Found:
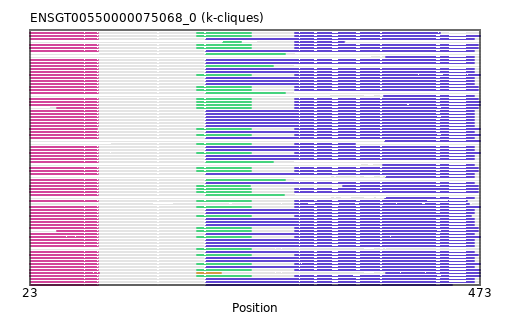
{"ENSTTRP00000003238": [["Domain_0", 23, 91, "PF08156"], ["Linker_0_2", 91, 190], ["Domain_2", 190, 244, "PF08060"], ["Linker_2_3", 244, 288], ["Domain_3", 288, 473, "PF01798"]], "ENSHGLP00100016718": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSMODP00000006385": [["Domain_0", 23, 91, "PF08156"], ["Linker_0_2", 91, 190], ["Domain_2", 190, 244, "PF08060"], ["Linker_2_3", 244, 288], ["Domain_3", 288, 471, "PF01798"]], "ENSAPLP00000008848": [["Domain_2", 216, 234, "PF08060"], ["Linker_2_3", 234, 288], ["Domain_3", 288, 337, "PF01798"]], "ENSFALP00000009336": [["Domain_0", 50, 91, "PF08156"], ["Linker_0_2", 91, 190], ["Domain_2", 190, 244, "PF08060"], ["Linker_2_3", 244, 288], ["Domain_3", 288, 471, "PF01798"]], "ENSPPAP00000008539": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSOGAP00000019309": [["Domain_3", 288, 462, "PF01798"]], "ENSACAP00000005642": [["Domain_0", 23, 91, "PF08156"], ["Linker_0_2", 91, 190], ["Domain_2", 190, 244, "PF08060"], ["Linker_2_3", 244, 288], ["Domain_3", 288, 433, "PF01798"]], "ENSCJAP00000039088": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSMMUP00000018256": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSEEUP00000001529": [["Domain_0", 23, 91, "PF08156"], ["Linker_0_2", 91, 190], ["Domain_2", 190, 244, "PF08060"], ["Linker_2_3", 244, 288], ["Domain_3", 288, 473, "PF01798"]], "ENSMICP00000015087": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSMPUP00000009042": [["Domain_0", 23, 91, "PF08156"], ["Linker_0_2", 91, 190], ["Domain_2", 190, 244, "PF08060"], ["Linker_2_3", 244, 288], ["Domain_3", 288, 471, "PF01798"]], "ENSCHIP00000027634": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSDORP00000021284": [["Domain_3", 377, 467, "PF01798"]], "ENSOGAP00000004183": [["Domain_0", 23, 91, "PF08156"], ["Linker_0_2", 91, 190], ["Domain_2", 190, 244, "PF08060"], ["Linker_2_3", 244, 288], ["Domain_3", 288, 460, "PF01798"]], "ENSTBEP00000009730": [["Domain_0", 23, 91, "PF08156"], ["Linker_0_2", 91, 190], ["Domain_2", 190, 244, "PF08060"], ["Linker_2_3", 244, 288], ["Domain_3", 288, 473, "PF01798"]], "ENSCAFP00000009947": [["Domain_0", 23, 91, "PF08156"], ["Linker_0_2", 91, 190], ["Domain_2", 190, 244, "PF08060"], ["Linker_2_3", 244, 288], ["Domain_3", 288, 471, "PF01798"]], "ENSPVAP00000016064": [["Domain_0", 23, 91, "PF08156"], ["Linker_0_2", 91, 190], ["Domain_2", 190, 244, "PF08060"], ["Linker_2_3", 244, 288], ["Domain_3", 288, 473, "PF01798"]], "ENSPCAP00000012021": [["Domain_0", 23, 91, "PF08156"], ["Linker_0_2", 91, 190], ["Domain_2", 190, 244, "PF08060"], ["Linker_2_3", 244, 288], ["Domain_3", 288, 464, "PF01798"]], "ENSMFAP00000041101": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSRNOP00000009563": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSSSCP00000021283": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSVPAP00000005727": [["Domain_0", 23, 92, "PF08156"], ["Linker_0_1", 92, 190], ["Domain_1", 190, 214, "PF08060"], ["Domain_3", 379, 473, "PF01798"]], "ENSSHAP00000020233": [["Domain_0", 23, 91, "PF08156"], ["Linker_0_2", 91, 190], ["Domain_2", 190, 244, "PF08060"], ["Linker_2_3", 244, 288], ["Domain_3", 288, 