Found:
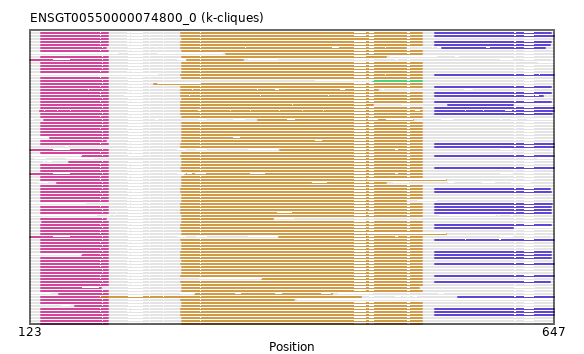
{"ENSCGRP00000009171": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSPANP00000029769": [["Domain_0", 123, 201, "PF02874"], ["Domain_1", 372, 515, "PF00006"]], "ENSOPRP00000001601": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 274], ["Domain_1", 274, 515, "PF00006"], ["Linker_1_3", 515, 528], ["Domain_3", 528, 606, "PF00306"]], "ENSHGLP00100014948": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSCAPP00000004649": [["Domain_1", 319, 515, "PF00006"]], "ENSCPOP00000016071": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 247], ["Domain_1", 247, 515, "PF00006"]], "ENSFCAP00000020709": [["Domain_0", 137, 201, "PF02874"], ["Domain_1", 351, 506, "PF00006"]], "ENSLAFP00000018666": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 274], ["Domain_1", 274, 515, "PF00006"], ["Linker_1_3", 515, 528], ["Domain_3", 528, 647, "PF00306"]], "ENSSSCP00000028820": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSCAFP00000033854": [["Domain_0", 134, 199, "PF02874"], ["Linker_0_1", 199, 274], ["Domain_1", 274, 499, "PF00006"], ["Linker_1_3", 499, 535], ["Domain_3", 535, 638, "PF00306"]], "ENSMODP00000024772": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 274], ["Domain_1", 274, 515, "PF00006"], ["Linker_1_3", 515, 528], ["Domain_3", 528, 643, "PF00306"]], "ENSGGOP00000011311": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSDNOP00000022596": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 274], ["Domain_1", 274, 515, "PF00006"], ["Linker_1_3", 515, 528], ["Domain_3", 528, 636, "PF00306"]], "ENSMLUP00000014760": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 274], ["Domain_1", 274, 515, "PF00006"], ["Linker_1_3", 515, 528], ["Domain_3", 528, 647, "PF00306"]], "ENSFCAP00000011162": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "MGP_PahariEiJ_P0087565": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSPTRP00000008686": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSCCAP00000016748": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSRNOP00000003965": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSODEP00000009826": [["Domain_0", 134, 199, "PF02874"], ["Linker_0_1", 199, 275], ["Domain_1", 275, 366, "PF00006"]], "ENSCSAP00000013290": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 274], ["Domain_1", 274, 471, "PF00006"]], "ENSMPUP00000001493": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 274], ["Domain_1", 274, 515, "PF00006"], ["Linker_1_3", 515, 528], ["Domain_3", 528, 644, "PF00306"]], "ENSOCUP00000005273": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 274], ["Domain_1", 274, 515, "PF00006"], ["Linker_1_3", 515, 528], ["Domain_3", 528, 644, "PF00306"]], "ENSPEMP00000014079": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSSSCP00000045621": [["Domain_0", 134, 194, "PF02874"], ["Linker_0_1", 194, 275], ["Domain_1", 275, 512, "PF00006"]], "ENSMOCP00000005096": [["Domain_0", 150, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 479, "PF00006"]], "ENSTTRP00000002011": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 274], ["Domain_1", 274, 515, "PF00006"], ["Linker_1_3", 515, 528], ["Domain_3", 528, 647, "PF00306"]], "ENSCJAP00000027524": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSCANP00000019980": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSCGRP00001017033": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSETEP00000008289": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 274], ["Domain_1", 274, 515, "PF00006"], ["Linker_1_3", 515, 528], ["Domain_3", 528, 647, "PF00306"]], "ENSTGUP00000015953": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 274], ["Domain_1", 274, 387, "PF00006"]], "ENSAMEP00000004389": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 274], ["Domain_1", 274, 515, "PF00006"], ["Linker_1_3", 515, 