Found:
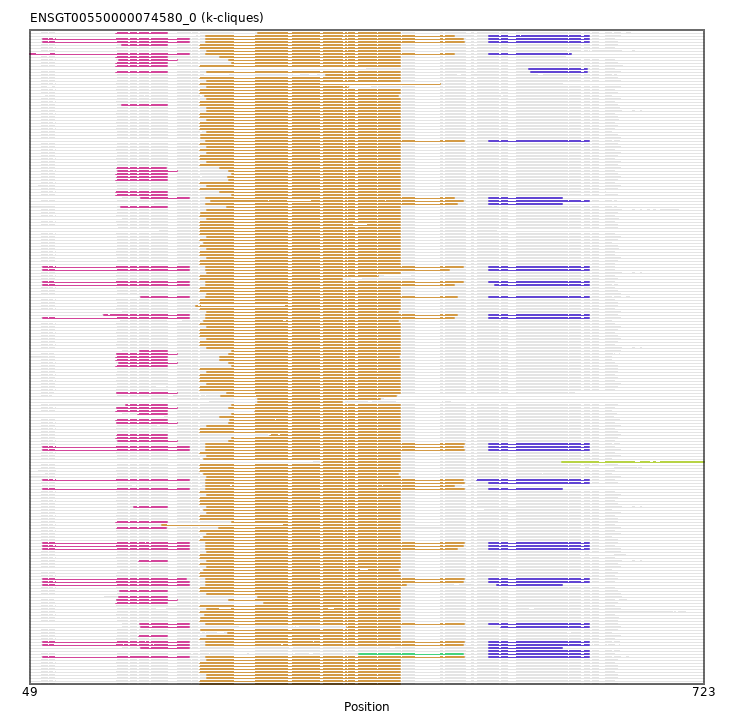
{"ENSP00000415178": [["Domain_1", 341, 419, "PF00650"], ["Linker_1_4", 419, 581], ["Domain_4", 581, 723, "PF10558"]], "ENSTSYP00000018627": [["Domain_0", 158, 186, "PF03765"], ["Linker_0_1", 186, 226], ["Domain_1", 226, 419, "PF00650"]], "MGP_PahariEiJ_P0030549": [["Domain_1", 219, 419, "PF00650"]], "ENSTBEP00000013979": [["Domain_0", 159, 208, "PF03765"], ["Linker_0_1", 208, 225], ["Domain_1", 225, 483, "PF00650"], ["Linker_1_3", 483, 508], ["Domain_3", 508, 608, "PF01105"]], "ENSETEP00000014777": [["Domain_1", 225, 476, "PF00650"], ["Linker_1_3", 476, 508], ["Domain_3", 508, 581, "PF01105"]], "ENSMFAP00000037928": [["Domain_1", 226, 419, "PF00650"]], "ENSOANP00000031344": [["Domain_1", 277, 361, "PF00650"]], "ENSPPAP00000026835": [["Domain_1", 226, 419, "PF00650"]], "ENSPANP00000045362": [["Domain_1", 226, 419, "PF00650"]], "ENSMLUP00000008657": [["Domain_0", 62, 208, "PF03765"], ["Linker_0_1", 208, 225], ["Domain_1", 225, 473, "PF00650"], ["Linker_1_3", 473, 508], ["Domain_3", 508, 608, "PF01105"]], "ENSFCAP00000011074": [["Domain_1", 220, 419, "PF00650"]], "ENSACAP00000009475": [["Domain_0", 135, 186, "PF03765"], ["Linker_0_1", 186, 277], ["Domain_1", 277, 419, "PF00650"]], "ENSMUSP00000019512": [["Domain_1", 226, 419, "PF00650"]], "ENSMODP00000011629": [["Domain_0", 159, 186, "PF03765"], ["Linker_0_1", 186, 251], ["Domain_1", 251, 419, "PF00650"]], "ENSCATP00000043718": [["Domain_1", 226, 419, "PF00650"]], "ENSMFAP00000036162": [["Domain_1", 219, 419, "PF00650"]], "ENSPANP00000036825": [["Domain_1", 223, 417, "PF00650"]], "ENSTSYP00000022253": [["Domain_1", 219, 419, "PF00650"]], "ENSCAPP00000005174": [["Domain_1", 226, 367, "PF00650"]], "ENSODEP00000003066": [["Domain_1", 219, 419, "PF00650"]], "ENSCJAP00000048834": [["Domain_1", 219, 419, "PF00650"]], "ENSMPUP00000012984": [["Domain_0", 135, 186, "PF03765"], ["Linker_0_1", 186, 251], ["Domain_1", 251, 419, "PF00650"]], "ENSTSYP00000018142": [["Domain_1", 247, 419, "PF00650"]], "ENSSSCP00000038806": [["Domain_1", 226, 418, "PF00650"]], "ENSMPUP00000012867": [["Domain_0", 138, 196, "PF03765"], ["Linker_0_1", 196, 248], ["Domain_1", 248, 419, "PF00650"]], "ENSMPUP00000012862": [["Domain_0", 135, 186, "PF03765"], ["Linker_0_1", 186, 239], ["Domain_1", 239, 419, "PF00650"]], "MGP_SPRETEiJ_P0026715": [["Domain_1", 219, 419, "PF00650"]], "ENSOPRP00000015242": [["Domain_0", 62, 208, "PF03765"], ["Linker_0_1", 208, 225], ["Domain_1", 225, 482, "PF00650"], ["Linker_1_3", 482, 508], ["Domain_3", 508, 608, "PF01105"]], "ENSVPAP00000000839": [["Domain_0", 160, 208, "PF03765"], ["Domain_3", 508, 581, "PF01105"]], "MGP_SPRETEiJ_P0026712": [["Domain_1", 226, 419, "PF00650"]], "ENSSSCP00000033603": [["Domain_1", 