Home
Query Results
"ENSGT00900000140789_0"
Found:
ENSGT00900000140789_0
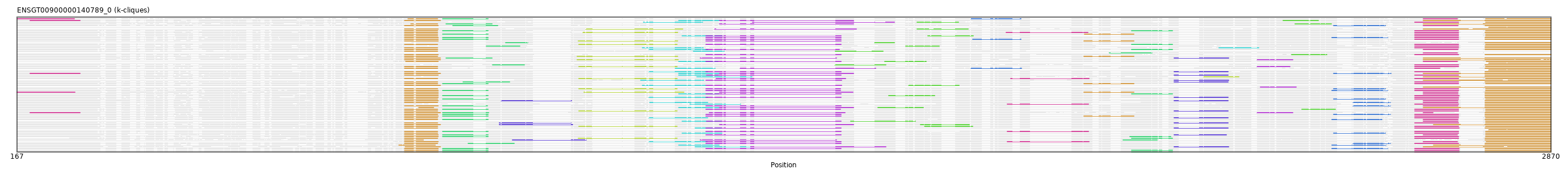
ENSOARP00000006528 (view gene)
ENSFDAP00000011250 (view gene)
ENSSBOP00000006474 (view gene)
ENSCANP00000030676 (view gene)
ENSPPYP00000021885 (view gene)
ENSETEP00000003436 (view gene)
ENSCCAP00000010123 (view gene)
ENSMLUP00000021226 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------GPEVVA
PF01391 // ENSGT00900000140789_0_Domain_15 (2358, 2421)
GPDGLPGRDGRPGQQG-EQGADGTP---GPMGPAGKRGNPGVA
GLPGAQGPPGFKGESGLPGQLGPPGKRGTEGGTGLPGNQGEP---GSKGQPGDSGQMGFP
GM-AGLFGPKASPPGDIGFKGIQGPRGPPGLM--------------------------G-
-KEGIIGPLGILGPSGRP-GPKG--DKGSQGNMGLQGPRGPPGPRGR-PGPPG-P-----
--PG--RPIHFQQDDLGAAFQTWMDTHGALRAE-VSSH-PDRLVLDQGGEIFKTLHYLSH
LIQS
PF01410 // ENSGT00900000140789_0_Domain_19 (2645, 2870)
IKTPLGTKENPARVCRDLMDCEQKMADGTYWVDPNLGCSSDTIEVSCNFTHGGQTC
LKPITASKVE------------------------------------------FAVSRVQM
NFLHLLSSEVTQHITIHCLNMTVWQEGPGQAPAKRAVRFRAWNGQIFEAGGQFRPEVSMD
GCKVQDGRWQQTLFTFRTQDPQQLPIVSVDNLPPASSGKQYRLEVGPACFL
ENSMUSP00000043816 (view gene)
ENSCAFP00000004923 (view gene)
ENSPANP00000018955 (view gene)
ENSPCOP00000003046 (view gene)
ENSMPUP00000008107 (view gene)
ENSRNOP00000010333 (view gene)
ENSPVAP00000011262 (view gene)
ENSPSIP00000018777 (view gene)
ENSMOCP00000000791 (view gene)
ENSCGRP00001021678 (view gene)
ENSJJAP00000017621 (view gene)
ENSMODP00000004868 (view gene)
ENSMNEP00000035197 (view gene)
ENSCHIP00000033038 (view gene)
ENSCLAP00000015376 (view gene)
------------------------------------------------------------
-----------GFLFAWILVSFACHLASTQGAPEDVDVLQRLGLSWTKA-GGG---QSPA
PPGIIPFPLGFIFTQRARLQAPTASVIPAALGTELALVLSLCSHRVNHAFLFAVRSRKYR
LQLGLQFLPGRTVVHLGPRRSVAFDLDVHDGRWHHLALELRSRTVTLVTACGQRRLPVPL
PFHRDSMLDPQGSFLFGKMSPHAVQFEGALCQFSVLPVTQVARNYCAHLRKQCGQADTYR
PQLGPLFPVDAGR---------PFALQSDLALLGLENLTTA---------TLALGARPVG
RGPTV-----------TV-------------PTKPLRTNPPDLHQHTTS----------P
PAK---LPASKA---------P-PSSSPAFPASPTR------ARPVAAQSLQ--------
------KVTATKIPKSLPTKPLAPSPSVAPTRSPQLTQKTAPRPFTRSAP-PTRKPVPPT
PH-PAPAKASGPTAKP-VQRNPVVPRLVPL-----------SARPPV----ASSS-----
--KSLPTVTQD----ESRLTSRASKLASTRASTHKPPQPAVLPSSSASSA---------S
TTVVAPALGTGTPRT-TLPTLAPGSVP-----SGNKKPTGSEASKKS------GPKNST-
--WK---PTPVKPGKAAGDA----PLND--------------------------------
----RTTRPSPSHW----RTTPAVVLAPA---RVLSSNP--RP-TSGSY----SFFHLVE
PTPFP-LLMGPPGPKGDCGLPGPPG----LPGLPGPPGPRGPRVICGET---SSPGDLVA
KGQKGDLGLSPGQAHDGAKGDMGLPGLSGNPGPPGRKGHKGYPGPAGHPGEQGQPGPEGS
PGAKGYPGRQ----------------------G
PF01391 // ENSGT00900000140789_0_Domain_2 (994, 1053)
LPGPVGDPGPKGSRGYIGIPGLFGL--
PGSDGERGLPGVPGKKGKMGRPGFPGDFGERGPPGLDGNPILFSF---------------
------------------------------------------------------------
FSQGDM-GVLGPIGYPGPKGMKGLMGS--VGEPGLKGDKG--------------------
------------------------------------------------------EQGVPG
VSGEPGFQGDKG---SQG--LPGVPGA---------------------------------
-----RGKPGPTVTSGLPGRKGFPGR--PGPDGS------KVRS--------------P-
----------------PEAWRAPQG-----------------------------------
--------PGLQPPIPIHPLLYCLQ-----------------------------------
------------------------------------------------------------
------------------------------------------------GEPGEPGRPGPM
GEQGLLGFIGLVGEPGIVGEKGEKATP---------------------------------
---DTLTVRLPFAWQGHPGMPGGVGIPGEPGPPG-----------------P
PF01391 // ENSGT00900000140789_0_Domain_7 (1733, 1792)
RGSR-GPP
GMRGA---KGRRGPRGPDGPAGEQ-GSRGQKGPAG-P-RGRPGPPGQQGTAGWRS-----
-------------------HLCL-SLQGIPGPSGPPGTKGLPGEP--VSSFPCLPD--ML
APGRLG---------------PPGAV---GEPGP-PGEAGMKGDLGPLGSPGEQGLIGQR
-------------GEPGLEGDSGPAGPDGLK-----------------------------
--------------------------------------------GDRGDPGPDGDRGEKG
QEGL-KGEDGPPGPPGITGIRQPPKLN---------------------------------
------------------------------------------------------------
-----------------------------------RKQGEDGKAEGPPGPPGDRVRHPLS
PGLDEVPGGGRPQRAPRSSVPFPAIVLVVQGPVGDR-GDRGEPGDPGYPVSRGGN-----
PGEWRGC----------------------------CFSAGPLLPREGLQGLPGPRGVVGR
Q----------GPEGAAGPDGLPGRDGRAGQQRGCQGKKGDTLRRLRHGPVPHSGVPPSP
DILRLQGTQ--NYFLGLPGQLGPPGKRGTEGGMGLPGSQGEP---GSKGQPGDSGEMGFP
GV-AGLFGPKG-PPGDLGFKGIQGPRGPPGLMVSSSFCAPAGTVPP------------GL
ASLQLVG---------------------------YLSPRLCPQNPGRKASL------GP-
SEAWAPQDSRYQQDDLGAAFQTWMDTNGALRSE-EYSY-PDRLVLDQGG
PF01410 // ENSGT00900000140789_0_Domain_18 (2630, 2707)
EIFKTLHYLSN
LIQSIKTPLGTKENPARVCRDLMDCEQKIADGTYWVDPNLGCASDTIEVSCNFTHGGQTC
LKPITAENSGT00900000140789_0_Linker_18_19 (2707, 2754)
SKVE------------------------------------------FPF01410 // ENSGT00900000140789_0_Domain_19 (2754, 2870)
AVSRVQM
NFLHLLSSEVTQHITIHCLNMTVWQEGPGHTPAKQAVRFRAWNGQVFEAGGQFRPEVSVD
DCKVHDGRWHRTLFTFRTQDPQQLPIVGVDNLPPASSGKQYRLEVGPACFL
ENSRBIP00000005850 (view gene)
ENSP00000348385 (view gene)
ENSHGLP00100024524 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------------------
PF01410 // ENSGT00900000140789_0_Domain_19 (2760, 2870)
M
NFLHLLSSEATHVITIHCLNTPRWTSAQTSG-PGLPVAFKGWNGQIFEENTLLEPEVLLD
DCKIQDGSWHKAKFLFHTPDPNQLPVIEVQKLPHLKTERKYYIESSSVCFL
ENSPPAP00000017942 (view gene)
ENSNGAP00000001460 (view gene)
ENSMICP00000013929 (view gene)
ENSOGAP00000014789 (view gene)
ENSFCAP00000036703 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------------------
