------------------------DYREVFRKH-
--------LKEK----------------YLKI-GVDRCYHRGK--------------HIE
TGLLVVRKTLYNRKATIGFY------------------------------------LIPN
CG--ALGMC----W--------------------------DWE--N----------MVTP
G--------RFDY-LIYVNCREV----------------A-ILLT-L-------V-IQVD
QSWTLFLYI---
PF05729 // ENSGT00900000140813_0_Domain_2 (793, 922)
-RRSLFILDGFPE-LQYSVSDQ------EE-D-----LSTNPLER-KP
-VETLLHSIMKTKLFPESSLLIT-AQPTTMKKLHSLL-----------------------
-KAYF----------------L--SQ-FSG---------ANAATRVLYDLRE-NEGLDIM
SSRPITSWMSHR-------------------------------KMECLNTLTGIS-VW--
-----------KGQICRWGLCSLA---AEGL-QN-QQ----VLFEVS-------------
--DLRRHRLGVYDTNCTFLNH-----FLKKVEG-VV-SVYTFLHFSFQEFLTAVFYTLK-
NGKIF----------------------Q-------------QY-EKGFSSLTIQFL----
----FGVLHKGKEKAVQTTFGKK---------------V--SPG-LRE-ELLKWTEKEIK
DK-----------------------------------PYRLQIEPIDLIIND--------
-----------------------------L-------------------RSIILLQLMYT
KM-DILAV--S----FSVK-SSHSHLS----------------V-----SLKCQHLLGF-
-EEE--------------------------AFMP--------------------------
---------PMLTISETFQ-----LHNLPIC---------PLHLLC--QPLHNP------
YC--K-IKDLKL---IFCHLT------ASYGRD---------------LALVFET-NQYL
TDLG-FVKN-TLEDS-GMKFLCEGLKQPN-------------LMR---SLVCSLKVVPAS
CG------------AL----ATVHGTNQWLTELEFSETKLEALALKLLRECLKDPNCK-V
QKLKLSASFLLGSSET-IGR---YLASVLIC-
PF13516 // ENSGT00900000140813_0_Domain_8 (1773, 1794)
NPNFTELDLSENLLGD-TGVKFLRDG--
LRH-SNCKVEKQ-D----------------------------------------------
------------------------LNTCYLT-----------------------------
----------------------------DGSCVYLFS-----------------------
--------------------------------FLQVSQTLKE---LF-VFANS-LRD---
-------TGVQHLCEGL----CLVNGITKNL-----------------------------
------------------------------------------------------------
------------------------------VLSECSLSPLCCESLTQV-LSSTRSLTRLL
L-INNKIEDLGLK-LLCEGLKQP--DCQ----L---------------------------
-----------------------KDLVL--WTC-----HLTGECCQHS------------
-------CNSL-YTNG
PF13516 // ENSGT00900000140813_0_Domain_15 (2357, 2378)
HLRVLELSASALGDEGMQL-FCEGLK-RP-SC-KLQTLR--LA-
--E-CHFTGACCG--------------------ALTSVLSR
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2465)
NENL-TLLDLSGNHLKGFG
VQMLCD--------ALLHPICK--LQTFY-------------------------------
------VDTDHLHEETFRK------IG----------ALN-VSKPGITW-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSSTOP00000026826 (view gene)
------------------------------------------------------------
------------------------------------------------------------
-------------PRCP-------------------GSGPCPLWTPRTP----------G
VRGRGHLAAYINPNR--AAAAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLAALEDLS--Q---EQLKRFRHKLRD------
------------APMDG-----------RSI-PWGR-LERAD-AMDLAEQLTHFYGPEPA
LDVARKT----LK-RADVRDVAARLK-AQRQQR---------L---------GPS-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------SS-ALLSVAEYKKKYREH-
--------VLRQ----------------HAKV--KERNARS------------VKINKRF
TKLLIAPETAALEEA--PLGLAE-------EPEP--------ERARRSDTHTFNRLFKRD
Q---EGQRP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYQ
G--------QVDF-AFFMPCHELLELSG------TRSLAD-LILD-QC-----PD-RGAP
VRQMLA-QP----QRLLFILDGADE-LPATGTA--------EAA-----PCTDPFEA-TS
-GTRVLGGLLSKALLPSARLLVT-TRTAAPGRLQGRL-C-----SPQ--CAEVRGFSDKD
KKKYF----------------Y--KF-FQD---------EWRAERAYRFVKE-NETLYGL
CFVPFVCWIVCTVLRQQLDLGQ-DL-S--RT-SKTTTSVYLLFIASVLSST-GADGS---
-----------WVQRELRKLCLLA---REGVLRR--K----AQFTEK-------------
--DLEKLELRGSKVQTLFLCKKEMPGVLKT-----E-VIFRFIDQSFQEFFAALSYMLE-
EEGAS-------------------G-APTGSVG-SLLRTDTEL--SGHLALTTRFL----
----FGLLSTERIRDIEHHFGC-----------------VVPER-VKE-DALRWVQGQGC
PR-----TASEGME------DTEE-L---------EEEGEEPNIPLELLYCLYETQE-EA
FVRQALSS---------------------L-------------------PELALERVRLS
RM-DLEVL--S----YCVR-CCPA-GQA-LRLV------------------NI----GLA
------AA----------------------------------------QEKK-K---KKS
----------------------------------------LVKR----------------
------LQGS----------------------------------------------RAST
KQ---LLGS-------PLRPLCEAMTDQQCG----LSSL--------TLS---------Y
C-----------------------------------------------------------
--------KLP---DA-ICR---DLSEALKV-APALTDLGLLHNRLS-EAGLRMLSEG--
-LAWPRCRVQTL------------------------------------------------
----------------------------RVQ-----------------------------
---------------LSGL---------PGALQSL-------------------------
------------------------------VGMLRQSPALTT---LD-LSGC-QLSDPTV
--T--------YMCAVL----QHPSCSLQTLRL--ASVELSEL-----------------
----------AKQELQAVKTAKP-------GLV----------------------I--IH
SE----------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSNGAP00000023365 (view gene)
ENSMNEP00000010656 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------MAESNSTDFD---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWYLENLS--D---KEFQSFKKYLAR------
---------K--ILDFK--------L--PQV-PLIN-LRA-T-KEELANLLPISYEGQHI
WNMLYSI----FL-MMRKEDHCMKI---IGRRN---------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------RNQEACKAL-
--------MRRK----------------ITLR--WENHTFGEFHYKFFCNVA--------
----------------------------------------------SDVFYVLQLAYDST
SC--YSAND----LN
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 930)
VFLMGDRASGKTVTINLA-----VMKWI--N---------GEMWQ
N--------MISY-VVHLTSHEINQMT-------DSSLAE-LIAK-DW-----PD-GQAP
IADILS-DP----KKLLFILEDVDN-LRFELNVN------EH-A-----LCSNSTQK-VP
-IPILLVSLLKRQMAPGCWFLIS-SRPTREENIKMFL-K-----TTD--CYVTLQFSNEK
RELYF----------------H--SF-FND---------RQRASTALMLVHD-DEILTGL
CRVAILCWITCTVLNRQMDKG-HDP-Q--HC-CETPTDLHAHFLADALTSKAGLT-AN--
----------QYHLGLLRRLCLLA---AGGL-FL-ST----LNFSAE-------------
--DLRCVGFTEV-DFSVLQAV----NILLPSST-HK-DRYKFIHLNVQEFCAAIAFLMAV
PNCLI-----------------P----SGSR-G--Y--KEKRE-QYCNFNQVFTFI----
----FGLLNENRRKILETTFGYQ---------------LPMVES-FRR-YSVEYMKHLGR
DQ-------------------------------------QKLTHHGSLFYCLFENQE-EE
FVKKVANT---------------------L-------------------KEVTLYLQSNK
DM---MVS--L----YCLE-YCCH-LRT-FKLS----------------VQR-----IFE
--YKEPLV-------------------------------------------RPTASQMKS
LVYWREIC-SLFCTMDTLWELHLFDSDLNDI---------SERILS--KALEHS------
SC--K-LRTLKLSY---------IST-ASGFEDLLKAL----------------ARNRSL
IHLS-LNCT---------------------------------------------------
------------------------------------------------------------
--------------------------------------------SISL-NMFSLLREI--
LDK-STCQIRHL-S----------------------------------------------
------------------------LMKCDLR-----------------------------
----------------------------ASECGEIAS-----------------------
--------------------------------LLVNGGSLRK---LT-LTNNP-LRN---
-------DGVNILCNAL----LHPNCTLTSL-----------------------------
------------------------------------------------------------
------------------------------ALVFCCLTEQCCSSLGRV-LLLGRTLKQLD
L-CVNHLQNYGVL-HVTFPLVFP--TCQ----L---------------------------
-----------------------EELYL--SGC-----FFTNDICQYI------------
-------AIVIA-T
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NETLRSLEIGSNSIEDAGMQL-LCGGLR-HP-DC-MLVNIG--LE-
--E-CMLTGACCE--------------------SLASVLT
PF13516 // ENSGT00900000140813_0_Domain_17 (2441, 2464)
TNKTL-ERLNILQNQLGDEG
VVKLLE--------SLILPECV--LQ--I-------------------------------
---IGLPLN-DVSAETRRM------VM----------TVK-ERKPNLIFLSETWSVKEGR
EIGVI-------------------------------------------------------
-------------------------------------------------------PASQP
GSV-----------------IPNSNLD---YM--FFKFPRMSTAMRMSSTASRQPL----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSGGOP00000023354 (view gene)
ENSPTRP00000019828 (view gene)
ENSMEUP00000004563 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------------ETLFDSD
AN--TSNAP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
TTVVLQGPAGIGKSTLARKV-----LQGWA--A---------GTLYP
G--------RFDY-IFYIGYQEARLLRE-------NSVDH-LITT-LC-GN-----NSAP
VSDILK-QP----QKLLFVLDGLDE-LHFLSAEE------SW--------------K-RS
-LDILLGDLIQKELLPQSTLLIT-VRPPTQEKLGPWL-M-----HPR--YVKVLGFSQKQ
KKEYF----------------Y--LY-FMN---------RDLAKKAFDFVQT-NKTISDE
CCAPLLCWIICCWMKRRMEQGK-SP-T--KA-LKNRTDIYMAYVSTFLPTEK----CLSK
----------PRQHVALRGLCSLA---TEGI-QR-QK----ILFTEG-------------
--DLRRHNLDGVDTS-AILYV----SGSQEGLG-GT-MLYSFRHLSFQEFFSAMFYLLE-
DEISGK---------------------ESQNLK-ELME-------RRH--QSVQFL----
----HELLKKENEKNLELKFNLR-----------------VSLP-LRK-EV-EIKKEDE-
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCJAP00000025340 (view gene)
ENSECAP00000017692 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------MVPSG----QL
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 326)
DFN---LQPLLERLD--Q---DELSQFKSLLRT------
---------L--SLQDE--------L--QHI-PQTE-VEEAS-GKQLAEILTNQCPRNWV
EMVTIQV----FD-KMNRTDLSERAK-DEFK--AL-KSLQ----ENK------PPSLG--
------KSSTDSPL----------------------------------------------
------------------------------------------------------------
-------------------------------------------H----YPIQKKG-WQE-
--------KIQK----------------FCQI--WEKNFWPGASENVHI-----------
---------------------------------------------VTQRYETLIPFCNPK
ML--TGPFP----HT
PF00004 // ENSGT00900000140813_0_Domain_2 (676, 854)
VVLHGPAGIGKTTLAKKL-----MLDWT--Q---------DNLAE
---------TFNA-AFYLSCKELNHKG-------ACTFAE-LISK-NW-----PD-LQDA
IPEIFA-QA----QKVLFIIDGFDE-LRVPSG------ALIH-D-----MCCDWQKQ-KP
-VPVLLGSLLKRKILPKATLLIT-TRPGTLRELRLLV-E-----QPL--FIEIEGFLELD
RKAYF----------------L--KH-FEE---------EVQALRAFDLMKS-NAALFHL
GSAPAVCWIVCTCMKLQMEKGE-DP-A--PT-CQTITSLFLHFLCSHCPGPF--------
------------LQVPVTALCLLA---AGGI-WT-QT----SMFDGE-------------
--DLERLGVKES-DFHPFLDK----NILQKGKD-CE-GCYSFIHLSVQQFLAALFYVLE-
REEEE-------------DGDSC--RWDIGGVQ-KLLSKEERL-KNPSLTHVGYFL----
----FGLLNEKRARELETTFGCQ---------------V--SLK-IKQ-ELLECRVKSTE
NK-----------------------------------PLS-VTEMQEVLYCLYESQE-ER
LVKDAMAH---------------------I-------------------KETSLHLKNKM
DI---IHA--S----FCLQ-HCRN-LQK-ISLQ----------------VEKG----VFL
-END-TAS----ES--------------------DTQVESC-GMW--DAASAFQND-EHV
LPFWMGLC-SIFGSNKNLIFLDISQSFLSTS---------SVKILC--EKIAS-A-----
TC--N-LQKVV--L-KNISPA--DA----YQ-NFCITF----------------GGHKTL
THLT-LQGN-DQDD--MFPSLCSVLRHPKCH----LQYL--------SLV---------S
C------S----------------------------------------------------
---------AT---TH-QWA---DLSSSLEI-NQSLTCLNLTANELLD-EDAKLLYMT--
LRH-PKCFLQRL-S----------------------------------------------
------------------------LENCHLT-----------------------------
----------------------------EASCKELSS-----------------------
--------------------------------ALIVNQRLTH---LC-LAKNA-LGD---
-------GGVKLLCEGL----SYPEC----------------------------------
--------------------------QLQ-TLV---------------------------
-------------------------------LWYCNITSDGCNHLSTL-LQQNSSLMHLD
L-GLNHIGIIGLK-FLCEALKKP--LCN----L---------------------------
-----------------------RCLWL--WGC-----AITPFSCEDL------------
-------SSAL-SSNQNLTTLDLGQNSLGYSGVKV-LCDALK-LQ-SC-PLQTLR--LK-
--I-DESDARIHK--------------------LLKEIKDSNPQLTIE------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPTRP00000019821 (view gene)
ENSFDAP00000002232 (view gene)
ENSPTRP00000019824 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------MAESDSTDFD---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWYLENLS--D---KEFQSFKKYLAR------
---------K--ILDFK--------L--PQF-PLIQ-M---T-KEELANVLPISYEGQYI
WNMLFSI----FS-MMRKEDLCRKI---IGRRN---------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------RNQEACKAV-
--------MRRK----------------FMLQ--WESHTFGKFHYKFFRDVS--------
----------------------------------------------SDVFYILQLAYDST
SY--YSANN----LN
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 930)
VFLMGERASGKTMVINLA-----VLRWI--K---------GEMWQ
N--------MISY-VVHLTSHEINQMT-------NSSLAE-LIAK-DW-----PD-GQAP
IADILS-DP----KKLLFILEDLDN-IRFELNVN------ES-A-----LCSNSTQK-VP
-IPVLLVSLLKRKMAPGCWFLIS-SRPTRGNNVKSFL-K-----EVD--CCTTLQLSKEK
REIYF----------------N--SF-FKD---------RQRASAALQLVHE-DEILVGL
CRVAILCWITCTVLKRQMDKG-RDF-Q--LC-CQTPTDLHAHFLADALTSEAGLT-AN--
----------QYHLGLLKRLCLLA---AGGL-FL-ST----LNFSGE-------------
--DLRCVGFTEA-DVSVLQAA----NILLPSST-HK-DRYKFIHLNVQEFCTAIAFLMAV
PNYLI-----------------P----SGSR-E--Y--KEKRE-QYSDFNQVFTFI----
----FGLLNANRRKILETSFGYQ---------------LPMVDS-FKW-YSVGYMKYLDR
DP-------------------------------------EKLTHHMPLFYCLYENRE-EE
FVKTIVDA---------------------L-------------------MEVTVYLRSDK
DM---MVS--L----YCLD-YCCH-LRT-LKLS----------------VQH-----IFQ
--NKEPLI-------------------------------------------RPTASQMKS
LVYWREIC-SLFYTMESLWELHIFDNDLNGI---------SERILS--KALEHS------
SC--K-LRTLKLSY---------VST-ASGFEDLLKAL----------------ARNRSL
TYLS-INCT---------------------------------------------------
------------------------------------------------------------
--------------------------------------------SISL-NMFSLLHDI--
LHE-PTCQISHL-S----------------------------------------------
------------------------LMKCDLR-----------------------------
----------------------------ASECEEIAS-----------------------
--------------------------------LLISGGSLRK---LT-LSSNP-LRS---
-------DGMNILCDAL----LHPNCTLISL-----------------------------
------------------------------------------------------------
------------------------------VLVFCCLTENCCSALGRV-LLFSPTLRQLD
L-CVNRLKNYGVL-HVTFPLLFP--TCQ----L---------------------------
-----------------------EELHL--SGC-----FFSSDICQYI------------
-------AIVIA-TNEKLRSLEIGSNKIEDAGMQL-LCGGLR-HP-NC-MLVNIG--LE-
--E-CMLTSACCR--------------------SLASVLTTNKTL-ERLNLLQNHLGNDG
VAKLLE--------SLISPDCV--LK--V-------------------------------
---VGLPLT-GLNTQTQQL------LM----------TVK-ERKPSLTFLSETWSLKEGR
EIGVT-------------------------------------------------------
-------------------------------------------------------PASQP
GSI-----------------IPNSNLD---YM--FFKFPRMSAAMRMSNTASRQPL----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSRBIP00000018090 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MSLDIQCEQ-LSD-ARWA----------------EL---------
----LPLL-------------------------------QQCQVV------RLDDC-GLT
EARCSDIS-SALRV
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1542)
NPALTELSLRGNELGDA---------GVHCVL--QGLQN-P-----
SC--K-IQKLS--L-QNCCLT--GA---GCG-VLSSTL----------------RS
PF13516 // ENSGT00900000140813_0_Domain_6 (1617, 1642)
LPTL
QELH-LSDN-LLGDA-GLQLLCEGLLDPQCR----LEKL--------QLE---------Y
C------N----------------------------------------------------
---------LS---AA-SCE---PLASVLRA-KPDFKELTVSNNDINE-AGVRVLCQG--
LKD-SPCQLEAL-K----------------------------------------------
------------------------LESCGIT-----------------------------
----------------------------SDNCRDLCS-----------------------
--------------------------------IVASK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
ASLRE---LA-LGSNK-LGD---
-------VGLAELCPGL----LHPSS----------------------------------
--------------------------SLR-TLW---------------------------
-------------------------------IWECGITAKGCGDLCRV-LRTKE
PF13516 // ENSGT00900000140813_0_Domain_13 (2215, 2237)
SLKELS
L-AGNELGDEGAR-LLCETLLEP--GCQ----L---------------------------
-----------------------ESLWV--KSC-----SFTAACCSHF------------
-------SSVL-TQ
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NKFLLELQISNNRLGDAGVQE-LCQGLG-QP-GS-VLRVLW--LG-
--D-CDVSDRSCS--------------------SLAATLLV
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2464)
NHSL-RELDLSNNCLGDAG
VLQLVE--------SVRQPGCL--LE--Q-------------------------------
---LVLYDI-YWSEAMEDR------LQ----------ALE-EEKPSLRVIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMFAP00000007813 (view gene)
------------------------------------------------------------
------------------------------------------------------------
-------------M-SDVNPPSD------------T-PIPFSSS-------------S-T
H---SSCNPPWTFSCYPGSPCENG---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
VMLYMRYVS--N---KELQWFKQFLLN------
---------E--L-RAG--------T--TPI-TWDQ-VKRAS-WAEVVHLLIERFPGRRA
WDVTSKI----FA-VMNCDKMCALVH-REINAILP-TLEPEDLNVG------E-------
------------------------------------------------------------
------------------------------------------------------------
-------TEV-------------------------------NLEEGES--GKIQQYKLN-
--------VIVK----------------CFPT--WDFMTWPGNQRDFFYQDV--------
----------------------------------------------HRHMEYLPCLLLPK
RP--RGRQP----K
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 928)
TVVIQGVPGIGKTTLAKTV-----MLEWA--S---------NRFYA
H--------KPWC-AFYFHCQEMNQTT-------EHSFSE-LIEH-KW-----PG-SQDL
VAKIMS-KP----DQLLLLLDGFEE-LTSTLI------NRPE-D-----LSEDWRQK-LP
-GSVLLSSLLSKRMLPEATLLIM-IRCTSWPTCKPLL-K-----RPS--LVTLPGFNTTE
KIKYF----------------Q--MY-FGY---------GE-TNRVLSFAME-NTVLFSM
CQVPVVCWMVCSGLKQQMERGN-NL-T--QA-CPNATSVFVRYISSLFPTRA-ENFSRKI
------------HQAQLEGLCHLA---ADGM-WH-RQ----WVFGKE-------------
--DLEEAKLDQM-GVAAFLGM----SVLRRIAG-EE-DRYTFTLLIFQEFFAALFYVLC-
FPQRL--------------QNFH--VLSHMNIQ-RLIASPR-G-SKSYLSHMGLFL----
----FGFLNGTCALAVEQSFRCK---------------V--SFN-NKR-KLLKVIPLLQK
CE-----------------------------------PPSPCSGVPQLFYCLHEIRE-EA
FVSQALNG---------------------Y-------------------HKAVLKIGNSK
DV---QVS--A----FCLK-RCEH-LQE-VELT----------------VTL-----NLK
-NVWKLSS----G-------------------------------SRPGS--EAPES-KEL
HCWWQDLC-SVFATNDKLEVLTLTNSALGPP---------FLKALA--AALKH-P-----
QC--K-LQKLL--L-RRVNST--ML---N-Q-DLIGVL----------------MGNEHL
RYLE-IQHV-EVESN-VVKLLCKALRSPRCR----LQCL--------RLE---------D
C------L----------------------------------------------------
---------AT---PR-IWT---DLGSNLQV-NRHLKTLILTKNSLEN-CGACY------
--L-SVAQLERL-S----------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------K
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2236)
SKMLTHLS
L-AENALKDEGVK-HIWNALSHL--RCP----L---------------------------
-----------------------QRLVL--RKC-----DLTFNCCQDM------------
-------ISAL-C
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
KNKTLKSLDLSFNSLKDDGVIL-LCEALK-NP-DC-TLQILE-----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSOARP00000003424 (view gene)
-----------------------------------------------------MPWRRAW
QPTPVF----------------------SPGESHAQRSLVGYSPWGRKESDTPEQLTLHP
SNR-----------SHWLRLPKT------------T-------------------FPF-C
L---SAQPCVRMREAKTAPFS
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 326)
SYG---LQWCFEQLG--K---EELQTFKALLKE------
---------H--ASEPA--------A--CSF-PLVQ-VDRAD-AASLASLLREHCRASLA
WKTSIGI----FE-KMRLFALSEMAR-DEMKKYPL-AETSEDSAPAKT--DQGPSMKEVP
ALA-SSKVSGTAYGGRQTRKAS-------------------------------AVSAG--
KS-GP------------VKGT-Q-------------------------------------
----IYFPGH-------------------------------PSALKREDPQHSRDYRIH-
--------VMTT----------------FSTR--LDTP-QHFEEF---------------
-----------------------------------AS-------ECPDA-HVLSGAFNPD
PS--GGFPG----L----SRP
PF05729 // ENSGT00900000140813_0_Domain_2 (682, 905)
PGVGKSMLARRL-----LLFWA--Q---------GDLYQ
G--------LFSY-VFLLRARDLQGSR-------ETSFAE-LISR-EW-----PD-APVP
VEKVLS-QS----KRLLIMVDGLEE-LELTLE--------DS-S-----LSANWVER-QP
-VAVLAHSLLKKVLLPECALLLT-VQDAGVQ---ALL-L-----SPP--YLWVGGLSVEN
RVQLL----------------L--GG-GKH---------GRQETRAWHAGTD-HQEVLDK
CQVPAGG----GPFPXHGEPEK-GL-P--VP-GHTLTGLYAAFVFQRLAPKD-AGRRALS
----------REERSALKGLCRLA---VDGV-WN-AK----FVFDGD-------------
--DLGVHGLKGP-ELSALQQA----SILLPDSR-CGRG-HAFSHLSLQEFFAALFYVLR-
GVEGD--------------GEGY--PLFPQSTK-SLMELRHVD-LNVHLVQMKRFL----
----FGLVSKEATRALETLLGCP---------------V--AP-VAKQ-CLLHWICLLGQ
HPA----------------------------------APAASPDLLEAFYCLFETQD-DE
FVRLALNG---------------------F-------------------QEVWLQLNRPM
DL---TVS--S----FCLR-RCQH-LRK-VRLD----------------VRE-----T-P
-EDESAEA----WP--------------------GA-----------PQ--GLKIK-TL-
DEHWEDLC-SVLGTHPNLRELDLSGSVLSKE---------AMKTLC--VKLRQ-P-----
AC--K-IQNLI--F-TGARV---TP---GLR-HLWMTL----------------LVNRNV
TRLD-LAGS-RLGEE-DVRAAGAALRHPHCA----LQTL--------RLD---------R
C------G----------------------------------------------------
---------LT---PA-SCR---VISQVLAT-SVSLKSLSLMGNKVAD-QGVKSLCDA--
LKV-APCTLQKL-I----------------------------------------------
------------------------LGSCGLA-----------------------------
----------------------------ATTCQDLAS-----------------------
--------------------------------ALIENQGLTH---LS-LSGDE-LGS---
-------EGMSLLCRAV----KLPSC----------------------------------
--------------------------GLQ-KLA---------------------------
-------------------------------LNACSLDVAGCGFLALA-LMSNRHLTHLS
L-SMNPLEDPGMN-LLCEVMMEP--SCP----L---------------------------
-----------------------QDLDL--VNC-----RLTAACCESL------------
-------SNVI-TRSRHLRSLDLAANALGDEGIVA-LCKGLK-QK--N-ALTRLG--LE-
--A-CGLTSEGCK--------------------ALSAALICSRHL-ASLNLMRNDLSPGG
MAMLCS--------AFMHPSSN--LQ--T-------------------------------
---IGLWKE-QYPARVRRL------LE----------QVQ-RLKPHVVISVAWYSEE---
--E---------------------------------------------------------
--------------------------------------------------SD-FQGEIE-
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCGRP00001001900 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------RSLF--
L-----------------SVAEDSRTHQEEDRE------PIPSDQGPDE-----------
--------GQ-------------------------------APGLHAGLGCDLQTYKAH-
--------MITK----------------LSTS--LEIC-Y--------------------
--------------------------------------------DHPEM-RVLSEAFKPD
Q---NTFRP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
HTVVLHGKPGIGKSALARSV-----ILGWA--Q---------DKLYQ
D--------I-SY-AFFFSPREMQWAE-------KSSLAQ-LISR-EW-----AD-SQAP
MTEIMS-QP----ERLLFVVDDFDD-LDSALPQE------DT-R-----LCEDWKDE-QP
-IHTLLYSLLKKTLLPQSFLLVT-TRDTGLQKLKSMV-V-----SPL--YILVEGLSATK
RTQLV----------------L--EN-VSD---------DHRRKQVIRSMLD-NQWLAEQ
CRVPSICSLVCKALKIQEKLGG-SC-T--PV-CQTLTSLYSTLVFHQLTPRE-PFRGCLS
----------PEERGTLMGLCRMA---AEGV-WS-KR----SVFYDN-------------
--DLKTYGLKES-EISALFGM----DVF-QASH-SAKRCYAFFHLSLQDFFAALHYVLE-
GLG-T--------------WNQS--CFVIKNLR-SLSELKGTH-FNAHLGGMKRFL----
----FGLVNKDTMRTLEVLLGYP---------------L--SP-VVNQ-VLQHWVFMLGQ
QAN----------------------------------DTS-PMDVLDAFRHVFESQD-KD
FVCSALKS---------------------F-------------------REVEIQVNQRT
DL---VVS--S----YCLQ-HCKN-LEA-LRVD----------------VSD-----IFS
-VEMNTEL----LW--------------------PD---LHLKPISKCC--MTQGK-PLI
MECWEDFC-SILGTHPKLKQLDLGSSILSEW---------AMKIMC--LKLKN-Q-----
AC--K-VQSLT--F-KNAEV---VS---GLR-HLWMTL----------------VSNRNL
KYLH-LGNT-LMEEN-DIKLACQALQHPKCS----LETL--------RLD---------S
C------A----------------------------------------------------
---------LT---PD-CYL---MLSNLLLS-TTSLRCLSLAGNKVTE-KSMRWLCDA--
LNS-SKCALQKL-I----------------------------------------------
------------------------LDSCELM-----------------------------
----------------------------PVSCHILAS-----------------------
--------------------------------ALCGNRTLTH---LC-LSDNS-LRT---
-------EEVRRLCRFM----SNPEC----------------------------------
--------------------------ALQ-RLI---------------------------
-------------------------------LKQCNLRGDAYGFLALM-LSH
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
NGKLTHLN
L-TMNPVEDGGMK-LLCDAIKKP--TLD--------------------------------
---------------------------------------------RDL------------
-------ASVI-TR
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NQHLKSLDLSSNALGDSGVIA-LCKALK-QN-NS-SLRRLG--LE-
--A-CGLTSDCCT--------------------ELSLALSC
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2465)
NSRL-TSLNLMKNDFSTPG
MLKLCS--------AFLHPTSN--LW--I-------------------------------
---IGLWKQ-QYYAKVRRQ------LE----------KVQ-FIKPHMVIDGDWYAFD---
--E---------------------------------------------------------
--------------------------------------------------DDRNWWKN--
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSSSCP00000047938 (view gene)
ENSNLEP00000013044 (view gene)
ENSOPRP00000012332 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------SEP--LAA
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
AREL---LLAALEDLS--Q---EQLKRFRHKLRD------
------------ARAGG-----------RGI-PWGR-LELAD-AMDLAEQLVHFYGPEPA
LDVARKT----LQ-RADVRDVAARLK-EQRRQR---------L---------GTG-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------SS-AMISVAEYRKKYREH-
--------VLRQ----------------HAKV--RERNARS------------VRINRRF
TKLLIAPEGTAGGD--VAPGTTD-------EPQE--------ERARRSDTRTFNRLFGRD
E---EGQRP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
LTVVLQGPAGIGKTMAAKKI-----LLDWA--A---------GKLYQ
G--------QVDF-AFFVPCRELLANPG------ARSLAD-LVLE-QC-----PD-RRAP
VPQMLA-QP----DRLLFILDGVDE-LPAPPGAL------EAAE-----PCTDPFEA-AS
-GARVLGGLLSKALLPAARLLVT-ARAAAPGRLR----------SPQ--CAEVYGFSDRD
RKKYF----------------Y--RF-FED---------ERGAARAYRLVKE-NQTLLAL
CFVPFVCCIVCTVLRQQLALGH-DP-A--HT-LRTATAVYLLFVTSFWTAA-GGA-----
-------------QAELRKLCGLA---REGTLER--R----ARFTEQ-------------
--DLGRLQLRGSPVQTLFLSKKELPGVLET-----E-VAYQFADLSFQEFFAALSYLLP-
DERAP-------------------ESVAARDLE-TLLPRDGRP--RAHLALTTRFL----
----FGLLSTERTREIERHFGC-----------------TVSAR-VKQ-DPLRWVQSQGQ
GC-----------QGLEAVEEPGEEKGEEADEEDEEEDGEELGEDL--NYXXXXXXX-XX
XXXXXXXX---------------------X-------------------XXXXXXXXXXX
XX-XXXXXXXX----XXXX-XXXX-XXXXXXXX------------------XX----XXX
------XX----------------------------------------XXXXXX---XXX
----------------------------------------XXXX----------------
------XXXX---X-----------------------------------------XXXXX
XXX--XXXX-------XXXXXXX-------------------------XX---------X
X-----------------------------------------------------------
--------XXX---------------------XXXXXXXXXXXXXXXXXXXXXXXXXX--
XXXXXXXXXXXXXX-XXX------------------------------------------
-------------------XXXXXXXXXXXX-----------------------------
---------------XXXX---------XXXXXXX-------------------------
-----------------------------XXXXXXXXXXXXXXXXXX-XXXXXXXXXXXX
XXXXXXXXXXXXXXXXX----XXXXXXXXXXXX--XXXXXXXX-----------------
----------XXXXXXXXXXXXX-------XLV----------------------I--VH
SALG-------------------GPSRPRWRISAL-------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMFAP00000004329 (view gene)
ENSCCAP00000037979 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---ESFFSDFG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 322)
-LLWYLKELR--K---EEFWKFKELLKQ------
---------E--PLKLE--------L--KPV-PWTE-LKKAS-KEDVAKLLDTHYPGKRA
WELTLNL----FL-QISRKDLWTKAQ-EEM-S----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------KLNPYRKQ-
--------IKEK----------------FQLI--WEKETCLHVPEH--------------
---------------------------------F-------YKESMKHECTVLNKAYSAA
------ATP----V
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVVEGPDGIGKTTLLRKV-----MLDWA--E---------GNLWK
D--------RFTF-VFFLTVYEMNSIT-------ETSLPE-LLSR-DW-----PES-SEK
IEDVFS-QP----ERILLIMDGFEQ-LKLDLD-H------TA-D-----LCDDWRQR-QP
-LPIILSSLLQKKMLPECSLLLA-LGQVGMRKNYFVL-E-----HPK--LIKLSGFTEPE
RKMYF----------------S--CF-FGE---------KNKALKVFNFVRE-TGPLFIL
CHNPFICWLVCTCMKYRLERGE-DL-E--IN-SQNTTSLHASFITNVFKAGS-QSSPPK-
-----------LNRARLKNLSALA---AEGL-WT-HT----FVFCRE-------------
--DLRRNGLSES-EGLMWVGM----KLLQRSG-----DCFTFVHVCIQEFCAAMFYLLK-
GPKDC-----------------P--NPAIGSIT-QLVRASVAQ-PQNFLTQMGVFV----
----FGISTEEIASMLETSFGFP---------------L--SKD-LQQ-EIAQCLKSFSQ
CE-----------------------------------ADREAIGFQELFSCLFETQE-KE
FVTQVMNF---------------------F-------------------EEVYIYIANIE
HL---VIA--S----FCVK-HCHN-LTT-LRLC----------------MEN-----IFP
-DDSGCVS-----------E-------------------H-N-------EK---------
LMYWRELC-SMFSNNKNFQTLDMENTSFDDA---------SLTILC--KALAQP------
VC--K-LQKLMLTF---------MSN-FGHDSELFRAV----------------LHNPHL
KSLS-LYGT-GLCRT-DVKYLCEALRHPMCK----IEEL--------ML-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------MYCCLT-----------------------------
----------------------------CVSCEVISE-----------------------
--------------------------------G---------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------------------LVRSKSLSLLD
L-GSNHLEDSAVA-LLCEALKHP--DCS----I---------------------------
-----------------------RELWL--MAC-----FLTSDSCKDI------------
-------ADVL-ICNEKLKILKLGHNEIGDAGVKQ-LCEALK-HP-NC-KLECLG--LQ-
--T-CRITHACCE--------------------DLAEALVASKTL-RSLNLDWITLDPEG
VVLLCG--------ALSHPDCA--LQ--M-------------------------------
---LGLQKS-AFDKESQKM------LM----------ALE-EKCPHLSISHEPWIEE---
--EYR-----IR------------------------------------------------
--------------------------------------------------GVIP------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCLAP00000024432 (view gene)
------------------------------------------------------------
------------------------------------------------------------
-------MEISEAV-----GIAE--------------------------------EEQ--
S---AHLHRAKMAGK----RKYHL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 413)
LVYYLDLLE--K---EELKEFQLLLQN------
---------K--TFLR--------IS--PTT-TAAQ-AKKIS-GREVALWLVAEYGEQQA
WDLAIKT----WE-QM--------------------------------------------
----------------------------------GLTV-------------LCDK--A--
---------RR--------------E------VFLTWGAR-GSEKKSLTQ--PSEK----
------S--------------------------------------VHSKG-IEPINFPK-
--------ISEK----------------SCPTQSR-------ETK---------YFHEKF
TQLLLLRTPHFRSKEY---L------AREMWIYES--------KVGQGHLIKIQDLFGPV
PG--TQEES----H
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
RVVLYGAAGIGKSTLARQV-----RGAWE--E---------GWLFR
D--------RFRY-VFYFNCRDLPQCQ-------VNSMEE-VIAA-SW-----AS-TAVP
IMDILS-SP----EQVLFILDNVDQ-PKLNLRFL------DS-KGLRLQAQPKLSHS-PQ
-VRLVLDHLLQSLLFVRAFFLIT-ARTTSLEMFPHSW-E-----KPR--WVEVLGFSEWG
RKDYF----------------K--KY-FTD---------ESQSIIAFSAVEP-NPLLLAQ
CLVPWVCWLVCTCLQQQIEQGE-VH-P--LT-SQTTTGLFLHYLSQVIP--------TA-
-----------LQEIQLRCLCSLA---AEGI-WR-RK----TLFSTN-------------
--ELSKHGLEGA-LITTFLKI----GFLQEH---PDTLSYSFTHQCFQEFFAALSCVLG-
N------SAH--------TDKYN--FSL-RWMT-QLLKEYRWP--ESSRIQILCFL----
----SSLLSDYGAKEMKKIFSCQ---------------L--LPR-RSW-ELLQIVRPHG-
--------------------------------------ILLSQYYLNFFHYLYETQD-EE
LVTHEMAH---------------------F-------------------QERRVCVRTDR
EL---LLV--T----FCVK-FCNH-VKW-LQLN----------------ESDL-------
------RK----------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------Q------------
------ERRR--------------------------------------------------
-------------------PGIAVSTWARLT-----------------------------
----------------------------DASWKVLFS-----------------------
--------------------------------VLQSTGSLEE---LA-LSGNP-LSL---
-------SAVQSLGETL----GQPR
PF13516 // ENSGT00900000140813_0_Domain_12 (2066, 2206)
CCLQTLR----------------------------
------------------------------------------------------------
-------------------------------LAGCGLTAKGCKHLENSGT00900000140813_0_Linker_12_13 (2206, 2213)
AFG-LSTPF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
SQTLTELE
L-DFNMLTDAGAQ-HLCLGLKHP--SCK----L---------------------------
-----------------------QRLRL--VHC-----GLTS------------------
------------------------------------------------------------
---------TCCQ--------------------DLASVLST
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2464)
SPSL-KELDLQQNELGSYG
VQLENSGT00900000140813_0_Linker_17_18 (2464, 2716)
LFE--------GLRNPTCR--LT--L-------------------------------
---LWLDLT-LLNE---EE------LR----------IGE-QEKPQLLISS-GQIPDTRI
PPEDPEEAEESYS-----------------TSSFMQQRPQ--------------------
-------------------------------------LGD-LHMEALD-IEDDFCDPTGP
LPIELVNK-DGSLWRPF13553 // ENSGT00900000140813_0_Domain_18 (2716, 2997)
VHFPMAGYYHWP---HTGLSFVVRREV-------MMAIQFCAWDQ
FLSIHDLQDTWMVAGPLFDIKAEQGA-VAAVYLPHFVALQRE--------HVDRSLFQVA
HFKEEGMLLDKPDRVEPHHTILENPTFSPMGVLLKIIPGARRFFRVTCTTLLYHHIHAEE
V-KFHLYLIPSDCTIRQAIDDEEKKFQFERIHKPPPTDPLYLGSRYIVSGS---------
GMLEIMPKELELCYRSPRQPQLFSEIYVGCLGSGIQLEMRYKKDETVVWEALLKPGENSGT00900000140813_0_Linker_18_19 (2997, 3052)
DLRP
AASLFPPSAIGQQRLSETTGDSGIPGTFCLVLEGREETGASASPPNVPASLPF00619 // ENSGT00900000140813_0_Domain_19 (3052, 3134)
HFVDQHREQ
LVARVTSVDSVLDKLLSEQVLSEEQYTSVRAEATKPSQMRKLFSFSPSWNSACKDKLYQA
LREIHPHLIMDLWEAGAKHSGDL-------------
ENSMFAP00000014076 (view gene)
ENSBTAP00000027232 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MAG-------EAR---SLEDLEEKK--K---QKLKELRLWL--------
-----------------PDEEPRLQS--PEV-VHAQ-PEKPR-SRKTMT-ILSVMKEVQH
ECMAIHT----KGPHVNEQGACTKTL-EETDLNSA---LY-SSFSH---SPSAPNLESPS
WPTSTKVLTFCKPKQP-----VLNPLK-----WSSSPLIFASGYRKEDLSPSIY--QG--
---------FPSSPY-HESPSQELPNDPTSTAVLRACEIPRHSCPEPREKEGPGTVWPLV
ETSGNSCTEVDTVENINSQNGRD-GIQ-PPLSLMPLLPPSTHLMFCFPCHRSRQRYQDQ-
--------RRET----------------PCSTWSW-------KNE---------DLHPKF
MQLLLLHTPYHKDYES---L------NRESWDHGV--------VEGQGHLIEVRDLFGSD
LG--SQEGP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
HTVILHGVAGIGKSTLARHI-----RRAWE--E---------GQLYR
N--------HFQH-VFYFNCRELATSK-------LTSLEE-LITK-DQ-----ST-TVAP
AGQIQS-QP----EQLLFILDGLDE-LECVSEEK------RA-E-----LCLHRSQQ-QS
-VQTLLGSLVEKTIFPEASLLIT-ARTAALQKFIPSL-K-----QPC--WVEVLGFSESS
RKDYF----------------Y--EY-FTE---------ESQATRAFSVVES-NQALLTM
CLTPLVSWLACTCLKQQMEAGE-EL-S--LK-SQTMTALYLRYLAQAIQ--------AQ-
-----------PLGTQLRGFCTLA---AEGI-CQ-GK----TLFSPG-------------
--DLKKHGLDGV-ISATPLKM----DVLQKH---PTSLNYSFIHLCFQEFFAAVSFALG-
D------IRD--------KSDHP--NST-GSME-KLVEVYGIN--NLFGAPTVHFL----
----SGLLSEQGAREMENIFQCK---------------L--SWE-RKRESLLRWAELELR
-----------------------------------R-----EHYSLQLFHCLYEIQD-EE
FLTQAMAH---------------------F-------------------QGMRVCIRTSM
EL---LVF--T----FCIR-FCSH-VWR-LQLN----------------EGRQ-------
------HPG---------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------ACRP--------------------------------------------------
-------------------TDTVLLSWVPAT-----------------------------
----------------------------EACWQVLCS-----------------------
--------------------------------VLQVTGNLKE---LD-LSRNI-LSR---
-------SAVQSLDEAL----RCPGCQLETLR----------------------------
------------------------------------------------------------
-------------------------------LASCSLTAESCKDIASG-LSTSQTLMQLE
L-SFNTLLDAGAE-HLCQGLRQP--TCK----L---------------------------
-----------------------QRLLL--AGC-----GLTS------------------
------------------------------------------------------------
---------GCCQ--------------------DLASMLRVSPSL-MELNLLQNDLDDLG
VRLLCE--------GLRHPSCQ--LI--C-------------------------------
---LWLDQT-QLSEEVTKM------LR----------ALE-RQKPQLLVSG-IWKPSLRI
LHEGPGGAEMSDT------------------SSLKRQRQASARQKNLKLA----------
----------AALSAPEGSSPQAAQ-VRLFCPSSPAPPED-EHMESLG-TEDDLWGPTGP
VATEAVDE-ERSLYRVHLPMAGFYHWP---NTGLRFEVRGPV-------TVEIEFCAWDQ
FLNRNFPQRSWLAAGPLFDIKAEPGA-VAAVFLPHFVAFRGN--------DVDISQFQVA
HFKEEGMLLEKPARVTPCYAVLENPSFSPMGVLLRMVQTALRFIPITSTVLLYHYLQPEE
V-TFHLYLIPSDCSIQKAIDNEEKMFQFVRIHKPPPLTPLYMGSRYIVSGS---------
EKLEILPEELELCYRSPRESQLFSEFYVRHLRSGIRLQIRSKKDGTVVWEALVKPGDLRR
AVTPVSP-----------------------------GLRASPDSPAALRL
PF00619 // ENSGT00900000140813_0_Domain_19 (3051, 3133)
RHFVDRHREQ
LVARVTSVDPILDKLH-GQVLSEEQYEGVRAETTAPGQMRRLFGFSRSWDWACKDRLYQA
LKETHPHLIMELWEKWGSEETRPGS-----------
ENSMAUP00000023846 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------MARA-NNPQEA---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWALTDLE--E---NSFKTLKFHLRD------
---------V--T------------Q--FHL-ARGE-LEGLS-PVDLASKLISMYGALEA
VRVVSRS----LW-AMNLMELVNYLS-QVC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LH--DYREIYREH-
--------VRC-----------------LE------ER---QDW----------GINSSC
NELLMVT-MSSPGSSGSPPC------SG-------LK-----W-EVGQV--RVEALFAPE
AE--SYSTS----PI
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 930)
VVMQGSAGTGKTTLARKL-----VQDWA--K---------GTLYP
G--------RFDY-VFYVSCREVVLMPK-------CDLPN-LICW-CC-GD-----NEAP
VTEILR-QP----ERLLFILDGYDE-LQK---------------------------S-SR
-AESVLRILIRRREVP-CSLLIT-TRTPALQSLEPML-G-----ERQ--HVRVLGFSEEE
RENYF----------------N--SY-FTD---------KEQARNVLEFVQS-NDILYKA
CQVPGICWVVCSWLERKMARGQ-EV-S--ET-PSNSTDIFTAYVSTFLPTDG-NEDSPDL
----------T-RHRVLRGLCSLA---AEGI-QH-QR----LLFEED-------------
--ILRKHSLDGSNLT-AFLDS----IDYRGGFG-IK-KLYSFRHISFQEFFYAMSFLVK-
EDQRQL----------------G--EATHREVA-KLLEQ---E-KCEEMTLSLQFL----
----FDMLKREGTLSMGLKFYFK-----------------IAPS-VRQ-DL-KYFKEQI-
E-------------------------------------TMKCNRPWDLEFSLYDSKI-KK
ITQGIHIK---------------------D-------------------ITFNIQRSG-E
KK-PGKRM--T----VLVA-SSID-----KGQV------------------QS----PLL
--------KT---NPS--T-------------------KKQQG-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------ASN----------------GKS---------R
-GTEE-PAG-KVR-NSR-MVST----EK-GHMEMRA---QEGGGV---E-RQED-GEG-Q
R-DEK-DGEMR---DQ-MNG---Y------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSRROP00000027247 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MAMTKA-RNPREA---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWALSDLE--E---NDFKKLKFYLRD------
---------M--TLSE--------------------------------ELLISKYGEKEA
VTVVLKG----LR-VMNLLELVDQLS-HIC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LH--DYREIYREH-
--------VHC-----------------IE------EW---QEA----------GVNGRY
SQVLLVA-KSSSESPES-AH------PI-------PE-----Q-ELDSV--MVEALFDSG
EK--PSLAP----S
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
LVVLQGSAGTGKTTLARKM-----VLDWA--T---------GSLYP
G--------RFDY-VFYVSCKEVVLLLE-------SKLEQ-LLFW-CC-GD-----NQAP
VTEILR-QP----EWLLFILDGFDE-LQRPFEEK------LK-K-----RG-----L-SP
-KESLLHLLIRRHTLPTCSLLIT-TRPLALRNLEPLL-I-----QAR--HVHILGFSEEE
RARYF----------------S--SY-FTD---------EKKADSAFDIVQK-NDILYKA
CQVPGICWVVCSWLKGQMERGK-AV-------------------STFLPPVD-DGGCSEF
----------S-RHRVLRSLCSLA---AEGI-QN-QR----FLFEEA-------------
--ELRKHNLDGPRLA-AFLSS----NDYQLGLA-IK-KFYSFRHISFQDFFHAMSYLVK-
EDQSRL----------------G--KESRRELQ-RLLEAKEQE-GNDEMTVTMQFL----
----LDISKKDSFLNLELKFCCR-----------------MSPC-LAQ-DL-KHFKEQM-
E-------------------------------------SMKHNRTWDLEFSLYEAKI-KN
LVKGIQMK---------------------D-------------------VSFKIKLSN-E
KK-SQSQN--S----FSVK-SSLS-HGP-KEEQ------------------KR----PSV
---RGQKEGK---DNI--A-------------------GTHKE-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------AST----------------GKG---------R
-GTEE-TDEEVRS-PKN--TYI--------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMLEP00000040542 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MAESNSDFD---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWYLENLS--D---KEFQSFKKYLAR------
---------K--ILDFK--------L--PQV-PLIN-LRA-T-KEELANLLPISYEGQHI
WNMLYSI----FL-MMRKEDHCMKI---IGRRN---------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------RNQEACKAL-
--------MRRK----------------LTPR--WENHTFGEFHYKFFCNVA--------
----------------------------------------------SDVFYVLQLAYDST
SC--YSTND----LN
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 930)
VFLMGDRASGKTMTINLA-----VMKWI--N---------GEMWQ
N--------MISY-VVHLTSHEINQMT-------DSSLAE-LIAK-DW-----PD-GQAP
IADILS-DP----KKLLFILEDVDN-LRFELNVN------EQ-A-----LCSNSTQK-VP
-IPILLVSLLKRQMAPGCWFLIS-SRPTREENIKMFL-K-----TTD--CYVTLQFSNEK
RELYF----------------H--SF-FND---------RQRASTALMLVHD-DEILTGL
CRVAILCWITCTVLNRQMDKG-HDP-Q--HC-CETPTDLHAHFLADALTSKAGLT-AN--
----------QYHLGLLRRLCLLA---AGGL-FL-ST----LNFSGE-------------
--DLRCVGFTEV-DVSVLQAV----NILLPSST-HK-DRYKFIHLNVQEFCAAIAFLMAV
PNCLI-----------------P----SGSR-G--Y--KEKRE-QYCNFNQVFTFI----
----FGLLNENRRKILETTFGYQ---------------LPMVDS-FRR-YSVEYMKHLGR
DQ-------------------------------------EKLTHHGSLFYCLFENQE-EE
FVKKVVNT---------------------L-------------------KEVTVYLQSNK
DM---MVS--L----YCLE-YCCH-LRT-FKLS----------------VQR-----IFE
--YKEPLV-------------------------------------------RPTASQMKS
LVYWREIC-SLFCTMDTLWELHLFDSDLNDI---------SERILS--KALEHS------
SC--K-LRTLKLSY---------IST-ASGFEDLLKAL----------------ARNRSL
IHLS-LNCT---------------------------------------------------
------------------------------------------------------------
--------------------------------------------SISL-NMFSLLREI--
LDK-STCQIRHL-S----------------------------------------------
------------------------LMKCDLR-----------------------------
----------------------------ASECGEIAS-----------------------
--------------------------------LLVNGGSLRK---LT-LTNNP-LRN---
-------DGVNILCNAL----LHPNCTLTSL-----------------------------
------------------------------------------------------------
------------------------------ALVFCCLTEKCCSSLGRV-LLLGRTLKQLD
L-CVNHLQNYGVL-HVTFPLIFP--TCQ----L---------------------------
-----------------------EELYL--SGC-----FFTNDICQYI------------
-------AIVIA-T
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NETLRSLEIGSNSIEDAGMQL-LCGGLR-HP-DC-MLVNIG--LE-
--E-CMLTGACCE--------------------SLASVLT
PF13516 // ENSGT00900000140813_0_Domain_17 (2441, 2464)
TNKTL-ERLNILQNQLGDEG
VVKLLE--------SLILPECV--LQ--I-------------------------------
---IGLPLN-DVSAETRRM------VM----------TVK-ERKPNLIFLSETWSVKEGR
EIGVI-------------------------------------------------------
-------------------------------------------------------PASQP
GSV-----------------IPNSNLD---YM--FFKFPRMSTAMRMSSTASRQPL----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
MGP_CAROLIEiJ_P0078148 (view gene)
ENSDNOP00000014238 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MAGG----I-K
PF02758 // ENSGT00900000140813_0_Domain_0 (203, 326)
LR---LAWYLELLS--K---EEMAEFQLQLPS------
---------K--AL-HE-------GS--SGV-TPAE-TGTAN-GMEVASRLVAQYGEQQA
WDLALHT----WQ-QMGLDRLYTQAR-AEANLGSHT------ALLT---SPSAPGLESPS
WPTSTEVLSLWGSKYL-----CSQDSD-PK------APKAGCSW-QVFPIDCSSWLGH--
---------ISSSPS-HTSPNQQSPTAPMSTAVLGGWVPPPQPSPDPREQAASQAKGLLT
EGSHKYVVQSRKRM--TGFSRESTGLSLPPWSRTEHQNVQ--KVTSGSQMGHLHHLEIS-
--------PNER----------------SQKSQSW-------KKE---------DFHQKF
TELILLHRTGPQVQEF---L------FTGSWPQEA---------KEQGHLIEVQDLFGPG
LG--TQEGP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
RTVLLMGAAGIGKSTLARQV-----RRAWE--D---------GQLYR
D--------RFQH-VFYLNCRELAQTK-------VVSLAE-LMA--DQ-----AA-PLAP
IGQILS-QP----TQLLLILDGFDE-PGWIFKKH------RS-E-----LCFLWSQQ-QP
-VHILLSSLLRKTLLPEASLLIT-AQTTSLLELLFFV-E-----QLH--WVEVLGFSEAG
RKEYF----------------Y--NY-FTD---------DSQATRAFSLVES-NQALLTM
CLVPWVSRLVCTCLKQQMERGE-EL-P--LT-SQTTTALCLHYLSQAFP--------AQ-
-----------PLRAQLKSLCSLA---AKST-EQ-GK----ALFSLC-------------
--YLKKQGFSKA-VISTFVKM----GVFQK----PTFLSFSFAHSGLQEFFAAIFYALG-
D-------EE--------RSHYP--KST-HDVR-KLLRVCGRQ--VVFGTLTTRFL----
----FGLAGEHGARELENIFNLK---------------P--SGE-LRG-ELLRWVETELR
-----------------------------------CGGFSKQPYRLELLLCLYEIQD-EE
FLTQVMAY---------------------F-------------------QEARVWIQTDV
DL---LVF--T----FCLQ-FCRN-VKR-LQLS------------------GQ-------
------H-G---------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------P------------
------AWRA--------------------------------------------------
-------------------HSVVLCMEVPIT-----------------------------
----------------------------DARWQILFS-----------------------
--------------------------------ILGVAGSLRE---LD-LSGNS-LSH---
-------SAVQGLCKAL----RHPQCHLETLR----------------------------
------------------------------------------------------------
-------------------------------L-DCGLGSRDVKGVRIQ-IRVGERGDEWD
H-KYSFFQDFDNKEEVDRVSGQE--PCK----L---------------------------
-----------------------HGVRL--VGC-----GLTS------------------
------------------------------------------------------------
---------SCCA--------------------DLASALGASPSL-RELDLQQNDLDAAG
VGLLCE--------GLRHPACP--LK--L-------------------------------
---LQLGQI-PWSDKVSQE------LR----------ALE-EEKPQLLVTS-RWKQSVAM
AKEDPDWGETSSE-----------------TSSLKRQRLGKGPSAAHRRGLGETGQESLE
LTTALPA--AEE------HSPQATL-AEPRWLAGPSLPED-LLTEPLA-TEDDFQGPTGL
VATQVVDE-EKGQYRVHFPMAGTYRWP---STRLCFVVRWAV-------TLEIEFCAWSQ
FLDGTSLQHGWMVAGPLFDIKAEPGA-VAAVHLPHFVNLEEG--------NVDISLFQVA
HIKEEGTLLEKPARVEPEHAVLESPSFSPMGVLLRVIHAALR-IPITSIVLLYHRRHPEE
V-VFHLYLVPSDCSIRKAIDDEEKMSQFVQLRKPPPLTPLYMGSRYVVSGS---------
ENLEMIPKELELCYRSPGQPQLFSEFYVGHLGSGIRLQVKDKKCGTMVWEALVKPGDLRT
AVTPLPP-----------------------------SPKASPPLPYAP-AL
PF00619 // ENSGT00900000140813_0_Domain_19 (3052, 3132)
HFVDRHREQ
LVARVTSVEPILDKLH-GWVLSEEQYERVRAEATTLDQMRKLFSFSRSWNLACKDRLYQA
LEEIHPHLTVELRKCDSHRDQEGLLTNSAAEV----
ENSLAFP00000001437 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------MASPA----Q
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
LDFN---LQPLLEELD--Q---DELGQFKSMLRS------
---------F--PLHKA--------FHQQEL-PHME-IEQAN-GKQLAEILIKNCHSYWI
EMAAIHG----FD-KMNRVDLSERAK-DELREAAL-KSLR----E---RK---PPSLG--
------KKKSWSLK-FA--------------------------W---WYFFFS-------
------------------------------------------------------------
------------------------------------------------KDKEDKFRITL-
---------KKK----------------FWEI--WKNNLNPGAAENFRV-----------
---------------------------------------------ITQRYKMLIPFCNPK
ML--TGPFP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
DTVVLRGPAGVGKTTLAKEL-----MRDWT--Q---------DNLAK
---------TFKY-IFYLSCKELNHIG-------PCTCAE-LITK-DY-----PE-LQHA
IPDVLA-QS----RKILIIIDGFDE-LRFPPG------ALIS-D-----ICGDWKKQ-KP
-VPVLLSSLFMKKMFPNATLLIT-TRPWALRELRFLV-E-----RPL--FVDIEGFLEVD
RREYF----------------L--KH-FQD---------EDQALRAFDLMRS-NTALFSL
GSAPMVCWVVCTCLKLQMDKGE-DP-T--PT-CQTTTSLFLRFICSQFMPA-PDSCPGQC
------------LQAPVKALCLLA---AEGV-WT-QT----SVFDGE-------------
--DLERLGVKEC-ELSPFLDK----NILQKVKG-CE-GCYSFIHLSVQQFFAAIFYVLG-
SEEEK----E--------DRESH--REDIGNIQ-KLFSKEERL-KNPSLPQVGYFL----
----FGLSNEKRTRELETTFGCQ---------------L--SID-VKQ-ELLRWKVKSN-
EN-----------------------------------KPFSVMGMKEVFYCLYESQE-EE
FVKDAMAH---------------------F-------------------KEISLKLKSEM
DV---VHA--S----FCLK-HCQN-LEK-ISLK----------------IEKG----IFL
-END-TSL----ES--------------------G------------SQVERSQKD-QRI
LPFWMDLC-SMFGSNKNPIFLEIDRSFLSDS---------SVRILC--EQMAS-A-----
TH--N-LQKVV--L-NDILPA--SA----YR-DFCLAF----------------SGQETL
THLT-LQGS-CQND--VLPLLCEVLRHPKCD----LQYL--------RLE---------S
C------S----------------------------------------------------
---------AT---TQ-QWG---DLFLVLEI-NQSLTYLNFTADELLD-EGVKLLCTT--
LRH-PRCSLQRL-S----------------------------------------------
------------------------LENCHLT-----------------------------
----------------------------EASCKDLSS-----------------------
--------------------------------ALIINQKLTH---LC-LAKNE-LGD---
-------GGVQLLCEGL----SYPDC----------------------------------
--------------------------KLQ-ALV---------------------------
-------------------------------LWNCGITDSGCNHLSKL-LQQKSSLTHLD
L-GLNHIGITGLN-ILCEALKEP--TCN----L---------------------------
-----------------------KHLRG--SSV-----SVVELHTQQLPVCQWRRAVLCS
LPIQMILVS----FNQGVYCPQRQQQSCYLR-------------------PAAPR--LN-
--I-NESDAQMQK--------------------LLEEVKGSNPQLTIENDKL------RF
---------------QKSPWSH--FS----------------------------------
----C-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSGGOP00000045325 (view gene)
MDQPEAPCS-----STASPQRAFVQSVPHQGLRFRPWCKAPSWGSLEIPPSGHVTCPGVW
KDRSRKGGAGEEGAPQSSDQRRPVPTSPSEGEEHQD------APTRVRGRGR--------
-------EKSGVTPRCPLPPPHG-----SRRCLARDSLRPCPRWVLRRQSRGRAGVTRGS
LPDDPRPPASSTGPR--LAVAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLAALEELS--Q---EQLKRFRHKLRD------
------------VGPDG-----------RSI-PWGR-LERAD-AVDLAEQLAQFYGPEPA
LEVARKT----LK-RADARDVAAQLQ-EQRLQR---------E---------GWA-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------ALLPYKKKYREH-
--------VLQL----------------HARV--KERNARS------------VKITKRF
TKLLIAPESAAPE-E--ALGPAE-------EPDP--------GRARRSDTHTFNRLFRRD
E---EGRRP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYQ
G--------QVDF-AFFMPCGELLERPG------TRSLAD-LILD-QC-----PD-RGAP
VPQMLA-QP----QRLLFILDGADE-LPALGGP--------EAA-----PCTDPFEA-AS
-GARVLGGLLSKALLPTALLLVT-TRAAAPGRLQGRL-C-----SPQ--CAEVRGFSDKD
KKKYF----------------Y--KF-FRD---------ERRAERAYRFVKE-NETLFAL
CFVPFVCWIVCTVLRQQLELGR-DL-S--RT-SKTTTSVYLLFITSVLSSAPVADGP---
-----------RLQGDLRNLCRLA---REGVLGR--R----AQFAEK-------------
--ELEQLELRGSKVQTLFLSKKELPGVLET-----E-VTYQFIDQSFQEFLAALSYLLE-
DGGVP-------------------R-TAAGGVG-TLLRGDAQP--HSHLVLTTRFL----
----FGLLSAERMRDIERHFGC-----------------MVSER-VKQ-EALRWVQGQGQ
GCPGVAPEVTEGAKGLEDTEEPEEE-----------EEGEEPNYPLELLYCLYETQE-DA
FVRQALCR---------------------L-------------------PELALQRVRFC
RM-DVAVL--S----YCVR-CCPA-GQA-LRLI------------------SC----RLV
------AA----------------------------------------QEK--K---KKS
----------------------------------------LGKR----------------
------LQAS---L--------------GGS------S----------------LCTCPS
LH---LEYS-------VCCLSCKSALAPPAG----WGPS--------W------------
------------------------------------------------------------
------------------------LQKHPFD-HPAVTSAPHLHQTLG-SSRARLLPEF--
-L----------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPPYP00000004673 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------DGTEYRIR-
--------IKEK----------------FCIT--WDKKSLAGKPEDFHHGIA--------
----------------------------------------------EKDRKLLEHLFDVD
VK--TGEQP----Q
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
VVVLQGAAGVGKTTLVRKA-----MLDWA--E---------GNLYQ
Q--------RFKY-VFYLNGREINQLK-------ERSFAQ-LISK-DW-----PS-TE-P
IEEIIS-QP----SSLLFIIDGFDE-LN-AFEEP------EF-A-----LCEDWTQE-HP
-VSFLMSSLLRKVMLPEASLLVT-TRLTTSKRLKQLL-K----N-HR--YVELLGMSEDA
REEYI----------------Y--QF-FED---------KRWAMKVFSSLKS-NEMLFSM
CQVPLVCWATCTCLKQQMEKGG-DV-T--LT-CQTTTALFTCYISSLFTPV-DGRSLSLP
------------NQAQLRRLCQVA---AKGI-WT-MT----YVFYRE-------------
--NLRRLGLTKS-DVSSFMDS----NIIKKDAE-YE-NCYVFTHLHV-EFFAAMFYMLK-
GNWEA--------------WNPS--CQPFEDLK-SLLQ-TSYK--DPQLTQMKCFL----
-----GLLNEDRVKQLERTFNCK---------------M--SLK-IKS-KLLQCMEVLGN
SD-----------------------------------YSPSQLGFLELFHYLYETQD-KA
FISQAMRY---------------------F-------------------PKVAINICEKI
DL---LVS--S----FCLK-HCRC-LRT-IRLS----------------VTG-----VFE
--K--KIL----KTSL-----PT--N------------------------TW---DGDRI
THCWQDLC-SVLHTNEHLRQLDLCHSNLDKS---------AMNILH--HELRHP------
NC--K-LQKLLLKF---------ITF-PDGCPDISTSL----------------IHNKNL
MHLD-LKGS-DIGDN-GVKSLCEALKHPECK----LQTL---------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSFDAP00000000941 (view gene)
ENSPPYP00000011689 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---ESFFS
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 328)
DFG---LLWYLKELR--K---EEFWKFKELLKQ------
---------P--LEKFE--------L--KPI-PWAE-LKKAS-KEDVAKLLDKHYPGKQA
WEVTLNL----FL-QINRKDLWTKAQ-EEMRN----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------KLNPYRKH-
--------MKEK----------------FQLI--WEKETCLHVPEH--------------
---------------------------------F-------YKETMKNESKELNDAYTAA
------ARR----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 929)
HTVVLEGPDGIGKTTLLRKV-----MLDWA--E---------GNLWK
D--------RFTF-VFFLNVYEMNGVA-------ETSLLE-LLSR-DW-----PES-SEK
IEDIFS-VP----ERILFIMDGFEQ-LKFNLQ-L------KA-D-----LSDDWRQR-QP
-MPIILSSLLQKKMLPESSLLIA-LGRLAMQKHYFML-R-----HPK--LIKLLGFTESE
KKSYF----------------S--YF-FGE---------KSKAVKVFNFVRE-SGPLFIL
CHNPFICWLVCTCVKQRLERGE-DL-E--IN-SQNTTSLYASFLTTVFRAGS-QSFPPK-
-----------VNRARLKSLCALA---AEGI-WT-YT----FVFSHG-------------
--DLRRNGLSES-EGLMWVDM----RLLQRRG-----DCFAFIHLCIQEFCAAMFYLLK-
RPKDD-----------------P--NPAIGSIT-QLVRASVVQ-PQTLLTQVGIFM----
----FGISTEEIVSMLETSFGFP---------------L--SKD-LKQ-EITQCLESLSQ
CE-----------------------------------ADREAIGFQELFIGLFETQE-KE
FVTKVMNF---------------------F-------------------EEVFIYIGNIE
HL---VIA--S----FCLK-HCRH-LTT-LRMC----------------VEN-----IFP
-DDSGCIS-----------E----------------------------------------
------------------------------------------------------------
-----------FTS---------V-Y-FGHDSELFKAV----------------LHNPHL
KLLS-LYGT-SLSQS-DIRHLCETLKHPMCK----IEEL--------IL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------GKCDIS-----------------------------
----------------------------SEACEDIAF-----------------------
--------------------------------VLACNSKLKH---LS-LVENP-LRD---
-------EGMTLLCEAL----KHPHCALERL-----------------------------
------------------------------------------------------------
------------------------------MLMYCCLTSVSCDSISQVL-LCSKSLSLLD
L-GSNALEDNGVA-SLCGALKHP--GCS----I---------------------------
-----------------------QELWL--MGC-----FLTSDSCKDI------------
-------AAVL-ICNEKLKTLKLGHNEIGDTGVRQ-LCAALQ-HP-NC-KLECLG--LQ-
--T-CPITRACCG--------------------DLAAALIACKTL-RSLNLDWITLDADA
VVVLCE--------ALSHPDCA--LQ--L-------------------------------
---LGLHKS-GFDEETQKI------LM----------SVEEK-IPHLTISHGPWIDE---
--EYK-----IR------------------------------------------------
--------------------------------------------------GVLL------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSSSCP00000042009 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MEPL--AAAAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLVALEDLS--Q---EQLKRFCHKLRD------