219, 419, "PF00650"]], "ENSDORP00000003434": [["Domain_1", 226, 419, "PF00650"]], "ENSPCAP00000012055": [["Domain_1", 225, 473, "PF00650"]], "ENSBTAP00000019231": [["Domain_0", 138, 196, "PF03765"], ["Linker_0_1", 196, 248], ["Domain_1", 248, 419, "PF00650"]], "ENSDORP00000003433": [["Domain_1", 219, 419, "PF00650"]], "ENSCGRP00000009385": [["Domain_1", 226, 419, "PF00650"]], "ENSCPOP00000011499": [["Domain_1", 226, 419, "PF00650"]], "ENSCAFP00000018795": [["Domain_0", 135, 186, "PF03765"], ["Linker_0_1", 186, 226], ["Domain_1", 226, 419, "PF00650"], ["Linker_1_3", 419, 550], ["Domain_3", 550, 606, "PF13897"]], "ENSP00000478755": [["Domain_1", 219, 419, "PF00650"]], "ENSOARP00000008171": [["Domain_0", 136, 196, "PF03765"], ["Linker_0_1", 196, 248], ["Domain_1", 248, 419, "PF00650"]], "ENSMLUP00000001976": [["Domain_0", 123, 208, "PF03765"], ["Linker_0_1", 208, 225], ["Domain_1", 225, 476, "PF00650"], ["Linker_1_3", 476, 508], ["Domain_3", 508, 608, "PF01105"]], "ENSMICP00000030131": [["Domain_1", 219, 419, "PF00650"]], "ENSTTRP00000001451": [["Domain_0", 62, 208, "PF03765"], ["Linker_0_1", 208, 225], ["Domain_1", 225, 483, "PF00650"], ["Linker_1_3", 483, 508], ["Domain_3", 508, 608, "PF01105"]], "ENSNGAP00000005593": [["Domain_1", 219, 419, "PF00650"]], "ENSMODP00000011810": [["Domain_0", 136, 196, "PF03765"], ["Linker_0_1", 196, 248], ["Domain_1", 248, 419, "PF00650"]], "ENSCGRP00001025165": [["Domain_1", 219, 419, "PF00650"]], "ENSCLAP00000016949": [["Domain_1", 219, 419, "PF00650"]], "ENSMOCP00000011049": [["Domain_1", 219, 419, "PF00650"]], "MGP_SPRETEiJ_P0026722": [["Domain_1", 219, 419, "PF00650"]], "ENSPVAP00000016719": [["Domain_0", 62, 208, "PF03765"], ["Linker_0_1", 208, 225], ["Domain_1", 225, 483, "PF00650"], ["Linker_1_3", 483, 508], ["Domain_3", 508, 608, "PF01105"]], "ENSCATP00000037579": [["Domain_1", 224, 417, "PF00650"]], "ENSMUSP00000065084": [["Domain_1", 219, 419, "PF00650"]], "ENSFCAP00000024232": [["Domain_1", 219, 419, "PF00650"]], "ENSSTOP00000012481": [["Domain_1", 220, 419, "PF00650"]], "ENSCGRP00001009665": [["Domain_1", 226, 419, "PF00650"]], "ENSMEUP00000000317": [["Domain_0", 62, 208, "PF03765"], ["Linker_0_1", 208, 225], ["Domain_1", 225, 482, "PF00650"], ["Linker_1_3", 482, 508], ["Domain_3", 508, 608, "PF01105"]], "ENSMNEP00000036801": [["Domain_1", 226, 419, "PF00650"]], "ENSMFAP00000024111": [["Domain_1", 219, 419, "PF00650"]], "ENSFDAP00000017956": [["Domain_1", 219, 419, "PF00650"]], "ENSHGLP00000022089": [["Domain_1", 219, 419, "PF00650"]], "ENSMAUP00000002141": [["Domain_1", 226, 419, "PF00650"]], "ENSCGRP00001012255": [["Domain_1", 219, 419, "PF00650"]], "ENSCHIP00000027753": [["Domain_1", 220, 419, "PF00650"]], "ENSPCOP00000017600": [["Domain_1", 