Home
Query Results
"ENSGT00640000091449_2"
Found:
ENSGT00640000091449_2
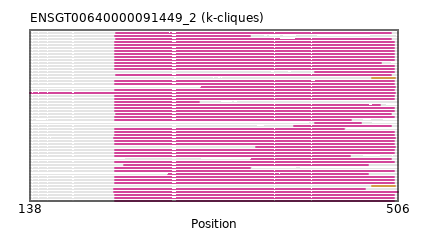
ENSOGAP00000016616 (view gene)
--------MRQVLLCNVCVTSQE-I-----RH-------HLHLPSILDKMPAPGALILLA
AV----SASGCLASPAHPDGFALGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FVFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 502)
SYLQEIGSGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGLCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHAHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPAHSA--
-------------PRPGTLSSPFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAQPWQPASAPPAPHANPSNPFYEA--------LSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLFP-NDWDPLDPGVPAPQAPQAPSEVPQ
LVSETWASPL-FPAP---RPFP-AQSSGSGSFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--ERGTAPWAEEEE-EEEE--GSSPGEDSSSIGGGPSRRGPLP
CPLCSREG-----ACSCLPLERGDAVAGWGGHPAL-GC----PHPPEDDSSLRAERGSLA
DLPLAPPTS------APPEFLDPLMGA-AAP----QYP---GRGPPPAPPPPPP------
--------------------------------------------------PPRVPADPAV
-----SPDPPSAVASPGSGLSSPGPKPGDSGYETETPFSPEGPFPGGGAA--EEEGVPRP
RAPPEPPDPGVPRPPPDPGPLLLP------------------------------------
------------------------------------GNREKPTFVVQVSTEQLLMSLRED
VTRNLLGEKGATARETGP-RKVGRGPGNREKVPGLNKDLTALGDGNQAPSL----S-LPV
NGTIVLENGGQRAPDIEGKAAIKEKEAIEEKAVENGGLGSPEREE----KALENGELTPP
RREEKV--L--------------------------ENGDLRSPEVGEKVVVNGGLTPPKS
GENVSENGGLRFPTNM-ERLPETGPWRVPGPWEKMPESGGPAPMIEE-PAPEISLEKAPA
PG-------TVASSWNGGETAPGPLGPAPKSGTLE--PGTERKAHETGGVLKAPGAGRLD
LGSGGRAPV-GTGMAPAGGPGSVVDVKAGWVDNMRQQPPLL---PPPPEAQLRRPE----
-PV---PQKAKPEVAP-EGEPGVPDSKA---------GGDMAPSGDGDP-----PKPERK
GPEMPR-LFLDLGPPQGNSEQIKA------------------------------------
------------------------KLSRLSLALPPL---TLTPFPGPSPRRPP--WEGAD
AG--AAGGEAGGAGAPGSAEEDGEDEDEDEEEDEE----AAAPDAAAGPRGSGRSRAAPV
PVVVSSADADAARPLRGLLKSPRGAEEPED------SELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQGPPEGDTDLSTPPAPPTPPHPATPGD--------------
---------------------GFPSNDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARAPDTRPAGPVEN----------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSHGLP00000008992 (view gene)
------MCQLC---V--GCVRLTPAS-WHVAHMSVPCRHHLHLPSILDKMPAPGALILLA
AV----SASGCLASPAHPDGFALGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 502)
SYLQEIGSGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGLCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYGDYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPSHSA--
-------------PRPGTLSSPFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAPAWQPASAPPAPHANPSNPFYEA--------LSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLFP-NDWDPLDPGLPAAQAPQAPSEVPQ
LVSETWASPL-FPAP---RPFP-AQSSASGSFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--ERGTAPWAEEE--EEEE--GSSPGEDSSSLGVGPSHRGPLP
CPLCSREG-----PCSCLPLERGDAVAGWGGHPAL-GC----PHPPEDDSSLRAERGSLA
DLPLAPPAS------APPEFLDPLMGA-AAP----QYP---GRGPPPAPPPPPP------
--------------------------------------------------PPRASAEPAV
-----TPDPPSAVASPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGEAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLLLP------------------------------------
------------------------------------GTREKPTFVVQVSTEQLLMSLRED
VTRNLLGDKGATPRETGL-RKAGRGPGNREKVLGLNRDLPVLGSGKKAPSL----S-LPV
NGVTVLGNGAQRAPDIEE------K------PAENGGLGSPEREV----KVLSNGELTPP
RREEKA--L--------------------------ENGGPRSPEAGEKVLANGGPIPPGI
EEKVSENGLLRFPRNT-ERLTETGPWRAPGPREKTPESGE--------PAPETPLERAPV
PS-------EVALLQNGLETAPGPLGPAPKIGMLD--PGKERRAHETGGVPRAPGAGRLD
LGSGGRAPV-GMGMAPGGGLGSGVDAKVGWVDSKKPQPLPPL---PPPEAHPRRPE----
-PV---SPRVRPEVAP-EGEPGVPDSRA---------GGDTALSGDGDP-----SKPERK
GPEMPR-LFLDLGPPQGNSEQIKA------------------------------------
------------------------KLSRLSLALPPL---TLSPFPGPGPRRPP--WEGAD
AG--AAGGEAGGAGSSGPAEEDGEDEDEDEEEE----------DAAAGPRGPERTRAAPV
PVVVSSGDADAARPLRGLLKSPRGADEPE-----DS-ELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQGPPEGDTDPSTPPAPPTPPHPATPGD--------------
---------------------GFPSNDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARAPDARSAGPVEN----------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSSTOP00000018152 (view gene)
--------ME--TSAPRVGVRAS-G-----RH-------HLHLPSILDKMPAPGALILLA
AV----SASGCLASPAHPDGFALGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLSLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 502)
SYLQEIGSGWFGKVILGE
LFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGLCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPSHSA--
-------------PRPGTLSSPFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAPPWQPASAPPAPHANPSNPFYEA--------LSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLFP-NDWDPLDPGVPAPQAPQAPSEVPQ
LVGPSAAGSPRATRP---RSAP-ATPR------------------------PGSP---PT
PR-----------------EPGEANFRSSSE-----------------------------
------H-------------------RAATDVPAG-GC----DKEP--------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------P---RG--QGGGERPREQRESPRP
EQGPD--SPGRREESPKPE-----------------------------------------
------------------------------------------------------------
----------------PP-RERG-------------------------------------
-------------------DSVGERG-----SESPGHGGEGEREE----KVLENGDLTPP
RKEEKA--L--------------------------ENGELRSPEAGEKLLVNGGLVSPKS
EEMVSENGSLSFPRNT-ERPPETGPWRAPGPREKTLESGGP-------PAPETSLEKALI
PG-------EVALPQNGLETAPGPLGPAPKSGTLE--PGTERRAHETGGVQRAPGAGRLD
LGSGGQAPV-GMGMAPGGGPGSGVDAKAGWVDSMRPQPL-P---PPPLEAQLRRPE----
-PA---PQRSKPEVAP-EGELGVPDSRA---------AGDTAPSGDGDP-----PKPERK
GPEMPR-LFLDLGPPQGNSEQIKG------------------------------------
------------------------EGFVGSLGATPS---GVR------------------
-G---AGGSRSG---------------------------------AAGPADPGATAGLR-
-SCLFPCPWLSWANVQTALPSPRGADESED------SELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQGPSEGDTDPSTPPAPPTPPHPATPGD--------------
---------------------GFPSNDSGFGGSFEWAEDFPLLPPPGPPLCFS-------
----------------------------------RFSVSPAL------------------
------ETPGPPARAPDARPAGPVEN----------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSCANP00000016788 (view gene)
MRQVLWLCNVC---E-----------------TSRETRHHLHLPAILDKMPAPGALILLA
AV----SASGCLASPAHPDGFALGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 502)
SYLQEIGSGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGLCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPAHSA--
-------------PRPGTLSSPFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAPAWQPASAPPAPHANPSNPFYEA--------LSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLFP-NDWDPLDP----------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------GGEPGAPDSRA---------GGDTAPSGDGDP-----PKPERK
GPEMPR-LFLDLGPPQGNSEQIKA------------------------------------
------------------------RLSRLSLALPPL---TLTPFPGKSTAGMG--AEP-K
AG-RIEQGAGRAWGEGGVWEETGQGNGAGLGRWEEALSVRAGPDSGARPGGRAEPRAGVG
PRVRAGLVMGAGC-------RGRGEAWRVRVSLEGKSGPRKIAGFVAFGGCGKWDVTVCP
LWL------PETPTNELSVQGPPEGDTDPSTPPAPPTPPHPATPGD--------------
---------------------GFPSNDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARAPDARPAGTIS--SALPLLCP-SLSPCVPLPTSCDHLSMSLPLSSQ
S-----LLSPLSVNRHPTQLW---NQLRPGDVHQTVVGRQA-----------GCPPPPSL
SFAFCG---DSHHPQLLPIFVPSLGSDPGLALAGS---------------------
ENSPCOP00000024449 (view gene)
--------------CNVCVTSQE-I-----RH-------HLHLPSILDKMPAPGALILLA
AV----SASGCLASPAHPDGFALGWAPLAPPYAVVLIS----------------------
-------C-SGLLA-FVFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 502)
SYLQEIGSGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGLCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPAHSA--
-------------PRPGTLSSPFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAQPWQPASAPPAPHANPSNPFYEA--------MSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLFP-NDWDPLDPGVPAPQAPQAPSEVPQ
LVSETWASPL-FPAP---RPFP-AQSSGSGSFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--ERGTAPWAEEEE-EEEE--GSSPGEDSSSLGGGPSRRGPLP
CPLCSREG-----ACSCLPLERGDAVAGWGGHPAL-GC----PHPPEDDSSLRAERGSLA
DLPLAPPSS------APPEFLDPLMGA-AAP----QYP---GRGPPPAPPPPPP------
--------------------------------------------------PPRAPVDSTV
-----SPDPPSAVASPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLPLP------------------------------------
------------------------------------GNREKPTFVVQVSTEQLLMSLRED
VTRNLLGEKGATARETGP-RKVGRGPGNREKVQGLHKDPTVLGNGKQAPSP----S-LPV
NGVTVLENGGQRA-------AIEEKA------AENGGLGSPEREE----KVLENGELTPP
RREEKV--L--------------------------ENGELRSPEAGEKVLVNGGLTPPKS
EEKVSENGGLRFPRNT-ERPPETGPWRVPGPWEKMPESGGPAPMTEE-PAPETSLERAPA
PG-------TVASSQNGGETAPGPLGPAPKNGTLE--PGTERKAHETGGVLRAPGAGRLD
LGSGGRALV-GTGMAPGGGPGSGVDAKAGWVDNMRPQPPLL---PPPLEAQQRRPE----
-PV---PPRARPEVAP-EGESGAPDSRA---------GGDTAPSGDGDP-----PKPERK
GPEMPR-LFLDLGPPQGNSEQIKA------------------------------------
------------------------KLSRLSLALPPL---TLTPFPGPGPRRPP--WEGAD
AG--AAGGEAGGAGAPGSAEEDGEDEDEDEEEDEE----AAASGVAAGPRGSGRARAAPV
PVVVSSADADAARPLRGLLKSPRGADEPED------SELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQAPPEGDTDLSTPPAPPTPPHPATPGD--------------
---------------------GFPSNDSGFGEGRGRVGGGVWQTEGAGPEQKRGRRVRAE
VWA-----------GDTRV-----------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------------
MGP_SPRETEiJ_P0082952 (view gene)
----------------------METS-SPRAAVRASGRRFLHLPSILDKMPAPGALILLA
AV----SASGCLASPAHPDGFALSRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVR-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPPHSDISTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 502)
SYLQEIGSGWFGKVILGE
VFSDYSPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGVCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGMSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWVPLRWAAPELLGELHGSFVL-V
DQSRESNVWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPSHSA--
-------------PRPGTLSSQFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAPPWQPASAPPAPHTNPSNPFYEA--------LSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLFP-NDWDPLDPGVPGPQAPQTPSEVPQ
LVSETWASPL-FPAP---RPFP-AQSSGSGGFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--EQGTAPWAEEEE----E--ESSPGEDSSSLGGGPSRRGPLP
CPLCSREG-----PCSCLPLERGDAVAGWGGHPAL-GC----PHPPEDDSSLRAERGSLA
DLPLVPPTS------APLEFLDPLMGA-AAP----QYP---GRGPPPAPPPPPP------
--------------------------------------------------PPRASAEPAA
-----SPDPPSALASPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLPLP------------------------------------
------------------------------------GSQEKPTFVVQVSTEQLLMSLRED
VTKNLLGDKGSTPGETGP-RKAGRSPANREKGPGPNRDLTSLVSRKKVPSR----S-LPV
NGVTVLENGKPGVPDMKE-------------------------------KVAENG-LESP
E---RA--L--------------------------VNGEPMSPEAGEKVLANGVLMSPKS
EEKVAENGVLRLPRNT-ERPPEIGPRRVPGPWEKTPETGGL--------VPETLLDRAPA
PC-------EAALPQNGLEMAPGQLGPAPKSGNPD--PGTEWRVHESGGAPRAPGAGKLD
LGSGGRALG-GVGTAPAGGPASAVDAKAGWVDNSRPLPPPP----QPLGAQQRRPE----
-PV---PLKARPEVAQ-EEEPGVPDNRL---------GGDMAPSVDEDP-----LKPERK
GPEMPR-LFLDLGPPQGNSEQIKA------------------------------------
------------------------KLSRLSLALPPL---TLTPFPGPGPRRPP--WEGAD
AG--AAGGEAGGAGAPGPAEEDGEDEDEDEEDE-----------EAAGSRGPGRTREAPV
PVVVSSADGDTARPLRGLLKSPRAADEPED------SELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQGTPEGDTEPSTPPAPPTPPHPTTPGD--------------
---------------------GFPNNDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARAPDARPAGPVEN----------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSBTAP00000015942 (view gene)
------------------------------------------------------------
-------------SPVPTDGFALGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLGLSRQHLS
PF00069 // ENSGT00640000091449_2_Domain_0 (224, 499)
YLQEIGSGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGVCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYK---------------XAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPQHSA--
-------------PRPGTLSSPFPLLD---GFPG--ADPDDVLTVTESSGGLNLECLWE-
KARRGAGR--GG-GPTWQPASAPPAPPSNPSNPFYEA--------LSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLFP-NDWDPLDPGVPAPQAPQAPSEVPQ
LVSETWASPL-FPAA---RPFP-AQSSASGSFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--ERGAAPWAEEE--EEEE--GSSPGEDSSSLGGGPSRRGPLP
CPLCSREG-----ACSCLPLERGDAVAGWGGHPAL-GC----PHPPEDDSSLRAERGSLA
DLPLNPPAS------APPEFLDPLMGA-AAP----QYP---GRGPPPAPPPPPP------
--------------------------------------------------PPRAPADPAV
-----SPDPSSAVASPGSGLSSPGPKPGDSGYETETPFSPEGAFPGAGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLPLP------------------------------------
------------------------------------GAREKPTFVVQVSTEQLLMSLRED
VTRNLLGEK-------GP-RKTGRGPGNRERALGPSPDPAVPESGKQAPSPNEGLS-LPV
NGVTVLENGGPRALGADE------K------VAENGDPGSPEREE----QVLANGELPPP
RREETV--L--------------------------ENGGPGPPEREERVLVNGGLTSPKI
EE-GSENGGLRLPRNP-ERLPETGPWRAPGPWEKMPESGAPAPTNGE-PAPETSLERAPA
PG-------AVALALNGGETAPGPAGPAPRSGALE--PGTERRAPETGGAPRAPGAGRLD
LGSGGQAPV-GTGMAPGGGPGSGVDAKAGWADSTRPQPPLP-----LLEAQPRRPE----
-PA---PQRVRPEAAS-EGEPGAPDSRA---------GGDTAPSGDGDP-----PKPERK
GPEMPR-LFLDLGPPQGNSEQIKA------------------------------------
------------------------KLSRLSLALPPL---TLTPFPGPGPRRPP--WEGAD
AG--AAGGEAGGAGAPGPSEEDGEDEDEEE------DEEAAAAGAAAGPRGPGRARAAPV
PVVVSSADADAARPLRGLLKSPRGADEPE-----DS-ELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQGPPEGDTDPSTPPAPPTPPHPATPGD--------------
---------------------GFPSNDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARAPDARPAGTVLQ----------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSP00000270238 (view gene)
MRQVLWLCNVC---V-----------------TARETRHHLHLPAILDKMPAPGALILLA
AV----SASGCLASPAHPDGFALGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 502)
SYLQEIGSGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGLCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPAHSA--
-------------PRPGTLSSPFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAPAWQPASAPPAPHANPSNPFYEA--------LSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLFP-NDWDPLDPGVPAPQAPQAPSEVPQ