------------APLDG-----------RSI-PRGR-LEGSD-AVDLAEQLIHFYGPELA
LEVARKT----LK-RADVRDVAAQLK-EQQLQR---------L---------GPR-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------SS-ALLSASEYKKKYREH-
--------VLRQ----------------HAKV--KERNARS------------VKISKRF
TKVLIAPET--LEHR--TLGPAE-------EPEP--------ERARRSDTHTFNRLFSRD
A---EGQRP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYH
G--------QVDF-AFFMPCRELLERSG------TCSLAD-LILE-QC-----PD-RSAP
VPQILA-QA----ERLLFILDGVEE-LPAPGPS--------EAV-----PCTDPLEA-AS
-GARVLGGLLSKVLLPAARLLVT-TRAAAPGRLQGRL-C-----SPQ--CAEVCGFSDKD
KKKYF----------------Y--KF-FRD---------ERRAEQAYRFVKE-NETLFSL
CFVPFVCWIVCTVLRQQLELGQ-DL-S--RT-SKTTTAVYLLFVASFLSSAPAADGP---
-----------QLQDELRKLCRLA---CEGVLGR--R----AQFSQK-------------
--DLERLELLHSQVQTPFLSKKELPGVLET-----E-VTYQFIDQSFQEFFAALSYLLE-
DEGSP-------------------R-APAGGVG-ALLRGDAEP--RCHLALTTRFL----
----FGLLSAERMRDIERHLGC-----------------SVSKR-VKQ-DVLRWVQEQGQ
GCPRAGPEGTKETQALQGAEEPEE------------EEDEELSYPLELLYCLYETQE-EA
FVRQALRG---------------------L-------------------PELALERVHFS
RM-DLAVL--S----YCIR-CCPV-GQA-LRLD------------------SC----RLA
------IR----------------------------------------QEKK-----KKS
----------------------------------------LVKR----------------
------LQGS---L--------------GGS------A-----------------SRATV
RK---PLGS-------PLRPLCEAMVDQQCG----LSSL--------TLS---------H
C-----------------------------------------------------------
--------KLP---DS-VCG---DLSEALRK-APALTELGLLHNGLS-EAGLRALSEG--
-LAWPECQVQTL------------------------------------------------
----------------------------RMQ-----------------------------
---------------QPGL---------QGTPQYV-------------------------
------------------------------AGLLQHSPALAT---LD-LSGC-QLSERMV
--T--------YLCAAL----RYPGCRLQTLRL--TSVELSER-----------------
----------SLQELQAVGTAKP-------GLV----------------------I--TH
EALG-------------------SRPKPPEGFDSAL------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPVAP00000005696 (view gene)
ENSMMUP00000031445 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MTSP----QLEWT---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 323)
LQTLLEQLN--E---DELKSFKSLLRA------
---------L--PLEDV--------L--QKA-PWSE-VEEAD-GKKLAEILIHTSSENWI
RSATVTV----LE-EMNLTELCK-MA-KSEM-----------------------------
------------------------------------------------------------
--LEAG---------------------------------QVQEIDNPQL-----------
-------GDA-------------------------------EEDSELEKPGEKEGRRNS-
---------VQK----------------QSLV--WKNTFWQGDIDSFHDDVI--------
-----------------------------------------------QRNQRFIPFLNPR
TP--RKLTP----Y
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVLHGPAGVGKTTLAKKC-----MLDWT--D---------CNLSP
---------TVRY-AFYLSCKELSRMG-------PCSFAE-LISK-DW-----PE-LQDD
IPHILA-QA----QRILFVVDGLDE-LKVPPG------ALIQ-D-----ICGDWEQQ-KP
-VPVLLGSLLKRKMLPKATLLVT-TRPRALRDLQLLV-Q-----QPI--YIRVEGFLEED
RRAYF----------------L--RH-FGD---------EDQAMRAFELMRS-NAALFQL
GSAPAVCWIVCTTLKLQMEKGE-DP-A--PT-CLTSTGLFLRFLCSQFPQG-------AQ
------------LRGALRALSLLA---AQGL-WA-QM----SVLHGE-------------
--DLESAGVQES-DLRPFLDR----DILRQDGV-AK-GCYSFIHLSFQQFLTALFYALE-
KE--E---EE--------DRDGH--AWDIGDVQ-KLLSEEERL-KNPDLIQVGHFL----
----FGLANEKRVKELEATFGCQ---------------M--SPE-IKQ-ELLRCKASLHA
NK-----------------------------------P-LSMTDLKEVLCCLYESQE-EE
LAKVVVAP---------------------L-------------------KKISINLTNTS
EM---MHC--S----FSLK-HCRD-LEK-VSLQ----------------IAKG----VFL
-ENY-VDF----EP--------------------DIEFE---------------------
------------SSNSNLKFLEVKQSFLSDS---------SVRILC--DHITR-S-----
TC--H-LQKVE--I-KNVTPD--TA----YR-DFCLAF----------------IGKRTL
THLT-LEGH-IEWERTMLLLLCDLLRNHKCN----LQYL--------RLG---------G
H------C----------------------------------------------------
---------AT---SE-QWV---DFFYVLKA-NQSLRHLHLSASVLLD-EGAKLLYKT--
MTH-PKHFLQML-S----------------------------------------------
------------------------LENCRLT-----------------------------
----------------------------EASCKDLAA-----------------------
--------------------------------VLVV
PF13516 // ENSGT00900000140813_0_Domain_11 (2017, 2054)
SQQLTH---LC-LAKNP-IGD---
-------TGVKFLCEGL----SYPEC----------------------------------
--------------------------KLQ-ALV---------------------------
-------------------------------LQQCSITKRGCRYLSEA-LQEACSLTNLD
L-SLNQIA-RGLW-ILCQALENP--NCN----L---------------------------
-----------------------KHLRL--WSC-----SLMPFYCQHL------------
-------GSAL-ISNQKLETLDLGQNHLWKSGIIQ-LFRVLR-QR-TG-SLKTLR--LK-
--T-YETNLEIKK--------------------LLEEVKERNPKLTIDCNAS------GA
---------------MAPACCD--FF----------------------------------
----C-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPPAP00000000338 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MAMAKA-RNPREA---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWALSDLE--E---NDFKKLKFYLRD------
---------M--TLSEG--------Q--PPL-ARGE-LEGLI-PVDLAELLISKYGEKEA
VKVVLQG----LK-VMNLLELVDQLS-HIC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LH--DYREVYREH-
--------VRC-----------------LE------EW---QEA----------GVNGRY
NQVLLVA-KPSSESPESLAC------PI-------PE-----Q-ELDSV--TVEALFDSG
EK--PSLAP----S
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
LVVLQGSAGTGKTTLARKM-----VLDWA--T---------GTLYP
G--------RFDY-VFYVSCKEVVLLLE-------SKLEQ-LLFW-CC-GD-----NQAP
VTEILR-QP----ERLLFILDGFDE-LQRPFEEK------LK-K-----RG-----L-SP
-KESLLHLLIRRHTLPTCSLLIT-TRPLALRNLEPLL-K-----QAR--HVHILGFSEEE
RARYF----------------S--SY-FTD---------EKQADRAFDIVQK-NDILYKA
CQVPGICWVVCSWLQGQMERGK-VV-L--ET-PRNSTDIFMAYVSTFLPPDD-DGGCSGL
----------S-RHRVLRSLCSLA---AEGI-QH-QR----FLFEEA-------------
--ELRKHNLDGPRLA-AFLSS----NDYQLGLA-IK-KFYSFRHISFQDFFHAMSYLVK-
EDQSRL----------------G--KESRREVQ-RLLEVKEQE-GNDEMTLTMQFL----
----LDISKKDSFSNLELKFCFR-----------------ISPC-LAQ-DL-KHFKEQM-
E-------------------------------------SMKHNRTWDLEFSLYEAKI-KN
LVKGIQMN---------------------N-------------------VSFKIKHSN-E
KK-SQSQN--L----FSVK-SSLS-HGP-KEEQ------------------KC----PSV
---HGQKEGK---DNI--A-------------------GTPKE-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------ASS----------------GKG---------R
-GTEE-TDEEVR--SPK-NTYI--------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPVAP00000014869 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------MALA-SN
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
PREA---LLQALNDLE--E---YDFKILKFHLRD------
---------G--TLLGG-----------PGL-ARGE-LEGLS-RVDLASRLISMYGTQEA
VKVVLKV----LK-VMNLLELVDQLS-HVC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LN--DYREIYREH-
--------VRC-----------------LE------ER---QEE----------GVDHSY
YQLLLVA-KPCAGSPI-------------------LG-----Q-EVDSV--TVEALFDPG
EK--PNQGP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
PTVVIQGSAGTGKTTLARKM-----VLDWA--T---------GALYP
G--------RFDY-VFYVSCREVTLLQE-------STLDQ-LLIW-CC-GD-----SKAP
VAEIRRQQP----ERLLFILDGYDE-LQRSYAER------LR-R-----PG-----P-SP
-VEDMLHRLIRREVLPLSSLLIT-TRPLALRNLEPLL-K-----QPR--HLHILGFSMEE
KERYF----------------R--SY-FTD---------EEQARKALDIARG-NDFLYKA
CQIPGICWVVCSWLKGQMERGG-VV-S--EM-PSDGTDIFMAYVSTFLPSSD-NRDCSKL
----------T-RDKVLKGLCSLA---AEGI-QH-QR----FLFEEA-------------
--DLEKHNLDGPSLA-AFLSS----IDYQEGLN-IK-KFYSFRHISFQEFFHAMSYLVK-
EDASQL----------------G--EGSLREVN-RLLDEKKQV-GSEEMTLSMQFL----
----LDISKKESSSNFELMFCFK-----------------VSPS-LKQ-DL-KNFKEQM-
S-------------------------------------SIKHKKTWDLELSLYESNM-KN
LLKSVQLS---------------------D-------------------VAFKMVHSK-N
KK-SPSRN--L----FSVK-TSLS-NKQ-KEEQ------------------KC----PAV
--------DK---NNI--A-------------------ETPME-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------ASN----------------GKD---------R
-GTKE-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMAUP00000005150 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------M---ASFFSDFG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 323)
-LMWYLQELN--K---KEFMKFKEVLKQ------
---------E--GLQCG--------L--QQI-PWME-VKKAS-REDLANLLVKYYE-KKA
WDVTLSI----FQ-NINRNDLSERAA-REI------------AG--------------QS
K---------------------------------------------------IYQA----
------------------------------------------------------------
----------------------------------------------------------H-
--------LKNK----------------LCQD--WSRKFYIPIQYF--------------
---------------------------------L-------KPEVTKDKVSYLQQLFISE
TT--EKR-P----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
HTVLLKGVSGVGKTLVLFQL-----MLAWS--E---------GLLFQ
D--------EFSY-IFYLCCEEVKQLK-------TESLAY-LMFR-EW-----SSP-SAS
IVEIVS-QP----AKLLFIIDSLEG-LNCDLTEP------EL-E-----LCDNWMEK-RP
-VNILLSSLLRRKMLPESSLLIA-GTPESFDQLEHRI-D-----YTD--MKTVTGFDENT
IRMCF----------------Q--EL-FQD---------ESTAQNVFRLVRE-KEQLFRM
CHIPTLCWAVATCLKNEKEKAR-DP-A--SV-CRCITSVYTAYILNLFIPKS-AHGPSK-
-----------KGQDLLRGLCSLA---AEGM-WT-DK----FVFNQE-------------
--DLGRNGIVSS-DISTLLNM----SVLLKSKD-SK-NFYKFHHPSIQEFCAAVFYLVK-
SHEDH-----------------P--SKDVQGIE-TLLFTFLSK-VRVHWIYLGCFI----
----FGLLHKSEQEKLQAFFGYQ---------------L--SQE-VEQ-KLCQSLEEISI
DED-----------------------------------LWEQVDDMKLFYCLFEMEN-EA
FVMQVMDF---------------------W-------------------QRIKFVAKNYS
DL---MVA--A----YCLR-NCFS-LQT-LSFS----------------IQD-----ILQ
-EVQ--DQ-----------S-----------------------------------S----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSTTRP00000005730 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------T----MKLDIQCEQ-LSD-ARWT----------------EL---------
----LPLI----QQ---------------------------YEVV------RLDDC-GLT
EVRCKDIG-SALRANP
PF00560 // ENSGT00900000140813_0_Domain_4 (1517, 1551)
SLTELSLRTNELGDS---------GVHLVL--QGENSGT00900000140813_0_Linker_4_5 (1551, 1566)
LQS-P-----
TC--KPF00560 // ENSGT00900000140813_0_Domain_5 (1566, 1617)
-IQKLN--L-QNCCLT--EA---GCR-VLPGVL----------------RSENSGT00900000140813_0_Linker_5_6 (1617, 1619)
LSPF00560 // ENSGT00900000140813_0_Domain_6 (1619, 1645)
TL
RELN-LSDN-PLGDA-GLQLLCEGLLDPQCH----LEKL--------QLE---------Y
C------N----------------------------------------------------
---------LT---AA-ACE---PLAAVLRA-TRDLKELMVSNNDLGE-AGIRELCRG--
LTD-SACQLEAL-K----------------------------------------------
----------------------LXXXXXXXXXXXXXXXXXXXXXXXX-------------
----------------------------XXX-XX-XX-----------------------
--------------------------------XXXXXXXXXX---XX-XXXXX-XXX---
-------XXXXX---XX----XXXXX----------------------------------
--------------------------XXX-XXX---------------------------
-------------------------------------XXX-------X-XXXXX--XXXX
X-XXXXXXXXXXX-XXXXXXXXX--XXX----X---------------------------
-----------------------XXXXXXXXXX-----XXXXXXXXXX------------
-------XXXX-XXXXXXXXXXXXXXXXXXXXXXX-XXXXXX-XX-XX-XXXXXX--XX-
--X-CDVTDGACG--------------------GLASLLLTNR
PF00560 // ENSGT00900000140813_0_Domain_17 (2444, 2465)
SL-RELDLSNNGLGDPG
VLQLLG--------SLEQPGCA--LE--Q-------------------------------
---LVLYDI-YWTEVVEER------LR----------ALE-GSKPGLRIIF---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPANP00000002473 (view gene)
ENSPANP00000001452 (view gene)
ENSMLUP00000012508 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---ESFF
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
SDIC---LLSYLKELR--K---NEFWKFKELLKQ------
---------E--PKQSD--------L--GPI-PWPE-LKRAS-KDDLAKLLCRHYPGKQA
WEVALRL----FP-QVNRRDLWAKAG-DENSG----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------QINPYKSH-
--------MRKK----------------FSDI--WQNETCLAVPES--------------
---------------------------------F-------HAESTKTEYERLSEAFA-A
DK--VGEHS---
PF05729 // ENSGT00900000140813_0_Domain_2 (673, 940)
-VTVALGSAKATGKTTLLRKV-----MLQWA--E---------GNLWK
G--------RFTF-VFFLNCCEMNIID-------KTSLVE-FLSK-DW-----PES-SEP
IHDVFS-RP----EKILFIIDGLED-LKVDMG-L------DD-D-----LCSGWEQP-GP
-VHVVLRSLLQRTLLPEASLLVS-LSGEWLRKFNYLL-R-----HPV--FVYLPGFSGHF
KWLYF----------------S--RF-FQD---------TDQADQAFSVVSG-HKPLFTL
CQYPLVCWLVCECLKKQLQRCQ-PL-G--VN-SQTTTCIYSSFLTNVFQPEC-TGWALG-
-----------HCHSRLKCLCALA---AEGT-WA-GR----FAFQPS-------------
--DLDENTVSEA-HALAWVRM----GLLRQNG-----SRWVFPHQSLQELCAALWYLLR-
LPREP-----------------Q--HPRICSAT-QLVTATMTE-APFYLKDTALFL----
----FGFATERVAGLLEAAWGFE---------------L--AAS-LRE-EITQCLQGLSQ
QVA----------------------------------RGELQINFQELFNCVFETQD-PG
FATQVMSM---------------------F-------------------PDIDVCISKPD
DL---MTY--A----CCLQ-YALN-LQT-LHLS----------------VED-----VFS
-EDSRGLL-----------D-------------------K-D-------RK---------
LSYWRALC-CVFTNSRDFQMLDLDNCIFNEA---------SQAILW--KALAQP------
TC--K-LQKLVYNF---------ASN-VGESADFFKAL----------------VHNPNL
TYLN-LHGT-NLSSE-EVAQLSEALRHPMCS----IQQL--------IMMESHFLLGSLK
CGHWVGRC------------------------------------KN--------------
---------------RSKYI---YIYILCLA----------------LSMSPALPCSP--
PCA--PCPLPAPAG----------------------------------------------
------------------------WGKCDIT-----------------------------
----------------------------DEACEDIAL-----------------------
--------------------------------LLVSSRKLKL---VS-LMENP-VMD---
-------SGL-LLSEAL----KHPDCALESL-----------------------------
------------------------------------------------------------
------------------------------LLTYCCLTSDACDYIAQA-LVRSTTLSLLD
L-GSNFLEDSGVK-LLCEALRQP--SCN----L---------------------------
-----------------------QQL----------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMNEP00000005715 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MAMTKA-RNPREA---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWALSDLE--E---NDFKKLKFYLRD------
---------M--TLSEG--------Q--PRL-ARGE-LEGLI-PVDLAELLILKYGEKEA
VKVVLKG----LR-VMNLLELVDQLS-HVC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LH--DYREIYREH-
--------VRC-----------------LE------EW---QEA----------GVNGRF
NQVLLVA-KSSSESPESPAH------PI-------PE-----Q-ELDSV--MVEALFDSG
EK--PSLAP----S
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
LVVLQGSAGTGKTTLARKM-----VLDWA--T---------GSLYP
G--------RFDY-VFYVSCKEVVLLLE-------SKLEQ-LLFW-CC-GD-----NQAP
VTEILR-QP----ERLLFILDGFDE-LQRPFEEK------LK-K-----RG-----L-SP
-KESLLHLLIRRHTLPTCSLLIT-TRPLALRNLEPLL-K-----QAR--HVHILGFSEEE
RARYF----------------S--SY-FTD---------EKKADSAFDIVQK-NDILYKA
CQVPGICWVVCSWLKGQMERGK-AV-L--ET-PRNSTDIFMAYVSTFLPPVD-DGGCSEF
----------S-RHRVLRSLCSLA---AEGI-QN-QR----FLFEEA-------------
--ELRKHNLDGPRLA-AFLSS----NDYQLGLA-IK-KFYSFRHISFQDFFHAMSYLVK-
EDQSRL----------------G--KESRREVQ-RLLEAKEQE-GNDEMTLTMQFL----
----LDILKKDSFLNLELKFCFR-----------------ISPC-LAQ-DL-KHFKEQM-
E-------------------------------------SMKHNRTWDLEFSLYEAKI-KN
LVKGIQMN---------------------D-------------------VSFKIKHSN-E
KK-SQSQN--S----FSVK-SSLS-HGP-KEKQ------------------KH----PSV
---HGQKEGK---DNI--A-------------------GTHKE-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------AST----------------GKG---------R
-GTEE-TDEEVRS-PKN--TYI----YT--------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSSSCP00000033039 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MNS---------------------------QKKRKDMEPA-----
--------CTWS------------------------------------------LLSE--
-----------------------------------------QK-----------------
----AASSSL-------------------------------TSSSWKELTDYREVFKKH-
--------LKEK----------------YLKI-GVDKCYHHGK--------------HID
SRLLVVKEHCISEKPSCGIP-------------------------QQDNFIEMEHVFDPD
EE--GSSSP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 928)
RTVVLQGCAGIGKTAVVHKF-----MFDWA--A---------GMVTP
G--------RFDY-VIYINCREISHFA-------NLSAAD-LITN-TF-----QD-VDGP
ILDVILTYP----EKLLFILDGFPE-LQYRVGDQ------DE-D-----LSANPQER-KP
-VETLLCSFVRKKLFPESSLLIT-ARPATMKKLHSLL-K-----QPL--QAEILWFTDAE
KRAYF----------------L--SQ-FPG---------ANAAMRVFHGLQE-NEGLDIM
SSLPIISWMICSVLQSQGDGDR-TL-M--RS-LQTMTDMYLFYFSKCLKTLIGIS-VW--
-----------KGQSCLWGLCSLA---AEGL-QN-QK----VLFEVS-------------
--DLRRHGIGVYDTSCTFLNH-----FLKKVKG-GG-SVYTFLHFSFQEFLTAVFYALK-
NDSSW---------------------MFSDQVGKTWQEIFQQY-GKGFSSLTIQFL----
----FGLLHKGKGKAVETTFGRK---------------V--SLG-LRE-ELLKWTEKEIK
DK-----------------------------------SSRLQIEPMDLFHCLYEIQE-EE
YAKRIIDD---------------------L-------------------GSIILLQPTYT
KM-DILAM--S----FCVK-SSHSHLS----------------V-----SLKCQHLLGF-
-EEEDRAS----------------------AFMP--------------------------
---------PASTFNQPFQ-----LCTLPVS---------PLHLFC--QALRNP------
YC--K-VKELKL---IFCHLT------SSYGRD---------------LSLVFET
PF13516 // ENSGT00900000140813_0_Domain_6 (1616, 1642)
-NQYL
TDLE-FVKN-TLEDS-GMKLLCEGLKQPNCI----LQTLR--------LYRCLI-PSA-S
CG------------AL----AAVLSTNQWLTELEFSETKLEASALKLLCEGLKDPNCK-V
QKLKLCASFLPGSSEA-VCK---YLASVLIC-
PF13516 // ENSGT00900000140813_0_Domain_8 (1773, 1796)
NPNLTELDLSENPLGD-TGVKHLCEG--
LRH-SNCKVEKL-D----------------------------------------------
------------------------LSTCSLT-----------------------------
----------------------------DASCVELSS-----------------------
--------------------------------FLQVSQTLKE---LF-VFANA-LGD---
-------TGVQHLCEGL----HHPSGTIQNL-----------------------------
------------------------------------------------------------
------------------------------VLSECSLSAACCESLAQV-LSSTRSLTRLL
L-INNKIEDLGLK-LLCEGLKQP--DCQ----L---------------------------
-----------------------KDLSL--WTC-----HLTGECCQDL------------
-------CDAL-Y
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
TNEHLRVLDLSDNALGDEGMQV-LCEGLK-HP-SC-KLQTLW--LA-
--E-CHLTDACCG--------------------ALASVLKR
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2465)
NENL-TLLDLSGNDLKDFG
VQMLCD--------ALIDPKCR--LQTFY-------------------------------
------IDTDHLHEETFRK------IE----------ALK-MSKPGITW-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCATP00000021782 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MNN---------------------------QKKRKYMELA-----
--------CTWS------------------------------------------LLSG--
-----------------------------------------EN-----------------
--------------------------------------------------DYWEIFRRH-
--------LKEK----------------YLKI-GVDKCYHHGK--------------HIE
SQLLLIKDHGISEETPCEIP-------------------------QQDHFIDMEHVFDPS
EE--GSSSS----R-------TVVLQG-CAGTG-----KQLWC--------------TSS
C-------
PF05729 // ENSGT00900000140813_0_Domain_2 (729, 870)
-LEIC-HIA-NLSAA----------------D-LITN-TF-----QD-INGP
ILDTILIYP----EKILFILDGFPE-LQGPVGDQ------EE-D-----LSVHPQER-KP
-VEGLLCSFVRKKLLPDSSLLIT-ARPTTMKL-HSVK-T-----TY--------------
---------------------------------------PGRDL-------T-NEDLDFM
SSLPIVSWMICSVLQSQGDGDR-TL-L--RS-LQTMTDVYLFYFSKCLKTLTGIS-VW--
-----------EGQSCLWGLCCLA---AEGL-QN-HQ----VLFAVS-------------
--DLRRHGIGVCDTNYIFLSR-----FLKKAEG-AV-SVYTFLHFSFQEFLTAVFHALK-
NDNSW---------------------IF-------------FF-LSSFSSLMMQFL----
----FGLLHKEKGKAVETTFGIK---------------L--SPG-LRE-GLLKWTEREIK
DK-----------------------------------ASRLQIEPVDLFHCL--------
------------------------------------------------------------
-------------------------YE----------------I-----QEEENHLLGF-
-EEEERAS----------------------TFVP--------------------------
---------LALTLNQSFQ-----LCSLPVS---------QLHLLC--QGLRNP------
CS--S-C--G---------------------RD---------------LSLVFET
PF13516 // ENSGT00900000140813_0_Domain_6 (1616, 1642)
-NQHL
TDLE-FVKN-TLEDS-GMKLLCEGLKQPNCQ----DNL-------------LFDTLILGV
NF------------YF----SAVLSTNQWLTELEFSETKLEASALKLLCEGLKDTNCK-L
QKLKLCASLLPEGSEA-VCR---YLSCVLIC-NPDLTELDPSENPLGD-IGVKYLCVG--
LRH-SNYKVEKL-D----------------------------------------------
------------------------LSTCHST-----------------------------
----------------------------DASCVELSS-----------------------
--------------------------------FLQVSQALKE---LF-VFADV-LGD---
-------TGVQHLCEGL----QHTKGIIENL-----------------------------
------------------------------------------------------------
-------------------------------LSEYSLSAACCEPLAQV-LSSTRSLTRLI
L-INNKIEDMGLK-LLCEGLKQP--DCH----L---------------------------
-----------------------KDLAL--WTC-----HLSGACCQDL------------
-------CNAL-Y
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
TNEHLRDLDFSDNALGDEGMRV-LCEGLK-CP-CC-KLQTLW--LA-
--E-CHLTDACCG--------------------ALASVLNRDE
PF13516 // ENSGT00900000140813_0_Domain_17 (2444, 2465)
NL-TLLDLSGNDLKDFG
VQMLCD--------ALIHPICK--LQTFY-------------------------------
------LDTDPLREDAFRK------ME----------VLK-MSKPGITCCLRECLI----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSSBOP00000025651 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----MSAD--M----QCVDIQCEE-LSD-ARWA----------------EL---------
----LPLL-------------------------------PRCQAV------RLDDC-GLS
EAQCEDVG-SALQA
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1542)
NSALTELSLRNNEVGDA---------GVRRVL--QALQS-S-----
PC--K-IQKLS--F-QSCYLT--VA---GCG-VLSGTL----------------RS
PF13516 // ENSGT00900000140813_0_Domain_6 (1617, 1642)
LPTL
QELH-LSDN-PMGDA-GLQLLCEGLLDPQCR----LEKL--------QLE---------Y
C------S----------------------------------------------------
---------LS---SA-GCE---PLASVLRA-KPD
PF13516 // ENSGT00900000140813_0_Domain_8 (1776, 1796)
FKELTVSNNDIDE-TGVRALCQG--
LKD-SPCQLETL-K----------------------------------------------
------------------------LEKCGVT-----------------------------
----------------------------SDNCRDLCS-----------------------
--------------------------------ILASK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
ASLQE---LA-LGNNK-LGD---
-------VGITELCPGL----LHPSS----------------------------------
--------------------------RLR-TLW---------------------------
-------------------------------IWECGLTAKSCVDLCRV-LRAKD
PF13516 // ENSGT00900000140813_0_Domain_13 (2215, 2237)
SLKELS
L-AGNELGDEGAQ-LLCESLLEP--GCQ----L---------------------------
-----------------------ESLWV--KSC-----SLTAACCSHF------------
-------SSVL-A
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
KNKFLLELQISENRLGDAGVQE-LCRGLG-QP-GS-VLRS-------
------------------------------------------------------------
------------------------------------------------------------
---LCLYDI-YWSLEMENR------LQ----------ALE-EEKPSLRIIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCAFP00000010243 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------MALT-RNP
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 324)
REA---LLWALSDLE--E---KNFKLFKFHLRD------
---------R--SLLEG--------R--LQL-ARGE-LEGLS-PVDLASRLISLYGALEA
VKVVVRV----LR-VMNLLEPVDQLS-HIC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LN--DYREIYREH-
--------VRCC----------------LE------ER---QEE----------SICGSH
SQLLLVA-TSSPGSPESSAC------PV-------LE-----Q-ELDSV--LVETLFDPG
EK--SYQAP----P
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLQGSAGTGKTTLARKM-----VLDWA--T---------GTLYP
G--------RFDY-VFYVSCREVVLLQE-------GVLDQ-LLFW-CC-GD-----NQAP
VREMLR-EA----ERLLFILDGFDE-LQRPFVRG------LK-S-----LR-----L-SP
-MEKVLHCLIRREVLSTCSLLIT-TRPLALQNLNPLL-K-----QSH--HIHILGFSEDE
RRRYF----------------S--SY-FMD---------EEQARNAFDIVRG-NDVLYKS
CQIPGICWVICSWIKEQMEKGR-EI-S--ET-PSNGTDIFMAYVSTFLPPSD-NEGCSKL
----------T-RHRVLKGLCSLA---AEGI-QH-QR----FMFEEA-------------
--DLRKHNLDGTRLA-AFLSS----MDYQEGLD-IK-KFYIFRHISFQEFFHAMSYLVK-
EDQSQL----------------G--KESRKEVK-RLLDEERQA-ENEEMTLSMHFL----
----LDILKKETSSNFELKFSLK-----------------ISPL-VKQ-DL-KHFKEQM-
K-------------------------------------SIKHKGAWDLEFSLNQSKI-RN
LVKGVQIS---------------------D-------------------VSLKMQRSN-K
KQ-PHDRN--S----FSVK-TSLI-NRW-KEKE------------------KC----PFV
--------DE---DNI--E-------------------KTQME-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------ASN----------------GKG---------R
-KMEK-QEMLSVG-AVG-REV-----EE-WDSRAEG---SGRLES---Q-GQ-E-DGG-R
QSLKK-MER---------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMNEP00000022637 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---ESFFSDFG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 322)
-LLWYLKELR--K---EEFWKFKELLKQ------
---------P--LEKFE--------L--KPI-PWAE-LKKAS-KEDVAKLLDKHYPGKQA
WEVTLHL----FL-QIDRKDLWTKAQ-EEMRN----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------KLNPYRKH-
--------MKEK----------------FQLI--WEKETCLPVPEH--------------
---------------------------------F-------YKETMKNKYKELNGAYTAA
------ARL----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
HTVVLEGSDGIGKTTLLRKV-----MLDWA--E---------GNLWK
D--------RFTF-VFFLNVYEMNGVT-------ETSLLE-LLSR-DW-----PES-SET
IEDIFS-QP----EGILFIMDGFEH-LKFNLK-F------KA-D-----LSDDWRRQ-QP
-TSIILSSLLQKKMLPECSLLIA-LGKLAMQKHYFML-Q-----HPK--LIKLSGFTESE
KKSYF----------------S--YY-FGE---------KNKALKVFNFVRD-SGPLFIL
CHNPFICWLVCTCVKQRLERGE-DL-E--IN-SQNTTSLYASFLTNVFKAGS-QSFVPK-
-----------VNRARLKNLCALA---AEGV-WT-YT----FVFCHE-------------
--DLRRNGLSES-EGLMWVDM----RLLQRRG-----DCFTFIHLSIQEFCAAMFYLLK-
RPKDN-----------------P--NSAIGSIT-QLVKATVIQ-PQTHLTQVGIFM----
----FGISAEEIVSMLETSFGFP---------------L--SKD-LQQ-EITQSLKSLSQ
CE-----------------------------------ADREAIGFQELFVSLFETQE-KE
FVTQVMNF---------------------F-------------------EEVFIYIGNIE
HL---VIA--S----FCLK-HCRG-LTT-LRMC----------------MEN-----IFP
-DDSGCAS-----------G-------------------F--------------------
------------------------------------------------------------
------------TS---------MLN-FGHDSELFKAV----------------LHNPHL
KLLS-LYGT-SLSHT-DVRHLCETLKHPMCK----IEEL--------ILG----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------KC-DIS-----------------------------
----------------------------SEDCEDVAS-----------------------
--------------------------------VLAC
PF13516 // ENSGT00900000140813_0_Domain_11 (2017, 2054)
NSTLKH---LS-LVENS-LRD---
-------EGMMLLCEAL----KHPHCALETL-----------------------------
------------------------------------------------------------
------------------------------MLMYCCLTCVSCDSISQVL-SCSK
PF13516 // ENSGT00900000140813_0_Domain_13 (2215, 2237)
SLSLLD
L-SSNALEDNGVA-SLCGALKHP--ACS----I---------------------------
-----------------------QEL----------------------------------
------------------------------------------------------C--LQ-
--T-CPITRACCG--------------------DLAAALVACKTL-RSLNLDWITLEPDA
VAVLCE--------ALSHPDCA--LQ--M-------------------------------
---LGLHKS-GFDEETQKM------LM----------SVEEKM-PHLTISHEPWIDE---
--EYK-----IR------------------------------------------------
--------------------------------------------------GVLL------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCHOP00000003344 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------ME---SSFF
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
SDYG---LMWYLEELR--K---GEFRKFKELLKQ------
---------E--ALQLG--------L--TQI-SWAE-VKKAT-REDLANLLIKHYEEQQA
WDLTFRI----FQ-KINRKDLCEKAK-RES------------TG--------------NT
K---------------------------------------------------MYQA----
------------------------------------------------------------
----------------------------------------------------------H-
--------VKKK----------------FNNI--WFRESITRIHEY--------------
---------------------------------F-------AQEVTLEEREYLELLFAPR
AN--GEE-P----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
RTVVLNGTQGIGKTTLLIKL-----MLAWA--K---------GTVYQ
N--------QFLY-VFFFCCREMKQLT-------VTSLAE-LISR-NW-----PTA-PVP
LAEIMS-QP----ERLLFIIDSFED-LKCALEEP------RS-A-----LCSDWMQQ-QP
-VEVVLGSLMTKKMLPESSLLIT-ATPACQWALRAGL-E-----RPE--FGALMGFDESN
RKLFF----------------S--HL-FQD---------RRGAMEAFNFVRE-NEQLFSM
CQTPLLCWIVCTCLKQEMERGR-DL-A--LS-CRRATSLYVSFMFNSFMPKG-ATCPLR-
-----------HSQGQLKGLCSLA---VEGM-WT-DT----FVFNEE-------------
--DLRRNGLADS-DIPALLGV----KALQKHGK-CE-NSYAFIHTCIQEFCAAMFYLLK-
NPSDH-----------------P--NPAVGHAE-ALLLTYR---RKAHWIYLGCFV----
----FGLLNEKEERRLEAFFGFQ---------------L--SPK-IKQ-QFLQCLKRFGE
H-E----------------------------------HLQGDINFMRLFYCLFEMQD-EA
FVRQVMNY---------------------F-------------------QEVNFSIVDNV
DM---MVS--V----YCVQ-HCCG-LRK-LQLS----------------IQN-----VFK
-EEN-KDS-----------P-----------------------------------XXXXX
XXXXXXXX-XXXXXXXXXXXXXXXXXXXXXX---------XXXXXX--XXXXXX------
XX--X-XXXXXLNH---------VSS-TGDSWIFFEAL----------------IHNPDL
KFLS-LSDI-RLSPN-EVKFLCDVLNQPTCN----IEEL--------LL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------TKCCLS-----------------------------
----------------------------ADDCEVFVS-----------------------
--------------------------------VLNSNKKLKH---LN-VAHNT-LG----
-------QGWDLLCEAL----CHPDCTLEAL-----------------------------
------------------------------------------------------------
------------------------------VLTYCGLNESCWENLSEV-LLCNK
PF00560 // ENSGT00900000140813_0_Domain_13 (2215, 2240)
SLVHLD
L-SANNLKDDGLK-LLCEALKHP--NGH----L---------------------------
-----------------------QSLCL--VKC-----LITADGCGDL------------
-------ASVL-TSNQ
PF00560 // ENSGT00900000140813_0_Domain_15 (2357, 2375)
QLRYLEIGCNGIEDVGVKL-LCEALT-HP-NC-HLKNLG--LD-
--A-CGLTGACCA--------------------DLSSALASSTTL-ERLSLTQNALDHDG
VVVLCE--------GLRHPDCA--LK--V-------------------------------
---LGLKKA-DFDEETQGL------LM----------AEE-ERNPYLTIRGN-CETTTRV
EIQ---------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCGRP00000022383 (view gene)
ENSMLEP00000012466 (view gene)
MDQPEDPGSRLRSLRTASPQRAFVQSVPHQRLRLRPWCKAPKLGLTK-RQPGHVTWPGVW
KNRSREGGAGEEGAPQSSGRRWPVPTSPSEGEEHQD------APTRVRGRWR--------
-------AKSGVTPCCPLPPPHG-----SS--LARGSLRPCPRWVLKRQSRGWAGVTRGP
LPDDPRPPVSSTGLR--LAVAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLAALEELS--Q---EQLKRFRHKLRD------
------------VGPDQ-----------RSI-PWGR-LERAD-AVDLAEQLAQFYGPEPA
LEVARKT----LK-RADARDVAAQLK-EQRLRR---------L---------GLG-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------SA-ALLSVSEYKKKYREH-
--------VLQL----------------HARV--KERNARS------------VKITKRF
TKLLIAPESAAPEEE--ALGPAE-------EPEP--------GRARRSDTHTFNRLFRGD
E---DGRRP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYQ
G--------QVDF-AFFMPCGELLERPG------TRSLAD-LILD-QC-----PD-RGAP
VPQMLA-QP----QRLLFILDGADE-LPALEGP--------EAA-----PCTDPFEA-AS
-GARVLGGLLSKALLPTALLLVT-TRAAAPGRLQGRL-C-----SPQ--CAEVRGFSDKD
KKKYF----------------Y--KF-FQD---------ERRAERAYRFVKE-NETLFAL
CFVPFVCWIVCTVLRQQLELGR-DL-S--RT-SKTTTSVYLLFIASVLSSAPVADGP---
-----------RVQGDLRNLCLLA---REGVLGR--R----AQFAEK-------------
--QLEQLELRGSKVQTLFLSKKELPGVLET-----E-VTYQFIDQSFQEFLAALSYLLE-
DEGVP-------------------R-TAAGGVG-TLLRGDSQP--HSHLVLTTRFL----
----FGLLSAERMRDIERHFGC-----------------VVSER-VKQ-EALRWVQGQGQ
GCPGVAPEVTEETKGLEDTEEPDE-----------EEEGEEPNYPLELLYCLYETQE-DA
FVRQALCR---------------------L-------------------PELALQGVRFC
RM-DVAVL--S----YCVR-CCPA-GQA-LRLI------------------SC----RLV
------AA----------------------------------------QEK--K---KKS
----------------------------------------LGKR----------------
------LQAS---L--------------GGS------S-----------------SRGTT
KQ---LPAS-------LLHPLFQTMTDPLCG----LTSL--------TLS---------H
C-----------------------------------------------------------
--------KVP---DA-VCR---DLSEALRA-APALTELGLLHNRLS-EAGLRMLSEG--
-LAWPQCRVQTV------------------------------------------------
----------------------------RVQ-----------------------------
---------------LPSS---------QQGLQYL-------------------------
------------------------------VGMLRQSPALTT---LD-LSGC-QLPAPMV
--T--------YLCAVL----QHQGCGLQTLRL--TSVELSEQ-----------------
----------SLQELQAVKTAKP-------DLV----------------------I--AH
PVLD-------------------GHPESP-KELVSTF-----------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPTRP00000061889 (view gene)
ENSCATP00000011069 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MSLDIQCEQ-LSD-ARWA----------------EL---------
----LPLL-------------------------------QQCQVV------RLDDC-GLT
EARCGDIS-SALRV
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1542)
NPALTELSLRSNELGDA---------GVHCVL--QGLQS-P-----
SC--K-IQKLS--L-QNCYLT--GA---GCG-VLSGTL----------------RS
PF13516 // ENSGT00900000140813_0_Domain_6 (1617, 1642)
LPTL
QELH-LSDN-LLGDA-GLQLLCEGLLDPQCR----LEKL--------QLE---------Y
C------N----------------------------------------------------
---------LS---AA-SCE---PLASVLRA-KPDFKELTVSNNDINE-AGVRVLCQG--
LKD-SPCQLEAL-R----------------------------------------------
------------------------LESCGIT-----------------------------
----------------------------SNNCRDLCS-----------------------
--------------------------------IVASK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
ASLRE---LA-LGSNK-LGD---
-------VGIAELCPGL----LHPSS----------------------------------
--------------------------SLR-TLW---------------------------
-------------------------------IWECGITAKGCADLCRV-LRTKE
PF13516 // ENSGT00900000140813_0_Domain_13 (2215, 2237)
SLKELS
L-AGNELGDEGAR-LLCETLLEP--GCQ----L---------------------------
-----------------------ESLWV--KSC-----SFTAACCSHF------------
-------SSVL-TQ
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NKFLLELQMSNNRLGDAGVQE-LCKGLG-QP-GS-VLRVLW--LG-
--D-CDMSDSSCS--------------------SLAATLLA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2464)
NHSL-RELDLSNNCLGDAG
VLQLVE--------SVRQPGCL--LE--Q-------------------------------
---LVLYDI-YWSEAMEDR------LQ----------ALE-EEKPSLRVIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPCOP00000031041 (view gene)
ENSOGAP00000005359 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MASG----A-HWQ---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 326)
LGYYLELLK--K---EELKEFQLQLSS------
---------E--ASFQG--------P--SSA-AAVP-PEKMS-GMEVASRLVAQYGQQQA
WDLALQT----WR-QMGLNSFCARAQ-AELALLSGDDL----PSLC---SPSIPNLESPN
PPSPAVMQESFDSRVLEKIGYLGYTQHSERRTSDQLTDTSRRHW-KENPYLPFYQ--A--
---------LPSSPD-QQSPSQESPNTPTSTAVLRGGKSPPQPNPEPRKQESPGAEWPLA
ETSEKYFTRIREKERKESRKEGRG---------SPEMPYGT--GPPSISNILGKRHQNQ-
--------KREH----------------LCFTQSW-------KNE---------DLHKKF
TQLLLLQRPHPRGLES---L------VIKKTQNPV--------VENLGHLIEIGDLFASG
LG--TQEEP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
RIVILHGAAGIGKSTLARQV-----QRGWE--E---------GRLYQ
D--------RFQH-VFYFSCAELAQSK-------VMSLAE-LITQ-AQ-----EA-PPAP
LQQVLS-RP----EQLLFILDGVDE-PRWVLWQE------ST-E-----FCQHWSQP-QQ
-VAMLLGSLLGKTILPEASFLIT-TRTVALQKLIPSL-K-----QPR--WVEVLGFSESS
RKEYF----------------Y--NY-FTD---------ENQALRALSLVES-NPAVWTM
CLVPWLSWLVCTCLKQQMERGE-DL-T--LT-TPTATGFCLRYLSQALP--------AQ-
-----------PLGHHLRGICSLA---AEGI-WR-KR----SLFRLH-------------
--DFGRHGLDGP-VISTFVTM----GVLRKY---PNSWGYCFIHRCFQEFFAAVSCALG-
D------EKE--------RGEHP--NSI-RGVG-GLLQAYKTH--DLSRAPTRRFL----
----FGLLSEQAGREMESIFTCQ---------------Q--APE-LKQ-ELVKWAKRESW
-----------------------------------VLRSPQQPGFLELFHCLYETQD-EE
FLTQVIGH---------------------F-------------------LASSVCVQTGM
EL---LVF--T----FCIK-FCHY-VKR-LQLH----------------EGGQ-------
------HRP-RWRPARVVL-----------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------FMQVPVT-----------------------------
----------------------------DACWQLLFS-----------------------
--------------------------------ILEVTGSLKE---LD-LSGNS-LSH---
-------SAVQSLSETL----RCPHCHLETLR----------------------------
------------------------------------------------------------
-------------------------------LASCGLTAEGCKDLVLG-LSTSETLTELE
L-SFNMLMDAGAE-HLCWGLRQQ--NCR----L---------------------------
-----------------------RRLQL--VGC-----GLTS------------------
------------------------------------------------------------
---------SCCR--------------------DLASVLSTNPSL-KKLDLQQNDLGDHG
VRLLCE--------GLRHPVCS--LT--L-------------------------------
---LWLDQA-PLGEELKEE------LR----------TLE-KEKPALLISS-RWEKPSVM
NPN-VGGEKTSN------------------MFSLKRQRRESGPQRSSIGGKGESRADH--
-----------CPAVPEGSSSQAVQ-MEPLSLSPPAPLRT-FHMEPPG-TEDEFLDPTGP
VAAEVVDIETRSLYRVHFPKSGTYDWH---NTGLHFVVRRAV-------TIEIEFCSWKQ
FLDGNMSQHSWMVAGPLLDIKAEPGSVAAV-YLPHFVDLQGG--------QVDVSLFQVA
HFKEEGMLLEKPAQVEPHYIILENPSFSPMGVLLRIFHAALPFLPITSMVLLYHQPQHKE
D-KFHLYLIPNDCCIRKAIDDEEKKFQFVRIHKPNLLTSLYMGSRYTMSGS---------
GKLEIIPKELELCYRSPGEAQMFSEFYVAHVGSGFRLKMRDKKHRTVVWKALVKPGDLRP
AASLGPP-----------------------------AAVALLSFPDTPALL
PF00619 // ENSGT00900000140813_0_Domain_19 (3052, 3133)
HFVDRHREP
LITRVTSVDSVLDKLH-GQVLNEEQYEQVRAEATRPDQMRKLFSFSKSWDQTCKDRLYQA
LKETHPHLIMDLFEGQMWLQRGLLPPRS--------
ENSOARP00000002238 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------RYRNT-
---------MEK----------------F-SV--WKDILWSGDDGGFHYTTT--------
-----------------------------------------------QRSPKLITYLNLQ
AP--TQSTP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TMVLHGPAGVGKTTLAKDL-----MLDWT--Q---------DDLAE
---------TSNS-AFYLSCKGLNHRG-------MCTFAE-LISA-NW-----PR-VQED
MPAILA-RA----QKVLFILDGFDE-LKVPSG------ALIH-D-----LCGDWKKQ-KP
-VPVLLGSLLKRKMLPKATLLIT-TRPGALQELRLLT-E-----QPL--FIEIEGFLEED
RKAYF----------------L--KH-FEE---------ESQALRAFDLMKS-NAALFQL
GSAPSVCWLVCTCLRQQMERGE-DP-A--AT-CRTTTALFLRFLCSRFPPP-RGGGPRRG
------------LQAPLQPLCLLA---AEGV-WT-RS----SVFDGE-------------
--DLRRLGVHPS-ALSPFLDG----NILQKSED-GE-ACYSFIHLSVQQFLAAMFYVLE-
PEEQEE---E--------GLGGC--WWHVGDVG-KLLSKAERL-KNPSLTHVGCFL----
----FGLCNERRAMELEATFGCL---------------V--STE-IKR-ELLKYTLMPHG
KK------------------------------------SFSVMDTKEVLSCLYESQE-EQ
LVKDAMAH---------------------V-------------------KEMSIHLTNTS
EM---MQS--S----FCLK-HCAK-LQK-ISLQ----------------VGKK----IFL
-END-PAL----KS--------------------D------------PQNQISQHD-HRS
LQLWADLC-SVLSSNENLNFLDVKESFLSHS---------SVRVLC--EQITH-I-----
TC--H-LQKGG--F-KMLHPARWSE----YE-ELEWLF----------------KGKKTL
THLV-LEGS-VHGDKMLLLLLCEILKHPRCN----LQYL--------RLG---------S
C------S----------------------------------------------------
---------DT---TW-QWA---DFSSALKV-NQSLKCLDLTASEFLD-AGVKLLCLT--
LRH-PKCVLQKL-S----------------------------------------------
------------------------LENCQLT-----------------------------
----------------------------EACCKELSS-----------------------
--------------------------------ALIVNQRLTH---LC-LAKND-LGD---
-------GGVKTLCEGL----SYPEC----------------------------------
--------------------------QLQ-TLV---------------------------
-------------------------------LSHCNINRHGCKYISKL-LQGDSSLTSLD
L-GFNPIA-TGLC-FLYEALKKP--NCN----L---------------------------
-----------------------KCLGL--WGC-----PITPFSCQHL------------
-------ASAL-VSSRSLETLDLGQDAWGQSGIVV-LLKALK-QN-HG-SLKMLR--LK-
--M-DKSTVEIQR--------------------LLKDVKENNPRLTIECNDA------RT
---------------TRSSCCD--FF----------------------------------
----C-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMNEP00000042967 (view gene)
ENSMLEP00000016844 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MNN---------------------------QKRRKYMELA-----
--------CTWS------------------------------------------LLSG--
-----------------------------------------GN-----------------
--------------------------------------------------DYWEIFRRH-
--------LKEK----------------YLKI-GVDKCYHHGK--------------HIE
SQLLLVKDHGISEETPCGIP-------------------------QQDHFIDMEHVFDPS
EE--GSSSS----R-------TVVLQGSVVHKF-----MFDWA--A---------GTVTP
G-------
PF05729 // ENSGT00900000140813_0_Domain_2 (729, 929)
-REIC-HIA-NLSAA----------------D-LITN-TF-----QD-INGP
ILDTILIYP----EKILFILDGFPE-LQDPVGDQ------EE-D-----LSVHPQER-KP
-VEGLLCSFVRKKLLPDSSLLIT-ARPTTMKL-HSLL-K-----QPI--QAEILRFTDTE
KTAYL----------------L--SQ-FSG---------ANTSM-------K-NEDLDFM
SSLPIVSWMICSVLQSQGDGDR-TL-L--RS-LQTMTDVYLFYFSKCLKTLTGIS-VW--
-----------EGQSCLWGLCCLA---AEGL-QN-HQ----VLFAVG-------------
--DLRRHGIGVCDTNYIFLSR-----FLKKAEG-AV-SVYTFLHFSFQEFLTAVFHALK-
NDNTE-------------------------------------------------------
---------------------------------------------------KMWQEIEIK
DK-----------------------------------ASRLQIEPVDLFHCL--------
------------------------------------------------------------
-------------------------YE----------------I-----QEEENHLLGF-
-EEEERAS----------------------TFVP--------------------------
---------LALTLNQSFQ-----LCSLPVS---------QLHLLC--QALRNP------
YC--K-IKDL---------------------RD---------------LSLVFET
PF13516 // ENSGT00900000140813_0_Domain_6 (1616, 1642)
-NQHL
TDLE-FVKN-TLEDS-GMKLLCEGLKQPNCQ----DNL-------------LFDTLILGV
NF------------YF----SAVLSTNQWLTELEFSETKLEASALKLLCEGLKDTNCK-L
QKLKLCASLLPEGSEA-VCR---YLSCVLIC-NPDLTELDPSENPLGD-IGVKYLCVG--
LRH-SNCKVEKL-D----------------------------------------------
------------------------LSTCHST-----------------------------
----------------------------DASCVELSS-----------------------
--------------------------------FLQVSQALKE---LF-VFANV-LGD---
-------TGVQHLCEGL----QHTKGIIENL-----------------------------
------------------------------------------------------------
-------------------------------LSEYSLSAACCEPLAQV-LSSTRSLTRLI
L-INNKIEDMGLK-LLCEGLKQP--DCH----L---------------------------
-----------------------KDLAL--WTC-----HLSGACCQDL------------
-------CNVL-Y
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
TNEHLRDLDFSDNALGDEGMRV-LCEGLK-CP-CC-KLQTLW--LA-
--E-CHLTDTCCG--------------------ALASVLNRDE
PF13516 // ENSGT00900000140813_0_Domain_17 (2444, 2465)
NL-TLLDLSGNDLKDFG
VQMLCD--------ALIHPICK--LQTFY-------------------------------
------LDTDPLREDAFRK------ME----------VLK-MSKPGITCCLRECLI----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSRNOP00000020393 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MALARA-NSPQEA---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWALNDLE--E---NSFKTLKFHLRD------
---------V--T------------Q--FHL-ARGE-LEGLS-QVDLASKLISMYGAQEA
VRVVGRS----LQ-AMNLMELVDYLN-QVC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LN--DYREIYREH-
--------VLC-----------------LE------ER---QDW----------GVNSSH
NKLLLVA-TSSSGSLRSPSC------SD-------LE-----Q-ELDPV--HVETLFAPE
AE--SYSTP----PI
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 930)
VVMQGSAGTGKTTLVKKL-----VQDWA--K---------GKLYP
G--------QFDY-VFYVSCREVVLLPK-------CDLPN-LICW-CC-GD-----DQAP
VTEILR-QP----ERLLFILDGYDE-LQK---------------------------S-SR
-AERVLHILMRRREVP-CSLLIT-TRPPALQSLEPML-G-----ERR--HVHVLGFSEEE
RETYF----------------C--SY-FMD---------KEQLRNALEFVQN-NAVLYKA
CQVPGICWVVCSWLKGKMARGQ-EV-S--ET-PSNSTDIFTAYVSTFLPTDG-NGDSSEL
----------T-RHKVLRGLCSLA---AEGM-RH-QR----LLFEED-------------
--VLRKHSLDGPSLT-AFLNC----TDYREGLG-IK-KLYSFCHISFQEFFYAMSFLVK-
EDQGQL----------------G--EATHKEVA-KLVDQ---E-NYEEVTLSLQFL----
----FDMLKTDNTLSLGLKFCFK-----------------IAPS-VRQ-DL-KHFKEQV-
V-------------------------------------AIKYNRSWDLEFSLYDSKI-KK
LKQGIQMK---------------------D-------------------VIFNVQHSD-E
KK-SGKKK--P----FSVT-SRFG-K--------------------------G----PIL
--------ET---GKS--T-------------------RKQKK-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------ASN----------------GKS---------R
-ETEE-PPAQDGG-KSR-LASR----GK-GNKEIND---K-DGGV---Y-GQED-GEG-Q
E-VKK-GGEIL---DE-MNG----------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMLUP00000022256 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------LSVGIKEKYQNQ-
--------RRES----------------SCTTQSW-------RKE---------YLHQKS
TQLLLLHRSQHRGYDF---L------LRSKKNHQL--------VDKQRHLMEIGDLFGPG
LS--TQKEP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
PTVVVHGVAGIGKSTLARQV-----RRAWE--E---------GWLFR
D--------LFKH-VFYFNCRELAQSE-------TVSLAE-LITK-DW-----AA-PDAP
IEQILS-QP----EQLLFILDNLDE-PKWDLKGK------SS-E-----LCPHWSQQ-QQ
-VHTLLGSLLKKTLLPTASLLIT-ARITDLRKLIPSL-K-----HPR--WVEVLGFSESA
RKEYF----------------F--QY-FTD---------EGQAIRAFTSVES-NPALFTM
CLMPLVSWLVCTCLKQQMEQKQ-PL-S--LT-SKTITALCLHYFFYVLQ--------AQ-
-----------SLGTNLKGFCSLA---AEGT-CQ-GK----ILFSPE-------------
--DFWKHGLDGD-NISTLLNM----GVLQEH---PTSRSYSFIHLCFQEFFAAMSYASG-
D------IVL--------KSGRF--ENM-KITP-DLRDAYDMR--DLFGKPTTCFL----
----FGLLSDQGMREMESIFKCS---------------L--SRE-RHR-SLLDFAKSEVL
-----------------------------------ENHLNLDAYSWHLLHCFYEIQD-ED
FMKNTF-T---------------------F-------------------YKMRICVQTDM
EL---LLL--T----FFFT-FCHH-IER-LQLN----------------ESGQ-------
------H-R---------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------Q------------
------AGRT--------------------------------------------------
-------------------SGAFLYSWGLFT-----------------------------
----------------------------DSWWQMFLS-----------------------
--------------------------------ILKVPG
PF00560 // ENSGT00900000140813_0_Domain_11 (2019, 2057)
HLRE---LD-LSGNS-LSC---
-------SAVQSLCDAL----KSPHCCLETLR----------------------------
------------------------------------------------------------
-------------------------------LVRCGL-----------------------
------------------------------------------------------------
----------------------------------------TY------------------
------------------------------------------------------------
---------NCCQ--------------------DLASMLGVSP
PF00560 // ENSGT00900000140813_0_Domain_17 (2444, 2463)
SL-TELDLRHNELDDLG
VELLCE--------GLRQPACQ--LK--H-------------------------------
---LRLYQT-QLSKEVKKM------LR----------LLQ-QEKPQLVISS-RRTTTLSV
PEESF-SQLLQKL-----------------S-----------------------------
----------------------------SLSTREPLGN---QPVESLT-MDNYFWGPTGP
VATEMVDK-HRSLYRVHFPVAGSYHWP---NTGLYFVVRGPV-------TIEIEFCAWDQ
FLDQIVPQHSWMVAGPLFDIKVKPGA-VAAVYLPHFIALQGQ--------HVDLSWFQVA
HIKEYGILMEKPARVEPHYIVLENPSFSPIGVLLRKIQRALS-IRVITNVLLYYCLQQEE
V-NFHLYLIPSDCTIQKAIDDEEKKFQFFQIRKPP-LTPLYLGSRYTVSGS---------
DWLEIIPEELELCYRSPGKPQLFSEFYVGHLGSGIWLQIKSKDDATVVWEALVKPGDLRP
AAPLV-----------------------------------SQAMTDAPTL
PF00619 // ENSGT00900000140813_0_Domain_19 (3051, 3135)
LHFVDQHRED
LVARVTSVDTVLDKLH-GQVLSEEQYERVRAEPTNPDKMRMLFSFSKSWDRACKDQLYQA
LKEIHPHLIIDLWEK---------------------
ENSSBOP00000021579 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------T----P--PG--------GPSSP---
------------------------------------------------------GN----
------------------------------------------------------------
---QS-TCLL-------------------------------EVSLATPKKDPQETYRDY-
--------VRRK----------------FRLM--EDRNARLGECV---------NLSHRY
TRLLLVKEHSNPMRAQQQLL------DTGREHARTVG--------HQTSPIKIETLFEPD
EE--RPEPP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
RTVVMQGAAGIGKSMLAHKV-----MLDWA--D---------RKLFQ
G--------RFDY-LFYINCRELNQSAA------ERSMRD-LISS-CW-----PE-PCAP
PRELVR-VP----ERLLFIIDGFDE-LKPSFHDP------QG-P-----WCLCWEEK-RP
-TELLLSSLIRKKLLPELSLLIT-TRPTALEKLHRLL-E-----HPR--HVEILGFSEAE
RKEYF----------------Y--KY-FQD---------AEQAGHVFNYVRD-NEPLFTM
CFVPLVCWVVCTCLQQQLEGGG-LL-R--QT-SRTTTEVYMLYLLSLMQPKPGT-PQL--
-----------QPPPNQRGLCSLA---ADGL-WN-QK----ILFEEQ-------------
--DLRRHGLHGA-DVSAFLNM----NIFQKDIN-CE-RYYSFIHLSFQEFFAAMYYLLD-
E--GE--------------R--G--AGPEQDVH-RLLAEYEFS-ERSFLALTIRFL----
----FGLLNEETRSHLEKSLCWK---------------V--SPH-IKT-RLLQWIQSKAQ
SA-----------------------------------GSTLQQGSLEFFSCLYEIQE-EE
FIQQALSH---------------------F-------------------QVIVVSNIA-S
KM-EHMVS--S----FCLK-NCRS-AQV-LHLY----------------GA------TYS
-ADREDRA--R-CS---------------------------------------AGT---H
TL------------------------L-------------------------V-------
QL-----------PERTVLLD-------TYSEHLASAL----------------CTNPNL
IELA-LYRN-ALGSR-GVKLLCQGLRHPNCK----LQNL--------RLK---------R
C-----------------------------------------------------------
--------GVS---SS-ACE---DLSAALIA-
PF13516 // ENSGT00900000140813_0_Domain_8 (1773, 1796)
NKNLTRLDLSGNGIGL-PGMTLLCEG--
LRH-PRCRLQMIQ-----------------------------------------------
------------------------LRKCQLE-----------------------------
----------------------------SGACQELAS-----------------------
--------------------------------VLS
PF13516 // ENSGT00900000140813_0_Domain_11 (2016, 2054)
TNPHLVE---LD-LTGNA-LED---
-------LGLRFLCQGL----RHPAC----------------------------------
--------------------------RLR-TLW---------------------------
-------------------------------LKICHLNAAACEGLAST-LSV
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
NQSLTELD
L-SLNDLGDPGAL-LLCEGLRHP--TCK----L---------------------------
-----------------------QTLRL--------------------------------
----------------------------------------------------------D-
--S-CGLTAKACK--------------------NLYFTLGI
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2465)
NQTL-TELYLTNNALGDTG
VRLLCK--------RLSHPGCK--LR--V-------------------------------
---LWLFGI-DLNKMTHSR------LA----------ALR-ATKPYLDIGC---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSGGOP00000010282 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MTSP----QLEWT---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 323)
LQTLLEQLN--E---DELKSFKSLLWA------
---------L--PLEDV--------L--QKT-PWSE-VEEAD-GKKLAEILVNTSSENWI
RNATVNI----LE-EMNLTELCKMAK--AEM-----------------------------
------------------------------------------------------------
--MEDG---------------------------------QVQEIDNPEL-----------
-------GDA-------------------------------EEDLELAKPGEKEGWRNS-
---------MEK----------------QSLV--WKNTFWQGDIDNFHDAVT--------
-----------------------------------------------LRNQRFIPFLNPR
TP--GKLTP----Y
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVLHGPAGVGKTTLAKKC-----MLDWT--D---------CNLSP
---------TVRY-AFYLSCKELSRMG-------PCSFAE-LISK-DW-----PE-LQDD
IPSILA-QA----QRILFVVDGLDE-LKVPPG------ALIQ-D-----ICGDWEKK-KP
-VPVLLGSLLKRKMLPQAALLVT-TRPRALRDLQLLA-Q-----QPI--YVRVEGFLEED
RRAYF----------------L--RH-FGD---------EDQAMRAFELMRS-NAALFQL
GSAPAVCWIVCTTLKLQMEKGE-DP-I--PT-CLTRTGLFLRFLCSRFPQG-------AQ
------------LRGALRTLSLLA---AQGL-WA-QM----SVFHRE-------------
--DLERLGVQES-DLRLFLDG----DILRQDRV-SK-GCYSFIHLSFQQFFAALFYALE-
KE--E---EE--------DRDGH--AWDIGDVQ-KLFSGEERL-KNPDLIQVGHFL----
----FGLANEKRAKELEATFGCR---------------M--SPD-VKQ-ELLQCKAHLHA
NK-----------------------------------P-LSVTDLKEVLGCLYESQE-EE
LAKVVVAP---------------------L-------------------REISIHLTNTS
EV---MHC--S----FSLK-HCQD-LQK-LSLQ----------------VAKG----VFL
-ENY-MDF----EL--------------------DIEFERC-TYL--TIPNWARQD-LRS
LRLWTDFC-SLFSSNSNLKFLEVKQSFLSDS---------SVRILC--DHVTR-S-----
TC--H-LQKVE--I-KNVTPD--TA----YR-DFCLAF----------------IGKKTL
THLT-LAGH-IEWERTMMLMLCDLLRNHKCN----LQYL--------RLG---------G
H------C----------------------------------------------------
---------AT---PE-QWT---EFFYVLKA-
PF13516 // ENSGT00900000140813_0_Domain_8 (1773, 1795)
NQSLKHLRLSANVLLD-EGAKLLYKT--
MTR-PKHFLQML-S----------------------------------------------
------------------------LENCRLT-----------------------------
----------------------------EASCKDLAA-----------------------
--------------------------------VLVVS
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
KKLTH---LC-LAKNP-IGD---
-------TGVKFLCEGL----SYPDC----------------------------------
--------------------------KLQ-TLV---------------------------
-------------------------------LQQCSITKLGCRYLSEA-LQEACSLTNLD
L-SINQIA-RGLW-ILCQALENP--NCN----L---------------------------
-----------------------KHLRL--WSC-----SLMPFYCQHL------------
-------GSAL-LSNQKLENLDLGQNHLWKSGIIK-LFGVLR-QR-TG-SLKTLR--LK-
--T-YETNLEIKK--------------------LLEEVKEKNPKLTIDCNAS------GA
---------------TAPPCCD--FF----------------------------------
----C-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCATP00000044950 (view gene)
ENSFDAP00000007526 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------MQSLCESL----RHPLCYLKSLW----------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------L--VGC-----GLTS------------------
------------------------------------------------------------
---------NCCQ--------------------DLASVLSASCSL-TELDLQQNDLGICG
VRLLCQ--------GLRHPTCQ--VT--L-------------------------------
---LWLDHT-L-SEEVTEE------LK----------ALK-EEKPQLLVCS-IWKPRVMI
PTEGRDGGKLGNS-----------------TSSFKRQRPQ--------------------
---------------SEEGSPQLLQ-MEPFLLPSPSPLGH-QHMEPLG-TEDDFLGPTGP
VLTEIIDR-ERNLCR
PF13553 // ENSGT00900000140813_0_Domain_18 (2716, 2997)
VHFPMAGSYHWP---STGLGLSVRKAV-------TIEIEFCAWDQ
FLGINDLQHTWMVAGPLFDIKAEHGA-VADVHLPHFVSLQEG--------TVDISLFQVA
HFKEEGMLLEKPARVEPHYTVLENPNFSPMGVLLRMIPAARRFIPIMSTTLLYHHLHAEE
V-TFHLYLIPSDCTIRKAIDDEEKKFKFVQIHKPPPMDTLYIGTRYTVSGS---------
RKLEIIPKELELCYRSPCEPQLFSEIYVANFGSGIQLQIRSKKDETLIWEALLKPGENSGT00900000140813_0_Linker_18_19 (2997, 3052)
DLRP
A---------------ATL--------VH------PVSLASPSPPNALALLPF00619 // ENSGT00900000140813_0_Domain_19 (3052, 3134)
HFIDQHREQ
LVAQVTSVDPVLDKLH-GQVLSEEQYESVRAEPTKPGQMRKLFGFSRSWNRACKDKVYQA
LKEIHPHLVMELWEKWGGGMGGL-------------
ENSMOCP00000017748 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MALAQA-SSPREA---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWALNDLE--E---NSFKTLKFHLRD------
---------M--T------------Q--FHL-ARGE-LEGLS-RVELASKLISMYGAQEA
VRVVSRS----LQ-AMNLMELVDYLG-QVC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LN--DYRKIYREH-
--------VRC-----------------LE------ER---PDW----------GVNSSR
NELLLVS-RSGPGSPGSPSR------SD-------LQ-----Q-ELDPI--RIEALFALE
AE--SYSAP----PI
