Found:
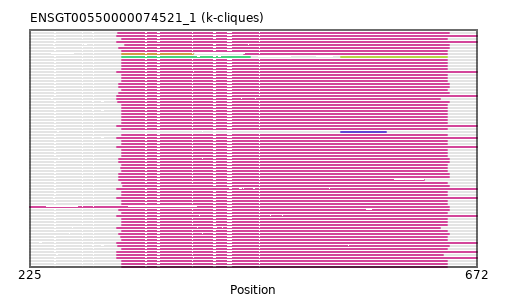
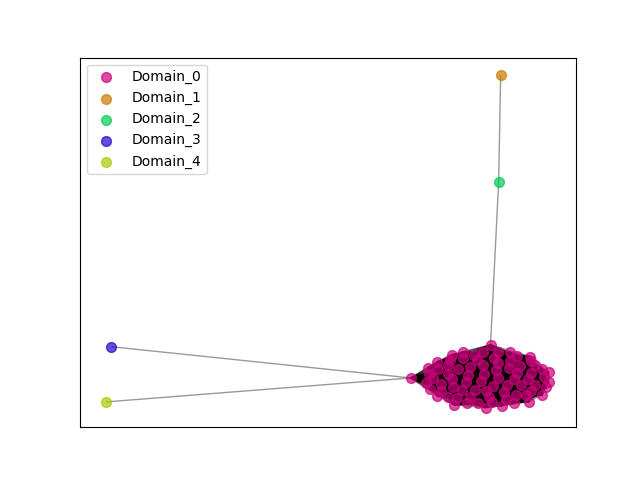
{"ENSCAPP00000006302": [["Domain_1", 317, 388, "PF04515"], ["Linker_1_0", 388, 439], ["Domain_0", 439, 642, "PF04515"]], "ENSCPOP00000022152": [["Domain_0", 317, 642, "PF04515"]], "ENSMLUP00000014044": [["Domain_0", 312, 672, "PF04515"]], "ENSCHOP00000012265": [["Domain_0", 312, 672, "PF04515"]], "ENSETEP00000005155": [["Domain_0", 312, 635, "PF04515"]], "ENSSSCP00000028533": [["Domain_0", 316, 642, "PF04515"]], "ENSCHIP00000005456": [["Domain_0", 317, 642, "PF04515"]], "ENSMEUP00000004312": [["Domain_3", 536, 581, "PF04515"]], "ENSSTOP00000011097": [["Domain_0", 317, 642, "PF04515"]], "ENSRBIP00000026110": [["Domain_0", 317, 642, "PF04515"]], "ENSCATP00000002029": [["Domain_2", 317, 445, "PF04515"], ["Linker_2_4", 445, 536], ["Domain_4", 536, 642, "PF04515"]], "ENSTSYP00000020559": [["Domain_0", 317, 642, "PF04515"]], "ENSPANP00000042891": [["Domain_0", 317, 642, "PF04515"]], "ENSCCAP00000039627": [["Domain_0", 317, 642, "PF04515"]], "ENSCSAP00000007522": [["Domain_0", 314, 644, "PF04515"]], "MGP_CAROLIEiJ_P0062403": [["Domain_0", 317, 642, "PF04515"]], "ENSMGAP00000007852": [["Domain_0", 312, 672, "PF04515"]], "ENSMOCP00000010004": [["Domain_0", 317, 642, "PF04515"]], "MGP_PahariEiJ_P0077539": [["Domain_0", 317, 642, "PF04515"]], "ENSOPRP00000009525": [["Domain_0", 312, 672, "PF04515"]], "ENSSBOP00000004612": [["Domain_0", 317, 642, "PF04515"]], "ENSDORP00000018016": [["Domain_0", 317, 642, "PF04515"]], "ENSOANP00000006665": [["Domain_0", 314, 644, "PF04515"]], "ENSGGOP00000038010": [["Domain_0", 317, 642, "PF04515"]], "ENSPTRP00000069665": [["Domain_0", 317, 642, "PF04515"]], "ENSCJAP00000067583": [["Domain_0", 317, 642, "PF04515"]], "ENSAPLP00000013559": [["Domain_0", 312, 672, "PF04515"]], "ENSCGRP00000004090": [["Domain_0", 317, 642, "PF04515"]], "ENSPPAP00000012185": [["Domain_0", 317, 642, "PF04515"]], "ENSCAFP00000004081": [["Domain_0", 314, 644, "PF04515"]], "ENSFCAP00000038138": [["Domain_0", 317, 642, "PF04515"]], "ENSFDAP00000019406": [["Domain_0", 317, 642, "PF04515"]], "ENSNGAP00000011282": [["Domain_0", 316, 642, "PF04515"]], "ENSPSIP00000011566": [["Domain_0", 314, 644, "PF04515"]], "ENSMMUP00000051775": [["Domain_0", 317, 642, "PF04515"]], "ENSP00000363852": [["Domain_0", 317, 642, "PF04515"]], "ENSODEP00000006112": [["Domain_0", 317, 642, "PF04515"]], "ENSMFAP00000026103": [["Domain_0", 317, 