471, "PF01798"]], "ENSSARP00000009048": [["Domain_2", 190, 244, "PF08060"]], "ENSSBOP00000013714": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSOPRP00000006879": [["Domain_0", 23, 91, "PF08156"], ["Linker_0_2", 91, 190], ["Domain_2", 190, 244, "PF08060"], ["Linker_2_3", 244, 288], ["Domain_3", 288, 473, "PF01798"]], "ENSOCUP00000012855": [["Domain_0", 23, 91, "PF08156"], ["Linker_0_2", 91, 190], ["Domain_2", 190, 244, "PF08060"], ["Linker_2_3", 244, 288], ["Domain_3", 288, 471, "PF01798"]], "ENSPCOP00000024928": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSBTAP00000025042": [["Domain_0", 23, 91, "PF08156"], ["Linker_0_2", 91, 190], ["Domain_2", 190, 244, "PF08060"], ["Linker_2_3", 244, 288], ["Domain_3", 288, 471, "PF01798"]], "ENSCCAP00000012117": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSMAUP00000025176": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSCPOP00000013801": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSPPYP00000012104": [["Domain_0", 23, 91, "PF08156"], ["Linker_0_2", 91, 190], ["Domain_2", 190, 244, "PF08060"], ["Linker_2_3", 244, 288], ["Domain_3", 288, 471, "PF01798"]], "ENSMNEP00000018564": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSCAPP00000007368": [["Domain_2", 199, 278, "PF01798"]], "ENSCGRP00000018331": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_2", 91, 199], ["Domain_2", 199, 266, "PF01798"]], "ENSCAPP00000012618": [["Domain_3", 379, 467, "PF01798"]], "ENSPTRP00000022562": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSMOCP00000021717": [["Domain_3", 376, 467, "PF01798"]], "ENSTSYP00000034586": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSXETP00000061034": [["Domain_0", 23, 91, "PF08156"], ["Linker_0_2", 91, 190], ["Domain_2", 190, 244, "PF08060"], ["Linker_2_3", 244, 288], ["Domain_3", 288, 473, "PF01798"]], "MGP_SPRETEiJ_P0056686": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 445, "PF01798"]], "ENSCGRP00001020345": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSCLAP00000020165": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "MGP_CAROLIEiJ_P0054577": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSMLEP00000004605": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSGALP00000048153": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSCANP00000030713": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSPSIP00000007373": [["Domain_0", 50, 91, "PF08156"], ["Linker_0_2", 91, 190], ["Domain_2", 190, 244, "PF08060"], ["Linker_2_3", 244, 288], ["Domain_3", 288, 471, "PF01798"]], "ENSSTOP00000011210": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSFDAP00000021835": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSNGAP00000021254": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_2", 91, 199], ["Domain_2", 199, 278, "PF01798"]], "ENSOARP00000006924": [["Domain_0", 23, 91, "PF08156"], ["Linker_0_2", 91, 190], ["Domain_2", 190, 244, "PF08060"], ["Linker_2_3", 244, 288], ["Domain_3", 288, 471, "PF01798"]], "ENSMGAP00000004512": [["Domain_3", 378, 473, "PF01798"]], "ENSLAFP00000001178": [["Domain_0", 23, 90, "PF08156"], ["Linker_0_2", 90, 190], ["Domain_2", 190, 244, "PF08060"], ["Linker_2_3", 244, 288], ["Domain_3", 288, 