Home
Query Results
"ENSGT00660000095541_0"
Found:
ENSGT00660000095541_0
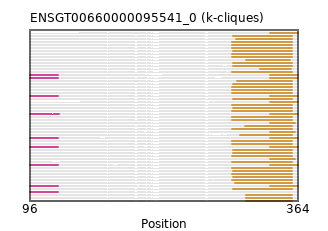
ENSP00000278886 (view gene)
------------------------------------------------------------
-------------------------MDEEENHYVSQLREVYSSCDTTGTGFLDRQELTQL
CLKLHLEQQLPVLLQTLLGNDHFARVNFEEFKEGFVAVLSSNAGVRPSDEDS-SSLESAA
SSAIPPKYVNGSKWYGRRSR--PELCDAATE-ARRV-P------EQQTQASLKSHLWRSA
SLESVESPKSDEEAESTKEAQNELFEAQGQ---LQTWDSEDFGSPQKSCSPSFDTPE
PF13499 // ENSGT00660000095541_0_Domain_1 (298, 358)
SQI
RGVWEELGVGSSGHLSEQELAVVCQSVGLQGLEKEELEDLFNKLDQDGDGKVSLEEFQLG
LFSHEPALLLESSTRVKPSKA----WSHYQ--VPEESGCHT-TTT-SSL-VSLCSSLRLF
SSIDDGSGFAFPDQVLAMWTQ-------EGIQ--NGREI----LQSLDFSVDEKVNLLEL
TWALDNELMTVDSAVQQAALACYHQELSYQQ--GQVEQLARER-DKARQDLE--------
-RAEKRNLEFVKEMDDCHSTLEQLTEKK-IKHLEQGYRERLS----LLRSEVEAERELFW
EQAHRQRAALEWDVG-RLQAEEAGLREKLTLALKENSRLQKEIVEVVEKLSDSERLALKL
QKDLEFVLKDK-L---EPQSAE-LLAQEERFAAVLKEYELKCR-DLQDRNDELQ----AE
LEGLWARLPKNRHSPSWSPDG--------RRRQLPGLG-PAGIS----------------
------------------------FLGNS--APVSIETELMMEQVKEHYQDLRT--QLET
KVNYYEREIAALKRNFEKERKDMEQARRREVSVLEGQKADLEELHEKSQEVIWGLQEQLQ
DTARGPEPEQ----MGL--APCCTQALCGLALRHHS-------HLQQIRR----------
------------------------------------------------------------
----------EAEAELSGELSGLGALPARRD--------------LTLELEE----PPQG
PLPRGSQRSEQLELER----------ALK-------------------------LQPCAS
------EKRAQMCVSLALEEEELELARGKRVDGPS-------------LEAEM-QALPKD
GL-VAGS-GQEGT-----RGLLPLRPGCGERPL----------------AWLAPGDGRES
EEAAGAGPRR-RQAQ---------DTE---ATQSPA--PAPAP-----------------
----ASHGPSERWSRMQ-----PCGVDGDIV-----PKEPEPFGAS--------------
-----------------AAGLEQPGARELPLLGTERDASQT------------QPRMWEP
PLRPAA---SCRGQAE---RLQAIQEERARS-----------------------------
----WSRG-------------TQEQASEQQARA-E----GALEPGCHK-HSVEVARRGSL
------PSHLQLADPQGS-------------------------WQEQLAAPEEGETKIAL
E---REKDDMETK--LLHLEDVVRALEK-----H--------------VDLRENDRLEFH
RLSE---ENTLLKN-------DLGRVRQELEA-----AESTHDAQRKEIEVLKKDKEKAC
SEMEV-LNRQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------NQNYKDQLSQLNVRVLQLGQEA-STHQAQN
EEHRVTIQMLTQSLEEVVRSGQQQ---------------SDQIQKLR--VELECLNQEHQ
SLQLPWSELTQTLEESQDQ-----------------------------------------
---------------------------------------------------VQGAHLRLR
QAQAQHLQEV--------RLVPQDRVAELHRLLSLQGE--------------QARRRLDA
Q-REEHEKQLKATEERVEEAEMILKNMEMLLQEKVDKLKEQFEKNTKSDLLLKELYVENA
HLVRALQATEEKQRGAEKQSRLLEEKVRALNKLVSRIAPAALSV----------------
---------------------------------------------------------
ENSRROP00000026175 (view gene)
------------------------------------------------------------
-------------------------MDEEENHYVSQLREVYSSCDTTGTGFLGRQELTQL
CLKLHLEQQLPVLLQTLLGNDHFARVNFEEFKEGFVAVLSSNADVRPSDEDS-SSLESAA
SSAIPPKYVNGSKWYGRRSR--PELCDTATE-ARHV-P------EQQTQASLKSQLWRSA
SLESVESLKSDEEADSTKESQNELFEAQGQ---LQTWDTEDFGSPQKSCSPSFDTPE
PF13499 // ENSGT00660000095541_0_Domain_1 (298, 358)
SQI
RGMWEELGVGSRGHLSEQELAVVCQSIGLRGLEKEELEDLFNKLDQDGDGKVSLEEFQLG
LFSHEPALLLESSTQVKLSKP----WSHYQ--VPEESGCHT-TTT-SSL-VSLCSSLRLF
SSIDDGSGFAFPDQILAMWTQ-------EGIQ--NGREI----LQSLDFSVDEKVNLLEL
TWALDNELMTVDSAVQQAALACYHQELSYQQ--GQVEQLVRER-DKARQDLE--------
-RAEKRNLEFVKEMDDCHSALEQLTEKK-IKHLEQGYRERLS----LLRSEVEAERELFW
EQAHKQRAALEWDMG-RLQAEEAGLREKLTLALKENSRLQKEIVEVAEKLSDSERLALKL
QKDLEFVLKDK-L---EPQSAE-LLAQEERFAAVLKEYELKCR-DLQDRNDELQ----AE
LEGLWARLPKNQHSPSWSPGG--------CRRQLSGLG-PAGIS----------------
------------------------FLGNS--VPVSIEVELMMEQVKEHYQDLRT--QLET
KVNYYEREIAALKRNFEKERKDMEQAHRREVSVLEGQKADLEERHEKSQEVIWGLQEQLQ
DTAHGPEPEW----MGL--APCCTQALCGLALQHQS-------HLRQIRR----------
------------------------------------------------------------
----------KAEAELSGELLGLRALPARRD--------------LTLELEE----PLQG
PLPHGSRRSEQLNLES----------ALK-------------------------LQPCAR
------EKGAQMCASLALEEEELELAGGKRVDGLS-------------QEAEMLGALPKE
GL-VAGS-GQEGA-----RGLLPLSPCYGERPL----------------AWLAPGDGGES
KEAAGAGSRC-RQAQ---------DTE---ATKSPDP--IPAP-----------------
----ASHGPSERWSHVQ-----PCGVDWGTV-----PEEPELFGVP--------------
------------------AGLEQPGSRELPLLGTEGDALQT------------QPRMWES
PLSPAA---SCRGQAE---RPQAIQEERGRS-----------------------------
----WSRG-------------TQGQASEEQARA-E----GALESACQE-HSVEVARRGSL
------PSHLQLADPRGS-------------------------GQEQLAAPEEGETKIAL
E---REKDDMETK--LLHLEDV-RALEK-----H--------------VDLRENDRLEFH
RLSE---ENALLKN-------DLGRVRQELEA-----AESTHNAQRKEIEVLKKDKEKAC
SEMEV-LNRQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------NQNYKDQLSQLNDRVLQLGQEA-STHQAQS
EEHRVTIQLLTQSLEEVACSGQQQ---------------SDQIQKLK--VELECLNQEYQ
SLQRSQSELTQTLEESQGQ-----------------------------------------
---------------------------------------------------VQGAHLRLR
QAQAQHSQEVR--------LVPQDRVAELQRLLNLQGE--------------QAGRRLDA
Q-REEHEKQLKATEERVEEVEMILKNMEMLLQEKVDELKEQFEKNRKSDLLLKELYVENA
HLVRALQATEEKQRDAEKQSRILEEKVRALNKLISRIAPAALSV----------------
---------------------------------------------------------
ENSMICP00000049940 (view gene)
------------------------------------------------------------
------------------------MDEEEESHYVSQLREVYSSCDTTGTGFLDREELTQL
CLKLHLEKQLPALLQTLLGNDHFARVNFEEFKEGFVAVLSSNAAVGPSEEES-STLETAA
SRAVPPKYVRGSKWYGRRSQ--PELCDAATK-ARCV-P------EQQTKANLKSQLRRSA
SLESVESLKSDEETESAKEPQNELFEAQGQ---LQTWGSEVFRSPQKSCSPSCDTPES
PF13499 // ENSGT00660000095541_0_Domain_1 (299, 358)
QV
QGIWEELGVGSSGHLNEQELAVVCQSVGLRGLQKEELKDLFNKLDRDGDGRVSLEEFQLG
LLSHEPTSLPESSTQVNLSRP----WSHYQ--VQEESCCHT-TTT-SSL-VSLCLGLRLF
SSIDDGTGFAFPEQVMAVWAQ-------EGIQ--NGREI----LQSLDFSVDEKVNLMEL
TWALDNELMTVDGAAQQAALACYRQELSYHQ--GQVEQLARER-DKARQDLE--------
-RVEKRNLEFVKEMDDCHSALEQLTEKK-IKHLEQGYRGRLN----LLRSEVEAERELFW
EQARRQRAALEHDVG-RLQAEEASLREKLTLALKENSRLQKEVVEVVEKLSDSEKLVLKL
QNDLELVLKDK-L---EPQSTE-LLAQEERFTAVLEEYEVKCR-DLQDCNDELQ----AQ
LEGLRAQLPASRQSSS---RG--------LGQRLSGQG-PAGIL----------------
------------------------LTGSS--APVSIETEIMMEQLKEHHRDLRI--QLET
KVNYYEREIEVMKRDFEKERKEMEQAHKREVGVLEEQKADLEMLHAKSQEVIQGLREQLQ
DLAHGPQPEQ----LGLALAPCCAQALSSLALRLEE-------ELHLRHRGQLQQIRREA
E--------------------------------------GELS--------RKLSWMEVQ
------------------------HAARCED--------------LSLQHQREKDELLQK
HLLQVSDLAEQLDQEKRRQAKREEEILTRCRKQQ----------------LK--LQELMS
------EEQAQICKSFALEKERLELAYRKQVEELA-------------QEVEALRALWKG
GLPVARS-EQESA-----HGLTPPRPGSGEQPP----------------ARLEPGDGGEM
GEPAGGGPGCVQAGCRDVAGQP-CCVD---GMQSPAAAPALVASRPSESLGVRENCQGLL
NTEEGASIPPERWSRVQ-----PHGVDRGAV-----PGESVPSARG--------------
-----------------PADLSQPEAQE-------GDVRPA------------QPQSWGP
QLSPAS---SGRGQAE---RPQAVLEERAGS-----------------------------
----RSRVLDSGG--------VQRPASEEQAGD-E------------------GARRGSL
------PSQ------------------------------------PQLAVPGEMEVDAAL
E---KEKNDVKTK--LLQLEDVVRALEK-----E--------------ADSREKDRIEFH
RLSE---ENTVLKN-------DLGRIQQELEA-----AESTNDAQRREIEALKRDKEKAC
FDMEE-LSKQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------SQKYKDQLSQLNHSVLQLREEA-STRQAQN
EESRTTIQLLTQSLEEAGRRGEQQ---------------SDQIQKLE--VELERVNQECQ
SLRLSQSELTKTLEESQDQ-----------------------------------------
---------------------------------------------------LQGAQQRLK
LAQSQHSQEVQQLQEQVGRLVPQDHVAKLQHLLSLQQTQQ--------------------
---EEHEKQLKATEERLGEAEVMLQHVEMLLQEKVEELKGQFEKNTKSNLLLKELYVENA
HLVKALQVTEQKQRGAEKKSRILEEKVQALNKLISKIAPAALSV----------------
---------------------------------------------------------
ENSOCUP00000005445 (view gene)
------------------------------------------------------------
--------------MARMDWACYGMDGEEESRYVLQLKELYSSCDTTGTGFLDRDELTQL
CRKLHLEKQLPVLLQTLFGQDHFARVNFEEFKEGFVAVLSSNAGVDPSDEES-SSLESVE
DLTVPPKYVSGSKWYGRRSQ--PELRDSE---AKYV-P------DQQAKASLKGQLCWSA
SLESVESLKSDEEAESAKEPQNELLEAQGQ---LETWDSEASGSPQKSCSPSFDTPGSRV
RGIWDALGVGSSGHLSKQELAVVCQSIGLQALDKE
PF00036 // ENSGT00660000095541_0_Domain_1 (336, 361)
ELEDLFNKLDRDGDGKVSLEEFQLGLFSHEPTSLPESCTLVKPSGP----WSHYQ--VPEESSCHT-PTA-SSL-VSVGSGPRLF
SNVDDGTGSAFPEQIITSWAQ-------EGIH--NGKEI----LQKQEAGTGQKVNLLEL
TWALDNELLTVDGVVQQAALACYRQELSYLQ--GQMEQLVQER-DKARQDLD--------
-RAEKRNLEFVKEMDDCHSALEQLTEKK-IKHLEQGYLERLS----LLRSEVEMERELVW
EQARRQKAALEQDVD-RLQVEEASLRQKLSLALKENSRLQKEIMEVVAKLSDSEKVALKL
QSDLELVLREK-L---EPQGTE-LLAQEERFAAVLKEYELKCR-DLQDHNDELQ----AE
LEGLRARLPESRHNPSRGPSG--------HRRRLPGRG-PAG------------------
----------------------ILFVGES--TPVSIETEIMMEQVKEHYQDLRV--QLET
KVSYYEREVEAMKRNFEKEKKEVELAHQLEVSALEEQRADLEALQAKAQEVIQGLQEQLR
DAAR------TPEPAQAGLAQCCAQALSGLA-------LRLEEELRLRHQDQLQQI----
----------------------------------RKEADLELS--------QKLSWLEA-
-----------------------QHATHCES--------------LALQHQHERDQLLQA
HLLQVHELTAQLDLEKERRQGREKEILVRCQKQQ----------------LK--LQEAMS
------EEQAQICKSFALEKEKLELTYRQQVQGLA-------------QEVQALQALLQG
RLTVAGV-EQEGA-----HGPAPLCPGIVERPP----------------AKWEPDAGGET
GELAGDGPGQSCCVDA---------------MQSPAAA----SASPLESISLRENSGGLL
DTEEVASVLSETESHVQ-----PCGTD------KG-------------------------
--A-------------------------------------------EADDLPAPPRMTEP
RLNS--------------------------------------------------------
-D--SLEEPAPAGNLGQ----------------ARG---GGNTLDTELQPCKGAAQ-REP
LLSHPG-----L----PDGQ--EQ-----------LAILG------------QTEDETV-
--LERERNDMKTK--LLQLEDVVRAL-------------------EAAADSRDHDRVEFQ
RLSE---ENTVLKT-------ELGRIQQELEA-----AENASEVQRKQMGVLKRDKEKAC
TEVEE-LNRQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------SQKYKDELSQLHHRVLQLEEEA-SAQQTQH
EKNCATIQLLTQRLEEAGHREAQQ---------------GEQLRTLE--GELERMTQECQ
SLRVSQSELAETLGESEQQ-----------------------------------------
---------------------------------------------------KINEFLKIM
QKHSASPSTV--PVGADRLLVPGARAALWQHLLGLQRVVGGQW-------------DTQQ
VDGRDVRNSWKLQRSGWGEVETRLRNVELLLQDKVRELTQQFERNTRSDLLLKELYVENA
HLVKALQVTEEKQRGAERRNLVLEEKVRALNRLVSRMAPAALAV----------------
---------------------------------------------------------
ENSCGRP00000003428 (view gene)
------------------------------------------------------------
------------------------MDNEEENHYVSQLRDVYRSCDTTGTGFLDQEELTQL
CTKLGLEEQLPALLQILLGNDRLARVNFEEFKDGFVAVLSSGSGVGPSDEDG-SSSESAT
SCAVPPKYVNGSKWYGRRSR--PELCDSATA-VKYG-S------EQQAKGSMKPPLRRSA
SLESVESLKSDEDAESAKEPQNELFEAQGQ---LRSWGCDVFGTSRKSCSPSFNTPE
PF13499 // ENSGT00660000095541_0_Domain_1 (298, 358)
NQV
QNIWHELGVGSSGHLDEQELAVVCRSIGLHGLEKQELEELFSKLDQDGDGRVSLEEFQLG
LFGHELPSLPESSSLVKPNRV----WSHYQ----EENGCHT-TTT-SSL-VSVCSGLRLF
SSVDDGSGFAFPEQIISAWAQ-------EGIQ--NGREI----LQSLDFGVDEKVNLLEL
TWALDNELLTVDGVIQQAALACYRQELNYHQ--GQVEQLVHER-DKVRQDLE--------
-KAEKRNLDFVREMDDCHSALEQLTEKK-IKHLEQEYRGRLS----LLRSEVEMERELFW
EQARRQRAVLEQDVG-RLQAEETSLREKLTLALKENSRLQKEIIEVVEKLSDSEKLVLRL
QSDLQFVLKDK-L---EPQNVE-LLAQEEKFTAILKEYELKCR-DLQDRNDELQ----TE
LEGLRMRLPRSRQSPSGTPGT--------HRRRLPGRG-PAGNL----------------
------------------------FVGDS--TPVSLETEIMVEQMKEHYQELRM--QLET
KVNYYEKEIEVMKRNFEKDKKEMEQAFQLEVSMLEGQKADLEALYAKSQEVILGLKEQLQ
DAARIPEPA------PASLAHCCAQALCTLAQRLEV-------EMHLRHQDQLQQIRR--
------------------------------------EAEEELN--------QKLSWLE--
----------------------AQHAACCES--------------LSLQHQCEKDQLLQT
HLQRVKDLAAQLDLEKGWREEREQEVLAHCRRQQ----------------LK--LQAVMS
------EEQARICRSFTLEKEKLEQTYREQVEGLV-------------QEADELRALLKN
GTTVVSD-QQERIPSSMTLGP-----DSRQQPP----------------AQQAVSPDGRT
RAPAEECPGQGRAKGTDLPGQP-CSIV---AM--PSPPPTLVPRRSLESLGVRDNHQCPL
SAEEGAVPK-----------------------------EPEPPARALI------------
-----------------------G--------------------QGQKLPLPTQPQMSEP
WLGPAA------------------------------------------------------
----LDRKPAPVG--------VQGQASERPTGNAE----GV--QETWLQYREETS-----
-RMRPSPPCSELSDSQEA-------------------------KFKHLAAPGETEAKVPL
VMSEREMNDVKT--KLLQLEDVVRALEK-----E--------------ADTRENYRTKFQ
RLSE---ENFVLKS-------DLGRIQLELET-----SVSRNETQRQEIEVLKRDKEQAC
FDLEE-LGTQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------AQRYKEELSQLNCRILQLEGDT-SALHTQK
EENRVAIQLLMQKLEEAGCREQQQ---------------GVQIQNLE--VELERVNEECQ
GLRLSQAELTETLKESQGQ-----------------------------------------
---------------------------------------------------LHSVQLTLE
NAQCQHNRIVQCLQEQMGQLVPGAHVAELQHLLSLEEE---------------AGRRLNA
Q-QEEYKQQLKAKEDQVEDAEARLRNVEWLLQEKVEELREQLEKNTRSDLLLKELYVENA
HLMKAVQLTEEKQRGAERKNCILEEKVRALNKLISKMAPASLSV----------------
---------------------------------------------------------
ENSTSYP00000027443 (view gene)
------------------------------------------------------------
-------------------------MDEEEDHYVAQLRKVYDSCDTTGTGFLDHEELTQL
CLKLHLEKQLPILLQTLLGDDHFARVNFEEFKEGFVALLSSNAGTGPLDDEG-NSLESAA
SCVVPPKYVSGSKWYGRRSR--PERCDTATE-ARHT-P------EQQTRASLRSQLRRSA
SLESVESLKSDEEAESTKEPQNESFEAQGQ---MQTWGLEAFGSPRKPCSPSFASPES
PF13499 // ENSGT00660000095541_0_Domain_1 (299, 358)
QV
RGIWEELGVGSSGHLSEQELAVVCQSVGLRGLKKEELEDLFNKLDQDGDGRVSLEEFQLG
LFSHEPTSLLDSSTQVKPSRP----WSHYQ--VPEETGGHTTTTT-SSL-VSLCSGLRLF
SSIDDGTGFAFPEQVLAMWAQ-------EGIP--NGSEI----LQSLDFSVDEKVNLLEL
TWALDSELMAVDGAIQQAALACYRQELSYHQ--EQTEQLARER-DKARQDLE--------
-RTEKRNLDFVKEMDDCHSALEQLTEKK-IQHLEQGYRGRLT----LLRSEVEAERELFW
EQARRQKAVLERDVG-RLQTEEASLRQKLTLALKENSRLQKEILEVVGKLSDSEKLVLKL
QGDLEFVLKDK-L---EPQSEE-FLVQEEQFAAVLREYELKCR-DLQDHNDELE----VA
LEGLRARLPENGHNLAGRS-------------PALGRA-AKGI-----------------
-----------------------SSAGDC--APVSVDTEIMMEQVKEHCHNLSI--QLET
KVNHYEREIEVIKRNFEKERKEMEQAHEREVHALEGQRADLEKLHEKSREVIQGLQEQLR
DTAPQP-------------GPCCARALSGLALQLEEELSL-------------RRARCGV
LSR-----YAAN--------------------QPE-------------------------
------------------------------------------------------------
--------------------------TPAWLRRM----------------AT--LTIL--
---------SSRCGPITMQ-----------------------------------------
------T-TEGCTPALAEMGT---------------------------------------
--------------------------------------PFLV------------------
-------GGSGEWPEHM-----GCGKGGLA------------------------------
------------------------------------------------------------
---------------L---LPPSFF-----------------------------------
--------PQAMG--------AQG----AQCFCSG----RYMP-----------------
--------------PPGD-------------------------GTT--------------
---------------SSPLSLGVY---------P--------------YFVNFSCRIEFH
RLAE---ENTVLKN-------DLGKTQQELEA-----AESANDTQRKEIEVLKKDKEKAC
SDMEE-LNKQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------NQKYKNELSQLKHRVLQLAEAA-STPQARE
EEHHVSIQQLTRSLEAARQQGVQQ---------------RDQIQKLE--AELECMNQECQ
SLRLSQSELMKTVRESRDQV----------------------------------------
----------------------------------------------------GDAHQRLR
LAQAQHAEESQ-------RLVPWERVAELQRLLHLQGEQ--------------AMRRLDA
-QREEHEKQLEATVTRLGEAETALQSAETLLQETMDELKAQLERNTKSDLLLKELYVENA
HLVKALQVTEEKQRGAEKKGRVLEEKVRALNKLIGKIAPAALSV----------------
---------------------------------------------------------
ENSPTRP00000083627 (view gene)
------------------------------------------------------------
-----------------------MGWMKKENHYVSQLRKSTGSCDTTRTGFLDRQTS---
-----PEQQLPVLLQTLLGNDHFASVNFEEFKEGFVAVLSSNAGVRPSDEDS-SSLESAA
SSAIPPKYVNGSKWYGRRSR--PELCDAATE-ARHV-P------EQQTQASLKSHLWRSA
SLESVESLKSDEEAESTKEAQNELFEAQGQ---LQTWDSEDFGSPQKSCSPSFDTPE
PF13499 // ENSGT00660000095541_0_Domain_1 (298, 358)
SQI
RGMWEELGVGSSGHLSEQELAVVCQSVGLQGLEKEELEDLFNKLDQDGDGKVSLEEFQLG
LFSHEPALLLESSTRVKPGKA----WSHYQ--VPEESGCHT-TTT-SSL-VSLCSSLRLF
SSIDDGSGFAFPDQVLAMWTQ-------EGIQ--NGREI----LQSLDFSVDEKVNLLEL
TWALDNELMTVDSAVQQAALACYHQELSYQQ--GQVEQLARER-DKARQDLE--------
-RAEKRNLEFVKEMDDCHSTLEQLTEKK-IKHLEQGYRERLS----LLRSEVEAERELFW
EQAHRQRAALEWDVG-RLQAEEAGLREKLTLALKENSRLQKEIVEVVEKLSDSERLALKL
QKDLEFVLKDK-L---EPQSAE-LLAQEERFAAVLKEYELKCR-DLQDRNDELQ----AE
LEGLWARLPKNRHSPSWSPDG--------RRRQLSGLG-PAGIS----------------
------------------------FLGNS--APVSIETELMMEQVKEHYQDLRT--QLET
KVNYYEREIAALKRNFEKERKDMEQARRREVSVLEGQKADLEELHEKSQEVIWGLQEQLQ
DTARGPEPEQ----MGL--APCCTQALCGLALRHQS-------HLQQIRR----------
------------------------------------------------------------
----------EAEAELSGEPSGLGALPARRD--------------LTLELEE----PPEG
PLPRGSQRSEQLELER----------ALK-------------------------LQPCVS
------EKRAQMCVLLALEEEELELARGKRVDGPS-------------REAEMLQALPKD
GL-VAGS-GREGA-----RGLLPLRPGCGERPL----------------AWLAPGDGRES
EEAAGAGPRGGRQAQ---------DTE---ATQSPA--PAPAP-----------------
----ASHGPSERWSRMQ-----PCGVDGDIV-----PKEPEPFGAS--------------
-----------------AAGLEQPGARELPLLGTERDASQT------------QPRMWEP
PLRPAA---SCRGRAE---RPQAIQEERARS-----------------------------
----WSRG-------------TQEQASEQQARA-E----GALEPGCHK-HSVEVARRGSL
------PSHLQVADPQGS-------------------------WQEQLAAPKEGETKIAL
E---REKDDMETK--LLHLEDVVRALEK-----H--------------VDLRENDRLEFH
RLSE---ENTLLKN-------DLGRVRQELEA-----AESTHDAQRKEIEVLKKDKEKAC
SEMEV-LNRQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------NQNYKDQLSQLNVRVLQLGQEA-STHQAQN
EEHRVTIQMLTQSLEEVVCSGQQQ---------------SDQIQKLR--VELECLNQEHQ
SLQLPWSELTQTLEESQDQ-----------------------------------------
---------------------------------------------------VQGAHLRLR
QAQAQHLQEVRQLQEQMGRLVPQDRVAELQRLLSLQGE--------------QARRCLDA
Q-REEHEKQLKATEERVEEAEMILKNMEMLLQEKVDKLKEQFEKNTKSDLLLKELYVENA
HLVRALQATEEKQRGAEKQSRLLEEKVRALNKLVSRIAPAALSV----------------
---------------------------------------------------------
ENSGGOP00000026802 (view gene)
------------------------------------------------------------