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 930)
VVMQGSAGTGKTTLVKKL-----VQDWA--K---------GTLYP
G--------RFDY-VFYVSCREVVLLPK-------CDLSS-LICW-CC-GD-----DQAP
VTEILK-QP----ERLLFVLDGYDE-LQK---------------------------S-SR
-AECVLHILIRRREVP-CSLLVT-TRPPALQSLEPML-G-----ERR--HVHVLGFSEEE
RKNYF----------------N--SY-FMD---------KEQARNALEFVQN-NDVLYKA
CQVPGICWVVCSWLKRKMAGGQ-EV-S--ET-PNNSTDIFTAYVSAFLPTDG-NGDSPEL
----------T-RHRVLRSLCSLA---AEGI-QH-QR----LLFEED-------------
--VLRKHSLDGASLT-AFLNS----IDYREGLG-VK-KLYSFRHVSFQEFFYAMSFLVK-
EDESQV----------------G--KATRREVA-KLVEQ---E-SSEEMTLSLQFL----
----SDMLKTDGTLSLGLKFYLR-----------------IAPS-VRQ-DL-KCFKEKI-
E-------------------------------------AIKYNRSWDLEFSLYESKI-KK
LTQGIQVK---------------------D-------------------VTFNIQHLD-E
KR-PDKER--L----FSVK-GSFG-----KGRV------------------QS----PLL
--------KT---NTS--M-------------------KKQKG-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------ASN----------------GKK---------R
-GAEE-SAQ-MVK-NSR-LAST----GK-GSTEIKG---QEGGGV---E-GRED-GEG-Q
R-VKK-DGELG---EP-MNG---Y-QRE--------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCCAP00000003221 (view gene)
ENSANAP00000022010 (view gene)
ENSCPOP00000020660 (view gene)
ENSOCUP00000004989 (view gene)
-----------------------------------------------------------M
RRRPLA------------------------GE-----SSKATAAWAWGGRRLISAIAVAP
GVRSTMAEERQVDPISSKLPKNL------------L-------------------SDQ-N
L---SSQPCVTKEDDQWVSFS
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 326)
NYG---LQWCLKELS--K---EELQTFKELLRK------
---------K--HSRST--------T--SAL-PWTE-VDNTN-TESLASLLHEYYERPTA
WTMAIDI----FE-EMKLLTLSEKAR-DEMKKHST-DTVPEDAAPVRT--DQGPSKEEAP
ET---------------------------------------------------SQARG--
QD-EAAAA---------ETQD-QVS-----------------------------------
-----------------------------------------------DDDTDRRDYKAH-
--------VKSK----------------FAIK--SDSH-NGSENA---------------
-----------------------------------AS-------DWPEM-QMLADIFNPD
Q---RGFRP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 928)
RTVVLHGRSGVGKSALARRV-----ILRWA--E---------GALYQ
D--------LFNY-VFFLHASDIKWRE-------ESSFVE-LLAR-EW-----PH-PQFP
ATEILS-RP----GRLLFILDSFDD-LDSAPGLC------AG-K-----LCTDWTER-RP
-RCALVRSLLKKALLPESSLLVT-VRDVGMEQLKSLI-V-----SPL--YLVVGGIPVQR
RIQLL----------------L--EH-IPS---------EHRKMQVLHSVVD-NHRLFDQ
CQVPAVCLLICSALELQEAVGK-SL-A--PT-CQTLTGLYATFVFHQPTPGA-ALQRCLS
----------YEERAVLKGLCHVA---VQGV-WS-RK----SVFSSD-------------
--DLRAHGLSAF-ELSSMFPM----SLLLQGVH-GERR-YAFVHLSLQDFFAALYYVLE-
ALEKE--------------TSSC--SLFFEAAQ-SWLELKQTG-IDSRLARVKHFL----
----FGLMSREVMRTLEIRLGCP---------------V--PQGVVKR-ELLHWVSLLGR
QAT----------------------------------AS-APADVIESFHCLFETQD-ED
FVLEALDS---------------------F-------------------QDVRLPISQRM
DL---IAS--S----FCLR-HCRH-LRR-IRVD----------------VKD-----LFL
-KDEFA-V----WP--------------------TI-----------PP--WMQTR-TLM
DECWENFC-SVLATHPSLRQLDLGSSILSDW---------AMKVLC--AKLRH-P-----
AC--R-IQTLI--L-KGAQV---AP---GLQ-HLWVTL----------------LTNRNI
KHLN-LGNT-LLKEE-DVKMACEALKHPNSL----LESL--------RLD---------S
C------G----------------------------------------------------
---------LT---QI-CYL---MISQVIHK-STSLKTLSIAGNKVTD-WGLKPLCDA--
LRL-SWCSLQRL-I----------------------------------------------
------------------------LENCGLT-----------------------------
----------------------------AVSCQVLAL-----------------------
--------------------------------ALTDNHSLTH---LC-LSNNN-PEH---
-------EGLSPLSRAI------PDS----------------------------------
--------------------------GLR-WLL---------------------------
-------------------------------LDQCNLDVAGCGYLAFA-LKDNRRLTHLS
L-NKNPLGDSGVK-LLYEVMREP--SCH----L---------------------------
-----------------------QDLEV--ADC-----RLTAACCGSL------------
-------SHVI-VRSKHLKSLDLAGNALGDTGMAA-LCEGLK-QR-TS-VLQRLG--LE-
--A-CGLTSDCCG--------------------ALLLALSHNQHL-TSLNLVRNNFSPNT
VKKLCS--------AFARPTSN--LR--I-------------------------------
---IGLWKE-QYPAQTQRL------LE----------ELQ-RLRPGLVVDGDWYSHG---
--E---------------------------------------------------------
--------------------------------------------------DDRYWWKD--
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSOGAP00000011980 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------MVSST----ELGFN--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 326)
-LKALLEKLD--Q---NELSEFKSLLKT------
---------F--AQKNE--------L--QEL-PQME-VDTAD-RKQLAEILLNHCPSYWV
DMVVIQA----FR-QMNRRDLWMKAK-NELREAAL-ASID----KNNNKN--EPLWTG--
------IE--WKEG-PE--------------------------Y---ENLEELLQH-LEK
QSER------------------------------------TEEEEVINL-----------
-------GKT-------------------------------KTFLEGEKPGKEDIYRSI-
--------VKKK----------------FWKH--WKNN-WPGLSENIHD-----------
---------------------------------------------VTQRDKMLIPFCDPT
ML--PGPFP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
HTVVLYGPAGVGKTTLAKKL-----MLDWA--E---------NNLNQ
---------VFKY-TFYLSCKELNHMG-------ACSFAE-LIFK-DW-----PE-WQDD
AAEVLA-QA----QKILFIIDGFEE-LRVPAG------ALIH-D-----ICGNWERE-MP
-VPVLLGSLLKRKMLPKASLLVT-TRPEALRELRLLV-E-----QPL--FIEMEGLLEPD
RRAYF----------------L--RQ-FAD---------QD-------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------TFGCR---------------M--FLQ-IRE-ELLKYRANFDE
NK-----------------------------------P-FSRTDMKELLYCLYESQD-EG
LVRGAMAP---------------------F-------------------KELSLHLNNKM
DV---IDS--S----FCLK-HCQN-LQK-LALR----------------VAKG----IFL
-END-TAL----EP--------------------H------------TQLERSQDD-QHI
LSFWTELC-SIFGSNENLIFLKISQSFLSTS---------SVRILC--EQIAS-A-----
TC--N-LQKVV--L-KNVSPA--DV----YQ-NFCFTF----------------IHNETL
TNLT-LQGN-DQDD--AFPSLCEILRHPKCN----LQYL--------SLT---------S
C------S----------------------------------------------------
---------AT---TQ-QWV---NLFLALET-SQSLMNLILSDNHLLD-KGVKLLLRA--
LTH-PKCCLERL-S----------------------------------------------
------------------------LENCYFT-----------------------------
----------------------------EACCKNLAS-----------------------
--------------------------------VLIVSVRLTH---LC-LANNN-LGD---
-------RGVKILCEGL----SYPEC----------------------------------
--------------------------RLQ-ILV---------------------------
-------------------------------LWHCNITTDGCHHLSKI-LQQKTSLTHLD
L-GLNHLGIAGLK-FLCEALKAP--LCN----L---------------------------
-----------------------RCLWL--WGC-----SITTFSCEDL------------
-------SAIL-KSNQNLSTLDLGQNSLGSSGVKM-LCKPLK-HQ-KC-PLQMLR--LK-
--M-DESDVEIQK--------------------LLKEIQATNPHLTIENDNH----DPRG
---------------KRPSSHY--FI----------------------------------
----V-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSNLEP00000012963 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------MAESDSTDFD---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWYLENLS--E---KEFQSFKKYLAR------
---------K--ILDFK--------L--PQV-PVIQ-M---T-KEELANVLPISYEGQYI
WNMLYSI----FL-MMRKEDLCRKI---IRRRN---------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------RNQETCKAL-
--------MRRK----------------FMLQ--WESHTFGEFHYKFFCDVT--------
----------------------------------------------SDVFYILQLAYDST
SC--YSAND----LN
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 930)
VFLVGDRASGKTMVIYLA-----VSRWI--K---------GEMWQ
N--------MISY-VVHLTSHEINQMP-------NSSLAE-LIAK-DW-----PE-GQAP
IADILS-DP----KKLLFIIEDLDN-VRFELNVN------ES-A-----LCSNSTQK-VP
-IPILLVSLLKRKMAPGCWFLIS-SRPTSKDNVKLFL-K-----ETD--CYTTLQLSNEK
REIYF----------------N--SF-FKD---------RQRASAALKLVHE-DEILVGL
CRVAILCWITCTVLNRQMDKG-RDF-Q--LC-CQTPTDLHAHFLADALTSEAGLT-AN--
----------QYHLGLLKRLCLLA---AGGL-FL-ST----LNFSGE-------------
--DLRCVGLTEV-DVSVLQAA----NILLPSST-HK-SRYKFIHLNVQEFCAAIAFLMAV
PNCLI-----------------P----SGGR-E--Y--KEKRE-QCSDFNQVFTFI----
----FGLLNENRRKILETSFGYQ---------------LPLVGS-FRW-YSVGYMKHSGX
XX-------------------------------------XXXXXXXXXXXXX--------
------XX---------------------X-------------------XXXXXXXXXXX
XX---XVS--L----YCLE-YCCH-LRT-LKLS----------------VQR-----IFQ
--NKEPLV-------------------------------------------RPTSSQMKS
LVYWREIC-SLFYTMESLRELHIFDNDLNDI---------SERILS--KALEHS------
SC--K-LRTLKLSY---------VST-ASGFEELLKAL----------------AHNRSL
TYLS-LNCT---------------------------------------------------
------------------------------------------------------------
--------------------------------------------SISV-NMFSLLHEI--
LDE-PTCQISHL-S----------------------------------------------
------------------------LMKCDLR-----------------------------
----------------------------ASECEEIAS-----------------------
--------------------------------LLVNGGSLRK---LT-LSNNP-LRS---
-------DGVNILCDAL----LHPNCTLISL-----------------------------
------------------------------------------------------------
------------------------------VLVFCCLTENCCSSLGRV-LLLSPTLKQLD
L-CVNHLKNYGVL-HVTFPLIFP--TCQ----L---------------------------
-----------------------EELHL--SGC-----FFSSDICQYI------------
-------AIVIA-T
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NEKLRSLEIGSNSIEDAGMQL-LCGGLR-HP-NC-MLVNIG--LE-
--E-CMLTSACCR--------------------SLASVLT
PF13516 // ENSGT00900000140813_0_Domain_17 (2441, 2464)
TNKTL-ERLNLLQNHLGNEG
VVKLLE--------SLIRPDCV--LK--V-------------------------------
---VGLPLT-GLSTQTQQL------LM----------TVK-ERKPNLIFLSETWSLKEGR
EIGVT-------------------------------------------------------
-------------------------------------------------------PASQP
GSI-----------------IPNSNLD---YM--FFKFPRMSAAMRTSNTASRQPL----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSDORP00000011074 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MAWAPA-KTPREA---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLLVLSDLE--E---NNFKILKFHLRD------
---------M--C------------Q--PHL-ARGE-LQGLS-PVDLASRLTLMYGAHEA
VRMVLRA----LK-TMHLLELVDQLS-HVC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LN--DYREIYREH-
--------VRC-----------------LE------EK---EAW----------GVNGSY
SQLLLVVAALNSKSPRSPPC------PN-------LE-----Q-ELGTVQVEVETLFSLG
EE--PCPSP----A
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
LVVLQGSAGTGKTTLVRKM-----MLDWA--T---------GTLYQ
D--------RFDY-VFYVSCREGVLFRT-------CDLDR-LVFW-CC-GD-----NQAP
VREILK-QP----ERLLFILDGFDE-LQR---------------------------S-SP
-MENVLHLLIQRKLLPTCSLLIT-TRPPALQSLEPML-D-----QRCHYHVHILGFSEEE
RKKYF----------------S--LF-FTD---------EGQARDALHRVST-NAVLYKA
CQVPGICWVVCSWLKRQMERGQ-LV-L--ET-PSNSTDIFTAYISTFLTTDDEDPNCSEL
----------T-RHKVLKNLCSLA---AEGI-QH-QR----FLFEEA-------------
--DLSRHNLHAPSLT-AFLNC----DDYQEGLD-MK-KFYSFRHISFQEFFYAMSFLVK-
EDPSQL----------------G--KETQREVE-RLVGIKEQE-KNEEITLSMQFL----
----LDILKCESPVNVGFKFFFK-----------------IAPP-VME-DL-KHFKEQM-
E-------------------------------------RIKHHRSWDLEFSLYDNKI-KK
LAKGIQLT---------------------D-------------------MVFTMGSNE-K
KS-KGMYI--N----YSV-----S-KRY-KEGQ------------------RE----SLM
--------GK---GNG--T-------------------RTQRG-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------TPN----------------TKI---------R
-GSEE-LGQQVRS-STL-AS-R----EK-GAREIEG---KKDGEV---E-RLED-EKA-K
IEIKK-DGE---------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSANAP00000015301 (view gene)
ENSCAFP00000040966 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------SQDKMT---ESFFS
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 328)
DFG---LLWYLEELK--K---EEFWKFKELLKQ------
---------E--PLQLG--------L--KPI-PWTE-LKKAS-REDLAKMLDKHYPGKQA
WDVTLNL----FL-QINRHDLWTKAQ-EEIRNLHQHSLVV--DD--------------FS
S-----------------------------------------------------------
------------------------------------------------------------
--------RD-------------------------------RSDSKYVILDKPSPYRNH-
--------MKEK----------------FRLI--WEKETCLPVPND--------------
---------------------------------F-------YKETIKHEYENLHAAYR-A
SQ--AEESS----P
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVLQGPEGIGKTTLLRKV-----MLEWA--E---------GNLWK
D--------RFTF-IFFLNGCEMNMIT-------ETSLVE-LISR-DL-----PLS-SEP
VEDILS-QP----ERILFIMDGFEE-LKFDLE-L------ST-P-----LCNDQRQR-QP
-MQIILSSLLQRKMLPESSLLIA-LGTEGMRKNYCLL-Q-----HPK--YITLPGFSEHE
RKLYF----------------Y--HF-FRE---------RNKALKAFSFVRG-NIPLFVF
CHNPLVCWLVCTCMKWQLEKGE-DL-E--IV-SESTTSLYTSFFISVFQSRN-ETCPPK-
-----------HSRTRLKGLCTLA---AMGV-WT-RM----FVFCHE-------------
--DLRRNGISES-DTLMWMGM----RILQRSG-----EYFTFTHMCLQEFCAAMFYVLK-
QPKDS-----------------P--NPAIGSVT-QLVTAGVSQ-VQSPLSRMITFL----
----FAFSTEKITNLLETSFGFL---------------L--SKE-LKQ-EITQCLKSLSQ
CD-----------------------------------PNQVAVNFQELFSGLFETHE-KG
FVAQVMDF---------------------F-------------------EEVNIYIGNTE
DL---VIS--A----FCLK-HCQN-LRT-LHLC----------------IEN-----VFS
-DDSGSII-----------K-------------------L-P-------TH--TSALKYI
SPFMQDFC-SVFTTNENFQMLDLDNCKFSEA---------SLAILC--KALAQP------
VC--K-LQNLLAYF---------LNL-PG-CQSVFSAI----------------LHNPHL
KHLN-LHGS-SLSHV-DVRQLCEMLKHPTCS----IEEL--------ML-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------GMCDIT-----------------------------
----------------------------AEACEEIAS-----------------------
--------------------------------VLVCNKKLKL---LS-LAENP-LRN---
-------EGMLMLCDAL----KHPDCVLETLLQG--NQMLTEGGCRADSVVLITPY----
------------------------------------SLLHS--CLCVFAAGVHALILLLF
---------------------SSDHGVGGITLMCCCLTSVACDYISQA-LLC
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2238)
NKSLSFLD
L-ESNFLKDDGVA-SLCEALKHP--NCH----I---------------------------
-----------------------EQLWL--ADC-----SLTSLCCKNL------------
-------SDVL-VCN
PF13516 // ENSGT00900000140813_0_Domain_15 (2356, 2379)
EKLKILKLGSNDIQDAGVKQ-LCEALK-HP-DC-KVEHLG--LD-
--M-CQLTTACCE--------------------ALASALTVCKSL-KSLNLHWISLDRDG
AVVLCE--------ALNHLDCA--LQ--Q-------------------------------
---LGLDKS-VFDKEIQML------LT----------AVE-EKNPHLTISHLLWINK---
--EYR-----IR------------------------------------------------
--------------------------------------------------DCNTWWWGL-
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSSBOP00000016349 (view gene)
ENSTGUP00000008037 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------------TTALFEPG
RD--GQTPQ-----
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
VVVLVGAPGMGKTTMVRKV-----MVEWG--E---------GTLHT
---------QFDF-VFCIDCKAITL-P------MEASVAD-LVSQ-CC-----PA-LQVP
AGKILD-DQ----KKILFIFDGFEA-LGFSLVQP------EA-E-----LSSDPREV-KP
-LELTLMSLLKKTVLPKASLLIT-MRPMALGSLRQCL-K-----GKH--YMEILGFSAAR
RKEYF----------------H--QY-FEN---------PRKADVAFRFVRG-NEILYSL
CVIPIMSWTICTILEQELCGKK-NL-L--EC-SKATTGMTMFYLSQLLKHRG-RDN-PQV
------------LQQFLLQLCSLA---AEGI-WK-HK----VLFEEK-------------
--EIKDYGLDQPGLLPYFLNE----RIPRKGED-NG-SVYAFTHLHLQEFFAALFYVLE-
DDGET--------------VSSS--GRLEKDVN-KLLESYK-S-RK-DLNVTVLFL----
----FGLVSQKSMEYMDKTFGCR---------------I--SPR-ARE-ELLEWLQGRHR
GLS--------------------------------HPSQALKISEMDTFHFLFEMNE-KS
FAESALGH---------------------F-------------------TDLDLHDTKLT
LY-DQMAL--S----FCIQ-HWAG-LGS-ITLQ----------------GCF-----FHW
-QDP---------------------------------------EA------ELD------
-----------------------------------------ASVPW--------------
-----------------------------------T------------------------
-----SWCP-RRDAL-PIHLFCRALRHRDSA----LQVL--------RLQ---------W
C------Q----------------------------------------------------
---------LS---ES-CCA---ELAALLAE-HPGLAHLEVADSSLGD-SGVRLLCEG--
LRT-AGSRLRIL-R----------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSRBIP00000011721 (view gene)
ENSACAP00000019163 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------------RKKYQRH-
--------IQDT----------------FSHI--PELNPRSGQKV---------SHHQRY
TELFLRGRKPDPEERGHELL------AMERKHQEMEA----HPERCPSL--GLEGLFGPD
AE--G-RSS----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
RTTVLFGPAGIGKTMALRKL-----MLDWA--S---------GESWP
A--------KFAY-AFYVSCGALNGSSE------TMSLVD-LIAN-SC-PP-----GTLA
MEDILM-HQ----DNLLIVLDAFDE-LNLGNLPS------E--G-----PSNDLHKR-KP
-VTDLVLGLLRRSSLP-NALLVA-TRPKAIGSLLQYL-R-----SAR--CVEVLGFHLAQ
RKEYF----------------Q--RF-FED---------REDGAQAFELVRS-NKTLSRM
CFLPASCSVICSIFSRTTQRDPLKD-I--PE-KATVTDIYVWLLFKFLGCHS--------
-------------LVNLEGLCRLA---KERL-LH-KT----AVFDEG-------------
--VLKEHGLGNASTHPG---------ILLQDCH-VT-TTYKFNHLGFQELLAALSFCLL-
DSDGNT-------------------I---SSFQ-ELEEVFGHQ-TESNFTLLLRFL----
----FGLCGKTRLDILQGNWGCR-----------------TS----SQ-ELGIWSEKEVK
H-----------------------------------HSFRSEEKLLELCHCLYETED-LL
FAKNIMRH---------------------I-------------------HDLDLRELLST
NL-DLAAV--A----FCLS-VSDS-LQS-LHLSDFELGPE--------------------
------RLGLLLPDAG--L---QELQL-QTRV--TSLV----------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------DAE----LQEL--------QLQ---------A
--------------SSEPMFVL----SL------FY--VKGIECLPYMCN-VIHPESPLG
FFLRLNRCGLT---VS-TCK---DLSSVVAA-NVSLTSLDLSENPLGD-CGVTELCQG--
LLD-SHCQLQSL-Q----------------------------------------------
------------------------LHGCHLT-----------------------------
----------------------------AAACEPLSAVLAS-------------------
------------------------------KP------SLVE---LN-LGGNP-LGD---
-------PGIHIFCKGM----KNPQC----------------------------------
--------------------------GLQ-RLK---------------------------
-------------------------------LHQCSLTGAACMDLASV-LETGPTLMELN
L-GDNSLGDDGVR-QLCLALKKP--DCK----L---------------------------
-----------------------RKL----------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMFAP00000038181 (view gene)
ENSCSAP00000016325 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---ASFFS
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 326)
DFG---LMWYLEELK--K---EEFRKFKEHLKQ------
---------M--TLHLE--------L--KQI-PWTE-VKKAS-REELANLLIKHYEEQQA
WNITLRI----FQ-KMDRKDLCMKVM-RER------------TG--------------YT
Q---------------------------------------------------TYQT----
------------------------------------------------------------
----------------------------------------------------------H-
--------AKRK----------------FSRL--WSSKSVTEIHLY--------------
---------------------------------F-------QEEVKQEECEHLDHLFAPK
ET--GKQ-P----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
RTVIIQGPQGIGKTTLLMKL-----LMAWS--D---------NKIFR
D--------RFLY-TFYFCCRELRELP-------AMSLAD-LISR-EW-----PDP-AAP
ITEIVS-QP----ERLLFVIDSFEE-LKDGLNEP------DS-A-----LCDDVMEK-RP
-VQVLLSSLLRKKMLPEASLLVA-VQPVCPKELRDRV-A-----ISE--IYEPRGFNESD
RLVYF----------------C--CF-FKD---------PKRAMEAFSLVRE-SEQLFSI
CQIPLLCWILCTSLKQEMQKGK-DL-T--LT-CQTTTSVYSCFIFNLFTPEG-AEGPTP-
-----------QSQHQLKALCSLA---AEGM-WT-DT----FEFRED-------------
--DLRRNGVVDT-DIPALLGT----KVLLKYGE-RE-SSYVFLHACIQEFCAALFYLLK-
SHLDH-----------------P--HPAVRSVQ-ELLIANFEKTRRAHWIFLGCFL----
----TGLLNKQEQQKLDAFFGFQ---------------L--SQE-IKQ-QLHQCLKSLGE
H-D----------------------------------DPLGQVDSLAIFYCLFEMQD-PA
FVKQAVNL---------------------L-------------------QKADFHIIDNA
DL---VVS--A----YCLK-YCSS-LKK-LSFS----------------VQN-----VFK
-KED-EHS-----------S-----------------------------------MSDYS
LVCWHHIC-SVLTTSGHLRELQVQDSTLSES---------TFVIWC--NQLRHP------
SC--R-LQRLGINN---------VSF-SGPSGQLFEML----------------FYQPDL
KYLS-FTLT-KLSRD-DVRSLCDALNSPAGN----VKEL--------AL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------VNCFLS-----------------------------
----------------------------PIDCEVLAC-----------------------
--------------------------------LLTNNKKLTY---LN-VSCND-LD----
-------AGLPLLCEAL----CNPDTVLVYL-----------------------------
------------------------------------------------------------
------------------------------MLAYCHLSEQCCEYISEM-LLHNKSVRYLD
L-GANVLKDEGLK-TLCEALKRP--DCR----L---------------------------
-----------------------DSLCM--VKC-----FITVAGCEDL------------
-------ASAL-VSNQNLKILEIGCNEIGDVGVKL-LCGALT-HP-NC-RLEVLG--LE-
--E-CGLTSACCK--------------------DLASVLTSSKTL-QQLNLTLNALDYAG
VVVLCE--------ALRHPECA--LQ--V-------------------------------
---LGLRKT-DFDEETQAL------LM----------AEE-ERNPNLTITDD-CDTVTRV
EI----------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSRBIP00000011121 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---ESFFSDFG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 322)
-LLWYLKELR--K---EEFWKFKELLKQ------
---------P--LEKFE--------L--KPI-PWAE-LKKAS-KEDVAKLLDKHYPGKQA
WEVTLNL----FL-QISRKDLWTKAQ-EEMRN----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------KLNPYRKH-
--------MKEK----------------FQHI--WEKETCLQVPEH--------------
---------------------------------F-------YKETMKNEYKELNGAYTAA
------ARL----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
HTVVLEGSDGIGKTTLLRKV-----MLDWA--E---------GNLWK
D--------RFTF-VFFLNVYEMNGVT-------ETSLLE-LLSR-DW-----PES-SET
IEDIFS-QP----ERILFILDGFEQ-LKFNLE-F------KA-D-----FSDDWRRR-QP
-TPIILSSLLQKKMLPESSLLIA-LGKLAMQKHYFTL-Q-----HPK--LIKLSGFTESE
KKSYF----------------S--YY-FGE---------KNKALKVFNFVRD-SGPLFIL
CHNPFICWLVCTCVKQRLERGE-DL-E--IN-SQNTTSLYASFLTNVFKAGS-QSFLPK-
-----------VNRARLKNLCALA---AEGV-WT-YT----FVFCHE-------------
--DLRRNGLSES-EGLMWVGM----RLLQRRG-----DCFTFVHLCIQEFCAAMFYLLK-
RPKDD-----------------P--NPAIGSIT-QLIKASMTQ-PQTHLTQVGVFT----
----FGISAEEIVSMLETSFGFP---------------L--SKD-LQQ-EITQCLKSLSQ
CE-----------------------------------ADREAIGFQELFVSLFETQE-KE
FVTQVMNF---------------------F-------------------EEVFIYIGNIQ
HL---AIA--S----FCLK-HCRR-LTT-LRMC----------------MES-----IFP
-DDSGCAS-----------G-------------------Y-N-------EK---------
LVYWRELC-SVFITNKNFQILDMENSSLDDA---------SLAILC--KALTQP------
VC--K-LRKLIFTS---------MLN-FGHDSELFKAV----------------LHNPHL
KLLS-LYGT-SLSHT-DVRHLCETLKHPMCK----IEEL--------IL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------GKCDIS-----------------------------
----------------------------SEACEDIAS-----------------------
--------------------------------VLAC
PF13516 // ENSGT00900000140813_0_Domain_11 (2017, 2054)
NSKLKH---LS-LVENS-LRD---
-------EGMMLLCEAL----KHPHCALETL-----------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------ML--MGC-----FLTSDSCKDI------------
-------AAVL-IC
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NEKLKTLKLGHNEIGDAGIRQ-LCAALK-HP-NC-KLEGLG--LQ-
--T-CPITRACCG--------------------DLAAALVACKTL-RSLNLDWITLEPDA
VAVLCE--------ALSHPDCA--LQ--M-------------------------------
---L--HKS-GFDEETQKM------LM----------SVEEKM-PHLTISHEPWIDE---
--EYK-----IR------------------------------------------------
--------------------------------------------------GVLL------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSOARP00000002124 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------MAPSA----ELG
PF02758 // ENSGT00900000140813_0_Domain_0 (203, 326)
FH---LQPPVEGLG--Q---DELSKFKSLLRD------
---------H--SLKEE--------L--QHL-LQME-VEEAS-RKQPADLLTARCPSFWV
EMVTIQ------G-AMNQMDLSERAK-DELRETAL-KLVQ----ENK------PPSFV--
------GMQRNKRPRLC--------------------------C---HPA---LQKFH-T
LTSSFV---------------------------------N---KEGNKR-----------
-------LGK-------------------------------PEFSSWKIRSKQDKYRNI-
--------LKKK----------------FCQC--W-KNFWPRLSEDVCI-----------
---------------------------------------------VTKRYETLIPFCNPK
ML--AGPFP----Y
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLHGPAGVGKTTLAKKL-----MLDWT--Q---------DDLAE
---------TSNS-AFYLSCKGLNHRG-------MCTFAE-LISA-NW-----PR-VQED
MPAILA-RA----QKVLFILDGFDE-LKVPSG------ALIH-D-----LCGDWKKQ-KP
-VPVLLGSLLKRKMLPKATLLIT-TRPGALQELRLLT-E-----QPL--FIEIEGFLEED
RKAYF----------------L--KH-FEE---------ESQALRAFDLMKS-NAALFQL
GSAPSVCWLVCTCLRQQMERGE-DP-A--AT-CRTTTALFLRFLCSRFPPPRG-GGPRRG
------------LQAPLQPLCLLA---AEGV-WT-RS----SVFDGE-------------
--DLRRLGVHPS-ALSPFLDG----NILQKSED-GE-ACYSFIHLSVQQFLAAMFYVLE-
PEEQE---EE--------GLGGC--WWHVGDVG-KLLSKAERL-KNPSLTHVGCFL----
----FGLCNERRAMELEATFGCL---------------V--STE-IKR-ELLKYTLMPHG
KK-----------------------------------SFS-VMDTKEVLSCLYESQE-EQ
LVKDAMAH---------------------V-------------------KEMSLRLKNKT
DV---VHS--S----FCLK-HCGN-LQK-LSLQ----------------VEEG----IFL
-DSD-SAL----ES--------------------GTQVER------------SQNE-QHM
LPLWMDLC-SVFESNRNLLFLDISQSFLSTS---------SVRILC--EKIAC-A-----
AS--S-LQKVV--L-KNISPA--DT----YR-NFCMAF----------------GGHKTL
THLT-LQGN-DQND--MLPPLCEVLRNPRCN----LKYL--------RLV---------S
C------S----------------------------------------------------
---------AS---AQ-QWA---DLSCCLRT-NQSLTCLNLTANEFLD-EGAKLLYMT--
LRY-PTCFLQRL-S----------------------------------------------
------------------------LENCHLT-----------------------------
----------------------------EAYCKDLSS-----------------------
--------------------------------ALIINQRLTH---LC-LAKNA-LGD---
-------RGVKLLCEGL----TYPEC----------------------------------
--------------------------QLQ-TLV---------------------------
-------------------------------LWCCNITSDGCIHLSTL-LQQNSSLTHLD
L-GLNHIGIIGLK-FLCEALKKP--LCK----L---------------------------
-----------------------RCLWL--WGC-----AITPFCCAEL------------
-------SSAL-RSNQNLITLDLGQNSLGSSGVNM-LCDALK-LQ-SC-PLQTLR--LK-
--I-DESDARIQK--------------------LLREMKESNPQLTIESDHR----DPKD
---------------NRPSSHD--FI----------------------------------
----F-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCSAP00000016321 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------MAESNST
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 325)
DFD---LLWYLENLS--D---KEFQSFKKYLAR------
---------K--ILDFK--------L--PQV-PLIN-LRA-T-KEELANLLPISYEGQHI
WNMLYSI----FL-MMRKEDHCMKI---IGRRN---------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------RNQEACKAL-
--------MRRK----------------ITLR--WENHTFGEFHYKFFCNVA--------
----------------------------------------------SDVFYVLQLAYDST
SC--YSASD----LN
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 930)
VFLMGDRACGKTMTINLA-----VMKWI--N---------GEMWQ
N--------MISY-IVHLTSHEINQMT-------DSSLAE-LIAK-DW-----PD-GQAP
IADILS-DP----KKLLFILEDVDN-LRFELNVN------EH-A-----LCSNSTQK-VP
-IPILLVSLLKRQMAPGCWFLIS-SRPTREENIKMFL-K-----TTD--CYVTLQFSNEK
RELYF----------------H--SF-FND---------RQRASTALMLVHD-NEILTGL
CRVAILCWITCTVLNRQMDKG-HDP-Q--HC-CETPTDLHAHFLADALTSKAGLT-AN--
----------QYHLGLLQRLCLLA---AAGL-FL-ST----LNFSGE-------------
--DLRCVGFTET-DVSVLQAV----NILLPSST-HK-DRYKFIHLNVQEFCAAVAFLMAV
PNCLI-----------------P----SGSR-G--Y--KEKRE-QYCNFNQVFTFI----
----FGLLNENRRKILETTFGYQ---------------LPMVDS-FRR-YSVEYMKHLGR
DQ-------------------------------------EKLTHHGSLFYCLFENQE-EE
FVKKVANT---------------------L-------------------KEVTVYLQSNK
DM---MVS--L----YCLE-YCCH-LRT-FKLS----------------VQR-----IFE
--YKEPLV-------------------------------------------RPTASQMKS
LVYWREIC-SLFCTMDTLWELHLFDSDLNDI---------SERILS--KALEHS------
SC--K-LRTLKLSY---------IST-ASGFEDLLKAL----------------ARNRSL
IHLS-LNCT---------------------------------------------------
------------------------------------------------------------
--------------------------------------------SISL-NMFSLLREI--
LDK-STCQISHL-S----------------------------------------------
------------------------LMKCDLG-----------------------------
----------------------------ASKCGEIAS-----------------------
--------------------------------LLVSGGSLRK---LT-LTNNP-LRN---
-------DGVNILCNAL----LHPNCTLTSL-----------------------------
------------------------------------------------------------
------------------------------ALVFCCLTEKCCSSLGRV-LLLGRTLKQLD
L-CVNHLQNYGVL-HVTFPLIFP--TCQ----L---------------------------
-----------------------EELYL--SGC-----FFTNDICQYI------------
-------AIVIA-TNETLRSLEIGSNSIEDAGMQL-LCGGLR-HP-DC-MLVNIG--LE-
--E-CMLTGACCE--------------------SLASVLTTNKTL-ERLNILKNQLGDEG
VVKLLE--------SLILPECG--LQ--I-------------------------------
---IGLPLN-DVSAETRRM------VM----------TVK-ERKPNLIFLSETWSVKEGR
EIGVI-------------------------------------------------------
-------------------------------------------------------PASQP
GSV-----------------IPNSNLD---YM--FFKFPRMSTPMGMSNTASRQPL----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSNLEP00000009988 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-MDQPEAPCSSTGPR--LAVAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLAALEELS--Q---EQLKRFRHKLRD------
------------VGPDG-----------RSI-PWGR-LERAD-AVDLAEQLAQFYGPEPA
LEVARKT----LK-RADARDVAAQLQ-EQRLQR---------L---------GLS-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------SV-ALLS------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------VSAPGRLQGRL-C-----SPQ--CAEVRGLSDKD
KKKYF----------------Y--KF-FQD---------ERRAERAYRFVKE-NETLFAL
CFVPFVCWIVCTVLRQQLEMGR-DL-S--RT-SKTTTSVYMLFIASVLSSAPVADGP---
-----------RVQGDLRNLCRLA---REGVLGR--R----AQFAEK-------------
--ELEQLELRGSKVQTLFLSKKELPGVLET-----E-VTYQFIDQSFQEFLAALSYLLE-
DGGVP-------------------W-TAAGGVG-TLLPGDAQP--HSHLVLTTRFL----
----FGLLSAERTRDIERHFGC-----------------MVSER-VKQ-EALRWVQGQGR
GCPGVAPEVTEGAKGLEDTEEPEEE-----------EEGEEPNYPLELLYCLYETQE-DA
FVRQALCR---------------------L-------------------PELALQRVRFC
RM-DVAVL--S----YCVR-CCPA-GQA-LRLI------------------SC----RLV
------AA----------------------------------------QEK--K---KKS
----------------------------------------LGKR----------------
------LQAS---L--------------GGS------S----------------SSRGTK
KQ---LPAS-------LLHPLFQAMTDPLCH----LSSL--------TLS---------H
C-----------------------------------------------------------
--------KVP---DA-VCR---DLSEALRA-APALTELGLLHNRLS-EAGLRMLSEG--
-LAWPQCRVQTV------------------------------------------------
----------------------------RVQ-----------------------------
---------------LPDP---------QRGLQYL-------------------------
------------------------------VGMLRQSPALTT---LD-LSGC-QLSAPMV
--T--------YLCALL----QHQGCGLQTL-----------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSTBEP00000011709 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------LGNSH--VS---------NPTVI---
--------CR-----------------------------------GEST-----------
-----------------------------------------------------EEDWTG-
------------------L-----------------LGYLSRISICKKKKDYCRKYRRH-
--------VRSR----------------FQCI--EDRNARLGESV---------NLNKRY
TRLRLVKEHPNRQEREHELL------AIGRT--KTWD------RP--VTSVKLEFLFD--
PDDEHVEPV----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
HTVVFQGAAGIGKTILARKI-----MLDWS--S---------GRLYQ
D--------RFDY-LFYIHCREVSLVT-------RRSLGD-LIAS-CC-----PD-SNPP
LGKIVS-KP----SRILFLMDGFDE-LQGAFDEH------EG-A-----LCTDWQRA-ER
-GDVLLSSLIRKKLLPEASLLIT-TRPVALEKLHHLL-D-----HPR--HVEIVGFSEAR
RKEYF----------------F--KY-FSD---------ESQAREAFSLIQE-NEVLFTM
CFIPLVCWIVCTGLRQQMESGK-SL-AQ--T-SKTTTAVYVFFLSSLLQPRGGSQEHH--
------------LSAHLRGLCSLA---ADGI-WN-QK----ILFEEC-------------
--DLRNHGLQKA-DISAFLRM----NLFQKEVD-CE-TFYSFIHMTFQEFFAAMYYLLE-
EEE---------EGVVRKPGSCS--KLPNRDVT-VLLENYGKF-EKGYLIFVVRFL----
----FGLVNQERTSYLEKKLSCK---------------I--SQQ-IRV-ELLKWIEAKAK
AK-----------------------------------KLPVHPAQLELFYCLYEMQE-ED
FVHRAMDH---------------------F-------------------PKIEINLSPRM
---DHVVS--S----FCIE-NCRS-VDS-LSLG----------------FSHN-----SP
KEEEE--------E------------------------------------E---------
---------------KDSQHPDMGQGVLPSP-----------------------------
------------------------------------------------------------
--------------------------QATCS----N-----------RLA----------
N-----------------------------------------------------------
--------YLT---SS-FCR---GLFSVLCT-NP
PF00560 // ENSGT00900000140813_0_Domain_8 (1775, 1801)
SLTELDLSDNTLGD-PGMRVLCET--LRH-PGCNIQRL-W----------------------------------------------
------------------------LGRCGLS-----------------------------
----------------------------EQCCFDISL-----------------------
--------------------------------VLSSSQ
PF00560 // ENSGT00900000140813_0_Domain_11 (2019, 2052)
KLVE---LD-LSDNA-LGD---
-------FGIRLLCVGL----THPLCSLKKL-----------------------------
--------------------------------W---------------------------
-------------------------------LVSCFLTSACCQDLASV-LSTNQSLTRLY
V-GENALGDSGVE-MLCEKVKHP--GCN----L---------------------------
-----------------------QKLGL--VNS-----GLTAACCSAL------------
-------SLAL-SGNQNLIHLYLRGNALGDVGVKR-LCEGLS-HP-NC-KLQFLE--LD-
--S-CNLTSRCCW--------------------DLCTLLTSTQ
PF00560 // ENSGT00900000140813_0_Domain_17 (2444, 2463)
SL-RKLSLGNNDLGDLG
VMMLCE--------VLKHGDCL--LQ--S-------------------------------
---LQLCEM-YFNYETKRA------LE----------ALQ-EERPELTLIFEPSW-----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPANP00000003150 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MTSP----QLEWT---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 323)
LQTLLEQLN--E---DELKSFKSLLRA------
---------L--PLEDV--------L--QQT-RWSE-VEEAD-GKKLAEILIHTSSENWI
RSATVTV----LE-EMNLTELCKMAK-AEM------------------------------
------------------------------------------------------------
--LEAG---------------------------------QVQEIDNPQL-----------
-------GDA-------------------------------GEDSELAKPGEKEGRRNS-
---------MQK----------------QSLV--WKNTFWQGDIDSFHDDVI--------
-----------------------------------------------LRHQRFIPFLNPR
TP--GKLTP----Y
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVLHGPAGVGKTTLAKKC-----MLDWT--D---------CNLSP
---------MVRY-AFYLSCKELSRMG-------PCSFAE-LISK-DW-----PE-LQDD
IPSILA-QA----QRILFVVDGLDE-LKVPPG------ALIQ-D-----ICGDWTQQ-KP
-VPVLLGSLLKRKMLPKATLLVT-TRPRALRDLQLLV-Q-----QPT--YIRVEGFLESD
RRAYF----------------L--RH-FGD---------EDQAMRAFELMRS-NAALFQL
GSAPAVCWIVCTTLKLQMEKGE-DP-A--PT-CLTSTGLFLRFLCSQFPQG-------AQ
------------LWGALRALSLLA---AQGL-WA-QM----SVLHGE-------------
--DLERVGVQES-DLRLFLDR----DILRQDRV-SK-GCYSFIHLSFQQFLTALFYALT-
-------------------GTGH--AWDIGDVQ-KLLSEEERL-KNPDLIQVGHFL----
----FGLANEKRVKELEPLLAGP---------------D--VTE-IKQ-ELLRCKAIFMS
CA-----------------------------------A-CTSL-----------------
-----------------------------------------------------RRRSWQR
NM---KHF--P----YSLK-QCRD-LEK-VSLQ----------------VAKG----VFL
-ENY-VDF----EP--------------------DIEFERC-SYV--TLPSWAQQD-LRS
LRLWTDFC-SLFSSNSNLKFLEVKQSFLSDS---------SVRILC--DHITR-S-----
TC--H-LQKVE--I-SLCHPR--WS----AVVQSCVFS----------------RGGIS-
------PCW-PGWSQTS-----DLRHEPP-------CLA--------QLG---------G
H------C----------------------------------------------------
---------AT---SE-QWV---DFFYVLKA-NQSLRHLHLSASVLLD-EGAKLLYKT--
MTH-PKHFLQML-S----------------------------------------------
------------------------LENCRLT-----------------------------
----------------------------EASCKDLAA-----------------------
--------------------------------VLVV
PF13516 // ENSGT00900000140813_0_Domain_11 (2017, 2054)
SQQLTH---LC-LAKNP-IGD---
-------TGVKFLCEGL----SYPEC----------------------------------
--------------------------KLQ-ALV---------------------------
-------------------------------LQQCSITKRGCRYLSEA-LQEACSLTNLD
L-SLNQIA-RGLW-ILCQALENP--NCN----L---------------------------
-----------------------KHLRL--WSC-----SLMPFYCQHL------------
-------GSAL-ISNQKLETLDLGQNHLWKSGIIQ-LFRVLR-QR-TG-SLKTLR--LK-
--T-YETNLEIKK--------------------LLEEVKEKNPKLTIDCNAS------GA
---------------TAPPCCD--FF----------------------------------
----C-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMOCP00000027502 (view gene)
ENSCAFP00000009685 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------VAVA
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 328)
RDL---LQATFEDLN--Q---EQLKRFRHKLRD------
------------ATLDG-----------RNI-PWGR-LDGLD-AMDLAAQLTHFYGPEPA
LDVTRKT----LK-RADMLDVAARLK-EQQLQR---------L---------GPS-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------SS-TLRSVSEYKKKYRDH-
--------VLQQ----------------HAKV--KERNARS------------VKINKRF
TKLLIAREGAALPDE--ALGCTK-----ELEPEP--------QRARRSDTHTFNRLFGRD
E---EGQRP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYH
G--------QVDF-AFFLPCRELLERPG------ACSLAD-LVLD-QC-----PQ-RKAP
LKPMLA-RP----ERLLFILDGADD-LPPLAP---------HAA-----PCTDPFQA-AD
-AARVLGGLLGKTLLPAARLLVT-ARTTAPGRLLDRL-C-----SPQ--CAEVCGFSDKD
KKKYF----------------H--KF-FRD---------ERLAERAYRFVKE-NETLFAL
CFVPFVCWIVCTVLRQQLELGQ-DL-S--RT-SKTTTSVYLLFIISVLSSAPAADAP---
-----------RMRAELRKLCRLA---REGTLGH--R----AQFAEK-------------
--DLERLELRGSKVQTLFLSKKELPGVLQT-----D-VTYQFMDQSFQEFFSALSYLLE-
EERDL-------------------R-ALL--------QG------ESHLTLTTRFL----
----FGLLNPDRVSDMQRHFGV-----------------AVPER-VKQ-DVLLWMQEQGQ
GRRRAAPEKTAGTDAAEDAEEPEE-------------EECEPNYPLELLYCLYETQE-DA
FVRQALRC---------------------L-------------------PELELERVRLS
RM-DLVIL--S----YCVR-CCPP-GQA-LRLV------------------SC----GLA
------PA----------------------------------------QK-K-----KKS
----------------------------------------LMKR----------------
------LHSS---L--------------GGS------S-----------------SQASK
RK---PQPS-------PLRPLFEAMTDEHCG----LNSL--------TLC---------R
C-----------------------------------------------------------
--------KLP---DS-VCH---DLSEALRE-APALTELGLLHNQLS-EAGLRVLLEG--
-LAWPQCRVQKL------------------------------------------------
----------------------------RVQ-----------------------------
---------------QPSL---------QEAPQYI-------------------------
------------------------------ISILRQSVA-ST---LY-LGDC-QLPGPVV
--T--------SLCAAL----QHPGCRLQTLTL--TSVELSKE-----------------
----------SLKELQAVKTAKP-------GLV----------------------I--IH
PALD-------------------SHPKPSKGLSDAL------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSFCAP00000018722 (view gene)
ENSMFAP00000017378 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---ESFFSDFG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 322)
-LLWYLKELR--K---EEFWKFKELLKQ------
---------P--LEKFE--------L--KPI-PWAE-LKKAS-KEDVAKLLDKHYPGKQA
WEVTLHL----FL-QIDRKDLWTKAQ-EEMRN----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------KLNPYRKH-
--------MKEK----------------FQLI--WEKETCLPVPEH--------------
---------------------------------F-------YKETMKNKYKELNGAYTAA
------ARL----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
HTVVLEGSDGIGKTTLLRKV-----MLDWA--E---------GNLWK
D--------RFTF-VFFLNVYEMNGVT-------ETSLLE-LLSR-DW-----PES-SET
IEDIFS-QP----EGILFIMDGFEH-LKFNLK-F------KA-D-----LSDDWRRQ-QP
-TSIILSSLLQKKMLPECSLLIA-LGKLAMQKHYFML-Q-----HPK--LIKLSGFTESE
KKSYI----------------S--YY-FGE---------KNKALKVFNFVRD-SGPLFIL
CHNPFICWLVCTCVKQRLQRGE-DL-E--IN-SQNTTSLYASFLTNVFKAGS-QSFVPK-
-----------VNRARLKNLCALA---AEGV-WT-YT----FVFCHE-------------
--DLRRNGLSES-EGLMWVDM----RLLQRRG-----DCFTFIHLCIQEFCAAMFYLLK-
RPKDN-----------------P--NSAIGSIT-QLVKATVIQ-PQTHLTQVGIFM----
----FGISAEEIVSMLETSFGFP---------------L--SKD-LQQ-EITQSLKSLSQ
CE-----------------------------------ADREAIGFQELFVSLFETQE-KE
FVTQVMNF---------------------F-------------------EEVFIYIGNIE
HL---VIA--S----FCLK-HCRG-LTT-LRMC----------------MEN-----IFP
-DDSGCAS-----------G-------------------Y-N-------EK---------
LVYWRELC-SVFITNKDFQILDMENSSLDNA---------SLAILC--KALTQP------
AC--K-LRKLIFTS---------MLN-FGHDSELFKAV----------------LHNPHL
KLLS-LYGT-SLSHT-DVRHLCETLKHPMCK----IEEL--------IYRYKPDDHSVVE
TKTIFFTH------------------------------------PSENCRFSLWNSI---
-PFRL----PC--FSQFSHQ---YISSYCRQ----------------CAPRVLSPANF--
KGF--FF-----------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------SFPPWAWFWS-----------------------------
------------------------------------------------------------
------------------------------FFVFIVLK------------NSRSFLHLWM
N--------------VCTGF----------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCLAP00000015171 (view gene)
ENSTTRP00000000282 (view gene)
------------------------------------------------------------
------------------------------------------------------------
-------------M-SDVSWD----------------SDSLTSF-------------S-P
S---SAHRLLSSSSSSSAFP
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
FKDG---VMLYMVYLS--K---ENLQRFKQLLVE------
---------E--SPRPG--------S--LQI-SWDQ-LKTAR-WAEVVHLLIESFPGRLA
WEVTHDI----FA-KMDQAELCLRVQ-MELNDILP-NLEPEALNPR------E-------
------------------------------------------------------------
------------------------------------------------------------
-------TQM-------------------------------NLEEDEA--DKLREYRLH-
--------LMNE----------------YSTL--RNSTAWPGNHMDFFYQDL--------
----------------------------------------------HREETFLPCLLLPR
KP--QGRQP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
KTVVLQGVAGVGKTSLAKKV-----MLEWA--E---------NKFYA
H--------KLWN-AFYFHCQEMDQLD-------EQSFAE-LIAR-KL-----SG-SRAL
ASKIIS-KA----DQLLLFFDGFEE-LTSTLI------DRPE-D-----LSEDWSQK-VP
-WSVLLTSLLSKRMLPGATLIIF-LRFTSWKNLNPFL-K-----CPS--LITLTGFSVPE
RSRYF----------------R--TY-FRN---------KREADEALSFVMG-NTILFSM
CQVPVICWMVCSCLKQQMERGA-NL-P--HT-FPNATAVFVHFLSSLFPTRV-RDLFNTT
------------HQEQLKGLCSLA---AEGM-WE-RR----WVFNKR-------------
--DLNLARLDET-DVETFLSA----NIFRRVVG-NR-DLYAFAHLSFQEFFAALLYVLC-
FPRRL--------------RNFH--VLDRFHVL-RLIAHPG-R-KRNHLAQMALFL----
----FGLLNDACALAVERVFGCK---------------V--SLG-NKK-KLLKVAAEPHE
CD-----------------------------------PPTPHHGVPQLFHCLHEIRE-EV
FVSQILSS---------------------Y-------------------RKATLVISRSK
DV---QVS--A----FCLK-FCRR-LQE-LELT----------------ITL-----IIT
-RASRLYP----DP-------------------------------LPAS--GPEGG-DKC
YLWWQDFC-SVFRTHESLEVLAVTNST-GPD---------SLKVLS--TALKH-P-----
HC--K-LQKLI--F-RHVSPL--VL---N-E-DLIRVL----------------VENQYL
RYLE-IQDT-EVRCQ-VMEFLCNALKHPQCF----LQCL--------RVE---------D
C------P----------------------------------------------------
---------FS---PR-NWA---DLARNLKN-NVHLKTLMLKRSSLER-FEPCHH-----
--L-PVAQLEKL-S----------------------------------------------
------------------------LENCDLT-----------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------SLSCKSLISS-LASNKNLTHLS
L-AKNALKDDGAK-QLWNALQCS--QYP----L---------------------------
-----------------------QRLVL--RNC-----ALTSECCPDM------------
-------ASAL-DKNK
PF00560 // ENSGT00900000140813_0_Domain_15 (2357, 2381)
NLRSLDLGFNSLKDDGLIL-LCQALM-NP-DS-GLQILE--LD-
--K-CLFTSVCCQ--------------------AMASVLLNNQTL-RYLDLSKNDIGFGG
ILHLRE--------AVRKRKWG--SE--V-------------------------------
---S------LYS-PASIG------II-----------------AFSPI--LLDFTA---
--EGV-------------------------------------------------------
-------------------------------------------RLSHV---NPSTW----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPPYP00000003332 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----MSLD--I----QSLDIQCEE-LSD-ARWA----------------EL---------
----LPLL-------------------------------QQCQVV------RLDDC-GLT
EARCKDIS-SALRVNPALAELNLRSNELGDA---------GVHCVL--QGLQS-P-----
SC--K-IQKLS--L-QNCCLT--GA---GCG-VLSSTL----------------RSLPTL
QELH-LSDN-LLGDV-GLQLLCEGLLDPQCR----LEKL---------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------Q---------------------------
-------------------------------IWECGITSKGCGDLCRV-LRAKESLKELS
L-AGNELGDEGAR-LLCETLLEP--CCQ----L---------------------------
-----------------------ESLWV--KSC-----SFTAACCSHF------------
-------SSVL-AQNKFLLELQISNNRLEDAGVRE-LCQGLG-QP-GS-VLRVLW--LA-
--D-CDVSDSSCS--------------------SLAATLLANH
PF00560 // ENSGT00900000140813_0_Domain_17 (2444, 2458)
SL-RELDLSNNCLGDAG
VLQLVE--------SVRQPGCL--LE--Q-------------------------------
---LVLYDI-YWSEEMEDR------LQ----------ALE-EDKPSLRVIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSTTRP00000000283 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------RVTFPTGPIC-FLPKNL------------L-------------------LQQ-N
L---SSQPCVKMGEAKLSSF
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
SSYG---LQWCFKQLH--K---EEFQIFKALLKE------
---------S--ASEPA--------A--CSF-PLVE-VDNAD-SEQLTSLLHEHYKESLA
WKISIDI----FE-KMSLSALSEMAR-DEMKKYLL-AEIPEHSTPTKT--DQGLSMEEVP
SH----------------------------------------------------------
------------------------------------------------------------
-------------------------------------------------RDDQQDYKSH-
--------VMRK----------------FSTK--LDAH-HSFEKF---------------
-----------------------------------AS-------DCPDV-HVLLGAFNPD
Q---GGFRP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
LTVVLHGKAGIGKSALARRI-----LLFWA--Q---------GELYQ
G--------MFSY-VFFLHAREVQRTR-------ECSFAE-LISR-QW-----PD-TQVQ
VEEILS-QP----ERLLFVVDGFED-LDLAFKDD------DS-N-----LSGDWAEK-QP
-VSVLMHSLLKKVLLPESSLIIT-VRDSGIEKLQSVL-L-----SPH--YLLVGGLSVEK
RMQLL----------------L--EH-MKH---------EHQKMQTLRSVVG-NHEVFDK
CQVSVVCSLVCEALNLQEAPGK-SL-S--IT-CQTLTGLYASFVFHQLAPKD-AVRSCLR
----------QEERVTLKGLCQMA---VEGV-WN-TK----FVFYSD-------------
--DLGIHGLKEP-ELSALFHT----SILLQGSR-CEEC-YMFLHLSLQEFFAALYYVLE-
GWEGE--------------GDPY--PLFTENVK-SLMELKQID-FNVHLVQMKRFL----
----FGLMSKEVMGALGTLLGSP---------------V--PL-VAKQ-RLLHWISLLGQ
HTN----------------------------------TT-APLDFLDAFYCLFETQD-DE
FVRSALNS---------------------F-------------------QEVWLPLNRPM
DL---IVS--S----FCFR-HCQY-LRK-IRLD----------------VRE-----V-P
-KHEFAES----WP--------------------SI-----------PQ--GLQIK-TH-
DEHWEHLC-SVLSTHPNLRQLDLSGSILSEE---------AMKTLC--IKLRQ-P-----
TC--K-IQNLI--F-TGAQI---TP---GLR-HLWLTL----------------I-NRNI
KYLD-LEST-HLKDR-DVMMACEAVKHPSCL----LESL--------RLD---------H
C------E----------------------------------------------------
---------LT---HT-CCQ---VIAQMLVT-SI
PF00560 // ENSGT00900000140813_0_Domain_8 (1775, 1801)
SLKSLSLAGNKVTD-QSIKSLCDA--LKV-TQCTLQKL-X----------------------------------------------
------------------------XXXXXXX-----------------------------
----------------------------XXXXXXXXX-----------------------
--------------------------------XXXXXXXXXX---XX-XXXXX-XXX---
-------XXXXXXXXXX----XXXXX----------------------------------
--------------------------XXX-XXX---------------------------
-------------------------------XXXXXXXXXXXXXXXXX-XXXXXXXXXXX
X-XXXXXXXXXXX-XXXXXXXXX--SCL----L---------------------------
-----------------------QDLEL--VKC-----RLTAACCKNL------------
-------SYVI-TRSQ
PF00560 // ENSGT00900000140813_0_Domain_15 (2357, 2381)
HLKSLDLAANALGDHGIAA-LCEGLQ----KN-SLRRLG--LE-
--T-CGLTSNGCE--------------------ALSSALSCNQHL-TSLNLMQNNFGPRG
MTKLCS--------AFAQPTSN--LQ--T-------------------------------
---IGLWKW-QYPARIRRL------LE----------ELE-RLKPHIVIGDGWYSFA---
--E---------------------------------------------------------
--------------------------------------------------DDRYWWKN--
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSAMEP00000011039 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------T----MSLDIQCQQ-LSD-ARWT----------------EL---------
----LPLL-------------------------------QQSEVV------RLDDC-GLT
EMRCKDIS-SALQANPSLTELSLCTNELGDA---------GVHLVL--QGLQS-P-----
TC--K-IRKLS--L-QNCCLT--KT---GCG-VLPDML----------------RSMP
PF00560 // ENSGT00900000140813_0_Domain_6 (1619, 1645)
TL
RELY-LNDN-PLGDA-GLQLLCSGLLDPQCH----LEKL--------QVE---------Y
C------N----------------------------------------------------
---------LT---AA-SCE---SLASALRA-KRHFKELAVSNNELGE-AGVRVLCRG--
LVD-SACQLEVL-K----------------------------------------------
------------------------LENCGLT-----------------------------
----------------------------SASCEDLCG-----------------------
--------------------------------VVASKP
PF00560 // ENSGT00900000140813_0_Domain_11 (2019, 2057)
SLQE---LD-LGDNK-LGD---
-------QGIATLCPRL----LHPSC----------------------------------
--------------------------QIR-VLW---------------------------
-------------------------------LWDCDITTAGCRDLCRV-LRAKESLKEMS
L-AGNALGDEGAR-LLCESLLEP--ACQ----L---------------------------
-----------------------QSLWV--KSC-----SFTAASCQHF------------
-------SSML-TRNT
PF00560 // ENSGT00900000140813_0_Domain_15 (2357, 2381)
CLVELQLSNNKLGDCGVQQ-LCQGLS-QP-GA-ALQVLW--LG-
--D-CDVAHSGCA--------------------SLASVLLVNRSL-RELDLSNNCMGEEG
ILRLME--------SVEQPGCV--LE--Q-------------------------------
---LVLYDI-YWSEEMDNS------LR----------ALE-GRKPSLRVIF---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMFAP00000006491 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MNN---------------------------QKKRKYMELA-----
--------CTWS------------------------------------------LLSG--
-----------------------------------------EN-----------------
--------------------------------------------------DYWEIFRRH-
--------LKEK----------------YLKI-GVDKCYHHGK--------------HIE
SQLLLVKDHGISEETPCGIP-------------------------QQDHFIDMEHVFDPS
EE--GSRMC----W-------DWET
PF05729 // ENSGT00900000140813_0_Domain_2 (686, 870)
AV-V-HKF-----MFDWA--A---------GTVTP
G--------REIC-HIA-NLSAA----------------D-LITN-TF-----QD-INGP
ILDTILIYP----EKILFILDGFPE-LQDPVGDQ------EE-D-----LSVHPQER-KP
-VEGLLCSFVRKKLLPDSSLLIT-ARPTTMKL-HSVK-T-----TY--------------
---------------------------------------PGRDL-------T-NEDLDFM
SSLPIVSWMICSVLQSQGDGDR-TL-L--RS-LQTMTDVYLFYFSKCLKTLTVIS-VW--
-----------EGQSCLWGLCYLV---AEGL-QN-HQ----VLFAVS-------------
--DLRRHGIGVCDTNCIFL-----------------------------------------
----------------------------------------------SFSSLMMQFL----
----FGLLHKGKGKAVETTFGIK---------------L--SPG-LRE-GLLKWTEREMK
DK-----------------------------------ASRLQIEPVDLFHCL--------
------------------------------------------------------------
-------------------------YE----------------I-----QEEENHLLGF-
-EEEERAS----------------------TFVP--------------------------
---------LALTLNRSFQ-----LCSLPVS---------QLHLLC--QALRNP------
YC--K-IKDL---------------------RD---------------LSLVFET
PF13516 // ENSGT00900000140813_0_Domain_6 (1616, 1642)
-NQHL
TDLE-FVKN-TLKDS-GMKLLCEGLKQPNCV----LQT-----------LRFFRRLISSS
CG------------AL----AAVLSTNQWLTELEFSETKLEASALKLLCEGLKDTNCK-L
QKLKLCASLLPEGSEA-VCR---YLSCVLIC-NPDLTELDPSENPLGD-IGVKYLCVG--
LRH-SNCKVEKL-D----------------------------------------------
------------------------LSTCHST-----------------------------
----------------------------DASCVELSS-----------------------
--------------------------------FLQVSQALKE---LF-L-----------
----------QHLCEGL----QHTKGITENL-----------------------------
------------------------------------------------------------
-------------------------------LSEYSLSAACCEPLAQV-LSSTRSLTRLI
L-INNKIEDMGLK-LLCEGLKQP--DCH----L---------------------------
-----------------------KDLAL--WTC-----HLSGACCQDL------------
-------CNAL-Y
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
TNEHLRDLDFSDNALGDEGMRV-LCEGLK-CP-CC-KLQTLW--LA-
--E-CHLTDACCG--------------------ALASVLNRDE
PF13516 // ENSGT00900000140813_0_Domain_17 (2444, 2465)
NL-TLLDLSGNDLKDFG
VQMLCD--------ALIHPICK--LQTFY-------------------------------
------LDTDPLREDAFRK------ME----------VLK-MSKPGI-CCLRECLI----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSEEUP00000003349 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MAAALA-HN
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
PREA---LLWALSDLE--E---NNFKVFKFHLRD------
---------Q--SLFGG--------Q--FQL-ARGE-LEGLN-RVDLASRLISICGAQEA
VKVVLKV----LK-IMNLMELVDQLS-HVC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LN--DYRHIYREY-
--------VRC-----------------LL------ER---WEG----------GVHKKY
NQLLLVT-KPSSGSPVSPVG------AA-------LE-----Q-ESDSD--TVKALFDPA
AE--ASQHP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
PLVVLQGSAGMGKTTLVRKM-----VQDWA--A---------GTLYQ
G--------QFDY-VFYVSCREVVLLPE-------GKLKQ-LLFW-CC-GD-----NQAP
VNEIVR-QP----ERLLFLLDGFDE-LQRPFAVG------LQ-K-----LG-----Q-SP
-MERLLTCLMKREVLPTCSLLVT-TRPLALQNLESLL-K-----KPR--HIHVLGFSKKE
REKYF----------------S--IY-FTD---------KEQARNAFAIVQG-SDVLYKA
CQVPGICWVVCSWLKGQMERGS-EI-S--EM-PGNDTDIFMAYVSTFLPPSG-NEGCPEA
----------S-RDKVLQGLCSLA---AEGI-QQ-QQ----FLFKEE-------------
--DVRKHGLDGPSLP-HFLNS----SDYYEGLD-TK-KFYSFRHIIFQEFFHAISYLVK-
KDKDQM----------------G--KGSCREVS-RLLDEKEEA-GGEEMTLSMHFL----
----LDISKKEKLSNLELKFCFK-----------------IP------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSEEUP00000012162 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MDSR----T
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
-EEK---LAQCLKEMK--K---EELRRFLHQLSH------
---------K--EPHS--------LS--TSE-VPSQ-LLKLD-AMEVASHLVSQYGEQKA
WDLALHT----WE-QMGLGDLRAKAQ-KEVILMPXXXXXX-XXXXX---XXXXXXXXXXX
XXXXXX-----XXXXX-----XXXXXX-----XX--XXXXXXXX-XXXXXXXXXX--X--
-----------XXXX----------XXXXXXXXXXXXX-------X-XXXXXX--XXXXX
XXXXXXXXXX---XXXXXXXXX-----------XXXXXXX-XXXXXXXXXXXXXIKYTR-
--------IRDY----------------PCPTQSW-------KDG---------NTQQNF
TQLLLLYRSHPGG------L------LRGSWHQGV--------VEEPGYLIKAIDLFVPE
LG--AQKEP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
YTVILHGEAASGKSTLAKQV-----R-AWG--K---------GHLYR
D--------HFQH-VFYFNCREVNDADE----FMPMTLAE-LLTE-DS-----AV-PVIS
IAKVLS-QP----EQLLFIPNSLDE--LWIWMKQ------RS-Q-----MCLHWSQQ-QP
-VYTLLGSFLEKTILPEASLLIT-ARNKALREFSPSL-K-----QPC---VEVLGFSESN
RNDYF----------------Y--KY-FTE---------ESQASRVLSVVES-NRA-LTL
CLLPWVSWLVCTCLKLQVDQEQ-DL-S--LM-CQTTTALCLQFLSQNLP--------DQ-
-----------SRGTQLKGLCFLA---AEGT-QP-GY----TMFTPL-------------
--DLTKHQLNDS-VISTFLNM----GVLQKH---STSLHYSFTHQCFQEFFAAMFCVLG-
D------NNE--------NTDPP--NSI-RTAT-RLLEIYGWY--DILGAATTHFL----
