Home
Query Results
"ENSGT00870000136417_1"
Found:
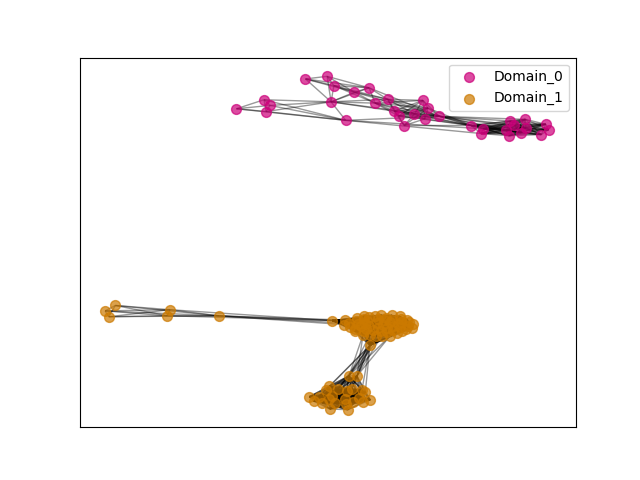
ENSGT00870000136417_1
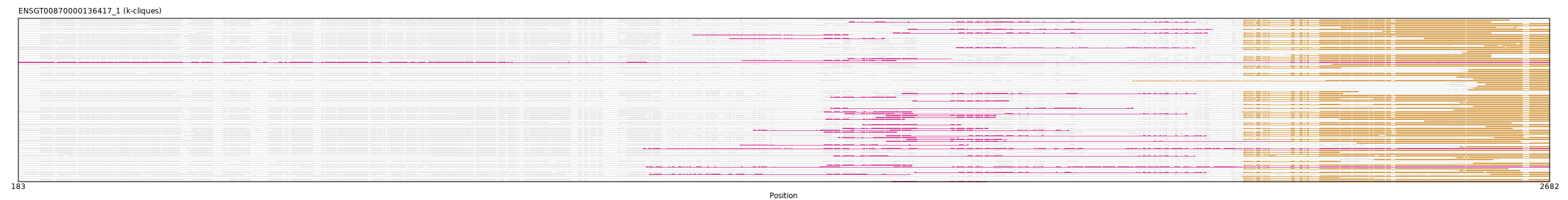
ENSPANP00000014781 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------MGNQDGKLKRSAGDALHEGGG-
--GGAEDALGPRDVETTKKGSGGKKALGKHGKGGG--G-GGGESGKKKS-KSDSRASVFS
-NLRIRKN-LSKGK-GAGGSREDVLD-SQALQTGELD-SAHSLL-TKTPDLSLSADETGL
SDTEC-ADPFEVTRPGGPGPAE-ARVRGRPVAEDVE-TAAGAQDGQRTSSGSDT-DIYSF
HSATEQEDLLSDIQQAIRLQQQQQQLQL--------------------QAPATARPPTAV
SPQPGAFLGLDRFLLGPSGG---------------A-GEAPGSPD-TEQALSALSDLPES
LAAEPRE-PQQPPSPGGLPAS--------E--------------------APSLPAAQPA
VADSPSSTAFPFP---ETGPGEE----AAGAPVRGAGDTDEEGEEDAFEDAPRGSPGEEW
APE-------------VGEDA-PQRLEEEPEEEAQGPDSPA-AASLPGSPAPSQRCFKPY
PLITPCYIKTTTRQLSSPNHSPSQSPNQSPRIK----RRPEPSLSRGSRTALASVAA-PA
K---KHR-ADGGLAGGLSRSADWTEELGART---PRAGGSAHLLERGVAAD-SGG--GVS
PALAAK-AS-GAPAAADGFQNVFTGRTLLEKLFSQQENGPPEEAEKFCSRIIAMGLLLPF
SDCFREPCN-QNAQTNAASFDQDQL---YTWAAVSQPTHSLDYSEGQFPRRVPSMGPPSK
PP---DEEHRLGDAETE----SQSAVSETPKKRSDAV--QK-----EVVDMKSE-GQANV
IQQLEQTIEDLRTKIAELERQYPALDTEVASGRQGLGNGVTASGDVCLEALRLEEKEGRH
QRIL-----------EAKSIQT---SPTEEG-G---VLTLP-PVG---AL-GHPPC----
---------------------PP----------GAESGPQTKFCSEISLIVSPRRISVQL
DSHQPTQSISQPPPP--PSLLWSAGQGQPG----------S-Q-P-P-H-S-SL-S-T-E
-F-E-T-S-H--E-R-S-V-S-S-A-F-K-N-S-C-H-I-P-S-P-PP-L-P-CT-ES-S
-S-S-M-P-G-L-G-M-V-P-PP-P-P-PL-P-G-M-T-V-PT-L-P-S-T-T-I-P-T-
S-SA-A-V-Y-R--M-R-QP---L-S---S-W-S-G-H-T-I--STY--R-GG-L--P-L
--A-P---------------------------------------------------HGSY
PLPIFGR--------------------LPLGPFYRAAG-TPPPLRA----------ATLH
TPRP--LY--PGR-AYPLRPLYPGRAYPLRPLYPER-GYLLRPLYPGQAYPLRPLYPGRA
YP--LRPLYPGRAYPLRPL-----------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------Y-P-E-WGYLLRPLYPGQ-
------------AYPLHPLYPGQAYPLRPLYPEPAYPLRPLYPEWG--------YLL---
------------------------------------------------------------
------------RMG--IPPAP-AP-----P-P----PPPGT--G-IPLP-----P----
----LLP--A-S-GPPLL-----------PQVGS----STLP------------------
T-P-QVC-G-FLP---P-PLPSGL--------------FG--------------------
--FGMNQDK---G-S-R-K--
PF02181 // ENSGT00870000136417_1_Domain_1 (2182, 2682)
-QPI-E-P-C-R-P-M-----KPLYWTR----I-Q-L-H
--SK-----------------------------------RDSSTS--------LIWEK--
-I---EEP-----------------SIDCHEFEELFSKTAVKERKKPISDTISKTKAK-Q
VVKLLSNKRSQAVGILMS-SLHLDMKDIQHAVVNLDNSVVDLETLQ-ALYENR-AQSDEL
EKIEKHGRSSKD--KENAKSLDK-------PEQFLYELSLIPNFSERVFCILFQSTFSES
ICSIHRKLELLQKLCETLKNGPGVMQVLGLVLAFGNYMNGGNKTRGQADGFGLDILPKLK
DVKSSDNSRSLLSYIVSYYLRNFD-EDAGKEQCLFPLPEPQDLFQASQMKFEDFQKDLRK
LKKDLKACEVEAGKVYQVSSKEHMQPFKENMEQFIIQAKIDQEAEENSLTETHKCFL---
--------ETTAYFFMKPKLGEKEVSPNAFFSIWHEFSSDFKDFWKKENKLLLQ----ER
VKEAEEVCRQKKGKSLYKIKPRHDSGIKAKISMKT-------------------------
--------------
ENSBTAP00000041415 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
V
PF02181 // ENSGT00870000136417_1_Domain_1 (2342, 2627)
VKLLSNKRSQ-VGISMS-SLHLDMKDTQHAVVNLDNSVVDLKTLQ-ALYENRYAQSVEL
EKTEKHIQSSKD--KENAKSLD-------KPEQFLYELSLIPNFQ-SVFCILFQSTFSET
ICSIRCKLELLQKLCETLKHGSRVMQVLGSVLGFGNYINGGNKTRGQADGFELDILPKLK
DVKSNDNSRSLLSHIMSYYICNFD-EDAGKEQCVFPLPESQDLFQASQMKCEDFQKDLRK
LKKDLRASKIDQE------------------------------AEENSLTE---------
------------------------------------------------------------
------------------------------------------------------------
--------------
ENSMAUP00000008930 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------MGNQDGKLKRSAGDASHEGGGG
G-GGAEDAAGPRDAETTKKASGSKKALGKHGKGGGG----SGETSKKKS-KSDSRASVFS
-NLRIRKN-LTKGK-GAGDSREDVLD-SQALPTGELD-SAHSIV-TKTPDLSLSAEETGL
SDTEC-ADPFEVIHPGGPRPAE-AGVGIQATAEDLG-TAAGAQDGQRTSSGSDT-DIYSF
HSATEQEDLLSDIQQ----QQQQQQQQQQKL-------------LL-QDSEEPAAPPTAI
SPQPGAFLGLDQFLLGPSSE---------------A-AK-----D-TEQPLPVRPDLPET
--IKSPV-PEHPPSSGGHLAS--------E--------------------TPGHETGHSA
VTDSLSSPAFTFP---DAGPREG----AAGAPVPEAGDTDEECEEDAFEDAPRGSPEEGW
VPE-------------VGEA--PERLEEEPEEGMRGPSVSA-VASLPGSPAPSPRCFKPY
PLITPCYIKTTTRQLSSPNHSPSQSPNQSPRIK----KRPEPSVSRSPRTALASAAA-PA
K---KHR-LEGSLAGGLSRSADWTEELGVRP---PGAGGSAHLLGRGATAD-DSG--GGS
PVLAAK-AP-GAPATADGFQNVFTGRTLLEKLFSQQENGPPEEAEKFCSRIIAMGLLLPF
SDCFREPCN-QNAGSSSAPFDQDQL---YTWAAVSQPTHSMDYSEGQFPRREPSTWPSSR
LP---DEEHSPKDVNA----EPQSSVLESPKKCSNSV--QQ-----EVCDVKSE-GQATV
IQQLEQTIEDLRTKIAELEKQYPALDLEGPRGLPGLENGFTASEEVGLDALRLHGDVAQP
PRTL-----------QAKSIQT---SPTEEG-G---ILTLA-SLK---ALPEGPPC----
---------------------PPAAQSGESAVLASASGPQTKFCSEISLIVSPRRISVQL
DAQPALQS-APPLPP--P------------------------------PDG--L-P---T
AL-E-T-S-H--K-H-S-V-F---SS----FQNSC-N-I-----PPAPPL-P-CTESSSF
VPGL---------G-M-A-I-PSPP---CL---SDITVPALPSTTAP-------------
-QPTSL---QGTE-M-LPAPP--------PPP----------------------------
------------------------------------------------------------
---SLPGLGIP--PPP-------------APPLPVVGIPPPPP-----------------
---------------------LPGVGIPPPHPLPGM-EIPPPP--------PPPPLPGVG
IP--PPPPLPGVGIPXXXXXXXXXXXXXXXXX----------------------------
----------------------XXXXXXXX---XXXXXXXXVGIPP----PPPLPGVGIP
----PPPPLPGV-GIP---PPPPLPGM-G-IP-PPPPLPGVGIPP-----------PPPL
PGVAIP--------PPPPL-P-G-V-GVPP--------------------------A---
---------------------------------------PPPLGAG--------------
------------------------------------------------------------
----------------------------------------IP--------------PP-P
----LLP--G-S-GPSLSS-----------QVG-------------SSTL-P--------
A-P-QVC-G-FLF---P-PLPTGIFG----------------------------------
--LGMNQDR---G-A-R-K--
PF02181 // ENSGT00870000136417_1_Domain_1 (2182, 2682)
-QPI-E-P-C-R-P-M-----KPLYWTR----I-Q-L-H
--SK-----------------------------------RETSPS--------LIWEK--
-I---EEP-----------------SIDCHEFEELFSKTAVKERKKPISDTISKTKAK-Q
VVKLLSNKRSQAVGILMS-SLHLDMKDIQHAVVNLDNSVVDLETLQ-ALYENR-AQSDEL
EKIEKHSRSSKD--KENAKSLD-------KPEQFLYELSLIPNFSERVFCILFQSTFSES
ICSIRRKLELLQKLCETLKNGPGVMQVLGLVLAFGNYMNAGNKTRGQADGFGLDILPKLK
DVKSSDNSRSLLSYIVSYYLRNFD-EDAGKEQCVFPLAEPQELFQASQMKFEDFQKDLRK
LKKDLKACEVEAGKVYQVSSEEHMQPFKEIMEQFISQAKIDQESQEAALTETHKCF----
-------LETTAYYFMKPKLGEKEVSPNVFFSVWHEFSSDFKDSWKKENKLILQ----ER
VKEAEEVCRQKKGKSLYKVKPRHDSGIINISMD---------------IC-------ERL
CPLLLGAGDGCTSS
ENSHGLP00000001950 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------HV
PF02181 // ENSGT00870000136417_1_Domain_1 (2531, 2682)
LLSYIVSYYLRNFE-EDAGKEQCVFPLPEPQEVFQASQMNFEDFQKDLRK
LKKDLKACETEAGKVYQVSSKEHMQPFKENMEQFIIQAKIDQEAKENSLTETHKCFL---
--------ETTAYFFMKPKIGEKEVSPNVFSSIWHEFSSDFKDFWKKENKLILQ----DR
VKEAEELCRQKKGKSLYKIKPRHDFGIKAKTSMKIQTGDRRMKMSHYNNF-------HKF
QLI-----------
ENSHGLP00000016533 (view gene)
ENSNGAP00000019809 (view gene)
ENSHGLP00100003278 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------------CSRAWAQF
SVPTIAEAFCHTLFHITFEI------------------------------------L
PF02181 // ENSGT00870000136417_1_Domain_1 (2578, 2682)
TRK
LKKDLKACETEAGKVYQVSSKEHMQPFKENMEQFIIQAKIDQEAEETSLTETHKCFL---
--------ETTAYFFMKPKIGEKEVSPNVFFSIWHEFSSDFKDFWKKENKLILQ----ER
VKEAEEVCRQKKGKSLYKIKPRHDSGIVSIQLFFIL------------------------
--------------
ENSTBEP00000014955 (view gene)
ENSSTOP00000016978 (view gene)
ENSPVAP00000004193 (view gene)
ENSTSYP00000030611 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------------------MS
KAVTIAEAFCHISF
PF02181 // ENSGT00870000136417_1_Domain_1 (2535, 2633)
HTIFEI---L-MRMLEEQCLFPLPEPQDLFQASQMKFEDFQKDLRK
LKKDLKACEVEAGKVYQVSSKEHMQPFKENMEQFIIQAKIDQEAEENSLTETHK------
------------------------------------------------------------
------------------------------------------------------------
--------------
ENSOPRP00000015241 (view gene)
ENSAMEP00000014813 (view gene)
ENSECAP00000022816 (view gene)
ENSHGLP00000020101 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------------------MS
KAVTIAEAFCHILFHIIFEI-LIR---------------TLEKN
PF02181 // ENSGT00870000136417_1_Domain_1 (2565, 2682)
SASQLKFEDFQKDLRK
LKKDLKASETEAGKVYQVSSKEHMQPFKENMEQFIIQTKIDQEAEETSLTETHKCFL---
--------ETTACFF-TPKIREKEVSPNVFFSIWHEFSSDFKDFWKKENKLILQ----ER
VKEAEEVCRQKKGKLLYKIKPRHDSRIKAKISMKT-------------------------
--------------
ENSRROP00000034007 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------DALGPRDVETTKKGSGGKKALGKHGKGGG--G-GGGESGKKKS-KADSRASVFS
-NLRIRKN-LSKGK-GAGGSREDVLD-SQALQTGELD-SAHSLL-TKTPDLSLSADETGL
SDTEC-ADPFEVTRPGGPGPAE-ARVGGRPVAEDVE-TAAGAQDGQRTSSGSDT-DIYSF
HSATEQEDLLSDIQQAIRLQQQQQQLQ-----------LQQQQQQQLQGAEEPAAPPTAV
SPQPGAFLGLDRFLLGPSGG---------------A-GEAPGSPD-TEQALSALSDLPES
LAAETRE-PQQPPSPGGLPAS--------E--------------------APSLPAAQPA
VTDSPSSTAFPFP---ETGPGAE----AAGAPVRGAGDTDEEGEEDAFEDAPRGSPGEEW
APE-------------VGEDA-PQRLEEEPEEEAQGPDSPA-AASLPGSPAPSQRCFKPY
PLITPCYIKTTTRQLSSPNHSPSQSPNQSPRIK----RRPEPSLSRGSRTALASVAA-PA
K---KHR-VDGGLAGGLSRSADWTEELGART---PRAGGSAHLLERGVPAD-SGG--GVS
PALAAK-AS-GAPAAADGFQNVFTGRTLLEKLFSQQENGPPEEAEKFCSRIIAMGLLLPF
SDCFREPCN-QNAQTNAASFDQDQL---YTWAAVSQPTHSLDYSEGQFPRRVPSMGPSSK
PP---DEEYRLEDAETE----SQSAVSETPKKRSDAV--QK-----EGVDMKSE-GQANV
IQQLEQTIEDLRTKIAELERQYPALDTEVASGRQGLGNGVTASGDVCLEALRLEEKEIRH
QRIL-----------EAKSIQT---SPTEEG-G---VLTLP-PVG---AL-GPPPC----
---------------------PP----------GAESGPQTKFCSEISLIVSPRRISVQL
DSHQPTQSISQPPPP--PSLLWSAGQGQPG----------S-Q-P-P-H-S-SL-S-T-E
-F-E-T-S-H--E-H-S-V-S-S-A-F-K-N-S-C-N-I-P-S-P-PP-L-P-CT-ES-S
-S-S-M-P-G-L-G-M-V-P-PP-P-P-PL-P-G-M-T-V-PT-L-P-S-T-A-I-P-P-
P-PP-L-P-G-TE-M-L-PP-P-P-P---P-L-P-G-V-G-I------------------
-----------------------------------------------------------P
PPPPLPGAGIP--SPVG-----I-------------------------------------
PP--------------------------------------------------LSPLPGVG
IP--PL-----GIPPSLPPVPRVGIPLPP-------------------------------
-----------------------PLPEAGI---PLPPQLPGVGIPP----LSPLLRAGIH
----PPPPLPGV-GTT---PPPLLPEA-G-IP-PLPPLPGTGIPP---------------
------------------------------------------------------------
----------------------------------------------------PPPLP---
------------------------------------------------------------
------------GMG--IPPAP-AP-----L-P----PPPGT--G-IPPP-----P----
----LLP--A-S-GPPLL-----------PQVGS----STLP------------------
T-P-QVC-G-FLP---P-PLPSGL--------------FG--------------------
--LGMNQDK---G-S-R-K--
PF02181 // ENSGT00870000136417_1_Domain_1 (2182, 2682)
-QPI-E-P-C-R-P-M-----KPLYWTR----I-Q-L-H
--SK-----------------------------------RDSSTS--------LIWEK--
-I---EEP-----------------SIDCHEFEELFSKTAVKERKKPISDTISKTKAK-Q
VVKLLSNKRSQAVGILMS-SLHLDMKDIQHAVVNLDNSVVDLETLQ-ALYENR-AQSDEL
EKIEKHGRSSKD--KENAKPWVGPFCLENANAVFLYELSLIPNFSERVFCILFQSTFSES
ICSIHRKLELLQKLCETLKNGSGVMQVLGLVLAFGNYMNGGNKTRGQADGFGLDILPKLK
DVKSSDNSRSLLSYIVSYYLRNFD-EDAGKEQCLFPLPEPQDLFQASQMKFEDFQKDLRK
LKKDLKACEVEAGKVYQVSSKEHMQPFKENMEQFIIQAKIDQEAEENSLTETHKCFL---
--------ETTAYFFMKPKLGEKEVSPNAFFSIWHEFSSDFKDFWKKENKLLLQ----ER
VKEAEEVCRQKKGKSLYKIKPRHDSGIKAKISMKT-------------------------
--------------
ENSPCOP00000013212 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------MGNQDGKLKRSAGDASHEGG--
---GAEDAVGPRDAEATKKGSGGKKALGKHGKGGGG---GGGEPGKKKS-KSDSRASVFS
-NLRIRKN-LSKGK-GSGGSREDVLD-TQALQTGELD-SAHSLL-TKTPDLSLSADETGL
SDTEC-ADPFEVTRVGGPGPAE-AGVGGRAVAEDLE-TAPGAQDGQRTSSGSDT-DIYSF
HSATEQEDLLSDIQQAIRLQQQQQQL-------------------LLQGSEEPAAPPTAT
SPQPGAFLGLDQFLRGPSGG---------------A-AEARDSVD-TEQALSALSDLPES
LVAEPSV-PEQPRTPGGLLSS--------E--------------------APGLPGAQPG
AEDSPSSTAFPFP---EAGPGES----AVGAPVQRAGDTDEECEEDAFEDASRGSPGEEW
GPE-------------VGEEA-AQRREGEPEEGARGPGTPT-AASLPGSPAPSPRCFKPY
PLITPCYIKTTTRQLSSPNHSPSQSPSQSPRIK----RRLEPSLSRGPRTALASAAA-PA
K---KHR-VEGGLAGGLSRSADWTEELGGRS---PQPGGSAHLLECGVATEGSGG--GVS
PASAAK-AS-GAPAAADGFQNVFTGRTLLEKLFSQQENGPPEEAEKFCSRIIAMGLLLPF
SDCFREPCN-QNAQSSSAPFDQDQL---YTWAAVSQPTHSLDYSEGQFPRRVPSAWPPSK
PP---EEEHRPMET------ESQSAVLETPKKGSDAI--QQ-----EVSDMKSE-GQATV
IQQLEQTIEDLRTKIAELEKQYPAMDMEVASGHQGVENGVAPSEDVCFEALRLGENDVRH
QRIL-----------QAKSIQT---SPTEEG-G---MLPP--------------------
-----------------------------------PPGNSTKFCSEISLIVSPRRISVQL
DAQQSIQGISQPPPP--PSLPWSDGQGQPGAR-----------PS---HPS--L-C---T