PF01410 // ENSGT00900000140789_0_Domain_19 (2760, 2870)
M
NFLHLLSSEATHIITIHCLNTPVWTSAQSSG-SGLPIGFKGWNGQIFEENTLLEPKVLSD
DCKIQDGSWHKAKFLFHTQDPNQLPVIEVQKLPHLKTERKYYIESSSVCFL
ENSNLEP00000020391 (view gene)
ENSTBEP00000013359 (view gene)
ENSCJAP00000033351 (view gene)
ENSPCAP00000009396 (view gene)
------------------------------------------------------------
-----------GFLFTWIIVSFACHLASTQGTPEDVDVLQRLGLSRTKV-AGG---RSPA
PLGVIPFQLGFIFTQQARLQAPTASVIPAALGTELALVLSLCSHRVNHAFLFAVRSRKRK
LQLGLQFLP
PF02210 // ENSGT00900000140789_0_Domain_0 (190, 278)
GKMVVHLGPRRSVAFDLDVHDGRWHHLALELRGRTVTLVTACGQRRVPVPL
PFHRDPALDPEGAFLFGKMSPRAVQFEGALCQFSIYPENSGT00900000140789_0_Linker_0_1 (278, 850)
VTQVAHNYCTHLKKQCGQADMYW
PQLGPLFPQDSGF---------SFTMQTNLPLLGLENLTTA---------ALALGSWPAG
QGAKV-----------TVAPITP-------T--KPLRTSTIGPHQHVV--VGGAAQTPLP
PAK---LLASQV-------------LPPGPPASWATTARTPHPAAPPAQISW--------
------KSTNTNIPKSVPAEPSIPSPSLVPVKSPRPTPKPVPPSFPESAS-STQKPVSPS
FPESAPASVTRPAVRL-TQRNPGTPRPPIP-----------SSRPLRPTA-VFSK-----
--KVFPTVTQS----KAKMTSHTSKPAPARTSIHKPPHPTITSLSPAPNLGSTRTPQPPA
MVA-RQISGTRTPRT-APPTLTPASSP-----SRSKKPTGSEATKKA------GPKSSS-
--RR---PAPLRPGKAARDA----LLNN--------------------------------
----PATLPKPR-----QQTTPAPALAPV---RFLSSSS--RP-TSSGY----SFFYLVG
PTPFS-LLMPF01391 // ENSGT00900000140789_0_Domain_1 (850, 913)
GPPGPKGDCGLPGPPG----LPGLPGPSGARGPRGPPGPYGNPGLPGLPGP
KGQKGDPGLSPGENSGT00900000140789_0_Linker_1_2 (913, 917)
KAYDPF01391 // ENSGT00900000140789_0_Domain_2 (917, 998)
GPKGDMGLPGLPGNPGPLGRKGLKGYPGPAGHPGEQGPPGPEGS
PGAKGYPGRQ----------------------GLPGPVGGPGPKGNRXXXXXXXXXXX--
XXXXXXXGLPGVPGKRGKMGRPGFPGDFGERGSPGLDGNPXXXXXXXXX-----------
---------------------------------------------------------XXX
XXXGDM-GSLGPIGYPGPKGMKGLMGN--VGEPGLKGDKG--------------------
------------------------------------------------------EQGVPG
VSGDPGFQGDKG---SQG--LPGFPGV---------------------------------
-----RGKPGPPGRIGDKGSLGFPGP--PGPEGF------PGDIGPPGDNGP-EGMKGKP
GARGLP
PF01391 // ENSGT00900000140789_0_Domain_6 (1387, 1626)
GPRGQL---GPEGDEGPMGPPGA--PGLE-------------------------
GQLGRKGFPGR-------PGPDGLK-----------------------------------
------------------------------------------------------------
------------------------------------------------GEPGNPGRPGPV
GEQXXXXXXXXXXXXXXXX------------------------------------XXGDR
GMMGPPGAPGPKGSMGHPGMPGSIGFPGEPGPPG-----------------PPGSR-GPS
GMRGA---KGRRGLRGPDGTAGEP-GSKVL-GPPG-P-QGRPGQPGQQGVAGERG-----
-------------------DAGPQGFPGIQGPSGPPGTKGLPGEP--GPQGPQGPIGPPG
EMGPKG---------------PPGAV---GEPGL-PGEAGMKGDLGPLGIPGEQGLIGQR
-------------GEPGLEGDSGPAGPDGLKXXXXXXXXXXXXXXXXXXXXXXXXXXXX-
----X------------XXXXXX------X--------------XXXXX-----XXXXXX
XXXX-XXX-------XXXXXXXXXXXX-----X-------------XXXXXXX-------
-----XX----XXXXXX--------------------XXXXXXXXXXXXXXXXXXXXXXX
XXXGH-KGILGPLGPP--GPKGEK-----------GEQGNDGKAEGSPGPPGD-------
-----------------------------RGPVGTR-GDRGQPGDPGYPXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXGPKD---KQGRAGAPGRRETQGLQRLPGPRGVVRR
Q----------
PF01391 // ENSGT00900000140789_0_Domain_15 (2352, 2415)
GPEGIAGPDGLPGRDGPAGQQG-EQGEDGDP---GPMGPAGKRGNPGVA
GLPGAQGPPGFKGESGLPGQLGPPGKRGTEGGMGLPGNQGEP---GSKGQPGDSGEMGFP
GM-AGLFGPKG-PPGDIGFKGIQGPRGSPGLMGXXXXXXXXX----------------XX
XXXXX---------------XXXXXXXXXXXXXELQVPRGPPGPRGR-PGPPVDPKRGPG
QMSS--LLFAQQQDHLGAAFQTWMDTDGALRTE-GHSH-ADQLVLDQGGEIFKTLHYLSN
LIQS
PF01410 // ENSGT00900000140789_0_Domain_18 (2645, 2662)
IKTPLGTKENPARVCRDENSGT00900000140789_0_Linker_18_19 (2662, 2754)
LMDCEQKMVDXXXXXXXXXXXXXXXXXXXXXXXXXXXX-
------------------------------------------XXXXXXXXXVLPF01410 // ENSGT00900000140789_0_Domain_19 (2754, 2870)
AISRVQM
NFLHLLSSEVTQHITIHCLNMTVWQESTGQTPAKQAVRFRAWNGQIFEAGGQFRPEVSMD
GCKVQDGHWHQTLFTFRTQDPQQLPIVGVDNLPPASSGKQYRLEVGPACFL
ENSECAP00000022951 (view gene)
-------------------------------------------GRGSQ---------GCK
SVVGRSAARGGGFLFTWILVSFACRLASTQGPPEDVDVLQRLGLSWTKA-AGG---RSPP
PPGVIPFQSGFIFTQRARLQAPTATVIPATLGAELALVLSLCSHRVNHAFLFAVRSRKHK
LQLGLQFVPGKTVVHLGPRRSVAFDLDMHDGRWHHLALELRGRTVTLVTACGQRRVPVPL
PFHRDPALDPEGSFLFGKMNPHAVQFEGALCQFSIHPVARVAHNYCAHLRKQCGQADMYR
PRLGPLLPQDAGS---------PFAFQTDLALLGLENLTTA---------APALGSRPAG
SGPWV-----------TVVPPTP-------S--KPQRTSTIDPQQRVT--VGSPAWTPLP
PAK---LSASEL----------LPLTSPASPASSS----------RSVQTLR--------
------KSTATKMPKSPPTEPTAPS-SVAPVQSPGSPQKAAPPSFTKSAR-PTKKPALPT
PH-PAPARVSRPTVKP-IQRNPGTPRPPPP-----------SGRPPPPAT-GSSK-----
--KLVPSVVQT----ETKKSSRASEPGPAHTGTRRPPQPTVLHPSPAPSPGSIRTTRPPA
-------------------TLALGSAP-----TGSKKSTGSEATRKA------RPKSGP-
--RK---PVPLRPGKAARDV----PSNP--------------------------------
----PTAGPSPRPPQPSPQTTPAPALAPV---RFPSSSP--RP-MSSGY----SFFHLAG
PTPFP-LLV
PF01391 // ENSGT00900000140789_0_Domain_1 (850, 908)
GPPGPKGDCGLPGPPG----LPGLPGPPGARGPRGPPGPYGNPGLPGPPGA
KGQKGDPGLSPGKAHDGAKGDMGLPGLSGNPGPLGRKGHKGYPGPAGHPGEQGQPGPEGS
PGAKGYPGRQ----------------------GLPGPVGDPGPKGSRVSGPWQVPHSLTS
SLSLPLQGLPGVPGKRGKMGRPGFPGDFGERGPPGLDGNPGE-LGLPGPPGVPGL-----
------------------------------------------------------------
--MGDM-GVLGPI
PF01391 // ENSGT00900000140789_0_Domain_4 (1154, 1331)
GYPGPKGMKGLMGS--MGEPGLKGDKG--------------------
------------------------------------------------------EQGVPG
VSGDTGFQGDKG---SQG--LPGFPGA---------------------------------