219, 419, "PF00650"]], "ENSPEMP00000009527": [["Domain_0", 153, 186, "PF03765"], ["Linker_0_1", 186, 226], ["Domain_1", 226, 419, "PF00650"]], "ENSSHAP00000000075": [["Domain_0", 138, 186, "PF03765"], ["Linker_0_1", 186, 277], ["Domain_1", 277, 419, "PF00650"]], "MGP_PahariEiJ_P0030556": [["Domain_1", 219, 419, "PF00650"]], "ENSGGOP00000004181": [["Domain_1", 219, 419, "PF00650"]], "ENSRNOP00000006053": [["Domain_0", 158, 186, "PF03765"], ["Linker_0_1", 186, 226], ["Domain_1", 226, 419, "PF00650"]], "ENSRROP00000037212": [["Domain_1", 219, 419, "PF00650"]], "ENSCHIP00000001738": [["Domain_1", 219, 419, "PF00650"]], "ENSPPAP00000042752": [["Domain_1", 219, 419, "PF00650"]], "ENSSHAP00000002162": [["Domain_0", 136, 196, "PF03765"], ["Linker_0_1", 196, 248], ["Domain_1", 248, 419, "PF00650"]], "ENSP00000255858": [["Domain_1", 226, 419, "PF00650"]], "ENSRBIP00000007362": [["Domain_1", 219, 419, "PF00650"]], "ENSPCAP00000006718": [["Domain_1", 225, 483, "PF00650"], ["Linker_1_3", 483, 508], ["Domain_3", 508, 608, "PF01105"]], "ENSMAUP00000019249": [["Domain_1", 219, 361, "PF00650"]], "ENSCCAP00000037619": [["Domain_1", 226, 419, "PF00650"]], "ENSDNOP00000016707": [["Domain_0", 136, 186, "PF03765"], ["Linker_0_1", 186, 247], ["Domain_1", 247, 419, "PF00650"]], "ENSAMEP00000007975": [["Domain_0", 62, 208, "PF03765"], ["Linker_0_1", 208, 225], ["Domain_1", 225, 482, "PF00650"], ["Linker_1_3", 482, 508], ["Domain_3", 508, 608, "PF01105"]], "ENSPCOP00000016540": [["Domain_1", 226, 419, "PF00650"]], "ENSANAP00000008065": [["Domain_0", 141, 186, "PF03765"], ["Linker_0_1", 186, 219], ["Domain_1", 219, 419, "PF00650"]], "ENSSTOP00000012473": [["Domain_1", 224, 419, "PF00650"]], "ENSANAP00000034365": [["Domain_1", 226, 419, "PF00650"]], "ENSECAP00000017411": [["Domain_0", 135, 186, "PF03765"], ["Linker_0_1", 186, 239], ["Domain_1", 239, 419, "PF00650"]], "ENSAPLP00000014471": [["Domain_0", 49, 208, "PF03765"], ["Linker_0_1", 208, 225], ["Domain_1", 225, 473, "PF00650"], ["Linker_1_3", 473, 508], ["Domain_3", 508, 590, "PF01105"]], "ENSAMEP00000007872": [["Domain_1", 225, 473, "PF00650"], ["Linker_1_3", 473, 508], ["Domain_3", 508, 608, "PF01105"]], "ENSRNOP00000006542": [["Domain_1", 219, 419, "PF00650"]], "ENSPEMP00000009048": [["Domain_1", 219, 419, "PF00650"]], "ENSOCUP00000012230": [["Domain_0", 136, 196, "PF03765"], ["Linker_0_1", 196, 248], ["Domain_1", 248, 419, "PF00650"]], "ENSSBOP00000005745": [["Domain_1", 219, 419, "PF00650"]], "ENSCSAP00000005693": [["Domain_0", 135, 186, "PF03765"], ["Linker_0_1", 186, 251], ["Domain_1", 251, 419, "PF00650"]], "ENSOGAP00000013208": [["Domain_0", 137, 196, "PF03765"], ["Linker_0_1", 196, 248], ["Domain_1", 248, 