QF-D-T-S-H--E-H-S-V-S---SS----LENSY-N-I-----PSPPPL-P-CTESSSS
MPGL---------G-M-V-T-PPSA---PL---PGMTVPAIPGAAIP-------------
-PPPPL---PGID-M-LPPPP--------PPL----------------------------
------------------------------------------------------PGVGIP
PPPPLPGVGIP--PAPPLPG
PF06346 // ENSGT00870000136417_1_Domain_0 (1521, 1717)
SGI----PPPPPLPGVGIPPPPPLPGS-----------GI
P----------------PPPPLPGTGIPPPPPLPGV-GIPPAPPLPGMGIPLPPPLPGVR
IP--PPPPLPGVGIPPPPPLPGTGIPP---------------------------------
-----------------------------------PPPLPGVGIPP----PPPLPGMGIP
----PPPPLPGV-GIP---PPPPLPGV-G-IP-PPPPLPGTGIPPPPPLPGVGIPPPPPP
SRYGNT--------PSPSS-T-W-M-GIPPPP--------PL-P-G-MGILPAPGPP---
---------------------------------------LPPPGT---------------
------------------------------------------------------------
--------------------------------------------G-IPPP-----PPP-P
----LLP--G-S-GPPLPA-----------QVG-------------SSTL-S--------
T-P-QVC-G-FLP---P-PLPSGLFG----------------------------------
--LGMSQDK---A-S-R-K---QPI-E-P-C-R-P-M-----KPLYWTR----I-Q-L-H
--SR-----------------------------------RDSSAS--------LIWEK--
-I---EEP-----------------SIDCHEFEELFSKTAVKERKKPISDTISKTKAK-Q
VVKLLSNKRSQAVGILMS-SLHLDMKDIQHAVVNLDNSVVDLETLQ-ALYENR-AQSDEL
EKIEKHGRSSKD--KKNPKSLD-------KPEQFLYELSLIPNFSERVFCILFQSTFSES
ICSIRRKLELLQKLCETLKNGPGVMQVLGLVLAFGNYMNSGNNTRGQADGFGLDILPKLK
DVKSSVSILC-IIYLVSDYLYNFL-ITLIH----------------------------IW
LCFDMFTCEVE
PF02181 // ENSGT00870000136417_1_Domain_1 (2592, 2682)
AEKVYQVSSKEHMQPFKENMEQFIIQAKIDQEAEENSLTETHKCFL---
--------ETVAYFFMKPKLGEKEVSPNVFFSIWHEFSSDFKDFWKKENKLILQ----ER
VKEAEEVCRQKKGKSLYKIKPRHDSGIVSI------------------------------
--------------
ENSRBIP00000014255 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------MGNQDGKLKRSAGDALHEGGG-
G-GGAEDALGPRDVETTKKGSGGKKALGKHGKGGG--G-GGGESGKKKS-KADSRASVFS
-NLRIRKN-LSKGK-GAGGSREDVLD-SQALQTGELD-SAHSLL-TKTPDLSLSADETGL
SDTEC-ADPFEVTRPGGPGPAE-ARVGGRPVAEDVE-TAAGAQDGQRTSSGSDT-DIYSF
HSATEQEDLLSDIQQAIRLQQQQQQL-----------QLQQQQQQQLQGAEEPAAPPTAV
SPQPGAFLGLDRFLLGPSGG---------------A-GEAPGSPD-TEQALSALSDLPES
LAAETRE-PQQPPSPGGLPAS--------E--------------------APSLPAAQPT
VTDSPSSTAFPFP---ETGPGAE----AAGAPVRGAGDTDEEGEEDAFEDAPRGSPGEEW
APE-------------VGEDA-PQRLEEEPEEEAQGPDSPA-AASLPGSPAPSQRCFKPY
PLITPCYIKTTTRQLSSPNHSPSQSPNQSPRIK----RRPEPSLSRGSRTALASVAA-PA
K---KHR-VDGGLAGGLSRSADWTEELGART---PRAGGSAHLLERGVPAD-SGG--GVS
PALAAK-AS-GAPAAADGFQNVFTGRTLLEKLFSQQENGPPEEAEKFCSRIIAMGLLLPF
SDCFREPCN-QNAQTNAASFDQDQL---YTWAAVSQPTHSLDYSEGQFPRRVPSMGPSSK
PP---DEEYRLEDAETE----SQSAVSETPKKRSDAV--QK-----EGVDMKSE-GQANV
IQQLEQTIEDLRTKIAELERQYPALDTEVASGRQGLGNGVTASGDVCLEALRLEEKEIRH
QRIL-----------EAKSIQT---SPTEEG-G---VLTLP-PVG---AL-GPPPC----
---------------------PP----------GAESGPQTKFCSEISLIVSPRRISVQL
DSHQPTQSISQPPPP--PSLLWSAGQGQPG----------S-Q-P-P-H-S-SL-S-T-E
-F-E-T-S-H--E-H-S-V-S-S-A-F-K-N-S-C-N-I-P-S-P-PP-L-P-CT-ES-S
-S-S-M-P-G-L-G-M-V-P-PP-P-P-PL-P-G-M-P-V-PT-L-P-S-T-A-I-P-P-
P-PP-L-P-G-TE-M-L-PP-G-H-T---F----------------------S-A--P-L
--P-G---------------------------------------------------AGVP
PPPLYPERNTP--FPTPTVPSGI----PLRWPLYRESAY---------------------
PSAP--CY--PER-PYPLRPLYPERH------------TPSGPLYP-ESH----------
-N--SGPLYPGRHTPSAPSTPRA-------------------------------------
----------------------------AY---PLPPPSTGAGIPP----PHPLYRKV--
-------------G--------------H-TP-LAPPLPGRHTP------------SGPL
PESSIP------------------------SA--------PL-P-D-QHYPLRPPYLRS-
------------AYPS-----------VPYYPERAYPPR-PSTRNGITSSAPRPSTW---
------------------------------------------------------------
------------YWD--SPPPP-FS-----P-P----PPPGT--G-IPPP-----P----
----LLP--A-S-GPPLL-----------PQVGS----STLP------------------
T-P-QVC-G-FLP---P-PLPSGL--------------FG--------------------
--LGMNQDK---G-S-R-K--
PF02181 // ENSGT00870000136417_1_Domain_1 (2182, 2682)
-QPI-E-P-C-R-P-M-----KPLYWTR----I-Q-L-H
--SK-----------------------------------RDSSTS--------LIWEK--
-I---EEP-----------------SIDCHEFEELFSKTAVKERKKPISDTISKTKAK-Q
VVKLLSNKRSQAVGILMS-SLHLDMKDIQHAVVNLDNSVVDLETLQ-ALYENR-AQSDEL
EKIEKHGRSSKD--KENAKPWVGPFCLENANAVFLYELSLIPNFSERVFCILFQSTFSES
ICSIHRKLELLQKLCETLKNGSGVMQVLGLVLAFGNYMNGGNKTRGQADGFGLDILPKLK
DVKSSDNSRSLLSYIVSYYLRNFD-EDAGKEQCLFPLPEPQDLFQASQMKFEDFQKDLRK
LKKDLKACEVEAGKVYQVSSKEHMQPFKENMEQFIIQAKIDQEAEENSLTETHKCFL---
--------ETTAYFFMKPKLGEKEVSPNAFFSIWHEFSSDFKDFWKKENKLLLQ----ER
VKEAEEVCRQKKGKSLYKIKPRHDSGIKAKISMKT-------------------------
--------------
ENSCJAP00000062816 (view gene)
MQRQAGGCVCGRDTMAPLNWNWASVLPHVKISSFLFNFSRGRSSSSEDRSASSSRQALRP
RTMHSIRTVEIKVPEIEETFFAPR----FREEPRGGRGGGGGGRGSRPELRPAQPLAGA-
---ALCTEHVRAPPAPSRSR-SRSRSDGSHGSSRALCKAAAD--ANGASRARVGLPSQRL
APQPPDSPGRLPRPGPGAEEGRDGLTARGAAAQQRDCTMGNQDGKLKRSAGDASHEGGGG
--GGTEDALGPRDVEATKKGSGGRKALGKHGKGGGG---GGGESGKKKS-KSDSRASVFS
-NLRIRKN-LSKGK-GAGGSREDVLD-SQALQTGELD-SAHSLL-TKTPDLSLSADETGL
SDTEC-ADPFEVTRPGVPGPAE-ARVGGRPIAEDVE-TAAGAQDGQRTSSGSDT-DIYSF
HSATEQEDLLSDIQQAIRLQQQQQL-------------QLQLQPQQLQGAEEPAAPPSAV
SPQPGAFLGLDRFLLGPSGG---------------T-REAPGSPD-TEQALSALSDLPAS
LAAEPSA-PQQPPSPGSLPAS--------E--------------------APSLPAAQPA
PRDSPSSTAFPFP---EGGPGEE----AAGAPARGAGDTDEEGEEDAFEDAPRGSPGEEW
APE-------------GGENA-PQRLGEEPEEGAQGPGVPV-AASLPGSPAPSQRCFKPY
PLITPCYIKTTTRQLSSPNHSPSQSPNQSPRIK----RRPEASLSQGSRTALASAAA-PA
K---KHR-ADGGLAGGLSRSADWTEELGAST---PGAGGSAHLLERGVAAD-RGG--GVS
PALTAK-AS-GAPAAADGFQNVFTGRTLLEKLFSQQENGPPEEAEKFCSRIIAMGLLLPF
SDCFREPCN-QNAQTNAASFDQDQL---YTWAAVSQPTHSLDYSEGQFPRRVPSMWPPSK
PP---EEEHKLKDAEAEDDGESQSAVSETPKTGLDAV--QQ-----EVIDMKSE-GQATV
IQQLEQTIEDLRTKIAELERQYPALDAEVASGRQGLENGVTASGDVCLEALRLEEKEVRH
QRIL-----------EAKSIQT---SPTEEG-G---VLTLP-PVD---AQPGGPPC----
---------------------SPGAER----------GPQTKFCSEISLIVSPRRISVQL
DSHQSAQSISQPPPP--PSLLWSDGQGQPGSQ-----------PP---HSS--L-S---T
EF-E-T-S-H--E-H-S-V-S---SAF-K---NSS-N-I-----PSPPPL-P-CTESSGS
-----M-P-G-L-G-M-V-P-PPPP---PL-P-G-MTA---PT-L-P-S---T-G----I
PPPPPL---PGTE-M-LPPP----------------------------------------
------------------------------------------------------------
-----------------------------PPPLPGVGIPPPPPLPGT-----------GI
PPPP--PL--PGV-GIPPPPPLPGAGIPPPPPLPGA-GIPPPPPLPGAGIPPPPPLP---
----------GAGVPPPPPLPGMGIPPA--------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------------PA-----
---------------PP-------------------------------------------
------------------------------------------------------------
--------------------------------P----PPLGT--G-I--------PPP-P
----LLP--A-S-GPPLLP-----------QVG-------------SSTL-P--------
T-P-QVC-G-FLP---P-PLPTGLFG----------------------------------
--LGMNQDK---G-S-R-K--
PF02181 // ENSGT00870000136417_1_Domain_1 (2182, 2682)
-QPI-E-P-C-R-P-M-----KPLYWTR----I-Q-L-H
--SK-----------------------------------RDSSTS--------LIWEK--
-I---EEP-----------------SIDCHEFEELFSKTAVKERKKPISDTISKTKAK-Q
VVKLLSNKRSQAVGILMS-SLHLDMKDIQHAVVNLDNSVVDLETLQ-ALYENR-AQSDEL
EKIEKHSRSSKD--KENAKSLD-------KPEQFLYELSLIPNFSERVFCILFQSTFSES
ICSIHRKLELLQKLCETLKNGPGVMQVLGLVLAFGNYMNGGNKTRGQADGFGLDILPKLK
DVKSSDNSRSLLSYIVSYYLRNLD-EDAGKEQCLFPLPEPQDLFQASQMKFEDFQKDLRK
LKKDLKACEVEAGKVYQVSSKEHMQPFKENMEQFISQAKIDQEAEENSLTETHKCFL---
--------ETTAYFFMKPKLGEKEVSPNVFFSIWHEFSSDFKDFWKKENKLILQ----ER
VKEAEEVCRQKKGKSLYKIKPRHDSGIKAKISMKT-------------------------
--------------
ENSPTRP00000072717 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------------------MS
RAVTIAEAFCHILFR
PF02181 // ENSGT00870000136417_1_Domain_1 (2536, 2682)
IISEI---L-MRMLEEQCLFPLPEPQDLFQASQMKFEDFQKDLRK
LKKDLKACEVEAGKVYQVSSKEHMQPFKENMEQFIIQAKIDQEAEENSLTETHKCFL---
--------ETTAYFFMKPKLGEKEVSPNAFFSIWHEFSSDFKDFWKKENKLLLQ----ER
VKEAEEVCRQKKGKSLYKIKPRHDSGIKAKISMKT-------------------------
--------------
ENSOANP00000016311 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------M
PF02181 // ENSGT00870000136417_1_Domain_1 (2478, 2682)
LKNGSGVMQVLGLVLALGNYMNGGNKARGQADGFGLDILPKLK
DVKSSDNSRSLLSYIVSYYLRNFD-EDAGKEQCTFPLPEPQDLFQASQMKLEDFQKDLRK
LKKDLRACETEAGKVYRTSLEEHLQPFKGSMEQFLSQAKIDQEAEENSLSEAHKCFL---
--------ETAAYFYMKPKMGEKEVSPNVFFSLWHEFSSDFKDSWKRENKLILQ----ER
------------------------------------------------------------
--------------
ENSHGLP00100004027 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------------------MS
KAVTIAEAFCHILFHIIFEI-LIR---------------TLEKN
PF02181 // ENSGT00870000136417_1_Domain_1 (2565, 2682)
SASQLKFEDFQKDLRK
LKKDLKASETEAGKVYQVSSKEHMQPFKENMEQFIIQTKIDQEAEETSLTETHKCFL---
--------ETTACFF-TPKIREKEVSPNVFFSIWHEFSSDFKDFWKKENKLILQ----ER
VKEAEEVCRQKKGKLLYKIKPRHDSRIKAKIRMKT-------------------------
--------------
ENSPTRP00000003647 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------MGNQDGKLKRSAGDALHEGG--
--GGAEDALGPRDVEATKKGSGGKKALGKHGKGGGG--GGGGESGKKKS-KSDSRASVFS
-NLRIRKN-LSKGK-GAGGSREDVLD-SQALQTGELD-SAHSLL-TKTPDLSLSADEAGL
SDTEC-ADPFEVTRPGGPGPAE-ARVGGRPIAEDVE-TAAGAQDGQRTSSGSDT-DIYSF
HSATEQEDLLSDIQQAIRLQQQQQQQ---QLQLQLQ---QQQQQQQLQGAEEPAAPPTAI
SPQPGAFLGLDRFLLGPSGG---------------A-GEAPGSPD-TEQALSALSDLPES
LAAEPRE-PQQPPSPGGLPVS--------E--------------------APSLPAAQPA
AKDSPSSTAFPFP---EAGPGEE----AAGAPVRGAGDTDEEGEEDAFEDAPRGSPGEEW
APE-------------VGEDA-PQRLEEEPEEEAQGPDAPA-AASLPGSPAPSQRCFKPY
PLITPCYIKTTTRQLSSPNHSPSQSPNQSPRIK----RRPEPSLSRGSRTALASVAA-PA
K---KHR-ADGGLAAGLSRSADWTEELGART---PRAGGSAHLLERGVASD-SGG--GVS
PALAAK-AS-GAPAAADGFQNVFTGRTLLEKLFSQQENGPPEEAEKFCSRIIAMGLLLPF
SDCFREPCN-QNAQTNAASFDQDQL---YTWAAVSQPTHSLDYSEGQFPRRVPSMGPPSK
PP---DEEHRLEDAETE----SQSAVSETPQKRSDAV--QK-----EVVDMKSE-GQATV
IQQLEQTIEDLRTKIAELERQYPALDTEVASGRQGLENGVTASGDVCLEALRLEEKEVRH
HRIL-----------EAKSIQT---SPTEEG-G---VLTLP-PVD---GLPGRPPC----
---------------------PP----------GAESGPQTKFCSEISLIVSPRRISVQL
DSHQPTQSISQPPPP--PSLLWSAGQGQPG----------S-Q-L-P-H---SI-S-T-E
-F-Q-T-S-H--E-H-S-V-S-S-A-F-K-N-S-C-N-I-P-S-P-PP-L-P-CT-ES-S
-S-S-M-P-G-L-G-M-V-P-PP-P-P-PL-P-G-M-T-V
PF06346 // ENSGT00870000136417_1_Domain_0 (1361, 1733)
-PT-L-P-S-T-A-I-P-Q-
P-PP-L-Q-G-TE-M-L-PP-P-P-P---P-L-P-R------------------------
----A---------------------------------------------------AGIP
PPPPLPGAGIP--PPPPLPGAGI----PPPPPLPGAGIPPPPPLPG----------AAGI
PLLR--PL--SRS-GNTPPPPLGAA-------------I---------------------
---------------PLH------------------------------------------
-------------------------------------PLPGAAIP-----LHPTSRSGIP
----PPPPLPGV-GIP---PPPPLPGA-G-IP-PPPPLPGVGIPPAPA------------
------------------------------------------------------------
-------------------------------------PPLPLPGTG--------------
------------------------------------------------------------
----------------------------------------------IPPP-----P----
----LLP--A-S-GPPLLPQ-----------VGSS-------------TL-A--------
T-P-QVC-G-FLP---P-PLPSGL--------------FG--------------------
--LGMNQDK---G-S-R-K---QPI-E-P-C-R-P-M-----KPLYWTR----I-Q-L-H
--SK-----------------------------------SAAVCY--------LKLKE--
-I---FFL----------------KYMLCYLV--------------PCPGAVPATREA-E
AGEWREPGRRTEIA-----PRHSSLGD--RARVGLKNK----------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------
ENSODEP00000022020 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------------ITR
AGVTIAEACCHISFHIIFEI------------------------------------L
PF02181 // ENSGT00870000136417_1_Domain_1 (2578, 2682)
MRK
LKKDLKACETEAGKVYQVSSQEHMQPFKENMEQFIIQAKIDQEAEETSLTETHKCFL---
--------ETTAYFFMKPKLGEKEVSPNVFFSIWHEFSSDFKDFWKKENKLILQ----ER
VKEAEEVCRQKKGKSLYKIKPRHDSGIVSIILFSAWAFPWL-------------------
--------------
ENSP00000318884 (view gene)
ENSTTRP00000010223 (view gene)
ENSCPOP00000024740 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------------IFLPRTI
-----AEAFC--------H
PF02181 // ENSGT00870000136417_1_Domain_1 (2540, 2682)
ILFHI-SRNLMKQCVFPLPEPQDLFQASQMKFEDFQKDLRK
LKKDLKACETEAGKVYQVSSEEHMQPFKENMEQFIVQAKIDQEAEENSLTETHKCFL---
--------ETTAYFFMKPKLGEKEVSPNVFFSIWHEFSSDFKDFWKKENKLILQ----ER
VKEAEEVCRQKKGKSLYKIKPRHDSGIVSIYSNF--------------------------
--------------
ENSVPAP00000001647 (view gene)