LVSETWASPL-FPAP---RPFP-AQSSASGSFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--ERGTAPWVEEEE-EEEE--GSSPGEDSSSLGGGPSRRGPLP
CPLCSREG-----ACSCLPLERGDAVAGWGGHPAL-GC----PHPPEDDSSLRAERGSLA
DLPMAPPAS------APPEFLDPLMG-AAAP----QYP---GRGPPPAPPPPPP------
--------------------------------------------------PPRAPADPAA
-----SPDPPSAVASPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLPLP------------------------------------
------------------------------------GPREKPTFVVQVSTEQLLMSLRED
VTRNLLGEKGATARETGP-RKAGRGPGNREKVPGLNRDPTVLGNGKQAPSL----S-LPV
NGVTVLENGDQRAPGIEE------K------AAENGALGSPEREE----KVLENGELTPP
RREEKA--L--------------------------ENGELRSPEAGEKVLVNGGLTPPKS
EDKVSENGGLRFPRNT-ERPPETGPWRAPGPWEKTPESWGPAPTIGE-PAPETSLERAPA
PS-------AVVSSRNGGETAPGPLGPAPKNGTLE--PGTERRAPETGGAPRAPGAGRLD
LGSGGRAPV-GTGTAPGGGPGSGVDAKAGWVDNTRPQPPPPPLP-PPPEAQPRRLE----
-PA---PPRARPEVAP-EGEPGAPDSRA---------GGDTALSGDGDP-----PKPERK
GPEMPR-LFLDLGPPQGNSEQIKA------------------------------------
------------------------RLSRLSLALPPL---TLTPFPGPGPRRPP--WEGAD
AG--AAGGEAGGAGAPGPAEEDGEDEDEDEEEDEEAA----APGAAAGPRGPGRARAAPV
PVVVSSADADAARPLRGLLKSPRGADEPE-----DS-ELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQAPPEGDTDPSTPPAPPTPPHPATPGD--------------
---------------------GFPSNDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARAPDARPAGPVE--N--------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSXETP00000054322 (view gene)
------------------------------------------------------------
--MSYFNPEKALAAPAENDGSSQGRVPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPDGEEFSGEYTPPAEETS-
-SSQSIPDVYILPLSEVSLPVSNTQAPKTELLKQTGFSRQD
PF00069 // ENSGT00640000091449_2_Domain_0 (222, 506)
LSYLQEIGNGWFGKVILGE
IFSDYTPAQVVVKELRVNASPLEQRKFLSEAQPYRSLQH----SNILQCLGLCTETIPFL
LIMEFCQLGDLKRYLRAQRKADGMTPDLLTCDLTTLQRMAYEITLGLMHLHKNNYIHSDL
ALRNCLLTSDLTVRIGDYGIS-HNNYKEDYYITPDRLWIPLRWVAPELLDEVHGNLVV-V
DQSKESNVWSLGVTMWELFEFGSQPYRHLSDEEVLTFVIKEQQMKLAKPRLKLPHSDYWY
EVMQSCWLPTEQRPSVEELHLQLTYLLNECSNQT----------EESFEWRWNALKPTKT
SGGPL---------HHSEEISAFPLLDNFNGGDGFHADMDDILTVTESSRGLNFEYMWE-
KARLGRWK--ATPQCQNYPPS--NGGLTVPANPFYESNHQHSRQSLETPSVVPVISARSP
S-VSSEYYIRLEEHTDRA---------SPIEPLFS-SDWDPLESERTSPEDPP--DEVPK
L-IDRWTPGNNNPFITVGRDLP-------------RYMNPFRGPADRDLSRRSWDSDTME
ERGAGETLAGDPESLAEFLDDRASPWDCEPSLMEDQ--SSESS---SSSTDNSNRRNMPS
CPLCSRGVMGGLQRCS-CAMLRGDVVIDWTAHVAM----HQDHYGLDDDSSLKVEQGSLS
ECSIPTRT----FGDEHQEFYDPLMGAAVKRIS-----------------------NSMA
NCSVIVPIEIPPLSTLAE------------------------------------------
--NIPSNTSSSDVCSPSSHYSSPGQKFGDSGYETENTFSPEFIFKDIKEE----------
-------------GGLRHVKLPLTPISESTSDLTLTEDTPEAHTTPGDEDSLRNLEIGTP
HRDSAYFSDYDNECEKHCRAAQQNLVAGSHNGFSKEEETTVPALVE-------------E
DTPNLEE---------AP-E---------------------------------------S
KDIEITETSCE---FTDEECS--------LQNASMDDIPMDSKEE----TLNCNGELNNS
SKENEPNHLLSENTNTPREKNNEKQTIIIDQRNEEDNTLKKQPESGFVVQEN--------
---VTRNVLSTFESSDIVNSCVLKDQAVLKLWSMETTAN---------PLSESLL-----
--------------KN------ESMNKQPSEEQLE--ESN----PRNSGSTN-------N
ME---------------SANECGNKEPMDII---KEEPEDPV-QQQQQEEAPR-------
-PEDKKDE-TKQET---FSETGIDLL--------------EQPVENC--------QDSSN
ETKEQAKLFLDFGSTNAGS-----------------------------------------
-------------------EAIKAKVARLSLSLPPL---NLQAFQNPTGRKSL--WDNNE
EWESPQ-----------TKEEDEEEDG---------FSF--QDQQRAGSESEG--DVAGI
PIVV--SETDDGRNLRSLLKSPKSADEAD-------DDLDRKRKMVSFFD-----DVTVY
LFD------QETPTNELSNQSGAEGDQIPHNPNVDPTI----------------------
--------------------DNF-SSTDGFSGSFEWDDDFPLMTQNSDTL----------
-------------------------------RFTRFTVSPAVDPHPVLETDVAPSAG---
-----------------------IEN----------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSMPUP00000003914 (view gene)
-------------------------------------------------MPGPGAFILLA
AV----SASGCLASPAHPDGFALGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLGLSRQHLS
PF00069 // ENSGT00640000091449_2_Domain_0 (224, 499)
YLQEIGSGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGVCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPP-----PPPRDGPFPWPWPPQHSA--
-------------PRPGTLSSPFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAPPWQPASAPPAPHANPSNPFYEA--------LSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLFP-NDWDPLDPGVPAPQASQAPSEVPQ
LVSETWASPL-FPAA---RPFP-AQSSASGSFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--ERGTAPWAEEEE-EEEE--GSSPGEDSSSLGAGPSRRGPLP
CPLCSREG-----ACSCLPLERGEAVAGWGGHPAL-GC----PHPPEDDSSLRAERGSLA
DLPLAPPTS------APPEFLDPLMGA-AAP----QYP---GRGPPPAPPPPPP------
--------------------------------------------------PPRVPADPAV
-----SPDPPSAVASPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGGGA--EEEG----
--PPEPPDPGAPRPPPDPGPHPLP------------------------------------
------------------------------------GTREKPTFVVQVSTEQLLMSLRED
VTRNLLGEKGANPRETGP-RKVGRGPGNREKAQGPSRDPTVLGSGKKAPSLNEDPS-LPM
NGVTVLENGGQRALGIEE------K------VAENGGPGTPEREE----KVLE-------
-----------------------------------KNGELRSPEREEKVLANGGLTPSKI
EEKVSENGGLRLPRNM-ERLPETGPRRAPGLWEKVPESGV--------PAPETSLERALE
PS-------AVALSRNGGETAPGPTGPAPRSGVLE--PGTERRAPETGGAPRAPGVGRLD
LGSGGRAPV-GTGMAPGGGLGSGVDAKAGWVDSTRPQLPPPL---PPSEAQPRRPE----
-PA---PQRARPEVASSEAEPGAPDSRA---------GGDTAPSTDGDP-----PKPERK
GPEMPR-LFLDLGPPQGNSEQIKV------------------------------------
------------------------RLGGLKLRCPWGPDSIGPSYPKPGPREFQ--TLPCP
TPSAPLPHAMGDLKTRNVFS--GRAPD--SRKISRDEQLRCFRGLW----EEGGTVTAPK
PIALRXADADAARPLRGLLKSPRGADEPE-----DS-ELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQGPPEGDTDPSTPPAPPTPPHPATPGD--------------
---------------------GFPSNDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARAPDARPAGTVLQ----------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSMOCP00000003426 (view gene)
----------------------METS-LPRAAVCASGRRYLHLPSILDKMPAPGALILLA
AV----SASGCLASPAHPDGFALSRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVR-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPAHSDISTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 502)
SYLQEIGSGWFGKVILGE
VFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGVCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGVSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWVPLRWAAPELLGELHGSFVL-V
DQSRESNVWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPSHSA--
-------------PRPGTLSSQFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAPPWQPASAPPAPHTNPSNPFYEA--------LSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLFP-NDWDPLDPGVPGPQAPQAPSEVPQ
LVSETWASPL-FPAP---RPFQ-TQSSGSGGFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--EQGTAPWAEEEE----E--ESSPGEDSSSLGGGPSRRGPLP
CPLCSREG-----PCSCLPLERGDAVAGWGGHPAL-GC----PHPPEDDSSLRAERGSLA
DLPLVPPTS------APPEFLDPLMGA-AAP----QYP---GRGPPPAPPPPPP------
--------------------------------------------------PPRAPAEPAA
-----SPDPPSALASPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLPLP------------------------------------
------------------------------------GSQEKPTFVVQVSTEQLLMSLRED
VTKNLLGDKGSTAGESGP-RKAWRNPANGEKDPGPNRDLTSLGNRKKVPSW----N-LPV
NGLTVLENGKPGAPDMKE-------------------------------KVVENG-LESP
ERGERA--L--------------------------VNGEPMPPEPGEKVLANGVLMSPKS
EEKVAENGVLRLPRNT-ERLPETGPRRAPGPWEKMPETGGL--------APETLLDQAPA
PC-------EVALPQNGLETAPGQLGLVPKSENPE--PGTEWRVHETGGALRAPGAGKLD
LGSGDRAQG-GVGMAPAGGPASGVDAKVGWVDNSRPLPPPP----QPLGAQQRRPE----
-PV---PLKARPEVAL-EDEPGVPDSRL---------GEDTAPSVDEDP-----PKLERK
GPEMPR-LFLDLGPPQGNSEQIKA------------------------------------
------------------------KLSRLSLALPPL---TLTPFPGPGPRRPP--WEGAD
AG--AAGGEAGGAGAPGPAEEDGEDEDEDEEDE-----------EAAGSRGPGRTREAPV
PVVVSSADADTARPLRGLLKSPRAADEPED------SELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQSTPEGDTDPSTPPAPPTPPHPATPGD--------------
---------------------GFPNNDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARAPDARPAGPVEN----------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSPPAP00000026690 (view gene)
MRQVLWLCNVC---V-----------------TARETRHHLHLPAILDKMPAPGALILLA
AV----SASGCLASPAHPDGFALGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 458)
SYLQEIGSGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGLCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRLAHPGLLACR----------
-------TPPRMVPASPLV-SHITHI----------------------------------
---------------------P------------------------FSEEGLSPPFLPAS
ATCRGTQA--GDGRAPFPSAAVSPLP----------------------------------
-----------------------------------PGPCQPLSMGIFPP--PLSPAQLPP
LNLYRLLSPL-LPGS---LPH---------------------------------------
--------------F---------------------------------------------
LGLCFPIS-----LGLCPPLSGSLS--------------------------L-SLSGSLS
---------------L--------------------------------------------
------------------------------------------------------------
-----SLLFFFFVADFCFCLSSPGPKPGDSGYETETPFSPEGAFPGGGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLPLP------------------------------------
------------------------------------GPREKPTFVVQVSTEQLLMSLRED
VTRNLLGEKGATARETGP-RKAGRGPGNREKVPGLNRDPTALGNGKQAPSL----S-LPV
NGVTVLENGDQRAPGIEE------K------AAENGALGSPEREE----KVLENGELTPP
RREEKA--L--------------------------ENGELRSPEAGEKVLVNGGLTPPKS
EDKVSENGGLRFPRNT-ERPPETGPWRAPGPWEKTPESWGPAPTIGE-PAPETSLERAPA
PS-------AVVSSRNGGETAPGPLGPAPKNGTLE--PGTERRAPETGGAPRAPGAGRLD
LGSGGRAPV-GTGTAPGGGPGSGVDAKAGWVDNTRPQPPPPPLP-PPPEAQPRRLE----
-PA---PPRARPEVAP-EGEPGAPDSRA---------GGDTAPSGDGDP-----PSPRGR
LGGRACLSFTRTGA--HQVSPLAA------------------------------------
------------------------RLSRLSLALPPL---TLTPFPGPGPRRPP--GEG--
----------------GVWGETGQGNGAGLGRWEEALSVRAGPGNGAGPGGRAEPRAGAG
PRVRAGLVT-------GAWSRGRGEAWRE-----SV-GLEG--------KRGY-------
---------WETPTNELSVQAPPEGDTDPSTPPAPPTPPHPATPGD--------------
---------------------GFPSNDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARAPDARPAGTIS--SALPPPSA-HLCPPM---FHCQLLVTIFQCPCL
SLPSLFSLPSQSTDTPPSSGT---SSDQATSIRRS-------------------------
--------------------------------------------------------
ENSNLEP00000042129 (view gene)
MRQVLWLCNVCV--------------------TSRETRHHLHLPAILDKMPAPGALILLA
AV----SASGCLASPAHPDGFALGRAPLAPPYAVAMAC----------------------
-------L-SALLS-STPL-LLS----------R-HQEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 459)
SYLQEIGSGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGLCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRLRCDLNPLHR--------KR
SLLLTCLIPE-------------------LTAQTGTP--PLSP-DVPICCCVPCAC-R--
-------------SLSASLYGHL--------------------PPTPASLGLNTQLPS--
-----------------PVTSSESLPSAVPSSPWISA--------PLFGSVFPSLSR---
-----------------------------------------------------------S
LSPSLWVSV-----P---LS----------------------------------------
----------------------------------------------------------FF
FFFFFLTK-----FCFCCPGWSAM-AQSWL--TAT-ST----SRVQADDSSLRAERGSLA
DLPMAPPAS------APPEFLDPLMGA-AAP----QYP---GRGPPPAPPPPPP------
--------------------------------------------------PPRAPADPAA
-----SPDPPSAVASPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLPLR------------------------------------
------------------------------------GPQEKPTFVVQVSTEQLLMSLRED
VTRNLLGEKGATARETGP-RKAGRGPGNREKVPGLNRDPTALGNGKQAPSL----S-LPV
NGVTVLENGDQRAPGIEEK------------AAENGALGSPEREE----KVLENGELTPP
RREEKA--L--------------------------ENGDLRSPEAGEKVLVNGGLTPPKS
EDKVSENGGLRFPRNT-ERPPETGPWRAPGPWEKTPESWGPAPTIGE-PAPETSLERAPA
PS-------AVASSRNGGETAPGPLGPAPKNGTLE--PGSERRAPETGGAPRAPGAGRLD
LGIQRKRTR-E-------GPS-------AFIF--LPTPPPP---SSCPCHQPRK------
--E---MPRCSPSVHKV---------------------GAEAQIVRGGL-----PEATQL
G-GRACLSFTGTGAH---------------------------------------------
-----------------QVSPLAARLSRLSLALPPL---TLTPFPGPGPRRPP--WEGRR
A------------GAPGPAEEDGEDEDEDEEEDEEAA----APGAAAGPRGPGRARAAPV
PVVVSSADADAARPLRGLLKSPRGADEPED------SELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQAPPEGDTDPSTPPAPPTPPHPATPGD--------------
---------------------GFPSNDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARAPDARPAGTISSALPPPLPISVPLCST------ANFL---------
------------------------------------------------------------
--------------------------------------------------------
ENSCGRP00001010660 (view gene)
----------------------METS-SPRAAVCASGRRYLHLPSILDKMPAPGALILLA
AV----SASGCLASPAHPDGFALSRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVR-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPVPAPQPAHSDISTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 502)
SYLQEIGSGWFGKVILGE
VFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGVCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGMSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWVPLRWAAPELLGELHGSFVL-V
DQSRESNVWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPSHSA--
-------------PRPGTLSSQFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAPSWQPASAPPAPHTNPSNPFYEA--------LSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLFP-NDWDPLDPGVPGPQASQAPSEVPQ
LVSETWASPL-FPAP---RPFQ-AQSSGSGGFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--EQGTAPWAEEEE----E--ESSPGEDSSSLGGGPSHRGPLP
CPLCSREG-----PCSCLPLERGDAVAGWGGHPAL-GC----PHPPEDDSSLRAERGSLA
DLPLVPPPS------APSEFLDPLMGA-AAP----QYP---GRGPPPAPPPPPP------
--------------------------------------------------PPRAPTEPAA
-----SPDPPSALASPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLPLP------------------------------------
------------------------------------GAQEKSTFVVQVSTEQLLMSLRED
VTKNLLGDKGSTPGETGP-RKAGRSPANREKGPGPNRDLTSLGSRKIVPSW----N-LPV
NGVTVLENGKPGAPDTKE-------------------------------KVAENG-LESP
ERGEKA--L--------------------------VNGEPMSPEAEEKVLANGVLMSPKI
EEKVAENGVLKLPRNM-ERPPETGPRRAPGPWEKMPETGGQ--------APEILLDQAPA
PC-------EAALPQNGLETVPGQLGPVPKSGNPE--PGTEWRVHETGGALRAPGAGKLD
LGSGGRAQG-GAGMAPAGGPASGVDAKAGWVDSSRPLPPPP----QPLGTKQRRPE----
-PV---PLKSRPEVAP-EEEPEVPDNRL---------GGDTALSVDEDP-----PKLERK
GPEMPR-LFLDLGPPQGNSEQIKA------------------------------------
------------------------KLSRLSLALPPL---TLTPFPGPGPRRPP--WEGAD
AG--AAGGEAVGAGAPGPAEEDGEDEDEDEEDE-----------EAAGSRGPGRTLEAPV