473, "PF01798"]], "ENSNLEP00000011184": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSDORP00000013082": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_2", 91, 199], ["Domain_2", 199, 278, "PF01798"]], "ENSCHOP00000001055": [["Domain_0", 23, 91, "PF08156"], ["Linker_0_2", 91, 190], ["Domain_2", 190, 244, "PF08060"], ["Linker_2_3", 244, 288], ["Domain_3", 288, 473, "PF01798"]], "ENSPANP00000016952": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSFCAP00000013404": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSODEP00000007660": [["Domain_3", 378, 467, "PF01798"]], "ENSECAP00000018952": [["Domain_0", 23, 91, "PF08156"], ["Linker_0_2", 91, 190], ["Domain_2", 190, 244, "PF08060"], ["Linker_2_3", 244, 288], ["Domain_3", 288, 471, "PF01798"]], "ENSGGOP00000002836": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSDNOP00000020005": [["Domain_0", 23, 91, "PF08156"], ["Linker_0_2", 91, 190], ["Domain_2", 190, 244, "PF08060"], ["Linker_2_3", 244, 288], ["Domain_3", 288, 471, "PF01798"]], "ENSHGLP00000022425": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSMOCP00000004868": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_2", 91, 199], ["Domain_2", 199, 266, "PF01798"]], "ENSMUSP00000099487": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSRROP00000004848": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSP00000370589": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSAMEP00000011901": [["Domain_0", 23, 91, "PF08156"], ["Linker_0_2", 91, 190], ["Domain_2", 190, 244, "PF08060"], ["Linker_2_3", 244, 288], ["Domain_3", 288, 473, "PF01798"]], "MGP_PahariEiJ_P0065556": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSNGAP00000020270": [["Domain_3", 379, 467, "PF01798"]], "ENSODEP00000007373": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_2", 91, 199], ["Domain_2", 199, 277, "PF01798"]], "ENSCATP00000014256": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSANAP00000031050": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSMGAP00000007622": [["Domain_2", 190, 244, "PF08060"], ["Linker_2_3", 244, 288], ["Domain_3", 288, 348, "PF01798"]], "ENSCSAP00000007129": [["Domain_0", 23, 91, "PF08156"], ["Linker_0_2", 91, 190], ["Domain_2", 190, 244, "PF08060"], ["Linker_2_3", 244, 288], ["Domain_3", 288, 471, "PF01798"]], "ENSMEUP00000002013": [["Domain_0", 23, 91, "PF08156"], ["Linker_0_2", 91, 190], ["Domain_2", 190, 244, "PF08060"], ["Linker_2_3", 244, 288], ["Domain_3", 288, 473, "PF01798"]], "ENSRBIP00000013371": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSPEMP00000007338": [["Domain_0", 24, 91, "PF08156"], ["Linker_0_3", 91, 199], ["Domain_3", 199, 467, "PF01798"]], "ENSOANP00000000804": [["Domain_0", 23, 91, "PF08156"], ["Linker_0_2", 91, 190], ["Domain_2", 190, 243, "PF08060"], ["Linker_2_3", 243, 336], ["Domain_3", 336, 471, "PF01798"]], "ENSETEP00000012933": [["Domain_0", 23, 91, "PF08156"], ["Linker_0_2", 91, 190], ["Domain_2", 190, 244, "PF08060"], ["Linker_2_3", 244, 288], ["Domain_3", 288, 473, "PF01798"]], "ENSMLUP00000021297": [["Domain_0", 23, 91, "PF08156"], ["Linker_0_2", 91, 190], ["Domain_2", 190, 244, "PF08060"], ["Linker_2_3", 244, 288], ["Domain_3", 288, 473, "PF01798"]]}