-----RGKPGPLGRVGDKGSLGFPGP--PGPEGF------PGDIGPPGDNGP-EGMKGKP
GARGLPGPRGHL---GPEGDEGPMGPPGA--PGLE-------------------------
GQPGRKGFPGR-------PGPDGLK-----------------------------------
------------------------------------------------------------
------------------------------------------------GEPGSPGRPGPV
GEQGLLGFIGLVGEPGIMGEK------------------------------------GDR
GMVGPPGVPGPKGSMGHPGMPGGVGTPGEPGPPG-----------------PPGSR-GPP
GMRGA---KGRRGPRGPDGPAGEQ-GSRGLKGPPG-P-QGRPGQPGQQGTAGERG-----
-------------------HSGPRGFPGIPGPSGPPGTKGLPGEP--GPQGPQGLIGPPG
EMGPKG---------------PPGAV---GEPGL-PGEAGMKGDLGPLGTPGEQGLIGQR
-------------GEPGLEGDSGSAGPDGLK-----------------------------
--------------------------------------------GDRGDPGPDGERGEKG
QEGL-K
PF01391 // ENSGT00900000140789_0_Domain_10 (2047, 2135)
GEDGPPGPPGITGIR-----------GPEGK-PGKQGEKGRTGAKGA-------
-----------KGYQGQLGEMGVPGDPGPPGTPGPKGSRGSLGPTGAPGRMGAQGEPGLA
GYEGH-KGVIGPLGPP--GPKGEK-----------GEQGEDGKAEGPPGPPGD-------
-----------------------------RGPVGDQ-GDRGEPGDPGYPGQEGVQGLRGN
PGQQGLPGHPGPRGRPGPKGSKGEEGPKG---KQGKAGAPGRRGIQGLQGLPGPRGVVGR
Q----------GPEGVAGPDGLPGRDGPVGQQG-EQGDDGDP---GPMGPAGKRGNPGVA
GLPGAQGPPGFKGESGLPGQLGPPGKRGTEGGTGLPGTQGEP---GSKGQPGDSGEMGFP
GM-AGLFGPKG-PPGDIGFKGIQGPRGPPGLM--------------------------G-
-KEGIIGPLGILGPSGRP-GPKG--DKGSQGDL-VSGWRGPPGPRGR-PGPPG-SPEGPG
YTPC--LLLAQQQDDLGAAFQTWMDTNGALRAE---------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------
ENSAPLP00000002291 (view gene)
ENSEEUP00000009875 (view gene)
ENSRROP00000009007 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------------------
PF01410 // ENSGT00900000140789_0_Domain_19 (2760, 2870)
M
NFLHLLSSEATHIITIHCLNTPRWTSTQTSG-PGLPIGFKGWNGQIFKVNTLLEPKVLSD
DCKIQDGSWHKATFLFHTQEPNQLPVTEVQKLPHLKTERKYYIESSSVCFL
ENSMFAP00000035402 (view gene)
ENSPEMP00000028693 (view gene)
ENSMLUP00000008345 (view gene)
ENSSSCP00000005883 (view gene)
ENSACAP00000013987 (view gene)
ENSFALP00000001341 (view gene)
ENSCATP00000020210 (view gene)
ENSTTRP00000005161 (view gene)
ENSOCUP00000003979 (view gene)
ENSBTAP00000001200 (view gene)
ENSHGLP00000023436 (view gene)
ENSPTRP00000036359 (view gene)
ENSTGUP00000003594 (view gene)
ENSCSAP00000008779 (view gene)
MGP_PahariEiJ_P0077769 (view gene)
ENSXETP00000047552 (view gene)
ENSRROP00000029393 (view gene)
ENSMEUP00000008207 (view gene)
-----------------------------------------------MEPE------SAP
VSRVRAAASRGGFFLAWIVFSSTCGLAFTQGDPEDVDVLQRLGLSGKKVW-------CPI
PPGVIPFQSGFIFTQRTRIEAPTHTIIPTTFGTELTLVLSLCSHRVNNAFLFAVRSIRRK
LQLGVQFIPGKIVLYLGQRRSLAFSYDVHDGRWHHLAISIRGQVVTLITSCGKHQIPMDL
HFKKDESLDPEGSFLFGRMNQHSVQFEGAVCQFNIYPSAKAAHNYCKYLKKQCRQADTYR
PLLPPLLPLLPQDP--------GPGLQTDVSLQSLRNLTTV---------MPPLASLPPG
KGAKI-----------TVVPSAL-------TSPQPMRTSTVDPWSSRSTPMSSLVWTTPV
TAKPLPLSSGKV---------SFPVLRPALT---TSQSRPPKPPIPAMMPVPPTS-----
------KASVVKTP-YIPIKLVTP-VTVVSAKRSHSMVKPVLTSPSKSFP-STMKPVLPA
SH-LIPAKAPHPPTAKPALQNSATAKTPLP-----------TARPVPPPIRSSFP-----
--ITTKTVPQGSRANMARSLPSTSKPVLASTGTAKLTSPSTKPSLPTSGYTLTQTPRPPV
KPV-FPTMGIKMPRT-IPPTPTSGLNS-----TASKKVAESTVKRKS------TPKNTP-
--WP---PSHPSPRK-AGDA----PLKN--------------------------------
----LTTQPIPKQSQPSHMAATI---LTQ---RLPSSSR--LT-HTDGY----LFFDPIG
PTLFP-LLLGPPGQKGDPGFPXXXXXXXXXXXXXXXXX----XXXXXXXXXXXXXXXXXX
XGQKGDPGLSPGKAYDGEKXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXGEPGPEGS
PGAKGYPGRQ----------------------GLPGPIGEPGPKGSRXXXXXXXXXXX--
XXXXXXXGPPGVHGKRGKMGRPGFPGDFGERGPPGLDGNPGE-LGAPGPPGVLGL-----
------------------------------------------------------------
--IGDM-GAIGPIGYPGPKGMKXXXXX--XXXXGLKGDKG--------------------
------------------------------------------------------DQGLPG
VPGDIGFQGDKG---SQG--MSGSPGA---------------------------------
-----RGKPGPPGKSGDKGPVGFPGP--PGPEGF------PGDI
PF01391 // ENSGT00900000140789_0_Domain_5 (1365, 1462)
GPPGQNGP-EGAKGKP
GARGLPGPRGEL---GQEGDEGPMGPPGS--PGLE-------------------------
GQLGRKGFPGR-------PGPDGSK-----------------------------------
------------------------------------------------------------
------------------------------------------------XXXXXXXXXXXX
XXXGLMGFIGLVGEPGIIGEK------------------------------------GGR
GAMGPPGAPGAKGSMXXXXXXXXXXXXXXXXXXG-----------------PPGTR-GPQ
GIRGA---KGRRGPRGPDGPAGEP-GSQGVKGPPG-T-QGKQGLDGQQXXXXXXX-----
-------------------XXXXXXXXGIPGPSGPPGAKGIPGEP--GPQGPQGLVGPLG
EIGPKG---------------PPGAL---GEPGL-PGEAGMKGDFGPLGIPGEQGLVGQR
-------------GETGLEGDGGPAGPDGLK-----------XXXXXXXXXX-XXXXXXX
XXXXXXXXXXXXXXX------X----------------------XXXX------XXXXXX
XXXX-XXXX------XXXXXXXXXXXXXX---X-------------XXXXXXX-------
-----XX----XXXXXXX-------------------XXXXXXXXXXXXXXXXXXXXXXX
XXXGH-KGLPGPIGPP--GPKGEKXXXXXXXXXX-----------XXXXXXXX-------
-----------------------------XVPVGDH-
PF01391 // ENSGT00900000140789_0_Domain_13 (2258, 2320)
GDRGEPGDPGYPGQEGVEGSRGK
AGQQGLPGHPGPRGQPGPKGSKGEEGQKG---KQGKRGEAGSRGMQGLMGQPGPRGVVGR
Q----------GQEGLPGQDGVPGKDGQAGEQG-EQGDDGEV---GPIGPAGRRGNPGVA
GLPGAQGPPGFKGESGLPGQLGPTGKRGTEGGTGLPGSQGEP---GSKGQPGDFGEMGFP
GI-AGLFGPKG--SGDVGFKGIQGPRGPPGLM--------------------------G-
-KEGVIGPLGILGPSGIP-GPKG--NKGSRGDLGLQGPRLGE------------------
--CI--SHLLQQQDDLGAAFQTWMDSSSALRTE-GYSH-SDLLVLDQGGEIFKTLHYLSS
LIQS
PF01410 // ENSGT00900000140789_0_Domain_19 (2645, 2870)
IKKPLGTKETPARICRDLMNCEQKMADGTYWIDPNLGCSSDTIEVTCNFTKGGQTC