-------------------------MDEEENHYVSQLREVYSSCDTTGTGFLDRQELTQL
CLKLHLEQQLPVLLQTLLGNDHFARVNFEEFKEGFVAVLSSNAGVRPSDEDS-SSLESAA
SSAIPPKCVNGSKWYGRRSR--PELCDAATE-ARRV-P------EQQTQASLKSHLWHSA
SLESVESLKSDEEAESTKEAQNELFEAQGQ---LQTWDSEDFGSPQKSCSPSFDTPE
PF13499 // ENSGT00660000095541_0_Domain_1 (298, 358)
SQI
RGMWEELGVGSSGHLSEQELAVVCQSVGLQGLEKEELEDLFNKLDQDGDGKVSLEEFQLG
LFSHEPALLLESSTRVKPSKA----WSHYQ--VPEESGCHT-TTT-SSL-VSLCSSLRLF
SSIDDGSGFAFPDQVLAMWTQ-------EGIQ--NGREI----LQSLDFSVDEKVNLLEL
TWALDNELMTVDSAVQQAALACYHQELSYQQ--GQVEQLARER-DKARQDLE--------
-RAEKRNLEFVKEMDDCHSTLEQLTEKK-IKHLEQGYRERLS----LLRSEVEVERELFW
EQAHRQRAALEWDVG-RLQAEEAGLREKLTLALKENSRLQKEIVEVVEKLSDSERLALKL
QKDLEFVLKDK-L---EPQSAE-LLAQEERFAAVLKEYELKCR-DLQDRNDELQ----AE
LEGLWARLPKNRHSPSWSPDG--------HRRQLSGLG-PAGIS----------------
------------------------FLGNS--APVSIETELMMEQVKEHYRDLRT--QLET
KVNYYEREIAALKRNFEKETKDMEQARRREVSVLEGQKADLEELHEKSQEVIWGLQEQLQ
DTARSPEPEQ----MGL--APCCTQALCGLDLQHQS-------HLQQIRK----------
------------------------------------------------------------
----------EAEAELSGEPSGLGALLARRD--------------LTLELEE----PPPG
PLPRGSQRSEQLELER----------ALK-------------------------LQPCAS
------EKRAQMCVSLALEEEELELARGKRVDGPS-------------REAEMLQALPKD
GL-VAGS-GREGA-----RGLLPLRPGCGERPL----------------PWLAPGDERES
EEAAGAGPRC-RQAQ---------DTE---ATQSPA--PAPAP-----------------
----ASHGPSERWSRMQ-----PCEVDGDIV-----PKEPEPFGAS--------------
-----------------AAGLEQPGARELPLLGTERDASQT------------QPRMWEP
PLRPAA---SCRGRAE---RPQAIQEERARS-----------------------------
----WSRG-------------TQEQASEQQARA-E----GAVEPGCHK-HSVEVARRGSL
------PSHLQLADPQGS-------------------------WQEQLAAPEEGETKIAL
E---REKDDMETK--LLHLEDVLRALEK-----H--------------VDLRESDRLEFH
RLSE---ENTLLKN-------DLGRVRQELEA-----AESTHDAQRKEIEVLKKDKEKAC
SEMEV-LNRQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------NQNYKDQLSQLNVRVLQLGQEA-STHKAQN
EEHRVTIQMLTQSLEEVVRSGQQQ---------------SDQIQKLR--VELECLNQEHQ
SLQLPWSELTQTLEESQDQ-----------------------------------------
---------------------------------------------------VQGAHLRLR
QAQAQHLQEVRQLQEQMGRLVPQDHVAELQRLLSLQGE--------------QARRRLDA
Q-REEHEKQLKATEERVEEAEMILKNMEMLLQEKVDKLKEQFEKNTKSDLLLKELYVENA
HLVRALQATEEKQRGAEKQSRLLEEKVRALNKLVSRIAPAALSV----------------
---------------------------------------------------------
ENSPPAP00000017533 (view gene)
------------------------------------------------------------
------MKLAGPWRRASDFQAC-YGMDEEENHYVSQLREVYSSCDTTGTGFLDRQELTQL
CLKLHLEQQLPVLLQTLLGNDHFARVNFEEFKEGFVAVLSSNAGVRPSDEDS-SSLESAA
SSAIPPKYVNGSKWYGRRSR--PELCDAATE-ARHV-P------EQQTQASLKSHLWRSA
SLESVESLKSDEEAESTKEAQNELFEAQGQ---LQTWDSEDFGSPQKSCSPSFDTPE
PF13499 // ENSGT00660000095541_0_Domain_1 (298, 358)
SQI
RGMWEELGVGSSGHLSEQELAVVCQSVGLQGLEKEELEDLFNKLDQDGDGKVSLEEFQLG
LFSHEPALLLESSTRVKPGKA----WSHYQ--VPEESGCHT-TTT-SSL-VSLCSSLRLF
SSIDDGSGFAFPDQVLAMWTQ-------EGIQ--NGREI----LQSLDFSVDEKVNLLEL
TWALDNELMTVDSAVQQAALACYHQELSYQQ--GQVEQLARER-DKARQDLE--------
-RAEKRNLEFVKEMDDCHSTLEQLTEKK-IKHLEQGYRERLS----LLRSEVEAERELFW
EQAHRQRAALEWDVG-RLQAEEAGLREKLTLALKENSRLQKEIVEVVEKLSDSERLALKL
QKDLEFVLKDK-L---EPQSAE-LLAQEERFAAVLKEYELKCR-DLQDRNDELQ----AE
LEGLWARLPKNRHSPSWSPDG--------RRRQLSGLG-PAGIS----------------
------------------------FLGNS--APVSIETELMMEQVKEHYQDLRT--QLET
KVNYYEREIAALKRNFEKERKDMEQARRREVSVLEGQKADLEELHEKSQEVIWGLQEQLQ
DTARGPEPEQ----MGL--APCCTQALCGLALQHQS-------HLQQIRRGSAWSSQTGC
CSA-----GTHCPVESIGPGLEEDGPSHHTGRATPSLTESHLS--------GRMRWLTPV
ISTLGGQGGREAEAELSGEPSGLGALPARRD--------------LTLELEE----PPEG
PLPRGSQRSEQLELER----------ALK-------------------------LQPCVS
------EKHAQMCVSLALEEEELELARGKRVDGPS-------------REAEMLQALPKD
GL-VAGS-GREGA-----RGLLPLRPGCGERPL----------------AWLAPGDGRES
EEAAGAGPRGGRQAQ---------DTE---ATQSPA--PAPAP-----------------
----ASHGPSERWSRMQ-----PCGVDGDIV-----PKEPEPFGAS--------------
-----------------AAGLEQPGARELPLLGTERDASQT------------QPRMWEP
PLRPAA---SCRGRAE---RPQAIQEERARS-----------------------------
----WSRG-------------TQEQASEQQARA-E----GALEPGCHK-HSVEVARRGSL
------PSHLQVADPQGS-------------------------WQEQLAAPKEGETKIAL
E---REKDDMETK--LLHLEDVVRALEK-----H--------------VDLRENDRLEFH
RLSE---ENTLLKN-------DLGRVRQELEA-----AESTHDAQRKEIEVLKKDKEKAC
SEMEV-LNRQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------NQNYKDQLSQLNVRVLQLGQEA-STHQAQN
EEHRVTIQMLTQSLEEVVRSGQQQ---------------SDQIQKLR--VELECLNQEHQ
SLQLPWSELTQTLEESQDQ-----------------------------------------
---------------------------------------------------VQGAHLRLR
QAQAQHLQEVRQLQEQMGRLVPQDRVAELQRLLSLQGE--------------QARRCLDA
Q-REEHEKQLKATEERVEEAEMILKNMEMLLQEKVDKLKEQFEKNTKSDLLLKELYVENA
HLVRALQATEEKQRGAEKQSRLLEEKVRALNKLVSRIAPAALSVFATRWSQILGAVSKEQ
LRLLELLVEGAYTHSRKGFTHWERQPSEKGGEAPSPGTLSVDSGMMVFIANAQSFSH
ENSSTOP00000021764 (view gene)
------------------------------------------------------------
-------------------------MDEEENHYVSQLREVYNSCDTTGTGFLDRDELTQL
CCKLHLEKQLPILLQTLLGSNHFARVNFEEFKEGFVAVLSSTVGVGPLDEES-SSLESAA
SCAVPPKYVSGSKWYGRRSR--PERWDSAAE-AKYV-P------EQQAKASLKSQLRRSA
SLESVESLKSDEEAESAKETQNELFEAQGQ---LQTWSSEVFGSSQKPCSPSLDSPERQV
PF13499 // ENSGT00660000095541_0_Domain_1 (301, 358)
RGIWEELGVGSSGHLNEQELAVVCQSIGLHGLEKEELEDLFNKLDQDGDGRVSLAEFQVG
LFSHEPASLLESSTLLKSSKP----WSHCQ--VPTENGCHPTPTT-SSL-VSVCSGLRFF
SNVDDGSGFAFPEQIITTWAQ-------EGIQ--NGREI----LQSLDFSMDEKVNLLEL
TWALDNELLTVDGVVQQAALACYRQELSYHL--TQVEQLVQER-DKARQDLE--------
-RAEKRNLEFVKEMDDCHSALEQLTEKK-IKNLEEGYRGRLS----LLRSEVEMEQELIW
EQACRQKAALEQDVS-RLRAEEASLHEKLTLALKENSRLQKEIVEVVEKLSDSEKLVLKL
QNDLEFVLKDK-L---EPQNME-LPSQEEQFAVLLKEYELKCR-DLQDCNDELQ----AE
LEGLRAQLPESWQSPSEDLGT--------HGQRVSGRG-PAGIL----------------
------------------------SVGDS--TPVNIETEIMMEQVKERYQDLKI--QLET
KVNYYEMEIEVMQRNFQKERKEMEQAHQLEVCTLESQKADLEALYTKSQEVILGLREQLQ
DATHGPEPG------QTGMEQCCAQVLCGLAQQLKE-------ELQLRHQNQLQQIRK--
------------------------------------EAEDVLT--------QKLSWLE--
----------------------AQHVARCER--------------LSLQHQSEKDRLLQT
HLWRLRELVAQLDLEKEQQGEREKEILTRCQKQQ----------------MK--LQAAMS
------EEQAQICRSMALEKEKLEHTYRKQVEGLA-------------QEAEALRALLND
GTTVTGS-EQGGIPGLT-----SLCLGGEQPLA----------------QLDLNTDGE-M
RELAGDTPGQRRTEGRDLPSQPRCVDT---T---QSPTPVLVSRESLKSFSLGENCQGPL
SAGEVASVPLEKRSY-----MQPCEGNKGAI-----AEEPVPSARA--------------
-----------------PMGLGQADAQELPVLATEGIAWLA---Q---------PQTLEP
WHSPAT------------------LEEQASS-----------------------------
----LSRDPAPES--------GPGQVFEGMSRGGD----DALE--TWLEHSQEAARWGSS
L------LHSKLPDPQQP-------------------------GLECLTALEETEAKMAL
E---KEKSDMRNKLL--QLEDVVRAL-----EKE--------------ADSRENDRIEFQ
RISE---ENMLLKN-------DLGKIQQELEA-----AESTNDAQRKEIEDLKRDKEKTC
LEMEE-LNIQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------IQKYKDELSQLNCRVLQLEGDA-STHQALK
EKNHIAIQLLRQRLEEARRREEQQ---------------DNQIKKLE--VELQHVNQECQ
NLRLSQSELKVSLEESQEQ-----------------------------------------
---------------------------------------------------LHSAHLRLK
AAQSQHAQEVQHLQEQKSQLVPRARVTELQHLLSVQEK--------------EAGRLLDA
-QREEHERQMKTKEGQAKEAEMCLQNVEWLLQEKVDELREQFKKNTKSDLLLKELYVENA
HLMKALQVTEEKQRGAESKNRILEEKVHALNKLINKIALASLSV----------------
---------------------------------------------------------
ENSCANP00000022283 (view gene)
------------------------------------------------------------
-------------------------MDEEENHYVSQLREVYSSCDTTGTGFLDRQELTQL
CLKLHLEQQLPVLLQTLLGNDHFARVNFEEFKEGFVAVLSSNAGVRPSDEDS-SSLESAA
SSAIPPKYVNGSKWYGRRSR--PELCDTTTE-ARHV-P------EQQTQTSLKSQLWRSA
SLDSVESLKSDEEAESAKEAQNELFEAQGQ---LQTWDTEDFGSPQKSCSPSFDTPE
PF13499 // ENSGT00660000095541_0_Domain_1 (298, 358)
SQI
RGMWKELGVGSRGHLSEQELAVVCQSVGLQGLEKEELEDLFNKLDQDGDGKVSLEEFQLG
LFSHEPALLLESSTQVKLSKP----WSHYQ--VPEESGCHT-TTT-SSL-VSLCSSLRLF
SSIDDGSGFAFPDQVLAMWTQ-------EGIQ--NGREI----LQSLDFSVDEKVNLLEL
TWALDNELMTVDSAVQQAALACYHQELSYQQ--GQVEQLVRER-DKARQDLE--------
-RAEKRNLEFVKEMDDCHSALEQLTEKK-IKHLEQGYRERLS----FLRSEVEAERELFW
EQAHKQRAALEWDVG-RLQAEEAGLREKLTLALKENSRLQKEIVEVAEKLSDSERLALKL
QKDLEFVLKDK-L---EPQSAE-LLAQEERFAAVLKEYELKCR-DLQDRNDELQ----AE
LEGLWARLPKNQHSPSWSPGG--------CRRQLSGLG-PAGIS----------------
------------------------FLGNS--VPVSIETELMMEQVKEHYQDLRT--QLET
KVNYYEREIAALKRNFEKERKDAEQARRCEVSVLEGQKADLEERHEKSQEVIWGLQEQLQ
DTAHGPEPEW----MGL--APCCTQALCGLALRHQS-------HLRQIRR----------
------------------------------------------------------------
----------EAEAELSGELLGLRALPARRD--------------LTLELEE----PLQG
PLPHGSRRSEQLNLES----------ALK-------------------------LQPCAS
------EKGAQMCASLALEEEELELAGGKQVDGLS-------------REAEMLGALPKE
GL-VAGS-GQEGA-----RGLLSLSPCYGERPL----------------AWLAPGDGGES
EEAAGAGFRC-RQAQ---------DAE---ATKSPGP--APAP-----------------
----ASHGPSERWSHVQ-----PCGVDWGTV-----PEEPELFGVP--------------
------------------AGLEQPGPRELPLLGTEGDASQT------------QPRMWES
PLSPAA---SCRGQAE---SPQAIQEERERS-----------------------------
----WSRG-------------TQGQASEEQARA-E----GALESAYQE-HSVEVARRGSL
------PPHLQLADRRGS-------------------------GQEQLAAPEEGETKIAL
E---REKDDMETK--LLHLEDVVRALEK-----H--------------VDLRENDRLEFH
RLSK---ENTLLKN-------DLGRVRQELEA-----AESTHNAQRKEIEVLKKDKEKAC
SEMEV-LNRQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------NQNYKDQLSQLNDRVLQLGQEA-STHQAQS
EEHRVTIQLLTQSLEEMACSGQQQ---------------SDQIQKLK--VELECLNQEYQ
SLQLSQSELTQTLEESQGQ-----------------------------------------
---------------------------------------------------VQGAHLRLR
QAQAQHSQEVR--------LVPQDRVAELQRLLNLQGE--------------QAGRRLDA
Q-REEHEKQLKATEERVEEAEMILKNMEMLLQEKVDELKEQFEKNTKSDLLLKELYVENA
HLVRALQATEEKQRDAEKQSRILEEKVRALNKLVSRIAPAALSV----------------
---------------------------------------------------------
ENSCGRP00001000566 (view gene)
------------------------------------------------------------
------------------------MDNEEENHYVSQLRDVYRSCDTTGTGFLDQEELTQL
CTKLGLEEQLPALLQILLGNDRLARVNFEEFKDGFVAVLSSGSGVGPSDEDG-SSSESAT
SCAVPPKYVNGSKWYGRRSR--PELCDSATA-VKYG-S------EQQAKGSMKPPLRRSA
SLESVESLKSDEDAESAKEPQNELFEAQGQ---LRSWGCDVFGTSRKSCSPSFNTPE
PF13499 // ENSGT00660000095541_0_Domain_1 (298, 358)
NQV
QNIWHELGVGSSGHLDEQELAVVCRSIGLHGLEKQELEELFSKLDQDGDGRVSLEEFQLG
LFGHELPSLPESSSLVKPNRV----WSHYQ----EENGCHT-TTT-SSL-VSVCSGLRLF
SSVDDGSGFAFPEQIISAWAQ-------EGIQ--NGREI----LQSLDFGVDEKVNLLEL
TWALDNELLTVDGVIQQAALACYRQELNYHQ--GQVEQLVHER-DKVRQDLE--------
-KAEKRNLDFVREMDDCHSALEQLTEKK-IKHLEQEYRGRLS----LLRSEVEMERELFW
EQARRQRAVLEQDVG-RLQAEETSLREKLTLALKENSRLQKEIIEVVEKLSDSEKLVLRL
QSDLQFVLKDK-L---EPQNVE-LLAQEEKFTAILKEYELKCR-DLQDRNDELQ----TE
LEGLRMRLPRSRQSPSGTPGT--------HRRRLPGRG-PAGNL----------------
------------------------FVGDS--TPVSLETEIMVEQMKEHYQELRM--QLET
KVNYYEKEIEVMKRNFEKDKKEMEQAFQLEVSMLEGQKADLEALYAKSQEVILGLKEQLQ
DAARIPEPA------PASLAHCCAQALCTLAQRLEV-------EMHLRHQDQLQQIRR--
------------------------------------EAEEELN--------QKLSWLE--
----------------------AQHAACCES--------------LSLQHQCEKDQLLQT
HLQRVKDLAAQLDLEKGWREEREQEVLAHCRRQQ----------------LK--LQAVMS
------EEQARICRSFTLEKEKLEQTYREQVEGLV-------------QEADELRALLKN
GTTVVSD-QQERIPSSMTLGP-----DSRQQPP----------------AQQAVSPDGRT
RAPAEECPGQGRAKGTDLPGQP-CSIV---AM--PSPPPTLVPRRSLESLGVRDNHQCPL
SAEEGAVPK-----------------------------EPEPPARALI------------
-----------------------G--------------------QGQKLPLPTQPQMSEP
WLGPAA------------------------------------------------------
----LDRKPAPVG--------VQGQASERPTGNAE----GV--QETWLQYREETS-----
-RMRPSPPCSELSDSQEA-------------------------KFKHLAAPGETEAKVPL
VMSEREMNDVKT--KLLQLEDVVRALEK-----E--------------ADTRENYRTKFQ
RLSE---ENFVLKS-------DLGRIQLELET-----SVSRNETQRQEIEVLKRDKEQAC
FDLEE-LGTQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------AQRYKEELSQLNCRILQLEGDT-SALHTQK
EENRVAIQLLMQKLEEAGCREQQQ---------------GVQIQNLE--VELERVNEECQ
GLRLSQAELTETLKESQGQ-----------------------------------------
---------------------------------------------------LHSVQLTLE
NAQCQHNRIVQCLQEQMGQLVPGAHVAELQHLLSLEEE---------------AGRRLNA
Q-QEEYKQQLKAKEDQVEDAEARLRNVEWLLQEKVEELREQLEKNTRSDLLLKELYVENA
HLMKAVQLTEEKQRGAERKNCILEEKVRALNKLISKMAPASLSV----------------
---------------------------------------------------------
ENSTTRP00000001543 (view gene)
------------------------------------------------------------
------------------------MDEEEESHYVS
PF00036 // ENSGT00660000095541_0_Domain_0 (96, 124)
QLRDVYSSCDTTGTGFLDREELTQL
CLKLHLEKQLPVLLQTLLGNDHFARVNFEEFKDGFVAVLSSQAGVGSSDEDS-GSLEPGA
PRAVPPKYVSGSKWYGRRSW--PEPCDAAGG-APCL-P------EQPTRPSVRSQLRRSA
SLESVESLKSDEEAESAKDPQNELFEAKGQ---LPTWGSEVFGSLRKPCSSSFDTPESQV
RGLWEELGVGSSGHLTEQELALLCQSVGLQGLEKEXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXX-XXXXXXXXXXXXXXX----XXXXX--XVPEEGGGQ-TTT-SSL-VSVSSGPRLL
CSVDDGAGFAFPEQIITLWAQ-------EGVH--NGREV----LQSLDFSVDEKVNLLEL
AWALENELMMVGGATQQAALACYRQELSFRQ--GQVEQMARER-DKARQDLE--------
-KAEQRHLEFVKETDDLHSALEQLAEEK-VRRLEQGYRERLS----LLRSEVEVERELSW
EQARRQRAALEEDLQ-RLRAEEASLRQKLSLALKENSRLQKEMIEVVEKLAESEKLVLKL
QNDLEFVLKDK-L---EPQSTE-LLTQEERFSEVLKEYKLKCR-DLQDCNDELQ----AA
LEGLRAQLPPSRHGQLP-----------------PGHG-PAXXXXXXXXX--XXXXXXX-
--------------XXXXXXXXXXXXXXX--XXX-------------------------X
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXEVGVLQGQKAELATLHQESQEAVKL-RDRLQ
V-----------------------RELERGHARSAQERDQLRRLQRVXXXXXXXXX----
----------------------------------XXXXXXXXXXXXXXXXXXXXXXXXX-
-----------------------XXXXXXXX--------------XXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXX------------XXXXXX----------------X----XXXXX
------XXXXXXXXXXXXXXXXXXXXXXXXXX-------------------XXXXXXXX-
----XXX-XXXXX-----XX-XXXXXXXXXXXXXXXXXXX--------XXXXXXXXXXXX
XXX--------XXXXXXXXX-------XXXXXXXXXXX----X------XXXXXXXXXX-
---XXXXXXXX-XXX-X-----XX-XX-----XXXXXXXXXXXX--XXXXXXXXXXXX--
XXX-------------------------------------------XXXXXXXX-----X
XXXXXX---------------------XXXXX----------------------------
-XXXXXXXXXXXXX--XXXXX--------------------XXXXXXXXXXXXXXXXXXX
XXXXXXX------XXXXXXX--XXXXXXXXX---X----XXX---XXXXXXXXXXXX-X-
--XXXXXXXXXXX--XXXX--------------------------XXXX----------X
XXXX---XXXXXXX-------XXXXXXXXXXX-----XXXXXXXXXEEVEFLKREKEKAC
SEMEE-LCTQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------SQKCKDELSQLNHRVLQLGEEA-STHQAQS
KKNHITVQLLTQRLEEAACREELQ---------------DNQIQKLE--LELEHVTQERQ
SLRLAQSQLREALEESRDQ-----------------------------------------
---------------------------------------------------LYGATTRLK
LAQSQH------LQEQMEQLVPRGRVAELQQLLEGERQAARQL---------------QV
EEWEAHEKQLKAAEERVEEVEMILKNVEALLQEKVRELKEQFEKNTRSDLLLKELYVENA
HLTKALQVTEEKQRGAEKKSRFLEERVRALNKLLSKIASASLSV----------------
---------------------------------------------------------
ENSSSCP00000007598 (view gene)
------------MFLVSLCLTSTERTWCPSQKPSCSPD---LGR-------MVGSWEPGR
L---VRAAARGYHCLPFPHAACDGMDEEEESHYVSKLRDLYHSCDTTGTGFLDREELTQL
CLKLHLEKQLPILLQTLLGNDHFARVNFEEFKDGFVAVLSSQASVSSSDEGS-VALESAA
PRAVPPKYVSGSKWYGRRSR--PEPCVSAGG-ASCL-P------EQPARPSVRNQLRRSA
SLESVESLKSDEEAESTKEPQIELFEAQGQ---LPAWS-EVFGNPRKPCSPSSDTPE
PF13499 // ENSGT00660000095541_0_Domain_1 (298, 358)
SRV
RDIWEELGVGSSGHLTEQELALVCQSVGLRGLQKEELEDLFNKLDRDGDGRVSLEEFQLG
LFNHGPPQLLESSILVKPNGS----WSHYQ--VSEEGGGQT---TTSSL-VSVCSGPRLF
CSVDDGTGFAFPEQIIALWAQ-------EGVH--NGREV----LQSLDFSVDEKVNLLEL
TWALENELLTVDGAAQQAALACYRQELSFCQ--GKVEQLARER-DKARQDLE--------
-KAEQRNLEFVKEMDDCHCTLEQLTEEK-IRRLEQGYRGRLS----LLRTEAEVERELFW
EQARRQRAALEGDLQ-HLRAEETSLREKLTLALKENSRLQKEIIEVVEKLSESEKLVLKL
QNDLEFVLRDK-L---EPQSTE-LLAQEERFSGVLKEYELKCR-DLQDHNDELQ----AA
LEGLRAQLPPSRHSLPHT-QL--------DGQLPPGHS-PAGIVT---------------
------------------------FVGDS--IPVSLETEIMLEQVKERYQDLEI--QLET
KVNYYEKEIEGMKRDFAEERQEMERAFKLEVSVLQDQKAKLVALHSESQEVVRGLRDRLQ
RIVELDQGHA-----------RLT---QE-RDRLE-------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------------------------------EALQRVRIELD
KLSE---ENTLLKN-------ELGRIQQELEA-----AKRTEDAQRKEIEVLKREKEKAC
SEMEE-LCTQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------S-----------------------------
----------------------------------------KQIQKLE--LELGHVTEERQ
SLRLAQSQLREALEESQDQ-----------------------------------------
---------------------------------------------------L-------S
LARSQHALEVQRLQEQVGQLVPQGRVAELQRLLDGERQAARQLRE-------EEGRRLET
Q-REAHEKQLKATEERVEEMEMILKNVEVLLQEKVGELKEQFEKNTRSDLLLKELYVENA
QLTKALQVTEERQRGAEKKSRFLEEKVRALNKLLSKIASAALSV----------------
---------------------------------------------------------
ENSCLAP00000016910 (view gene)
------------------------------------------------------------
MGKPPRLH----CGETAELPAC-YGMDEEDNYYISQLKDVYNSCDATGTGFLDREELTQL
CRKLHLEKQLPALLQTLLGDDLIARVTFEEFKDGLVAVLSSKAGGDPGREEA-GSLQSAA
SCAVPPKYVSGSKRYGRRSQ--PERRISATE-ATCG-P------EQQARAGLKSQLRRAA
SLESVESLKSDEEAESPREPQDELFEAQGQ---LQTWGSEVFGNAQKS----SASPES
PF13499 // ENSGT00660000095541_0_Domain_1 (299, 358)
QV
QGIWEQLGVGRRGHLDEQELAVVCRSIGLRGLTEEELKDLFHKLDQDGDGRVSLAEFQLG
LCRHEPTLHPES-TLVKPSRP----WSHCQGQIPEESGCHTATTTSSSL-ASMSCSLHLF
SSIDDGSGFALPEQVIATWAQ-------EGIH--SGREI----LQSLDFSLDEKVDLLEL
TWALDNELLTVDSVIQQAALACYRQELSYHQ--EQAEQLVQER-NKARQDLE--------
-RAERRSLELVREMDDSHTALEQLSERR-IERLEQDYRGRLS----LLRAEVEAEREQIW
EQACRRSAVLEKDLG-HLRAEEASLRHRLGLATKENNRLQQEILEVVGKLSDSEKLVLKL
QGDLEFVLREK-L---EPQGME-LRAQEERFAAVLKEYELKCR-DLQDQNDELQ----AK
LESLRARPPGS--RSWQCPA------------GEPGCR-PAGML----------------
