Found:
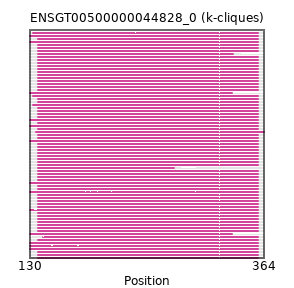
{"ENSSTOP00000031730": [["Domain_0", 138, 358, "PF06456"]], "ENSGGOP00000004971": [["Domain_0", 138, 358, "PF06456"]], "MGP_SPRETEiJ_P0084707": [["Domain_0", 138, 358, "PF06456"]], "MGP_CAROLIEiJ_P0081327": [["Domain_0", 138, 358, "PF06456"]], "ENSCAFP00000009675": [["Domain_0", 138, 358, "PF06456"]], "ENSLAFP00000012887": [["Domain_0", 130, 358, "PF06456"]], "ENSCANP00000015180": [["Domain_0", 138, 358, "PF06456"]], "ENSCPOP00000024250": [["Domain_0", 138, 358, "PF06456"]], "ENSBTAP00000004638": [["Domain_0", 138, 358, "PF06456"]], "ENSPVAP00000002281": [["Domain_0", 130, 358, "PF06456"]], "ENSMNEP00000039993": [["Domain_0", 138, 358, "PF06456"]], "ENSEEUP00000001857": [["Domain_0", 130, 332, "PF06456"]], "ENSAPLP00000008923": [["Domain_0", 130, 358, "PF06456"]], "ENSPANP00000006167": [["Domain_0", 138, 358, "PF06456"]], "ENSCGRP00001005186": [["Domain_0", 138, 358, "PF06456"]], "ENSPCAP00000014861": [["Domain_0", 130, 358, "PF06456"]], "ENSTGUP00000015146": [["Domain_0", 143, 358, "PF06456"]], "ENSHGLP00100016124": [["Domain_0", 138, 358, "PF06456"]], "ENSP00000484121": [["Domain_0", 138, 358, "PF06456"]], "ENSPPYP00000004061": [["Domain_0", 138, 358, "PF06456"]], "ENSRROP00000025527": [["Domain_0", 138, 358, "PF06456"]], "ENSGALP00000043736": [["Domain_0", 133, 358, "PF06456"]], "ENSAMEP00000017247": [["Domain_0", 130, 358, "PF06456"]], "ENSPCOP00000001824": [["Domain_0", 138, 358, "PF06456"]], "ENSMUSP00000120387": [["Domain_0", 138, 358, "PF06456"]], "ENSCCAP00000009547": [["Domain_0", 138, 358, "PF06456"]], "ENSRBIP00000031517": [["Domain_0", 138, 358, "PF06456"]], "ENSSHAP00000009185": [["Domain_0", 138, 358, "PF06456"]], "ENSMEUP00000007185": [["Domain_0", 130, 358, "PF06456"]], "ENSACAP00000010034": [["Domain_0", 133, 358, "PF06456"]], "ENSTTRP00000012190": [["Domain_0", 130, 358, "PF06456"]], "ENSDNOP00000017827": [["Domain_0", 138, 358, "PF06456"]], "ENSOCUP00000014560": [["Domain_0", 138, 358, "PF06456"]], "ENSMMUP00000060653": [["Domain_0", 138, 358, "PF06456"]], "ENSHGLP00000010421": [["Domain_0", 138, 358, "PF06456"]], "ENSNGAP00000001247": [["Domain_0", 138, 358, "PF06456"]], "ENSJJAP00000001527": [["Domain_0", 138, 358, "PF06456"]], "ENSOARP00000019533": [["Domain_0", 138, 358, "PF06456"]], "ENSPEMP00000002113": [["Domain_0", 138, 358, "PF06456"]], "ENSCSAP00000000668": [["Domain_0", 138, 358, "PF06456"]], "ENSFDAP00000002343": [["Domain_0", 138, 358, "PF06456"]], "ENSTSYP00000029934": [["Domain_0", 138, 358, "PF06456"]], "ENSVPAP00000009985": [["Domain_0", 130, 358, "PF06456"]], "ENSCJAP00000046338": [["Domain_0", 138, 358, "PF06456"]], "ENSOANP00000024054": [["Domain_0", 138, 274, "PF06456"]], "ENSCHIP00000014537": [["Domain_0", 138, 358, "PF06456"]], "ENSMOCP00000013482": [["Domain_0", 138, 358, "PF06456"]], "ENSDORP00000026532": [["Domain_0", 138, 358, "PF06456"]], "ENSMLEP00000006065": [["Domain_0", 138, 358, "PF06456"]], "ENSNLEP00000044149": [["Domain_0", 138, 358, "PF06456"]], "ENSMGAP00000016219": [["Domain_0", 136, 364, "PF03114"]], "ENSMLUP00000015752": [["Domain_0", 130, 358, "PF06456"]], "ENSOPRP00000007283": [["Domain_0", 130, 358, "PF06456"]], "ENSPTRP00000005766": [["Domain_0", 138, 358, "PF06456"]], "ENSFCAP00000004279": [["Domain_0", 138, 358, "PF06456"]], "ENSCLAP00000023659": [["Domain_0", 138, 358, "PF06456"]], "ENSXETP00000035859": [["Domain_0", 130, 358, "PF06456"]], "ENSMICP00000001922": [["Domain_0", 138, 358, "PF06456"]], "ENSCGRP00000007184": [["Domain_0", 138, 358, "PF06456"]], "ENSRNOP00000025159": [["Domain_0", 138, 358, "PF06456"]], "ENSANAP00000026890": [["Domain_0", 138, 358, "PF06456"]], "ENSMODP00000023176": [["Domain_0", 138, 358, "PF06456"]], "ENSMFAP00000012786": [["Domain_0", 138, 358, "PF06456"]], "ENSTBEP00000011739": [["Domain_0", 130, 332, "PF06456"]], "MGP_PahariEiJ_P0018138": [["Domain_0", 138, 358, "PF06456"]], "ENSCAPP00000002588": [["Domain_0", 138, 333, "PF06456"]], "ENSMPUP00000005568": [["Domain_0", 138, 358, "PF06456"]], "ENSSSCP00000015554": [["Domain_0", 138, 358, "PF06456"]], "ENSECAP00000017458": [["Domain_0", 138, 358, "PF06456"]], "ENSPPAP00000039585": [["Domain_0", 138, 358, "PF06456"]], "ENSODEP00000011858": [["Domain_0", 138, 358, "PF06456"]], "ENSSBOP00000026363": [["Domain_0", 138, 358, "PF06456"]], "ENSFALP00000008509": [["Domain_0", 133, 358, "PF06456"]], "ENSMAUP00000011752": [["Domain_0", 138, 358, "PF06456"]], "ENSOGAP00000002497": [["Domain_0", 138, 358, "PF06456"]], "ENSCATP00000022911": [["Domain_0", 138, 358, "PF06456"]]}