Found:
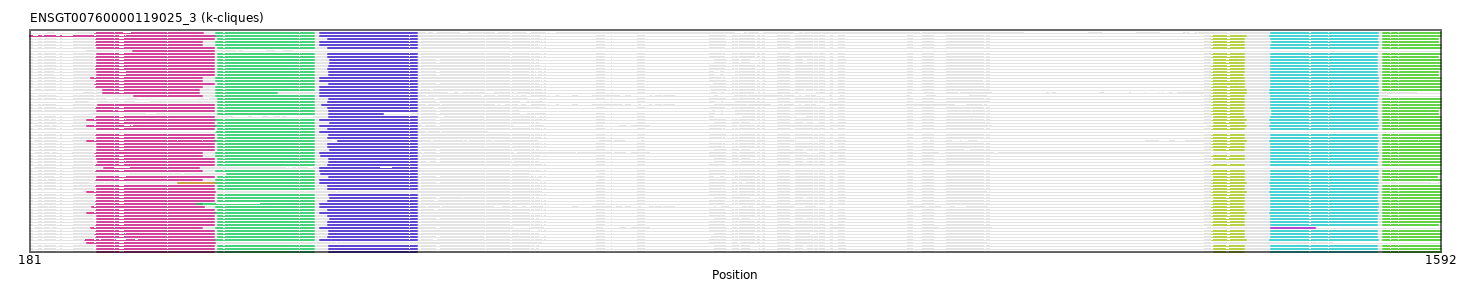
{"ENSEEUP00000014365": [["Domain_0", 254, 350, "PF00047"], ["Linker_0_2", 350, 367], ["Domain_2", 367, 465, "PF00193"], ["Linker_2_3", 465, 471], ["Domain_3", 471, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1397, "PF07974"], ["Linker_4_5", 1397, 1421], ["Domain_5", 1421, 1529, "PF00059"], ["Linker_5_7", 1529, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSFDAP00000018463": [["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 479], ["Domain_3", 479, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSODEP00000013541": [["Domain_0", 248, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 480], ["Domain_3", 480, 568, "PF00193"], ["Linker_3_4", 568, 1365], ["Domain_4", 1365, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1588, "PF00084"]], "ENSBTAP00000020965": [["Domain_0", 247, 353, "PF07686"], ["Linker_0_2", 353, 367], ["Domain_2", 367, 465, "PF00193"], ["Linker_2_3", 465, 471], ["Domain_3", 471, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1423], ["Domain_5", 1423, 1529, "PF00059"], ["Linker_5_7", 1529, 1534], ["Domain_7", 1534, 1589, "PF00084"]], "ENSP00000331210": [["Domain_0", 247, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 479], ["Domain_3", 479, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSLAFP00000026161": [["Domain_0", 238, 366, "PF07686"], ["Linker_0_2", 366, 367], ["Domain_2", 367, 465, "PF00193"], ["Linker_2_3", 465, 471], ["Domain_3", 471, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1397, "PF00008"], ["Linker_4_5", 1397, 1421], ["Domain_5", 1421, 1529, "PF00059"], ["Linker_5_7", 1529, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSCJAP00000011200": [["Domain_0", 247, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 479], ["Domain_3", 479, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSPPAP00000033220": [["Domain_0", 247, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 479], ["Domain_3", 479, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSRBIP00000022692": [["Domain_0", 248, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 479], ["Domain_3", 479, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSOARP00000006609": [["Domain_0", 247, 353, "PF07686"], ["Linker_0_2", 353, 367], ["Domain_2", 367, 465, "PF00193"], ["Linker_2_3", 465, 471], ["Domain_3", 471, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1529, "PF00059"], ["Linker_5_7", 1529, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSHGLP00000005824": [["Domain_0", 248, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 480], ["Domain_3", 480, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1394, "PF00008"], ["Linker_4_5", 1394, 1423], ["Domain_5", 1423, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1589, "PF00084"]], "ENSMGAP00000004468": [["Domain_2", 367, 465, "PF00193"], ["Linker_2_3", 465, 471], ["Domain_3", 471, 568, "PF00193"]], "ENSTSYP00000025023": [["Domain_0", 247, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 479], ["Domain_3", 479, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSANAP00000026375": [["Domain_0", 247, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 479], ["Domain_3", 479, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1589, "PF00084"]], "ENSOPRP00000000562": [["Domain_1", 329, 366, "PF07686"], ["Linker_1_2", 366, 367], ["Domain_2", 367, 465, "PF00193"], ["Linker_2_3", 465, 471], ["Domain_3", 471, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1397, "PF00008"], ["Linker_4_5", 1397, 1421], ["Domain_5", 1421, 1529, "PF00059"]], "ENSPCOP00000022740": [["Domain_0", 247, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 480], ["Domain_3", 480, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSCSAP00000015864": [["Domain_0", 242, 353, "PF07686"], ["Linker_0_2", 353, 367], ["Domain_2", 367, 465, "PF00193"], ["Linker_2_3", 465, 471], ["Domain_3", 471, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1529, "PF00059"], ["Linker_5_7", 1529, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSGALP00000061057": [["Domain_0", 249, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 473], ["Domain_3", 473, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSMNEP00000003179": [["Domain_0", 248, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 