Found:
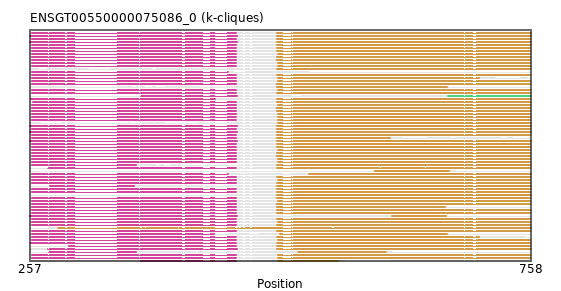
{"ENSACAP00000000550": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 758, "PF02913"]], "ENSPANP00000040480": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 757, "PF02913"]], "ENSCAFP00000039899": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 757, "PF02913"]], "ENSMEUP00000011609": [["Domain_0", 257, 464, "PF01565"], ["Linker_0_1", 464, 504], ["Domain_1", 504, 758, "PF02913"]], "ENSTSYP00000032458": [["Domain_0", 296, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 757, "PF02913"]], "ENSCANP00000037642": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 757, "PF02913"]], "ENSCAPP00000007724": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 505], ["Domain_1", 505, 757, "PF02913"]], "ENSJJAP00000004500": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 505], ["Domain_1", 505, 757, "PF02913"]], "ENSGGOP00000013937": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 757, "PF02913"]], "ENSTGUP00000006082": [["Domain_0", 258, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 757, "PF02913"]], "ENSFDAP00000011239": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 505], ["Domain_1", 505, 757, "PF02913"]], "ENSPPYP00000014198": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 672, "PF02913"]], "ENSCHIP00000002924": [["Domain_1", 504, 757, "PF02913"]], "ENSMFAP00000041430": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 757, "PF02913"]], "ENSAPLP00000013839": [["Domain_0", 257, 464, "PF01565"], ["Linker_0_1", 464, 504], ["Domain_1", 504, 758, "PF02913"]], "MGP_CAROLIEiJ_P0019277": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 505], ["Domain_1", 505, 757, "PF02913"]], "ENSCATP00000044719": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 757, "PF02913"]], "ENSRBIP00000035300": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 757, "PF02913"]], "ENSPCOP00000007114": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 757, "PF02913"]], "MGP_SPRETEiJ_P0020298": [["Domain_0", 368, 463, "PF01565"], ["Linker_0_1", 463, 505], ["Domain_1", 505, 565, "PF02913"]], "ENSMUSP00000108502": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 505], ["Domain_1", 505, 757, "PF02913"]], "ENSAMEP00000017324": [["Domain_0", 257, 464, "PF01565"], ["Linker_0_1", 464, 504], ["Domain_1", 504, 758, "PF02913"]], "ENSNLEP00000009450": [["Domain_0", 259, 363, "PF01565"], ["Domain_1", 504, 757, "PF02913"]], "ENSMLEP00000035702": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 617, "PF02913"]], "ENSRROP00000012626": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 757, "PF02913"]], "ENSCLAP00000002761": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 505], ["Domain_1", 505, 706, "PF02913"]], "MGP_PahariEiJ_P0074580": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 505], ["Domain_1", 505, 757, "PF02913"]], "ENSPVAP00000006630": [["Domain_0", 257, 464, "PF01565"], ["Domain_1", 619, 673, "PF02913"]], "ENSP00000315351": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 757, "PF02913"]], "ENSECAP00000015745": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 757, "PF02913"]], "ENSCGRP00001002344": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 505], ["Domain_1", 505, 757, "PF02913"]], "ENSMAUP00000007294": [["Domain_1", 505, 757, "PF02913"]], "ENSRNOP00000071036": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 505], ["Domain_1", 505, 757, "PF02913"]], "ENSMLUP00000018146": [["Domain_0", 257, 464, "PF01565"], ["Linker_0_1", 464, 504], ["Domain_1", 504, 758, "PF02913"]], "ENSOPRP00000013979": [["Domain_0", 277, 361, "PF01565"], ["Domain_1", 504, 758, "PF02913"]], "ENSEEUP00000011221": [["Domain_0", 368, 464, "PF01565"], ["Domain_2", 675, 758, "PF02913"]], "ENSSARP00000006720": [["Domain_1", 285, 758, "PF02913"]], "ENSMMUP00000024402": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 757, "PF02913"]], "ENSFCAP00000013253": [["Domain_0", 260, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 757, "PF02913"]], "ENSPCAP00000009828": [["Domain_1", 504, 758, "PF02913"]], "ENSMOCP00000023432": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 505], ["Domain_1", 505, 757, "PF02913"]], "ENSCHIP00000021763": [["Domain_0", 259, 455, "PF01565"]], "ENSSBOP00000009301": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 757, "PF02913"]], "ENSMGAP00000003044": [["Domain_0", 257, 464, "PF01565"], ["Linker_0_1", 464, 504], ["Domain_1", 504, 758, "PF02913"]], "ENSDORP00000004503": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 505], ["Domain_1", 505, 757, "PF02913"]], "ENSPPAP00000000182": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 757, "PF02913"]], "ENSNGAP00000026074": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 505], ["Domain_1", 505, 757, "PF02913"]], "ENSPTRP00000063874": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 757, "PF02913"]], "ENSMNEP00000006989": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 757, "PF02913"]], "ENSCCAP00000019436": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 757, "PF02913"]], "ENSOCUP00000016021": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 757, "PF02913"]], "ENSVPAP00000010114": [["Domain_0", 276, 363, "PF01565"], ["Domain_1", 525, 613, "PF02913"]], "ENSANAP00000020162": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 757, "PF02913"]], "ENSDNOP00000018708": [["Domain_1", 504, 674, "PF02913"]], "ENSCPOP00000000006": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 505], ["Domain_1", 505, 757, "PF02913"]], "ENSTTRP00000005436": [["Domain_0", 275, 464, "PF01565"], ["Linker_0_1", 464, 504], ["Domain_1", 504, 758, "PF02913"]], "ENSGALP00000052543": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 757, "PF02913"]], "ENSFALP00000003347": [["Domain_0", 258, 442, "PF01565"], ["Linker_0_1", 442, 504], ["Domain_1", 504, 757, "PF02913"]], "ENSMODP00000010716": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 757, "PF02913"]], "ENSODEP00000014643": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 505], ["Domain_1", 505, 757, "PF02913"]], "ENSSTOP00000018895": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 505], ["Domain_1", 505, 757, "PF02913"]], "ENSOARP00000018894": [["Domain_0", 259, 455, "PF01565"], ["Linker_0_1", 455, 536], ["Domain_1", 536, 758, "PF02913"]], "ENSOANP00000001326": [["Domain_0", 275, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 757, "PF02913"]], "ENSCSAP00000002059": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 757, "PF02913"]], "ENSPEMP00000024832": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 505], ["Domain_1", 505, 757, "PF02913"]], "ENSSHAP00000018979": [["Domain_0", 259, 441, "PF01565"], ["Linker_0_1", 441, 507], ["Domain_1", 507, 674, "PF02913"]], "ENSHGLP00100026019": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 505], ["Domain_1", 505, 757, "PF02913"]], "ENSMICP00000015031": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 757, "PF02913"]], "ENSPSIP00000010327": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 757, "PF02913"]], "ENSSSCP00000058694": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 706, "PF02913"]], "ENSCJAP00000020619": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 757, "PF02913"]], "ENSCGRP00000017856": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 505], ["Domain_1", 505, 757, "PF02913"]], "ENSMPUP00000000188": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 757, "PF02913"]], "ENSOANP00000032889": [["Domain_1", 602, 676, "PF02913"]], "ENSBTAP00000003690": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 758, "PF02913"]], "ENSOGAP00000020239": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 504], ["Domain_1", 504, 757, "PF02913"]], "ENSHGLP00000004264": [["Domain_0", 259, 463, "PF01565"], ["Linker_0_1", 463, 505], ["Domain_1", 505, 757, "PF02913"]]}