ENSMODP00000008810 (view gene)
ENSMFAP00000045272 (view gene)
ENSFCAP00000020057 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------MGNQDGKLKRSAGDALHEGG--
--SSAEDAVGPRDVEATKKGSGGKKALGKHGKGGGGG--GGGEPGKKKS-KSESRASVFS
-NLRIRKN-LSKGK-GTSGSREDVLD-SQALQIGELD-SAHSLV-TKTPDLSLSADETGL
SDTEC-ADPFEVTRPSGSGPTD-VGVGGRVVLGDLE-AAAGVQDGQRTSSGSDT-DIYSF
HSATEQEDLLSDIQQAIRQQQQQQQQQQQQQQQQQ----QQQQLLLL------QAAPTAA
SPQPGAFLGLDHYLLGTSSA---------------A------------------------
--GEPRV-PEQPPPPGA---L--------A--------------------VPGLPVGQPA
AADSPSSPAFRFPESS--PAEEG----SARSPGPGVGDTDEEGEEDTFEDAPRDSPGEAW
GLE-------------AGEDP--PRPGAEPEGEPGRPGALA-AASLPGSPAPSPRCFKPY
PLITPCYIKTTTRQLSSPNHSPSQSPSQSPRIK----RRLESSLSRGPRAALVSAAA-PP
K---KHR-ADGGLTCGLSRSADWTEELGSRA---PPAGGSAHLPAREAASE-GGG--GVP
PGLAAK-V-PGAQAAADGFQNVFTGRTLLEKLFSQQENGPPEEAEKFCSRIIAMGLLLPF
SDCFREPCD-QNAQSSSAPFDQDQL---YTWAAVSQPTHSLDYTEGQFPRRVPPVWPPST
PP---DEEHRSKDVETE----SQSAVLETPKTCSDAV--QQ-----EAFDMKSE-GQATV
IQQLEQTIEDLRTKIAELEKGYPALDVEMARGA--------PLEDVCLEALRLGEKDVRH
QRIL-----------QAKSIQT---SPTEEG-G---IPILP-SVN---TLPECPHH----
---------------------PPGAESGDSPVPSSPSGPQTKFCSEISLIVSPRRISVQL
DTHQPTL-----PPP-SPSLLKSDSQRQSGSQ-----------PS---HPSLH------T
EFET---S-H--E-H--SV-F---SSFE----NSH-N-V-----QPSPPL--PCTESSSS
MPGVGMAA-------------PPPPLL--------------PGTTRP-------------
-------------------------------V----------------------------
------------------------------------------------------LPMGIP
PPPPLPGVGIP--PPPPLPGVGI----PPPPPLPGVGIPP--------------------
-----------------------------------------PPPLPGVGIPPPPLLPGEG
IP--PPPPLPGVGVPPPPPLPGMGIPPVPA------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------T-L----PTSGT--G-IPPP-----P----
----LLP--G-S-GPPLPP-----------QVG-------------SSTL-S--------
A-P-QVC-G-FLP---P-PLPAGLFG----------------------------------
--LGLNQDR---G-S-R-K--
PF02181 // ENSGT00870000136417_1_Domain_1 (2182, 2341)
-QPV-E-P-C-R-P-M-----KPLYWTR----I-Q-L-H
--SK-----------------------------------RDSSAS--------LIWEK--
-I---EEP-----------------SIDCHEFEELFSKTAMKERKKPISDTITKTKAKQ-VSICVFSEIGQYFGHKYELSLEKKNETVFWAV----------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------
ENSPCAP00000005171 (view gene)
ENSCATP00000045295 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------MGNQDGKLKRSAGDALHEGGG-
G-GGTEDALGPRDVETTKKGSGGKKALGKHGKGGG--G-GGGESGKKKS-KSDSRASVFS
-NLRIRKN-LSKGK-GAGGSREDVLD-SQALQTGELD-SAHSLL-TKTPDLSLSADETGL
SDTEC-ADPFEVTRPGGPGPAE-ARVRGRPVTEDVE-TAAGAQDGQRTSSGSDT-DIYSF
HSATEQEDLLSDIQQAIRLQQQQQQQ------LQLQLQQQQQQQQQLQGAEEPAAPPTAV
SPQPGAFLGLDRFLLGPSGG---------------A-GEARGSPD-TEQALSALSDLPES
LAAEPRE-PQQPPSPGGLPAS--------E--------------------APSLPAAQPA
VTDSPSSTAFPFP---ETGPGEE----AAGAPVLGAGDTDEEGEEDAFEDAPRGSPGEEW
APE-------------VGEDA-PQRLKEETEEEAQGPDSPA-AASLPGSPAPSQRCFKPY
PLITPCYIKTTTRQLSSPNHSPSQSPNQSPRIK----RRPEPSLSRGSRTALASVAA-PA
K---KHR-ADGGLAGGLSRSAYWTEELGART---PRAGGSAHLLERGVAAD-SGG--GVS
PALAAK-AS-GAPAAADGFQNVFTGRTLLEKLFSQQENGPPEEAEKFCSRIIAMGLLLPF
SDCFREPCN-QNAQTNAASFDQDQL---YTWAAVSQPTHSLDYSEGQFPRRVPSMGPPSK
PP---DEEHRLGDAETE----SQSAVSETPKKRSDAV--QK-----EVVDMKSE-GQANV
IQQLEQTIEDLRTKIAELERQYPALDTEVASGRQGLGNGVTASGDVCLEALRLEEKEGRH
QRIL-----------EAKSIQT---SPTEEG-G---VLTLP-PVG---AL-GHPPC----
---------------------PP----------GAESGPQTKFCSEISLIVSPRRISVQL
DSHQPTQSISQPPPP--PSLLWSAGQGQPG----------S-Q-P-P-H-S-SL-S-T-E
-F-E-T-S-H--E-H-S-V-S-S-A-F-K-N-S-C-N-I-P-S-P-PP-L-P-CT-ES-S
-S-S-M-P-G-L-G-M-V-P-PP-P-P-PL-P-G-M-T-V-PT-L-P-S-T-A-I-P-P-
P-PP-L-P-G-TE-M-L-PP-P-P-R---L-L-P--------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------GIPP---PPPLP---
------------------------------------------------------------
------------GMG--IPPAP-AP-----P-P----PPPGT--G-IPPP-----P----
----LLP--A-S-GPPLL-----------PQVGS----STLP------------------
T-P-QVC-G-FLP---P-PLPSGL--------------FG--------------------
--LGMNQDK---G-S-R-K--
PF02181 // ENSGT00870000136417_1_Domain_1 (2182, 2682)
-QPI-E-P-C-R-P-M-----KPLYWTR----I-Q-L-H
--SK-----------------------------------RDSSTS--------LIWEK--
-I---EEP-----------------SIDCHEFEELFSKTAVKERKKPISDTISKTKAK-Q
VVKLLSNKRSQAVGILMS-SLHLDMKDIQHAVVNLDNSVVDLETLQ-ALYENR-AQSDEL
EKIEKHGRSSKD--KENAKSLDK-------PEQFLYELSLIPNFSERVFCILFQSTFSES
ICSIHRKLELLQKLCETLKNGPGVMQVLGLVLAFGNYMNGGNKTRGQADGFGLDILPKLK
DVKSSDNSRSLLSYIVSYYLRNFD-EDAGKEQCLFPLPEPQDLFQASQMKFEDFQKDLRK
LKKDLKACEVEAGKVYQVSSKEHMQPFKENMEQFIIQAKIDQEAEENSLTETHKCFL---
--------ETTAYFFMKPKLGEKEVSPNAFFSIWHEFSSDFKDFWKKENKLLLQ----ER
VKEAEEVCRQKKGKSLYKIKPRHDSGIKAKISMKT-------------------------
--------------
ENSCCAP00000028876 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------MGNQDGKLKRSAGDASHEGG--
G-GGTEDALGPRDVEATKKGSGGKKALGKHGKGGGGG---GGESGKKKS-KSDSKASVFS
-NLRIRKN-LSKGK-GAGGSREDVLD-SQALQTGELD-SAHSLL-TKTPDLSLSADETGL
SDTEC-ADPFEVTRPGVPGPAE-ARVGGRPIAEDVE-TAAGAQDGQRTSSGSDT-DIYSF
HSATEQEDLLSDIQQAIRLQQQQQLQ-------------LQLQPQQLQEAEEPAAPPSAV
SPQPGAFLGLDRFLLGPSGV---------------A-REATGSPD-TEQALSALSDLPAS
LAAESPA-PQQPPSPGSLPAS--------E--------------------APSLPAAQPA
PRDSPSSTAFPFP---EAGPGEE----AAGAPARGAGDTDEEDEEDAFEDAPRGSPGEEW
APE-------------VGEDA-PQRLGEEPEEGAQGPGAPA-AASLPGSPAPSQRCFKPY
PLITPCYIKTTTRQLSSPNHSPSQSPNQSPRIK----RRPEASQSQGSRTALASAAA-PA
K---KHR-VDGGLAGGLSRSADWTEELGART---PGAGGSAHLLERGVAAD-RGG--GVS
PALAAK-AS-GAPAAADGFQNVFTGRTLLEKLFSQQENGPPEEAEKFCSRIIAMGLLLPF
SDCFREPCN-QNAQTNAASFDQDQL---YTWAAVSQPTHSLDYSEGQFPRRVPSMWPPSK
PP---DEEHELKDAEAE----SQSAVSETPKKRLDAV--QQ-----EVIDMKSE-GQATV
IQQLEQTIEDLRTKIAELERQYPAPDTEGASGRQGLENGVTASGDVCLEALRLEEKEVRH
QRIL-----------EAKSIQT---SPTEEG-G---VLTLP-PVD---AQPGGPPC----
---------------------SP----------GAERGPQTKFCSEISLIVSPRRISVQL
NSHQSAQSTSQPPQP--PSLLWSDGQGQPG----------S-Q-P-P-H-S-SL-S-T-E
-F-E-T-S-H--E-H-S-V-S-S-A-F-K-N-S-S-N-I-P-S-P-PP-L-P-CT-ES-S
-G-S-M-A-G-L-G-M-V-P-PA
PF06346 // ENSGT00870000136417_1_Domain_0 (1344, 1596)
-P-P-PL-P-G-M-T-A-PT-L-P-S-T-G-I-P-P-
P-PP-L-P-G-TE-M-L-PP-P-P-P---P-L-P-G-V-G-I--PP------P-P--P-L
--P-G---------------------------------------------------VGIP
PPPPLPGMGIP--PPPPLPGAGL----PPPPPLPGAGI----------------------
PPPP--PL--PGM-GIPPPP-----------PLPGV-GIPPPPPLPG-GIPPPPPLPGAG
IP--PPPPLPGTGIPPPPPLPGAGIPPPP-------------------------------
-----------------------------------PPPLPGAGIPP----PP--------
----PAPPLPGV-GIP---PPPPLPGT-G-IP-PPPPLPGMGIPPAPA------------
--PPPP----------------------------------PL-G-T--------------
---------------------------------------------GI---PPPPLLPA--
------------------------------------------------------------
---------------------S--GPP---------------------------------
----LLP--Q---------------------VGS----STLP----T-------------
P---QVC-G-FLP---P-PLP----T-----G-----FFG--------------------
--LGMNQDR---G-S-R-K---QPI-E-P-C-R-P-M-----KPLYWTR----I-Q-L-H
--SK-----------------------------------RDSGTS--------LIWEK--
-I---EEP-----------------SIDCHEFEELFSKTAVKERKKPISDTISKTKAK-Q
VVKLLSNKRSQAVGILMS-SLHLDMKDIQHAVVNLDNSVVDLETLQ-ALYEN--------
------------------------------------------------------------
----------------
PF02181 // ENSGT00870000136417_1_Domain_1 (2477, 2682)
TLKNGPGVMQVLGLVLAFGNYMNGGNKTRGQADGFGLDILPKLK
DVKSSDNSRSLLSYIVSYYLRNFD-EDAGKEQCLFPLPEPQDLFQASQMKFEDFQKDLRK
LKKDLKACEVEAGKVYQVSSKEHMQPFKENMEQFISQAKIDQEAEENSLTETHKCFL---
--------ETTAYFFMKPKLGEKEVSPNVFFSIWHEFSSDFKDFWKKENKLILQ----ER
VKEAEEVCRQKKGKSLYKIKPRHDSGIKAKISMKT-------------------------
--------------
ENSACAP00000003593 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------MGNQDGKLKKNAGDAH---DGG
-----EDASGPRDADGTKKGAGSKKGLGKHSKGN--------ESGKKKG-KSESRGSVFS
-NLRIRKN-LSKAKDGNSSSREDVLE-SQASQTGELD-SAHSIL-TKTPDISISADEGGL
SDTDV-E-HFEHTN------------------ETRP-AAAEPQDGQRTSSGSDT-DIYSF
HSATEQEDLLSDIQQAIRLQLQQQQGAINS--------------GTEQ--HGTKTMGLHM
ANSQAAPFDFERFLSLSS-D----------------KDSSPEM----EQPVSAKSEITDY
IPASD-IESGVKEVTNGIGHSCSSVFDVQEERR---------------------------
-------SVFNFP---SPIGESE---TEVSAFEFSGSEREEEDIANTSHNQVEGAKERK-
-RS-------------ISAS--REALDDGFSAT-IKNGTLS-SNDVSSSPMHSSKGLKPF
PPINPCYIKTTTRQLSSPNHSPCLSPSQSPLFK----RRRESTQRKENVPTKKQR-----
---------ASSLAGPFSRSADWTVTLSTQTPERGRQMTRLARAAQAS----ASK-GVPP
QPALPA--PEGVPAEYGEFSDVFLGRTLLEKLSCQQENAPQEEAEKICSWIIAMGLLLPF
SDCFREPCGGQSAQQSAPTFDQDQL---YTWAAVNQPTHSSDYIEGRFPGRIPSVWPPPK
PP---DEEQRIKHKEPDP--TLS-----PLKGIQSCLIGIQ-----EDVIIKSE-ERASE
IQRLEQTIQDLKIKIAELEKQFPASEVLVSSSNQ-RESGQTTCEELFSAGLEEQELQTLP
KTI------------TAKSVQT---SPTDDC-L---IHTMP-SAS---TLPQAPP-----
--------------------LLKGNVSQGPTQLLPTLEPQTTFCSELSLVLSPKQISLQL
DDVQEKLTGLQ--ISGPDANFPPLLQEQ---P-----------VS---STALS------T
ESFT--CS----E-A--SPS--------LSSHPVLES---------HPGL--SDTGVSPS
LPGAGVPF---------V--MP-----------------PSPS-----------------
-----------SG-VPLPLAPFSS---------G--H-----------------------
--------------------------------------------ALSSLPAPLLPSGGIP
PPPPLPGMDMAPPPPPPLPAAVMPTEIPPPPPLPDMGVP---LPTS--------------
--------------------------------------------LPGATVPLAPSLPGMS
MP-PAPPPLPDMEVPQPPPLPWTGFSSPHL------------------------------
------------------------LPGTGIPP-PPPPPPPGIGVSL---HPPHLPGMGIP
P---APPPLPGM-GVPLPPPPPPLPGMW--GPPPPPPLPGMGGPPP--------------
---------P--PPP-PPL-P-G-M-GGPPPPP---P-PPPL-P-G-MG-GPPPP-----
-----------------PPLPGM-------------------------------------
------------------------------------------------------------
--------------D--G-P-P---P-P--P-P--L-P--G-A-G--P-P--P--P-P-P
--P-L-P--G-M-G--V-P-P--P-P-P----------------I--S-L-P--H--G--
-----S--G-F---L-P-P--P-L--P--S-G--L--F----------------------
-VMGMNQ-E-K-G-S-R-K--
PF02181 // ENSGT00870000136417_1_Domain_1 (2182, 2616)
-HAI-E-P-S-R-P-M-K-P---LYWTR----I-Q-L-H
--N-----------------------------------KRDSSAS--------LVWEK--
-I---EEP-----------------SIDYHEFEELFSKTAVKERKKPISDTITKTKNK-Q
VVKLLSNKRSQAVGILMS-SLHLDMKDIQHAVVNLDNSVVDLETLQ-ALYENR-AQSDEL
EKIEKHGKSSKE--KENAKSLD-------KPEQFLYELSLIPNFSERVFCILFQSTFSES
ISSIRSKLELLQKLCETLKSGSGVMRVLGLVLAFGNYMNGGNRTRGQADGFGLDILPKLK
DVKSSDNSRSLLSYIVSYYLRNFD-EDAGKEQCIFPLPEPQDLFQASQLKFEEFQKDLRK
LKKDLKACETEAGKVCQFSLEEHIQPFKDNMEKFISQ-----------------------
------------------------------------------------------------
------------------------------------------------------------
--------------
ENSSARP00000011242 (view gene)
ENSOCUP00000003468 (view gene)
ENSOGAP00000000107 (view gene)
ENSAPLP00000014376 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------MGNQDGKLRKNTSDVH---EGG
-----EDASGPKDTDGTKKVSGSRRGLGKHAKGS--------ESAKKKS-KSESRASVFS
-NLRIRKN-LSKAKDGNSSSREDVLE-SQALQTGELD-STHSIV-TKTPDISISADEGGL
SDTDA-E-HFEIRN------------------ETVP-AAAETQDGQRTSSGSDT-DIYSF
HSAAEQEDLLSDIQQAIRLQQ--QQGAINV--------------GTED--LVTKTMGRHM
ANSQAAPLDFERFLSITTTD----------------KESSPDT----VKAFPTASEIAEN
VPVSCATERELKEAVNGTGSPENA------------------------------------
--SETHSFTLRSP---TSVADSETHIAEVQAIDLQGSGT-EDGISDAGHDHAAETQEGK-
-HT-------------LSDS--QENLHNGLSTE-IKDGILS-SEGTAGSPMLGSRCFKPY
-LINPCYIKTTTRQLSSPNHSPSPSPSQSPLFT----RRQESFHKKEKVPIKKQR-----
---------SASLAGLFSRSADWTEEVSNHKYEITQKVGSAGYLEHKGFREKFQT-ERVP
LTHSRK--SSGVQASAETFPNVFSGRTLLEKLFSQQENAPQEEAEKLCSRIIAMGLLLPF
TDCFREPCN-QSAQQSTPTFDQDQL---YTWAAVSQPTHSSDYAEGSFPRRIPSVWPPPK
PP---DEEQGLKITETDS--ELKSTTLETEKKFIEDVQ--Q-----DDFEVKSE-ERASV
IQQLEQTIEDLRTKIAELEKQFPATEVQPAGREQENEHAHTVCREHSIVNLQTNREEYVP
RTVS-----------QARSVQT---SPTEDY-L---THTMP-SAN---ALPQPPSP----
-------------------LHLEGGK---------TLELQPTFCSEISLILSPKLISVPL
DGSQSVQTTSQSPLPGPKVNLSFAFEGETKFS-----------SA---HQPLS------T
GNVT--CS----E-H--TLS--------LPPEPTCSI---------HPSL--SDTELAAP
VTGVHGPS---------I--TA-----------------LMPVAE--------------V
PPPPPL---PGAG-LPPSLPGPAM---------P--H-----------------------
--------------------------------------------LDHSLPGIVIPPPQLL
P------------PPSPLPSGEIA-ACPPAPPLPCTGA----------------------
--------------------------------------------LPGAGVPLPPPSWGIL
PPPPPPPPLPGTAIPPQ-PPPL--------------------------------------
-------------------------PGLGMSTLPTAPPLPGAGVPL---LPPPLPRGDIP