-------------------------SDNS--SPVSLEMEIMVERMRERCQELRT--QLEA
KVNSYEGEIEAMKNSFEKERMELQQARQREVAVLEAQRADLEALCAKSQEVILGLREQLW
DAAGRSESS------GAGLTSCCAQALCDLAQWLDQ-------HTRRQHQGELQQIR---
-----------------------------------REAAEELS--------WMLSQLE--
----------------------AQHDVRCKS--------------LALRHQHERDWLQQA
HLR------------------WEEDVFARCQEQQ----------------QK--LQAALG
------KEQAQMCRSFALERKRLERAHREQVGGLV-------------REAEALRALLQG
GTTVTIGREQKGAPG-----PSSPCPGIH----------------------------GGT
GELA-----------GDRPGQP-CCVD-------------AVSRGPPERLDHGQSSWVLL
DAKEATSVPCEEGSHVQ-----PPGQSKGAN-----PNEPAPSARAVVSPRQ--------
-----------------PDVQE------LPSWGTE-------------------------
------------------------------------------------------------
---------------------------------GD----GALQ--TWLQHH-----PG--
----PLPPHPELPNPAG---------------------------------------ETEA
EME-REKNDMKTK--LLQLEDVVRALEREADS------------------K-ENDRVELQ
RLSE---ENSLLKN-------DLERVQQELGA-----TEHVNDKQRQEIEALKRDKEKAC
SEMDE-LSMQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------NQKYKNEVSQLSHRVLQLEGVA-STHQAQN
EENLAAAQLLTQRLEEAGHREEQQ---------------CAQIQKLE--AELQHLSQECQ
SLRLAWSELTENQREGRDQEEE--------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------RQ-DTREG-------QGEEAERLLREEL----ERLEKSTRSSLLLKELYVENA
HLMKALQVAEQKQWGAEKQSCILEEKVRALSRLISKLAPASLSV----------------
---------------------------------------------------------
ENSEEUP00000014019 (view gene)
------------------------------------------------------------
-------------------------MDGEENHYVS
PF00036 // ENSGT00660000095541_0_Domain_0 (96, 124)
KLREVYNSCDTTGTGFLDQEELTRL
CLKENSGT00660000095541_0_Linker_0_1 (124, 336)
LHLEKQLPLLLQTLLGNDHFARVNFEEFKDGFVAVLSATAGADPSDDDSCSSLESAA
PGAVPPKYMSGSKWYGRRSQPEPCSGGPARL-PD-------------LQAGLRSQPHRCT
SLESVESLKSDEETESSKEPQNELFEAQGQ---LPGWGAEVFSSPQKSRRISFDAAESQV
RSLWAELGVGSSGHLTQQELVLVCRNIGLQALEKEPF00036 // ENSGT00660000095541_0_Domain_1 (336, 364)
ELEDLFNKLDQDGDGRVSLEEFQLG
LFNHGPNSLPEFSIPVRPARP----WSHWQ--VPEEGSCHT-ATT-SSL-VSLYSCPRLF
CSLDDGSGFAFPEQLVALWAQ-------EGIQ--NGMEV----LQSLDFSVDEKVNLWEL
AWALENELTAL-GATQQAALAXXXXXXXXXX--GQVEQMVRER-DKARQDLE--------
-KAEKRNLEFVKEMDDCHSALEQLTEQK-IQRLEQGYRGQLS----LLRSEVEVERELLW
EQARRQRATLDKDLG-RLRAEESGLREKLTLALKENSRLQKEILEVVEKLSESEKLVLKL
QADLEFVLKEK-A---RPQSTE-LLVQEQRFSEILTEYERKCR-DLQDRNDELQ----AA
LEGLQTQLPQIPRS---LQQP--------DGQLLPGHS-PTGPLQ---------------
------------------------CVSSA--APVSIETEIKMEQVKEHCQDLKI--QLET
KVNYYEREMQMLKRAFEKEREEMEQAFRLEVCVLEDQKADLETLHAESQELIHTLREQLQ
DALSMELEQR----------LEC----RGWRQNLAQDKEQL--------KEQLQRVRR--
------------------------------------EAEAERH--------QRLVWTEA-
----------------------RQ------------------------------EEMLQR
HLPA--------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----ASGLAEKWSQVH-----LHGADGGVI-----LEEPEPSTQGPAH-----------
-----------------------------LSWPAAQELLVGADRRAQILPASGE------
------------------------------------------------------------
------------------------------------------------------------
---------PRLSPPTSG-------------------------GGE---------AQVVL
----QRENEMK--AKLLQLEDVVRAIERA-------------------SDCKESHRI-ED
RFSE---ENALLKS-------ELEKIQQDLEA-----AERSCEAQRKEIPANRRDKKEAC
SAIEE-XXXX--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------SQKWKAELSQISHRVLQLGEEA-SMQQTRN
EENHMTIQLLTRRLEEARCKGQLQ---------------DDQIQKLE--RELEGVTQECQ
SLKVLRSKQREALEESWEQXXXXXXXXXXXXXXXXXXXXX--------------------
---------------------------------------------------XXXXX----
------------------XX---------------------XXXXXXXXXX-XXXXXXXX
XXXEELGRQLKVTEERAEEVAMALKNVEVLLQEKVGELREQFEKSTKSDVLLRGLYVENA
HLMRALQLAEEKQHSAEKKTRILEEKVQALNKLVSNISVASLCV----------------
---------------------------------------------------------
ENSANAP00000023813 (view gene)
------------------------------------------------------------
-------------------------MDEEENHYVSQLREVYSSCDTTGTGFLDREELTQL
CLKLHLEQQVPVLLQTLLGNNHFGRVNFEEFKEGFVTVLSLNAGVRPSDEES-SSLESAA
SCAIPPKYVSGSKWYGRRSQ--PELCDAATE-ARHV-P------EQQTQTSLKSQLRRLA
SLESVESLKSDEEAESAKEAQNELFEAQGQ---LQTWDSEDFGSPQKSCSPSFDTPEG
PF13499 // ENSGT00660000095541_0_Domain_1 (299, 358)
QI
RGMWEELGVGSSGHLSEQELAVVCQSVGLRGLKKEELEDLFNKLDQDGDGRVSLEEFQLG
LFSHEPTLLLESCTQVKPSRP----WSHYQ--VPEETGCHT-TTT-SSL-VSLCSSLRLF
SSIDDGSGFAFPEQVLAVWTQ-------EGIQ--NGGKI----LQSLDFSVDEKVNLLEL
TWALDNELMTVDNAIQQAALACYRQELSYQQ--GQVEQLVRER-DKARQDLE--------
-MAEKRNLEFVKEMDDCHSALEQLTEKK-IKHLEQGYRERLS----LLRSEVEAEQELFW
EQAHKQRAVLERDMG-RLQAEEAGLREKLTLALKENSRLQKEIVEVVEKLSDSERLSLKL
QKDLEFVLKDKL----EPQSAE-LLAQEERFTAVLKEYELKCR-DLQDRNDELQ----AE
LEGLRTRLPESRHSPSWGPGG--------RGRILL----FACVS----------------
------------------------FLGNS--APVSIETELMVEQVKEHYQDLRT--QLET
KVNYYEREIAALKRNFEKERKDMEQACRHEVSVLEGQKADLEELNEKSQEVIQGLQEQLQ
DAARSPKLEW----MGL--APCCTQALCDLALRHQS-------HLRQIRR----------
------------------------------------------------------------
----------EAKAEVSGEPSGLGALPACED--------------LTLGLEE----PPQG
FLSRGSWRPEQLELKR----------PQWP-------------------------QSRAS
------RVRAQMCASLALEEDGWELARVKRVDRLP-------------REAEMLRTLPKD
GL-VAES-GREGT-----HGPLPLCPGCGERLL----------------AWPALGNGGES
GEAAGPGPGR-RQVQ---------DKD---AVQSPAP--APAP-----------------
----ASHRPSEK-SRVQ-----PLEVDGGTD-----SEEPKTFCTG--------------
-----------------TAGLEQPGAQELPLLGTEGNALPA------------QPGMWEP
PLNLAT---LRRGQAE---SPQAVQEEPAQS-----------------------------
----WSRG-------------Q---ASEGQARA-E----GALEPRCHQ-HSVEVTRRRSS
------LSPFQLSDPRGA-------------------------GQEHLAVLGEKEAEIVL
E---REKDDMETK--LLHLEDVVRALEK-----H--------------VDLRENDRLEFH
RLTE---ENTLLKN-------NLGRVRQELEA-----AESTHDAQRKEIEVLKKDNEKAC
SEMEA-LSRQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------NQNYKDQLSQLNDRVLQLGQE---------
--------LLTQSLEEVACSWQQQ---------------SDQIQKLE--VELERLNQECQ
SMRLSQAELTQTLEESQSQ-----------------------------------------
---------------------------------------------------VQGAHLRLR
QAQAQHSQ----------ELVPRDHVAELQRLLRLQGE--------------RAGRRLDA
Q-REEHEKQLKATEERAEEAEMILKNMEMLLQEKVDELKEQFEKNTKSDLLLKELYVENA
HLVRALQATEEKQRGAEKQSRILEEKVRALNKLVSRIAPAALSV----------------
---------------------------------------------------------
ENSPANP00000017187 (view gene)
------------------------------------------------------------
-------------------------MDEEENHYVSQLREVYSSCDTTGTGFLDRQELTQL
CLKLHLEQQLPVLLQTLLGNDHFARVNFEEFKEGFVAVLSSNAGVRPSDEDS-SSLESAA
SSAIPPKYVNGSKWYGRRSR--PELCDTATE-ARHV-P------EQQTQASLKSQLWRSA
SLESVESLKSDEEAESTKEAQNELFEAQGQ---LQTWDTEDFGSPEKCCSPSFDTPE
PF13499 // ENSGT00660000095541_0_Domain_1 (298, 358)
SQI
RGMWEELGVGSHGHLSEQELAVVCQSVGLRELEKEELEDLFNKLDQDRDGKVSLEEFQLG
LFSHEPALLLESSTQVKVSKP----WSHYQ--VPEESGCHT-TTT-SSL-MSLCSSLRLF
SSIDDGSGFAFPDQVLAMWTQ-------EGIQ--NGREI----LQSLDFSVDEKVNLLEL
TWALDNELMTVDSAVQQAALACYHQELSYQQ--GQVEQLVRER-DKARQDLE--------
-RAEKRNLEFVKEMDDCHSALEQLTEKK-IKHLEQGYRERLS----LLRSEVEAERELFW
EQAHRQRAALEWDVG-RLQAEEAGLREKLTLALKENSRLQKEIVEVAEKLSDSERLALKL
QKDLEFVLKGQGL---EPQSAESPWPREGRFAAVLKEYELKCRYDLQDRNDELQ----AE
LEGLWARLPKNQHSPSWSPGG--------CRRQLSGLG-PAGIS----------------
------------------------FLGNS--VPVSIETELMMEQVKEHYQDLRT--QLET
KVNHYEREIAALKRNFEKERKDMEQARRREVSVLEGQKADLEELHKKSQEVIWGLQEQLQ
DTAHDPEPER----MGL--APCCTQALCGLALRHQS-------HLRQIRR----------
------------------------------------------------------------
----------EAHAELSGELLGLRALPARRD--------------LTLELEE----PLQG
PLPRGSRRSEQLNLES----------ALN-------------------------L-ECAS
------EKGAQMCASLALE-EELELAGGKRVDGLS-------------REAEMLGALPKE
GL-VAGS-GREGA-----RGLLPLSPGYGERPL----------------AWLTPGDGGES
EEAAGAGSRC-RQAQ---------DTE---ATKSPAPAPAPAP-----------------
----ASHGPSERWSHVQ-----PCGVDGGTV-----PEEPELFGVP--------------
------------------VGLKQPGPRELPLLGTEGDASQT------------QPRMWES
PLSPAA---SCRGQAE---KPQAIQEERARS-----------------------------
----WSRG-------------TQGQASEEQARA-E----GALESACQE-HGVEVARRGSL
------PSHFQLSDPRAS-------------------------RQEQLAAPEEGETKTAL
E---REKDDMETK--LLHLEDVVRALEK-----H--------------VDLRENDRLEFH
RLSE---ENALLKN-------DLGRVRQELEA-----AESTHNAQRKEIEVLKKDKEKAC
SEMEV-LTRQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------NQNYKDQLSQLNDRVLQLGQEA-STHQSQS
EEHRVTIQLLTQSLEEVACSGQQQ---------------SDQIQKLK--VELECLNQEYQ
SLQLSQSELTQALEESQGQ-----------------------------------------
---------------------------------------------------VQGAHLRLR
QAQAQHSQEVR-------RLVPQDRVAELQRLLSLQGE--------------QAGRRLDA
Q-REEHEKQLKATEERVEEAEMILKNMEMLLQEKVNELKEQFEKNTKSDLLLKELYVENA
HLVRALQATEEKQRGAQKQSRILEEKVRALNKLVSRIAPAALSV----------------
---------------------------------------------------------
ENSCATP00000008377 (view gene)
------------------------------------------------------------
-------------------------MDEEENHYVSQLREVYSSCDTTGTGFLDRQELTQL
CLKLHLEQQLPVLLQTLLGNDHFARVNFEEFKEGFVAVLSSNAGVRPSDEDS-SSLESAA
SSAIPPKYVNGSKWYGRRSR--PELCDTATE-ARHV-P------EQQTQASLKSQLWRSA
SLESVESLKSDEEAESTKEAQNELFEAQGQ---MQTWDTEDFGSPQKSCSPSFDTPE
PF13499 // ENSGT00660000095541_0_Domain_1 (298, 358)
SQI
RGMWEELGVGSRGHLSEQELAVVCQSVGLRGLEKEELEDLFNKLDQDGDGKVSLEEFQLG
LFSHEPALLLECSTQVKLSKP----WSHYQ--VPEESGCHT-TTT-SSL-VSLCSSLRLF
SSIDDGSGFAFPDQT--PWTR-------EGAE--R-CLL----GESLDFSVDEKVNLLEL
TWALDNELMTVDSAVQQAALACYHQELSYQQ--GQVEQLVRER-DKARQDLE--------
-RAEKRNLEFVKEMDDCHSALEQLTEKK-IKHLEQGYRERLS----LLRSEVEAERELFW
EQAHRQRAALEWDVG-RLQAEEAGLREKLTLALKENSRLQKEIVEVAEKLSDSERLALKL
QKDLEFVLKDK-L---EPQSAE-LLAQEERFAAVLKEYELKCR-DLQDRNDELQ----AE
LEGLWARLPKNQHSPSWSPGG--------CTRQLSGLG-PAGIS----------------
------------------------FLGNS--VPVSIETELMMEQVKEHYQDLGT--QLET
KVNHYEREIAALKRNFEKERKDMEQARRREVSVLEGQKADLEELHKKSQEVIWGLQEQLQ
DTAHDPEPER----MGL--APCCTQALCGLALRHQS-------HLRQIRR----------
------------------------------------------------------------
----------EAQAELGGELLGLRALPARRD--------------LTLELQE----PLQG
PLPRGSRRSEQLNLES----------ALN-------------------------L-ECAS
------EKGAQMCASLALK-EELELAGGKRVDGLS-------------REAEMLGALPKE
GL-VAGS-GREGA-----RGLLPLSPGYGERPL----------------AWLTPGDGGES
EEAAGAGSRC-KQAQ---------DTE---ATKSPAP--APAP-----------------
----ASHGPSERWSHVQ-----PCGVDGGTV-----PEEPELFGVP--------------
------------------VGLEQPGPRELPLLGTEGDASQT------------QPRMWES
PLSPAA---SCRGQAE---KPQAIQEERARS-----------------------------
----WSRG-------------TQGQASEEQARA-E----GALESACQE-HGVEVARRGSL
------PSHLQLADPRAS-------------------------RQEQLAAPEEGETKTAL
E---REKDDMKTK--LLHLEDVVRALEK-----H--------------VDLRENDRLEFH
RLSE---ENALLKN-------DLGRVRQELEA-----AESTHNAQRKEIEVLKKDKEKAC
SEMEV-LTRQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------NQNYKDQLSQLNDRVLQLGQEA-STHQAQS
EEHRVTIQLLTQSLEEVACSGQQQ---------------SDQIQKLK--VELECLNQEYQ
SLQLSQSELTQTLEESQGQ-----------------------------------------
---------------------------------------------------VQGAHLRLR
QAQAQHSQEVR-------RLVPRDRVAELQRLLSLQGE--------------QAGRRLDA
Q-REEHEKQLKATEERVEEAEMILKNMEMLLQEKVDELKEQFEKNTKSDLLLKELYVENA
HLVRALQATEEKQRGAEKQSRILEEKVRALNKLVSRIAPAALSV----------------
---------------------------------------------------------
ENSCJAP00000021371 (view gene)
------------------------------------------------------------
-------------------------MDEEENHYVSQLREVYSSCDTTGTGFLDREELTQL
CLKLHLEQQLPVLLQTLLGNNHFARVNFEEFKEGFVTVLSSNAGVRPSDKDS-SSLESAA
SCAIPPKYVSGSKGYGRRSR--PELFDAATE-ARHV-P------EQQTQASLKSQLRRSA
SLESVESLKSDEEAESAKEAQNELFEAQGQ---LQTWDSEDFGSPQKSCSPSFDTPEG
PF13499 // ENSGT00660000095541_0_Domain_1 (299, 358)
QI
QGMWEELGVGSSGHLSKQELAVVCQSVGLRGLKKEELEDLFNKLDQDGDGRVSLEEFQLG
LFSHEPALLLESCTQVKPSRP----WSHYQ--VPEETGCHT-TTT-SSL-VSLCSSLCLF
SSIDDGSGFAFPEQVLAVWTQ-------EGIQ--NGGEI----LQSLDFSMDEKVNLLEL
TWALDNELMTVDNAIQQAALACYRQELSYQQ--GQVEQLVKER-DKVRQDLE--------
-MAEKRNLEFVKEMDDCHSALEQLTEKK-IKHLEQGYRERLS----LLRSEVEAEQELFW
EQARKQRAVLEQDVG-RLQAEEAGLREKLTLALKENSRLQKEIVEVVGKLSDSERLSLKL
QKDLEFVLKDKL----EPQGAE-LLAQEERFTAVLKEYELKCR-DLQDRNDELQ----AE
LEGLRTRLPESRHSPSWGPGG--------RGRVLL----FACIS----------------
------------------------FLGNS--APVSIETELMVEQVKEHYRDLRT--QLEA
KVNYYEREIAALKRNFEKERKDMEQACRHEVSVLEGQKADLEELNEKSQEVIQGLQEQLQ
DAARGPKLEW----VWL--APGCTQALCGLALWHQS-------HLRQIR-----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------------LKFH
RLTE---ENALLKN-------NLGRVRQELEA-----AENAHNAQRKEIEVLKKDNEKAC
SETEA-LSRQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------NQNYKDQLSQLNDRVLQLGQEA-STHQTQN
EEHRVTIQLLTQSLEEVACSWQQQ---------------SDQIQKLE--VELERLNQECQ
SMRLSQAELTQTLEESQGQ-----------------------------------------
---------------------------------------------------VQGAHLRLR
QAQAQHSQ----------ELVPRDHVAELQRLLRLQGE--------------RAGRSLDA
Q-REEHEKQLKATEERAEEAEMILKNMEMLLQEKLDELKEQFEKNTKCDLLLKQLYVENA
HLARALQATEEKQRGAEKQSRILEEKVRALNKLVSRIAPAALSV----------------
---------------------------------------------------------
MGP_CAROLIEiJ_P0055140 (view gene)
------------------------------------------------------------
------------------------MDNEDENHYVSRLRDVYSSCDTTGTGFLDQEELTQL
CTKLGLEEQLPALLHILLGDDHLARVNFEEFKEGFVAVLSSGSGVEPSDEEG-SSSESAT
SCAVPPKYMSGSKWYGRRSL--PELGDSATE-IKYG-S------EQQTKGIVKPPLRRSA
SLESVESLKSDEDAESAKEPQNELFEAQGQ---LRSWGCEVFGTLRKSCSPSFSTPENLV
QGIWHELGIGS
PF13499 // ENSGT00660000095541_0_Domain_1 (312, 358)
SGHLNEQELAVVCRSIGLHSLEKQELEELFSKLDQDGDGRVSLAEFQLG
LFGHEPPSLPASSSLIKPNRL----WSHYQ----EESGCHT-TTT-SSL-VSVCSGLRLF
SSVDDGSGFAFPEQVISAWAQ-------EGIQ--NGREI----LQSLDFSVDEKVNLLEL
TWALDNELLTVDGVIQQAALACYRQELSYHQ--GQVDQLVQER-DKARQDLE--------
-KAEKRNLDFVREMDDCHSALEQLTEKK-IKHLEQEYRGRLS----LLRSEVEMERELFW
EQARRQRAVLEQDVG-RLQAEETSLREKLTLALKENSRLQKEIIEVVEKLSDSEKLVLRL
QSDLQFVLKDK-L---EPQSME-LLAQEEQFTAILNDYELKCR-DLQDRNDELQ----AE
LEGLRLRLPRSRQSPSGTPGT--------HRRRIPGRG-PADNL----------------
------------------------FVGES--TPVSLETEIMVEQMKEHYQELRM--QLET
KVNYYEKEIEVMKRNFEKDKKEMEQAFQLEVSILEGQKADLEALYAKSQEVILGLKEQLQ
DAARSPEPA------PAGLAHCCAQALCTLAQRLEV-------EMHLRHQDQLLQIRR--
------------------------------------EAEEELN--------QKLSWLE--
----------------------AQHAACCES--------------LSLQHQCEKDQLLQT
HLQRVKDLAAQLDREKGWREEREQEVLAHCRRQQ----------------LK--LQAVMS
------EEQARICRSFTLEKEKLEQAYREQVEGLV-------------QEADVLRALLKN
GTTVVSD-QQERTPSSMSLGP-----DSRQQPT----------------ARQAVSPDGRT
GAPA-EWPGPGKAEGRDFPGQL-CSID---AM--PSPTPTLLSRRSSENLGVRDNHQRPL
NAEEGAIPK-----------------------------EPEPSARTLT------------
-----------------------G--------------------QGQKLPLPVYPQMLEP
SLGTTA------------------------------------------------------
----LDRKAASVG--------VPGQASEGPVGDGE----GV--QEAWLQLRGEAA-----
-RMRPSLPCSELPNPQET--------------------------------------TVM-
--SESEMKDVKT--KLLQLEDVVRALEK--------------------ADSRESYRAELQ
RLSE---ENLVLKS-------DLGKTQLELET-----SESRNEVQRQEIEVLKRDKEQAC
FDLEE-LSTQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------TQKYKDEMSQLNYRVLQLEGEP-SGLHTQK
EENHSAIQVLMRKLEEAGCREEQQ---------------GDQIQNLK--MELERVNEECQ
YLRLSQAELTESLEESRGQ-----------------------------------------
---------------------------------------------------LYSVQLRLE
AAQSQHGRIVQHLQEQMSQLVPGARVAELQHLLNVKEE---------------EAGRLSS
Q-QEEYRQQLKAREDQVEDAEARLRNVEWLLQEKVEELRKQFEKNTRSDLLLKELYVENA
HLMKAVQLTEEKQRGAEKKNCVLEEKVRALNKLISKMAPASLSV----------------
---------------------------------------------------------
ENSMMUP00000013021 (view gene)
------------------------------------------------------------
-------------------------MDEEENHYVSQLREVYSSCDTTGTGFLDRQELTQL
CLKLHLEQQLPVLLQTLLGNDHFARVNFEEFKEGFVAVLSSNAGVRPSDEDS-SSLESAA
SSAIPPKYVNGSKWYGRRSR--PELCDTATE-ARHV-P------EQQTQASLKSQLWRSA
SLESVESLKSDEEAESTKEAQNELFEAQGQ---LQTWDTEDFGSPQKSCSPSFDTPE
PF13499 // ENSGT00660000095541_0_Domain_1 (298, 358)
SQI
RGMWEELGVGSRGHLSEQELAVVCQSVGLRGLEKEELEDLFNKLDQDGDGKVSLEEFQLG
LFSHEPVLLLECSTQVKLSKP----WSHYQ--VPEESGCHT-TTT-SSL-VSLCSSLRLF
SSIDDGSGFAFPDQVLAMWTQ-------EGIQ--NGREI----LQSLDFSVDEKVNLLEL
TWALDNELMTVDSAVQQAALACYHQELSYQQ--GQVEQLVRER-DKARQDLE--------
-RAEKRNLEFVKEMDDCHSALEQLTEKK-IKHLEQGYRERLS----LLRSEVEAERELFW
EQAHRQRAALEWDVG-RLQAEEAGLREKLTLALKENSRLQKEIVEVAEKLSDSERLALKL
QKDLEFVLKDK-L---EPQSAE-LLAQEERFAAVLKEYELKCR-DLQDRNDELQ----AE
LEGLWARLPKNQHSPSWSPGG--------CRRQLSGLG-PAGIS----------------
------------------------FLGNS--VPVSIETELMMEQVKEHYEDLRT--QLET
KVNHYEREIAALKRNFEKERKDMEQARRREVSVLEGQKADLEELHKKSQEVIWGLQEQLQ
DTAHDPEPER----MGL--APCCTQALCGLALRHQS-------HLRQIRR----------
------------------------------------------------------------
----------EAQAELSGELLGLRALPARRD--------------LTLELEE----PLQG
PLPRGSRRSEQLNLES----------ALN-------------------------L-ECAS
------EKGAQMCASLALK-EELELAGGKRVDRLS-------------REAEMLGALPKE
GL-VAGS-GREGA-----RGLLPLSPGYGERPL----------------AWLTPGDGGES
EEAAGAGSRC-RQAQ---------DTE---ATKRPAP--APAP-----------------
----ASHGPSERWSHVQ-----PCGVDGGTV-----PEELELFGVP--------------
------------------VGLEQPGSRELPLLGTEGDASQT------------QPRMWES
PLSPAA---SCRGQAE---KPQSIQEERARS-----------------------------
----WSRG-------------TQGQASEEQARA-E----GALESACQE-HGVEVARRGSL
------PSHLQLADPGAS-------------------------RQEQLAAPEEGETKTAL
E---REKDDMETK--LLHLEDVVRALEK-----H--------------VDLRENDRLEFH
RLSE---ENALLKN-------DLGRVRQELEA-----AESTHNAQRKEIEVLKKDKEKAC
SEMEV-LTRQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------NQNYKDQLSQLNDRVLQLGQEA-STHQAQS
