Home
Query Results
"ENSGT00730000111382_0"
Found:
ENSGT00730000111382_0
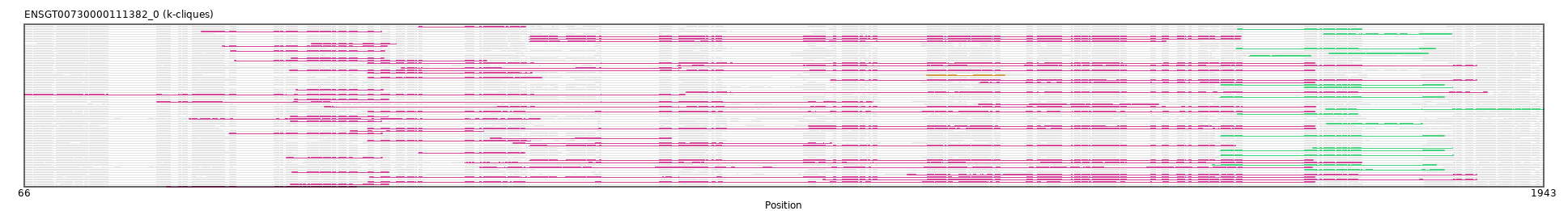
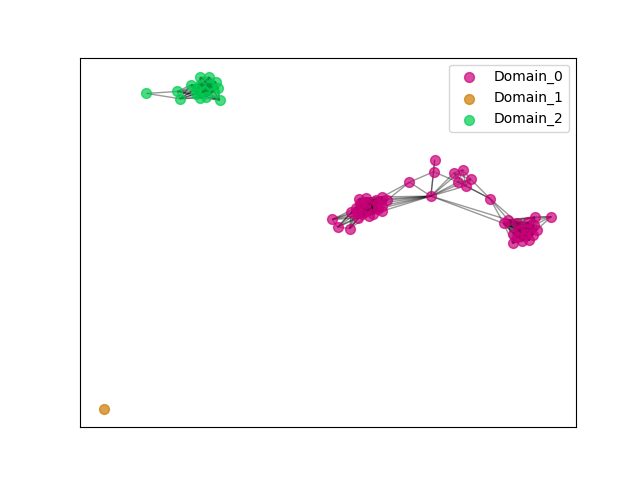
ENSECAP00000012329 (view gene)
ENSPANP00000002769 (view gene)
MA-EARKRRELLPLIYHHLLRAGYVRAAR-------EVKEQSGQKCFLAQPVTLLDIYTH
WQQTSELGRKRKAEE-DAALQAKKTRVSDPISTSESSEE-E---EEAEAETAKATPRLAS
TNSSVLGADLPSSLKEKAKAETEK-AGKTGNSMPHPATGKTVTNLLSGK-----------
------------------------------------------------SPRKSAEPSANT
TLVSET---------EEEG-SVPAFGA-----AAKPGMVSAGQADSSSEDTSSSSDETDV
EGKPSVKPAQVKA-----SSVSTKESP---------------------------------
-------------ARKAAPA-PGKV---GDVTPQVRGGALPPA-----------KRAKKP
EEESESSEE-GSESEEEAPAGTPS-QVKTS---EKILQVRAAS---TAAKGTPGKGA---
---------TPAPPGKA--------GAVAFQTKAGKP-------EEDSESSSEES---SD
SEEETPA-AK--ALLQA-------------------------------------------
---------KASGKTPQVRAA------SAP--AKESPRKGAAPAPPGKTGPAVAKAQTGK
--QEEDSQSSSEESDSE-K-----EA----PVQTKPSGKTSQVRAASASAKE-SPRKRAA
PPPP----------RKTGPAATQAQAGKQEEDSGSSSEE-SDSDREAPAAM----NAA--
------------------------------------------------------------
---------QVKPLGKNPQVKPAST--------MGTGPLGKGPGPVLPGKVGPTTPSAQV
GKWEDS-DSS----SEE--SSDSDDGE---------------------------------
------------------------------------------------------------
-------VPTAVAPAQEKSLGKVLQAKPASGPAKG---------PPQK-------AGPVA
IQVKAEKPMEDSESS--EESSDSADS-EET---PAAMTA---------------------
----------------------------------------AQAKPALKIPQTKACPKKTN
--T---ASAKVTPVRVGTQAPRKAGTVTSPVGSS-PAVAGGTQRPAEDSSSSEESDSEEE
-K-TGPAVTMG--------------------------------QAKSVGKG-----LQVK
AASVPVKGSLGQGTAPVLPGKTGPTVT-----------QV--KAEMQEDS-ESSEEESD-
SE--EAAAPSAQVKTSVKKTQAKANPAAVRASPAKGTI-SAPRKVVT-------------
----------------AAAQAKQ-------RSPAKAKPP----------VRNLQNSTV-L
---VRGP--------ASVPPVGKAVAAA-AQVQTGP-------
PF03546 // ENSGT00730000111382_0_Domain_2 (1544, 1820)
EEDSGSSEEESDSEEE-
TETPAQAKPS--------------------------------------------------
--------------------------GKTPQVRAALAPAKES---PRK--GAAPAPPGKT
GPSA--TQAGKQ-DGSGSS-SEE-SNSDGEAPAAATS-----------------------
----------------------------------------------------AQKDSNSK
PARSKTLAPAPPEGNTEGSSESSEEELPLTQVI-KPPLIFV----DPNRSPAGPA---AT
PVQAQAASTPRKSRASESTARSS--SSESEDEDVIPATQCLTP-G-IRTNVVTVPTAHPR
IAPKAS-MTGASSSKES---SRISDGKKQEGPATQVSKKNPASLPLTQAALKVLAQKASE
AQPPVARTQPSCGADSAVGTLPATSPQSTPVQAKGTNKLRKPKLPEGQQAT---------
KAPGSSDDSEDSS-NSSSGSEEDAE---GPQVAKSAHTLVGPTPSRTETLVEETAAESSE
DDVVAPSQSLLSGYVTPGLTPA---NSQASKATPKPYSSPSVS-STLAAKDDPD------
------GKQEAKPQQA--AGMLS------------------------------PKTGGKE
AASGTTPQKSRKPKKGAGN--------PQASTLALQSNITQCLLGQPWPLNEAQVQASVV
KVLTELLEQERKKVVDATKESS------RKG--WESRKRKLSGDQPAARTPRSKKKKKLG
AGEGGEAS-VSPEKTSTTSKGKAKRDKASDDVKEKKGKGSL-GSQGAKDEPEKELQKGIG
KVEGGDQSNPK---SKKEKKKSDKRKKDKEKKEKK--KKAKKSSTKDSESPSQ--KKKKK
KKKTAEQTV---------------------------------------------------
--
ENSMFAP00000029393 (view gene)
MA-EARKRRELLPLIYHHLLRAGYVRAAR-------EVKEQSGQKCFLAQPVTLLDIYTH
WQQTSELGRKRKAEE-DAALQAKKTRVSDPISTSESSEE-E---EEAEAETAKATPRLAS
TNSSVLGADLPSSLKEKAKAETEK-AGKTGNSMPHPATGKTVTNLLSGK-----------
------------------------------------------------SPRKSAEPSANT
TLVSET---------EEEG-SVPAFGA-----AAKPGMVSAGQADSSSEDTSSSSDETDV
EGKPSVKPAQVKA-----SSVSTKESP---------------------------------
-------------ARKAAPA-PGKV---GDVTPQVRGGVLPPA-----------KRAKKP
EEESESSEE-GSESEEEAPAGTPS-QVKTS---EKILQVRAAS---TAAKGTPGKGA---
---------TPAPPGKA--------GAVAFQTKAGKP-------EEDSESSSEES---SD
SEEETPA-AK--ALLQA-------------------------------------------
---------KASGKTPQVGAA------SAP--AKESPRKGAAPAPPGKTGPAVAKAQTRK
--QEEDSQSSSEESDSE-E-----EA----PVQTKPSGKTSQVRAASASAKE-SPRKGAA
PAPP----------RKTGPAATQAQAGKQEEDSGSSSEE-SDSDREAPAAM----NAA--
------------------------------------------------------------
---------QVKPLGKNPQVKPAST--------MGTGPLGKGPGPVLPGKAGPTTPSAQV
GKWEDS-DSS----SEE--SSDSDDGE---------------------------------
------------------------------------------------------------
-------VPTAVAPAQEKSLGKVLQAKPASGPAKG---------PPQK-------AGPVA
IQVKAEKPMEDSESS--EESSDSADS-EET---PAAMTA---------------------
----------------------------------------AQAKPALKIPQTKACPKKTN
--T---ASAKVTPVRVGTQAPRKAGTVTSPVGSS-PAVAGGTQRPAEDSSSSEESDSEEE
-K-TGPAVTMG--------------------------------QAKSVGKG-----LQVK
AASVPVKGSLGQGTAPVLPGKTGPTVT-----------QV--KAEMQEDS-ESSEEESD-
SE--EAAAPSAQVKTSVKKTQAKANPAAVRASPAKGTI-SAPGKVVT-------------
----------------AAAQAKQ-------RSPAKAKPP----------VRNLQNSTV-L
---VRGP--------ASVPPVGKAVAAA-AQVQTGP-------
PF03546 // ENSGT00730000111382_0_Domain_2 (1544, 1820)
EEDSGSSEEESDSEEE-
TETPAQAKPS--------------------------------------------------
--------------------------EKTPQVRAALAPAKES---PRK--GAAPAPPGKT
GPSA--TQAGKQ-DGSGSS-SEE-SNSDEEAPAAATS-----------------------
----------------------------------------------------AQKDSNSK
PARSKTLAPAPPEGNTEGSSESSEEELPLTQVI-KPPLIFV----DPNRSPAGPA---AT
PVQAQAASTPRKSRASESTARSS--SSESEDEDVIPATQCLTP-G-IRTNVVTVPTAHPR
IAPKAS-MTGASSSKES---SRISDGKKQEGPATQ-------------------------
-------------ADSAVGTLPATSPQSTPVQAKGTNKLRKPKLPEGQQAT---------
KAPGSSDDSEDSS-NSSSGSEEDAE---GPQVAKSAHTLVGPTPSRTETLVEETAAESSE
DDVVAPSQSLLSGYVTPGLTPA---NSQASKATPKPYSSPSVS-STLAAKDDPD------
------GKQEAKPQQA--AGMLS------------------------------PKTGGKE
AVSGTTPQKSRKPKKGAGN--------PQASTLALQSNITQCLLGQPWPLNEAQVQASVV
KVLTELLEQERKKVVDATKESS------RKG--WESRKRKLSGDQPAARTPRSKKKKKLG
AGEGGEAF-VSPEKTSTTSKGKAKRDKASDDVKEKKGKGSL-GSQGAKDEPEKELQKGIG
KVEGGDQSNPK---SKKEKKKSDKRKKDKEKKEKK---KAKKSSTKDSESPSQ--KKKKK
KKKTAEQTV---------------------------------------------------
--
ENSHGLP00100016341 (view gene)
MA-EARKRRELLPLIYQHLLHAGYVRAAR-------EVKEQSGQKSFLAQPFTLLDIYTH
WQQTSEIGQKRKAED-DAALQAKKTRVSDPISSSESSEEEEEEEEEAEAKAAKTTPRPAS
TNSSAMGMVLPSSMKEKAKAKTRN-TSKTVNSVPHLASGKAVVHLLSGK-----------
------------------------------------------------SPKKSAELSGST
VLVSET---------EEEG-SIQALEA-----AAKPGTVSAGQADNSNKDTSSSSDETDV
EVKSSVETSQVRP-----SLTLAKESS---------------------------------
-------------ASKAAPS-PGKV---GGVARQARGGALTPA-----------GSAVKP
EEEPP--------------TGAPS-QVKAS---EKILQVRLAP---ASAKGTPGKQ----
--------AT
PF03546 // ENSGT00730000111382_0_Domain_0 (491, 705)
CALPGKG--------GPLATQAKARRP-------EQDSESSS--E-GESD
SEVETPP-VM--TALQA-------------------------------------------
---------KLSGKTLQVRAA------SAP--AKESPRKGHPPATPGKTGPAATQAQVGK
QEEDSDSSS--EESDSD-GETPAAQTLTTNPSQVKPLGKTPQVTAASAPAKG-SPRKEVP
PAYPGKTGP----------AGTQAQVGKQEEDSDSSSEE-SDSDEETPAAQTLTTNPS--
------------------------------------------------------------
---------QVKPLGKTPQVTAASA--------PAKGSPRKEVPPAYPGKTGPAGTQAQV
GKQEEDSDSS----SEE---SDSDGETPAAQTLTTNPSQVKPLGKTPQVTAASAAAKGSP
RKEVPPAYPGKTGPAGTQAQVGKQEEDS-----------------------DSSSEESDS
DGETPGAQTLTTNSSQAKPSGKILQVRPASGPTKGLLGKGAAPKPPQK-------SGPVA
IQVKAEKSKDDSESS--EESSDSE----EEA--PAAVTPA--------------------
-----------------------------------------QAKPELKMPQTKASPSKSI
LLTP--TSAKGAPVRVGTPAPWKAGTVASA---SSSAVPRGAQRPKDNSSSSEEESEGKE
A---TPAAAVA--------------------------------QAKSLGKG-----LRGQ
AASAPTMGPSGQGATPVPPGKTRPTVA-----------QV--RAEAHESS-ESSEEESDD
SE-VGAVVTPAQAKAPGKTPQAKANPASTKASTDKEAA-WTPAKLVT-------------
----------------AVVQAKP-------GSMAKGKSP----------VRAPKNSTIS-
---ARGQ--------ASVPASGKAVTAA-AQSQKGPTQESE--EKDLESSEESSESEEE-
--LPTQAKPS--------------------------------------------------
--------------------------GKTLQVRAATAPAKES---SRK--GPSPTTPGKT
GPAGTQAQVGKQEEDSDSSSE--ESDSDGETPAAQTLSTNPSQV----------------
--------------------------------------------------LSLQKNINSK
TVGSNTLTPAPREKSPEGSSESSEEELPSSQVI-KTPLIFV----DPNRSPAGPA---AT
PTQAQAASPPRKARASSSTAGSS---SEGEDEDVIPATQSSTLA--VRKEVVTVPTASPR
MAPRAS------SSLES---SRVSPGQKQEAAAPQVTKRNLASLPLTQAALKVLAQKASE
GQP-------SSEAGSDAGTLPVTCPQSNPVQARVTNKLRKPKPSEGQQTTVTTSGHLKA
TAPGSSDNSDDSS-DNSSGSEEDAE---EPLMTKSAPGLAGPSPSKRETLVEETTAESSE
DEVVVPSQSLLSGFVTPGLTMA---NNRASKATARQNLHPSLS-SPPPTKDDPA------
------GKQAAESQHI--AGAVS------------------------------PKPGRKE
AASGPTPQKPRKPKKGPLS--------TSASLQTLQSNITQHLQGQPWPLNEAQVQASVV
KVLTELLEQERKKAEDTARESS------RKG-----RKRKLSGDQVASRAPKGKKKKQLA
AGEGREDA-ASSEKAPRTSKGKSKRDKASGDLK---EEKGS-GSGGSKDKPDGEL----L
KVESGNQNDSK---SKKEKKKSNKKKKDKEKKEK---KKGKKPSAKDSDSPFQ--KKKKK
KKKTAEQTV---------------------------------------------------
--
ENSAMEP00000004999 (view gene)
MA-EARKRRELLPLIYQHLLQAGYVRAAR-------EVKEQSGQKSFLAQPVTLMDIYTH
WQQTSEIGQKRKAEE-DAALQAKKTRVSDPISSSESSEEED----DAEAEAAKATPRLAS
TNSLVAGSVLPSSAKEKDKAKTKK-AGKTVNSTPHPASGKAVAHLLSGR-----------
------------------------------------------------SPRKSAGPSANT
ILVSET---------EEEG-SVPALGT-----TAKPGMASANQADSSSEETSSSSDETYV
EMKPSVKPPQVKA-----SPAPAKDSL---------------------------------
-------------GKGAAPA-PGKA---GDVTPQARGGALTPT-----------SKAKEA
EEDSESSED-ESGSEDGALAEPPR-QAKGS---EKIVQVRAAS---GPVKGTPGKGA---
---------TPAPPGKA--------GPAAAQAKAGKP-------EEDSESSSEEE---SD
SEEEAPA-AK--
PF03546 // ENSGT00730000111382_0_Domain_0 (553, 685)
TPLQT-------------------------------------------
---------KPSGKTPQVKAA------SVS--VKESPRKGAPPVPAGKAGPVAAQVRK--
GEE--DPESSTEESGSE-EEAPATVP----PAKSPA--QRARLR-------------ACP
--PS----------PHLGPCALHLPS---------SPFVSPRTQSPVGPCPDPVFCYS--
------------------------------------------------------------
---------QAKPLGQNAQVRAASG--------S---------VKGPPPKVGLAAI--PV
GKQEEDSESS----SEE--ESDSKWEAPA-------------------------------
------------------------------------------------------------
-------------QAQAKSSGKVLQVRAASGPAKG---------PPQK-------AGPVA
TQGKAEVSKDDSDSS--EEESDSE----EEA--PAALAA---------------------
----------------------------------------AQAKPAMRTPQTKASPKKGT
PITP--APAKVPPVRVDTPAPWKAGAATSPACTSSPAMARGTQRPED-SSSSEESESEED
P---APAPAVG--------------------------------QAKPVGKG-----VTVK
AAPTPTKGPSGQGTAPAPPGKAGPAMA-----------LV--KAEVQADSSESSEDDSDS
E---EAARAPAQVKASVKTPQSKTNPAPPRAASAKAAA-SAPGKVVP-------------
----------------AAAQAKQ-------QSPAKVKPP----------ARTPQSNAVS-
---ARGQ--------GSVPAVGKAVATA-AQAQPGPVRSPQ--E-DSESSEEESDSDGE-
--ALTQAKPS--------------------------------------------------
--------------------------GHTPQVRTASAPNKGF---SRK--GAAAAAPRKT
GATA--SQAGKLEEDSESS-SEEESDSEEEA-----------------------------
------------------------------------------------------------
----------------------PAQDVEGSQVI-KPPLIFV----DPNRSPAGPA---AT
ATQAPAANTPRKARASESTARSS--SSESEDEDVIPATQCSTP-A-SRPNATVL-SAHAR
PAVRAS-AVGASGSEASG---RASEGKKQEAPVAQVTKRSPARLPLTQAALKVLAQKASE
AQPPAARTPSSSGVDGALGTLPAMSPQSAPVQAKGTNKLRKPVVPAVQRATDTPAGHAKA
KASDTSNDSADGS-GSSSGSEEVAD---GPRMAPSAHRPVGPAPSTTETLVEETTAESSE
DEAVAPSQSLLSGYVTPGPTLA---NSQASKATPRPDHNPSAC-STSATTDAPD------
------GKQELEPRHV--AGTVS------------------------------PKTSRR-
-EANPTPQKPRKAKKEAGS--------PQASMLALQNDISRRLLSEPWPLNEAQVQASVV
KVLTELLQ-ERKKAADATKESG------RKG--QLGRKRKLSGDQTAARAPKSKKKKQLV
AGGGGEGG-GSPEKALRTPKAKAKRDGASGDIKEKKEKESP-GSQGAKEKPEGEP--GTL
KVEGGDQGNPK---SKKEKKKSDKKKKDKEKKEKK--KKVKKASTKDPDSPFQ--KKKKK
KKKTAEQTV---------------------------------------------------
--
ENSODEP00000007198 (view gene)
ENSPCOP00000011729 (view gene)
MA-EARKRRELLPLIYHHLLRAGYVRAAR-------EVKEQSGQKILLAQPVTLLDIYTH
WQQTSELGQKRKAEA-DAALLAKKTRVSDPISSSESSEE-E---EEAEAETAKATPRLAC
TNSSAMGADLLSSMKEKAKAKTET-AGKMGNSMPNPASMKTVAHLLSGK-----------
------------------------------------------------SSKKAGEPSANT
VLASET---------EEE-GSVLAFGA-----AAKPGVVSAGQADSSSEDTSSSSDETDA
EVIAVTRSWKGCS-----QEREGKDSG---------------------------------
-------------AAWRTLSPEGII---R-------------------------------
--------E-FS-ETGEQLLFLFY-QVKAS---EKILQIRAAS---APAKGTPGKGP---
---------
PF03546 // ENSGT00730000111382_0_Domain_0 (490, 691)
TLAPPGKA--------GPIATQAKAGKP-------EEDSESSSEEE---SD
SEKETPA-AK--APLQG-------------------------------------------
---------RPSEKTPQVRAA------SAP--AKESPRKGAVAAPPGKTGPAAAQAQAGK
--QEEDSGSSSEESDSD-GEAPAADT----LHGPRPASECLLWAF--------------L
PSVPS---------ACAAPFACLLPV---------S----SRTFSLWAPCPDLVLCHS--
------------------------------------------------------------
---------QAKPVGKNPQVRPAST--------LGVRPSGKGTIPVLPGKAGTAPPFAQV
GKQEDPTSS-----SEE--SSDSEGEAPTAVAP----A----------------------
------------------------------------------------------------
---------------QAKPSGKILQVRPASSPAKG-----TVPVGPQK-------AGPVA
TLVKAERSKTDSESS--EESSDSE----EEA--PAAVPP---------------------
----------------------------------------AQAKPALKTPQTKASPRKGT
PSTL--ASAKTPSVRVGTPAPWKAGA-----------VAPPAQRPEEDSSSSEESESEEE
E--AAPVAAVG--------------------------------QAKSVGKG-----LQAK
AASVPTTGPSGQGTAPVLPGKTGLAVT-----------QV--KAEAQEDS-ESSEEESD-
SE--DAATAPAQVKASVKMPLAKASPAATRVSSAKGAT-SVPGKGVA-------------
----------------AAAQTKQ-------GSTAKAKPP----------ARTLQNSAVSV
----RAQ--------APVPSVGNAGATA-AQAQTGP----R--EEDSESS-EESD--SE-
EEAPAPAKPL--------------------------------------------------
--------------------------GKTLQVRAASAPAKES---PRK--GAAPGTPGKT
GPAAAQAQVGKREEDSESS-SEE-SDGEGKAPAAVTPA----QKNGN-------------
----------------------------------------------------------LK
TGRSKTLAPAPPDKKAAGSSESSDEELPASQVI-KTPLIFV----DPNRSPAGPA---AT
PAQAQAASTPRKARASESTAKSS--SSESEDEDMIPATQCLTP-A-IRANVT-VSATHPR
TTPKAS-AAGATNSKES---SRVSDGKKQEGPATQ-------------------------
-------------VDSAIGTFPVTGSQSTPVQTGVTSNPRKPKLSDSQQAK---------
-APGSSDDSEESS-GSSLGMEEDAK---GPQMAKSAHLPVGPATSSRETLVEETT-ESSE
DEVVAPSQSLLSGYVTPGLTSA---NSQASKATRKPISNPSVS-STPAIKDDPD------
------GKQQAEPQQS--AGITS------------------------------LKTGKKE
ATSDTTSQKPRKPKKGAGN--------LQASTLALQNDITQRLLGEAWPLSEAQVQASVA
KVLAELLEQEKRKATDAIKESG------KKG-----HKRKLSGDQLATKAPKSKKKKQLA
TGEGGEGT-VSPEKASVTSKGKAKKDRASGDVKEKKGKGSS-GSQGAKDKPKGELPKGVA
KVEDGDQSDPK---TKKEKKKSDKKKKDKEKKEKK--KKAKKTSTKDPELPFQ--KKKKK
KVE---------------------------------------------------------
--
ENSMLEP00000022799 (view gene)
MA-EARKRRELLPLIYHHLLRAGYVRAAR-------EVKEQSGQKCFLAQPVTLLDIYTH
WQQTSELGRKRKAEE-DAALQAKKTRVSDPISTSESSEE-E---EEAEAETAKATPRLAS
TNSSVQGADLPSSLKEKAKAETEK-AGKTGNSMPHPATRKTVTNLLSGK-----------
------------------------------------------------SPRKSAEPSANT
TLVSET---------EEEG-SVPAFGA-----AAKPGMVSAGQADSSSEDTSSSSDETDV
EGKPSVKPAQVKA-----SSVSTKESP---------------------------------
-------------ARKAAPA-PGKV---GDVTPQVRGGALP
PF03546 // ENSGT00730000111382_0_Domain_0 (402, 509)
PA-----------KRAKKP
EEESESSEE-GSESEEEAPAGTPS-QVKTS---EKILQVRAAS---TAAKGTPGKGA---
---------TPAPPGKA--------GAVAFQTKAGKP-------EEDSESSSEES---SD
SEEETPA-AK--ALLQA-------------------------------------------
---------KASGKTPQVGAA------SAP--AKESTRKGAAPAPPGKTGPAVAKAQTGK
--QEEDSQSSSEESDSE-E-----EA----PVQTKPSGKTSQVRAASASAKE-SPRKGAA
PAPP----------RKTGPAATQAQAGKQEEDSGSSSEE-SDSDREAPAAM----NAA--
------------------------------------------------------------
---------QVKPVGKNPQVKPAST--------MGTGPLGKGPGPVLPGKVGPTTPSAQV
GKWEDS-DSS----SEE--SSDSDDGE---------------------------------
------------------------------------------------------------
-------VPTAVAPAQEKSLGKVLQAKPASGPAKG---------PPQK-------AGPVA
IQVKAEKPMEDSESS--EESSDSADS-EET---PAAMTA---------------------
----------------------------------------AQAKPALKIPQTKACPKKTN
--T---ASAKVTPVRVGTQAPRKAGTVTSPVGSS-PAVAGGTQRPAEDSSSSEESDSEEE
-K-TGPAVTVG--------------------------------QAKSVGKG-----LQVK
AASVPVKGSLGQGTAPVLPGKTGPTVA-----------QV--KAEMQEDS-ESSEEESD-
SE--EAAAPSAQVKTSVKKTQAKANPAAVRASPAKGTI-SAPGKVVT-------------
----------------AAAQAKQ-------RSPAKAKPP----------VRSLQNSTV-L
---VRGP--------ASVPPVGKAVAAA-AQVQTGP-------EEDSGSSE-ESDSEEE-
TETPAQAKPS--------------------------------------------------
--------------------------GKTPQVRAALAPAKES---PRK--GAAPAPPGKT
GPSA--TQAGKQ-DGSGSS-SEE-SNSDGEAPAAATS-----------------------
------------------------------------------------------------
-----------------------------AQVI-KPPLIFV----DPNRSPAGPA---AT