----FGLLSEQGMRNLEDIFNCT---------------L--TLE-KKW-ELLQWAQL---
------------------------------------VQSFLWPYSLQLLYCLYEIQD-EK
FVTQAMAH---------------------F-------------------QGKWMLFQTDI
NL---LVF--T----FCTK-FCHH-IQR-LQLK----------------MGGQ-------
------Y-R---------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------Q------------
------AWKP--------------------------------------------------
-------------------PGIILFTGAPVT-----------------------------
----------------------------DVWWQSLFS-----------------------
--------------------------------ITGIMGSLKD---LD-LSGNF-LSQ---
-------SAVQSLCVAL----TFPLCHLQILW----------------------------
------------------------------------------------------------
-------------------------------LDSCGL-----------------------
------------------------------------------------------------
----------------------------------------RS------------------
------------------------------------------------------------
---------SCCQ--------------------DLVSALSSSPSL-TELYLRQNDLGDFG
MRLICE--------GLRHTFCQ--LT--F-------------------------------
---LWLDQA-QLSEKVTEL------LS----------ILE-EEKPQLHISG-RRKPHVIS
PTKGSGGSERNDS-----------------TTSLSQQRPESLLV----------------
-------------------SSQLA---QVECLSPLTSSED-LHIEPVR-AEDNFWSPMRP
VTTEMVDR-ERSLYRVHFPIPGSYHWP---NIGLHFVVKGSV-------TIEIEFCDWKQ
FLDKIVPQENWMVAGPLFDIKAEPGT-VAALYLPHFVSFQGG--------HVDTSQFRVA
HFKMEGMVLEQPTRVVPCYTVLENPGFSPMGVLLRMFPVTRNFIPITTIVLLYHKPHPQD
V-TFHLYLIPNDGAIQKAIDNDESLSQFVRIRKPPPLMPLHVGSHYTVSAS---------
QQLHILP-ELKLCYRSPREAQLFSEIYVDQMGSGIILKIKSKKDGTTVWETLVRPGDLRP
AGALDPP-----------------------------FPSVSASTPDARNV
PF00619 // ENSGT00900000140813_0_Domain_19 (3051, 3135)
MRFLEQNRNN
LVTRVTTVDPLLDKLL-GPVLSEEEYKMVQAEDTTQKQMRRLFSFSPAWDLACKNCFFQA
LQETHPHLIGELLEMGGGLLTRGG------------
ENSCLAP00000004444 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MSLEICHEQ-LSD-ARWM----------------EL---------
----LPLI-------------------------------QQHQVV------RLEFC-DLT
EGRCRDIS-SALWA
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1542)
NPTLTELSLNHNELGDA---------GVHLVL--QGLQS-P-----
TC--K-IQSLS--L-QYCGLT--ET---GCG-VLSSLL----------------PSM
PF13516 // ENSGT00900000140813_0_Domain_6 (1618, 1642)
PTL
RELY-LSGN-SLGDG-GLLLLENSGT00900000140813_0_Linker_6_7 (1642, 1650)
SKGLQDAQPF13516 // ENSGT00900000140813_0_Domain_7 (1650, 1766)
CH----LEKL--------QLE---------Y
C------N----------------------------------------------------
---------LS---DA-SCE---PLASVLKA-KSNIKELVVSNNDIHE-AGMRTLLRG--
LKD-SACLLETL-W----------------------------------------------
------------------------LESSGVT-----------------------------
----------------------------AANCKDLCD-----------------------
--------------------------------VMAARPFLQD---LE-LGDSR-LGD---
-------AGVATLCSWL----LHPSC----------------------------------
--------------------------RLR-KLW---------------------------
-------------------------------LWECDITAKGCRDLSQV-LR
PF13516 // ENSGT00900000140813_0_Domain_13 (2212, 2237)
TNQNLKALS
L-MLNRLGDEGAQ-LLCEALQEP--TCQ----L---------------------------
-----------------------ECLWV--KEC-----SFTAACCPYF------------
-------RAVL-A
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
QNKFLTELQLGENNLGDTGVQE-LCQGLG-QP-GS-VLQVLG--LV-
--D-CKLTNSSCS--------------------SLASVLLA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2464)
CHSL-RVLDLSNNGLGDPG
ALQLLE--------SLRQPACA--LD--C-------------------------------
---LAVFDV-YLSSGVWDQ------LE----------ALE-KEKPSLRIIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSANAP00000000245 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MAGG----A-WGR---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LACYLELLK--K---EELKKFQLLLTN------
---------K--VHSR--------GS--LGE-TPAQ-PEKTS-GMQVASHLVAQYGEQQA
WDLAIHT----WE-QMGLRSLCAQAQ-EEAG-------HS-LSFPY---SVSEPSAGSPN
QPTSTAVLRSSIPEL------QGCTQGSKRSILRHLPDTSGHRW-REITSSLLYQ--A--
---------PLSSPD-YGSPSHESPNAPISTAELGSWESPLQSSPMPREQEAPGIQWPLD
ETLGNYYTGETLLSTY-----M------PRWFAVSIPLSL-T---SGISPHVIPPQPSQ-
--------PLES----------------NCSKWSW-------KNE---------DLSQKF
TQLLLLQRPHPRSHEC---L------IRRRLLHNV--------EEDQGRLIEIRDLFHPG
LD--AQ-EP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
RIVILQGAAGIGKSTLARRV-----REAWE--R---------GQLYR
D--------CFQH-VFYFSCRELAQSK-------VLSLAE-LIGK-DW-----TA-TQAP
IRQILS-RP----ERLLFILDGVDE-PKWVLKEQ------SS-E-----LCLHWSQP-QP
-ADALLGSLLGKTLLPKASFLIT-ARTTVLQSLIPSL-E-----QAR--WVEVLGFSESS
RKEYF----------------Y--KY-FTD---------ERQASRAFRLVKS-NKELWAL
CLVPWVSWLACTCLMQQMKQKE-EL-T--LT-SKTTTTLCLHYFSQALQ--------AQ-
-----------PLGSQLRGFCSLA---AEGI-WQ-RK----TLFSPD-------------
--DLKKHGLDEA-IISSLLKM----GILQEH---PIPLSYSFIHLCFQEFFAAMSCALE-
D------KKE--------RGEHS--NHI-MGLG-KLLEAYGTH--DLFRAPTTRFL----
----LGLLSKQGAREMDNIFDCR---------------L--SQE-RKR-RLLQCAQF---
------------------------------------LQPPLQSHSLEFLHCLYETQG-KR
FLTQMMAH---------------------F-------------------QEVCMCVETDM
EL---LVC--T----FCIK-FCHD-VKK-LQLI----------------DSRQ-------
------R-G---------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------P------------
------AWSL--------------------------------------------------
-------------------DRVVLFRWVTAT-----------------------------
----------------------------DAYWQILFS-----------------------
--------------------------------VLKVTGSLKE---LD-LSGNL-LSL---
-------SAVQSLCETL----RYPRCL
PF13516 // ENSGT00900000140813_0_Domain_12 (2068, 2206)
LETLR----------------------------
------------------------------------------------------------
-------------------------------LAGCGLTAEGCKDLENSGT00900000140813_0_Linker_12_13 (2206, 2212)
ASG-LRPF13516 // ENSGT00900000140813_0_Domain_13 (2212, 2237)
TNQTLTEVE
L-SFNMLTDAGAK-HLCQGLRRP--SCK----L---------------------------
-----------------------QRLQL--VSC-----GLTS------------------
------------------------------------------------------------
---------GCCQ--------------------ALASVLSA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2465)
SPSL-KELDLQQNDLDDVG
VQLLENSGT00900000140813_0_Linker_17_18 (2465, 2716)
CK--------GLRRPTCK--LT--R-------------------------------
---LGLDQT-TLSDETRQE------LR----------TLM-QEKPRIQIFPMRRKPSTMI
PNEGLDTGEMSNS-----------------TSSLKRQRLRSE------------------
----CG--ADHCLAVSEGSVPRITQ-QELACLPSPASQGD-LHTKPLG-TDDDFWGPTGP
VATEVVDK-ERSLYRPF13553 // ENSGT00900000140813_0_Domain_18 (2716, 2995)
VHFPVAGTYHWP---NMGLCFVVREAV-------TTEIEFCVWDK
LLGEINQQHSWMVAGPLLDIKAEPGA-IEAVHLPHFVALQGG--------HMDTSLFQVA
HFKEEGMLLEKPAKVELHHVVLEDPSFSPMGVLLRVIHNALRIFPVTSMVLLYHRIHPEE
V-TFHLYLIPSDCSIRKAIDDEETKFQFVRLHKPPPLTPLYMGSRYPSGERPPMQDEQGW
QLNSLPLQELELCYRSPGEPQLFSEFYVGHLGSGIRLQMKNKTDETLVWETLVKENSGT00900000140813_0_Linker_18_19 (2995, 3052)
PEPVCS
-------------------------------------LPALPSPLDAPELLPF00619 // ENSGT00900000140813_0_Domain_19 (3052, 3134)
HFVDQYREQ
LVARVTSVEPVLDNLL-GHVLSQEQYERVLAEDTRPSKMRKLFSFSRSWDRSCKDRLYQA
LKETHPYLIMELWEKGSKKGLLPLSI----------
ENSCATP00000017631 (view gene)
ENSFDAP00000020144 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---SPFFADFG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 322)
-LMWYLEELK--K---KEFMKFKELLRQ------
---------E--TPQLG--------L--GQI-PWGE-VKRAS-REELANLLLRHYEGEQA
WNVTFRV----FQ-KLGRKDLCAKAV-RES------------SG--------------NV
K---------------------------------------------------IYQA----
------------------------------------------------------------
----------------------------------------------------------H-
--------MREK----------------FCSF--LAKESIPMVGEH--------------
---------------------------------F-------NLNVTQEKCKFLELLFEP-
----GKQ-T----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
RTLCLHGIQGVGKTTFLAKI-----IVAWS--K---------GFIYQ
D--------RFSY-IFYFCCKELKRLP-------AVSLME-LIAR-AW-----PTA-SAP
VSEIMA-HP----EKVLFMVDDFEE-LGSDLREP------ES-E-----LCGDWAEA-HP
-PKLLLSSLLRKKMVPECFLLVT-LSPKARQGLEDRL-K-----DPD--IRVLTGFNECD
MKIFI----------------S--QI-FPD---------MYRALDAFAVILD-NHQIFSL
CQLPVLCWMACVALKWEMDQGN-DV-K--LT-CQRRTALYTSFLLNLFTPQG-AAGPSW-
-----------QGQAWLQSLCSLA---VQGM-WT-DT----LVFGER-------------
--DLVRNGIGSS-DLPALLEL----RVLLLCQE-AE-ASYRFLHSSVQEFCAALFYLLK-
SPGHH-----------------P--HTAVQGTQ-ELLLTYLQK-SGEHWVFVGCFL----
----FGLLHERERQKLDAFFGFQ---------------L--SGE-MKW-QLYRCLQDLAK
ITG----------------------------------KLRRETTFLALFYALFEMQE-EA
FTDQVMDL---------------------M-------------------SELTFSVFHET
DL---KVS--A----YCLK-RCSN-LKK-LSLS----------------AHS-----VFK
-NEERGQS-----------S-----------------------------------VSSTE
LLSWHQVC-SVLTTNRDLQVLQVKDSTLNEP---------ALLTLC--HQLRQP------
NC--L-LENLFLNN---------ITF-HCEDWALFEVF----------------THNPNL
KYLN-LSNT-LLGCQ-EMQSLYSMLSHPACN----LEKL--------LL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------MNCKLS-----------------------------
----------------------------ADDCKVLAS-----------------------
--------------------------------ILIESRKLRH---LN-LSCNY-LN----
-------KGARALCKAL----CHPACTLELL-----------------------------
------------------------------------------------------------
------------------------------VLALCKLRAWCWDYLADV-LLLSKSLVHLD
L-SINSLRDEGLQ-VLCEALRHP--VCR----L---------------------------
-----------------------QTLCL--MKC-----SITVGGCRDL------------
-------ASVL-TSSQSLRTLFITENSIEDAGMKM-LCWALA-RP-SC-LLETLG--VK-
--D-CGLTSACCE--------------------DLATVLTSSKTL-SMLNLIGNPLGHSG
VALLCQ--------GLRHPECA--LQ--V-------------------------------
---LGLNKA-EFGEETRAL------LE----------AEE-ERNPDLIIKES-W------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCAFP00000003671 (view gene)
ENSPEMP00000009823 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MSLDIQCEH-LSD-ARWT----------------EL---------
----VPLI-------------------------------QQYQVV------RLDDC-GLT
EVRCKDIS-SAIQA
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1543)
NPTLTELSLCTNELGDA---------GVSLVL--QGLQK-P-----
TC--K-IQKLS--L-QNCSLT--EA---GCG-VLPNVL----------------RS
PF13516 // ENSGT00900000140813_0_Domain_6 (1617, 1642)
LPTL
RELH-LSDN-PLGDA-GLKLLCEGLLDPQCR----LEKL--------QLE---------Y
C------N----------------------------------------------------
---------LT---AT-SCE---PLASVLRV-KPDFKEIVLSNNDLHE-AGIQMLCQG--
LKA-SACQLESL-K----------------------------------------------
------------------------LESCGIT-----------------------------
----------------------------SANCKDLCD-----------------------
--------------------------------VVASK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
ASLRQ---LD-LGSNK-LGN---
-------AGIAALCSGL----LLPSC----------------------------------
--------------------------RLR-TLW---------------------------
-------------------------------LWECDITAEGCKDLCRV-LRAKE
PF13516 // ENSGT00900000140813_0_Domain_13 (2215, 2237)
SLKELS
L-AGNELRDEGAQ-LLCESLLEP--GCQ----L---------------------------
-----------------------ESLWV--KTC-----SLTAASCSHF------------
-------CSVL-T
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
KNRSLLELQMSSNPLGDLGVLE-LENSGT00900000140813_0_Linker_15_16 (2378, 2391)
CKALG-QP-DT-VPF13516 // ENSGT00900000140813_0_Domain_16 (2391, 2436)
LRVLW--LG-
--D-CDVTDSGCS--------------------SLENSGT00900000140813_0_Linker_16_17 (2436, 2441)
ASVLLPF13516 // ENSGT00900000140813_0_Domain_17 (2441, 2465)
TNRSL-RELDLSNNCMGDSG
VLQLME--------SLKQPSCA--LQ--Q-------------------------------
---LVLYDI-YWTGEVEDQ------LR----------ALE-EERPSLRIIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSSBOP00000032399 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------MAMA-RNPREA---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWALSDLD--E---KDFKKLKFYLRD------
---------M--TWSEG--------Q--APL-ARGE-LEGLI-AVDLAELLISKYGEKEA
VKVVFEG----LK-AMNLLELVDQLS-HIC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LH--DYREIYREH-
--------VLC-----------------LE------EW---QEV----------GVNGRY
NQVLLVA-KPSSESQESPAC------PIT------PE-----Q-ELDSV--MVEALFDSG
EK--PSLAP----S
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
LVVLEGSAGTGKTTLSRKM-----VLDWA--A---------GTLYP
G--------QFDY-VFYVSCKEVVLLPE-------SKLEE-LVFW-CC-RD-----NQAP
VAEILR-QP----ERLLFILDGFDE-LQRPFEEK------LK-K-----RG-----S-SP
-KEGLLHLLIRRCTLPACSLLIT-TRPPALRNLRPLL-K-----QAR--HVHILGFSEEE
RGRYF----------------S--SY-FTD---------EKQADHAFEIVQK-NDILYKA
CQVPGICWMVCSWLKQQMERGE-AV-L--ET-PRNSTDIFMAYVSTFLLPDD-DGGCSEF
----------S-RHRVLMSLCSLA---AEGI-QH-QR----FLFEEA-------------
--ELRKYNLDGPRLA-AFLSS----NDYQLGLA-IK-KFYSFRHISFQEFFYAMSFLVK-
EDQSRL----------------G--KESHREVQ-RLLEVKKQE-GNDEMTLTMQFL----
----LDISKKDSVSNLELKFCFR-----------------ISPC-IAQ-DL-KHFKEQL-
E-------------------------------------SIKQKRTWDLEFSLYEAKT-KN
LIKGIQMN---------------------N-------------------VSLKIAHSN-E
KK-SQSQN--P----FSIK-TS-----H-GEKQ------------------KC----PLV
--------GK---DNI--A-------------------RTQKE-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------AST----------------GKG---------R
-GTEE-PDEEYQK-N----TYI--------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSLAFP00000022644 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------SSS----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
RTVVLQGCAGIGKTAVVHKF-----MFDWA--A---------GVVTP
G--------RFDY-LIYINCREISHIA-------NISAAD-LIAD-TF-----ED-INGP
VLDIILTYP----EKLLFILDGFPE-LQYSVGEQ------EE-D-----LNANPQEK-KP
-VETLLCSFVRKKLFPESSLLIT-ARPTAMKKLHSLL-K-----QPI--QVEILWFTEAE
KRAYF----------------L--SQ-FSG---------ANAAMRVFYGLRE-NEGLDIM
SSLPIVSWMICSVLQSQGDGDR-AL-M--RS-LQTMTDVYLFYFSKCLKTLTGIS-IW--
-----------KGQSCLWGLCSLA---AEGL-RN-KR----VQFEVN-------------
--DLRRHGVGVFDTNCIFLNH-----FLEKVER-GV-NVYTFLHFSFQEFLTAVFYALK-
NDNSW---------------------MFLDQVGKTWQEIFQQY-GKGFSSLMIQFL----
----FGLLHKGSGKTVEATFGRK---------------I--SPR-LRE-ELLKWTETEIK
DK-----------------------------------TSRLQLEQIDLFHCLYEIQE-EG
YAKRIMDD---------------------L-------------------QSIILLQPTYT
KM-DLLAM--S----YCVK-SSHNNLS----------------V-----SLKCQHLLGF-
-EDEFYF-----------------------------------------------------
---------------------------------------------C--EHLII-------
--------------------------------S---------------LCITFEL-YKDV
KDLE-FVKN-TLEDS-GMKLLCEGLKQPNSV----LHKL---------------------
------------------------------------------------------------
-------SFLPESSEA-VCK---YLASVLMC-NS
PF00560 // ENSGT00900000140813_0_Domain_8 (1775, 1805)
NLTELDLSENPLGD-TGVKYLCEG--
LRH-SNCKVEKL-D----------------------------------------------
------------------------LSTCSLT-----------------------------
----------------------------DASCVELSS-----------------------
--------------------------------FLQMNQTLKE---LF-VFANA-LGD---
-------TGVQHLCEGL----RHAKGIVENL-----------------------------
------------------------------------------------------------
------------------------------VLSECSLSATCCEPLAQV-LSSTRSLTRLL
L-INNKIEDLGLK-LLCEGLKQP--HCQ----L---------------------------
-----------------------KDLAL--WTC-----HLTGACCQDL------------
-------CNAL-YTNE
PF00560 // ENSGT00900000140813_0_Domain_15 (2357, 2381)
HLRDLDLSDNALGDEGMQV-LCEGLK-HP-CC-KLQTLW--LA-
--E-CHLTDVCCG--------------------ALASVLNRNE
PF00560 // ENSGT00900000140813_0_Domain_17 (2444, 2475)
NL-TLLDLSGNDFRDLG
VQMLCD--------ALIHPIC---------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMAUP00000022693 (view gene)
ENSCHOP00000009422 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MRSA----Q
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
LDFN---SRHLLEQFD--Q---DELDKFNSLLSF------
---------L--PLPSE--------L--QQI-PHME-LEGAD-GKQLAEILINNCPSCWV
EAVAILA----FD-KMSQMDLSEKAK-DLRXXX-X-----XXXXX---------------
------------------------------------------------------------
----------------------------------------------XXX-----------
XX-------X-------------------------------XTMWNTESKDNEEIQLHL-
--------EKEK----------------------QELSQVQGEYV---------KILGGA
KESLQRQKQGRDNKYRSILR----KKFWKWKNWP-GAS--EHFPIVTQGYERLITFSYPK
ML--EGSFP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 941)
YTVVLQESAGLGKAMLAKKL-----MLDWE--DNLRQ----------
---------TFKY--TYYLCKEFNEVGT------C-TFAE-LISK-AW-AE-----LQDT
ITGILT-PP----QK--VIIIDFDE-LTVPIV-A------VI-N-----DISDWKIQ-NP
-VCILLGSLLKRQVFPKATLLITMK-PVLR-QLQ-LL-K-----K----PIEIEGFSEFD
RKPYFL---------------K--H--F-K---------DADALQDFNLVRS-NADMFNK
GLVPV-VWIICSCLKVQKENGG-DI-I--LT-CQTSTSLFLHYLCIQFTPAHLQS----P
----------------VKSPCLLT---CQGI-WT-QA----LVFDGG-------------
--RLRSLGEEEGELC-PFMDK----NTLQMGKY-SE-GC-YFNHLSVQQLLVAMFVLLS-
KEEQH-----------------V---------E-TCR-WDGNV-ENASLTQVGYFF----
----FDLLNKKFE--LETTFNCQ-----------------ISMV-IKQ-ELLKCKGKS-N
E-----------------------------------NKQFSSVREMKELFCLYETQE-EE
CVKDVMVH---------------------T-------------------KEMY-LCLK-N
KV-DVIHS--S----FCLQ-HCQN-LQK-SLH-----------------VEKG----IFL
------ENGTVLESNP--------------------------------QVKRSQND-QYI
IPFWMDLC-SMF------------------------------------------------
------------------------------------------------------------
-------------------------DAN--------------------------------
--------------------NN-------MLFLDV-SESFLCNSVRTLCEIITSP-HN-L
Q--KVVLRVSP---LD-AYQ---DFCHAFRG-KECLTLLTLQGNDQND-KP----LPE--
VRH-PKYKLQYL-R----------------------------------------------
------------------------LECCSAT-----------------------------
----------------------------TQQCSDFSS-----------------------
--------------------------------TLKINQSLTY---LN-LTSNE-LLD---
-------EGVKL-YMIL----SNPKCSL--------------------------------
------------------------------QML---------------------------
-------------------------------LEGCYLIEACSEDLISS-FIVQKRLTHLY
L-AKNDLRDEVVK-LLCEGLCYP--DLQ----S---------------------------
-----------------------QVLWC----C-----NIMEAGYNHP------------
--------KLL-QQKSRLTHLDMRLIYIRITGLTSL-CKAL-KNT-FC-NLKYLL--LR-
--SS---------------FKPFSCI----------ELCSSTKSF-FYLVLCQNNLGHNG
IKILFK--------ALRLQTCL--LQ--F-------------------------------
---RLMINE-SD-TEILRM------LK----------EIR-KSNAQLIIESGNPDTK---
--KYR-----HSS-----------------------------------------------
--------------------------------------------------QD-FLF----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCGRP00001002994 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------MARA-NSPQEA---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWALKDLE--E---NSFKMLKFHLRD------
---------M--T------------Q--FHL-ARGE-LEGLS-RVDLASKLISMYGAPEA
VRVVSRS----LQ-AMNLMELVNYLS-QVC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LH--DYREIYREH-
--------VRC-----------------LE------ER---QDW----------GVNSSR
NELLMVA-TSSPGSPGSPPC------SD-------LQ-----Q-ELGPV--SVEALFTPE
AE--SYSTL----PI
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 930)
VVMQGSAGIGKTTLVKRL-----VQDWA--K---------GILYP
G--------QFDY-VFYVSCRDVVLLPK-------CDLPN-LICW-CC-GD-----NQAP
VTEILR-QP----ERLLFILDGYDE-LQK---------------------------S-SR
-AESVLHILIRRREVP-CSLIIT-TRPPALQSLEPML-G-----ERR--HVHVLGFSEQE
RENYF----------------N--SY-FMD---------KEQARNTLEFVQN-NDVLYKA
CQVPGICWVVCSWLERKMARGQ-EV-S--ET-PSNSTDIFTAYVSTFLPTDG-NEDSSGL
----------T-RHRVLRSLCSLA---AEGI-QH-QR----LLFEEN-------------
--ILRKHSLDGPSLT-AFLNS----IDYREGFS-IK-KLYSFRHISFQEFFYAMSFLVK-
EDQNQQ----------------G--EATRREVA-KLLEQ---E-KYEDMTLSLQFL----
----FDMLKIEGTLSLGLTFYFK-----------------IAPS-VRR-EL-NYFKERI-
Q-------------------------------------TIKCNRSWDLEFSLYDSKI-KK
LTQGIHMK---------------------D-------------------ITVNIQRSG-E
KK-PGKRK--T----VTVE-SSLG-----KGQV------------------QS----PLL
--------KT---NNS--T-------------------EKQKG-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------ASN----------------GKS---------R
-ETEE-PTG-KVR-NSR-LADT----ER-GNMEMKG---QEGGGV---E-RQED-GEG-Q
R-VTK-DGEMR---DQ-MNG---Y------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSSBOP00000027355 (view gene)
ENSSARP00000007120 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MDNI----L
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
-QQR---LAQYLNLLR--K---DELNEFLSWLPE------
---------M--ILIR--------CS--STE-TLTQ-PKTRE-GTEVASKMITCHGEIKA
WKKALQT----WE-KMGLSSLRAKAE-ADTDLIPXXXXXX-XXXXX---XXXXXXXXXXX
XXXXXXXXXXXXXXXX------------------XXXXXXXXXX---XXX-XXXX--X--
----------XXXXX----------XXXX--XXXXXXXXX-XXXXXXXXXXXXXXX--XX
XXXXXXXXXX--XXXXX-XXXXX----------XXXXXXX---XXXXXXXXXXALRQRG-
--------QQES----------------SSLIHT-------CRNC---------TYYTNF
TQLRLLQLXXXXXXXX---X------XXXXXXXXXXX----XXXXXXXXXXXXXXXXXXX
XX--XX-X-----XXXXXXXXXXXXXXXXXXXX-----XXXXX--X---------XXXXX
X--------XXXX-XXXXXXXXXXXXX-----------XX-XXXX-XX-----XX-XXXX
XXX-XX-XX----XXXXXXXXXXXX-XXXXXXXX------XX-X-----XXXXXXXX-XX
XXXXXXXNFLENTNLPE-SIVIP-VRSTNMNGFIPP--K-----PSR--YVEVLRYSEFG
RKNYF----------------Y--KY-FTE---------ENQACRAISLVES-NQALWTM
CFIPWVSWLVCTCLKQQMDQGQ-IL-S--LT-CKTTTALCLQFLSQTLP--------AQ-
-----------SLGMQMRALCSLA---DKEN-WQ-TK----INVTIF-------------
--TCRKYWLDET-TKSTLLQM----GVLQQCQPCSTPLGYRFTHRYFQEFFKAMSIILR-
I------CED--------TSDIF--YPR-KDIG-MLLKNYFTK--DMFMSPTMCFL----
----FGFLSNRGKQEVERIFHCD---------------L--ALE-KKW-RVMCWGPS---
-------------------------------------SKPFLPYSQHLFYCLYEIQD-QE
FLTEVMDK---------------------F-------------------QGTKIFIQTDI
EF---LVV--T----FCIK-FWHY-VMR-LQVN----------------ENGH-------
------G-----------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------P------------
------IWIA--------------------------------------------------
-------------------PDVILLTRAPVT-----------------------------
----------------------------DNCWRILFS-----------------------
--------------------------------ISGLMGCLKE---LN-LSGNF-LSC---
-------SAVQSLCKAL----SFPNCNLRILF----------------------------
------------------------------------------------------------
--------------------------------VDYNMTAEDCKDLAS--LSTIHIITEPE
L-SNHMLIDAVVQ-NFHQSLRQP--NCH----L---------------------------
-----------------------R-LLL--DNC-----GLEL------------------
------------------------------------------------------------
---------SSCQ--------------------YLTSMLRTS-NL-RRLSLEQNDLGSQG
LRMFCK--------ELRHPSCK--LQ--T-------------------------------
---LWLDQX-XXXXXXXXX------XX----------XXX-XXXXXXXXXX-XXIPPQMT
PKMDDEENTMKDD-----------------AYLLKQQRRKSE------------------
-----------------GSSSQVL--RRPLCLSSPEPLGD-LHMKSLG-TDDYFWGPTGP
VATEIVDR-ERNLYRIHLPMAGFFYWP---HMGLHFVVRGPV-------TIEIEFCAWDQ
FLDKTVLQNNWMVAGPLFDIKAEPGT-VEAVYLPHFVAIRXXXXXXXXXXXXXXXXXXXX
XXXXXXXX--------XXXXXXXXXXXXXXXXXXXXXXXXXX-XXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXAVDDEEKRFQFVRIYKPPPIRPLYMGSHFTVSGS---------
EVNEIIPKXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXDLKP
RVPQFPP-----------------------------FSNVSPLHLGEQAL
PF00619 // ENSGT00900000140813_0_Domain_19 (3051, 3135)
KHFLDKNREK
LVSRVTSVDSILDKLI-GTALNEEQYEKIRAKTTRHDQMCMLFLFSQSWSPANRYSFYCA
LKEIHPHLIGELWESCSQALQTSR------------
ENSSBOP00000017054 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MAGG----A-WGR---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LACYLELLK--E---EDLKKFQLLLTN------
---------K--VHSR--------GS--LGE-SPAQ-PEKMS-GMQVASHLVAQYGEQQA
WDLAIHT----WE-QMGLRSLCAQAQ-EEAG-------HS-SSFPY---SLSEPSLGSPN
RPTSTAVLRSWIPEL------QGCTQGSERSVLRHLPDMSGHRW-REITSSLLYQ--A--
---------PLSSPD-YGSPSHESPNAPTSTAELGSWESPLHPSPVPREQEAPGTQWPLD
EMLGNCYTGIRER------------------------------------------DQNL-
--------WRES----------------PCSTWSW-------KNE---------DLNQKF
TQLLLLQRPHPRSHEC---L------VRGRLLHNV--------EEDQGHLIEIGDLFGPG
LD--TQ-EP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
RIVILQGAAGIGKSTLARWV-----REAWG--R---------GQLYR
D--------CFQH-VFYFSCRELAQSK-------LLSLAE-LIGK-DW-----TA-TQAP
IRQILS-SP----EQLLFILDGVDE-PKWVLKEQ------SP-E-----LCLHWSQP-QP
-ADALLGSLLGRSILPKASFLIT-ARTTALQSLIPSL-E-----QAH--WVEVLGFSESS
RKEYF----------------Y--KY-FTD---------KRQANRAFRLVKS-NKELWAL
CLVPWVSWLACTCLMQQMKQKE-EL-T--LT-SKTTTTLCLHYFSQALR--------AQ-
-----------PLGSQLLGFCSLA---AEGI-WQ-RK----TLFSPD-------------
--DLKKHGLDGA-IISSLLKM----GILQEH---PIPLSYSFIHLCFQEFFAAMSCALE-
D------KKG--------RSEHS--NHI-VGLG-KLLEAYGTH--DLFRAPTTRFL----
----LGLLSKRGTREMENIFDCQ---------------L--SQE-RKR-RLLQCAQF---
------------------------------------LQPSLQSHSLEFLHCLYETQG-KS
FLTQMMAH---------------------F-------------------QEVCMCVETDM
EL---LVC--T----FCIK-FCHD-VKK-LQLI----------------EGRQ-------
------H-G---------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------P------------
------AWSL--------------------------------------------------
-------------------DRVVLFRWVPAT-----------------------------
----------------------------DAYWQILFS-----------------------
--------------------------------VLKITGSLKE---LD-LSGNL-LSL---
-------SAVQSLCETL----RYP
PF13516 // ENSGT00900000140813_0_Domain_12 (2065, 2206)
CCLLETLR----------------------------
------------------------------------------------------------
-------------------------------LAGCGLTAEGCKNLENSGT00900000140813_0_Linker_12_13 (2206, 2213)
ASG-LRTPF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
SQTLTEVE
L-SFNALTDAGAK-HLCRGLRQP--SCK----L---------------------------
-----------------------QQLQL--VSC-----GLTS------------------
------------------------------------------------------------
---------GCCR--------------------ALASVLSA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2465)
SPSL-KELDLQQNDLGDVG
VQLLENSGT00900000140813_0_Linker_17_18 (2465, 2716)
CK--------GLRHPTCK--LT--R-------------------------------
---LGLDQT-TLSDETRQE------LR----------ALE-HEKSQL-LLSYTRKPTVMI
PNEGLDTGEMSNS-----------------TSSLKRRRLRSETVSSYVAQADLEHL-DWS
NPPAKA--FHSAGM-TKKSIPRIAQ-QELACLPSPASHED-LHMKPLG-TDDDFWGPTGP
VATEVVDK-ERSLYRPF13553 // ENSGT00900000140813_0_Domain_18 (2716, 2997)
VHFPVAGTYRWP---NMGLCFVVREAV-------TTEIEFCVWDK
LLGEINQQHSWMVAGPLLDIKAEPGA-IKAVHLPHFVALQGG--------HMDTSLFQVA
HFKEEGMLLEKPAKVELHHIVLEDPSFSPMGVLLRVIHNALRIFPITSMVLLYHRIHPEE
V-TFHLYLIPSDGSIRKAIDDEEMKFQFVRLHKPPPLTPLYMGSRYAVSGSEE-------
--LEIIPEELELCYRSPGEPQLFSEFYVGHLGSGIRLQMKNKTDETLVWEALVKPGENSGT00900000140813_0_Linker_18_19 (2997, 3052)
DLTP
ATTLVPP-----------------------------ALKALPSPLDAPGLLPF00619 // ENSGT00900000140813_0_Domain_19 (3052, 3134)
HFVDQYREQ
LVARVTSVEPVLDNLL-GHVLSQEQYERVLAENTRPSKMRELFSFSQSWDRSCKDRLYQA
LKETHPYLIMELWEKGSKKGLLPLGI----------
ENSPANP00000008559 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MSLDIQCEQ-LSD-ARWA----------------EL---------
----LPLL-------------------------------QQCQVV------RLDDC-GLT
EARCGDIS-SALRV
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1542)
NPALTELSLRSNELGDA---------GVHCVL--QGLQS-P-----
SC--K-IQKLS--L-QNCYLT--GA---GCG-VLSGTL----------------RS
PF13516 // ENSGT00900000140813_0_Domain_6 (1617, 1642)
LPTL
QELH-LSDN-LLGDA-GLQLLCEGLLDPQCR----LEKL--------QLE---------Y
C------N----------------------------------------------------
---------LS---AA-SCE---PLASVLRA-KPDFKELTVSNNDINE-AGVRVLCQG--
LKD-SPCQLEAL-K----------------------------------------------
------------------------LESCGIT-----------------------------
----------------------------SNNCRDLCS-----------------------
--------------------------------IVASK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
ASLRE---LA-LGSNK-LGD---
-------VGIAELCPGL----LHPSS----------------------------------
--------------------------SLR-TLW---------------------------
-------------------------------IWECGITAKGCADLCRV-LRTKE
PF13516 // ENSGT00900000140813_0_Domain_13 (2215, 2237)
SLKELS
L-AGNELGDEGAR-LLCETLLEP--GCQ----L---------------------------
-----------------------ESLWV--KSC-----SFTAACCSHF------------
-------SSVL-TQ
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NKFLLELQISNNRLGDAGVQE-LCKGLG-QP-GS-VLRVLW--LG-
--D-CDMSDSSCS--------------------SLAATLLA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2464)
NHSL-RELDLSNNCLGDAG
VLQLVE--------SVRQPGCL--LE--Q-------------------------------
---LVLYDI-YWSEAMEDR------LQ----------ALE-EEKPSLRVIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPSIP00000011293 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------LLPAA--TASK---MA
PF02758 // ENSGT00900000140813_0_Domain_0 (210, 326)
EELEDLE--K---KDFKRFRHKLSD------
---------F--RYE-G--------K--RKV-PRGK-LENAD-EMDVANILVEFYGEEKA
VTVTIEI----FD-KIGLRNSSVKLQ-EEREDWGK-INAGLIVPGV-------SCLYH--
-----AALQA--------------------------------------------------
------------------------VCVGV-------------------L-----------
------SSAS-------------------------------DGGNCSLKLDYKAKYKEH-
--------IREK----------------YQLI--EDRNNRVGECV---------ELNTRY
TKLLIVKEHRDQKEKSEIIA-------------------------KRASSTKLETLFEAS
GS--RQTPL----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
RTIVLQGAAGIGKTMLTKKL-----MVDWA--Y---------GKLYA
G--------KFDY-IFYINSREMNCIS------GPRSVAD-LILN-NC-----PE-RNAP
IQDILM-NQ----EKLLFIIDGFDV-FKFSFDQE------ES-S-----LCTDPCQT-KP
-VEIMLSSLLRKAILPKSYLLIT-TRPTALDKLRQCL-K-----FPH--FATILGFSEEE
RKEYF----------------R--KF-FAN---------VSQAKQALDFVKE-NETLFTM
CFAPIVCWILCTVMKQQMESGE-DL-A--KS-SKTTTSVYMLFLSSLLDNHS-N-S-KEV
------------VHADLRKLCSLA---AYGI-LN-NK----ILFEEE-------------
--DMKKHGLDGSDIQSLFLTK----DVFQKDIS-FE-SVYSFIHLSFQEFFAALFYLLM-
KDAEK--------------GRDS--MTVQKDVN-EILKKYGNS-RDKSLTLTVRFL----
----FGLLNKERVNHIEKIFGCK---------------I--SLD-IKQ-NLLKWFGVVAK
ASC--CLT---------------------------VKSIQEKRFQLELFHCLYEIHE-EE
FVKSAMDH---------------------F-------------------QEIGLNNSNFT
KI-EHMVL--S----FCIK-NCRN-GQS-LYLN----------------ACT-----FGV
-EEQEKEK----IK-------------------------DKHESN---------------
----------------------------------------SQRLL---------------
-N--M-HAACG--L-RCCHLT--SA---CCE-DLVSAL----------------KTNP
PF00560 // ENSGT00900000140813_0_Domain_6 (1619, 1634)
SL
KELD-LSGN-KLGEA-GIKLLCKGLKHPNCN----VQKL--------ELR---------G
N------S----------------------------------------------------
---------VT---PT-CLG---DLAAVLTK-NQSLTELDLSYSKWGD-LGVKQLYKA--
LKH-PSCQLQKI-K----------------------------------------------
------------------------LDCCLLT-----------------------------
----------------------------SACCNDLAS-----------------------
--------------------------------ALGTNETLME---LD-LRNNE-LRD---
-------SGVKHLCGGL----KQDRC----------------------------------
--------------------------KIQ-KLV---------------------------
-------------------------------LSNCYVTGA--------------------
------------------------------------------------------------
-------------------------------------------CCGAL------------
-------SAIL-SKSKSLQELDLCYNKLGDAGLKV-LCEGLK-QP-AC-KLQKLR--LT-
--G-CGLTPACCE--------------------DLLVVISTNQSL-TELNINHDALGVSG
MKQLRD--------GLKSTSCK--LQ--T-------------------------------
---LLLSQV-LLNHTNPLR------TT----------ITE-----EHDLQ----------
------------------------------------------------------------
-----------------------------------------------------QWHETSM
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMICP00000046583 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MSLDIQCEQ-LSD-ARWT----------------EL---------
----LPLI-------------------------------QQYQVV------RLDDC-GLT
EVRCQDIS-SALRA
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1542)
NPALTELSLRSNELGDA---------GVHLVL--QGLQS-P-----
TC--K-IQKLS--L-QNCCLT--EA---GCG-VLPAVL----------------RSV
PF13516 // ENSGT00900000140813_0_Domain_6 (1618, 1642)
PTL
RELH-LSDN-PLGDG-GLRLLCEGLLDPRCY----LERL--------QLE---------Y
C------N----------------------------------------------------
---------LT---AA-SCE---PLASVLRA-KPDFKELMLSNNDIGE-AGVRVVCQG--
LKD-SACQLESL-K----------------------------------------------
------------------------LESCGVT-----------------------------
----------------------------AANCRDLCG-----------------------
--------------------------------ILASK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
ASLKE---LD-LGSNQ-LGD---
-------AGLAELCPGL----LQPSS----------------------------------
--------------------------RLR-TLW---------------------------
-------------------------------LWECGITAEGCRELCRV-LRAKE
PF13516 // ENSGT00900000140813_0_Domain_13 (2215, 2237)
SLKELS
L-AGNELGDAGAR-LLCESLLDP--SCQ----L---------------------------
-----------------------ESLWV--KTC-----SLTTACCPHF------------
-------SSVL-A
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2374)
QNKSLLELQMSSNKLGDAGVHE-LSQGLR-QP-GS-VLRVLW--LG-
--D-CEVTDGGCG--------------------SLASTLLA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2464)
NRSL-RELDLSNNCMGDPG
VLQLIE--------SLRQPACA--LE--Q-------------------------------
---LVLYDI-YWSEEMDNQ------LR----------ALE-EHKPSLRIIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMLUP00000008807 (view gene)
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------TSS----------------
----SSSTSTLPSLPSFTSP
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 332)
FKSG---VMRYMLYLS--Q---EDLERFKQLLVR------
---------E--QPKAG--------S--TQI-TWDQ-VRTAR-WGEVVHLLTESFPGHLA
WDMANHI----FA-QMDKTQLCYLTQ-LELQELLP-HLEPTGPPPR------R-------
------------------------------------------------------------
------------------------------------------------------------
-------ALR-------------------------------TLEEEYSALAMGQRYREQ-
--------VLDH----------------YFHP--VDGLAWPKNQADFFYQDI--------
----------------------------------------------LPHQRFLPCLFLPC
RT--HGRPP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
RTVVLRSVAGAGKTTLAHQV-----MLEWA--G---------RTFYP
H--------KAWC-AFYVHCRHVVQDE-------EQSLCG-LLSR-QW-----PQ-AGAL
AAKALA-RP----EQLLLILDGFEV-LALPLA------HRRK-D-----LRADWSRK-LP
-VPILLASLLSGKMLPQASLLVL-LRFSSWRVLKPLL-S-----CPS--VLTLSGFTAPE
RTRYL----------------R--RY-FGD---------GRAADKAVDFLRG-SAIPSSM
CQAPAVCWLVCLCLKEPMERGA-DL-F--LA-FPNAAALFARHLSNALVAAV-GGLPGTL
------------LQECLQGACSLA---AEGM-WA-AR----WVFGEK-------------
--ELERAQLGPE-AVDALLRA----HVLQSVVR-GE-ARYTFVLLSFQEFFAALWYVLC-
FPGRF--------------PEFQ--KLDRAQVR-RLLAQPS-R-RRNDLAHMALFL----
----FGLFNETCARAVGRSFRCE---------------V--SLG-NKE-VLCRTALW--Q
DR-----------------------------------LPTKHHGLPLLFYCLHETRE-EA
FLAQVLQP---------------------C-------------------KQATLMIAKHR
DL---QVC--A----FCLR-CCQH-LRE-MQLI----------------LTL-----TIH
-KAIVTSN----PD--------------------SLLD------SSLPH--RPEGT-NWR
LCWWEELC-MALAL-QDLEALSLTNSVIELD---------AMQALS--AALKS-P-----
RC--Q-LHTLR--V-TTTMFP--LL---CPP-HSFASV----------------RAEEDL
DHLC-VCLP-LTGGP-EVKELPQGMESPFFP----LKTV--------WLE---------G
C------S----------------------------------------------------
---------VT---PG-SML---EFAKDLSS-NLKLKTLMVRRIYLKD-EGACS------
--L-SKVQWERL-A----------------------------------------------
------------------------LECCDYT-----------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------LLSYRSLILP-LSSNHRLTHLS
L-AGNALRHEGAK-DLWSALQKA--KCP----L---------------------------
-----------------------QRLVL--ERC-----QLTSDCCQAV------------
-------ASLL-CRHPSLRHLDLRKNDIRLRGAEL-LYQTF-------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSGALP00000045145 (view gene)
ENSPPAP00000012732 (view gene)
ENSNGAP00000018646 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------------MLRG-AR
LVPRVF----------------LSPPPGPL-------------------NKWQSF---RT
RR-RHCIP--T----MSLDIQCEQ-LSD-ARWA----------------EL---------
----LPLI-------------------------------QQYQVV------RLEDC-GLT
EVRCKDIS-SVLQA
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1542)
NPTLTELSLRTNELGNA---------GVHLVL--QGLQN-S-----
TC--K-IQKLS--L-QNCGLT--EA---GCG-VLTGPL----------------RS
PF13516 // ENSGT00900000140813_0_Domain_6 (1617, 1642)
LTTL
RELH-LNDN-PLGDA-GLQLLENSGT00900000140813_0_Linker_6_7 (1642, 1650)
CKAFLDPQPF13516 // ENSGT00900000140813_0_Domain_7 (1650, 1766)
CH----LERL--------QLE---------Y
C------N----------------------------------------------------
---------LT---AT-SCE---PLATVLRA-KPDFKELVLSNNDLHE-AGVHTLCQG--
LKD-SACQLELL-K----------------------------------------------
------------------------LENCGIT-----------------------------
----------------------------SANCRDLCD-----------------------
--------------------------------VVGSK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
ASLQE---LG-LGCNK-LGN---
-------AGIATLCPGL----LHPNC----------------------------------
--------------------------RLR-TLW---------------------------
-------------------------------LWECDITAEGCKDLCHV-LRT
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
KQSLKELS
L-ASNELGDEGAR-LLCDSLLEP--GCQ----L---------------------------
-----------------------ESLWV--KSC-----SLTAACCSHF------------
-------CSVL-T
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
QNRSLLELQMSNNPLGDSGVQE-LCKALG-QP-GT-VLRVLW--LG-
--D-CDVSDIGCR--------------------SLAPILLA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2465)
SRSL-RELDLSNNCMGDSG
VLQLME--------SIKQPSCV--LQ--Q-------------------------------
---LVLYDI-YWSEEVEDQ------LR----------ALE-EERPSLRIIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSOCUP00000009034 (view gene)
------------------------------------------------------------
-----------------------------------------------------------M
DVR----VFLTI---SAIEGK-R------------SESGVWT------G------HWS-E
F---NLYPLIVMAMARA-CNPQ
PF02758 // ENSGT00900000140813_0_Domain_0 (203, 325)
KA---LLGALSDLE--E---NSFKILKFHLRD------
---------M--TMVEG--------H--RQL-TRGQ-LEGLS-RVDLASRLILMYGAQEA
VKVVLSA----LK-VMNLMELVDQLS-LVC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LD--DYREIYREH-
--------VRC-----------------LE------ER---QEA----------GVNSSY
RQLLLVA-KPSPESPGSPGC------PI-------SE-----Q-ELDSV--MVEALFAPG
EK--PYAAP----S
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
VVVLQGSAGTGKTTLARKM-----LLDWA--T---------GTLYP
G--------RFDY-VFYVSCREVVLLPG-------RKLAQ-LLFW-CC-GD-----NEAP
AAEILK-QP----ERLLFILDGFDE-LQRPLGDQ------LK-S-----RR-----S-SP
-AENMLHLLIRRKVLPTCSLLIT-TRPSALRNLEPLL-D-----QPC--HVHVLGFSEGE
RIKYF----------------S--SY-FMD---------EKQARKAFDFVQG-NDILYKA
CQVPGICWMVCSWLKKKMERGQ-AV-S--ET-PSNSTDIFTAYVSTFLPADD-KGGCSEL
----------T-RHRVLTGLCSLA---SEGI-RC-QK----LLFEEA-------------
--DLQRYNLDGPRLA-DFLSS----NDYQEGRD-SK-KFYSFLHISFQEYFHAMSYLMK-
EDHSQL----------------G--KESCREVE-KLLEAE-----DQEMTLSMQFL----
----LDISKNQSFSNLELKLCFK-----------------ISPF-VTQ-EL-KYFREQM-
E-------------------------------------SIKCNRSWDLEFALYGSRI-KK
LAKSVHMS---------------------D-------------------ISFKLKHSN-K
TK-PHGDT--S----FSVK-TSLN-HGD-IEKQ------------------KC----PAV
--------RQ---DNM--A-------------------ETQKE-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------ISN----------------GKG---------R
-DTEE-LDKEVGG-SRM-GSRK----KM-A-KELKG---QKVVRL---E-RQEE-EED-R
--VKR-MER---------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSVPAP00000011582 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------MLPSAQL
PF02758 // ENSGT00900000140813_0_Domain_0 (198, 328)
-GFHLQPLLE------QLH--QD---ELSKFKALLRT------
---------L--SLKDE--------F--QHI-SLTE-VEEAD-GKQLAEILTNRCPSCWV
ETVTIQV----FD-KMNRTDLSERAK-DEFREASL-KSL---LENK------PPSVXXXX
----XXXXXXXXXX-----------------------X-XXX-------X----------
--XXX----------XXXXXXX----XXX-------------------X-----------
------XXXX-------------------------------XXXXXXXXXXKQDKYRNT-
--------LKKK----------------F-------------------------------
-------GQIGKNLRPGPSE------N-------VHI--------APQRCEALIPFCNPK
ML--AGPFP----HTAVLQGPAGAGRTTPAERP-----TLGWP--R---------GSPAE
T--------S-GP-ASRLRRGALGRGG-------ARTFA---------------------
-------------------------------------------E-----VGSRRRAG-RL
-PPLLRGCSG---------------------ALRAEL-Q-----AA--------------
------------------------------------------------------------
------------------------------------------------------------
--------------------GPLA---ARGA-WS-TP----VSGG-Q-------------
--APRRLGVD-SNG-------------SDENRD-QG-GCHAFLRLTARKLPAAAFQCLR-
KEAEE--------------XXX------XXXXX-XXXXXXXXX-XXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXX---------------X--X-------XXXXXXXXXXX
XX-----------------------------------XXSSAADTTEVLYCLCESQD-EQ
LVKDAVAQ---------------------V-------------------KEISVHLTSPS
EV---MRS--S----FCL-KHCQN-LEK-VSLQ----------------VD---------
----KGIF----LE-------------------------NDAALKPDTQAERSLND-QHI
LPFWVDLC-SVFDSNRNLTFLDISQSFLSTS---------SVRVLC--EKIAS-A-----
AS--S-LQKVI--L-KNLSP---AD---AYR-NICLAL----------------GGLETL
THLT-FQGN-DQ-DD-LLPPLCEVLRHPKCN----LQYL--------RLA---------S
C------S----------------------------------------------------
---------AT---TE-QWA---TLSSCLKI-SQSLTCLNLTANEFPG-EAAKGLCRT--
LSH-PKCFLQRL-S----------------------------------------------
------------------------LENCHLT-----------------------------
----------------------------EANCKELSS-----------------------
--------------------------------ALIVSQRLTH---LC-LAKNA-LGD---
-------RGVKLLCEGL----SYPEC----------------------------------
--------------------------QLQ-TLV---------------------------
-------------------------------LWCCSIASGGCFHLSKL-LRQNSRLTHLD
L-GLNHIGITGLK-FLCESLKKP--LCN----L---------------------------
-----------------------RCLWL--WGC-----AISPVCCADL------------
-------SSAL-QSNQNLITLDLGQNFLGYSGIKM-LCDALK-LQ-SC-PLRTVR--LK-
--I-DESVFRIRK--------------------LLEEIKESNPQL-TIESDHQG------
----PK--------HSRPSSRD--FI--F-------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCHIP00000030097 (view gene)
------------------------------------------------------------
-----------------------------MGA------------SQEDQGPD--------
-WVSYHLLLLFENF--------P------------LPTT----------------GWR-S
P---APQSRDKMA---DSFFSDFG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 322)
-LLWYLEELK--K---EEFWKFKELLKQ------
---------E--PLKFK--------L--KPI-PWTE-LKKAS-RENVSKLLSKHYPGKLA
WDVTLSL----FL-QISRDDLWRKAR-NEIRQ----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------TVNPYRSH-
--------MKQK----------------FQVL--WEKEACLLVPED--------------
---------------------------------F-------YRETTKSEYGFLNAVYLAA
LQ--PGEPS----P
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVILHGPEGIGKTTLLRKV-----MLEWA--K---------GNLWR
D--------RFSF-VFLLTGREMNGVT-------DMSLVE-LLSR-DW-----PES-SEP
IEDIFS-QP----ERILFILDGVEE-LKFDLD-C------NT-D-----LCEDWEEP-HS
-MQVVLRSLLQKQMLPECSLLLA-LSKTGMRKNHSLL-K-----HIK--CIFLLGFSEHQ
RKLYF----------------S--HY-FQE---------NDASSRAFSFVRE-KRSLFVL
CQSPFLCWLVCTSLKCQLDKGE-DL-E--LD-SETITGLYVSFFTKVFKSGS-ETCPLK-
-----------QRRACLKSLCTLA---AEGM-WT-RT----FLFCPE-------------
--DLRRNGVSES-DTLMWLDM----KLLHRSG-----DCLAFIHTCIQEFCAAMFYMFK-
RPKDP-----------------P--HSVIGNVT-QLITRAVSE-HYSRLSWTAVFL----
----FVFSTERMTNRLETSFGFP---------------L--SKE-IKQ-EITQSLDTLSQ
YD-----------------------------------PNNVMVSFQALFNCLFETQD-PE
FVAQVVNF---------------------F-------------------KDIDIYIGTKE
EL---IIC--A----ACLR-HCHS-LQK-FHLC----------------MEH-----VFP
-DESGCIS-----------N-------------------T-I-------EK---------
LTLWRDVC-SAFTASEDFEMLNLENCQFDEA---------SLAVLC--RTLSQP------
VC--K-LRKFVCNF---------ASN-LANSLELFKAI----------------LHNPHL
KHLN-LYGS-SLSHM-DAKQLCEALKHPMCN----IEEL--------ML-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------GKCDIT-----------------------------
----------------------------GEACEDIAS-----------------------
--------------------------------VIIHNKKLKL---LS-MCENA-LKD---
-------DGVQVLCEAL----KNPDCALEAL-----------------------------
------------------------------------------------------------
------------------------------LLSHCCFTSAACDHLSQV-LLG
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
NRSLTFLD
L-GSNVLKDEGVT-TLCESLRHP--SCN----L---------------------------
-----------------------QELWL--MNC-----YFTSVCCVDI------------
-------ATVL-I
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
HSEKLTTLKLGNNKIYDAGAKQ-LCKALK-HP-KC-KLEHLG--LE-
--A-CELSPASCE--------------------DLASALTTCKSL-SCVNLEWITLDYDG
AAVLCE--------ALVSLECS--LQ--V-------------------------------
---LGLNKS-SYDEEIKMM------LT----------QVE-EMNPNLIISHHLWTHD---
--EGR-----LR------------------------------------------------
--------------------------------------------------GVLV------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSETEP00000012003 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------------LRLC-GLT
EARCQELG-ASLQANPGLQELCVDFNEIGDG---------GLSLLL--QVLLG-P-----
TC--K-IQKLX--X-XXXXXX--X----XXX-XXXXXX----------------XXXXXX
XXXX-XXXX-XXXXX-XXXXX---X-XXXXX-----XXX--------XXX--X-XXXXXX
X------X----------------------------------------------------
---------XX---XX-XXX---XXXXVLKA---------RPLKELVL-SGISACLQE--
LCQ-AASNLETL-K----------------------------------------------
------------------------LVQCGIT-----------------------------
----------------------------AADCKGLSS-----------------------
--------------------------------MLAASETLQE---LH-LGENE-LGS---
-------EAMALLCSGM----LSPSS----------------------------------
--------------------------RLR-VLG---------------------------
-------------------------------LWECCISAEGCRELSRV-LGAKE
PF00560 // ENSGT00900000140813_0_Domain_13 (2215, 2240)
SLRELN
L-TYNELGDTGVA-LLCEAMREP--GCR----L---------------------------
-----------------------ESLRM--RSC-----SLTAACCTDL------------
-------GAML-VQNK
PF00560 // ENSGT00900000140813_0_Domain_15 (2357, 2381)
HLRELQLSSNNLGDNGVQL-LCQGLG-QP-GT-TLRQL------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSNGAP00000019327 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---ESDFSPYD---WV
PF02758 // ENSGT00900000140813_0_Domain_0 (210, 322)
HYLKELK--D---EEFLKFKELLRE------
---------E--PEKFQ--------L--QPI-SWSK-LQKAS-KDELAELLDEHYPGQQA
WEMIRGL----FL-QLNRKDLCTKVQ-EQMAS----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------KPNKYKQH-
--------MKKK----------------FQLI--WDNEGCRQIPES--------------
---------------------------------F-------YKQIIKAEYEALKDAFSEP
-----G--A----I
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 928)
TMVVEGPEGQGRTAFLRKV-----MLDWA--E---------GNLWK
D--------RFTF-VFFIYVYELNSMS-------ETSLVE-LISR-EW-----PGS-LEM
INDIFS-KP----ERILFILDGFDG-LKFDLE-V------RT-N-----LCDDWRQR-QP
-IQIVLSSLLQKRMLPDSSLILE-LGKQSLPKIYYLL-Q-----FPR--SIFMTGLPKEL
TEPYF----------------Q--HF-FCS---------KDKGSEAFRYLKTDISPFLEL
CRNLRMCWMACSCLKWQYDRGD-KY-K--IV-ADTDSSTYTSFVVSSFKSRT-ANCPSK-
-----------QNRDRLRKLCTLA---AEGM-WH-RT----FVFSTG-------------
--DLRKIGITES-DEAMWLGM----RFLLKRG-----NCFVFHNHTLQAYFAALFYFLR-
QTKDK-----------------P--NPAIGSLP-ELLREVYVH-FQTHWLQIGMFL----
----FGISTERSITLLQQSIGFL---------------P--SKE-IRQ-EVIKYLKSLGQ
HKH----------------------------------IDKV-IRSKDLLPTLLEVQE-GR
FETQVMDL---------------------F-------------------EELTTNVFDSE
TL---LEA--S----NCLD-HCNK-IRK-LHLC----------------VHNN----GV-
----------------------------------------------------CYLHCRAH
IHFWGKLC-SVFS--KNFQALELDSCNFNET---------SFKILC--EALSSP------
VC--K-LQSLTCSF---------MSD-IGNGSLLFQTI----------------AKNSCL
KYLN-LYGT-KLSCH-SVGSLCQVLQDPSCK----VEDL--------LL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------GKCSIT-----------------------------
----------------------------SPSCRAIAS-----------------------
--------------------------------VVLCN-KVKR---LS-LVENP-VKN---
-------KGVRLLCEAL----KQPTCALETL-----------------------------
------------------------------------------------------------
------------------------------LLSYCCLTFVACGHIYEA-LLSNKSLSLLD
L-ASNHLEDNGII-SLCEALKSP--NCP----L---------------------------
-----------------------QALWL--SGC-----SLTLECCEDL------------
-------SAVL-VCNKRLKTLKLGNNYLVDAGVQL-LFEALR-NP-NC-KLQSLG--ID-
--M-CYLSNDCCE--------------------DLVSTLTTCKTL-KSLNLAWITLDHDR
LVMLCE--------ALLHKSCT--LK--V-------------------------------
---L--------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSRNOP00000068986 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------M---ASFSSDFS---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LVWYLKELN--Q---KEFMKFKDFLIQ------
---------E--ILELK--------L--KQI-SWTE-VKKAS-REGLSNLLLKCYGENQA
LDMTFKI----LQ-KINRKDLTKKAT-REIADSSTLGLTD--SE--------------NS
K---------------------------------------------------LYRE----
------------------------------------------------------------
----------------------------------------------------------H-
--------LKKK----------------LSHN--CSKMFNINIQDF--------------
---------------------------------I-------LETVIQDDYDTFENLLVSK
GD--EKK-P----H
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
MVFLKGMAGIGKTLMLKNV-----MLAWA--K---------GLVFQ
N--------KFSY-TFYFCCQDVKQLK-------TASLAE-LISR-EW-----PIS-SAP
IEEILS-QP----EKLLFIIDSLEG-MEWDLTKQ------ES-E-----LCGNCTEK-QP
-VSLLLSSLLRRKMLPESSFLIS-ATPETFEKMEDRI-R-----CTD--VKTATAFNERS
IEICF----------------H--RL-FQD---------RNTAQKAFSLVRE-NEQLFTL
CQAPLLCWMVATCLKKEIEKGK-DP-V--SI-CRHITSLYTTYIFNSFIPQS-AKYPSK-
-----------KSQDQLQGLCSLA---AEGM-WT-DT----FVFSEE-------------
--ALRRNGIMDS-DIPTLLDI----GMLGKIRE-FE-NSYLFLHPSVQEICAAIFYLLK-
SHGVH-----------------P--SQDVKSVE-TVLFMFLKK-VKTQWILWGSFI----
----FGLLHKSEQEKLAVFFGYQ---------------L--SQK-IRH-KLYQCLETING
NVE-----------------------------------LQEQIDGMKLFYCLFEMED-DA
FLVKAVNC---------------------M-------------------QQINFVANYYS
DF---FVA--A----YCLK-RCST-LKK-LSFS----------------TQN-----ALN
-EEL--EQ-----------Q-----------------------------------YRERL
LICWNDIC-SVFVRSKNIQTLQIKDTNFSEP---------TFRVLY--ESLKYP------
SF--P-LSKLVANN---------VHF-FGDNHMFFEL-----------------I
PF13516 // ENSGT00900000140813_0_Domain_6 (1616, 1641)
QNHNL
QYLD-LSCS-SLSPN-GVKLLFDVLNQAECN----IEKL--------MV-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------AHCKLS-----------------------------
----------------------------PDDCKIFGS-----------------------
--------------------------------ILMSSKTLKI---LN-LASNN-LN----
-------QGISSLCEGL----CHPNCVLEYL-----------------------------
------------------------------------------------------------
------------------------------VLANCSLSEQCWDYLSDV-LR
PF13516 // ENSGT00900000140813_0_Domain_13 (2212, 2233)
KNKTLNHLD
I-SCNDLKDKGIK-ILCKALTLP--DCF----M---------------------------
-----------------------KSLW---------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMUSP00000104293 (view gene)
ENSOCUP00000018965 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------MEPA-----
--------CTWS------------------------------------------LLSE--
-----------------------------------------KK-----------------
----ATSSSL-------------------------------ALLSWKEFTDYREAFQKH-
--------LKQK----------------YQKI-GVDKCYYHGK--------------HIR
PRLLLVKEHGISEKTPCGIP-------------------------QQDHFIELEHVFGPN
EE--SCSSS----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
RTVVLQGCAGIGKTAVVHKF-----MFDWA--A---------GMVTP
D--------RFDY-LIYVNCREISRTA-------NLSAAD-LITN-SF-----QD-LKGP
ILDTVLLYP----EKLLFILDGFPE-LQYPVSDQ------EE-D-----LSVNPQER-KP
-VESLLCSFVSKKLFPESSLLIT-ARPTAMKKLHSLL-K-----QPI--QAEILWFTDTE
KRAYF----------------L--SR-FSG---------ANAAMKVFYTLRE-NEGLDIM
SSLPIVSWMICSVLQSQGEGDR-TL-L--KS-LQTMSDVYLFYFSKCLKTLTGVS-VW--
-----------KGQSCLWGLCSLA---AEGL-QK-HQ----VLFAAS-------------
--DLRRHGIGVYDANCAFLSH-----FLKKAEG-GV-NVYTFLHFSFQEFLTAVFYALK-
NDSSW---------------------MFFDQVGKTWQEIFQQY-GKGFSSLLIRFL----
----FGLLHRGKGHTVETTFGRK---------------V--SPG-LRE-ELLKWTEREIQ
DK-----------------------------------SSRLQIEPVDLFHCLYEIQE-EE
YTKKIIDD---------------------L-------------------QSIILLQPTYT
KM-DMLAM--S----FCVK-SSHSHLS----------------V-----SLKCQHILGF-
-QEEEQAS----------------------ALTP--------------------------
---------PALTLNQSFQ-----LYNVPVS---------RLHLLC--QALQNP------
YC--K-IKDLKL---IFCHLT------ASCGRD---------------LSSVFET-NQYL
TDLE-FVKN-TLEDS-GLKLLCEGLKRPNCI----LQKLR--------LYRCLI-SPA-S
CG------------AL----AAVLSTNQWLTELEFSETKLEASALKLLCEGLKDPNCK-L
QKLKLCASLLPESSEA-VCK---YLASVLIC-NPHLTELDLSENPLGD-TGVKYLCEG--
LRH-ANCKVEKL-D----------------------------------------------
------------------------LSTCSLT-----------------------------
----------------------------DASCVELSS-----------------------
--------------------------------FLQVSQTLQE---LF-VFANV-LGD---
-------TGVRRLCEGL----LHAKGMIKNL-----------------------------
------------------------------------------------------------
------------------------------VLSECSLSAACCEPLAQV-LSSTRSLTRLL
L-INNKIEDLGLK-LLCEGLKQP--DCQ----L---------------------------
-----------------------KDLAL--WTC-----HLTGACCQDL------------
-------CNAL-YTNEHLRDLDLSDNALGDEGMQV-LCEGLK-HP-YC-KLQTLW--LA-
--E-CHLTDACCG--------------------ALASVLSRNE
PF00560 // ENSGT00900000140813_0_Domain_17 (2444, 2461)
NL-TLLDLSGNHLRDFGVQMLCD--------ALIHPVCK--LQTFY-------------------------------
------IDTDHLHDDTFRK------ME----------ALK-RSKPGITW-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCHIP00000014734 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MKLDIHCEQ-LSD-ARWT----------------EL---------
----LPLI-------------------------------QQYEVV------RLDDC-GLT
EVRCKDIG-SALQA
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1543)
NPSLTELSLRTNELGDG---------GVLLVL--QGLQS-P-----
TC--K-IQKLS--L-QNCCLT--EA---GCR-VLPGAL----------------RSL
PF13516 // ENSGT00900000140813_0_Domain_6 (1618, 1642)
