Found:
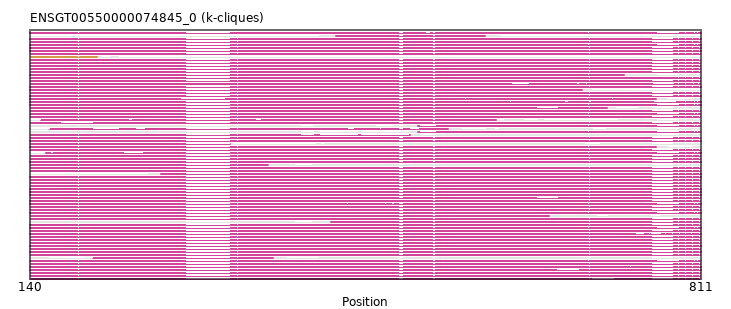
{"ENSGALP00000016108": [["Domain_0", 141, 810, "PF00995"]], "ENSAMEP00000005408": [["Domain_0", 140, 811, "PF00995"]], "ENSVPAP00000010860": [["Domain_0", 140, 811, "PF00995"]], "ENSCANP00000027939": [["Domain_0", 141, 810, "PF00995"]], "ENSSTOP00000008023": [["Domain_0", 203, 383, "PF00995"]], "ENSMAUP00000003319": [["Domain_0", 141, 340, "PF00995"]], "ENSMICP00000037480": [["Domain_0", 141, 810, "PF00995"]], "ENSLAFP00000005129": [["Domain_0", 140, 811, "PF00995"]], "ENSOANP00000022110": [["Domain_0", 140, 811, "PF00995"]], "ENSMEUP00000005738": [["Domain_0", 140, 766, "PF00995"]], "ENSNGAP00000015138": [["Domain_0", 141, 810, "PF00995"]], "ENSRNOP00000039132": [["Domain_0", 141, 810, "PF00995"]], "ENSGGOP00000010735": [["Domain_0", 141, 810, "PF00995"]], "ENSACAP00000014540": [["Domain_0", 140, 811, "PF00995"]], "ENSPTRP00000075506": [["Domain_0", 141, 810, "PF00995"]], "ENSHGLP00100024585": [["Domain_0", 141, 810, "PF00995"]], "ENSTGUP00000012189": [["Domain_0", 140, 810, "PF00995"]], "ENSOPRP00000002516": [["Domain_0", 140, 811, "PF00995"]], "MGP_PahariEiJ_P0082354": [["Domain_0", 141, 810, "PF00995"]], "ENSFALP00000006053": [["Domain_0", 140, 811, "PF00995"]], "ENSPCOP00000026396": [["Domain_0", 141, 659, "PF00995"]], "ENSHGLP00000012366": [["Domain_0", 160, 558, "PF00995"]], "ENSCGRP00000013300": [["Domain_0", 141, 810, "PF00995"]], "ENSOGAP00000015311": [["Domain_0", 140, 810, "PF00995"]], "ENSDNOP00000032135": [["Domain_0", 140, 692, "PF00995"]], "ENSSHAP00000017417": [["Domain_0", 140, 811, "PF00995"]], "ENSRROP00000018084": [["Domain_0", 141, 810, "PF00995"]], "ENSCHIP00000029070": [["Domain_0", 141, 810, "PF00995"]], "ENSMODP00000021910": [["Domain_0", 271, 811, "PF00995"]], "MGP_SPRETEiJ_P0033727": [["Domain_0", 141, 810, "PF00995"]], "ENSTTRP00000008654": [["Domain_0", 140, 811, "PF00995"]], "ENSMLUP00000000504": [["Domain_0", 140, 811, "PF00995"]], "ENSCAFP00000018746": [["Domain_0", 140, 811, "PF00995"]], "ENSCATP00000026326": [["Domain_0", 141, 810, "PF00995"]], "ENSTSYP00000017686": [["Domain_0", 141, 810, "PF00995"]], "ENSECAP00000003971": [["Domain_0", 140, 811, "PF00995"]], "ENSPCAP00000005388": [["Domain_0", 140, 811, "PF00995"]], "ENSMNEP00000023189": [["Domain_0", 141, 810, "PF00995"]], "ENSOCUP00000006775": [["Domain_0", 140, 811, "PF00995"]], "MGP_CAROLIEiJ_P0032239": [["Domain_0", 141, 810, "PF00995"]], "ENSCCAP00000029242": [["Domain_0", 141, 