PVQAQAASTPRKSRASESTARSS--SSESEDEDVIPATQCLTP-D-IRTNVVTVPTAHPR
IAPKAS-MTGASSSKES---SRISDGKKQEGPATQVSKKNPASLPLTQAALKVLAQKASE
AQPPVARTQPSRGADSAVGTLPATSPQSTPVQAKGTNKLRKPKLPEGQQAT---------
KAPGSSDDSEDSS-NSSSGSEEDAE---GPQVAKSAHT-LGPTPSRTETLVEETAAESSE
DDVVAPSQSLLSGYVTPGLTPA---NSQASKATPKPYSSPSVS-STLAAKDDPD------
------GKQEAKPQQA--AGMLS------------------------------PKTGGKE
AASGTTPQKSRKPKKGAGN--------PQASTLALQSNITQCLLGQPWPLNEAQVQASVV
KVLTELLEQERKKVVDATKESS------RKG--WESRKRKLSGDQPAARTPRSKKKKKLG
AGEGGEAS-VSPEKTSTTSKGKAKRDKASDDVKEKKGKGSL-GSQGAKDEPEKELQKGIG
KVEGGDQSNPK---SKKEKKKSDKRKKDKEKKEKK--KKAKKSSTKDSESPSQ--KKKKK
KKKTAEQTV---------------------------------------------------
--
ENSCJAP00000066463 (view gene)
MA-EARKRRELLPLIYHHLLRAGYVRAAR-------AVKEQSGQKCFLAQPVTLLDIYTH
WQQTSELGRKRKAEE-DAALQAKKTRVSDPISTSESSEE-E---EEAEAETTKATPRLTS
TNSSVMGADLTSSMKEKAKAETEK-AGKTGNSMPHPATGKMVAHLLSGK-----------
------------------------------------------------SPRKSAEPSANT
TLVSET---------KKEL-SNPAFGA-----AAKPGMVSTGQAGSSSEDTSSSSDETDV
EGKPSVKPAQVKA------S
PF03546 // ENSGT00730000111382_0_Domain_0 (321, 511)
VSTKESP---------------------------------
-------------ARKAVPA-LGKV---GDVTPQVSGGALPPS-----------RRAKKP
EEESESSEE-GSESEEEAPAGTPS-QVKAS---EKILQVRAAS---APAKGTSGKVA---
---------APAPTGKA--------GPLASQSKTGKP-------EEDSESSSVES---SD
SEEEISA-AK--VLLQA-------------------------------------------
-----------SAKTPQVRAV------SAP--AKESPRKGAA------------KAQAVK
--QEKDSESSSEESDSE-E-----EA----PARVKPSGKTPQVRAALAPTKE-SPRKGAA
PTPPGKTVT----------AAAQAQVGNGEEDSGSSSEE-SDSDEEAPAAM----STA--
------------------------------------------------------------
---------QVKPLGKNPQVRPAST--------VGTGPLGKGTGPVLPGKV---------
AKKEEELESS----SEE--SSDSDDGE---------------------------------
------------------------------------------------------------
-------VPIAVAPAQEKSLGKILQAKPASVPAKGPLGRGAAPVPPQE-------AGPVA
IQVKAEKPTEDSESS--EESSDSEEA-PAA------VTA---------------------
----------------------------------------AQAKPALKAPQTKACPKKTS
--TT--VSAKVAPVQVGAQASRKAGSATS-AGSS-PAVAWGAQRPAEDSSSSEESDSEEE
-K-TGPAVTTG--------------------------------QAKSVGKG-----LQVK
AASVPVKGPLGQGTAPLLPGKTQPSLT-----------QV--KAEMQEDS-ESSEEESD-
SE--EAATTPAQVKTSVTKNQAKANPAATRASSAKGTI-AAPGKVVS-------------
----------------ATAQAKQ-------RSPAKVKPQ----------ARNPQNSAV-L
---VRGP--------ASVPPVGKAVA------QTGP-------EEDSGSSEEESDSEEET
MEMPAQMKPS--------------------------------------------------
--------------------------GKNPQVRAALAPAKES---PRK--GAGPAPPGKT
GPLT--TQAGRQKDDSGSS-SEE-SDSDGEALAAVTSA----QVL---------------
---------------------------------------------------SPQKGGNSK
PARSKTLVPTPPERNMEESSESSEEELPLTQVI-KPPLIFV----DPNRSPAGPA---AT
PAQAQAPSTPRKARASESTARSS--SSESEDEDMISATQCLTP-A-IRTNVVTVPTAHPR
IAPKAS-TARASSNKES---SRISDGKKQEGPATQVSKKNPTSLPLTQAALKVLAQKASE
AQPPVARTQPSSGVDSAVGTFPATSPQSTPVQARGTNKLRKPKLPDGQQAA---------
KAPGSSDDSEDSS-DSSSGSEEDAE---GPQVAKSAHRLVGPTPSRTETLVEETTAESSE
DDVVAPSQSLLSGYVTPGLTLA---NSQASKATPKPDSNLSVS-SALATKDDPD------
------SKQEAKLQQA--ADTLS------------------------------PKTGGKE
AVSDTRPQKSQKPKKGAGI--------PQASTLALQSNIAQRLLGQPWPLSEAQVQASVM
KVLTELLEQEKKKAADVTKESS------RKG--WESRKRKLSGDQPAARN----KKKKLG
AGEGGEAS-VSPEKASMTSKGKAKRDKASGGIKEKKGKGPL-GSQGVKDKPEEELQKGMG
KVEGGDQSNPK---SKKEKKKSDKRKKDKEKRGKK--KKAKKASTKDPESLSQ--KKKKK
KKKTPEQTV---------------------------------------------------
--
ENSSTOP00000008058 (view gene)
MA-EARKRQELLPLIYQHLVQAGYVRAAR-------AVKEQSSQKKFPVQPFTLLDIYTH
WQLTSELGRKQKAEK-DAALQAKKTRVSDPVSSSESSEEEE---DEAEAETVKATPRLAS
TNVSAVGAHLPSSMKEKAKAKTKK-AGKTVNSVPHPPSAKAVVHLLSGK-----------
------------------------------------------------SPKKSTEPSANA
TLVSET---------EEES-SVPGHRA-----TAKPGTVSAGPADSSSEDTSSSSDETDM
EVETSGKPAQVKA-----SPAAPKESP---------------------------------
-------------AKRAAPR-PGKM---GDVTPQVR
PF03546 // ENSGT00730000111382_0_Domain_0 (397, 516)
ESALTPA-----------QGAKKP
KEELESSEE-SSESDEEVPAGTPN-QVKAT---EKTLQARAAP---ASAKGIPGKGA---
---------TTVTPGKA--------GPVATQTKVGKP-------KEESESSSEEE---SD
SEEDNPA-AT--ALLQA-------------------------------------------
---------KPSEKSPQVRAV------STP--AKESPRKGAPPVPPGKTGPAAIQASAGK
--PEEDSDSSSEDSDSE-RT----PV----------------------------------
---------------------------------------------AMTLTV----NPA--
------------------------------------------------------------
---------QAKSLGKNLQMKPAST--------VVQGPLGKGISPVVPAA--------QV
KKVEKASESS----SEE--SDSEEEA----------------------------------
------------------------------------------------------------
---------PPATLAQAKLSGKTTPVRPASGSAKGHLGKVAAPAPPQK-------AGPGV
SQVKAERSKEDSESS--ETSSDSEE----EA--SAAVTP---------------------
----------------------------------------AQAKPALKTPQTKASPKKGT
PITP--VSSKIPPVHVGTPAPWKAGTMTS--SSS-PAVAWSTQKPEEDSSSSSEEESEEE
-GKSACVAAVA--------------------------------QVKSVGKG-----LQAR
AALVPTKAPSGQGTAPAPPRKTGPSVS-----------QV--RAQDSEDS-ESSEEESD-
SE--EEAATPAPVKALGNTVQAKASPAPIRASSAKGT--SAPGKVLT-------------
----------------AVAPAKQ-------GSPAKGKPS----------ARTPQNSTVT-
---MKGQ--------ASVVTTGKAVAGA-AQTQKGSVGRAE--EEDSESSEEESDSEEEE
VAPAQAKPLE--------------------------------------------------
--------------------------NKLQQVRNASGPAKGS---PMK--GAPPAPPKSA
-SAQ--APPGKQEDDSDSS-SEE-SDSEGETPAAVTP-----------------------
----------------------------------------------------AQKDGTSK
TARGKALTSGP-TENAEVSSDSSGEELPSSQVI-KPPLIFV----DPNRSPAGPP---AT
PAQAQAANTLRKARVSDGTARHS--SSESEDEDLIPATQSSTP-A-IKTNVI-VPTAHGR
TTPRA------NSREES---SRTSESKKQEAAATQVAKRNSASLPLTQAALKVLAHKANE
AQPP-ARTQSSSEVDSAVGALTVTHPQSSPVQNRVNNKLKQPKPPAGQQATATPSGHSKA
KAPGGSDDSEDSS-DSSSGSE-DTR---GPQTAKSIPRLVGPTPSKKETLVEETTEGSSE
DEVVAPSQSLLSGYVTPGLTLA---NSQASKVTPKPDSSPLVS-SALAKKDDPD------
------GKLES---QQ--AGTVS------------------------------LKTGKKE
ATSGATSE---KPRKEAPS--------STDSTQGLQSNIAQHFQGAPWPLSEAQVQASVM
KVLKELLEQERKKATDAAKDSS------RKG-----RKRKLSGDQSATRAPKSKKKKQLV
SEEGGESA-VSSEKASRTSKGK-KRDRASGNTKEKKGKGPS-GSHGARDQPEGEL--GMV
KVEGRDQSDPK---SKKEKKKSDKSKKDKEKKEKK--KKAKKASTKDPDSLLQ--KKKKK
KKKTAEQTV---------------------------------------------------
--
ENSMUSP00000135639 (view gene)
ENSFCAP00000029727 (view gene)
ENSANAP00000019635 (view gene)
MA-EARKRRELLPLIYHHLLRAGYVRAAR-------EVKEQSGQKCFLAQPVTLLDIYTH
WQQTSELGRKRKAEE-DAALQAKKTRVSDPISTSESSEE-E---EEAEAETAKATPRLAS
TNSSVMGVDLTSSMKEKAKAETEK-AGKTGNSMPHPATGKVVAHL-SGK-----------
------------------------------------------------SPRKSAEPSANT
TLVSET---------KEEV-SIPAFGA-----AAKPGIVSAGQADSSSEDTSSSSDETDV
EGKPSVKPAQVKA------SVSTKESP---------------------------------
-------------ARKAVPA-LGKV---GDVTPQVRGGALPPS-----------RRAKKP
EEESESSEE-GSESEEEAPAGTPS-QVKAS---EKILQVRAAS---APAKGTPGKVA---
---------APAPPGKA--------GPIASHAKAGKP-------EEDSESSSEES---SG
SEEETSA-AK--LLLQA-------------------------------------------
-----------SAKTPQVRAA------SAP--AKESPRKGAAPAPPGKTGPAAAKAQAVK
--REEDSESSSEESDSE-E-----EA----PAQAKPSGKTPQVRAALAPAKE-SPRKGAA
LTPPGK----------TGPAATQAQAGKREEDSGSSSEE-SDSDEEAPAAM----STA--
------------------------------------------------------------
---------QEKSLGKILQAKPASG--------PAKGPLGR-------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------GVPPGPPQE-------AGPVA
IQVKAEKPTEDSESS--EESQLETEF-RNLL--QPLFVS---------------------
----------------------------------------PQAKPALKASQTKACPEKTN
--TT--ASAKVAPVQVGAPAPRKAGTATS-AGSS-PAVAWSAQRPAEDSSSSEESDSEEE
-K-TGPTVTMG--------------------------------QAKSVGKG-----LQVK
AASVPVKGPLGRGTAPVLPGKTQPSVT-----------QV--KAEMQEDS-ESSEEESD-
SE--KAAATPAQVKTSVKKTQAKANPAAARASSVKGTI-SAPGKVVS-------------
----------------AAAQAKQ-------RSPAKASRV----------RNHRRYGGLGQ
---TGGP--------LTDPSLE-----------KGP-------AWAVGPER-NCFHLQYH
YPPA
PF03546 // ENSGT00730000111382_0_Domain_2 (1565, 1718)
IQMKPS--------------------------------------------------
--------------------------GKSPKVRAALVPAKES---PRK--GAGPAPPGKT
GPLT--AQAGRQKDDSGSS-SEE-SDSDGEAPAAVTSA----------------------
------------------------------------------------------------
------------------------------QVI-KPPLIFV----DPNRSPAGPA---AT
PVQAQAASTPRKARASESTARSS--SSESEDEDMISATQCLTP-A-IRTNVVTVPTAHPR
IAPKAS-TARDSSNKES---SRISDGKKQEGPATQVSKKNPASLPLTQAALKVLTQKASE
AQPPVARTQPSSGVDSAVGTLPATSPQSTPVQARGTNKLRKPKLPDGQQAA---------
KAPGSSDDSEDSS-DSSSGSEEDAE---GPQVAKSAHRL-GPTPSKMETLVEETTAESSE
DDVVAPSQSLLSGYVTPGLTLA---NSQTSKATPKPDSNPSVS-STLATKDDPD------
------SKQEAKLQQV--ADTLS------------------------------PKTGGKE
AVSGTKTQKSQKPKKGAGN--------PQASTPALQSNIAQRLVGQPWPLSEAQVQASVM
KVLTELLEQEKKKAADVTKESS------RKG--WESRKRKLSGDQPAARN----KKKKLG
AGEGGEAS-VSPEKASMTSKGKAKRDKASGGIKEKKEKGSL-GSQGAKDKPEEELQKGMG
KVEGGDQSNPK---SKKEKKKSDKRKKDKEKKEKK--KKAKKASTKDPESLSQ--KKKKK
KVE---------------------------------------------------------
--
ENSCANP00000002555 (view gene)
ENSMPUP00000000440 (view gene)
ENSP00000367028 (view gene)
MA-EARKRRELLPLIYHHLLRAGYVRAAR-------EVKEQSGQKCFLAQPVTLLDIYTH
WQQTSELGRKRKAEE-DAALQAKKTRVSDPISTSESSEE-E---EEAEAETAKATPRLAS
TNSSVLGADLPSSMKEKAKAETEK-AGKTGNSMPHPATGKTVANLLSGK-----------
------------------------------------------------SPRKSAEPSANT
TLVSET---------EEEG-SVPAFGA-----AAKPGMVSAGQADSSSEDTSSSSDETDV
EGKPSVKPAQVKA-----
PF03546 // ENSGT00730000111382_0_Domain_0 (319, 508)
SSVSTKESP---------------------------------
-------------ARKAAPA-PGKV---GDVTPQVKGGALPPA-----------KRAKKP
EEESESSEE-GSESEEEAPAGTRS-QVKAS---EKILQVRAAS---APAKGTPGKGA---
---------TPAPPGKA--------GAVASQTKAGKP-------EEDSESSSEES---SD
SEEETPA-AK--ALLQA-------------------------------------------
---------KASGKTSQVGAA------SAP--AKESPRKGAAPAPPGKTGPAVAKAQAGK
--REEDSQSSSEESDSE-E-----EA----PAQAKPSGKAPQVRAASAPAKE-SPRKGAA
PAPP----------RKTGPAAAQVQVGKQEEDSRSSSEE-SDSDREALAAM----NAA--
------------------------------------------------------------
---------QVKPLGKSPQVKPAST--------MGMGPLGKGAGPVPPGKVGPATPSAQV
GKWEEDSESS----SEE--SSDSSDGE---------------------------------
------------------------------------------------------------
-------VPTAVAPAQEKSLGNILQAKPTSSPAKG---------PPQK-------AGPVA
VQVKAEKPMDNSESS--EESSDSADS-EEA---PAAMTA---------------------
----------------------------------------AQAKPALKIPQTKACPKKTN
--TT--ASAKVAPVRVGTQAPRKAGTATSPAGSS-PAVAGGTQRPAEDSSSSEESDSEEE
-K-TGLAVTVG--------------------------------QAKSVGKG-----LQVK
AASVPVKGSLGQGTAPVLPGKTGPTVT-----------QV--KAEKQEDS-ESSEEESD-
SE--EAAASPAQVKTSVKKTQAKANPAAARAPSAKGTI-SAPGKVVT-------------
----------------AAAQAKQ-------RSPSKVKPP----------VRNPQNSTV-L
---ARGP--------ASVPSVGKAVATA-AQAQTGP-------EEDSGSSEEESDSEEE-
AETLAQVKPS--------------------------------------------------
--------------------------GKTHQIRAALAPAKES---PRK--GAAPTPPGKT
GPSA--AQAGKQ-DDSGSS-SEE-SDSDGEAPAAVTS-----------------------
------------------------------------------------------------
-----------------------------AQVI-KPPLIFV----DPNRSPAGPA---AT
PAQAQAASTPRKARASESTARSS--SSESEDEDVIPATQCLTP-G-IRTNVVTMPTAHPR
IAPKAS-MAGASSSKES---SRISDGKKQEGPATQVSKKNPASLPLTQAALKVLAQKASE
AQPPVARTQPSSGVDSAVGTLPATSPQSTSVQAKGTNKLRKPKLPEVQQAT---------
KAPESSDDSEDSS-DSSSGSEEDGE---GPQGAKSAHTL-GPTPSRTETLVEETAAESSE
DDVVAPSQSLLSGYMTPGLTPA---NSQASKATPKLDSSPSVS-STLAAKDDPD------
------GKQEAKPQQA--AGMLS------------------------------PKTGGKE
AASGTTPQKSRKPKKGAGN--------PQASTLALQSNITQCLLGQPWPLNEAQVQASVV
KVLTELLEQERKKVVDTTKESS------RKG--WESRKRKLSGDQPAARTPRSKKKKKLG
AGEGGEAS-VSPEKTSTTSKGKAKRDKASGDVKEKKGKGSL-GSQGAKDEPEEELQKGMG
TVEGGDQSNPK---SKKEKKKSDKRKKDKEKKEKK--KKAKKASTKDSESPSQ--KKKKK
KKKTAEQTV---------------------------------------------------
--
ENSNGAP00000007791 (view gene)
MA-EARKRRELLPLIYQHLLQAGYVRAAR-------EVKEQSGQKSFPTQPVTLLDIYTH
WQQSSELGQKQKAEN-DAALQAKKSRVSDPISSSESSEEEEE--AEAETAKASKTPRPTP
ANSSAVAADLPSSMKEK--AKTKK-ASQMVNSVLCPASGKSAVHLLSGK-----------
------------------------------------------------SPKKLAEPSANT
VLVSET---------EEEG-SVQALGA-----TANPGMLSAGQVSSSSEDTSSSSDETDV
EVKPSIKPAHTKA-----LPTPAKEHP---------------------------------
--------------A--VPG-PGKL---GDVTPQLRGGTLTAA-----------GRTADQ
EEESESSEDDSES-EDEAPAGVPT-QVSTG---E---------------------GALAP
AKQSSQKGALPTIPGKT--------GPAAAQTKR--P-------EDSDSSSEESD---SD
NRKTPAA-AT--QTPS--------------------------------------------
---------------P-------------------------AQVRPGDLVS---------
---------FLERPAADSASLPPIRS----------------------------------
------------------------------------------------FFPDRAHCDS--
------------------------------------------------------------
---------QVKPLGKTPQVRPAST--------VGLGSSGKGTHMAPAGKAGSAAPWAQV
GKLEENLESS----SEEEEESGNEGA----------------------------------
------------------------------------------------------------
------------------PLGKA-----------------AAPILPHK-------VEPVA
TQVKAERSKEDSEST--EES----TDSEEETA-PAATA----------------------
---------------------------------------PA----QVGS-Q-----NRST
PAST--V--------------VAPVTAPSQASLSSPALARGIQRPEVDSSSESLSESESE
A----AVPAS--------------------------------VGGKPMGKG-----PQGR
A-------ALAQGITPVPPGRSGPEVS-----------QV--RAKAQENS-QSSEEESS-
SEE--EVETPVQAKPLGKLPQAKASPSPTKVSSVKEPT-STPGKAAG-------------
----------------VV-------------APAKQGTL----------S----KATVI-
---QPGP--------GSAQA-------------------GG-----RLSHKDLPGSPSS-
--SN
PF03546 // ENSGT00730000111382_0_Domain_2 (1565, 1713)
AAAKPV--------------------------------------------------
--------------------------VKTSQIRAASAPAKES---PQK--GAHLGPSRKK
GS-AAQAQTRKPE-DSESS--SEESDSDREMPT---------------------------
------------------------------------------------------------
-------------------------------VI-KSPLIFV----DPNRSPAGPAGLAAT
PARTQAMSALRKARASDSTVQSS--SSESEDEDVIPATQSSIP-A-IRANVITVPAAPSR
TVPQAS------RSKQS---VQMSKGKKPKAA----------------------------
----------ATQVDSAMGTLPVTPPQSTPIQSRATNKLKNPKLPDKQQAPA---SHP--
KAPRSSDDSEDSS-DTSSGSEEDPK---GPQLTKSAPR---PAPSQKETVVEETPAESSE
DEVVAPSQSLFSGYVTPGLTLA---NSQTSNATLKPVSNPLVS-SVPLTKNDQD------
------GKQKAKSQPA--AHIMS------------------------------PKTGREE
ASSC-TPQKRRKSKKDTG---------LAASAEALQSSITQ--LDQPWPLSKAQVQASVV
KVLTELLEQEKKKATSAIKESS------KKA--QVAQKRKLSGDQPGAGAPKSKKRKQLV
AGEGGEGA-ASPEKASRTSKGKSK-----QDGK---EKGSP-GSRGVKDRPKS--E-SRM
KVEGGDHNDPK---SKKEKKKSNKNKKDKEK--K---KKGKKTSAKDPDSPFQ--KKKKK
KKKTTEPVL---------------------------------------------------
--
ENSCATP00000016431 (view gene)
ENSBTAP00000020259 (view gene)
MA-EARKRRELLPLIYQHLLQAGYVRAAR-------EVKEQSGQKIFLSQPVTLLDIYTH
WQQTSEIGRKRKAEE-DAALQAKRSRVSDPISSSESSEEEE-EEEEAEAKTTKPTPRLAA
TNSSVTGTVSSSNVKEKAKAKTTK-ASKTVNSTTHPAPGKAVAHLLTGK-----------
------------------------------------------------SPQKTSGPSANA
VLVSET---------EEEG-SVSALRT-----ATTPGMASADQA
PF03546 // ENSGT00730000111382_0_Domain_0 (285, 507)
DSSSEDTTSSSDETDL
EAKAAVKPLQVRA-----SATAVKESP---------------------------------
-------------RKSAAPT-PGKA---GVVTPQVRGGAVTPA-----------SRAKKP
EEVSESSEE-SDESEEAAPAGTPS-QVKAL---EKTVPIRVAP---SPAKGTPGKGA---
---------TPAPPGKA--------GPL------GKP-------EEDSESSS-EE---SD
SEEETQA-VK--TPVQVGPGERAPELAPTPGSLLRVDSPTPRAPATHRATWSLWSSTPSA
LLFPRVHHAKPSGKIPQVKAA------STPKIQVFFPKKGSPPTSPGKAGPAATQAQPRK
QEEEESSSSSSKESDSE-GEVPTAMQ----Q-----------------------------
-----------------------------------------------------TTSLA--
------------------------------------------------------------
---------QAKPLEKNSQVRAASA--------TGAGPSAKGTVPALPQKARSVAA--QD
GKQE-DSESS----SEE--ESDSEEEMQKAGTS----AQ---------------------
------------------------------------------------------------
----------------AQSSGKALQIRPASGPTKR---------PPQK-------AGPAT
TQVKVERAEEDSESS--EEESDSE----DEA--PVAKTP---------------------
----------------------------------------AQAKSAVKTSQIKASPRKGT
PITP--SSARVPPVQVGTPAPRTAAAVSTPTCASSPAIVRHTQKPGEDSSSSEESESEEE
T---APAAAGG--------------------------------QMKSIGKT-----LPVK
AASPATKGPSGKGTTPGPPGKAGPAVA-----------QV--KAEVQEDS-ESSE-ESDN
