Found:
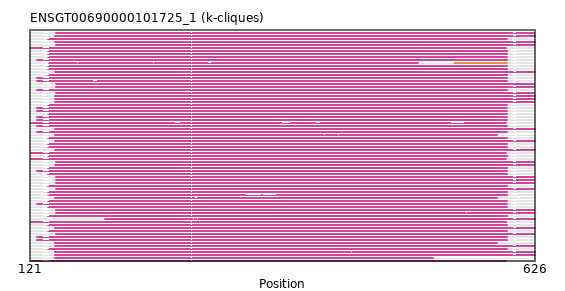
{"ENSCLAP00000009565": [["Domain_0", 128, 598, "PF00501"]], "ENSFDAP00000009284": [["Domain_0", 128, 598, "PF00501"]], "ENSTTRP00000001497": [["Domain_0", 146, 626, "PF00501"]], "ENSTBEP00000002461": [["Domain_0", 146, 588, "PF00501"]], "ENSCPOP00000001398": [["Domain_0", 128, 598, "PF00501"]], "ENSSBOP00000018471": [["Domain_0", 139, 598, "PF00501"]], "ENSAMEP00000019025": [["Domain_0", 146, 626, "PF00501"]], "ENSCSAP00000009726": [["Domain_0", 147, 625, "PF00501"]], "ENSHGLP00000001837": [["Domain_0", 128, 598, "PF00501"]], "ENSPVAP00000012807": [["Domain_0", 196, 626, "PF00501"]], "ENSMUSP00000033634": [["Domain_0", 139, 598, "PF00501"]], "ENSRROP00000025331": [["Domain_0", 139, 598, "PF00501"]], "ENSDNOP00000026696": [["Domain_0", 147, 625, "PF00501"]], "ENSECAP00000014924": [["Domain_0", 147, 625, "PF00501"]], "ENSANAP00000039845": [["Domain_0", 139, 598, "PF00501"]], "ENSHGLP00100009494": [["Domain_0", 122, 597, "PF00501"]], "ENSCJAP00000013688": [["Domain_0", 139, 598, "PF00501"]], "ENSOARP00000019273": [["Domain_0", 147, 625, "PF00501"]], "ENSSSCP00000056573": [["Domain_0", 128, 598, "PF00501"]], "ENSGGOP00000029666": [["Domain_0", 139, 598, "PF00501"]], "ENSCGRP00000003674": [["Domain_0", 128, 598, "PF00501"]], "ENSPPYP00000023091": [["Domain_0", 147, 625, "PF00501"]], "ENSEEUP00000000174": [["Domain_0", 146, 626, "PF00501"]], "ENSOANP00000018121": [["Domain_0", 147, 625, "PF00501"]], "ENSMAUP00000008254": [["Domain_0", 128, 598, "PF00501"]], "ENSPANP00000007465": [["Domain_0", 139, 598, "PF00501"]], "ENSFCAP00000033611": [["Domain_0", 139, 598, "PF00501"]], "ENSPSIP00000019638": [["Domain_0", 147, 625, "PF00501"]], "ENSDORP00000013834": [["Domain_0", 128, 598, "PF00501"]], "ENSMICP00000013231": [["Domain_0", 139, 598, "PF00501"]], "ENSCGRP00001014725": [["Domain_0", 139, 598, "PF00501"]], "ENSSARP00000000601": [["Domain_0", 146, 626, "PF00501"]], "ENSPCAP00000011048": [["Domain_0", 146, 588, "PF00501"]], "ENSPPAP00000018085": [["Domain_0", 139, 598, "PF00501"]], "ENSPTRP00000038175": [["Domain_0", 139, 598, "PF00501"]], "ENSBTAP00000050572": [["Domain_0", 147, 625, "PF00501"]], "ENSNLEP00000017493": [["Domain_0", 139, 598, "PF00501"]], "ENSMOCP00000018252": [["Domain_0", 122, 597, "PF00501"]], "ENSTGUP00000004473": [["Domain_0", 146, 625, "PF00501"]], "ENSTSYP00000010351": [["Domain_0", 139, 598, "PF00501"]], "ENSNGAP00000007181": [["Domain_0", 128, 598, "PF00501"]], "ENSMFAP00000044718": [["Domain_0", 139, 598, "PF00501"]], "ENSCCAP00000029622": [["Domain_0", 139, 598, "PF00501"]], "ENSSTOP00000004738": [["Domain_0", 128, 598, "PF00501"]], "ENSCATP00000045898": [["Domain_0", 139, 598, "PF00501"]], "ENSHGLP00100002563": [["Domain_0", 128, 598, "PF00501"]], "ENSETEP00000000496": [["Domain_0", 146, 626, "PF00501"]], "ENSCHIP00000000112": [["Domain_0", 139, 598, "PF00501"]], "ENSOGAP00000013291": [["Domain_0", 147, 625, "PF00501"]], "ENSMLEP00000034679": [["Domain_0", 139, 598, "PF00501"]], "ENSMNEP00000042308": [["Domain_0", 139, 598, "PF00501"]], "ENSGALP00000013111": [["Domain_0", 128, 598, "PF00501"]], "ENSFALP00000000180": [["Domain_0", 146, 625, "PF00501"]], "ENSMPUP00000003946": [["Domain_0", 146, 625, "PF00501"]], "ENSRNOP00000026057": [["Domain_0", 122, 597, "PF00501"]], "ENSOCUP00000004338": [["Domain_0", 147, 625, "PF00501"]], "ENSJJAP00000018749": [["Domain_0", 128, 598, "PF00501"]], "ENSAPLP00000012207": [["Domain_0", 146, 626, "PF00501"]], "ENSMODP00000014342": [["Domain_0", 147, 625, "PF00501"]], "ENSCAFP00000026704": [["Domain_0", 121, 597, "PF00501"]], "ENSACAP00000012627": [["Domain_0", 147, 625, "PF00501"]], "ENSXETP00000027173": [["Domain_0", 146, 524, "PF00501"]], "ENSMLUP00000005927": [["Domain_0", 146, 626, "PF00501"]], "MGP_SPRETEiJ_P0096659": [["Domain_0", 122, 597, "PF00501"]], "ENSMEUP00000009441": [["Domain_0", 146, 588, "PF00501"]], "ENSMGAP00000006613": [["Domain_0", 146, 626, "PF00501"]], "ENSCHOP00000003421": [["Domain_0", 146, 626, "PF00501"]], "ENSCANP00000035732": [["Domain_0", 139, 598, "PF00501"]], "ENSVPAP00000000472": [["Domain_0", 146, 626, "PF00501"]], "ENSODEP00000018066": [["Domain_0", 139, 598, "PF00501"]], "ENSPEMP00000017828": [["Domain_0", 128, 598, "PF00501"]], "ENSLAFP00000016511": [["Domain_0", 146, 626, "PF00501"]], "ENSMMUP00000005601": [["Domain_0", 122, 597, "PF00501"]], "ENSP00000339787": [["Domain_0", 139, 598, "PF00501"]], "ENSCGRP00001008714": [["Domain_0", 140, 509, "PF00501"], ["Linker_0_1", 509, 545], ["Domain_1", 545, 598, "PF00501"]], "ENSPCOP00000024774": [["Domain_0", 139, 598, "PF00501"]], "ENSSHAP00000001925": [["Domain_0", 147, 625, "PF00501"]]}