EEHRVTIQLLTQSLEEVACSGQQQ---------------SDQIQKLK--VELECLNQEYQ
SLQLSQSELTQTLEESQGQ-----------------------------------------
---------------------------------------------------VQGAHLRLR
QAQAQHSQEVR-------RLVPQDRVAELQRLLSLQGE--------------QAGRRLDA
Q-REEHEKQLKATEERVEEAEMILKNMEMLLQEKVDELKEQFEKNTKSDLLLKELYVENA
HLVRALQATEEKQRGAEKQSRILEEKVRALNKLVGRIAPAALSV----------------
---------------------------------------------------------
ENSMEUP00000001891 (view gene)
------------------------------------------------------------
------------------------------------------------------------
-------------------------VNFEEFKEGFVAILSSDIDVSTSDEDS-SYLESAE
PAEVQPKYVNGAKWYGRRSR--PEVQNPESE-NKFL-Q------EQQINANVKSPLRRST
SLESVESLKSDEEAESAKELQNETFETQQA----QTWN-EIFGKSPKSPNACLDVTENQV
RGIWEGLGVGSSGYLDEEELATVCKNIGLQELEKE
PF00036 // ENSGT00660000095541_0_Domain_1 (336, 364)
ELEDLFNKLDQDGDGRVSFKEFQLG
VFSHGPCSVPASSTPIKPNNH----RPHHQ--VYEENGCRT-STT-SSL-LSICVDVHLF
SSIDDGTGFAVPEQVIAVWAQ-------EGIK--NGKEI----LQNLDFNMEEKVNLLEL
TWAFDNELMTG-GGIQQAALASYRHELCYLQ--GQAEQTVKER-DKLKHDLE--------
-KLEKRNLEFVKEMDDCHIAMEHLNESK-IKHLEEDYRGKLS----LLRSEIEMEREMLW
QQVNRQRAKFEADIE-YLQGEEICLQEKLTLALKENSRLQKEIIEVVEKLTASEKLVSKL
QDDLEFVLKDK-L---EPANPE-LFSQEERFAEVIKEYELKCR-DLSDRNDELQ----TE
LEGLRSQLNDSKHHHSRSPEK---------------------------------------
--------------------DITFIGDDH--VPVSIETEIMVEQLKEHYQDLKI--KLET
KVKYYEREIELMKKHFEKERKAIEQGFKVEISELEEQKAHLEELNVKSQELIEGLKDQIQ
KSTQS------------------------------------QELERLKHQNELQLM----
----------------------------------RTEAEMELN--------QKLSWTEA-
-----------------------QHAKE--------------------------------
--------------------------------------------------LK--LEERMN
------EEHARICKMFVIEKEKVEFTYRMQIERLT-------------HEAEKRWETETQ
----LER-EKDE---------MKIKLIQLE------------------------------
------------------------------------------------------------
---EVVQIL---------------------------------------------------
----------------------------------------------------------EK
ETYSRA---------------------NDXXX----------------------------
-XXXXXXXXXX--------------------------------------------X-XXX
XXXXXX-----------XXX------------------XXXX------X--X------X-
--X--------XX---XXX-----XX----------------------------------
---------------------------------------------------XXXXXXXXX
XXXXX-XXXX--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------TEKHKNEISQLNIQISQLTDEV-SVLQAEN
QSSQHTVQQLTQRLTEAEHQKKEK---------------XXXXXXXX--XXXXXXXXXXX
XXXXXXXXXXXXXX----------------------------------------------
----------------------------------------------------XXX--XXX
XXXXXXXX-X--XXXXXXXX-XXXXXX----------XXXXXXXXXXXXXX-XXXXXXXX
XXXEQHEERLKKMEERVEEVEMNLKNMEILLQEKVEQLKEQFAKNAKSDLLLKDLYVENA
HLMKALQITEQKQKGAEKKNFVLEEKIIALNKLINKITPASLSV----------------
---------------------------------------------------------
ENSCAFP00000006567 (view gene)
------------------------------------------------------------
------------------------MDEEEENHYVSQLREVYSSCDSTGTGFLDREELTQL
CLKLHLEKQLPILLQTLLGNDHFARVNFEEFKEGFVALLSSNAGAGPSDEDS-SSLESTT
SRAVPPKYVSGCKWYGRWSRPEPKPCGSATG-VKCL-P------EQQPWTSRRGPLRRSA
SLESVESLKSDEEAESAKEPQNELFEARGQ---LPAWGSEVFECLHKPSSLSLDTPESRV
R
PF13499 // ENSGT00660000095541_0_Domain_1 (302, 358)
GIWEELGVGSSGHLNEQELAIVCQSIGLQGLEKEELEDLFNKLDQDGDGRVSLEEFQLG
LFNHGPTSLPGSSTLSKPSMS----WSHYQ--VLEEGGCQT-TTT-SSL-VSVCSGLRLL
GSIDDGTGCAFPEQLIALWAQ-------EGIQ--NGREI----LQSLDFSVDEKVNLLEL
TWALENELMMVGGATQQAALACYRQELSFRQ--GQVEQMARER-DKARQDLE--------
-RAEKRNLEFVQEMDDCHSALEQLTEKK-IQHLEHGYRERLS----LLRSEVERERELFW
EQTCRQRAVLEKDLE-RLQAEEASLREKLTLAQKENSRLQKEIVEVVEKLSESEKLVLKL
QNDLDFVLKDK-L---EPQSIE-LLAQEERFAEVLKEYELKCR-DLQDRNDELL----AA
LEGLRAQLPSSWHGRPHA-QP--------EEQLLPGHG-PAGIS----------------
------------------------FIGDS--IPVNLETEIMMEQVKEHYQALQI--QLET
KVNSYEKEMEAMRRDFAKEREELQQAFRLEVRVLECRRADLEVLLGKSQEFIRGLQDQLQ
QDTRGPLPE------QAGLGQCCVHTLSGLVGRLAPEQGPPEQEPRQRHWNELQQVRK--
------------------------------------EAKAVLN--------YELLQTE--
----------------------AQQAANYKD--------------LVLQPQQEKEEVLQT
SLLQVHEVTEQHDLEKERREGRGKETLLRYGSQK----------------LK--LQEPVG
------KEQVRLCKSSTLEKEKRELACRKQMGSLV-------------WETDMLQACLRD
QPAVAGC-EQEGT-----VGPMPLCAGRREQLP----------------AWLGPGDGQ--
--------GRGQAGCRDGPDQHQGHVD---GRQSPALTPALAHRGPLEGLGLREDGHGVL
NAEEVASAPPDRWAEVH-----PCELDRRVV-----LEELVPPVGG--------------
---------------------SAERAGKLS-WGLA-------------------------
------------------------------------------------------------
----------PAG--------SQGE-----AQCGE----RVLEQGLQDHRG--KATVGTL
ISQTH------LTGPQAA-------------------------GQDQREALQEEEADSGL
Q---REKDHMKTK--LLQLEDVVWTLEK-----E--------------AESRENNRIELD
RLSE---ENSWLKN-------ELRKIQQELEA-----AERTKDAQRKEIEVLRRDKEKTC
SEVEE-LNKQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------SQKCKDEVSQLSHKVLQLGEEA-SIHQAQD
EKNRVTIQLLTHRLEEAGQQEELQ---------------SEQIQKLE--LELEHMNQECQ
SLRLSQSQLRETLEESQDQVGMLEHLECPSS-----------------------------
----------------VFKR------------------------------RVQR------
---------QSRLDSYQQGTRTAEDLHIL----------GNFHSEECKL------LICNL
FWQEEREKELKDTKEHLEEVGIVLKNVEMLLQEKVAQLKEQFEKNTKSDLLLKELYVENA
HLTKALQVTEEKLRGAEKKNRILEEKVRALNRLVSMLASASLSV----------------
---------------------------------------------------------
ENSHGLP00100017905 (view gene)
------------------------------------------------------------
-------------------------MDEEDNHYISQLKDVYNSCDSTGTGFLDQEELTEL
CRKLHLEKQLPALLQTLLGDDHIARVNFEEFKEGLVTVLSSKAGGNPISEEA-GSLQSAA
SYTVPPKYVSGSKWYGRWSQ--PEQRASATK-ARCG-P------EQQARAGLKSQLHHSM
SLESVESLKSNEEAESPREPQDELFEAQGQ---LQNWGSEVFGSPQKSCSPLYASSE
PF13499 // ENSGT00660000095541_0_Domain_1 (298, 358)
SQV
QDIWEQLGVGSRGHLDEQELAVVCQSIGLHGLEKEELKDLFHKLDQDGDGRVSLEEFQLG
LCSHKPSQIPESSTLVKPSRP----WSHCQGQVPEESGCHTATTT-SSL-VSMSCGLRLF
SSIDDGSGFSLPEQVIATWAQ-------EGIH--SGREI----LQSLDFSLDEKVDLLEL
TWALDNELLTVDSVIQQAALACYRQELSYHQ--EQVEQLVQER-DKARQDLE--------
-RAERRSLELVREMDDSHTTLEQLTEQK-IVCLEQDYRGRLN----LLRAEVEAERELIW
EQACRQSAMLEQDLG-HLRAEEASLRHRLGLASKENSRLQQEILEVVGKLSDSEKLVLKL
QGDLEFMLREK-L---ETQGME-LQAQEEQSAAVLKEYELKCR-DLQDQNDELQ----AE
LESLQARLPQS--QSWQCPA------------GDAGHH-PAGVY----------------
-------------------------MDDS--SPVSLETEILVEQMREHCQELRT--QLEA
KVNSYEREIEAMKNNFEKERMELEQARQQEVTVLEARRADLEVLCATSQEVILGLREQLR
AAAGGPEAS------WAAMTSCCAPVLCDLAQQLDK-------HMWQQHQGELRQIR---
-----------------------------------QEAAEELS--------RM----L--
----------------------SQHDIHCKS--------------LVLRHQLEKDRLQQV
HLR------------------REEEVLVRCQEQQ----------------QR--LQVALG
------EEQAQMCRSFALERKRLERAHREQVGSLV-------------QEAEALRVLLQG
GAAATTDKEQQGAPI-----PMSPCLDIH----------------------------GGT
WEPV-----------GDGPGQP-CCTD-------------AVSRGLPESLDHEQSCWGLM
DAEETASVPFEKGPPIQ-----APGQNKGAD-----PEEPAPSARATVSPSW--------
-----------------PDVQE------LPSRGTG-------------------------
------------------------------------------------------------
---------------------------------ED----GALQ--TWLQHH-----SG--
----P-GTTPEFPNPAG---------------------------------------ETEA
DMVEREKNDMKTK--LLQLEDVVRALKREADS------------------R-ENDREELQ
RLSE---ENGLLKN-------DLERIQQKLGA-----AEHMSDMQRQEIEALKRDKEKVC
FEMDKQLSIQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------NLKYKNEVSQLNCRVLQLEGVT-STHQAQN
EENLAAAQLLTQRLEEVGRREEQQ---------------CAQIQKLE--TELEHMSQQCQ
RLKLVRSELTESREEGQDQEEQ--------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------EQLDTRAG-------PAVEVELLLREKL----DQLEKTTRSGLLLKELYVENA
HLTKALQVAEQKQWGAEKQKRILEEKVRALNTLISKLAPAALSV----------------
---------------------------------------------------------
ENSFCAP00000005012 (view gene)
------------------------------------------------------------
------------------------MGEEEENHYVSQLREVYSSCDTTGTGFLDREELTQL
CLKLHLEKELPILLQTLLGNDHFARVNFEEFKEGFVAVLSSNAGAGPSDEDS-SSLESTS
SRAVPPKYVSGSKWYGRWSCPEPEPCGSATG-AKCL-P------EQQARATGRGQLRRSA
SLESVESLKSDEEAESAKEPQNELFEAQGQ---LPAWGSEVFRCLQKPCSLSLDTPESRV
RG
PF13499 // ENSGT00660000095541_0_Domain_1 (303, 358)
IWEKLGVGSNGHLSEQELAVVCQSIGLQALEKEELEDLFNKLDQDGDGRVSLEEFQLG
LFNHGPTSLPESSTAIKPSMS----WSHHQ--VLEEGSCQT-ATT-SSL-VSVCSGLRLL
CSIDDGTGFAFPEQLITLWAQ-------EGIR--NSREI----LQSLGFSVDEKVNLLEL
TWALENELMMVGGATQQAALACYRQELSFRQ--GQVEQMARER-DKARQDLE--------
-KAEKRNLEFVQEMDDCHSALEHLTEKK-IQHLEQGYRERLS----LLRSEVERERELLW
EQTCRQTAVLEKDLE-RLRGEEANLRGKLTLALKENSRLQKEIIEVVEKLSESEKLVLKL
QNDLEFVLKDK-L---EPQSIE-LLAQEERFVEVLKEYELKCR-DLQDHNDELQ----TA
LEGLRAQLPPSWRGRPHA-QP--------EERLLSEHG-PTGITS---------------
------------------------FVGDC--IPVSLETEIMMEQVKEHYQDLKI--QLET
KVNSYEREMEVMRRDFAKEREEMQQAFRLEVRVLECRRADLEVLLMKSQELIRGLQDQLQ
QASHSPLTE------RAVLGQCCAQALSGPAQRLAQEQDPLKQEQRQRHRKELQRVRMLY
YIT-----HEEM--------------------DKRKEAEVELN--------HKLSRTEA-
----------------------LQQTAHCKD--------------LALQPQQEKEEQKH-
-LLQVHEVIEQHDLEKEWREGKGKEILPLCGRQQ----------------LK--LQELTG
------EEQMQPCRSSALEKERLELACRNQMGKLM-------------QEEDMLQACLKD
GPAVAGG-DQEGM-----GGPMPLCAGRREQSL----------------VWQEPGAGH--
--------GHGQARHRDVPGRYLCHMD---PRQSPALDPALACRGSLENLGLREDGQGVL
DLEEVASAPPGRWLQVH-----QCGVDKKAV-----LELVPPAI----------------
----------------GPSGLNQPEAQELL-WKTEGDASPA---QPPTSG-------R--
------------GETE---RPPAILERAG-N-----------------------------
----LSGGLAPMG--------SQGQALEEQAWCGN----STLEQGLQHS-SD-KAKEGTL
FSHLH------LADPQGA-------------------------RQEQLEVLREEEANMAL
E---QEKDGMKTK--LLQLEYVVWALEK-----E--------------ADARENNRIKLD
RLSE---ENTLLKN-------ELGRIQQELEA-----AEKTNDAQRKEIEILKRDKEKAC
SEMEE-LNKQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------SQKCKDEVSQLNHKILQLGEEA-SVHQAQD
EKNLINIQLLTQRLEEAGHQEELQ---------------SELIQKLE--LELEHMNQECQ
SLRLSQSQLRETLEESQN------------------------------------------
--------------------------------------------------QLHGANLRLR
LAQSQHSEEVHQLQEQMQWLVPQDHMTELQQLLEEERQATQQLRDECILQK-EKGRRLET
QWE-EHEKELKDSEERVEEVGRALKNVDMLLQEKVAELKEQFEKNTKSHLLLKDLYVENA
HLTKALQVTEEKLRDAEKKNCILEEKVRALNRLISKIASASLSV----------------
---------------------------------------------------------
ENSMPUP00000015671 (view gene)
------------------------------------------------------------
------------------------MDEEEENHYVSQLREVYSSCDTTGTGFLDREELTQL
CLKLHLEKQLPILLQTLLGNDHFARVNFEEFKEGFVAVLSSSAGVGSSDEDS-SSLESTA
SRAVPPKYVSGSKWYGRWSRPEPEPGGPAVG-AKFL-P------EQQAWTSGRGQLRRSA
SLESVESLKSDEEAESAKEPQNELFEARGQ---LPAWGSEVFGCLQKPCSPSLDTPESRV
RGIWEELGVGSSGHLSEQELAIVCESIGLQGLEKE
PF00036 // ENSGT00660000095541_0_Domain_1 (336, 361)
ELKDLFNKLDQDGDGRVSFEEFQLGLFNHEPTSLPESSTPVKPSMS----WSHYQ--VLEEGGCPT-ATT-SSL-VSVCSGLRLL
CSIDDGTGFAVPEQLIALWAQ-------EGIP--NGREI----LQSLDFNVDEKVNLMEL
TWALENELMMVGGATQQAALACYREELTFRQ--GQVEQMAKER-DKARQDLE--------
-KAEKRNLEFVQEMDDCHSALEQLTEKK-IQHLEQGYRERLS----LLRSEVERERELFW
EQTCKQRAMLEKDLE-RLRAEEASLREKLTLALKENSRLQKEIIEVVEKLLESEKLVLKL
QNDMDFVLKDK-L---EPQSTE-LLAQEERFAEVLKEYECKCR-DLQDRNDELQ----AA
LEGLQARLPPSWHSQPHA-QP--------EEWLLSGHG-PAGWNDEVYDN--LVVTSGQR
GKKGCVDHLEPLLPAAHSPGPTWPEPGDS--IPVSLETEIMVEQVKEHCQDLKI--QLET
KVNSYEREMEAMRRDFMKEREEMQQSFRLEIHMLECRRADLEVLLRKSQELIRGLQDQLQ
QVTRGPLPE------RADLGQCCMQALSGPAGLLAQEQDLLEQEQHQRHRTELPRIRK--
------------------------------------EAEVVWN--------YELSRMQ--
----------------------AQQAAHCKL--------------LVLQPLQEKEEVLL-
-S-------PLLQVHEEQRQGRGKEMYPHWRSQL----------------LK--LQELTG
------REQGELCKPFALEEERLELAWR--------------------------------
------------------------------------------------------------
---------NGQTRGRDVPGQLLCHVN---AGQSPA----LAHGGPLESFGLREDGQHVL
TAKELASAPPEWLAQVY-----LCGVDREEA-----NMALEREKDDMKTKLLQLEDVVWA
LEKEADLRLRLIHALSQKPGILETRLFQML-WHMALDIFLA---TLEMEGFPG--SCYGY
WHARHS---HVMPAPS---RPRPILEHDHGCVVRPAVQLRHQLRTRSPH-LLEIPLLDV-
LANSYVGVSKPQS--------T-LSSQFHHPWNAE----GALCQRAWSTKCM-RKLKGRV
AETVYISPRERRLDCRGC-------------------------REDGIFAG-----KYPT
R---REHQSVDQPLATTPSILALWPLVRKHMGPP--------------SKPTPERRIELD
RLSE---ENELLKN-------ELGRIQQELEA-----AERTNDAQRKEIEVLKRNQEKSC
SEVEE-LNKQ-------EGKKFFLEDLRVASPSAPSLAQMLYNFITVCVHGQDTE-----
---QFKTERLEGLWQLLPNHSITVTQISPMVNFLEHHTIKDDSLNHQCHSDSAPGGSVTR
GHQEDHMAGDKASDLRNHSIRQTPQPSHGSIWQTPSEEVLKGRDPQILRHIIVRGCWRLR
RGSGAHALPAALESGTRLLKIIIGNNHERNSQKCRDEVSQLNEKVLQQGEVA-SPHQAQN
-ENCIAIQLLTQRLEEVGQWEELQVSINAHLEEGLKSQQGEHIQKLE--LELKHMNQECQ
SLRLSQSQLRDTLEESQDQHTRFSRYREEGGELVVLDNLKGSLRSCLEQFEPLGTRNQPL
SQAEDTLLRQSFCYMSWFPLLCDLEEDPQNHISENSHEQSHQPCLGFIPWQLHRANLGLG
LAQSQHSEEVHRLQEQMQGLVPRDHVAELQQLLEEERQVAWRFREECLLQK-EKGRRLET
QWQEEHEKELKDTEEQVEEMGMVLKNVEILLQEKVAELKEQFEKNTKSDLLLKELYVENA
HLMKALQVTEEKLQGAQKRNHILEEKVRALNRLISKIASASLSV----------------
---------------------------------------------------------
ENSPCAP00000003101 (view gene)
------------------------------------------------------------
------------------------MDEEEENPYVL
PF00036 // ENSGT00660000095541_0_Domain_0 (96, 124)
QLKEVYSSCDTTGTGFLDREELTQL
CLKENSGT00660000095541_0_Linker_0_1 (124, 336)
LHMEKQLPVLLQTLLGSDHLARVNFQEFKEGFVAVLSSNPSDGPSDEEN-SSSEAAV
SWAVLPKYMKGSKWYGRRTR--PEICVET---EARH-S------DQQAQASLNSQLCHAT
SPQSVEXXXXXXXXXXXXXXXXXXXXXXXQ---LQVCYAEGLGSPQELSSPSFDAPESQA
RAIWEEIGVGSSGYLSTQELALVCQRIGLQRLEKEPF00036 // ENSGT00660000095541_0_Domain_1 (336, 364)
ELEDLFDKLDQDGDGKVSLEEFQLG
LFSHGSTSFPESSTPVQPSIP----WSHYQ--XXXXXXXXX-XXX-XXX-XXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXX-------XXXX--XXXXX----XXNLDFGVDEKVNLLEL
SWAFDNELMMVTGIIQQTALASYRQELTYIQ--GQVEQVTRER-DKVKQDLE--------
-EARKRNLELVTEMDDCHSAIEELAEQK-IKHLEQGYHRQLA----LLRSEMEMERELLW
QQTSRQRARLEEDLG--LTQEKTCLREKLALALKENSQLQKEIFEVVKKHLKSEKLVLKL
QSELEFVLKDK-L---ESQSTX-XXXXXXXXXXXXXXXXXXXX-DLQDHNDELQ----AA
LENLWTRLHKNQCPVQC------------SVRWLPPADAPEXXXXXXXXX--XXXXXXX-
--------------XXXX----XXXXXXX--XXXXXXXXXXX-----------------X
XXXXXXXXX------------------------------------------X-X-X--XX
XXX--------XXXXXX----XXXXXXXXXX--XX---XXXXXXXXXXX-----XX----
----------------------------------XXXXXXXXX--------XXXXXX---
-------------------------XXXXXXXXXXXXXXXXXXXXXXXXXXXXXEEMLQS
HLLQVRNMMAQLDEK------------------A----------------WK--SRPNAN
------EEQAWICKSFALEKGKV-LTYRKQVEGLA-------------ET-QMLKARLKE
KTAAAHE----S----------GLPPY----RW--------------------------S
RGNRRERRGPGGAEAMGVLGWYSW-YSCTDTIQSPASISTLTSGASSEM-----------
----WSCIGPPGVNGPV-----PLTSPAGLKQPKG-------------------------
------------------------------------------------------------
QEHPLI---------------------VDGDN----------------------------
-LHKELALEEP-------------------------------------------------
-------------------------ETLQAS------RGHQVGWQEEKA--IQREAETA-
--LKREKIVGKTG---LLLEDMVGVL-------------------VKNANSKEKNXXXXX
XXXX---XXXXXXX-------XXXXXXXXXXX-----XXXXXXXXXKEIEILKKDKDQAC
VEMET-LNKQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------CQKYKDQLFQLNHRILKLGKEI-SAHEARD
EKNQVAIQQLTQSLEVASHQKEEQ---------------RDQIQNLK----LEHVSRECQ
YLHLSRSQLGEVLEEIGQX-----------------------------------------
---------------------------------------------------XXXXXXXXX
XXXXXXXXXXXXXXX-------XXXXXXX----------------XXXXXX-XXXXXXXX
XXXEEYEKQLTATKEWMKEVETILKNMEKLLHKNVVELKDQXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXKQREAEEKNVILEEKTRALNKLIGKIAPASLSV----------------
---------------------------------------------------------
MGP_SPRETEiJ_P0057281 (view gene)
------------------------------------------------------------
------------------------MDNEEENHYVSRLRDVYSSCDTTGTGFLDQEELTQL
CTKLGLEEQLPALLHILLGDDRLARVNFEEFKEGFVAVLSSGSGVEPSDEEG-SSSESAT
SCAVPPKYMSGSKWYGRRSL--PELGDSATA-TKYG-S------EQQAKGSVKPPLRRSA
SLESVESLKSDEDAESAKEPQNELFEAQGQ---LRSWGCEVFGTLRKSCSPSFSTPENLV
QGIWHELGIGS
PF13499 // ENSGT00660000095541_0_Domain_1 (312, 358)
SGHLNEQELAVVCQSIGLHSLEKQELEELFSKLDQDGDGRVSLAEFQLG
LFGHEPPSLPASSSLIKPNRL----WSHQ-----EESGCHT-TTT-SSL-VSVCSGLRLF
SSVDDGSGFAFPEQVISAWAQ-------EGIQ--NGREI----LQSLDFSVDEKVNLLEL
TWALDNELLTVDGVIQQAALACYRQELSYHQ--GQVDQLVQER-DKARQDLE--------
-KAEKRNLDFVREMDDCHSALEQLTEKK-IKHLEQEYRGRLS----LLRSEVEMERELFW
EQARRQRAVLEQDVG-RLQAEETSLREKLTLALKENSRLQKEIIEVVEKLSDSEKLVLRL
QSDLQFVLKDK-L---EPQSME-LLAQEEQFTAILNDYELKCR-DLQDRNDELQ----AE
LEGLRLRLPRSRQSPSGTPGT--------HRRRIPGRG-PADNL----------------
------------------------FVGES--TPVSLETEIMVEQMKEHYQELRM--QLET
KVNYYEKEIEVMKRNFEKDKKEMEQAFQLEVSVLEGQKADLEALYAKSQEVILGLKEQLQ
DATQSPEPA------PAGLAHCCAQALCTLAQRLEV-------EMHLRHQDQLLQIRR--
------------------------------------EAEEELN--------QKLSWLE--
----------------------AQHAACCES--------------LSLQHQCEKDQLLQT
HLQRVKDLAAQLDLEKGRREEREQEVLAHCQRQQ----------------LK--LQAVMS
------EEQARICRSFTLEKEKLEQTYREQVEGLV-------------QEADVLRALLKN
GTTVVSD-QQERTPSSTSLGP-----DSRQQPT----------------ARQAVSPDGRT
GAPA-EWPGPEKAEGRDFPGQL-CSID---AM--PSPTPTLLSRRSSENLGVRDNHQRPL
NAEEGAIPK-----------------------------EPEPSARPLT------------
-----------------------G--------------------QGHKLPLRVHPQMLEP
SSGTTA------------------------------------------------------
----LDRKAASVG--------VQGQASEGPLGDGE----GV--QEAWLQFRGEAT-----
-RMRPSLPCSELPNPQEA--------------------------------------TVMP
AMSESEMKDVKT--KLLQLEDVVRALEK--------------------ADSRESYRAELQ
RLSE---ENLVLKS-------DLGKIQRELET-----SESKNEVQRQEIEVLKRDKEQAC
CDLEE-LSTQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------TQKYKDEMSQLNCRVLQLEGEP-SGLHTQK