E---EAAATPAQVKTSVKTPQTKANSSATRVASAKAAA-PAPGKGGL-------------
----------------AVAQAKV-------GSPAKVKPP----------ARTPQSSAVS-
---GRGQ--------VSVPAVGKAAAAA-AQAQPGPVKGSQ--E-DSESSEEESDSEGE-
--APPQGKPS--------------------------------------------------
--------------------------GKTPQVRTASAPTKAS---PKK--GAAPVPPGKA
GPPA--TQAKRQEEDSESS-SEEESDSDGDVPA---------------------------
------------------------------------------------AAPPA-------
------------------------------QVI-KTPLIFV----DPNRSPAGPA---AT
PAQAQATGTLRKAQALESTARSS---SESEDEDVIPATQCPTP-A-TRTSVVTVPTAPPK
AALRAS-VVGAPS---SEDTSQGSEGKKQEASAT---QVTKRNLPLTQAALKVLAQKASE
AQSPAAKTPSSSGTDSAVGTLSTASPQSTPTQVRMANKLRKPEVPTAQQATATPSGHPTA
KAPATS---DDSN-SSSSGSEDDAK---GSQAAPSAHRP-GPAPSRRETLVEETTTESSD
DEVVAPSQSLLSGFVTPGLTPA---GSKTSKAAPKPDASPSVS-STPATRDAPE------
------GKQEVEPQQA--AGTMS------------------------------PKTGRK-
-EAGATPQKPG---KGLGS--------PPASMLALQSNITQRLLSEPWPLSEAQVQASVA
KVLTELLEQERKKAMDTAKEGS------RKG--RLGHKRKLSEDQTAPKAPKNKKKKQLA
AADGGEHV-VSSEKAPRTVKGKSKKDRASGDVKEKKEKESP-GSQGAKEKPVGEL--GTP
KGDGGDHGNLK---MKKEKKKSDKKKKDKEKKEKK--KKAKKASTKDPGSPSQ--KKKKR
KKKTAEQTV---------------------------------------------------
--
ENSPCAP00000013361 (view gene)
---------ELLPLIYQHLLQAGYVRAAR-------EVKEQSGQKSFLAQSVTLLDIYTH
WQQTSEAGQKRKAEN-DAALQAKKTRVSDPITSSESSDEEEE--AEAQIANAKKTPRLVS
TNSLVVGADLPSSMKEQAKAKTKK-ASKMVNSMPHPATGKAVTPHLSGK-----------
------------------------------------------------SPKKLAVPL-NT
NLVSET---------EEEG-SSV-LSV-----TAKPGVGD--PADSSSEDTSSSSGETDI
KAKSSRKMPPAKAPPIQASPVLSKEPP---------------------------------
-------------GVRAAPA-PAKV---GDVTAQVKGGPVTTA-----------SSAKKS
EEESESSEE-SE-SE----EETPS-QVKTV---GKTVHVRTAS---APAKGPPGKGG---
---------ATASPVKA--------GA--AQAKAGKR-------GQDSESSSEEE---SD
SKEETST-AV--APFQV-------------------------------------------
---------KPSGKVPQARAA------SAP--AKASPAKGA
PF03546 // ENSGT00730000111382_0_Domain_0 (642, 865)
ATAPPGKIGPAAAP-----
------------------------------------------------------------
-------------------------TGKPEEDSESSSEDESDSGEETSAAKTPTGSTA--
------------------------------------------------------------
---------LAKPSGKTSQARATSA--------LGTGPSGKGAVSAPVGKAASVVPQAQV
VKTEEDSESG----SEE--ESDSD-GE---------------------------------
------------------------------------------------------------
-------ASAAVTPVQVKPSGKTIQVRPASVPTKA---------PPQK-------TGPVA
TQVKAERPRADSDSS--EEESDSE----EET--PAAVAA---------------------
----------------------------------------AQAKPAVKTPQTKASPRRST
PITS--ASAKLPPVHVGTPAPWKAGTVTSAACTASPAVARGTQRPEKESSSSDGSDSEEE
E--MASMATGG--------------------------------QAKSVGKS-----LPGK
AASAPVKAHSGPATTPVLPGKAVLT-A-----------AV--KTETCEDS-ESSEEDSD-
SDEAPTAVNAAXMKASVKTPQTKANPASSRVSTVKGAA-STPGKVVT-------------
----------------GGVQTKL-------VSPAKVKSP----------VTTSQSSAT-S
---VQGQ--------ASVPAMGKAVAIA-AQAKTGS-------EEDSESSE-ESDSEEET
---PAQVKPS--------------------------------------------------
--------------------------GKAPEVKAASASTKGS---PGK--RAAPAPPGKT
DPAA--AQSGKPEEDSESS-SEEESDSAGEAPSAMTPT----------------------
------------------------------------------------------------
------------------------------QVI-KPPLIFV----DPNRSPAGPA---AT
PAPAQAANTPRKAQALESTARSS--SSESEDEDVIPATQCSIPPV-VRTSVVA-------
-----------SSSEES---RGVAEGKQQKMPATQVTKKNPVTLPLTQAALKVLAQKASE
TQPPTARMQPSSWVGSTLGKSPVLSPQSIPVPARGTSKLKNSKLPEAQPAT---------
RASGTSEDSEDSG-DSSSGSDKDVE---GAPRAKSTHKP-GPAPEGKETLVEETT-ESSE
DEVVEPSQSLLSGYVIPGVTPA---SSQASKATPRPGLNPSVS-STPATKDALA------
------GKLDTEPQQA--VATMS------------------------------AKTXXXX
XXXXXXXXXXXXXXXXXXX--------XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXX------XXX--XXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXX-XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX-XXXXXXXXXXXXXXXXXX
XXXXXXXXXXX---XXXXXXXXXXXKKDKEKKEKK--KKTKKASTKDSDSLSQ--KKKKK
KKKTVAEQSV--------------------------------------------------
--
ENSMOCP00000007313 (view gene)
ENSPTRP00000068671 (view gene)
MA-EARKRRELLPLIYHHLLRAGYVRAAR-------EVKEQSGQKCFLAQPVTLLDIYTH
WQQTSELGRKRKAEE-DAALQAKKTRVSDPISTSESSEE-E---EEAEAETAKATPRLAS
TNSSVLGADLPSSMKEKAKAETEK-AGKTGNSMPHPATGKTVANLLSGK-----------
------------------------------------------------SPRKSAEPSANT
TLVSET---------EEEG-SVLAFGA-----AAKPGMVSAGQADSSSEDTSSSSDETDV
EGKPSVKPAQVKA-----SSVSTKESP---------------------------------
-------------ARKAAPA-PGKV---GDVTPQVKGGALPPA-----------KKAKKP
EEESESSEE-GSESEEEAPAGTRS-QVKAS---EKILQVRAAS---APAKGTPGKGA---
---------TPAPHGKA--------GAVASQTKAGKP-------EEDSESSSEES---SD
SEEETPA-AK--ALLQA-------------------------------------------
---------KASGKTSQVGAA------SAP--AKESPRKGAAPAPPGKTGPAVAKAQAGK
--REEDSQSSSEESDSE-E-----EA----PAQAKPSGKAPQVRAASAPAKE-SPRKGTA
PAPP----------RKTGPAAAQVQVGKQEEDSGSSSEE-SDSDREALAAM----NAA--
------------------------------------------------------------
---------QVKPLGKSPQVKPAST--------MGMGPLGKGAGPVPPGKVGPATPSAQV
GKWEEDSESS----SEE--SSDSSDGE---------------------------------
------------------------------------------------------------
-------VPTAVAPAQEKSLGNILQAKPASSPAKG---------PPQK-------AGPVA
VQVKAEKPMDNSESS--EESSDSADS-EEA---PAVMTA---------------------
----------------------------------------AQAKPALKIPQTKACPKKTN
--TT--ASAKVAPVRVGTQAPRKAGTATSPAGSS-PAVAGGTQRPAEDSSSSEESDSEEE
-K-TGLAVTVG--------------------------------QAKSVGKG-----LQVK
AASVPLKGSLGQGTAPVLPGKTGPTVT-----------QV--KAEKQEDS-ESSEEESD-
SE--EAAASPAQVKTSVKKTQAKANPAAARAPSAKGTI-SAPGKVVT-------------
----------------AAAQVKQ-------RSPSKVKPP----------VRNPQNSTV-L
---ARGP--------ASVPSVGKAVATA-ARAQTGP------
PF03546 // ENSGT00730000111382_0_Domain_2 (1543, 1820)
-EEDSGSSEEESDSEEE-
VETLAQAKPS--------------------------------------------------
--------------------------GKTPQIRAALAPAKES---PRK--GAAPTPPGKT
GPSA--AQAGKQ-DDSGSS-SEE-SDSDGEAPAAVTS-----------------------
----------------------------------------------------AQKDSNSK
PARSKTLAPAPPERNTEGSSESSEEELPLTQVI-KPPLIFV----DPNRSPAGPA---AT
PAQAQAASTPRKGRASESTARSS--SSESEDEDVIPATQCLTP-G-IRTNVVTMPTAHPR
IAPKAS-MAGASSSKES---SRISDGKKQEGPATQVSKKNPASLPLTQATLKVLAQKASE
AQPPVARTQPSSGVDSAVGTLPATSPQSSSVQAKGTNKLRKPKLPEVQQAT---------
KAPESSDDSEDSS-DSSSGSEEDAE---GPQGAKSAHTL-GPTPSRTETLVEETAAESSE
DDVVAPSQSLLSGYMTPGLTPA---NSQASKATPKLDSSPSVS-STLAAKDDPD------
------GKQEAKPQQA--AGMLS------------------------------PKTGGKE
AASGTTPQKSRKPKKGAGN--------PQASTLALQSNIAQCLLGQPWPLNEAQVQASVV
KVLTELLEQERKKVVDTTKESS------RKG--WESRKRKLSGDQLAARTPRSKKKKKLG
AREGGEAS-VSPEKTSTTSKGKAKRDKASGDVKEKKGKGSL-GSQGAKDEPEEELQKGMG
KVEGGDQSNPK---SKKEKKKSDKRKKDKEKKEKK--KKAKKASTKDSESPSQ--KKKKK
KKKTAEQTV---------------------------------------------------
--
ENSOCUP00000003342 (view gene)
MA-EARKRRELLPLIYQHLLQAGYVRAAR-------EVKEQSGQKTFLTQPVTLLDIYTH
WQQTSELGQKRKAED-DAALQAKKTRVSDPISSSESSEE-E---EEAEALLAKTTPRPAS
VNSSVMAVTLPSGVKEKAKAQSKA--AKAVNSVPHPASGKALAHLLTGK-----------
------------------------------------------------SARKSGEPSANT
VLASET---------EEEG-GVPAPAP-----TAKPGLVSASQADSSSEDTSSSSDETDV
EVTPSAKPAPIRA-----PPVPAQEPA---------------------------------
-------------VRRAAPA-PGKV---GAVTPQAKGGALTLA-----------ARARKP
EEESESSEE-SD-SEEEAPAGRPS-QVNAS---TKVLQVRPSS---TTAKGTPGKVV---
---------TPAPPRKA--------GLVAPQAKAGRP-------EEDSGSSSEAS---SD
SEEETPA-AR--TPLQA-------------------------------------------
---------KPPGKTPQVRAA------FAP--AKGSPRKGTLPVAPGKTGPAATQAQVGK
--LEVDSESSSKESDSD-EETPAAMG----V-----------------------------
-----------------------------------------------------TVSPA--
------------------------------------------------------------
---------QVKPSGKNSWVRPAPT--------VGAGPSGKGTGLAPPGKAAPAAPLTQV
VKPEDESDSS----SEE--ESDSEGETTPAVSA---------------------------
------------------------------------------------------------
--------------TQAKPPGKILQVRPASGPTKGPLGKDVAMAPSQK-------LGPGA
TQVKAERAKENSESS--EESSDSV----EEA--PTATTP---------------------
----------------------------------------AQAKPALKT-QAKASPKKGT
PV-TPA-NTKVAPMRVGTPAPWKAGTATSPATASSPAVAWGARRPQEVSSSSEESESEEE
T---APAA-----------------------------------TVTSVGKG-----LQAK
AAP---A------TTPLLPGKMGPAVQ------------I--KAKAHEDS-ESSEEESD-
SA--QAASTPAQVQASARTPQTKASAAATKASSAEGVT-SPPGKVVA-------------
----------------AAAQAKQ-------GSPAKVKLP----------VRAPQNSALGK
AVATTGP--------AQKGPV------------GRP----Q--EEDSESSEESSD--SE-
EEAPAQVKPL--------------------------------------------------
--------------------------GKTPQVRAASAPAKES---PRK--GVPL
PF03546 // ENSGT00730000111382_0_Domain_2 (1675, 1793)
VPPGKA
GPAAAQAQLGKKE-DSESS-SEE-SDSGREAPAAMTPA----QVKPAVKNPQVKASPV--
-----KGALGAPTSVKASTAK---ADTPAPWKAKQASPAKV---------LSPRKDSNSK
TPRSKTLTPAPPEKKAQESSESSDEEWPSSQVI-KPPLIFV----DPNRSPAGPA---AA
PAQAQAAGAQRKTRASESSAQ-S--SSDTEDEDVIPATQSLAP-A-YRTS-V-VPTGHPR
TGLGA------SGSEES---SRVSEGKKQETPAPQVAKRNPASLPLTQAALKVLAQKASE
AQPPRARTQSSSGVDGTVGTLPVTSPQSAAVQAKVTSNLRKS---KGQQTTAPPLGHPKA
KAPGSSDDSEDSS-NSSSGSEEGAE---GPQLTKPTPRPAGSVPAGKETLVEETS-ESSE
DEVVAPSQSLLSGYVAPGLTPA---NSQVSKSTPRPDSNPLVS-STPATKDDLD------
------GKQEAEPQQA--VGTVS------------------------------PKAGRKQ
ATSGTTPQKPRKPKKGAPG----------TQASILPSSITQRLLGEPWPLSEAQVQASVV
KVLTELLEQEKKKALDASKESS------KKG-----RKRKLVVDQLAPKAPKSKKKKPAA
AGECREGA-DSQEKASRTSKGKAKKDKASGDGQEKKGKGPF-GSPGARDKSEGEL--GTA
KVEGGDQSNSK---SKKEKKKSDKKKKDKEKKEKK--KKAKRASAKDPESPFQ--KKKKK
KKV-AEQAV---------------------------------------------------
--
ENSCPOP00000002489 (view gene)
MA-EARKRRELLPLIYQHLLHAGYVRAAR-------EVKEQSGQKSFLAQPFTLLDIYTH
WQQTSELGQKRKAED-DAALQAKKTRVSDPLSSSESSDEEEE-DEEAEARTAKAAPRPAS
TNSLARGVDLPSSMKEKAKAQARS-TSKTMNSMPHLAPGKAAGHFLSGK-----------
------------------------------------------------SPKKSAQSSGSI
ILVSET---------EEEG-SIRALGA-----TAIPGTVSADQVDNSSKDSSSSSDETDV
EVKSSGKAAQVRP-----SPAPAKESS---------------------------------
-------------ANKVAPS-PRKV---GGLVPQAGGGALTPA-----------GSAKVL
KEDPGNSKE--SESEEEP----PS-QVKVS---EKILPVRAAS---ASTKGTPGKG----
--------ATCALPGKV--------GPVTGQAKVGRP-------EQEQDSGSSSE-EESH
SEVEMPA-AT--TAPQA-------------------------------------------
---------KSAGKTSQVRAA------TAP--AKESPRKGAPPACPGKAGPAATQTQVGR
QAEDSVSSSSSEESDSD-TETPA-------------------------------------
-------------------------------------------------PQAPVAKPV--
------------------------------------------------------------
---------QLKPLGKTPQVRAAPA--------PAKESPRKGVPPAPPAKTGPAATQAQM
GKRKRDSDSS--------------------------------------------------
----------------------------------------------------SSSEESDS
DGETPAPRIPATSPAQAKPSGKNLQVRPASAPTKEPLGKGAAPKPPQK-------SAPVA
IQVKTEMSKEDSDSSDSDESSDSE----EEA--PAAAPA---------------------
----------------------------------------AQEKPALKTPQAKASPRKSA
LLTP--TSAKGPPVRVGTPAPWKAGTMTSA---PAPALPPGTQRPENDSLSSE-ESEAEG
A---TPAAAVA--------------------------------QTK------------QQ
AASAPTKGPAGQGVTPAPPGKTRPTIAAPAGKARPTVTHV--RAEAHESS-ESSEEDSDD
SE-AAAVMTPPQVKASGKTPQAKASPASTTASAVKGAA-SAPGKLVT-------------
----------------PVAQGKR-------GPMAKGKSP----------VRAPQNGAIP-
---ARGQ--------ASVSAVEKAVTAA-AQA-----------EKDSESSEESSESEEK-
--VPAQVKPTGKTPA---
PF03546 // ENSGT00730000111382_0_Domain_2 (1579, 1655)
-SAPAKEPPRKGAPPAPIGKTVPTAT--QVGRQEENSVSSSS
SEESDSDTETPAPQAPTTNPAQLKPLGKTPQVRAAPAPGKES---PRK--GAPPAPPAKT
GLAATQAQVGRQEEDSDSSSSSEESDSDGETPAPRIPATSPAQLKPSGKTPQVKAAPTPA
KESPRKGVPPAPLGKTGHVGG-----VKQEDSDSSSGEESDSDRETLAPQTPAQKNITSK
TARSSTVTPAPPEKNPEGSSESSDEELPPSQVI-KPPLIFV----DPNRSPAGPA---VT
PAQAQAASAPRKARAPSSTAGSS--SSESEDEDVIPATQSSAPAVR---TDATMPTTSTR
TAPRAS------ASEES---SRASEDKKQEVAASQVTRKNPASLPLTQAALKVLA-----
----------QKEAGSAAGTLPVTQPQSTAVQARVANKLRQTKLSEGQQNTAIPSGCPKA
EAPGSSDDSEDSS-DDSSENEEDAE---GPQKAKSDPRLAGPPLSKRETLVEETTAESSE
DEVVAPSQSLLSGYVTPGLTMA---NNQASKATAGRESNPTIS-SASRTKD-PA------
------GKP-AAESQP--AGIMS------------------------------PKPGRKE
AASGPTTQKPRKPKKRPAS--------TEASLQALQSSITQRLLGQPWPLSEAQVQASVV
KVLTELLEQQRQKATDATKESS------RKG-----QKRKLSGNQAASKAPKSKKKKQLA
AGELKEGA-ASSEKASRTSKGKSKRGKASGDIKEK-EKGAP-GSGGAMAKPEGEL----P
KVEGGDQNDPK---TKKEKKKSSKKRKDKEKKEKK--KKAKKASAKDCDSPFQ--KKKKK
KKTTAEPTV---------------------------------------------------
--
MGP_SPRETEiJ_P0048783 (view gene)
MA-EARKRRELLPLIYHHLLQAGYVRAAR-------EVKEQSGQKSFLTQPVTLLDIYTH
WQQTSELGQKQKAED-DETLQAKKSRVSDPVSSSESSEQEKE--EEAATERAKATPRPTP
VNSAT--AALPSKVKEK--GKTKT-ANKTVNSVSHPGSGKTVVHLLSGK-----------
------------------------------------------------SPKKSAEPLANT
V
PF03546 // ENSGT00730000111382_0_Domain_0 (242, 476)
LASET---------EEEG-NAQALGP-----TAKSGTVSAGQGSSSSEDSSISSDETDV
EVKSPAKPAQAKA-----SAAPAKDPP---------------------------------
--------------ARTAPG-PTKL---GNVAPTPAKPARAAAA-AAA-AAVAA-AAA-A
AEESESSEEDSDS-EDEAPAGLPS-QVKAS---GKGPHVRAAS---VSAKGISGKVPISA
AKGISGKGPILATPGKT--------GPAATQAKAERP-------EKDSETSSEDD---SD
SEDEMPV-T--VNTPQA-------------------------------------------
---------RTSGKSPWARGT------SAP--AKESSQKGAPAVTPGKARPVAA--QAGK
PE---AK--SSEESESDSGETPAA------------------------------------
--------------------------------------------------ATLTTSPA--
------------------------------------------------------------
---------KVKPLGKSSQVRPVST--------VTPGSSGKGANLPCPGKVGSAALKVQM
VKKEDASESS----SEELD---SDEPVS--------------------------------
------------------------------------------------------------
-------------PAKAKPPGKK-----------------ASPALPQK-------VGPVA
TQVKTDRGKDHSGSS--EES----PDSEEEA-----------------------------
----------------------------------APAASAAQAKPALEK-QMKASSRKGT
PASA--TGASTS-------SHCKAGAVTSSASLSSPALAKGTQRSDVDSSSESESE--GA
A----PSTPR--------------------------------VQGKSGGKG-----LQGK
A-------ALGQGMAPVHTQKTGPS--------------V--KAMAQEDS-ESLEEDSS-
IEE--EDETPAQATPLGRLPQAKANPPPTKTPPA-----SASGKA---------------
----------------VA-------------APTKGKPP-------------VPNSTVS-
---ARGQ--------RSVPAVGKAGAPA-TQAQKGPVTGAG--EDSESSSKEESDSEEE-
--TPAQIKPV--------------------------------------------------
--------------------------GKTSQVRAASAPAKES---PKK--GAHPGTSGKT
GSSATQAQTGKTE-DSDSS--SEESDSDTEMPS----A----------------------
------------------------------------------------------QDDISQ
PARGKASGPASPEKSIEGSSESSDEDLPSGQAI-KSPPVSV----NRNSGPAVPA---PT
PEEVQAVNTTKK-A-SGTTAQSSSSESEDGDEDLIPATQPSTY-A-LRTSVT-TPAALSR
AASQPS------KSEQS---SRMPKGKKAKAAA---------------------------
----------SAQTSSAVETLPMMPPQSAPIQPKATNKLGKSKLPEKQQLAP---GYP--
KAPRSSE---DSS-DTSSEDEEDAK---RPQMPKSAHRL-DPDPSQKETVVEETPTESSE
DEMVAPSQSLLSGYMTPDLTVA---NSQASKATPRPDSNPLAS-SAPATKDNPD------
------GKQKSKSQHS--ADTAL------------------------------PKTGRKE
ASSGSTPQKPKKPKKITSS--------SPAPTQTLPNSITQRLLEQAWPLSEAQVQASVV
KVLTELLEQERLKATEAIKESG------KKS-----QKRKLSGD-LEAGAPKNNNKKEQP
V--PRASA-VSPEKAPMTSKAKSKLDKGSAGGK---GKGSP-GPQGAKEKPDGGEL-LGI
KLESGEQSDPK---SKSKKKKSLKKKKDKEKKEK---KKGKKSLAKDSASPIQ--KKKKK
KKKSVEPAV---------------------------------------------------
--
ENSPVAP00000005127 (view gene)
MA-EARKRRELLPLIYQHLLQAGYVRAAR-------EVKEQSAQ-KFPTQPVTLLDIYTH
WQ-TSVIGQKRKAEE-DAALQAKKTRVSDPISSSESSEEE----EEAEAETTKAAPRLTS
INSSAVEAVLPSSMKEKAKAKTKN-VNKTVNSTPHPASGKTVAHLLSGK-----------
------------------------------------------------SPRKSSGPSANT
ILVSET---------E-ES-SVPALGT-----TAKPGMASAGQVASSSEDTSSSSDETDI
EXXXXXXXXXXXXXXXXXXXXXXXXXX---------------------------------
-------------XXXXXXX-XXXX---XXXXXXXX-----XX-----------XXXXXX
XXXXXXXXX-XXXXXXXXXXXXXX-XAKAS---EKIVQVRAAS---GPSKGTLAKGG---
---------TPAPPGKA--------GPATAPAKAGRQ-------GEDSESSSE-D---SD
SEEEAPA-AK--
PF03546 // ENSGT00730000111382_0_Domain_0 (553, 684)
TPLQA-------------------------------------------
---------KPSGKTLQVKAA------SAT--AKESPRKGGTPVPPGKAVPAAAQAKAGR
--QEEDSESSSEESDSE-EEAPAAKT----PLQAKPSGKTPQVKAASATAKE-SPRKGAT
PAAPGKAGP----------AAAQAKAGRQEEDSESSSEE-SDSEGAAPAAVPVTTNPA--
------------------------------------------------------------