479], ["Domain_3", 479, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSCATP00000003078": [["Domain_0", 248, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 479], ["Domain_3", 479, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSMAUP00000008756": [["Domain_0", 248, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 481], ["Domain_3", 481, 568, "PF00193"], ["Linker_3_4", 568, 1365], ["Domain_4", 1365, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSMMUP00000022489": [["Domain_0", 248, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 479], ["Domain_3", 479, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSCPOP00000032780": [["Domain_0", 248, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 480], ["Domain_3", 480, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1394, "PF00008"], ["Linker_4_5", 1394, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1589, "PF00084"]], "ENSFCAP00000006092": [["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 480], ["Domain_3", 480, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSMEUP00000008413": [["Domain_0", 238, 366, "PF07686"], ["Linker_0_2", 366, 367], ["Domain_2", 367, 465, "PF00193"], ["Linker_2_3", 465, 471], ["Domain_3", 471, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1397, "PF00008"], ["Linker_4_5", 1397, 1421], ["Domain_5", 1421, 1529, "PF00059"], ["Linker_5_7", 1529, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSMFAP00000009523": [["Domain_0", 248, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 479], ["Domain_3", 479, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSPANP00000010953": [["Domain_0", 248, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 479], ["Domain_3", 479, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSMICP00000026117": [["Domain_0", 247, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 480], ["Domain_3", 480, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSTTRP00000013753": [["Domain_0", 236, 366, "PF07686"], ["Linker_0_2", 366, 367], ["Domain_2", 367, 465, "PF00193"], ["Linker_2_3", 465, 471], ["Domain_3", 471, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1397, "PF00008"], ["Linker_4_5", 1397, 1421], ["Domain_5", 1421, 1529, "PF00059"], ["Linker_5_7", 1529, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSMUSP00000088491": [["Domain_0", 248, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 480], ["Domain_3", 480, 568, "PF00193"], ["Linker_3_4", 568, 1365], ["Domain_4", 1365, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSCHIP00000008519": [["Domain_0", 247, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 480], ["Domain_3", 480, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSAMEP00000015680": [["Domain_0", 181, 366, "PF07686"], ["Linker_0_2", 366, 367], ["Domain_2", 367, 465, "PF00193"], ["Linker_2_3", 465, 471], ["Domain_3", 471, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1397, "PF07974"], ["Linker_4_5", 1397, 1421], ["Domain_5", 1421, 1529, "PF00059"], ["Linker_5_7", 1529, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSCGRP00000006381": [["Domain_0", 248, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 481], ["Domain_3", 481, 568, "PF00193"], ["Linker_3_4", 568, 1365], ["Domain_4", 1365, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "MGP_CAROLIEiJ_P0058331": [["Domain_0", 248, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 480], ["Domain_3", 480, 568, "PF00193"], ["Linker_3_4", 568, 1365], ["Domain_4", 1365, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSCAPP00000001112": [["Domain_0", 284, 365, "PF07686"]], "ENSMLUP00000018314": [["Domain_0", 238, 366, "PF07686"], ["Linker_0_2", 366, 367], ["Domain_2", 367, 465, "PF00193"], ["Linker_2_3", 465, 489], ["Domain_3", 489, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1397, "PF00008"], ["Linker_4_5", 1397, 1421], ["Domain_5", 1421, 1529, "PF00059"], ["Linker_5_7", 1529, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSPSIP00000009237": [["Domain_0", 243, 355, "PF07686"], ["Linker_0_2", 355, 367], ["Domain_2", 367, 465, "PF00193"], ["Linker_2_3", 465, 471], ["Domain_3", 471, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1394, "PF00008"], ["Linker_4_5", 1394, 1422], ["Domain_5", 1422, 