EESHGAIQVLMKKLEEAGCREEQQ---------------GDQIQNLK--IELERVNEECQ
YLRLSQAELTESLEESRGQ-----------------------------------------
---------------------------------------------------LYSVQLRLE
AAQSQHGRIVQRLQEQMSQLVPGARVAELQHLLNVKEE---------------EARRLSA
Q-QEEYRQQLKAREDQVEDAEARLRNVEWLLQEKVEELRKQFEKNTRSDLLLKELYVENA
HLMKAVQLTEEKQRGAEKKNCVLEEKVRALNKLISKMAPASLSV----------------
---------------------------------------------------------
ENSCHIP00000024306 (view gene)
------------------------------------------------------------
------------------------MDKEEENHYVSQLRDLYSSCDTTGTGFLDREELTQL
CLKLHLEKQLPVLLHTLLGNNQFARVNFEEFKDGFVAVLSSQAGLASSDEDS-GSLESVA
SQAVPPKYVSGSKWYGRRSQ--PEPCGTAGG-APGL-P------EQPARPSARSQLRRSA
SLESVESLKSEEEADSAKEPQNELFEAQGQ---LPTWGSEVFGSPRKPHSHSFDTPESQV
WGLWEELGVGS
PF13833 // ENSGT00660000095541_0_Domain_1 (312, 356)
SGYLTEEELALVCQSIGLQGLAKEELEDLFNKLDQDGDGRVSLEELQLG
LFNHGSPRPLELAPLVKPSGS----WSHYQ--VPEESVGQT---TTSSL-VSVCSGPRLF
CSVDDGSGFAFPEQIISLWAQ-------EGVH--NGREV----LQSLDFSVDEKVNLLEL
TWALENELMMVGGAAQQAALACYRQELSFCQ--EQVEQMARER-DKARQDLE--------
-KAEQRNLEFVKETDDLHSALEQLAEEK-VRRLEQGYRGRLS----LLRSEVEGERELFW
EQARRQRAGLEEDLR-RLQAEETSLREKLTLALKENSRLQKEMIEVVEKLSDSEKLVLKL
QNDLEFVLKDQ-L---EPRSTE-LLAQEERFSDILKEYELKCR-DLQDRNDELQ----AA
LEGLQVKAAPSW-----------------HGRLSPGHG-PAGTVT---------------
------------------------FVGDS--APASIETEIMLEQLKERYQELKI--QLET
KVNDHERKMEVTKRAFAEERRQLERAFQLEVGVLQGQKAELVTLQVESQEVVRGLREQLQ
RRAELEQGHA-----------HRL---SQERARLQ---------------EALQR-----
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------LRIKLE
KLSE---ENTLLKN-------ELGRIRQELEA-----SERTEAAQRKEIEVLKRDKEKAC
SEMEE-LCTQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------SQKCRDELSQLNHRVLQLGEEA-STHQTQS
EKNHVTVQLLTHRLEEAVLREELQ---------------GNQIQKLE--LELDRVTQDRQ
NLRLAQSQLREALEESRDQ-----------------------------------------
---------------------------------------------------LYGATQRLR
LARSQHAQRVQQLREEMAQLVPAGRVAELQRLLEGERQGARRLQE-------E-------
A-QEVHEKQLKAAEERVEEVEMILKNMEVLLREKVGELKEQFEKNTRSDLLLKELYVENA
HLAKALQVTEEKQRGAEKKSRFLEEKVSALNKLLSKIATASLAV----------------
---------------------------------------------------------
ENSMFAP00000003111 (view gene)
------------------------------------------------------------
------MEFCV-DSRNKRKLAC-YGMDEEENHYVSQLREVYSSCDTTGTGFLDRQELTQL
CLKLHLEQQLPVLLQTLLGNDHFARVNFEEFKEGFVAVLSSNAGVRPSDEDS-SSLESAA
SSAIPPKYVNGSKWYGRRSR--PELCDTATE-ARHV-P------EQQTQASLKSQLWRSA
SLESVESLKSDEEAESTKEAQNELFEAQGQ---LQTWDTEDFGSPQKSCSPSFDTPE
PF13499 // ENSGT00660000095541_0_Domain_1 (298, 358)
SQI
RGMWEELGVGSRGHLSEQELAVVCQSIGLRGLEKEELEDLFNKLDQDGDGKVSLEEFQLG
LFSHEPALLLECSTQVKLSKP----WSHYQ--VPEESGCHT-TTT-SSL-VSLCSSLRLF
SSIDDGSGFAFPDQVLAMWTQ-------EGIQ--NGREI----LQSLDFSVDEKVNLLEL
TWALDNELMTVDSAVQQAALACYHQELSYQQ--GQVEQLVRER-DKARQDLE--------
-RAEKRNLEFVKEMDDCHSALEQLTEKK-IKHLEQGYRERLS----LLRSEVEAERELFW
EQAHRQRAALEWDVG-RLQAEEAGLREKLTLALKENSRLQKEIVEVAEKLSDSERLALKL
QKDLEFVLKDK-L---EPQSAE-LLAQEERFAAVLKEYELKCR-DLQDRNDELQ----AE
LEGLWARLPKNQHSPSWSPGG--------CRRQLSGLG-PAGIS----------------
------------------------FLGNS--VPVSIETELMMEQVKEHYEDLRT--QLET
KVNHYEREIAALKRNFEKERKDMEQARRREVSVLEGQKADLEELHKKSQEVIWGLQEQLQ
DTAHDPEPER----MGL--APCCTQALCGLALRHQS-------HLRQIRR----------
------------------------------------------------------------
----------EAQAELSGELLGLRALPARRD--------------LTLELEE----PLQG
PLPRGSRRSEQLNLES----------ALN-------------------------L-ECAS
------EKGAQMCASLALE-EELELAGGKRVDWLS-------------REAEMLGALPKE
GL-VAGS-GREGA-----RGLLPLSPGYGERPL----------------AWLTPGDGGES
EEAAGAGSRC-RQAQ---------DTE---ATKRPAP--APAP-----------------
----ASHGPSERWSHVQ-----PCGVDGGTV-----PEELELFGVP--------------
------------------VGLEQPGPRELPLLGTEGDASQT------------QPRMWES
PLSPAA---SCRGQAE---KPQSIQEERARS-----------------------------
----WSRG-------------TQGQASEEQARA-E----GALESACQE-HGVEVARRGSL
------PSHLQLADPRAS-------------------------RQEQLAAPEEGETKTAL
E---REKDDMETK--LLHLEDVVRALEK-----H--------------VDLRENDRLEFH
RLSE---ENALLKN-------DLGRVRQELEA-----AESTHNAQRKEIEVLKKDKEKAC
SEMEV-LTRQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------NQNYKDQLSQLNDRVLQLGQEA-STHQAQS
EEHRVTIQLLTQSLEEVACSGQQQ---------------SDQIQKLK--VELECLNQEYQ
SLQLSQSELTQTLEESQGQ-----------------------------------------
---------------------------------------------------VQGAHLRLR
QAQAQHSQEVRQLQEQMSRLVPQDRVAELQRLLSLQGE--------------QAGRRLDA
Q-REEHEKQLKATEERVEEAEMILKNMEMLLQEKVDELKEQFEKNTKSDLLLKELYVENA
HLVRALQATEEKQRGAEKQSRILEEKVRALNKLVGRIAPAALSV----------------
---------------------------------------------------------
ENSMLEP00000000025 (view gene)
------------------------------------------------------------
-------------------------MDEEENHYVSQLREVYSSCDTTGTGFLDRQELTQL
CLKLHLEQQLPVLLQTLLGNDHFARVNFEEFKEGFVAVLSSNAGVRPSDEDS-SSLESAA
SSAIPPKYVNGSKWYGRRSR--PELCDTATE-ARHV-P------EQQTQASLKSQLWRSA
SLESVESLKSDEEAESTKEAQNELFEAQGQ---LQTWDTEDFGSPQKSCSPSFDTPE
PF13499 // ENSGT00660000095541_0_Domain_1 (298, 358)
SQI
RGMWEELGVGSRGHLSEQELAVVCQSVGLRGLEKEELEDLFNKLDQDGDGKVSLEEFQLG
LFSHEPALLLECSTQVKLSKP----WSHYQ--VPEESGCHT-TTT-SSL-VSLCSSLRLF
SSIDDGSGFAFPDQVLAMWTQ-------EGIQ--NGREI----LQSLDFSVDEKVNLLEL
TWALDNELMTVDSAVQQAALACYHQELSYQQ--GQVEQLVRER-DKARQDLE--------
-RAEKRNLEFVKEMDDCHCALEQLTEKK-IKHLEQGYRERLS----LLRSEVEAERELFW
EQAHRQRAALEWDVG-RLQAEEAGLREKLTLALKENSRLQKEIVEVAEKLSDSERLALKL
QKDLEFVLKDK-L---EPQSAE-LLAQEERFAAVLKEYELKCR-DLQDRNDELQ----AE
LEGLWARLPKNQHSPSWSPGG--------CRRQLSGLG-PAGIS----------------
------------------------FLGNS--VPVSIETELMMEQVKEHYQDLRT--QLET
KVNHYEREIAALKRNFEKERKDMEQARRREVSVLEGQKADLEELHKKSQEVIWGLQEQLQ
DTAHDPEPER----MGL--APCCTQALCGLALRHQS-------HLRQIRR----------
------------------------------------------------------------
----------EAQAELSGELLGLRALPARRD--------------LTLELQE----PLQG
PLPRGSRRSEQLNLES----------ALN-------------------------L-ECAS
------EKGAQMCASLALK-EELELGGGKRVDGLS-------------REAEMLGALPKE
GL-VAGS-GREGA-----RGLLPLSPGYGERPL----------------AWLTPGDGGES
EEAAGAGSRC-RQAQ---------DTE---ARKSLAP--APAP-----------------
----ASHGPSERWSHVQ-----PCGVDGGTV-----PEEPELFGVP--------------
------------------VGLEQPGPRELPLLGTEGDASQT------------QPRMWES
PLSLAA---SRRGQAE---KPQAIQEERARS-----------------------------
----WSRG-------------TQGQASEDQARA-E----GALESACQE-HGVEVARRGSL
------PSYLQLADPRAS-------------------------RQEQLAAPEEGETKTAL
E---REKDDMETK--LLHLKDVVRALEK-----H--------------VDLRENDRLEFH
RLSE---ENALLKN-------DLGRVRQELEA-----AESTHNAQRKEIEVLKKDKEKAC
SEMEV-LTRQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------NQNYKDQLSQLNDRVLQLGQEA-STHQSQS
EEHRVTIQLLTQSLEEVACSGQQQ---------------SDQIQKLK--VELECLNQEYQ
SLQLSQSELTQTLEESQGQ-----------------------------------------
---------------------------------------------------VQGAHLRLR
QAQAQHSQEV--------RLVPQDRVAELQRLLSLQGE--------------QAGRRLDA
Q-REEHEKQLKATEERVEEAEMILKNMEILLQEKVDELKEQFEKNTKSDLLLKELYVENA
HLVRALQATEEKQRGAEKQSRILEEKVRALNKLVSRIAPAALSV----------------
---------------------------------------------------------
ENSFDAP00000004647 (view gene)
------------------------------------------------------------
MKKKP--------KRPLDCPAC-YGMDEDDNHYVSQLKDVYSSCDSTGTGFLDREELTQL
CRKLHLEKQLPALLHTLLGDNHVTRVNFEEFKEGLVAVLSSKAGGNSISEET-GSLQSAA
SNAVPPKYVSGSKWYGRRSQ--PEQRASATK-ARCG-P------EQQTRAGLKSQLRRFM
SLESVESLKSDEEAESPREPQDELFEAQGQ---LQTWGSEVFGSPQKSCSPSYPSSES
PF13499 // ENSGT00660000095541_0_Domain_1 (299, 358)
QV
QIIWEQLGVGSRGHLDEQELAFVCRSIGLHGLKKEELKDLFHKLDQDGDGRVSLAEFQLG
LCSHKPMLLPESSTPVKPSWP----WSHCQVQVPEESGCHTATTT-SSL-VSMSCGLRLF
SSIDDGNGFALPEQIITTWAQ-------EGIH--SGRKI----LQSLDFSLDEKVDLLEL
TWALDNELLTVDSTIHQAALACYRQELSYHQ--EQAEQLVQER-NKARQDLE--------
-RAERRSLELVREMDDSHAALEQLTEQK-MARLEQDYRGRLS----LLRAEVEAERELIW
EQACRQSAILEQDLD-HLRAEEAGLRHRLGLAAKENSRLQQEILEVVRKLSDSEKLVLKL
QGDLEFVLREK-L---EPQGVE-LQAQEERSAAVLKEYKLKCR-DLQDQNDELQ----AE
LESLRVQLPQSRSRSWQRPA------------GHAGHR-AAGVF----------------
-------------------------VDNS--SPVSLETEIMVEQMREHCQELRT--QLEA
KVNSYEREIEAMKNNFEKERLELQHAQRQEVAVLEAQRADLEVLCAKSQEVILGLQEQLR
DSAGGPELS------RAGLTSCCAPVLCDLAQRLDE-------HVRWQHQGELRQIR---
-----------------------------------QEAAEELS--------RTLSRLE--
----------------------AQHDIHCKS--------------LVLRHRREKDRLQQA
HLQ------------------REEEVLAHCQEQQ----------------QR--LQAALG
------EEQAEMCRSFALERKRLERAHREQVGGLV-------------QEAEALRALLRG
GAIATTEKEQQETPS-----SVSPCLGTH----------------------------GGT
QEPV-----------GDGPGQA-CCMD-------------TVSRGPLESLDHEQSCLGLL
DAEEAASVSCEKRPHMQ-----PPGQNEGTD-----PEEPPPSARATVSLSW--------
-----------------PDVQE-------PSGDAG-------------------------
------------------------------------------------------------
---------------------------------ED----GILQ--ITLQHH-----PG--
----SLPPHPEFPHPAG---------------------------------------ETEA
DMVEKEKNDMKIK--LLQLEDIVRALEREADL------------------R-ENDRVVLQ
RLSE---ENGLLKN-------DLERIQQKLGA-----AEHMSDLQRQEIEALKRDKEKVC
FEMDE-LSIQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------NQKYKNEVSQLNCRVLQMEGVA-STHQAQN
EENLAAARLLTQKLEEVGRREEQQ---------------CAQIQKLE--VELEHMNQQCQ
SLKLARSELTENHEEGQDQQKQ--------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------EQLNTRAR-------PAVEVEQLLWEKL----DQLEKSTRSGLLLKELYVENA
HLTKALQVAEQKQWGAEKQNRILEEKVRALNMLISKLAPAALSV----------------
---------------------------------------------------------
ENSMUSP00000105522 (view gene)
------------------------------------------------------------
------------------------MDNEEENHYVSRLRDVYSSCDTTGTGFLDQEELTQL
CTKLGLEEQLPALLHILLGDDRLARVNFEEFKEGFVAVLSSGSGVEPSDEEG-SSSESAT
SCAVPPKYMSGSKWYGRRSL--PELGDSATA-TKYG-S------EQQAKGSVKPPLRRSA
SLESVESLKSDEDAESAKEPQNELFEAQGQ---LRSWGCEVFGTLRKSCSPSFSTPENLV
QGIWHELGIG
PF13499 // ENSGT00660000095541_0_Domain_1 (311, 358)
SSGHLNEQELAVVCRSIGLHSLEKQELEELFSKLDQDGDGRVSLAEFQLG
LFGHEPPSLPASSSLIKPNRL----WSHYQ----EESGCHT-TTT-SSL-VSVCSGLRLF
SSVDDGSGFAFPEQVISAWAQ-------EGIQ--NGREI----LQSLDFSVDEKVNLLEL
TWALDNELLTVDGVIQQAALACYRQELSYHQ--GQVDQLVQER-DKARQDLE--------
-KAEKRNLDFVREMDDCHSALEQLTEKK-IKHLEQEYRGRLS----LLRSEVEMERELFW
EQARRQRAVLEQDVG-RLQAEETSLREKLTLALKENSRLQKEIIEVVEKLSDSEKLVLRL
QSDLQFVLKDK-L---EPQSME-LLAQEEQFTAILNDYELKCR-DLQDRNDELQ----AE
LEGLRLRLPRSRQSPAGTPGT--------HRRRIPGRG-PADNL----------------
------------------------FVGES--TPVSLETEIMVEQMKEHYQELRM--QLET
KVNYYEKEIEVMKRNFEKDKKEMEQAFQLEVSVLEGQKADLEALYAKSQEVILGLKEQLQ
DAAQSPEPA------PAGLAHCCAQALCTLAQRLEV-------EMHLRHQDQLLQIRQ--
------------------------------------EAEEELN--------QKLSWLE--
----------------------AQHAACCES--------------LSLQHQCEKDQLLQT
HLQRVKDLAAQLDLEKGRREEREQEVLAHCRRQQ----------------LK--LQAVMS
------EEQARICRSFTLEKEKLEQTYREQVEGLV-------------QEADVLRALLKN
GTTVVSD-QQERTPSSMSLGP-----DSRQQPT----------------ARQAVSPDGRT
GAPA-EWPGPEKAEGRDFPGQL-CSID---AM--PSPTPTLLSRRSSENLGVRDNHQRPL
NAEEGAIPK-----------------------------EPEPSARTLT------------
-----------------------G--------------------QGQKLPLPVHPQMLEP
SLGTTA------------------------------------------------------
----LDRKAASVG--------VQGQASEGPVGDGE----GV--QEAWLQFRGEAT-----
-RMRPSLPCSELPNPQEA--------------------------------------TVMP
AMSESEMKDVKI--KLLQLEDVVRALEK--------------------ADSRESYRAELQ
RLSE---ENLVLKS-------DLGKIQLELET-----SESKNEVQRQEIEVLKRDKEQAC
CDLEE-LSTQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------TQKYKDEMSQLNCRVLQLEGEP-SGLHTQK
EENHGAIQVLMKKLEEAGCREEQQ---------------GDQIQNLK--IELERVNEECQ
YLRLSQAELTESLEESRSQ-----------------------------------------
---------------------------------------------------LYSVQLRLE
AAQSQHGRIVQRLQEQMSQLVPGARVAELQHLLNVKEE---------------EARRLSA
Q-QEEYRQQLKAREDQVEDAEARLRNVEWLLQEKVEELRKQFEKNTRSDLLLKELYVENA
HLMKAVQLTEEKQRGAEKKNCVLEEKVRALNKLISKMAPASLSV----------------
---------------------------------------------------------
ENSTBEP00000010686 (view gene)
------------------------------------------------------------
------------------------MDEEDENHYVS
PF00036 // ENSGT00660000095541_0_Domain_0 (96, 124)
LLKDVYNSYDTTGTGFLDREELTQL
CLKENSGT00660000095541_0_Linker_0_1 (124, 336)
LHLEQQPPDLLQILFGDDLFSRVNFEEFKEGFIAVLSSNAGVSPSDAES-GSLESAT
SHAVPPKYVRGSKWYGRRSQPE--LRDSAE--AT-CGL------EQQTKASLRSQLRSSA
SLESVESLKSDEEAESAREPQDELFGAQDQ---LRSWGSEIFGSPQKSSSPSFDTPESQV
RGIWEELGVGSSGHLNEQELAVVCQSIGLQGLRQEPF00036 // ENSGT00660000095541_0_Domain_1 (336, 364)
XXXDLFNKLDQDGGGRVSLQEFQLG
LFSHDPASLLESSTSVKPDRP----WSHFQ--XXXXXXXXXXXXX-XXX-XXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXX-------XXX---XXXXX----XXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX--GQVEQLTRER-DKARQDLE--------
-RAEKRNLQFVKEMDDCHSALEQLTEKK-IXXXXXXXXXXXX----XXXXXXXXXXXXXX
XXXXXXXXXXXXXXX-XXXXXXXXXXXXXXXXXXENSRLQKEIVEVMGKLSDSEKLVRKL
QNDLEFVLKDK-A---VLPSPE-LVAQEERFAPVLKEYELKCL-XXXXXXXXXX----XX
XXXXXXXXXXXXXXXXXX--------------XXXXXX-XXXXXX--X----X-------
---------------------XXXXXXXX--XXXXXXXXXXXXXXXXXXXXXXX--XXXX
XVNYYEREIEAVKRSSEKERRDMEQAHRLEVRVLEDQKADLEVLHTKSQEVIRGLREQLQ
DTTRSPEPV------GPGRAQCCVQVLSGLALRHQR---------------QLQQMXXXX
XXX-----XXXX--------------------X---------------------------
------------------------XXXXXXX--------------XXXXXXXXXXXLLQE
HLLQVSEMAEQLGLEK-----------------A----------------LK--LQERMS
------EEQAQICRSLTLEKQKLELSYRKQVQGLT-------------QEAEALRALLAG
T-----------------HSPAPLCPGGGEQSP----------------AQLELNNGGQP
EELAEGGLGRGQVK---------CVD----TVQSPVSAPTPT------------------
-----SHGTSERWSHGH-----PCGVHRGTA-----PEDPVPSASGPGS-----------
--------------------LSQPAGPELL-LGVQADDMS--------------------
------------PQPQ---NSEPQSRS--------AASG---------------------
--RGEAKSQEPAC--------KQGQAWEEQAH-SE----NTLEPGLQHH--REAARRGPS
L------PHPGLTDPWG--------------------------------AEQEAKAQMAL
E---REKEDMKTKL--LHLEDVVWALEKEADSRE--------------NEXXXXXX----
-XXX---XXXXXXX-------XXXXXXXXXXX-----XXXXXXXXXKDMEVLKRDKEKAC
FDMEA-LSEQ-------XXXXXXXXXXXXXXXXXXXXXXXXXXXXX--------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------XXXXXXXXXXXXXXXXXXX-XX-XXXXXXX
-XXXXXXXXXXX----XXXXXXXX---------------XXXX--X--------XXXXXX
XXXXXXXXXXXXXXXXXXX-----------------------------------------
-----------------X---------------------------------XXXXXX---
---------X--------XX------------------XXXXXXXXXXXXX-XXXXXXXX
XXXEEHENQRKASEERVEAVGALLKNVEVLLQEKAADLREQFEKNAKSCLLITELYVENG
HLMKALQITEEKQRHAEKENRLLEEKVHALNKLVGKMAAASLSV----------------
---------------------------------------------------------
ENSETEP00000008000 (view gene)
------------------------------------------------------------
-------------------------MGEEENPYVL
PF00036 // ENSGT00660000095541_0_Domain_0 (96, 124)
QLKDVYNSCDTTGTGYLDQLELTQL
CHKENSGT00660000095541_0_Linker_0_1 (124, 336)
LHLDKQLPILLQTLFGSDHFARESFEEFKEGFVAALSSNPGDDPSDDSE-SNSLEAL
SQAVLPKYVSGPKWYGRRTR--PELCTETEVR--RA-A------EKQGQSSLSSQLHRAA
SLQSVESLKSDEEPDGANEPQNELFEAQGQ---LQVWSPL------ELSSPSFRTPEIQA
QIIWEELGVGSSGFLSMQELTLVCQTIGLQDLEKEPF00036 // ENSGT00660000095541_0_Domain_1 (336, 364)
EMEDLFNKLDQDGDGRVSLEEFQLS
LFSHTPNSLPKSSTPIELSGP----WTHYQ--ATEESSCRT-TT--SSL-VSICSDLHLF
SSIDDGTGFAYPEQVVAMWAQ-------EGIQ--NGKEI----LQSLEFSVDEKLNLLDL
AWAIDNKLMMVTGVIQQAALASYHQELSYLQ--GQMKQMVKER-DKARQDLE--------
-KAEKRNLEFVTEMDDCHSAMEQLTERR-IXXXXXXXXXXXX----XXXXXXXXXXXXXX
XXXXXXXXXXXXXXX-XXXXXXXXX---XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXX-XXXXXPDGIT-LFSQ-EQFMEILKEYEFKCR-XXXXXXXXXX----XX
XXXXXXXXXXXXXXXX-------------------------------XXX--XXXXXXX-
--------------XXXXXXXFFTCVRAP--TPVNLETEIMMEQVKEHYQDLKN--QLET
KVNYYKQEIELMKKSFEKEKREMEHMFKSEVSMLESQRAALEAHHAQSQDALCSLQGQLR
EMECGPIAP------------TSLPPLS---------------GQNLRHQHGLQQIXXXX
XXX--------XXXX----XXXXXXXX-------XXXXXXXXX----XXX-XXXXXXXXX
----------------------XXXXXXXXX--------------XXX------------
XXXXXXXXXXXXXXX------------XXXXXXX----------------XX--XXXXXX
------XXXXXXXXXXXXXXXXXXXXXXXXXXXX-----------------XXXXXXXXX
XXX-XXX-XXXXX-----XXXXXXXXXXXXXXX----------------XXX---XXXXX
XXX--------XXXXXXXXXXXXX---XXXXXXXXXXX----X------XXXXXXXXXX-
---XXXXXXXXXXXXXX-----XXXXXXX---XXX--------------XXXXXXXXX--
XXX-------------------------------------------X-X-----X---XX
XXXXXX---------------------XXXXX----------------------------
-XXXXXXXXXXXX-XXX----------------------XXX--XXXXXXXXXX-XXXXX
XXXXXX----------XXXX-----XXXXXX---XXXXXXXXXXXXXXXXXXXXKVETV-
--LEREKSDMKTR--LLQLEDMLLAM-------------------EKETESRENNKIELQ
RLSE---ENTLLKN-------ELGRIQQELKV-----TERTNDVQRKEIDILKKEKYQAC
LEMEM-INNQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------XXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXX----X---------------X-XXXXXX----XXXXXXXXX
XXXXXXXXXXXXXX----------------------------------------------
----------------------------------------------------XXXXXXXX
XXXXXXXXXX--XXXXXXXX-XXXXXX-------XXXXXXXXXXXXXXXXX-XXXXXXXX
XXXEIHEKQLNATEKQVDKVEIILKNVEMLLQEKVDELKDQ-------------------
------------------------------------------------------------
---------------------------------------------------------
ENSCPOP00000004536 (view gene)
------------------------------------------------------------
-------------MCTTGWPAC-YGMDEEDSYYISQLKDVYNSCDTTGTGFLDLEELIQL
CRKLHLDKQLPALLQTLLGDDLNARVTFEEFKEGLVTVLSSKANGDLGSKEA-GSSQAAA
VCAVPPKYVSGSKRYGRWSQ--PEWRASGAE-ATRG-P------EQQARAGLKGQLCHAA