---------QAKPLGKNSQVRAASA--------MAMEPSGKGTVPASPGKAGSAATLAQA
GKQENSE--------------SSSKGA---------------------------------
------------------------------------------------------------
-------VPAALAVARAKSSGKGLQIRPASGPTKD---------TPQK-------TGPAA
TQVKIERSKEDSESS--EEESDSE----GEA--SAAMTP---------------------
----------------------------------------AQAKLAVKTPQAKASPRKGI
PITP--ASAKVPPVRVGTPAPWKAGTANSPACTSSPAVARSTQRPEEDSSSSEESESEEE
---TAPATAGG--------------------------------QVKSVRKS-----LPAK
AASAPTKGPSGQGTTPVLPGKAAPSVA-----------QV--KVRLXXXX-XXXXXXXXX
XXXXXXXXXXXXVKTSVKTPQTKANPASSRVASAKGAA-SAPGKVVS-------------
----------------AAAQAKR-------GSLAKVKPP----------ARNSQTNAV-S
---VKGQ--------VSMPAMGKAVAGA-AQAQPKPV------KEDSDSSEEESDSEGEA
---PVQVKPS--------------------------------------------------
--------------------------GKTMKIRTALVPTMGS---PRK--GIAPAASGKI
GPTA--TQARKQ-EDSESS-SEEESDNEKEAPGAVTPA----------------------
------------------------------------------------------------
------------------------------QVI-KTPLIFI----ELNRSPAVPA---AT
PTPAQAANTPKKARASESTARSS--SSESEDEDVIPATQCLAP-G-IKTNMVAVPTAHPK
TALKTS-IVGASSNEES---SRVLKGKKQEAPATQVTKRNLAHLPLTQAALKVLAQKASE
AQPPAARTPSSSGIDSALGILPAASPHSTPSQAKVTNKLRTPEVPVVPRAT---------
KASGTSDDS-DSS-GSSLGSKEDAK---RPQMAPLAPRL-GPAPSRKETLVEETTAESSD
DEVVAPSQXXXXXXXXXXXXXX---XXXXXXXXXXXXXXXXXX-XXXX--XXXX------
------XXXXXXXXXX--XXXXX------------------------------XXXXXXR
QEASTTPQKPQKPKKGAGN--------PQASTLALHSDLTRRLLSEPWPLSEAQVQASVV
KVLTELLEQERKKAMDAAKESS------KKV--RAGHKRKLSGDQAVVKASKSKKKKQLV
VXXXXXXX-XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXX---XXXXXXXXXXXKKDKEKKEKK--KKSKKVPTKDPDSEFQ--KKKKK
KKKTAEQTV---------------------------------------------------
--
ENSSHAP00000021147 (view gene)
ENSRNOP00000045755 (view gene)
MA-EARKRRELLPLIYHHLLQAGYVRAAR-------EVKEQSGQKNFLTHPVTLLDIYTH
WQQTSELGLKQKAEN-DETLQAKKSRVSDPISSSESSEEEE---EEAETGTAKATPRPTP
VNSTA--AALPSNMKQK--AKTKT-ANKTVNSVLPPASGKTAVQLLSGK-----------
------------------------------------------------SPKKSAEPLASI
VLASET---------EEEG-SAQVLGP-----TAKPGMVSAGQASSSSEDTSSSSDETDV
EVKPSTK-PQAKA-----PVTPAKDPP---------------------------------
--------------ARTASG-PTKL---G
PF03546 // ENSGT00730000111382_0_Domain_0 (390, 508)
DVTPASARA-----------------AAT-K
AAESESSEEDSES-EDEAPAGLPS-QVTTP---GKGLHVRAAS---VSAKG---------
---MSRKGPISATPEKT--------GPITPQAKTGRP-------QKDMETSSEDD---SD
SEDEMPV-T--VTTPQA-------------------------------------------
---------RPSGKSPQVRGT------SAP--AKESSQKGAPAVTPGKTRPVAAQAQAGK
PE---TR--SSEESESDSGETPAA------------------------------------
--------------------------------------------------GTLTTSPA--
------------------------------------------------------------
---------KVKPLGKSPQVRPAST--------ANLGSSRKGANPPCLGKVGSAVLKVQT
GKEEEDSESS----SEESD---SDGAVN--------------------------------
------------------------------------------------------------
-------------AAKAKPPGRK-----------------ATPALPRK-------TGPVA
TQVKTDKGKDHAESS--EES----TDSEEEA-----------------------------
----------------------------------APAASAAQAKPAPIK-QMKASPRKGT
AAST--TGASAS-------SPRKAGTKTSSASLSSLALPKGTQKPDVDSSSEWESE--GA
A----PGTTG--------------------------------VQGKSGGKG-----LQGR
A-------ASGQGVAPLHAQKTGPSGA-----------QV--KATAQEDS-ESSEEESS-
SEE--EDETPAQVMALGRLPPAKANPPPTKTPLA-----SASGKAAA-------------
----------------VV-------------PPPKGKAP----------ASTVQNSTIS-
---ARGQ--------RAVPATGKAGAPA-TQAQKGPMAGTG--EDSESSSEEESDSEEE-
--TPAQVKPV--------------------------------------------------
--------------------------GKTSQVRAASAPVKES---PNK--GAYSGTSRKT
GPPATQAQTGKTE-DSESS--SEESDSDREIPPAITPA----------------------
------------------------------------------------------QDGISQ
STRSKLSGLAPPEKSTEESSESSDEDLPSGQAI-KSPPISV----NPNRGPAAPV---PT
PEQHQAVNTRKA-QASGSTAQSS--SSESEDEDMIPATQPPTL-A-IRTNVT-TPTALSQ
TAAQPS------KSEQS---SRMPKGKKPKTA----------------------------
----------STQISSAMEALPVTLPQSTPAQSKTTNKLGDPKLAEKQQLTP---GYP--
KAPRSSE---DSS-DTSSESEEDAK---RPQMSKSSQRL-DPDASQKETVVEETPTESSD
DEMVAPSQSLLSGYVTPGLTVA---NSQPSKATPRPDANPLVS-SAPATKDNPD------
------GKQKSKSQDS-TADTTL------------------------------RKTGEKE
ASSGSTPQKPKKPKKSTLS--------SPAPAQTLPNSITQRLLEQPWPLSEAQVQASVM
KVLTELLEQERQKATEAIKESG------KKG-----QKRKLSGDQVEAGAPKNKKKKQQL
A--AGTSA-GSPEKASRTSKAKSKLNKGSAGGK---GKGSP-VPQGAKEKPEG--K-LGI
KLESGEQSDPK---SKKEKKKSSKKRKKEKRPQP-------RT------LPHR--SRRRK
RRRRRQPSLLCEGQAAHR----TESWQSVTIPRPLTSG-HCELGLDFLICPPSAEGDGWR
CC
ENSMLUP00000012077 (view gene)
ENSMICP00000035903 (view gene)
MA-EARKRRELLPLIYHHLLRAGFVRAAR-------EVKEQSGQKILLAQPVTLLDIYTH
WQQTSELGQKRKAEV-DAELLAKKTRVSDPISSSESSEE-----EEAEAETAKATPRLAC
TNSSAMGADLLSSMKEKAKTKTET-ASKMGNSMPNPASMKTVSHLLSGK-----------
------------------------------------------------SPKRSGEPSANT
VLASET---------EEEGGSVPVFGA-----AAKPGTVSAGQADSSSEDTSSSSDETDV
EAKPSVKPAQV-------------------------------------------------
----------------------------K-------------------------------
--------A-LS----------------IP---AKELPVRR-A---AP---APGKVG---
---------DVTPQVRA--------GALTPARTTKKP-------GEESESS-EES---SE
SE-EEAP-AG--TPSQV-------------------------------------------
---------RPSEKTPQVRAA------SAP--AKESPRKGAAPATPGKTGPAAT--QVGT
--QEEDSESSSEESDSD-GEAPAAVS----LAQVR--LESRLWAF--------------L
PSVPS---------ACTAPFALLLPV---------S----SHTCSLWAPSPDLVLCHS--
------------------------------------------------------------
---------QAKPLGKNPQVRPAST--------VGVRPSGKGAIPALPGKAGTTAPFAQV
GKQEDPTSS-----SEE--SSDSEGEAPTVGAP----AQVRPHLSCQALLS---------
LSVQSPTWPPPACPLSALR----LRLPS-----------------------QSL---CLH
TSTVWAPLLILCVSFQAKPLGKILQVRPAPSPAKG-----TVPVGPQK-------AGPVA
TLVKAERSKEDSESS--EESSDSE----EEA--PAAMPS---------------------
----------------------------------------AQAKPALKTPQTKASPRKGA
PSTL--ASAKTPSARGGTPAPWKAGA-----------VAPPAQRPEEDSSSSEESESEEE
E--RAPVAAVG--------------------------------QAKSVGKG-----LQAK
AA----SGPSGPGTAPVLPGRMGLAVT-----------QV--KAEAQEDS-ESSEE----
----DKAMTPAQVKALVKACLAKANPAATRVSSAKG-T-SAPGKGVA-------------
----------------VTAQTKQ-------GSTAKTKPP----------GRTLQNSAVSV
----RAQ--------APMPSVGKAGATA-AQAQTGP----G--EEDSDSSEEESD--SE-
EEAPAPAKPL--------------------------------------------------
--------------------------
PF03546 // ENSGT00730000111382_0_Domain_2 (1647, 1830)
GKNLQVRAASAATKES---PRK--GAASVPPRKT
GPAATQAQAGKQG-DSESS-SEE-SDSDGEVPAAVTP-----------------------
------------------------------------------------------------
-----------------------------AQVI-KTPLIFV----DPNRSPAGPA---AT
PAQAQAASTPGKARASESTAKSS--SSESEDEDVIPATQCLTP-A-IRANVT-ASATHPR
TNPKAN-IAGASNSKES---SRGSDGKRQQGPATQVTKRNRASLPLTQV---------LE
SQPPIGRTQASSGVDSAVGTFPVMGSQSIPVQTGVTSNPRKPKLPDSQQAT---------
-TAPLD--DSEDS-DSSSGSEEDAE---GPQMAKSAQR-PGPATSSRETLVEETT-DSSE
DEVVAPSQSLLSGYVTPGLTPA---NSQASKATPRPDSNPSVS-SAPAIKDDPD------
------GKREAEPQQA--AGITS------------------------------PKTGKKE
ATSGATSQKPQKPKKGAGN--------PQASTLALQNDITQRLLGESWPLSEAQVQASVA
KVLAELLEQEKKKATDATKESS------KKG-----RKRKPSGDQLATKAPKSKKKKQLA
AGEGGEGA-VSPEKASVTSKGKAKKDKSSGDVKEKKEKGSP-GSQGGKDKPKGELPKGAV
KVEGGDQSSPK---TKKEKKKSDKKKKDKDKKEKK--KKAKKTSTKDPELPFQ--KKKKK
KVE---------------------------------------------------------
--
ENSTBEP00000003838 (view gene)
ENSCGRP00000012633 (view gene)
------------------------------------------------------------
--------------------MMHHCKPSSPISSSESSEEEE---EEAETKTAKAIPRPTS
VNSIATAADLPSSMKEKAQAKTKT-ANKMVNSVQQPASGKAVVHLLSGK-----------
------------------------------------------------SPKKSAEPLANT
VLASET---------EEEG-SVPALGA-----TAKPGVLSTGQASSSSEDTSSSSDETDV
EVKVSVKPAQAKV-----SPAPAKEPP---------------------------------
--------------ARTAPC-PTKS---GSGMXXX----XAAP-----------GRTA-H
PF03546 // ENSGT00730000111382_0_Domain_0 (421, 525)
AEESESSEEDSES-EDEPPAGMPS-QVKAP---GKVLHGRAAS---VYAKE---------
---VSGKGPTSAPPEKT--------RPIVTPAKAGKP-------EKETESSSEDS---SD
SEDEEPV-AV-MTTPQV-------------------------------------------
---------RPSGKNPQVKST------STP--AKGSSQKGAPPVTSGKAGPAAAQAQAGK
PE---ESDSSSEGSESDSGKTPAA------------------------------------
--------------------------------------------------VTLSTSSA--
------------------------------------------------------------
---------QVKPLGKSQQVRPAST--------VSLGPSGKGARPPCPGKTGSAVLGVQM
GKVEEDSESS----SEDS----DDGAVT--------------------------------
------------------------------------------------------------
-------------PAKAKALGKR-----------------ATPALPQK-------AGPVA
AQVKIERRKEDSESS--EESSEESTDSEEEA-----------------------------
---------------------------------APAASAPAQAKPDLEK-QTKASPRKGT
PATP--ASAMVS-------SMRV---ST------------------APSSEEGSGA--AP
------GTTR--------------------------------VQGKSVGKG-----LQGK
G-------DLGQGMAPVHPERPGPTVA-----------HV--RITAQEDS-EDSEEESS-
SEE---DETSAQAKPLGKPLQAKAIPPPTKASPAKRPT-SASGKV---------------
-------------------------------SPSKGKPP----------MSTVQNSALS-
---AR------------VPATGKAGAPA-VQAQKGLVAGAG--QDSESSSEEESDSEEE-
--APAQTKPV--------------------------------------------------
--------------------------GKTSQVRAASAPAKES---PGK--GPHPGATKKT
LPTSAQAQTGKTE-GSESS--SEESDSDGEMPSAIIPA----------------------
------------------------------------------------------QGGASQ
PAKSRASAPAAPEKSIEDSSESSDEELPSSQVI-KPPLISV----NPNRSP---Q-----
-----AVSNPRKAQASGTMAQSS--SSESEDEDMIPDTQPSTH-A-TKANVVTVPTSLPR
TAPQPS------KSEQS---SRMPKGKKPKAA----------------------------
----------STQISSDVKTLPVTPPQSTPVQSKTTNKPRKPKLPAKQQPPS---TYR--
SS---SS---DSS-DTSSGSEEDAK---RPQVATSAPRL-DPHLSQKETVVEETPDESGE
DELVAPSQSLLSGYVTPGATVA---NSQASKATSRPDSNPLVS-SAPVTKDNPD------
------GKQKAKSQD--AADTVF------------------------------PKTGRKE
TSSDTTPPKPKKAKKSTWT--------SATPTQALQNSITQCLREQSWSLSEAQVQASVV
KVLAELLEQERQKATEAIKESG------KKG-----KKRKLSGDQLAAGAPESKKK--AS
-------------GTS---KTKSKLDEVNADGK---GKGSS-GSQGAKAKPES--E-LGL
KVESGQQNDTK---NKKEKKKSSKKKKSKDKKGK---KKGKKTSAKDPDSQIK--KKKKK
KK-TAEPAV---------------------------------------------------
--
ENSDNOP00000009653 (view gene)
MA-EARKRRELLPLIYQHLLHAGYVRAAR-------EVKEQSGQKSFLAQPVTLLDIYTH
WQQTSEVGRKRKAED-DAALQAKKARVSDPVSSSESSEEEEE--AEAQAAGARMTPRLVS
TNSSGIGTDLPSSVKEKAQAKTKK-ASSTVNSTPRPASGKAVVHLLSGK-----------
------------------------------------------------SPKKSAESLTNT
TLVPET---------EEEG-SVPALGT-----LAKPGIASAGQADSSSEDTSSSSDETDV
EVKPSMKPPQVK------------E
PF03546 // ENSGT00730000111382_0_Domain_0 (326, 637)
SP---------------------------------
-------------GRKAAPA-PGKA---GPATPQVRGGAPTPA-----------SRARKP
GEESESSTE-ESESEEEAPTGTAG-KVKAT---GKSFQIKSAS---VPAKGTPGKGA---
---------TPAPPEKT--------GPAATQAKVGGS-------EDGSDSSSEEA---SD
SEGAPPP-AM--ARLQV-------------------------------------------
---------RPGLASRRAGAL------EAP--AAPGA--GCGRPPPSDPGAPCARSSSC-
--------LSAHPPRDA-LL----IL----SLQAKPSGKAPQLRGASVPTKG-ALARGAT
PAPPGKAGPAAVQ--------AKAQAGKLEEDSESSSEEESDSAEEAPAAEALTTSPA--
------------------------------------------------------------
---------QAEPSVRNSQ--------------VRQGRQGKGPPQC-SRESGVCSPLGQG
GKEDSDSEGK----AR--------QGR---------------------------------
------------------------------------------------------------
-------GAEPCAPAQVRPRP-----------RRPPLGPSSGPFLPQH-------QEPCT
LPRNLAKPSGKTIQV--RP----ASG-GAFP--SLSPVS---------------------
----------------------------------------PQAKPATKTPQTKASPRKGT
PVTP--TPARVPPAPGGTPAPRTAGAATSPACTSSLTAARGAQRPEEDSSSSEEETESEE
-E--TAPAAVG--------------------------------QAKSVGKG-----LQVK
AAPAPAKGTSAQQTTPGPPGKAVPAAT-----------QV--KTETD--S-ESSEEESD-
SEEAPAAATPAQVKASVKIPQAKANPASTRASSGKGAV-SAPGKVVT-------------
----------------PAVQAKQ-------GSPAKGKPP----------VRTPQSSAT-L
---AKGQ--------GSVPAVGKAVATA-GQAQAGAVRRPE--EEDSESSEEDSDSEEEA
---LAQAEIS--------------------------------------------------
--------------------------GKNPQVRATSASIKGS---PGK--VATPAPPGKA
GHAA--AQVGKLEEDSESS-SEEESDSEEEPPKAAAP-----------------------
----------------------------------------------------AQKEGNSK
TARSKSLALAPREKSAEGSWE-SEEELSSGQVI-KPPLIFV----DPNRSPAGPA---AT
PAQAPAASTPRKAWASESTARSS--SSESEDEDVIPATQCPTPPV-IRTSMVTVPAVHPR
TAPRAR-AAGTSSSEES---SGESEGKKKEAPATQVTKKNPASLPLTQAALKVLAQKASE
AQPPSARTQSSSEVDSALGKLPVLSPQSTAVQT-STTKLRKPKLPEAQQPAAAPLGCPKA
KASGTSDDSEDSS-DSSSGSEEPAK---GTQTAKAAARLVGPAPSRKETLVEETTAESSE
DEVVEPSQSLLSGYVIPGSTPA---DSQASKATARPDTNPSIS-STTATRDAPG------
------GKPEVGHQQA--ATTTS------------------------------PKKGKKE
AALGTKPQKPRKPKKGAVN--------PQASTLALQSDIAQRIRSAPWPLSEAQVQASVA
KVLAELLEQERKVAVGAVKESS------KKA--RVDRKRKLSEDQPAAKTPKNKKKKQPA
AGEGRDGA-DSPEQAPRTSKEKSKRDKASGDIQEKKGKGSP-SSRGAKEKPGGEL--GVG
KVEGGDQGNPK---SRKEKKRSDKKKKDKEKKEKK---KVKKASTKDTDLPSQ--KKKKK
KKKTAEQTV---------------------------------------------------
--
ENSCAFP00000031314 (view gene)
MA-EARKRRELLPLIYQHLLQAGYVRAAR-------EVKEQSGQKSFLTQPVTLMDIYTH
WQQTSEVGQKRKAEE-DAALLAKKTRVSDPISSSESSEEEEE--EETEAEATKTTPRLAS
TNSSVPGSVLPSSTKEKGVAKTNK-ASKMVNSTPHPASAKAVAHILSGR-----------
------------------------------------------------SPRKSAGPSANT
ILVSET---------EEEG-SVPALGT-----TAKPGMASANQADSSSETSSS-SDETDV
------------------------------------------------------------
------------------------------------------------------------
-------------------------EVKAS---EKIVQAKAAS---GPVKGTPGKGA---
---------TPAPPGKA--------GPSAAQAKTEKP-------KEDSDS-SEED---SD
SEEEPPA-AK--TPLQV-------------------------------------------
---------KPSGKTPQVKAA------SAS--AKESPRKGVPPVPPGKVGPAAGQAKKGA
GEE--DPDSSTEESDSE-EEAPTAVP----PTRSPV--QCPCSED---------PSVGLS
--SP----------SAPGPHALHLPP---------SPPVSPPKQSPVGPCPDPVFCYS--
------------------------------------------------------------
---------QAKS-----------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------SGKIPQVRAASGPAKG---------PPQK-------AGPAA
TQAKAKMSKDDSESS--EEESESE----EEA--PAAMAP---------------------
----------------------------------------VQAKSTVKTPQTKASPKKGT
PITP--IPAKAPPVRVGTPAPWKARAEASPACASSPAMARGAQKPEA-SSSSEESESEEE
T---APAPAAG--------------------------------QAKPVGKG-----VTVK
AASTPTKRPSGQGTALAPPGKAGPATA-----------PV--KTEVQEDSSESSEEESGS
EEAEEAAAAPAQVKTAVKTPQSKANQAPTRATSAKGAA-SAPGKVVS-------------
----------------AAVQIKQ-------ESPAKVKPP----------ARTSQSNAVS-
---VRGQ--------GAVPAVGKAVATA-AQAQPGPVKNPE--E-DSESSEEESDSDGE-
--APTPVKPS--------------------------------------------------
--------------------------GKTPQVGTASAPSKGL---SRK--
PF03546 // ENSGT00730000111382_0_Domain_2 (1671, 1829)
GATAAPPRKT
GAVA--TQAGKPEEDSESS-SEEESDSEEE----A-PA----QVKPGIKPPQTKASLL--
-----KGVSGTPASAEASTTR---VDNSAPQKAR--------------PAPPAKKEGSSK
TAKSKAQAPAPPEKNAEGSSESSDDELPATQVI-KPPLIFV----DPNRSPAGPA---AT
ATQAPAADTPRKAQASESTARSS--SSESEDEDVIPATQCSVP-A-GRTNVTPL-TAHPR
PAVRAS-TVGASGGEASG---RVSEGKKQEAPITQVTKRNPAHLPLTQAALKVLAQKASE
AQPPAARTPSSSGVDHALGTLPVMSPQITPVQAKMTNKLKKAEVPAVERATATPVGHPKA
KASETSNDSEDSS-GSSSGSEEDAD---GPQK---AHRRVGPAPSATETLVEETTAESSE
DEVVAPSQSLLSGYVTPGPTLA---NSQASKATPRPDLNPSAS-STSISKDAPD------
------GKQEVEPQHV--AGTVS------------------------------PKTSRR-
-EADATPQKPRKPKKEAGS--------PQASTLALQSDISRRLLSEPWPLNEAQVQASVV
KVLTELLEQERKKAADAAKESN------RKG--RVGRKRKLSGDQTAARVPKSKKKKQLV
AGGGGEGAAGSPEKALRTPKGKAKRDGASSDIKEKKEKESP-GSLEAKEKPEGEP--GTL
KVEGGDQGNPK---IKKEKKKSDKKKKDKEKKEKK--KKAKKASTKDPDSLFQ--KKKKK
KKKTAEQTV---------------------------------------------------
--
ENSGGOP00000041929 (view gene)
--------------------------------------------KCFLAQPVTLLDIYTH
WQQTSELGRKRKAEE-DAALQAKKTRVSDPISTSESSEE-E---EEAEAETAKATPRLAS
TNSSVLGADLPSSMKEKAKAETEK-AGKTGNSMPHPATGKTVANLLSGK-----------
------------------------------------------------SPRKSAEPSANT
TLVSET---------EEEG-SVPAFGA-----AAKPGMVSAGQADSSSEDTSSSSDETDV
EGKPSVKPAQVKA-----SSVSTKESP---------------------------------
-------------ARKAAPA-PGKV---GDVTPQVKGGALPPA-----------KRAKKP
EEESESSEE-GSESEEEAPAGTRS-QVKAS---EKILQVRAAS---APAKGTPGKGA---