810, "PF00995"]], "ENSP00000390783": [["Domain_0", 141, 810, "PF00995"]], "ENSNLEP00000019191": [["Domain_0", 141, 810, "PF00995"]], "ENSFDAP00000010847": [["Domain_0", 141, 810, "PF00995"]], "ENSMFAP00000043153": [["Domain_0", 141, 810, "PF00995"]], "ENSPPYP00000006502": [["Domain_0", 140, 810, "PF00995"]], "ENSHGLP00000003117": [["Domain_0", 151, 606, "PF00995"]], "ENSACAP00000015607": [["Domain_0", 446, 595, "PF00995"]], "ENSMMUP00000025874": [["Domain_0", 141, 378, "PF00995"]], "ENSCPOP00000006514": [["Domain_0", 141, 810, "PF00995"]], "ENSHGLP00100020674": [["Domain_0", 141, 520, "PF00995"]], "ENSANAP00000016037": [["Domain_0", 141, 810, "PF00995"]], "ENSPVAP00000011217": [["Domain_0", 140, 811, "PF00995"]], "ENSPPAP00000039762": [["Domain_0", 441, 810, "PF00995"]], "ENSBTAP00000043412": [["Domain_0", 140, 811, "PF00995"]], "ENSHGLP00100013798": [["Domain_0", 528, 810, "PF00995"]], "ENSSARP00000013096": [["Domain_0", 140, 811, "PF00995"]], "ENSRBIP00000023769": [["Domain_0", 141, 810, "PF00995"]], "ENSFCAP00000022249": [["Domain_0", 141, 717, "PF00995"]], "ENSMLEP00000032515": [["Domain_0", 141, 810, "PF00995"]], "ENSCGRP00001011991": [["Domain_0", 141, 810, "PF00995"]], "ENSAPLP00000015410": [["Domain_0", 140, 811, "PF00995"]], "ENSCLAP00000017896": [["Domain_0", 141, 810, "PF00995"]], "ENSPANP00000017745": [["Domain_0", 141, 810, "PF00995"]], "ENSCJAP00000011215": [["Domain_0", 141, 810, "PF00995"]], "ENSODEP00000006194": [["Domain_0", 141, 810, "PF00995"]], "ENSMPUP00000005335": [["Domain_0", 140, 811, "PF00995"]], "ENSCSAP00000011837": [["Domain_0", 140, 811, "PF00995"]], "ENSSSCP00000037872": [["Domain_0", 141, 810, "PF00995"]], "ENSMOCP00000019173": [["Domain_0", 141, 810, "PF00995"]], "ENSETEP00000003239": [["Domain_0", 140, 811, "PF00995"]], "ENSSBOP00000030250": [["Domain_0", 141, 810, "PF00995"]], "ENSPSIP00000012442": [["Domain_0", 140, 811, "PF00995"]], "ENSOARP00000005739": [["Domain_0", 140, 811, "PF00995"]], "ENSCAPP00000000230": [["Domain_1", 141, 207, "PF00995"]], "ENSHGLP00000003651": [["Domain_0", 141, 810, "PF00995"]], "ENSHGLP00000004431": [["Domain_0", 528, 810, "PF00995"]], "ENSEEUP00000012419": [["Domain_0", 140, 766, "PF00995"]], "ENSPEMP00000013224": [["Domain_0", 141, 810, "PF00995"]], "ENSMGAP00000013045": [["Domain_0", 156, 811, "PF00995"]], "ENSCHOP00000001871": [["Domain_0", 140, 734, "PF00995"]], "ENSMUSP00000021335": [["Domain_0", 141, 810, "PF00995"]], "ENSDORP00000012608": [["Domain_0", 141, 810, "PF00995"]]}