Found:
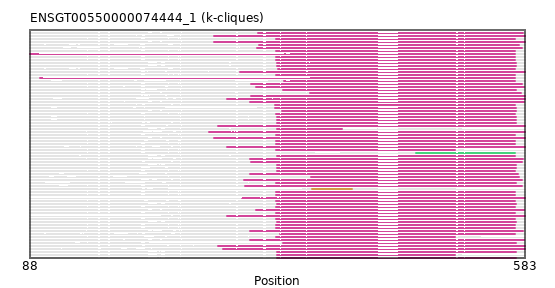
{"ENSMLEP00000018936": [["Domain_0", 334, 573, "PF00777"]], "ENSRROP00000032583": [["Domain_0", 334, 573, "PF00777"]], "ENSRBIP00000023586": [["Domain_0", 334, 573, "PF00777"]], "ENSCAPP00000000925": [["Domain_0", 88, 573, "PF00777"]], "ENSCGRP00001014189": [["Domain_0", 335, 574, "PF00777"]], "ENSPTRP00000040768": [["Domain_0", 334, 573, "PF00777"]], "ENSAPLP00000008049": [["Domain_0", 272, 583, "PF00777"]], "MGP_SPRETEiJ_P0047168": [["Domain_0", 335, 574, "PF00777"]], "ENSGGOP00000040821": [["Domain_0", 334, 573, "PF00777"]], "ENSTSYP00000006363": [["Domain_0", 341, 574, "PF00777"]], "ENSSBOP00000011029": [["Domain_0", 334, 573, "PF00777"]], "ENSANAP00000014802": [["Domain_0", 334, 573, "PF00777"]], "ENSMEUP00000007228": [["Domain_0", 267, 583, "PF00777"]], "ENSMNEP00000026540": [["Domain_2", 474, 573, "PF00777"]], "ENSCANP00000013028": [["Domain_0", 334, 573, "PF00777"]], "ENSJJAP00000007019": [["Domain_0", 335, 574, "PF00777"]], "ENSCHIP00000011655": [["Domain_0", 336, 573, "PF00777"]], "ENSPANP00000007584": [["Domain_0", 334, 573, "PF00777"]], "ENSCHOP00000007763": [["Domain_0", 298, 583, "PF00777"]], "ENSMAUP00000022245": [["Domain_0", 335, 400, "PF00777"]], "ENSCSAP00000013100": [["Domain_0", 309, 580, "PF00777"]], "ENSNGAP00000005725": [["Domain_0", 335, 574, "PF00777"]], "ENSSTOP00000015426": [["Domain_0", 334, 513, "PF00777"]], "ENSMUSP00000025000": [["Domain_0", 335, 574, "PF00777"]], "ENSMGAP00000015716": [["Domain_0", 272, 583, "PF00777"]], "MGP_PahariEiJ_P0044783": [["Domain_0", 335, 513, "PF00777"]], "ENSTGUP00000010013": [["Domain_0", 308, 582, "PF00777"]], "ENSHGLP00000025076": [["Domain_0", 335, 574, "PF00777"]], "ENSCCAP00000001952": [["Domain_0", 334, 573, "PF00777"]], "ENSCAFP00000003057": [["Domain_0", 315, 580, "PF00777"]], "ENSFDAP00000017000": [["Domain_0", 333, 574, "PF00777"]], "ENSOPRP00000004272": [["Domain_1", 370, 410, "PF00777"]], "ENSTTRP00000010007": [["Domain_0", 276, 583, "PF00777"]], "ENSECAP00000009700": [["Domain_0", 368, 579, "PF00777"]], "ENSODEP00000012476": [["Domain_0", 334, 574, "PF00777"]], "ENSAMEP00000014079": [["Domain_0", 272, 583, "PF00777"]], "ENSPVAP00000001714": [["Domain_0", 285, 583, "PF00777"]], "ENSCATP00000043844": [["Domain_0", 334, 573, "PF00777"]], "ENSDORP00000011749": [["Domain_0", 341, 574, "PF00777"]], "ENSPSIP00000013325": [["Domain_0", 314, 582, "PF00777"]], "ENSFCAP00000025500": [["Domain_0", 333, 573, "PF00777"]], "ENSNLEP00000013152": [["Domain_0", 335, 573, "PF00777"]], "ENSCJAP00000056613": [["Domain_0", 334, 573, "PF00777"]], "ENSHGLP00100019673": [["Domain_0", 335, 574, "PF00777"]], "ENSCGRP00000004748": [["Domain_0", 335, 574, "PF00777"]], "ENSOCUP00000006679": [["Domain_0", 302, 580, "PF00777"]], "ENSOANP00000017654": [["Domain_0", 308, 576, "PF00777"]], "ENSXETP00000022803": [["Domain_0", 281, 583, "PF00777"]], "ENSCLAP00000013428": [["Domain_0", 98, 573, "PF00777"]], "ENSSHAP00000015986": [["Domain_0", 308, 581, "PF00777"]], "ENSMICP00000042863": [["Domain_0", 334, 573, "PF00777"]], "ENSPCAP00000000261": [["Domain_0", 300, 583, "PF00777"]], "ENSOGAP00000015085": [["Domain_0", 303, 580, "PF00777"]], "ENSP00000386942": [["Domain_0", 334, 573, "PF00777"]], "ENSMMUP00000002397": [["Domain_0", 334, 513, "PF00777"]], "ENSCPOP00000014080": [["Domain_0", 342, 573, "PF00777"]], "ENSFALP00000003738": [["Domain_0", 308, 582, "PF00777"]], "ENSMLUP00000011021": [["Domain_0", 285, 583, "PF00777"]], "ENSRNOP00000071786": [["Domain_0", 335, 574, "PF00777"]], "ENSEEUP00000007989": [["Domain_0", 309, 583, "PF00777"]], "ENSMODP00000003179": [["Domain_0", 308, 581, "PF00777"]], "ENSPCOP00000013982": [["Domain_0", 334, 573, "PF00777"]], "ENSMOCP00000026046": [["Domain_0", 335, 573, "PF00777"]], "ENSMFAP00000003580": [["Domain_0", 334, 573, "PF00777"]], "ENSETEP00000011445": [["Domain_0", 285, 583, "PF00777"]], "ENSDNOP00000022270": [["Domain_0", 314, 582, "PF00777"]], "ENSSSCP00000008692": [["Domain_0", 338, 573, "PF00777"]], "ENSGALP00000035783": [["Domain_0", 340, 574, "PF00777"]], "ENSMPUP00000010734": [["Domain_0", 309, 580, "PF00777"]], "ENSBTAP00000000937": [["Domain_0", 317, 577, "PF00777"]], "MGP_CAROLIEiJ_P0045276": [["Domain_0", 335, 574, "PF00777"]], "ENSLAFP00000003891": [["Domain_0", 276, 583, "PF00777"]], "ENSPEMP00000021822": [["Domain_0", 335, 574, "PF00777"]], "ENSPPAP00000025736": [["Domain_0", 334, 573, "PF00777"]], "ENSOARP00000022521": [["Domain_0", 369, 577, "PF00777"]], "ENSACAP00000002385": [["Domain_0", 316, 581, "PF00777"]]}