PSL
RELH-LSDN-PLGDA-GLRLLCEGLLDPQCR----LEKL--------QLE---------Y
C------N----------------------------------------------------
---------LT---AA-SCE---PLAAVLRA-TRD
PF13516 // ENSGT00900000140813_0_Domain_8 (1776, 1796)
LKELVVSNNDIGE-AGVQVLCRG--
LAE-SACQLETL-K----------------------------------------------
------------------------LENCGLT-----------------------------
----------------------------AANCKDLCG-----------------------
--------------------------------IVASQA
PF13516 // ENSGT00900000140813_0_Domain_11 (2019, 2054)
SLKD---LD-LGSNR-LGD---
-------AGLAELCPGL----LSPSS----------------------------------
--------------------------QLR-TLW---------------------------
-------------------------------LWECDLSVSGCRDLCRI-LQAK
PF13516 // ENSGT00900000140813_0_Domain_13 (2214, 2237)
ETLKELS
L-AGNGLGDEGAQ-LLCESLLQP--GCQ----L---------------------------
-----------------------ESLWV--KSC-----GFTAACCQHF------------
-------GSVL-TQ
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2377)
NKRLLELQLSSNPLGDAGVHV-LCQALG-QP-GT-VLRVLW--VG-
--D-CELTNSSCG--------------------GLASLLLA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2464)
SRSL-RELDLSNNGLGDPG
VLQLLG--------SLEQPACG--LE--Q-------------------------------
---LVLYDI-YWTEAVDER------LR----------AVE-ESKPGLRIIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSNLEP00000013036 (view gene)
MGP_SPRETEiJ_P0080669 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------M---ASLFSDFG---F
PF02758 // ENSGT00900000140813_0_Domain_0 (209, 319)
IWYWKELN--K---IEFMYFKELLAL------
---------E--MLNMG--------L--KQI-SWTE-VKEAS-REDLAILLVKHCGRNQA
WNTTFRV----FQ-MIGRNVINNRAT-GEI------------AA--------------HS
T---------------------------------------------------IYRA----
------------------------------------------------------------
----------------------------------------------------------H-
--------LKEK----------------LTRD--CSRKFNISIQNF--------------
---------------------------------F------------QDEYDHLENLLVPN
VT--ENN-P----K
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
MVVLQGVAGIGKTILLKNL-----MIVWS--E---------GLVFQ
N--------KFSY-IFYFCCHDVKQLQ-------TASLAD-LISR-EW-----PSP-SAP
MEEILS-QP----EKLLFIIDSLEG-MEWNFTQQ------DS-Q-----LCYNCMEK-QP
-VNVLLSSLLRKKILPESSLLIS-TSCETFKDLKDWI-E-----YTN--VRTITGFKENN
INMCF----------------H--SL-FED---------RNRAQEAFSLIRE-NEQLFTV
CQAPVVCYMVATCLKNEIESGK-DP-V--SI-CRRTTSLYTTHILNLFIPHD-AQNPSN-
-----------DSEDLLDNLCFLA---AEGM-WT-DI----SVFNEE-------------
--ALRRNGIMDS-DIPTLLDI----GILEQSRE-SE-NSYIFLHPSVQEFCAAMFYLLH-
EDTAH-----------------S--CQAVDFIE-TLLFTFLKK-IKKQWVFLGCFF----
----FGLLHETEQEKLEAFFGYH---------------L--SEK-LRQ-QLFLWLELVLD
TLH----------------------------------PDVKKINTMKFFYCLFEMEE-EV
FVQSAMNC---------------------R-------------------EQIAVVVKDYS
DF---IVA--A----YCLS-HGSA-LTD-FSIS----------------AQN-----VLN
-EEQ--GQ-----------R-------------------------------------GKL
LVLWHQIC-SVFQRNKDIKILRIEDTIFNEP---------VFKIFY--SYLKNS------
SC--I-LKTLVAIN---------VSF-LCDKRLFLEL-----------------IQSYNL
EELY-LRGT-FLSHS-DVEMLCDILNQAECN----IRI----------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------L-----------------------------
------------------------------------------------------------
------------------------------DLANCSLCEHCWDYLSEV-LR
PF13516 // ENSGT00900000140813_0_Domain_13 (2212, 2237)
QNKSLRHLD
I-SYNNLKDEGLK-ALCRALTLP--NSV----L---------------------------
-----------------------QS-----------------------------------
-----------------------------------------------------LS--LE-
--A-CQLTGACCK--------------------DLASTFTQYKCL-RWINLAKNSLGFSG
LIVLCK--------AMKDQTST--LY--E-------------------------------
---LKLRMA-DFDSESQEF------LL----------SEM-ERNKILNIENG-V------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSGGOP00000016379 (view gene)
ENSRROP00000014783 (view gene)
MDQPEAPCSRLRSLRTASSQRAFVESVPHQRLRLRPWCKAPKLGLTK-RQPGHVTWPGVW
KNRSRKGGAGEEGAPQSSGRRWPVPTSPSEGEEHQD------APTRVRGRRR--------
-------AKSGVTPHCPLPPPHG-----SS--LARGSLRPCARWVLERQSRGGR----GS
PEDDPRPPVSSTGLR--LAVAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLAALEELS--Q---EQLKRFRHKLRD------
------------VGPDG-----------RSI-PWGR-LERAD-AVDLAEQLTQFYGPEPA
LEVARKT----LK-RADARDVAAQLQ-EQRLRR---------L---------GLG-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------SV-ALLSVSEYKKKYREH-
--------VLQL----------------HARV--KERNARS------------VKITKRF
TKLLIAPESAAPEEE--ALGPAE-------EPEP--------GRARRSDTHTFNRLFRGD
E---EGRQP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYQ
G--------QVDF-AFFMPCGELLERPG------TRSLAD-LILD-QC-----PD-RGAP
VPQMLA-QP----QRLLFILDGADE-LPALEGP--------EAA-----PCTDPFEA-AS
-GARVLGGLLSKALLPTALLLVT-TRAAAPGRLQGRL-G-----SPQ--CAEVRGFSDKD
KKKYF----------------Y--KF-FQD---------ERRAERAYRFVKE-NETLFAL
CFVPFVCWIVCTVLRQQLELGR-DL-S--RT-SKTTTSVYLLFIASVLSSAPVADGP---
-----------RVQGDLRNLCRLA---REGVLGR--R----AQFAEK-------------
--QLEQLELRGSKVQTLFLSKKELPGVLET-----E-VTYQFIDQSFQEFLAALSYLLE-
DEGVP-------------------R-TAAGGVG-TLLRGDSQP--HSHLVLTTRFL----
----FGLLSAERMRDIERHFGC-----------------VVSER-VKQ-EALRWVQGQGQ
GCPGMAPEVTEETKGLEDTEEPEEE----------EEEGEEPNYPLELLYCLYETQE-DA
FVRQALCR---------------------L-------------------PELALQGVRFC
RM-DVAVL--S----YCVR-CCPA-GQA-LRLI------------------SC----RLV
------TA----------------------------------------QEK--K---KKS
----------------------------------------LGKR----------------
------LQAS---L--------------GGS------S-----------------SRGTT
KQ---LPAS-------LLHPLFQAMTDPLCR----LTSL--------TLS---------H
C-----------------------------------------------------------
--------KVP---DA-VCR---DLSEALRA-APALTELGLLHNRLS-EAGLRMLSEG--
-LAWPQCRVQTV------------------------------------------------
----------------------------RVQ-----------------------------
---------------LPSS---------QQGLQYL-------------------------
------------------------------VGMLRQSPALTT---LD-LSGC-QLPAPMV
--T--------YLCAIL----QHQGCGLQTLS--LTSVELSEQ-----------------
----------SLQELQAVKRVKP-------DLV----------------------I--AH
PALD-------------------GHPEPP-KEVISTF-----------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSCJAP00000032660 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----DG---------------------------------QVEEVDNPEL-----------
-------GDL-------------------------------EEDLESAKPGEKEGWRNV-
---------TDE----------------QSLV--SRNTFWRGDSDNFHDHVT--------
-----------------------------------------------QRNQRFIPFLNPR
RP--TQPAP----Y
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVLHGPAGVGKTTLAKKC-----MLDWK--D---------YNLGP
---------KARY-AFYLSCKGLSRLG-------PCSFAE-LIAR-DQ-----PE-LQDD
IPDILA-QA----QRILIVVDGLDE-LRVPPG------ALIY-D-----ICGDWLKP-KP
-VPVLLGSLLKRKMLPKAALLVT-TRPRSLRDLQLLA-E-----KPI--YIRVEGFLEED
RRAYF----------------L--RH-FGE---------EDQATRAFELMRS-NAALFQL
GSAPAVCWMVCTSLKLQMENGE-DP-A--TT-CHTRTGLFLHFLCSRFPKG-------AQ
------------LGGTLRALSILA---AQGL-WA-QM----SVFHRE-------------
--DLESFGVQES-DLRPFLDR----GILRQDRD-SK-DCYSFLHLSFQQFFTALFYALE-
KE------EE--------DRDGH--AWDIGDVQ-RLLSGEERV-QNPDLIQAGHFL----
----FGLANEKRAAELETTFGCV---------------M--SRE-IKQ-ELLRCRANLHV
KK-----------------------------------P-SSMAGLRELLCCLYESQD-GE
LAKAVMAP---------------------F-------------------REISVHLTSKS
EM---VQC--Y----FSLK-HCPD-LEK-LSLQ----------------VAKG----VFL
-ENY-VDL----EP--------------------DLELE---------------------
------------SLNSNLKFLEVRQSFLSES---------SVRVLC--DHIVR-N-----
TC--H-LQKAE--I-KNVTPD--SA----YR-DFCLAF----------------IGKPTL
THLT-LEGH-IEWERTMLLLMCDLFRTPRGN----LQYL--------RFG---------G
H------S----------------------------------------------------
---------AT---PE-QWA---DFSFLLKA-
PF13516 // ENSGT00900000140813_0_Domain_8 (1773, 1795)
NQSLRHLHLSANLLLD-EGAKLLYET--
IGH-PTNLLQKL-S----------------------------------------------
------------------------LENCRLT-----------------------------
----------------------------EASCKELAN-----------------------
--------------------------------VLIV
PF13516 // ENSGT00900000140813_0_Domain_11 (2017, 2054)
NQHLTH---LC-LAKNP-IGD---
-------RGVELLCEGL----SYPDC----------------------------------
--------------------------KLQ-TLV---------------------------
-------------------------------LQQCNITKRGCRPISKV-LQGVCSLTDLD
L-SLNHIG-CGLW-ILCKALENP--DCN----L---------------------------
-----------------------KHLRL--WSC-----SLLPFYCQHL------------
-------GSAL-ISNQKLETLDLGQNHLWKSGIIK-LLGVLK-QR-PG-SLKTLR--LK-
--T-YDTSLEIEK--------------------LLEEVKGKNPKLTIDCDAS------GE
---------------AAPGCCD--SF----------------------------------
----C-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSDORP00000027616 (view gene)
ENSPPYP00000004039 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MADSSSSFFP
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 328)
DFG---LLLYLEELN--K---EELNTFKLFLK-------
---------E--TMEPE--------H--GLT-PWTE-VKKAR-REDLANLMKKYYPGEKA
WSVSLKI----FG-KMNLKDLCERAK-EEINWSAQ-NIGPDDAKA-GE------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------TQ-DQEAVLGDGTEYRIR-
--------IKEK----------------FCIT--WDKKSLAGKPEDFHHGIA--------
----------------------------------------------EKDRKLLEHLFDVD
VK--TGEQP----Q
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
VVVLQGAAGVGKTTLVRKA-----MLDWA--E---------GNLYQ
Q--------RFKY-VFYLNGREINQLK-------ERSFSQ-LISK-DW-----PS-TEGP
IEEIIS-QP----SSLLFIIDSFDE-LNFAFEEP------EF-A-----LCEDWTQE-HP
-VSFLMSSLLRKVMLPEASLLVT-TRLTTSKRLKQLL-K----N-HC--YVELLGMSEDA
RQEYI----------------Y--QF-FED---------KRWAMKVFSSLKS-NEMLFSM
CQVPLVCWATCTCLKQQMEKGG-DV-T--LI-CQTTTALFTCYISSLFTPV-DGRSPSLP
------------NQAQLRRLCQVA---AKGI-WT-MT----YVFYRE-------------
--NLRRLGLTKS-DVSSFMDS----NIIKKDAE-YE-NCYVFTHLHVQEFFAAMFYMLK-
GNWEA--------------WNPS--CQPFEDLK-SLLQSTSYK--DPQLTQMKCFL----
----FGLLNEDRVKQLERTFNCK---------------M--SLK-IKS-KLLQCMEVLGN
SD-----------------------------------YSPSQLGFLELFHYLYETQD-KA
FISQAMRY---------------------F-------------------PKVAVNICEKI
HL---LVS--S----FCLK-HCRC-LRT-IRLS----------------VTG-----VFE
--K--KIL----KTSL-----PT--N------------------------TW---DGDRI
THCWQDLC-SVLHTNEHLRQLDLCHSNLDKS---------AMNILH--HELRHP------
NC--K-LQKLLLES---------CNLTVFCCLNISNAL----------------IRSQSL
IFLN-LSTN-NLLDD-GVQLLCEALRHPKCY----LERL--------SLE---------S
CN----------------------------------------------------------
---------LT---VF-CCL---NISNALIR-SQSLIFLNLSTNNLLD-DGVQLLCEA--
LRH-PKCYLERL-S----------------------------------------------
------------------------LESCGLT-----------------------------
----------------------------EAGCEDLSL-----------------------
--------------------------------ALISNKRLTH---LC-LADNV-LGD---
-------GGIKLMSDAL----QHAQC----------------------------------
--------------------------TLK-SLV---------------------------
-------------------------------LRRCHFTSLSSEYLSTS-LLHNKSLTHLD
L-GSNWLQDNGVK-LLCDVFRHP--SCN----L---------------------------
-----------------------QDLEL--MGC-----VLTNACCLDL------------
-------ASVI-LINPNLRSLDLGNNDLQDDGVKI-LCDALR-HP-NC-NIQRLE--LE-
--Y-CGLTSLCCQ--------------------DLSSALICNKRL-IKMNLTQNTLGYEG
IVKLYK--------VLKSPKCK--LQ--V-------------------------------
---LGLCKE-AFDEEAQKL------LE----------AVG-VSNPHLIIKPDCNYHN---
--E---------------------------------------------------------
--------------------------------------------------EDVSWWRCF-
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
MGP_SPRETEiJ_P0080593 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------------DKQKEE-
--------WKTR----------------YTAK--WKKNFWPKCNKEIYV-----------
---------------------------------------------ATESYKTLLALCNPK
I---ETPFA----H
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 931)
AIVLHGPPGSGKTTMAKQL-----MLEWS--E---------SKQAQ
---------IFSC-AFYISCREVNNTK-------PCTFAH-LLSM-DN-----PS-WRDC
VIRDLI-LG----KEFLFVVDGFDE-LTFPAG------ALIR-D-----LCGDWNTV-KP
-VEVLLGSLLKRKMAPHATLLVT-TRTQSLRQIFVMM-D-----QPL--LVETLGFLEQE
KQEYF----------------Q--KY-FEDEEGEEEDKGEGKALRALKEVRC-NADLYQM
ASLPTACGIFCLCLELRMKKGE-DL-S--LT-CQTYTSMFLNFLCEVFSPETCEGHLNKE
------------FQILFKKICILA---ANSL-LE-QV----PILCEE-------------
--DFLTLNLNLN-NLHPIVCR----HIFFKDSS-ST-HCLSFICLGIQQLLAAIIFVQE-
LGQES-----------------K--GVSKYSIQ-NMLSREARL-KNPDLSGLLPFV----
----FGLLNETRIQELKTTFGCQ---------------I--STE-VKR-KFLECE--SGE
NK-----------------------------------PLLLLMNMQEILSCLYESQE-EG
FVKEAMVL---------------------F-------------------EDISLHLKTST
DL---IHA--S----FCLK-NSQN-LQT-MSLK----------------VEKA----VFP
-ENV-AAL----ES--------------------TAKHQR------------SPDE-QRM
LTFWTDFC-DTFNSNEKLVFLDIHESFLNSS---------ALEILC--EKLPS-A-----
SC--C-LQKVV--L-KNISPD--DA----YE-KLCLIF----------------NGYKTI
SHLI-LQGG-NLDS--MHHSLCEVLKNPACN----LKFL--------SLG---------S
C------S----------------------------------------------------
---------TA---AQ-KWD---DFFPVLKV-
PF13516 // ENSGT00900000140813_0_Domain_8 (1773, 1796)
NQSLIYLDLTGNNLLD-KSAKLLCNI--
WKE-PKCILQRV-S----------------------------------------------
------------------------LENCQLT-----------------------------
----------------------------EACCKDLSS-----------------------
--------------------------------ILMV
PF13516 // ENSGT00900000140813_0_Domain_11 (2017, 2054)
SRTLTH---LS-LANNK-LGD---
-------NGVKNLCESL----SFTEC----------------------------------
--------------------------NLQ-TLV---------------------------
-------------------------------LWSCNITNTGCHYLSKM-LKQTL
PF13516 // ENSGT00900000140813_0_Domain_13 (2215, 2237)
SLKHLD
L-GLNRIGTKGAK-FLCEALKNP--KSK----L---------------------------
-----------------------KSLWL--CGC-----SITPLNCQDF------------
-------SETL-R
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
SNKSLNTLDLSQNVLGTDAIKT-FCEALK-LQ-IC-PLQMLR--LK-
--F-DEAKPSIQN--------------------LIQEMKASHPQLRISSDQV----DLKN
-----------------PPLPH--FI----------------------------------
----F-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
MGP_SPRETEiJ_P0080597 (view gene)
MGP_CAROLIEiJ_P0081454 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MALARA-NSPQEA---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWALKDLE--E---NSFKTLKFHLRD------
---------V--T------------Q--FHL-ARGE-LESLS-QVDLASKLISMYGAQEA
VRVVSRS----LR-AMNLMELVDYLN-QVC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LN--DYREIYREH-
--------VRC-----------------LE------ER---QDW----------GVNSSH
NKLLLMA-TSSSGGRRSPSC------SD-------LE-----Q-ELDPV--DVKTLFAPE
AE--SYSTP----PI
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 930)
VVMQGSAGTGKTTLVKKL-----VQDWS--K---------GKLYP
G--------QFDY-VFYVSCREVVLLPK-------CDLPN-LICW-CC-GD-----DQAP
VTEILR-QP----GRLLFILDGYDE-LQK---------------------------S-SR
-AERVLHILMRRREVP-CSLLIT-TRPPALQSLEPML-G-----ERR--HVLVLGFSEEE
RETYF----------------S--SC-FTD---------KEQLKNALEFVQN-NAVLYKA
CQVPGICWVVCSWLKKKMARGQ-EV-S--ET-PSNSTDIFTAYVSTFLPTDG-NGDSSEL
----------T-RHKVLKSLCSLA---AEGM-QH-QR----LLFEEE-------------
--VLRKHGLDGPSLT-AFLNC----IDYRKGLG-IK-KFYSFRHISFQEFFYAMSFLVK-
EDQSQQ----------------G--EATHKEVA-KLVDP---E-NHEEVTLSLQFL----
----FDMLKTDGTLSLGLKFCFR-----------------IAPS-VRQ-DL-KHFKEQI-
E-------------------------------------AIKYKRSWDLEFSLYDSKI-KK
LTQGIQMK---------------------D-------------------VILNVQHLD-E
KK-SDKKK--S----VSVT-SSFS-----SGKV------------------QS----PFL
--------GN---GKS--T-------------------RKQKK-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------ASH----------------GKS---------R
-GTEE-PVQ-GVR-NHR-LTSR----EK-GHMEMND---KEDGGV---E-GQED-EEG-Q
T-LKK-DGEMI---DK-MNG----------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSECAP00000002676 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----SSSS----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 928)
RTVVLQGCAGTGKTAVVHKF-----MFDWA--A---------GTVTP
G--------RFDY-LIYVNCREISHIA-------NLSAAD-LITN-TF-----QD-INGP
ILDVTLMYP----EKLLFILDGFPE-LQHTIGDQ------EE-D-----LSANPQER-KP
-VETLLCSFVRKKLFPESSLLIT-ARPTTMKKLHSLL-K-----QPI--QAEILWFTDAE
KRAYF----------------L--SQ-FLG---------ANTAMRVFYDLRE-SEGLDIM
SSLPIISWMICSVLQSQGDDDR-TL-M--RS-LQTMTDVYLFYFSKCLKTLTGVS-VW--
-----------KGQSCLWGLCSLA---AKGL-QN-QQ----VLFEVS-------------
--DLRRHGIGVYDTNCTFLNH-----FLKKAEG-GV-NVYTFLHFSFQEFLTAVFYALK-
NDSSW---------------------MFFDQVGKTWQEIFQQY-GKGFSSLTIQFL----
----FGLLHKGKGKALETTFGRK---------------V--SPG-LRE-ELLKWTEKEIK
DK-----------------------------------SSRLQIEPMDLFHCLYEIQE-EE
YAKR--------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSDNOP00000011474 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------------WAYKHH-
--------VITK----------------FSAD--LDVI-LPFKES---------------
-----------------------------------VT-------CWPQM-QILVGAFHPD
E---SGSQP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
HTVVLHGKSGTGKSVLARRV-----MLSWA--Q---------GQLYK
D--------MFSY-VFFLHAREIRWLE-------EGSFAE-LISR-EW-----PG-SQAP
VLEIMS-QP----EKLLFILDSFDD-LGFTLKD--------E-K-----VCRDWAEK-QP
-VPVLLHSLLKKVLLPKSSLMIT-VRDVGIGSLKSVV-V-----FPR--YLLVGGISVEQ
RIQMI----------------S--RQ-VKD---------EPQKSQILKSVMD-NDQLFDN
CQVFAVCSLVSVA--LQQAARN-DF-S--PV-CQTLTGLYATFAFHQLTPQS-QVQCCLN
----------QRERAILKALCRMA---LEGV-WH-MK----FVFYSD-------------
--DLVLYGLKES-ELSTLFHV----KVLAQDVL-DKGS-YTFFHLSFQEFCAALYYVLQ-
GIERE--------------RDQF--PLSIGNAR-SLGEFRQTG-FSPHLLQMKRFL----
----FGLMNKKALRKLEAVLGRP---------------I--PL-AIKQ-ELLQWISLLAQ
ERK----------------------------------AP-SSLDILDSFYCLFETQD-EE
FVQLAVLP---------------------L-------------------QEVWLPIYQKM
DL---LVS--S----FCLR-YCQH-LKK-IRLN----------------VKE-----IFL
-KDEVTEA----QP--------------------LT-----------AN--GSPGQ-TL-
AAWWQDFC-SVLGSLAGLTLLDLSSCVLSEW---------AMRSLC--AKLRL-R-----
TS--R-LQHLV--L-KDAQI---TL---GLQ-HLWATL----------------ITNHNL
KYLN-LENT-RLKDQ-DIEAVCVVLKHRNCF----LESL--------RLN---------F
C------G----------------------------------------------------
---------LT---HA-CGL---MLSKALIL-CPSLKSLSLTGNNMTD-VGLQPLCEA--
LKY-PTCNLQKL-A----------------------------------------------
------------------------LGHCGLT-----------------------------
----------------------------ESTCQSLAM-----------------------
--------------------------------VFIISQSLTH---LY-LAGND-LGN---
-------DGVKALCRFM----KHASC----------------------------------
--------------------------YLQ-RLI---------------------------
-------------------------------LTQCNLHSTSCGFLAFA-LMSNRSLTHLN
L-NSNPLQDDGIK-LLCEVMGEP--CCH----L---------------------------
-----------------------QDLEL--VEC-----SLTSACCESL------------
-------SSVI-SRSSHLQSLDLSFNDLGDGGLIT-LCQGLT-TE-TS-HLRRLG--LE-
--A-CGLSSSCCE--------------------AFSSALCRNQQL-TSLNLMRNVFSTAG
LEKLCM--------AFACS--K--LQ--R-------------------------------
---VGICKV-LFALKQKRL------LE----------KVQ-RIKPNFVIDENWYDVD---
--G---------------------------------------------------------
--------------------------------------------------DDRYWWQN--
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMOCP00000027176 (view gene)
ENSCGRP00001013748 (view gene)
ENSMLEP00000024895 (view gene)
ENSMMUP00000011259 (view gene)
ENSCAFP00000003638 (view gene)
ENSOGAP00000021129 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---SSFFS
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 328)
DFG---LMWYLEELN--K---KEFKKFKERFKQ------
---------E--TLKLG--------L--KQI-SWTE-VKKSS-RQELANLLIQHYEAQQA
WGLTLRI----FH-SMARQDLCDKVQ-RER------------AE--------------HK
K---------------------------------------------------IYRA----
------------------------------------------------------------
-------------------------------------------------YV---------
---------KNK----------------FSSL--CTSKFITDFFEC--------------
---------------------------------F-------GEKVRPEELEYLHLLFAAK
EN--GQP-S----H
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 929)
TTVLQGAEGIGKTTLLMKL-----LLGWA--E---------GSLYE
D--------RFLY-AFYFCCRELRQLK-------ETSLAD-LMSG-NW-----PYPSPAP
IAEIVS-HP----EKLLFIIDSFEQ-LKCDLSEP------ES-D-----LCSDWMKQ-QP
-VRVLLSGLLRKKMLPEASLLIA-VTPMCPAELQARL-V-----CPE--LITLSGFSEEE
IEFYF----------------R--RL-FPS---------RSRAAEAFRFLRE-NAQVYSL
CHTPRVCWVIGTCMKQVMEKGR-SL-A--PI-CRRLTSLYASFILNLFTGED-ASCPSP-
-----------ESQVQLRGLCSLA---VEGM-WT-DE----FEFNDE-------------
--ALRRNGIADS-DVPALLDT----NIFLKCT---E-SSYTFAHTSIQEFCAAMFYLLR-
--RDH-----------------P--NPAVRCRE-TLLATHLKKVKKAHWTYWVRFL----
----FGFLKEKEQEEVDTFFGFQ---------------P--SQE-IRQ-EFHQILKNLGE
HEG-----------------------------------LPGQTDFLALFYCLFELED-TT
FVREAVNF---------------------L-------------------QEINFSINNDL
DL---VVC--A----YCLE-YCSG-LRR-LSFS----------------VPK-----VIE
-EDGQGCA-----------S-------------------------------------GPS
VTHWHHIF-SVFTTNEHLKELEITDTDFCQS---------VLVILC--NQLRHP------
TC--H-LEKLQLNR---------VSL-SGGSSVFFEAL----------------SQSPDL
RYLN-LHNV-NLSRD-DMEKFCKVLSLSMCN----IEEL--------TL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------SGCFLR-----------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------N-PCWDLLHEA-LLTKKSLCSLD
L-SASNLHDKGLN-VLSVALRHP--DCC----L---------------------------
-----------------------RSLGL--ANC-----SFTAVGCGDL------------
-------ATVL-TSNRKLTSLQIGLNKVGDMGAKV-LCEALA-RP-SC-HLEILG--FK-
--D-CGLTSACCR--------------------DLYSALTHSRTL-RVLDLTLNTLDYSG
VAVLCE--------ALKYPECP--LW--V-------------------------------
---LGLPKT-RFDEKTEML------LM----------AVE-ESNPHLTIIGF-W------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSRNOP00000019808 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---MD-AAGASCSSS--EAVAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 321)
LLAALQDLS--Q---EQLKRFRHKLRD------
------------APLDG-----------RSI-PWGR-LEHSD-AVDLTDKLIEFYAPEPA
VDVTRKI----LK-KADIRDVSLRLK-EQQLQR---------L---------GSS-----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------S-ALLTVSEYKKKYREH-
--------VLRQ----------------HAKV--KERNARS------------VKINKRF
TKLLIAPGSGAGEDE--LLGTSG-------EPEP--------ERARRSDTHTFNRLFRGN
DD--EGPRP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLQGPAGIGKTMAAKKI-----LYDWA--G---------GKLYH
S--------QVDF-AFFMPCGELLERPG------TRSLAD-LILE-QC-----PD-RTAP
VRRILA-QP----HRLLFILDGADE-LPTLAAP--------EAT-----PCRDPFEA-TS
-GLRVLSGLLSQELLPSARLLVT-SRNATLGRLQGRL-C-----SPQ--CAEVRGFSDKD
KKKYF----------------F--KF-FRD---------ERKAERAYRFVKE-NETLYAL
CFVPFVCWIVCTVLLQQMELGR-DL-S--RT-SKTTTSVYLLFITSMLKSA-GTNGP---
-----------RVQGELRMLCRLA---REGILKH--Q----AQFSEK-------------
--DLERLKLQGSQVQTMFLSKKELPGVLET-----V-VTYQFIDQSFQEFLAALSYLLD-
AEGAP-------------------G-NSAGSVQ-MLVNSDAGL--RGHLALTTRFL----
----FGLLSTERIRDIGNHFGC-----------------VVPGR-VKQ-DTLRWVQGQSQ
PK--VATVGAEKKDELKDEEAE--EEEE-----EEEEEEEELNFGLELLYCLYETQE-DD
FVRQALSS---------------------L-------------------PEMVLERVRLT
RM-DLEVL--S----YCVQ-CCPD-GQA-LRLV------------------SC----GLV
------AA----------------------------------------KEKKKK---KKS
----------------------------------------FMNR----------------
------LKGS----------------------------------------------QSTG
KQ---PPAS-------LLRPLCEAMITQQCG----LSIL--------TLS---------H
C-----------------------------------------------------------
--------KLP---DA-VCR---DLSEALKV-APSLRELGLLQNRLT-EAGLRLLSQG--
-LAWPKCKVQTL------------------------------------------------
----------------------------RIQ-----------------------------
---------------MPGL---------QEVIHYL-------------------------
------------------------------VIVLQQSPVLTT---LD-LSGC-QLPGTVV
--E--------PLCSAL----KHPKCGLKTLSLT--SVELTEN-----------------
----------PLRELQAVKTLKP-------DLA----------------------I--IH
SKLG-------------------THPQPLKG-----------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPCAP00000008349 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------MAAPSY
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 330)
TDSD---LLWYLEKLN--D---KELQSVKEHYK-------
------------HLEFR--------V--PEI-PWFQ-LMQAQ-QEDLDKILTTSYEAHIT
WNMMFSI----FQ-KISRQDLCDKI---DARRK---------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------RNKEIYKDR-
--------IREN----------------LLLQ--WEDSISTEVNDRFYESFA--------
----------------------------------------------PEIFHRLETTFDRR
GN--WPLGN----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
LNMLLIGNEAAGKTMLTKMA-----MSLWA--R---------GMMWK
D--------TFSY-VIRLTSHEINQMV-------VSSFVE-LLSK-DW-----PD-SQAP
VGDILS-DS----QKLLFILEDLDN-IGLNLNIE------ES-L-----LCSDSRKQ-VP
-ILLLLVSLLKRKMMPGSSFLIS-TRPRNQDVIDTLM-K-----EKG--CSTNLNFTDEI
RQKYF----------------T--IF-FQD---------SRRAMIALKFVHE-NEVLVDL
CSVPILSWIVCTTLNQQLGTA-DDV-Q--LR-FRTSSDIHAHFLTKMLTSEAHV--PQ--
----------QHQQVLLQRLCWLA---VNGI-LQ-EK----LEFTDQ-------------
--DLRSVELTEA-DVAVLRTT----HILKPSSN-YK-DHCVFIHRCVQEFCAAIAYMRPL
IFGPF-----------------P----SGTN----V--RDRRS-ECQDFSLIMAFI----
----FGLLNKKTRNILEISFDCQ---------------LF-SDM-HRQ-YFLENMQLLGA
NP-------------------------------------RAMEHHLPLFYCLFENQE-EE
FVKQIMGF---------------------F-------------------TEATVVIRENK
DL---MVS--L----YCLK-NCHP-LQR-LRLC----------------IQG-----IFK
--NSA----------------------------------------------NPTSSEMRS
LHHWTGLC-SLLYTKGNLQELEICNSNLDKV---------SETILC--RAVRHP------
SC--Q-LQKLKLTY---------LTI-DIRFEELFKTV----------------AHNQNL
IHLS-LTCV---------------------------------------------------
------------------------------------------------------------
--------------------------------------------PMSQ-KIFSLMHEV--
LAN-PMCNIQRL-S----------------------------------------------
------------------------LIKCDLE-----------------------------
----------------------------TSECERIAS-----------------------
--------------------------------LLEN-EKLKQ---LN-LSHNE-LSD---
-------KALETLCNGV----LHPHCALESL-----------------------------
------------------------------------------------------------
------------------------------VXXXXXXXXXXXXXXXXX-XXXXXXXXXXX
X-XXXXXXXXXXX-XXXXXXXX--------------------------------------
-----------------------XX-X---XX------XXXX-XXXXX------------
-------XXXXX---XX-XXXXXXXXXXXXXXXXX-XXXXXX-XX-XX-XXXXXX--XX-
--X---XXX--------------------------XXXXXXXXXX-XXXXXXXXXXXXXX
XXXXXX----------XXXXXX--XX--X-------------------------------
---XXXXXX-X-------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
XXX-----------------XXXXXXX---XX--XXXXPKVYRSTRSA--ADPQSLL---
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMAUP00000022945 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------------MLRG-AW
LTLRVF----------------LESPPGPL-------------------NTWQSV---WT
RR-LHCVP--T----MSLDIQCEQ-LSD-ARWT----------------EL---------
----LPLI-------------------------------QQYQVV------RLDDC-GLT
EVRCKDIS-SAIQA
PF13516 // ENSGT00900000140813_0_Domain_4 (1515, 1542)
NPTLTELSLCTNELGDA---------GVCLVL--QGLQN-P-----
TC--K-IQKLS--L-QNCSLT--EA---GCR-VLPDVL----------------RS
PF13516 // ENSGT00900000140813_0_Domain_6 (1617, 1642)
LPTL
RELH-LSDN-PLGDA-GLKLLCEGLLDPQCR----LEKL--------QLE---------Y
C------N----------------------------------------------------
---------LT---AT-SCE---PLASVFRV-KPDFKEIVLSNNDLHE-AGVQMLCQG--
LKD-SACQLESL-K----------------------------------------------
------------------------LENCGIT-----------------------------
----------------------------SANCKYLCD-----------------------
--------------------------------VVASK
PF13516 // ENSGT00900000140813_0_Domain_11 (2018, 2054)
ASLQQ---LD-LSSNK-LGN---
-------AGIAALCSGL----LLPSC----------------------------------
--------------------------RIR-TLW---------------------------
-------------------------------LWECDITAEGCKDLCRV-LRT
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
KPSLKELS
L-AGNELKDEGAQ-LLCESLLEP--GCQ----L---------------------------
-----------------------ESLWI--KTC-----SLTAACCSHL------------
-------CSVL-T
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2378)
KNRSLLELQMSSNPLGDSGVLE-LENSGT00900000140813_0_Linker_15_16 (2378, 2391)
CKALG-QP-DI-VPF13516 // ENSGT00900000140813_0_Domain_16 (2391, 2436)
LRVLW--LG-
--D-CDVTDNGCS--------------------SLENSGT00900000140813_0_Linker_16_17 (2436, 2442)
ATVLLAPF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2462)
NRSL-RELDLSNNCMGDTG
VLRLIE--------SLRQPSCA--LQ--Q-------------------------------
---LVLYDI-YWTEEVEDQ------LR----------ALE-EERPSLRVIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
MGP_PahariEiJ_P0045396 (view gene)
ENSMAUP00000020106 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------ME------ETTGYG---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 321)
LNLCLQMLS--N---AEFLRFKSLYKE------
---------V--QEQFK--------L--KPMIPWTT-IEKSS-RKDMETLLHTHYP-GLA
WAFAARL----FD-CINRQDLFRMTE-KLMRGKQ--------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------------RPYREI-
--------MKTI----------------FNYI-------WTKENN---------------
----------------IYLK------S-----SDFKV-------SIKRQYSALKEVFDD-
-----QLKP----V
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 905)
TLVVLGESGIGKSTFLRRA-----MLDWA--S---------GDLWN
N--------RFHY-VFFFSLISLNNTT-------ELSLAQ-LLLS-KL-----SE-SSEA
LDDVLS-NP----RRILFILEGFDY-LNFDLELQ-------T-D-----LCSDWKKS-MP
-TQIVLRSLLQKVMLPESSLILE-LGPQSEPKIYPLL-R-----YPR--KIILGGLLDSA
IRLYC----------------L--LF-FE----------VNEGVQVFNYIAA-NEPLFCI
CKNPHLCWVVCSTIKWKCARRE-EA-I--LP-FDTDSALYASFILSSFRSKY-DKCLPKE
------------NRARLKTLCTLA---VEGM-WK-RV----FVFNSE-------------
--DLRRNGIC-ESELTVWLQM----NFLNTR----G-DCFVFYSPTLQWYFGALFYFLR-
EGKEK-----------------H--NPIIGSLP-QLLSESYAH-SETQLFMPGMFV----
----LGIATENVAAMLNPHFGLI---------------P--SKA-MRQ-EIFKCFRS---
-----------------LSQ----------------GEGREKLILLRLFDGLLENQD-ER
FETQVLDL---------------------F-------------------EEMTVHIGSHN
EL---STA--T----IYL-PKSKK-LKK-LHMH----------------IE---------
----HRIF----SD--------------------------VYKPEDSDLEDFIKGS-RV-
NNEWREIC-SIFQ---NLKVLDLDSCNFNET---------AIQNLC--LSISD-P-----
KM--P-LNAFKLES-MSCSFM--TN--FGDG-ALFPTL----------------LRLPHL
KSLN-LYGT-RLSSD-VVGKMCSVLL----------------------------------
------------------------------------------------------------
------------------------------------------------------------
-TC-PTCRVEEL-L----------------------------------------------
------------------------LGKCDIS-----------------------------
----------------------------SEDCGVIAT-----------------------
--------------------------------SLT-KCKVKH---LS-LVENP-LKN---
-------KGVMLLCQIL----KHPNC----------------------------------
--------------------------VLE-TLM---------------------------
-------------------------------LSHCCLTFIACGHLYED-ILCNKFLSLLD
L-GSNFLEDAGVN-ILCEALKKP--VCT----L---------------------------
-----------------------QELWL--TGC-----SLTSACCERI------------
-------SAVL-SCQKNLKTLKLGNNNIQDTGVQQ-LCKALR-NP-EC-KLQCL------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSBTAP00000056032 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------V--WKDILWSGDDGGFHYNIT--------
-----------------------------------------------QRSPKLITYLNPQ
AP--TQSTP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TMVLYGPAGVGKTTRAKEL-----MLDWM--Q---------DDLAE
---------TSNS-AFYLSCKGLNHRR-------TCTFAE-LISA-NW-----PH-VQED
IPAILA-QA----QKVLLILDGFDE-LKVPSG------ALIH-D-----ICGDWKKQ-KP
-VPVLLGSLLKRKMLPKATLLIT-TRPGALRELRLLT-E-----QPL--FIEMEGFLEED
RKAYF----------------L--KH-FEE---------ESQALRAFDLMKN-NAALFQL
GSAPSVCWMVCTCLRQQMERGE-DP-A--AT-CRTTTALFLRFLCGRFTPP-HGGGPRRG
------------LQAPLKPLCLLA---AEGV-WT-QS----SVFDGE-------------
--DLRRLGVDPS-ALCPFLDG----NILQKSED-GE-ACYSFIHLSVQQFLAAMFYVLE-
PEEQEE---E--------GLGGR--RWHIGDVG-KLLSKEERL-KNPSLTHVGYFL----
----FGLCNKRRAMELETTFGCL---------------V--STE-IKR-ELLKYTLMPHG
KK------------------------------------SFSVTDTKEVLSCLYESQE-EQ
LVKDAMAH---------------------V-------------------KEMSIHLMNTS
EM---MQS--S----FCLK-HCAN-LQK-ISLQ----------------VGKK----IFL
-END-PAL----KS--------------------D------------PQNETSQHD-HHS
LQLWADLC-SVFSSNENLNFLDVKESFLSHS---------SVRILC--EQITH-V-----
TC--H-LQKVV--I-KNVSPA--SA----YR-DFCLAF----------------IGKKTL
THLV-LEGS-VRGDKILLLLLCEILKHPRCN----LQYL--------RLG---------S
C------S----------------------------------------------------
---------DT---TQ-QWA---DFSSALKV-NQSLKCLDLTASEFLD-EGVKLLCLT--
LRH-PKCVLQKL-S----------------------------------------------
------------------------LENCQLT-----------------------------
----------------------------QACCKELSS-----------------------
--------------------------------ALIVNQRLTH---LC-LAKND-LGD---
-------GGVKILCEGL----RYPEC----------------------------------
--------------------------QLQ-TLV---------------------------
-------------------------------LRHCNINRHGCKYISKL-LQGDSSLTSLD
L-GFNPIA-TGLC-FLYEALKKP--NCN----L---------------------------
-----------------------KCLGL--WGC-----SITPFSCQHL------------
-------ASAL-VSSRSLETLDLGQNAWGQSGIVV-LLKALK-QN-HG-SLKTLR--LK-
--M-DKSTVEIQR--------------------LLKDVKENNPRLTIECNDA------RT
---------------TRSSCCD--FF----------------------------------
----S-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMICP00000018849 (view gene)
ENSMFAP00000038232 (view gene)
ENSMLUP00000013077 (view gene)
ENSODEP00000001162 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------MEF-----VLI-L----CV------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------CVCFFLIGLGVRTEYKLR-
--------IKEK----------------FLIL---DKNSLLGEPFHFYYGAS--------
----------------------------------------------Q-QQKLLECLFQVN
AR--GDEQL----Q
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLRGAAGVGKTTLMRKA-----VLQWA--E---------ENLYQ
H--------KFTY-VFYLSGKEISQEK-------EQSFAQ-LISK-DW-----PS-TEIP
IEKILS-QP----SNLLFIIDGFDE-LNFAFEEP------EF-A-----LCGDWTQV-HP
-VSFLISSLLRKVMLPKSSLWVT-VRHTASNRL-----------NQC--YVDLKGLSQDQ
RKEYI----------------Y--QL-FED---------ETWAMNAFDLIRK-NEVLFNM
CHVPSVCRLICNCLKQQMETNN-DI-T--KI-CQTATAVFTCYLSSLFSPR-DEYLPTLP
------------NEVQLRSLCCLA---AKGL-WT-MT----HVLYKN-------------
--ELRKHGLTTY-DTSVFLDM----NVLQKDID-YD-NCYIFTNLHIQEFFAAMFYILR-
SDWED--------------REHS--SQFFEDLN-LLLESKSDC--DPNLTQLKCFL----
----FGLLNEDLLKQLEETLKCQ---------------M--SLK-IKW-EILQWIKVLGD
RD-----------------------------------ISPSDTVFLDLFHYLYETQD-EA
FVAQAMGN---------------------F-------------------SNVVINICREA
HL---LVS--S----FCLK-HCRC-LQT-LKLD----------------VV---------
------MI----RSSP-----AA--E------------------------T---------
IQCWQNLF-SVLNTNEHLTELDLCHSNLDEL---------IMKILH--QELRNP------
NC--K-LQRLLLRF---------VAF-PDGYQDVFSFL----------------THKQNL
MHLD-LKGS-NTEDD-GVKLLCEALKHPECK----LQNL--------GLE---------A
CN----------------------------------------------------------
---------LT---SV-CCQ---DIAKALIK-
PF13516 // ENSGT00900000140813_0_Domain_8 (1773, 1796)
SKSLVFLNLSTNTLHD-DGVKFLCEA--
LGH-PECYLRRL-S----------------------------------------------
------------------------LESCGLT-----------------------------
----------------------------EVSCEDLSS-----------------------
--------------------------------ALIR
PF13516 // ENSGT00900000140813_0_Domain_11 (2017, 2054)
NKSLTH---LC-LADNS-VGD---
-------RGIKLLSAAL----KHPEC----------------------------------
--------------------------TLQ-SLV---------------------------
-------------------------------LRRCHFTPISSEYLSSS-LL
PF13516 // ENSGT00900000140813_0_Domain_13 (2212, 2237)
YNKSLIHLD
L-GSNDLQDDGLA-VLCNVFQQS--NSN----L---------------------------
-----------------------QDLEL--MGC-----VLTTACCPDL------------
-------ASAI-L
PF13516 // ENSGT00900000140813_0_Domain_15 (2354, 2374)
NSPNLLNLDLGYNDLQDEGVKI-LCEALR-NP-NC-SIQSLG--LE-
--Y-CGLTSLCCQ--------------------DLSSALLSNQRL-IKINLAQNSLGHEG
LRTLCE--------VLSCPECE--LQ--V-------------------------------
---LRLCKD-GFDKELQRL------LA----------TVR-VSNPKLAINF--SC-----
--N---------------------------------------------------------
--------------------------------------------------ESGYWWWRF-
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMLUP00000014116 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------K
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
MDFN---LRPLLEQLE--L---EELSQFKSLLRT------
---------L--PPQDE--------L--QHV-SQTE-VDEAN-GQQLAEILTTRCPVFWV
EMVTIQV----FD-KMNRTDLSKRAK-DELREAAL-KS------LQKNKV---PLSGG--
------CGGHRLVP-LI--------------------------D---ACLEFSSCARELT
QEPGGS--------------------------------------VTNPE-----------
-------EVT-------------------------------EGLSPWRELGIRWLLSNL-
---------TST----------------SGPI--WKQNLWPEASEKLHR-FT--------
-----------------------------------------------QRYERLVPFCNPN
ML--DGPFP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
HTVVLQGAAGVGKTTLAKKW-----MLDWA--Q---------DHLPG
---------TFHS-AFYLSCKELGRQA-------PCTFAE-LL-K-KC-----PD-AQEV
GPQVLA-QV----------------------------------T-----ICGNWEKP-KP
-VSVLLGSLLMRKLLPKATLLVT-TRPEALGELRLLV-D-----QPL--FIEVEGFLEAD
RRGYF----------------L--KH-FEQ---------QDQALRAFEGMKG-SPALFHM
GAAPAVCWVVCTCLKGQLEKGE-DP-A--PT-CRTPTSLFLRFLCNQCSPVAPGSGPAPH
------------LPTALQALSLLA---ARGI-WA-QT----SLFDCE-------------
--DLGQLGVSES-DLRPFLDG----KVLQADAD-CE-GCYDFLHLSVQQFLAALFYLLD-
SAEPG-------------DAGGW--APDIGNVR-ALLSKEERL-RNPSLTRVGYFL----
----FGLANQERARELEATFGCR---------------V--SLD-VRQ-ELLRVTRSRD-
-W-----------------------------------SFSSRADTRELMCCLYESQE-EA
LVREALAP---------------------I-------------------TELALHLQSER
DL---VHA--S----FCLR-HGQA-LRR-LWLQ----------------VEKG----IFL
-END-GAS----EP--------------------G------------TWEERSQSD-RHL
LPFWADLC-SVFGANKNLVFLDISRSFLSTS---------SVRLLC--EKISS-V-----
PC--S-LQKVV--L-RNVSPA--DA----YR-NFCVIL----------------GGYETL
THLT-LEGN-DQ--NNMLPILCGVLLHPKCH----LQYL--------RLV---------S
C------S----------------------------------------------------
---------AT---AQ-QWA---DLASSLET-NHSLTCLNLTANEFQD-EGARLLYMT--
LRH-PKCILQRL-S----------------------------------------------
------------------------LENCHLT-----------------------------
----------------------------AAYCKELTS-----------------------
--------------------------------TLMVNQRLTH---LC-LAKND-LGD---
-------HGVRLLCEGL----SYPEC----------------------------------
--------------------------RLQ-TLV---------------------------
-------------------------------LFHCSITSDGCIIL-AL-LQESSSLTHLD
L-GLNHIGTTGVK-FLCEALKKP--LCH----L---------------------------
-----------------------RCLWL--WGC-----AITPFSCADL------------
-------SSAL-SSNQHLITLDLGQNSLEYSGIKM-LGDALK-LQ-SC-PLQKLR--LK-
--I-DHSDAQIQK--------------------LLEEIKEGNPKLTIERDNE----EPRN
---------------NRPSSAD--FI----------------------------------
----F-------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSRROP00000024058 (view gene)
------------------------------------------------------------
--------------------------------------------MKVAQGLEL-A-ADLL
RDPLLNLVILSTGPTCSVLPKNP------------L-------------------SPQ-N
L---SSQPCLKMEGDKSLTFSSYG--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 323)
-LQWCLHELD--K---EEFQTFKELLKK------
---------K--SSEST--------T--CSI-PQFE-IENAN-VECLALLLHEYYGASLA
WATSISI----FE-NMNLPTLSEKAR-DDMKRHS-----PEDLEPMTT--GQGPSKEEVP
G-----ISQTVQQESA--TAAE-------------------------TKEQEISQTME--
QE-GATAA---------ETEEQEISQTMEQEGA---------------------------
-----T---A-------------------------------AETEEQEYGDDKWDYKSH-
--------VVTK----------------FAVK--EDVR-CGFEKT---------------
------------------------------------A-------DWPEM-QMLADAFNSD
Q---WGFRP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 925)
HTVVLHGKSGIGKSALARRI-----VLCWA--R---------GGLYQ
G--------MFSY-VFFLPVREMQQKK-------ESSVAE-FISR-EW-----PD-SQAP
VTEIMS-RP----ERLLFIIDGFDD-LG-SVLNS------DT-K-----LCKDWAER-QP
-PFILIQSLLRKVLLPESFLMVT-VRDVGIEKLKSEV-V-----SPR--YLFVRGISAEQ
RIHLL----------------L--ER-GIG---------EHQQTQGLRAIMN-NRKLFDQ
CQVPAVGSLICVALQLQDVVGE-SI-A--PF-GQTLTGLYATFVFHQLTPQG-VVRRCLN
----------LEERVVLKRFCRMA---VEGV-WN-MK----SVFDGD-------------
--DLMVHGLGES-ELRTLFHM----NILLRDSR-CE-EFYTFFHLSLQDFCAALYYVLE-
GLEIE--------------PALC--PLFVEKTK-RSMELQQAG-FHIHLLWMKRFL----
----FGLMSEDVRRPLEVLLGCP---------------V--PL-GVKQ-KLLHWVSLLGQ
QPN----------------------------------AA-APGDTLDAFHCLFETQD-KE
FVRLALNG---------------------F-------------------QEVWLPINQNL
DL---IAS--S----FCLQ-HCPY-LRK-IRMD----------------VKG-----IFP
-RDELAEA----WP--------------------VV-----------PQ--WMQGK-TLM
EEQWEDFC-SVLSTHPHLRQLDLGSSILTER---------AMKTLC--AKLRH-S-----
TC--K-IQTLM--F-RNAQI---AP---GLQ-HLWRTL----------------IANRNL
KCLN-LGGT-HLKEE-DVRMACEALKHPKCL----LESL--------RLD---------R
C------G----------------------------------------------------
---------LT---HA-CYL---KISQILTT-
PF13516 // ENSGT00900000140813_0_Domain_8 (1773, 1796)
SPSLKSLSLAGNEVTD-QGVTPLENSGT00900000140813_0_Linker_8_10 (1796, 1807)
SDA--
LRV-SQPF13516 // ENSGT00900000140813_0_Domain_10 (1807, 1956)
CTLQKL-T----------------------------------------------
------------------------LEDCGIT-----------------------------
----------------------------ATGCQSLENSGT00900000140813_0_Linker_10_11 (1956, 2016)
AS-----------------------
--------------------------------ALVPF13516 // ENSGT00900000140813_0_Domain_11 (2016, 2054)
NNRSLTH---LC-LSNND-LGN---
-------EGVNLLCRSM----RLPHC----------------------------------
--------------------------SLR-RLM---------------------------
-------------------------------LNRCNLDTAGCGFLALA-LMG
PF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
NAWLTHLS
L-SMNPVEDNGMK-LLCEVMREP--SCH----L---------------------------
-----------------------QDLEL--VKC-----HLTAACCESL------------
-------SCVI-SR
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
SRHLKSLDLTDNALGDGGVTA-LCEGLK-QK-KS-VLTRLG--LK-
--A-CGLTSDCCE--------------------ALSL-ALSC
PF13516 // ENSGT00900000140813_0_Domain_17 (2443, 2465)
NRHLTSLNLVQNNFSPKG
MMKLCS--------AFACPTSN--LQ--I-------------------------------
---IGLWKW-QYPVQIKKL------LE----------EVQ-LLKPRIVIDGSWHSFD---
--E---------------------------------------------------------
--------------------------------------------------DDRYWWKN--
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMUSP00000050252 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MALARA-NSPQEA---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWALNDLE--E---NSFKTLKFHLRD------
---------V--T------------Q--FHL-ARGE-LESLS-QVDLASKLISMYGAQEA
VRVVSRS----LL-AMNLMELVDYLN-QVC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LN--DYREIYREH-
--------VRC-----------------LE------ER---QDW----------GVNSSH
NKLLLMA-TSSSGGRRSPSC------SD-------LE-----Q-ELDPV--DVETLFAPE
AE--SYSTP----PI
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 930)
VVMQGSAGTGKTTLVKKL-----VQDWS--K---------GKLYP
G--------QFDY-VFYVSCREVVLLPK-------CDLPN-LICW-CC-GD-----DQAP
VTEILR-QP----GRLLFILDGYDE-LQK---------------------------S-SR
-AECVLHILMRRREVP-CSLLIT-TRPPALQSLEPML-G-----ERR--HVLVLGFSEEE
RETYF----------------S--SC-FTD---------KEQLKNALEFVQN-NAVLYKA
CQVPGICWVVCSWLKKKMARGQ-EV-S--ET-PSNSTDIFTAYVSTFLPTDG-NGDSSEL
----------T-RHKVLKSLCSLA---AEGM-RH-QR----LLFEEE-------------
--VLRKHGLDGPSLT-AFLNC----IDYRAGLG-IK-KFYSFRHISFQEFFYAMSFLVK-
EDQSQQ----------------G--EATHKEVA-KLVDP---E-NHEEVTLSLQFL----
----FDMLKTEGTLSLGLKFCFR-----------------IAPS-VRQ-DL-KHFKEQI-
E-------------------------------------AIKYKRSWDLEFSLYDSKI-KK
LTQGIQMK---------------------D-------------------VILNVQHLD-E
KK-SDKKK--S----VSVT-SSFS-----SGKV------------------QS----PFL
--------GN---DKS--T-------------------RKQKK-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------ASN----------------GKS---------R
-GAEE-PAP-GVR-NRR-LASR----EK-GHMEMND---KEDGGV---E-EQED-EEG-Q
T-LKK-DGEMI---DK-MNG----------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSLAFP00000000878 (view gene)
ENSCCAP00000008804 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MTSS----QLEYT---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LQTLLEQLS--E---GELGTFKSILWA------
---------F--PLEDV--------L--QKT-PWSE-VEEAN-GKKLAEILVNTSSECWI
RNATVKV----FE-ELNLPELSKMAK--AEL-----------------------------
------------------------------------------------------------
--LEDG---------------------------------QVEDADNHEL-----------
-------GDS-------------------------------DEDLELAKPGEKEGWRNV-
---------MDE----------------QSLV--QRNSFWRGDSDNFHDHVT--------
-----------------------------------------------QRNQRFIPFLNPR
RP--TQPSP----Y
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVLHGPAGVGKTTLAKKC-----MLDWK--D---------NNLGP
---------KARY-AFYLSCKELSRLG-------PCSFAE-LIAK-DQ-----PE-LQDD
IPNILA-QA----QRILFVVDGLDE-LRVPPG------ALIH-D-----ICGDWLKP-KP
-VPVLLGSLLKRKMLPKAALLVT-TRSWALRDLRPLM-E-----QPI--YIRVEGFLEED
RRAYF----------------L--RH-FGE---------EDQATRAFELMRS-NAALFQL
GSAPAVCWMVCMSLKLQMENGE-DP-A--PT-CHTRTGLFLHFLCSRFPKG-------AE
------------LGRTLRALSLLA---AQGL-WA-QM----SVFHAE-------------
--DLESLGVQES-DLRPFLDR----GILRQDGD-SK-DCYSFLHLSFQQFFTALFYALE-
KE------E---------EEDGH--AWDIGDVQ-KLLSGEERL-QNPDLIQAGHFL----
----FGLANEQRAAELETTFGCV---------------M--SRE-IKQ-ELLRCKANLHV
KK-----------------------------------P-SSMAGLKELLCCLYESQD-DE
LAKVVMAP---------------------F-------------------KEISIHLTNTS
EM---MQC--Y----FSLK-HCPD-LEK-LSLQ----------------VAKG----VFL
-END-VDF----DL--------------------GFELE---------------------
------------SSNSNLKFLEVRQSFLSES---------SVRVLC--DHIAR-N-----
TC--H-LQKAE--I-KNVTPD--TA----YR-DFCLAL----------------IGKTTL
THLT-LEGH-IEWERTMLLLMCDLFRSPKGN----LQYL--------RFG---------G
H------S----------------------------------------------------
---------AT---PE-QWD---DFSLILKA-NQSLRRLHLSASLLLD-EGAKLLYET--
VGH-PTSLLQKL-S----------------------------------------------
------------------------LENCRLT-----------------------------
----------------------------EASCKELAD-----------------------
--------------------------------VLIL
PF13516 // ENSGT00900000140813_0_Domain_11 (2017, 2054)
SQHLTH---LC-LAKNP-IGD---
-------KGVQLLCEGL----SYPDC----------------------------------
--------------------------KLQ-NLV---------------------------
-------------------------------LQQCNITKHGCRALSEV-LQGA
PF13516 // ENSGT00900000140813_0_Domain_13 (2214, 2228)
CNLTDLD
L-SFNHIG-CGLW-ILCQALENP--DCN----L---------------------------
-----------------------KHL----------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSOGAP00000016316 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------TLSW-------KNE---------DLHKKF
TQLLLLQRPHPRSLKS---L------VRRKRQVPI--------MENQGHLIEIRDLFGPG
LG--AQEEP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 929)
RIVILHGAAGIGKSTLAREV-----QRGWE--E---------GQLYQ
D--------RFQH-VFYFSCAELAQSK-------VMSLAD-LITQ-AQ-----EA-PPAS
LQQVLS-RP----EQLLFILDGVDE-PRWVLQQQ------NA-E-----FCQHWSQP-QQ
-VAMLLGSLLGKTILPEASFLIT-TRTVALQQLIPSL-K-----QPR--WVEVLGFSESS
RKEYF----------------Y--SY-FTV---------KSQALRALSLVES-NPAVWTM
CLMPWLSWLVCTCLKQQMERGE-DL-T--LT-TMTVTGLYWCYLSQALP--------AQ-
-----------PLRPHLRSICSLA---AEGI-RR-KR----SLFSPG-------------
--DLRRHGLNGR-IISTFVTM----GVLLKH---PNSRVYSFIHRCFQEFLAAVSYVLG-
D------EK---------RGEHC--NSI-RGVR-GLPQAHKTH--VLSRAPTRRFL----
----FGLLGEQAGREMESIFTCQ---------------R--APE-LKQ-ELLQWAKREAC
-----------------------------------VLQSPQLPGFLELFYCLYETQD-EE
FVTQVMVH---------------------F-------------------QGSSVCVQTGM
EL---LVF--T----FCIK-FCHH-MKR-LQLN----------------KDLV-------
------VGI---------------------------------------------------
------------------------------------------------------------
N-----------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------LSLC----SNVYFMQIPVT-----------------------------
----------------------------DACWQPLFS-----------------------
--------------------------------ILEVTG
PF00560 // ENSGT00900000140813_0_Domain_11 (2019, 2037)
SLKE---LD-LSGNS-LSH---
-------SAVQSLRKTL----RRPRCHLETLR----------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------L--VSC-----GLTF------------------
------------------------------------------------------------
---------SCCK--------------------DLASVLSTSPSL-KELDLRWNNLGDLG
VRQLCE--------GLRHPTCS--LT--L-------------------------------
---LWLDQA-FLGEKMKEE------LR----------TLE-KEKPALLISS-RWERGPRT
PSG---------------------------WCRLRAEPRD-----RQIS-----------
------------------------------RSKSAKVSGT-FHMEPPG-TEDEFLGPMGP
VAAEVVDIETRSLYRWEKVSFTTYIYRHCDKDGYHFKMERSKTSLFL-AGLNSQFCTYTL
LINNHSITS-VTLPLP-----YRQGEETTPPVSHHYMMLESSIT------FQDSAKISMA
HTQHHIIFIQTKQLISEHLTINEKTSETKIKVLLTKLDSVRALNPCLSILLLYRR-----
--------PPNDCSIQKAIDDEEKKFQFVRIHKPNLLTSLYMGSRYSVSGS---------
GKLEMIPNELELCYRSPGEAQMFSEFYVAHMESGFRLQMRDKKHRTVVWKALVKPDKLSP
MLPR------------------------------------LSGQTEAPDSSHREEP
PF00619 // ENSGT00900000140813_0_Domain_19 (3057, 3132)
HKHQ
LVARVTSVDSVLDKLY-GQVLSEEQYKQVRAEVTRPDQMRKLFSFSKSWDQACKDRLYQA
LKETHPHLIMDLL-----------------------
ENSOCUP00000014204 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---SSFFS
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 328)
DFG---LIWYLGELR--R---EELAKFKELLRL------
---------E--TSQLG--------I--QQI-PWAK-VKVAS-REVLADLLTKHYGEQQA
WDMTLRI----FH-TMGREDLCERAR-RES------------AAG--------QSLLKAD
K---------------------------------------------------MYQD----
------------------------------------------------------------
----------------------------------------------------------H-
--------VRST----------------FPHV--WSRDSLFRSHE---------------
---------------------------------F-------EWKLTQEERETLKFVLAPT
DT--GRQ-F----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
HTVVLQGSRGIGKTALLRKL-----MLAWS--D---------GSIYQ
D--------RFSY-VFYLCCREVRQLA-------STSLAS-LISR-DW-----PHH-STP
LADVVS-QP----ERLLFLVDNVEE-MSGDMAGP------PS-D-----LCSHWLEP-RP
-GKVLLRSLLAKSLLPAAFLLLA-VTPQCRGELLSGL-Q-----DPS--VKTVLGFSRNS
KKLYF----------------G--SL-FSN---------KRRALEAFQLVNR-NDRLFSM
CQNPILCWLVGTSMKQDMDRGR-DV-A--LT-CQSVTSTFISFIFNAFTPNG-AAYPTQ-
-----------QSQEQLKNLCSLA---AEGT-WT-HI----QFFNQE-------------
--VLGRNGIGDT-AIPTLLDT----KVL-RCRA-PE-NLYGFPHKALLQFCTALFYLLQ-
HPSAH-----------------P--NPAVQCTR-TLLLTVLNR--NRSSILLGLFL----
----FGLLNERERQKLDAFFGSN---------------L--SRQ-RKE-ELEQLLQNISG
C-A----------------------------------DFHPQVDFLVLFYCLSEIQD-DV
FAGQVMAC---------------------F-------------------QELNLTFLSSS
DL---GMA--A----SYLK-HCYN-LRK-LCFS----------------IQA-----LFR
-GED-PRT-----------Y-----------------------------------RTRQN
FLYWSQIC-SVLASSEHLQELQVVDSSFTEP---------TLVIFC--HQLRQP------
GC--R-LQKLMMNK---------VTM-TCEKLAFFEVF----------------THNPGL
KYLR-LNRM-ALSWD-DMKLLREALNHPMCG----VEGL--------LP-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------ASRGLA-----------------------------
----------------------------PDDCRVLAW-----------------------
--------------------------------VLFSGEVSPG---LG-GSTS--------
-------------VSPT----HRRPQTSENM-----------------------------
------------------------------------------------------------
------------------------------ALTSCHLSEAAWELLAQV-LISNCRLLYLD
V-SINILGDAGLK-QFCDALRHP--SCR----L---------------------------
-----------------------RSLRV--FEC-----LITAAGCGDL------------
-------AQAL-VSSQDLRCLQIGSNAIGDAGVKL-LCEALV-QP-SC-HLETLG--LQ-
--I-CGLSSACCE--------------------DLAYALTGSRTL-QDLDLSCNALDHNG
LAVLLA--------AVSHPGCA--LK--V-------------------------------
---LRLKHM-ELGEETQAL------LT----------AVQ-ERNPYLRVISQ--------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSOPRP00000009298 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------MA----G
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
ARCK---LAQYLEELE--E---LDLKKFKMHLED------
---------Y--PPGQG--------C--VPL-PKGQ-VARAD-PMDLATLMTKVNGEERA
WAMALWI----FM-AINRRDLYEKAK-RDEPAWXXXXXXXXXX---------XXXXX---
--------XX-----------------------------------XXXX-----------
-----------------------------------------------------XXXXXX-
------------------X-----------------XXXXXXXXX--XXXXX-XXXXXX-
--------XXXX----------------XX-X--XXXXXXXXXXX---------XXXXXX
XXXXXXXXXXXXXXXXXXXX------XXXXX--XXXX------XX--XXXXXXXXXXXXX
XXXXXXXXX----XXXXXXXXXXXXXXXXXXXX-----XXXXX--X---------XXXXX
X-------
PF05729 // ENSGT00900000140813_0_Domain_2 (729, 940)
-XXXX-XXXXHCRELSLGP-------HRSPGK-LIAG-CC-----PA-PGRP
VGQMLS-GP----ARVLFLIDGFDE-LQGAFDEC------PG-A-----SCCDWRAS-VR
-RDILLGSLIRKQLLPEASLLIT-TRPVALEKLQHLL-E-----RPR--HVAVLCFSEAQ
RSEY-----------------F--KF-FPE---------EAQAREAFRLLQE-NEVLLSM
CCIPLVCWIVCTGLRQQMSQD--------QT-CKTTTAVYTCFLSSLLQARGGGREPG--
------------LPSHLRGLCSLA---ADGI-WN-QK----ILFEEW-------------
--DLRRHGLREA-EVSAFLSM----SLFQKEVD-CE-AFYSFVHLTFQEFFAAMYYMLE-
EEEGA--------GTASPGRGGS--ELPPRDVR-VLLENYGRF-EKGYLIFVVRFL----
----FGLVNQDRAAYLEKKLGCS---------------V--SRQ-VRP-ELLRWIEAKAE
AK-----------------------------------KLQVQPSQLELFYCLYETQE-DT
FLRAAMGH---------------------F-------------------PRIELSLSTRM
---DHVVA--S----FCIQ-NCHS-VES-LSLG----------------LLHS-----SS
KEEGEEEE-----E------------------------------------EE---E----
---------------EGWLAAEGRHMCLR-P-----------------------------
------------------------------------------------------------
--------------------------PAACP----S-----------RSV---------S
------------------------------------------------------------