LRPITASKVD------------------------------------------FAIGRVQM
NFLHLLSSEVIQHITIHCLNMSVWQEATGQVSPKQAVRFRAWNGQIFEAGGQFEPEVSSD
DCKXQDGRWHQTLFTFRTQDPHQLPIVSVYNLPPATPGKQYRLEVGPACFL
ENSGGOP00000019773 (view gene)
ENSANAP00000038025 (view gene)
MGP_SPRETEiJ_P0065114 (view gene)
ENSDNOP00000022128 (view gene)
------------------------------------------MNTNAVRRPRKD---RPA
RLAPRDPPVSRGFLFSWIIVSFVCPLASTQGAPEDVDVLRQLGLGGTKAAGGRG---SAA
PPGVIPFPSGFIFTQRARLQAPTGAVLPAALGTELALVLSLCSHRVNHAFLFAVRSRKRK
LQLGLQFLPGQTVVHLGPRRSVAFDLDVHDGRWHHLALELRGRTVTLVTGCGRRRVPVQL
PFRRDSALDPDGTFLFGKMTPHAVQFEGALCQFSIYPVTQVAHDYCTHLRKQCGQADPSR
PQRGPFFL---RDSG------TLFAFRTGPALLGLGNLTTA---------TPAPGARPAG
GGAGG-----------TATP------------TKPLGTNGRDPPRA------------LS
PSR---LSTSK-------------VLPAVAPTSPRG------------------------
-------------------SPSTPQPAAPPXG-----------------------PPPPS
-------------------------------------------SRLPPSTTGSA------
---------PNRVPTVAPTATGAGKPGPARTSTHRPPQPPVLPLSPVPSPGSARTPRPLA
TAG-PAASGARTSRT-ASPPVAPISAP-----TGSKKPPGAEATKKA------GPESSP-
--WK---PGPLGLGKAASEA----PLDR--------------------------------
----PTARPGPRPPQPRGQSTPAPALAPA---PVLASSP--GPTGS-----ASSFLRLGG
PTPFP-LLM
PF01391 // ENSGT00900000140789_0_Domain_1 (850, 908)
GPPGPKGDCGLPGAPG----LPGLPGPPGARGPRGPPGPYGNPGLPGLPGA
KGQKGDPGLSPGKAHDGAKGDLGLPGLAGNPGPLGRKGYKGYPGPAGHPGEQGQPGPEGS
PGAKGYPGRQ----------------------GLPGPVGDPGPKGSRGYIGLPGLFGL--
PGSDGERGPPGVPGKRGKMGRPGFPGDFGERGPPGLDGNPGE-LGLPGPPGVPGL-----
------------------------------------------------------------
--IGDM-GPLGPVGYPGPKGMKGLMGN--VGGPGLKGDKGEQGVPGVSWSEGVSGRPRAP
PGEGTDPGALTSRARLLR--APAWRWLCCSLPPGGPVLTQPRCLPPGEPLRLAASPLGPH
APQLLGIQGAALPAGSPGSRVPGPPGSHSCSSLGVGGSRAGPDSHLWSVRGSRREQRAHT
LPGPLLPLPGVLGWSSELSSPGWA-------SHSVSSQ
PF01391 // ENSGT00900000140789_0_Domain_5 (1359, 1413)
GFPGDIGPPGDNGP-EGVKGKP
GAQGLPGPRGQL---GPEGDEGPMGPPGV--PENSGT00900000140789_0_Linker_5_7 (1413, 1609)
GLELMLRDRNHQRGSQTSQDKRQSSLLQ
GQPGRKGFPGR-------PGPDGVKGWEGVIKKELARGRRMILKLWVAPLSGFPRNPLSG
LDFPSLPEQGLWAKFRNGQILRGLGHGQGSRGLLTPLVHFPFGGRSPHICPGPSGFRHPG
SSGQSSARLESRTRGPDSMAPGAAWAGAACCSENGGFRWGLNLVAWGAPF01391 // ENSGT00900000140789_0_Domain_7 (1609, 1693)
GEPGNPGRPGPV
GEQGLPGFIGLVGEPGIVGEKGLAY---------------------------SPGPHGDR
GMMGPPGAPGPKGSMGHPGMPGGVGTPGEPGPPA-----------------SSGPVLGSS
AHQGFL--GAFKGPRGPDGTAGEP-GSRGLKGPPG--PQGRPGQPGQQGVAGERG-----
-------------------DVGERGFPGIQGPSGPPGTKGLPGEP--GPQGPQGPLGPPG
EMGAKGTPGLGPRSSCIFLQGPPGAV---GEPGL-PGEAGMKVSWDERGEKGVGPGCGLS
-------------GVPGWPGAPAP------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------
ENSBTAP00000056025 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------------------
PF01410 // ENSGT00900000140789_0_Domain_19 (2760, 2870)
M
NFLHLLSSEATHIITIHCLNTPVWTSPQTSG-SGLTLGFKGWNGQIFEENTLLEPKVLSN
DCKIQDGSWHKAKFLFHTQDPNQLPVIEVQKLPHLKTERKYYIESSSVCFL
MGP_CAROLIEiJ_P0062640 (view gene)
ENSDORP00000007684 (view gene)
ENSMLEP00000009837 (view gene)
ENSCPOP00000007274 (view gene)
ENSODEP00000024872 (view gene)
ENSDNOP00000032430 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------------------PQ
PF01391 // ENSGT00900000140789_0_Domain_11 (2092, 2167)
GA-------
-----------KGYQGQLGEMGVPGDPGSPGTPGPKGSRGSMGPTGAPGRMGAQGEPGLA
GYDGH-KGIVGPLGPP--GPKGEK-----------GEQGEDGRAEGPPGPPGD-------
-----------------------------RGPVGDR-GDRGDPGDPGYRGQEGVQGARGK
PGQQGLPGHPGPRGRPGPKGSKGEEGPKG---KQGRAGAPGRRGVQGLQGLPGPRGVVGR
Q----------GPEGVAGPDGLPGRDGRAGEQG-EQGDDGDP---GPLGPAGKRGNPGVA
GLPGAQGPPGFKGENGLPGQLGPPGKRGTEGGTGLPGNQGEP---GSKGQPGDIGEMGFP
GI-SGLFGPKG-PPGDIGFKGIQGPRGPPGLT--------------------------G-
-REGIIGPLGILGPSGLP-GPKG--DKGSRGDLGLQGPRGPPGPRGQ-PGPPGPP-GSPI
Q---------FQRDDLGAVFQTWMDTNGAFTPE-VYGH-PERLGLDQGGEIFKTLHYLSN
LIQS
PF01410 // ENSGT00900000140789_0_Domain_18 (2645, 2705)
IKTPLGTRENPARVCRDLMDCEQKMVDGTYWVDPNLGCSSDTIEVSCNFTHGGQTC
LKPITAS------------------------------------------KV---------
------------------------------------------------------------
---------------------------------------------------
ENSSHAP00000017379 (view gene)
ENSCGRP00000014778 (view gene)
ENSMGAP00000003226 (view gene)
ENSMAUP00000006878 (view gene)
ENSAMEP00000013505 (view gene)
ENSSTOP00000009048 (view gene)
ENSOPRP00000005214 (view gene)
ENSCAPP00000006605 (view gene)
ENSLAFP00000024867 (view gene)
ENSCHOP00000001916 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------GAPG----LPGLPGPPGARGPRXXXXXXXXXXXXXXXXX
XGQKGDPGLSPGKAHDGAKGNVGLPGLPGNPGPPGRKXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXX----------------------XXXXXXXXXXXXXXXXXXXXXXXXXX--
XXXXXXX
PF01391 // ENSGT00900000140789_0_Domain_2 (1028, 1067)
GPPGVPGKRGKMGRPGFPGDFGERGPPGLDGNPGE-LG-PGPPGVPGL-----
------------------------------------------------------------
--IXXX-XXXXXXXXXXXXXXXXXXXX--XXXXXXXXXXG--------------------
------------------------------------------------------EQGVPG
VSGDTGFQGDKXXXXXXXXXX---------------------------------------
----XXXXXXXXXXXXXXXXXXXXXX--XXXXGF------PGDIGPPGDNGP-EGEKGKP
GARGLPGPRGQL---GPEGDEGPLGPPGV--PGLE-------------------------
GQPGRKGFPGR-------PGPDGVK-----------------------------------
------------------------------------------------------------
------------------------------------------------XXXXXXXXXXXX
XXXGLLGFIV--LEPGIXXXX------------------------------------
PF01391 // ENSGT00900000140789_0_Domain_7 (1678, 1713)
