Found:
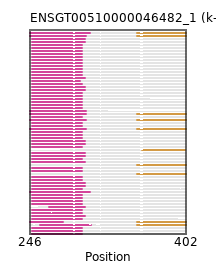
{"ENSMOCP00000008740": [["Domain_0", 248, 298, "PF01448"]], "ENSCATP00000032048": [["Domain_0", 248, 298, "PF01448"]], "ENSHGLP00000019850": [["Domain_0", 248, 298, "PF01448"]], "ENSMICP00000007917": [["Domain_0", 248, 298, "PF01448"]], "ENSOGAP00000019317": [["Domain_0", 248, 301, "PF01448"]], "ENSCGRP00001012842": [["Domain_0", 248, 298, "PF01448"]], "ENSOARP00000008920": [["Domain_0", 248, 301, "PF01448"]], "ENSXETP00000014376": [["Domain_0", 256, 307, "PF01448"], ["Linker_0_1", 307, 353], ["Domain_1", 353, 402, "PF00249"]], "ENSP00000264819": [["Domain_0", 248, 298, "PF01448"]], "ENSECAP00000006571": [["Domain_0", 248, 301, "PF01448"]], "ENSSBOP00000009811": [["Domain_0", 248, 298, "PF01448"]], "ENSMNEP00000025814": [["Domain_0", 248, 298, "PF01448"]], "ENSTGUP00000000607": [["Domain_0", 248, 301, "PF01448"]], "ENSPCOP00000014892": [["Domain_0", 248, 298, "PF01448"]], "ENSPTRP00000075430": [["Domain_0", 248, 298, "PF01448"]], "ENSMUSP00000127387": [["Domain_0", 248, 298, "PF01448"]], "ENSCJAP00000061886": [["Domain_0", 248, 298, "PF01448"]], "ENSCPOP00000021225": [["Domain_0", 248, 298, "PF01448"]], "ENSDORP00000008446": [["Domain_0", 248, 298, "PF01448"]], "ENSCJAP00000026666": [["Domain_0", 248, 298, "PF01448"]], "ENSODEP00000016325": [["Domain_0", 248, 298, "PF01448"]], "ENSPPAP00000014054": [["Domain_0", 248, 298, "PF01448"]], "ENSSHAP00000001189": [["Domain_0", 265, 301, "PF01448"]], "ENSOPRP00000011602": [["Domain_1", 353, 402, "PF00249"]], "ENSPVAP00000015175": [["Domain_0", 246, 306, "PF01448"]], "ENSAPLP00000001259": [["Domain_0", 246, 302, "PF01448"], ["Linker_0_1", 302, 353], ["Domain_1", 353, 402, "PF00249"]], "ENSCAFP00000029202": [["Domain_0", 248, 301, "PF01448"]], "ENSMODP00000008240": [["Domain_0", 248, 301, "PF01448"]], "ENSFDAP00000014145": [["Domain_0", 248, 298, "PF01448"]], "ENSTTRP00000003542": [["Domain_0", 246, 279, "PF01448"], ["Linker_0_1", 279, 353], ["Domain_1", 353, 402, "PF00249"]], "ENSMGAP00000002152": [["Domain_0", 246, 302, "PF01448"], ["Linker_0_1", 302, 353], ["Domain_1", 353, 402, "PF00249"]], "ENSAMEP00000002731": [["Domain_0", 246, 306, "PF01448"], ["Linker_0_1", 306, 353], ["Domain_1", 353, 402, "PF00249"]], "ENSCCAP00000007663": [["Domain_0", 248, 298, "PF01448"]], "ENSMLEP00000012276": [["Domain_0", 248, 298, "PF01448"]], "ENSGGOP00000019654": [["Domain_0", 248, 298, "PF01448"]], "ENSMLUP00000014598": [["Domain_0", 246, 302, "PF01448"], ["Linker_0_1", 302, 353], ["Domain_1", 353, 402, "PF00249"]], "ENSCLAP00000011439": [["Domain_0", 248, 298, "PF01448"]], "ENSCGRP00000012723": [["Domain_0", 248, 298, "PF01448"]], "ENSPCAP00000009088": [["Domain_1", 353, 402, "PF00249"]], "ENSPSIP00000003684": [["Domain_0", 248, 301, "PF01448"]], "ENSMPUP00000010052": [["Domain_0", 248, 301, "PF01448"]], "MGP_SPRETEiJ_P0024844": [["Domain_0", 248, 298, "PF01448"]], "MGP_CAROLIEiJ_P0023695": [["Domain_0", 248, 298, "PF01448"]], "ENSMMUP00000057890": [["Domain_0", 248, 298, "PF01448"]], "ENSMFAP00000008537": [["Domain_0", 248, 298, "PF01448"]], "ENSPANP00000009228": [["Domain_0", 248, 298, "PF01448"]], "ENSBTAP00000026341": [["Domain_0", 248, 301, "PF01448"]], "ENSCHIP00000019121": [["Domain_0", 248, 298, "PF01448"]], "ENSRROP00000024518": [["Domain_0", 248, 298, "PF01448"]], "ENSCANP00000038496": [["Domain_0", 248, 298, "PF01448"]], "MGP_PahariEiJ_P0086848": [["Domain_0", 248, 298, "PF01448"]], "ENSPPYP00000010403": [["Domain_0", 248, 301, "PF01448"]], "ENSTSYP00000023730": [["Domain_0", 248, 298, "PF01448"]], "ENSCSAP00000009304": [["Domain_0", 248, 301, "PF01448"]], "ENSLAFP00000018639": [["Domain_0", 246, 302, "PF01448"]], "ENSGALP00000001660": [["Domain_0", 248, 298, "PF01448"]], "ENSMEUP00000009303": [["Domain_0", 246, 302, "PF01448"], ["Linker_0_1", 302, 353], ["Domain_1", 353, 402, "PF00249"]], "ENSRNOP00000068538": [["Domain_0", 248, 298, "PF01448"]], "ENSSTOP00000010974": [["Domain_0", 248, 298, "PF01448"]], "ENSNLEP00000000742": [["Domain_1", 358, 401, "PF00249"]], "ENSHGLP00100012408": [["Domain_0", 248, 298, "PF01448"]], "ENSDNOP00000021790": [["Domain_0", 248, 296, "PF01448"]], "ENSFALP00000014500": [["Domain_0", 248, 301, "PF01448"]], "ENSFCAP00000028522": [["Domain_0", 248, 298, "PF01448"]], "ENSOCUP00000021502": [["Domain_0", 248, 301, "PF01448"]], "ENSSSCP00000014888": [["Domain_0", 248, 298, "PF01448"]], "ENSRBIP00000005406": [["Domain_0", 248, 298, "PF01448"]], "ENSCAPP00000013071": [["Domain_0", 248, 298, "PF01448"]]}