PVVVSSVDADTARPLRGLLKSPRAADEPED------SELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQGTPEGDTDPSTPPAPPTPPHPATPGD--------------
---------------------GFPNNDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPTRAPDARPAGPVEN----------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSCLAP00000012167 (view gene)
----------------------METS-APRAGARASGRHHLHLPSILDKMPAPGALILLA
AV----SASGCLASPAHPDGFALGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPAHSDMTTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 489)
SYLQEIGSGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGLCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFVV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAHSPTCSL-----------SGPPGPRP--PPPPRDGPFPWPWPPSHSA--
-------------PRPGTLSSQFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAPAWQPASAPPAPHTNPSNPFYEA--------LSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLFP-NDWDPLDPGVPAAQAPQAPSEVPQ
LVSETWASPL-FPAP---RPFP-AQSSASGSFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--ERGTAPWAEEE---EEE--GSSPGEDSSSLGVGPSRRGPLP
CPLCSREG-----ACSCLPLERGDAVAGWGGHPAL-GC----PHPPEDDSSLRAERGSLA
DLPLAPPAS------APAEFLDPLMGA-AAP----QYP---GRGPPPAPPPPPP------
--------------------------------------------------PPRAPSEPAA
-----APDPPSAEASPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGEAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLPLP------------------------------------
------------------------------------GAREKPAFVVQVDTEQLLVSLREG
VTRSLLGDS----RETGP-GKAGRGPGNREKALGLNRDLPVLGGGKKAPSL----S-LPV
NGVTVLGSGAQRAPDIEE------K------PAENGGLGSPEREV----KALANGE----
-----------------------------------------SPEAGEKVLANGGPIPPEI
EEKVSENGLLRFPRNT-ERPTETGPWRAPGPREKTPESGGP--------APETPLERAPA
PS-------EVASPQNGLETAPGPPGPAPKSGMLD--PGTERRAHETGGVPRAPGAGRLD
LGSGGRAPV-GVAMAPGGGPGSGVDAKAGWADSTRPQPLPP---LPPPEAQPRRPE----
-PV---PPRVRPEVAS-EGEPGVPDSKA---------GGDTAPSGDGDL-----SKPERK
GPEMPR-LFLDLGPPQGNSEQIKA------------------------------------
------------------------KLSRLSLALPPL---TLSPFPGPGPRRPP--WEGR-
------------------------------------------------------TRAAPV
PVVVSSADADAARPLRGLLKSPRGADEPED------SELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQGPPEGDTDPSTPPAPPTPPHPATPGD--------------
---------------------GFPSNDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARAPDARPAGTVNSISAPPDVA--------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSSBOP00000026538 (view gene)
--KCLWLCNVC---V-----------------TSRETWVFAIH----------GAMIPGS
RD----LKLGTQPSPVPTDGFALGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 502)
SYLQEIGSGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGLCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPAHSA--
-------------PRPGTLSSPFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAPAWQPASAPPAPHANPSNPFYEA--------LSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLF-PNDWDPLDPGVPAPQAPQAPSEVPQ
LVSETWASPL-FP----------AQSSASGSFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--ERGTAPWVEEEE-EEEE--GSSPGEDSSSLGGGPSRRGPLP
CPLCSREG-----ACSCLPLERGDAVA---------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------GLSSPGPKPGDSGYETETPFSPEGAFPGGGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLLLP------------------------------------
------------------------------------GPREKPTFVVQVSTEQLLMSLRED
VTRNLLGEKGATARETGP-RKAGRGPGNREKAPGLSRDPTALGNGKQAPSP----S-LPV
NGVTVLENGDQRALDIEK------K------AAENGALASPEREE----KVLENGELTPP
RR-EKA--L--------------------------ENGELRSPEAGEKVLVNGGLTPPKS
EDKVSENGGLRFPRNT-ERPPETGPWRAAGPWEKKPESWGPAPTIGE-PAPETSLERAPA
LS-------AVGSSRNGGETAPGPLGPAPKNGTLE--PGTERRAPETGGAPRVPGAGRLD
LGSGGRAPV-GTGTAPGGGPGSGVDAKVGWVDNTRPQPPPPPLP-PPPEVHPRRPE----
-PV---PPRTRPEVAS-EGEPGDPDSRA---------GGDTAPSGDGDP-----PKPERK
GPEMPR-LFLDLGPPQGNSEQIKG------------------------------------
------------------------EEAALN------------------------------
--------------------------------------VRAGPGNGAGPGGRTEPRAGVG
PGAGAGLVM-------GAGSRGRGEAWRV-----RVRSPEILAAFAAFGGCSKWDMTVCP
LWFPPNLPSQETPTNELSVQGPPEGDTDPSTPPAPPTPPHPATTPG--------------
--------------------DGFPSNDSGFGEGGAYR------TGGGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARAPDARPAGPVE--N--------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSMLEP00000004546 (view gene)
MRQVLWLCNVC---E-----------------TSRETRHHLHLPAILDKMPAPGALILLA
AV----SASGCLASPAHPDGFALGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 307)
SYLQEIGSGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGLCVETLPFL
LIMEFCQLVRRQIKWKGWSQCEGAGPGR-----------GW---RAAPALTSSHSSRSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPAHSA--
-------------PRPGTLSSPFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAPAWQPASAPPAPHANPSNPFYEA--------LSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLFP-NDWDPLDPGVPAPQAPQAPSEVPQ
LVSETWASPL-FPAP---RPFP-AQSSASGSFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--ERGTAPWK---------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------PTFVVQVSTEQLLMSLRED
VTRNLLGEKGATARETGP-RKAGRGPGNREKVPGLNRDPTALGNGKQAPSP----S-LPV
NGVTVLENGDQRAPGIEE------K------AAENGALGSPEREE----KVLENGELTPP
RREEKA--L--------------------------ENGELRSPEAGEKVLVNGGLTPPKS
EDKVSENGGLRFPRNT-ERPPETGPWRAPGPWEKTPESWGPAPTIGE-PAPETSLERAPA
PS-------AVASSRNGGETAPGPLGPAPKNGTLE--PGTERKAPETGGAPRAPGAG---
------------------------------------------------------------
-------------------------SRA---------GGDTAPSGDGDP-----PKPERK
GPEMPR-LFLDLGPPQGNSEQIKA------------------------------------
------------------------RLSRLSLALPPL---TLTAP----------------
------------------------------------------------------------
---------------------------------------EILTSFVAFGGCGKWDVTVCP
LWL------PETPTNELSVQGPPEGDTDPSTPPAPPTPPHPATPGD--------------
---------------------GFPSNDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARAPDARPA---------------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSMICP00000041476 (view gene)
--------ME--ASAPRVGVRAA-G-----RH-------HLHLPSILDKMPAPGALILLA
AV----SASGCLASPAHPDGFALGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FVFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 502)
SYLQEIGSGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGLCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPAHSA--
-------------PRPGTLSSPFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAQPWQPASAPPAPHGNPSNPFYEA--------MSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLFP-NDWDPLDPGVPAPQAPQAPSEVPQ
LVSETWASPL-FPAP---RPFP-AQSSGSGSFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--ERGNAPWAEEEE-EEEE--GSSPGEDSSSLGGGPSRRGPLP
CPLCSREG-----ACSCLPLERGDAVAGWGGHPAL-GC----PHPPEDDSSLRAERGSLA
DLPLNPPAS------APPEFLDPLMGA-AAP----QYP---GRGPPPAPPPPPP------
--------------------------------------------------PPRAPVDSAV
-----SPDPPSAVASPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGP--LP------------------------------------
------------------------------------GNQEKPTFVVQVSTEQLLMSLRED
VTRNLLGEKGAAGRETGP-RKAGRGPGNREKVPGLNKDPTVLANGKQAPSL----S-LPV
NGVTVLENGGQRAPDIEAKAAIAEKA------AENGGLGSPEREE----KALENGELTPP
RREEKA--L--------------------------ENGELRSPEAGQKVLVNGGLTPPKS
AEKVLENGGLRFPRST-ERPPETGPWRVPGPWEKMPESGGPAPTTEE-PAPETSLERAPA
PG-------AAASSRNGGETAPGPLGPAPKNGTLE--PGTERKARETGGAPRAPGAGRLD
LGSGARALV-GTGMAPGGGPGSGVDAKAGWADNMRPQP-LL---PPPPETQPRRPE----
-PV---PPRARPEGAP-EGESGAPDSRP---------GGDTAPSGDGDP-----PKPERK
GPEMPR-LFLDLGPPQGNSEQIKA------------------------------------
------------------------KLSRLSLALPPL---TLTPFPGPGPRRPP--WEGAD
AG--AAGGEAGGAGAPGSAEEDGEDEDEDEEEDEE----AAASGAAAGPRGPGRARAAPV
PVVVSSADADAARPLRGLLKSPRGADEPED------SELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQGPPEGDTDLSTPPAPPTPPHPATPGD--------------
---------------------GFPSNDSGFGEGRGRVGGGAWRGPRG-----RGRRAGRE
LGL-----------GARVSEGLTCRLSVPAAPRRQFRVGGGFPPPPPARAPPVLLPLLRL
ACAGDPGAPRPGPRRPARRPRGELIPQRPDPAAPPEEG---LRVESSVDDGATAT-----
------------------------T----ADAASGEAPEAGPSPS---------------
-PTMCQTGGPGPLPPS-----PPDGS-----------PDPPDPLGAK---------
ENSMAUP00000014952 (view gene)
----------------------METS-SPRAAVCASGRRYFHLPSILDKMPAPGALILLA
AV----SASGCLASPAHPDGFALSRAPVAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVR-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPAHSDISTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 502)
SYLQEIGSGWFGKVILGE
VFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGVCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGMSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWVPLRWAAPELLGELHGSFVL-V
DQSRESNVWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPSHSA--
-------------PRPGTLSSQFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAPPWQPASAPPAPHTNPSNPFYEA--------LSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLFP-NDWDPLDPGVPGPQGSQGPSEVPQ
LVSETWASPL-FPAP---RPFQ-AQSS-SGGFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--EQGTAPWAEEEE----E--ESSPGEDSSSLGGGPSRRGPLP
CPLCSREG-----PCSCLPLERGDAVAGWGGHPAL-GC----PHPPEDDSSLRAERGSLA
DLPLVPPTS------APSEFLDPLMGA-AAP----QYP---GRGPPPAPPPPPP------
--------------------------------------------------PPRAPAEPAA
-----SPDPPSALASPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLPPP------------------------------------
------------------------------------GSQEKPTFVVQVSTEQLLMSLRED
VTKNLLGDKGSTPGETGP-RKAGRSPANREKGPGPNRDLTSLGSRKIVPSW----N-LPV
NGVTVLENGKAGAQDMKE-------------------------------KVAENG-LDSP
ERGEKA--L--------------------------VNGEPMSPEAGEKVLANGVLMSPKI
EEKVAENGVLRLPRNM-ERPPETGPRRAPGPWEKMPETGDL--------DPEIQLDQAPA
PC-------EAALPQNGLETVPGQLGPVPKSENPG--PGTEWKVHETGGAPRVPGAGKLD
LGNGGRAQG-GAATAPAGGPASGVDAKAGWVDSSRPLPP------PPLGTQQRRPE----
-PV---PLKARPEVGP-EEEPGVPDSRL---------GGDTALSGDEDP-----PKLERK
GPEMPR-LFLDLGPPQANSEQIKA------------------------------------
------------------------KLSRLSLALPPL---TLTPFPGPGPRRPP--WEGAD
AG--AAGGEAGGAGAPGPAEEDGEDEDEDEEDE-----------EAAGSRGPGRTREAPV
PVVVSSADADTARPLRGLLKSPRAADEPED------SELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQGPPEGDTDPSTPPAPPTPPHPATPGD--------------
---------------------GFPNNDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPTRAPDARPAGPVEN----------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSDNOP00000030849 (view gene)
MRARLWLCHLC---LRAC---------------VFACCHHLHLPPILDKMPASGALILLA
AV----SASGCLASPAHPDGFALGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLGLSRQHLSYLQEIGSGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGLCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSQG
GEEGRVRGGRGGGRVGGI---ERGGVREGRVPGGH---VVWPWGAGGLGXELHGTFMV-V
DQ
PF07714 // ENSGT00640000091449_2_Domain_0 (423, 502)
SRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPP---PPPPRDGPFPWPWPAAHPP--
-------------PRPGTLSSPFPLLD---GFPG--TDPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGPPPWQPATAPPAHQTKPSNPFYEA--------LXPSQ----------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------------------------DPS-LPA
NGATVLENGAQVAPGTEE------K------AAANGGPGSPGREE----KVLENGELTPP
RREAKL--L--------------------------ENGGPKSPEATEKVLVNGGLTPPKS
EE-ASENGGPRLPRNA-ERPPETGPWRAPGPWEKMPESGGPAPTIRG-PAPETSPARVPE
PG-------AA--PPSGAETAPGTSGPAPRSGAPE--AGTERRAPATGGALRAPAAGRLD
PGSGGRVPV-GSGTAPGGGPGSGVDAKAGWADSTRPPPLPPP--EKPPETPPRRPE----
-PA---PLRARPEVAP-EGDPGVPDSRA---------AGDTAPSGDGDP-----PKLERK
GPEMPR-LFLDLGPPQGNSEQIKA------------------------------------
------------------------KLSRLSLALPPL---TLTPFPGPGPRRPP--WEGAD
AG--AAGGEAGGAGAPGPAEEDGEDEDEEEDEDEE-----AAAGAAAGPRGPRRARAAPV
PVVVSSADADAARPLRGLLKSPRGADEPE------DSELERKRKMVSFHG-----DVTVY
LFDQ--------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSGGOP00000010451 (view gene)
MRQVLWLCNVC---V-----------------TARETRHHLHLPAILDKMPAPGALILLA
AV----SASGCLASPAHPDGFALGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 502)
SYLQEIGSGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGLCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFMV--
--SRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPAHSA--
-------------PRPGTLSSPFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAPAWQPASAPPAPHANPSNPFYEA--------LSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLFP-NDWDPLDPGVPAPQAPQAPSEVPQ
LVSETWASPL-FPAP---RPFP-AQSSASGSFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--ERGTAPWVEEEE-EEEE--GSSPGEDSSSLGGGPSRRGPLP
CPLCSREG-----ACSCLPLERGDAVAGWGGHPAL-GC----PHPPEDDSSLRXXXXXXX
XXXXXXXXX------XXXXXXXXXXXXXXXX----XXX---XXXXXXXXXXXXX------
--------------------------------------------------XXXXPADPAA
-----SPDPPSAVASPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLPLP------------------------------------
------------------------------------GPREKPTFVVQVSTEQLLMSLRED
VTRNLLGEKGATARETGP-RKAGRGPGNREKVPGLNRDPTALGNGKQAPSL----S-LPV
NGVTVLENGDQRAPGIEE------K------AAENGALGSPEREE----KVLENGELTPP
RREEKA--L--------------------------ENGELRSPEAGEKVLVNGGLTPPKS
EDKVSENGGLRFPRNT-ERPPETGPWRAPGPWEKTPESWGPVPVIGELPAPETSLERAPA
PS-------AVVSSRNGGETAPGPLGPAPKNGTLE--PGTERRAPETGGAPRAPGAGRLD
LGSGGRAPV-GTGTAPGGGPGSGVDAKAGWVDNTRPQPPPPPLP-PPPEAQPRRLE----
-PA---PPRARPEVAP-EGEPGAPDSRA---------GGDTAPSGDGDP-----PKPERK
GPEMPR-LFLDLGPPQGNSEQIKA------------------------------------
------------------------RLSRLSLALPPL---TLTPFPGPGPRRPP--WEGAD
AG--AAGGEAGGAGAPGPAEEDGEDEDEDEEEDEEAA----APGAAAGPRGPGRARAAPV
PVVVSSADADAARPLRGLLKSPRGADEPE-----DS-ELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQAPPEGDTDPSTPPAPPTPPHPATPGD--------------
---------------------GFPSNDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARAPDARPAGTIS--SALPPPLP-I---SVPLCSTANFL---------
------------------------------------------------------------
--------------------------------------------------------
ENSCAPP00000014635 (view gene)
-----WEQRGAILFLRGCKLQLPQGXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XX----XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 502)
SYLQEIGNGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRYEGLSASWVLECPCLGLCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPSHSA--