-------CYPT---SG-LCR---SLFSVLGS-NP
PF00560 // ENSGT00900000140813_0_Domain_8 (1775, 1801)
SLVELDLSDNSLGD-PGMRVLCEA--LQH-PSCGIRRL-W----------------------------------------------
------------------------LGRCGLS-----------------------------
----------------------------HQCCFDLSL-----------------------
--------------------------------VLSGSQ
PF00560 // ENSGT00900000140813_0_Domain_11 (2019, 2064)
KLAE---LD-LSDNT-LGD---
-------FGVRLLCVGL----RHLSCQLQKL-----------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSLAFP00000009291 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-MEEPRASCSSPGQR--AAA
PF02758 // ENSGT00900000140813_0_Domain_0 (201, 328)
AREL---LLAAFEDLS--Q---EQLKRFRHKLRN------
------------APEGS-----------RGI-PWGR-LEGAD-AIDLVERLVEFYGPEPA
LEVAHKA----LK-RSDVRDVAARLK-EQRLQQ---------L---------GPG-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------PS-AQLSVSEYKKKYRQH-
--------VLQL----------------HAKV--KERNARS------------VKITKRF
TKLLIAPESPDPEDE--TLGLAE-------EPEP--------RRARRSDTDTFNRLFSRD
E---EGGRP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
LTVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYH
G--------QVDF-AFFLPCRELLERQG------DSSLAD-LILD-QC-----PD-RGAP
VRQILA-HP----ERLLFILDGADE-LFAQGSA--------EAA-----PCADPFEA-TS
-GAQVLGALLSKALLPEARLLVT-TRAAPPGGLHRHL-R-----SPQ--CAEVRGFSAKD
KKKFF----------------Y--KF-FRD---------ERRAERAYRFVKE-NETLFAL
CFVPFVCWIVCSVLRQQLKSSQ-DL-S--RT-SKTTTSVYLLFIVSLLGSAPPANGA---
-----------YVQGELRKLCRLA---REGVLGR--R----AQFAEK-------------
--DLERLELSDSVVQTLFLNKKEMVGVLEA-----E-VTYQFVDQSFQEFFAALSYLLE-
EEGAP-------------------E-APAGGVG-ALLRRDAEL--RGHLTLTTRFL----
----FGLLSTERMRDIERHFGC-----------------LVSER-VKQ-DALLWVQGQGR
ST-----AAPEGTEGTEGLEASEEPE---------EEEYEELNHPLELLYCLYETQE-DA
FVRQALRG---------------------L-------------------PELALERVCFS
RM-DLAVL--S----YCVQ-CCPE-GQA-LRLL------------------RC----QLS
------ML----------------------------------------QEKK-K---KRS
----------------------------------------LVKR----------------
------LQGS---L--------------GSR------N----------------S-GASS
RQ---PPAS-------PLRALCEAMNDQHCG----LSRL--------T------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSNGAP00000017481 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----PSEV---------TTSEENVCLSPNSQGRVE----THVVQAGPKL-------YVNF
---TESKTSF-------------------------------FCPLLPDHGGDQQDYKAH-
--------MVAK----------------LKTS--VEVH-H--------------------
--------------------------------------------DSPDI-QILLDAFKPD
Q---KGFQP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 928)
HTVVLHGKSGIGKSALARRI-----VLGWA--Q---------GELYQ
D--------MFSY-IIVLSTREMKWTE-------ESSFAK-LIAR-EW-----PN-SQAP
VAQIMS-QP----ERLLFVVDSFDD-LDSTTIHD------DT-R-----LCGDWNRE-QL
-VSILMYSLLKKALLPESFLMIL-TRDTGLEKLKSAV-V-----SPQ--YILVEGMSCKE
RTQLL----------------L--SH-IAS---------DHRKAQVFHSVMD-NHQLFDQ
CQIPSVCLLVCEALQLQERLGR-SC-A--PC-HQTLTGLYTTFVFHQLTPRD-PFQSCLS
----------QEERVSLMGLCRMA---VEGV-WN-MR----SVFYSN-------------
--DLRSHGLKES-ELRALFHM----NILLHED--RGERCFTFFHLSLQDFCAALYYVLE-
GLE-R--------------WDYY--FLFIKNLR-GITELQQTG-SNAHLLRMKQFL----
----FGLVNKDTMKALELLLGRP---------------V--SP-AVKQ-KLWRWVSLLGQ
QVN----------------------------------TTSTQVDVLNAFHYLYETQD-KE
FVSLSLNS---------------------F-------------------QEVWLTVSRRM
DL---MVS--S----FCLQ-HCQN-LRK-IRVD----------------VKD-----IFS
-VDKTAEL----CP--------------------MA-----------PQ--RTERK-ALI
VEWWENFC-SVLGSHPNLEQLHLGSSILSEW---------AMKTLC--IQLRH-P-----
AC--K-IQNLT--F-KSAEV---AP---GLQ-YLWMTL----------------ISNRNL
KYLN-LGST-LLREE-DVKMACNALRHPKCF----LETL--------RLD---------S
C------E----------------------------------------------------
---------LS---NT-CYP---MVSQLLNS-STSLKSLSLARNKVAE-DSMKSLCDA--
LMS-SKCGLQRL-I----------------------------------------------
------------------------LDSCDLK-----------------------------
----------------------------SASFHVLSS-----------------------
--------------------------------ALLSNRKLTH---LC-LSNND-LGT---
-------EEMRLLCQSL----RHPGC----------------------------------
--------------------------TLQ-RLI---------------------------
-------------------------------LNDCNLRGQAYGFLALT-LM
PF13516 // ENSGT00900000140813_0_Domain_13 (2212, 2237)
NNTQLTHLS
L-TMNPVEDSGMK-LLCEAIREP--TCH----L---------------------------
-----------------------QDLEL--VHC-----QLTGNCCKDL------------
-------ACVI-TR
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NKYLRSLDLGANALGDSGVVA-LCEGLK-QR-DS-HLRRLG--LE-
--A-CRLTSDCCQ--------------------ELSSALSC
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2465)
NQHL-ISLNLVRNDFSTSG
MMKLCS--------AFMHPTSN--LW--M-------------------------------
---IGLWKK-EYHAQVRRQ------LE----------ELQ-LLKPHMMIKGDWYSFD---
--E---------------------------------------------------------
--------------------------------------------------DDRCWWKN--
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPCOP00000013304 (view gene)
ENSOGAP00000020794 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------V---SSFFSDFG---R
PF02758 // ENSGT00900000140813_0_Domain_0 (209, 328)
IWYLGELK--K---K--KKCKEHFKQ------
---------E--TLKLG--------F--KQI-SWTE-VKKSS-REELANQLIQQYEDQQA
WNLTLRI----FH-RMSRQDLCDKVQ-RER------------AE--------------HR
K---------------------------------------------------IYRGNW--
--AV------------------------V-----T-------------------------
----------------------------------------------YHSLVRS---TGH-
--------QKSDCSRRCLAGPGW-------------------------------------
-----------------------------------------G--------------FEST
TL--GVHGW----CLSRLSYRSRA
PF05729 // ENSGT00900000140813_0_Domain_2 (685, 929)
GKVTFLMKL-----VLGWA--E---------GSLYQ
D--------RFLY-LFYFC-QEPRQLK-------ETSLAN-LISR-NW-----P--SPAP
ITEIMS-HP----ETFLFFIYSFEE-LKCDLSEP------ES-D-----LCSDWMKQ-QL
-VWALMRGLLRKKMLLEASLLIT-VTPVCLA----QL-V-----CPE--LI--TGFSEGK
IELYS----------------R--HL-FPS---------RNGTVEAF-YLRE-NTGVFHV
PPPPLICWV-GTCTKQVMEKGR-DV-A--PI-CRHLTSLYASFILNLFTCKD-ACRPSL-
-----------QSQVQLRGLCSLA---VEGM-WT-NT----FEFNDK-------------
--ALRRNGIADS-DVPALLDT----NIFLKCT---E-SSYTFVHTSVQEFCAAMFYLLR-
--CDH-----------------P--NPAVRCRE-MLLADHLEKVNKACWIYWGKFS----
----FWFK------KVDTVFCFQ---------------P--SQE-IRQ-QFHQILKNLGE
REG-----------------------------------LR-QTDFLALFCCLFEMEN-KA
FVREAVNF---------------------L-------------------QDINFSINSDL
DL---VIC--A----YCLK-DCSG-LRR-LSFS----------------VSN-----VTE
-KS------------------------------------------------------SHN
VIPWHHIF-SVFTTNEHLKELEITNTDS-------------ANILC--NQLRHP------
TG--P-LEKLQLNR---------VSL-AGGSSVFLEVL----------------SHSSDL
KDLN-LHNV-NLSRD-DTEMFCKVLSSSMCN----TEEL--------VL-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------SGCFLR-----------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------N-PCWDLLCEA-LLSKKSL-ALD
L-SASNLHNKGLN-VLCAALRHP--DCC----L---------------------------
-----------------------RSLGL--ANC-----SFTAVDCPDL------------
-------ASVL-TSNQKLTSLQIGLNKIGDVGVKV-LCEALT-HP-NC-HLEILG--LR-
--D-CGLTGA-CR--------------------DLYSALSNSGTL-RMLDLTINTLDHRG
-AVLCE--------ALRYPGCP--LW--V-------------------------------
---LGLPKT-RVDEETQML------LT----------VVE-ERNPYLTIIGI-W------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSECAP00000016221 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------CRLT------ASCGRD---------------LSLVFET-NQCL
TNLE-FVKN-TLEDS-GVKLLCQGLKHSNCI----LQTLR--------LYRCLI-SPA-S
CE------------AL----AAVLSTSQWLTELEVSETKLETSAFKFLCEGLKDPNCK-L
QKLN----ILPESSEA-VCK---YLASVLIC-NPHLTELDLSENPLGD-TGVKYLCEG--
LRH-SNCKVEKL-D----------------------------------------------
------------------------LSTCHLT-----------------------------
----------------------------DASCVELSS-----------------------
--------------------------------FLQVSQTLKE---LI-VFANA-LGD---
-------TGVQHLCGGL----RHTKGIIENL-----------------------------
------------------------------------------------------------
------------------------------VLSECSLSAACCESLAQV-LRSTWSLTRLL
L-INNKIEDLGLK-LLCEGLKQP--DCQ----L---------------------------
-----------------------KDLAL--WTC-----NLTGECCQDL------------
-------CNAL-YTNEQLRVLDLSDNALGDEGMQV-LCEGLK-QP-SC-KLQTLW--LA-
--Q-CHLTDACSG--------------------ALASVLNRNE
PF00560 // ENSGT00900000140813_0_Domain_17 (2444, 2461)
NL-TLLDLSGNDFKDFGVQMLCD--------ALIHPICK--LQE---------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPCOP00000001237 (view gene)
ENSCCAP00000039719 (view gene)
ENSTSYP00000032706 (view gene)
ENSANAP00000041367 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-MDQSDAPCSSTGPR--FAVAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LQAALEDLS--Q---EQLKRFRHKLRD------
------------VGSDG-----------RSI-PWGR-LERAD-ATELAEQLAQFYGPEPA
LEVARKT----LK-RADARDVAAQLQ-EQRLQR---------L---------GPG-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------PA-ALLSVSEYKKKYREH-
--------VLQR----------------HARV--KERNARS------------VKITKRF
TKLLIAPESAAP---------SE-------EPEP--------QRARRSDTHTFNRLFRGD
E---EGRRP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 930)
TVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYQ
E--------QVDF-AFFLPCGELLERPG------TRSLAD-LILD-QC-----PD-RGAP
LRRMLA-QP----ERLLFILDGADE-LPAPGGP--------EAA-----PCTDPFEA-AS
-GARVLGALLNKALLPSALLLVT-TRAAAPGRLQGRL-C-----SPQ--CAEVRGFSDKD
KKKYF----------------Y--KF-FGN---------ERRAERAYRFVKE-NETLFAL
CFVPFVCWIVCTVLHQQLELGR-DL-S--RT-SKTTTSVYLLFVASVLSSAPAADRP---
-----------RVQGDLRKLCRLA---RGGVLER--R----AQFAEE-------------
--ELEQLELRDSKVQTQFFSKKERPGVLET-----E-VTYQFLDQSFQEFLAALSYLLE-
DESVP-------------------G-TAAGGVG-TLLRGDAQP--HSHLVLTTRFL----
----FGLLSAERMGDIQRHFGC-----------------RVSER-VKQ-EALRWVQG--Q
GCPGAAPEVTEGAKGLEDTEEPEEEE------------EEEPNYSLELLYCLYETQE-DA
FVRQALSS---------------------L-------------------PELALQRVRFS
RM-DVAVL--S----YCVR-CCHS-GQA-LRLS------------------SC----RLV
------AT----------------------------------------QEK--K---KKS
----------------------------------------LRKR----------------
------LQAS---L--------------GGS------S----------------SSQATA
KQ---LPAS-------LLHPLFQAMTDPLCR----LSSL--------TLS---------H
C-----------------------------------------------------------
--------KLP---DA-VCR---DLSEALRA-APMLTELSLLHNGLS-EAGLRMLSEG--
-LAWPQCQVQTF------------------------------------------------
----------------------------RVQ-----------------------------
---------------LSGP---------QRGLQYL-------------------------
------------------------------VPMLRESPALTT---LD-LSGC-QLPAPMV
--T--------YLCAVL----QHPGCSLQTLSL--ASVELSEE-----------------
----------SLQELRAAKRAKP-------NLV----------------------I--AH
PALD-------------------GHPEPP-KELSSIF-----------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMUSP00000075794 (view gene)
ENSPVAP00000013249 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------MSLDIHFEQ-LSD-ARWT----------------EL---------
----LPLI-------------------------------QQHQVV------RLIDC-GIT
EGRCKDIS-SALGDNP
PF00560 // ENSGT00900000140813_0_Domain_4 (1517, 1551)
TLTELNLCNNELGDA---------GMRLLL--QGLHS-P-----
TC--K-IQKLS--V-QNCGLT--EA---GCG-VLPGAL----------------RSVP
PF00560 // ENSGT00900000140813_0_Domain_6 (1619, 1645)
TL
RELY-LSNN-PLGDA-GLQLL-DGLLDPQCH----IERL--------XXX---------X
X------X----------------------------------------------------
---------XX---XA-SCE---PLAAVLRA-KREFKELMVSNNDLGD-AGVRALCRG--
LAD-SASPLESL-R----------------------------------------------
------------------------LEGCNLT-----------------------------
----------------------------PASCQDLGS-----------------------
--------------------------------IVASKA
PF00560 // ENSGT00900000140813_0_Domain_11 (2019, 2057)
SLCT---LE-LGDNK-LGD---
-------AGIAKLCPGL----LSPSS----------------------------------
--------------------------QLR-TLW---------------------------
-------------------------------LWECDITASGCRDLCRV-LKAKGSLKELS
V-AGNAVGDEGAQ-LLCESLLAP--SCH----L---------------------------
-----------------------ESLWA--KSC-----GFTAACCQHF------------
-------SAML-ARNT
PF00560 // ENSGT00900000140813_0_Domain_15 (2357, 2381)
RLLELQLSGNSLGDAGVQQ-LCQGLG-QP-GA-VLRVLC--LG-
--D-CEVTNNGCS--------------------SLASLLLANH
PF00560 // ENSGT00900000140813_0_Domain_17 (2444, 2475)
SL-CELDLSNNCMSDMG
VLQLAE--------SLQQPGCM--LE--K-------------------------------
---LVLFDT-YWMEDTDNR------LR----------ALE-ESKPSLKIIS---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSAMEP00000018180 (view gene)
ENSDORP00000010033 (view gene)
ENSCSAP00000016317 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------MA---ESFFS
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 328)
DFG---LLWYLKELR--K---EEFWKFKELLKQ------
---------P--LEKFE--------L--KPI-PWAE-LKKAS-KEDVAKLLNKHYPGKQA
WEVTLHL----FL-QIDRKDLWTKAQ-EEIRN----------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------KLNPYRKH-
--------MKEK----------------FQLI--WEKETCLPIPEH--------------
---------------------------------F-------YKETMKNKYKELNGAYTAT
------ARL----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 929)
HTVVLEGSDGIGKTTLLRKV-----MLDWA--E---------GNLWK
D--------RFTF-VFFLNVYEMNGVT-------ETSLLE-LLSR-DW-----PES-SET
IEDIFS-QP----ERILFIMDGFEQ-LKFNLT-F------KA-D-----LSDDWRPR-QP
-PPIILSSLLQKKMLPESSLLIA-LGKLAMQKYYFML-Q-----HPK--LIKLSGFTESE
KKSYF----------------S--YY-FGE---------KNKALKVFNFVRD-NGPLFIL
CHNPFICWLVCTCVKQRLERGE-DL-E--IN-SQNTTSLYASFLTNVFKAGS-QSFVPK-
-----------VNRARLKNLCALA---AEGV-WT-YT----FVFCHE-------------
--DLRRNGLSES-EGLMWVGM----RLLQRRG-----DCFTFVHLCIQEFCAAMFYLLK-
RPKDD-----------------P--NPAIGSIT-QLVKATVIQ-PQTHLTQVGIFM----
----FGISAEEIVSMLETSFGFP---------------L--SKD-LQQ-EITQSLKSLSQ
CE-----------------------------------ADREAIGFQELFVSLFETQE-KE
FVTEVMNF---------------------F-------------------EEVFIYIGNIE
HL---VIA--S----FCLK-HCRG-LMT-LRMC----------------MEN-----IFP
-DDSGCAS-----------S-------------------Y-N-------EK---------
LVYWRELC-SVFITNKNFQILDMENSSLDDA---------SLAILC--KALTQP------
VC--K-LRKLIFTS---------MLN-FGHDSELFKAV----------------LHNPHL
KLLS-LYGT-SLSHT-DVRHLCETLKHPMCK----IEEL--------INLKLKY------
-------V------------------------------------NDHKCDCLIWQVT---
-ALKV----L---------G---YMCAMCRQ----------------RPTSVILLCRC--
CCL--LFIVIVFLR----------------------------------------------
------------------------LGKCDIS-----------------------------
----------------------------SEDCEDVAS-----------------------
--------------------------------VLTCNSTLKH---LS-LVENS-LRD---
-------EGMLLLCEAL----KHPPCALETL-----------------------------
------------------------------------------------------------
------------------------------MLTYCCLTCVSCDSISQVL-LCNKSLSLLD
L-SSNALEDEGVA-SLCGALKHP--ACS----I---------------------------
-----------------------QELWL--------------------------------
----------------------------------------------------------Q-
--T-CPITRACCG--------------------DLAAALVACKTL-RSLNLDWITLEPDA
VAVLCE--------ALSHPDCA--LQ--M-------------------------------
---LGLHKS-GFDEETQKM------LM----------SVEEKV-PHLTISHEPWIDE---
--EYK-----IR------------------------------------------------
--------------------------------------------------GVLL------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSPANP00000023154 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------MAESNSTDFD---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWYLENLS--D---KEFQSFKKYLAR------
---------K--ILDFK--------L--PQV-PLIN-LRA-T-KEELANLLPISYEGQHI
WNMLYSI----FL-MMRKEDHCMKI---IGRRN---------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------RNQEACKAL-
--------MRRK----------------LTLQ--WENHTFGEFHYKFFCNVA--------
----------------------------------------------SDVFYVLQLAYDST
SC--YSAND----LN
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 930)
VFLMGDRASGKTMTINLA-----VMKWI--N---------GEMWQ
N--------MISY-VVHLTSHEINQMT-------DSSLAE-LIAK-DW-----PD-GQAP
IADILS-DP----KKLLFILEDVDN-LRFELNVN------EQ-A-----LCSNSTQK-VP
-IPILLVSLLKRQMAPGCWFLIS-SRPTREENIKMFL-K-----TTD--CYVTLQFSNEK
RELYF----------------H--SF-FND---------RQRASTALMLVHD-DEILTGL
CRVAILCWITCTVLNRQMDKG-HDP-Q--HC-CETPTDLHAHFLADALTSKAGLT-AN--
----------QYHLGLLRRLCLLA---AGGL-FL-ST----LNFSGE-------------
--DLRCVGFTEV-DVCVLQAV----NVLLPSST-HK-DRYKFIHLNVQEFCAAIAFLMAV
PNCLI-----------------P----SGSR-G--Y--KEKRE-QYCNFNQVFTFI----
----FGLLNENRRKILETTFGYQ---------------LPMVDS-FRR-YSVEYMKHLGR
DQ-------------------------------------EKLTHHGSLFYCLFENQE-EE
FVKKVVNT---------------------L-------------------KEVTVYLQSNK
DM---MVS--L----YCLE-YCCH-LRT-FKLS----------------VQR-----IFE
--YKEPLV-------------------------------------------RPTASQMKS
LVYWREIC-SLFCTMDTLWELHLFDSDLNDI---------SERILS--KALEHS------
SC--K-LRTLKLSY---------IST-ASGFEDLLKAL----------------ARNRSL
IHLS-LNCT---------------------------------------------------
------------------------------------------------------------
--------------------------------------------SISL-NMFSLLREI--
LDK-STCQIRHL-S----------------------------------------------
------------------------LMKCDLR-----------------------------
----------------------------ASECGEIAS-----------------------
--------------------------------LLVNGGSLRK---LT-LTNNP-LRN---
-------DGVNILCNAL----LHPNCTLTSL-----------------------------
------------------------------------------------------------
------------------------------ALVFCCLTEKCCSSLGRV-LLLGRTLKQLD
L-CVNHLQNYGVL-HVTFPLIFP--TCQ----L---------------------------
-----------------------EELYL--SGC-----FFTNDICQYI------------
-------AIVIA-T
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NETLRSLEIGSNSIEDAGMQL-LCGGLR-HP-DC-MLVNIG--LE-
--E-CMLTGACCE--------------------SLASVLT
PF13516 // ENSGT00900000140813_0_Domain_17 (2441, 2464)
TNKTL-ERLNILQNQLGDEG
VVKLLN--------SLILPECV--LQ--I-------------------------------
---IGLPLN-DVSAETQRM------VM----------AVK-ERKPNLIFLSETWSVKEGR
EIGVI-------------------------------------------------------
-------------------------------------------------------PASQP
GSV-----------------IPNSNLD---YM--FFKFPRMSTAMRMSSTASRQPL----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMMUP00000010478 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------MAESNSTDFD--
PF02758 // ENSGT00900000140813_0_Domain_0 (207, 322)
-LLWYLENLS--D---KEFQSFKKYLAR------
---------K--ILDFK--------L--PQV-PLIN-LGA-T-KEQLANLLPISYEGQHI
WNMLYSI----FL-MMRKEDHCMKI---IGRRN---------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------RNQEACKAL-
--------MRRK----------------ITVR--WENHTFGEFHYKFFCNVA--------
----------------------------------------------SDVFYVLQLAYDST
SC--YSAND----LN
PF05729 // ENSGT00900000140813_0_Domain_2 (676, 930)
VFLMGDRASGKTMTINLA-----VMKWI--N---------GEMWQ
N--------MISY-VVHLTSHEINQMT-------DSSLAE-LIAK-DW-----PD-GQAP
IADILS-DP----KKLLFILEDVDN-LRFELNVN------EH-A-----LCSNSTQK-VP
-IPILLVSLLKRQMAPGCWFLIS-SRPTREENIKMFL-K-----TTD--CYVTLQFSNEK
RELYF----------------H--SF-FND---------RQRASTALMLVHD-NEILTGL
CRVAILCWITCTVLNRQMDKG-HDP-Q--HC-CETPTDLHAHFLADALTSKAGLT-TN--
----------QYHLGLLRRLCLLA---AGGL-FL-ST----LNFSGE-------------
--DLRCVGFTEV-DFSVLQAV----NILLPSST-HK-DRYKFIHLNVQEFCAAIAFLMAV
PNCLI-----------------P----SGSR-G--Y--KEKRE-QYCNFNQVFTFI----
----FGLLNENRRKILETTFGYQ---------------LPMVES-FRR-YSVEYMKHLGR
DQ-------------------------------------QKLTHHGSLFYCLFENQE-EE
FVKKVVNT---------------------L-------------------KEVTLYLQSNK
DM---MVS--L----YCLE-YCCH-LRT-FKLS----------------VQR-----IFE
--YKEPLV-------------------------------------------RPTASQMKS
LVYWREIC-SLFCTMDTLWELHLFDSDLNDI---------SERILS--KALEHS------
SC--K-LRTLKLSY---------IST-ASGFEDLLKAL----------------ARNRSL
IHLS-LNCT---------------------------------------------------
------------------------------------------------------------
--------------------------------------------SISL-NMFSLLREI--
LDK-STCQIRHL-S----------------------------------------------
------------------------LMKCDLR-----------------------------
----------------------------ASECGEIAS-----------------------
--------------------------------LLVNGGSLRK---LT-LTNNP-LRN---
-------DGVNILCNAL----LHPNCTLTSL-----------------------------
------------------------------------------------------------
------------------------------ALVFCCLTEQCCSSLGRV-LLLGRTLKQLD
L-CVNHLQNYGVL-HVTFPLIFP--TCQ----L---------------------------
-----------------------EELYL--SGC-----FFTNDICQYI------------
-------AIVIA-T
PF13516 // ENSGT00900000140813_0_Domain_15 (2355, 2378)
NETLRSLEIGSNSIEDAGMQL-LCGGLR-HP-DC-MLVNIG--LE-
--E-CMLTGACCE--------------------SLASVLT
PF13516 // ENSGT00900000140813_0_Domain_17 (2441, 2464)
TNKTL-ERLNILQNQLGDEG
VVKLLE--------SLILPECV--LQ--I-------------------------------
---IGLPLN-DVSAETRRM------VM----------TVK-ERKPNLIFLSETWSVKEGR
EIGVI-------------------------------------------------------
-------------------------------------------------------PASQP
GSV-----------------IPNSNLD---YM--FFKFPRMSTAMRMSSTASRQPL----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSOANP00000018610 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------TTMA-PAP
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 325)
RDK---LLLSLKELR--E---EELRTFKFKLLG------
---------I--PLQDG--------Y--FHV-PRGE-VDGLK-PVELADLLILYYGEKYA
VMVVLEV----LK-AMHMNGVVETLG-QDT------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------WE--DSRETYRER-
--------MKKS----------------LE------R----AEE----------SLPGAQ
EEE-----GPAPVAEGNGSS------GI-------GE-----K-EVHSI--GLETLLDPE
EE--GTDSP----RT
PF00004 // ENSGT00900000140813_0_Domain_2 (676, 871)
VVLHGEAGIGKTTLAKKL-----MLDWA--S---------GDFYP
D--------TFDY-TFLVSCREINLMAE-------KSLAE-LICH-CY-GD-----GHAP
VTEVLK-RP----ERLLFIIDGLDE-LKYSLEEE------GE-E-----PGSELRDK-QP
-VQTLLSSLVRKKLLPESSLLIT-TRPRALEKLQPLL-E-----DPR--YVEILGFSEVE
REEYF----------------F--KF-FTD---------ENKAKKAFNFVQR-NENLSSL
CSIPLVCWFSCTCLKQQMVRGE-PL-T--HT-SKTITDVYMSYISIFLQPDRA---RAKQ
----------P-AYPTLKRLCSLA---ADGI-RN-RK----ILFQED-------------
--DLRQHSLDQADIA-AFLSK----ANFRQESK-GE-TFYSFIHYSFQEFFSALYYMLE-
GD--------------------G--NESNRDVN-ALLEKQEQT-RSDGSALTLRFL----
----FDFLSQETVSTLEIKFSYK-----------------IPPK-LQK-EL-KCIKARV-
K-------------------------------------SLRHFRT---------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSTSYP00000033223 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------MA----S-RTQ---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LAHFLELLK--K---EELKEFQLQLPS------
---------K--VL-SG-------AS--SGS-IPAQ--PEMS-GLEVASCLVAQYGEKQA
WNMAFNI----WE-KMGLKLVCAQAQ-AEGAGEIPPDLSC-EVGLG---SPN--------
QPTSTAVLGILGPGSL-----ESPQDSEKRRILRWPPDTSGNNW-REIKPLPLFE--A--
---------LSSFPV-YQSPSQESPNTPTSTAVLRGWASPHRPSPDPRSQEAPKPRWLLA
DTPESCCAEKK----------------------------S--EAGKQRCPRKVEGYQKQ-
--------MREN----------------PCSTQSW-------KNE---------DLHQEF
TQLLLLHRPHPRGHRP---L------VRGSWHQDV--------LRDQGHLIEIGDLFGPS
LG--SQEEP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 940)
RVVILQGAAGTGKSTLARQV-----RRAWE--E---------GHLYR
D--------RFQH-VFYFCCRQLAKSK-------EVSLAE-LIIK-DW-----VA-PLAP
IGQILS-QP----EQLLFILDGVDE-LRCVLEKQ------SS-K-----LCLHWSQL-QP
-LHELLASLLGRTTLPGASLLIT-TRTAALQKLIPSS-E-----QPR--WVEVLGFSESS
RKEYF----------------Y--KY-FTD---------ESQAIRAFSLVES-NRALWTL
CLVPWVSWLVCTCLKQQMEQGE-EL-T--LT-AQTITALCLRYFSQALQ--------AQ-
-----------SLGPQLRGFCSLA---AEGI-WR-GT----TLFSPA-------------
--DLRKHRLDEA-IIFTFLKV----GVLQKH---PTSLSFSFTHLCFQEFFAAMSYALG-
D------KE---------RGEHP--NHI-RGVE-KLLEAYGRH--DLLKTLTMRFL----
----FGLLGEQGMRDMENIFHCQ---------------L--PRE-RKW-GLLRWPS----
-------------------------------------QIPLLPRSLELLHCLYETQD-ED
FLTQAMAH---------------------F-------------------QGTRICVQTDM
EL---LVF--T----FCIK-FRHR-VKR-LQLN----------------EGGQ-------
------H-E---------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------P------------
------VWRP--------------------------------------------------
-------------------PGIVLFKWIPVT-----------------------------
----------------------------DAYWHVLFS-----------------------
--------------------------------ILKITGSLKE---LD-LSGNS-LNC---
-------SAVQSLCETL----RHPR
PF13516 // ENSGT00900000140813_0_Domain_12 (2066, 2206)
CHLETLQ----------------------------
------------------------------------------------------------
-------------------------------LADCSLTAEGCKDLENSGT00900000140813_0_Linker_12_13 (2206, 2213)
ASA-LSTPF13516 // ENSGT00900000140813_0_Domain_13 (2213, 2237)
SQTLTKLE
L-SFNVLTDAGVE-HLCQGLRQP--SCR----L---------------------------
-----------------------QQLQL--VSC-----GLTT------------------
------------------------------------------------------------
---------SCCQ--------------------DLASVLSA
PF13516 // ENSGT00900000140813_0_Domain_17 (2442, 2465)
SPSL-TELDLQQNDLGNLG
VKLLENSGT00900000140813_0_Linker_17_18 (2465, 2716)
CE--------GLRQPTCQ--LT--L-------------------------------
---LWLDQT-PLSDKVREE------LR----------AVE-EERPLLLISS-IWKPSVMT
PNEGPDGEEMDSN-----------------RSSLKRQRQESGNLKGA-------------
-DHCPA--VPEG------SSPQVAQ-VESFCLSFPASPGN-LHVESLG-TKDEFWGPMGP
VATEVVDE-ERSLHRPF13553 // ENSGT00900000140813_0_Domain_18 (2716, 2997)
VHFPSAGSYHWS---NTGLCFVVREAV-------TIKMEFCAWGQ
FLGRTISRHSWMVAGPLLDIKAEPGAAVAAVYLPHFVALQGG--------HVDLSLFQVA
HFKEEGMLLEKPARVEPHYIVLENPSFSPVGVLLRMIHAALRFIPITSIVLLYHHLRPEE
V-TFHLYLIPSDCSIQKAIDDEEKKFQFVRIHKPPPLTPLYMGTRYTVSGS---------
RKLEIIPKELELCYRSPGEAQLFSEFYVGHLGSGIRLQMRDKKEGAVVWKTLVKPGENSGT00900000140813_0_Linker_18_19 (2997, 3052)
DLRP
AAALVPP-----------------------------APIGSPSPLDAPAMLPF00619 // ENSGT00900000140813_0_Domain_19 (3052, 3134)
HFIDRHREQ
LVARVTSVDPLLDKLY-GQVLSEEQYERVRAEATKPGQMRKLLSFSRSWDQACKDQLYQA
LKETHPHLIMDLWEKWSGQGGV--LPLSC-------
ENSNGAP00000012423 (view gene)
ENSCJAP00000021914 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------MAMA-RNPRNA---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLWALSDLE--E---KDFKKLKFYLRD------
---------M--TLSEC--------Q--APL-ARGE-LEGLI-AVDLAELLIAKYGEKEA
VKVVFEG----LK-AMNLLELVDQLS-HIC------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------LH--DYREIYREH-
--------VRC-----------------LE------EW---QEV----------GVNGRY
NQVLLVA-KPSAESPESPAC------PIT------LE-----Q-ELDSI--VVEALFDSG
EK--PSLAP----S
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
LVVLEGSAGTGKTTLSRKM-----VLDWA--T---------GTLYP
G--------QFDY-VFYVSCKEVVLLLE-------SKLEE-LLFW-CC-GD-----NQAP
VTEILR-QP----ERLLFILDGFDE-LQRPFEEK------LK-------RG-----L-SS
-KESLLHLLIRRCILPTCSLLIT-TRPLALRNLGPLL-K-----QAR--HVHILGFSEEE
RGRYF----------------S--SY-FTD---------EKQADHAFEIVQK-NDILYKA
CQVPGICWMVCSWLKEQMEKGE-AV-L--ET-PRNSTDIFMAYVSTFLLPDD-DGDCSEF
----------S-RHRVLMSLCSLA---AEGI-QH-QR----FLFEED-------------
--ELRKHNLDGPRLA-AFLSS----NDYQLGRA-IK-KFYSFRHISFQEFFYAMSFLVK-
KDQSQL----------------G--KESGREVQ-RLLEVKKQE-GNDEMTLTMQFL----
----LDISKKDSISNLELKFCFR-----------------ISPC-IAQ-DL-KHFKEQW-
E-------------------------------------SIKQKRTWDLEFSLYKAKV-KT
LAKGIQMN---------------------E-------------------VSFKIAHSN-E
KK-SQSQN--S----FSVK-TSLS-RGQ-KEKQ------------------KC----PLV
--------GK---DNK--A-------------------GTQKE-----------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------AST----------------GKG---------R
-GSEE-PDEKHQK-N----TYI--------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
MGP_SPRETEiJ_P0084791 (view gene)
ENSOPRP00000012868 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------
PF02758 // ENSGT00900000140813_0_Domain_0 (270, 328)
PL-PKGQ-VARAD-PMDLATLMTKVNGEERA
WAMALWI----FM-AINRRDLYEKAK-RDEPAWRS-----NPAAAS------NPIVI---
------------------------------------------------C-----------
---------------------------------------QGELTEESSM-----------
GVL-GY-LTR-------------------------------I-SFCHRKKDYCRKYRRH-
--------VLSR----------------FQCI--EDRNARLGESV---------NLNRRY
TRLRLAKEHPSRQEREQQLV------AIGQTPVR----------DSPASPVRLEMLFEAD
EE--CPGPV----RTVVLQGTXXXXXXXXXXXX-----XXXXX--X---------XXXXX
X--------XXXX-XXXXXXXXXXXXXXX----XXXXXXX-XXXX-XX-XX-----XXXX
XXXXXX-XX----XXXXXXXXXXXX-XXXXXXXX------XX-X-----XX----XX-XX
-XXXXXXXXXXXXXXXXXXXXXX-XXXXXXXXXXXXX-X-----XXX--XXXXXXXXXXX
XXXXX----------------X--XX-XXX---------EEAQARGFRLLQE-NEVLLSM
CCIPLVCWIVCTGLRRRMSQDQ--------T-CKTTTAVYTCFLSSLLQARGGG---REP
----------G-LPSHLRGLCSLA---ADGI-WN-QK----ILFEEW-------------
--DLRRHGLREAEVS-AFLSM----SLFQKEVD-CE-AFYSFVHLTFQEFFAAMYYMLE-
EEEGAGTASP--------GRGGS--ELPPRDVR-VLLENYGRF-EKGYLIFVVRFL----
----FGLVNQDRAAYLEKKLGCS-----------------VSRQ-VRP-ELLRWIEAKAE
A-----------------------------------KKLQVQPSQLELFYCLYETQE-DT
FLRAAMGH---------------------F-------------------PRIELSLS--T
RM-DHVVA--S----FCIQ-NCHS-VES-LSLGLL-------------------------
-HSSSKEEGEEEEEEE--EEGWLAAEGRHMCLRPPAACPSXXXXXXXXXXXXXXXX-XXX
XXXXXXXX-XX------------------XX---------XX------------------
-----------------------------------XXX----------------XXXXXX
--------------X-XXXXXXXXXXXXXXX-----XXX--------XXX---------X
-XXXX-XXX-XXX-XXX-XXXX----XX-XXXXXXX---XXXXXXXXXXXXXXXXXXX-X
XXXXLVSCCLT---ST-CCP---DLASVLDT-NHSLTGLYLGENALGD-DGVSLLCEK--
LKH-PQCSLRKL-G----------------------------------------------
------------------------LVSSGLT-----------------------------
----------------------------SVCCAAISAALGS-------------------
------------------------------SK------
PF00560 // ENSGT00900000140813_0_Domain_11 (2019, 2057)
SLTH---LY-LRGNA-LGD---
-------NGLKLLCQGL----LHPSC----------------------------------
--------------------------RLQ-MLE---------------------------
-------------------------------LDSCGLSSHCCWDLCRL-LTSSRCLQKLS
L-GSNDLGDLGVV-MLCEVLRQP--GCP----L---------------------------
-----------------------QRL----------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSLAFP00000010734 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLHGKSGTGKSALIKRI-----MLHWA--K---------GQLYQ
G--------IFSY-VFFLHAREMQHMR-------EGSFAE-LISM-EW-----PD-SCAP
VTEIMS-QP----ERLLFVIDGFDD-LDFALQDC------SE-K-----LCENWAEK-QP
-VSVLMCSLLKKVLLPESSLLIT-VTDIGIKKLTPVV-V-----SPH--YLFVGGIPVEL
RTQFI----------------L--QG-TVD---------EQQKMPILTSVVT-NTQLFDE
CQVYNMCWLASVALQLQHVAER-DA-P--PT-CQTFTSLCATFMFHQFKPED-DLWCCLN
----------GEEKAVLKNLCQMA---VEGV-WN-MK----LVFYSG-------------
--DLGVHRLKEL-ELSALFHM----NILPQDVL-CEGC-YTFFHHSLQEFCAALYYVLE-
GLERK--------------HDPS--PLCIENAR-SPMEIRQVG-FNSHLLQMKRFL----
----FGLMNKELVRKLEALLGHG---------------V--PL-VIKE-ELLQWVSLLGE
QAS----------------------------------TT-SPLDFLDAFHCLFETQN-EE
FVRLALGS---------------------F-------------------QEVWLPINQQM
DL---LVS--S----FCLQ-HCQY-LQR-IRLD----------------IKE-----IFS
-TGPRTT-----------------------------------------------PK-AHV
TVFWQNLC-MVLSTHETLRLLDLSGSVLNKW---------AMRILC--SKLRR-P-----
TC--K-IQTLI--F-KNAQI---TL---GLQ-HLWTTL----------------I-NQEL
KNLY-LESM-HLSNE-DVEAVCGALKHPHCL----LESL--------RLD---------R
C------G----------------------------------------------------
---------LT---YT-SCL---ELSQVLVV-SASLSSLSLAGNKVTD-EGLKTLCSV--
LNF-TRSNLKKL-I----------------------------------------------
------------------------LGNCGLT-----------------------------
----------------------------AAGCGDLAL-----------------------
--------------------------------ALFTNQNLTH---LC-LSDND-LGS---
-------EGMQLLCGAI----KNPNC----------------------------------
--------------------------GLK-RLI---------------------------
-------------------------------LSQCNLDVTSCGFLAFA-LMKNRRLTHLT
L-SMNPLEDNGIK-MLCEVMKEP--SCY----L---------------------------
-----------------------QDLEL--VRC-----HLTSACCEDL------------
-------SCVI-ARNPHLKSLDLAYNALGDSGAAL-LCKGLK-HK-QS-SLTRLG--LE-
--A-CGLTSTCCE--------------------ELSLAIYHNQSL-TSLNLMQNLLDPVG
ISKLCS--------ASACPKSN--LQ--I-------------------------------
---I--------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSRROP00000040567 (view gene)
ENSDORP00000013746 (view gene)
ENSP00000445135 (view gene)
ENSDNOP00000024216 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-MGEPEAS-GSRERR--DAAA
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 328)
REL---LLAALEDLS--E---EQLKRFRLKLWD------
------------APAGG-----------RSI-PRGR-LEHAD-AVDLVERLAEFYGPEPA
LDVARKA----LK-KSDVRDVAARLK-EQRLQR---------L---------GPS-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------PS-PPLSVSDYKKLFREH-
--------VQQQ----------------HAKV--KERNARS------------VKISKRF
TKLLIAPESAALEDE--ALGPEAGLGARGWSPEP--------VRARRSDTDTFNRLFSRD
A---EGGRP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLRGPAGIGKTMAAKKI-----LYDWA--A---------GKLYH
G--------QVDF-AFFMPCREVLGRPG------ACSLAD-LILE-QC-----PD-RGAP
VRQMLA-QA----ERLLFILDGADE-LP---PA--------EAA-----PCTDPFEA-AG
-GARVLGGLLSKALLPSSRLLVT-SRATAPERLQGRL-R-----SPQ--CAEVRGFSHKD
KKKYF----------------Y--KF-FQD---------EWRAERAYRFVKE-NETLYGL
CFVPFVCWIVCSVLRQQLQRRQ-DL-S--RT-SKTTTAVYLLFISSMLSSAPATGGQ---
-----------QVQGELRKLCRLA---RKGLLGH--R----AQFSEK-------------
--DLEQLELRGSSVQMLFLSKKELPGVLEN-----E-VAYQFMDQSFQEFFAALSYLLE-
EEGAP-------------------G-APADSVR-TLLCSEVEM--RGHLALTTRFL----
----FGLLNTERIRDIERHFGC-----------------VVSER-VKQ-EALLWVQGQGQ
GVTVAVPEKPEGPEGLNSSEPLEE--D-------EEDENEELNHPLELLYCLYETQE-DA
FVRQALRS---------------------L-------------------PELALERVRFS
RM-DLVVL--S----YCVR-CCPP-GQV-LRLL------------------GC----GLG
------TA----------------------------------------QEKK-K---KKS
----------------------------------------LAKR----------------
------LRGS---L--------------GGS------G----------------SSRA-T
GQ---HPVS-------PLRPLCEAMTDQQCG----LSSL--------TLS---------G
C-----------------------------------------------------------
--------KLP---NT-VCR---DLSEALKV-APALRELGLLRIGLS-EAGMRMLCEG--
-LAWPRCQVQTL------------------------------------------------
----------------------------RVQ-----------------------------
---------------LPGR---------QEVLQYL-------------------------
------------------------------AGLLRQTPALAT---LD-LSGC-QLPGPIL
--T--------ALCAAL----QHPRCSLQSLSL--PSKELSEQ-----------------
----------ELQELKAVKSAKP-------GLV----------------------I--MH
PALD-------------------RPLEPPEGVSSVL------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
ENSMNEP00000010301 (view gene)
MDQPEDPGSRLRSLRTASPQRAFVESVPHQRLRLRPWCKAPKLGLTK-RQPGHVTWPGVW
KNRSRKGGAGEEGAPQSSGRRWPVPTSPSEGEEHQD------APTRVRGRRR--------
-------AKSGVTPRCPLPPPHG-----SS--LARGSLRPCPRWVLKRQSRGWAGVTRGP
LPDDPRPPVSSTGLR--LAVAREL---
PF02758 // ENSGT00900000140813_0_Domain_0 (208, 322)
LLAALEELS--Q---EQLKRFRHKLRD------
------------VGPDR-----------RSI-PWGR-LERAD-AVDLAEQLAQFYGPEPA
LEVARKT----LK-RADARDVAAQLK-EQRLRR---------L---------GPG-----
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------SA-ALLSVSEYKKKYREH-
--------VLQL----------------HARV--KERNARS------------VKITKRF
TKLLIAPESAAPEEE--ALGPAE-------EPEP--------GRARRSDTHTFNRLFRGD
E---DGRRP----L
PF05729 // ENSGT00900000140813_0_Domain_2 (675, 940)
TVVLQGPAGIGKTMAAKKI-----LYDWA--A---------GKLYQ
G--------QVDF-AFFMPCGELLERPG------TRSLAD-LILD-QC-----PD-RGAP
VPQMLA-QP----QRLLFILDGADE-LPALEGP--------EAA-----PCTDPFEA-AS
-GARVLGGLLSKALLPTALLLVT-TRAASPGRLQGRL-C-----SPQ--CAEVRGFSDKD
KKKYF----------------Y--KF-FQD---------ERRAERAYRFVKE-NETLFAL
CFVPFVCWIVCTVLRQQLELGR-DL-S--RT-SKTTTSVYLLFIASVLSSAPVADGP---
-----------RVQGDLRNLCRLA---REGVLGR--R----AQFAEK-------------
--QLEQLELRGSKVQTLFLSKKELPGVLET-----E-VTYQFIDQSFQEFLAALSYLLE-
DEGVP-------------------R-TAAGGVG-TLLRGDAQP--HSHLVLTTRFL----
----FGLLSAERMRDIERHFGC-----------------VVSER-VKQ-EALRWVQGQGQ
GCPGVAPEVTEETKGLEDTEEPEE-----------EEEGEEPNYPLELLYCLYETQE-DA
FVRQALCR---------------------L-------------------PELALQGVRFC
RM-DVAVL--S----YCVR-CCPA-GQA-LRLI------------------SC----RLV
------AA----------------------------------------QEK--K---KKS
----------------------------------------LGKR----------------
------LQAS---L--------------GGS------S-----------------SRGTT
KQ---LPAS-------LLHPLFQTMTDPLCR----LTSL--------TLS---------H
C-----------------------------------------------------------
--------KVP---DA-VCR---DLSEALRA-APALTELGLLHNRLS-EAGLRMLSEG--
-LAWPQCRVQTV------------------------------------------------
----------------------------RVQ-----------------------------
---------------LPSS---------QQGLQYL-------------------------
------------------------------VGMLRQSPALTT---LD-LSGC-QLPAPMV
--T--------YLCAVL----QHQGCGLQTLRL--TSVELSGQ-----------------
----------SLQELQAVKRAKP-------DLV----------------------I--AH
PVLD-------------------GHPESP-KELVSTF-----------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
MGP_SPRETEiJ_P0029372 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------------------------ME---
------------------------------------ESPPKQKSNTK-------------
---VAQHE---------------------GQQDLNTTRHMNVELKHRPKLERHLKLVPSN
ED---AEI--R----FALW-VNAG-KMT-PQIH----------------WL---------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------EEAG--------------------------------------------------
-------------------KQQGRYRWTPIT-----------------------------
----------------------------NASCEILFY-----------------------
--------------------------------NLKFTRNLEG---LD-LSGNS-LRY---
-------SVVQSLCNTL----RYPGCQLKTLW----------------------------
------------------------------------------------------------
-------------------------------LVKCGL-----------------------
------------------------------------------------------------
----------------------------------------TS------------------
------------------------------------------------------------
---------RYCS--------------------LLASVFGACSSL-IELDLQLNYLGDDG
VRMLCE--------GLRNPVCN--LS--I-------------------------------
---LWLDLS-SLSARVITE------LR----------TLE-EKNPKLYIRS-IWMPHMMV
PTENMDEEDIL--------------------TSFKQQRQESGE-----------------
-----------------------------------------KPMEILG-TEEDFWGPTGP
VATELVDR-VRNLYR
PF13553 // ENSGT00900000140813_0_Domain_18 (2716, 2997)
VQLPMAGSYHCP---STGLHFVVTRAV-------TIEIEFCAWSQ
FLDKTPLQQSHMVVGPLFDIKAEQGA-VTAVYLPHFVSLKANFTLQQGYEWASTFDFKVA
HFQEHGMVLETPDRVKPGYTVLKNPSFSPMGVVLKIFPAARHFIPITSITLIYYRLNLEE
V-TLHLYLVPNDCTIQKAIDDEEMKFQFVRINKPPPVDNLFIGSRYIVSGS---------
ENLEITPKELELCYRSSKEFQLFSEIYVGNMGSEIKLQIKNKKHMKFIWEALLKPGDLRP
ALPRIA-----------------------------------QALNDAPSLLHFMDQHREQ
LVA---------------------------------------------------------
------------------------------------
ENSMMUP00000004772 (view gene)
ENSSHAP00000007616 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------AEG--V
PF02758 // ENSGT00900000140813_0_Domain_0 (202, 326)
RCQ---LARHLEDLT--P---DELKKFKLYLKD------
---------H--SS-----------G--LGI-PRGR-VDGVD-STEMAEILVSSRGEQEA
WQLALSI----WK-KMGLRELWEQAS-KVRPLRNNNCYSYTILNLLCQ----TSSAFNQY
----LGK-----------------------------------------------------
------------------------LNEEI-------------------Q-----------
------ITRA-------------------------------KK-----RMDYINKYRER-
--------MRKK----------------FQLM--RDRNSRPGEHE---------LFHHRF
TELLLLRGYRYKEQKHHELL------VHGWEHATVMQ-------ER-GQLTRVSTLFDPD
QK--IGIPP----
PF05729 // ENSGT00900000140813_0_Domain_2 (674, 930)
HTVVLQGAAGIGKTTLASKV-----MLDWA--E---------GNFFQ
E--------RFNY-VFYLTCRELNPLEG-----KIISFAD-LIAN-DW-----PG-PQAP
MTEIMS-QP----ERLLFIIDGLDE-LKFPCNEH------RY-D-----LCKDWKQQ-RP
-VPILLSSLLRKTLLPEACLLLT-TRLTALGKFCPLL-E-----SPR--HVEILGFSVQH
REEYF----------------C--KF-FRD---------KHLGEKIFHLVKG-NGTLLTM
CSVPLMNWIVCTCLKQQMEKGQ-DP-V--QA-LKTTTALYMCYLSNLIIPDD-----KNF
------------IPQHLRGLCRLA---AEGI-WD-RK----LLFEEE-------------
--DLRRNTLETADV-SAFLDM----NIFQKDSG-CE-NCYSFIHLSFQEFFAAMFYVMS-
TEEEG--------------MKSS--DSFIPGVR-ELLEECSGF-NASFVVLVVRFL----
----FGFLNVETARELERKFRCR---------------M--SPE-IKS-QLLQWAQGESR
RNY--------------------------------KRSQFSDIYFPEWYSHLYETQD-AE
FVTQVLVN---------------------I-------------------QEINVD--IVY
PS-DALSA--V----FCIK-QCHG-AQR-LFLQ----------------IGT-----YFN
-T-----V--------------------------------KY------------------
--------------KQDPRRND-------NP---------TQHFLCIYTDLLY-S-----
CC--K-----K--L-EHFKVA--YS---CCQ-YLFSAL--------------------SL
KSLH-FKRI-FFEDD-GMKLPCETLGKKNCQ----LQSL--------KIS---------H
C------Q----------------------------------------------------
---------GF---GV-NIL---DFFSAFVN-NESLRILELSFICLKE-DEVKLLCNV--
LTK-QDCKLHAL-S----------------------------------------------
------------------------LRS---------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------CDLTTTGYQNLFSA-LSSNMSLKNLD
L-SRNSFEETEIK-SLCKTLENQ--NCK----L---------------------------
-----------------------EKLRL--TYC-----KLSPASCCCL------------
-------SSNL-MKNESLKDLCLSRNCVGDCAVKL-LCKALG-NQ-NC-KLHTLR--LS-
--G-CNLTATCLQ--------------------DILSVICSSDKL-KKLDLSRNYFGDEG
MKLLCE--------ALQKQDCK--LQ--R-------------------------------
---LNLSRC-PLSEESERA------WT----------SLN-SRKSFHITW----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------
{"ENSOGAP00000021901": [["Domain_2", 674, 930, "PF05729"]], "ENSSARP00000012840": [["Domain_2", 674, 940, "PF05729"], ["Domain_6", 1619, 1645, "PF00560"]], "ENSCCAP00000028558": [["Domain_4", 1515, 1542, "PF13516"], ["Domain_6", 1617, 1642, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2215, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSSBOP00000005889": [["Domain_2", 674, 929, "PF05729"], ["Domain_8", 1773, 1796, "PF13516"], ["Linker_8_10", 1796, 1805], ["Domain_10", 1805, 1955, "PF13516"], ["Domain_15", 2359, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSCGRP00000019004": [["Domain_4", 1514, 1542, "PF13516"], ["Domain_6", 1617, 1642, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Linker_15_16", 2378, 2391], ["Domain_16", 2391, 2436, "PF13516"], ["Linker_16_17", 2436, 2441], ["Domain_17", 2441, 2464, "PF13516"]], "ENSCCAP00000023046": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSFCAP00000013894": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 658, "PF14484"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2373, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSCSAP00000016257": [["Domain_2", 675, 930, "PF05729"]], "ENSBTAP00000054745": [["Domain_0", 202, 326, "PF02758"], ["Domain_2", 675, 929, "PF05729"]], "ENSMOCP00000010558": [["Domain_2", 674, 930, "PF05729"], ["Domain_6", 1616, 1641, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2441, 2465, "PF13516"]], "ENSCSAP00000007255": [["Domain_17", 2444, 2458, "PF00560"]], "ENSP00000309767": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSCLAP00000011529": [["Domain_2", 675, 940, "PF05729"], ["Domain_17", 2442, 2465, "PF13516"], ["Linker_17_18", 2465, 2716], ["Domain_18", 2716, 2997, "PF13553"], ["Linker_18_19", 2997, 3052], ["Domain_19", 3052, 3134, "PF00619"]], "ENSCSAP00000016258": [["Domain_0", 202, 326, "PF02758"], ["Domain_2", 674, 930, "PF05729"]], "ENSNGAP00000025650": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2373, "PF13516"], ["Domain_17", 2441, 2465, "PF13516"]], "ENSBTAP00000055476": [["Domain_2", 674, 930, "PF05729"]], "ENSCGRP00001007377": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSPANP00000000631": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 719, 940, "PF05729"]], "ENSBTAP00000048061": [["Domain_0", 203, 326, "PF02758"], ["Domain_2", 675, 930, "PF05729"]], "ENSETEP00000006872": [["Domain_0", 198, 332, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "MGP_SPRETEiJ_P0086559": [["Domain_4", 1515, 1542, "PF13516"], ["Domain_6", 1617, 1642, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Linker_15_16", 2378, 2390], ["Domain_16", 2390, 2436, "PF13516"], ["Linker_16_17", 2436, 2442], ["Domain_17", 2442, 2464, "PF13516"]], "ENSMLUP00000015241": [["Domain_2", 674, 940, "PF05729"], ["Domain_15", 2357, 2405, "PF00560"]], "ENSMLUP00000014098": [["Domain_11", 2019, 2057, "PF00560"], ["Domain_15", 2357, 2381, "PF00560"]], "MGP_CAROLIEiJ_P0090776": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"]], "ENSP00000478516": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"], ["Linker_17_18", 2465, 2716], ["Domain_18", 2716, 2997, "PF13553"], ["Linker_18_19", 2997, 3052], ["Domain_19", 3052, 3134, "PF00619"]], "ENSBTAP00000017623": [["Domain_0", 202, 326, "PF02758"], ["Domain_2", 675, 905, "PF05729"]], "ENSMNEP00000029873": [["Domain_4", 1515, 1542, "PF13516"], ["Domain_6", 1617, 1642, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2215, 2237, "PF13516"], ["Domain_15", 2356, 2378, "PF13516"], ["Domain_17", 2442, 2464, "PF13516"]], "ENSMFAP00000021717": [["Domain_4", 1515, 1542, "PF13516"], ["Domain_6", 1617, 1642, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2215, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2464, "PF13516"]], "ENSPCAP00000002371": [["Domain_2", 674, 940, "PF05729"], ["Domain_14", 2252, 2355, "PF00560"]], "ENSJJAP00000023655": [["Domain_2", 675, 930, "PF05729"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Linker_11_12", 2054, 2126], ["Domain_12", 2126, 2206, "PF13516"], ["Linker_12_13", 2206, 2212], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2374, "PF13516"]], "ENSMPUP00000006858": [["Domain_2", 675, 930, "PF05729"]], "ENSCANP00000022391": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_18", 2716, 2997, "PF13553"], ["Linker_18_19", 2997, 3052], ["Domain_19", 3052, 3134, "PF00619"]], "ENSCLAP00000000175": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSSSCP00000023003": [["Domain_11", 2016, 2050, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2443, 2465, "PF13516"]], "ENSRNOP00000046459": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_11", 2017, 2048, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSPPAP00000034190": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_11", 2016, 2052, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2374, "PF13516"]], "ENSPCAP00000003709": [["Domain_2", 674, 940, "PF05729"], ["Domain_11", 2019, 2057, "PF00560"]], "ENSMPUP00000008374": [["Domain_0", 202, 328, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSPPAP00000011890": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 658, "PF14484"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_15", 2355, 2373, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSCANP00000024527": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 929, "PF05729"], ["Domain_13", 2213, 2236, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2441, 2462, "PF13516"]], "ENSPTRP00000019830": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 929, "PF05729"], ["Domain_13", 2214, 2236, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2441, 2465, "PF13516"]], "ENSMLEP00000015777": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"]], "ENSMOCP00000012017": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSMLEP00000025857": [["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1617, 1633, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Linker_13_14", 2237, 2247], ["Domain_14", 2247, 2348, "PF13516"], ["Linker_14_15", 2348, 2354], ["Domain_15", 2354, 2378, "PF13516"]], "MGP_SPRETEiJ_P0084839": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 676, 930, "PF05729"]], "ENSSTOP00000015184": [["Domain_4", 1515, 1542, "PF13516"], ["Domain_6", 1617, 1642, "PF13516"], ["Linker_6_7", 1642, 1650], ["Domain_7", 1650, 1766, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2464, "PF13516"]], "ENSCCAP00000024896": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1617, 1633, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Linker_13_14", 2237, 2247], ["Domain_14", 2247, 2348, "PF13516"], ["Linker_14_15", 2348, 2354], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSCGRP00000011757": [["Domain_2", 675, 930, "PF05729"], ["Domain_11", 2019, 2054, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"], ["Linker_17_18", 2465, 2716], ["Domain_18", 2716, 2997, "PF13553"], ["Linker_18_19", 2997, 3052], ["Domain_19", 3052, 3133, "PF00619"]], "ENSMUSP00000139357": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSMLEP00000032802": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSPTRP00000005809": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSOGAP00000016874": [["Domain_2", 674, 940, "PF05729"]], "ENSGGOP00000023539": [["Domain_4", 1515, 1542, "PF13516"], ["Domain_6", 1617, 1642, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2215, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2464, "PF13516"]], "ENSCJAP00000064897": [["Domain_2", 675, 930, "PF05729"]], "MGP_CAROLIEiJ_P0078152": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSMLUP00000000748": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSCGRP00000002480": [["Domain_0", 209, 323, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_13", 2213, 2237, "PF13516"]], "ENSMEUP00000000871": [["Domain_0", 204, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_13", 2215, 2236, "PF00560"], ["Domain_15", 2357, 2375, "PF00560"]], "ENSMAUP00000020881": [["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Linker_13_14", 2237, 2247], ["Domain_14", 2247, 2348, "PF13516"]], "ENSCAFP00000003831": [["Domain_0", 203, 326, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_11", 2017, 2055, "PF13516"], ["Domain_13", 2213, 2238, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"]], "ENSMOCP00000000825": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_13", 2212, 2233, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSNLEP00000012957": [["Domain_0", 210, 323, "PF02758"], ["Domain_2", 781, 930, "PF05729"]], "ENSCATP00000031257": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSANAP00000003104": [["Domain_4", 1515, 1542, "PF13516"], ["Domain_6", 1618, 1642, "PF13516"], ["Linker_6_7", 1642, 1650], ["Domain_7", 1650, 1765, "PF13516"], ["Domain_13", 2215, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSCSAP00000008835": [["Domain_0", 208, 328, "PF02758"], ["Domain_2", 675, 940, "PF05729"], ["Domain_11", 2019, 2037, "PF00560"]], "ENSCCAP00000005912": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_12", 2068, 2206, "PF13516"], ["Linker_12_13", 2206, 2212], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_17", 2442, 2464, "PF13516"], ["Linker_17_18", 2464, 2716], ["Domain_18", 2716, 2997, "PF13553"], ["Linker_18_19", 2997, 3052], ["Domain_19", 3052, 3134, "PF00619"]], "ENSSSCP00000024653": [["Domain_0", 209, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1617, 1633, "PF13516"], ["Domain_11", 2019, 2054, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Linker_13_14", 2237, 2247], ["Domain_14", 2247, 2348, "PF13516"], ["Linker_14_15", 2348, 2354], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2441, 2461, "PF13516"]], "ENSMLUP00000019706": [["Domain_2", 674, 940, "PF05729"]], "ENSOGAP00000019290": [["Domain_0", 203, 325, "PF02758"], ["Domain_2", 783, 930, "PF05729"]], "ENSMLEP00000028379": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_11", 2016, 2036, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSRNOP00000066377": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 675, 931, "PF05729"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2214, 2237, "PF13516"]], "ENSMUSP00000066841": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_11", 2016, 2048, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSETEP00000012779": [["Domain_0", 194, 332, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_8", 1775, 1801, "PF00560"], ["Domain_11", 2019, 2064, "PF00560"], ["Domain_15", 2357, 2385, "PF00560"], ["Domain_17", 2444, 2463, "PF00560"]], "ENSHGLP00100023958": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_6", 1618, 1642, "PF13516"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2374, "PF13516"]], "ENSRROP00000033846": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_11", 2016, 2036, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSPTRP00000039083": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_8", 1773, 1795, "PF13516"], ["Domain_11", 2020, 2054, "PF13516"], ["Domain_13", 2214, 2237, "PF13516"], ["Domain_15", 2355, 2377, "PF13516"]], "ENSCATP00000028404": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 929, "PF05729"], ["Domain_13", 2213, 2236, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2441, 2465, "PF13516"]], "MGP_CAROLIEiJ_P0077856": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSPPYP00000011653": [["Domain_0", 202, 326, "PF02758"], ["Domain_2", 675, 930, "PF05729"]], "ENSCJAP00000034052": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2373, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "MGP_CAROLIEiJ_P0077858": [["Domain_3", 1227, 1408, "PF05729"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2356, 2378, "PF13516"], ["Domain_17", 2442, 2464, "PF13516"]], "ENSSBOP00000006129": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 675, 930, "PF05729"]], "ENSPANP00000023613": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 