Found:
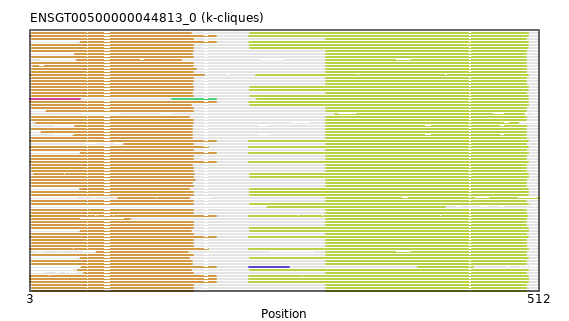
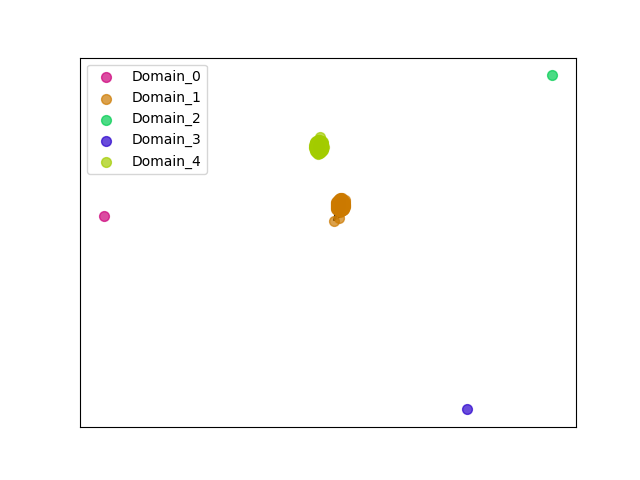
{"ENSMNEP00000039623": [["Domain_1", 5, 166, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSNGAP00000021598": [["Domain_1", 5, 165, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSMFAP00000040668": [["Domain_1", 5, 166, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSCJAP00000027829": [["Domain_1", 5, 166, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSCCAP00000022929": [["Domain_1", 50, 154, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSHGLP00100020901": [["Domain_1", 46, 165, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSRBIP00000021595": [["Domain_1", 5, 166, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSCHIP00000033487": [["Domain_1", 5, 166, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSANAP00000014162": [["Domain_1", 5, 166, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSTBEP00000013274": [["Domain_1", 54, 189, "PF00875"], ["Linker_1_3", 189, 222], ["Domain_3", 222, 262, "PF03441"], ["Linker_3_4", 262, 391], ["Domain_4", 391, 446, "PF03441"]], "ENSAPLP00000008083": [["Domain_1", 53, 189, "PF00875"], ["Linker_1_4", 189, 222], ["Domain_4", 222, 501, "PF03441"]], "ENSOGAP00000001085": [["Domain_1", 5, 166, "PF00875"], ["Linker_1_4", 166, 223], ["Domain_4", 223, 501, "PF03441"]], "ENSCPOP00000027151": [["Domain_1", 5, 165, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSHGLP00000009964": [["Domain_1", 5, 165, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSCAPP00000014244": [["Domain_1", 48, 165, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSSSCP00000035060": [["Domain_1", 5, 165, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSTSYP00000022858": [["Domain_1", 57, 166, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSMGAP00000013551": [["Domain_1", 53, 189, "PF00875"], ["Linker_1_4", 189, 222], ["Domain_4", 222, 501, "PF03441"]], "ENSCSAP00000015109": [["Domain_1", 5, 166, "PF00875"], ["Linker_1_4", 166, 223], ["Domain_4", 223, 501, "PF03441"]], "ENSPPYP00000005604": [["Domain_1", 5, 166, "PF00875"], ["Linker_1_4", 166, 223], ["Domain_4", 223, 501, "PF03441"]], "ENSRNOP00000009124": [["Domain_1", 5, 166, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSMAUP00000013083": [["Domain_1", 97, 166, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSHGLP00000002633": [["Domain_1", 9, 166, "PF00875"], ["Domain_4", 299, 491, "PF03441"]], "ENSEEUP00000006465": [["Domain_0", 3, 53, "PF00875"], ["Linker_0_2", 53, 145], ["Domain_2", 145, 189, "PF00875"], ["Linker_2_4", 189, 230], ["Domain_4", 230, 501, "PF03441"]], "ENSCATP00000027012": [["Domain_1", 5, 166, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSCGRP00001001622": [["Domain_1", 5, 169, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSBTAP00000051134": [["Domain_1", 5, 166, "PF00875"], ["Linker_1_4", 166, 223], ["Domain_4", 223, 501, "PF03441"]], "ENSTTRP00000012157": [["Domain_1", 3, 189, "PF00875"], ["Linker_1_4", 189, 222], ["Domain_4", 222, 501, "PF03441"]], "ENSHGLP00100006166": [["Domain_1", 14, 165, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSMUSP00000020227": [["Domain_1", 5, 168, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSGALP00000020598": [["Domain_1", 5, 162, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSGGOP00000025197": [["Domain_1", 5, 166, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSRROP00000022771": [["Domain_1", 5, 166, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSSTOP00000025979": [["Domain_1", 5, 165, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSFDAP00000016170": [["Domain_1", 19, 166, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSFDAP00000021037": [["Domain_4", 308, 497, "PF03441"]], "ENSTGUP00000011352": [["Domain_1", 51, 164, "PF00875"], ["Linker_1_4", 164, 223], ["Domain_4", 223, 501, "PF03441"]], "ENSDORP00000008183": [["Domain_1", 5, 165, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSP00000008527": [["Domain_1", 5, 166, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "MGP_CAROLIEiJ_P0024322": [["Domain_1", 5, 166, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSPVAP00000001499": [["Domain_1", 3, 189, "PF00875"], ["Linker_1_4", 189, 222], ["Domain_4", 222, 501, "PF03441"]], "ENSPANP00000012029": [["Domain_1", 5, 166, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSCCAP00000024672": [["Domain_1", 5, 166, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSCHOP00000009859": [["Domain_1", 3, 177, "PF00875"], ["Linker_1_4", 177, 