-------------PRPGTLSSQFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAPAWQPASAPPAPHANPSNPFYEA--------LSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLFP-NDWDPLDPGVPAAQAPQPPSEVPQ
LVSETWASPL-FPAP---RPFQ-AQSSASGSFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--ERGNAPWAEEE--EEEE--GSSPGEDSSSLGVGPGRRGPLP
CPLCSREG-----ACSCLPLERGDAVAGWGGHPAL-GC----PHPPEDDSSLRAERGSLA
DLPLVPPTS------APTEFLDPLMGA-AAP----QYP---GRGPPPAPPPPPP------
--------------------------------------------------PPRAPAEPAA
-----TPDPPSAVASPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGEAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLPLP------------------------------------
------------------------------------GTREKPTFVVQVSTEQLLMSLRED
VTRNLLGDKGVTSRETGP-RKVGRGPGNREKVLGLDRDLPVLGGGKKAPSL----S-LPV
NGVTVLGNGAQRAPDIEEK------------PAENGGLGSPEKEV----KVLTNGELMPP
RR-EKA--L--------------------------ENGELRSPEAGEKVLANGGPIPPEI
EEKVSENGLLSFPRNT-ERPTETGPWRAPGPQEKMHESGGP--------APETLLERAPV
PG-------EVASPQNGLETSPGPLGPAPKSGILD--PGMERRAHETGGVPRAPGAGRLD
LGSGGRAPV-GVGMVPGGGPGSGVDAKAEWVDNMRQQPLPP---LPPPEAQPRRPE----
-PV---PPRIRPEAVS-EGEPGVPDSRA---------GGDTAPRGDGDL-----SKPERK
GPEMPR-LFLDLGPPQGNSEQIKGEEVWGGGETGALC--------RGCGPIWAQILTFSL
PYSEVLGRDVCCSVTQFPCLSRTGDQFRMIVR---V---E------LGPRAQA-------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSCATP00000000765 (view gene)
MRQVLWLCNVC---E-----------------TSRETRHHLHLPAILDKMPAPGALILLA
AV----SASGCLASPAHPDGFALGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 502)
SYLQEIGSGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGLCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPAHSA--
-------------PRPGTLSSPFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAPAWQPASAPPAPHANPSNPFYEA--------LSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLFP-NDWDPLDPGVPAPQAPQAPSEVPQ
LVSETWASPL-FPAP---RPFP-AQSSASGSFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--ERGTAPWVEEEE-EEEE--GSSPGEDSSSLGGGPSRRGPLP
CPLCSREG-----ACSCLPLERGDAVAGWGGHPAL-GC----PHPPEDDSSLRAERGSLA
DLPMAPPAS------APPEVLDPLMG-AAAP----QYP---GRGPPPAPPPPPP------
--------------------------------------------------PPRAPADPAA
-----SPDPPSAVASPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLPLP------------------------------------
------------------------------------GPREKPTFVVQVSTEQLLMSLRED
VTRNLLGEKGATARETGP-RKAGRGPGNREKVPGLNRDPTALGNGKQAPSS----S-LPV
NGVTVLENGDQRAPGIEE------K------AAENGALGSPEREE----KVLENGELTPP
RREEKA--L--------------------------ENGELRSPEAGEKVLVNGGLTPPKS
EDKVSENGGLRFPRNT-ERPPETGPWRAPGPWEKTPESWGPAPTIGE-PAPETSLERAPA
PS-------AVASSRNDGETAPGPLGPAPKNGTLE--PGTERKAPETGGAPRAPGAGRLD
LGSGGRAPV-GTGTAPGGGPGSGVDAKAGWVDYTRPQPPPPPLP-PPPEAQPRRLE----
-PA---PPRARPEVAP-EGEPGAPDSRA---------GGDTAPSGDGDP-----PKPERK
GPEMPR-LFLDLGPPQGNSEQIKA------------------------------------
------------------------RLSRLSLALPPL---TLTPFPGPGPRRPP--WEGAD
AG--AAGGEAGGAGAPGPAEEDGEDEDEDEEEDEEAA----ASGAAAGPRGPGRARAAPV
PVVVSSADADAARPLRGLLKSPRGADEPE-----DS-ELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQGPPEGDTDPSTPPAPPTPPHPATPGD--------------
---------------------GFPSNDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARAPDARPAGTIS--SNTEARKE-VRS----------WDSKRQLLGL-
------------------------------------------------------------
--------------------------------------------------------
ENSOCUP00000019567 (view gene)
------------------------------------------------------------
-------------SPVSTDGFAMGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 452)
SYLQEIGSGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGLCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFESGRSSIANLSDEE---------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSACAP00000016312 (view gene)
-------------------------------------------------MLRQGPFLLLA
ASMSYFNPEKALAAPSENDGL---SHPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPDGEEYSGEYTPPAEETS-
-SSQSIPDVYILPLSEVSLPSSNQQMPKTDLVKHLGLSRQHLS
PF00069 // ENSGT00640000091449_2_Domain_0 (224, 499)
YLQEIGNGWFGKVILGE
IFSDFTPAQVVVKELRVNAGPLEQRKFISEAQPYRL-QH----PNILQCLGLCTETIPYL
LIMEFCQLGDLKRYLRAQRKADGMTPDLLTRDLSTLQRMAYEITLGVLHLHKNNYIHSDL
ALRNCLLTSDLTVRVGDYGLS-HNNYKEDYYITPDRLWIPLRWVAPELLDEMHGNVIV-V
DQTKESNVWSMGVTIWELFEFGSQPYRHLSDEEVLTFVVKEQQMKLAKPRLKLPYSDYWY
EVMQSCWLPASHRPTMEELHLQVSYLLNERSNAQ---------AEESFEWRWSALKPKRS
SSPPAASSSSARHRRSSEEISAYPLLD---GFHG--GDMDDILTVTESSGGLNFEYMWE-
KARLGRWK--GPASAPPEYPSSNGGLAVPGSNPFYEQASQR-GHSLETPSVVPVISARSP
S-VSSEYYIRLEDHGSGEGPVCAPSPGSPPQPLFP-SDWDCPGASYRPQEQPTSNPFLPS
SGGD--SNPFRADSEIQETSLTETPTTGGESFLAESLLTVY---RELEDQQRSWDAEMME
ERGAGETLAGDPSEFQG--S---PSWEQEPSLMEDQ--SSGSSTDGGGSSIGGGRRNLLP
CPLCTRVR------CT-CAIPRGDVVSGWSNHGPPLQRPHRDSYGTEDDSSLKVERGSLS
ECPMLPPRDCDGEGEAHSEFFDPLMGTVVKTYELQDYPKQRGGGSTRIAPGSPFPAMSSG
CCNVIVPVEIPPLSTLAESNDEETIAILPDVIDGFSTGPVTLTLSPAAPDRQRGTLDSVD
SLDIPSNTSSSDVCSPASHYSSPGQKFGDSGYETENTFSPEFIFKEDPPP----------
LAPPEEEEE--TEGDEDSGKLPTTSISQSTSDLTLSVSEEGV---ELEEEVGRSLGPTTP
HRDSAYFSDGEAESLEVLAP--PAPAAGSEEGKSIEKTTLDGGFVVQVCKEQLLVTLHEN
VTRNLLCSQKKPVPGFRP-V-------------------------------------APL
KDQAI-------------------------------------------------------
------------------------------------------------------------
----------------------------LRLWPLEPEEGQEETEEEE------EVEEED-
-------------KKNHKKDVTDSQQP----EIME--DKEKEE---------VVEEEEVD
MENKEKHQEEVNGELEGIKPGC--------EEEERKEKEKPS----QIEIQSVAGSSVVE
LPDVHKDSSVATEQPPKEQEPGLPGNQ---------------------------------
------------------------------------------------------------
-----------------AGEDIKVKLSRLSLSLPPL---ALQAFPPPGGGRKGPFWEGAA
LGEGPVLGGREAA----MAAEDEEDDDAEE--DE--------EGGLAGSGGRG------V
PIVV--SESDDAHNLRGLLKSPRSAEEAA-------AELDRKRKMVSFFD-----DVTVY
LFD------QETPTNELSSQGAPDGDAPSPGST--------LEPGL--------------
--------------------DPFP-AADGFSGSFEWDDDFPLMAQNPSYVSMVSDPSTVK
PSLPAMSSPVPLAPALLPSAKRTESGLVDALRFSRFTVSPALEPHLSPSCDT--------
-------ELGPT--------ANPIEN----------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSPTRP00000019283 (view gene)
MRQVLWLCNVC---V-----------------TARETRHHLHLPAILDKMPAPGALILLA
AV----SASGCLASPAHPDGFALGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 502)
SYLQEIGSGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGLCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPAHSA--
-------------PRPGTLSSPFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAPAWQPASAPPAPHANPSNPFYEA--------LSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLFP-NDWDPLDPGVPAPQAPQAPSEVPQ
LVSETWASPL-FPAP---RPFP-AQSSASGSFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--ERGTAPWVEEEE-EEEE--GSSPGEDSSSLGGGPSRRGPLP
CPLCSREG-----ACSCLPLERGDAVAGWGGHPAL-GC----PHPPEDDSSLRAERGSLA
DLPMAPPAS------APPEFLDPLMG-AAAP----QYP---GRGPPPAPPPRRH------
--------------------------------------------------L---------
-----------LGPPPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLPLP------------------------------------
------------------------------------GPREKPTFVVQVSTEQLLMSLRED
VTRNLLGEKGATARETGP-RKAGRGPGNREKVPGLNRDPTALGNGKQAPSL----S-LPV
NGVTVLENGDQRAPGIEE------K------AAENGALGSPEREE----KVLENGELTPP
RREEKA--L--------------------------ENGELRSPEAGEKVLVNGGLTPPKS
EDKVSENGGLRFPRNT-ERPPETGPWRAPGPWEKTPESWGPAPTIGE-PAPETSLERAPA
PS-------AVVSSRNGGETAPGPLGPAPKNGTLE--PGTERRAPETGGAPRAPGAGRLD
LGSGGRAPV-GTGTAPGGGPGSGVDAKAGWVDNTRPQPPPPPLP-PPPEAQPRRLE----
-PA---PPRARPEVAP-EGEPGAPDSRA---------GGDTAPSGDGDP-----PKPERK
GPEMPR-LFLDLGPPQGNSEQIKA------------------------------------
------------------------RLSRLSLALPPL---TLTPFPGPGPRRPP--WEGAD
AG--AAGGEAGGAGAPGPAEEDGEDEDEDEEEDEEAA----APGAAAGPRGPGRARAAPV
PVVVSSADADAARPLRGLLKSPRGADEPE-----DS-ELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQAPPEGDTDPSTPPAPPTPPHPATPGD--------------
---------------------GFPSNDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARAPDARPAGTIS--SALPPPLP-ISV---PLCSTANFL---------
------------------------------------------------------------
--------------------------------------------------------
ENSSSCP00000020665 (view gene)
-------------------------------------------------MPAPGALILLA
AV----SASGCLASPAHPDGFALGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMSTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 502)
SYLQEIGSGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGVCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPC-----A--
-------------PRPGTLSSPFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAPAWQPASAPPAPHANPSNPFYEA--------LSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLFP-NDWDPLDPGVPAPQAAQAPSEVPQ
LVSETWASPL-FPAA---RPFP-AQSSASGSFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--ERGAAPWAEEEE--EEE--GSSPGEDSSSIGGGPSRRGPLP
CPLCSREG-----ACSCLPLERGDAVAGWGGHPAL-GC----PHPPEDDSSLRAERGSLA
DLPLNPPAS------APPEFLDPLMGA-AAP----QYP---GRGPPPAPPP---------
-------------------------------------------------------RHPAV
-----SPDPPSAVASPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLPLP------------------------------------
------------------------------------GAREKPTFVVQVSTEQLLMSLRED
VTRNLLGEKGAAAHETGP-RKAARGSGNREKAPGPSRDPVVLDSGKQ------GLS-LPV
NGVTVLENGPQRALGIEE------K------VAENGGPRSPEREE----QVL--------
------------------------------------NGDPGSPEREEQVLANGGLTLPKI
EE-VSENGGLTLPRNP-ERLPETGPWRAPGPWEKNPESG---------PVPETSLERAPA
PG-------AVALALNGGETAPGPTGPAPRSGVLE--PGTERRAPETGGAPRAPGAGRLD
LGSGARAPV-GMGMAPGGGLGSGVDAKAGWVDSMRPQPPL-----PPPEAQPRRPE----
-PM---LQRARQEVVP-EGEPGVPDSKA---------GGDMAPSGDADP-----PKPERK
GPEMPR-LFLDLGPPQGNSEQIKA------------------------------------
------------------------KLSRLSLALPPL---TLTPFPGPGPRRPP--WEGAD
AG--APGGEAGGAGAPGPAEEDGEDEDEDEEEDEE----AAAAGAAAGPRGPGRARAAPV
PVVVSSTDADAARPLRGLLKSPRGADEPE------DSELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQGPPEGDTDPSTPPAPPTPPHPATPGD--------------
---------------------GFPSNDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARAPDARPAGTVENGSGTLPLVSNSKHSPR----H-------------
------------------------------------------------------------
--------------------------------------------------------
ENSOANP00000030592 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------------MV-V
DQ
PF07714 // ENSGT00640000091449_2_Domain_0 (423, 469)
SRESNVWSLGVTLWELFEFGSQPYRHLSDDEVLAYVIRQQHVQLARPRLKLPYGDYW-
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSLAFP00000024940 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------
PF00069 // ENSGT00640000091449_2_Domain_0 (309, 503)
GDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPTHTA--
-------------PRSGTLSSPFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARR--------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSPCAP00000003280 (view gene)
-VRWMLLCSVC---VTSKKXXXXXXX-XXXXXXX----------------XXXXXXXXXX
XX----XXXXXXXXXXXXXGFALGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLGLSRQHLSYLQEIGSGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYXXXXX----XXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXDL
ALR
PF00069 // ENSGT00640000091449_2_Domain_0 (364, 503)
NCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPSHTA--
-------------PRPGTLSSPFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAPPWQPASAPPAPHTNPSNPFYEA--------LSTPSVLPVISARSP
S-VGSEYYIRLEEHGSP------------PEPLFP-NDWDPLDPGVPAPQPPQAPSEVPQ
LVSETWASPL-FPAP---RSFP-AQASASGGFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--ERGTAPWAEEE--EEEE--GSSPGEDSSSLGGGASRRGALP
CPLCSSEG-----ACSCLPLERGDAVVGWGGHPAL-GC----PHLPRDDSFLRAERGSLG
DLPLAPAAS------APPEFLGPLMGA-AAL----QYP---WRGPPPGSPPPPP------
--------------------------------------------------PLRGPYRPAA
-----FPEPPTVVASPXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX--XXXXXXXX
XXXXX--------------XXXXX------------------------------------
------------------------------------XXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXX-XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX-XXX
XXXXXXXXXXXXXXXXXXXXXXXXX------XXXXXXXXXXXXXX----XXXXXXXXXXX
XXXXXX--X--------------------------XXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXX-XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XX-------XXXXXXXXXXXXXXXXXXXXXXXXXX--XXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXX-XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX----
-XX---XXXXXXXXXXXXXXXXXXXXXX---------XXXXXXXXXXXX-----XXXXXX
XXXXXXXXXXXXXXXXXXXXXXXX------------------------------------
------------------------KLSRLSLALPPL---TLTPFPGPGPRRPP--WXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX----XXX--XXXXXXX-----X-XXXXX
XXXXXXXXX-----X-XXXXXXXXXXXXX-----XXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXETPTNELSVQGPPEGDTDPSTPPAPPTPPHPTTPGD--------------
---------------------GFPSNDSGFGGSIEWAEDFSLLLSPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGLAWAL--DAPAGPVEN----------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSCSAP00000015206 (view gene)
-------------------------------------------------MPAPGALILLA
AV----SASGCLASPAHPDGFALGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 502)
SYLQEIGSGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGLCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPAHSA--
-------------PRPGTLSSPFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAPAWQPASAPPAPHANPSNPFYEA--------LSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLFP-NDWDPLDPGVPTPQAPQAPSEVPQ
LVSETWASPL-FPAP---RPFP-AQSSASGSFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--ERGTAPWVEEEE-EEEE--GSSPGEDSSSLGGGPSRRGPLP
CPLCSREG-----ACSCLPLERGDAVAGWGGHPAL-GC----PHPPEDDSSLRAERGSLA
DLPMAPPAS------APPEFLDPLMG-AAAP----QYP---DP-----------------
----------------------------------------------------------AA
-----SPDPPSAVASPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLPLP------------------------------------
------------------------------------GPREKPTFVVQVSTEQLLMSLRED
VTRNLLGEKGATARETGP-RKAGRGPGNREKVPGLNRDPTALGNGKQAPSP----S-LPV