---------
PF03546 // ENSGT00730000111382_0_Domain_0 (490, 693)
TPAPPGKA--------GAVASQTKAGKP-------EEDSESSSEES---SD
SEEETPA-AK--ALLQA-------------------------------------------
---------KASGKTSQVGAA------SAP--AKESPRKGAAPAPPGKTGPAVAKAQAGK
--REEDSQSSSEESDSE-E-----EA----PAQAKPSGKAPQVRAASAPAKE-SPRKGAA
PAPP----------RKTGPAAAQVQVGKQEEDSGSSSEE-SDSDREALAAM----NAV--
------------------------------------------------------------
---------QVKPLGKSPQVKPAST--------MGMGPLGKGAGPVPPGKVGPATPSAQV
GKWEEDSESS----SEE--SSDSSDGE---------------------------------
------------------------------------------------------------
-------VPTAVAPAQEKSLGNILQAKPASSPAKG---------PPQK-------AGPVA
VQVKAEKPMDNSESS--EESSD---S-AEA---PAAMTA---------------------
----------------------------------------AQAKPALKIPQTKACPKRTN
--TT--ASAKVAPVRVGTQAPRKAGTATSPAGSS-PAVAGGAQRPAEDSSSSEESDSEEE
-K-TGLAVTVG--------------------------------QVKSVGKG-----LQVK
AASVPVKGSLGQGTAPVLPGKTGPTVT-----------QV--KAEKQEDS-ESSEEESD-
SE--EAAASPAQVKTSVKKTQAKANPAAARAPSAKGTI-SAPGKVVT-------------
----------------AAAQAKQ-------RSPSKVKPP----------VRNPQNSTV-L
---ARGP--------ASVPSVGKAVATA-AQAQTGP-------EEDSGSSEEESDSEEE-
AETLAQAKPS--------------------------------------------------
--------------------------GKTPQIRAALAPAKES---PRK--GAAPTPPGKT
GPSA--AQAGKQ-DDSGSS-SEE-SDSDGEAPAAVTS-----------------------
------------------------------------------------------------
-----------------------------AQVI-KPPLIFV----DPNRSPAGPA---AT
PAQAQAASTPRKARASESTARSS--SSESEDEDMIPATQCLTP-G-IRTNVVTMPTAHPR
IAPKAS-MAGASSSKES---SRISDGKKQEGPATQVSKKNPASLPLTQAALKVLAQKASE
AQPPVARTQPSSGVDSAVGTLPATSPQSTSIQAKGTNKLRKPKLPEVQQAT---------
KAPESSDDSEDSS-DSSSGSEEDAE---GPQGAKSAHTL-GPTPSRTETLVEETAAESSE
DDVVAPSQSLLSGYMTPGLTPA---NSQASKATPKLDSSPSVS-STLAAKDDPD------
------GKQEAKPQQA--AGMLS------------------------------PKTGGKE
AASGTTPQKSRKPKKGAGN--------PQASTLALQSNITQCLLGQPWPLNEAQVQASVV
KVLTELLEQERKKVVDTTKESS------RKG--WESRKRKLSGDQPAARTPRSKKKKKLG
AGEGGEAS-VSPEKTSTTSKGKAKRDKASGDVKEKKGKGSL-GSQGAKDEPEEELQKGMG
KVEGGDQSNPK---SKKEKKKSDKRKKDKEKKEKK--KKAKKASTKDSESPSQ--KKKKK
KK-TAEQTV---------------------------------------------------
--
ENSOANP00000003653 (view gene)
-------------------------------------------QKTFPTQPVTLLDIYTH
WERTSEEAKKRKADELDENLQAKKVRVSDPVSSSESSEVEEE--YTAK--GMKTTRPLLA
KSVTLMPEKNLAVTKDRAKGKNKKVTGKAVNSISPPTPNKGPAPL--GK-------PGLA
TLAQ----------------------------------ALSPRSN--PVSKNQAADSSSS
--SSDS---------EAEG-AASVDRLHP
PF03546 // ENSGT00730000111382_0_Domain_0 (270, 703)
RP-VAGPVAR---KPESSSSGASGDE--TDI
EVIS---AFSSCS------VLPS-------------------------------------
----------------ASPP-------WCWR-GSCTGKPVTASPQAKAGTPVRIAGDGRP
EESSESSEESE--SEEEMPASQG--QVKAA---GRTPLVSSVPVSPVSVKGSQEKATAPA
L-------------RKP--------VTQPSQAKVGTPNVATKAGVESSESSE-----ESE
SEEEAPA-PE----GKV-------------------------------------------
---------SQARA-P------AGRGA----VTAPASTKGAAATHPQAQA---KKVGSR-
--KPEESSESSESSESEEAS--AS----KGSLQVKAAGRTPRALSVPVTPISFTGTHERV
----SASVA-TRPQAKAGTPIQASGAGRAEESS--ESSEESESEEEAPAVQGPVCSP---
------------------------------------------------------------
----------------------YA---------PRLQTSEPEASGPPAKIVTPTAKAVGA
GKPGESSESS--------EESESEEETPASQG------QVKAAGRTPR------------
---------AVSVPVTPISVTGTHERVSTSVATQLRTKAGTPVQAEKAEKSSESSEESES
EEEAPAVRG---------------KVGITPGSARKPQGRVSTPAAGNRVAATAPMAKVAT
PTTKASGGGKPGESSESSEESESE----EEA-----------------------------
------------------------------------PASQGQVKPAVTTSQVSVTRGSAK
KPQG--RASTPAAGKAMAATSLQAKVVTPT------AKAVGAGKPGESSESSEESESEEE
A----PASQGQ-----------------------------------ASGAGRAEKS----
--------------------------------------------------SESSE-ESES
EE--EAPTVRGQKFQGKASTPAAGKGVATK---------ST---RAKVVTSTDASGGGKP
GESSESSEESESEEETPASQGQVRLVETEV-------APLV------KPGR---------
SLSSLARV---T--TQPQAKAGTPIKAS-----G-----AGRAEKSS-ESSEESESEEEA
LAVQGPPQGR--------------------------------------------------
--------------------------AST--------------PAAGKAMAATPLQAKVV
TPTAKAAGGGKPGES---SESSEESESEEETPASQGQKRLKSI-----------------
--------------------------------------------------L----CVE--
----------ECCLHIPSGMMVERQRPPPTETV-NGSLAGSPAPRLPGSTPKSTP---TV
PAKA---------QPPE----------------TLP--RGPPKPE-VGKTAAANDPLGV-
--IVPASSATDDSS-------DGGSSEDDGKPTAVKPPVG--------------------
-TGRQHPLPPSLRR------EEKHLDTQACACSAETRV------V---------------
----------------------GPSA----A----SAW-LLLCPPCRPS--CARPRAAQV
PPFLDPLPRICSTVRWN-------------ERPGGHHLCTAVP-LQPSPL-----PPPKG
GGKGEGGTL-----PV--RGR---------------------------------------
-------------EDKGGCSSPVAARAGAESS------GDGELDSEEEALATSRAQASVV
KVLTDLLQQEKKKATKIIKAAKSPKVKEGKK-QKSSQKRKLAGEDGDTKATGNKKQKL-A
TEDSQEGE-AKAGK-GGRTK-KGKKEKGTGDAQEKRVRGAS-GTKKSKEKSGDDL--DPT
KVRSEDAGDTKTKKEKKEKKKSDKKDKTP----KDK-KKRKKSLSKDSDSSV--LKKKKK
KKNKYAQKTP--------------------------------------------------
--
ENSCGRP00001007261 (view gene)
MA-EARKRRELLPLIYHHLLQAGYVRAAR-------EVKEQSGQKSFLTQPTTLLDIYTH
WQQTSELGQKQKAKD-DASLQAKQSRVSDPISSSESSEEEE---EEAETKTAKAIPRPTS
VNSIATAADLPSSMKEKAQAKTKT-ANKMVNSVQQPASGKAVVHLLSGK-----------
------------------------------------------------SPKKSAEPLANT
VLASET---------EEEG-SVPALGA-----TAKPGVLSTGQASSSSEDTSSSSDETDV
EVKVSVKPAQ
PF03546 // ENSGT00730000111382_0_Domain_0 (311, 514)
AKV-----SPAPAKEPP---------------------------------
--------------ARTAPC-PTKS---GSGMPRLQEGTPAAP-----------GRTA-H
AEESESSEEDSES-EDEPPAGMPS-QVKAP---GKVLHGRAAS---VSAKE---------
---VSGKGPTSAPPEKT--------RPIVTPAKAGKP-------EKETESSSEDS---SD
SEDEEPV-AV-MTTPQV-------------------------------------------
---------RPSGKNPQVKST------STP--AKGSSQKGAPPVTSGKAGPAAAQAQAGK
PE---ESDSSSEGSESDSGKTPAA------------------------------------
--------------------------------------------------VTLSTSSA--
------------------------------------------------------------
---------QVKPLGKSQQVRPAST--------VSLGPSGKGARPPCPGKTGSAVLGVQM
GKVEEDSESS----SEDS----DDGAVT--------------------------------
------------------------------------------------------------
-------------PAKAKALGKR-----------------ATPALPQK-------AGPVA
AQVKIERRKEDSESS--EESSEESTDSEEEA-----------------------------
---------------------------------APAASAPAQAKPDLEK-QTKASPRKGT
PATP--ASAMVS-------SMRV---ST------------------APSSEEGSGA--AP
------GTTR--------------------------------VQGKSVGKG-----LQGK
G-------DLGQGMAPVHPERPGPTVA-----------HV--RITAQEDS-EDSEEESS-
SEE---DETSAQAKPLGKPLQAKAIPPPTKASPAKRPT-SASGKV---------------
-------------------------------SPSKGKPP----------MSTVQNSALS-
---AR------------VPATGKAGAPA-VQAQKGLVAGAG--QDSESSSEEESDSEEE-
--APAQTKPV--------------------------------------------------
--------------------------GKTSQVRAASAPAKES---PGK--GAHPGATKKT
GPTSAQAQTGKTE-GSESS--SEESDSDGEMPSAIIPA----------------------
------------------------------------------------------QGGASQ
PAKSRASAPAAPEKSIEDSSESSDEELPSSQVI-KPPLISV----NPNRSP---A---AT
LSQVQAVSNPRKAQASGTMAQSS--SSESEDEDMIPDTQPSTH-A-TKANVVTVPTSLPR
TAPQPS------KSEQS---SRMPKGKKPKAA----------------------------
----------STQISSDVKTLPVTPPQSTPVQSKTTNKPRKPKLPAKQQPPS---TYR--
SS---SS---DSS-DTSSGSEEDAK---RPQVATSAPRL-DPHLSQKETVVEETPDESGE
DELVAPSQSLLSGYVTPGATVA---NSQASKATSRPDSNPLVS-SAPVTKDNPD------
------GKQKAKSQD--AADTVF------------------------------PKTGRKE
TSSDTTPPKPKKAKKSTWT--------SATPTQALQNSITQCLREQSWSLSEAQVQASVV
KVLAELLEQERQKATEAIKESG------KKG-----KKRKLSGDQLAAGAPESKKK--AS
-------------GTS---KTKSKLDEVNADGK---GKGSS-GSQGAKAKPES--E-LGL
KVESGQQNDTK---NKKEKKKSSKKKKSKDKKGK---KKGKKTSAKDPDSQIK--KKKKK
KK-TAEPAV---------------------------------------------------
--
ENSCCAP00000031848 (view gene)
ENSTTRP00000012360 (view gene)
ENSSBOP00000020200 (view gene)
MA-EARKRRELLPLIYHHLLRAGYVRAAR-------EVKEQSGQKCFLAQPVTLLDIYTH
WQQTSELGRKRKAEE-DAALQAKKTRVSDPISTSESSEE-E---EEAEAETTKATPRPPS
TNSSVMGADLTSSMKEKAKAETEK-ASKTGNSMPHPATGKMVAHLLSGK-----------
------------------------------------------------SPRKSVEPSANT
KLVSET---------KEEV-SIPAFGA-----AAKPGMVSAGQADSSSEDTSSSSDETDV
EGKPSVKPAQVKA------SVSTKESP---------------------------------
-------------ARKAVLA-LGKV---GDVTPQVREGALPPS-----------RRAKKP
EEESESSEE-GSESEEEAPARTPS-QVKAS---EKITQVRTAS---ALPKGTPGKVA---
---------APAPPGKA--------GPTAPQAKAGKP-------EEDSESSSEES---SD
SEEETSA-AK--VLLQA-------------------------------------------
-----------SAKTPQVKAA------SAP--AKESPRKGAAPAPPGKMGPAAAKAQAVK
--REEDSESSSEESDSE-E-----EA----PAQAKPSGKTPQVRAALAPAKE-SPRKGAA
PTPPGKTGPAPTPPRKTGPAAAQAQAAKREEDSGSSSEE-SDSDEEAPAAM----STA--
------------------------------------------------------------
---------QVKPLGKNPQVRPAST--------VGTGPLGKGAGPVPPGKV---------
GKREEDSESS----SEE--SSDSDDGE---------------------------------
------------------------------------------------------------
-------VPRAVAPAQEKSLGKILQAKPASGPAK---GRGAAPVPPQE-------AGPVA
IQVKAEKPTEDSESS--EESSDSEKA-PAA------VTA---------------------
----------------------------------------AQAKPALKAPQTKTCPKKTN
--TT--ASAKAAPVQVGARAPGKAGTATS-AGSS-PAVAWGAQRPAEDSSSSEESDSEEE
-K-TGPTVTTG--------------------------------QAKSVGKG-----LQAK
AASLPVKGLLGQGTTPVLPGKTQPSVT-----------QV--KAEMQEDS-ESSEEESD-
SE--EADGTPAQVKTSVKKTQDKANPAAARASSAKGTI-SAPGKVVS-------------
----------------AAAQAKQ-------RSPAKVKPQ----------ARNPQNSAV-L
---VRGP--------ASVPPVGKAVA------Q
PF03546 // ENSGT00730000111382_0_Domain_2 (1534, 1810)
TEP-------EEDSGSSEEESDSEEET
TEMPAQMKPS--------------------------------------------------
--------------------------GKSPQVRAALVPAKES---PSK--GAGLAPPGKT
GPLT--AQAGRQKDDSGSS-SEE-SDSDGEAPAAVTS-----------------------
----------------------------------------------------AQKDGNSK
PARSKTLVPAPPKRNMEGSSESSEEELPLTQVI-KTPLIFV----DPNRSPAGPA---AT
PAQAQAASTPRKARASESTARSS--SSESEDEDMVSATQCLTP-A-IRTNAVTVPTTHPR
TAPKAS-TARASSNKEY---SRISDGKKQEGPATQVLKKNPASLPLTQAALKVLAQKASE
AQPPVARTQPSSGVDSAVGTLPATSPQSTPVQARGTNKLRKPKLPDGQQAA---------
KAPGSSDDSEDSS-DSSSGSEEDAA---GPQVAKSAHRLVGPNPSRMETLVEETTAESSE
DDVVAPSQSLLSGYVTPGLTLA---NSQASKATPKPDSNPSVS-STLATRDDPD------
------SKQEAELQQA--TDTLS------------------------------PKTGGKE
AVSGTKPQKSQKPKKGAGN--------PQTSTLALQSNIAQRLMGQPWPLSEAQVQASVM
KVLTELLEQEKKKAADVTKESS------RKG--WESRKRKLSGDQPAARN----KKKKLG
AGEGGEAS-VSPEKASMTSKGKAKRDKASDGIKEKKGKGSS-GSQGAKDKPEEELQKGMG
KVEGGDQSNPK---SKKEKKKSDKRKKDKEKKEKK--KKAKKASTKDPESLSQ--KKKKK
KKKTPEQTV---------------------------------------------------
--
ENSTSYP00000014474 (view gene)
ENSMLUP00000008094 (view gene)
ENSMODP00000031401 (view gene)
------------------------------------------------------------
------------------------------------------------------------
-----------------------MEMEQAL-----------RTPQ--GN-----------
-----------------------------------------------------SALVRGN
SVASIP---------TKVS-QEKVVTPSPGRLVTKPLTTKDKNPEETSESSDESESDEDA
PQSQGQAQEVKMP------HHLS-------------------------------------
----------------EVPG-------RVGKTTPTTGKAVTPSSKPNLGVPAKAAMTQKP
EETSESSEESE--SEEENAKLQR--------------QVKSVPASGTIA--SPGKGALPV
P-------------GKS--------VTSNLQAKAPQHE----SPETSSESSE-----ESE
SDEDASA-PQ----GQE-------------------------------------------
---------KSSMKTPQASATPATRTPTPSTLTKVSPRKGTLQAKTGPPS---KTSEVN-
--LPETGSDTSEESESEDNV--IV-----PQRQGKSSGKASQGNATPSKGRSLAPGKPMT
LVPPPRMGP-STPSPRAGTSAMASKPGSSETSS--ESSEESESEEEILVPQEKKKSPVKV
LQVNAIVTKVAPVPPQGPLGKEALSTTGKSGTLASGVEISSSTETSGSESSEGSDESESE
EEKTPAPKTQVKPSEKAQAQVSPAPLPKSPLEKGVLNSPEKAAMVTPSAKMAPQAKGVGP
GRSQASSGCS--------GESDSEGETPTSQG------QAKPVVKALQINA---------
-------SPRKLTPLTPISTKGIQGRVST-------------------------------
------------------------------------------PAPWKSG--------ILP
LQAKAPGPERPAETSGSSEESES-----EEI-----------------------------
------------------------------------PASQGQAKPTLKAPQIDTPPGKGT
PVTP--VPTKCSQGRVSTPAPWKSGTVTPQ------PKVPGPGR
PF03546 // ENSGT00730000111382_0_Domain_0 (1245, 1467)
PAETSGSSDESESEEE
I----PAIQAK-----------------------------------PAVKIPQANTPSRK
GVSTSTVSTQDSQGKAPSPLKSGTSSLQSKGEV----PVKKAWLGRQEENLESSE-DSES
DD--SILGPQGQAKPAVKTSPGLGGEKGTT--------ASTPEKSATTIQTT-KVDSGSS
EESSCSSDESESDVVIPVSQKQVKPSVIDKGADKAVMVPVTMVEETSSESSEDTESEETE
ILTSMGQVKTVMRTPQVSAASGQSAS-S-----SMEPERPTRPEECSESSEEESDSEDEI
IVCQGQVKPT--------------------------------------------------
--------------------------VKTSEIPAASAEKKGSIPASGK--SATPSPQNQA
TPQAKDTRTEISEES---SDRSDESESEVEIPVSQQQVSSRKKDEKAV------A-----
---------------------------------------SKNNMGKVSPISPVKQLAS--
----------PCTIRGQKSESNSEQLS-PAQVI-KTPLIFV----DPNRSPAGQA---AI
PMKA--KTDSRNRQVSESREQSS---SESGDEDVIPDTQYPNLPT-VKNAVMKASPLYQR
AGSTPSTSKPEESSS-------DSASEHQGTGTS--------------------------
--------------------------QTTCVKSKTARRDKKSKLTGGIQAIAVQDKSSTT
TS------------NSTRNEKENPEA----VRAKQTQN-LGLSPKDKIPVKTENPEDSSE
EEIIEPSQSILSGYEFPRPSLPTSINAQLEKMPAKQGITRVVS-ADPALQASSKGRPQEG
SGDSEKEKQKVEQPHE--TGRDS---------------------KTSVSKLQATKKGSK-
--PEAVDYKPSSPKEVDICESLLRTKPPSNLPSLVQLQSSGCIENLGFEDAFFKSKAFRL
RMLTDLPEKKKKNT-------ESPKLAKG---KKSKSKRKLTEEGPVAKAPKNKKKKL-M
GEEPKEGE-ALQEKTPASPK-EEKREKASGDAKEKKPKEVQ-SSKKKKEKPEGNV--IS-
VKEYGDAPDTKPKKEKKHKKKSDKKKKDKDKKDEKK-KKKKKSISKDPDSASVSLKKEKK
KKKKVKQVDG--EELNNRIFLTTS------------------------------------
--
ENSOGAP00000004130 (view gene)
ENSOARP00000007663 (view gene)
MA-EARKRRELLPLIYQHLLQAGYVRAAR-------EVKEQSGQKIFLSQPVTLLDIYTH
WQQTSEIGRKRKAEE-DAALQAKRTRVSDPISSSESSEEEEEEEEEAEAKTTKPTPRLAA
TNSSVTGTVSSSNVKEKAKAKTMK-ASKTVNSTTHPAPGKAVAHLLTGK-----------
------------------------------------------------SPQKTTGPSANA
VLECGC---------WLAL-S---PLL-----VYPSGMASADQADSSSEDTTSSSDETDL
EVKAAVKPLQVRA-----SATAVKESP---------------------------------
-------------RKSAAPT-PGKA---GVVTPQ
PF03546 // ENSGT00730000111382_0_Domain_0 (395, 507)
VKGGAVTPA-----------SRAKKP
EEVSESSEE-SDESEEEAPSPEPA-QVKAL---EKTTPIRVAP---SPAKGTPGKGA---
---------TPAAPGKA--------GPL------GKP-------EEDSESSS-EE---SD
SEEETQA-VK--TPVQA-------------------------------------------
---------KPSGKIPQVKAA------SAAPTQVLSPKKGAPPAPAGKAGPAATQAQPRK
QEKEESSSSSSEESDSE-GEAPTATR----Q-----------------------------
-----------------------------------------------------TTSLA--
------------------------------------------------------------
---------QAKPLEKNSQVRAASA--------TGAGPSAKGAVPAPPQKARSVAA--QD
GKQE-DSESS----SEE--ESDSEEEMQRAGTS----AQVSPLPVRF-------------
---ISPAQCRKPLPF---------------------------------------------
-PHLHSLDR-VTLSISAQSSGKALQVRPSSGPTKG---------PPQK-------AGPAT
TQVKVERAEEDSESS--EEESDSE----DEA--PVAKTP---------------------
----------------------------------------AQAKSTVKTSQIKASPRKGT
PITP--TSARVPPVQVGTPSPRTAATVSTPTCASSPAIVRHTQKPGEDSSSSEESESEEE
T---APAATGG--------------------------------QMKSVGKS-----LPVK
AASASTKGPSGKGTTPGPPGKAGPPVA-----------QV--KAEVQEDS-ESSE-ESDS
E---EAAAAPAQVKTSVKTPQTKANPSATRVASAKAAA-PAPGKGGL-------------
----------------AVAQAKV-------GSPAKVTPA----------ARAPQSSAVS-
---GSSQ--------VSMPAVGKAAAAA-AQAQPGPVGGSQ--E-DSESSEEESDSEGE-
--APPQAKPS--------------------------------------------------
--------------------------GKTPQVKTASAPTKAC---PKK--GAAPVPPGKA
GPPA--AQARRQEEDSESS-SEEESDSDGDVPA---------------------------
------------------------------------------------AAPPAQKEGSSK
TATSKTLTPASLEKKAEESSTSSDEDLPSNQVI-KTPLIFV----DPNRSPASPA---AT
PTQAQATGTPRKAQALESTARSS---SESEDEDVIPATQCPTP-A-TRTSVVTVPTAPPK
AALRAS-VVGAPS---SEDTSQGSEGKKQEASAT---QVTKRNLPLTQAALKVLAQKANE
AQPPAAKTPSSSGTDSAVGTLSTVSPQSTPTQVRMANKLRKPEVPAAQRATATPSGHPTP
KAPAAS---DDSN-SSSSGSEDEAK---GSQAAPSAHRP-GSAPSRRETLVEETTTESSD
DEVVAPSQSLLSGFVTPGLTPA---GSKTSKATPKPDTSPSVS-STPATRDAAE------
------GNQEVEPQQA--AGTMS------------------------------PKTGRK-
-EAGATPQKPG---KGPGS--------TPASMLALQSNITQRLLSEPWPLSEAQVQASVA
KVLTELLEQERKKAVETAKEGN------RKG--RLGHKRKLSEDQTAPKAPKSKKKKQLA
AADSGEHV-VSSEKAPRTVKGKSKKDRASGDAKEKKEKESP-GSQGAKEKPVGEL--GTP
KSDGGDHGNLK---IKKEKKKSDKKKKDKEKKEKK--KKTKKASTKDPGSPSQ--KKKKR
