Found:
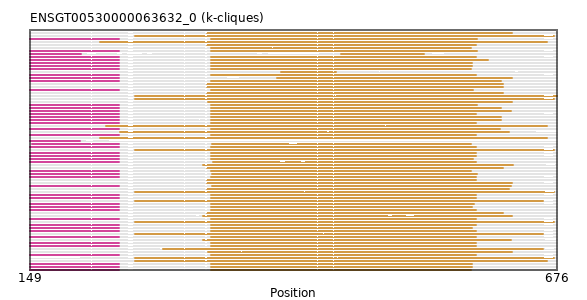
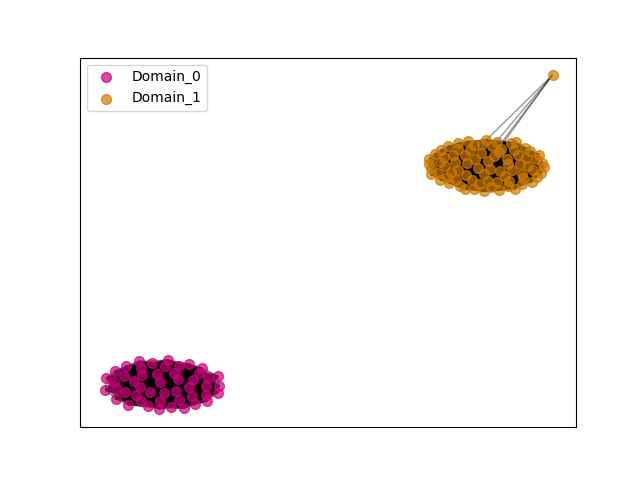
{"ENSBTAP00000017718": [["Domain_1", 326, 595, "PF03071"]], "ENSMNEP00000026385": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 595, "PF03071"]], "ENSEEUP00000007289": [["Domain_1", 254, 676, "PF03071"]], "ENSMLEP00000029550": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 595, "PF03071"]], "ENSANAP00000017536": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 596, "PF03071"]], "ENSCHIP00000013528": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 590, "PF03071"]], "ENSTTRP00000012503": [["Domain_1", 254, 673, "PF03071"]], "ENSPVAP00000012907": [["Domain_1", 254, 673, "PF03071"]], "ENSOPRP00000014611": [["Domain_1", 254, 673, "PF03071"]], "ENSSARP00000010993": [["Domain_1", 254, 662, "PF03071"]], "ENSJJAP00000018753": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 595, "PF03071"]], "MGP_CAROLIEiJ_P0063752": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 591, "PF03071"]], "ENSMLUP00000003421": [["Domain_1", 254, 673, "PF03071"]], "ENSMGAP00000011013": [["Domain_1", 219, 666, "PF03071"]], "ENSTGUP00000008474": [["Domain_1", 327, 631, "PF03071"]], "ENSPEMP00000019180": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 591, "PF03071"]], "ENSP00000361060": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 595, "PF03071"]], "ENSMFAP00000000634": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 595, "PF03071"]], "ENSMICP00000039652": [["Domain_0", 151, 238, "PF15711"], ["Linker_0_1", 238, 331], ["Domain_1", 331, 590, "PF03071"]], "ENSPCAP00000007674": [["Domain_1", 254, 662, "PF03071"]], "ENSSHAP00000002998": [["Domain_1", 322, 630, "PF03071"]], "ENSCANP00000003611": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 596, "PF03071"]], "ENSMICP00000027578": [["Domain_0", 149, 199, "PF15711"]], "ENSPSIP00000013254": [["Domain_1", 322, 631, "PF03071"]], "ENSXETP00000009518": [["Domain_1", 254, 666, "PF03071"]], "ENSCAFP00000006400": [["Domain_1", 330, 590, "PF03071"]], "MGP_SPRETEiJ_P0066248": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 590, "PF03071"]], "ENSSTOP00000030409": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 595, "PF03071"]], "ENSMMUP00000040259": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 595, "PF03071"]], "ENSCPOP00000021630": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 620, "PF03071"]], "ENSNLEP00000020172": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 595, "PF03071"]], "ENSPANP00000024281": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 595, "PF03071"]], "ENSGGOP00000002327": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 595, "PF03071"]], "ENSCGRP00001013069": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 591, "PF03071"]], "ENSSSCP00000045060": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 595, "PF03071"]], "ENSETEP00000008845": [["Domain_1", 254, 673, "PF03071"]], "ENSCLAP00000009454": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 396], ["Domain_1", 396, 631, "PF03071"]], "ENSOARP00000002347": [["Domain_1", 326, 595, "PF03071"]], "ENSMEUP00000010883": [["Domain_1", 239, 628, "PF03071"]], "ENSMOCP00000009789": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 592, "PF03071"]], "ENSRROP00000044832": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 