NGVTVLENGDQRAPGIEEKAGIEQK------AAENGALGSPEREE----KVLENGELTPP
RREEKA--L--------------------------ENGELRSPEAGEKVLVNGGLTPPKS
EDKVSENGGLRFPRNT-ERPPETGPWRAPGPWEKTPESWGPAPTIGE-PAPETSLERAPA
PS-------AVASSRNGGETAPGPLGPAPKNGTLE--PGTERKAPETGGAPRAPGAGRLD
LGSGGRAPV-GTGTAPGGGPGSGVDAKAGWVDYTRPQPPPPPLP-PPPEAQPRRLE----
-PA---PPRARPEVAP-EGEPGAPDSRA---------GGDTAPSGDGDP-----PKPERK
GPEMPR-LFLDLGPPQGNSEQIKA------------------------------------
------------------------RLSRLSLALPPL---TLTPFPGPGPRRPP--WEGAD
AG--AAGGEAGGAGAPGPAEEDGEDEDEDEEEDEEAA----ASGAAAGPRGPGRARAAPV
PVVVSSADADAARPLRGLLKSPRGADEPE-----DS-ELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQGPPEGDTDPSTPPAPPTPPHPATPGD--------------
---------------------GFPSNDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARAPDARPAGTIS--S--------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSCJAP00000000681 (view gene)
--KCLWLCNVC---V-----------------TSRETRHHLHLPAILDKMPAPGALILLA
AV----SASGCLASPAHPDGFALGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 502)
SYLQEIGSGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGLCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPAHSA--
-------------PRPGTLSSPFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAPAWQPASAPPAPHANPSNPFYEA--------LSTPSVLPVISARSP
S-VGSEYYIRLEEHGSP------------PEPLF-PNDWDPLDPGVPAPQAPQAPSEVPQ
LVSETWASPL-FP----------AQSSASGSFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--ERGNAPWVEEEE-EEEE--GSSPGEDSSSLGGGPSRRGPLP
CPLCSREG-----ACSCLPLERGDAIAGWGGHPAL-GC----PHPPEDDSSLRAERGSLA
DLPMAPPAS------APPEFLDPLMGA-AAP----QYP---GRGPPPAPPPPPP------
--------------------------------------------------PPRVPADPAA
-----SPDPPSAVASPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLLLP------------------------------------
------------------------------------GPREKPTFVVQVSTEQLLMSLRED
VTRNLLGEKGATARETGP-RKAGRGPGSREKAPGLSRDPTALGNGKQAPSP----S-LPV
NGVTVLENGDQRAPDIEE------K------AAENGALASPEREE----KVLENGELTPP
RREEKV--L--------------------------ENGELRSPEAGEKVLVNGGLTPPKS
EDKVSENGGLRFPRNT-ERPPETGPWRAAGPWEKKPESWGPAPTIGE-PAPETSLERAPA
LS-------AVGSSRNGGETAPGLLGPAPKNGTLE--PGTERRAPETGGAPRVLGAGRLD
LGSGGRALV-GTGTAPGGGPGSGVDAKVGWVDNTRPQPPPPPLP-PPPEVHPRRPE----
-PA---PPRTRPEVAS-EGEPGAPDSRT---------GGDTAPSGDGDP-----SKPERK
GPEMPR-LFLDLGPPQGNSEQIKA------------------------------------
------------------------RLSRLSLALPPL---TLTPFPGPGPRRPP--WEGAD
AA--AAGGEAGGAGSSGPAEEDGEDEDEDEEEDEEAA----AAGAGAGPQGPGRARAAPV
PVVVSSVDADAARPLRGLLKSPRGADEPE-----DS-ELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQGPPEGDTDPSTPPAPPTPPHPATPGD--------------
---------------------GFPSNDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARAPDARPAGPVE--N--------------------------------
------------------------------------------------------------
--------------------------------------------------------
MGP_PahariEiJ_P0016477 (view gene)
----------------------MEAS-SPRAAVRASGRRFLHLPSILDKMPAPGALILLA
AV----SASGCLASPAHPDGFALSRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVR-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPPHSDISTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 502)
SYLQEIGSGWFGKVILGE
VFSDYSPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGVCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGMSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWVPLRWAAPELLGELHGSFVL-V
DQSRESNVWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPSHSA--
-------------PRPGTLSSQFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAPPWQPASAPPAPHTNPSNPFYEA--------LSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLFP-NDWDPLDPGVPGPQAPQTPSEVPQ
LVSETWASPL-FPAP---RPFP-AQSSGSGGFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--EQGTAPWAEEEE----E--ESSPGEDSSSLGRGPSRRGPLP
CPLCSREG-----PCSCLPLERGDAVAGWGGHPAL-GC----PHPPEDDSSLRAERGSLA
DLPLVPPTS------APLEFLDPLMGA-AAP----QYP---GRGPPPAPPPPPP------
--------------------------------------------------PPRASAEPAA
-----SPDPPSALASPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLPLP------------------------------------
------------------------------------GSQEKPTFVVQVSTEQLLMSLRED
VTKNLLGDKGSTPGETGP-RKAGRSPANREKGPGPSRDLTSLV-RKKVPSR----S-LPV
NGVTVLENGKPGVPDVKE-------------------------------KVAENG-LESP
ERGERA--L--------------------------VNGEPMSPEAGEKVLANGVLMSPKS
EEKVAENGVLRLPRNT-ERPPEIGPRRVPGPWEKTPETGGL--------APETLLDRAPA
SC-------EAALPQNGLEMAPGQLGPAPKSGNPD--PGTEWRVHESGGAPRAPGAGKLD
LGSGGRAPG-GVGTAPAGGPASAVDAKAGWVDNLRPLPPPP----QPLGAQQKRPE----
-PV---PLKARPEAAQ-EEEPGVPDNRL---------GGDMAPSVDEDP-----LKPERK
GPEMPR-LFLDLGPPQGNSEQIKA------------------------------------
------------------------KLSRLSLALPPL---TLTPFPGPGPRRPP--WEGAD
AG--AAGGEAGGAGAPGPAEEDGEDEDEDEEDE-----------EAAGSRGPGRTREAPV
PVVVSSADGDTARPLRGLLKSPRAADEPED------SELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQGNPEGDTEPSTPPAPPTPPHPTTPGD--------------
---------------------GFPNNDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARAPDARPAGPVEN----------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSOARP00000012709 (view gene)
-------------------------------------------------TGRGGARNLGA
RG----VKTGAQPSPVPTDGFALGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLGLSRQHLSYLQEIGSGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGVCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHRWQ
PGSH-------RAELGRGGRARGLRTQEDYYLTPERLWIPL
PF00069 // ENSGT00640000091449_2_Domain_0 (402, 499)
RWAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPPPPPP--PPPPRDGPFPWPWPPQHSA--
-------------PXPA-------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------------DPAV
-----SPDPSSAVASPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLPLP------------------------------------
------------------------------------GAREKPTFVVQVSTEQLLMSLRED
VTRNLLGEK-------GP-RKTGRGPGNRERALGPSRDPPVLESGKQAPSLNEGLS-LPV
NGVTVLENGVPRALGADE------K------VAENGDPGSPEREE----QVLANGELPPP
RREETV--L--------------------------ENGGPGPPEREERVLVNGGLTPPKI
EE-GSENGGLRLPRNP-ERLPETGPWRAPGPWEKMPESGAPAPTNGE-PAPETSLERAPA
PG-------AVALALNGGETAPGPAGPAPRSGAPE--PGTERRAPETGGAPRAPGAGRLD
LGSGGQAPV-GTGMAPGGGPGSGADAKAGWADSTRPQPPLP-----PLEPQPRRPE----
-PA---PPRIRPEVAS-EGEPGPPDSRA---------GGDTAPSGDGDP-----PKPERK
GPEMPR-LFLDLGPPQGNSEQIKA------------------------------------
------------------------KLSRLSLALPPL---TLTPFPGPRQRA---------
-------------------------------------------------TXPGRARAAPV
PVVVSSADADAARPLRGLLKSPRGADEPE-----DS-ELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQGPPEGATDPAPRPPPLL-----------------------
-------------------------SLPTLGGSFEWAEDFPLLPPPGPPPPA--------
--------------------------------------SPPP------------------
------KPRGPPARAPDARPAGTVLQ----------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSOPRP00000006442 (view gene)
-------------------------------------------------MPAPSALILFA
AV----SASGCLASPAHPDGFAMGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FK-FENPEGENCSGENPPPAKETF-
-SSESLPXXXXXXXXXXXXXXXXXXXXXXXXXXXXXLSRQH
PF00069 // ENSGT00640000091449_2_Domain_0 (222, 503)
LSYLQEIGSGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGLCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSXXXXXXXXXXXXXXXXXXXXXXXXXXX-----
-------------XXXXXXXXXXXXXX---XXXX--XXXXXXXXXXXXXXXXXXXXXXX-
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX--------XXXXXXXXXXXXXXX
X-XXXXXXXXXXXXXXX------------XXXXXX-XXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXX----X---XXXXXXXXXX-----------------XXXXX---XX
XXXXXXXXXXXXXXXXX--XXXXXXXXXXXXXXX---XXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXX-----XXXXXXXXXXXXXXXXXXXXXX------XXXXXXXXXXXXXXXXX--
XXXXXXXXX------XXXXXXXXXXXXXXXXXX--------XXXXXXX--XX--------
------------------------------------------XXXXXXXXXXXXXXXXXX
-----XXXXXXXXXXXXXXXXXXXXXPGDSGYETETPFSPERAFPGEGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLPHP------------------------------------
------------------------------------GTREKPNFVVQVSTEQLLMSLRED
VTRNLLGDKGATPRETGP-RKAGRGLGNRERVPGLPRDLPAPGSGKPAPSLPH-----SV
NGETALETGAQRAPRAEEK------------AAENGPLGSPEREE----TALQNGELTPP
RREETL--A--------------------------ENGPLGSPEREETALQNGELTPPRR
EETLVENGAPRLLRSM-EKPVETGPRRAPGPWGEMPENGEPVPEAGL--------ETAPT
AS-------TADSPQSGSQTGHGPLGPVPRSGTPG--PGTERRALETGGAL-RAGAGRLD
PGSGGRAPV-GTGTALG-----GGLESGV---------XXXX-----XXXXXXXXXXXX-
----XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXX-XXXXXXXXXXXXXXXXX------------------------------------
------------------------XXXXXXXXXXXX---XXXXXXGPGLWRSP--CEVAD
VG--AAEGETAGSGSSGPAEG-GREDEEVE-----------AEVEAAGPRGPGRARAAPV
PVVVSSADADAARPLRGLLKSPRAADEPED------SELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQGPPEADTDPSTPPAPPTPPHPATPGD--------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSMMUP00000031546 (view gene)
MRQVLWLCNVC---E-----------------TSRETRHHLHLPAILDKMPAPGALILLA
AV----SASGCLASPAHPDGFALGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 488)
SYLQEIGSGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGLCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYA---Y
DILQSCARPSATL-------------------------------WYPFPLPWPRAPGT--
-------------ACPGD-----PLLRHSPATGD--SDPDDVLTVYGEHREPPASRPVG-
EGRRVAGA--GWGDSAWRRVP-PGPPRQNPSNPFYEA--------LSTPSVLPVISGREP
A-S---NYIRAGGATAPL-----------LSRLFPQRTGDPLDPGVPAPQAPQAPSEVPQ
LVSETWASPL-FPAP---RPFP-AQSSASGSFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--ERGTAPWVEEEE-EEEE--GSSPGEDSSSLGGGPSRRGPLP
CPLCSREG-----ACSCLPLERGDAVAGWGGHPAL-GC----PHPPEDDSSLRAERGSLA
DLPMVKDQR------HIKP----------------Q------------------------
------------------------------------------------------------
------RWGKSMAENPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLPLT------------------------------------
------------------------------------GPREKPTFVVQVSTEQLLMSLRED
VTRNLLGEKGATARETGP-RKAGRGPGNREKVPGLNRDPTALGNGKQAPSP----S-LPV
NGVTVLENGDQRAPGIEE------K------AAENGALGSPEREE----KVLENGELTPP
RREEKA--L--------------------------ENGELRSPEAGEKVLVNGGLTPPKS
EDKVSENGGLRFPRNT-ERPPETGPWRAPGPWEKTPESWGPAPTIGE-PAPETSLERAPA
PS-------AVASSRNGGETAPGPLGPAPKNGTLE--PGTERKAPETGGAPRAPGAGRLD
LGSGGRAPV-GTGTAPGGGPGSGVDAKAGWVDYTRPQPPPPPLP-PPPEAQPRRLE----
-PA---PPRARPEVAP-EGEPGAPDSRA---------GGDTAPSGDGDP-----PKPERK
GPEMPR-LFLDLGPPQGNSEQIKA------------------------------------
------------------------RLSRLSLALPPL---TLTPFPGPGPRRPP--WEGAD
AG--AAGGEAGGAGAPGPAEEDGEDEDEDEEEDEEAA----ASGAAAGPRGPGRARAAPV
PVVVSSADADAARPLRGLLKSPRGADEPE-----DS-ELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQGPPEGDTDPSTPPAPPTPPHPATPGD--------------
---------------------GFPSNDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARAPDARPAGTIS--SALPLLCP-SLSPYVPLPTSCDHLSMSLPLSSQ
S-----LLSPLSVI-RHPTQLWNQLR--PGDVHQTVVGRQAGCPQP-LSLSFAFCGDSPH
PQ-------------LLPIFVPSLGSDSGLALAGS---------------------
ENSCHIP00000016066 (view gene)
MRSSGPASAER---DATQGLRDHREV-RAQGGRPCFRRHHLHLPAILDKMPAPGALILLA
AV----SASGCLASPAHPDGFALGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 502)
SYLQEIGSGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGVCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPQHSA--
-------------PRPGTLSSPFPLLD---GFPG--ADPDDVLTVTESSGGLNLECLWE-
KARRGAGR--GGGAPTWQPASAPPAPHANPSNPFYEA--------LSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLFP-NDWDPLDPGVPPPQAPQAPSEVPQ
LVSETWASPL-FPAA---RPFP-AQSSASGSFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--ERGAAPWAEEEE-EEEE--GSSPGEDSSSLGGGPSRRGPLP
CPLCSREG-----ACSCLPLERGDAVAGWGGHPAL-GC----PHPPEDDSSLRAERGSLA
DLPLNPPAS------APPEFLDPLMGA-AAP----QYP---GRGPPPAPPPPPP------
--------------------------------------------------PPRAPADPAV
-----SPDPSSAVASPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLPLP------------------------------------
------------------------------------GAREKPTFVVQVSTEQLLMSLRED
VTRNLLGEK-------GP-RKTGRGPGNRERALGPSRDPPVPESGKQAPSPNEGLS-LPV
NGVTVLENGVPRALGADE------K------VAENGDPGSPEREE----QVLANGELPPP
RREETV--L--------------------------ENGGPGPPEREERVLVNGGLTPPKI
EE-GSENGGLRLPRNP-ERLPETGPWRAPGPWEKMPESGAPAPTNGE-PAPETSLERAPA
PG-------VVALALNGGETAPGPAGPAPRSGAPE--PGTERRAPETGGAPRAPGAGRLD
LGSGGQAPV-GTGMAPGGGPGSGADAKAGWADSTRPQPPLP-----PLEAQPRRPE----
-PA---PPRVRPEVAS-EGEPGAPDSRA---------GGDTAPSGDGDP-----PKPERK
GPEMPR-LFLDLGPPQGNSEQIKA------------------------------------
------------------------KLSRLSLALPPL---TLTPFPGPGPRRPP--WEGAD
AGA--AGGEAGGAGAPGPSEEDGEDEDEEE------DEEAAAAGAAAGPRGPGRARAAPV
PVVVSSADADAARPLRGLLKSPRGADEPE-----DS-ELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQGPPEGDTDPSTPPAPPTPPHPATPGD--------------
---------------------GFPSNDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARAPDARPAGPVEN----------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSECAP00000011401 (view gene)
------------------------------------------------------------
---------GPQPSPVPTDGFAPGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLGLSRQHLS
PF00069 // ENSGT00640000091449_2_Domain_0 (224, 499)
YLQEIGSGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGVCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGEVHGSFLV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQVTNLLSERPPRPSPP--PPTPRKGPF------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSPVAP00000008969 (view gene)
--------------------------------------------------------VKQV
LW----LCNGCVTSKEXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
PF00069 // ENSGT00640000091449_2_Domain_0 (232, 476)
XXXXVILGE
IFSDYSPAQVVVKELRASAGALEQRKFISEAQPYRSLQH----PNVLQCLGVCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX--XXXXXXXXXXXXX-----
-------------XXXXXXXXXXXXXX---XXXX--XXXXXXXXXXXXXXXXXXXXXXX-
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX--------XXXXXXXXXXXXXXX
X-XXXXXXXXXXXXXXX------------XXXXXX-XXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXX----X---XXXXXXXXXX-----------------XXXXX---XX
XXXXXXXXXXXXXXXXX--XXXXXXXXXXXXXXX---XXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXX-----XXXXXXXXXXXXXXXXXXXXXX------XXXXXXXXXXXXXXXXX--