GDR
GMMGPPGVPGPKGSMGHPGMPGGVGTPGEPGPPX-----------------XXXXX-XXP
GMRGT---KRVRXXXXXXXXXXXX-XXXXXXXXXX--XXXXXXXXXXXGTAGERG-----
-------------------DSGPRGFPGIQGPSGPPGTKGLPGEP--XXXXXXXXXXXXX
XXXXXX---------------XXXXX---XXXXX-XXXXXXXXXXXXXXXXXXXXXXXXX
-------------XXXXXXXXXXXXXXXXXX-----------------------------
--------------------------------------------GDRGDLGPDGEHGEKG
QEGL-MGEDGPPGPPGITGIR-----------XXXXX-XXXXXXXXXXXXXGA-------
-----------KGYQGQLGEMGIPGDPGPPGTPGPKGSRGSLGPMGAPGRMGAQGEPGLA
GYDXX-XXXXXXXXXXXX---XXX-----------XXXXXXXXXXXXXXXXXX-------
-----------------------------XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXGPKG---KQGKAGAPGRRGIQGLQGLPGPRGVVGR
Q----------GSEGITGPDGLPGRDGQAGQQG-EQGDDGDP---GPLGPAGKRGNPGVA
GLPGAQGPPGSKGESGLPGQLGPPGKRGTEGGTGLPGNQGEP---VSKGQPGDAGEMGFP
GI-SGLFGPKX-XXXXXXXXXXXXXXXXXXXX--------------------------G-
-KEGIIGPLGTLGPSGLP-GPKG--NRGSRGDLGLQGPRGSPGPRGR-PGPPVXX-XX--
------XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX-XXXXXXXXXXXXXXXXXXXXX
XXXXXXXXX--------XXXXXXXXXXXXXX-XXXXX----XXXXXXXXXXXXX------
X--XXXX----------------XXXXXXXXXXXXXXXXXXXXXXXXXXXVEF
PF01410 // ENSGT00900000140789_0_Domain_19 (2754, 2870)
AVSRVQM
NFLHLLSSEVTQHITIHCLNMTVWQEGTGQIPAKQAVRFRAWNGQIFEAGGQFKPEVSVD
GCKVQDGHWHQTLFTFRTQDPQQLPIVSVDNLPPASSGKHYRLEVGPACFL
ENSTSYP00000022850 (view gene)
{"ENSOARP00000006528": [["Domain_1", 850, 909, "PF01391"], ["Domain_3", 1020, 1144, "PF01391"], ["Domain_12", 2206, 2301, "PF01391"], ["Domain_18", 2645, 2706, "PF01410"], ["Linker_18_19", 2706, 2754], ["Domain_19", 2754, 2870, "PF01410"]], "ENSFDAP00000011250": [["Domain_2", 1005, 1061, "PF01391"], ["Domain_7", 1608, 1698, "PF01391"], ["Domain_18", 2630, 2707, "PF01410"], ["Linker_18_19", 2707, 2754], ["Domain_19", 2754, 2870, "PF01410"]], "ENSSBOP00000006474": [["Domain_1", 850, 908, "PF01391"], ["Domain_5", 1339, 1409, "PF01391"], ["Domain_17", 2487, 2578, "PF01391"], ["Linker_17_18", 2578, 2630], ["Domain_18", 2630, 2707, "PF01410"], ["Linker_18_19", 2707, 2755], ["Domain_19", 2755, 2870, "PF01410"]], "ENSCANP00000030676": [["Domain_1", 850, 908, "PF01391"], ["Linker_1_2", 908, 917], ["Domain_2", 917, 997, "PF01391"], ["Domain_11", 2131, 2203, "PF01391"], ["Domain_18", 2630, 2707, "PF01410"], ["Linker_18_19", 2707, 2755], ["Domain_19", 2755, 2870, "PF01410"]], "ENSPPYP00000021885": [["Domain_1", 850, 908, "PF01391"], ["Linker_1_2", 908, 917], ["Domain_2", 917, 997, "PF01391"], ["Domain_17", 2484, 2572, "PF01391"], ["Linker_17_18", 2572, 2645], ["Domain_18", 2645, 2706, "PF01410"], ["Linker_18_19", 2706, 2754], ["Domain_19", 2754, 2870, "PF01410"]], "ENSETEP00000003436": [["Domain_1", 850, 913, "PF01391"], ["Domain_4", 1154, 1332, "PF01391"], ["Domain_15", 2352, 2415, "PF01391"], ["Domain_19", 2645, 2870, "PF01410"]], "ENSCCAP00000010123": [["Domain_1", 850, 908, "PF01391"], ["Domain_5", 1339, 1409, "PF01391"], ["Domain_6", 1381, 1619, "PF01391"], ["Domain_7", 1772, 1852, "PF01391"], ["Domain_18", 2630, 2707, "PF01410"], ["Linker_18_19", 2707, 2755], ["Domain_19", 2755, 2870, "PF01410"]], "ENSMLUP00000021226": [["Domain_15", 2358, 2421, "PF01391"], ["Domain_19", 2645, 2870, "PF01410"]], "ENSMUSP00000043816": [["Domain_1", 850, 909, "PF01391"], ["Linker_1_2", 909, 917], ["Domain_2", 917, 997, "PF01391"], ["Domain_7", 1703, 1784, "PF01391"], ["Domain_18", 2630, 2708, "PF01410"], ["Linker_18_19", 2708, 2754], ["Domain_19", 2754, 2870, "PF01410"]], "ENSCAFP00000004923": [["Domain_1", 850, 909, "PF01391"], ["Domain_4", 1169, 1340, "PF01391"], ["Linker_4_6", 1340, 1396], ["Domain_6", 1396, 1646, "PF01391"], ["Domain_7", 1753, 1843, "PF01391"], ["Domain_19", 2645, 2870, "PF01410"]], "ENSPANP00000018955": [["Domain_1", 850, 908, "PF01391"], ["Domain_4", 1157, 1330, "PF01391"], ["Domain_12", 2206, 2301, "PF01391"], ["Domain_18", 2630, 2707, "PF01410"], ["Linker_18_19", 2707, 2755], ["Domain_19", 2755, 2870, "PF01410"]], "ENSPCOP00000003046": [["Domain_1", 850, 908, "PF01391"], ["Linker_1_2", 908, 917], ["Domain_2", 917, 997, "PF01391"], ["Domain_6", 1381, 1619, "PF01391"], ["Domain_17", 2487, 2587, "PF01391"], ["Linker_17_18", 2587, 2630], ["Domain_18", 2630, 2707, "PF01410"], ["Linker_18_19", 2707, 2754], ["Domain_19", 2754, 2870, "PF01410"]], "ENSMPUP00000008107": [["Domain_1", 850, 909, "PF01391"], ["Domain_5", 1333, 1403, "PF01391"], ["Domain_6", 1381, 1619, "PF01391"], ["Domain_11", 2131, 2203, "PF01391"], ["Domain_18", 2645, 2706, "PF01410"], ["Linker_18_19", 2706, 2754], ["Domain_19", 2754, 2870, "PF01410"]], "ENSRNOP00000010333": [["Domain_1", 850, 908, "PF01391"], ["Domain_5", 1362, 1408, "PF01391"], ["Domain_6", 1381, 1619, "PF01391"], ["Domain_12", 2206, 2301, "PF01391"], ["Domain_18", 2630, 2708, "PF01410"], ["Linker_18_19", 2708, 2754], ["Domain_19", 2754, 2870, "PF01410"]], "ENSPVAP00000011262": [["Domain_1", 850, 913, "PF01391"], ["Domain_3", 1017, 1146, "PF01391"], ["Domain_6", 1405, 1641, "PF01391"], ["Domain_7", 1759, 1845, "PF01391"], ["Domain_19", 2663, 2870, "PF01410"]], "ENSPSIP00000018777": [["Domain_5", 1339, 1408, "PF01391"], ["Domain_6", 1382, 1619, "PF01391"], ["Linker_6_7", 1619, 1636], ["Domain_7", 1636, 1750, "PF01391"], ["Domain_18", 2645, 2706, "PF01410"], ["Linker_18_19", 2706, 2755], ["Domain_19", 2755, 