SLESVESLKSDEEAESPREPQEELFEAQGQ---LQTWGSEVFGSIQKSCSSSCASPESRI
QGIWEQLGVGSRGHLGEQELAVVCH
PF13833 // ENSGT00660000095541_0_Domain_1 (326, 357)
SIGLLGLAEEELKDLFHKLDQDGDGRVSLAEFQLG
LCHHKPMLLPESPL----LKP----WSHSQGQVPEESGCHTAATT-SSL-ASVSCGLRLF
SSIDDGSGFALPEQVIATWAQ-------EGIR--SGREI----LQSLDFSLEEKVDLLEL
TWALDNELLTVDSVIQQAALACYRQELSYLQ--EQVEQLARER-DKARQDLE--------
-RAERRSLELAREMDDSHTALEQLTERR-IQCLEQDYQGRLS----LLRAEVAAEQEQVW
EQACRQNAALEQDLG-LLRAEESRLRHRLGLAAKENSRLQQEILEVVGKLSDSEKLVLKL
QGDLEFILKEK-L---EPQGME-LRGQEERFEAVLKEYELKCR-DLQDQNDELQ----AK
LERLRMRPPGN--GSQQCPA------------GEPRHH-SAGMF----------------
-------------------------SNNS--RPVSLETEIMVEEMREHCQELRT--QLEA
KVNAYEGEIKAMKNSFEKERMELEQVRQQEVTALEAQRADLEALCAKSQEVILGLREQLQ
EAAGSPKSV------RAGPTSCCAQALCDLTHWLEL-------RTQRQYQGELRQIR---
-----------------------------------QQAAAELS--------WMLSRLE--
----------------------AQHDTCYKS--------------LVPRHQCEKDWLRQV
LLQ------------------REAEVHVRCQEQQ----------------QR--LQAALG
------KEQVQMCRCFALERKRLEHAHCEQLEHLV-------------QEAEVLQALLRG
GAAATIGREWKGAPS-----HSSLCPGTP----------------------------GGP
EKLV-----------GDWPGQP-CCVD-------------AVSQQPPESLNHEQNCWGLL
DLEEVVSDPCEKGSLRW-----PPGQSEGPD-----SEKQAPSARATVSPSQ--------
-----------------PDVRE------QPSQGVA-------------------------
------------------------------------------------------------
---------------------------------GN----DALQ--TWLQHH-----PG--
----LLLLHPELPNQTG---------------------------------------ETEA
EIVER-ETGMKTK--LLQLEDVVQAFEREADL------------------R-ESDRVELQ
RLSA---ANSLLKS-------DLERIQQELGA-----AEHMSNQQRREIEVLKRDKEKAC
SEMDE-LSIQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------NQKYKNEVLQLNCRVLQLEEVA-SAHQAQN
EENLAAAQLLTQRLQEAARQEEQQ---------------CAQIRKLE--AELLHLREEGQ
SVRPAQLGLTEDQEEGRTQEEE--------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------PR-DTMEG-------QAGEMEQLLQEKL----DWLEKSARSGLLLKELYVENA
QLTKALQVAEQKQWGAEKQNRILEEKVHALGTLISKLAPASLSV----------------
---------------------------------------------------------
ENSPVAP00000013573 (view gene)
------------------------------------------------------------
------------------------MDEEEENHYVS
PF00036 // ENSGT00660000095541_0_Domain_0 (96, 124)
QLKEVYSSCDTTGTGFLDREELTQL
CLKENSGT00660000095541_0_Linker_0_1 (124, 336)
LHLEKQLPIILQTLLGNDHFARVNFEEFKEGFVAMLSSNAGFGPFDEDS-NSLES--
--AVPPKYVSGSKWYGRRSQ--PEPCDSATG-AKCL-P------EQQARATVRSQVRRSA
SLESVESLKSDEEAESAQEPQNDLFETQGQ---LPTWDSEVFGSPQKSCSPFFDTPESQV
RGIWEELGVGRSGHLSEQELAVVCQSIGLQGLKKEPF00036 // ENSGT00660000095541_0_Domain_1 (336, 364)
ELEDLFNKLDQDQDGRVSLEEFQLG
LSNYGPTSLLKSSTPVKLSRS----WSHYQ--VLEEGDCHT-ATT-SSL-VSMCSGPRLF
CSVDNGTGFAFPEQLFALWAQ-------EGIQ--NGREV----LQSLDFSVDEKVNLLEL
TWALENELMMVHGATQQAALACYRQELSFCQ--GQVEQMARER-DKARKDLE--------
-KAEKRNLEFVKEMDDCHLALEQLTEKK-IKHLEQGYRERLC----LLRSEVEMERELFW
EQACRQRAALEKDLE-HLQAEDASLREKLTLALKXXXXXXXXXXXXXXXXXXXXXXXXXX
QNDLEFVLK-K-L---EPQSIE-LLAQEERFMEVLKEYELKCR-DLQDRSDELQ----AA
LESLQAQLPQSWRSHQ-C-KP--------NGQLFPRHG-RAGS-----------------
------------------------LLGDS--TPVSIETEIMMEQVKEHYQDLKI--QLET
KVNYYEREIEVIKRDFAKEKKEMEQAFRLEFSVLEDQKADLESLHVKSQEVIKGLQDQLQ
SAAHGPELE------RIEMEQRYTQALSGLAQR---------------HRHELQQIRK--
------------------------------------KVEVELN--------QKLSWMK--
----------------------AQQATCYKN--------------LSLQYQQEKKELLQK
HLLQVREMTELDL-EKKQ------------------------------------KPQLLM
------SEEQALCRSFALEKEKLELTYRKQVESLA-------------WEAEMLT-LLKD
GPAVAGG-EQDST-----GGPIPLNAGGMLAMM----------------P----------
------------------------------SSPTPTLAARC-----LRRASATALVRSVL
DPKKVGFVPTGRWLKVH-----PCGVVGGAV-----RENLLPYAGG--------------
-----------------RAGPSHPEAQQLL-LGTEGDASNT---QPQRV-----------
----------VWGKTE---RQQAILEEQAGNP----------------------------
----------SGG--------TLGQASEEQAQCSK----STLEGGYHSRN---EARRGTL
SS-----HHHRLTGPQAA-------------------------LQKQLKTLEEAEAKTPV
G---KKKNDVKIRLL---QLENVWALEKEANS-------------------RENNXXXXX
XXXX---XXXXXXX-------XXXXXXXXXXX-----XXXXXXXXXKEIEVLKRDKEKAC
SEVEE-LNKQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------SQKCKDELTQLNHRVLQLEEEV-STQQARN
EKNCITIQVLTQRLEEAGCRVELQ---------------GDQIQKLE-------HLQKCQ
SLRLSEPQLETF-E----------------------------------------------
------------------------------------------------------------
----------------------------------------------------------ES
QDPEEHKKQLKDTEEQVEEMEMILKNLEMLLQEKVSELGEQFEKNTKSDLLLKDLYVENA
HLIKALQVTEEKQRRAEKKNRILEEKVRALNKLISKITSASLSV----------------
---------------------------------------------------------
ENSPEMP00000011154 (view gene)
------------------------------------------------------------
------------------------MDSEEEDHYVSQLRDVYSSCDTTGTGFLDQEELTQL
CTKLGLEEQLPALLQMLLGNDRLARVNFEEFKEGFVAVLSSGSGVGPSDDEG-SSSESAT
SCAVPPKYMRGSKWYGRRSR--PELCDSATA-TKYG-S------EQQPKGGVKPPLRRSA
SLESVESLKSDEDAESTKEPQNELFEAQGQ---LRSWGCEVFGTPRKSCSPSFNTPEN
PF13499 // ENSGT00660000095541_0_Domain_1 (299, 358)
QV
QSIWHELGVGSSGHLNEQELAVVCQSIGLHRLEKQELEELFSKLDQDGDGRVSLAEFQLG
LFGHEPPSLPESSSPVKPNGP----WSHYQ----EENGCHT-ATT-SSL-VSVCSGLRLF
SSVDDGSGFAFPEQVISAWAQ-------EGIQ--NGREI----LQSLDFSVDEKVNLLEL
TWALDNELLTVDGVIQQAALACYRQELSYHQ--GQVEQLVHER-DKARQDLE--------
-KAEKRNLDFVREMDDCHSALEQLTEKK-IKHLEQEYRGRLS----LLRSEVEMERELFW
EQARRQRAVLEQDVD-RLQAEETSLREKLTLALKENSRLQKEIIEVVEKLSDSEKLVLRL
QSDLQFVLKDK-L---EPQSME-LLAQEEQFTAILKEYELKCR-DLQDRNDELQ----TE
LEGLRVRLPRSRQSPSGTPGT--------HRRRLPGRG-PAGNL----------------
------------------------FMGDS--TPMSLETEIMVEQMKEHYQELRM--QLET
KVNYYEKEIEVMKRNFEKDKKEMEQAFQLEVSVLEGQKADLEALYAKSQEVILGLKEQLQ
DAARSPEPA------AAGLAHCCAQALCTLAQRLEV-------EMHLRHQEQLQQIRK--
------------------------------------EAEEELN--------QKLSWLE--
----------------------AQHAACCES--------------LSLQHQCEKDQLLQT
HLQRVKDLAAQLDLEKGWREEREQEVLAHCRRQQ----------------WK--LQAVMS
------EEQARICRSFTLEKEKLEQTYREQVEGLA-------------READALRALLKD
GTTVVSD-QQERTPSSTSLGP-----DSRPQPP----------------VQQAVSPDGRT
GVPAEEWPGQERAKGSELPGQP-CNIV---AM--PRPTPTLVSRRSSESLGVRDNHQGPL
SAEEGTVPK-----------------------------EPELPARTLP------------
-----------------------G--------------------QGQKLPLPTQPQMLEP
WLGPTA------------------------------------------------------
----LDRKPAPVG--------VQGQASEEPTGGTE----GV--QETWLQFRGEAS-----
-RMRLSLPCSELPDPQEA-------------------------KFKHLAILGETEAKVPS
VMSESEMNDVKT--KLLQLEDVVRALEK-----E--------------ADSRENYRPSFE
VMFL---ICF---------------------------------FNRQEIEVLKTDKEQAC
FDLEE-LSTQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------TQKYKDELSQLNCRILQLEGDT-SALHTQK
EENRVAIQLLMQKLEEAGCREEQQ---------------GVQIQNLE--VELERVNEECQ
GLRLSQAELTERLKESQDQ-----------------------------------------
---------------------------------------------------LHSAQLSLE
AAQSQHGRIVKHLQEQMSQLVPRAHVAELQHLLNLEEE---------------AGRRLNA
Q-QEEYRQRLKAKEDQVEDAEARFRNVEWLLQEKVEELREQLEKNTRSDLLLKELYVENA
HLMKAVQLTEEKQRGAERKNCVLEEKVRALNKLISKMAPASLSV----------------
---------------------------------------------------------
ENSCCAP00000031453 (view gene)
------------------------------------------------------------
-------------------------MDEEENHYVSQLREVYSSCDTTGTGFLDREELTQL
CLKLHLEQQLPVLLQTLLGNNHFARVNFEEFKEGFVSVLSSNAGVRPSDEDS-SSLESAA
SCAIPPKYVSGSKWYGRRSR--PELCDAATE-ARHV-P------EKQTQASLKSQLRRSA
SLESVESLKSDEEAESAKEAQNELFEAQGQ---LQTWDSEDFGSPQKSCSPSFDTPEG
PF13499 // ENSGT00660000095541_0_Domain_1 (299, 358)
QI
RGMWEELGVGSSGHLSEQELAVVCQSVGLRGLKKEELEDLFNKLDQDGDGRVSLEEFQLG
LFSHEPALLLESCTQVKPSRP----WSHYQ--VPEETGCHT-TTT-SSL-VSLCSSLRLF
SSIDDGSGFAVPEQVLALWTQ-------EGIQ--NGREI----LQSLDFSVDEKVNLLEL
TWALDNELMTVDSAVQQAALACYRQELSYQQ--GQVEQLVGER-DKARQDLE--------
-MAEKRNLEFVKEMDDCHSALEQLTEKK-IKHLEQGYRERLS----LLRSEVEAEQELFW
EQAHKQRAMLEQDMG-RLQAEEAGLREKLTLALKENSRLQKEIVEVVEKLSDSERLSLKL
QKDLEFVLKDKL----EPQSAE-LLAQEERFTAVLKEYELKCR-DLQDRNDELQ----AE
LEGLRTQLPESRHSPSRGPGG--------RGRVLL----FACIS----------------
------------------------FLGNS---PVSIETELMVEQVKEHYQDLRT--QLET
KVNYYEREIAALKRNFEKERKDMEQACRREVSVLEGQKADLEELNEKSQEVIQGLQEQLQ
DAARSPRLAW----MGL--APCCTQAPCGLALRHQS-------HLRQIRR----------
------------------------------------------------------------
----------EAEAEMSGEPSRLGALPACED--------------LTLGLEE----PPQG
FLPRWSRRPEQLELKR----------PQW-------------------------PQSRAS
------REHAQMCASLALEEDGWELARVKRADRPP-------------REAEMPWALPKD
GL-VAGR-GREGA-----HGPPPLCPRCGEWLL----------------AWPATGNGGGS
GEAAGAGPGR-RQDQ---------DKD---AVQSPAS--APAP-----------------
----ASHRPSDK-SPVQ-----PRGVDGGTV-----PEEPEPFCTG--------------
-----------------TAGLEQPGAQELPLLGTEGNALPA------------QPGMWEP
PLNLAA---LCRGQAE---SPQAVQEEPARS-----------------------------
----WSRG-------------Q---ASEGQARA-E----GALEPGCHQ-HRMEVTRRRSS
------PSPFQLTDPRGT-------------------------GQEPLAVPGEKEAEIAL
E---REV-DMETK--PLHLEEVVRALEK-----H------------------ENDRLEFQ
RLTE---ENTLLKN-------NLGRVRQELEA-----AESTHDAQRKEIEVLKKDSEKVC
SEMEA-LNRQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------NQKYKDQLSQLNDRVLRLGQEA-STHQTQN
EEHRVTIQLLTQSLEEVACSWQQQ---------------SDQIQKLE--VELERLNQECQ
SMRLSQAELTQTLEESQGQ-----------------------------------------
---------------------------------------------------VQGAHLRLR
QAQAQHWQ----------ELVPRDHVAELQRLLHLQGE--------------RAGRRLDA
Q-REEHEKQLKATEERAEEAEMILKNMEMLLQEKADELKEQFEKNTKSDLLLKELYVENA
HLVRALQATEEKQRGAEKQSRILEEKVRALNKLVSRIAPAALSV----------------
---------------------------------------------------------
ENSDORP00000009766 (view gene)
------------------------------------------------------------
------------------------MDDEEENHYVSRLREVYCSCDSTGTGFLDREELTQL
CHKLHLEEQLPALLQTLLGDDHFARVNFEEFKEGFVAVLSSNAGVSSSDEGS-SSLESAA
SCAVPPKYVNGSKWYGRRSQ--PELWDSVTE-AKYSGA------EQQAKASQKQQLRRSA
SLESVESLKSDEETDSAKEPQNELFEAQGQ---LQNWGSEVFGIPRKSCNLS-NTPE
PF13499 // ENSGT00660000095541_0_Domain_1 (298, 358)
NQV
REIWEELGVGSSGHLNEQELAVVCQSIGLHGLEKEELEDLFNKLDQDGDGRVSLEEFQLG
LFSHEPASLPESSTPIKPSRP----WSHYQ-----ENGCHTTTTT-SSL-VSVCSGLCFF
SSVDDGSGFAFPEQLIAAWAQ-------EGIQ--NGREI----LKSLDFNVEEKVNLLEL
TWALDNELLTVDGVIQQAALACYRQELSYHQ--GQVEQLVRER-DKVRQDLE--------
-RVEKRNLEFVKEMDDCHSALEQLTEKK-VKHLEQEYRGRLS----LLRSEVEMERELFW
EQARRQRSVLEQDVS-RLRAEEVSLREKLTLSLKENSRLQKEIVEVVEKLSDSEKLVLKL
QSDLEFVLKDK-L---EPQNTE-LLAQEERFAAVLKEYERKCR-DLQDQNDELQ----SK
LEGLQAQLHESWPSPSGELST--------RGRRLPGRG-PAGIL----------------
------------------------FVGDS--TPVSIETEIMLEQVKEHYQDLKI--QLEA
KVNYYERDIEVMKKNFEKEKREMKQAHQVEVSTLEGQKTDLEALYTKSQEVIQGLQEQLQ
DKMYSPELA------KTGLAHCCAQELCDLARRLEE-------EMNLRHQNQLQQIRR--
------------------------------------EAGNELS--------QKLSWLE--
----------------------AQHAAYCES--------------LSLQHQCEKDKLLQT
HLRRANELAIQLDLEKGRQEEREKEILARCRRQQ----------------LK--LQAVMN
------EEQAQICRSFTLEKEKLERTYRNQVERLA-------------REAEVLRALLKN
GNAKVSN-ESERP-----CSPIPLCPDSGDQLL----------------VQMAPCADRDM
GVSVGHWPSQGQTQGQ------PCNVY---AIQSPSPTQM--SRSLSEHPSLQENYQGPP
SAGTI-------------------------------------------------------
-----------------------------------------------LEGLP---PS---
F--RAS---ASPGQPD---TQELLLEEQAGCLIRDPISLGSQE-----------------
----------------------------GLARGSK----GALETQLQLD--GEAASTGLL
LT------CPKLPDPQEV-------------------------TA----------AETLS
E---REKHDVKIK--LLHLEDVVRALE-----KD--------------ANSRENDRIEFR
RLSE---ENTLLQT-------DLRKIQQELEA-----AENTNGAQRKEIEVLKRDKEKAC
FEMEK-LNTQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------IQNYKDELSQINSRVLQLEGDT-STHQAWK
EKRHVAIQLLMQRLEEASQREQQQ---------------DKQIQKLE--VELEHMNQECQ
NLKLSQSELTESLQETQEQ-----------------------------------------
---------------------------------------------------LYSAHRRLE
AAQGQHEQKVQCLQEQMAQLVPGARVAELQHLLSLQKE--------------EARRKLDS
QWE-EHQQQLRSREEQMREAEMRLQNVEWLLQEKVEELKEQFEKNSKSDLLLKELYVENS
HLMKALQVTEEKQLCAERKSRILEEKVRALNKLISKIAPASLSV----------------
---------------------------------------------------------
ENSRBIP00000029549 (view gene)
MLWGKKENISRSIFTGPGRLLPSMVVHCPGVRPFHRTTGHSRGRGAHPSQPLLSEEEAGQ
KQSSPRLATTG-VPSQDRTRAC-YGMDEEENHYVSQLREVYSSCDTTGTGFLGRQELTQL
CLKLHLEQQLPVLLQTLLRNDHFARVNFEEFKEGFVAVLSSNADVRPSDEDS-SSLESAA
SSAIPPKYVNGSKWYGRRSR--PELCDTATE-ARHV-P------EQQTQASLKSQLWRSA
SLESVESLKSDEEADSTKESQNELFEAQGQ---LQTWDTEDFGSPQKSCSPSFDTPE
PF13499 // ENSGT00660000095541_0_Domain_1 (298, 358)
SQI
RGMWEELGVGSRGHLSEQELAVVCQSIGLRGLEKEELEDLFNKLDQDGDGKVSLEEFQLG
LFSHEPALLLESSTQVKLSKP----WSHYQ--VPEESGCHT-TTT-SSL-VSLCSSLRLF
SSIDDGSGFAFPDQVLAMWTQ-------EGIQ--NGREI----LQSLDFSVDEKVNLLEL
TWALDNELMTVDSAVQQAALACYHQELSYQQ--GQVEQLVRER-DKARQDLE--------
-RAEKRNLEFVKEMDDCHSALEQLTEKK-IKHLEQGYRERLS----LLRSEVEAERELFW
EQAHKQRAALEWDMG-RLQAEEAGLREKLTLALKENSRLQKEIVEVAEKLSDSERLALKL
QKDLEFVLKDK-L---EPQSAE-LLAQEERFAAVLKEYELKCR-DLQDRNDELQ----AE
LEGLWARLPKNQHSPSWSPGG--------CRRQLSGLG-PAGIS----------------
------------------------FLGNS--VPVSIEVELMMEQVKEHYQDLRT--QLET
KVNYYEREIAALKRNFEKERKDMEQARRREVSVLEGQKADLEERHEKSQEVIWGLQEQLQ
DTAHGPEPEW----MGL--APCCTQALCGLALQHQS-------YLRQIRR----------
------------------------------------------------------------
----------EAEAELSRELLGLRALPARRD--------------LTLELEE----PLQG
PLPHGSRRSEQLNLES----------ALK-------------------------LQPCAR
------EKGAQMCASLALEEEELELAGGKRVDGLS-------------QEAEMLGALPKE
GL-VAGS-GQEGA-----RGLLPLSPCYGERPL----------------AWLAPGDGGES
KEAAGAGSRC-RQAQ---------DTE---ATKSPDP--APVP-----------------
----ASHGPSERWSHVQ-----PCGVDWGTV-----PEEPELFGVP--------------
------------------AGLEQPGSRELPLLGTEGDALQT------------QPRMWES
PLSPAA---SCRGQAE---RPQAIQEERGRS-----------------------------
----WSRG-------------TQGQASEEQARA-E----GALESACQE-HSVEVARRGSL
------PSHLQLADPRGS-------------------------GQEQLAAPEEEETKIAL
E---REKDDMETK--LLHLEDV-RALEK-----H--------------VDLRDNDRLEFH
RLSE---ENALLKN-------DLGRVRQELEA-----AESTHNAQRKEIEVLKKDKEKAC
SEMEV-LNRQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------NQNYKDQLSQLNDRVLQLGQEA-STHQAQS
EEHRVTIQLLTQSLEEVTCSGQQQ---------------SDQIQKLK--VELECLNQEYQ
SLQRSQSELTQTLEESQGQ-----------------------------------------
---------------------------------------------------VQGAHLRLR
QAQAQHSQEVQQLQEQMSRLGHQDRVAELQRLLNLQGE--------------QAGRRLDA
Q-REEHEKQLKATEERVEEVEMILKNMEMLLQEKVDELKEQFEKNRKSNLLLKELYVENA
HLVRALQATEEKQRDAEKQSRILEEKVRALNKLISRIAPAALSV----------------
---------------------------------------------------------
ENSSBOP00000017104 (view gene)
------------------------------------------------------------
-------------------------MDEEENHYVSQLREVYSSCDTTGTGFLDREELTQL
CLKLHLEQQLPVLLQTLLGNNHFARVNFEEFKEGFVAVLSSNAGVRPSDEDR-SSLESAA
SCAIPPKYVSGSKWYGRRSR--PELCDAATE-ARHV-P------EQQTQASLKSQLRRSA
SLESVESLKSDEEAESAKEAQNELFEAQGQ---LQTWDSEDFGSPQKSCSPSFDTPEG
PF13499 // ENSGT00660000095541_0_Domain_1 (299, 358)
QI
RGMWEELGVGSSGHLSKQELAVVCQSVGLQGLKKEELEDLFNKLDQDGDGRVSLEEFQLG
LFSHEPALLLESCTQVKPIRP----WSHYQ--VPEETGCHT-TTT-SSL-VSLCSSLRLF
SSIDDGSGFAFPEQVLAVWTQ-------EGIQ--NGGEI----LQSLDFSVDEKVNLLEL
TWALDNELMTVDNAIQQAALACYRQELSYQQ--GQVEQLVRER-DKVRQDLE--------
-MAEKRNLVFVKEMDDCHSALEQLTEKK-IKHLEQGYRERLS----LLRSEVEAEQELFW
EQAHKQRAVLERDMG-RLQAEEAGLREKLTLALKENSRLQKEIVEVVEKLSDSERLSLKL
QKDLEFVLKDKL----EPQSAE-LLAQEERFTAVLKEYELKCR-DLQDCNDELQ----AE
LEGLRTRLPESRHSPSWGPGG--------RGRQL------SGLS----------------
------------------------PAGSS--APVSIETELMVEQVKEHYQDLRT--QLET
KVNYYEREIAALKRNFEKERKDMEQACRREVSVLEGQKADLEELNEKSQEVIQGLREQLQ
DAARSPKLEW----MGL--APCCTQALCDLALRHQS-------HLRQIRR----------
------------------------------------------------------------
----------EVEVEMSGEPSRLGALPACEY--------------LTLALEE----PPQG
FLPLGSRRPEQLELER----------PQWAERGP----------------PK--PQSRAS
------RDHAQMCASLALEEDGWELARVKRVDRLP-------------REAEMLLALPKD
GL-VAGS-GREGA-----HGPPPLCPGCGERLL----------------AWPAAGNGRES
GEAAGAGPGR-GQAQ---------DKD---AVQSPTP--IPAP-----------------
----ASYRPSEK-SRVR-----PRGVDGGNV-----PEEPDSFYTG--------------
-----------------TAGLEQPGAQELPLLGTEGNALPS------------QPGMWEP
PLSLDT---LRRGQAE---SPHAVQEEPARS-----------------------------
----WSRV-------------CP-VGSQGQARA-E----GALEPGCYQ-HSMEVTR-RSS
------PSPFQLTDPRGA-------------------------GQEHLAVPGEKEAEIAL
E---REKDDMETK--FLHVEDVVRALEK-----H--------------VDLRENDRLEFH
RLTE---ENTLLKN-------NLGSVRQELEA-----AESTHDAQRKEIEVLKKDNEKAC
SEMEA-LNRQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------NQNYKDRLSQLNDRVLQLGQEA-STHQAQN
EEHRVTIQLLTQSLEEVACSWQQQ---------------SDQIQKLE--VELERLNQECQ
SMRLSQAELTQTLEESQSQ-----------------------------------------
---------------------------------------------------VQGAHLRLR
QAQAQHSQEVRQLQEQMGQLVPRDHVVELQHLLRLQGE--------------RAGRHLDA
Q-REEHEKQLKATEERAEEAEMILKNMEMLLQEKVDELKEQFEKNTKSDLLLKELYVENA
HLVRALQATEEKQRGAEKQNRVLEEKVRALNKLVSRIAPAALSV----------------
---------------------------------------------------------
ENSMAUP00000025222 (view gene)
------------------------------------------------------------
------------------------MDSEEENHYVSQLRDVYSSCDTTGTGFLDQEELTQL
CTKLGLEEQLPALLQILLGNDRLARVNFEEFKDGFVAVLSSGSGVGPSDEEG-SSSESAT
SCAVPPKYVSGSKWYGRRSR--PELCDSATA-VKYG-S------EQQAKGSMKPPLRRSA
SLESVESLKSDEDAESAKEPQNELFEAQGQ---LRSWGCDVFGTPRKSCSPSFNTPEN
PF13499 // ENSGT00660000095541_0_Domain_1 (299, 358)
QV
QSIWHELGVGSSGHLNEQELAVVCRSIGLHGLEKQELEELFSKLDQDGDGRVSLEEFQLG
LFGHELPSLPESSSLVKPNRP----WSHYQ----EENGCHT-TTT-SSL-VSVCSGLRLF
