Found:
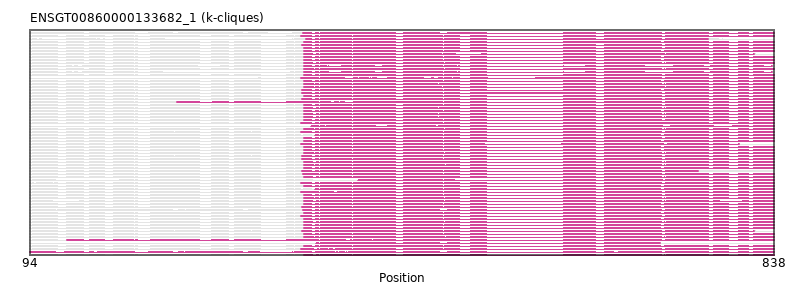
{"ENSCGRP00000015760": [["Domain_0", 368, 836, "PF00443"]], "ENSETEP00000012762": [["Domain_0", 241, 838, "PF00443"]], "ENSCLAP00000020018": [["Domain_0", 368, 838, "PF00443"]], "ENSPTRP00000051643": [["Domain_0", 368, 838, "PF00443"]], "ENSGGOP00000007337": [["Domain_0", 368, 838, "PF00443"]], "ENSMLUP00000004133": [["Domain_0", 365, 803, "PF00443"]], "ENSSSCP00000043152": [["Domain_0", 368, 838, "PF00443"]], "ENSMUSP00000106936": [["Domain_0", 368, 838, "PF00443"]], "ENSMFAP00000031487": [["Domain_0", 379, 838, "PF00443"]], "ENSLAFP00000013671": [["Domain_0", 365, 838, "PF00443"]], "ENSCATP00000023880": [["Domain_0", 368, 838, "PF00443"]], "ENSOCUP00000002071": [["Domain_0", 367, 838, "PF00443"]], "MGP_SPRETEiJ_P0022236": [["Domain_0", 368, 838, "PF00443"]], "ENSNGAP00000015238": [["Domain_0", 368, 838, "PF00443"]], "ENSHGLP00100008567": [["Domain_0", 368, 838, "PF00443"]], "ENSMODP00000006496": [["Domain_0", 366, 838, "PF00443"]], "ENSLAFP00000020052": [["Domain_0", 375, 838, "PF00443"]], "ENSMNEP00000020708": [["Domain_0", 368, 838, "PF00443"]], "ENSANAP00000020209": [["Domain_0", 370, 772, "PF00443"]], "ENSPPAP00000041271": [["Domain_0", 368, 838, "PF00443"]], "ENSCHIP00000019016": [["Domain_0", 368, 838, "PF00443"]], "ENSCJAP00000009687": [["Domain_0", 368, 838, "PF00443"]], "ENSCGRP00000006872": [["Domain_0", 368, 838, "PF00443"]], "ENSCPOP00000013808": [["Domain_0", 368, 838, "PF00443"]], "ENSMEUP00000004036": [["Domain_0", 365, 838, "PF00443"]], "ENSXETP00000038347": [["Domain_0", 94, 838, "PF00443"]], "ENSPEMP00000019311": [["Domain_0", 368, 838, "PF00443"]], "ENSCCAP00000012365": [["Domain_0", 368, 838, "PF00443"]], "ENSCAFP00000018832": [["Domain_0", 368, 838, "PF00443"]], "ENSCSAP00000015400": [["Domain_0", 366, 838, "PF00443"]], "ENSCAPP00000002233": [["Domain_0", 368, 818, "PF00443"]], "ENSPANP00000005868": [["Domain_0", 379, 838, "PF00443"]], "ENSSTOP00000005733": [["Domain_0", 368, 838, "PF00443"]], "ENSMPUP00000014153": [["Domain_0", 366, 838, "PF00443"]], "ENSDNOP00000022893": [["Domain_0", 366, 838, "PF00443"]], "ENSAMEP00000016389": [["Domain_0", 365, 838, "PF00443"]], "ENSPEMP00000016198": [["Domain_0", 368, 838, "PF00443"]], "ENSODEP00000014366": [["Domain_0", 368, 838, "PF00443"]], "ENSPCOP00000000523": [["Domain_0", 368, 838, "PF00443"]], "ENSOGAP00000008979": [["Domain_0", 422, 838, "PF00443"]], "ENSRROP00000033874": [["Domain_0", 368, 838, "PF00443"]], "ENSTGUP00000016381": [["Domain_0", 380, 724, "PF00443"]], "ENSMMUP00000029798": [["Domain_0", 368, 838, "PF00443"]], "ENSTTRP00000008720": [["Domain_0", 365, 838, "PF00443"]], "ENSTSYP00000026009": [["Domain_0", 368, 838, "PF00443"]], "ENSCHOP00000011665": [["Domain_0", 365, 838, "PF00443"]], "ENSACAP00000004141": [["Domain_0", 367, 838, "PF00443"]], "ENSECAP00000014073": [["Domain_0", 366, 838, "PF00443"]], "ENSMAUP00000007684": [["Domain_0", 368, 838, "PF00443"]], "ENSMLEP00000016446": [["Domain_0", 368, 838, "PF00443"]], "ENSOARP00000010328": [["Domain_0", 366, 762, "PF00443"]], "ENSJJAP00000004584": [["Domain_0", 368, 838, "PF00443"]], "ENSFCAP00000009119": [["Domain_0", 368, 838, "PF00443"]], "ENSANAP00000041874": [["Domain_0", 368, 838, "PF00443"]], "ENSMICP00000006603": [["Domain_0", 368, 838, "PF00443"]], "ENSPCAP00000012592": [["Domain_0", 365, 838, "PF00443"]], "ENSPPYP00000000711": [["Domain_0", 366, 838, "PF00443"]], "ENSSHAP00000019394": [["Domain_0", 366, 818, "PF00443"]], "ENSSBOP00000021840": [["Domain_0", 368, 838, "PF00443"]], "ENSCGRP00001024861": [["Domain_0", 368, 836, "PF00443"]], "ENSFDAP00000019602": [["Domain_0", 368, 838, "PF00443"]], "ENSOPRP00000002971": [["Domain_0", 365, 838, "PF00443"]], "ENSBTAP00000029184": [["Domain_0", 366, 838, "PF00443"]], "ENSDORP00000001581": [["Domain_0", 368, 838, "PF00443"]], "ENSPVAP00000005228": [["Domain_0", 365, 838, "PF00443"]], "ENSHGLP00000024472": [["Domain_0", 368, 838, "PF00443"]], "ENSNLEP00000017080": [["Domain_0", 368, 838, "PF00443"]], "ENSPSIP00000005405": [["Domain_0", 367, 838, "PF00443"]], "ENSP00000356981": [["Domain_0", 368, 838, "PF00443"]], "ENSRNOP00000055192": [["Domain_0", 368, 838, "PF00443"]], "ENSCANP00000013721": [["Domain_0", 368, 838, "PF00443"]], "ENSMOCP00000004277": [["Domain_0", 368, 838, "PF00443"]], "ENSTBEP00000014315": [["Domain_0", 131, 838, "PF00443"]], "ENSRBIP00000032862": [["Domain_0", 368, 838, "PF00443"]], "ENSCGRP00001011847": [["Domain_0", 368, 838, "PF00443"]]}