1529, "PF00059"], ["Linker_5_7", 1529, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSOANP00000020921": [["Domain_0", 255, 350, "PF00047"], ["Domain_3", 471, 568, "PF00193"]], "MGP_SPRETEiJ_P0060661": [["Domain_0", 248, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 480], ["Domain_3", 480, 568, "PF00193"], ["Linker_3_4", 568, 1365], ["Domain_4", 1365, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "MGP_PahariEiJ_P0069292": [["Domain_0", 248, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 480], ["Domain_3", 480, 568, "PF00193"], ["Linker_3_4", 568, 1365], ["Domain_4", 1365, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSGGOP00000022275": [["Domain_0", 247, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 479], ["Domain_3", 479, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSJJAP00000019350": [["Domain_0", 247, 365, "PF07686"], ["Domain_3", 480, 568, "PF00193"], ["Linker_3_4", 568, 1365], ["Domain_4", 1365, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSOCUP00000025883": [["Domain_2", 378, 465, "PF00193"], ["Linker_2_3", 465, 472], ["Domain_3", 472, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1529, "PF00059"], ["Linker_5_7", 1529, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSETEP00000014603": [["Domain_0", 254, 350, "PF00047"], ["Linker_0_2", 350, 367], ["Domain_2", 367, 428, "PF00193"], ["Linker_2_3", 428, 471], ["Domain_3", 471, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1397, "PF07974"], ["Linker_4_5", 1397, 1421], ["Domain_5", 1421, 1529, "PF00059"]], "ENSRNOP00000025496": [["Domain_0", 248, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 481], ["Domain_3", 481, 568, "PF00193"], ["Linker_3_4", 568, 1365], ["Domain_4", 1365, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSHGLP00100016079": [["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 480], ["Domain_3", 480, 534, "PF00193"], ["Linker_3_4", 534, 1363], ["Domain_4", 1363, 1394, "PF00008"], ["Linker_4_5", 1394, 1423], ["Domain_5", 1423, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1589, "PF00084"]], "ENSOGAP00000015052": [["Domain_0", 247, 353, "PF07686"], ["Linker_0_2", 353, 367], ["Domain_2", 367, 465, "PF00193"], ["Linker_2_3", 465, 471], ["Domain_3", 471, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1529, "PF00059"], ["Linker_5_7", 1529, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSSBOP00000038341": [["Domain_0", 247, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 479], ["Domain_3", 479, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1589, "PF00084"]], "ENSECAP00000015557": [["Domain_0", 247, 353, "PF07686"], ["Linker_0_2", 353, 367], ["Domain_2", 367, 465, "PF00193"], ["Linker_2_3", 465, 471], ["Domain_3", 471, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1529, "PF00059"], ["Linker_5_7", 1529, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSNGAP00000008229": [["Domain_0", 249, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 480], ["Domain_3", 480, 568, "PF00193"], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSMLEP00000040761": [["Domain_0", 248, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 479], ["Domain_3", 479, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSVPAP00000009282": [["Domain_0", 238, 366, "PF07686"]], "ENSMODP00000033648": [["Domain_0", 246, 353, "PF07686"], ["Linker_0_2", 353, 367], ["Domain_2", 367, 465, "PF00193"], ["Linker_2_3", 465, 471], ["Domain_3", 471, 568, "PF00193"], ["Domain_5", 1422, 1529, "PF00059"], ["Linker_5_7", 1529, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSFALP00000003443": [["Domain_0", 285, 353, "PF07686"], ["Linker_0_2", 353, 367], ["Domain_2", 367, 465, "PF00193"], ["Linker_2_3", 465, 471], ["Domain_3", 471, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1529, "PF00059"]], "ENSMPUP00000004089": [["Domain_0", 247, 353, "PF07686"], ["Linker_0_2", 353, 367], ["Domain_2", 367, 465, "PF00193"], ["Linker_2_3", 465, 472], ["Domain_3", 472, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1529, "PF00059"], ["Linker_5_7", 1529, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSRROP00000031636": [["Domain_0", 248, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 479], ["Domain_3", 479, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSPCAP00000005141": [["Domain_0", 