Found:
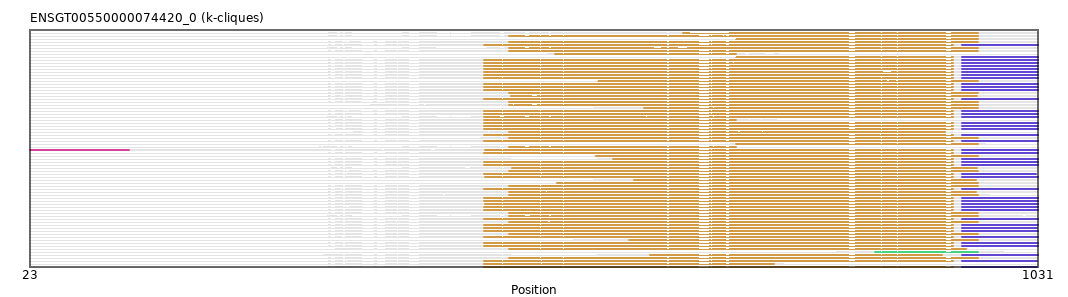
{"ENSMMUP00000023495": [["Domain_1", 606, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSMEUP00000005342": [["Domain_1", 502, 971, "PF00501"]], "ENSFCAP00000003460": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSCSAP00000016433": [["Domain_1", 502, 971, "PF00501"]], "ENSSBOP00000016708": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSAPLP00000001385": [["Domain_1", 502, 971, "PF00501"]], "ENSCAPP00000005013": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSTGUP00000017288": [["Domain_2", 868, 971, "PF00501"]], "ENSCGRP00000008777": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSMPUP00000008223": [["Domain_1", 502, 971, "PF00501"]], "ENSAMEP00000007889": [["Domain_1", 502, 971, "PF00501"]], "ENSMLUP00000002483": [["Domain_1", 589, 971, "PF00501"]], "ENSMODP00000041023": [["Domain_1", 505, 970, "PF00501"]], "ENSCAFP00000040553": [["Domain_1", 729, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSSTOP00000003787": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "MGP_PahariEiJ_P0045785": [["Domain_1", 477, 767, "PF00501"]], "ENSDORP00000008370": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSTSYP00000022115": [["Domain_1", 643, 935, "PF00501"], ["Linker_1_3", 935, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSCHOP00000004007": [["Domain_1", 591, 971, "PF00501"]], "ENSGALP00000051180": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSP00000263093": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSODEP00000001202": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSPSIP00000011431": [["Domain_1", 502, 971, "PF00501"]], "ENSMAUP00000010188": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSMNEP00000032338": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSRNOP00000026574": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1030, "PF13193"]], "MGP_SPRETEiJ_P0080726": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1030, "PF13193"]], "ENSPTRP00000019940": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSPCOP00000012983": [["Domain_1", 478, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSSARP00000001405": [["Domain_1", 502, 971, "PF00501"]], "ENSMICP00000029409": [["Domain_0", 23, 122, "PF00651"], ["Linker_0_1", 122, 478], ["Domain_1", 478, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSSHAP00000003445": [["Domain_1", 622, 971, "PF00501"]], "ENSSSCP00000051828": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSANAP00000016200": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSLAFP00000002286": [["Domain_1", 502, 938, "PF00501"]], "ENSCLAP00000000120": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSOANP00000018887": [["Domain_1", 627, 969, "PF00501"]], "ENSACAP00000011619": [["Domain_1", 502, 971, "PF00501"]], "ENSCJAP00000000459": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSFALP00000009413": [["Domain_1", 637, 971, "PF00501"]], "ENSPPAP00000036014": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSFDAP00000018055": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSMGAP00000006338": [["Domain_1", 502, 729, "PF00501"]], "ENSTTRP00000001871": [["Domain_1", 502, 971, "PF00501"]], "ENSECAP00000014705": [["Domain_1", 502, 971, "PF00501"]], "ENSMLEP00000010604": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSOCUP00000025341": [["Domain_1", 502, 971, "PF00501"]], "ENSMFAP00000013478": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSGGOP00000048749": [["Domain_1", 477, 729, "PF00501"]], "ENSNGAP00000004855": [["Domain_1", 478, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSCCAP00000014826": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSPEMP00000017108": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSBTAP00000020170": [["Domain_1", 502, 971, "PF00501"]], "ENSOPRP00000015552": [["Domain_1", 502, 971, "PF00501"]], "ENSPPYP00000011775": [["Domain_1", 502, 971, "PF00501"]], "ENSMUSP00000032539": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1029, "PF13193"]], "ENSHGLP00100025189": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSCPOP00000011353": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSCHIP00000001178": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSCAFP00000039755": [["Domain_1", 548, 729, "PF00501"]], "ENSCANP00000015650": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSOARP00000004141": [["Domain_1", 550, 971, "PF00501"]], "ENSHGLP00000006311": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "MGP_CAROLIEiJ_P0077425": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1030, "PF13193"]], "ENSPANP00000011813": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSMOCP00000004940": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSMGAP00000001838": [["Domain_1", 729, 971, "PF00501"]], "ENSRROP00000041040": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSPVAP00000013176": [["Domain_1", 502, 971, "PF00501"]], "ENSOGAP00000017610": [["Domain_1", 502, 969, "PF00501"]], "ENSCATP00000002733": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSCGRP00001013527": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSDNOP00000013903": [["Domain_1", 504, 970, "PF00501"]], "ENSRBIP00000006913": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSJJAP00000003742": [["Domain_1", 477, 946, "PF00501"], ["Linker_1_3", 946, 955], ["Domain_3", 955, 1031, "PF13193"]], "ENSACAP00000010446": [["Domain_1", 676, 971, "PF00501"]], "ENSETEP00000008005": [["Domain_1", 502, 971, "PF00501"]], "ENSTBEP00000003438": [["Domain_1", 502, 959, "PF00501"]], "ENSACAP00000021681": [["Domain_1", 729, 971, "PF00501"]]}