2870, "PF01410"]], "ENSMOCP00000000791": [["Domain_1", 850, 909, "PF01391"], ["Linker_1_2", 909, 917], ["Domain_2", 917, 997, "PF01391"], ["Domain_6", 1381, 1618, "PF01391"], ["Domain_17", 2484, 2582, "PF01391"], ["Linker_17_18", 2582, 2630], ["Domain_18", 2630, 2707, "PF01410"], ["Linker_18_19", 2707, 2755], ["Domain_19", 2755, 2870, "PF01410"]], "ENSCGRP00001021678": [["Domain_1", 850, 909, "PF01391"], ["Linker_1_2", 909, 917], ["Domain_2", 917, 997, "PF01391"], ["Domain_6", 1381, 1619, "PF01391"], ["Domain_8", 1851, 1936, "PF01391"], ["Domain_18", 2630, 2707, "PF01410"], ["Linker_18_19", 2707, 2754], ["Domain_19", 2754, 2870, "PF01410"]], "ENSJJAP00000017621": [["Domain_1", 850, 908, "PF01391"], ["Domain_5", 1280, 1384, "PF01391"], ["Domain_6", 1381, 1619, "PF01391"], ["Domain_12", 2206, 2301, "PF01391"], ["Domain_18", 2630, 2707, "PF01410"], ["Linker_18_19", 2707, 2754], ["Domain_19", 2754, 2870, "PF01410"]], "ENSMODP00000004868": [["Domain_0", 167, 269, "PF02210"], ["Linker_0_1", 269, 850], ["Domain_1", 850, 909, "PF01391"], ["Domain_4", 1166, 1342, "PF01391"], ["Linker_4_6", 1342, 1405], ["Domain_6", 1405, 1640, "PF01391"], ["Domain_10", 2047, 2135, "PF01391"], ["Domain_18", 2645, 2706, "PF01410"], ["Linker_18_19", 2706, 2754], ["Domain_19", 2754, 2870, "PF01410"]], "ENSMNEP00000035197": [["Domain_1", 850, 908, "PF01391"], ["Domain_4", 1157, 1330, "PF01391"], ["Linker_4_6", 1330, 1381], ["Domain_6", 1381, 1618, "PF01391"], ["Domain_17", 2487, 2578, "PF01391"], ["Linker_17_18", 2578, 2630], ["Domain_18", 2630, 2707, "PF01410"], ["Linker_18_19", 2707, 2755], ["Domain_19", 2755, 2870, "PF01410"]], "ENSCHIP00000033038": [["Domain_1", 850, 909, "PF01391"], ["Domain_4", 1156, 1331, "PF01391"], ["Linker_4_6", 1331, 1381], ["Domain_6", 1381, 1619, "PF01391"], ["Domain_10", 2047, 2135, "PF01391"]], "ENSCLAP00000015376": [["Domain_2", 994, 1053, "PF01391"], ["Domain_7", 1733, 1792, "PF01391"], ["Domain_18", 2630, 2707, "PF01410"], ["Linker_18_19", 2707, 2754], ["Domain_19", 2754, 2870, "PF01410"]], "ENSRBIP00000005850": [["Domain_1", 850, 908, "PF01391"], ["Domain_4", 1157, 1330, "PF01391"], ["Domain_7", 1766, 1851, "PF01391"], ["Domain_18", 2630, 2707, "PF01410"], ["Linker_18_19", 2707, 2755], ["Domain_19", 2755, 2870, "PF01410"]], "ENSP00000348385": [["Domain_1", 850, 908, "PF01391"], ["Linker_1_2", 908, 917], ["Domain_2", 917, 997, "PF01391"], ["Domain_6", 1381, 1618, "PF01391"], ["Domain_16", 2431, 2490, "PF01391"], ["Domain_18", 2630, 2707, "PF01410"], ["Linker_18_19", 2707, 2755], ["Domain_19", 2755, 2870, "PF01410"]], "ENSHGLP00100024524": [["Domain_19", 2760, 2870, "PF01410"]], "ENSPPAP00000017942": [["Domain_1", 850, 908, "PF01391"], ["Domain_5", 1280, 1384, "PF01391"], ["Domain_12", 2206, 2301, "PF01391"], ["Domain_18", 2630, 2707, "PF01410"], ["Linker_18_19", 2707, 2755], ["Domain_19", 2755, 2870, "PF01410"]], "ENSNGAP00000001460": [["Domain_1", 850, 909, "PF01391"], ["Domain_5", 1280, 1384, "PF01391"], ["Domain_6", 1381, 1619, "PF01391"], ["Domain_12", 2206, 2301, "PF01391"], ["Domain_18", 2630, 2707, "PF01410"], ["Linker_18_19", 2707, 2754], ["Domain_19", 2754, 2870, "PF01410"]], "ENSMICP00000013929": [["Domain_1", 850, 908, "PF01391"], ["Linker_1_2", 908, 953], ["Domain_2", 953, 1035, "PF01391"], ["Domain_12", 2206, 2301, "PF01391"], ["Domain_18", 2630, 2707, "PF01410"], ["Linker_18_19", 2707, 2754], ["Domain_19", 2754, 2870, "PF01410"]], "ENSOGAP00000014789": [["Domain_1", 850, 909, "PF01391"], ["Linker_1_2", 909, 917], ["Domain_2", 917, 996, "PF01391"], ["Domain_6", 1381, 1619, "PF01391"], ["Domain_17", 2521, 2587, "PF01391"], ["Linker_17_18", 2587, 2645], ["Domain_18", 2645, 2706, "PF01410"], ["Linker_18_19", 2706, 2754], ["Domain_19", 2754, 2870, "PF01410"]], "ENSFCAP00000036703": [["Domain_19", 2760, 2870, "PF01410"]], "ENSNLEP00000020391": [["Domain_1", 850, 908, "PF01391"], ["Linker_1_2", 908, 917], ["Domain_2", 917, 997, "PF01391"], ["Domain_17", 2487, 2578, "PF01391"], ["Linker_17_18", 2578, 2630], ["Domain_18", 2630, 2707, "PF01410"], ["Linker_18_19", 2707, 2755], ["Domain_19", 2755, 2870, "PF01410"]], "ENSTBEP00000013359": [["Domain_1", 850, 883, "PF01391"], ["Domain_3", 1040, 1170, "PF01391"], ["Linker_3_6", 1170, 1371], ["Domain_6", 1371, 1611, "PF01391"], ["Domain_11", 2116, 2176, "PF01391"], ["Domain_19", 2754, 2870, "PF01410"]], "ENSCJAP00000033351": [["Domain_1", 850, 908, "PF01391"], ["Domain_4", 1157, 1330, "PF01391"], ["Linker_4_6", 1330, 1381], ["Domain_6", 1381, 1619, "PF01391"], ["Domain_11", 2131, 2203, "PF01391"], ["Domain_18", 2630, 2707, "PF01410"], ["Linker_18_19", 2707, 2755], ["Domain_19", 2755, 2870, "PF01410"]], "ENSPCAP00000009396": [["Domain_0", 190, 278, "PF02210"], ["Linker_0_1", 278, 850], ["Domain_1", 850, 913, "PF01391"], ["Linker_1_2", 913, 917], ["Domain_2", 917, 998, "PF01391"], ["Domain_6", 1387, 1626, "PF01391"], ["Domain_15", 2352, 2415, "PF01391"], ["Domain_18", 2645, 2662, "PF01410"], ["Linker_18_19", 2662, 2754], ["Domain_19", 2754, 2870, "PF01410"]], "ENSECAP00000022951": [["Domain_1", 850, 908, "PF01391"], ["Domain_4", 1154, 1331, "PF01391"], ["Domain_10", 2047, 2135, "PF01391"]], "ENSAPLP00000002291": [["Domain_1", 861, 908, "PF01391"], ["Linker_1_2", 908, 923], ["Domain_2", 923, 1004, "PF01391"], ["Domain_6", 