SSVDDGSGFAFPEQVISTWAQ-------EGIQ--NGREI----LQSLDFSVDEKVNLLEL
TWALDNELLTVDGVIQQAALACYRQELNYHQ--GQVEQLMHER-DKARQDLE--------
-KAEKRNLDFVREMDDCHSALEQLTEKK-IKHLEHEYKGRLS----LLRSEVEMERELFW
EQARRQRAVLEQDVG-RLQAEETSLREKLTLALKENSRLQKEIIEVVEKLSDSEKLVLRL
QSDLQFVLKDK-L---EPQNME-LLAQEEKFTAMLKEYELKCR-DLQDRNDELQ----TE
LEGLRMRLPRSRQSPSRTPGT--------HRRRLPRRG-PTGNL----------------
------------------------FVGDC--TSVSLETEIMVEQMKEHYQELRM--QLES
KVNYYEKEIEVMKRNFEKDKKEMEQAFQLEVSMLEGQKADLEALYAKSQEVILGLKEQLQ
DAARSPEPA------AAGLAHCCAQALCTLAQRLEV-------EMDLRHQDQLQQIRR--
------------------------------------EAEEELN--------RKLSWLE--
----------------------AQHAACCES--------------LSLQHQCEKDQLLQT
HLQRVKDLAAQLDLEKGWREEREQEVLEHCRRQH----------------LK--LQAVMS
------EEQARICRSFTLEKEKLEQAYREQVEGLV-------------QEADALRALLKN
GTSVVSD-QQERTPSSTPRGP-----DSRQQPP----------------PQQAVSPDGRT
GAPAEECPDQRRAKGSNLPGQP-CSIV---AM--PSPTPTLVPTRSLESLGVRDNHQGPL
SAEEGPIPK-----------------------------ESELPARALI------------
-----------------------G--------------------HEQKLPLPTQPQMMEP
WLGPTA------------------------------------------------------
----LDRKPAPVG--------VQEQASERPNRDAE----GV--QETWLQFREEAS-----
-RMRPSLPCSELPDPQ--------------------------------------EAKVPS
VMSESEMDDVKT--KLLQLEDVVRALER-----E--------------ADARENCRAEFQ
RLSE---ENFVLKS-------DLGRIQLELET-----SERRNETQRQEIEVLKKDKEQAR
FDLEE-LGTQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------AQKYKDELSQLNCRILQLEGDT-SALHTQK
EENRVAIQLLMQKLEEAGCREEQQ---------------GVQIQNLE--IELERVNEECQ
GLRLSQAELTENLKESQSQ-----------------------------------------
---------------------------------------------------LHSVQLTLE
AAQSQHDQIVQCLQEQMGHLVPGAHVAELQHLLSLEEE---------------AGRRLNA
Q-QEEYKQQLKAKEDQVEDAEARLRSVEWLLQEKVEELREQLEKNTRSDLLLKELYVENA
HLMKAVQLTEEKQRGTERKNCVLEEKVRALNKLISKMAPASLSV----------------
---------------------------------------------------------
ENSAMEP00000010995 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------QVTFEELKEGLVAVLSSNAGAGPLDGDS--SLESAA
SRDVPPKYVSGSKRYGRWSRPEPEPCGSATE-ANCS-P------EQQARTTGRSQLRRSA
SLESVESLKSDEEAESAKEPQNELFEARGQ---LPAWGSEVFGCLQKPCSPSLDTPESRV
RGIWEELGVGSSGHLNEQELAVVCQSIGLQGLQKE
PF00036 // ENSGT00660000095541_0_Domain_1 (336, 364)
ELEDLFNKLDQDGDGRVSLEEFQLG
LFNHGPTSLPESSTPVKPSMS----WSHYQ------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------------
ENSCAPP00000012747 (view gene)
------------------------------------------------------------
-------------------------MDEEDSYYISQLKDVYNSCDTTGTGFLDLEELIQL
CRKLHLDKQLPALLQTLLGDDLNARVTFEEFKEGLVTVLSSKANGDPDSKEA-GSSQAAA
VCAVPPKYVSGSKRYGRWSQ--PEWRASGAE-ATRG-P------EQQARAGLKGQLCHAA
SLESVESLKSDEEAESPREPQEELFEAQGQ---LQTWGSEVFGSIQKSCSSSCASPE
PF13499 // ENSGT00660000095541_0_Domain_1 (298, 358)
SRI
QGIWEQLGVGSRGHLDEQELAVVCHSIGLLGLAEEEFKDLFHKLDQDGDGRVSLAEFQLG
LCHHKPMLLPESPL----LKP----WSHSQ-----ESGCHTATTT-SSL-ASVSCGLRLF
SSIDDGSGFALPEQVIATWAQ-------EGIR--SGREI----LQSLDFSLEEKVDLLEL
TWALDNELLTVDSVIQQAALACYRQELSYLQ--EQVEQLARER-DKARQDLE--------
-RAERRSLELAREMDDGHTALEQLTERR-IQCLEQDYQGRLS----ILRAEVAAEREQVW
EQACRQHAALEQDLG-LLRAEESRLRHRLGLAAKENSRLQQEILEVVGKLSDSEKLVLKL
QGDLEFILKEK-L---EPQAMD-LRGQEERFEAVLKEYELKCR-DLQDQNDELQ----AL
LVTRLS-----------CHV------------V--TFR-TSGL-----------------
---------------------------------------------------ALL--LISL
QVNAYEGEIKAMKNSFEKERMELEQVRQREVAALEAQRADLEALCAKSQEVILGLREQLQ
EAAGSPKSV------RAGPTSCCAQALCDLTHWLEL-------RTQRQYQGELRQIRSAL
SLS-----G--------------------------QEAAAELS--------RMLSRLE--
----------------------AQHDTCYKS--------------LVPRHQCEKDWLRQV
LLQ------------------REAEVHVRCQEQQ----------------QR--LQAALG
------KEQVQMCRCFALERKRLEHAHCEQLEHLV-------------QEAEVLQALLRG
GAAATIGREWKGAPS-----HSSLCPGTP----------------------------GGP
EKLV-----------GDWPGQP-CCVD-------------AVSQQPPESLNHEQNCWGLL
DLEEVVSDPCEKGSLRW-----PPGQSEGPD-----SEKQAPSARATVTGVL--------
-----------------LGPLSEQSAYLCPSNGPAVLVAVL-------------------
------------------------------------------------------------
------------G--------RLGR----GTEAAC----GAVG--RDLHHR-----SG--
----MSWG--WFSSRCC-------------------------------------------
----RLQGQFAFL--LIACFKTSSGLNAEAVVLM--------GQNYSSVYRVSTVTVTFQ
QLHR---ATGTT------------LPQSTLGLPSSSLSRWEATWSSREIEVLKRDKEKAC
SEMDE-LTIQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------NQKYKNEVLQLKCRVLQLEGVA-SAHQAQN
EENLAAAQLLTQRLQEAARQEEQQ---------------CAQIRKLE--AELVHLREEGQ
SVRPAQLGLTEDQEEGRTQV----------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------GLEKSARSGLLLKELYVENA
QLTKALQVAEQKQWGAEKQNRILEEKVHALGTLISKLAPASLSV----------------
---------------------------------------------------------
ENSMLUP00000000827 (view gene)
------------------------------------------------------------
------------------------MDEEEEDHYVS
PF00036 // ENSGT00660000095541_0_Domain_0 (96, 125)
QLREVYSSCDTTGTGFLDREEL-QL
CLKLENSGT00660000095541_0_Linker_0_1 (125, 336)
HLEKQLPILLQTLLGNDHFARVNFEDFKEGFVAVLSSSAGVGSSDEDS-SSLESAA
SHAVPPKYVSGSKWYGRRSR--PELCDSATR-TKCL-P------EQQAKASVKSQIRRSA
SLESVESLKSEEEAESAKGPQNELFEAKGQ---LPLWGSEAFGNTQKSCSPFSDTPESQV
RAIWEELGVGSSGHLNEQELALVCQSIGLQGLKKEPF00036 // ENSGT00660000095541_0_Domain_1 (336, 364)
ELEDLFNKLDQDQDGRVSLAEFQRG
LFNHGPTSLLESSSPVKLSRS----WSHSQ--VLEEGGCHT-TTT-SSL-VSLCSGPRLF
CSVDNGTGFAFPEQLIALWAQ-------EGIH--NGKEV----LQSLDFSVDEKVNLLEL
TWALENELMMIHGATQQAALSCYRQELRFCQ--GQVEQMVRER-DKARQDLE--------
-KAEKRNLEFVKEMDDCHSALEQLTEKK-IKHLEQEYRERLN----LLRSEVEMERELFW
EQACRQRTMLERDLE-RLQAEEASLREKLTLALKENSRLQKEIIEVVEKLSESEKLVLKL
QTDLEFVLKDK-L---EPQSTE-LLAQEEHFEEVLKEYKLKCR-DLQDSNDELQ----AA
VEGLRAQLPQSWRGQPHA-QP--------NGRLLSRHG-RAGIIS---------------
------------------------FLGDS--TPVSLETEIMMEQVKEHYQDLKI--QLET
KVNCYEREIEMMKKDFANKRKEMEQAFQHEVSVLEDQKANLEKLHVKSQEVIQVLQDQLQ
SASQGPELERTFELKRAEMEQCYAQALSGLAQQLAREKEQLEAELRRRHQQELQQFRK--
------------------------------------EAKVELN--------PKLTRME--
----------------------AQLASHCKN--------------LFTQHE-QKEEMPQK
HLLHVREMTQ-LDLEKEQREERGREILTRTQQQ---------------------LQAWMS
------EEQAQLCKSFAL---RLELTFRKQVLALV-------------SPALVLSIVLER
QVSVLLG-QQQRRPSTTGLGTVPQQALRLLQPV----------------PQGHPKVGR--
--------WPGDVR---------LSAR---SRE------------P----------REMV
SYLKKA-SPPE-------------------------------------------TDHSWA
QREHLGDL---LHVLRKQ-RPCHS----------------N---QVPLLNKPS--PKHSS
YRTPAG---QGWSLPL---SPSLFL---------------------------SVSLLHY-
K---LIFVS------------N---QNKKQSECVG----DGMGQPSAYLTSR-KLEVDAL
LPSPYLP----LSPPP---------------------------LHTHIPALSEDFFK--N
V---AFKTGINSLV---SNDGIIY------AESL--------------ASSESKGRIKLD
RLSE---ENTVLKN-------ELGRMQQELES-----TERTNDAQRKEIELLKRDKEKAC
SEMEE-LNKQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------SQKCNDELLQLNHRVLQLGEEA-SSQQAHI
ENNGITIQLLTQRLEEAGRREEVQ---------------GDQIKKLE--LELELINQECQ
SLRLSWSQLKETLEESRNQQ----------------------------------------
-----------------HELLCDLEDSPQDH-----------------------------
------TDDVHQLQKQMEQSLPVGCVAELRQLLEWEQRAAQQLCQECSLQ----------
-QEEEHKKQLKDTEEQVEEVEMILKNMEMLLQEKVGELGEQFEKNTKSDLLLKELYVENA
HLMKALQVTEEKQRGAEKENRILEEKVRALNKLICRITSASFSV----------------
---------------------------------------------------------
ENSPPYP00000012053 (view gene)
------------------------------------------------------------
-------------------------MDEEENHYVSQLREVYSSCDTTGTGFLDRQELTQL
CLKLHLEQQLPVLLQTLLGNDHFARVNFEEFKEGFVAVLSSNAGVRPSDEDS-SSLESAA
SSAIPPKYVNGSKWYGRRSW--PELCDAATE-ARRV-P------EQQTQASLKSHLWRSA
SLESVESLKSDEEAESTKEAQNELFEAQGQ---LQTWDSEDFGSPQKSCSPSFDTPESQI
RGMWEELGVGGSGHLSEPELGVVCQSVGLQGFQKE
PF00036 // ENSGT00660000095541_0_Domain_1 (336, 361)
ELEDLFNKLDQDGDGKVSLEEFQLGLFSHEPALLLESSTQVKPSKA----WSHYQ--VPEESGCHT-TTT-SSL-VSLCSSLRLF
SSIDDGSGFAFPDQVLAMWTQ-------EGIQ--NGREI----LQSLDFSVDEKVNLLEL
TWALDNELMTVDSAIQQAALACYHQELSYQQ--GQVEQLARER-DKVRQDLE--------
-RAEKRNLEFVKEMDDCHSALEQLTEKK-IKHLEQGYRERLS----LLRSEVEAERELFW
EQAHRQRAALEWDVG-RLQAEEAGLREKLTLALKENSRLQKEIVEVVEKLSDSERLALKL
QKDLEFVLKDK-L---EPQGAE-LLAQEERFAAVLKEYELKCR-DLQDCNDELQ----AE
LEGLWARLPENRHSRSWSPGG--------RRRQLSGLG-PAGIS----------------
------------------------FLGNS--APVSIETELMMEQVKEHYQDLRT--QLET
KVNYYEKEIAALKRNFEKERKDMEQARRREVSVLEGQKADLEELHEKSQEVIRGLQEQLQ
DTARGPEPEQ----MGL--APCCTQALCGLALRHQS-------HLQQIRR----------
------------------------------------------------------------
----------EAEAELSGEPSGLGALPARRD--------------LTLELEE----PPQG
PLPRGSRRSEQLELER----------ALK-------------------------LQPCAS
------EKRAQMCASLALEEEELELARRKRVDELS-------------REAEMLRALPKD
GL-VAGS-GQEGA-----RGLLPLRLGCGERPL----------------AWLAPGDGGES
EAAAGARPRR-RQAQ---------DTE---ATQSPA--PAPAP-----------------
----ASHGPSERWSRVQ-----PCGVDGDTV-----PEEPEPFGTS--------------
-----------------AAGLEQPGARELALLGTEGDASQT------------QPWMWEP
LLRPAA---SCRGQVE---RPQAIQEERARS-----------------------------
----WSRG-------------TQGQASEQQARA-E----GALEPGCHK-HSVEVARRGSL
------PSHLQLADPRGS-------------------------WQEQLAAPEEGETEIAL
E---REKDDMETK--LLHLEDVVRALEK-----H--------------VDLRENDRLEFH
RLSE---ENTLLKN-------DLGKVRQELEA-----AESTHDAQRKEIEVLKKDKEKAC
SEMEV-LNRQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------NQNYKDQLSQLNVRVLQLGQES-STHQALN
EEHRVTIQLLTQSLEEVVHSGQQQ---------------SDQIQKFR--VELECLNQEHQ
SLQLPQSELTQTLEERQGQ-----------------------------------------
---------------------------------------------------VQGAHLRLR
QAQAQHLQEVRQLQD---------CVAELQHLLSLQGE--------------QAGRRLDA
Q-REEHEKQLKATEERVEEAEMILKNMEMLLQEKVDKLKEQFEKNTKSNLLLKELYVENA
HLVRALQATEEKQRGAEKQSRVLEEKVRALNKLVSRIAPAALSV----------------
---------------------------------------------------------
ENSMNEP00000021366 (view gene)
------------------------------------------------------------
-------------------------MDEEENHYVSQLREVYSSCDTTGTGFLDRQELTQL
CLKLHLEQQLPVLLQTLLGNDHFARVNFEEFKEGFVAVLSSNAGVRPSDEDS-SSLESAA
SSAIPPKYVNGSKWYGRRSR--PELCDTATE-ARHV-P------EQQTQASLKSQLWRSA
SLESVESLKSDEEAESTKEAQNELFEAQGQ---LQTWDTEDFGSPQKSCSPSFDTPE
PF13499 // ENSGT00660000095541_0_Domain_1 (298, 358)
SQI
RGMWEELGVGSRGHLSEQELAVVCQSVGLRGLEKEELEDLFNKLDQDGDGKVSLEEFQLG
LFSHEPALLLECSTQVKLSKP----WSHYQ--VPEESGCHT-TTT-SSL-VSLCSSLRLF
SSIDDGSGFAFPDQVLAMWTQ-------EGIQ--NGREI----LQSLDFSVDEKVNLLEL
TWALDNELMTVDSAVQQAALACYHQELSYQQ--GQVEQLVRER-DKARQDLE--------
-RAEKRNLEFVKEMDDCHSALEQLTEKK-IKHLEQGYRERLS----LLRSEVEAERELFW
EQAHRQRAALEWDVG-RLQAEEAGLREKLTLALKENSRLQKEIVEVAEKLSDSERLALKL
QKDLEFVLKDK-L---EPQSAE-LLAQEERFAAVLKEYELKCR-DLQDRNDELQ----AE
LEGLWARLPKNQHSPSWSPGG--------CRRQLSGLG-PAGIS----------------
------------------------FLGNS--VPVSIETELMMEQVKEHYEDLRT--QLET
KVNHYEREIAALKRNFEKERKDMEQARRREVSVLEGQKADLEELHKKSQEVIWGLPEQLQ
DTAHDPEPER----MGL--APCCTQALCGLALRHQS-------HLRQIRR----------
------------------------------------------------------------
----------EAQAELSGELLGLRALPARRD--------------LTLELEE----PLQG
PLPRGSRRSEQLNLES----------ALN-------------------------L-ECAS
------EKGAQMCASLALE-EELELAGGKRVDRLS-------------REAEMLGALPKE
GL-VAGS-GREGA-----RGLLPLSPGYGERPL----------------AWLTPGDGGES
EEAAGAGSRC-RQAQ---------DTE---ATKRPAP--APAP-----------------
----ASHGPSERWSHVQ-----PCGVDGGTV-----PEELELFGVP--------------
------------------VGLEQPGSRELPLLGTEGDASQT------------QPRMWES
PLSPAA---SCRGQAE---KPQSIQEERARS-----------------------------
----WSRG-------------TQGQASEEQARA-E----GALESACQE-HGVEVARRGSL
------PSHLQLVDPRAS-------------------------RQEQLAAPEEGETKTAL
E---REKDDMETK--LLHLEDVVRALEK-----H--------------VDLRENDRLEFH
RLSE---ENALLKN-------DLGRVRQELEA-----AESTHNAQRKEIEVLKKDKEKVC
SEMEV-LTRQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------NQNYKDQLSQLNDRVLQLGQEA-STHQAQS
EEHRVTIQLLTQSLEEVACSGQQQ---------------SDQIQKLK--VELECLNQEYQ
SLQLSQSELTQTLEESQGQ-----------------------------------------
---------------------------------------------------VQGAHLRLR
QAQAQHSQEVRQLQEQMSRLVPQDRVAELQRLLSLQGE--------------QAGRRLDA
Q-REEHEKQLKATEERVEEAEMILKNMEMLLQEKVDELKEQFEKNTKSDLLLKELYVENA
HLVRALQATEEKQRGAEKQSRILEEKVRALNKLVSRIAPAALSV----------------
---------------------------------------------------------
MGP_PahariEiJ_P0066124 (view gene)
------------------------------------------------------------
------------------------MDNEEENHYVSRLRDVYNSCDTTGTGFLDQEELTQL
CTKLGLEEQLPALLHILLGDDRLARVNFEEFKEGFVAVLSSGSGVEPSDEEG-SSSESAT
SCAVPPKYMSGSKWYGRRSL--PELGDSATA-TKYG-L------EQQVKGSVKPPLRRSA
SLESVESLKSDEDAESAKEPQNELFEAQGQ---LRSWGCEVFGTLRKSCSPSFSTPENLV
QGIWHELGIGS
PF13499 // ENSGT00660000095541_0_Domain_1 (312, 358)
SGHLNEQELAVVCRSIGLHSLEKQELEELFSKLDQDGDGRVSLAEFQLG
LFGHEPPSLPASSSLIKPNRL----WSHYQ----EESGCHT-TTT-SSL-VSVCSGLRLF
SSVDDGSGFAFPEQVISAWAQ-------EGIQ--NGREI----LQSLDFSVDEKVNLLEL
TWALDNELLTVDGVIQQAALACYRQELSYHQ--GQVEQLVQER-DKARQDLE--------
-KAEKRNLDFVREMDDCHSALEQLTEKK-IKHLEQEYRGRLS----LLRSEVEMERELFW
EQARRQRAVLEQDVG-RLQAEETSLREKLTLALKENSRLQKEIIEVVEKLSDSEKLVLRL
QSDLQFVLKDK-L---EPQSME-LLAQEEQFTAILNDYELKCR-DLQDRNDELQ----AE
LEGLRVRLPRSQQSPSGTPGT--------HRRRIPGRG-PADNL----------------
------------------------FVGES--TPVSLETEIMMEQMKEHYQERRM--QLET
KVNYYEKEIDVMKRNFEKDKKEMEQAFQLEVSVLEGQKADLEALYAKSQEVILGLKEQLQ
DAARSPEPA------PAGLAHCCAQALCTLAQRLEV-------EMHLRHQDQLLQIRR--
------------------------------------EAEEELN--------QKLSWLE--
----------------------AQHAACCES--------------LSLQHQCEKDQLLQT
HLQRVKDLAAQLDLEKGRREEREQEVLAHCRRQQ----------------LK--LQAVMS
------EEQARICRSFTLEKEKLEQTYREQVEVLV-------------QEADVLRALLKN
GTTVVSD-QQERTPSSMSLGP-----DSRQQPT----------------ARQAVSPDGRT
GAPA-EWPGPGKAEGRDFPGQL-CSID---AI--PSPTPTLLSRRSSENLGVRDNHQGPL
SAEEGAIPK-----------------------------ELEPSARTLT------------
-----------------------G--------------------QDQKLPLPVHPQMLEL
RLGTAV------------------------------------------------------
----LDRKPASVG--------VQGQASEGTIGDGD----GV--QEAWLQFRGEAA-----
-RMRPSLPCSELPNPQEA--------------------------------------TVTP
AVSESEMKDVKT--KLLQLEEVVRALEK--------------------ADSRESYRAELQ
RLSE---ENLVLKS-------DLGKIQLELET-----SESRNEVQRQEMEVLKRDKEQAL
LDLEE-LSTQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------TQKYKDEMSQLNCRILQLEGES-SGFHTQK
EESHTAIQVLMKKLEEAGCREEQQ---------------GDQIQNLK--IELECVNEECQ
RLRLSQAELTENLEESRGQ-----------------------------------------
---------------------------------------------------LHSVQLRLE
AAQSQHGRIVQRLQEQMSQFVPGARVAELRHLLNVREE---------------EAGRLSA
Q-QEEYRQQLKAREEQVEDAEARLRNVEWLLQEKVEELRKQLEKSTRSDLLLKELYVENA
HLMKAVQLTEEKQRGAEKKNCVLEEKVRALNKLISKMAPASLSV----------------
---------------------------------------------------------
ENSRNOP00000061689 (view gene)
------------------------------------------------------------
------------------------MDNEEENHYVSRLRDVYSSCDTTGTGFLDQEELTQL
CTKLGLEEQLPALLHILLGDGRLARVNFEEFKEGFVAVLSSATGVEPSDEEG-SSSESAT
SCAVPPKYMSGSKWYGRRSL--PELGDSATT-TKCG-S------EQQAKGSVKPPLRRSA
SLESVESLKSDEDAESPKEPQNELFEAQGQ---LRSWGCEVFGTPRKSCSPSFNTPEN
PF13499 // ENSGT00660000095541_0_Domain_1 (299, 358)
QV
QGIWHELGVGSSGHLNEQELAVVCRSIGLHGLEKQELEELFSKLDRDGDGRVSLAEFQLG
LFGHEPPSLPASSSLIKPNGP----WSHYQ----EESGCHT-TTT-SSL-VSVCSGLRLF
SSVDDGSGFAFPEQVISAWAQ-------EGIQ--NGREI----LQSLDFNVDEKVNLLEL
TWALDNELLTVDGVIQQAALACYRQELNFHQ--GQVEQLVQER-DKARQDLE--------
-KAEKRNLDFVREMDDCHSALEQLTEKK-IKHLEQEYRGRLS----LLRSEVEMERELFW
EQARRQRAVLEQDVG-RLQAEETSLREKLTLALKENSRLQKEIIEVVEKLSDSEKLVLRL
QSDLQFVLKDK-L---EPQSME-LLAQEEQFTAILNDYELKCR-DLQDRNDELQ----AE
LEGLRVRLPRSRQSPSGTPGT--------HRRWTPGRG-PADNL----------------
------------------------FVGES--IPVSLETEIKMQQMKENYQELRM--QLET
KVNYYEKEIEVMKRNFEKDKKEMEQAFQLEVSVLEGQKADLETLYAKSQEVILGLKEQLQ
DAARSPEPA------PAGLAPCCAQALCTLAQRLGV-------EMHLRHQDQLLQIRR--
------------------------------------EAEEELN--------QKLSWLE--
----------------------AQHAACCES--------------LSLQHQCEKDQLLQT
HLQRVKDLAAQLDLEKGWREEREQEVLAHCRRQQ----------------LK--LQAD-E
------EEQARICRSFTLEKEKLEQTYREQVEGLV-------------QEADVLRALLKN
GTTVVSD-QQERIPGSMYPGP-----DSRQQPP----------------TWQTVSPDGRT
GAPA-EWPGPGRADGRDLPGQL-CSLD---AV--PSPTPTLLSRRSSESLDVRDNHQGPL
SAEEGAVPK-----------------------------EPEPSARTLT------------
-----------------------G--------------------QDQKLPLPIQPQMLEP
WLGPAA------------------------------------------------------
----VDRKPDSVR--------VQGQASEGPTGDDK----GV--QETPLQLRGETA-----
-RMRPSLPYSELPNPQEA--------------------------------------KVMS
VMSESEMNDVKT--KLLQLEDVVRALEK--------------------ADSRESYRAELQ
RLSE---ENSVLKS-------DLGKIQLELGT-----SESRNEVQRQEIEVLKRDKEQAC
FDLEE-LSTQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------TQKYKDEMSQLNCRILQLEGDS-SGLHTQK
EENHAAIQVLMKKLEEAECREKQQ---------------GDQIKHLK--IELERVNEECQ
RLRLSQAELTGSLEESQGQ-----------------------------------------
---------------------------------------------------LHSVQLRLE
AAQSQHDRIVQGLQEQMSQLVPGARVAELQHLLSLREE---------------EAERLNA
Q-QEEYKQQLKAREDQVEEAEARLHNVEWLLQEKVEELRKQFEKNTRSDLLLKELYVENA
HLMKAVQLTEEKQRGAEKKNCVLEEKVRALNKLISKMAPASLSV----------------
---------------------------------------------------------
ENSNLEP00000019042 (view gene)
------------------------------------------------------------
-------------------------MGEEENHYVSQLREVYSSCDTTGTGFLDRQELTQL
CLKLHLEQQLPILLQTLLGNDHFARVNFEEFKEGFVAVLSSNAGVRPSDEDS-SSLESGA
SSAIPPKYVNGSKWYGRRSR--PELCDAATE-ARHV-P------EQQTQASLKSHLWRSA