AP----PPLPGA-GAPLPPPPPPLPGAGAPLPPPPPPLPGAGAPLPPP--------PPPL
PGAGA--PLP--PPP-PPL-P-G-A-GAPLP-P---P-PPPL-P-G-AG-IAPPP-----
---------------PPPPLP---------------------------------------
------------------------------------------------------------
------G-A--G--I--A-P-P---P-P--P-P--L-P--G-L-G--M-P--PL-A-P-P
--P-L-P--G-E-A--P-P-L--P-P-P-V--Q-G--I-A--Y-P--S-V-P--Q--V--
-----G--G-F---L-P-P--P-L--P--S-G--L--F----------------------
-AMGMNQ-E-K-G-S-R-K--
PF02181 // ENSGT00870000136417_1_Domain_1 (2182, 2682)
-HVI-E-P-S-R-P-M-K-P---LYWTR----I-Q-L-H
--S-----------------------------------KRDSSAS--------LVWEK--
-I---EEP-----------------SIDYHEFEELFSKTAVKERKKPISDTITKTKTKQQ
VVKLLSNKRSQAVGILMS-SLHLDMRDIQHAVVNLDNSVVDLETLQ-ALYENR-AQSDEL
EKIEKHSKASKE--KENAKSLD-------KPEQFLYELSLIPNFSERVFCILFQSTFSES
ICSIHRKLELLQKLCETLKNGSGVMQVLGLVLAFGNYMNGGNRTRGQADGFGLDILPKLK
DVKSSDNSRSLLSYIVSYYLRNFD-EDAGKEQCIFPLPEPQDLFQASQMKFEDFQKDLRK
MKKDLRVCETEAAKVYQLSLEEHLQPFKDSMEQFISQAKLDQENEEKSLTEAHKSFL---
--------ETAAYFCMKPKMGEKEVSPHSFFSIWHEFSSDFKDFWKKENKLILQ----ER
VKEAEEVCRQKKGKSLYNIRPKHDSGIKAKISMKI-------------------------
--------------
ENSMUSP00000030039 (view gene)
ENSLAFP00000000880 (view gene)
ENSGGOP00000001003 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------MGNQDGKLKRSAGDALHEGG--
--GGAEDALGPRDVEATKKGSGGKKALGKHGKGGGG-G-GGGESGKKKS-KSDSRASVFS
-NLRIRKN-LSKGK-GAGGSREDVLD-SQALQTGELD-SAHSLL-TKTPDLSLSADEAGL
SDTEC-ADPFEVTRPGGPGPAE-ARVGGRPIAEDVE-TAAGAQDGQRTSSGSDT-DIYSF
HSATEQEDLLLDIQQAIRLQQQQQQQL----QLQLQQ-QQQQQQQQLQGAEEPAAPPTAI
SPQPGAFLGLDRFLLGPSGG---------------A-GEAPGSPD-TEQALSALSDLPDS
LAAEPRE-PQQPPSPGGLPVS--------E--------------------APSLPAAQPA
AKDSPSSTAFPFP---EAGPGEE----AAGAPVRGAGDTDEEGEEDAFEDAPRGSPGEEW
APE-------------VGEDA-PQRLGEEPEEEAQGPDAPA-AASLPGSPAPSQRCFKPY
PLITPCYIKTTTRQLSSPNHSPSQSPNQSPRIK----RRPEPSLSRGSRTALASVAA-PA
K---KHR-ADGGLAAGLSRSADWTEELGART---PRAGGSAHLLERGVASD-SGG--GVS
PALAAK-AS-GAPAAADGFQNVFTGRTLLEKLFSQQENGPPEEAEKFCSRIIAMGLLLPF
SDCFREPCN-QNAQTNAASFDQDQL---YTWAAVSQPTHSLDYSEGQFPRRVPSMGPPSK
RP---DEEHRLEDAETE----SQSAVSETPKKRSDAV--QK-----EVVDMKSE-GQATV
IQQLEQTIEDLRTKIAELERQYPALDTEVASGRQGLENGVTASGDVCLEALRLEEKEVRH
HRIL-----------EAKSIQT---SPTEEG-G---VLTLP-PVD---GLPGRPPY----
---------------------PP----------GAESGPQTKFCSEISLIVSPRRISVQL
DSHQPTQSISQPPPP--PSLLWSAGQGQPG----------S-Q-P-P-H-S--L-S-T-E
-F-Q-T-S-H--E-H-S-V-S-S-A-F-K-N-S-C-N-I-P-S-P-PP-L-P-CT-ES-S
-S-S-M-P-G-L-G-M-V-P-PP-P-P-PL-P-G-M-T-V-PT-L-P-S-T-A-I-P-Q-
P-PP-L-P-G-TE-M-L-PP-P-P-P---P-L-P-G-A-G-I--P------PP-P--P-L
--P-G---------------------------------------------------AGIL
PLP---GAGIP--P----------------------------------------------
--PP--PL--PGA-GIPPPP-----------------------PL-----------PGA-
------------AIP---------------------------------------------
----------------------------------PPP-----------------------
------------------------------------------------------------
-----------------------------------------P-L-G-AGIPPPPPLPGA-
------------GIPPPPPLPGVGIPPPPPLPGVGIPPPPPLPGAGIPP---PPPLP---
------------------------------------------------------------
------------GMG--IPPAP-AP-----P-L----PPPGT--G-IPPP-----P----
----LLP--A-S-GPPLL-----------PQVGS----STLP------------------
T-P-QVC-G-FLP---P-PLPSGL--------------FG--------------------
--LGMNQDK---G-S-R-K--
PF02181 // ENSGT00870000136417_1_Domain_1 (2182, 2682)
-QPI-E-P-C-R-P-M-----KPLYWTR----I-Q-L-H
--SK-----------------------------------RDSSTS--------LIWEK--
-I---EEP-----------------SIDCHEFEELFSKTAVKERKKPISDTISKTKAK-Q
VVKLLSNKRSQAVGILMS-SLHLDMKDIQHAVVNLDNSVVDLETLQ-ALYENR-AQSDEL
EKIEKHGRSSKD--KENAKSLDK-------PEQFLYELSLIPNFSERVFCILFQSTFSES
ICSIRRKLELLQKLCETLKNGPGVMQVLGLVLAFGNYMNGGNKTRGQADGFGLDILPKLK
DVKSSDNSRSLLSYIVSYYLRNFD-EDAGKEQCLFPLPEPQDLFQASQMKFEDFQKDLRK
LKKDLKACEVEAGKVYQVSSKEHMQPFKENMEQFIIQAKIDQEAEENSLTETHKCFL---
--------ETTAYFFMKPKLGEKEVSPNAFFSIWHEFSSDFKDFWKKENKLLLQ----ER
VKEAEEVCRQKKGKSLYKIKPRHDSGIKAKISMKT-------------------------
--------------
ENSCAFP00000038900 (view gene)
ENSNLEP00000004370 (view gene)
ENSANAP00000019455 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------MGNQDGKLKRSAGDASHEGG--
G--GTEDALGPRDVEATKKGSGGKKALGKHGKGGGGG---GGESGKKKS-KSDSRASVFS
-NLRIRKN-LSKGK-GAGGSREDVVD-SQALQTGELD-SAHSLL-TKTPDLSLSADETGL
SDTEC-ADPFEVTRPGVPGPAK-ARVGGRPIAEDVE-TAAGAQNGQRTSSGSDT-DIYSF
HSATEQEDLLSDIQQAIRLQQQQQLQ-------------LQLQPQQLQGAEEPAAPPSAV
SPQPGAFLGLDRFLLGPSGG---------------A-REAPGSPD-TEQALSALSDLPAS
LASEPPA-PQQPPSPGSLPAS--------E--------------------APSLPAAQPA
PRDSPSSTAFPFP---EAGPGEE----AARAPARGAGDTDEEGEEDAFEDAPRGSPGEEW
APE-------------VGEDA-PQRLGEEPEEGAQGPGVPA-AASLPGSPALSQRCFKPY
PLITPCYIKTTTRQLSSPNHSPSQSPNQSPRIK----RRPEASLSQGSRTALASAAA-PA
K---KHR-ADGGLAGGLSRSADWTEELGART---PGAGGSAHLLERGVAAD-RGG--GVS
PALAAK-AS-GAPAAADGFQNVFTGRTLLEKLFSQQENGPPEEAEKFCSRIIAMGLLLPF
SDCFREPCN-QNAQTNAASFDQDQL---YTWAAVSQPTHSLDYSEGQFPRRVPSMWPPSK
AP---DEEHKLKDAEAE----SQSAVSETPKKRLDAV--QQ-----EVIDMKSE-GQATV
IQQLEQTIEDLRTKIAELERQYPALDTEGASGRQGLENGVTASGDVCLEALRLEEKEVRH
QRIL-----------EAKSIQT---SPTEEG-G---VRTLP-PVD---AQPGGPPC----
---------------------SP----------GAERGPQAKFCSEISLIVSPRRISVQL
DSHQSAQSISQPPPP--PSLLWSDGQGQPG----------S-Q-L-P-H-S-SL-A-T-E
-F-E-T-S-H--E-H-S-V-S-S-A-F-K-N-S-----I-P-S-P-PP-L-P-CT-ES-S
-G-S-M-P-G-L-G-M-V-P-PP-S-P-PL-P-G-M-T-A-PT-L-P-S-T-G-I-P-P-
P-PP-L-P-G-TE-M-L-PP-P-P-P---P-L-P-G-V-G-I--PP------P-P--P-L
--P-G---------------------------------------------------VGIP
PPPPLPGMGLP--PPPPLPGVGI----PPPPPLPGMGI----------------------
PPPP--PL--PGV-GILPSH-----------FLEWA--YPSS-PLQEWNTSSA-------
-------SSTRSGNTPSTP-----------------------------------------
------------------------STRSGN---TSPSPTSRRNTTPSAHIPTQSGTGNTL
----PPPPLPGA-GIP---PPPPLPGA-G-IP-PPPPLPGMGIPPAPA------------
--PPPP----------------------------------PL-G-T--------------
---------------------------------------------GI---PPPPLLPAS-
------------------------------------------------------------
------------------------GPP---------------------------------
----LLP--Q---------------------VGS----STLP----T-------------
--P-QVC-G-FLP---P-PLPTGL--------------FG--------------------
--FGMNQDK---G-S-R-K--
PF02181 // ENSGT00870000136417_1_Domain_1 (2182, 2682)
-QPI-E-P-C-R-P-M-----KPLYWTR----I-Q-L-H
--SK-----------------------------------RDSGTS--------LIWEK--
-I---EEP-----------------SIDCHEFEELFSKTAVKERKKPISDTISKTKAK-Q
VVKLLSNKRSQAVGILMS-SLHLDMKDIQHAVVNLDNSVVDLETLQ-ALYENR-AQSDEL
EKIEKHGRSSKD--KENAKLRVGPFCLENANAVFLYELSLIPNFSERVFCILFQSTFSES
ICSIRRKLELLQKLCETLKNGPGVMQVLGLVLAFGNYMNGGNKTRGQADGFGLDILPKLK
DVKSSDNSRSLLSYIVSYYLRNFD-EDAGKEQCLFPLPEPQDLFQASQMKFEDFQKDLRK
LKKDLKACEVEAGKVYQVSSKEHMQPFKENMEQFISQAKIDQEAEENSLTETHKCFL---
--------ETTAYFFMKPKLGEKEVSPNVFFSIWHEFSSDFKDFWKKENKLILQ----ER
VKEAEEVCRQKKGKSLYKIKPRHDSGIKAKISMKT-------------------------
--------------
ENSGALP00000050709 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------MGNQDGKLKKNAGDVH---EGG
-----EDASGPKDTDGTKKVSGSKKGLGKHAKGS--------ESGKKKG-KSDSRASVFS
-NLRIRKN-LSKAKDGNSSSREDVLE-SQALPAGELD-STHSII-TKTPDISISADEGGL
SDTDA-E-HFEIRN------------------ETVP-VAAETQDGQRTSSGSDT-DIYSF
HSATEQEDLLSDIQQAIRLQQ-QQQGAINI--------------GTED--LVTKTMGRHM
ANSQAAPLDFERFLSVTTAD----------------KESSPDA----VEAFPAVSEIAED
VPVSCATERERKEAVNGTGSPGNGLFEPQERKQLPEEQLTAGVDAVACELEGDLNRTEIE
NGSEARSSTIHPP---TSAADSEAKIAEVQAVDFQGSVT-EDGASDVGRDYAAGTQEGK-
-RT-------------PSAS--QEDLHNGLSME-MKNGILP-SEGTAGSPMFGSRCFKPY
-LINPCYIKTTTRQLSSPNHSPSPSPSQSPLFT----RRQESFHKKEKVPIKKQR-----
---------SASLAGLFSRSADWTEEVSKHKSEITQKVGSAGYLEHKGFSEKFQT-ERVP
LTHSRK--SSGVQASTEAFPNVFSGRTLLEKLFSQQENAPQEEAEKLCSRIIALGLLLPF
TDCFREPCN-QSAQQSTPTFDQDQL---YTWAAVSQPTHSSDYIEGSFPRRIPSAWPPPK
PP---DEEQRLKIPDAES--ESKSTVLEAEKEHTEDVQQVQ-----DDLEVKSE-ERASV
IQKLEQTIEDLRTKIAELEKQFPATEVQATEKEQENEHVHTACREFSNVTLQTHGKETVP
RTVS-----------QARSVQT---SPTEDY-S---VHAMP-SAN---TLPLPPSP----
-------------------LHLESSKSEEPSEKLPALELQPTFCSEISLILSPKLISVPL
DASQSVQTASQSPLPGPKANLSLAFEGETEFS-----------AP---HQLIG------I
RNVT--CS----E-H--SLS--------LPPEPTCSI---------PSSL--STTAHTAP
VPGVHGSS---------V--TA-----------------PMPVAG--------------V
PLPPPL---PGSG-LPLPFAGPAM---------P--H-----------------------
--------------------------------------------LDHSLPDVTTAPQQLL
PPPP------PPPPPPPLPSGEIP-VLPPAPSLPCTGVPPPPLPPP--------------
--------------------------------------------LPGAGVSLP-PGWSIP
---PPPPPLPAAGVPPP-LLPL--------------------------------------
-------------------------PGLGMP--PTAPPLSGVGEPP---LPPPLPGGAIP
S---------------FPPPPPPLPGAGVSSPQPP-------------------------
------------PPP-PPL-P-G-A-G--VP-P---P-PPPL-P-G-AG-VP--P-----
---------------PPPPLP---------------------------------------
------------------------------------------------------------
------G-A--G--V----P-P---P-P--P-P--L-P--G-W-G--V-P--PA-A-P-P
--P-L-P--G-E-T--A-P-L--P-P-P-A--Q-G--S-A--Y-T--A-V-P--Q--V--
-----G--G-F---L-P-P--P-L--P--S-G--L--F----------------------
-AMGMNQ-E-K-G-S-R-K--
PF02181 // ENSGT00870000136417_1_Domain_1 (2182, 2682)
-HVI-E-P-S-R-P-M-K-P---LYWTR----I-Q-L-H
--S-----------------------------------KRDSSAS--------LVWEK--
-I---EEP-----------------SIDYHEFEELFSKTAVKERKKPISDTITKTKTK-Q
VVKLLSNKRSQAVGILMS-SLHLDMRDIQHAVVNLDNSVVDLETLQ-ALYENR-AQSDEL
EKIEKHSKASKE--KENAKSLD-------KPEQFLYELSLIPNFSERVFCILFQSTFSES
ICSIHRKLELLQKLCETLKNGSGVMQVLGLVLAFGNYMNGGNRTRGQADGFGLDILPKLK
DVKSSDNSRSLLSYIVSYYLRNFD-EDAGKEQCIFPLPEPQDLFQASQLKFEDFQKDLRK
MKKDLRVCETEAAKVYQLSLEEHLQPFKDSMEQFISQAKIDQENEEKSLTEAHKSFL---
--------ETAAYFCMKPKMGEKEVSPHSFFNIWHEFSSDFKDFWKKENKLILQ----ER
VKEAEEVCRQKKGKSLYNIRPKHDSGIKAKISMKI-------------------------
--------------
ENSHGLP00000014867 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------ES-KAVKIAECVFPL
PF02181 // ENSGT00870000136417_1_Domain_1 (2558, 2682)
PEPQELFQASQMKFEDFQKDLRK
LKKDLKACETEAGKVYQVSSKEHTQAFKENMEQFIIQVKIDQEPEETSLTETHKCFS---
--------ETTAYFFMKPKIGEKEVSPDAFFSIWHEFSSDFKDFWKKENKLILQ----ER
IKEAEQVCRQKKGKSLYKIKPRHDSRIKAKISMKT-------------------------
--------------
ENSTGUP00000010919 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------PPPPPLPGAAVPPPPP--------P--L
PSLGF--A----PPP-PLL-P-G-A-G--LP-P---P-PPPL-P-G-GG-IPPPP-----
---------------P--------------------------------------------
------------------------------------------------------------
-------------------------P-P--P-P--L-P--G-A-A--V-P--PP-S-P-P
--P-P----------------------------------P--L-P--A-F-P--Q--G--
-----C--G-F---L-P-P--P-L--P--S-G--L--F----------------------
-AMGMNQ-E-K-G-S-R-K--
PF02181 // ENSGT00870000136417_1_Domain_1 (2182, 2614)
-HVI-E-P-T-R-P-M-K-P---LYWTR----I-Q-L-H
--S-----------------------------------KTDSSAS--------LVWEK--
-I---EEP-----------------SIDYHEFEELFSKTAVKERKKPISDTITKRKTK-Q
VVKLLSNKRSQAVGILMS-SLHLDMKDIQRAVVNLDNSVVDLETLQ-ALYENR-AQSDEL
EKIEKHSKASKE--KENAKSLD-------KPEQFLYELSLIPNFSERVFCILFQSTFSES
IGSIHRKLELLQKLSANLVFVEKIMRVLGLVLAFGNYMNGGNRTRGQADGFGLDILPKLK
DVKSSDNSRSLLSYIVSYYLRNFD-EDAGREQCIFPLPDPQDLFQASQLKFDDFQKDLRK
MKKDLRACETEAAKVYQLSLEEHLQPFKDNMEQF--------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------
ENSBTAP00000056407 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------AQSVGL
GKIEKHDRS
PF02181 // ENSGT00870000136417_1_Domain_1 (2410, 2682)
SKD--KENAKSLD-------KPEQFLYELSLIPNFSERVFCILFQSTFSES
ICSIRCKLELLQKLCETLRHDSGVMQVWGLVLAFGNYMNGGNNTQGQADGFGLDILLKLK
DVKSRDNSRSLLSYIVSYDLRNFD-EDAGKEQCVFPLPKPQDLFQASQMKFEHFQKDLRK
LKKDLRACEVEAGKVYQVPSKEHIQPFKENMEQFIIQAKIDQKAEENSLTETHKCFL---
--------ETTAYFSMRPKIGEKEVSPNVFFGIWHEFSSHFKDFWKNENKLILCVFSPSR
VKEAEEVCRQKKGKSLYKIKPRHDSGIKAKKSMKT-------------------------
--------------
ENSMNEP00000008627 (view gene)
ENSDORP00000026256 (view gene)
ENSJJAP00000020675 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------MGNQDGKLKRSAGDAPPEGG--
---GAEDAAGPRDAETTKKGSASKKALGKHHGKGTG--AGGGESGKKKS-KSDSRASVFS
-NLRIRKN-LSKGK-GAGDSREDALD-SQALQVGELD-SAHSVV-TKTPDLSLSAEETGL
SDTEC-ADPFEVTHAGGPRPAE-AGVRVQAVAEDLE-TAAGTQDGQRTSSGSDT-DIYSF