419, "PF00650"]], "MGP_CAROLIEiJ_P0025397": [["Domain_1", 219, 419, "PF00650"]], "ENSHGLP00000024731": [["Domain_1", 226, 419, "PF00650"]], "ENSVPAP00000004515": [["Domain_2", 378, 482, "PF00650"], ["Linker_2_3", 482, 508], ["Domain_3", 508, 608, "PF01105"]], "ENSFDAP00000016111": [["Domain_1", 219, 419, "PF00650"]], "ENSPTRP00000056683": [["Domain_1", 219, 419, "PF00650"]], "ENSCCAP00000008473": [["Domain_1", 219, 419, "PF00650"]], "ENSGGOP00000004193": [["Domain_1", 226, 419, "PF00650"]], "ENSMMUP00000007654": [["Domain_1", 223, 417, "PF00650"]], "ENSPEMP00000003810": [["Domain_1", 219, 419, "PF00650"]], "ENSMLEP00000034814": [["Domain_1", 226, 417, "PF00650"]], "ENSFDAP00000021693": [["Domain_1", 219, 419, "PF00650"]], "ENSOANP00000023299": [["Domain_0", 136, 196, "PF03765"], ["Linker_0_1", 196, 246], ["Domain_1", 246, 419, "PF00650"]], "ENSCAFP00000018721": [["Domain_0", 135, 186, "PF03765"], ["Linker_0_1", 186, 219], ["Domain_1", 219, 419, "PF00650"]], "ENSMLEP00000026390": [["Domain_1", 215, 419, "PF00650"]], "ENSOARP00000007802": [["Domain_0", 145, 186, "PF03765"], ["Linker_0_1", 186, 251], ["Domain_1", 251, 419, "PF00650"]], "ENSBTAP00000051657": [["Domain_0", 135, 186, "PF03765"], ["Linker_0_1", 186, 251], ["Domain_1", 251, 419, "PF00650"]], "ENSDORP00000007911": [["Domain_1", 219, 419, "PF00650"]], "ENSNGAP00000006378": [["Domain_1", 226, 419, "PF00650"]], "ENSFDAP00000006183": [["Domain_1", 226, 419, "PF00650"]], "ENSPPYP00000013065": [["Domain_1", 181, 419, "PF00650"]], "ENSPPYP00000013064": [["Domain_0", 135, 186, "PF03765"], ["Linker_0_1", 186, 251], ["Domain_1", 251, 419, "PF00650"]], "ENSCHOP00000001704": [["Domain_1", 225, 483, "PF00650"], ["Linker_1_3", 483, 508], ["Domain_3", 508, 608, "PF01105"]], "ENSPANP00000013127": [["Domain_1", 219, 419, "PF00650"]], "ENSECAP00000017690": [["Domain_0", 135, 186, "PF03765"], ["Linker_0_1", 186, 251], ["Domain_1", 251, 419, "PF00650"]], "ENSCHIP00000012417": [["Domain_1", 220, 419, "PF00650"]], "ENSMEUP00000003030": [["Domain_0", 62, 208, "PF03765"], ["Linker_0_1", 208, 225], ["Domain_1", 225, 473, "PF00650"], ["Linker_1_3", 473, 514], ["Domain_3", 514, 608, "PF01105"]], "ENSAMEP00000008121": [["Domain_0", 62, 208, "PF03765"], ["Linker_0_1", 208, 225], ["Domain_1", 225, 483, "PF00650"], ["Linker_1_3", 483, 508], ["Domain_3", 508, 608, "PF01105"]], "ENSHGLP00100019883": [["Domain_1", 219, 419, "PF00650"]], "ENSCJAP00000019384": [["Domain_1", 219, 419, "PF00650"]], "ENSMNEP00000009185": [["Domain_1", 219, 419, "PF00650"]], "ENSPPAP00000039352": [["Domain_1", 219, 419, "PF00650"]], "ENSMICP00000021725": [["Domain_1", 219, 419, "PF00650"]], "ENSTSYP00000000999": [["Domain_1", 220, 419, "PF00650"]], "ENSPTRP00000024534": [["Domain_1", 219, 