528], ["Domain_3", 528, 647, "PF00306"]], "ENSFALP00000000453": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 274], ["Domain_1", 274, 515, "PF00006"], ["Linker_1_3", 515, 528], ["Domain_3", 528, 645, "PF00306"]], "ENSOARP00000008911": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 274], ["Domain_1", 274, 515, "PF00006"], ["Linker_1_3", 515, 528], ["Domain_3", 528, 644, "PF00306"]], "ENSGALP00000056479": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSECAP00000020278": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 274], ["Domain_1", 274, 515, "PF00006"], ["Linker_1_3", 515, 528], ["Domain_3", 528, 644, "PF00306"]], "ENSMEUP00000002629": [["Domain_1", 274, 515, "PF00006"], ["Linker_1_3", 515, 528], ["Domain_3", 528, 647, "PF00306"]], "ENSODEP00000007791": [["Domain_1", 275, 515, "PF00006"]], "ENSMGAP00000000549": [["Domain_0", 175, 201, "PF02874"], ["Linker_0_1", 201, 274], ["Domain_1", 274, 515, "PF00006"], ["Linker_1_3", 515, 528], ["Domain_3", 528, 647, "PF00306"]], "ENSP00000262030": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSPSIP00000003495": [["Domain_0", 175, 201, "PF02874"], ["Linker_0_1", 201, 274], ["Domain_1", 274, 515, "PF00006"], ["Linker_1_3", 515, 528], ["Domain_3", 528, 644, "PF00306"]], "ENSMMUP00000001377": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSVPAP00000003740": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 274], ["Domain_1", 274, 515, "PF00006"], ["Linker_1_3", 515, 528], ["Domain_3", 528, 647, "PF00306"]], "MGP_CAROLIEiJ_P0025093": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSOANP00000005132": [["Domain_1", 274, 515, "PF00006"], ["Linker_1_3", 515, 528], ["Domain_3", 528, 645, "PF00306"]], "ENSPCAP00000009968": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 274], ["Domain_1", 274, 515, "PF00006"], ["Linker_1_3", 515, 528], ["Domain_3", 528, 647, "PF00306"]], "ENSNGAP00000021992": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSCAFP00000000202": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 274], ["Domain_1", 274, 515, "PF00006"], ["Linker_1_3", 515, 528], ["Domain_3", 528, 643, "PF00306"]], "ENSSHAP00000003002": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 274], ["Domain_1", 274, 515, "PF00006"], ["Linker_1_3", 515, 528], ["Domain_3", 528, 643, "PF00306"]], "ENSMMUP00000012433": [["Domain_0", 123, 201, "PF02874"], ["Linker_0_1", 201, 278], ["Domain_1", 278, 515, "PF00006"]], "ENSEEUP00000002149": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 274], ["Domain_1", 274, 466, "PF00006"], ["Linker_1_3", 466, 541], ["Domain_3", 541, 606, "PF00306"]], "ENSFDAP00000014471": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSETEP00000015281": [["Domain_0", 133, 201, "PF02874"], ["Linker_0_1", 201, 274], ["Domain_1", 274, 515, "PF00006"], ["Linker_1_3", 515, 528], ["Domain_3", 528, 647, "PF00306"]], "ENSNLEP00000026054": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 499, "PF00006"]], "ENSSTOP00000012435": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSMICP00000043808": [["Domain_0", 162, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSMOCP00000006064": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSSBOP00000038965": [["Domain_0", 134, 201, "PF02874"], ["Domain_1", 355, 515, "PF00006"]], "ENSCHIP00000007152": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSSTOP00000019713": [["Domain_0", 152, 201, "PF02874"], ["Linker_0_1", 201, 288], ["Domain_1", 288, 425, "PF00006"]], "ENSOGAP00000003156": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 274], ["Domain_1", 274, 515, "PF00006"], ["Linker_1_3", 515, 528], ["Domain_3", 528, 644, "PF00306"]], "ENSMUSP00000026459": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSRROP00000025118": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSMNEP00000020632": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 