Found:
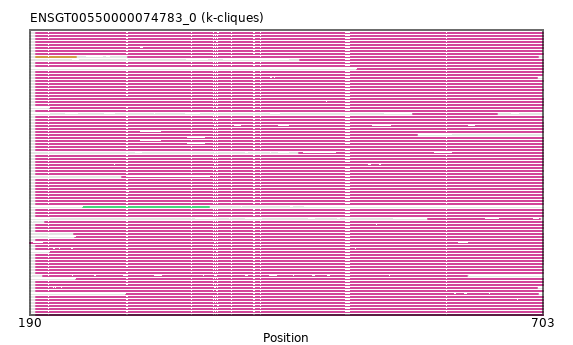
{"ENSHGLP00100002521": [["Domain_0", 196, 703, "PF00118"]], "ENSHGLP00000009843": [["Domain_0", 196, 703, "PF00118"]], "ENSHGLP00100015403": [["Domain_0", 196, 703, "PF00118"]], "ENSAMEP00000008610": [["Domain_0", 196, 703, "PF00118"]], "ENSBTAP00000049380": [["Domain_0", 196, 703, "PF00118"]], "ENSGGOP00000036928": [["Domain_0", 196, 703, "PF00118"]], "ENSMMUP00000044579": [["Domain_0", 196, 703, "PF00118"]], "ENSPSIP00000018899": [["Domain_0", 210, 703, "PF00118"]], "ENSCAFP00000036154": [["Domain_0", 196, 702, "PF00118"]], "ENSCATP00000031533": [["Domain_0", 196, 703, "PF00118"]], "ENSDNOP00000029189": [["Domain_0", 196, 702, "PF00118"]], "ENSHGLP00000005698": [["Domain_0", 196, 703, "PF00118"]], "ENSHGLP00100000103": [["Domain_0", 196, 703, "PF00118"]], "ENSCHIP00000018445": [["Domain_0", 196, 702, "PF00118"]], "ENSCGRP00000020780": [["Domain_0", 517, 703, "PF00118"]], "ENSP00000286788": [["Domain_0", 196, 703, "PF00118"]], "MGP_PahariEiJ_P0029495": [["Domain_0", 196, 703, "PF00118"]], "ENSHGLP00100026726": [["Domain_0", 196, 703, "PF00118"]], "ENSSTOP00000024099": [["Domain_0", 196, 703, "PF00118"]], "ENSGGOP00000006209": [["Domain_0", 196, 703, "PF00118"]], "ENSJJAP00000001862": [["Domain_0", 459, 703, "PF00118"]], "MGP_CAROLIEiJ_P0042137": [["Domain_0", 196, 703, "PF00118"]], "ENSRNOP00000002169": [["Domain_0", 196, 703, "PF00118"]], "ENSCANP00000013312": [["Domain_0", 196, 703, "PF00118"]], "ENSSHAP00000004777": [["Domain_0", 203, 627, "PF00118"]], "ENSCJAP00000060195": [["Domain_0", 196, 703, "PF00118"]], "ENSVPAP00000011543": [["Domain_0", 196, 703, "PF00118"]], "ENSCSAP00000005424": [["Domain_0", 196, 703, "PF00118"]], "ENSPPYP00000012641": [["Domain_0", 196, 703, "PF00118"]], "ENSMUSP00000026704": [["Domain_0", 196, 703, "PF00118"]], "ENSMNEP00000018042": [["Domain_0", 196, 702, "PF00118"]], "ENSCAPP00000014847": [["Domain_1", 196, 236, "PF00118"], ["Linker_1_0", 236, 288], ["Domain_0", 288, 698, "PF00118"]], "ENSMLUP00000000334": [["Domain_0", 196, 703, "PF00118"]], "ENSCATP00000024568": [["Domain_0", 460, 703, "PF00118"]], "ENSPVAP00000003385": [["Domain_0", 196, 703, "PF00118"]], "MGP_SPRETEiJ_P0044033": [["Domain_0", 196, 703, "PF00118"]], "ENSHGLP00000014087": [["Domain_0", 196, 577, "PF00118"]], "ENSHGLP00000019361": [["Domain_0", 196, 703, "PF00118"]], "ENSCHOP00000002459": [["Domain_0", 196, 697, "PF00118"]], "ENSTGUP00000013990": [["Domain_0", 196, 703, "PF00118"]], "ENSSSCP00000021676": [["Domain_0", 196, 703, "PF00118"]], "ENSXETP00000043707": [["Domain_0", 196, 703, "PF00118"]], "ENSOARP00000015560": [["Domain_0", 196, 703, "PF00118"]], "ENSPEMP00000028330": [["Domain_0", 190, 703, "PF00118"]], "ENSOANP00000031259": [["Domain_2", 243, 369, "PF00118"]], "ENSFCAP00000026497": [["Domain_0", 196, 702, "PF00118"]], "ENSTSYP00000030778": [["Domain_0", 