Found:
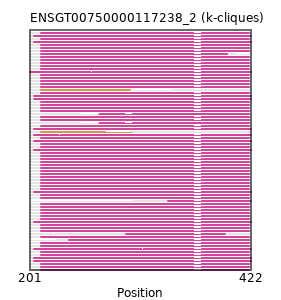
{"ENSPCAP00000007548": [["Domain_0", 339, 422, "PF00566"]], "ENSSBOP00000010121": [["Domain_0", 212, 420, "PF00566"]], "ENSMFAP00000034209": [["Domain_0", 212, 420, "PF00566"]], "ENSPTRP00000001657": [["Domain_0", 212, 420, "PF00566"]], "ENSHGLP00000007233": [["Domain_0", 212, 420, "PF00566"]], "ENSEEUP00000003528": [["Domain_0", 205, 422, "PF00566"]], "ENSCAFP00000029885": [["Domain_0", 212, 420, "PF00566"]], "ENSCAPP00000011120": [["Domain_0", 212, 398, "PF00566"]], "ENSTTRP00000000075": [["Domain_0", 205, 422, "PF00566"]], "ENSBTAP00000040066": [["Domain_0", 212, 420, "PF00566"]], "ENSPVAP00000006545": [["Domain_0", 205, 422, "PF00566"]], "ENSCSAP00000016554": [["Domain_0", 212, 420, "PF00566"]], "ENSOPRP00000001373": [["Domain_0", 205, 422, "PF00566"]], "ENSCANP00000028923": [["Domain_0", 212, 420, "PF00566"]], "ENSOCUP00000001554": [["Domain_0", 212, 420, "PF00566"]], "ENSANAP00000008191": [["Domain_0", 212, 420, "PF00566"]], "ENSOANP00000001627": [["Domain_0", 212, 420, "PF00566"]], "ENSP00000440826": [["Domain_0", 212, 420, "PF00566"]], "ENSTBEP00000008139": [["Domain_0", 205, 422, "PF00566"]], "ENSNGAP00000004574": [["Domain_0", 212, 420, "PF00566"]], "ENSCCAP00000032121": [["Domain_0", 212, 420, "PF00566"]], "ENSMGAP00000008358": [["Domain_0", 205, 422, "PF00566"]], "ENSCLAP00000012383": [["Domain_0", 212, 420, "PF00566"]], "ENSAPLP00000015686": [["Domain_0", 205, 422, "PF00566"]], "ENSPANP00000005502": [["Domain_0", 212, 420, "PF00566"]], "ENSOARP00000017642": [["Domain_0", 212, 420, "PF00566"]], "ENSRBIP00000017632": [["Domain_0", 212, 420, "PF00566"]], "ENSVPAP00000011343": [["Domain_0", 205, 422, "PF00566"]], "ENSRROP00000033803": [["Domain_0", 212, 420, "PF00566"]], "MGP_PahariEiJ_P0032697": [["Domain_0", 212, 420, "PF00566"]], "ENSCGRP00000019090": [["Domain_0", 212, 420, "PF00566"]], "ENSSARP00000009764": [["Domain_0", 297, 396, "PF00566"]], "ENSHGLP00100000613": [["Domain_0", 212, 420, "PF00566"]], "ENSMUSP00000108261": [["Domain_0", 212, 420, "PF00566"]], "ENSJJAP00000000497": [["Domain_0", 212, 420, "PF00566"]], "ENSDORP00000003933": [["Domain_1", 212, 301, "PF00566"]], "MGP_SPRETEiJ_P0072221": [["Domain_0", 212, 420, "PF00566"]], "ENSMAUP00000018407": [["Domain_1", 212, 302, "PF00566"]], "MGP_CAROLIEiJ_P0069560": [["Domain_0", 212, 420, "PF00566"]], "ENSSSCP00000020009": [["Domain_0", 212, 420, "PF00566"]], "ENSHGLP00100013492": [["Domain_0", 270, 420, "PF00566"]], "ENSTSYP00000026885": [["Domain_0", 212, 420, "PF00566"]], "ENSPEMP00000017616": [["Domain_0", 212, 420, "PF00566"]], "ENSCPOP00000006238": [["Domain_0", 212, 420, "PF00566"]], "ENSFDAP00000023376": [["Domain_0", 212, 420, "PF00566"]], "ENSMOCP00000008596": [["Domain_0", 212, 420, "PF00566"]], "ENSRNOP00000058347": [["Domain_0", 212, 420, "PF00566"]], "ENSETEP00000013747": [["Domain_0", 205, 422, "PF00566"]], "ENSOGAP00000011866": [["Domain_0", 212, 420, "PF00566"]], "ENSMLEP00000017937": [["Domain_0", 212, 420, "PF00566"]], "ENSODEP00000008122": [["Domain_0", 212, 420, "PF00566"]], "ENSHGLP00000006268": [["Domain_0", 270, 420, "PF00566"]], "ENSAMEP00000010444": [["Domain_0", 205, 422, "PF00566"]], "ENSPPYP00000001337": [["Domain_0", 212, 420, "PF00566"]], "ENSMMUP00000039738": [["Domain_0", 212, 420, "PF00566"]], "ENSCHIP00000032477": [["Domain_0", 212, 420, "PF00566"]], "ENSNLEP00000028595": [["Domain_0", 212, 420, "PF00566"]], "ENSPSIP00000009589": [["Domain_0", 212, 420, "PF00566"]], "ENSPCOP00000005389": [["Domain_0", 212, 420, "PF00566"]], "ENSCJAP00000032538": [["Domain_0", 212, 420, "PF00566"]], "ENSTGUP00000006220": [["Domain_0", 212, 420, "PF00566"]], "ENSFCAP00000034761": [["Domain_0", 212, 420, "PF00566"]], "ENSCHOP00000005166": [["Domain_0", 201, 422, "PF00566"]], "ENSMPUP00000004123": [["Domain_0", 212, 420, "PF00566"]], "ENSSTOP00000013614": [["Domain_0", 212, 420, "PF00566"]], "ENSCATP00000029810": [["Domain_0", 212, 420, "PF00566"]], "ENSMNEP00000003326": [["Domain_0", 212, 420, "PF00566"]], "ENSMICP00000049850": [["Domain_0", 212, 420, "PF00566"]], "ENSDNOP00000027105": [["Domain_0", 212, 420, "PF00566"]], "ENSFALP00000001819": [["Domain_0", 212, 420, "PF00566"]], "ENSMODP00000014766": [["Domain_0", 212, 420, "PF00566"]], "ENSECAP00000021285": [["Domain_0", 212, 420, "PF00566"]], "ENSACAP00000004356": [["Domain_0", 212, 420, "PF00566"]], "ENSCGRP00001024535": [["Domain_0", 212, 420, "PF00566"]], "ENSPPAP00000021790": [["Domain_0", 212, 420, "PF00566"]], "ENSSHAP00000008826": [["Domain_0", 240, 420, "PF00566"]], "ENSMEUP00000012660": [["Domain_0", 205, 422, "PF00566"]], "ENSGGOP00000027762": [["Domain_0", 212, 420, "PF00566"]], "ENSMLUP00000014355": [["Domain_0", 205, 422, "PF00566"]], "ENSLAFP00000015376": [["Domain_0", 205, 422, "PF00566"]]}