539, "PF00006"]], "ENSOCUP00000024969": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 274], ["Domain_1", 274, 515, "PF00006"], ["Linker_1_3", 515, 528], ["Domain_3", 528, 644, "PF00306"]], "ENSMFAP00000035499": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSCATP00000027918": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 479, "PF00006"]], "ENSDORP00000008682": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSXETP00000063929": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 274], ["Domain_1", 274, 515, "PF00006"], ["Linker_1_3", 515, 528], ["Domain_3", 528, 647, "PF00306"]], "ENSCHOP00000004746": [["Domain_1", 274, 515, "PF00006"], ["Linker_1_3", 515, 528], ["Domain_3", 528, 647, "PF00306"]], "ENSPVAP00000007200": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 274], ["Domain_1", 274, 515, "PF00006"], ["Linker_1_3", 515, 528], ["Domain_3", 528, 647, "PF00306"]], "ENSODEP00000026847": [["Domain_0", 135, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSTSYP00000023801": [["Domain_0", 168, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSJJAP00000011897": [["Domain_0", 134, 199, "PF02874"], ["Linker_0_1", 199, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSBTAP00000017710": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 274], ["Domain_1", 274, 515, "PF00006"], ["Linker_1_3", 515, 528], ["Domain_3", 528, 644, "PF00306"]], "MGP_SPRETEiJ_P0026379": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSSARP00000008566": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 274], ["Domain_1", 274, 515, "PF00006"], ["Linker_1_3", 515, 528], ["Domain_3", 528, 647, "PF00306"]], "ENSCSAP00000001350": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 274], ["Domain_1", 274, 515, "PF00006"], ["Linker_1_3", 515, 528], ["Domain_3", 528, 644, "PF00306"]], "ENSACAP00000016489": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 274], ["Domain_1", 274, 515, "PF00006"], ["Linker_1_3", 515, 528], ["Domain_3", 528, 644, "PF00306"]], "ENSPPAP00000028635": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSPANP00000004994": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 539, "PF00006"]], "ENSMMUP00000035315": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSHGLP00000008347": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSRBIP00000012778": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSDNOP00000008750": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 274], ["Domain_1", 274, 515, "PF00006"], ["Linker_1_3", 515, 528], ["Domain_3", 528, 644, "PF00306"]], "ENSMLEP00000020721": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSTSYP00000009143": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSJJAP00000005891": [["Domain_0", 143, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSPPYP00000005318": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 274], ["Domain_1", 274, 515, "PF00006"], ["Linker_1_3", 515, 528], ["Domain_3", 528, 644, "PF00306"]], "ENSPCOP00000010115": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSANAP00000030171": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 275], ["Domain_1", 275, 515, "PF00006"]], "ENSCLAP00000016921": [["Domain_0", 134, 201, "PF02874"], ["Domain_2", 466, 515, "PF00006"]], "ENSCATP00000031545": [["Domain_0", 123, 201, "PF02874"], ["Linker_0_1", 201, 280], ["Domain_1", 280, 336, "PF00006"]], "ENSMICP00000040907": [["Domain_1", 275, 515, "PF00006"]], "ENSTBEP00000002148": [["Domain_0", 134, 201, "PF02874"], ["Domain_1", 193, 454, "PF00006"], ["Domain_3", 551, 647, "PF00306"]], "ENSPSIP00000007185": [["Domain_0", 134, 201, "PF02874"], ["Linker_0_1", 201, 274], ["Domain_1", 274, 515, "PF00006"], ["Linker_1_3", 515, 528], ["Domain_3", 528, 644, "PF00306"]], "ENSMFAP00000022686": [["Domain_0", 123, 201, "PF02874"], ["Domain_1", 373, 515, "PF00006"]]}