Found:
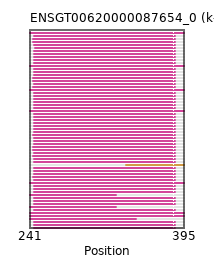
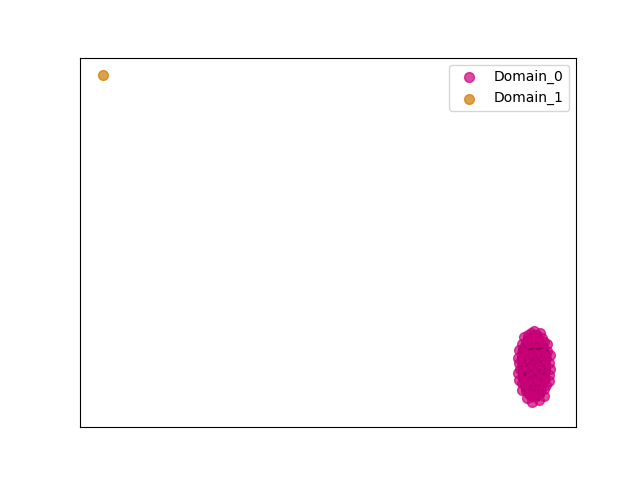
{"ENSMAUP00000015773": [["Domain_0", 245, 386, "PF01569"]], "ENSRNOP00000022625": [["Domain_0", 245, 386, "PF01569"]], "ENSMLEP00000007690": [["Domain_0", 245, 386, "PF01569"]], "ENSCAPP00000014721": [["Domain_0", 246, 386, "PF01569"]], "ENSMFAP00000027583": [["Domain_0", 245, 386, "PF01569"]], "ENSXETP00000018013": [["Domain_0", 241, 347, "PF01569"]], "ENSOCUP00000007711": [["Domain_0", 244, 386, "PF01569"]], "ENSMICP00000027315": [["Domain_0", 245, 386, "PF01569"]], "ENSP00000359204": [["Domain_0", 245, 386, "PF01569"]], "ENSFCAP00000016974": [["Domain_0", 245, 386, "PF01569"]], "ENSHGLP00000001930": [["Domain_0", 245, 386, "PF01569"]], "ENSBTAP00000025329": [["Domain_0", 244, 386, "PF01569"]], "MGP_CAROLIEiJ_P0060183": [["Domain_0", 245, 386, "PF01569"]], "ENSJJAP00000016701": [["Domain_0", 245, 386, "PF01569"]], "MGP_PahariEiJ_P0071120": [["Domain_0", 245, 386, "PF01569"]], "ENSTBEP00000010512": [["Domain_0", 241, 327, "PF01569"]], "ENSCLAP00000008004": [["Domain_0", 245, 386, "PF01569"]], "ENSMNEP00000006718": [["Domain_0", 245, 386, "PF01569"]], "ENSPANP00000011938": [["Domain_0", 245, 386, "PF01569"]], "ENSCGRP00001010930": [["Domain_0", 245, 386, "PF01569"]], "ENSANAP00000021997": [["Domain_0", 245, 386, "PF01569"]], "MGP_SPRETEiJ_P0062574": [["Domain_0", 245, 386, "PF01569"]], "ENSRROP00000018833": [["Domain_0", 245, 386, "PF01569"]], "ENSCAFP00000029711": [["Domain_0", 244, 386, "PF01569"]], "ENSCSAP00000016528": [["Domain_0", 244, 386, "PF01569"]], "ENSCCAP00000022140": [["Domain_0", 245, 386, "PF01569"]], "ENSCPOP00000005685": [["Domain_0", 245, 386, "PF01569"]], "ENSMPUP00000003227": [["Domain_0", 244, 386, "PF01569"]], "ENSOGAP00000004831": [["Domain_0", 244, 386, "PF01569"]], "ENSNLEP00000013928": [["Domain_0", 245, 386, "PF01569"]], "ENSPTRP00000001706": [["Domain_0", 245, 386, "PF01569"]], "ENSCATP00000021753": [["Domain_0", 245, 386, "PF01569"]], "ENSFDAP00000021550": [["Domain_0", 245, 386, "PF01569"]], "ENSSARP00000011580": [["Domain_0", 241, 327, "PF01569"]], "ENSOARP00000018957": [["Domain_0", 244, 386, "PF01569"]], "ENSMUSP00000052306": [["Domain_0", 245, 386, "PF01569"]], "ENSCGRP00000000038": [["Domain_0", 245, 386, "PF01569"]], "ENSSSCP00000055216": [["Domain_0", 245, 386, "PF01569"]], "ENSDORP00000027239": [["Domain_0", 245, 386, "PF01569"]], "ENSAMEP00000009637": [["Domain_0", 241, 395, "PF01569"]], "ENSPEMP00000005836": [["Domain_0", 245, 386, "PF01569"]], "ENSSBOP00000035768": [["Domain_0", 245, 386, "PF01569"]], "ENSDNOP00000034684": [["Domain_0", 244, 386, "PF01569"]], "ENSCANP00000035401": [["Domain_0", 245, 386, "PF01569"]], "ENSEEUP00000008009": [["Domain_0", 241, 395, "PF01569"]], "ENSNGAP00000016234": [["Domain_0", 245, 386, "PF01569"]], "ENSCHIP00000006143": [["Domain_0", 245, 386, "PF01569"]], "ENSPCAP00000004257": [["Domain_1", 337, 395, "PF01569"]], "ENSPPYP00000001305": [["Domain_0", 244, 386, "PF01569"]], "ENSVPAP00000000944": [["Domain_0", 241, 395, "PF01569"]], "ENSPPAP00000007057": [["Domain_0", 245, 386, "PF01569"]], "ENSPCOP00000018338": [["Domain_0", 245, 386, "PF01569"]], "ENSRBIP00000032585": [["Domain_0", 245, 386, "PF01569"]], "ENSPVAP00000014882": [["Domain_0", 241, 395, "PF01569"]], "ENSECAP00000004000": [["Domain_0", 244, 386, "PF01569"]], "ENSGGOP00000013151": [["Domain_0", 245, 386, "PF01569"]], "ENSHGLP00100007288": [["Domain_0", 245, 386, "PF01569"]], "ENSTSYP00000015118": [["Domain_0", 245, 386, "PF01569"]], "ENSLAFP00000008693": [["Domain_0", 241, 395, "PF01569"]], "ENSCJAP00000066369": [["Domain_0", 245, 386, "PF01569"]], "ENSCHOP00000001797": [["Domain_0", 241, 395, "PF01569"]], "ENSSTOP00000000702": [["Domain_0", 245, 386, "PF01569"]], "ENSMMUP00000027675": [["Domain_0", 245, 386, "PF01569"]], "ENSTTRP00000011194": [["Domain_0", 241, 395, "PF01569"]], "ENSODEP00000006361": [["Domain_0", 245, 386, "PF01569"]], "ENSMOCP00000000731": [["Domain_0", 245, 386, "PF01569"]]}