HSATEQEDLLSDIQQAIRLQQQKLQL--------------------QDSGEEPAAPPAAT
SPQPGAFLGLDQFLLGSSGG---------------APDPG----LDTGQAPSTRVDLPET
TP--KPPAPERPPSPAGRLAS--------E--------------------APS------A
VTDSLSCAAFAFP---DAGAGEG----AAG--VPGDGDTDEEVEEDAFEDATRGSPGEEW
GPE-------------VGDAP--RRREEEAQERTRGHEVPA-VTSLPGSPAPSPRCFKPY
PLITPCYIKTTTRQLSSPNHSPSASPNQSPRIK----KRPEPSASRSPRTASPPAAA-QA
G---KQR-PEGGLAAGLSRSADWTEELGART---PGPGGSAHLLGSGEAADG--G--GGS
PALATK-AS-GAPAAADGFQNVFTGRTLLEKLFSQQENGPPEEAEKLCSRIIAMGLLLPF
GDCFREPRQ-RGAGSRSAPFDQDQL---YTWAAVSQPTHSMDYNEGQFPRRAPSVWPLSR
HP---DEEHSPKDVETE----LQASALESTEKGSSAI--LQ-----DVSDVKSE-GQATV
IQQLEQTIEDLRTKIAALEKQYPALDREGASCSQGSDNRRRGLDDVTLESLRGGGKDKLP
ERIL-----------EAKSIQT---SPTEEG-R---SLTPS-PLN---ALPEGLPC----
---------------------LPSTESGEPVVPPSPSGPQTKFCSEISLIVSPRRISVQL
DSHQSKQSISQPPPP--PFLPGSDGQGKPGLQ-----------PS---YPS--I-H---T
EFET---S-H---VH--TV---S-SS----FENSCNI-------PLTPPL--PCTESSAP
MPGLGME-------------TPPPP---CLS---DKTVPDLPSTTV--------------
----------------LPPP-PQP---------------G--------------------
------------------------------------------------------SEMLPP
SPPPLPELGIS--SPPPFPGVGI----PPPPPLPGMGILPPPP-----------------
-----------------PPPPLPGMGI-----------------------PPPPPLPGMG
IP--PPPPLPGVGILPPPPLPGMGI-----------------------------------
-----------------------------------------------------------P
----PPPPLPGM-GIP---PPPPLPGV-G-IP-PPPPLPGVGXXPPPPLPGVGIPPPPPL
PGLGIP--------PPPPL-P-G-M-GVPPAP----------------------G-----
------------------------------------------------------------
------------------------------------------------------------
---------------------------P--S-P----PLPGT--G-IPPP-----PPP-P
----LPP--G-S-GPSLSE------------AG-------------SGTF----------
TAP-QVC-G-FTI---P-PLPVGLFG----------------------------------
--LGINQDR---G-A-R-K--
PF02181 // ENSGT00870000136417_1_Domain_1 (2182, 2369)
-QPI-E-P-C-R-P-M-----KPLYWTR----I-Q-L-H
--SK-----------------------------------RDSSTS--------LIWEK--
-I---EEP-----------------SIDCHEFEELFSKTAVKERKKPISDTISKTKSK-Q
VVKLLSNKRSQAVGILMS-SLHLDMKDIQH------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------
ENSMLEP00000032851 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------------------MS
RAVTIAEAFCHILFR
PF02181 // ENSGT00870000136417_1_Domain_1 (2536, 2682)
IISEI---L-MRMLEEQCLFPLPEPQDLFQASQMKFEDFQKDLRK
LKKDLKACEVEAGKVYQVSSKEHMQPFKENMEQFIIQAKIDQEAEENSLTETHKCFL---
--------ETTAYFFMKPKLGEKEVSPNAFFSIWHEFSSDFKDFWKKENKLLLQ----ER
VKEAEEVCRQKKGKSLYKIKPRHDSGIKAKISMKT-------------------------
--------------
ENSEEUP00000014216 (view gene)
ENSCHIP00000032612 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------KGSFPRESQLLGHHLNLLMKNT
G------------------LRM-----LTQEVCGMKSEEP------SIGSSKTHP
PF02181 // ENSGT00870000136417_1_Domain_1 (2576, 2682)
KDLRK
LKKDLRACEVEAGKVY--------QPFKENMEHFLIQAKIDQEADENSLTETPKCFL---
--------ETTAYFFMRPKIGKKEVSPNVFFSIWHEFSSDFKDFGKKENKLILQ----ER
VKEAEEVCRQKKGKSLYKIKLRHDSGIKAKISMKT-------------------------
--------------
ENSMPUP00000015581 (view gene)
ENSSHAP00000015245 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------------------F
YVSLLINQRNQSPSRLAE-YLLVDWIIILTLCIRENVNVIDFISLQKGDHIKR-A
PF02181 // ENSGT00870000136417_1_Domain_1 (2396, 2589)
QTDEL
EKIEKHGRSCKE--KENAKSLD-------KPEQFLYELSLIPNFSERVFCILFQSTFSES
ICSIRRKLELLQKLCETLKNGSGVMQVLGLVLAFGNYMNGGNKTRGQADGFGLDILPKLK
DVKSSDNSRSLLSYIVSYYLRNFD-QDAGKEQCVFPLPEPQDLFQASQMKFEDFQKDLRK
LKKDLRGNRIQCSPIIHLYN----------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------
ENSJJAP00000016692 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------------VSLPR--
---TTAEAFCR-------ILFLII-CDAG
PF02181 // ENSGT00870000136417_1_Domain_1 (2550, 2682)
KEQCVFPLPEPQELFQATQMKFEDFQKDLRK
LKKDLKACEVEAGKVYQVSSEEHMQPFRENMEQFISQAKIDQESQETALTETHKCFL---
--------ETTAYYFMKPKLGEKEVSPNVFFSIWHEFSSDFKDFWKKENKLILQ----ER
VKEAEEVCRQKKGKSLYKIKPRHDSGIVIALISMLGI-----------------------
--------------
ENSXETP00000035438 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------MGNQDAKLKKCSGDVQEGAE--
--------AGPKDGENTKK--GGKK-----------------TLGKKRS-KSESRSSMFS
-NLRIRKT-LSKARDGACGSKEDVLGS-QALHTEELD-SACSVL-TKTPDISISADEAGL
SDTD--ADHFEIPHDVPVAA-----------------AKCDAQEGQRTSSGSDT-DIYSF
HSATEQDDLLSDIQQAIRLQQATCDSDTTN--------------LLSKVMGG------TL
TNSHTLFLDAAEHLVAPEKENVQLAGPCPPAPIAQGQQGLTGTGD-TGLGLTEVSDPTQ-
---EPPLDEQAT-----------AQIHIQ-------------V--------STPV-----
TSKE----------------GEN------------AAGLQSDTDNETI--RLEGSNGDLA
VHSIFYESCQSKDTADMSIH--NQRLNGSTDLADCAM---N-STPTTPQLATNHSGVKPY
PPINPCYIKTTTRQLSSPNHSPFASPSHSPQLY----RKQGQTFKHES------------
A---IRKKRSCSLVGNISRSADWTEELIKDQNKPLRKGNSSDFLLFGANGKGAESL---P
S--DSR--KSSIQASASSFPNVFTGRTLLEKLFNQQKNA-PEEAEKLCSQIIAVGLLLPF
SDCFREQCS-KSAPQIPSTFDQDQL---YTWAAVSQPTHSLDYLEGRFPRRIQAAWPPVT
KATVREEEHQSHDSEVEN----KAKEESVPKKCKKRL--DQ-----KEIDVENKSRKVEI
IQQLEQTIEDLRTKLAEREKQNGQANISAWEQI-KPSTE---PSTEVEDEWHISLKK---
-------------CVEGKSVQT---SPV-EELQE-----VP-SLSI---LAQLDSAARN-
-------------------KYM----KQLPS--SLELPSHLTYCSELSVVLSPKPNSQQI
DSNIFTDILASALQTGSVSLPMSDSSDH-KHI----------------------------
-------------------------------------------------L----------
-------------------------------------------SR--------------I
PTPPPL---PFVS--------INN---------L--E-----------------------
--------------------------------------------N----------ASQML
PDKH--------------------------------------------D-----------
-LGP-----------------CTT-HIPPPPP-P----APPSSFIPGIGVPPPPPLPSNL
Y-FDSQHPLPNG----------I-------------------------------------
-------------------------SILN---APPPPP-------P---PPPLPPDIGMA
----SAA---GF-PVP---PPP-----------APPLPPGMGVSPSLPSSIVGI------
---G-S-R-P--T-S-P-S-M-G-N-Y-RPP-P-P-P-PP-L-P-V-AF-L-G-S-----
---------------S-PS-P-L-------------------A----------CS-G--P
-A-PP-P-------------------------------L---------------------
---------P-G-SV-P-I-----------------------------------------
------P-P-----R-F-L------A-S-HV-Q-G-S----------V-L-P-S-F-GC-
A-P-A-L-G-C-L-T-P-PL-P-------A-G-L---F-A-F------------------
G-L-N-Q-D-K-G-Y-R-K
PF02181 // ENSGT00870000136417_1_Domain_1 (2180, 2682)
-A-P-I-E-P-S-K-P-M-K-P-L-Y-W-T-R--I-E-L-H
-G-K----------------------------------R-D-S-N-I-P-L--V-W-E-A
-V-S-E-P-K--V--------------DFHELENLFSKTAVKERKKPISDTITKTKAK-Q
VVKLLSNKRSQAVGILMS-SLHLDMKDIQHAVLKMDYSVVDLETLQ-ALFENR-AVPEEQ
EKIDKHVKASKT--KDQSKPLD-------KPEQFLYELTTIPNFTERVFCILSHSTISES
TSSLRRKLELLQKVCETLREGP-VLRVLGLVLAFGNYMNGGNRTRGQADGFALDILPKLK
DVKSNDNSRNLLSYIVSYYLRHFD-ENAGKEDCVFTLPEPQDLFQTSQMKFEDFQKDLRK
TKKDFQACETEADKVYQKSLEEHLQPFKDNMEVFLSKAKIELEDTEKFLTEVHSRFL---
--------ETTSYFCVKPKIGEKEVSPNSFFSIWHEFTTDFKDFWKKENKLILQ----ER
LKEAEEVYKQKKEKTSIIVKPKHESGIKAKLSLRS-------------------------
--------------
ENSSSCP00000049388 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------------------VS
LPRTIAEAFCHISCPIIFFL-QRV-SHNLFFICVFPL
PF02181 // ENSGT00870000136417_1_Domain_1 (2558, 2682)
PEPQDLFQASQMKFEDFQKDLRK
LKKDLRACEVEAGKVYQVSSQEHIQPFKENMEQFIIQAKIDQEAEENSLTETHKCFL---
--------ETTAYFFMRPKIGEKEVSPNVFFSIWHEFSSDFKDFWKKENKLILQ----ER
VKEAEEVCRQKKGKSLYKIKPRHDSGIKAKISMKT-------------------------
--------------
MGP_CAROLIEiJ_P0021418 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------C-G-FLF---P-PLPTGLFG----------------------------------
--LGMNQDR---V-A-R-K--
PF02181 // ENSGT00870000136417_1_Domain_1 (2182, 2395)
-QPI-E-P-C-R-P-M-----KPLYWTR----I-Q-L-H
--SK-----------------------------------RDSSPS--------LIWEK--
-I---EEP-----------------SIDCHEFEELFSKTAVKERKKPISDTISKTKAK-Q
----------------------------------------------------R-AQSDEL
EKIEKHSRSSKD--KENAKSLD-------KPEQFLYELSLIPNFSERVFC----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------
MGP_PahariEiJ_P0076664 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------MGNQDGKLKRSAGDAFHEGG--
---GAEDAAGPRDTETTKKASGSKKALGKHGRGGGG----SGETSKKKS-KSDSRASVFS
-NLRIRKN-LTKGK-GACDSREDVLD-SQALPIGELD-SAHSIV-TKTPDLSLSAEETGL
SDTEC-ADPFEVIHPGASRPAE-AGVGIQATAEDLE-TAAGAQDGQRTSSGSDT-DIYSF
HSATEQEDLLSDIQQAIRLQQQQQQQQQQKL-------------LL-QDSEEPAAPPTAI
SPQPGAFLGLDQFLLGPRSE---------------A-EK-----D-TVQARPVRPDLPET
---KSPV-PEHPPSSGSHLAS--------E--------------------TPGYATAPSA
VTDSLSSPAFTFP---EAGPGDG----ATGVPVAGTGDTDEECEEDAFEDAPRGSPGEEW
VPE-------------VEEA--SQRMEEEPEEGMREPITSA-VASLPGSPAPSPRCFKPY
PLITPCYIKTTTRQLSSPNHSPSQSPNQSPRIK----KRPDPSVSRSPRTALASAAA-PA
K---KHR-LEGGLTGGLSRSADWTEELGVRT---PGAGGSVHLLGRGATAD-DSG--GGS
PVLAAK-AP-GAPATADGFQNVFTGRTLLEKLFSQQENGPPEEAEKFCSRIIAMGLLLPF
SDCFREPCN-QNSGSSSAPFDQDQL---YTWAAVSQPTHSMDYSEGQFPRREPSMWPSSK
LP---DEEPSSKDIDT----EPQSSILESPKKCSDGI--QQ-----EVFDVKSE-GQATV
IQQLEQTIEDLRTKIAELEKQYPSLDLEGPRGLSGLENGLTATADVSLDALGLHGKVAQP
PRTL-----------EAKSIQT---SPTE-------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--EGMNQDR---V-A-R-K--
PF02181 // ENSGT00870000136417_1_Domain_1 (2182, 2682)
-QPI-E-P-C-R-P-M-----KPLYWTR----I-Q-L-H
--SK-----------------------------------RDSSPS--------LIWEK--
-I---EEP-----------------SIDCHEFEELFSKTAVKERKKPISDTISKTKAK-Q
VVKLLSNKRSQAVGILMS-SLHLDMKDIQHAVVNLDNSVVDLETLQ-ALYENR-AQSDEL
EKIEKHSRSSKD--KENAKSLD-------KPEQFLYELSLIPNFSERVFCILFQSTFSES
ICSIRRKLELLQNLCETLKNGPGVMQVLGLVLAFGNYMNAGNKTRGQADGFGLDILPKLK
DVKSSDNSRSLLSYIVSYYLRNFD-EDAGKEQCVFPLAEPQELFQASQMKFEDFQKDLRK
LKKDLKACEVEAGKVYQVSSEEHMQPFKENMEQFISQAKIDQESQETALTETHKCF----
-------LETTAYYFMKPKLGEKEVSPNVFFSVWHEFSSDFKDAWKKENKLILQ----ER
VKEAEEVCRQKKGKSLYKVKPRHDSGIKAKISM---------------KT----------
--------------
ENSCGRP00001005121 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------MGNQDGKLKRSAGDASHEGGG-
--GGAEDAAGPRDAETTKKASGSKKALGKHGKGGGG---S-GETSKKKS-KSDSRASVFS
-NLRIRKN-LTKGK-GAGDSREDVLD-SQALPIGELD-SAHSIV-TKTPDLSLSAEETGL
SDTEC-ADPFEVIHPGGPRPAE-AGVGVQATAEDLG-TAAGAQDGQRTSSGSDT-DIYSF
HSATEQEDLLSDIQQQQ-----------------------QQQKLLLQDSEEPAAPPTAI
SPQPGAFLGLDQFLLGPSSE---------------AAKD-------TEQAQPVRPDLPET
T---KSPVPEHPPSSGAHLAS--------E--------------------TPGYATAHSA
VTDSLSSPAFTFP---EAGPGEV----AAGTPVPEAGDTDEECGEDAFEDAPRGSPEEEW
VPE-------------VEEA--PQRLEEEPEEGMRRPSVTA-VASLPGSPAPSPRCFKPY
PLITPCYIKTTTRQLSSPNHSPSQSPNQSPRIK----KRPEPSVSRSPRTALASAAA-PA
K---KHR-LEGSLAGGLSRSADWTEELGVHT---PGAGGSVHLLGRGSTAD-DSG--GGS
PVLASK-AP-GASATADGFQNVFTGRTLLEKLFSQQENGPPEEAEKFCSRIIAMGLLLPF
SDCFREPCN-QNAGSSSAPFDQDQL---YTWAAVSQPTHSMDYSEGQFPRREPSMWPPSR
LP---DEEHSPKDVDT----EPQSSILESPKKCSNGV--QQ-----EVFDVKSE-GQATV
IQQLEQTIEDLRTKIAELEKQYPALDLEGPRGLPGLENGFTASEEVGLDALRLHGRVAQP
PRTL-----------EAKSIQT---SPTEEG-G---ILTLA-PLK---ALPGGPPC----
---------------------PPAAQSGESAALASPAGPQTKFCSEISLIVSPRRISVQL
DAQPALQSASQLQPP--P-----DGQGQ---------------PS---CPS--L-H---P
EL-E-T-S-P--E-H-S-V-F---SSF-E---NSC-N-V-----PPAPPL-P-CTESSSL
-----M-P-D-L-G-M-A-I-PPPP---CL-S-D-VTV---PA-L-P-S---T-T----A
PQPTSL---QGTE-M-LPAP--------PP------------------------------
------------------------------------------------------------
-----------------------------PPPLPGLGLPPP-------------------
--PP--PL--PGV-GIPPPPPLPGVGIPPPPPLPGV-GIPPPPP----------------
--------LPGVAIPPPPPLPGMGAPPA--------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------P----PPPGA--G-I--------PPP-P
----LLP--G-S-GPSLSS-----------QAG-------------SSTL-P--------
A-P-PVC-G-FLF---P-PLPTGIFG----------------------------------
--LGMNQER---G-A-R-K--
PF02181 // ENSGT00870000136417_1_Domain_1 (2182, 2682)
-QPI-E-P-C-R-P-M-----KPLYWTR----I-Q-L-H
--SK-----------------------------------RDSSPS--------LIWEK--
-I---EEP-----------------SIDCHEFEELFSKTAVKERKKPISDTISKTKAK-Q
VVKLLSNKRSQAVGILMS-SLHLDMKDIQHAVVNLDNSVVDLETLQ-ALYENR-AQSDEL
EKIEKHSRSSKD--KENAKSLD-------KPEQFLYELSLIPNFSERVFCILFQSTFSES
ICSIRRKLELLQKLCETLKNGPGVMQVLGLVLAFGNYMNAGNKTRGQADGFGLDILPKLK
DVKSSDNSRSLLSYIVSYYLRNFD-EDAGKEQCVFPLAEPQELFQASQMKFEDFQKDLRK
LKKDLKACEVEAGKVYQVSSEEHMQPFKENMEQFISQAKIDQESQEAALTETHKCFL---
--------ETTAYYFMKPKLGEKEVSPNVFFSVWHEFSSDFKDSWKKENKLILQ----ER
VKEAEEVCRQKKGKSLYKVKPRHDSGIKAKISMKT-------------------------
--------------
ENSETEP00000010185 (view gene)