SLESVESLKSDEEAKSTKEAQNELFEAQGE---LQTWDSEDFGSPKKSCSPSFDTPE
PF13499 // ENSGT00660000095541_0_Domain_1 (298, 358)
SQI
RGMWEELGVGSSGHLSEQELAVVCQSVGLQGLKKEELEDLFNKLDQDGDGKVSLEEFQLG
LFSHEPVLLLESSTQVKPSKA----WSHYQ--VPEESGCHT-TTT-SSL-VSLCSSLRLF
SSIDDGSGFAFPDQVLAMWTQ-------EGIQ--NGREI----LQSLDFSVDEKVNLLEL
TWALDNELMTVDRAVQQAALACYHQELSYQQ--GQVEQLARER-DKARQDLE--------
-RAEKRNLEFVKEMDDCHSALEQLTEKK-IKHLEQGYRERLS----LLRSEVEAERELFW
EQAHRQRAALEWDVG-RLQAEEAGLREKLTLALKENSRLQKEIVEVVEKLSDSERLALKL
QKDLEFVLKDKAS---PFADS---LP--PEGRACWAEWGWRGRDDLQDRNDELH----AE
LEGLWARLPENRHSPSWSPGG--------RRRQLSGLG-PAGIS----------------
------------------------FLGNS--APVSIETELMMEQVKEHYQNLST--QLET
KVNYYEREIAALKRNFEKERKDMEQAHRREVSVLEGQKANLEELHEKSHEVIRGLQEQL-
DTARCPEPER----MGL--APCCTQALCGLALRHQS-------HLWQIRR----------
------------------------------------------------------------
----------EAEAELSGEPSGLGALLARRD--------------LTLELEE----PLQG
PLPRGSRRSEQLELER----------ALK-------------------------LQPCAS
------EKRAQMCASLALE-EELELARGKRVDGLS-------------REAEMLWALPKD
RL-VAGS-GREGA-----RGLLPLRPGCGERPL----------------AWLAPGDGGES
EEAAGAGPHR-RQAQ---------DTE---ATQSPA--LAPAP-----------------
----ASHGPSERWSRVQ-----PCGVDGDTV-----SEEPEPFGAS--------------
-----------------AAGLEQPGARELPLLGTEGDASQT------------QPWMWEP
PLRPAT---SCRGQPE---RPQAIQEERARS-----------------------------
----WSRG-------------TQGQALEEQARA-E----GDLEPGCHK-HSVEVARRESL
------PSHLQLADPRGS-------------------------WQEQLAAPEEGETEIAL
E---REKDDMETK--LLHLEDVVRALEK-----H--------------VDLRENDRSHHC
TSAWATKAKLCLKKKLQKLSQLLGRLRQENRL-----N------------LR----GRGF
S-EPR-SHHC--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------TPNYKDQLSQLNVRVLQLGQEA-STHQAQN
EEHRVTIQLLTESLEEVVRSRQQQ---------------SDQIQKLR--VELECLNQEHQ
SLQLPRSELTQTLEESQGQ-----------------------------------------
---------------------------------------------------VQGAHLRLR
QAQAQHLQ-------EMGRLVPRDRVAELQRLLSLQGE--------------QAGRRLDA
Q-REEHEKQLKATEERVEEAEMMLKNMEMLLQEKVDKLKEQFEKNTKSDLLLKELYVENA
HLVRALQATEEKQRGAEKQSRVLEEKVRALNKLVSRIAPAALSV----------------
---------------------------------------------------------
ENSLAFP00000010637 (view gene)
------------------------------------------------------------
------------------------MDEEEENHYVL
PF00036 // ENSGT00660000095541_0_Domain_0 (96, 124)
QLKEFYSSCDTTGTGYLDREELTQL
CLKENSGT00660000095541_0_Linker_0_1 (124, 336)
LHLEKQLPILLQTLLGSDRFARVNFQEFKDGFVAVLSSNPSHGPSGEEN-SSLEPAV
SQAVVPKYVNGSKWYGRRTR--PELYTCGTE-DGHS-A------DQQAQASLSSQLCHAT
SLQSVESFKSDEEADSAREPQSELFEAQGQ---LRAWSPKVLGSPQELSSPTFDTPQSQA
QVIWEELGVGSSGYLSAQELAVVCQSIGLQRLEKEPF00036 // ENSGT00660000095541_0_Domain_1 (336, 364)
ELEDLFKKLDRDGDGKVSLAEFQLG
LFSHGPASLPESSTPIQLSRP----WPDFQ--AVEESGCRT-TTT-SSL-VSICSGLRLF
SSIDDGTGFASPKQVVTLWAQ-------EGIQ--NGKEI----LQNLDFSVDEEVDLLEL
SWAFDNELMMVTGVTQQAALASYRQELTYLQ--GQMEQMVKER-DKARQDLE--------
-KAEKRNLEFVTEMDDCHSAMEQLAERK-IKHLEQGYHGRLA----LLRSEMEMERELLW
QQTSRQRARLEEDLA-ALRREEACLREKLALALKENSRLQKEIFEVVEKHSESEKLVLKL
QNDLEFVLKDK-L---EPQSTE-LLSQEERFTEILKEYEVKCK-DLQDHNDELQ----AA
LEGLQAWLHKTQRGPPPSPCGGACGA----------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------HA-------LLSGGERWKEELRLRHQHELQQV----
----------------------------------RKEAEAELS--------QKLLWMEA-
-----------------------QQATLREH--------------LSRQHQREEEDMLQT
HLLQVRDVAVQLDSEKGWRDEREKEILAQCRKQQ----------------LK--LEELMS
------EEEAHICKAFALEKGKLELTHRKQVEGLI-------------QETQMLRPALKE
GTAAADH-ERERR-----PPPGLLGPAEV----------------------------GEE
AGPAGGRPGPGGAEAMGIPGWPHS---CAGPVQSPTSIPALRSKGPSASFSIRDGHPGLL
YAVEMACVPSEKPV--------PLPSNRA-----G--------------QSWQSSEQR--
QPK-------------------------------------------A-------P-----
EQLWLV---------------------MDGDN----------------------------
-------------------------------------------------------LHGVP
ALGRSL-------------------SILEGEHGAGPTETGRQEWE---GAIQRQEAETV-
--LEREKIEMKTR--LLQLEDVVRAL-------------------EKEADTRENNRIELQ
RLSE---QNALLAD-------ELERIQQELGA-----AERTNDAQRKEIEVLKKDKDQAR
FEMET-LNKQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------SQKYKDQLSQLNHRTLQLDREVNNAYRAQN
EKNEVTIQLLTQRLEGAGHQEEEW---------------G-QLRDLE--TRTARHVAEAD
IGGAAQGPKRHAL-----------------------------------------------
---------------------------------------------------------ARH
VARGGHWRGSSGTRKCMLGLVRTAKTWDWGHLSHS---------------Q-AEKPEHES
SQQEEHEKQLTATKEQVEEVELILKNVEMLLQEKVDELKDQFEKSTKSDLLLKELYVENA
HLMKALQVTEEKQRGAEKNNLILEEKIRALNKLISKIAPASLSV----------------
---------------------------------------------------------
ENSOPRP00000009399 (view gene)
------------------------------------------------------------
-------------------------MEEEESHYIL
PF00036 // ENSGT00660000095541_0_Domain_0 (96, 124)
QLRAVYSSCDTTGTGFLDRDELTQL
CRKENSGT00660000095541_0_Linker_0_1 (124, 336)
LQLEKQLPVLLQTLFGSDPFARXXXXXXXXXXXXXXXXXXXX-----XXXXXXXXXX
XXXVPPKYVNGSKWYGRRSQ--PELWDPVTE-AKRV-P------EPQAKGNLKSQLQRST
SLESVEXXXXXXXXXXXXXXXXXXXXXXXQ---LEAWGSEVLGGSQKSCSPSLDSPESQV
RAIWDKLGVGNSGHLSRQELAVVCQSIGLRTLDTEPF00036 // ENSGT00660000095541_0_Domain_1 (336, 364)
ELEDLFNKLDRDGDGRVSLEEFQLG
LFSHVPPSLTETCSVVEPSRP----WCHCQ--VPKESSCHT-PTA-SSL-VSVASGLLLF
SSIDDGTGSAFPEQIIATLAQ-------EGIH--NGREI----LQXXXXXXXXXXXXXXL
SWADECT---VD-GDQQAALACYRQELSFLQ--GRMEQLVQER-DKARQDLD--------
-RAEKRNLEFVKEVDDCHSTLEQLTEKK-IKHLEQGYLERLN----LLRSEVEMERELLW
EQARKQKATLEQDVA-RLLAEEAXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXX-----XXXXXX-XXXXXXXXXXXXXXXXXXXXXDLQDRNDELQ----AE
LEGLRARLPESQHSSAQGPSG--------HRRQLPGRG-PAXXXXXXXXX--XXXXX---
-------------------------------------XXXX--XXXXXXXXXXXXXXXXX
XVNYYEREVETMKRNFEKERKEVELAHQQEISALEEQRADLEALQAKAQEVIQGLQEQLR
DVARTPEPT------QAGLGQCCAQALSGLALRHQDQLQQMXXXXXXXXXXXXXXXXXXX
XXX---------------------------------XX-XXXX--------XXXXXXX--
----------------------XXXXXXXXX--------------XXXXXX----XXXXX
XXX------X----XX----------XXXXXX-X----------------XX--XXXXXX
------X--XXXXXXXX----XXXXXXX--XXXXX--------------X--XXX-XXXX
X--XXXX-----------XXXXXXXXXXXXXXX----------------XXXXXGADGE-
-----------TEEPREGPGHS-CCAD---TVQSPAAAPILAS-----------------
------RVPSERWSCVQ-----SCGMDKRGN-----LEDPVPPDRAPESL----------
---------------SGPAGLEPTC------PGAEANDLAA------------SSRMTEP
QLSGQ---------VE---RMRTALEEA--------------------------------
----QEGVP--------------------QARDSN----GALDPARQPP-------KGAA
T-REPSFSHAGLPD-----------------------------GQEQLAMLGQTEAEGTA
E---REKRDVQAK--LVQLEGIVCALEAA-------------------AASRDAERIEFQ
RLSE---ENVLLKK-------ELGRVQQELEA-----AENASDAYXXXXXXXXXXXXX--
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------XXXXXXXXXXXGQKYEDELSQLHSRVLQLEEGA-SAHHTWR
EKNHVTVALLTQGLEEAGEREGQQV-------------QGEQIQKLE----LERMNQECQ
RLRLSQSELTETLEESQPQ-----------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------------------------------------------------------
ENSHGLP00000000366 (view gene)
------------------------------------------------------------
-------------------------MDEEDNHYISQLKDVYNSCDSTGTGFLDQEELTEL
CRKLHLEKQLPALLQTLLGDDHIARVNFEEFKEGLVTVLSSKAGGNPISEEA-GSLQSAA
SYTVPPKYVSGSKWYGRWSQ--PEQRASATK-ARCG-P------EQQARAGLKSQLHHSM
SLESVESLKSNEEAESPREPQDELFEAQGQ---LQTWGSEVFGSPQKSCSPLYASSE
PF13499 // ENSGT00660000095541_0_Domain_1 (298, 358)
SQV
QDIWEQLGVGSRGHLDEQELAVVCQSIGLHGLEKEELKDLFHKLDQDGDGRVSLEEFQLG
LCSHKPSQIPESSTLVKPSRP----WSHCQGQVPEESGCHTATTT-SSL-VSMSCGLRLF
SSIDDGSGFSLPEQVIATWAQ-------EGIH--SGREI----LQSLDFSLDEKVDLLEL
TWALDNELLTVDSVIQQAALACYRQELSYHQ--EQVEQLVQER-DKARQDLE--------
-RAERRSLELVREMDDSHTTLEQLTEQK-IVCLEQDYRGRLN----LLRAEVEAERELIW
EQACRQSAMLEQDLG-HLRAEEASLRHRLGLASKENSRLQQEILEVVGKLSDSEKLVLKL
QGDLEFMLREK-L---ETQGME-LQAQEEQSAAVLKEYELKCR-DLQDQNDELQ----AE
LESLQARLPQS--QSWQCPA------------GDAGHH-PAGVY----------------
-------------------------VDDS--SPVSLETEILVEQMREHCQELRT--QLEA
KVNSYEREIEAMKNNFEKERMELEQARQQEVTVLEARRADLEVLCATSQEVILGLREQLR
AAAGGPEAS------WAAMTSCCAPVLCDLAQQLDK-------HMWQQHQGELRQIR---
-----------------------------------QEAAEELS--------RM----L--
----------------------SQHDIHCKS--------------LVLRHQLEKDRLQQV
HLR------------------REEEVLVRCQEQQ----------------QR--LQVALG
------EEQAQMCRSFALERKRLERAHREQVGSLV-------------QEAEALRVLLQG
GAAATTDKEQQGAPI-----PMSPCLDIH----------------------------GGT
WEPV-----------GDGPGQP-CCTD-------------AVSRGLPESLDHEQSCWGLM
DAEETASVPFEKGPPIQ-----APGQNKGAD-----PEEPAPSARATVSPSW--------
-----------------PDVQE------LPSRGTG-------------------------
------------------------------------------------------------
---------------------------------ED----GALQ--TWLQHH-----SG--
----P-GTTPEFPNPAG---------------------------------------ETEA
DMVEREKNDMKTK--LLQLEDVVRALKREADS------------------R-ENDREELQ
RLSE---ENGLLKN-------DLERIQQKLGA-----AEHMSDMQRQEIEALKRDKEKVC
FEMDEQLSIQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------NLKYKNEVSQLNCRVLQLEGVT-STHQAQN
EENLAAAQLLTQRLEEVGRREEQQ---------------CAQIQKLE--TELEHMSQQCQ
RLKLVRSELTESREEGQDQEEQ--------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------EQLDTRAG-------PAVEVELLLREKL----DQLEKTTRSGLLLKELYVENA
HLTKALQVAEQKQWGAEKQKRILEEKVRALNTLISKLAPAALSV----------------
---------------------------------------------------------
ENSPCOP00000015224 (view gene)
------------------------------------------------------------
------------------------MDEEEENHYVSQLREVYSSCDTTGTGFLDREELTQL
CLKLHLEKQLPVLLQTLLGNDHFARVNFEEFKEGFVAVLSSNAAVGPSEEDS-STLDTAA
SRAVPPKYMRGSKWYGRWSQ--PEVCDAAIK-ARYV-P------EQQTKANLKSQLRRSA
SLESVESLKSDEETESTKEPQNELFEAQGQ---LQTWGSEVFGSPQKSRSPSFDTPES
PF13499 // ENSGT00660000095541_0_Domain_1 (299, 358)
QV
QGIWEELGVGSSGHLNEQELAVVCRSVGLRGLEKEELEDLFHKLDQDGDGRVSLEEFQLG
LLSHEPTSLPESSTQVNLSRP----WSHYQ--VPEESGCHT-TTT-SSL-VSLCLGLRLF
SSIDDGTGFAFPEQVMAVWAQ-------EGIQ--NGGEI----LQSLDFSVDEKVNLMEL
TWALDNELMTVDGAAQQAALACYRQELSYHQ--GQVEQLARER-DKARQDLE--------
-RVEKRNLEFVKEMDDCHSALEQLTEKK-IKHLEQGYRGRLN----LLRSEVETERELFW
EQARRQRAALERDVG-RLQAEEASLREKLTLALKENSRLQKEIVEVVEKLLDSEKLVLKL
QNDLELVLKDK-L---EPQSTE-LLAQEERFTAVLKEYEVKCR-DLQDRNDELQ----AE
LEGLRARLAPSRQNSS---RG--------LGQRLSGQG-PAGIL----------------
------------------------LTGSS--APVSIETEIMMEQLKEHHRDLRI--QLET
KVNYYEREIEVMKRDFEKERKEMEQAHKHEVGVLEEQKADLEVLHAKSQEVIQGLREQLQ
DLAHGPAPEQ----LGLALAPCCTQALSSLALRHRS---------------QL-------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------------QQIRIEFH
RLSE---ENTLLKN-------DLGSIQQELKA-----AESTNDAQRREIEVLKRDKEKAC
FDVEE-LSKQ-------V------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------NQKYRDQLSQLNHSVLQLGEEA-STHQAQN
EENRITIQLLTQSLEEAGRRGEQQ---------------SDQIQKLE--VELERLNQECQ
SLRLSQSELTKTLEESQDQ-----------------------------------------
---------------------------------------------------LQGAQLRLK
LAQSQHSQE-------MGRLVPRDDVAKLQHLLSLQRTQARL--D--PWSS-T-------
---KKHEKQLKATEERVGEAEMMLKHMEMLLQEKAEELKGQFEKNTKSNLLLKELYVENA
HLVKALQVTEEKQRGAEKKSRILEEKVQALNKLISKIAPAAMSV----------------
---------------------------------------------------------
ENSODEP00000019318 (view gene)
------------------------------------------------------------
-------------------------MDEGDNYYVSQLKDIYSSCDATGTGFLDREELTQL
CQKLHLQKQLPALLKILLGDNPSARVTFEEFKEGLVAVLSSKAGGDLSSEEA-GSLQSAA
SSAVPPKYVSGSKRYGHRSQ--PERRTSATE-ATYG-S------EQLARAGLKRQHCRSA
SLESVESLKSDEEAESPREPQDELFEAQGN---S--W---------PR----ICCPLPLV
QGIWE
PF13499 // ENSGT00660000095541_0_Domain_1 (306, 358)
QLGVGCRGHLDEQELAVVCRSIGLHASLVQELKDLFHKLDQDGDGRVSLAEFQLG
LCHHEPMLLLES-SLGKPSRP----WSHCQ-----ESGCHTATTTTSSL-ASMSCGLRLF
SSVDDGSGFALPEQVIAIWAQ-------EGIH--SGREI----LQSLDFSLDEKVDLLEL
TWALDNELLTVDSIIQQAALACYRQELSYHQ--EQAEQLVQER-DKARQDLE--------
-QAERRSLELAKEMDDSHTALEQLTERR-IERLEQDYRGRLS----LLRAEAEAEREQIW
EQSCRRSAVLETDLG-HLRAEEASLRHRLGLATKENSRLQQEILEVVGKLSDSEKLVMKL
QGDLEFVMREK-L---EPQGLE-LRAQEERFTAILKEYELKCR-GLQDQNDELQ----AK
LESLR--APGS--RSWQRPA------------GEAESR-PASRS----------------
-----------------------PQGEDS--SPVSLEMEIMVERLRQHCQELRT--QLEA
KVNSYEGEIEAMKNNFEKERMDLEQARQREVAVLEAQRADLEALCAKSQEVILGLREQLW
DAAGRPEPS------RAQLTSCCAQALCDLARRECE-------VW-----DPRFLPR---
-----------------------------------WEAAEELS--------RMLSQLE--
----------------------AQHDIHCKS--------------LALRHQREKDWLRQA
HLR------------------WEEEVFTRCQEQQ----------------QR--LQAALG
------EEQAQMCRSFALERKRLARAHREQVGDLV-------------REAEALRALLQG
GATVIVGREQNGAPS-----PLSLSPGTH----------------------------RET
GEPV-----------GEQPGQP-CCVD-------------TVSRGPPESPDPGQSCWVLL
DAKEATSVPCEEGSHVR-----PPGQSKGAN-----PEEPAPSARATVSPRQ--------
-----------------PDSQE------LPLWGAE-------------------------
------------------------------------------------------------
---------------------------------GD----STLP--TSLQHQ-----PE--
----PLLPYPELPNPGG---------------------------------------ETEA
EMA-ERENDMKTK--LLQLEDVVRALERETDL------------------R-ENDRVELQ
RLSE---ENTLLKT-------DLERIQQELGA-----TEHMNDEQRLEIEALKRDKEKTC
FDMDA-LNIQ--------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------NQKYQNEVAQLSCRVLQLEEAA-ATHQARN
EENLAAAQRLTQRLEEARCREEQQ---------------CVRIQKLE--AELEHVSEECQ
SLRLVRLEQMEKRKEGWDQVRV--------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------CG-SWR-L-------LGQQGRVWASPGG----LWLEKSTRCGLLLEELYVENA
HLMKALQVAEQKQWGAEKQNRILEEKVGALSTLVGRLAPAALSV----------------
---------------------------------------------------------
{"ENSP00000278886": [["Domain_1", 298, 358, "PF13499"]], "ENSRROP00000026175": [["Domain_1", 298, 358, "PF13499"]], "ENSMICP00000049940": [["Domain_1", 299, 358, "PF13499"]], "ENSOCUP00000005445": [["Domain_1", 336, 361, "PF00036"]], "ENSCGRP00000003428": [["Domain_1", 298, 358, "PF13499"]], "ENSTSYP00000027443": [["Domain_1", 299, 358, "PF13499"]], "ENSPTRP00000083627": [["Domain_1", 298, 358, "PF13499"]], "ENSGGOP00000026802": [["Domain_1", 298, 358, "PF13499"]], "ENSPPAP00000017533": [["Domain_1", 298, 358, "PF13499"]], "ENSSTOP00000021764": [["Domain_1", 301, 358, "PF13499"]], "ENSCANP00000022283": [["Domain_1", 298, 358, "PF13499"]], "ENSCGRP00001000566": [["Domain_1", 298, 358, "PF13499"]], "ENSTTRP00000001543": [["Domain_0", 96, 124, "PF00036"]], "ENSSSCP00000007598": [["Domain_1", 298, 358, "PF13499"]], "ENSCLAP00000016910": [["Domain_1", 299, 358, "PF13499"]], "ENSEEUP00000014019": [["Domain_0", 96, 124, "PF00036"], ["Linker_0_1", 124, 336], ["Domain_1", 336, 364, "PF00036"]], "ENSANAP00000023813": [["Domain_1", 299, 358, "PF13499"]], "ENSPANP00000017187": [["Domain_1", 298, 358, "PF13499"]], "ENSCATP00000008377": [["Domain_1", 298, 358, "PF13499"]], "ENSCJAP00000021371": [["Domain_1", 299, 358, "PF13499"]], "MGP_CAROLIEiJ_P0055140": [["Domain_1", 312, 358, "PF13499"]], "ENSMMUP00000013021": [["Domain_1", 298, 358, "PF13499"]], "ENSMEUP00000001891": [["Domain_1", 336, 364, "PF00036"]], "ENSCAFP00000006567": [["Domain_1", 302, 358, "PF13499"]], "ENSHGLP00100017905": [["Domain_1", 298, 358, "PF13499"]], "ENSFCAP00000005012": [["Domain_1", 303, 358, "PF13499"]], "ENSMPUP00000015671": [["Domain_1", 336, 361, "PF00036"]], "ENSPCAP00000003101": [["Domain_0", 96, 124, "PF00036"], ["Linker_0_1", 124, 336], ["Domain_1", 336, 364, "PF00036"]], "MGP_SPRETEiJ_P0057281": [["Domain_1", 312, 358, "PF13499"]], "ENSCHIP00000024306": [["Domain_1", 312, 356, "PF13833"]], "ENSMFAP00000003111": [["Domain_1", 298, 358, "PF13499"]], "ENSMLEP00000000025": [["Domain_1", 298, 358, "PF13499"]], "ENSFDAP00000004647": [["Domain_1", 299, 358, "PF13499"]], "ENSMUSP00000105522": [["Domain_1", 311, 358, "PF13499"]], "ENSTBEP00000010686": [["Domain_0", 96, 124, "PF00036"], ["Linker_0_1", 124, 336], ["Domain_1", 336, 364, "PF00036"]], "ENSETEP00000008000": [["Domain_0", 96, 124, "PF00036"], ["Linker_0_1", 124, 336], ["Domain_1", 336, 364, "PF00036"]], "ENSCPOP00000004536": [["Domain_1", 326, 357, "PF13833"]], "ENSPVAP00000013573": [["Domain_0", 96, 124, "PF00036"], ["Linker_0_1", 124, 336], ["Domain_1", 336, 364, "PF00036"]], "ENSPEMP00000011154": [["Domain_1", 299, 358, "PF13499"]], "ENSCCAP00000031453": [["Domain_1", 299, 358, "PF13499"]], "ENSDORP00000009766": [["Domain_1", 298, 358, "PF13499"]], "ENSRBIP00000029549": [["Domain_1", 298, 358, "PF13499"]], "ENSSBOP00000017104": [["Domain_1", 299, 358, "PF13499"]], "ENSMAUP00000025222": [["Domain_1", 299, 358, "PF13499"]], "ENSAMEP00000010995": [["Domain_1", 336, 364, "PF00036"]], "ENSCAPP00000012747": [["Domain_1", 298, 358, "PF13499"]], "ENSMLUP00000000827": [["Domain_0", 96, 125, "PF00036"], ["Linker_0_1", 125, 336], ["Domain_1", 336, 364, "PF00036"]], "ENSPPYP00000012053": [["Domain_1", 336, 361, "PF00036"]], "ENSMNEP00000021366": [["Domain_1", 298, 358, "PF13499"]], "MGP_PahariEiJ_P0066124": [["Domain_1", 312, 358, "PF13499"]], "ENSRNOP00000061689": [["Domain_1", 299, 358, "PF13499"]], "ENSNLEP00000019042": [["Domain_1", 298, 358, "PF13499"]], "ENSLAFP00000010637": [["Domain_0", 96, 124, "PF00036"], ["Linker_0_1", 124, 336], ["Domain_1", 336, 364, "PF00036"]], "ENSOPRP00000009399": [["Domain_0", 96, 124, "PF00036"], ["Linker_0_1", 124, 336], ["Domain_1", 336, 364, "PF00036"]], "ENSHGLP00000000366": [["Domain_1", 298, 358, "PF13499"]], "ENSPCOP00000015224": [["Domain_1", 299, 358, "PF13499"]], "ENSODEP00000019318": [["Domain_1", 306, 358, "PF13499"]]}