1405, 1641, "PF01391"], ["Domain_16", 2419, 2483, "PF01391"], ["Domain_19", 2645, 2870, "PF01410"]], "ENSEEUP00000009875": [["Domain_1", 850, 913, "PF01391"], ["Linker_1_2", 913, 923], ["Domain_2", 923, 1004, "PF01391"], ["Domain_6", 1371, 1611, "PF01391"], ["Domain_12", 2206, 2302, "PF01391"], ["Domain_19", 2645, 2870, "PF01410"]], "ENSRROP00000009007": [["Domain_19", 2760, 2870, "PF01410"]], "ENSMFAP00000035402": [["Domain_1", 850, 908, "PF01391"], ["Domain_4", 1157, 1330, "PF01391"], ["Linker_4_6", 1330, 1399], ["Domain_6", 1399, 1627, "PF01391"], ["Domain_9", 1918, 2056, "PF01391"], ["Domain_18", 2630, 2707, "PF01410"], ["Linker_18_19", 2707, 2755], ["Domain_19", 2755, 2870, "PF01410"]], "ENSPEMP00000028693": [["Domain_1", 850, 909, "PF01391"], ["Linker_1_2", 909, 917], ["Domain_2", 917, 997, "PF01391"], ["Domain_6", 1381, 1619, "PF01391"], ["Domain_10", 2047, 2135, "PF01391"], ["Domain_18", 2630, 2707, "PF01410"], ["Linker_18_19", 2707, 2754], ["Domain_19", 2754, 2870, "PF01410"]], "ENSMLUP00000008345": [["Domain_1", 850, 913, "PF01391"], ["Domain_5", 1269, 1380, "PF01391"], ["Linker_5_6", 1380, 1381], ["Domain_6", 1381, 1620, "PF01391"], ["Domain_7", 1738, 1827, "PF01391"]], "ENSSSCP00000005883": [["Domain_1", 850, 909, "PF01391"], ["Linker_1_2", 909, 917], ["Domain_2", 917, 997, "PF01391"], ["Domain_11", 2128, 2200, "PF01391"], ["Domain_18", 2630, 2707, "PF01410"], ["Linker_18_19", 2707, 2755], ["Domain_19", 2755, 2870, "PF01410"]], "ENSACAP00000013987": [["Domain_0", 168, 268, "PF02210"], ["Linker_0_1", 268, 856], ["Domain_1", 856, 908, "PF01391"], ["Linker_1_2", 908, 917], ["Domain_2", 917, 997, "PF01391"], ["Domain_8", 1848, 1936, "PF01391"], ["Domain_18", 2645, 2705, "PF01410"], ["Linker_18_19", 2705, 2755], ["Domain_19", 2755, 2870, "PF01410"]], "ENSFALP00000001341": [["Domain_1", 852, 909, "PF01391"], ["Domain_5", 1333, 1402, "PF01391"], ["Domain_6", 1381, 1619, "PF01391"], ["Linker_6_7", 1619, 1696], ["Domain_7", 1696, 1769, "PF01391"], ["Domain_18", 2645, 2705, "PF01410"], ["Linker_18_19", 2705, 2756], ["Domain_19", 2756, 2870, "PF01410"]], "ENSCATP00000020210": [["Domain_1", 850, 908, "PF01391"], ["Linker_1_2", 908, 917], ["Domain_2", 917, 997, "PF01391"], ["Domain_10", 2048, 2135, "PF01391"], ["Domain_18", 2630, 2707, "PF01410"], ["Linker_18_19", 2707, 2755], ["Domain_19", 2755, 2870, "PF01410"]], "ENSTTRP00000005161": [["Domain_1", 840, 904, "PF01391"], ["Domain_5", 1333, 1404, "PF01391"], ["Domain_17", 2484, 2579, "PF01391"], ["Domain_19", 2663, 2870, "PF01410"]], "ENSOCUP00000003979": [["Domain_1", 850, 908, "PF01391"], ["Domain_5", 1282, 1384, "PF01391"], ["Domain_17", 2521, 2587, "PF01391"], ["Linker_17_18", 2587, 2645], ["Domain_18", 2645, 2706, "PF01410"], ["Linker_18_19", 2706, 2754], ["Domain_19", 2754, 2870, "PF01410"]], "ENSBTAP00000001200": [["Domain_1", 850, 909, "PF01391"], ["Linker_1_2", 909, 935], ["Domain_2", 935, 1014, "PF01391"], ["Domain_17", 2487, 2578, "PF01391"], ["Linker_17_18", 2578, 2645], ["Domain_18", 2645, 2706, "PF01410"], ["Linker_18_19", 2706, 2754], ["Domain_19", 2754, 2870, "PF01410"]], "ENSHGLP00000023436": [["Domain_1", 850, 908, "PF01391"], ["Domain_5", 1327, 1397, "PF01391"], ["Linker_5_6", 1397, 1441], ["Domain_6", 1441, 1680, "PF01391"], ["Domain_8", 1848, 1937, "PF01391"], ["Domain_18", 2630, 2674, "PF01410"], ["Linker_18_19", 2674, 2754], ["Domain_19", 2754, 2870, "PF01410"]], "ENSPTRP00000036359": [["Domain_1", 850, 908, "PF01391"], ["Domain_3", 1017, 1143, "PF01391"], ["Domain_12", 2206, 2301, "PF01391"], ["Domain_18", 2630, 2707, "PF01410"], ["Linker_18_19", 2707, 2755], ["Domain_19", 2755, 2870, "PF01410"]], "ENSTGUP00000003594": [["Domain_1", 850, 909, "PF01391"], ["Domain_5", 1280, 1383, "PF01391"], ["Domain_6", 1381, 1619, "PF01391"], ["Domain_9", 1912, 2056, "PF01391"], ["Domain_18", 2645, 2705, "PF01410"], ["Linker_18_19", 2705, 2756], ["Domain_19", 2756, 2870, "PF01410"]], "ENSCSAP00000008779": [["Domain_1", 850, 908, "PF01391"], ["Domain_5", 1280, 1384, "PF01391"], ["Domain_6", 1381, 1616, "PF01391"], ["Domain_11", 2131, 2203, "PF01391"], ["Domain_18", 2645, 2706, "PF01410"], ["Linker_18_19", 2706, 2754], ["Domain_19", 2754, 2870, "PF01410"]], "MGP_PahariEiJ_P0077769": [["Domain_1", 850, 909, "PF01391"], ["Linker_1_2", 909, 917], ["Domain_2", 917, 997, "PF01391"], ["Domain_11", 2131, 2203, "PF01391"], ["Domain_18", 2630, 2708, "PF01410"], ["Linker_18_19", 2708, 2754], ["Domain_19", 2754, 2870, "PF01410"]], "ENSXETP00000047552": [["Domain_1", 851, 913, "PF01391"], ["Domain_5", 1362, 1459, "PF01391"], ["Linker_5_6", 1459, 1460], ["Domain_6", 1460, 1698, "PF01391"], ["Domain_12", 2206, 2302, "PF01391"], ["Domain_19", 2645, 2870, "PF01410"]], "ENSRROP00000029393": [["Domain_1", 850, 908, "PF01391"], ["Linker_1_2", 908, 917], ["Domain_2", 917, 997, "PF01391"], ["Domain_6", 1381, 1619, "PF01391"], ["Domain_9", 1912, 2055, "PF01391"], ["Domain_18", 2630, 2707, "PF01410"], ["Linker_18_19", 2707, 2755], ["Domain_19", 2755, 2870, "PF01410"]], "ENSMEUP00000008207": [["Domain_5", 1365, 1462, "PF01391"], ["Domain_13", 2258, 2320, "PF01391"], ["Domain_19", 2645, 2870, "PF01410"]], "ENSGGOP00000019773": [["Domain_1", 850, 908, "PF01391"], ["Domain_4", 1157, 1330, "PF01391"], ["Domain_15", 