KKKTAEQTV---------------------------------------------------
--
MGP_CAROLIEiJ_P0046834 (view gene)
ENSRBIP00000018158 (view gene)
MA-EARKRRELLPLIYHHLLRAGYVRAAR-------EVKEQSGQKCFLAQPVTLLDIYTH
WQQTSELGRKRKAEE-DAALQAKKTRVSDPISTSESSEE-E---EEAEAETAKATPRLAS
TNSSVLGADLPSSLKEKAKAETEK-ASKTGNSMPHPATGKTVTNLLSGK-----------
------------------------------------------------SPRKSAEPSANT
TLVSET---------EEEG-SVPAFGA-----AAKPGMVSAGQADSSSEDTSSSSDETDV
EGKPSVKPAQVKA-----SSVSTKESP---------------------------------
-------------ARKAAPA-PGKV---GDVTPQVRGGALPPA-----------KRAKKP
EEESESSEE-GSESEEEAPAGTPS-QVKAS---EKNLQVRAAL---TPAKGTPGKGA---
---------TPAPPGKA--------GAVAFQTKAGKP-------EEDSESSSEES---SD
SEEETPA-AK--ALLQA-------------------------------------------
---------KASGKTPQVGAA------SAP--AKESPRKGAAPAPPGKTGPAVAKAQTGK
--QEEDSQSSSEESDSE-E-----EA----PAQTKPSGKTSQVRAASASAKE-SPRKGAA
PAPP----------RKTGPAATQAQAGKQEDDSGSSSEE-SDSDREAPAAM----NAA--
------------------------------------------------------------
---------QVKPLGKNPRVKPAST--------MGTGPLGKGAGPVLPGKVGPTTPSAQV
GKWEDS-DSS----SEE--SSDSDDGE---------------------------------
------------------------------------------------------------
-------VPTAVAPAQEKSLGKVLQAKPASGPAKG---------PPQK-------AGPVA
IQVKAEKPMEDSESS--KESSDSVDS-EEA---PAAVTA---------------------
----------------------------------------AQAKPALKIPQTKACPKKTN
--T---ASAKVTPVRVGTQAPRKAGTAPSPVGSS-PAVAGGTQRPAEDSSSSEESDSEEE
-K-TGPAVTMG--------------------------------QAKSVGKG-----LRVK
AASVPVKGSLGQGTAPVLPGKTGPTVT-----------QV--KAEMQEDS-ESSEEESD-
SE--EAAATSAQVKTSVKKTQAKANPAAVRASPAKGTI-SAPGKVVT-------------
----------------AAAQAKQ-------RSPAKAKPP----------VRNLQNSTV-L
---VKGP--------ASVPPVGKAVAAA-AQVQTGP-------
PF03546 // ENSGT00730000111382_0_Domain_2 (1544, 1831)
EEDSGSSEEESDSEEE-
TEMPAQAKPS--------------------------------------------------
--------------------------GKTPQVRAALAPAKES---PRK--GAAPAPPGKT
GPSA--TQEGKQ-DDSGSS-SEE-SNSDGEAPAAATS-----------------------
------------------------------------------------------------
-----------------------------AQVI-K-PLIFV----DPNRSPAGPA---AT
PVQAQAASTPRKSRASESTARSS--SSESEDEDVIPATQCLTP-G-IRTNVVTVPTAHPR
IAPKAS-MTGASSSKES---SRISDGKKQEGPATQVSKKNPASLPLTQAALKVLAQKASE
AQPPVARTQPSSGADSAVGTLPATSPQSTPVQAKGTNKLRKPKLPEGQQAT---------
KAPGSSDDSEDSS-DSSSGSEEDAE---GPQVAKSAHTLG-PTPSRTETLVEETAAESSE
DDVVAPSQSLLSGYVTPGLTPA---NSQASKATPKPYSSPSVS-STLATKDDPD------
------GKQKAKPQQA--AGMLS------------------------------PKTGGKE
AASGTTPQRSRKPKKGAGN--------PQASTLALQSNITQCLLGQPWPLNEAQVQASVV
KVLTELLEQERKKVVDATKESS------RKG--WESRKRKLSGDQPAARTPRSKKKKKLG
AREGGEAS-VSPEKTSTTSKGKAKRDKASDDVKEKKGKGSV-GSQGAKDEPEKELQKGMG
KVEGGDQSNPK---SKKEKKKSDKRKKDKEKKEKK---KAKKSSTKDSESPSQ--KKKKK
KKKTAEQTV---------------------------------------------------
--
ENSCLAP00000015410 (view gene)
MA-EARKRRELLPLIYQHLLHAGYVRAAR-------EVKEQSGQKNLLAQPFTLLDIYTH
WQQTSELGQKRKAED-DAALQAKKTRVSDPLSSSESSDEEEE---EAEAKTAKATPRPAS
TNSSAIGVDLPSSMKEKAKAKTKS-ASKTVNSLPHLASGKAVVHLLSGK-----------
------------------------------------------------SPKKSAEPSGSI
IVVSET---------EEEG-SIQALAA-----TAKPGTVSAGQADS--KDTSSSSDETDV
EVKSSVK-AQVRP-----PPAPTKES----------------------------------
-------------SSKVAPR-PGRV---GGVTPQARGGALTPA-----------GSAMKP
KEDSESSKE--SESEEEPSVGTPS-QVKAS---EKSLQVRGAS---APAKGTPGKGVTPA
---PPRKGTTCALSGKA--------GPTATQAKAGRP-------EQDSESSSKE------
------------------------------------------------------------
------------------------------------------------------------
------------SSDSE-GETP--------------------------------------
----------------------------------------------------AAVTAL--
------------------------------------------------------------
---------QVKPSGKTPQLKASSG--------PAKESPRKGVPPAPPGKTGPTATPPQA
GKEEEEEDSS--------------------------------------------------
----------------------------------------------------SSCEESDS
DGETPASRLPITNPVQADALGKNLQVRPASGPTQGPLGKGAAPKPPQK-------SAPVA
IQVKAEGSQEDSDSS--DESSDSE----EEA--PAAVAP---------------------
----------------------------------------AQAKPALKTPQTKGSSRKGT
LLTP--TSAKGPPVQVGTPAPWKAGTVTSA---IAPAMPQGTQSPESDSSSSEEESEGEE
A---TPAAAVA--------------------------------QTKSLGKG-----LQQQ
AASATTKGPSGQGATPVLPGKTRPTIV-----------QV--RTEAHEES-ESE-EDCD-
---DSEAATPPQAKASGKTPQAKANLASTKASTVKGAA-SPCGKLVT-------------
----------------PVAQGKP-------GPMAKGKSP----------VRAPQNSTAS-
---ARGQ--------ASVPAVEKAATAA-AQA-----------KEDSESSDESSESKEE-
--VPTQVKPSGKTPQPRAASGPAKESPRKGIPPAPSGKTGPAATPPQVGKQEEDSD-SSS
SEESGSDGEVPALRTPTMNPVQRKPPGKTPQLRAASTPAKES---SRK--GAPPAPP
PF03546 // ENSGT00730000111382_0_Domain_2 (1678, 1800)
GRT
GLAATPAQAGKQEEDSDSS-SSEESDSDGETPAPQTPNMNPV-QKPSGKTPQLRAASAPA
KESPRKGAPPAPPGKIGHAAIQAQVGKQAEDSGSSSSEELDSDGETPASGTPAQKNIASK
TARSSALTPAPPEKNPEGSSESSDEELPSSQVI-KPPVIFV----DPNRSPAGPA---VT
PTQAQATSTPRNAQASSSTAGSS--SSESEDEDVIPATQSSTPAGR---AHVPVPAASTR
TAPRAS------TSEES---SQVSEGKKQA------------------------A-----
----------VSQAGSATGMLPVTRPQSTPVQAKAANTLRQPKLSEGQQNTATPSARPKA
KAPASSD-SEDSS-DNSSGSEEDAE---RPQ--MSAPRLAGPPLSKRETLVEETTAESSE
DEVVAPSQSLLSGYVTPGLTTA---KNRALKTTARRESNPEAS-SAPPTKGDPA------
------SEQATESEHT--VGTVS------------------------------LKPGRKE
AVSDPTPKKPRKPKKKAAS--------TEASLQVLQNSITQHLLGQPWPLNEAQVQASVA
KVLSELLQA-----TDTARESS------RKG-----RKRKLLGDQVASKAPKGKKKKQLA
AVEGREGA-ASSEKAARTSKGKSRGDKASGDTK---EKGSP-GSGGAKARPEGEL----L
KVEGGDQNDPK---SKKEKKKSNKKKKDKEKKEK---KKAKKASAKDCDSPFQ--KKKKK
KKIAEPPTTV--------------------------------------------------
--
ENSNLEP00000012588 (view gene)
MA-EARKRRELLPLIYHHLLRAGYVRAAR-------EVKEQSGQKCFLAQPVTLLDIYTH
WQQTSELGRKRKAEE-DAALQAKKTRVSDPISTSESSEE-E---EEAEAETAKATPRLAS
TNSSVLGADLPSSMKEKAKAGTEK-AGKTGNSMPPQANGKTVAHLLSGK-----------
------------------------------------------------SPTKSAEPSANN
TLVSSL---------SP--------LR-----VRSSGMVSAGQADSSSEDTSSSSDETDV
EGKPSVKPAQVKA-----SSVSTKESP---------------------------------
-------------ARKAAPA-PGKM---GDVTPQ
PF03546 // ENSGT00730000111382_0_Domain_0 (395, 515)
VKGGALPPA-----------KRAKKP
EEESESSEE-GSESEEEAPAGTPS-QVKAS---EKILQVRAAS---APAKGTPGKGA---
---------TPAPPGKA-----------------GKP-------EEDSESSSEES---SD
SEEETPA-AK--ALLQA-------------------------------------------
---------KASGKTSQVGAA------STP--AKESARKGAAPAPPGKTGPAVAKAQAGK
--REEDSQSSSEESDSE-E-----AP----PAQAKPSGKAPQVRAASAPAKE-SPRKGAA
PAPP----------RKTGPAAAQAQAGKQEEDSGSSSEE-SDSDREAPAAM----NAT--
------------------------------------------------------------
---------QVRLEAALHGLCPFLN--------VC-------------------------
--------------------THYPGLP---------------------------------
------------------------------------------------------------
-------LPILCISFQEKSLGKILQAKPASSSAKG---------PPQK-------AGPVA
IQVKAEKPMEDSESS--EESSDSADS-EEA---PAAMTA---------------------
----------------------------------------PQAKPALKIPQTKACPKKTN
--TT--ASAKVAPVRVGTQAPRKAGAATSPAGSC-PAVAGGTQRPAEDSSNSEESDSEEE
-K-TGLAVTMG--------------------------------QAKSVGKG-----LQVK
AASVPVKGSLGQGTAPVLPGKTGPTVT-----------QV--KAEMQEDS-ESSEEESD-
SE--EAAASPAQVKTSVKKTQAKANPAAARASSAKGTI-SAPGKVVT-------------
----------------AAAQAKQ-------RSPSKVKPP----------VRNPQNSTV-L
---ARGP--------ASVPPVGKAVAAA-AQAQTRP-------EEDSGSSEEESDSEEE-
AETPAQAKPS--------------------------------------------------
--------------------------RKTPQIRAALAPAKQS---PRK--GAAPTPPGKT
GPSA--AQAGKQ-DDSGSS-SEE-SDSDGEAPAAVTS-----------------------
------------------------------------------------------------
-----------------------------AQVI-KPPLIFV----DPNRSPAGPA---AT
PAQAQAASTPRKARASESTARSS--SSESEDEDVIPATQCLTP-G-IRTNVVTMPTAHPR
IAPKAS-MAGASSSKES---SQISDGKKQEGPATQVSKKNPASLPLTQAALKVLAQKASE
AQPPVARTQPSSGVDSAVGTLPATSPQSTSVRAKGTNKLRKPKLPEVQQAT---------
KAPGSSDDSEDSS-DSSSGSEEDAE---GPQGAKSAHTL-GPTPSRTETLVEETAAESSE
DDVVAPSQSLLSGYMTPGITPA---NSQASKATPKLDSSPSVS-STLAAKDDPD------
------GKQEAKPQQA--AGMLS------------------------------PKTGGKE
AASGTTPQKSRKPKKGAGN--------PQASTLALQSNIAQCLLGQPWPLNEAQVQASVV
KVLTELLEQERKKVVDTTKESC------RKG--WESRKRKLSGDQPAARTPRSKKKKKLG
AGEGGEAS-VSPEKTSMTSKGKAKRDKASGDVKEKKGKGSL-GSQGAKDEPEEELQKGMG
KVEGGDQSNPK---GKKEKKKSDKRKKDKEKKEKK--KKAKKASTKDSESPSQ--KKKKK
KVESSWGVSGQKTDPNPRPLLHTQTWFLPPFPFSGPSGGRQEL-----------------
--
ENSCSAP00000007758 (view gene)
MA-EARKRRELLPLIYHHLLRAGYVRAAR-------EVKEQSGQKCFLAQPVTLLDIYTH
WQQTSELGRKRKAEE-DAALQAKKTRVSDPISTSESSEE-E---EEAEAENAKATPRLAS
TNSSVLGADLPSSLKEKAKAETEK-AGKTGNSMPHPATGKTVTNLLSGK-----------
------------------------------------------------SPRKSAEPSANT
TLVSET---------EEEG-SVPAFGA-----AAKPGMVSAGQADSSSEDTSSSRDETDV
EGKPSVKPAQVKA-----SSVSTKESP---------------------------------
-------------ARKVAPA-PGKV---GDVTPQV
PF03546 // ENSGT00730000111382_0_Domain_0 (396, 510)
RGGALPTA-----------KRAKKP
EEESESSEE-GSESEEEAPIGTPS-QVKAS---EKILQVRAAS---TPAKGTPGKGA---
---------TAAPPGKA--------GAVAFQTKAGKS-------EEDSESSSEES---SD
SEEETPA-AK--ALLQA-------------------------------------------
---------KASGKTPQVGAA------SAP--AKESTRKGAAPAPPGKTGPAVAKAQTGK
--QEEDSQSSSEDSDSE-E-----EA----PAQTKPSRKTSQVRAAPASAKE-SPRKGAA
SAPP----------RKTGPAATQAQAGKQEEDSGSSSEE-SDSDREAPAAM----NAA--
------------------------------------------------------------
---------QVKPLGKNPQVKPAST--------MGTGPLGKGAGPVLPGKVAPTTPSAQV
GKWEDS-DSS----SEE--SSDSDDGE---------------------------------
------------------------------------------------------------
-------VPTAVAPAQEKSLGKVLQAKPASGPAKG---------PPQK-------AGPVA
IQVKAEKPMEDSESS--EESSDSADS-EET---PAAMTT---------------------
----------------------------------------AQAKPALKIPQTKACAKKTN
--T---ASAKVTPVRVGTQAPRKSGTATSPVGSS-PAVAGGTQRPAEDSSSSEESDSEEE
-K-TGPAVTMG--------------------------------QAKSVGKG-----LQVK
AASVPVKGSLGQGTTPVLPGKTGPTVT-----------QV--KAEMQEDS-ESSEEESD-
SE--EAAAPSAQVKTSVKKTQAKANPAAVRASPAKGTI-SAPGKVVT-------------
----------------AAAQAKQ-------RSPAKAKPP----------VRNLQNSTV-L
---VRGP--------ASVPPVGKAVAAA-AQVQTGP-------EEDSGSSEEESDSEEE-
TETPAQAKPS--------------------------------------------------
--------------------------GKTPQVRAALAPAKES---PRK--GAAPAPPGKT
GPLA--TQVGKQ-DDSGSS-SEE-SNSDGEAPAAATS-----------------------
----------------------------------------------------AQKDSNSK
PARSKTLAPAPPKGNTEGSSESSEEELPLTQVI-KPPLIFV----DPNRSPAGPA---AT
PVQAQAASTPRKSRASESTARSS--SSESEDEDVIPATQCLTP-G-IRTNVVTVPTAQPR
IAPKAS-MTGASSSKES---SQISDGKKQEGPATQVSKKNPASLPLTQAALKVLTQKASE
AQPPVARTQPSRGADSAVGTLPATSPQSTPVQAKGTNKLRKPKLPEGQQAT---------
KAPGSSDDSEDSS-HSSSGSEEDAE---GPQVAKSAHTLGGPTPSRTETLVEETAAESSE
DDVVAPSQSLLSGYVTPGLTPA---NSQASKAIPKPYSSPSVS-STLAAKDDPD------
------GKQEAKPQQA--AGMLS------------------------------PKTGGKE
AASGTTPQKSRKPKKGAGN--------PQASTLALQSNITQCLLGQPWPLNEAQVQASVV
KVLTELLEQERKKVVDATKESS------RKG--WESRKRKLSGDQPAARTPRSKKKKKLG
AGEGGEAS-ISPEKTSTTSKGKAKRDKASDDVKEKKGKGSL-GSQGAKDEPEKELQKGMG
KVEGGDQSNPK---SKKEKKKSDKRKKDKEKKEKK--KKARKSSTKDSESPSQ--KKKKK
KKKTAEQTV---------------------------------------------------
--
MGP_PahariEiJ_P0039419 (view gene)
MA-EARKRRELLPLIYYHLLQAGYVRAAR-------EVKEQSGQKSFLAQPITLLDIYTH
WQQTSELGQKQKAEN-DETLQAKKSRVSDPVSSSESSEEEEE--E----EEAKATPRPTP
VNSAA--AALPSKVKEK--AKTKT-ASKTVNSAPSPGSGKTVVHLLSGK-----------
------------------------------------------------SAKKPAEPLANT
VLASET---------EEEG-SAQALGP-----TAKSGTVSAGQASSSSEDTSISSDETDV
EVRSSAKPPQAKA-----LATPAKNPP---------------------------------
--------------ARTAPG-PTKL---GNVVPT
PF03546 // ENSGT00730000111382_0_Domain_0 (395, 516)
PAKPARAAAA-AGA-AATA--AAA-A
AEASESSEEDSDS-EDEAPAGLPS-QVKAS---GKGPHVRTAS---VTAKG---------
---ISGKGPISATPGKT--------GPIATQAKAGRP-------EKDMETSSEDD---SD
SEDEMPV-A--VTTPQA-------------------------------------------
---------RPSRNSPRARGT------SAP--AKESSQKGAPAVTPGKAKPVAA--QAGK
PE---AK--SSEESESDSGETPAA------------------------------------
--------------------------------------------------ATLTTSPA--
------------------------------------------------------------
---------KVKPLGKSSQVRPVST--------VTPGSSRKGANPPCPGKAGSAALKVQV
GKKEEDSWSS----SEESD---NDGSVS--------------------------------
------------------------------------------------------------
-------------PAKAKPPGKM-----------------ASPVL-QK-------AGPVA
TQVKTDGGKDHSESS--EEW----TDSEEEA-----------------------------
----------------------------------APAASAAQAKPALGK-QMKASLRKGT
SASA--TGASTS-------SHCKAGTVTSSANLSSLALAKGTQKPHVYSSSDSE-S--GA
A----PSTPT--------------------------------VQGKSGGKG-----LQGK
A-------ALGQGMAPVHTQKTGPS--------------V--KAMAQEDS-ESSE-DSS-
SEE--EDETPAQATPLGKLPQIKANPPPTKTPPA-----SASGKA---------------
----------------VA-------------TPTKGKPP----------ARTVPNSTVS-
---ARGQ--------QSVPAVGKTGAPA-TQAQKGPVAGAG--EDSESSSEEESDSEEE-
--TPPQIKPA--------------------------------------------------
--------------------------GKTSQVRAASAPAKEP---TKK--GAHPVTSGKT
GSSATQAQTGKAE-DSDSS--SEESDSDAEMPSAAIPA----------------------
------------------------------------------------------QDDISQ
PARGKASGPAPPEKSIEGSSESSDEDLPSSQAI-KSPPVSV----NRNRGPAVPT---PT
PEGVQAVNTTKK-A-LGATAQSS---SESEDEDMIPATQPSTH-A-ITTSG-MTPAALSQ
AASQPS------KSEQS---SRTPKGKKAKAAA---------------------------
----------PTQTSSAMETLPVTPPQSAPVQPKATNKLGKPKLPEKQQLAP---GYP--
KAPGSSE---DSS-DTSSEGEEDAK---RPQMSKSAHRL-DPDPSQKETVVEETPTESSE
DEMVAPSQSLLSGYVTPSLTVA---NSQPSKANPRPDSDPLVS-SAPATKDNPD------
------GKQKSESQHS--AGTAF------------------------------PKTGRKD
ATSGLTPQKPKKPKKNTSS--------SPAPAQTLPNSITQRLLEQAWPLSEAQVQASVV
KVLTELLEQERQKATEAIKESG------KKG-----QKRKLSGD-LEAGVPKNKKKQQQP
V--PGASP-VFPEKVPTSSKAKSKLDKGSAGGK---GKGSP-GPQGAKEKPD-GEL-LGI
KLESGEQSDPK---SKKEKKKSGKKKKDKEKKEK---KKGKKTPTKDPASPVQ--KKKKK
KKKTAEPAV---------------------------------------------------
--
ENSPPYP00000017847 (view gene)
MA-EARKRRELLPLIYHHLLRAGYVRAAR-------EVKEQSGQKCFLAQPVTLLDIYTH
WQQTSELGRKRKAEE-DAALQAKKTRVSDPISTSESSEE-E---EEAEAETAKATPRLAP
TNSSVLGADLPSSMKEKAKAETEK-AGKTGNSVPHPATGKTVANLLSGK-----------
------------------------------------------------SPRKSAEPSANT
TLVSET---------EEEG-SVPAFGA-----AAKPGMVSAGQADSSSEDTSSSSDETDV
EGKPSVKPAQVKA-----SSVSTKESP---------------------------------
-------------ARKAAPA-PGKV---GNVTLQVKGGALPPA-----------KRAKKP
EEESESSEE-GSESEEEAPAGTPS-QVKAS---EKILQVRAAS---APAKGTPGKGA---
---------TPAPPGKA--------GAVASQTKAGKP-------EEDSESSSEES---SD
SEEETPA-AK--ALLQA-------------------------------------------
---------KASGKTSQVGAA------SAP--AKESPRKGAAPAPPGKTGPAVAKAQAGK
--REEDLQSSSEDSDSE-EE----AA----PAQAKPSGKASQVRAASALAKE-SPRKGAA
PAPP----------RKTGPAAAQVQAGKQEEDSGSSSEE-SDSDREAPAAM----NAA--
------------------------------------------------------------
---------QVKPLGKSPQVKSAST--------MGMGPLGKGAAPVPPGKVGPTTPSAQV
GKWEEESESS----SEE--SSDSSDGE---------------------------------
------------------------------------------------------------
-------VPTAVAPAQEKSLGKILQAKPASSPAKG---------PPQK-------AGPVA
IQVKAEKPMEDSESS--EESSDSADS-EEA---PAAMTA---------------------
----------------------------------------AQAKPALKIPQTKACPKKTN
--TT--ASAKVAPVRVGTQAPRKAGTVTSPAGSS-PAVAGGTQRPAEDSSSSEESDSEEE
-K-TGLAVTVG--------------------------------QAKSVGKG-----LQVK
AASVPVKGSLGQGTAPVLPGKTGPTVT-----------QV--KAEMQEDS-ESSEEESD-
SE--EAAASPAQVKTSVKKTQAKANPAAARASSAKGTI-SAPGKVVT-------------
----------------AAAQAKQ-------RSPSKVKPP----------VRNPQNSTV-L
---ARGP--------TCVPPVGKAVATA-VQAQTGP-------EEDSGSSEEESDSEEE-
AETPAQAKPS--------------------------------------------------
--------------------------GKTPQIRAALP
PF03546 // ENSGT00730000111382_0_Domain_2 (1658, 1830)
PAKES---PRK--GAAPTPPGKT
GPSA--AQAGKQ-DDSGSS-SEE-SDSDGEAPAAVTS-----------------------
------------------------------------------------------------
-----------------------------AQVI-KPPLIFV----DPNRSPAGPA---AT
PAQAQAASNPRKARASESTARSS--SSESEDEDVIPATQCLTP-D-SRTNVVTMPTAHPR