595, "PF03071"]], "ENSCSAP00000016787": [["Domain_1", 326, 622, "PF03071"]], "ENSTSYP00000009695": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 595, "PF03071"]], "ENSHGLP00000002900": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 620, "PF03071"]], "ENSODEP00000012305": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 331], ["Domain_1", 331, 630, "PF03071"]], "ENSPTRP00000001193": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 595, "PF03071"]], "ENSHGLP00100015764": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 620, "PF03071"]], "ENSMPUP00000012863": [["Domain_1", 326, 622, "PF03071"]], "ENSMUSP00000112751": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 590, "PF03071"]], "ENSCHOP00000007130": [["Domain_1", 400, 455, "PF03071"]], "ENSCAPP00000000930": [["Domain_0", 149, 200, "PF15711"], ["Domain_1", 460, 543, "PF03071"]], "ENSRNOP00000035825": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 591, "PF03071"]], "ENSOCUP00000004522": [["Domain_1", 326, 631, "PF03071"]], "ENSAPLP00000007607": [["Domain_1", 219, 666, "PF03071"]], "ENSMOCP00000017436": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 332], ["Domain_1", 332, 595, "PF03071"]], "ENSOGAP00000003139": [["Domain_1", 326, 628, "PF03071"]], "ENSTBEP00000007910": [["Domain_1", 282, 662, "PF03071"]], "ENSMODP00000002564": [["Domain_1", 322, 632, "PF03071"]], "ENSFALP00000009913": [["Domain_1", 327, 631, "PF03071"]], "ENSPCOP00000023012": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 593, "PF03071"]], "ENSECAP00000019774": [["Domain_1", 326, 622, "PF03071"]], "ENSCJAP00000039220": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 595, "PF03071"]], "ENSSBOP00000011390": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 595, "PF03071"]], "ENSNGAP00000018874": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 331], ["Domain_1", 331, 596, "PF03071"]], "ENSDNOP00000003073": [["Domain_1", 326, 622, "PF03071"]], "ENSPPAP00000015371": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 595, "PF03071"]], "ENSLAFP00000023852": [["Domain_1", 225, 666, "PF03071"]], "ENSMAUP00000023814": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 619, "PF03071"]], "ENSCCAP00000005292": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 607, "PF03071"]], "ENSACAP00000002475": [["Domain_1", 326, 631, "PF03071"]], "ENSAMEP00000014622": [["Domain_1", 254, 673, "PF03071"]], "ENSFDAP00000020192": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 620, "PF03071"]], "ENSCATP00000024303": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 595, "PF03071"]], "ENSCGRP00000012488": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 591, "PF03071"]], "ENSGALP00000048138": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 630, "PF03071"]], "ENSRBIP00000006132": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 595, "PF03071"]], "ENSPPYP00000001624": [["Domain_1", 326, 622, "PF03071"]], "ENSDORP00000022054": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 592, "PF03071"]], "MGP_PahariEiJ_P0078857": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 591, "PF03071"]], "ENSFCAP00000000513": [["Domain_0", 149, 238, "PF15711"], ["Linker_0_1", 238, 330], ["Domain_1", 330, 596, "PF03071"]]}