------------------------------------------------------------
--MHSIHTVETEVPQIEETCLLPL----FSVEPLGGSGEGDPPPGRRLGL-PAQPLAGGG
RGAWRSTRHVRAVPAAGSSR-SR---R----KGGSLWKAKPVAAENKAIRAPGGLGSRRL
T-QHSNLLQSLLPPKSGGKEGHDGLTSGGA-ARRPACNMGNQDGKLKRSTGDASHEGAGA
S----EDALGPTDVEAARKGSGVKKALSKHGKGGSGG--GSGEPGKKKS-KSDSRASVFS
-NLRIRKN-LSRA-KSTAGSRGDVLD-GQALQTEELD-SAQSLV-TKTPDLSLSADETGL
S-DFECVDPFEVTGAGRPGPAEAEIGGQ-AVAEDVVSAA-GAQDGQRTSSGSDT-DIYSF
HSATEQEDLLSDIQQAIRLQQQQQQQQQQQ--------------KQ--GSGETTVPPTDT
PPQPGASLSLDQFLLGFSVR----------------TEEALGNDN-SEQTLHTPSYLPEN
C--EPPLASSASP-------------HSE-------------G--------P--------
LPVTQSVGAFPFP---ETQVGEV----VTKSLVLEAVDTDEDGEEDAFDDARQGSPGENG
GQE-------------MGEA--PQGLGQEAE---GQVPTPE-TASLPGSPAPSPRCFKPY
RMITPCYIKTTTRQLSSPNHSPSQSPSQSPRIQ----RRLESSLIRGQRSALATTAP-QG
K---KHQ-VDGGLGAGLSLSADWTDELSTC---EPKDGDSADLLERGTISDGVSHSSRVS
LALASK--SSGSQAAADGFQNVFTGRTLLEKLFSQQENGPPEEAEKFCSRIIAMGLLLPF
SDCFREPCD-QNIQSSSAPFDQDQL---YTWAAVSQPTHSLDYAEEQFPRRIPSAWPLSK
LP---DEDKRPQDAEIES----QSAVLETPEKCLDVL--QQ-----DIVDMKSD-GQANV
IQRLEQTIEDLRSKIAELEKQYPAVDMEVAIHRHGAVNEGVAAEDVCLEALRFKEKDTSH
HRILE-----------AKSIQT---SPTGES-G----------------LE---------
---------------------TVTSAECLPPAPGPESGLQTTFCSEISLIVSPRLIAMQL
DTPQSMPSISQP--PPPPSCLSSDAQGQP-------------------------------
------------------------------------------------------------
------------------------------------------------------------
-GSQPS---HG-------------------------------------------------
-----------------------------------------------YLSSDFETSHGDT
V------------FSVSENSCSI----PPPPPLPC-------------------------
-TMPS------------SSLPGLSLMMPPLPPFPAMAV----PVLHSTVIPPPPPLPGTE
MLPPSPPPLPGAGMYLLPLMPLI-----P-------------------------------
-----------------------PVSIT----------------EA---PPLFPPAVGIP
----PTPPLAGL-GMT---XXXXXXXXX--XXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX----
---------------XXXXXXXX----------------X--X---------XXXXXXXX
XXXXXXXX------------------------------XX--------------------
---------XXXXXXXXXXXXXXXXXXX--XXX-XX--XXXXXXXXXXXXXXX-XXXXXX
----X-XXXXXXXXXXXXX-----XXXXXXXXXXXXXX---------XXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXX------XXXXXX--XXXXXX-----------------
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXX---------------------------------XXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
PF02181 // ENSGT00870000136417_1_Domain_1 (2327, 2682)
XXXXXXXXXXXX-X
VVKLLSNKRSQAVGILMS-SLHLDMKDIQHXXXXXXXXXXXXX-XXXXXXXXR-AQSDEL
EKIEKHGRSSKD--KENTKSLD-------KPEQFLYELSLIPNFSERVFCILFQSTFSES
ICSIHRKLELLQKLCETLKNGPGVMKILGLVLAFGNYMNGGNKTRGQADGFGLDILPKLK
DVKSSDNSRSLLSYIVSYFLRNFD-EDAGKEQCVFPLPEPQDLFQASQMKFEDFQKDLRK
LKKDLKVCEE-AGKVYQXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXFL---
--------ETTAYFFMKPKIGEKEVSPNVFFSIWHEFSSDFKDFWKKENKLILQ----ER
VIEAGRPGRQKKGXXXXXXXXXXXXXXKAKISMKT-------------------------
--------------
ENSRNOP00000070887 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------MGNQDGKLKRSAGDASHEGGG-
--GGAEDAAGPRDAETTKKASGSKKALGKHGKGGGG----SGETSKKKS-KSDSRASVFS
-NLRIRKN-LGKGK-GAGDSREDVLD-SQALPIGELD-SAHSIV-TKTPDLSLSAEETGL
SDTEC-ADPFEVIHPGGSRPAE-AGVGIQAIAEDLE-TAAGAQDGQRTSSGSDT-DIYSF
HSATEQEDLLSDIQQAIRLQQQQQQQ----------------QKLLLQDSEEPAAPPTAI
SPQPGAFLGLDQFLLGTSSE---------------AKKD-------TVQAPPVRPDLPET
---TKSLVPEHPPSSGSHLAS--------E--------------------TPGYATAHSA
VTDTLSSPAFTFPEVPEAGPGEG----AAGVPPAGTGDTDEECEEDAFEDAPRGSPGEEW
APE-------------VEEAS--QRLEEEPEEGMRGPIIPA-VASLPGSPAPSPRCFKPY
PLITPCYIKTTTRQLSSPNHSPSQSPNQSPRIK----KRPDPSVSRSPRTALASAAT-PA
K---KHR-LEGGLVGGLSRSADWTEELGART---PGAGGSVHLLGRGATAD-DSG--GGS
PVLAAK-AP-GAPATADGFQNVFTGRTLLEKLFSQQENGPPEEAEKFCSRIIAMGLLLPF
SDCFREPCN-QNAGSSSAPFDQDQL---YTWAAVSQPTHSMDYSEGQFPRREPSTWRPSK
LP---DEEPSPKDIDTE----PQSSILESPKKCSNGV--QQ-----EVFDVKSE-GQATV
IQQLEQTIEDLRTKIAALEKQYPSLDLEG----PRLENGLTASADVSLDALGLHGKVAHP
PRTL-----------EAKSIQT---SPTEED-R---ILTLP-PPK---APPEGLPG----
---------------------PPAAESGESALLTSSSGPQTKFCSEISLIVSPRRISVQL
DAQPAMQSTSQLPPP--PSLLGADSQGQPSQP------------S--LHPE---------
-L-E-T-S-H--E-H-S-V-F---SSFEN-------N---P--DPAPPS---ALYESSSF
MPGLAQ---------------PIPHLP-CLSD---Y------------------------
------------N-M-LPLPSPTA-----PALST------------YQ------------
----------------------------------------------------IRYFCPVP
PLPPPPGVGIP--SPTSLPGMGI----PPLLPSFSLG-------------------MWNS
LPSPSSPLESVRNTGLPHPSLFPGNRSP-PTPLPRSREIPRPPPLHFPGAM---------
EY-----------ILPPFSLPS--------------------------------------
----------------------GYSPI---------------------------------
-----------I-PLP---SPSSLPGV-G-TT-THT------------------------
---------------VLNF-P-G-M-GIPPPS--------LY-L-E-IVTYP--------
-------------------------------------PPLCSYPW---------RLPFLP
------------------------------------------------------------
---------P-SLVADQVFVG---STPA--P-L-----PPGA--G-NLP-----------
SILPQLP--G-S-GLPLSS-----------QDG-------------SSTL-P--------
AAP-QGC-G-FLF---P-PLPTGLFG----------------------------------
--LGMNQDR---V-A-R-K--
PF02181 // ENSGT00870000136417_1_Domain_1 (2182, 2339)
-QPI-E-P-C-R-P-M-----KPLYWTR----I-Q-L-H
--SK-----------------------------------RDSSPS--------LIWEK--
-I---EEP-----------------SIDCHEFEELFSKTAVKERKKPISDTISKTKAKQ-
V-----------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------
ENSFALP00000013764 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--MS------PLPPPPHLPSGEIP-TLPPAPLLPCPGFPPPPLPPP--------------
--------------------------------------------LPGAGVPSPLPGWGIP
PP-PPPPPLPGAGVPPP-PPPL--------------------------------------
-------------------------PGLGLP--PPAPPLPGAGLPP---PPPPLPGGGIP
PPPPPPPPLPGV-AVPPPPPPPPLPGVAVPPPPPPPPLPGVAVPPPPP--------PPPL
PGVAV--PPP--PPP-PPL-P-G-V-AVPPP-P---P-PPPL-P-G-VA-VPPPP-----
---------------PPPPLPGV-----------AVPPPP--PLPGVA------------
--------------------------------------V---P---P---P--P--P---
L--P--G-A--G--V----P-P---P-P--P-P--L-P--G-L-G--M-P--PP-A-P-P
--P-L-P--G-S-A--P-P-L--P-A-S-V--Q-G--G-S--Y-P--A-V-P--Q--G--
-----C--G-F---L-P-P--P-L--P--S-G--L--F----------------------
-AMGMNQ-E-K-G-S-R-K--
PF02181 // ENSGT00870000136417_1_Domain_1 (2182, 2682)
-HVI-E-P-S-R-P-M-K-P---LYWTR----I-Q-L-H
--S-----------------------------------KRDSSAS--------LVWEK--
-I---EEP-----------------SIDYHEFEELFSKTAVKERKKPISDTITKTKTKQQ
VVKLLSNKRSQAVGILMS-SLHLDMKDIQHAVVNLDNSVVDLETLQ-ALYENR-AQSDEL
EKIEKHSKASKE--KENAKSLD-------KPEQFLYELSLIPNFSERVFCILFQSTFSES
IGSIRRKLELLQKLCETLKNESGVMRVLGLVLAFGNYMNGGNRTRGQADGFGLDILPKLK
DVKSSDNSRSLLSYIVSYYLRNFD-EDAGREQCIFPLPDPQDLFQASQLKFDDFQKDLRK
MKKDLRACETEAAKVYQLSLEEHLQPFKDNMEQFISQAKIDQENEEKSLTEAHKSFL---
--------ETTAYFCMKPKMGEKEVSPHSFFSIWHEFSSDFKDFWKKENKLILQ----ER
VKEAEEVCRQKKGKSLYNIRPKHDSGIKAKISMKI-------------------------
--------------
ENSFDAP00000012870 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------ESRPVMTTA-EDA
PF02181 // ENSGT00870000136417_1_Domain_1 (2549, 2682)
GKEQCVFPLPEPQDLFQASQMKFEDFQKDLRK
LKKDLKACETEAGKVYQVSSKEHMQPFKENMERFITQAKIDQEAEETSLTETYKCFL---
--------ETTAYFFMKPKIGEKEVSPNVFFSIWHEFSSDFKDFWKKENKLILQ----ER
VKEAEEVCRQKKGKSLYKIKPRHDSGIVSVPFSFSLVISLVFEPCLH-----SMHKF--S
NFPLKF--------
ENSPEMP00000028348 (view gene)
ENSMOCP00000012110 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------MGNQDGKLKKSAGDASHEGG--
G-GGAEDAAGPRDAETTKKASGSKKALGKHGKGGGG----GGESSKKKS-KSDSRASVFS
-NLRIRKN-LTKGK-GAGDSREDVLD-SQALPVGELD-SAHSIV-TKTPDLSLSAEETGL
SDTEC-ADPFEVIHPGGPRPAE-AGVGIQAPAEDLE-TAAGAQDGQRTSSGSDT-DIYSF
HSATEQEDLLSDIQQAIRLQQQQQQQ--------------QQQKLLLQDSEEPAAPPTAI
SPQPGAFLGLDQFLLGPSSE---------------A-RKDA------DQTQPVRPDLPET
T--KSPV-TEHPPSSGGHLAS--------E--------------------TPGYATAHSA
GTDSLSSPAFTFP---EPESREG----ATGAPAPEAGDTDEECEEDAFEDAPRGSPEEEW
VPE-------------KGESS-Q-RPEEEPEEGIRGPSVSA-VASLPGSPAPSPRCFKPY
PLITPCYIKTTTRQLSSPNHSPSQSPNQSPRIK----KRPEPSVSRSPRTALASAAA-PA
K---KHR-LEGSLTGGLSRSADWTEELGVRT---PGAGGSVHLLGRGATAD-ESG--GES
PVLTAK-AP-GAAATADGFQNVFTGRTLLEKLFSQQENGPPEEAEKFCSRIIAMGLLLPF
SDCFREPCN-QNAGASSAPFDQDQL---YTWAAVSQPTHSMDYNEGRFPRREPSRL----
-P---DEKHSPKDVDAE----PQSSILESPKKCPNGV--QK-----ELYDVKSE-GQATV
IQQLEQTIEDLRTKIAELEKQYPALDLEGPRGLPGLENGFPDSAEVGLDALRLHGKVAQL
PRTL-----------QAKSIQT---SPTEEG-G---ILTLA-PLM---SRPEGPPC----
---------------------PPAAEIGESAVLPSPSGPQTKFCSEISLIVSPRRISVQL
EAQPTMQNTSQLHPL-PPSLSGSDG----Q----------E-L-P-L-H-P-SL-H-T-A
-L-E-T-S-H--E-H-S-V-L-S-S-L-E-N-S-R-N-I-P-P-A-PP-L-P-CT-ES-S
-S-F-M-P-G-L-G-M-A-I-PP-T-P-CL-S-D-I-T-V-PA-L-P-S-T-T-A-P-Q-
S-TS-L-Q-G-TE-M-L-PA-P-P-PP-PP-L-P-G-L-G-I--PP------P-P--P-L
--P-G---------------------------------------------------VEIP
PPPPLPGVEIR--PPPPLPGVGI----PPPPPLPGIGIPPPPPLPG----------I-GI
PPPP--PL--PGV-GIPPPPPL-----------PGM-GIPPPPPLPGMGIPPPPPLPGVG
IP-------------PPPPLPGVGIPPPP-------------------------------
-----------------------PLPXXXX---XPP---------------PPLPGVGIP
----PPPPLPGV-GIP---PPPPLPGV-A-IP-PPPPLPGMGVPPAPPPPGA--------
------------------------------------------------------------
---------------------------------------------G--------------
------------------------------------------------------------
----------------------------------------------IPPP-----P----
----LLP--G-S-GPSLSSQ-----------VG-------------SSSL-P--------
A-P-QVC-G-FLF---P-PLPTGLFG----------------------------------
--LGMSQDR---G-A-R-K---QPI-E-P-C-R-P-M-----KPLYWTR----I-Q-L-H
--SK-----------------------------------RDSSPS--------LIWEK--
-I---EEP-----------------SIDCHEFEELFSKTAVKERKKPISDTISKTKAK-Q
VVKLLSNKRSQAVGILMS-SLHLDMKDIQHAVVNLDNSVVDLETLQ-ALYEN--------
------------------------------------------------------------
------------------------------------------------------------
----
PF02181 // ENSGT00870000136417_1_Domain_1 (2525, 2682)
-DNSRSLLSYIVSYYLRNFD-EDAGKEQCVFPLAEPQELFQASQMKFEDFQKDLRK
LKKDLKACEVEAGKVYQVSSEEHMQPFKENMEQFISQAKIDQESQETALTETHKCFL---
--------ETTAYYFMKPKIGEKEVSPNVFFSVWHEFSSDFKDSWKKENKLILQ----ER
VKEAEEVCRQKKGKALYKVKPRHDSGIKAKISMKT-------------------------
--------------
ENSPPYP00000000074 (view gene)
ENSCGRP00000000755 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------MGNQDGKLKRSAGDASHEGGG-
--GGAGG-----------KASGSKKALGKHGKGGGG---S-GETSKKKS-KSDSRASVFS
-NLRIRKN-LTKGK-GAGDSREDVLD-SQALPIGELD-SAHSIV-TKTPDLSLSAEETGL
SDTEC-ADPFEVIHPGGPRPAE-AGVGVQATAEDLG-TAAGAQDGQRTSSGSDT-DIYSF
HSATEQEDLLSDIQQQQ-----------------------QQQKLLLQDSEEPAAPPTAI
SPQPGAFLGLDQFLLGPSSE---------------AAKD-------TEQAQPVRPDLPET
T---KSPVPEHPPSSGAHLAS--------E--------------------TPGYATAHSA
VTDSLSSPAFTFP---EAGPGEV----AAGTPVPEAGDTDEECGEDAFEDAPRGSPEEEW
VPE-------------VEEA--PQRLEEEPEEGMRRPSVTA-VASLPGSPAPSPRCFKPY
PLITPCYIKTTTRQLSSPNHSPSQSPNQSPRIK----KRPEPSVSRSPRTALASAAA-PA
K---KHR-LEGSLAGGLSRSADWTEELGVHT---PGAGGSVHLLGRGSTAD-DSG--GGS
PVLASK-AP-GASATADGFQNVFTGRTLLEKLFSQQENGPPEEAEKFCSRIIAMGLLLPF
SDCFREPCN-QNAGSSSAPFDQDQL---YTWAAVSQPTHSMDYSEGQFPRREPSMWPPSR
LP---DEEHSPKDVDT----EPQSSILESPKKCSNGV--QQ-----EVFDVKSE-GQATV
IQQLEQTIEDLRTKIAELEKQYPALDLEGPRGLPGLENGFTASEEVGLDALRLHGRVAQP
PRTL-----------EAKSIQT---SPTEEG-G---ILTLA-PLK---ALPGGPPC----
---------------------PPAAQSGESAALASPAGPQTKFCSEISLIVSPRRISVQL
DAQPALQSASQLQPP--P-----DGQGQ---------------PS---CPS--L-H---P
EL-E-T-S-P--E-H-S-V-F---SSF-E---NSC-N-V-----PPAPPL-P-CTESSSL
-----M-P-D-L-G-M-A-I-PPPP---CL-S-D-VTV---PA-L-P-S---T-T----A
PQPTSL---QGTE-M-LPAP--------PP------------------------------
------------------------------------------------------------
-----------------------------PPPLPGLGLPPP-------------------
--PP--PL--PGV-GIPPPPPLPRPPP--PPASPCR-PPSPSPS----------------
--------SPWIAIPPPPPLPGMGAPPA--------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------P----PPPGA--G-I--------PPP-P
----LLP--G-S-GPSLSS-----------QAG-------------SSTL-P--------
A-P-PVC-G-FLF---P-PLPTGIFG----------------------------------
--LGMNQER---G-A-R-K--
PF02181 // ENSGT00870000136417_1_Domain_1 (2182, 2682)
-QPI-E-P-C-R-P-M-----KPLYWTR----I-Q-L-H
--SK-----------------------------------RDSSPS--------LIWEK--
