Found:
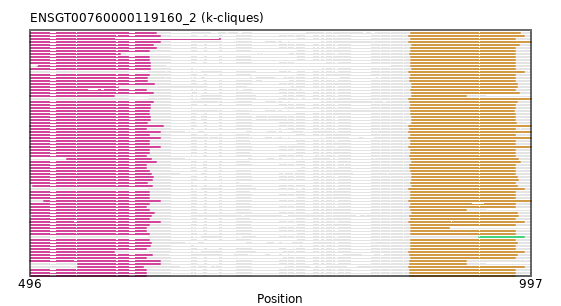
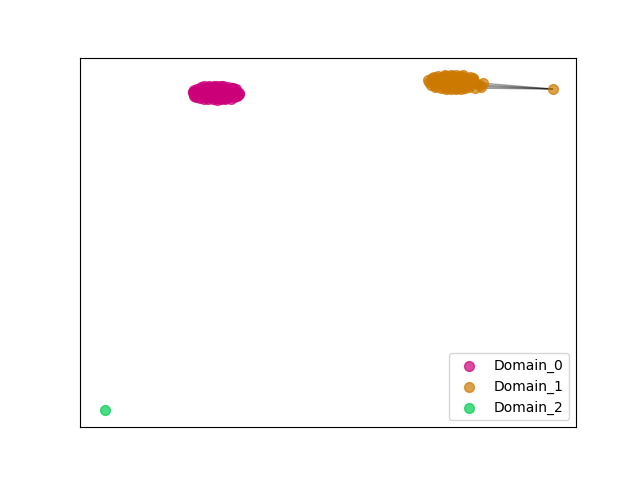
{"ENSMEUP00000010457": [["Domain_0", 496, 626, "PF00068"], ["Linker_0_1", 626, 875], ["Domain_1", 875, 997, "PF00068"]], "ENSOCUP00000010355": [["Domain_0", 497, 618, "PF00068"], ["Linker_0_1", 618, 878], ["Domain_1", 878, 984, "PF00068"]], "ENSMMUP00000047562": [["Domain_0", 497, 615, "PF00068"], ["Linker_0_1", 615, 877], ["Domain_1", 877, 981, "PF00068"]], "ENSHGLP00100007997": [["Domain_0", 497, 616, "PF00068"], ["Linker_0_1", 616, 878], ["Domain_1", 878, 981, "PF00068"]], "ENSLAFP00000004256": [["Domain_0", 496, 629, "PF00068"], ["Linker_0_1", 629, 875], ["Domain_1", 875, 997, "PF00068"]], "MGP_SPRETEiJ_P0040142": [["Domain_0", 497, 612, "PF00068"], ["Linker_0_1", 612, 876], ["Domain_1", 876, 981, "PF00068"]], "ENSRNOP00000046821": [["Domain_0", 497, 612, "PF00068"], ["Linker_0_1", 612, 877], ["Domain_1", 877, 981, "PF00068"]], "ENSPPYP00000021191": [["Domain_0", 497, 620, "PF00068"], ["Linker_0_1", 620, 878], ["Domain_1", 878, 983, "PF00068"]], "ENSBTAP00000037179": [["Domain_0", 497, 619, "PF00068"], ["Linker_0_1", 619, 878], ["Domain_1", 878, 985, "PF00068"]], "ENSRBIP00000008749": [["Domain_0", 497, 615, "PF00068"], ["Linker_0_1", 615, 877], ["Domain_1", 877, 981, "PF00068"]], "ENSOARP00000007552": [["Domain_0", 497, 619, "PF00068"], ["Linker_0_1", 619, 878], ["Domain_1", 878, 983, "PF00068"]], "ENSANAP00000036656": [["Domain_0", 497, 686, "PF00068"], ["Linker_0_1", 686, 877], ["Domain_1", 877, 981, "PF00068"]], "ENSMOCP00000003915": [["Domain_0", 497, 612, "PF00068"], ["Linker_0_1", 612, 876], ["Domain_1", 876, 981, "PF00068"]], "ENSAMEP00000013676": [["Domain_0", 496, 626, "PF00068"], ["Linker_0_1", 626, 875], ["Domain_1", 875, 990, "PF00068"]], "ENSPCAP00000008696": [["Domain_0", 510, 626, "PF00068"], ["Linker_0_1", 626, 875], ["Domain_1", 875, 997, "PF00068"]], "ENSFALP00000002704": [["Domain_0", 497, 619, "PF00068"], ["Linker_0_1", 619, 876], ["Domain_1", 876, 983, "PF00068"]], "ENSMICP00000003280": [["Domain_0", 497, 615, "PF00068"], ["Linker_0_1", 615, 877], ["Domain_1", 877, 981, "PF00068"]], "ENSTSYP00000003172": [["Domain_0", 497, 612, "PF00068"], ["Linker_0_1", 612, 877], ["Domain_1", 877, 981, "PF00068"]], "ENSMNEP00000019017": [["Domain_0", 497, 615, "PF00068"], ["Linker_0_1", 615, 877], ["Domain_1", 877, 981, "PF00068"]], "ENSTTRP00000012769": [["Domain_0", 496, 