IAPKAS-MAGASSSKES---SRISDGKKQEGPATQVSKKNPASLPLTQAALKVLAQKASE
AQPPVAKTQPSSGVDSAVGTLPATSPQSTSVQAKGTNKLRKPKLPEVQQAT---------
KAPGSSDDSEDSS-DSSSGSEEDAE---GPQGAKSAHTL-GPTPSRTETLVEETAAESSE
DDVVAPSQSLLSGYMTPGLTPA---NSQASKATPKLDSSPSVS-STLAAKDDPD------
------GKQEAKPQQA--AGMLS------------------------------PKTGGKE
AAAGTTPQKSRKPKKGAGN--------PQASTLALQSNIAQCLLGQPWPLNEAQVQASVV
KVLTELLEQERKKVVDATKESS------RKG--WESRKRKLSGDQPAARTPRSKKKKKLG
AGEGGEAS-VSPEKTSTTSKGKAKREKATGDVKEKKGKGSL-GSQGAKDEPEEELQKGMG
KVEGGDQSNPK---SKKEKKKSDKRKKDKEKKEKK--KKAKKASTKDSESPSQ--KKKKK
KKKTAEQTV---------------------------------------------------
--
ENSCHIP00000003682 (view gene)
MA-EARKRRELLPLIYQHLLQAGYVRAAR-------EVKEQSGQKIFLSQPVTLLDIYTH
WQQTSEIGRKRKAEE-DAALQAKRTRVSDPISSSESSEEEE-EEEEAEAKTTKPTPRLAA
TNSSVTGTVSSSNVKEKAKAKTTK-ASKTVNSTTNPAPGKAVAHLLTGK-----------
------------------------------------------------SPQKTTGPSANA
VLVSET---------EEEG-SVSALRT-----AATPGMASADQADSSSEDTTSSSDETDL
EVKAAVKPLQVRA-----SAAAVKESP---------------------------------
-------------RKSAAPT-PGKA---GVVTPQVKGGAVTPA-----------SRAKKP
EEVSESSEE-SDESEEEAP-------VKAL---EKTTPIRVAP---SPAKGTPGKGA---
---------TPAAPGKA--------GPL------GKP-------EEDSESSS-EE---SD
SEEETQA-VK--TPVQA-------------------------------------------
---------KPSGKIPQVKAA------SAAPTQVLSPKKGAPPAPAGKAGPAATQAQPRK
QEKEESS-SSSEESDSE-GEAPTATR----Q-----------------------------
-----------------------------------------------------TTSLA--
------------------------------------------------------------
---------QAKPLEKNSQVRAASA--------TGAGPSVKGTVPAPPQKARSVAA--QD
GKQE-DSESS----SEE--ESDSEEEMQRAGTS----AQ---------------------
------------------------------------------------------------
----------------AQSSGKALQVRPSSGPTKG---------PPQK-------AGPAT
TQVKVERAEEDSESS--EEESDSE----DEA--PVAKTP---------------------
----------------------------------------AQAKSAVKTSQIKASPRKGT
PITP--TSARVPPVQVGTPSPRTAATVSTPTCASSPAIVRHTQKPGEDSSSSEESESEEE
T---APAAAGG--------------------------------QMKSVGKS-----LPVK
AASASTKGPSGKGTTPGPPGKAGPPVA-----------QV--KAEVQEDS-ESSE-ESDS
E---EAAAASAQVKTSVKTPQTKANPSATRVASAKAAA-PAPGKGGL-------------
----------------AVAQAKV-------GSPAKVTPA----------ARDPQNSAVS-
---GSGQ--------VSMPAVGKAAAAA-AQAQPGPVGGSQ--E-DSESSEEESDSEGE-
--
PF03546 // ENSGT00730000111382_0_Domain_2 (1563, 1809)
APPQAKPS--------------------------------------------------
--------------------------GKTPQVKTASAPTKAS---PKK--GAAPVPPGKA
GPPA--AQARRQEEDSESS-SEEESDSDGDAPA---------------------------
------------------------------------------------AAPPAQKEGSSK
TATSKTLAPASLEKKAEESSTSSDEDLPSNQVI-KTPLIFV----DPNRSPASPA---AT
PTQAQATGTPRKAQALESTARSS---SESEDEDVIPATQCPTP-A-TRTSVVTVPTAPPK
AALRAS-VVGAPS---SEDTSQGSEGKKQEASAT---QVTKRNLPLTQAALKVLAQKASE
AQPPAAKTPSSSGTDSAVGTLSTASPQSTPTQVRMANKLRKPEVPAAQRATATPSGHPTP
KAPATS---DDSN-SSSSGSEDEAK---GSQAAPSAHRP-GLAPSRRETLVEETTTESSD
DEVVAPSQSLLSGFVTPGLTPA---GSKTSKATPKPDTSPSVS-STPATRDAPE------
------GNQEVEPQQA--AGTMS------------------------------PKTGRK-
-EAGATPQKPG---KGPGS--------TPASMLALQSNITQRLLSEPWPLSEAQVQASVA
KVLTELLEQERKKAVETAKEGN------RKG--RLGQKRKLSEDQTAPKAPKSKKKKQLA
AADGGEHV-VSSEKAPRTVKGKSKKDRASGDVKEKKEKESP-GSQGAKEKPVGEL--GTP
KGDGGDHGNLK---TKKEKKKSDKKKKDKEKKEKK--KKTKKASTKDPGSPSQ--KKKKR
KKKTAEQTV---------------------------------------------------
--
ENSSSCP00000015382 (view gene)
MA-EARKRRELLPLIYQHLLQAGYVRAAR-------EVKEQSGQKSFLTQPVTLLDIYTH
WQQTSELGRKRKAEE-DAALQAKKTRVSDPISSSESSEEEEEEEEAAEARNAKATPRPAS
TNSSVPGPVLPSSIKDEGAAKTKK-ASQTGNPTARAASGKPVAQLLGQK-----------
------------------------------------------------SPRKAAGPSANA
VLASET---------EEEG-SVSALRT-----AAKPGMTAASQAHSSSEDTSSSSDETDV
EGKPLVAPAQVKA-----SAATAQEPP---------------------------------
-------------RKRAPPT-PGKA---GAGTPQGRGGTLTPA-----------SRAKKP
EEVSESSEE-ESESDDQAPASTPS-QTKAS---EK-TQVRAAS---GPARGAPGKGA---
---------TPAPPGRA--------GPAAARDKVVKP-------EEGSESSSEEASSSSD
SEEEAPA-AK--TPPQA-------------------------------------------
---------KPPGKVPGVRAT------SAAP-AKESPRKGAPPAPASRAAQA--------
GQREEDSESSSEESDAG-GQAAAAAP----QTSSPA------------------------
------------------------------------------------------------
------------------------------------------------------------
---------LVKPMGQNSHGRAPRA--------L----AGKGAVPALPGQAGQ-------
--Q--DSGSS----SEDSDSGDSEEEA---------------------------------
------------------------------------------------------------
-------------ATTAKPSEKVLPARTASGPSTG---------PPQK-------ARPAA
TQVKAERSESSEE-----SSTDSE----EEA--PAAAAA---------------------
----------------------------------------AQAKPAVKTPQTKASPRKGG
PVTP--ASAKAAPVRVGTPAPWKAGATT-----ASPATAKAAQRPEADSSSSETSESEEE
E---EEAP----------------------------------------------------
------------------------------------------------------------
-----PAAAAGRMKNSVTAPQTKATPAVPRVASAKVAA-SAPGKGAP-------------
----------------AGAQATL-------GSPAKVRPP----------ARTPQGRPGA-
---GKGP--------GAVPAVGKAATTA--APQPGPVGGAE--D-DSESSDGDSDSEEA-
--APAQAKPT--------------------------------------------------
--------------------------R
PF03546 // ENSGT00730000111382_0_Domain_2 (1648, 1820)
KTPQVRAASAPSKAS---PRK--GAAPAPAGKT
GPPA--TQAGKPTEDSESS-SEESSGSDGEAPPAQ-GG----GCSPASGRP---------
---------------------------------------------------PAATA----
-SAGKAEQKKTKEEKAEGPSASSDDELPASQVI-KTPLIFV----DPNRSPAGPA---AT
LAQAQATSAGRKVQASESTAGSS---SESEDEDVVPATQCSAP-A-LRTSSATAPPVPPR
TAPRAS-SGKE--------PGQAAEGRKPEAPATQVTKRSPADLPLTQAALKVLAQKASE
GRPAAARTPSSCATD-GLGTLPTTRPQSAPAQTRGANELRKPGVPAAP-----PSKRPKA
KASGTSEDSEDGSSSSSSGSEDDAA---GPRVAPGAPGP-GAAPARKETLVEETTAESSD
DEVVAPSQSLLSGYVTPGLTPG---TSQASKTTPKPAPSPSVS-SPPATKDALG------
------GKRGAEAPRA--AGTTS------------------------------PKSGRG-
-EAGATPQRPR---EGPES--------PQASRLALQHDIARRLLGEPWPLSEAHVQASVA
KVLTELLEQERRKAADAAREGR------RK-----GRKRKLSEDQTAAKAPKSKKKKQLA
AGEGGERA-VSPDKAPRTCQGKPKRDRAGGASKEKQEKEAP-GSQGAKEKLEGEP--GMV
KGEGGDQGAPR---SKKEKKKPDKKKKDKEKREKK--KKAKKASTKAPDSPCQ--KKKKK
KKKTAEQTV---------------------------------------------------
--
ENSMNEP00000029270 (view gene)
MA-EARKRRELLPLIYHHLLRAGYVRAAR-------EVKEQSGQKCFLAQPVTLLDIYTH
WQQTSELGRKRKAEE-DAALQAKKTRVSDPISTSESSEE-E---EEAEAETAKATPRLAS
TNSSVLGADLPSSLKEKAKAETEK-AGKTGNSMPHPATGKTVTNLLSGK-----------
------------------------------------------------SPRKSAEPSANT
TLVSET---------EEEG-SVPAFGA-----AAKPGMVSAGQADSSSEDTSSSSDETDV
EGKPSVKPAQVKA-----SSVSTKESP---------------------------------
-------------ARKAAPA-PGKV---GDVTPQVRGGA
PF03546 // ENSGT00730000111382_0_Domain_0 (400, 516)
LPPA-----------KRAKKP
EEESESSEE-GSESEEEAPAGTPS-QVKTS---EKILQVRAAS---TAAKGTPGKGA---
---------TPAPPGKA--------GAVAFQTKAGKP-------EEDSESSSEES---SD
SEEETPA-AK--ALLQA-------------------------------------------
---------KASGKTPQVGAA------SAP--AKESPRKGAAPAPPGKTGPAVAKAQTGK
--QEEDSQSSSEESDSE-E-----EA----PVQTKPSGKTSQVRAASASAKE-SPRKGAA
PAPP----------RKTGPAATQAQAGKQEEDSGSSSEE-SDSDREAPAAM----NAA--
------------------------------------------------------------
---------QVKPLGKNPQVKPAST--------MGTGPLGKGPGPVLPGKVGPTTPSAQV
GKWEDS-DSS----SEE--SSDSDDGE---------------------------------
------------------------------------------------------------
-------VPTAVAPAQEKSLGKVLQAKPASGPAKG---------PPQK-------AGPVA
TQVKAEKPMEDSESS--EESSDSADS-EET---PAAMTA---------------------
----------------------------------------AQAKPALKIPQTKACPKKTN
--T---ASAKVTPVRMGTQAPRKAGTVTSPVGSS-PAVAGGTQRPAEDSSSSEESDSEEE
-K-TGPAVTMG--------------------------------QAKSVGKG-----LQVK
AASVPVKGSLGQGTAPVLPGKTGPTVT-----------QV--KAEMQEDS-ESSEEESD-
SE--EAAAPSAQVKTSVKKTQAKANPAAVRASPAKGTI-SAPGKVVT-------------
----------------AAAQAKQ-------RSPAKAKPP----------VRNLQNSTV-L
---VRGP--------ASVPPVGKAVAAA-AQVQTGP-------EEDSGSSEEESDSEEE-
TETPAQAKPS--------------------------------------------------
--------------------------EKTPQVRAALAPAKES---PRK--GAAPAPLGKT
GPSA--TQAGKQ-DGSGSS-SEE-SNSDGEAPAAATS-----------------------
----------------------------------------------------AQKDSNSK
PARSKTLAPAPPEGNTEGSSESSEEELPLTQVI-KPPLIFV----DPNRSPAGPA---AT
PVQAQAASTPRKSRASESTARSS--SSESEDEDMIPATQCLTP-G-IRTNVVTVPTAHPR
IAPKAS-MTGASSSKES---SRISDGKKQEGPATQVSKKNPASLPLTQAALKVLAQKASE
AQPPVARTQRSRGADSAVGTLPATSPQSTPVQAKGTNKLRKPKLPEGQQAT---------
KAPGSSDDSEDSS-NSSSGSEEDAE---GPQVAKSAHTLVGPTPSRTETLVEETAAESSE
DDVVAPSQSLLSGYVTPGLTPA---NSQASKATSKPYSSPSVS-STLAAKDDPD------
------GKQEAKPQQA--AGMLS------------------------------PKTGGKE
AVSGTTPQKSRKPKKGAGN--------PQASTLALQSNITQCLLGQPWPLNEAQVQASVV
KVLTELLEQERKKVVDATKESS------RKG--WESRKRKLSGDQPAARTPRSKKKKKLG
AGEGGEAF-VSPEKTSTTSKGKAKRDKASDDVKEKKGKGSL-GSQGAKDEPEKELQKGIG
KVEGGDQSNPK---SKKEKKKSDKRKKDKEKKEKK--KKAKKSSTKDSESPSQ--KKKKK
KKKTAEQTV---------------------------------------------------
--
ENSETEP00000006388 (view gene)
------------------------------------------------------------
---TSVVGQKQKAEE-DAALLARKSRVSDPVSTSESSEDE----EEAEAKTANATPSLVS
TNSSVVRENLPSSEKEPAAANAKG-ASKMVNSVPHPVSGKAVAHPLSGK-----------
------------------------------------------------SPKKLAVPSANT
TLVSET---------EEEG-SVQPLKA-----AAKPGAVD--PADSSSDDTSSSSDETDV
EXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXVKLPGKSPPAKATPAPARGGTGTPR------------PAERP
EQEADSSEEESSESEE------EL-QVKAP---EKKLQVRAAK---HS------------
-------------PTHA--------GP--TQAKAGKP-------EQDSESG
PF03546 // ENSGT00730000111382_0_Domain_0 (532, 877)
SEEE---SS
SEEETAV-TR--A--PT-------------------------------------------
---------KPSGKAPQVRAA------SAP--TKGSPAKGAGPAQPGKSGPAAAQ-----
------------------------------------------------------------
--------------------------GRPEEDSGSSSEEDSDSEGPTPAAV----ASV--
------------------------------------------------------------
---------QAKPPGRNLQARPPTA--------LAAGPSGKGAILVPPGKAGSGVSRAQV
GRPEQTSESDSPSS----------EE----------------------------------
------------------------------------------------------------
-------APTVRTPIQAMPSGKSPQVRPASGPTKG---------PPQK-------SAPLA
TEVKTEGPKEESESS--EEESDSEESDSEEA--PAATTP---------------------
----------------------------------------AQAKPATKISQTKASPRKGP
PITPA-SQGKTPPVRVGTPTPWKAGVVASPACSASPGVARGTQKPEEESSSSEESESEPE
V---APAATAK--------------------------------QVKSVGKS-----LPGK
AVPAPSKGPSAPGSTPVPPGKAGLAAT-----------EI--KKESSEDS-EDSEEEPG-
------TKTPAPVKAPG-RTLTKASPASPRAPTVKGIA-AAPGKALA-------------
----------------AGVQAKL-------ASPAKARLP----------PPPGRSLPT-S
AASARSQ--------VSAPAVEKSVAGA-AQAKPGAARGSQ--EEDSESSKQQSSE----
EEAPTQVKPS--------------------------------------------------
--------------------------GKAPQARAAAAPTKGS---PAK--GTVPAWPGKS
RPAA--SPAEKLEEDSGSS-SEE-SDREAPAVVAGTP-----------------------
------------------------------------------------------------
-----------------------------AQVI-KPPLIFV----DPNRSPAGPA---AT
PSPAPPASAPRKA--SESSARSS--SSESEDEDVVPATQCSTPPV-MRTNVVPMPTTRPT
AAPRAS-LAGASSSEES---RGVARGKQQKAPATQXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXX----------XXXXXXXX---XXXXXXXXXXXXXXXX-XXXXXX---------
-XXXXX--XXXXX-X-----X-XXX---XXXXXXXXXXX-XXX-XXXXXXXXXXX----X
XXXXXXXXXXXXXXXXXXXXXX---XX-------------XXX-XXXXXXXXXX------
------XXXXXXXXXX--XXXXX------------------------------XXXXXXX
RREDTKPQKPRKPSKASAS--------PPASTLALQNDIAQRLLGQPWPLSEAHVQASVA
XXXXXXXXXXXXXXXXXXXXXX------XXX--XXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXX-XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX-XXXXXXXXXXXXXXXXXX
XXXXXXXXXXX---XXXXXXXXXXXKKDKGKKEKK--KKTKKASTKDHSSPPQ--KKKKK
KKKTPEQTV---------------------------------------------------
--
ENSLAFP00000017111 (view gene)
ENSRROP00000020240 (view gene)
ENSEEUP00000013352 (view gene)
ENSSARP00000010981 (view gene)
ENSOPRP00000007676 (view gene)
MA-EARKRRELLPLIYQHLLQAGYVRAAR-------EVKEQSGQKSFLTQPVTLLDIYTH
WQQTSELGRKKKAEE-DAALQAKKMRVSDPVSSSESSEE-D---EEVEALAAKATPRPTA
VNSSVTGVDLPASMKEKAKVQAQTRADKVVNSVPHSASKKALAHLLTGK-----------
------------------------------------------------SAQKSAEPSMNT
VLASET---------EEEA-GGPAPAP-----AAMPGLVSAGQADSSSEDTSSSSDETDM
EVKPTQVKAPL--------PAPAQEST---------------------------------
-------------VKRTTPA-PGKV---GARPTQARGGALTPA-----------AETQKT
QKDSESSEE-SD-SEEEPPTGRPG-QVMAS---GKVLLAGSAS---AS
PF03546 // ENSGT00730000111382_0_Domain_0 (469, 633)
AKGTPRKAA---
---------TPAAPG-----------------KTGRP-------EEDSDSSSEAS---SS
DSEETPA-AV--TPLEV-------------------------------------------
---------KPSGKTPQARA------------AKGSPRKGAPPVAPSRIGPAATQAQVGK
--QKEVSESSSEESDSD-EETPVT------Q-----------------------------
-----------------------------------------------------TMNPA--
------------------------------------------------------------
---------QPKPLGKNSQVRPAPA--------VGVGPSGKGTGLVPPGKARPAAPLTQV
AKPKEDSDSSSSSSSEE--ESYSNGEVVPAVSA---------------------------
------------------------------------------------------------
--------------AQVKPPGKGLQVI--LSPTQGPLGKVAAPVPSQK-------PGPGP
GA-----PKEDSESS--EESSDSE----EEA--RTAPPP---------------------
----------------------------------------TQAKLAQKT-QPKASPKKGT
PAPMTPTSTKAPA---------------MQVSSSSPAVAQGAQRPTEISSSSEESESDEE
T--TPA----A--------------------------------TAKSVGKG-----LQAR
ATPATSTGPLGQGTTPMLPGKTGPASA-----------QL--KAKAHK---ESSEEGSD-
SE--GAASTTTQVKASAKAPQARVGTASARAAAAQGVT-SPPRKVIT-------------
----------------PAAQAKQ-------GSPAKGKPP----------MKTAQNSAVGK
AATTVAQ--------ANKVPV------------GRP----Q--ED-SESSEESSDSEEE-
EEAPAQVKLL--------------------------------------------------
--------------------------GKTPQLRAALAPAKES---PRK--GTPLAPSGKA
VPVPAQTQLGKKEEDSESS-SED-SDSDGETPMAVTPA----------------------
------------------------------------------------------------
------------------------------QVI-KPPLIFV----DPNRSPAGPA---AT
PAQAQTAGAQRKTRASESTTR-S--SSESEDDDVIPATQSLAP-A-IRTN-V-LATGHPR
TGHSA------SGSE-C---SRASEDKKQEAPATQITKRNPASLPLTQAALKVLAQKASQ
AQSPH--AQSSSGVDGATGTVPVTGLQSNSLQARVANKLGKS---KGQQ---------AT
KVPRSSEHSEDSS-SSSSGSEEGAE---GPPPAKSAPKP-GAVPAGKEILVEETS-ESSD
DEVVAPSQSLLSGYVTPGLTPA---NSQVSKAIPRPDPKPLVF-STPTTKDNPN------
------GKQEAEPPRG--VDATS------------------------------PKKGRKE
PASGTTPQKPRKPKKGTLG----------AQASVLQSSITQRLLNEHWPLSEAQVQASVV
KVLTELLEQEKKKAMDATKETS------KKG-----HKRKLSGDQPAPK---GKKKKLLE
AGANGENA-GSPEKASRTSKGKTKKDKASSDSK-KKGKGSS-GSQGAKGKPEEEL--GTV
KAEEEDQSNSK---SKKEKKKSDKRKKDKEKKEKK--KKAKKASAKDPESPFQ--KKKKK
KKKAAEQVQAV-------------------------------------------------
--
ENSMPUP00000013688 (view gene)
MA-EARKRRELLPLIYQHLLQAGYVRAAR-------EVKEQSGQKSFLTQPVTLMDIYTH
WQQTSEVGQKRKAEE-DAALQAKKTRVSDPISSSESSEEEEE--EEAEAEAAKATPRLAA
TNSSAAGPVLPSGAKEKAKAKTKK-ASKMMNSTPHPASGKAVAHLLSGR-----------
------------------------------------------------SPRKSAEPSANT
VLVSET---------EEEG-GVPALGT-----TAKPGMASANQVDSSSEETSSSSDETYV
EMKPSVKPPQVKA-----SPVPAKDSL---------------------------------
-------------GRGTAPT-PGKA---GDVTPQVRGGALTSA-----------NKAKVP
EEDSESSEE-ESESEEGAPAEPPC-QAKAS---NTVVQVRAAL---GPVKGTSGKGA---
---------APAPPGKA--------GPAAAQATAGKP-------EEDSDSSSEEE---SD
SEEEAPA-AK--TPLQA-------------------------------------------
---------KSSGKTPQVKGA------SAS--AKESPRKVAPALPPGKAGPVAAQVGKGK
GED--NAESSTEESDSE-EEAPAAVP----PAKSPA------------------------
------------------------------------------------------------
------------------------------------------------------------
---------QAKPLGQNSQVRAASG--------P---------VKGPPQKLGPAAT--PV
GKQEEDSESS----SEE--ESDSEAEAPAQLRA----RSRLPQTTYT-------------
---HTD----SACPF---------------------------------------------
-AMLSTPSPILCVSLQAKSSG-KIQVRPASGPAKG---------PPQK-------AGSVT
TQGKAKVSKDDSESS--E-ESDSD----EKV--PAAP-----------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------APSGQGTAAAPPGKAGLPVA-----------PVKVKAELPEDSSESSEEDSDS
E---ETAKAPTQMKASMKTPQNKANPAPTRAASAKGAA-SAPGKGVM-------------
----------------AAAQAKQ-------ESPAK-------------------SNAVS-