419, "PF00650"]], "ENSPTRP00000024535": [["Domain_1", 226, 419, "PF00650"]], "ENSFALP00000007218": [["Domain_0", 140, 186, "PF03765"], ["Linker_0_1", 186, 246], ["Domain_1", 246, 419, "PF00650"]], "ENSRROP00000029714": [["Domain_1", 219, 419, "PF00650"]], "ENSOCUP00000008314": [["Domain_0", 135, 186, "PF03765"], ["Linker_0_1", 186, 249], ["Domain_1", 249, 419, "PF00650"]], "ENSDNOP00000028749": [["Domain_0", 135, 186, "PF03765"], ["Linker_0_1", 186, 248], ["Domain_1", 248, 419, "PF00650"]], "ENSVPAP00000001235": [["Domain_3", 508, 608, "PF01105"]], "ENSOCUP00000000673": [["Domain_1", 239, 419, "PF00650"]], "ENSCATP00000013467": [["Domain_1", 219, 419, "PF00650"]], "ENSSARP00000011309": [["Domain_0", 62, 208, "PF03765"], ["Linker_0_1", 208, 225], ["Domain_1", 225, 425, "PF00650"], ["Linker_1_3", 425, 516], ["Domain_3", 516, 581, "PF01105"]], "ENSBTAP00000002561": [["Domain_0", 135, 186, "PF03765"], ["Linker_0_1", 186, 239], ["Domain_1", 239, 419, "PF00650"]], "ENSETEP00000005191": [["Domain_0", 160, 208, "PF03765"], ["Linker_0_1", 208, 225], ["Domain_1", 225, 473, "PF00650"], ["Linker_1_3", 473, 508], ["Domain_3", 508, 581, "PF01105"]], "ENSODEP00000002587": [["Domain_1", 226, 419, "PF00650"]], "ENSRBIP00000003489": [["Domain_1", 226, 419, "PF00650"]], "ENSPANP00000013135": [["Domain_1", 219, 419, "PF00650"]], "ENSCJAP00000019542": [["Domain_1", 226, 419, "PF00650"]], "ENSSARP00000011292": [["Domain_0", 62, 208, "PF03765"], ["Linker_0_1", 208, 225], ["Domain_1", 225, 482, "PF00650"], ["Linker_1_3", 482, 508], ["Domain_3", 508, 608, "PF01105"]], "ENSSHAP00000005975": [["Domain_0", 135, 186, "PF03765"], ["Linker_0_1", 186, 283], ["Domain_1", 283, 419, "PF00650"]], "ENSSSCP00000047139": [["Domain_1", 219, 419, "PF00650"]], "ENSMOCP00000010549": [["Domain_1", 219, 419, "PF00650"]], "ENSRROP00000001525": [["Domain_1", 226, 419, "PF00650"]], "ENSCCAP00000008955": [["Domain_0", 141, 186, "PF03765"], ["Linker_0_1", 186, 219], ["Domain_1", 219, 419, "PF00650"]], "ENSCGRP00000009749": [["Domain_1", 219, 419, "PF00650"]], "ENSPVAP00000016723": [["Domain_0", 62, 208, "PF03765"], ["Linker_0_1", 208, 225], ["Domain_1", 225, 476, "PF00650"], ["Linker_1_3", 476, 508], ["Domain_3", 508, 608, "PF01105"]], "ENSCGRP00000015175": [["Domain_1", 219, 419, "PF00650"]], "ENSCAPP00000002381": [["Domain_1", 219, 459, "PF00650"]], "ENSTBEP00000014836": [["Domain_0", 160, 208, "PF03765"], ["Domain_1", 366, 419, "PF00650"], ["Linker_1_3", 419, 520], ["Domain_3", 520, 608, "PF01105"]], "ENSCSAP00000005475": [["Domain_0", 136, 185, "PF03765"], ["Linker_0_1", 185, 239], ["Domain_1", 239, 419, "PF00650"]], "ENSMMUP00000055549": [["Domain_1", 219, 419, "PF00650"]], "ENSCANP00000027706": [["Domain_1", 226, 419, "PF00650"]], "ENSMUSP00000003681": [["Domain_1", 