626, "PF00068"], ["Linker_0_1", 626, 876], ["Domain_1", 876, 932, "PF00068"]], "ENSPVAP00000009372": [["Domain_0", 496, 626, "PF00068"], ["Linker_0_1", 626, 875], ["Domain_1", 875, 990, "PF00068"]], "ENSSARP00000007645": [["Domain_2", 945, 990, "PF00068"]], "ENSTBEP00000011674": [["Domain_0", 497, 580, "PF00068"], ["Linker_0_1", 580, 875], ["Domain_1", 875, 990, "PF00068"]], "ENSP00000262283": [["Domain_0", 497, 615, "PF00068"], ["Linker_0_1", 615, 877], ["Domain_1", 877, 981, "PF00068"]], "ENSSSCP00000006364": [["Domain_0", 497, 616, "PF00068"], ["Linker_0_1", 616, 877], ["Domain_1", 877, 981, "PF00068"]], "ENSCGRP00000008298": [["Domain_0", 497, 616, "PF00068"], ["Linker_0_1", 616, 877], ["Domain_1", 877, 981, "PF00068"]], "ENSPSIP00000006978": [["Domain_0", 498, 617, "PF00068"], ["Linker_0_1", 617, 876], ["Domain_1", 876, 984, "PF00068"]], "ENSCJAP00000008090": [["Domain_0", 497, 615, "PF00068"], ["Linker_0_1", 615, 877], ["Domain_1", 877, 981, "PF00068"]], "ENSOANP00000012878": [["Domain_0", 497, 617, "PF00068"], ["Linker_0_1", 617, 878], ["Domain_1", 878, 981, "PF00068"]], "ENSMLUP00000005956": [["Domain_0", 496, 626, "PF00068"], ["Linker_0_1", 626, 875], ["Domain_1", 875, 997, "PF00068"]], "ENSPTRP00000035206": [["Domain_0", 497, 615, "PF00068"], ["Linker_0_1", 615, 877], ["Domain_1", 877, 981, "PF00068"]], "ENSXETP00000059896": [["Domain_0", 544, 609, "PF00068"], ["Linker_0_1", 609, 875], ["Domain_1", 875, 990, "PF00068"]], "ENSJJAP00000017394": [["Domain_0", 497, 613, "PF00068"], ["Linker_0_1", 613, 878], ["Domain_1", 878, 982, "PF00068"]], "ENSACAP00000010594": [["Domain_0", 497, 622, "PF00068"], ["Linker_0_1", 622, 877], ["Domain_1", 877, 986, "PF00068"]], "ENSFDAP00000009142": [["Domain_0", 497, 616, "PF00068"], ["Linker_0_1", 616, 876], ["Domain_1", 876, 981, "PF00068"]], "ENSMLEP00000021256": [["Domain_0", 497, 615, "PF00068"], ["Linker_0_1", 615, 877], ["Domain_1", 877, 981, "PF00068"]], "ENSECAP00000012376": [["Domain_0", 497, 619, "PF00068"], ["Linker_0_1", 619, 878], ["Domain_1", 878, 985, "PF00068"]], "ENSAPLP00000003985": [["Domain_0", 496, 626, "PF00068"], ["Linker_0_1", 626, 875], ["Domain_1", 875, 997, "PF00068"]], "ENSSTOP00000009967": [["Domain_0", 497, 612, "PF00068"], ["Linker_0_1", 612, 877], ["Domain_1", 877, 981, "PF00068"]], "ENSCLAP00000001790": [["Domain_0", 497, 613, "PF00068"], ["Linker_0_1", 613, 876], ["Domain_1", 876, 981, "PF00068"]], "ENSNGAP00000007404": [["Domain_0", 