---GRSQ--------GSMPAVGKAVATA-APAQPGPVKSPQ--EEDSESSEEESDSDGE-
--VPTQVKPA--------------------------------------------------
--------------------------GKSPQVRTASAPSKEL---SRK--GAA
PF03546 // ENSGT00730000111382_0_Domain_2 (1674, 1943)
VAPPRKT
GAIA--TQAGKPEEDSESS-SEEESDSEEEA-----------------------------
---------------------------------------------------P--------
-----------------------------AQVI-KPPLIFV----DPNRSPAGPA---AT
ATHAPAINTPRKARASESTARSS--SSGSEDEDVIPATQCSTP-A-SRTSTSVL-TAQPR
PAVRAN-AVGTGSGAVSG---RMSEVSSSSPTALC-------------------------
---------ASTGPLLPQGTL--------PVPAKGTNKVRKPEVPALQRATATPTGHTKA
KASDTSSDSNDSS-GSSSGSEEDAD---RPRMAPSAPRL-GPAPSTTETLVEETTAESSE
EEVVAPSQSLLSGYVTPGPTLA---NSQASKATPKPDLNPSSASSTSATKDAPD------
------SKQEVEPQHV--AGAVS------------------------------PKTSRK-
-EANPTPQKPRKPKKEAGN--------PQASVLALQSDISRRLLSEPWPLNEAQVQASVV
KVLTELLEQERKKAADTAKETG------RKG--RVGRKRKLSGDQTAARVPKNKKKKQLV
AGEGGEGA-GSPEKALRTPKGKAKKDGASGNTKEK-EKEAS-GSRGAKEKPEGEP--GTL
KVEVGDQGNPK---SKKEKKKSDKKKKDKEKKEKK--KKVKKASTKDPDSPFQ--KKKKK
KKKTAEQTV---------------------------------------------------
--
ENSPPAP00000030386 (view gene)
ENSPEMP00000003182 (view gene)
MA-EARKRRELLPLIYHHLLQAGYVRAAR-------EVKEQSGQKSFLTQPITLLDIYTH
WQQTSELGQKQKAKD-DVALQAKKFRVSDPVSSSESSEEEE---EEAEPKTAKASKNPSC
QV------------LCGVGAKTKT-VNKMVNSVLPPASGKAVVHLLSGK-----------
------------------------------------------------SPKKSAEPSANT
VLASET---------EEEG-SVQALGA-----TAKPGMLSAGQASSSSDDTSSSSDETDV
EVKLSVKPAPAKV-----APAPAKEPP---------------------------------
--------------ARTAPG-PTKL---GSVTPRPQAGTPAAP-----------GRTA-D
TEESESSEEDSESEEDEPPAGMPS-QVKAP---GKVPQAGAAS---VSA-K---------
---ISGKGPISAPPEKT--------GPTAAQAKAGRP-------EKDTESSSEDD---SD
SEEEEEE-PIDVSTPQV-------------------------------------------
---------KSAGPTYPFL-----------------------------------------
-----LIRL
PF03546 // ENSGT00730000111382_0_Domain_0 (670, 1063)
SSEESGIDMGKTPAA------------------------------------
--------------------------------------------------APLSTSPA--
------------------------------------------------------------
---------QMKPLGKSPQARPASA--------VSPGSSGKG-----PGKAGSAALKVQT
GKAEEDSESS----SEESES---DRALT--------------------------------
------------------------------------------------------------
-------------PAKVSPRRAS---------A-------PEPVWPQ-------------
--S---------------------------YV-GSGWQGGQEE-----------------
----------------------TDVACESRRPLQPSSALSPQAKPALGK-QAKASPRKGT
PASA--TVA----------PV----------------------------------L--VT
A----PGATR--------------------------------VQGTSVGKG-----LQGK
A-------ASGQGTASVHPGRTGPPVA-----------QV--RATAQEDS-SGDSEEES-
SSEDEDETPPAQAKPLGKLPQAKANPPPTKASPAKGTV-VAP------------------
-----------------------------------GKAP----------GSAGQNSALS-
---ARGQ--------RSVAATTKAGSPATTQAQKGPVAGAGKAS--E-SSSEESDSEEE-
--APAQVKPV--------------------------------------------------
--------------------------GKTSQVRAASAPAKAS---PGK--GAHPGATKKT
GPTAAQAQKRKME-DSESS--SEESDSDGEMPSADGP-----------------------
----------------------------------------------------------SQ
PAKSKASAPAPPEKNGEESSESSDEELPSAQVI-KSPLISV----DPNRSPAGPA---AT
PAQVQAVNTPRKAQASGTTAQSS--SSESEDEDMIPATQPTTH-A-IRTNV-TVPTSLPR
TAPQPS------KSEQS---GRVPKGKKPKAA----------------------------
----------STQISSAVETPSVTHPQSTPVQARAANKLRKPKLAEKQQPPP---SYP--
RPSRSPS---NSS-DSSSESEADAK---RPQMAKAAPRL-DPDPSQKEIVVEETPDESSE
DELVAPSQSLLSGYVTPGVTVA---NSQASKATPKQDSNPLVS-SAPATKDNVD------
------GKQKGKPQNSVAADTAS------------------------------PKTGRKE
ASPGATPQKPKKAKKSTQT--------PAAPTQDLQNSITQRLREQPWPLSEAQVQASVM
KVLTELLEQERQKATEAIKESG------KKG-----QKRKLSGDQLAAGTPKSKKKK-QL
V--AGAGA-ITPEKTSRTSKAKAKPDRADADGK---GKESP-GTRGAKEKPES--E-LGL
KVESGEQGDAK---SKKEKKKSGKKKKDKEKKEK---KKGKKTSVKDPDSLVK--KKKKK
KKKTAETAM---------------------------------------------------
--
ENSHGLP00000011017 (view gene)
MA-EARKRRELLPLIYQHLLHAGYVRAAR-------EVKEQSGQKSFLAQPFTLLDIYTH
WQQTSEIGQKRKAED-DAALQAKKTRVSDPISSSESSEEEEEEEEEAEAKAAKT------
--SKTMGMVLPSSMKEKA--KTRN-TSKTVNSVPHLASGKAVVHLLSGK-----------
------------------------------------------------SPKKSAELSGST
VLVSET---------EEEG-SIQALEA-----AAKPGEKPPSLGA---------------
-----------KP-----KL----------------------------------------
------------------------------------------------------------
------------------------------------CFVKLWD--ILGFKG---------
------------------------------------------------------------
------------------------------------------------------------
--------------VPPRQAA------SAP--AKESPRKGHPPATPGKTGPAATQAQVGK
QEEDSDSSS--EESDS--------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
DGETP----------AAKPSGKILQVRPASGPTKGLLGKGAAPKPPQK-------SGPVA
IQVKAEKSKDDSESS--EESSDSE----EEA--PAAVTPAQVRPGRGGLEPPSHCPTWSW
AALQSECGFWPEGRIAWGLLSAIDGVSGSRSLSHPLSSLS
PF03546 // ENSGT00730000111382_0_Domain_1 (1181, 1277)
PQAKPELKMPQTKASPSKSI
LLTP--TSAKGAPVRVA--------------------LPRGAQRPKDNSSSSEEESEGKE
A---TPAAAVAQVRPVSACPSSGLAPQALWTPVRPFPLSHSSFQAKSLGKG-----LRGQ
AASAPTMGPSGQGATPVPPGKTRPTVA-----------QV--RAEAHESS-ESSEEESDD
SE-VGAVVTPAQAKAPGKTPQAKANPASTKASTDKEAA-WTP------------------
-----------------------------------GKSP----------VRAPKNSTIS-
---ARGQ--------ASVPASGKAVTAA-AQSQKGPTQESE--EKDLESSEESSESEEE-
--LPTQLKPL--------------------------------------------------
--------------------------GKTPQVKAASATANGS---PRK--GAPPAPPGKT
GPAAIQAQVGKWDRLPS----------------ANT-LHMLLQV----------------
--------------------------------------------------LSLQKNINSK
TVGSNTLTPAPREKSPEGSSESSEEELPSSQVI-KTPLIFV----DPNRSPAGPA---AT
PTQAQAASPPRKARASSSTAGSS---SEGEDEDVIPATQSSTLGECSLKEVVTVPTASPR
MAPRAS------SSLES---SRVSPGQKQEAAAPQ-------------------------
-------------AGSDAGTLPVTCPQSNPVQARVTNKLRKPKPSE--------------
-APGSSDNSDDSS-DNSSGSEEDAE---EPLMTKSAPGL-GPSPSKRETLVEETTAESSE
DEVVVPSQSLLSGFVTPGLTMA---NNRASKATARQNLHPSLS-SPPPTKDDPA------
------GKQAAESQHI--AGAVS------------------------------PKPGRKE
AASGPTPQKPRKPKKGPLS--------TSASLQTLQSNITQHLQGQPWPLNEAQVQASVV
KVLTELLEQERKKAEDTARESS------RKG-----RKRKLSGDQVASRAPKGKKKKQLA
AGEGREDA-ASSEKAPRTSKGKSKRDKASGDLK---EEKGS-GSGGSKDKPDGEL----L
KVESGNQNDSK---SKKEKKKSNKKKKDKEKKEK---KKGKKPSAKDSDSPFQ--KKKKK
KKKTAEQTV---------------------------------------------------
--
ENSMMUP00000060903 (view gene)
MA-EARKRRELLPLIYHHLLRAGYVRAAR-------EVKEQSGQKCFLAQPVTLLDIYTH
WQQTSELGRKRKAEE-DAALQAKKTRVSDPISTSESSEE-E---EEAEAETAKATPRLAS
TNSSVLGADLPSSLKEKAKAETEK-AGKTGNSMPHPATGKTVTNLLSGK-----------
------------------------------------------------SPRKSAEPSANT
TLVSET---------EEEG-SVPAFGA-----AAKPGMVSAGQADSSSEDTSSSSDETDV
EGKPSVKPAQVKA-----SSVSTKESP---------------------------------
-------------ARKAAPA-PGKV---GDVTPQVRGGALPPA-----------KRAKKP
EEESESSEE-GSESEEEAPAGTPS-QVKTS---EKILQVRAAS---TAAKGTPGKGA---
---------TPAPPGKA--------GAVAFQTKAGKP-------EEDSESSSEES---SD
SEEETPA-AK--ALLQA-------------------------------------------
---------KASGKTPQVGAA------SAP--AKESPRKGAAPAPPGKTGPAVAKAQTGK
--QEEDSQSSSEESDSE-E-----EA----PVQTKPSGKTSQVRAASASAKE-SPRKGAA
PAPP----------RKTGPAATQAQAGKQEEDSGSSSEE-SDSDREAPAAM----NAA--
------------------------------------------------------------
---------QVKPLGKNPQVKPAST--------MGTGPLGKGPGPVLPGKAGPTTPSAQV
GKWEDS-DSS----SEE--SSDSDDGE---------------------------------
------------------------------------------------------------
-------VPTAVAPAQEKSLGKVLQAKPASGPAKG---------PPQK-------AGPVA
IQVKAEKPMEDSESS--EESSDSADS-EET---PAAMTA---------------------
----------------------------------------AQAKPALKIPQTKACPKKTN
--T---ASAKVTPVRVGTQAPRKAGTVTSPVGSS-PAVAGGTQRPAEDSSSSEESDSEEE
-K-TGPAVTMG--------------------------------QAKSVGKG-----LQVK
AASVPVKGSLGQGTAPVLPGKTGPTVT-----------QV--KAEMQEDS-ESSEEESD-
SE--EAAAPSAQVKTSVKKTQAKANPAAVRASPAKGTI-SAPGKVVT-------------
----------------AAAQAKQ-------RSPAKAKPP----------VRNLQNSTA-L
---VRGP--------ASVPPVGKAVAAA-AQVQTGP------
PF03546 // ENSGT00730000111382_0_Domain_2 (1543, 1820)
-EEDSGSSEEESDSEEE-
TETPAQAKPS--------------------------------------------------
--------------------------EKTPQVRAALAPAKES---PRK--GAAPAPPGKT
GPSA--TQAGKQ-DGSGSS-SEE-SNSDGEAPAAATS-----------------------
----------------------------------------------------AQKDSNSK
PARSKTLAPAPPEGNTEGSSESSEEELPLTQVI-KPPLIFV----DPNRSPAGPA---AT
PVQAQAASTPRKSRASESTARSS--SSESEDEDVIPATQCLTP-G-IRTNVVTVPTAHPR
IAPKAS-MTGASSSKES---SRISDGKKQEGPATQ-------------------------
-------------ADSAVGTLPATSPQSTPVQAKGTNKLRKPKLPEGQQAT---------
KAPGSSDDSEDSS-NSSSGSEEDAE---GPQVAKSAHTLVGPTPSRTETLVEETAAESSE
DDVVAPSQSLLSGYVTPGLTPA---NSQASKATPKPYSSPSVS-STLAAKDDPD------
------GKQEAKPQQA--AGMLS------------------------------PKTGGKE
AVSGTTPQKSRKPKKGAGN--------PQASTLALQSNITQCLLGQPWPLNEAQVQASVV
KVLTELLEQERKKVVDATKESS------RKG--WESRKRKLSGDQPAARTPRSKKKKKLG
AGEGGEAF-VSPEKTSTTSKGKAKRDKASDDVKEKKGKGSL-GSQGAKDEPEKELQKGIG
KVEGGDQSNPK---SKKEKKKSDKRKKDKEKKEKK---KAKKSSTKDSESPSQ--KKKKK
KKKTAEQTV---------------------------------------------------
--
ENSMAUP00000000729 (view gene)
MA-EARKRRELLPLIYHHLLQAGYVRAAR-------EVKEQSGQKSFLTQPTTLLDIYTH
WQQTSELGQKQKAQD-DASLQAKQSRVSDPISSSESSEEEE---EEAETKTAKAIPRPTS
VNSTAPAADLPSNKKEK--AKTKA-ANKMVNSVQHPAPGKAMVHLLSGK-----------
------------------------------------------------SPKKSAEPLANT
VLASET---------EEEG-SVPALGA-----TAKPGVLSTGQASSSSEDTSSSSDETDV
EVKLSVKPAQAKP-----SVAPAKEPP---------------------------------
--------------ARTAPV-PTKS---GSVTSRLQEGTPAAP-----------GRTA-H
AEESESSEEDSES-EDEPPAGMPS-QVKAP---GKV-HAKAAS---VSAKE---------
---VSGKGPTSAPPEKT--------RPTVTQVKAGRP-------EE-TESSSEDS---SD
SEDEEPV-AV--TAPQV-------------------------------------------
---------KPSGKKPQVKST------SSP--AKGSSQKRAPLVTPGKAGPAAAQARAGQ
PE---DSDSSSEESESDSEKTPAA------------------------------------
--------------------------------------------------VTLSTSPA--
------------------------------------------------------------
---------QVKPLGKSQQVRPAS--------------TGKGAHPPCPGKAGSAALGVQM
GKVEEDSESS----SEDS----DDGAVT--------------------------------
------------------------------------------------------------
-------------PAKAKALGKR-----------------ATPALLQK-------AEPVA
AQVKIKRRKEDSESS--E----ESTDSEEEA-----------------------------
---------------------------------APAAPAAAQAKPALGK-QTTASPRKGT
PATP--ASAMVT-------SVGV---ST-----------------
PF03546 // ENSGT00730000111382_0_Domain_0 (1246, 1660)
-SPSSKEGSGA--AP
------GITK--------------------------------VQGKSVGKG-----LQGR
D-------ALGQG-------RPGPTVA-----------HV--RITTQEDS-EDSEEEAS-
SEE---DETSAQTTPSGKPLQAKANPPPTKASPAKRPV-SASGKVNT-------------
----------------VV-------------APAKGKPP----------VSTVQNSAVS-
---AR------------VPATGKAGPPA-VQAQKGPVAGVG--QDSESSSEEESESEEE-
--AAAQTKPV--------------------------------------------------
--------------------------GKTSQVRAASAPAKES---PGK--GAHPGAAKKT
GPTATQAQTGKTE-DSESS--SEESDSDEEMPSAIIPA----------------------
------------------------------------------------------QGATSQ
PARSRASAPAAPEKNIEESSESSSEELPSAQVI-KSPLISA----NPNRSPAGPA---AT
LSQVQAVNIPRKAQASGTVAQSS--SSESEDEDMIPATQPSTQ-A-TKTNVATVPTSLPR
MASQPG------KSEPS---SQMPKGKKPKAA----------------------------
----------STQISGAVQTLPVTPPQSTPVQSKTTNKPRKPKLPAKQQPPS---SYP--
SS---SS---DSS-DSSSGSEEDAK---RPQMAKSAPRL-DPDLSQKETVVEETPAESSE
DELVAPSQSLLSGYVTPEVTVA-----QASKATPRPDSNPLVS-SAPATKDNPD------
------GKQRSKSQD--ATDTMF------------------------------PKTGRKE
TSSETTPPKAKKAKKSTWT--------SATPTQALQNSISQCLREQAWPLSEAQVQASVV
KALTELLEQERQKATAAIKESG------KKG-----HKRKLSGDQLAAGAPERKRK--AS
-------------GTS---KTKSKLAEVNADGK---EKGSP-GSQGAKEKSKS--E-PGS
QVESGEQNDAK---NKKEKKKSSKKKKGKDKKGK---KKGKKTSAKDPDSQIK--KKKKK
KK-TAEPAV---------------------------------------------------
--
{"ENSECAP00000012329": [["Domain_0", 490, 1660, "PF03546"]], "ENSPANP00000002769": [["Domain_2", 1544, 1820, "PF03546"]], "ENSMFAP00000029393": [["Domain_2", 1544, 1820, "PF03546"]], "ENSHGLP00100016341": [["Domain_0", 491, 705, "PF03546"]], "ENSAMEP00000004999": [["Domain_0", 553, 685, "PF03546"]], "ENSODEP00000007198": [["Domain_0", 1035, 1660, "PF03546"]], "ENSPCOP00000011729": [["Domain_0", 490, 691, "PF03546"]], "ENSMLEP00000022799": [["Domain_0", 402, 509, "PF03546"]], "ENSCJAP00000066463": [["Domain_0", 321, 511, "PF03546"]], "ENSSTOP00000008058": [["Domain_0", 397, 516, "PF03546"]], "ENSMUSP00000135639": [["Domain_0", 491, 1660, "PF03546"]], "ENSFCAP00000029727": [["Domain_0", 394, 1660, "PF03546"]], "ENSANAP00000019635": [["Domain_2", 1565, 1718, "PF03546"]], "ENSCANP00000002555": [["Domain_0", 690, 1569, "PF03546"]], "ENSMPUP00000000440": [["Domain_0", 437, 1661, "PF03546"]], "ENSP00000367028": [["Domain_0", 319, 508, "PF03546"]], "ENSNGAP00000007791": [["Domain_2", 1565, 1713, "PF03546"]], "ENSCATP00000016431": [["Domain_0", 690, 1568, "PF03546"]], "ENSBTAP00000020259": [["Domain_0", 285, 507, "PF03546"]], "ENSPCAP00000013361": [["Domain_0", 642, 865, "PF03546"]], "ENSMOCP00000007313": [["Domain_0", 230, 1114, "PF03546"]], "ENSPTRP00000068671": [["Domain_2", 1543, 1820, "PF03546"]], "ENSOCUP00000003342": [["Domain_2", 1675, 1793, "PF03546"]], "ENSCPOP00000002489": [["Domain_2", 1579, 1655, "PF03546"]], "MGP_SPRETEiJ_P0048783": [["Domain_0", 242, 476, "PF03546"]], "ENSPVAP00000005127": [["Domain_0", 553, 684, "PF03546"]], "ENSSHAP00000021147": [["Domain_0", 629, 1657, "PF03546"]], "ENSRNOP00000045755": [["Domain_0", 390, 508, "PF03546"]], "ENSMLUP00000012077": [["Domain_0", 66, 882, "PF03546"]], "ENSMICP00000035903": [["Domain_2", 1647, 1830, "PF03546"]], "ENSTBEP00000003838": [["Domain_0", 1157, 1860, "PF03546"]], "ENSCGRP00000012633": [["Domain_0", 421, 525, "PF03546"]], "ENSDNOP00000009653": [["Domain_0", 326, 637, "PF03546"]], "ENSCAFP00000031314": [["Domain_2", 1671, 1829, "PF03546"]], "ENSGGOP00000041929": [["Domain_0", 490, 693, "PF03546"]], "ENSOANP00000003653": [["Domain_0", 270, 703, "PF03546"]], "ENSCGRP00001007261": [["Domain_0", 311, 514, "PF03546"]], "ENSCCAP00000031848": [["Domain_0", 690, 1476, "PF03546"]], "ENSTTRP00000012360": [["Domain_0", 1053, 1860, "PF03546"]], "ENSSBOP00000020200": [["Domain_2", 1534, 1810, "PF03546"]], "ENSTSYP00000014474": [["Domain_0", 493, 1660, "PF03546"]], "ENSMLUP00000008094": [["Domain_0", 884, 1873, "PF03546"]], "ENSMODP00000031401": [["Domain_0", 1245, 1467, "PF03546"]], "ENSOGAP00000004130": [["Domain_0", 490, 1661, "PF03546"]], "ENSOARP00000007663": [["Domain_0", 395, 507, "PF03546"]], "MGP_CAROLIEiJ_P0046834": [["Domain_0", 491, 1660, "PF03546"]], "ENSRBIP00000018158": [["Domain_2", 1544, 1831, "PF03546"]], "ENSCLAP00000015410": [["Domain_2", 1678, 1800, "PF03546"]], "ENSNLEP00000012588": [["Domain_0", 395, 515, "PF03546"]], "ENSCSAP00000007758": [["Domain_0", 396, 510, "PF03546"]], "MGP_PahariEiJ_P0039419": [["Domain_0", 395, 516, "PF03546"]], "ENSPPYP00000017847": [["Domain_2", 1658, 1830, "PF03546"]], "ENSCHIP00000003682": [["Domain_2", 1563, 1809, "PF03546"]], "ENSSSCP00000015382": [["Domain_2", 1648, 1820, "PF03546"]], "ENSMNEP00000029270": [["Domain_0", 400, 516, "PF03546"]], "ENSETEP00000006388": [["Domain_0", 532, 877, "PF03546"]], "ENSLAFP00000017111": [["Domain_0", 1062, 1860, "PF03546"]], "ENSRROP00000020240": [["Domain_0", 690, 1658, "PF03546"]], "ENSEEUP00000013352": [["Domain_0", 873, 1860, "PF03546"]], "ENSSARP00000010981": [["Domain_0", 611, 1718, "PF03546"]], "ENSOPRP00000007676": [["Domain_0", 469, 633, "PF03546"]], "ENSMPUP00000013688": [["Domain_2", 1674, 1943, "PF03546"]], "ENSPPAP00000030386": [["Domain_0", 690, 1562, "PF03546"]], "ENSPEMP00000003182": [["Domain_0", 670, 1063, "PF03546"]], "ENSHGLP00000011017": [["Domain_1", 1181, 1277, "PF03546"]], "ENSMMUP00000060903": [["Domain_2", 1543, 1820, "PF03546"]], "ENSMAUP00000000729": [["Domain_0", 1246, 1660, "PF03546"]]}