925, "PF05729"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSRBIP00000006660": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2212, 2232, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSPTRP00000005475": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSCHOP00000001173": [["Domain_2", 674, 940, "PF05729"]], "ENSPTRP00000093283": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_8", 1773, 1795, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_15", 2354, 2377, "PF13516"]], "ENSMEUP00000011516": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSRBIP00000014522": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSAMEP00000003008": [["Domain_2", 676, 940, "PF05729"], ["Domain_8", 1775, 1805, "PF00560"], ["Domain_17", 2444, 2475, "PF00560"]], "ENSMICP00000005760": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSRROP00000035661": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1617, 1633, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Linker_13_14", 2237, 2247], ["Domain_14", 2247, 2348, "PF13516"], ["Linker_14_15", 2348, 2354], ["Domain_15", 2354, 2378, "PF13516"]], "ENSCANP00000035667": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 680, 940, "PF05729"]], "ENSPCOP00000021743": [["Domain_0", 207, 323, "PF02758"], ["Domain_6", 1616, 1640, "PF13516"], ["Domain_8", 1774, 1795, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSMLUP00000000546": [["Domain_2", 674, 940, "PF05729"], ["Domain_11", 2019, 2057, "PF00560"], ["Domain_19", 3051, 3135, "PF00619"]], "ENSOPRP00000007037": [["Domain_9", 2019, 2132, "PF00560"], ["Domain_15", 2357, 2381, "PF00560"]], "ENSCATP00000001010": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 676, 930, "PF05729"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2441, 2464, "PF13516"]], "ENSMICP00000046163": [["Domain_2", 674, 929, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "MGP_PahariEiJ_P0019911": [["Domain_4", 1515, 1542, "PF13516"], ["Domain_6", 1617, 1642, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2215, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2464, "PF13516"]], "ENSMAUP00000007100": [["Domain_0", 208, 321, "PF02758"], ["Domain_2", 676, 929, "PF05729"]], "ENSACAP00000017210": [["Domain_2", 675, 930, "PF05729"]], "ENSMFAP00000020036": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 676, 930, "PF05729"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2441, 2464, "PF13516"]], "ENSCSAP00000016343": [["Domain_0", 202, 326, "PF02758"], ["Domain_2", 674, 922, "PF05729"]], "ENSCGRP00001015691": [["Domain_0", 208, 321, "PF02758"], ["Domain_2", 676, 905, "PF05729"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSMUSP00000152151": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_13", 2212, 2233, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSMNEP00000023610": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 658, "PF14484"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2373, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSHGLP00000021761": [["Domain_2", 675, 890, "PF05729"]], "ENSCANP00000035810": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 925, "PF05729"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2443, 2465, "PF13516"]], "ENSRROP00000004574": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_8", 1773, 1795, "PF13516"], ["Domain_13", 2214, 2237, "PF13516"], ["Domain_15", 2355, 2377, "PF13516"]], "ENSRNOP00000040262": [["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1617, 1642, "PF13516"], ["Domain_8", 1772, 1796, "PF13516"], ["Domain_11", 2019, 2054, "PF13516"]], "ENSMMUP00000012994": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 928, "PF05729"], ["Domain_13", 2213, 2236, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2441, 2465, "PF13516"]], "ENSPVAP00000000490": [["Domain_0", 204, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_11", 2019, 2052, "PF00560"], ["Domain_15", 2357, 2385, "PF00560"]], "ENSCJAP00000021695": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 658, "PF14484"], ["Domain_8", 1772, 1796, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2212, 2233, "PF13516"], ["Domain_15", 2357, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSMICP00000050110": [["Domain_0", 210, 322, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_8", 1773, 1795, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_15", 2354, 2377, "PF13516"]], "ENSMPUP00000010362": [["Domain_2", 674, 930, "PF05729"]], "ENSAMEP00000017920": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_8", 1775, 1798, "PF00560"], ["Domain_11", 2019, 2064, "PF00560"], ["Domain_17", 2444, 2463, "PF00560"]], "ENSCSAP00000000724": [["Domain_0", 202, 328, "PF02758"], ["Domain_2", 674, 930, "PF05729"]], "ENSRNOP00000038641": [["Domain_0", 209, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSPSIP00000006673": [["Domain_0", 208, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSCJAP00000059395": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 674, 926, "PF05729"], ["Domain_8", 1773, 1795, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSGGOP00000004737": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 657, "PF14484"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSMPUP00000012314": [["Domain_2", 675, 940, "PF05729"]], "ENSMAUP00000016758": [["Domain_0", 210, 323, "PF02758"], ["Domain_2", 675, 931, "PF05729"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2017, 2051, "PF13516"], ["Domain_13", 2214, 2237, "PF13516"]], "ENSPPAP00000002657": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 657, "PF14484"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSGGOP00000008087": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1617, 1633, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Linker_13_14", 2237, 2252], ["Domain_14", 2252, 2348, "PF13516"], ["Linker_14_15", 2348, 2354], ["Domain_15", 2354, 2378, "PF13516"]], "ENSCGRP00001001791": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSTBEP00000005214": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSANAP00000039831": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 926, "PF05729"], ["Domain_8", 1773, 1795, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSRROP00000007398": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"]], "ENSPPAP00000014381": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSMNEP00000040976": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_11", 2016, 2052, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2374, "PF13516"]], "ENSOPRP00000003739": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSMNEP00000015393": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 930, "PF05729"]], "ENSMPUP00000003790": [["Domain_2", 687, 929, "PF05729"]], "ENSSBOP00000008549": [["Domain_2", 674, 928, "PF05729"], ["Domain_13", 2214, 2236, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2456, "PF13516"]], "ENSVPAP00000002974": [["Domain_2", 691, 940, "PF05729"], ["Domain_19", 3051, 3135, "PF00619"]], "ENSMMUP00000031438": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_8", 1773, 1795, "PF13516"], ["Domain_13", 2214, 2237, "PF13516"], ["Domain_15", 2355, 2377, "PF13516"]], "ENSETEP00000004560": [["Domain_0", 198, 332, "PF02758"], ["Domain_8", 1775, 1805, "PF00560"], ["Domain_11", 2019, 2057, "PF00560"], ["Domain_13", 2215, 2237, "PF00560"]], "ENSETEP00000004566": [["Domain_0", 208, 332, "PF02758"], ["Domain_2", 774, 940, "PF05729"]], "ENSPTRP00000080469": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 925, "PF05729"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSMOCP00000002645": [["Domain_4", 1515, 1542, "PF13516"], ["Domain_6", 1618, 1642, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSGGOP00000016043": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 676, 930, "PF05729"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2441, 2464, "PF13516"]], "ENSCHOP00000007146": [["Domain_0", 201, 419, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_13", 2215, 2240, "PF00560"]], "ENSCPOP00000007980": [["Domain_6", 1618, 1642, "PF13516"], ["Linker_6_7", 1642, 1650], ["Domain_7", 1650, 1766, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2464, "PF13516"]], "ENSRNOP00000029443": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 676, 930, "PF05729"]], "ENSAMEP00000004746": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSANAP00000025829": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1617, 1633, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSCLAP00000001546": [["Domain_2", 674, 928, "PF05729"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSP00000301295": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_11", 2016, 2036, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSCGRP00000007759": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 675, 931, "PF05729"], ["Domain_11", 2017, 2051, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSCHIP00000029121": [["Domain_2", 675, 930, "PF05729"], ["Domain_11", 2017, 2054, "PF13516"]], "ENSPCAP00000003633": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_15", 2357, 2381, "PF00560"], ["Domain_17", 2444, 2475, "PF00560"]], "ENSRBIP00000003237": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_8", 1773, 1795, "PF13516"], ["Domain_13", 2214, 2237, "PF13516"], ["Domain_15", 2355, 2377, "PF13516"]], "ENSMICP00000044792": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 674, 929, "PF05729"], ["Domain_8", 1774, 1795, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2377, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSMLUP00000020634": [["Domain_2", 674, 940, "PF05729"], ["Domain_11", 2019, 2057, "PF00560"], ["Domain_17", 2444, 2463, "PF00560"]], "ENSPPAP00000019584": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_11", 2016, 2036, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSCAFP00000015730": [["Domain_0", 202, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_8", 1775, 1796, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2379, "PF13516"], ["Domain_17", 2443, 2466, "PF13516"]], "ENSOCUP00000002291": [["Domain_0", 203, 330, "PF02758"], ["Domain_2", 676, 930, "PF05729"]], "ENSMICP00000045945": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSPCOP00000013110": [["Domain_4", 1515, 1543, "PF13516"], ["Domain_6", 1618, 1642, "PF13516"], ["Linker_6_7", 1642, 1650], ["Domain_7", 1650, 1766, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2215, 2237, "PF13516"], ["Domain_15", 2354, 2377, "PF13516"], ["Domain_17", 2442, 2464, "PF13516"]], "ENSLAFP00000022456": [["Domain_2", 674, 940, "PF05729"], ["Domain_13", 2215, 2240, "PF00560"]], "ENSGGOP00000011186": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"], ["Linker_17_18", 2465, 2716], ["Domain_18", 2716, 2997, "PF13553"], ["Linker_18_19", 2997, 3052], ["Domain_19", 3052, 3134, "PF00619"]], "ENSEEUP00000014333": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSNGAP00000019592": [["Domain_2", 675, 929, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_13", 2212, 2236, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSCHOP00000007242": [["Domain_2", 674, 941, "PF05729"]], "ENSSBOP00000010225": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 930, "PF05729"]], "ENSMLUP00000021000": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_8", 1775, 1801, "PF00560"], ["Domain_11", 2019, 2064, "PF00560"]], "ENSTSYP00000003797": [["Domain_2", 676, 930, "PF05729"], ["Domain_13", 2214, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2441, 2464, "PF13516"]], "ENSCHOP00000003625": [["Domain_0", 201, 328, "PF02758"], ["Domain_8", 1775, 1801, "PF00560"], ["Domain_11", 2019, 2056, "PF00560"], ["Domain_15", 2357, 2381, "PF00560"]], "ENSOCUP00000012880": [["Domain_0", 202, 328, "PF02758"], ["Domain_2", 675, 930, "PF05729"]], "ENSOPRP00000011804": [["Domain_2", 674, 940, "PF05729"], ["Domain_11", 2019, 2057, "PF00560"]], "ENSPVAP00000011856": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_4", 1517, 1550, "PF00560"], ["Domain_12", 2127, 2208, "PF00560"], ["Domain_15", 2357, 2381, "PF00560"]], "ENSSARP00000006107": [["Domain_4", 1517, 1551, "PF00560"], ["Linker_4_5", 1551, 1566], ["Domain_5", 1566, 1617, "PF00560"], ["Linker_5_6", 1617, 1619], ["Domain_6", 1619, 1645, "PF00560"], ["Domain_15", 2357, 2381, "PF00560"], ["Domain_17", 2444, 2476, "PF00560"]], "ENSMLEP00000009094": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"], ["Linker_17_18", 2465, 2716], ["Domain_18", 2716, 2997, "PF13553"], ["Linker_18_19", 2997, 3052], ["Domain_19", 3052, 3134, "PF00619"]], "ENSMLUP00000017003": [["Domain_2", 674, 940, "PF05729"], ["Domain_11", 2019, 2057, "PF00560"], ["Domain_19", 3051, 3135, "PF00619"]], "ENSPPYP00000008863": [["Domain_0", 208, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_19", 3052, 3133, "PF00619"]], "ENSEEUP00000008107": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 686, 940, "PF05729"]], "ENSMMUP00000049609": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 674, 925, "PF05729"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2443, 2465, "PF13516"]], "ENSSTOP00000012288": [["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2052, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2374, "PF13516"]], "ENSOARP00000003372": [["Domain_2", 675, 929, "PF05729"]], "ENSMUSP00000045077": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 675, 931, "PF05729"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2215, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSECAP00000014445": [["Domain_2", 675, 940, "PF05729"]], "ENSPSIP00000012294": [["Domain_0", 202, 326, "PF02758"], ["Domain_2", 675, 929, "PF05729"]], "ENSMLEP00000022240": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_11", 2016, 2052, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2374, "PF13516"]], "ENSOARP00000003407": [["Domain_0", 208, 326, "PF02758"], ["Domain_2", 675, 930, "PF05729"]], "ENSTSYP00000021583": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 905, "PF05729"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2441, 2465, "PF13516"]], "ENSMICP00000049318": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 676, 930, "PF05729"], ["Domain_11", 2017, 2051, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSRNOP00000031864": [["Domain_11", 2017, 2036, "PF13516"], ["Domain_13", 2218, 2233, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSCJAP00000010235": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 930, "PF05729"]], "ENSCGRP00000000203": [["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Linker_13_14", 2237, 2247], ["Domain_14", 2247, 2348, "PF13516"]], "ENSOCUP00000021941": [["Domain_0", 202, 328, "PF02758"], ["Domain_2", 675, 783, "PF05729"]], "ENSMGAP00000003606": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSRNOP00000043888": [["Domain_2", 674, 930, "PF05729"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2356, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSCAFP00000022816": [["Domain_0", 202, 324, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_11", 2019, 2055, "PF13516"], ["Linker_11_12", 2055, 2065], ["Domain_12", 2065, 2207, "PF13516"], ["Domain_17", 2446, 2465, "PF13516"], ["Linker_17_18", 2465, 2715], ["Domain_18", 2715, 2998, "PF13553"], ["Linker_18_19", 2998, 3052], ["Domain_19", 3052, 3133, "PF00619"]], "ENSCATP00000010211": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 657, "PF14484"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2212, 2232, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSSBOP00000005801": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSMLUP00000007905": [["Domain_2", 674, 940, "PF05729"], ["Domain_15", 2357, 2381, "PF00560"]], "ENSPANP00000002923": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_11", 2016, 2052, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2374, "PF13516"]], "ENSOANP00000014644": [["Domain_0", 205, 326, "PF02758"], ["Domain_2", 674, 930, "PF05729"]], "ENSMLUP00000022275": [["Domain_2", 674, 940, "PF05729"], ["Domain_11", 2019, 2057, "PF00560"], ["Domain_17", 2444, 2463, "PF00560"], ["Domain_19", 3051, 3135, "PF00619"]], "ENSSBOP00000016702": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 926, "PF05729"], ["Domain_8", 1773, 1795, "PF13516"], ["Linker_8_10", 1795, 1807], ["Domain_10", 1807, 1956, "PF13516"], ["Linker_10_11", 1956, 2016], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"]], "ENSPANP00000033792": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 929, "PF05729"], ["Domain_13", 2213, 2236, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2441, 2465, "PF13516"]], "ENSMICP00000037714": [["Domain_0", 210, 320, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"], ["Linker_17_18", 2465, 2716], ["Domain_18", 2716, 2997, "PF13553"], ["Linker_18_19", 2997, 3052], ["Domain_19", 3052, 3134, "PF00619"]], "ENSOARP00000008143": [["Domain_0", 202, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSAMEP00000003461": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSPTRP00000069850": [["Domain_4", 1515, 1542, "PF13516"], ["Domain_6", 1618, 1642, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2215, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2464, "PF13516"]], "ENSMLUP00000022171": [["Domain_2", 779, 940, "PF05729"], ["Domain_11", 2019, 2057, "PF00560"], ["Domain_13", 2215, 2240, "PF00560"]], "ENSGGOP00000002851": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 929, "PF05729"], ["Domain_13", 2214, 2236, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2441, 2465, "PF13516"]], "ENSAMEP00000003422": [["Domain_2", 674, 1048, "PF05729"]], "ENSMODP00000022770": [["Domain_2", 674, 929, "PF05729"]], "ENSMPUP00000008675": [["Domain_6", 1620, 1638, "PF00560"], ["Domain_17", 2444, 2458, "PF00560"]], "ENSLAFP00000011752": [["Domain_2", 674, 940, "PF05729"], ["Domain_12", 2127, 2208, "PF00560"], ["Domain_15", 2357, 2385, "PF00560"]], "ENSCATP00000024116": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"]], "ENSCAFP00000010008": [["Domain_0", 202, 326, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_6", 1617, 1643, "PF13516"], ["Domain_8", 1774, 1797, "PF13516"], ["Domain_11", 2017, 2051, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"]], "ENSMUSP00000081819": [["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_8", 1772, 1796, "PF13516"], ["Linker_8_10", 1796, 1806], ["Domain_10", 1806, 1956, "PF13516"], ["Linker_10_11", 1956, 2018], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"]], "ENSCATP00000004222": [["Domain_1", 536, 658, "PF14484"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_15", 2355, 2373, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSNGAP00000012000": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 657, "PF14484"], ["Domain_8", 1772, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2441, 2465, "PF13516"]], "ENSCJAP00000015134": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 674, 930, "PF05729"]], "ENSPEMP00000026823": [["Domain_0", 210, 322, "PF02758"], ["Domain_2", 675, 930, "PF05729"]], "ENSRROP00000045869": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2212, 2232, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSAMEP00000004163": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_6", 1619, 1644, "PF00560"]], "ENSTBEP00000004281": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "MGP_CAROLIEiJ_P0034749": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_13", 2212, 2233, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSMICP00000046552": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_13", 2213, 2236, "PF13516"], ["Domain_15", 2355, 2377, "PF13516"]], "ENSDNOP00000034243": [["Domain_2", 674, 930, "PF05729"]], "ENSOGAP00000006687": [["Domain_0", 202, 326, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSNLEP00000028046": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSPEMP00000002485": [["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_8", 1772, 1796, "PF13516"], ["Linker_8_10", 1796, 1806], ["Domain_10", 1806, 1949, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"]], "ENSCGRP00000011966": [["Domain_0", 208, 325, "PF02758"], ["Domain_2", 676, 905, "PF05729"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSGGOP00000008682": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 658, "PF14484"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2373, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSCATP00000029059": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSOANP00000018896": [["Domain_2", 755, 930, "PF05729"]], "ENSTTRP00000014925": [["Domain_2", 674, 940, "PF05729"]], "ENSPANP00000019578": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"], ["Linker_17_18", 2465, 2716], ["Domain_18", 2716, 2997, "PF13553"], ["Linker_18_19", 2997, 3052], ["Domain_19", 3052, 3134, "PF00619"]], "ENSMLEP00000031413": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 658, "PF14484"], ["Domain_8", 1772, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2373, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSCGRP00001016160": [["Domain_2", 675, 930, "PF05729"], ["Domain_11", 2019, 2054, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"], ["Linker_17_18", 2465, 2716], ["Domain_18", 2716, 2996, "PF13553"], ["Linker_18_19", 2996, 3052], ["Domain_19", 3052, 3133, "PF00619"]], "ENSTBEP00000005770": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSODEP00000019883": [["Domain_0", 208, 322, "PF02758"]], "ENSECAP00000016749": [["Domain_0", 203, 325, "PF02758"], ["Domain_2", 676, 940, "PF05729"]], "ENSPEMP00000011414": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 676, 930, "PF05729"]], "ENSCGRP00001023035": [["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Linker_13_14", 2237, 2247], ["Domain_14", 2247, 2348, "PF13516"]], "ENSPTRP00000014740": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"], ["Linker_17_18", 2465, 2716], ["Domain_18", 2716, 2997, "PF13553"], ["Linker_18_19", 2997, 3052], ["Domain_19", 3052, 3134, "PF00619"]], "ENSMPUP00000006824": [["Domain_0", 203, 326, "PF02758"], ["Domain_2", 675, 930, "PF05729"]], "ENSCANP00000026970": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_11", 2016, 2052, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2374, "PF13516"]], "ENSBTAP00000002954": [["Domain_0", 202, 326, "PF02758"], ["Domain_2", 674, 930, "PF05729"]], "ENSPTRP00000003728": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 657, "PF14484"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSHGLP00000014368": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_13", 2214, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"]], "ENSANAP00000003731": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 675, 930, "PF05729"]], "ENSJJAP00000024566": [["Domain_4", 1515, 1542, "PF13516"], ["Domain_6", 1617, 1642, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Linker_15_16", 2378, 2391], ["Domain_16", 2391, 2436, "PF13516"], ["Linker_16_17", 2436, 2442], ["Domain_17", 2442, 2465, "PF13516"]], "ENSMFAP00000000969": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSVPAP00000002585": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_4", 1517, 1550, "PF00560"], ["Domain_15", 2357, 2381, "PF00560"]], "ENSMMUP00000045983": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2373, "PF13516"]], "ENSNLEP00000021218": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2374, "PF13516"]], "ENSTTRP00000012845": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSP00000291971": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 929, "PF05729"], ["Domain_13", 2214, 2236, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2441, 2465, "PF13516"]], "ENSCGRP00000010646": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSPEMP00000002722": [["Domain_2", 675, 930, "PF05729"], ["Domain_17", 2442, 2465, "PF13516"], ["Linker_17_18", 2465, 2716], ["Domain_18", 2716, 2948, "PF13553"], ["Linker_18_19", 2948, 3051], ["Domain_19", 3051, 3133, "PF00619"]], "ENSRROP00000025388": [["Domain_2", 675, 930, "PF05729"]], "ENSCAFP00000003989": [["Domain_0", 207, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_8", 1773, 1797, "PF13516"], ["Domain_11", 2018, 2055, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2373, "PF13516"], ["Domain_17", 2443, 2466, "PF13516"]], "ENSMLUP00000016674": [["Domain_19", 3051, 3135, "PF00619"]], "ENSRBIP00000031628": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_11", 2016, 2036, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "MGP_PahariEiJ_P0034731": [["Domain_11", 2017, 2054, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"], ["Linker_17_18", 2465, 2716], ["Domain_18", 2716, 2948, "PF13553"]], "MGP_PahariEiJ_P0034732": [["Domain_2", 675, 930, "PF05729"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"], ["Linker_17_18", 2465, 2716], ["Domain_18", 2716, 2997, "PF13553"], ["Linker_18_19", 2997, 3052], ["Domain_19", 3052, 3132, "PF00619"]], "ENSSBOP00000014848": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_8", 1773, 1795, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_15", 2354, 2377, "PF13516"]], "ENSSSCP00000057427": [["Domain_18", 2716, 2997, "PF13553"], ["Linker_18_19", 2997, 3052], ["Domain_19", 3052, 3133, "PF00619"]], "ENSMMUP00000014119": [["Domain_4", 1515, 1542, "PF13516"], ["Domain_6", 1618, 1642, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2215, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2464, "PF13516"]], "ENSPPAP00000037748": [["Domain_4", 1515, 1542, "PF13516"], ["Domain_6", 1618, 1642, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2215, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2464, "PF13516"]], "ENSSTOP00000009174": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_8", 1772, 1791, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2441, 2465, "PF13516"]], "ENSECAP00000013016": [["Domain_0", 202, 326, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSSSCP00000058159": [["Domain_4", 1515, 1542, "PF13516"], ["Domain_6", 1617, 1642, "PF13516"], ["Domain_8", 1776, 1795, "PF13516"], ["Domain_11", 2019, 2054, "PF13516"], ["Domain_13", 2214, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Linker_15_16", 2378, 2389], ["Domain_16", 2389, 2436, "PF13516"], ["Linker_16_17", 2436, 2442], ["Domain_17", 2442, 2464, "PF13516"]], "ENSNLEP00000013011": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSPCOP00000023924": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSAPLP00000009811": [["Domain_0", 295, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_11", 2019, 2057, "PF00560"]], "ENSSSCP00000003594": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_13", 2212, 2236, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2441, 2457, "PF13516"]], "ENSSBOP00000019275": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1617, 1633, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Linker_13_14", 2237, 2247], ["Domain_14", 2247, 2348, "PF13516"], ["Linker_14_15", 2348, 2354], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSOGAP00000015737": [["Domain_0", 202, 328, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSFCAP00000018243": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_8", 1775, 1796, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2356, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSCAFP00000040841": [["Domain_2", 674, 930, "PF05729"], ["Domain_8", 1773, 1797, "PF13516"], ["Domain_15", 2358, 2379, "PF13516"], ["Domain_17", 2443, 2465, "PF13516"]], "ENSSSCP00000003593": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 676, 930, "PF05729"], ["Domain_11", 2017, 2051, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2443, 2464, "PF13516"]], "ENSRBIP00000018340": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSMPUP00000006685": [["Domain_0", 202, 326, "PF02758"], ["Domain_2", 674, 930, "PF05729"]], "ENSCAPP00000001320": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_8", 1776, 1796, "PF13516"], ["Domain_11", 2019, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2356, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSPANP00000007375": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2373, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSRBIP00000027142": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 929, "PF05729"], ["Domain_13", 2213, 2236, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSOCUP00000014512": [["Domain_0", 203, 323, "PF02758"], ["Domain_2", 675, 929, "PF05729"]], "ENSNLEP00000012949": [["Domain_0", 209, 323, "PF02758"], ["Linker_0_2", 323, 529], ["Domain_2", 529, 930, "PF05729"], ["Domain_8", 1773, 1795, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_15", 2354, 2377, "PF13516"]], "ENSMFAP00000035233": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_11", 2016, 2036, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSPPAP00000033490": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_8", 1773, 1795, "PF13516"], ["Domain_11", 2020, 2054, "PF13516"], ["Domain_13", 2214, 2237, "PF13516"]], "ENSMLEP00000013154": [["Domain_4", 1515, 1542, "PF13516"], ["Domain_6", 1617, 1642, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2215, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2464, "PF13516"]], "MGP_CAROLIEiJ_P0083139": [["Domain_4", 1515, 1542, "PF13516"], ["Domain_6", 1617, 1642, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Linker_15_16", 2378, 2389], ["Domain_16", 2389, 2436, "PF13516"], ["Linker_16_17", 2436, 2442], ["Domain_17", 2442, 2464, "PF13516"]], "ENSMICP00000048054": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_8", 1772, 1795, "PF13516"], ["Domain_11", 2017, 2053, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"]], "ENSPEMP00000002348": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_8", 1772, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSAPLP00000008960": [["Domain_4", 1517, 1551, "PF00560"], ["Linker_4_5", 1551, 1566], ["Domain_5", 1566, 1597, "PF00560"], ["Domain_8", 1775, 1801, "PF00560"]], "ENSECAP00000018270": [["Domain_0", 202, 328, "PF02758"], ["Domain_2", 674, 930, "PF05729"]], "ENSDNOP00000026801": [["Domain_0", 202, 324, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSECAP00000018272": [["Domain_17", 2444, 2462, "PF00560"]], "ENSMNEP00000038446": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"]], "ENSOARP00000001012": [["Domain_0", 202, 328, "PF02758"], ["Domain_2", 675, 930, "PF05729"]], "ENSDNOP00000020663": [["Domain_0", 202, 328, "PF02758"], ["Domain_2", 674, 891, "PF05729"]], "ENSMMUP00000033280": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"]], "ENSMNEP00000026032": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 925, "PF05729"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2443, 2465, "PF13516"]], "ENSPEMP00000014889": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSMMUP00000046788": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSPSIP00000017030": [["Domain_0", 202, 326, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSMUSP00000015866": [["Domain_2", 674, 930, "PF05729"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2356, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSMUSP00000113095": [["Domain_0", 209, 319, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_13", 2212, 2237, "PF13516"]], "ENSMPUP00000011174": [["Domain_0", 202, 326, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_19", 3052, 3133, "PF00619"]], "ENSRBIP00000037431": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_17", 2443, 2465, "PF13516"], ["Linker_17_18", 2465, 2716], ["Domain_18", 2716, 2997, "PF13553"], ["Linker_18_19", 2997, 3052], ["Domain_19", 3052, 3134, "PF00619"]], "ENSNLEP00000012932": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 674, 929, "PF05729"]], "MGP_SPRETEiJ_P0086520": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSTSYP00000030146": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 658, "PF14484"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2214, 2237, "PF13516"], ["Domain_15", 2354, 2372, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSMFAP00000014158": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_11", 2016, 2052, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2374, "PF13516"]], "ENSGGOP00000026379": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 674, 929, "PF05729"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"]], "ENSTSYP00000012813": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 930, "PF05729"]], "ENSP00000327763": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSAMEP00000003541": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSCGRP00001022670": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 657, "PF14484"], ["Domain_8", 1772, 1796, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSPVAP00000008200": [["Domain_0", 201, 328, "PF02758"]], "ENSMOCP00000012739": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 676, 928, "PF05729"]], "ENSRBIP00000003387": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_11", 2016, 2051, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2374, "PF13516"]], "ENSNGAP00000001161": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"]], "ENSP00000466285": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 676, 930, "PF05729"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2441, 2464, "PF13516"]], "ENSTSYP00000028812": [["Domain_2", 674, 929, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSRROP00000041080": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 676, 930, "PF05729"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2441, 2464, "PF13516"]], "ENSMODP00000015678": [["Domain_0", 207, 324, "PF02758"], ["Domain_2", 676, 930, "PF05729"]], "ENSMMUP00000009423": [["Domain_6", 1616, 1642, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2444, 2465, "PF13516"]], "ENSCANP00000020630": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 674, 930, "PF05729"]], "ENSCATP00000004673": [["Domain_2", 675, 930, "PF05729"], ["Domain_11", 2017, 2054, "PF13516"]], "ENSRNOP00000047770": [["Domain_0", 209, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_13", 2212, 2237, "PF13516"]], "ENSRROP00000041840": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_11", 2016, 2051, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"]], "ENSRROP00000044374": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_17", 2443, 2465, "PF13516"], ["Linker_17_18", 2465, 2716], ["Domain_18", 2716, 2997, "PF13553"], ["Linker_18_19", 2997, 3052], ["Domain_19", 3052, 3134, "PF00619"]], "ENSHGLP00100021899": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 839, "PF05729"]], "ENSPTRP00000019737": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_8", 1773, 1795, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_15", 2354, 2377, "PF13516"]], "ENSJJAP00000003663": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_8", 1772, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2441, 2465, "PF13516"]], "ENSMNEP00000042530": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 657, "PF14484"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2212, 2232, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSOCUP00000008333": [["Domain_0", 207, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSPVAP00000004083": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_6", 1619, 1644, "PF00560"]], "ENSLAFP00000005807": [["Domain_0", 201, 330, "PF02758"], ["Domain_2", 676, 940, "PF05729"]], "ENSMOCP00000021229": [["Domain_2", 675, 928, "PF05729"], ["Domain_6", 1617, 1642, "PF13516"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"]], "ENSMICP00000008087": [["Domain_0", 208, 320, "PF02758"], ["Domain_2", 675, 929, "PF05729"], ["Domain_13", 2215, 2235, "PF13516"], ["Domain_15", 2358, 2378, "PF13516"], ["Domain_17", 2441, 2465, "PF13516"]], "ENSPANP00000034802": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSCCAP00000018679": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 926, "PF05729"], ["Domain_8", 1773, 1795, "PF13516"], ["Linker_8_10", 1795, 1807], ["Domain_10", 1807, 1956, "PF13516"], ["Linker_10_11", 1956, 2016], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSOANP00000022985": [["Domain_2", 675, 930, "PF05729"]], "ENSRBIP00000010509": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 676, 930, "PF05729"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2441, 2464, "PF13516"]], "ENSOCUP00000004733": [["Domain_0", 202, 328, "PF02758"], ["Domain_2", 674, 929, "PF05729"]], "ENSSHAP00000002191": [["Domain_0", 202, 326, "PF02758"], ["Domain_2", 676, 862, "PF00004"]], "ENSCANP00000022505": [["Domain_4", 1515, 1542, "PF13516"], ["Domain_6", 1617, 1642, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2215, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2464, "PF13516"]], "ENSP00000375063": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 925, "PF05729"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSCGRP00001014679": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 676, 898, "PF05729"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"]], "ENSFALP00000011314": [["Domain_2", 676, 930, "PF05729"]], "ENSECAP00000021583": [["Domain_0", 202, 326, "PF02758"], ["Domain_2", 674, 930, "PF05729"]], "ENSMLUP00000021929": [["Domain_6", 1619, 1645, "PF00560"], ["Domain_8", 1775, 1794, "PF00560"], ["Domain_11", 2019, 2057, "PF00560"], ["Domain_13", 2215, 2235, "PF00560"], ["Domain_15", 2357, 2375, "PF00560"], ["Domain_17", 2444, 2463, "PF00560"]], "ENSECAP00000006832": [["Domain_2", 674, 930, "PF05729"], ["Domain_19", 3052, 3133, "PF00619"]], "ENSACAP00000020972": [["Domain_0", 209, 326, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSP00000433999": [["Domain_4", 1515, 1542, "PF13516"], ["Domain_6", 1618, 1642, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2215, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2464, "PF13516"]], "ENSMUSP00000104155": [["Domain_2", 675, 930, "PF05729"], ["Domain_17", 2442, 2465, "PF13516"], ["Linker_17_18", 2465, 2716], ["Domain_18", 2716, 2997, "PF13553"], ["Linker_18_19", 2997, 3052], ["Domain_19", 3052, 3133, "PF00619"]], "ENSMFAP00000034555": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_11", 2017, 2054, "PF13516"]], "ENSMUSP00000104158": [["Domain_2", 675, 930, "PF05729"], ["Domain_17", 2443, 2465, "PF13516"], ["Linker_17_18", 2465, 2716], ["Domain_18", 2716, 2997, "PF13553"], ["Linker_18_19", 2997, 3052], ["Domain_19", 3052, 3132, "PF00619"]], "ENSCATP00000003382": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 925, "PF05729"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2443, 2465, "PF13516"]], "MGP_SPRETEiJ_P0028307": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 657, "PF14484"], ["Domain_8", 1772, 1796, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2441, 2458, "PF13516"]], "ENSBTAP00000006201": [["Domain_0", 202, 328, "PF02758"], ["Domain_2", 675, 930, "PF05729"]], "ENSSSCP00000059045": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 658, "PF14484"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSCHIP00000026281": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 905, "PF05729"], ["Domain_8", 1775, 1796, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSCANP00000019338": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSPPAP00000039657": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2215, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"]], "ENSOCUP00000000403": [["Domain_0", 202, 326, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSOCUP00000019453": [["Domain_0", 210, 326, "PF02758"], ["Domain_2", 676, 931, "PF05729"]], "ENSP00000299481": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_11", 2016, 2052, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2374, "PF13516"]], "ENSMMUP00000031420": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_11", 2016, 2036, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSCSAP00000007374": [["Domain_0", 202, 328, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSNLEP00000007510": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2373, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSCJAP00000055581": [["Domain_2", 676, 930, "PF05729"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_15", 2357, 2378, "PF13516"], ["Domain_17", 2441, 2464, "PF13516"]], "ENSSHAP00000003923": [["Domain_2", 674, 805, "PF05729"]], "ENSCATP00000010619": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_8", 1773, 1795, "PF13516"], ["Domain_13", 2214, 2237, "PF13516"], ["Domain_15", 2355, 2377, "PF13516"]], "ENSCATP00000025317": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2358, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"], ["Linker_17_18", 2465, 2716], ["Domain_18", 2716, 2997, "PF13553"], ["Linker_18_19", 2997, 3052], ["Domain_19", 3052, 3134, "PF00619"]], "ENSOARP00000017438": [["Domain_0", 202, 326, "PF02758"], ["Domain_2", 674, 930, "PF05729"]], "ENSSARP00000006776": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_11", 2019, 2057, "PF00560"], ["Domain_17", 2444, 2476, "PF00560"]], "ENSNLEP00000010087": [["Domain_4", 1515, 1542, "PF13516"], ["Domain_11", 2018, 2053, "PF13516"], ["Domain_13", 2215, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2464, "PF13516"]], "ENSCCAP00000038497": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 658, "PF14484"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2373, "PF13516"]], "ENSNGAP00000014801": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSFCAP00000001890": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"]], "ENSCAPP00000012130": [["Domain_6", 1618, 1642, "PF13516"], ["Linker_6_7", 1642, 1650], ["Domain_7", 1650, 1766, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2464, "PF13516"]], "ENSMLUP00000017257": [["Domain_2", 674, 940, "PF05729"], ["Domain_6", 1619, 1645, "PF00560"], ["Domain_8", 1775, 1794, "PF00560"], ["Domain_11", 2019, 2052, "PF00560"], ["Domain_13", 2215, 2235, "PF00560"], ["Domain_15", 2357, 2375, "PF00560"]], "ENSCANP00000036520": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 657, "PF14484"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSMNEP00000020184": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_17", 2446, 2465, "PF13516"], ["Linker_17_18", 2465, 2716], ["Domain_18", 2716, 2997, "PF13553"], ["Linker_18_19", 2997, 3052], ["Domain_19", 3052, 3134, "PF00619"]], "ENSGGOP00000007590": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_8", 1773, 1795, "PF13516"], ["Domain_11", 2020, 2054, "PF13516"], ["Domain_13", 2214, 2237, "PF13516"], ["Domain_15", 2355, 2377, "PF13516"]], "ENSTSYP00000002782": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_8", 1773, 1796, "PF13516"], ["Linker_8_10", 1796, 1807], ["Domain_10", 1807, 1955, "PF13516"], ["Linker_10_11", 1955, 2016], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2374, "PF13516"]], "ENSVPAP00000004578": [["Domain_0", 201, 328, "PF02758"], ["Domain_6", 1619, 1645, "PF00560"]], "ENSMLUP00000008763": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_6", 1619, 1645, "PF00560"], ["Domain_8", 1775, 1798, "PF00560"], ["Domain_15", 2357, 2380, "PF00560"]], "ENSOGAP00000010273": [["Domain_17", 2444, 2457, "PF00560"]], "ENSECAP00000015250": [["Domain_0", 207, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSFDAP00000022072": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_4", 1514, 1528, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"]], "ENSCCAP00000002303": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 657, "PF14484"], ["Domain_8", 1772, 1796, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2212, 2233, "PF13516"], ["Domain_15", 2357, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSLAFP00000001775": [["Domain_6", 1619, 1645, "PF00560"], ["Domain_8", 1775, 1801, "PF00560"], ["Domain_17", 2444, 2476, "PF00560"]], "ENSPTRP00000005793": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_11", 2016, 2052, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2374, "PF13516"]], "ENSMMUP00000007829": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 657, "PF14484"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSCANP00000036805": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 930, "PF05729"]], "ENSOCUP00000023322": [["Domain_2", 675, 929, "PF05729"]], "ENSMEUP00000004119": [["Domain_2", 674, 940, "PF05729"], ["Domain_6", 1619, 1645, "PF00560"], ["Domain_8", 1775, 1801, "PF00560"], ["Domain_17", 2444, 2476, "PF00560"]], "ENSMLUP00000019239": [["Domain_0", 201, 330, "PF02758"], ["Domain_2", 684, 940, "PF05729"]], "MGP_PahariEiJ_P0018223": [["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_8", 1772, 1796, "PF13516"], ["Domain_11", 2019, 2054, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"]], "ENSOPRP00000010415": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 879, "PF05729"], ["Domain_11", 2019, 2057, "PF00560"], ["Domain_13", 2215, 2237, "PF00560"]], "ENSHGLP00100012129": [["Domain_2", 674, 930, "PF05729"], ["Domain_13", 2214, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"]], "ENSMUSP00000072895": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 676, 940, "PF05729"]], "ENSGALP00000011167": [["Domain_4", 1514, 1546, "PF13516"], ["Domain_6", 1618, 1642, "PF13516"], ["Linker_6_7", 1642, 1649], ["Domain_7", 1649, 1765, "PF13516"], ["Linker_7_8", 1765, 1773], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2214, 2237, "PF13516"], ["Domain_15", 2355, 2374, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSOPRP00000011835": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_11", 2019, 2056, "PF00560"], ["Domain_15", 2357, 2381, "PF00560"]], "ENSRNOP00000051298": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "MGP_CAROLIEiJ_P0083101": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSCHIP00000027263": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_8", 1775, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2212, 2232, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2441, 2465, "PF13516"]], "ENSPCOP00000009760": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 658, "PF14484"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2051, "PF13516"], ["Domain_17", 2441, 2465, "PF13516"]], "ENSRROP00000044645": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 658, "PF14484"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSCHIP00000016479": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 657, "PF14484"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSCGRP00001004006": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 675, 931, "PF05729"], ["Domain_11", 2017, 2051, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSPPYP00000011692": [["Domain_0", 202, 325, "PF02758"], ["Domain_2", 676, 930, "PF05729"]], "ENSPPYP00000011693": [["Domain_0", 202, 326, "PF02758"], ["Domain_2", 674, 930, "PF05729"]], "ENSPPYP00000011694": [["Domain_0", 202, 326, "PF02758"], ["Domain_2", 675, 930, "PF05729"]], "ENSPPYP00000011695": [["Domain_0", 202, 326, "PF02758"], ["Domain_2", 674, 925, "PF05729"]], "ENSLAFP00000017981": [["Domain_0", 204, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_13", 2215, 2237, "PF00560"], ["Domain_15", 2357, 2381, "PF00560"]], "ENSOGAP00000011492": [["Domain_2", 675, 940, "PF05729"]], "ENSMODP00000025242": [["Domain_0", 202, 328, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSBTAP00000001428": [["Domain_0", 202, 328, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSJJAP00000002893": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSCAFP00000041931": [["Domain_1", 574, 658, "PF14484"], ["Domain_8", 1773, 1797, "PF13516"], ["Domain_11", 2018, 2055, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2373, "PF13516"], ["Domain_17", 2443, 2466, "PF13516"]], "ENSJJAP00000013786": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2356, 2373, "PF13516"], ["Domain_17", 2441, 2465, "PF13516"]], "ENSTBEP00000013080": [["Domain_0", 201, 328, "PF02758"]], "ENSODEP00000019410": [["Domain_4", 1515, 1542, "PF13516"], ["Domain_6", 1618, 1642, "PF13516"], ["Linker_6_7", 1642, 1650], ["Domain_7", 1650, 1766, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2464, "PF13516"]], "ENSECAP00000021325": [["Domain_2", 675, 930, "PF05729"]], "ENSOPRP00000002921": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSSHAP00000001069": [["Domain_0", 208, 324, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSPANP00000004746": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 657, "PF14484"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2212, 2232, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSPPYP00000011607": [["Domain_0", 207, 328, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSCSAP00000005252": [["Domain_0", 202, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSCGRP00000016706": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 739, "PF14484"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2373, "PF13516"], ["Domain_17", 2441, 2465, "PF13516"]], "ENSMPUP00000009935": [["Domain_0", 207, 325, "PF02758"], ["Domain_2", 674, 930, "PF05729"]], "ENSCCAP00000000787": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 930, "PF05729"]], "ENSMODP00000014409": [["Domain_2", 675, 930, "PF05729"], ["Domain_19", 3052, 3134, "PF00619"]], "ENSTBEP00000012634": [["Domain_2", 686, 930, "PF05729"]], "ENSFCAP00000020543": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 657, "PF14484"], ["Domain_8", 1775, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2441, 2465, "PF13516"]], "ENSMPUP00000012189": [["Domain_0", 202, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSPPAP00000004041": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 676, 930, "PF05729"]], "ENSMICP00000009458": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 940, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_11", 2016, 2052, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2374, "PF13516"]], "ENSPTRP00000072999": [["Domain_2", 675, 930, "PF05729"]], "ENSMLEP00000006325": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 925, "PF05729"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"]], "MGP_SPRETEiJ_P0081462": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_13", 2213, 2237, "PF13516"], ["Linker_13_14", 2237, 2303], ["Domain_14", 2303, 2348, "PF13516"], ["Linker_14_15", 2348, 2354], ["Domain_15", 2354, 2378, "PF13516"]], "ENSSHAP00000000660": [["Domain_2", 675, 930, "PF05729"]], "ENSMGAP00000009467": [["Domain_4", 1517, 1551, "PF00560"], ["Linker_4_5", 1551, 1566], ["Domain_5", 1566, 1597, "PF00560"], ["Domain_8", 1775, 1801, "PF00560"], ["Domain_17", 2444, 2475, "PF00560"]], "MGP_SPRETEiJ_P0081466": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSFDAP00000016473": [["Domain_4", 1515, 1542, "PF13516"], ["Domain_6", 1618, 1642, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2464, "PF13516"]], "ENSMFAP00000026016": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_15", 2355, 2373, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "MGP_PahariEiJ_P0019874": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSPPYP00000000034": [["Domain_0", 202, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSMLUP00000021206": [["Domain_2", 674, 940, "PF05729"], ["Domain_11", 2019, 2057, "PF00560"], ["Domain_17", 2444, 2463, "PF00560"], ["Domain_19", 3051, 3135, "PF00619"]], "ENSCHOP00000002517": [["Domain_2", 674, 940, "PF05729"], ["Domain_19", 3051, 3135, "PF00619"]], "ENSDNOP00000005182": [["Domain_2", 674, 940, "PF05729"]], "ENSPCOP00000010092": [["Domain_2", 674, 929, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "MGP_PahariEiJ_P0045529": [["Domain_2", 675, 