497, 612, "PF00068"], ["Linker_0_1", 612, 876], ["Domain_1", 876, 981, "PF00068"]], "ENSOPRP00000014234": [["Domain_1", 875, 990, "PF00068"]], "ENSCHIP00000012540": [["Domain_0", 497, 616, "PF00068"], ["Linker_0_1", 616, 878], ["Domain_1", 878, 981, "PF00068"]], "ENSNLEP00000006946": [["Domain_0", 497, 615, "PF00068"], ["Linker_0_1", 615, 877], ["Domain_1", 877, 981, "PF00068"]], "ENSCCAP00000026553": [["Domain_0", 497, 615, "PF00068"], ["Linker_0_1", 615, 877], ["Domain_1", 877, 981, "PF00068"]], "ENSCAPP00000006420": [["Domain_0", 497, 586, "PF00068"], ["Linker_0_1", 586, 876], ["Domain_1", 876, 981, "PF00068"]], "ENSRNOP00000045370": [["Domain_0", 497, 612, "PF00068"], ["Linker_0_1", 612, 877], ["Domain_1", 877, 915, "PF00068"]], "ENSVPAP00000001322": [["Domain_0", 544, 626, "PF00068"], ["Linker_0_1", 626, 876], ["Domain_1", 876, 932, "PF00068"]], "ENSCAFP00000001544": [["Domain_0", 497, 622, "PF00068"], ["Linker_0_1", 622, 877], ["Domain_1", 877, 981, "PF00068"]], "ENSRROP00000040772": [["Domain_0", 497, 615, "PF00068"], ["Linker_0_1", 615, 877], ["Domain_1", 877, 981, "PF00068"]], "ENSDORP00000012907": [["Domain_0", 497, 612, "PF00068"], ["Linker_0_1", 612, 877], ["Domain_1", 877, 981, "PF00068"]], "ENSPEMP00000014372": [["Domain_0", 497, 612, "PF00068"], ["Linker_0_1", 612, 876], ["Domain_1", 876, 981, "PF00068"]], "ENSPPAP00000022956": [["Domain_0", 497, 615, "PF00068"], ["Linker_0_1", 615, 877], ["Domain_1", 877, 932, "PF00068"]], "ENSOGAP00000011511": [["Domain_0", 499, 618, "PF00068"], ["Linker_0_1", 618, 878], ["Domain_1", 878, 983, "PF00068"]], "ENSGGOP00000001057": [["Domain_0", 497, 615, "PF00068"], ["Linker_0_1", 615, 877], ["Domain_1", 877, 981, "PF00068"]], "MGP_PahariEiJ_P0041717": [["Domain_0", 497, 612, "PF00068"], ["Linker_0_1", 612, 876], ["Domain_1", 876, 981, "PF00068"]], "ENSPANP00000004957": [["Domain_0", 497, 615, "PF00068"], ["Linker_0_1", 615, 877], ["Domain_1", 877, 981, "PF00068"]], "ENSP00000254627": [["Domain_0", 497, 615, "PF00068"], ["Linker_0_1", 615, 877], ["Domain_1", 877, 981, "PF00068"]], "ENSHGLP00000022533": [["Domain_0", 497, 616, "PF00068"], ["Linker_0_1", 616, 878], ["Domain_1", 878, 981, "PF00068"]], "ENSCGRP00001001056": [["Domain_0", 504, 616, "PF00068"], ["Linker_0_1", 616, 877], ["Domain_1", 877, 981, "PF00068"]], "ENSMGAP00000013519": [["Domain_0", 496, 626, "PF00068"], ["Linker_0_1", 626, 875], ["Domain_1", 