229], ["Domain_4", 229, 501, "PF03441"]], "ENSPEMP00000021870": [["Domain_1", 5, 166, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSMODP00000001926": [["Domain_1", 7, 167, "PF00875"], ["Linker_1_4", 167, 223], ["Domain_4", 223, 501, "PF03441"]], "ENSOPRP00000015117": [["Domain_4", 240, 418, "PF03441"]], "ENSNLEP00000008191": [["Domain_1", 5, 166, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSECAP00000013000": [["Domain_1", 5, 166, "PF00875"], ["Linker_1_4", 166, 223], ["Domain_4", 223, 501, "PF03441"]], "ENSCAFP00000002620": [["Domain_1", 5, 166, "PF00875"], ["Linker_1_4", 166, 223], ["Domain_4", 223, 501, "PF03441"]], "ENSPCAP00000010567": [["Domain_1", 3, 189, "PF00875"], ["Linker_1_4", 189, 222], ["Domain_4", 222, 501, "PF03441"]], "ENSFALP00000013984": [["Domain_1", 5, 164, "PF00875"], ["Linker_1_4", 164, 223], ["Domain_4", 223, 501, "PF03441"]], "ENSOARP00000019119": [["Domain_1", 5, 166, "PF00875"], ["Linker_1_4", 166, 223], ["Domain_4", 223, 501, "PF03441"]], "ENSMMUP00000028723": [["Domain_1", 5, 165, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSMOCP00000010857": [["Domain_1", 5, 166, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSHGLP00000002808": [["Domain_1", 48, 165, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "MGP_PahariEiJ_P0088341": [["Domain_1", 5, 166, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSXETP00000059100": [["Domain_1", 3, 189, "PF00875"], ["Linker_1_4", 189, 222], ["Domain_4", 222, 501, "PF03441"]], "ENSAMEP00000002891": [["Domain_1", 3, 189, "PF00875"], ["Linker_1_4", 189, 222], ["Domain_4", 222, 501, "PF03441"]], "ENSMLEP00000027970": [["Domain_1", 5, 166, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSFCAP00000024940": [["Domain_1", 5, 166, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSJJAP00000003123": [["Domain_1", 5, 165, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSMPUP00000012065": [["Domain_1", 5, 166, "PF00875"], ["Linker_1_4", 166, 223], ["Domain_4", 223, 501, "PF03441"]], "ENSCLAP00000018813": [["Domain_1", 5, 166, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSVPAP00000007946": [["Domain_1", 3, 189, "PF00875"], ["Linker_1_4", 189, 222], ["Domain_4", 222, 501, "PF03441"]], "ENSPSIP00000015791": [["Domain_1", 53, 164, "PF00875"], ["Linker_1_4", 164, 223], ["Domain_4", 223, 501, "PF03441"]], "ENSPTRP00000009157": [["Domain_1", 5, 166, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSLAFP00000020605": [["Domain_1", 3, 189, "PF00875"], ["Linker_1_4", 189, 222], ["Domain_4", 222, 501, "PF03441"]], "ENSSBOP00000000276": [["Domain_1", 69, 161, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSETEP00000006713": [["Domain_1", 3, 189, "PF00875"], ["Linker_1_4", 189, 222], ["Domain_4", 222, 501, "PF03441"]], "ENSSHAP00000016769": [["Domain_1", 53, 162, "PF00875"], ["Linker_1_4", 162, 223], ["Domain_4", 223, 501, "PF03441"]], "MGP_SPRETEiJ_P0025504": [["Domain_1", 5, 166, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSMICP00000027430": [["Domain_1", 5, 166, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSODEP00000016702": [["Domain_1", 5, 166, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSOCUP00000011809": [["Domain_1", 5, 166, "PF00875"], ["Linker_1_4", 166, 223], ["Domain_4", 223, 501, "PF03441"]], "ENSMLUP00000014840": [["Domain_1", 3, 189, "PF00875"], ["Linker_1_4", 189, 222], ["Domain_4", 222, 501, "PF03441"]], "ENSSBOP00000018535": [["Domain_1", 5, 166, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSODEP00000004259": [["Domain_1", 91, 162, "PF00875"], ["Domain_4", 299, 512, "PF03441"]], "ENSDNOP00000009091": [["Domain_1", 5, 166, "PF00875"], ["Linker_1_4", 166, 223], ["Domain_4", 223, 501, "PF03441"]], "ENSCANP00000002163": [["Domain_1", 5, 166, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSPPAP00000003201": [["Domain_1", 5, 166, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSPCOP00000004921": [["Domain_1", 5, 103, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSCGRP00000013734": [["Domain_1", 6, 166, "PF00875"], ["Domain_4", 299, 499, "PF03441"]], "ENSACAP00000015006": [["Domain_1", 5, 164, "PF00875"], ["Linker_1_4", 164, 223], ["Domain_4", 223, 501, "PF03441"]], "ENSSARP00000009775": [["Domain_1", 3, 181, "PF00875"], ["Linker_1_4", 181, 222], ["Domain_4", 222, 501, "PF03441"]], "ENSOANP00000008782": [["Domain_1", 53, 162, "PF00875"], ["Linker_1_4", 162, 223], ["Domain_4", 223, 501, "PF03441"]], "ENSMEUP00000013409": [["Domain_1", 3, 181, "PF00875"], ["Linker_1_4", 181, 222], ["Domain_4", 222, 501, "PF03441"]]}