-I---EEP-----------------SIDCHEFEELFSKTAVKERKKPISDTISKTKAK-Q
VVKLLSNKRSQAVGILMS-SLHLDMKDIQHAVVNLDNSVVDLETLQ-ALYENR-AQSDEL
EKIEKHSRSSKD--KENAKSLD-------KPEQFLYELSLIPNFSERVFCILFQSTFSES
ICSIRRKLELLQKLCETLKNGPGVMQVLGLVLAFGNYMNAGNKTRGQADGFGLDILPKLK
DVKSSDNSRSLLSYIVSYYLRNFD-EDAGKEQCVFPLAEPQELFQASQMKFEDFQKDLRK
LKKDLKACEVEAGKVYQVSSEEHMQPFKENMEQFISQAKIDQESQEAALTETHKCFL---
--------ETTAYYFMKPKLGEKEVSPNVFFSVWHEFSSDFKDSWKKENKLILQ----ER
VKEAEEVCRQKKGKSLYKVKPRHDSGIKAKISMKT-------------------------
--------------
ENSCHIP00000007126 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------MGNQDAKPKRSAGDVPHGGG--
---GGEDAAVPREAEGTKKAGGGKKAPGRHGKGGG-----GAEPGRKKS-RSDPRASVFS
-NLRIRKT-LSKGR-GTGGSREDVLD-PQALQAGELD-SAHSLV-TKTPDLSLSADETGL
SDTEC-ADPFEVTRPGGPSLA---RVGGPAVAEYSE-TAAGAQDGQRTSSGSDT-DLYSF
HSAAEQEDLLSDIQQAIRLQQLQQQS---------------------PGPESPPVSLPSP
APLPGALPGLDRPPPGPSAE---------------A-------------ALSAPLD----
---------------------------------------------------------RPA
AAEPPAATAFASP---GSVPG-----------ERGAADTDEEGEEDAFEDAPRDSPGVEC
GLA-------------AGEAP-R-TPGATREEGAPKPVFPV-TASPPRSPAHSPRRPRAR
PLLAPCYVRTTTRQLSSPGPSPSPSPSHSPRVQ----RRPESSLGRGSGATAAPAAA-PA
R---KPR-------AGRSRSADWTAELG-RA---PGAGVSAHLVQRGAGGL-QGV--FAG
------------ENASICIQRIKGRRTLLEKLFSQQENGPPEEAEKFCSRIIAMGLLLPF
SDCFREPCG-QNAQSSSVPFDQDRL---YTWAAVSQPTHSLDYVEGQFPRRVPAAWPPSK
PP---DEEQRPKDADTD----SQSAVLETPKKCSDAA--QQ-----EVCDMKSE-GQATV
IQRLEQTIEDLKTKIAELEKQYPALDT-------------------GLEALKLGEKDVGY
ERIV-----------QAKSIQT---SPMEDG-G---VLARP-PGG---APLVGPL-----
----------------------PLTAHGESAGLPLPLGPQTMFCSEISLVVSPRRISVQL
DAQPSLQ-----PPP-PASLPWSDSHGQAG----------S-Q-P-S-H-P--L-H-T-G
-F-E-T-S-Q--E-H-S-A-F-P-S-S-G-N-G-C-D-I-P-T-A-PP-L-P-PE--T-S
-P-L-M-P-G-V-G-T-E-A-SP-A-P-LT-P-G-A-P-G-PA-L-P-S-P-A-G-L-S-
P-PP-C-L-G-PE-M-L-PP-P-P-L---P-L-P-G-V-G-I--PP--------------
------------------------------------------------------------
-APPLPGVGIP--PAPPLPGVGI----PPAPPLPGVGI----------------------
PPAP--PL--PGV-GIPPAPPLPGVGIPPAPPLPGV-GIPPAPLFPGWGSLPPPLFPGWG
SL--PPPLFPGWGSLPPRLFPGWGSL----------------------------------
---------------------------------------------P--------------
---------PRM-GIP---PTPPLPGV-G-IS-PPPPLSSEGIPPAPPPP----------
----------------PPP-P-G-V-----------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------G-IPPP-----LTP-P
P----LP--T-A-GSGLP--------LP-RQVG-------------SSTL-P--------
T-P-QVC-G-FLP---P-PLPLGLFG----------------------------------
--FGMNQEK---G-S-R-K--
PF02181 // ENSGT00870000136417_1_Domain_1 (2182, 2682)
-QPV-E-P-C-R-P-M-----KPLYWTR----I-Q-L-H
--SR-----------------------------------RDSSAS--------LIWEK--
-I---EEP-----------------SIDCHEFEELFSKSTVKERKKPISDTITKTKAK-Q
VVKLLSNKRSQAVGILMS-SLHLDMKDIQHAVVNLDNSVVDLETLQ-ALYENR-AQSDEL
EKIEKHGRSSKD--KENAKSLD-------KPEQFLYELSLIPNFSERVFCILFQSTFSES
ICSIRRKLELLQKLCETLKHGSGVMQVLGLVLAFGNYMNGGNNTRGQADGFGLDILPKLK
DVKSSDNSRSLLSYIVSYYLRNFD-EDAGKEQCVFPLPEPQDLFQASQMKFEDFQKDLRK
LKKDLRACEVEAGKVYQVSSKEHIQPFKENMEQFIIQGKFCKDAAL------PSLFL---
--------ETTAYFFMRPKLGEKEVSPNVFFGIWHEFSSDFKDFWKKENKLILQ----ER
VKEAEEVCRQKKGKSLYKIKPRHDSGIVSIYSVF--------------------------
--------------
ENSMICP00000028946 (view gene)
ENSMMUP00000010836 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------------------MS
RAVTIAEAFCHILF----------------QQCLFPL
PF02181 // ENSGT00870000136417_1_Domain_1 (2558, 2682)
PEPQDLFQASQMKFEDFQKDLRK
LKKDLKACEVEAGKVYQVSSKEHMQPFKENMEQFIIQAKIDQEAEENSLTETHKCFL---
--------ETTAYFFMKPKLGEKEVSPNAFFSIWHEFSSDFKDFWKKENKLLLQ----ER
VKEAEEVCRQKKGKSLYKIKPRHDSGIKAKISMKT-------------------------
--------------
ENSCLAP00000016505 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------SRYIEMTIA-ED
PF02181 // ENSGT00870000136417_1_Domain_1 (2548, 2682)
AGKEQCVFPLPEPQDLFQASQMKFEDFQKDLRK
LKKDLKACETEAGKVYQVSSKEHMQPFKENMEQFIIQAKIDQEAEEASLTETHKCFL---
--------ETTAYFFMKPKIGEKEVSPNVFFSIWHEFSSDFKDFWKKENKLILQ----ER
VKEAEEVCRQKKGKSLYKIKPRHDSGIVSI------------------------------
--------------
ENSSSCP00000049308 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------MGNQDAKPKRSAGDALHGGGG-
--GGGEDAAGPRDAEGTKKGGGGKKAPGRHGKGGGG-----GEPGRKKS-KSDPRASVFS
-NLRIRKT-LTKGR-GAGGSREDVLE-PQALQAGELD-SAHSLV-TKTPDLSLSADDTGL
SDTEC-ADPFEVTRPRGPGSAE-A--GGPAVAENSE-TAAGAQDGQRTSSGSDT-DLYSF
HSATEQEDLLSDIQQAIRLQQLQQQP---------------------QSPESPAASPPSA
PYQFGALLGLERPLPGPSAE---------------QTPS---------------------
------ARPDQPPAPDLL------------------------------------------
----SASAALPSP---ESAPGEP----SAGVPLRGAGDTDEEGEEDAFEDAPRGSPGAEC
DLA-------------EREAL--QRPGVPREEGAPRPGAPT-AASLPGSPARSPRCLRSR
PLLAPCYVRTTTRQLSSPGPSPSASPSHSPRVQ----RRPESSLGRDSGAVTAPAAARAR
K-----------PRAGRSRSADWTAELG-RA---PGVGGSAHLPQRGAGGLQDSE---GR
PGLRAR-GPGDLPMLLPNDRVSPAGRTLLEKLFSQQENGPPEEAEKFCSRIIAMGLLLPF
SDCFREPCS-QNAQSSSAPFDQDQL---YTWAAVSQPTHSLDYVEGQFPRRVPSAWPPPK
PP---DEEHRPKDADTE----SQSAVLETPKKCSDAV--QQ-----EVCDMKSE-GPATV
IQQLEQTIEDLRTKIAELEKQYPPADMEG-------------TVDICLEALRLGEKDVQH
QRIL-----------QAKSIQT---SPVEEG-G---TLTLT-SVS---ALPECLPH----
---------------------TPWAESGNSAVPSSPSRPQTKFCSEISLVVSPRRISVQL
DAPQSLQ-----PSP-PPSLPWSVGQGQAGSR-----------PS--LH----------T
EFET---S-H--E-H--SV-FPS-------SENSQNL-------PLTPPL--PCIESSSA
MPGLGMAT-------------PP-----SLSPPG-TLGPAL-------------------
------------------------------------------------------------
------------------------------------------------------PSTAVP
SPPPLPGVGIP--PAPPLPGMGI----PPPPPLPGVGIPPWEYLLP--------PSSWNG
N---------------PHSPAFARSGNTS------------CPPSSWNGNPPSPTFARSG
NT--SCPPSSWNGNPPSPAFARSG------------------------------------
----------------------------------------------------------NT
----SCPPSSWN-GNP---PSP-------------TFARRVGIPPAPPLPGMGIPPPPPL
PGMGIP--------PAPPL-P-G-M-GIPPPP--------PL-P-G-MGIPPAPPLP---
---------------------------------VGVPPPPPLPGMGIPA-----------
------------------------------------------------------------
--------------------------PP--P-P----PPPGT--G-ILLP-----PPP-P
----LLP--G-S-GLPLPP-----------QVG-------------SSTL-P--------
T-P-QVC-G-FLP---P-PLPTGLFG----------------------------------
--LGMNQDK---G-S-R-K--
PF02181 // ENSGT00870000136417_1_Domain_1 (2182, 2341)
-QPI-E-P-C-R-P-M-----KPLYWTR----I-Q-L-H
--SK-----------------------------------RDSSAS--------LIWEK--
-I---EEP-----------------SIDCHEFEELFSKTTVKERKKPISDTITKTKAK-QVSTHVFFEIRQSFGHKCELSLEKSFEIVSQAV----------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------
ENSFCAP00000021688 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------VFLPRTIA-EDAG
PF02181 // ENSGT00870000136417_1_Domain_1 (2550, 2682)
KEQCVFPLPEPQDLFQASQMKFEDFQKDLRK
LKKDLKACEVEAGKVYQVSSKEHIQPFKENMEQFIIQAKIDQEAEENSLTETHKCFL---
--------ETTAYFFMKPKIGEKEVSPNVFFSIWHEFSSDFKDFWKKENKLILQ----ER
VKEAEEVCRQKKGKSLYKIKPRHDSGIKAKISMKT-------------------------
--------------
ENSDNOP00000033153 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------MGNQDGKLKRSAGDGPHEAG--
---GAEDAAGPRDADAARKGGAGKKAPGRHGKGGA------GEAGRRKS-KADSRASVFS
-NLRLRKS-LARGR-GAGGSREDVLD-AQPPPGGELD-SAHSLA-TRTPDLSLSADEAGL
SDTEG-ADPFEGARPGRRGPAE-SGGGARAAAEDLD-AGAGAQDGQRTSSGSDT-DIYSF
HSAAEQEDLLSDIQQAIRLQQQQQQQQQQQQQQQQQQQQQQQQQQQQQGSQEG-TAPPAA
SPQTGAFLCLDRLLLGPLGQ---------------A-AEAPGSPE-PAPA--APSERPGS
PAAELPA-PERPLPPGAR-DP--------E--------------------AARPPAAQPA
AADSPWPTAFPFP---EGAPGQG----AAAATAPGAADTDEDGEEEAFEDAPRGSPGEAW
VEG-------------RAEAMQPEAGVFLAVALAECGCPAN-ALELPQSPFEDGRCLSPS
PFICPGNLINIVKQIMWPSRWPSRWPSRSPACPSRAPRRRASSKCRGPGGPGLTHAGSSP
RVHVAEA-QQAGLYGKLAHAECWPAQSAYLR---RRAGPRRELMQQDDVTK-RDT--EVS
PVDCADWASWGSTTSKGMPGSHFTSSTLIKHLGGHQDNIPPPSGNSFCGTLAALGKSLPS
NSCFLEGPP-RHSSFEAALLEQDQL---YTWAAVSQPTHSLDYTEGQFPRRIPSVWPPSK
PP---DEEHRPKAVET----ESQSAVLESPKKCSDAV--QQ-----EVFDMKSE-GQATV
IQQLEQTIEDLRTKIAELEKQYPAVDAEAAARRPGVLNAPMTPPDVSLEELRLGEKEARQ
QRIL-----------QARSIQT---SPTEEG-G---VPAAP-AAN---ALPERPPC----
---------------------PTGAEGGGPAVPTSAPGPPTKFCSEISLLVSPRLISVQL
DARQSPESTPQAPPP--PPPPAPGAEGLGPHE-----------VS---------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------TYILPIPPISGS-LV---------------------
------------------------------------------------------------
--------------------------------------------YL----FISLRSCSIC
----P--SMLGC-F---------------------TQHKGLKIHP-----------C---
-------------------------------------------------------YH---
---------------------------------------PCVCSVFV-------------
------------------------------------------------------------
--------------------------------L----K--------APLV-----FRA-P
----L------------LP-----------EAG-------------SSSL-P--------
T-P-QVC-G-FLP---P-PLPAGLFG----------------------------------
--LGTNQEK---G-S-R-K--
PF02181 // ENSGT00870000136417_1_Domain_1 (2182, 2586)
-QPI-E-P-C-R-P-M-----KPLYWTR----I-Q-L-H
--SK-----------------------------------RDSTAS--------LIWEK--
-I---EEP-----------------SIDCHEFEELFSKTAVKERKKPISDTITKTKAK-Q
VVKLLSNKRSQAVGILMS-SLHLDMKDIQHAVVNLDNSVVDLETLQ-ALYENR-AQSDEL
EKIEKHGRSSKD--KENAKSLD-------KPEQFLYELSLIPNFSERVFCILFQSTFSES
ICSIRRKLELLQKLCETLKNGPGVMQVLGLVLAFGNYMNGGNKTRGQADGFGLDILPKLK
DVKSSDNSRSLLSYIVSYYLRNFD-EDAGKEQCVFPLPEPQDLFQASQMKFEDFQKDLRK
LKKDLK------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------
ENSCANP00000020293 (view gene)
ENSMLUP00000004690 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------C-G-FLP---P-PLPTGWFG----------------------------------
--LGMSQDR---G-S-R-K--
PF02181 // ENSGT00870000136417_1_Domain_1 (2182, 2682)
-QPI-E-P-C-R-P-M-----KPLYWTR----I-Q-L-H
--SK-----------------------------------RDSGAS--------LIWEK--
-I---EEP-----------------SIDCHEFEELFSKTAVKERKKPISDTISKTKAK-Q
----------------------------------------------------R-AQSDEL
EKIEKHGRSSKD--KESAKSLD-------KPEQFLYELSLIPNFSERVFCILFQSTFSES
IGTIRRKLELLQKLCETLKNGPGVMQVLGLVLAFGNYMNGGNKTRGQADGFGLDILPKLK
DVKSSDNSRSLLSYIVSYYLRNFD-EDAGKEQCVFPLPEPQDLFQASQMKFEDFQKDLRK
LKKDLKACEVEAGKVYQVSPEEHIQPFKENMERFIIQAKIDQEAEENSLTETHKWFL---
--------ETTAYFFMKPKLGEKEVSPNVFFSIWHEFSSDFKDFWKKENKLILQ----ER
VKEAEEVCRQKKGKSLYKIKPRHDSGIKAKISMKT-------------------------
--------------
ENSHGLP00100020910 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------ES-KAVKIAECVFPL
PF02181 // ENSGT00870000136417_1_Domain_1 (2558, 2682)
PEPQELFQASQMKFEDFQKDLRK
LKKDLKACETEAGKVYQVSSKEHTQAFKENMEQFIIQVKIDQEPEETSLTETHKCFS---
--------ETTAYFFMKPKIGEKEVSPDAFFSIWHEFSSDFKDFWKKENKLILQ----ER
IKEAEQVCRQKKGKSLYKIKPRHDSRIKAKISMKT-------------------------
--------------
ENSBTAP00000046424 (view gene)
ENSCSAP00000005791 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------MGNQDGKLKRSAGDALHEGG--
G-GGAEDALGPRDVETTKKGSGGKKALGKHGKGGGG---GGGESGKKKS-KSDSRASVFS
-NLRIRKN-LSKGK-GAGGSREDVLD-SQALQTGELD-SAHSLL-TKTPDLSLSADEAGL
SDTEC-ADPFEVTRPGGPGPAE-ARVRGRPVAEDVE-TAAGAQDGQRTSSGSDT-DIYSF
HSATEQEDLLSDIQQAIRLQQQQQQL---QLQLQ-----QQQQQQQLQDAEEPAAPPTAV
SPQPGAFLGLDRFLLGPSGG---------------A-GEAPGSPD-TEQALSALSDLPES
LAAEPRE-PQQPPSPGGLPAS--------E--------------------APSLPAAQPA
VTDSPSSTAFPFP---ETGPGEE----AAGAPVRGAGDTDEEGEEDAFEDAPRGSPGEEW
APE-------------VGEDA-PQRLEEEPEEEAQGPDSPA-AASLPGSPAPSQRCFKPY
PLITPCYIKTTTRQLSSPNHSPSQSPNQSPRIK----RRPEPSLSRGSRTALASVAA-PA
K---KHR-ADGGLAGGLSRSADWTEELGART---PRAGGSAHLLERGVAAD-SGG--GVS
PALAAK-AS-GAPAAADGFQNVFTGRTLLEKLFSQQENGPPEEAEKFCSRIIAMGLLLPF
SDCFREPCN-QNAQTNAASFDQDQL---YTWAAVSQPTHSLDYSEGQFPRRVPSMGPPSK
PP---DEEHRFEDAETE----SQSAVSETPKKRSDAV--QK-----EVVDMKSE-GQANV
IQQLEQTIEDLRTKIAELERQYPALDTEVASGHQGLGNGVTASGDVCLEALRLEEKEGRH
QRIL-----------EAKSIQT---SPTEEG-G---VLTLP-PVG---AL-GHPPC----
---------------------PP----------GAESGPQTKFCSEISLIVSPRRISVQL
DSHQPTQSISQPPPP--PSLLWSAGQGQPGS-----------QPP---HSS--L-S---T
EF-E-T-S-H--E-H-S-V---S-SAF-K---NSC-N-I-P----SPPPL-P-CTESSSS
-----M-P-G-L-G-M-V---PPPP-P-PL-P-G-MTV---PT-L-P-------------
------------S-TAIPPP---------PPL-P-----GTE------------------
--------------------------------------------------------MLPP
PPPPLPGVGIP--PPPPLPGAGI----PPPPG---KGLSSGSNSSHLAAEPLLFLIKWSL