XXXXXXXXX------XXXXXXXXXXXXXXXXXX--------XXXXXXXXXXX--------
------------------------------------------XXXXXXXXXXXXXXXXXX
-----XXXXXSAVASPGSGLSSPGPKFGDSGYETETPFSPEGAFPGGGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDLGPLLLS------------------------------------
------------------------------------GTREKPTFVVQVSTEQLLMSLRED
VTRNLLGEKAATPQETGP-RKAAGGPGNREKAPGPSSDPAVLGSGKKAPGLNEDPSSLPV
NGATVLENGERRVPGVEEK------------VAENGDLGSPERDE----EVLENGELTPP
RRKETV--L--------------------------ENGELRSPEREETVLVNGGLTPPKM
EEMVSENGGLRLSRNT-ERLPETGSWRASGPWGKMCGSGDPAPETSM--------ERVLT
PSVLALPQISV--------TAPGPTGQAPRSGGLE--PGSER----------RLGPGRLD
LGSGGXXXX-XXXXXXXXXXXXXXXXXXX---------XXXX-----XXXXXXXXX----
------XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXX-XXXXXXXXXXXXXXXXX------------------------------------
------------------------KLSRLSLALPPL---TLTPFPGPGPRRPP--WEGAD
AG--AAGGEAGGAGAPGPPEEDGEDEDEDEEEEDEE-----AAGAAAGRRGSGRARAAPV
PVVVSSADADAARPLRGLLKSPRGADEPED------SELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQGPSEGDTDPSTPPAPPTPPHPVTPGD--------------
---------------------GFPSNDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPTRAPDARPAGPVEN----------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSANAP00000038485 (view gene)
-------CNVC---V-----------------TSRETRHHLHLPAILDKMPAPGALILLA
AV----SASGCLASPTHPDGFALGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 358)
SYLQEIGSGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGLCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHRWE
GRGAAGGATNG---KG-----------ERM-------------EAADEGGRMTGGHKIGM
NQGG-----------------------------------------------------SGG
GGARGEWEEPSEREAGSDEEESE------------SG--XXXXXXXXXPWPWPPAHSA--
-------------PRPGTLSSPFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAPAWQPASAPPAPHANPSNPFYEA--------LSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLF-PNDWDPLDPGVPAPQAPQAPSEVPQ
LVSETWASPL-FP----------AQSSASGSFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--ERGTAPWVEEEE-EEEE--GSSPGEDSSSLGGGPSRRGPLP
CPLCSREG-----ACSCLPLERGDAVAGWGGHPAL-GC----PHPPEDDSSLRAERGSLA
DLPMAPPAS------APPEFLDPLMGA-AAP----QYP---GRGPPPAPPPPPP------
--------------------------------------------------PPRVPADPAA
-----SPDPPSAVASPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLLLP------------------------------------
------------------------------------GPREKPTFVVQVSTEQLLMSLRED
VTRNLLGEKGATARETGP-RKAGRGPGNREKAPGLSRDPTALGNGKQAPSP----S-LPV
NGVTVLENGDQR------------K------AAENGALASPEREE----KVLENGELTPP
RREEKA--L--------------------------ENGELRSPEAGEKVLVNGGLTPPKS
EDKVSENGGLRFPRNT-ERPPETGPWRAAGPWEKKPESWGPAPTIGE-PAPETSLERAPA
LS-------AVGSSRNGGETAPGPLGPAPKNGTLE--LGTERRAPETGGAPRVPGAGRLD
LGSGGRAPV-GTGTVPGGGPGSGVDAKVGWVDNTRPAPPAPTCAPPRHHAHPRRPE----
-PA---PPRTRPEVAS-EGEPGAPDSRA---------GGDTAPSGDGDP-----PKPERK
GPEMPR-LFLDLGPPQGNSEQIKA------------------------------------
------------------------RLSRLSLALPPL---TLTPFPGPGPRRAP--WEGAD
AG--AAGGEAGGAGSSGPAEEDGEDEDEDEETGEERR----PSGAAAACGAPVRARAAPV
PVVVSSADADAARPLRGLLKSPRGADEPE-----DS-ELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQGPPEGDTDPSTPPAPPTPPHPATPGD--------------
---------------------GFPSNDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARAPDARPAGIMFKASALPLLCP-SLCLYVPLPISCDHL---------
------------------------------------------------------------
--------------------------------------------------------
ENSEEUP00000009799 (view gene)
-------------------------------------------------MPAPGALVFLA
AL----SSSCCLASPAHPDGFALGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPAHSXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX--XXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXX-XXXX--X-XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
PF07714 // ENSGT00640000091449_2_Domain_1 (480, 503)
Y
DILQSCWRPPAQRPSASELQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPQHSA--
-------------PRPGTLSSPFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGGPAWQPASAPPAPHANPSNPFYEA--------LSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLFP-NDWDPLDPGVPAPQAPQGPSEVTQ
LVSETWASPLFPGAR---ALFR-AQSSTXXXXX-----------------XXXXX---XX
XXXXXXXXXXXXXXXXX--XXXXXXXXXXXXXXXXX--XXXXXXXXXXXXXXXXXXXXXX
XXXXXXXG-----TCSCLPLERGDAVAGWGGHPAL-GC----PHPPEFYSSSRAERRSLG
NMSLNGPWT---------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
----------------------------------------------------------PK
KAPE--------------------------------------------L-----------
--KCSGSSPLR-----------------------------------------------SA
TS-------GGAPSWAGGGTMPGALGPAP--GILE--PGTERRTPETGGAPRALGAGRLD
FGSGGRPPA-GTGTGPGGGLGSGGDVKAVWADSSRPPPPPP-----LPAAQPRRPE----
-PAV--PPRARPEGAP-EGEPGAPDSRA---------DVDTAPSGDVDP-----PKPERK
GPEMPR-LFLDLGPPQGNSEQIKXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
------------------------XXXXXXXXXXXX---XXXX------------XXXXX
XX---X-----------XX---------XXXXX-----------XXX--XXXXXXXXXXX
-XXXXXXXX-----XXXXXXXXXXXXXXX-------XXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXETPTNELSVQGPPEGDSDPSTPPAPPTPPHPTTPGD--------------
---------------------GFPSNDSGF------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSCPOP00000012298 (view gene)
----------------------------METSAPRRGRHHLHLPSILDKMPAPGALILLA
AV----SASGCLASPAHPDGFALGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 502)
SYLQEIGNGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGLCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFMV-V
DQSPESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPSHSA--
-------------PRPGTLSSQFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAPAWQPASAPPAPHANPSNPFYEA--------LSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLFP-NDWDPLDPGVPAAQAPQPPSEVPQ
LVSETWASPL-FPAP---RPFQ-AQSSASGSFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--ERGNAPWAEEEE--EEE--GSSPGEDSSSLGVGPGRRGPLP
CPLCSREG-----ACSCLPLERGDAVAGWGGHPAL-GC----PHPPEDDSSLRAERGSLA
DLPLVPPTS------APTEFLDPLMGA-AAP----QYP---GRGPPPAPPPPPP------
--------------------------------------------------PPRAPAEPAA
-----TPDPPSAVASPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGEAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLPLP------------------------------------
------------------------------------GTREKPTFVVQVSTEQLLMSLRED
VTRNLLGDKGVTSRETGP-RKVGRGPGNREKVLGLDRDLPVLGGGKKAPSL----S-LPV
NGVTVLGNGAQRAPDIEE------K------PAENGGLGSPEKEV----KVLANGELMPP
RRE-KA--L--------------------------ENGELRSPEAGEKVLANGGPIPPEI
EEKVSKNGLLSFPRNT-ERPTETGPWRAPGPQEKMHESGGP--------APETLLERAPV
PG-------EVASPQNGLETSPGPLGPAPKSGILD--PGMERRAHETGGVPRAPGA----
--------------------GSGVDAKAEWVDNMRQQPLPP---LPPPEAQPRRPE----
-PV---PPRIRPEAVS-EGEPGVPDSRA---------GGDTAPRGDGDL-----SKPERK
-----------------------A------------------------------------
------------------------KLSRLSLALPPL---TLSPFPGSGPAATPVGWRGPR
RD---LAGKAGGAGSSGPPE-DGEDEDETRRRDE--------------AAGAARTRAAPV
PVVVSSADADAARPLRGLLKSPRGADESE------DSELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQGPPEGDTDPSTPPAPPTPPHPATPGD--------------
---------------------GFPSNDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARAPDARPAGPVEN----------------------------------
------------------------------------------------------------
--------------------------------------------------------
MGP_CAROLIEiJ_P0079612 (view gene)
----------------------METS-SPRAAVRASGRRFLHLPSILDKMPAPGALILLA
AV----SASGCLASPAHPDGFALSRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVR-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPPHSDISTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 502)
SYLQEIGSGWFGKVILGE
VFSDYSPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGVCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGMSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWVPLRWAAPELLGELHGSFVL-V
DQSRESNVWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPSHSA--
-------------PRPGTLSSQFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAPPWQPASAPPAPHTNPSNPFYEA--------LSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLFP-NDWDPLDPGVPGPQAPQTPSEVPQ
LVSETWASPL-FPAP---RPFP-AQSSGSGGFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--EQGTAPWAEEEE----E--ESSPGEDSSSLGGGPSRRGPLP
CPLCSREG-----PCSCLPLERGDAVAGWGGHPAL-GC----PHPPEDDSSLRAERGSLA
DLPLVPPTS------APLEFLDPLMGA-AAP----QYP---GRGPPPAPPPPPP------
--------------------------------------------------PPRASAEPAA
-----SPDPPSALASPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLPLP------------------------------------
------------------------------------GSQEKPTFVVQVSTEQLLMSLRED
VTKNLLGDKGSTPGETGP-RKAGRSPANREKGPGPNRDLTSLVSRKKVPSR----S-LPV
NGVTVLENGKPGVPDMKE-------------------------------KVTENG-LESP
ERGERA--L--------------------------VNGEPMSPEAGEKVLANGVLMSPKS
EEKVAENGVLRLPRNT-EKPPEIGPRRVPGPWEKTPETGGL--------APETLLDRAPA
PC-------EAALPQNGLEMAPGQLGPAPKSGNPD--PGTEWRVHESGGAPRAPGAGKLD
LGSGGRALG-GVGTAPAGGPASAVDAKAGWVDNSRPLPPPP----QPLGAQQRRPE----
-PV---PLKTRPEVAQ-EEEPGVPDNRL---------GGDMAPSVDEDP-----LKLERK
GPEMPR-LFLDLGPPQGNSEQIKA------------------------------------
------------------------KLSRLSLALPPL---TLTPFPGPGPRRPP--WEGAD
AG--AAGGEAGGAGAPGPAEEDGEDEDEDEEDE-----------EAAGSRGPGRTREAPV
PVVVSSADGDTARPLRGLLKSPRAADEPED------SELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQGTPEGDTEPSTPPAPPTPPHPTTPGD--------------
---------------------GFPSNDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARAPDARPAGPVEN----------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSMUSP00000112592 (view gene)
-------------------------------------------------MPAPGALILLA
AV----SASGCLASPAHPDGFALSRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVR-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPPHSDISTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 502)
SYLQEIGSGWFGKVILGE
VFSDYSPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGVCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGMSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWVPLRWAAPELLGELHGSFVL-V
DQSRESNVWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPSHSA--
-------------PRPGTLSSQFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAPPWQPASAPPAPHTNPSNPFYEA--------LSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLFP-NDWDPLDPGVPGPQAPQTPSEVPQ
LVSETWASPL-FPAP---RPFP-AQSSGSGGFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--EQGTAPWAEEEE----E--ESSPGEDSSSLGGGPSRRGPLP
CPLCSREG-----PCSCLPLERGDAVAGWGDHPAL-GC----PHPPEDDSSLRAERGSLA
DLPLVPPTS------APLEFLDPLMGA-AAP----QYP---GRGPPPAPPPPPP------
--------------------------------------------------PPRASAEPAA
-----SPDPPSALASPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLPLP------------------------------------
------------------------------------GSQEKPTFVVQVSTEQLLMSLRED
VTKNLLGDKGSTPGETGP-RKAGRSPANREKGPGPNRDLTSLVSRKKVPSR----S-LPV
NGVTVLENGKPGVPDMKE-------------------------------KVAENG-LESP
EKEERA--L--------------------------VNGEPMSPEAGEKVLANGVLMSPKS
EEKVAENGVLRLPRNT-ERPPEIGPRRVPGPWEKTPETGGL--------APETLLDRAPA
PC-------EAALPQNGLEMAPGQLGPAPKSGNPD--PGTEWRVHESGGAPRAPGAGKLD
LGSGGRALG-GVGTAPAGGPASAVDAKAGWVDNSRPLPPPP----QPLGAQQRRPE----
-PV---PLKARPEVAQ-EEEPGVPDNRL---------GGDMAPSVDEDP-----LKPERK
GPEMPR-LFLDLGPPQGNSEQIKA------------------------------------
------------------------KLSRLSLALPPL---TLTPFPGPGPRRPP--WEGAD
AG--AAGGEAGGAGAPGPAEEDGEDEDEDEEDE-----------EAAGSRDPGRTREAPV
PVVVSSADGDTVRPLRGLLKSPRAADEPED------SELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQGTPEGDTEPSTPPAPPTPPHPTTPGD--------------
---------------------GFPNSDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARAPDARPAGPVEN----------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSMNEP00000021300 (view gene)
MRQVLWLCNVC---E-----------------TSRETRHHLHLPAILDKMPAPGALILLA
AV----SASGCLASPAHPDGFALGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 502)
SYLQEIGSGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGLCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPAHSA--
-------------PRPGTLSSPFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAPAWQPASAPPAPHANPSNPFYEA--------LSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLFP-NDWDPLDPGVPAPQAPQAPSEVPQ
LVSETWASPL-FPAP---RPFP-AQSSASGSFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--ERGTAPWVEEEE-EEEE--GSSPGEDSSSLGGGPSRRGPLA
CPLCSREG-----ACSCLPLQRGDAVAGWGGHPAL-GC----PHPPEDDSSLRAERGSLA
DLPMAPPAS------APPEFLDPLMG-AAAP----QYP---GRGPPPAPPPPPP------
--------------------------------------------------PPRAPADPAA
-----SPDPPSAVASPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLPLT------------------------------------
------------------------------------GPREKPTFVVQVSTEQLLMSLRED
VTRNLLGEKGATARETGP-RKAGRGPGNREKVPGLNRDPTALGNGKQAPSP----S-LPV
NGVTVLENGDQRAPGIEE------K------AAENGALGSPEREE----KVLENGELTPP
RREEKA--L--------------------------ENGELRSPEAGEKVLVNGGLTPPKS
EDKVSENGGLRFPRNT-ERPPETGPWRAPGPWEKTPESWGPAPTIGE-PAPETSLERAPA
PS-------AVASSRNGGETAPGPLGPAPKNGTLE--PGTERKAPETGGAPRAPGAGRLD
LGSGGRAPV-GTGTAPGGGPGSGVDAKAGWVDYTRPQPPPPPLP-PPPEAQPRRLE----
-PA---PPRARPEVAP-EGEPGAPDSRA---------GGDTAPSGDGDP-----PKPERK
GPEMPR-LFLDLGPPQGNSEQIKA------------------------------------
------------------------RLSRLSL--------------------------GAD
AG--AAGGEAGGAGAPGPAEEDGEDEDEDEEEDEEAA----ASGAAAGPRGPGRARAAPV
PVVVSSADADAARPLRGLLKSPRGADEPE-----DS-ELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQGPPEGDTDPSTPPAPPTPPHPATPGD--------------
---------------------GFPSNDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARAPDARPAGTIS--SNTEARKE-VRS----------WDSKRQLLGL-
------------------------------------------------------------
--------------------------------------------------------
ENSTTRP00000007455 (view gene)
MRQVPMLCNGY---VTSEXXXXXXXX-XXXXXX-----------------XXXXXXXXXX
XX----XXXXXXXXXXXXXGFALGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLGLSRQH
PF07714 // ENSGT00640000091449_2_Domain_0 (222, 473)
LSYLQEIGSGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGVCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEITRGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYGDYXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX--X----XXXXXXXXXXXXXX--
-------------XXXXXXXXXXXXXXXXXXXXX--XXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXX--XXXXXXXXXXXXXXXXXXXXXXXXXXX--------XXXXXXXXXXXXXXX
X-XXXXXXXXXXXXXXXX-----------XXXXXXXXXXXXXXXXXXXX--XXXXXXXXX
XXXXXXXXXX-XXXX---XXXX-XXXXXXXXXX-----------------XXXXX---XX
XXXXXXXXXXXXXXXXX--XXXXXXXXXXXXXXXXX--XXXXXXXXXXXXX--XXXXXXX
XXXXXXXX-----XXXXXXXXXXXXXXXXXXXXXX-XX----XXXXXXXXXXXXXXXXXX
XXXXXXXXX------XXXXXXXXXXXXXXXX----XXX---XXXXXXXXXPPPP------
--------------------------------------------------PPRAPADPAV
-----SPDPPSAVASPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLPLP------------------------------------
------------------------------------GAREKPTFVVQVSTEQLLMSLRED
VTRKLLGEKGAAARETGP-RRAARGPGNREKAPGPSRGPTVLDSGKQAPSPQEGPS-LPV
NGVTVLENGEQRALGVEE------K------VAENGSPGSPEREE----QVLANGELPPP
RREEKV--L--------------------------ENGGLGSPAREEQVLVNGGLTPPKI
EE-VSENGGLRLPRNP-ERLPETGPWRAPGPWEKMPESGAPAPMNGE-PAPETSLERALA
PG-------AVVLALNGGETAPGPTGPALRSGVLE--PGTERRAPETGGAPRAPGAGRLD
LGSGVRAPV-GMGMAPGGGLGSGVDAKAGWVDSTRPQPPPP-----PLEAQPTRPE----
-PA---PQRARPEAAS-EGEPGVPDSRA---------GGDTAPNEDGDP-----PKPERK
GPEMPR-LFLDLGPPQGNSEQIKA------------------------------------
------------------------KLSRLSLALPPL---TLTPFPGPGPRRPP--WEGVD
AG--AAGGEAGGAGASGPSEEDGEDEDEDEEEDE----EAAAAGALAGPRGPGRARAAPV
PVVVSSADTDAARPLRGLLKSPRGADEPE-----DS-ELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQGPPEGDTDPSTPPAPPTLPHSATPGD--------------
---------------------GFPSNDSGFGAGX------FPLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARAPDARPAGPVEN----------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSMEUP00000003310 (view gene)
-------------------------------------HHHLPLPSILDKMPSPSPLVLLA
------LSASCLATPAHPDGLFSGQGPLAPPYAVVLIS----------------------
-------C-SGLLA-FV
PF00069 // ENSGT00640000091449_2_Domain_0 (138, 503)
FL-LLTCL-CCKRGDVG-FKEFENPEGEECSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPXXXXXXXXXXLGTPLGLSRQHLSYLQEIGSGWFGKXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX----XXXXXXXXXXXXXXXXX
XXXXXXXXGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEISRGLAHLHAHNYVHSDL
ALRNCLVTSDLTVRIGDYGLS-HSNYKEDYYLTPERLWIPLRWAAPELLGEIHGTFMV-V
DQTKESNVWSLGVTLWELFEFGAQPYRHLSDEEVLAFVIRQQHVKLARPRLKLPYGDYWY
DVLQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPPPPPPPPRDAPFPWPWPAP-----
-------------PHQGALSSPFPLLD---GFPG--SDPDDVLTVTESSHGLNLECLWE-
KARRGAGRGAGGGPAPWQPASAPPAPQAVPSNPFYEA--------LTTPSVLPVISARSP
S-VGSEYYIRLEDHGSP------------PEPLFP-NDWDPLDPGVPAPQTNAAPAEVPQ
LVAETWASPLFPPPR----S---FSSKASGSFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--ERGTAPWGEEEEEEE---GEESSLGEGSSSGGRRSRRGPLP
CPLCSHEG-----PCSCLPLQRGEMVSGWGGHPAL------GPQPPEDDSSLRAERGH--
TDPLPPPSS------APPXXXXXXXXXXXXXXX--------X-XXXXX------------
-------------------------------------------XXXXXXXXXXXXXXXXX
-----XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXTPFSPEGAFPGGGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLP--------------------------------------
------------------------------------------GFLVQVSTEQLLLSLRED
VTRNVLGERAAPAREGPSGPRLARVPGNREKVLGQNGELMSPGSREKVPENGEQ--RPPG
SEDRVLENGAQRPPGSQDR------------VLENGAQRPPGSLE----TLLENGGQTPP
GSRETM--V--------------------------ENGLQGPPRSGEKPQENGVQRLPGS
GEKVSENGGQRPPGNR-EKVLENGGLRPPENGKKIPEFGEPAPGIQE--------ERALG
NGEKFLERGSASPREPGQ---DPGTEPVPRSGGPA--RGSGKRAP--------PGPGRLD
PENGLQALG-GQGKAPDAGPAGGGVGGAA---------PPGE-----MRLEPRRPELAP-
----PGAAAPAGDAG-D---PGAPV-DGAV-ET-GG-DGD-PPH-LG-RKG-PE-MP-R-
L-------FLDLGPPPGSSEQIKA------------------------------------
------------------------KLSRLSLALPPL---SLTPFPGPGPRRPP--WEAGD
AG--VPGAESGGLGALGLVED-GEDEEEEEDDGAAV-----GEGEEAAARGPGRGRAAPV
PIVVSSADGDRSRPLRGLLKSPRGADEAEE-----SLELDRKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQAPPEGDPDPSTPPAPPTPPHPPSPGD--------------
---------------------GFP--DSGFGGSFEWGEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARPPDAVCPA--------------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSPEMP00000004536 (view gene)
----------------------METS-SPRAAVCASGRRYLHLPSILDKMPAPGALILLA
AV----SASGCLASPAHPDGFALSRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVR-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPAHSDISTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 502)
SYLQEIGSGWFGKVILGE
VFSDYTPAQVVVKELRASAGPMEQRKFISEAQPYRSLQH----PNVLQCLGVCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGMSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWVPLRWAAPELLGELHGSFVL-V
DQSRESNVWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPSHSA--
-------------PRPGTLSSQFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAPPWQPASAPPAPHTNPSNPFYEA--------LSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLFP-NDWDPLDPGVPGPQAPQAPSEVPQ
LVSETWASPL-FPAP---RPFQ-AQSSGSGGFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--EQGTAPWTEEEE----E--ESSPGEDSSSLGGGPSRRGPLP
CPLCSREG-----PCSCLPLERGDAVAGWGGHPAL-GC----PHPPEDDSSLRAERGSLA
DLPLVPPTS------APSEFLDPLMGA-AAP----QYP---GRGPPPAPPPPPP------
--------------------------------------------------PPRAPAEPAA
-----SPDPPSALASPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLPLP------------------------------------
------------------------------------ESQEKPTFVVQVSTEQLLMSLRED
VTKNLLGDKGSTPGETGP-RKAGRSPANREKGLDPNRDLTSLGNRKKVPSW----S-LPV
NGVTVLETGKPGALDVKE-------------------------------KVAENG-LESP
ERGERA--L--------------------------VNGEPMSPEAEEKVLANGVLMSPKS
EEKVAENGVLRLPRNT-ERPPETGPRRAPGPWEKMPETGGL--------APETLLDQAPA
PC-------EAVLPQNGLETAPGQLGPVPKNGNPE--PGTEWRVHETGGALRVPGAGKLD
LGSGGRAQG-GVGMAPAGGPASGVDAKAGWVDSSRPLPPPP----PPLGVQQRRPE----
-PA---PPKARPEVAP-EEEPGVPDSRL---------GGDTAPSVDEDP-----PKPERK
GPEMPR-LFLDLGPPQGNSEQIKA------------------------------------
------------------------KLSRLSLALPPL---TLTPFPGPGPRRPP--WEGAD
AG--AAGGEAGGAGAPGPAEEDGEDEDEEDEDE-----------EAAGSRGPGRTREAPV
PVVVSYADADTARPLRGLLKSPRAADEPED------SELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQGTPEGDTDPSTPPAPPTPPHPTTPGD--------------
---------------------GFPNNDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARAPDARPAGPVEN----------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSTBEP00000009604 (view gene)
------------------------------------------------------------
-------------------GFALGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPVPAPQPSHSXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX----XXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXX-XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX-X
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
PF07714 // ENSGT00640000091449_2_Domain_1 (480, 503)
Y
DILQSCWRPPAQRPSAADLQLQLTYLLSERPPRPPPP--TRPHRYGPFPWPWPPTHSA--
-------------PRPGTLSSPFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAPPWQPASAPPAPHANPSNPFYEA--------LSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLFP-NDWDPLDPGVQAPQAPQAPSEVPQ
LVSETWASPL-FPAP---RPFP-AQSSASGSFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--ERGNAPWAEEEE----E--EGSSPEDSSSLGGGPSRRGPLP
-PLCSRGP------CSCLEG---D-AAGWGGPALY-------PHPPEDDSSLRAERGSLA
DLPLAPPTS------APPEFLDPLMGAAAPQ-----Y---PGRGPPPAPPPPPPTSR---
-----------------------------------------------------GPTDTGV
-----FPDPPSGVARPGSGLSAPGP-PGDGGFQTKTPISPESTFQVGGVTQ--KVGCPGL
PGSPETSELK---------ALPIP---TEPSPSRLP------------------------
------------------------------------ERRKKQKLIQ---RKS-LPLSPEK
AWLWLPLGKGANTLETGP-LKAGRGLGNREKAPGLNRDPTVLGSGKKAPML----S-LPM
NGVMV-ENGAQRVPGSEEK------------AAENGGLGSPEREE----KVLENGELTPP
RREEKV--L--------------------------ENGELRSPEAEEKVLANGGLTPTKS
EEKVSENGGPRFPRNM-ERPPETGPWRAPGHWEKMPESGGPVPTIGE-PAPETSLERAPT
PPD------TAASPRNGGVTAPGPLGPAPRSAMPEPETGTERRAHETGGVPRAPGAGRLD
LGSGQAP--VGIGTAPGGGPASRVDAKAGWVDTR-PQPALPLPRAPPPEXXXXXXXXXXX
XXX----X-XXXXXXX-------XXX----------------------------------
--XXXX-XXXXXXX--XXX--XXX-------------------------------XXXXX
XXXXXXXXXXXXXXXXXXXXXXXXKLSRLSLALPPL---TLTPFPGPGPRRPP--WEGAD
AG--AAGGEAGGAGAPGPAEEDGEDEDEDEEEDEEAA----AAGEAAGPRGPGRARAAPV
PVVVSSADADAARPLRGLLKSPRTDQSEDE----------IVREEISFHRV-------VY
RFXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX---XXXXX---XXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXPVEN----------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSRROP00000014300 (view gene)
MRQVLWLCNVC---E-----------------TSRETWVFAI-------RVSPACLCVSS
AV----PASLC-PSPVPADGFALGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 502)
SYLQEIGSGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGLCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPAHSA--
-------------PRPGTLSSPFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAPAWQPASAPPAPHANPSNPFYEA--------LSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLFP-NDWDPLDPGVPAPQAPQAPSEVPQ
LVSETWASPL-FPAP---RPFP-AQSSASGSFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--ERGTAPWVEEEE-EEEE--GSSPGEDSSSLGGGPSRRGPLH
ETPFSPEG-----AFP--GGGRGDAVAGWGGHPAL-GC----PHPPEDDSSLRAERGSLA
DLPMAPPAS------APPEFLDPLMG-AAAP----QYP---GRG----------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
--------------------------------------------------L----S-LPV
NGVTVLENGDQTAPGIEE------K------AAENGALESPETEE----KVLENGELTPP
RREEKA--L--------------------------ENGELRSPEAGEKVLVNGGLTPPKS
EDKVSENGVLRFPRNT-ERLPETGPWRAPGPWEKTPESWGPAPTIGE-PAPETSLERAPA
PS-------AVASSRNGGETAPGPLGPAPKNGTLE--PGTERRAPETGGAPRAPGAGRLD
LGSGGRAPV-GTGTGPGGGPGSGVDAKAGWVDYTRPQPPPPPLP-PPPEAQPRRLE----
-PA---PPRARPEVAP-EGEPGAPDSRA---------GGDTAPSGDGDP-----PKPERK
GPEMPR-LFLDLGPPQGNSEQIKA------------------------------------
------------------------RLSRLSLALPPL---TLTPFPGPGPRRPP--WEGAR
AG--PRGEGG-------VWEETGQGNGAGLGRWEEAL----RPDSWARPGGRAEPRAGVG
PRV-----------RAGLVM-GAGCRGPR-----------KIAGFVAFGGCDKWDVTVCP
LWL------PETPTNELSVQGPPEGDTDPSTPPAPPTPPHPATPGD--------------
---------------------GFPSNDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARAPDARPAGTIS--SALPLLCP-SLSPYVPLPTSCDHLSMSLPLSSQ
S-----LLSPLSVNRHPTQLW---NQLRPGDVHRTVVGRQA-----------GCPPPPSL
SFAFCG---DSPHPQLLPIFVPSLGSDSGLALAGS---------------------
ENSPANP00000014733 (view gene)
MRQVLWLCNVC---E-----------------TSRETRHHLHLPAILDKMPAPGALILLA
AV----SASGCLASPAHPDGFALGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 502)
SYLQEIGSGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGLCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPAHSA--
-------------PRPGTLSSPFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAPAWQPASAPPAPHANPSNPFYEA--------LSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLFP-NDWDPLDPGVPAPQAPQAPSEVPQ
LVSETWASPL-FPAP---RPFP-AQSSASGSFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--ERGTAPWVEEEE-EEEE--GSSPGEDSSSLGGGPSRRGPLP
CPLCSREG-----ACSCLPLERGDAVAGWGGHPAL-GC----PHPPEDDSSLRAERGSLA
DLPMAPPAS------APPEFLDPLMG-AAAP----QYP---GRGPPPAPPPPPP------
--------------------------------------------------PPRAPADPAV
-----SPDPPSAVASPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLPLP------------------------------------
------------------------------------GPREKPTFVVQVSTEQLLMSLRED
VTRNLLGEKGATARETGP-RKAGRGPGNREKVPGLNRDPTALGNGKQAPSP----S-LPV
NGVTVLENGDQRAPGIEE------K------AAENGALGSPEREE----KVLENGELTPP
RREEKA--L--------------------------ENGELRSPEAGEKVLVNGGLTPPKN
EDKVSENGGLRFPRNT-ERPPETGPWRAPGPWEKTPESWGPAPTIGE-PAPETSLERAPA
PS-------AVASSRNGGETAPGPLGPAPKNGTLE--PGTERKAPETGGAPRAPGAGRLD
LGSGGRAPV-GTGTAPGGGPGSGVDAKAGWVDYTRPQPPPPPLP-PPPEAQPRRLE----
-PA---PPRARPEVAP-EGEPGAPDSRA---------GGDTAPSGDGDP-----PKPERK
GPEMPR-LFLDLGPPQGNSEQIKA------------------------------------
------------------------RLSRLSLALPPL---TLTPFPGPGPRRPP--WEGAD
AG--AAGGEAGGAGAPGPAEEDGEDEDEDEEEDEEAA----ASGAAAGPRGPGRARAAPV
PVVVSSADADAARPLRGLLKSPRGADEPE-----DS-ELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQGPPEGDTDPSTPPAPPTPPHPATPGD--------------
---------------------GFPSNDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARAPDARPAGTIS--SALPLLCP-SLSPDVPLPTSCSFNVPASLFPV-
S-----SLSPLSQQTPHPALEPAQTRRCPSD-GRRQAGRLSPTPEPQFRLLWGFPPPPTL
AH-LCPISGLRLRSGTCRILIPPLGNTVLRGRQGEKNPECSKPAGVNQSREDTAGG
ENSRBIP00000012589 (view gene)
MRQVLWLCNVC---E-----------------TSRETRHHLHLPAILDKMPAPGALILLA
AV----SASGCLASPAHPDGFALGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 358)
SYLQEIGSGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGLCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHRW-
-------------------------------------------AGRG-------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------T--AGGA----------------------------------------------
------------------------------------------------------------
----------------------------------------------------------TK
GKGEGG------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
---------EAAGEGPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLPLP------------------------------------
------------------------------------GPREKPTFVVQVSTEQLLMSLRED
VTRNLLGEKGATARETGP-RKAGRGPGNREKVPGLNRDPTALGNGKQAPSL----S-LPV
NGVTVLENGDQTAPGIEE------K------AAENGALESPETEE----KVLENGELTPP
RREEKA--L--------------------------ENGELRSPEAGEKVLVNGGLTPPKS
EDKVSENGVLRFPRNT-ERLPETGPWRAPGPWEKTPESWGPAPTIGE-PAPETSLERAPA
PS-------AVASSRNGGETAPGPLGPAPKNGTLE--PGTERRAPETGGAPRAPGAGRLD
LGSGGRAPV-GTGTGPGGGPGSGVDAKAGWVDYTRPQPPPPPLP-PPPEAQPRRLE----
-PA---PPRARPEVAP-EGEPGAPDSRA---------GGDTAPSGDGDP-----PKPERK
GPEMPR-LFLDLGPPQGNSEQIKG------------------------------------
------------------------EEAPKAGRIEQ----------GA---GRA--WGE--
---------------GGVWEETGQGNGAGLGRWEEAL----RPGSGAGPGGRAEPRAGVG
PRVRA-----------GLVM-GAGCRG-----------PRKIAGFVAFGGCGKWDVTVCP
LWL------PETPTNELSVQGPPEGDTDPSTPPAPPTNDSGFGEXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXTGFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARAPDARPAGTIS--SALPLLCP-SLSPYVPLPTSCDHLSMSLPLSSQ