931, "PF05729"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2215, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSOGAP00000013513": [["Domain_0", 203, 325, "PF02758"], ["Domain_2", 676, 930, "PF05729"]], "ENSFCAP00000029963": [["Domain_5", 1564, 1594, "PF13516"], ["Linker_5_6", 1594, 1617], ["Domain_6", 1617, 1642, "PF13516"], ["Linker_6_7", 1642, 1650], ["Domain_7", 1650, 1766, "PF13516"], ["Linker_7_8", 1766, 1774], ["Domain_8", 1774, 1796, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2215, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSCSAP00000016337": [["Domain_0", 202, 326, "PF02758"], ["Domain_2", 675, 930, "PF05729"]], "ENSCSAP00000016339": [["Domain_0", 202, 325, "PF02758"], ["Domain_2", 675, 928, "PF05729"]], "ENSNLEP00000027320": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1617, 1633, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSCJAP00000044881": [["Domain_4", 1515, 1542, "PF13516"], ["Domain_6", 1617, 1642, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2215, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"]], "ENSOGAP00000016459": [["Domain_2", 675, 940, "PF05729"]], "ENSETEP00000006696": [["Domain_2", 674, 940, "PF05729"], ["Domain_19", 3051, 3135, "PF00619"]], "ENSEEUP00000003038": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 894, "PF05729"], ["Domain_15", 2357, 2381, "PF00560"]], "ENSMNEP00000036266": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1617, 1633, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSCCAP00000031788": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_11", 2016, 2053, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2374, "PF13516"]], "ENSGGOP00000000443": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_11", 2016, 2052, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2374, "PF13516"]], "ENSMICP00000002549": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 676, 940, "PF05729"]], "ENSANAP00000020408": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_4", 1514, 1529, "PF13516"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2374, "PF13516"]], "ENSMOCP00000005221": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 658, "PF14484"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2355, 2377, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSCHIP00000021210": [["Domain_2", 674, 930, "PF05729"], ["Domain_18", 2716, 2997, "PF13553"], ["Linker_18_19", 2997, 3051], ["Domain_19", 3051, 3134, "PF00619"]], "ENSNLEP00000005171": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 657, "PF14484"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2212, 2232, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSP00000375653": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2373, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSCLAP00000024372": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_15", 2354, 2374, "PF13516"]], "ENSPVAP00000008198": [["Domain_0", 208, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSCLAP00000000911": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_8", 1776, 1792, "PF13516"], ["Domain_11", 2017, 2050, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2356, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSRNOP00000069711": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2373, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSDNOP00000027235": [["Domain_0", 202, 326, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSPANP00000008010": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_11", 2016, 2036, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSPANP00000008011": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"]], "ENSPPYP00000011655": [["Domain_0", 207, 326, "PF02758"], ["Domain_2", 675, 930, "PF05729"]], "ENSP00000467123": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_8", 1773, 1795, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_15", 2354, 2377, "PF13516"]], "ENSTBEP00000006180": [["Domain_11", 2019, 2057, "PF00560"], ["Domain_15", 2357, 2381, "PF00560"]], "ENSRBIP00000027983": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 925, "PF05729"], ["Domain_8", 1773, 1796, "PF13516"], ["Linker_8_10", 1796, 1807], ["Domain_10", 1807, 1956, "PF13516"], ["Linker_10_11", 1956, 2016], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"]], "ENSMEUP00000005340": [["Domain_4", 1517, 1551, "PF00560"], ["Domain_8", 1775, 1801, "PF00560"]], "ENSMAUP00000023936": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "MGP_SPRETEiJ_P0080288": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 658, "PF14484"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2377, "PF13516"], ["Domain_17", 2442, 2461, "PF13516"]], "ENSMUSP00000104024": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSCATP00000008305": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_11", 2016, 2036, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSPCAP00000005877": [["Domain_2", 674, 942, "PF05729"]], "ENSPEMP00000021727": [["Domain_2", 674, 930, "PF05729"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2441, 2465, "PF13516"]], "ENSMMUP00000031417": [["Domain_0", 265, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1617, 1633, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Linker_13_14", 2237, 2247], ["Domain_14", 2247, 2348, "PF13516"], ["Linker_14_15", 2348, 2354], ["Domain_15", 2354, 2378, "PF13516"]], "ENSSTOP00000030969": [["Domain_2", 675, 940, "PF05729"], ["Domain_18", 2716, 2996, "PF13553"], ["Linker_18_19", 2996, 3052], ["Domain_19", 3052, 3133, "PF00619"]], "ENSBTAP00000051793": [["Domain_2", 699, 930, "PF05729"]], "ENSPVAP00000016707": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSP00000343891": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1617, 1633, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSPTRP00000038834": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_11", 2016, 2036, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSCANP00000031685": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_15", 2354, 2377, "PF13516"]], "ENSCCAP00000025568": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 676, 930, "PF05729"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2213, 2232, "PF13516"], ["Domain_15", 2357, 2378, "PF13516"], ["Domain_17", 2441, 2464, "PF13516"]], "ENSANAP00000013563": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSGGOP00000047842": [["Domain_4", 1515, 1542, "PF13516"], ["Domain_6", 1617, 1642, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2215, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"]], "ENSFDAP00000005705": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 795, "PF05729"]], "ENSMODP00000017539": [["Domain_2", 675, 929, "PF05729"]], "ENSSBOP00000017295": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2377, "PF13516"]], "ENSFCAP00000018506": [["Domain_0", 208, 323, "PF02758"]], "ENSCANP00000025648": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 676, 930, "PF05729"], ["Domain_6", 1617, 1633, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Linker_13_14", 2237, 2247], ["Domain_14", 2247, 2348, "PF13516"], ["Linker_14_15", 2348, 2354], ["Domain_15", 2354, 2378, "PF13516"]], "ENSCCAP00000012304": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_8", 1773, 1795, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2377, "PF13516"]], "ENSODEP00000001307": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 657, "PF14484"], ["Domain_8", 1776, 1792, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2356, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSTTRP00000012884": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_8", 1775, 1801, "PF00560"], ["Domain_11", 2019, 2064, "PF00560"], ["Domain_15", 2357, 2375, "PF00560"], ["Domain_17", 2444, 2463, "PF00560"]], "MGP_CAROLIEiJ_P0077022": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 658, "PF14484"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2377, "PF13516"], ["Domain_17", 2442, 2461, "PF13516"]], "ENSSSCP00000003586": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSCGRP00001005030": [["Domain_4", 1514, 1542, "PF13516"], ["Domain_6", 1617, 1642, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Linker_15_16", 2378, 2391], ["Domain_16", 2391, 2436, "PF13516"], ["Linker_16_17", 2436, 2441], ["Domain_17", 2441, 2464, "PF13516"]], "MGP_PahariEiJ_P0018264": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 676, 930, "PF05729"]], "ENSHGLP00000021086": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_6", 1618, 1642, "PF13516"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2374, "PF13516"]], "ENSP00000331857": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 674, 929, "PF05729"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2215, 2237, "PF13516"], ["Domain_15", 2357, 2378, "PF13516"]], "ENSLAFP00000028929": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_6", 1619, 1644, "PF00560"]], "ENSRNOP00000074961": [["Domain_4", 1515, 1542, "PF13516"], ["Domain_6", 1617, 1642, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Linker_15_16", 2378, 2391], ["Domain_16", 2391, 2436, "PF13516"], ["Linker_16_17", 2436, 2442], ["Domain_17", 2442, 2464, "PF13516"]], "ENSMLUP00000019501": [["Domain_0", 201, 330, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSGGOP00000008815": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSCAFP00000003649": [["Domain_0", 203, 326, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_13", 2214, 2238, "PF13516"], ["Domain_15", 2355, 2374, "PF13516"], ["Domain_17", 2442, 2461, "PF13516"]], "ENSVPAP00000007865": [["Domain_0", 204, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_15", 2357, 2378, "PF00560"]], "ENSOARP00000002811": [["Domain_2", 674, 930, "PF05729"], ["Domain_19", 3051, 3133, "PF00619"]], "ENSBTAP00000001675": [["Domain_0", 202, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSMUSP00000147928": [["Domain_4", 1515, 1542, "PF13516"], ["Domain_6", 1617, 1642, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Linker_15_16", 2378, 2389], ["Domain_16", 2389, 2436, "PF13516"], ["Linker_16_17", 2436, 2442], ["Domain_17", 2442, 2464, "PF13516"]], "ENSPPAP00000030933": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"], ["Linker_17_18", 2465, 2716], ["Domain_18", 2716, 2997, "PF13553"], ["Linker_18_19", 2997, 3052], ["Domain_19", 3052, 3134, "PF00619"]], "ENSODEP00000023419": [["Domain_2", 674, 940, "PF05729"], ["Domain_18", 2716, 2996, "PF13553"], ["Linker_18_19", 2996, 3052], ["Domain_19", 3052, 3134, "PF00619"]], "ENSCJAP00000020787": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"], ["Domain_11", 2018, 2054, "PF13516"], ["Linker_11_12", 2054, 2068], ["Domain_12", 2068, 2206, "PF13516"], ["Linker_12_13", 2206, 2212], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"], ["Linker_17_18", 2465, 2716], ["Domain_18", 2716, 2997, "PF13553"], ["Linker_18_19", 2997, 3052], ["Domain_19", 3052, 3134, "PF00619"]], "ENSPPAP00000010743": [["Domain_2", 675, 930, "PF05729"], ["Domain_8", 1773, 1795, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_15", 2354, 2377, "PF13516"]], "ENSRNOP00000004280": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_8", 1772, 1796, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2441, 2458, "PF13516"]], "ENSJJAP00000000928": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_11", 2017, 2036, "PF13516"], ["Domain_13", 2212, 2233, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"]], "ENSMLEP00000020588": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2212, 2232, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSOANP00000018607": [["Domain_2", 674, 930, "PF05729"]], "ENSCSAP00000016111": [["Domain_0", 207, 328, "PF02758"], ["Domain_2", 674, 930, "PF05729"]], "ENSBTAP00000056628": [["Domain_2", 674, 940, "PF05729"]], "ENSCANP00000018943": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_11", 2016, 2036, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSAMEP00000004586": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSPSIP00000012133": [["Domain_0", 203, 328, "PF02758"], ["Domain_2", 675, 930, "PF05729"]], "ENSPEMP00000020353": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_8", 1772, 1796, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSPCOP00000019391": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_11", 2016, 2052, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2374, "PF13516"]], "ENSCAFP00000034020": [["Domain_6", 1617, 1643, "PF13516"], ["Linker_6_7", 1643, 1650], ["Domain_7", 1650, 1767, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2215, 2238, "PF13516"], ["Domain_17", 2442, 2462, "PF13516"]], "ENSTBEP00000002286": [["Domain_2", 674, 940, "PF05729"], ["Domain_11", 2019, 2057, "PF00560"], ["Domain_19", 3051, 3135, "PF00619"]], "ENSPCAP00000008455": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 692, 940, "PF05729"]], "ENSCCAP00000039880": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 930, "PF05729"]], "ENSFCAP00000016408": [["Domain_2", 675, 929, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSOARP00000000498": [["Domain_0", 208, 326, "PF02758"], ["Domain_2", 674, 930, "PF05729"]], "ENSANAP00000026326": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 657, "PF14484"], ["Domain_8", 1772, 1796, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2212, 2233, "PF13516"], ["Domain_15", 2357, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSMFAP00000041029": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSANAP00000022958": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSPCOP00000019540": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 676, 940, "PF05729"]], "ENSOGAP00000013522": [["Domain_0", 202, 328, "PF02758"], ["Domain_2", 675, 929, "PF05729"]], "ENSMUSP00000046503": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_11", 2017, 2036, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSOGAP00000013525": [["Domain_0", 202, 326, "PF02758"], ["Domain_2", 675, 930, "PF05729"]], "ENSCGRP00000003281": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 676, 930, "PF05729"]], "ENSCJAP00000031587": [["Domain_2", 675, 930, "PF05729"]], "ENSPPAP00000037285": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1617, 1633, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Linker_13_14", 2237, 2252], ["Domain_14", 2252, 2348, "PF13516"], ["Linker_14_15", 2348, 2354], ["Domain_15", 2354, 2378, "PF13516"]], "ENSCANP00000021141": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 658, "PF14484"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2373, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSNLEP00000011124": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 674, 929, "PF05729"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_17", 2442, 2464, "PF13516"], ["Linker_17_18", 2464, 2716], ["Domain_18", 2716, 2997, "PF13553"], ["Linker_18_19", 2997, 3052], ["Domain_19", 3052, 3134, "PF00619"]], "ENSETEP00000016431": [["Domain_0", 208, 344, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSANAP00000015859": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 676, 930, "PF05729"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2213, 2232, "PF13516"], ["Domain_15", 2357, 2378, "PF13516"], ["Domain_17", 2441, 2464, "PF13516"]], "ENSECAP00000009788": [["Domain_0", 202, 326, "PF02758"], ["Domain_2", 674, 930, "PF05729"]], "ENSEEUP00000001468": [["Domain_2", 674, 940, "PF05729"]], "ENSCANP00000033003": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 676, 930, "PF05729"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2441, 2464, "PF13516"]], "ENSETEP00000010989": [["Domain_2", 674, 940, "PF05729"]], "ENSOGAP00000017957": [["Domain_0", 203, 325, "PF02758"], ["Domain_2", 675, 930, "PF05729"]], "ENSHGLP00100018962": [["Domain_6", 1618, 1642, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2464, "PF13516"]], "ENSTBEP00000003854": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSSSCP00000038303": [["Domain_2", 674, 930, "PF05729"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"]], "ENSMUSP00000098707": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 657, "PF14484"], ["Domain_8", 1772, 1796, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2441, 2458, "PF13516"]], "ENSCGRP00000001860": [["Domain_2", 674, 930, "PF05729"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSSSCP00000003558": [["Domain_2", 675, 930, "PF05729"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2213, 2228, "PF13516"]], "ENSPCOP00000029322": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSANAP00000031681": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2214, 2228, "PF13516"]], "ENSOGAP00000020003": [["Domain_2", 674, 929, "PF05729"], ["Domain_8", 1775, 1792, "PF00560"]], "ENSPVAP00000010051": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 729, 940, "PF05729"]], "ENSP00000337383": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 657, "PF14484"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSHGLP00000006766": [["Domain_6", 1618, 1642, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2464, "PF13516"]], "ENSMICP00000014662": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"]], "MGP_CAROLIEiJ_P0081408": [["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_8", 1772, 1796, "PF13516"], ["Linker_8_10", 1796, 1806], ["Domain_10", 1806, 1956, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"]], "ENSMNEP00000023257": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 928, "PF05729"], ["Domain_13", 2213, 2236, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2441, 2465, "PF13516"]], "ENSOGAP00000007576": [["Domain_0", 215, 328, "PF02758"], ["Domain_2", 675, 926, "PF05729"]], "ENSPANP00000045277": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_8", 1773, 1795, "PF13516"], ["Domain_13", 2214, 2237, "PF13516"], ["Domain_15", 2355, 2377, "PF13516"]], "ENSOGAP00000001153": [["Domain_2", 674, 819, "PF05729"]], "ENSTSYP00000003410": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"]], "ENSCPOP00000004339": [["Domain_2", 674, 930, "PF05729"], ["Domain_6", 1617, 1642, "PF13516"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_15", 2354, 2374, "PF13516"]], "ENSSBOP00000000646": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 676, 930, "PF05729"], ["Domain_13", 2213, 2232, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2441, 2464, "PF13516"]], "ENSRNOP00000044415": [["Domain_0", 208, 321, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSOPRP00000013263": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSSTOP00000011399": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2050, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"]], "ENSPPAP00000017303": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 925, "PF05729"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSMLEP00000024232": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 674, 930, "PF05729"]], "ENSSARP00000010938": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_7", 1655, 1768, "PF00560"], ["Domain_12", 2127, 2208, "PF00560"], ["Linker_12_13", 2208, 2215], ["Domain_13", 2215, 2243, "PF00560"], ["Domain_15", 2357, 2385, "PF00560"]], "ENSANAP00000007054": [["Domain_4", 1515, 1542, "PF13516"], ["Domain_6", 1617, 1642, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2215, 2232, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSCHIP00000006950": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2377, "PF13516"]], "ENSPEMP00000019983": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 930, "PF05729"]], "ENSMLUP00000005806": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_13", 2215, 2243, "PF00560"], ["Domain_15", 2357, 2381, "PF00560"]], "ENSAMEP00000007804": [["Domain_2", 682, 940, "PF05729"]], "ENSFCAP00000010670": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2374, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSFCAP00000025751": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSPSIP00000007091": [["Domain_0", 207, 326, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSHGLP00100026435": [["Domain_2", 674, 940, "PF05729"], ["Domain_11", 2019, 2054, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"], ["Linker_17_18", 2465, 2716], ["Domain_18", 2716, 2996, "PF13553"], ["Linker_18_19", 2996, 3052], ["Domain_19", 3052, 3134, "PF00619"]], "ENSAMEP00000004644": [["Domain_0", 204, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_11", 2019, 2052, "PF00560"], ["Domain_15", 2357, 2385, "PF00560"]], "ENSMUSP00000083106": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 675, 925, "PF05729"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSTTRP00000002365": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSDNOP00000018711": [["Domain_0", 206, 328, "PF02758"], ["Domain_2", 674, 930, "PF05729"]], "MGP_PahariEiJ_P0041372": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSOARP00000009163": [["Domain_0", 202, 326, "PF02758"]], "ENSRBIP00000022982": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2373, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSAMEP00000004761": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_15", 2357, 2381, "PF00560"]], "ENSAMEP00000004763": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_6", 1619, 1645, "PF00560"], ["Linker_6_7", 1645, 1655], ["Domain_7", 1655, 1769, "PF00560"], ["Domain_13", 2215, 2240, "PF00560"], ["Linker_13_14", 2240, 2252], ["Domain_14", 2252, 2355, "PF00560"]], "ENSCGRP00000003634": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSRNOP00000034130": [["Domain_2", 675, 930, "PF05729"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_17", 2443, 2465, "PF13516"], ["Linker_17_18", 2465, 2716], ["Domain_18", 2716, 2997, "PF13553"], ["Linker_18_19", 2997, 3052], ["Domain_19", 3052, 3133, "PF00619"]], "ENSPPYP00000003320": [["Domain_0", 202, 328, "PF02758"], ["Domain_2", 744, 940, "PF05729"]], "ENSRROP00000000525": [["Domain_4", 1515, 1542, "PF13516"], ["Domain_6", 1618, 1642, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2215, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2464, "PF13516"]], "ENSCHIP00000015005": [["Domain_2", 793, 922, "PF05729"], ["Domain_8", 1773, 1794, "PF13516"], ["Domain_15", 2357, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSSTOP00000026826": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSNGAP00000023365": [["Domain_2", 675, 930, "PF05729"], ["Domain_18", 2716, 2996, "PF13553"], ["Linker_18_19", 2996, 3052], ["Domain_19", 3052, 3133, "PF00619"]], "ENSMNEP00000010656": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 676, 930, "PF05729"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2441, 2464, "PF13516"]], "ENSGGOP00000023354": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 925, "PF05729"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSPTRP00000019828": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1617, 1633, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Linker_13_14", 2237, 2252], ["Domain_14", 2252, 2349, "PF00560"], ["Linker_14_15", 2349, 2354], ["Domain_15", 2354, 2378, "PF13516"]], "ENSMEUP00000004563": [["Domain_2", 674, 940, "PF05729"]], "ENSCJAP00000025340": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_4", 1514, 1529, "PF13516"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2374, "PF13516"]], "ENSECAP00000017692": [["Domain_0", 202, 326, "PF02758"], ["Domain_2", 676, 854, "PF00004"]], "ENSPTRP00000019821": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 674, 929, "PF05729"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2215, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"]], "ENSFDAP00000002232": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2356, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSPTRP00000019824": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 676, 930, "PF05729"]], "ENSRBIP00000018090": [["Domain_4", 1515, 1542, "PF13516"], ["Domain_6", 1617, 1642, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2215, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2464, "PF13516"]], "ENSMFAP00000007813": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 928, "PF05729"], ["Domain_13", 2213, 2236, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSOARP00000003424": [["Domain_0", 202, 326, "PF02758"], ["Domain_2", 682, 905, "PF05729"]], "ENSCGRP00001001900": [["Domain_2", 674, 930, "PF05729"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSSSCP00000047938": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 657, "PF14484"], ["Domain_8", 1775, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2212, 2232, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2441, 2465, "PF13516"]], "ENSNLEP00000013044": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 925, "PF05729"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSOPRP00000012332": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSMFAP00000004329": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1617, 1633, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Linker_13_14", 2237, 2247], ["Domain_14", 2247, 2348, "PF13516"], ["Linker_14_15", 2348, 2354], ["Domain_15", 2354, 2378, "PF13516"]], "ENSCCAP00000037979": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 675, 930, "PF05729"]], "ENSCLAP00000024432": [["Domain_0", 208, 413, "PF02758"], ["Domain_2", 675, 940, "PF05729"], ["Domain_12", 2066, 2206, "PF13516"], ["Linker_12_13", 2206, 2213], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_17", 2442, 2464, "PF13516"], ["Linker_17_18", 2464, 2716], ["Domain_18", 2716, 2997, "PF13553"], ["Linker_18_19", 2997, 3052], ["Domain_19", 3052, 3134, "PF00619"]], "ENSMFAP00000014076": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_8", 1773, 1795, "PF13516"], ["Domain_13", 2214, 2237, "PF13516"], ["Domain_15", 2355, 2377, "PF13516"]], "ENSBTAP00000027232": [["Domain_2", 674, 930, "PF05729"], ["Domain_19", 3051, 3133, "PF00619"]], "ENSMAUP00000023846": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 676, 930, "PF05729"]], "ENSRROP00000027247": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSMLEP00000040542": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 676, 930, "PF05729"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2441, 2464, "PF13516"]], "MGP_CAROLIEiJ_P0078148": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_13", 2213, 2237, "PF13516"], ["Linker_13_14", 2237, 2303], ["Domain_14", 2303, 2348, "PF13516"], ["Linker_14_15", 2348, 2354], ["Domain_15", 2354, 2378, "PF13516"]], "ENSDNOP00000014238": [["Domain_0", 203, 326, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_19", 3052, 3132, "PF00619"]], "ENSLAFP00000001437": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSGGOP00000045325": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSPPYP00000004673": [["Domain_2", 675, 930, "PF05729"]], "ENSFDAP00000000941": [["Domain_2", 674, 940, "PF05729"], ["Domain_18", 2716, 2997, "PF13553"], ["Linker_18_19", 2997, 3052], ["Domain_19", 3052, 3134, "PF00619"]], "ENSPPYP00000011689": [["Domain_0", 202, 328, "PF02758"], ["Domain_2", 674, 929, "PF05729"]], "ENSSSCP00000042009": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSPVAP00000005696": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_8", 1775, 1798, "PF00560"], ["Domain_11", 2019, 2064, "PF00560"], ["Domain_15", 2357, 2375, "PF00560"]], "ENSMMUP00000031445": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_11", 2017, 2054, "PF13516"]], "ENSPPAP00000000338": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSPVAP00000014869": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSMAUP00000005150": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSTTRP00000005730": [["Domain_4", 1517, 1551, "PF00560"], ["Linker_4_5", 1551, 1566], ["Domain_5", 1566, 1617, "PF00560"], ["Linker_5_6", 1617, 1619], ["Domain_6", 1619, 1645, "PF00560"], ["Domain_17", 2444, 2465, "PF00560"]], "ENSPANP00000002473": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1617, 1633, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Linker_13_14", 2237, 2247], ["Domain_14", 2247, 2348, "PF13516"], ["Linker_14_15", 2348, 2354], ["Domain_15", 2354, 2378, "PF13516"]], "ENSPANP00000001452": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_8", 1773, 1795, "PF13516"], ["Domain_13", 2214, 2237, "PF13516"], ["Domain_15", 2355, 2377, "PF13516"]], "ENSMLUP00000012508": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 673, 940, "PF05729"]], "ENSMNEP00000005715": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSSSCP00000033039": [["Domain_2", 674, 928, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSCATP00000021782": [["Domain_2", 729, 870, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2444, 2465, "PF13516"]], "ENSSBOP00000025651": [["Domain_4", 1515, 1542, "PF13516"], ["Domain_6", 1617, 1642, "PF13516"], ["Domain_8", 1776, 1796, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2215, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSCAFP00000010243": [["Domain_0", 202, 324, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSMNEP00000022637": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2215, 2237, "PF13516"]], "ENSCHOP00000003344": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_13", 2215, 2240, "PF00560"], ["Domain_15", 2357, 2375, "PF00560"]], "ENSCGRP00000022383": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_8", 1772, 1796, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"]], "ENSMLEP00000012466": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSPTRP00000061889": [["Domain_0", 210, 323, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2373, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSCATP00000011069": [["Domain_4", 1515, 1542, "PF13516"], ["Domain_6", 1617, 1642, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2215, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2464, "PF13516"]], "ENSPCOP00000031041": [["Domain_0", 210, 319, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_13", 2214, 2237, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"], ["Linker_17_18", 2465, 2810], ["Domain_18", 2810, 2997, "PF13553"], ["Linker_18_19", 2997, 3052], ["Domain_19", 3052, 3133, "PF00619"]], "ENSOGAP00000005359": [["Domain_0", 208, 326, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_19", 3052, 3133, "PF00619"]], "ENSOARP00000002238": [["Domain_2", 675, 940, "PF05729"]], "ENSMNEP00000042967": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_11", 2016, 2036, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSMLEP00000016844": [["Domain_2", 729, 929, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2444, 2465, "PF13516"]], "ENSRNOP00000020393": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 676, 930, "PF05729"]], "ENSMLUP00000022256": [["Domain_2", 674, 940, "PF05729"], ["Domain_11", 2019, 2057, "PF00560"], ["Domain_17", 2444, 2463, "PF00560"], ["Domain_19", 3051, 3135, "PF00619"]], "ENSSBOP00000021579": [["Domain_2", 674, 940, "PF05729"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSGGOP00000010282": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_8", 1773, 1795, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"]], "ENSCATP00000044950": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_11", 2016, 2052, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2374, "PF13516"]], "ENSFDAP00000007526": [["Domain_18", 2716, 2997, "PF13553"], ["Linker_18_19", 2997, 3052], ["Domain_19", 3052, 3134, "PF00619"]], "ENSMOCP00000017748": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 676, 930, "PF05729"]], "ENSCCAP00000003221": [["Domain_2", 674, 929, "PF05729"], ["Domain_7", 1616, 1744, "PF13516"], ["Linker_7_8", 1744, 1773], ["Domain_8", 1773, 1796, "PF13516"], ["Linker_8_10", 1796, 1805], ["Domain_10", 1805, 1955, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSANAP00000022010": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 658, "PF14484"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSCPOP00000020660": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 657, "PF14484"], ["Domain_8", 1776, 1796, "PF13516"], ["Domain_11", 2019, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2356, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSOCUP00000004989": [["Domain_0", 202, 326, "PF02758"], ["Domain_2", 674, 928, "PF05729"]], "ENSOGAP00000011980": [["Domain_0", 207, 326, "PF02758"], ["Domain_2", 674, 930, "PF05729"]], "ENSNLEP00000012963": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 676, 930, "PF05729"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2441, 2464, "PF13516"]], "ENSDORP00000011074": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSANAP00000015301": [["Domain_2", 675, 929, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_8", 1773, 1796, "PF13516"], ["Linker_8_10", 1796, 1805], ["Domain_10", 1805, 1955, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSCAFP00000040966": [["Domain_0", 202, 328, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_13", 2213, 2238, "PF13516"], ["Domain_15", 2356, 2379, "PF13516"]], "ENSSBOP00000016349": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_4", 1514, 1529, "PF13516"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2374, "PF13516"]], "ENSTGUP00000008037": [["Domain_2", 675, 930, "PF05729"]], "ENSRBIP00000011721": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1617, 1633, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Linker_13_14", 2237, 2247], ["Domain_14", 2247, 2348, "PF13516"], ["Linker_14_15", 2348, 2354], ["Domain_15", 2354, 2378, "PF13516"]], "ENSACAP00000019163": [["Domain_2", 674, 930, "PF05729"]], "ENSMFAP00000038181": [["Domain_2", 674, 808, "PF05729"], ["Domain_8", 1773, 1796, "PF13516"], ["Linker_8_10", 1796, 1809], ["Domain_10", 1809, 1956, "PF13516"], ["Linker_10_11", 1956, 2016], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2443, 2465, "PF13516"]], "ENSCSAP00000016325": [["Domain_0", 202, 326, "PF02758"], ["Domain_2", 674, 930, "PF05729"]], "ENSRBIP00000011121": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"]], "ENSOARP00000002124": [["Domain_0", 203, 326, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSCSAP00000016321": [["Domain_0", 202, 325, "PF02758"], ["Domain_2", 676, 930, "PF05729"]], "ENSNLEP00000009988": [["Domain_0", 208, 322, "PF02758"]], "ENSTBEP00000011709": [["Domain_2", 674, 940, "PF05729"], ["Domain_8", 1775, 1801, "PF00560"], ["Domain_11", 2019, 2052, "PF00560"], ["Domain_17", 2444, 2463, "PF00560"]], "ENSPANP00000003150": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_11", 2017, 2054, "PF13516"]], "ENSMOCP00000027502": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 657, "PF14484"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSCAFP00000009685": [["Domain_0", 202, 328, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSFCAP00000018722": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1617, 1633, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Linker_13_14", 2237, 2247], ["Domain_14", 2247, 2348, "PF13516"], ["Linker_14_15", 2348, 2354], ["Domain_15", 2354, 2378, "PF13516"]], "ENSMFAP00000017378": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 674, 930, "PF05729"]], "ENSCLAP00000015171": [["Domain_0", 208, 412, "PF02758"], ["Domain_2", 675, 940, "PF05729"], ["Domain_12", 2066, 2206, "PF13516"], ["Linker_12_13", 2206, 2213], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_18", 2716, 2997, "PF13553"], ["Linker_18_19", 2997, 3052], ["Domain_19", 3052, 3134, "PF00619"]], "ENSTTRP00000000282": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_15", 2357, 2381, "PF00560"]], "ENSPPYP00000003332": [["Domain_17", 2444, 2458, "PF00560"]], "ENSTTRP00000000283": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_8", 1775, 1801, "PF00560"], ["Domain_15", 2357, 2381, "PF00560"]], "ENSAMEP00000011039": [["Domain_6", 1619, 1645, "PF00560"], ["Domain_11", 2019, 2057, "PF00560"], ["Domain_15", 2357, 2381, "PF00560"]], "ENSMFAP00000006491": [["Domain_2", 686, 870, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2444, 2465, "PF13516"]], "ENSEEUP00000003349": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSEEUP00000012162": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_19", 3051, 3135, "PF00619"]], "ENSCLAP00000004444": [["Domain_4", 1515, 1542, "PF13516"], ["Domain_6", 1618, 1642, "PF13516"], ["Linker_6_7", 1642, 1650], ["Domain_7", 1650, 1766, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2464, "PF13516"]], "ENSANAP00000000245": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_12", 2068, 2206, "PF13516"], ["Linker_12_13", 2206, 2212], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"], ["Linker_17_18", 2465, 2716], ["Domain_18", 2716, 2995, "PF13553"], ["Linker_18_19", 2995, 3052], ["Domain_19", 3052, 3134, "PF00619"]], "ENSCATP00000017631": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1617, 1633, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Linker_13_14", 2237, 2247], ["Domain_14", 2247, 2348, "PF13516"], ["Linker_14_15", 2348, 2354], ["Domain_15", 2354, 2378, "PF13516"]], "ENSFDAP00000020144": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 674, 930, "PF05729"]], "ENSCAFP00000003671": [["Domain_0", 203, 325, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1617, 1633, "PF13516"], ["Domain_11", 2021, 2054, "PF13516"], ["Domain_13", 2213, 2238, "PF13516"], ["Linker_13_14", 2238, 2247], ["Domain_14", 2247, 2349, "PF13516"], ["Linker_14_15", 2349, 2355], ["Domain_15", 2355, 2379, "PF13516"]], "ENSPEMP00000009823": [["Domain_4", 1515, 1543, "PF13516"], ["Domain_6", 1617, 1642, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2215, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Linker_15_16", 2378, 2391], ["Domain_16", 2391, 2436, "PF13516"], ["Linker_16_17", 2436, 2441], ["Domain_17", 2441, 2465, "PF13516"]], "ENSSBOP00000032399": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSLAFP00000022644": [["Domain_2", 674, 940, "PF05729"], ["Domain_8", 1775, 1805, "PF00560"], ["Domain_15", 2357, 2381, "PF00560"], ["Domain_17", 2444, 2475, "PF00560"]], "ENSMAUP00000022693": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_8", 1772, 1796, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSCHOP00000009422": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 941, "PF05729"]], "ENSCGRP00001002994": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 676, 930, "PF05729"]], "ENSSBOP00000027355": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_8", 1772, 1796, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2212, 2233, "PF13516"], ["Domain_15", 2357, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSSARP00000007120": [["Domain_0", 201, 328, "PF02758"], ["Domain_19", 3051, 3135, "PF00619"]], "ENSSBOP00000017054": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_12", 2065, 2206, "PF13516"], ["Linker_12_13", 2206, 2213], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"], ["Linker_17_18", 2465, 2716], ["Domain_18", 2716, 2997, "PF13553"], ["Linker_18_19", 2997, 3052], ["Domain_19", 3052, 3134, "PF00619"]], "ENSPANP00000008559": [["Domain_4", 1515, 1542, "PF13516"], ["Domain_6", 1617, 1642, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2215, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2464, "PF13516"]], "ENSPSIP00000011293": [["Domain_0", 210, 326, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_6", 1619, 1634, "PF00560"]], "ENSMICP00000046583": [["Domain_4", 1515, 1542, "PF13516"], ["Domain_6", 1618, 1642, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2215, 2237, "PF13516"], ["Domain_15", 2354, 2374, "PF13516"], ["Domain_17", 2442, 2464, "PF13516"]], "ENSMLUP00000008807": [["Domain_0", 201, 332, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSGALP00000045145": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 676, 930, "PF05729"], ["Domain_6", 1618, 1642, "PF13516"], ["Domain_9", 1773, 2378, "PF13516"], ["Domain_12", 1773, 2378, "PF13516"]], "ENSPPAP00000012732": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 929, "PF05729"], ["Domain_13", 2214, 2236, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2441, 2465, "PF13516"]], "ENSNGAP00000018646": [["Domain_4", 1515, 1542, "PF13516"], ["Domain_6", 1617, 1642, "PF13516"], ["Linker_6_7", 1642, 1650], ["Domain_7", 1650, 1766, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSOCUP00000009034": [["Domain_0", 203, 325, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSVPAP00000011582": [["Domain_0", 198, 328, "PF02758"]], "ENSCHIP00000030097": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSETEP00000012003": [["Domain_13", 2215, 2240, "PF00560"], ["Domain_15", 2357, 2381, "PF00560"]], "ENSNGAP00000019327": [["Domain_0", 210, 322, "PF02758"], ["Domain_2", 675, 928, "PF05729"]], "ENSRNOP00000068986": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1616, 1641, "PF13516"], ["Domain_13", 2212, 2233, "PF13516"]], "ENSMUSP00000104293": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 658, "PF14484"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2377, "PF13516"], ["Domain_17", 2442, 2461, "PF13516"]], "ENSOCUP00000018965": [["Domain_2", 674, 930, "PF05729"], ["Domain_17", 2444, 2461, "PF00560"]], "ENSCHIP00000014734": [["Domain_4", 1515, 1543, "PF13516"], ["Domain_6", 1618, 1642, "PF13516"], ["Domain_8", 1776, 1796, "PF13516"], ["Domain_11", 2019, 2054, "PF13516"], ["Domain_13", 2214, 2237, "PF13516"], ["Domain_15", 2355, 2377, "PF13516"], ["Domain_17", 2442, 2464, "PF13516"]], "ENSNLEP00000013036": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 929, "PF05729"], ["Domain_8", 1773, 1803, "PF13516"], ["Domain_13", 2214, 2236, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2441, 2465, "PF13516"]], "MGP_SPRETEiJ_P0080669": [["Domain_0", 209, 319, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_13", 2212, 2237, "PF13516"]], "ENSGGOP00000016379": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_11", 2016, 2036, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSRROP00000014783": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSCJAP00000032660": [["Domain_2", 675, 930, "PF05729"], ["Domain_8", 1773, 1795, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"]], "ENSDORP00000027616": [["Domain_4", 1515, 1543, "PF13516"], ["Domain_6", 1618, 1642, "PF13516"], ["Linker_6_9", 1642, 1650], ["Domain_9", 1650, 2436, "PF13516"], ["Domain_11", 1650, 2436, "PF13516"], ["Linker_11_17", 2436, 2442], ["Domain_17", 2442, 2464, "PF13516"]], "ENSPPYP00000004039": [["Domain_0", 202, 328, "PF02758"], ["Domain_2", 675, 930, "PF05729"]], "MGP_SPRETEiJ_P0080593": [["Domain_2", 675, 931, "PF05729"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2215, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "MGP_SPRETEiJ_P0080597": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_11", 2017, 2036, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "MGP_CAROLIEiJ_P0081454": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 676, 930, "PF05729"]], "ENSECAP00000002676": [["Domain_2", 674, 928, "PF05729"]], "ENSDNOP00000011474": [["Domain_2", 674, 930, "PF05729"]], "ENSMOCP00000027176": [["Domain_2", 675, 930, "PF05729"], ["Domain_17", 2441, 2465, "PF13516"], ["Linker_17_18", 2465, 2716], ["Domain_18", 2716, 2996, "PF13553"], ["Linker_18_19", 2996, 3051], ["Domain_19", 3051, 3132, "PF00619"]], "ENSCGRP00001013748": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2373, "PF13516"], ["Domain_17", 2441, 2465, "PF13516"]], "ENSMLEP00000024895": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 929, "PF05729"], ["Domain_13", 2213, 2236, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2441, 2465, "PF13516"]], "ENSMMUP00000011259": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_11", 2016, 2053, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2374, "PF13516"]], "ENSCAFP00000003638": [["Domain_0", 202, 328, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_11", 2017, 2053, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2379, "PF13516"]], "ENSOGAP00000021129": [["Domain_0", 202, 328, "PF02758"], ["Domain_2", 675, 929, "PF05729"]], "ENSRNOP00000019808": [["Domain_0", 208, 321, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSPCAP00000008349": [["Domain_0", 201, 330, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSMAUP00000022945": [["Domain_4", 1515, 1542, "PF13516"], ["Domain_6", 1617, 1642, "PF13516"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Linker_15_16", 2378, 2391], ["Domain_16", 2391, 2436, "PF13516"], ["Linker_16_17", 2436, 2442], ["Domain_17", 2442, 2462, "PF13516"]], "MGP_PahariEiJ_P0045396": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 658, "PF14484"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2377, "PF13516"]], "ENSMAUP00000020106": [["Domain_0", 208, 321, "PF02758"], ["Domain_2", 675, 905, "PF05729"]], "ENSBTAP00000056032": [["Domain_2", 675, 940, "PF05729"]], "ENSMICP00000018849": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 657, "PF14484"], ["Domain_8", 1772, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2441, 2465, "PF13516"]], "ENSMFAP00000038232": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 657, "PF14484"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSMLUP00000013077": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_8", 1775, 1801, "PF00560"], ["Domain_15", 2357, 2375, "PF00560"], ["Domain_17", 2444, 2463, "PF00560"]], "ENSODEP00000001162": [["Domain_2", 675, 940, "PF05729"], ["Domain_8", 1773, 1796, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2354, 2374, "PF13516"]], "ENSMLUP00000014116": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSRROP00000024058": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 674, 925, "PF05729"], ["Domain_8", 1773, 1796, "PF13516"], ["Linker_8_10", 1796, 1807], ["Domain_10", 1807, 1956, "PF13516"], ["Linker_10_11", 1956, 2016], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2443, 2465, "PF13516"]], "ENSMUSP00000050252": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 676, 930, "PF05729"]], "ENSLAFP00000000878": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_8", 1775, 1801, "PF00560"], ["Domain_11", 2019, 2064, "PF00560"], ["Domain_15", 2357, 2381, "PF00560"], ["Domain_17", 2444, 2465, "PF00560"]], "ENSCCAP00000008804": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2214, 2228, "PF13516"]], "ENSOGAP00000016316": [["Domain_2", 674, 929, "PF05729"], ["Domain_11", 2019, 2037, "PF00560"], ["Domain_19", 3057, 3132, "PF00619"]], "ENSOCUP00000014204": [["Domain_0", 202, 328, "PF02758"], ["Domain_2", 674, 930, "PF05729"]], "ENSOPRP00000009298": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 729, 940, "PF05729"], ["Domain_8", 1775, 1801, "PF00560"], ["Domain_11", 2019, 2064, "PF00560"]], "ENSLAFP00000009291": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"]], "ENSNGAP00000017481": [["Domain_2", 674, 928, "PF05729"], ["Domain_13", 2212, 2237, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSPCOP00000013304": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 657, "PF14484"], ["Domain_8", 1772, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2441, 2465, "PF13516"]], "ENSOGAP00000020794": [["Domain_0", 209, 328, "PF02758"], ["Domain_2", 685, 929, "PF05729"]], "ENSECAP00000016221": [["Domain_17", 2444, 2461, "PF00560"]], "ENSPCOP00000001237": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 676, 930, "PF05729"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2464, "PF13516"]], "ENSCCAP00000039719": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 674, 929, "PF05729"], ["Domain_13", 2214, 2236, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSTSYP00000032706": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 657, "PF14484"], ["Domain_8", 1772, 1796, "PF13516"], ["Domain_11", 2016, 2053, "PF13516"], ["Domain_13", 2212, 2232, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2441, 2465, "PF13516"]], "ENSANAP00000041367": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 930, "PF05729"]], "ENSMUSP00000075794": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_11", 2017, 2036, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"]], "ENSPVAP00000013249": [["Domain_4", 1517, 1551, "PF00560"], ["Domain_6", 1619, 1645, "PF00560"], ["Domain_11", 2019, 2057, "PF00560"], ["Domain_15", 2357, 2381, "PF00560"], ["Domain_17", 2444, 2475, "PF00560"]], "ENSAMEP00000018180": [["Domain_0", 201, 328, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_11", 2019, 2057, "PF00560"], ["Domain_17", 2444, 2463, "PF00560"], ["Domain_19", 3051, 3135, "PF00619"]], "ENSDORP00000010033": [["Domain_0", 208, 323, "PF02758"], ["Linker_0_1", 323, 536], ["Domain_1", 536, 657, "PF14484"], ["Domain_8", 1772, 1796, "PF13516"], ["Domain_11", 2016, 2054, "PF13516"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2441, 2465, "PF13516"]], "ENSCSAP00000016317": [["Domain_0", 202, 328, "PF02758"], ["Domain_2", 674, 929, "PF05729"]], "ENSPANP00000023154": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 676, 930, "PF05729"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2441, 2464, "PF13516"]], "ENSMMUP00000010478": [["Domain_0", 207, 322, "PF02758"], ["Domain_2", 676, 930, "PF05729"], ["Domain_15", 2355, 2378, "PF13516"], ["Domain_17", 2441, 2464, "PF13516"]], "ENSOANP00000018610": [["Domain_0", 202, 325, "PF02758"], ["Domain_2", 676, 871, "PF00004"]], "ENSTSYP00000033223": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_12", 2066, 2206, "PF13516"], ["Linker_12_13", 2206, 2213], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"], ["Linker_17_18", 2465, 2716], ["Domain_18", 2716, 2997, "PF13553"], ["Linker_18_19", 2997, 3052], ["Domain_19", 3052, 3134, "PF00619"]], "ENSNGAP00000012423": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_8", 1772, 1796, "PF13516"], ["Domain_11", 2017, 2051, "PF13516"], ["Domain_15", 2354, 2372, "PF13516"]], "ENSCJAP00000021914": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "MGP_SPRETEiJ_P0084791": [["Domain_2", 675, 930, "PF05729"], ["Domain_6", 1616, 1642, "PF13516"], ["Domain_8", 1772, 1796, "PF13516"], ["Linker_8_10", 1796, 1806], ["Domain_10", 1806, 1956, "PF13516"], ["Linker_10_11", 1956, 2018], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2212, 2237, "PF13516"]], "ENSOPRP00000012868": [["Domain_0", 270, 328, "PF02758"], ["Domain_11", 2019, 2057, "PF00560"]], "ENSLAFP00000010734": [["Domain_2", 675, 940, "PF05729"]], "ENSRROP00000040567": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 929, "PF05729"], ["Domain_13", 2213, 2236, "PF13516"], ["Domain_15", 2354, 2378, "PF13516"], ["Domain_17", 2441, 2462, "PF13516"]], "ENSDORP00000013746": [["Domain_0", 208, 323, "PF02758"], ["Domain_2", 674, 930, "PF05729"], ["Domain_8", 1772, 1795, "PF13516"], ["Domain_11", 2017, 2054, "PF13516"], ["Domain_13", 2216, 2231, "PF13516"], ["Domain_15", 2355, 2377, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"]], "ENSP00000445135": [["Domain_0", 207, 323, "PF02758"], ["Domain_2", 675, 930, "PF05729"], ["Domain_8", 1773, 1795, "PF13516"], ["Domain_11", 2020, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_15", 2355, 2377, "PF13516"]], "ENSDNOP00000024216": [["Domain_0", 202, 328, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "ENSMNEP00000010301": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 675, 940, "PF05729"]], "MGP_SPRETEiJ_P0029372": [["Domain_18", 2716, 2997, "PF13553"]], "ENSMMUP00000004772": [["Domain_0", 208, 322, "PF02758"], ["Domain_2", 674, 940, "PF05729"], ["Domain_11", 2018, 2054, "PF13516"], ["Domain_13", 2213, 2237, "PF13516"], ["Domain_17", 2442, 2465, "PF13516"], ["Linker_17_18", 2465, 2716], ["Domain_18", 2716, 2997, "PF13553"], ["Linker_18_19", 2997, 3052], ["Domain_19", 3052, 3134, "PF00619"]], "ENSSHAP00000007616": [["Domain_0", 202, 326, "PF02758"], ["Domain_2", 674, 930, "PF05729"]]}