219, 419, "PF00650"]], "ENSTTRP00000001453": [["Domain_0", 62, 208, "PF03765"], ["Linker_0_1", 208, 225], ["Domain_1", 225, 482, "PF00650"], ["Linker_1_3", 482, 508], ["Domain_3", 508, 608, "PF01105"]], "ENSANAP00000010770": [["Domain_1", 219, 419, "PF00650"]], "ENSCAFP00000018793": [["Domain_3", 548, 606, "PF13897"]], "ENSGGOP00000012366": [["Domain_1", 219, 419, "PF00650"]], "ENSGGOP00000038187": [["Domain_1", 223, 419, "PF00650"]], "ENSHGLP00100020250": [["Domain_1", 226, 419, "PF00650"]], "ENSMNEP00000016759": [["Domain_1", 219, 419, "PF00650"]], "ENSETEP00000006650": [["Domain_1", 230, 482, "PF00650"], ["Linker_1_3", 482, 508], ["Domain_3", 508, 608, "PF01105"]], "MGP_PahariEiJ_P0030562": [["Domain_1", 226, 419, "PF00650"]], "ENSPCAP00000014092": [["Domain_0", 62, 208, "PF03765"], ["Linker_0_1", 208, 225], ["Domain_1", 225, 483, "PF00650"], ["Linker_1_3", 483, 508], ["Domain_3", 508, 581, "PF01105"]], "ENSCLAP00000017913": [["Domain_1", 219, 419, "PF00650"]], "ENSCANP00000017421": [["Domain_1", 219, 419, "PF00650"]], "ENSOGAP00000011529": [["Domain_0", 136, 186, "PF03765"], ["Linker_0_1", 186, 289], ["Domain_1", 289, 419, "PF00650"]], "ENSHGLP00100023058": [["Domain_1", 219, 419, "PF00650"]], "ENSJJAP00000004868": [["Domain_1", 219, 419, "PF00650"]], "ENSCSAP00000005567": [["Domain_0", 137, 196, "PF03765"], ["Linker_0_1", 196, 248], ["Domain_1", 248, 419, "PF00650"]], "ENSSBOP00000021287": [["Domain_0", 139, 186, "PF03765"], ["Linker_0_1", 186, 219], ["Domain_1", 219, 419, "PF00650"]], "ENSNGAP00000006256": [["Domain_1", 219, 419, "PF00650"]], "ENSMOCP00000017542": [["Domain_1", 226, 419, "PF00650"]], "ENSMMUP00000025455": [["Domain_1", 219, 419, "PF00650"]], "MGP_CAROLIEiJ_P0025387": [["Domain_1", 226, 419, "PF00650"]], "ENSPCAP00000005153": [["Domain_0", 62, 208, "PF03765"], ["Linker_0_1", 208, 225], ["Domain_1", 225, 482, "PF00650"], ["Linker_1_3", 482, 496], ["Domain_3", 496, 608, "PF01105"]], "ENSP00000385695": [["Domain_1", 223, 419, "PF00650"]], "ENSFCAP00000008017": [["Domain_1", 226, 419, "PF00650"]], "ENSMGAP00000015268": [["Domain_0", 160, 208, "PF03765"], ["Linker_0_1", 208, 225], ["Domain_1", 225, 476, "PF00650"], ["Linker_1_3", 476, 508], ["Domain_3", 508, 608, "PF01105"]], "ENSCPOP00000013148": [["Domain_1", 219, 419, "PF00650"]], "ENSRBIP00000018380": [["Domain_1", 219, 419, "PF00650"]], "ENSLAFP00000024694": [["Domain_0", 62, 208, "PF03765"], ["Linker_0_1", 208, 225], ["Domain_1", 225, 468, "PF00650"], ["Linker_1_3", 468, 508], ["Domain_3", 508, 608, "PF01105"]], "ENSCANP00000003187": [["Domain_1", 345, 419, "PF00650"]], "ENSOCUP00000000399": [["Domain_0", 157, 186, "PF03765"], ["Linker_0_1", 186, 251], ["Domain_1", 251, 