TLLP--RLECSGVISAHCNLPLPGSSDSSAS-ASQVAGITGAGPCDQAGL---------K
L--------LTSGDPPTLDSQSAGMTGPT-------------------------------
-----------------------FCLPEGM---PVTTLFAGMGIRHVAQAGLVLLGSGNP
----PASASQSA-GIR---SPQPTPLP-A-TP-NSVPLWTCCIQPGAIRARCGAPRRIHV
PWWFAA--------PINPS-S-A-L-GISPNA--------I-------CLPVPHPLTGPS
AFPNTPSLKSGSVILPVLSLPGFGITSASLVPAI--------------------------
------------------------------------------------------------
--------------------------------------------H-LPQP-----PGN-P
PTSASQS--T-------------------RIAG--MSHPPCR----AEKL-Q--------
T------PG-LSL---P-APPILLFIPAA--GDPSLPLCTDHAAQAK-RIVWCQPDSLEI
SLVGMNLEL---D---------VPY-D-P-A-I-P-L-----LGIYP-------------
------------------------------WATRAKLHLR
PF02181 // ENSGT00870000136417_1_Domain_1 (2261, 2586)
KKKKK--------SIWSK--
-V---QSG----------------QLDLQVEFREMFKEKTKNSLQTLKKPTILNTLGN-D
VVKLLSNKRSQAVGILMS-SLHLDMKDIQHAVVNLDNSVVDLETLQ-ALYENR-AQSDEL
EKIEKHGRSSKD--KENAKSLD-------KPEQFLYELSLIPNFSERVFCILFQSTFSES
ICSIHRKLELLQKLCETLKNGPGVMQVLGLVLAFGNYMNGGNKTRGQADGFGLDILPKLK
DVKSSDNSRSLLSYIVSYYLRNFD-EDAGKEQCLFPLPEPQDLFQASQMKFEDFQKDLRK
LKKDLK------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------
MGP_SPRETEiJ_P0022480 (view gene)
ENSMGAP00000012134 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------G-F---L-P-P--P-L--P--S-G--L--F----------------------
-AMGMNQ-E-K-G-S-R-K--
PF02181 // ENSGT00870000136417_1_Domain_1 (2182, 2682)
-HVI-E-P-S-R-P-M-K-P---LYWTR----I-Q-L-H
--S-----------------------------------KRDSSAS--------LVWEK--
-I---EEP-----------------SIDYHEFEELFSKTAVKERKKPISDTITKTKTK-Q
----------------------------------------------------R-AQSDEL
EKIEKHSKASKE--KENAKSLD-------KPEQFLYELSLIPNFSERVFCILFQSTFSES
ICSIHRKLELLQKLCETLKNGSGVMQVLGLVLAFGNYMNGGNRTRGQADGFGLDILPKLK
DVKSSDNSRSLLSYIVSYYLRNFD-EDAGKEQCIFPLPEPQDLFQASQLKFEDFQKDLRK
MKKDLRVCETEAAKVYQLSLEEHLQPFKDSMEQFISQAKIDQENEEKSLTEAHKSFL---
--------ETAAYFCMKPKMGEKEVSPHSFFNIWHEFSSDFKDFWKKENKLILQ----ER
VKEAEEVCRQKKGKSLYNIRPKHDSGIKAKISMKI-------------------------
--------------
ENSOARP00000003377 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------QDQL---YTWAAVSQPTHSLDYAEGQFPRRVPAAWPPSK
PL---DEEQRPKDADTD----SQSAVLETPKKCSDAA--QQIKFFWEVCDMKSE-GQATV
IQRLEQTIEDLKTKIAELEKQYPALDT-------------------GLEALKLGEKDVGY
ERIV-----------QAKSIQT---SPMEDG-G---VLARP-PGG---APLVGPL-----
----------------------PLTAHGESAGLPLSSGPQTMFCSEISLVVSPRRISVQL
DAQPSLH-----PPP-PASLPWSDSHGQAG----------S-Q-P-S-H-P--L-H-T-G
-F-E-T-S-Q--E-H-S-A-F-P-Y-S-G-N-G-C-D-I-P-T-A-PP-L-P-PE--T-S
-P-L-M-P-G-V-G-A-E-A-SP-A-P-LT-P-S-A-P-G-PA-L-P-S-P-A-G-L-S-
P-PP-R-L-G-PE-M-L-PP-P-P-L---P-L-P-G-V-G-I--PP--------------
------------------------------------------------------------
-APPLPTMGIP--PGIPP------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------PPPLLPSEGIPP---AP----------
----------------PPP-P-G-V-----------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------R-IPPP-----LTP-P
P----PP--T-A-GSGLP--------LP-GQVG-------------SSTL-P--------
T-P-QVC-G-FLP---P-PLPLGLFG----------------------------------
--FGMNQEK---G-S-R-K--
PF02181 // ENSGT00870000136417_1_Domain_1 (2182, 2619)
-QPV-E-P-C-R-P-M-----KPLYWTR----I-Q-L-H
--SR-----------------------------------RDSSAS--------LIWEK--
-I---EEP-----------------SIDCHEFEELFSKSTVKERKKPISDTITKTKAK-Q
VVKLLSNKRSQAVGILMS-SLHLDMKDIQHAVVNLDNSVVDLETLQ-ALYENR-AQSDEL
EKIEKHGRSSKD--KENAKSLD-------KPEQFLYELSLIPNFSERVFCILFQSTFSES
ICSIRRKLELLQKLCETLKHGSGVMQVLGLVLAFGNYMNGGNNTRGQADGFGLDILPKLK
DVKSSDNSRSLLSYIVSYYLRNFD-EDAGKEQCVFPLPEPQDLFQASQMKFEDFQKDLRK
LKKDLRACEVEAGKVYQVSSKEHIQPFKENMEQFIIQGKF--------------------
------------------------------------------------------------
------------------------------------------------------------
--------------
ENSPSIP00000013896 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------MS
PF02181 // ENSGT00870000136417_1_Domain_1 (2368, 2682)
IFFSVVNLDNSIVDLETLQ-ALYENR-AQSDEL
EKIEKHGKSSKE--RENAKSLD-------KPEQFLYELSLIPNFSERVFCILFQSTFSES
ICSIHRKLELLQKLCEMLKSGAGVMQVLGLVLAFGNYMNGGNRTRGQADGFGLDILPKLK
DVKSSDNSRSLLSYIVSYYLRNFD-EDAGKEQCIFPLPEPQDLFQASQMKFEDFQKDLRK
LRKDLRACETEAGKVYQVSLEEHIQPFKDNMEKFINQAKIEQENEEKSLTEAHKSFL---
--------ETAAYFCMKPKMGEKEVSPHAFFSIWHEFSSDFKDLWKKENKLILQ----EK
VKEAEEVCRQKKGKSLYNIKPRHDSGIVRMDCIIIIIIIIISLLTFFLFILFVFHMYIIL
TYLVIYF-------
ENSCHOP00000004598 (view gene)
ENSSBOP00000028002 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---TIAEAFC
PF02181 // ENSGT00870000136417_1_Domain_1 (2531, 2682)
HILFRIISEI---L-MR---EQCLFPLPEPQDLFQASQMKFEDFQKDLRK
LKKDLKACEVEAGKVYQVSSKEHMQPFKENMEQFISQAKIDQEAEENSLTETHKCFL---
--------ETTAYFFMKPKLGEKEVSPNVFFSIWHEFSSDFKDFWKKENKLILQ----ER
VKEAEEVCRQKKGKSLYKIKPRHDSGIKAKISMKT-------------------------
--------------
{"ENSPANP00000014781": [["Domain_1", 2182, 2682, "PF02181"]], "ENSBTAP00000041415": [["Domain_1", 2342, 2627, "PF02181"]], "ENSMAUP00000008930": [["Domain_1", 2182, 2682, "PF02181"]], "ENSHGLP00000001950": [["Domain_1", 2531, 2682, "PF02181"]], "ENSHGLP00000016533": [["Domain_1", 2001, 2682, "PF02181"]], "ENSNGAP00000019809": [["Domain_0", 1583, 1777, "PF06346"], ["Linker_0_1", 1777, 2182], ["Domain_1", 2182, 2682, "PF02181"]], "ENSHGLP00100003278": [["Domain_1", 2578, 2682, "PF02181"]], "ENSTBEP00000014955": [["Domain_0", 1208, 2682, "PF02181"]], "ENSSTOP00000016978": [["Domain_0", 1503, 1641, "PF06346"], ["Domain_1", 2182, 2682, "PF02181"]], "ENSPVAP00000004193": [["Domain_0", 1203, 2682, "PF02181"]], "ENSTSYP00000030611": [["Domain_1", 2535, 2633, "PF02181"]], "ENSOPRP00000015241": [["Domain_0", 1383, 1897, "PF06346"], ["Linker_0_1", 1897, 2215], ["Domain_1", 2215, 2682, "PF02181"]], "ENSAMEP00000014813": [["Domain_0", 1539, 2103, "PF06346"], ["Linker_0_1", 2103, 2182], ["Domain_1", 2182, 2586, "PF02181"]], "ENSECAP00000022816": [["Domain_0", 1364, 1615, "PF06346"], ["Domain_1", 2182, 2682, "PF02181"]], "ENSHGLP00000020101": [["Domain_1", 2565, 2682, "PF02181"]], "ENSRROP00000034007": [["Domain_1", 2182, 2682, "PF02181"]], "ENSPCOP00000013212": [["Domain_0", 1521, 1717, "PF06346"], ["Domain_1", 2592, 2682, "PF02181"]], "ENSRBIP00000014255": [["Domain_1", 2182, 2682, "PF02181"]], "ENSCJAP00000062816": [["Domain_1", 2182, 2682, "PF02181"]], "ENSPTRP00000072717": [["Domain_1", 2536, 2682, "PF02181"]], "ENSOANP00000016311": [["Domain_1", 2478, 2682, "PF02181"]], "ENSHGLP00100004027": [["Domain_1", 2565, 2682, "PF02181"]], "ENSPTRP00000003647": [["Domain_0", 1361, 1733, "PF06346"]], "ENSODEP00000022020": [["Domain_1", 2578, 2682, "PF02181"]], "ENSP00000318884": [["Domain_0", 1498, 1616, "PF06346"], ["Domain_1", 2182, 2682, "PF02181"]], "ENSTTRP00000010223": [["Domain_0", 1646, 2121, "PF06346"], ["Linker_0_1", 2121, 2182], ["Domain_1", 2182, 2682, "PF02181"]], "ENSCPOP00000024740": [["Domain_1", 2540, 2682, "PF02181"]], "ENSVPAP00000001647": [["Domain_0", 1213, 1637, "PF06346"], ["Domain_1", 2586, 2682, "PF02181"]], "ENSMODP00000008810": [["Domain_0", 1498, 1641, "PF06346"], ["Domain_1", 2180, 2682, "PF02181"]], "ENSMFAP00000045272": [["Domain_0", 1509, 1615, "PF06346"], ["Domain_1", 2182, 2682, "PF02181"]], "ENSFCAP00000020057": [["Domain_1", 2182, 2341, "PF02181"]], "ENSPCAP00000005171": [["Domain_0", 1600, 2121, "PF06346"], ["Linker_0_1", 2121, 2182], ["Domain_1", 2182, 2639, "PF02181"]], "ENSCATP00000045295": [["Domain_1", 2182, 2682, "PF02181"]], "ENSCCAP00000028876": [["Domain_0", 1344, 1596, "PF06346"], ["Domain_1", 2477, 2682, "PF02181"]], "ENSACAP00000003593": [["Domain_1", 2182, 2616, "PF02181"]], "ENSSARP00000011242": [["Domain_0", 1513, 2103, "PF06346"], ["Linker_0_1", 2103, 2182], ["Domain_1", 2182, 2682, "PF02181"]], "ENSOCUP00000003468": [["Domain_0", 1561, 1721, "PF06346"], ["Linker_0_1", 1721, 2182], ["Domain_1", 2182, 2682, "PF02181"]], "ENSOGAP00000000107": [["Domain_0", 1528, 1765, "PF06346"], ["Linker_0_1", 1765, 2182], ["Domain_1", 2182, 2621, "PF02181"]], "ENSAPLP00000014376": [["Domain_1", 2182, 2682, "PF02181"]], "ENSMUSP00000030039": [["Domain_0", 1599, 1778, "PF06346"], ["Linker_0_1", 1778, 2182], ["Domain_1", 2182, 2682, "PF02181"]], "ENSLAFP00000000880": [["Domain_0", 1624, 2105, "PF06346"], ["Linker_0_1", 2105, 2182], ["Domain_1", 2182, 2344, "PF02181"]], "ENSGGOP00000001003": [["Domain_1", 2182, 2682, "PF02181"]], "ENSCAFP00000038900": [["Domain_0", 1611, 2123, "PF06346"], ["Linker_0_1", 2123, 2182], ["Domain_1", 2182, 2586, "PF02181"]], "ENSNLEP00000004370": [["Domain_0", 1501, 1629, "PF06346"], ["Domain_1", 2339, 2682, "PF02181"]], "ENSANAP00000019455": [["Domain_1", 2182, 2682, "PF02181"]], "ENSGALP00000050709": [["Domain_1", 2182, 2682, "PF02181"]], "ENSHGLP00000014867": [["Domain_1", 2558, 2682, "PF02181"]], "ENSTGUP00000010919": [["Domain_1", 2182, 2614, "PF02181"]], "ENSBTAP00000056407": [["Domain_1", 2410, 2682, "PF02181"]], "ENSMNEP00000008627": [["Domain_0", 1509, 2002, "PF06346"], ["Linker_0_1", 2002, 2182], ["Domain_1", 2182, 2682, "PF02181"]], "ENSDORP00000026256": [["Domain_0", 1537, 1704, "PF06346"], ["Domain_1", 2182, 2682, "PF02181"]], "ENSJJAP00000020675": [["Domain_1", 2182, 2369, "PF02181"]], "ENSMLEP00000032851": [["Domain_1", 2536, 2682, "PF02181"]], "ENSEEUP00000014216": [["Domain_0", 183, 2682, "PF02181"]], "ENSCHIP00000032612": [["Domain_1", 2576, 2682, "PF02181"]], "ENSMPUP00000015581": [["Domain_0", 1532, 2090, "PF06346"], ["Linker_0_1", 2090, 2182], ["Domain_1", 2182, 2682, "PF02181"]], "ENSSHAP00000015245": [["Domain_1", 2396, 2589, "PF02181"]], "ENSJJAP00000016692": [["Domain_1", 2550, 2682, "PF02181"]], "ENSXETP00000035438": [["Domain_1", 2180, 2682, "PF02181"]], "ENSSSCP00000049388": [["Domain_1", 2558, 2682, "PF02181"]], "MGP_CAROLIEiJ_P0021418": [["Domain_1", 2182, 2395, "PF02181"]], "MGP_PahariEiJ_P0076664": [["Domain_1", 2182, 2682, "PF02181"]], "ENSCGRP00001005121": [["Domain_1", 2182, 2682, "PF02181"]], "ENSETEP00000010185": [["Domain_1", 2327, 2682, "PF02181"]], "ENSRNOP00000070887": [["Domain_1", 2182, 2339, "PF02181"]], "ENSFALP00000013764": [["Domain_1", 2182, 2682, "PF02181"]], "ENSFDAP00000012870": [["Domain_1", 2549, 2682, "PF02181"]], "ENSPEMP00000028348": [["Domain_0", 1633, 1795, "PF06346"], ["Linker_0_1", 1795, 2182], ["Domain_1", 2182, 2682, "PF02181"]], "ENSMOCP00000012110": [["Domain_1", 2525, 2682, "PF02181"]], "ENSPPYP00000000074": [["Domain_0", 1600, 2260, "PF06346"], ["Linker_0_1", 2260, 2262], ["Domain_1", 2262, 2634, "PF02181"]], "ENSCGRP00000000755": [["Domain_1", 2182, 2682, "PF02181"]], "ENSCHIP00000007126": [["Domain_1", 2182, 2682, "PF02181"]], "ENSMICP00000028946": [["Domain_0", 1643, 1799, "PF06346"], ["Linker_0_1", 1799, 2182], ["Domain_1", 2182, 2682, "PF02181"]], "ENSMMUP00000010836": [["Domain_1", 2558, 2682, "PF02181"]], "ENSCLAP00000016505": [["Domain_1", 2548, 2682, "PF02181"]], "ENSSSCP00000049308": [["Domain_1", 2182, 2341, "PF02181"]], "ENSFCAP00000021688": [["Domain_1", 2550, 2682, "PF02181"]], "ENSDNOP00000033153": [["Domain_1", 2182, 2586, "PF02181"]], "ENSCANP00000020293": [["Domain_0", 1284, 1537, "PF06346"], ["Domain_1", 2182, 2682, "PF02181"]], "ENSMLUP00000004690": [["Domain_1", 2182, 2682, "PF02181"]], "ENSHGLP00100020910": [["Domain_1", 2558, 2682, "PF02181"]], "ENSBTAP00000046424": [["Domain_0", 1635, 2132, "PF06346"], ["Linker_0_1", 2132, 2182], ["Domain_1", 2182, 2682, "PF02181"]], "ENSCSAP00000005791": [["Domain_1", 2261, 2586, "PF02181"]], "MGP_SPRETEiJ_P0022480": [["Domain_0", 1609, 1763, "PF06346"], ["Linker_0_1", 1763, 2182], ["Domain_1", 2182, 2682, "PF02181"]], "ENSMGAP00000012134": [["Domain_1", 2182, 2682, "PF02181"]], "ENSOARP00000003377": [["Domain_1", 2182, 2619, "PF02181"]], "ENSPSIP00000013896": [["Domain_1", 2368, 2682, "PF02181"]], "ENSCHOP00000004598": [["Domain_0", 1714, 2103, "PF06346"], ["Linker_0_1", 2103, 2180], ["Domain_1", 2180, 2682, "PF02181"]], "ENSSBOP00000028002": [["Domain_1", 2531, 2682, "PF02181"]]}