642, "PF04515"]], "ENSMPUP00000007089": [["Domain_0", 314, 644, "PF04515"]], "ENSSARP00000002785": [["Domain_0", 312, 635, "PF04515"]], "ENSCLAP00000021946": [["Domain_0", 317, 642, "PF04515"]], "ENSJJAP00000008666": [["Domain_0", 317, 642, "PF04515"]], "ENSTTRP00000003265": [["Domain_0", 312, 672, "PF04515"]], "ENSEEUP00000000847": [["Domain_0", 312, 672, "PF04515"]], "ENSMICP00000037936": [["Domain_0", 317, 642, "PF04515"]], "ENSOGAP00000004946": [["Domain_0", 318, 644, "PF04515"]], "ENSBTAP00000009435": [["Domain_0", 320, 644, "PF04515"]], "ENSLAFP00000000155": [["Domain_0", 312, 672, "PF04515"]], "ENSPCAP00000003993": [["Domain_0", 312, 672, "PF04515"]], "ENSTBEP00000014506": [["Domain_0", 312, 672, "PF04515"]], "ENSOARP00000007829": [["Domain_0", 314, 644, "PF04515"]], "ENSOCUP00000005149": [["Domain_0", 314, 619, "PF04515"]], "ENSRROP00000038588": [["Domain_0", 317, 642, "PF04515"]], "ENSPPYP00000021811": [["Domain_0", 225, 644, "PF04515"]], "ENSGALP00000024856": [["Domain_0", 317, 642, "PF04515"]], "ENSAMEP00000015966": [["Domain_0", 312, 672, "PF04515"]], "ENSXETP00000002007": [["Domain_0", 312, 672, "PF04515"]], "ENSSHAP00000014009": [["Domain_0", 314, 644, "PF04515"]], "ENSMUSP00000099975": [["Domain_0", 317, 642, "PF04515"]], "ENSECAP00000002912": [["Domain_0", 314, 644, "PF04515"]], "ENSMLEP00000022743": [["Domain_0", 317, 642, "PF04515"]], "ENSFALP00000005098": [["Domain_0", 313, 644, "PF04515"]], "ENSANAP00000019470": [["Domain_0", 317, 642, "PF04515"]], "ENSMNEP00000042284": [["Domain_0", 317, 642, "PF04515"]], "ENSCANP00000029044": [["Domain_0", 317, 642, "PF04515"]], "ENSPEMP00000007716": [["Domain_0", 317, 642, "PF04515"]], "ENSNLEP00000043519": [["Domain_0", 317, 642, "PF04515"]], "ENSCGRP00001025445": [["Domain_0", 317, 642, "PF04515"]], "ENSMODP00000020345": [["Domain_0", 314, 644, "PF04515"]], "ENSVPAP00000009081": [["Domain_0", 312, 638, "PF04515"]], "ENSHGLP00100001685": [["Domain_0", 317, 642, "PF04515"]], "ENSDNOP00000030094": [["Domain_0", 314, 644, "PF04515"]], "ENSRNOP00000071535": [["Domain_0", 317, 642, "PF04515"]], "ENSTGUP00000000348": [["Domain_0", 313, 644, "PF04515"]], "ENSPVAP00000016113": [["Domain_0", 312, 672, "PF04515"]], "ENSHGLP00000007746": [["Domain_0", 317, 642, "PF04515"]], "MGP_SPRETEiJ_P0064870": [["Domain_0", 317, 642, "PF04515"]], "ENSACAP00000008303": [["Domain_0", 313, 644, "PF04515"]], "ENSMAUP00000013992": [["Domain_0", 317, 642, "PF04515"]]}