196, 703, "PF00118"]], "ENSODEP00000003800": [["Domain_0", 588, 700, "PF00118"]], "ENSFDAP00000018228": [["Domain_0", 196, 703, "PF00118"]], "ENSMGAP00000015356": [["Domain_0", 196, 703, "PF00118"]], "ENSTGUP00000016447": [["Domain_0", 286, 698, "PF00118"]], "ENSMODP00000026257": [["Domain_0", 196, 703, "PF00118"]], "ENSMOCP00000003885": [["Domain_0", 196, 703, "PF00118"]], "ENSETEP00000008817": [["Domain_0", 196, 703, "PF00118"]], "ENSJJAP00000024903": [["Domain_0", 196, 703, "PF00118"]], "ENSPPAP00000006734": [["Domain_0", 196, 703, "PF00118"]], "ENSPCOP00000025804": [["Domain_0", 235, 703, "PF00118"]], "ENSLAFP00000001930": [["Domain_0", 196, 703, "PF00118"]], "ENSNGAP00000016653": [["Domain_0", 196, 703, "PF00118"]], "ENSNLEP00000041939": [["Domain_0", 196, 703, "PF00118"]], "ENSCCAP00000024216": [["Domain_0", 196, 703, "PF00118"]], "ENSCPOP00000032203": [["Domain_0", 196, 702, "PF00118"]], "ENSGALP00000058407": [["Domain_0", 196, 702, "PF00118"]], "ENSFALP00000006596": [["Domain_0", 210, 702, "PF00118"]], "ENSSHAP00000012971": [["Domain_0", 236, 703, "PF00118"]], "ENSPANP00000028436": [["Domain_0", 196, 702, "PF00118"]], "ENSECAP00000009680": [["Domain_0", 196, 703, "PF00118"]], "ENSMEUP00000015214": [["Domain_0", 196, 703, "PF00118"]], "ENSSBOP00000029971": [["Domain_0", 196, 703, "PF00118"]], "ENSTBEP00000006887": [["Domain_0", 196, 697, "PF00118"]], "ENSAPLP00000004553": [["Domain_0", 196, 703, "PF00118"]], "ENSCLAP00000006238": [["Domain_0", 196, 702, "PF00118"]], "ENSMFAP00000021489": [["Domain_0", 196, 702, "PF00118"]], "ENSPTRP00000092452": [["Domain_0", 196, 703, "PF00118"]], "ENSMPUP00000001036": [["Domain_0", 196, 702, "PF00118"]], "ENSMAUP00000019260": [["Domain_0", 196, 703, "PF00118"]], "ENSOPRP00000015720": [["Domain_0", 196, 703, "PF00118"]], "ENSRBIP00000025585": [["Domain_0", 196, 703, "PF00118"]], "ENSSARP00000007612": [["Domain_0", 196, 703, "PF00118"]], "ENSODEP00000002478": [["Domain_0", 196, 703, "PF00118"]], "ENSANAP00000004521": [["Domain_0", 196, 703, "PF00118"]], "ENSMICP00000024314": [["Domain_0", 196, 702, "PF00118"]], "ENSMLEP00000023919": [["Domain_0", 282, 703, "PF00118"]], "ENSBTAP00000018917": [["Domain_0", 196, 703, "PF00118"]], "ENSOGAP00000004175": [["Domain_0", 205, 703, "PF00118"]], "ENSPEMP00000026841": [["Domain_0", 196, 703, "PF00118"]], "ENSTTRP00000006361": [["Domain_0", 196, 703, "PF00118"]], "ENSDORP00000002864": [["Domain_0", 196, 702, "PF00118"]], "ENSOCUP00000002283": [["Domain_0", 196, 703, "PF00118"]], "ENSCGRP00001001545": [["Domain_0", 196, 703, "PF00118"]], "ENSRROP00000013276": [["Domain_0", 196, 702, "PF00118"]], "ENSEEUP00000010426": [["Domain_0", 196, 703, "PF00118"]], "ENSACAP00000001856": [["Domain_0", 196, 703, "PF00118"]], "ENSPCAP00000014243": [["Domain_0", 234, 703, "PF00118"]], "ENSFDAP00000014014": [["Domain_0", 573, 657, "PF00118"]]}