2352, 2410, "PF01391"], ["Domain_18", 2630, 2707, "PF01410"], ["Linker_18_19", 2707, 2755], ["Domain_19", 2755, 2870, "PF01410"]], "ENSANAP00000038025": [["Domain_5", 1271, 1375, "PF01391"], ["Linker_5_6", 1375, 1463], ["Domain_6", 1463, 1713, "PF01391"], ["Linker_6_7", 1713, 1753], ["Domain_7", 1753, 1826, "PF01391"], ["Domain_18", 2630, 2707, "PF01410"], ["Linker_18_19", 2707, 2755], ["Domain_19", 2755, 2870, "PF01410"]], "MGP_SPRETEiJ_P0065114": [["Domain_1", 850, 909, "PF01391"], ["Linker_1_2", 909, 917], ["Domain_2", 917, 997, "PF01391"], ["Domain_11", 2131, 2203, "PF01391"], ["Domain_18", 2630, 2708, "PF01410"], ["Linker_18_19", 2708, 2754], ["Domain_19", 2754, 2870, "PF01410"]], "ENSDNOP00000022128": [["Domain_1", 850, 908, "PF01391"], ["Domain_5", 1359, 1413, "PF01391"], ["Linker_5_7", 1413, 1609], ["Domain_7", 1609, 1693, "PF01391"]], "ENSBTAP00000056025": [["Domain_19", 2760, 2870, "PF01410"]], "MGP_CAROLIEiJ_P0062640": [["Domain_1", 850, 909, "PF01391"], ["Linker_1_2", 909, 917], ["Domain_2", 917, 997, "PF01391"], ["Domain_6", 1381, 1619, "PF01391"], ["Domain_17", 2484, 2582, "PF01391"], ["Linker_17_18", 2582, 2630], ["Domain_18", 2630, 2708, "PF01410"], ["Linker_18_19", 2708, 2754], ["Domain_19", 2754, 2870, "PF01410"]], "ENSDORP00000007684": [["Domain_1", 850, 909, "PF01391"], ["Domain_6", 1381, 1619, "PF01391"], ["Domain_16", 2413, 2475, "PF01391"], ["Domain_18", 2630, 2707, "PF01410"], ["Linker_18_19", 2707, 2754], ["Domain_19", 2754, 2870, "PF01410"]], "ENSMLEP00000009837": [["Domain_1", 850, 908, "PF01391"], ["Linker_1_2", 908, 917], ["Domain_2", 917, 997, "PF01391"], ["Domain_10", 2047, 2135, "PF01391"], ["Domain_18", 2630, 2707, "PF01410"], ["Linker_18_19", 2707, 2755], ["Domain_19", 2755, 2870, "PF01410"]], "ENSCPOP00000007274": [["Domain_1", 850, 908, "PF01391"], ["Domain_5", 1269, 1376, "PF01391"], ["Domain_14", 2285, 2355, "PF01391"], ["Domain_18", 2630, 2707, "PF01410"], ["Linker_18_19", 2707, 2754], ["Domain_19", 2754, 2870, "PF01410"]], "ENSODEP00000024872": [["Domain_5", 1353, 1442, "PF01391"], ["Domain_9", 1912, 2055, "PF01391"], ["Domain_18", 2630, 2707, "PF01410"], ["Linker_18_19", 2707, 2754], ["Domain_19", 2754, 2870, "PF01410"]], "ENSDNOP00000032430": [["Domain_11", 2092, 2167, "PF01391"], ["Domain_18", 2645, 2705, "PF01410"]], "ENSSHAP00000017379": [["Domain_1", 850, 909, "PF01391"], ["Linker_1_2", 909, 917], ["Domain_2", 917, 997, "PF01391"], ["Domain_6", 1381, 1619, "PF01391"], ["Domain_12", 2206, 2298, "PF01391"], ["Domain_18", 2645, 2705, "PF01410"], ["Linker_18_19", 2705, 2755], ["Domain_19", 2755, 2870, "PF01410"]], "ENSCGRP00000014778": [["Domain_1", 850, 909, "PF01391"], ["Linker_1_2", 909, 917], ["Domain_2", 917, 997, "PF01391"], ["Domain_6", 1381, 1619, "PF01391"], ["Domain_17", 2484, 2582, "PF01391"], ["Linker_17_18", 2582, 2630], ["Domain_18", 2630, 2707, "PF01410"], ["Linker_18_19", 2707, 2754], ["Domain_19", 2754, 2870, "PF01410"]], "ENSMGAP00000003226": [["Domain_5", 1266, 1377, "PF01391"], ["Linker_5_6", 1377, 1381], ["Domain_6", 1381, 1620, "PF01391"], ["Domain_12", 2206, 2302, "PF01391"], ["Domain_19", 2645, 2870, "PF01410"]], "ENSMAUP00000006878": [["Domain_1", 850, 909, "PF01391"], ["Domain_5", 1333, 1402, "PF01391"], ["Domain_6", 1382, 1619, "PF01391"], ["Domain_12", 2206, 2301, "PF01391"], ["Domain_18", 2630, 2707, "PF01410"], ["Linker_18_19", 2707, 2754], ["Domain_19", 2754, 2870, "PF01410"]], "ENSAMEP00000013505": [["Domain_0", 190, 278, "PF02210"], ["Linker_0_1", 278, 850], ["Domain_1", 850, 913, "PF01391"], ["Domain_5", 1333, 1404, "PF01391"], ["Linker_5_6", 1404, 1405], ["Domain_6", 1405, 1641, "PF01391"], ["Domain_16", 2398, 2460, "PF01391"], ["Domain_19", 2645, 2870, "PF01410"]], "ENSSTOP00000009048": [["Domain_1", 850, 908, "PF01391"], ["Domain_4", 1156, 1330, "PF01391"], ["Domain_11", 2131, 2203, "PF01391"], ["Domain_18", 2630, 2707, "PF01410"], ["Linker_18_19", 2707, 2754], ["Domain_19", 2754, 2870, "PF01410"]], "ENSOPRP00000005214": [["Domain_1", 875, 901, "PF01391"], ["Domain_5", 1333, 1404, "PF01391"], ["Linker_5_6", 1404, 1405], ["Domain_6", 1405, 1641, "PF01391"], ["Linker_6_7", 1641, 1684], ["Domain_7", 1684, 1764, "PF01391"], ["Domain_19", 2645, 2870, "PF01410"]], "ENSCAPP00000006605": [["Domain_1", 850, 908, "PF01391"], ["Domain_4", 1164, 1331, "PF01391"], ["Domain_9", 1910, 2054, "PF01391"], ["Domain_18", 2630, 2707, "PF01410"], ["Linker_18_19", 2707, 2754], ["Domain_19", 2754, 2870, "PF01410"]], "ENSLAFP00000024867": [["Domain_0", 190, 278, "PF02210"], ["Linker_0_1", 278, 850], ["Domain_1", 850, 913, "PF01391"], ["Domain_5", 1333, 1404, "PF01391"], ["Linker_5_6", 1404, 1405], ["Domain_6", 1405, 1641, "PF01391"], ["Domain_17", 2487, 2588, "PF01391"], ["Linker_17_19", 2588, 2645], ["Domain_19", 2645, 2870, "PF01410"]], "ENSCHOP00000001916": [["Domain_2", 1028, 1067, "PF01391"], ["Domain_7", 1678, 1713, "PF01391"], ["Domain_19", 2754, 2870, "PF01410"]], "ENSTSYP00000022850": [["Domain_1", 850, 908, "PF01391"], ["Linker_1_2", 908, 962], ["Domain_2", 962, 1043, "PF01391"], ["Domain_6", 1381, 1619, "PF01391"], ["Domain_17", 2521, 2587, "PF01391"], ["Linker_17_18", 2587, 2630], ["Domain_18", 2630, 2707, "PF01410"], ["Linker_18_19", 2707, 2754], ["Domain_19", 2754, 2870, "PF01410"]]}

