Found:
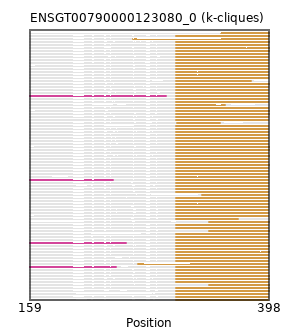
{"ENSBTAP00000005542": [["Domain_1", 305, 397, "PF01588"]], "ENSCHIP00000002356": [["Domain_1", 305, 397, "PF01588"]], "ENSNLEP00000038982": [["Domain_1", 305, 397, "PF01588"]], "ENSPVAP00000010026": [["Domain_1", 305, 398, "PF01588"]], "ENSCHIP00000020449": [["Domain_1", 305, 397, "PF01588"]], "MGP_PahariEiJ_P0071489": [["Domain_1", 305, 397, "PF01588"]], "ENSOARP00000009777": [["Domain_1", 305, 397, "PF01588"]], "ENSRNOP00000015213": [["Domain_1", 305, 397, "PF01588"]], "ENSSBOP00000039335": [["Domain_1", 267, 397, "PF01588"]], "ENSMNEP00000011688": [["Domain_1", 305, 397, "PF01588"]], "ENSFDAP00000001495": [["Domain_1", 305, 397, "PF01588"]], "ENSCJAP00000043263": [["Domain_1", 305, 397, "PF01588"]], "ENSCAFP00000009470": [["Domain_1", 305, 398, "PF01588"]], "ENSODEP00000014345": [["Domain_1", 305, 397, "PF01588"]], "ENSGALP00000017152": [["Domain_1", 305, 397, "PF01588"]], "ENSHGLP00100018028": [["Domain_1", 305, 397, "PF01588"]], "ENSMICP00000009285": [["Domain_1", 305, 397, "PF01588"]], "ENSETEP00000000227": [["Domain_1", 305, 398, "PF01588"]], "ENSMEUP00000006483": [["Domain_1", 305, 398, "PF01588"]], "MGP_SPRETEiJ_P0062969": [["Domain_1", 305, 397, "PF01588"]], "ENSPPAP00000030806": [["Domain_1", 305, 397, "PF01588"]], "ENSGGOP00000019960": [["Domain_1", 305, 397, "PF01588"]], "ENSPPYP00000016731": [["Domain_0", 159, 255, "PF04880"], ["Linker_0_1", 255, 305], ["Domain_1", 305, 397, "PF01588"]], "ENSMODP00000025782": [["Domain_0", 159, 242, "PF04880"], ["Linker_0_1", 242, 305], ["Domain_1", 305, 397, "PF01588"]], "ENSJJAP00000014403": [["Domain_1", 305, 397, "PF01588"]], "ENSANAP00000031351": [["Domain_1", 262, 397, "PF01588"]], "ENSECAP00000004623": [["Domain_1", 305, 397, "PF01588"]], "ENSCAFP00000016229": [["Domain_1", 305, 398, "PF01588"]], "ENSOARP00000021097": [["Domain_1", 305, 397, "PF01588"]], "ENSTBEP00000003803": [["Domain_1", 305, 398, "PF01588"]], "ENSMFAP00000026819": [["Domain_1", 305, 397, "PF01588"]], "ENSSTOP00000007327": [["Domain_1", 305, 397, "PF01588"]], "ENSMLUP00000008701": [["Domain_1", 305, 398, "PF01588"]], "ENSCHIP00000025630": [["Domain_1", 305, 380, "PF01588"]], "ENSLAFP00000010564": [["Domain_1", 305, 398, "PF01588"]], "ENSMOCP00000004497": [["Domain_1", 305, 397, "PF01588"]], "ENSMPUP00000008882": [["Domain_1", 305, 397, "PF01588"]], "ENSOARP00000009218": [["Domain_1", 331, 397, "PF01588"]], "ENSPTRP00000083697": [["Domain_1", 305, 397, "PF01588"]], "ENSCGRP00000020427": [["Domain_1", 305, 397, "PF01588"]], "ENSSSCP00000022764": [["Domain_1", 305, 397, "PF01588"]], "ENSODEP00000011178": [["Domain_1", 305, 397, "PF01588"]], "ENSMUSP00000029663": [["Domain_1", 305, 397, "PF01588"]], "ENSPANP00000000086": [["Domain_1", 305, 397, "PF01588"]], "ENSEEUP00000004792": [["Domain_1", 