875, 997, "PF00068"]], "MGP_CAROLIEiJ_P0038354": [["Domain_0", 497, 612, "PF00068"], ["Linker_0_1", 612, 876], ["Domain_1", 876, 981, "PF00068"]], "ENSSBOP00000031077": [["Domain_0", 497, 615, "PF00068"], ["Linker_0_1", 615, 877], ["Domain_1", 877, 981, "PF00068"]], "ENSCSAP00000006251": [["Domain_0", 497, 620, "PF00068"], ["Linker_0_1", 620, 878], ["Domain_1", 878, 983, "PF00068"]], "ENSMFAP00000032089": [["Domain_0", 497, 615, "PF00068"], ["Linker_0_1", 615, 877], ["Domain_1", 877, 981, "PF00068"]], "ENSCPOP00000008104": [["Domain_0", 497, 613, "PF00068"], ["Linker_0_1", 613, 876], ["Domain_1", 876, 981, "PF00068"]], "ENSGALP00000045769": [["Domain_0", 497, 615, "PF00068"], ["Linker_0_1", 615, 877], ["Domain_1", 877, 980, "PF00068"]], "ENSMODP00000001477": [["Domain_0", 533, 617, "PF00068"], ["Linker_0_1", 617, 877], ["Domain_1", 877, 984, "PF00068"]], "ENSMPUP00000001401": [["Domain_0", 497, 622, "PF00068"], ["Linker_0_1", 622, 878], ["Domain_1", 878, 986, "PF00068"]], "ENSPCOP00000005998": [["Domain_0", 497, 615, "PF00068"], ["Linker_0_1", 615, 877], ["Domain_1", 877, 981, "PF00068"]], "ENSMUSP00000062865": [["Domain_0", 497, 612, "PF00068"], ["Linker_0_1", 612, 876], ["Domain_1", 876, 981, "PF00068"]], "ENSSHAP00000006542": [["Domain_0", 497, 617, "PF00068"], ["Linker_0_1", 617, 877], ["Domain_1", 877, 983, "PF00068"]], "ENSODEP00000000565": [["Domain_0", 497, 613, "PF00068"], ["Linker_0_1", 613, 877], ["Domain_1", 877, 981, "PF00068"]], "ENSCHOP00000006753": [["Domain_1", 875, 990, "PF00068"]], "ENSFCAP00000036449": [["Domain_0", 497, 616, "PF00068"], ["Linker_0_1", 616, 877], ["Domain_1", 877, 981, "PF00068"]], "ENSTGUP00000012909": [["Domain_0", 497, 618, "PF00068"], ["Linker_0_1", 618, 876], ["Domain_1", 876, 983, "PF00068"]], "ENSETEP00000006868": [["Domain_1", 875, 997, "PF00068"]], "ENSDNOP00000025229": [["Domain_0", 497, 584, "PF00068"], ["Linker_0_1", 584, 878], ["Domain_1", 878, 983, "PF00068"]], "ENSCANP00000026750": [["Domain_0", 497, 615, "PF00068"], ["Linker_0_1", 615, 877], ["Domain_1", 877, 981, "PF00068"]], "ENSEEUP00000004997": [["Domain_0", 496, 580, "PF00068"], ["Linker_0_1", 580, 878], ["Domain_1", 878, 932, "PF00068"]], "ENSCATP00000012496": [["Domain_0", 497, 615, "PF00068"], ["Linker_0_1", 615, 877], ["Domain_1", 877, 981, "PF00068"]], "ENSMAUP00000015281": [["Domain_0", 497, 612, "PF00068"], ["Linker_0_1", 612, 876], ["Domain_1", 876, 981, "PF00068"]]}