419, "PF00650"]], "ENSPTRP00000055691": [["Domain_1", 223, 419, "PF00650"]], "ENSNLEP00000004656": [["Domain_1", 219, 419, "PF00650"]], "ENSCATP00000035033": [["Domain_1", 219, 419, "PF00650"]], "ENSODEP00000003001": [["Domain_1", 219, 419, "PF00650"]], "ENSOGAP00000011606": [["Domain_0", 135, 186, "PF03765"], ["Linker_0_1", 186, 251], ["Domain_1", 251, 419, "PF00650"]], "ENSP00000215812": [["Domain_1", 219, 419, "PF00650"]], "ENSSARP00000000988": [["Domain_0", 62, 205, "PF03765"], ["Linker_0_1", 205, 225], ["Domain_1", 225, 483, "PF00650"], ["Linker_1_3", 483, 508], ["Domain_3", 508, 608, "PF01105"]], "ENSOPRP00000015269": [["Domain_0", 62, 208, "PF03765"], ["Linker_0_1", 208, 225], ["Domain_1", 225, 483, "PF00650"], ["Linker_1_3", 483, 508], ["Domain_3", 508, 608, "PF01105"]], "ENSSTOP00000019020": [["Domain_1", 219, 419, "PF00650"]], "ENSPPAP00000007251": [["Domain_1", 223, 419, "PF00650"]], "ENSPCOP00000002693": [["Domain_1", 219, 419, "PF00650"]], "ENSMLEP00000030159": [["Domain_1", 219, 419, "PF00650"]], "ENSCLAP00000014603": [["Domain_1", 226, 419, "PF00650"]], "ENSCHIP00000002286": [["Domain_1", 226, 419, "PF00650"]], "ENSOANP00000025029": [["Domain_1", 240, 415, "PF00650"]], "ENSOARP00000008498": [["Domain_0", 135, 186, "PF03765"], ["Linker_0_1", 186, 274], ["Domain_1", 274, 419, "PF00650"]], "ENSPPYP00000023838": [["Domain_0", 136, 185, "PF03765"], ["Linker_0_1", 185, 238], ["Domain_1", 238, 419, "PF00650"]], "ENSNLEP00000004711": [["Domain_1", 219, 419, "PF00650"]], "MGP_CAROLIEiJ_P0025390": [["Domain_1", 219, 419, "PF00650"]], "ENSMAUP00000003751": [["Domain_1", 219, 397, "PF00650"]], "ENSRROP00000020037": [["Domain_1", 223, 417, "PF00650"]], "ENSMMUP00000048635": [["Domain_1", 226, 419, "PF00650"]], "ENSSBOP00000032956": [["Domain_1", 226, 419, "PF00650"]], "ENSMODP00000011824": [["Domain_0", 135, 186, "PF03765"], ["Linker_0_1", 186, 239], ["Domain_1", 239, 418, "PF00650"]], "ENSOANP00000028450": [["Domain_1", 277, 399, "PF00650"]], "ENSGALP00000048837": [["Domain_1", 219, 419, "PF00650"]], "ENSHGLP00000026076": [["Domain_1", 219, 419, "PF00650"]], "ENSLAFP00000004319": [["Domain_0", 62, 208, "PF03765"], ["Linker_0_1", 208, 225], ["Domain_1", 225, 482, "PF00650"], ["Linker_1_3", 482, 508], ["Domain_3", 508, 608, "PF01105"]], "ENSOPRP00000015226": [["Domain_1", 225, 483, "PF00650"], ["Linker_1_3", 483, 508], ["Domain_3", 508, 608, "PF01105"]], "ENSXETP00000035215": [["Domain_0", 62, 208, "PF03765"], ["Linker_0_1", 208, 225], ["Domain_1", 225, 483, "PF00650"], ["Linker_1_3", 483, 508], ["Domain_3", 508, 608, "PF01105"]], "ENSPVAP00000016722": [["Domain_0", 62, 208, "PF03765"], ["Linker_0_1", 208, 225], ["Domain_1", 225, 482, "PF00650"], ["Linker_1_3", 482, 508], ["Domain_3", 508, 608, "PF01105"]]}