305, 398, "PF01588"]], "ENSMMUP00000044008": [["Domain_1", 305, 397, "PF01588"]], "ENSOPRP00000009529": [["Domain_1", 305, 367, "PF01588"]], "ENSFDAP00000020028": [["Domain_1", 305, 397, "PF01588"]], "ENSPCOP00000000497": [["Domain_1", 305, 397, "PF01588"]], "ENSMLEP00000032907": [["Domain_1", 305, 397, "PF01588"]], "ENSAPLP00000005622": [["Domain_1", 305, 398, "PF01588"]], "ENSAMEP00000018487": [["Domain_1", 305, 398, "PF01588"]], "ENSOANP00000004417": [["Domain_1", 305, 397, "PF01588"]], "ENSOGAP00000001866": [["Domain_1", 305, 397, "PF01588"]], "ENSCHOP00000000552": [["Domain_1", 305, 398, "PF01588"]], "ENSXETP00000057929": [["Domain_1", 305, 398, "PF01588"]], "ENSDORP00000026799": [["Domain_1", 305, 354, "PF01588"]], "ENSOPRP00000014798": [["Domain_1", 338, 398, "PF01588"]], "ENSPCAP00000014446": [["Domain_1", 338, 398, "PF01588"]], "ENSFALP00000012892": [["Domain_1", 305, 397, "PF01588"]], "ENSSHAP00000000891": [["Domain_0", 159, 245, "PF04880"], ["Linker_0_1", 245, 305], ["Domain_1", 305, 397, "PF01588"]], "ENSCHIP00000004196": [["Domain_1", 305, 397, "PF01588"]], "ENSOCUP00000023259": [["Domain_1", 305, 397, "PF01588"]], "ENSOARP00000010015": [["Domain_1", 305, 397, "PF01588"]], "ENSTGUP00000004034": [["Domain_1", 305, 398, "PF01588"]], "ENSCGRP00001010824": [["Domain_1", 305, 397, "PF01588"]], "ENSP00000378191": [["Domain_1", 305, 397, "PF01588"]], "ENSFCAP00000011958": [["Domain_1", 305, 397, "PF01588"]], "ENSCLAP00000005574": [["Domain_1", 305, 397, "PF01588"]], "ENSCPOP00000006068": [["Domain_1", 305, 397, "PF01588"]], "ENSDNOP00000019294": [["Domain_1", 305, 397, "PF01588"]], "ENSTSYP00000012648": [["Domain_1", 305, 397, "PF01588"]], "ENSCCAP00000028085": [["Domain_1", 305, 397, "PF01588"]], "ENSFCAP00000029113": [["Domain_1", 306, 349, "PF01588"]], "ENSDORP00000004694": [["Domain_1", 305, 397, "PF01588"]], "ENSMOCP00000001453": [["Domain_1", 305, 397, "PF01588"]], "ENSRBIP00000010056": [["Domain_1", 305, 397, "PF01588"]], "ENSMGAP00000005879": [["Domain_1", 305, 398, "PF01588"]], "ENSPSIP00000007164": [["Domain_1", 305, 398, "PF01588"]], "ENSPEMP00000002927": [["Domain_1", 305, 397, "PF01588"]], "ENSCATP00000021750": [["Domain_1", 305, 397, "PF01588"]], "ENSRROP00000015190": [["Domain_1", 305, 397, "PF01588"]], "ENSTTRP00000007360": [["Domain_1", 338, 398, "PF01588"]], "ENSCAPP00000009628": [["Domain_1", 305, 397, "PF01588"]], "ENSCSAP00000000344": [["Domain_0", 159, 295, "PF04880"], ["Linker_0_1", 295, 305], ["Domain_1", 305, 397, "PF01588"]], "MGP_CAROLIEiJ_P0060560": [["Domain_1", 305, 397, "PF01588"]], "ENSVPAP00000003664": [["Domain_1", 305, 398, "PF01588"]], "ENSACAP00000021668": [["Domain_1", 350, 398, "PF01588"]], "ENSCANP00000027250": [["Domain_1", 305, 397, "PF01588"]], "ENSHGLP00000005280": [["Domain_1", 305, 397, "PF01588"]]}