S-----LLSPLSVNRHPTQLW---NQLRPGDVHRTVVGR-----------QAGCPPPPSL
SFAFCGDSPH---PQLLPIFVPSLGSDSGLALAGS---------------------
ENSSARP00000005953 (view gene)
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-----------------------------------
PF00069 // ENSGT00640000091449_2_Domain_0 (276, 476)
SLQH----PNVLQCLGVCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDMRTLQRMGLEIARGLAHLHAHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNY-EDYYLTPERLWIPLRWAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX-----
-------------XXXXXXXXXXXXXX---XXXX--XXXXXXXXXXXXXXXXXXXXXXX-
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX--------XXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXX------------XXXXXX-XXXXXXXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXX----X---XXXXXXXXXX-----------------XXXWC---TE
S-GRDGTLDGVSPEYMG--KKRSAPSAKEKVMYDNQKEVSFPGGDSSSQGGSPSHRGPLH
CPLCSREG-----AYSCLPLERREPVVGWWGHPVF------GPPPPENSSREKRDSQA--
DLPLAPPFS------APPEFLDPFMGAAAPQYP--------GRGPPP-------------
-------------------------------------------APPPPPPPPRVPADPAV
-----SPEPLSAVASPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPRPAP------------------------------------
------------------------------------GVREKPAFMVQVSTEQLLVSLRED
VTRNLLGDKRPNSRETGPWK--------ASRAPEASRDPSVPGSGKRAPSL-----SPPV
NGLTVSENGGQRALGLEEK------------VVENGGLGSPEREE----KVLENGELTPP
RKEK-P--L--------------------------ANGELRSPERDEKVLVNGELIPPKI
EETVSENGGLRLIRNT-ERPPETGPWRAPGPWEETTENGGPAPETTL--------QRAPT
PGTAALARNSG-------ETAPGPPGPAPRSGVLE--PGTEGRAPETGGAPKAPGAGRLD
LGSGGRALV-GMGTAPGGGLGSGVDAKAGWADSTRPLPPPPL-----PEGPPRRPEPAP-
----PKARP-EGP---PEGEPGFPDSR-------A--GGDTAPSGDGA----P-RPPERK
GPEMPR-LFLDLGPPQGNSEQIKX------------------------------------
------------------------XXXXXXXXXXXX---XXXXXXXXXXXXXX--XXXXX
XX--XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX-----XXXXXXXXXXXXXXXXXXX
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXX-----XXXXXXXXXXXXXXX-----XXXXX
XXX------XETPTNELSVQGPPEGDTDPSTPPAPPTPPHPATPGD--------------
---------------------GFPSNDSGFGGSFEWAXXXXXXXXXXXXXXXXX------
-----------------------------------XXXXXXX------------------
------XXXXXXXXXXXXXXXXPVEN----------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSCCAP00000039983 (view gene)
-------------------------------------------------MPAPGALILLA
AV----SASGCLASPAHPDGFALGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 502)
SYLQEIGSGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGLCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPAHSA--
-------------PRPGTLSSPFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAPTWQPASAPPAPHANPSNPFYEA--------LSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLF-PNDWDPLDPGVPAPQAPQAPSEVPQ
LVSETWASPL-FP----------AQSSASGSFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--ERGTAPWVEEEEEEEEE--GSSPGEDSSSLGGGPSRRGPLP
CPLCSREG-----ACSCLPLERGDAVAGWGGHPAL-GC----PHPPEDDSSLRAERGSLA
DLPMAPPAS------APPEFLDPLMGA-AAP----QYP---GRGPPPAPPPPPP------
--------------------------------------------------PPRVLADPAA
-----SPDPPSAVASPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLLLP------------------------------------
------------------------------------GPREKPTFVVQVSTEQLLMSLRED
VTRNLLGEKGATARETGP-RKAGRGPGNREKAPGLSRDPTALGNGKQAPSP----S-LPV
NGVTVLENGDQRAPDIEKAGT-EEK------AAENGALASPEREE----KVLENGELTPP
RREEKA--L--------------------------QNGELRSPEAGEKVLVNGGLTPPKS
EDKVSENGGLRFPRNT-ERPPETGPWRAAGPWEKKPESWGPAPTIGE-PAPETSLERAPA
LS-------AVGSSPNGGETAPGPLGPAPKNGTLE--PGTERRAPETGGAPRVPGAGRLD
LGSGGRAPV-GTGTAPGGGPGSGVDAKVGWVDNMRPQPPPPPLP-PPPEAHPRRPE----
-PA---PPRTRPEVAS-EGEPGAPDSRA---------GGDTAPSGDGDP-----PKPERK
GPEMPR-LFLDLGPPQGNSEQIKA------------------------------------
------------------------RLSRLSLALPPL---TLTPFPGPGPRRPP--WEGAD
AG--AAGGEAGGAGSSGPAEEDGEDEDEDEEEDEEAA----ASGAAAGPRGPGRARAAPV
PVVVSSADADAARPLRGLLKSPRGADEPE-----DS-ELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQGPPEGDTDPSTPPAPPTPPHPATPGD--------------
---------------------GFPSNDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARAPDARPAGPVE--N--------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSMFAP00000017404 (view gene)
MRQVLWLCNVC---E-----------------TSRETRHHLHLPAILDKMPAPGALILLA
AV----SASGCLASPAHPDGFALGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 502)
SYLQEIGSGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGLCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPAHSA--
-------------PRPGTLSSPFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAPDWQPASAPPAPHANPSNPFYEA--------LSTPSVLPVISARSP
S-VS--------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------SDPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLPLT------------------------------------
------------------------------------GPREKPTFVVQVSTEQLLMSLRED
VTRNLLGEKGATARETGP-RKAGRGPGNREKVPGLNRDPTALGNGKQAPSP----S-LPV
NGVTVLENGDQRAPGIEE------K------AAENGALGSPEREE----KVLENGELTPP
RREEKA--L--------------------------ENGELRSPEAGEKVLVNGGLTPPKS
EDKVSENGGLRFPRNT-ERPPETGPWRAPGPWEKTPESWGPAPTIGE-PAPETSLERAPA
PS-------AVASSRNGGETAPGPLGPAPKNGTLE--PGTERKAPETGGAPRAPGAGRLD
LGSGGRAPV-GTGTAPGGGPGSGVDAKAGWVDYTRPQPPPPPLP-PPPEAQPRRLE----
-PA---PPRARPEVAP-EGEPGAPDSRA---------GGDTAPSGDGDP-----PKPERK
GPEMPR-LFLDLGPPQGNSEQIKA------------------------------------
------------------------RLSRLSLALPPL---TLTPFPGPGPRRPP--WEGAD
AG--AAGGEAGGAGAPGPDE--------DEEEDEEAA----ASGAAAGPRGPGRARAAPV
PVVVSSADADAARPLRGLLKSPRGADEPE-----DS-ELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQGPPEGDTDPSTPPAPPTPPHPATPGD--------------
---------------------GFPSNDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARAPDARPAGTIS--SNTKARKE-VRS----------WDSKRQLLGL-
------------------------------------------------------------
--------------------------------------------------------
ENSPPYP00000011408 (view gene)
-------------------------------------------------MPAPGALILLA
AV----SASGCLASP---------------------------------------------
------------------------------------------------------------
----------------------------------------------------AHPVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGLCVETLPFL
LIMEFCQL--------------------------------------------------
PF00069 // ENSGT00640000091449_2_Domain_0 (359, 499)
DL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPAHSA--
-------------PRPGTLSSPFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--G-------------------------------------------------
------------------------------------------------------------
------------------------------------------------------------
-------------------------WVEDE-EEEEE--GSSPGEDSSSLGGGPSRRGPLP
CPLCSREG-----ACSCLPLERGDAVAGWGGHPAL-GC----PHPPEDDSSLRAERGSLA
DLPMAPPAS------APPEFLDPLMGA-AAP----QYP---GRGPPPAPPPPPP------
--------------------------------------------------PPRAPADPAA
-----SPDPPSAVASPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGGAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLPLP------------------------------------
------------------------------------GSREKPTFVVQVSTEQLLMSLRED
VTRNLLGEKGATARETGP-RKAGRGPGNREKVPGLNRDPTALGNGKQAPSL----S-LPV
NGVTVLENGDQRAPGIEE------K------AAENGALGSPEREE----KVLENGELTPP
RREEKA--L--------------------------ENGELRSPEAGEKVLVNGGLTPPKS
EDKVSENGGLRFPRNT-ERPPETGPWRAPGPWEKTPESWGPAPTIGE-PAPETSLERAPA
PS-------AVASSRNGGETAPGPLGPAPKNGTLE--PGTERRAPETGGAPRAPGAGRLD
LGSGGRAPV-GTGTAPGGGPGSGVDAKAGWVDNTRPQPPPPPLP-PPPEAQPRRLE----
-PA---PPRARPEVAP-EGEPGAPDSRA---------GGDTAPSGDGDP-----PKPERK
GPEMPR-LFLDLGPPQGNSEQIKA------------------------------------
------------------------RLSRLSLVLPPL---TFTPFP-PGPRRPP--WEGAD
AG--AAGGEAGGAGAPGPAEEDGEDEEEDEEE----DEEAAAPGAAAGPRGPGRARAAPV
PVVVSSADADAARPLRGLLKSPRGADEPED------SELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQAPPEGDTDPSTPPAPPTPPHPATPGD--------------
---------------------GFPSNDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPARAPDARPAGPVEN----------------------------------
------------------------------------------------------------
--------------------------------------------------------
ENSODEP00000024052 (view gene)
-------------------------------------------------MPAPGAFILLA
AV----SASGCLASPAHPDGFALGRAPLAPPYAVVLIS----------------------
-------C-SGLLA-FIFL-LLTCL-CCKRGDVG-FKEFENPEGEDCSGEYTPPAEETS-
-SSQSLPDVYILPLAEVSLPMPAPQPSHSDMTTPLGLSRQHL
PF07714 // ENSGT00640000091449_2_Domain_0 (223, 502)
SYLQEIGNGWFGKVILGE
IFSDYTPAQVVVKELRASAGPLEQRKFISEAQPYRSLQH----PNVLQCLGLCVETLPFL
LIMEFCQLGDLKRYLRAQRPPEGLSPELPPRDLRTLQRMGLEIARGLAHLHSHNYVHSDL
ALRNCLLTSDLTVRIGDYGLA-HSNYKEDYYLTPERLWIPLRWAAPELLGELHGTFMV-V
DQSRESNIWSLGVTLWELFEFGAQPYRHLSDEEVLAFVVRQQHVKLARPRLKLPYADYWY
DILQSCWRPPAQRPSASDLQLQLTYLLSERPPRPPPP--PPPPRDGPFPWPWPPSHSA--
-------------PRPGTLSSQFPLLD---GFPG--ADPDDVLTVTESSRGLNLECLWE-
KARRGAGR--GGGAPAWQPASAPPAPHTNPSNPFYEA--------LSTPSVLPVISARSP
S-VSSEYYIRLEEHGSP------------PEPLFP-NDWDPLDPGVPATQAPQAPSEVPQ
LVSETWASPL-FPAP---RPFP-AQSSASGSFL-----------------LSGWD---PE
GRGAGETLAGDPAEVLG--ERGTAPWAEE---EEEE--GSSPGEDSSSLGVGPSRRGPLP
CPLCSREG-----ACSCLPLERGDAVAGWGGHPAL-GC----PHPPEDDSSLRAERGSLA
DLPLAPPAS------APPEFLDPLMGA-AAP----QYP---GRGPPPAPPPPPP------
--------------------------------------------------PPRAPSEPAA
-----TPDPPSAVASPGSGLSSPGPKPGDSGYETETPFSPEGAFPGGEAA--EEEGVPRP
RAPPEPPDPGAPRPPPDPGPLPVP------------------------------------
------------------------------------GTREKPTFVVQVSTEQLLISLRED
VTRNLLGDKGATPRETGP-RKAGRGPGNREKVLGLNRDLPVLGGGKKAPSL----S-LPM
NGVTVLG--------------------------NNGGLGSPEREV----KVLVNGELTPP
RK-EKA--L--------------------------QNGELRSPEAGEKVLANGGPIPPEI
-EEVSENGLLRFPRNT-ERPTETGPWRAPGPREKMPESGGP--------APETLLERAPA
PS-------EVALPQNGLETVPGPLGPAPKSGMLD--PGTERRAHETGGAPKAPGAGRLD
LGSGGRALV-GVAMAPGGGPGSGVDVKAGWVDSTRPQPLPPLPPLPPPEAQQRRPE----
-PV---PPRIRPEVAS-EGEPGVPDSKA---------GGDTAPSGDGDP-----SKPERK
GPEMPR-LFLDLGPPQGNSEQIKD--RWGGQPAGLREKLGSRPRVLGSAAPWEKALEWDR
GGPASVSPT---AHALFPAA----KLSRLSLALPPL---TLSPFPGPGPRRPP--WEGAD
AG--AAGGEAGGAGSPGPAEEDGEDEDEDEEEDEA----A----------GAARTRAAPV
PVVVSSADADAARPLRGLLKSPRGADEPED------SELERKRKMVSFHG-----DVTVY
LFD------QETPTNELSVQGPPEGDTDPSTPPAPPTPPHPATPGD--------------
---------------------GFPSNDSGFGGSFEWAEDFPLLPPPGPPLCFSR------
-----------------------------------FSVSPAL------------------
------ETPGPPTRAPDARPADWV------------------------------------
------------------------------------------------------------
--------------------------------------------------------
{"ENSOGAP00000016616": [["Domain_0", 223, 502, "PF07714"]], "ENSHGLP00000008992": [["Domain_0", 223, 502, "PF07714"]], "ENSSTOP00000018152": [["Domain_0", 223, 502, "PF07714"]], "ENSCANP00000016788": [["Domain_0", 223, 502, "PF07714"]], "ENSPCOP00000024449": [["Domain_0", 223, 502, "PF07714"]], "MGP_SPRETEiJ_P0082952": [["Domain_0", 223, 502, "PF07714"]], "ENSBTAP00000015942": [["Domain_0", 224, 499, "PF00069"]], "ENSP00000270238": [["Domain_0", 223, 502, "PF07714"]], "ENSXETP00000054322": [["Domain_0", 222, 506, "PF00069"]], "ENSMPUP00000003914": [["Domain_0", 224, 499, "PF00069"]], "ENSMOCP00000003426": [["Domain_0", 223, 502, "PF07714"]], "ENSPPAP00000026690": [["Domain_0", 223, 458, "PF07714"]], "ENSNLEP00000042129": [["Domain_0", 223, 459, "PF07714"]], "ENSCGRP00001010660": [["Domain_0", 223, 502, "PF07714"]], "ENSCLAP00000012167": [["Domain_0", 223, 489, "PF07714"]], "ENSSBOP00000026538": [["Domain_0", 223, 502, "PF07714"]], "ENSMLEP00000004546": [["Domain_0", 223, 307, "PF07714"]], "ENSMICP00000041476": [["Domain_0", 223, 502, "PF07714"]], "ENSMAUP00000014952": [["Domain_0", 223, 502, "PF07714"]], "ENSDNOP00000030849": [["Domain_0", 423, 502, "PF07714"]], "ENSGGOP00000010451": [["Domain_0", 223, 502, "PF07714"]], "ENSCAPP00000014635": [["Domain_0", 223, 502, "PF07714"]], "ENSCATP00000000765": [["Domain_0", 223, 502, "PF07714"]], "ENSOCUP00000019567": [["Domain_0", 223, 452, "PF07714"]], "ENSACAP00000016312": [["Domain_0", 224, 499, "PF00069"]], "ENSPTRP00000019283": [["Domain_0", 223, 502, "PF07714"]], "ENSSSCP00000020665": [["Domain_0", 223, 502, "PF07714"]], "ENSOANP00000030592": [["Domain_0", 423, 469, "PF07714"]], "ENSLAFP00000024940": [["Domain_0", 309, 503, "PF00069"]], "ENSPCAP00000003280": [["Domain_0", 364, 503, "PF00069"]], "ENSCSAP00000015206": [["Domain_0", 223, 502, "PF07714"]], "ENSCJAP00000000681": [["Domain_0", 223, 502, "PF07714"]], "MGP_PahariEiJ_P0016477": [["Domain_0", 223, 502, "PF07714"]], "ENSOARP00000012709": [["Domain_0", 402, 499, "PF00069"]], "ENSOPRP00000006442": [["Domain_0", 222, 503, "PF00069"]], "ENSMMUP00000031546": [["Domain_0", 223, 488, "PF07714"]], "ENSCHIP00000016066": [["Domain_0", 223, 502, "PF07714"]], "ENSECAP00000011401": [["Domain_0", 224, 499, "PF00069"]], "ENSPVAP00000008969": [["Domain_0", 232, 476, "PF00069"]], "ENSANAP00000038485": [["Domain_0", 223, 358, "PF07714"]], "ENSEEUP00000009799": [["Domain_1", 480, 503, "PF07714"]], "ENSCPOP00000012298": [["Domain_0", 223, 502, "PF07714"]], "MGP_CAROLIEiJ_P0079612": [["Domain_0", 223, 502, "PF07714"]], "ENSMUSP00000112592": [["Domain_0", 223, 502, "PF07714"]], "ENSMNEP00000021300": [["Domain_0", 223, 502, "PF07714"]], "ENSTTRP00000007455": [["Domain_0", 222, 473, "PF07714"]], "ENSMEUP00000003310": [["Domain_0", 138, 503, "PF00069"]], "ENSPEMP00000004536": [["Domain_0", 223, 502, "PF07714"]], "ENSTBEP00000009604": [["Domain_1", 480, 503, "PF07714"]], "ENSRROP00000014300": [["Domain_0", 223, 502, "PF07714"]], "ENSPANP00000014733": [["Domain_0", 223, 502, "PF07714"]], "ENSRBIP00000012589": [["Domain_0", 223, 358, "PF07714"]], "ENSSARP00000005953": [["Domain_0", 276, 476, "PF00069"]], "ENSCCAP00000039983": [["Domain_0", 223, 502, "PF07714"]], "ENSMFAP00000017404": [["Domain_0", 223, 502, "PF07714"]], "ENSPPYP00000011408": [["Domain_0", 359, 499, "PF00069"]], "ENSODEP00000024052": [["Domain_0", 223, 502, "PF07714"]]}

