Found:
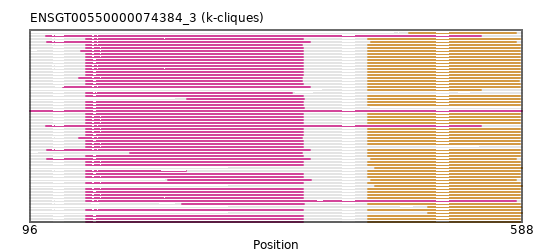
{"ENSHGLP00000025022": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSMUSP00000050730": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSCLAP00000020737": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSACAP00000015089": [["Domain_1", 475, 582, "PF00083"]], "ENSPVAP00000007032": [["Domain_1", 441, 588, "PF00083"]], "ENSCPOP00000012593": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSSBOP00000029111": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSMFAP00000043297": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSCAFP00000016465": [["Domain_0", 152, 368, "PF00083"], ["Linker_0_1", 368, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSPTRP00000084838": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSMLEP00000003533": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSCGRP00001010605": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSMLUP00000006565": [["Domain_0", 112, 547, "PF07690"]], "ENSCANP00000020647": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSAMEP00000017438": [["Domain_0", 112, 547, "PF07690"]], "ENSP00000479104": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSOARP00000018469": [["Domain_0", 113, 376, "PF07690"], ["Linker_0_1", 376, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSJJAP00000015344": [["Domain_0", 152, 370, "PF00083"]], "ENSOGAP00000015798": [["Domain_0", 113, 376, "PF07690"], ["Linker_0_1", 376, 437], ["Domain_1", 437, 582, "PF00083"]], "ENSPPYP00000005619": [["Domain_0", 234, 377, "PF07690"], ["Linker_0_1", 377, 437], ["Domain_1", 437, 582, "PF00083"]], "ENSECAP00000020797": [["Domain_0", 129, 376, "PF07690"], ["Linker_0_1", 376, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSRROP00000034059": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSSSCP00000048514": [["Domain_0", 152, 582, "PF00083"]], "ENSPCAP00000014979": [["Domain_1", 441, 588, "PF00083"]], "MGP_CAROLIEiJ_P0069955": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSLAFP00000025268": [["Domain_0", 96, 588, "PF00083"]], "ENSPCOP00000025917": [["Domain_0", 152, 252, "PF00083"], ["Domain_1", 434, 586, "PF07690"]], "ENSANAP00000003091": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSGGOP00000014555": [["Domain_0", 253, 370, "PF00083"], ["Linker_0_1", 370, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSMOCP00000015617": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSRBIP00000017405": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSCATP00000026315": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSTTRP00000003834": [["Domain_1", 494, 588, "PF00083"]], "ENSBTAP00000029564": [["Domain_0", 113, 376, "PF07690"], ["Linker_0_1", 376, 437], ["Domain_1", 437, 582, "PF00083"]], "ENSRNOP00000000874": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSMPUP00000003413": [["Domain_0", 145, 368, "PF00083"], ["Linker_0_1", 368, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSHGLP00100020794": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSCSAP00000015183": [["Domain_0", 145, 368, "PF00083"], ["Linker_0_1", 368, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSMNEP00000032805": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSEEUP00000007504": [["Domain_1", 434, 547, "PF07690"]], "ENSTBEP00000011381": [["Domain_1", 494, 587, "PF00083"]], "ENSCGRP00000009121": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSPPAP00000010180": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "MGP_PahariEiJ_P0058191": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSCCAP00000036434": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSNGAP00000017424": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSCHIP00000027226": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "MGP_SPRETEiJ_P0072631": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSMMUP00000059380": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSTSYP00000020445": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSODEP00000010763": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSPEMP00000020897": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSFDAP00000006052": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSMICP00000034093": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSSTOP00000021409": [["Domain_0", 248, 370, "PF00083"], ["Linker_0_1", 370, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSOCUP00000013950": [["Domain_0", 196, 368, "PF00083"], ["Linker_0_1", 368, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSCAPP00000013338": [["Domain_0", 147, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSMAUP00000014728": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSDORP00000008546": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSCJAP00000028557": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]], "ENSNLEP00000008296": [["Domain_0", 152, 369, "PF00083"]], "ENSFCAP00000021774": [["Domain_0", 152, 358, "PF00083"]], "ENSDNOP00000004104": [["Domain_0", 152, 368, "PF00083"], ["Linker_0_1", 368, 434], ["Domain_1", 434, 585, "PF07690"]], "ENSPANP00000001231": [["Domain_0", 152, 369, "PF00083"], ["Linker_0_1", 369, 434], ["Domain_1", 434, 586, "PF07690"]]}