238, 366, "PF07686"], ["Domain_4", 1363, 1397, "PF07974"], ["Linker_4_5", 1397, 1421], ["Domain_5", 1421, 1529, "PF00059"], ["Linker_5_7", 1529, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSNLEP00000015444": [["Domain_0", 247, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 479], ["Domain_3", 479, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSPVAP00000007085": [["Domain_0", 238, 366, "PF07686"], ["Linker_0_2", 366, 367], ["Domain_2", 367, 465, "PF00193"], ["Linker_2_3", 465, 471], ["Domain_3", 471, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1397, "PF00008"], ["Linker_4_5", 1397, 1421], ["Domain_5", 1421, 1529, "PF00059"], ["Linker_5_7", 1529, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSCAFP00000024595": [["Domain_0", 247, 353, "PF07686"], ["Linker_0_2", 353, 367], ["Domain_2", 367, 465, "PF00193"], ["Linker_2_3", 465, 471], ["Domain_3", 471, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1529, "PF00059"], ["Linker_5_7", 1529, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSDORP00000009823": [["Domain_0", 246, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 480], ["Domain_3", 480, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSPTRP00000051705": [["Domain_0", 247, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 479], ["Domain_3", 479, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSPPYP00000000820": [["Domain_0", 247, 353, "PF07686"], ["Domain_2", 348, 465, "PF00193"], ["Linker_2_3", 465, 471], ["Domain_3", 471, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1529, "PF00059"], ["Linker_5_7", 1529, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSCANP00000016346": [["Domain_0", 248, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 479], ["Domain_3", 479, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSDNOP00000017416": [["Domain_0", 247, 353, "PF07686"], ["Linker_0_2", 353, 367], ["Domain_2", 367, 465, "PF00193"], ["Linker_2_3", 465, 471], ["Domain_3", 471, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1529, "PF00059"], ["Linker_5_7", 1529, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSSSCP00000043407": [["Domain_0", 247, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 480], ["Domain_3", 480, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSPEMP00000016621": [["Domain_0", 248, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 481], ["Domain_3", 481, 568, "PF00193"], ["Linker_3_4", 568, 1365], ["Domain_4", 1365, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSCGRP00001000523": [["Domain_0", 248, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 481], ["Domain_3", 481, 568, "PF00193"], ["Linker_3_4", 568, 1365], ["Domain_4", 1365, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSCLAP00000004360": [["Domain_0", 248, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 480], ["Domain_3", 480, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1589, "PF00084"]], "ENSACAP00000004819": [["Domain_0", 246, 354, "PF07686"], ["Linker_0_2", 354, 367], ["Domain_2", 367, 465, "PF00193"], ["Linker_2_3", 465, 471], ["Domain_3", 471, 568, "PF00193"], ["Domain_5", 1422, 1529, "PF00059"], ["Linker_5_7", 1529, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSMOCP00000000164": [["Domain_0", 248, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 481], ["Domain_3", 481, 568, "PF00193"], ["Linker_3_4", 568, 1365], ["Domain_4", 1365, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSSTOP00000006851": [["Domain_0", 246, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 480], ["Domain_3", 480, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1592, "PF00084"]], "ENSSHAP00000018302": [["Domain_0", 242, 353, "PF07686"], ["Linker_0_2", 353, 367], ["Domain_2", 367, 465, "PF00193"], ["Linker_2_3", 465, 471], ["Domain_3", 471, 568, "PF00193"], ["Domain_6", 1422, 1466, "PF00059"]], "ENSCCAP00000038667": [["Domain_0", 247, 365, "PF07686"], ["Linker_0_2", 365, 369], ["Domain_2", 369, 465, "PF00193"], ["Linker_2_3", 465, 479], ["Domain_3", 479, 568, "PF00193"], ["Linker_3_4", 568, 1363], ["Domain_4", 1363, 1395, "PF00008"], ["Linker_4_5", 1395, 1422], ["Domain_5", 1422, 1528, "PF00059"], ["Linker_5_7", 1528, 1534], ["Domain_7", 1534, 1589, "PF00084"]]}