Found:
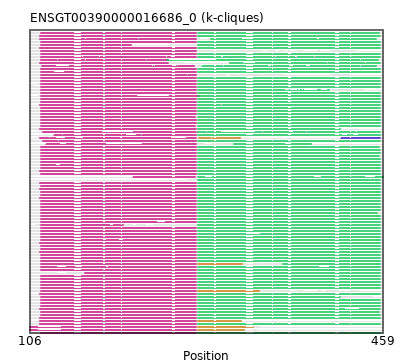
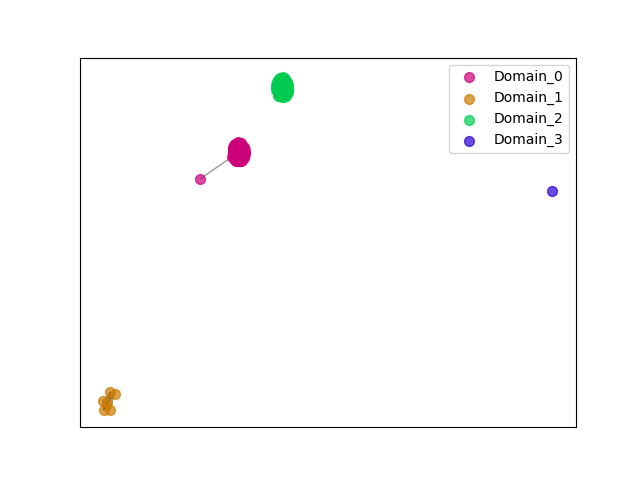
{"ENSMPUP00000015894": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 453, "PF02866"]], "ENSOCUP00000012316": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSFCAP00000039521": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSGGOP00000011503": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSRBIP00000008270": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSODEP00000021545": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSMICP00000033834": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSCAPP00000000418": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSCAFP00000037554": [["Domain_0", 118, 270, "PF00056"], ["Linker_0_2", 270, 278], ["Domain_2", 278, 452, "PF02866"]], "ENSMAUP00000014712": [["Domain_0", 209, 271, "PF00056"], ["Linker_0_2", 271, 274], ["Domain_2", 274, 459, "PF02866"]], "ENSOGAP00000008680": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSETEP00000010139": [["Domain_0", 116, 272, "PF00056"], ["Linker_0_1", 272, 275], ["Domain_1", 275, 316, "PF02866"], ["Linker_1_3", 316, 417], ["Domain_3", 417, 456, "PF02866"]], "ENSFDAP00000016595": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSJJAP00000016044": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSBTAP00000012454": [["Domain_0", 117, 272, "PF00056"]], "ENSSTOP00000014485": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSLAFP00000004400": [["Domain_0", 116, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSPCAP00000003672": [["Domain_0", 116, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSMLUP00000016235": [["Domain_0", 116, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSPEMP00000016650": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSCAFP00000036399": [["Domain_0", 117, 244, "PF00056"], ["Domain_2", 243, 454, "PF02866"]], "ENSMNEP00000016303": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSFALP00000010040": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 455, "PF02866"]], "ENSNLEP00000037554": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSCAFP00000038247": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSVPAP00000003285": [["Domain_0", 116, 272, "PF00056"], ["Linker_0_1", 272, 274], ["Domain_1", 274, 317, "PF02866"]], "ENSCHIP00000029021": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSMUSP00000019323": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "MGP_SPRETEiJ_P0074109": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSP00000327070": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSSBOP00000030859": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 416, "PF02866"]], "ENSANAP00000034558": [["Domain_0", 117, 207, "PF00056"], ["Domain_2", 274, 456, "PF02866"]], "ENSACAP00000001455": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 455, "PF02866"]], "ENSNGAP00000002265": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "MGP_CAROLIEiJ_P0071363": [["Domain_0", 106, 272, "PF00056"], ["Linker_0_1", 272, 274], ["Domain_1", 274, 329, "PF02866"]], "ENSCANP00000023188": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSMOCP00000004215": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSEEUP00000011301": [["Domain_0", 116, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 416, "PF02866"]], "ENSODEP00000016572": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSCCAP00000002380": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 405, "PF02866"]], "ENSRNOP00000001958": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSCATP00000027982": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSMMUP00000052500": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSPPYP00000019586": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_1", 272, 274], ["Domain_1", 274, 318, "PF02866"], ["Linker_1_2", 318, 359], ["Domain_2", 359, 456, "PF02866"]], "ENSCJAP00000030058": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSHGLP00000027096": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSETEP00000012378": [["Domain_0", 119, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSETEP00000002733": [["Domain_0", 119, 273, "PF00056"], ["Linker_0_2", 273, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSCPOP00000009110": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSOPRP00000008336": [["Domain_0", 116, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSCHOP00000012027": [["Domain_0", 116, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSCGRP00001025623": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSANAP00000001638": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSSSCP00000025866": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSAPLP00000004131": [["Domain_0", 116, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSCAFP00000038344": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSGALP00000003012": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 455, "PF02866"]], "ENSTSYP00000024464": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSECAP00000020738": [["Domain_0", 117, 271, "PF00056"], ["Linker_0_2", 271, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSXETP00000019672": [["Domain_0", 116, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSCCAP00000012898": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSMFAP00000011294": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSETEP00000016522": [["Domain_0", 122, 272, "PF00056"], ["Linker_0_2", 272, 310], ["Domain_2", 310, 387, "PF02866"]], "ENSPANP00000009603": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "MGP_PahariEiJ_P0059591": [["Domain_0", 106, 272, "PF00056"], ["Linker_0_1", 272, 274], ["Domain_1", 274, 320, "PF02866"]], "ENSDNOP00000028022": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSSHAP00000015668": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSTGUP00000004737": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 455, "PF02866"]], "ENSMEUP00000014768": [["Domain_0", 116, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSHGLP00100004236": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSCANP00000028103": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSMGAP00000003476": [["Domain_0", 116, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSCSAP00000014159": [["Domain_0", 117, 271, "PF00056"], ["Linker_0_2", 271, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSPVAP00000003211": [["Domain_0", 161, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSMLEP00000023430": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSCAFP00000019927": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSAMEP00000013516": [["Domain_0", 116, 272, "PF00056"], ["Linker_0_2", 272, 287], ["Domain_2", 287, 452, "PF02866"]], "ENSMAUP00000013207": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSPCOP00000020316": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSRROP00000000457": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSOARP00000014638": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSSARP00000010435": [["Domain_0", 116, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSMEUP00000009162": [["Domain_2", 274, 456, "PF02866"]], "ENSCGRP00000019203": [["Domain_0", 117, 275, "PF00056"], ["Domain_2", 274, 456, "PF02866"]], "ENSCAFP00000036577": [["Domain_0", 116, 244, "PF00056"], ["Linker_0_2", 244, 306], ["Domain_2", 306, 456, "PF02866"]], "ENSCAFP00000042888": [["Domain_0", 117, 272, "PF00056"]], "ENSRROP00000015266": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_1", 272, 274], ["Domain_1", 274, 335, "PF02866"]], "ENSMODP00000017108": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSRROP00000012775": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSTBEP00000002753": [["Domain_0", 116, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSRBIP00000011313": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSDORP00000008993": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSPTRP00000033039": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSOANP00000002188": [["Domain_0", 117, 199, "PF00056"], ["Domain_2", 274, 456, "PF02866"]], "ENSCAFP00000036265": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSETEP00000007264": [["Domain_0", 130, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSPPAP00000021260": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSPSIP00000018233": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSTTRP00000014063": [["Domain_0", 116, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSAMEP00000011416": [["Domain_0", 116, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]], "ENSCLAP00000009181": [["Domain_0", 117, 272, "PF00056"], ["Linker_0_2", 272, 274], ["Domain_2", 274, 456, "PF02866"]]}