Found:
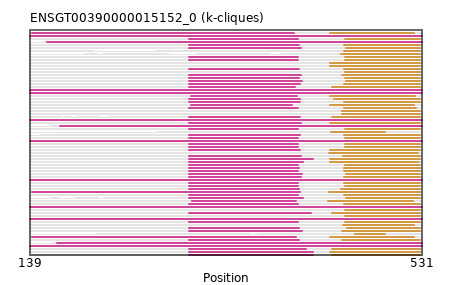

{"ENSSSCP00000002553": [["Domain_0", 298, 408, "PF01494"], ["Linker_0_1", 408, 456], ["Domain_1", 456, 529, "PF01494"]], "ENSOPRP00000002315": [["Domain_0", 139, 531, "PF01494"]], "ENSGALP00000016557": [["Domain_0", 300, 401, "PF01494"], ["Linker_0_1", 401, 439], ["Domain_1", 439, 523, "PF01494"]], "ENSDNOP00000011977": [["Domain_0", 298, 411, "PF01494"], ["Linker_0_1", 411, 455], ["Domain_1", 455, 529, "PF01494"]], "ENSDORP00000026899": [["Domain_0", 298, 409, "PF01494"], ["Linker_0_1", 409, 453], ["Domain_1", 453, 529, "PF01494"]], "ENSHGLP00000009092": [["Domain_1", 451, 529, "PF01494"]], "ENSTSYP00000005160": [["Domain_0", 298, 408, "PF01494"], ["Linker_0_1", 408, 451], ["Domain_1", 451, 529, "PF01494"]], "ENSFALP00000006518": [["Domain_0", 300, 406, "PF01494"], ["Linker_0_1", 406, 439], ["Domain_1", 439, 524, "PF01494"]], "ENSMLEP00000040043": [["Domain_0", 298, 407, "PF01494"], ["Linker_0_1", 407, 454], ["Domain_1", 454, 529, "PF01494"]], "ENSCHIP00000000872": [["Domain_0", 298, 408, "PF01494"], ["Linker_0_1", 408, 453], ["Domain_1", 453, 529, "PF01494"]], "ENSMFAP00000039814": [["Domain_0", 298, 407, "PF01494"], ["Linker_0_1", 407, 454], ["Domain_1", 454, 529, "PF01494"]], "ENSMOCP00000006806": [["Domain_0", 298, 409, "PF01494"], ["Linker_0_1", 409, 439], ["Domain_1", 439, 529, "PF01494"]], "ENSSTOP00000027073": [["Domain_0", 298, 411, "PF01494"], ["Linker_0_1", 411, 451], ["Domain_1", 451, 529, "PF01494"]], "MGP_SPRETEiJ_P0034330": [["Domain_0", 298, 422, "PF01494"], ["Linker_0_1", 422, 441], ["Domain_1", 441, 529, "PF01494"]], "ENSANAP00000027619": [["Domain_0", 298, 407, "PF01494"], ["Linker_0_1", 407, 454], ["Domain_1", 454, 529, "PF01494"]], "ENSJJAP00000009331": [["Domain_0", 298, 409, "PF01494"], ["Linker_0_1", 409, 441], ["Domain_1", 441, 529, "PF01494"]], "ENSPPYP00000006796": [["Domain_0", 298, 412, "PF01494"], ["Linker_0_1", 412, 455], ["Domain_1", 455, 529, "PF01494"]], "ENSSHAP00000003982": [["Domain_1", 452, 523, "PF01494"]], "ENSTGUP00000012453": [["Domain_0", 141, 405, "PF01494"], ["Linker_0_1", 405, 439], ["Domain_1", 439, 523, "PF01494"]], "ENSSBOP00000024705": [["Domain_0", 298, 407, "PF01494"], ["Linker_0_1", 407, 454], ["Domain_1", 454, 529, "PF01494"]], "ENSFCAP00000017346": [["Domain_0", 298, 409, "PF01494"], ["Linker_0_1", 409, 443], ["Domain_1", 443, 529, "PF01494"]], "ENSMLUP00000007939": [["Domain_0", 139, 531, "PF01494"]], "ENSODEP00000021774": [["Domain_0", 298, 411, "PF01494"], ["Linker_0_1", 411, 455], ["Domain_1", 455, 529, "PF01494"]], "ENSCLAP00000004962": [["Domain_1", 454, 529, "PF01494"]], "ENSMODP00000008175": [["Domain_1", 438, 527, "PF01494"]], "ENSOCUP00000005878": [["Domain_0", 298, 407, "PF01494"], ["Linker_0_1", 407, 453], ["Domain_1", 453, 529, "PF01494"]], "ENSACAP00000015933": [["Domain_0", 141, 403, "PF01494"], ["Linker_0_1", 403, 439], ["Domain_1", 439, 523, "PF01494"]], "ENSRROP00000045643": [["Domain_1", 454, 529, "PF01494"]], "ENSPEMP00000023279": [["Domain_0", 141, 409, "PF01494"], ["Linker_0_1", 409, 438], ["Domain_1", 438, 529, "PF01494"]], "ENSHGLP00100016899": [["Domain_1", 451, 529, "PF01494"]], "ENSEEUP00000000735": [["Domain_0", 139, 531, "PF01494"]], "ENSCCAP00000029064": [["Domain_0", 298, 407, "PF01494"], ["Linker_0_1", 407, 454], ["Domain_1", 454, 529, "PF01494"]], "ENSBTAP00000027093": [["Domain_0", 298, 408, "PF01494"], ["Linker_0_1", 408, 453], ["Domain_1", 453, 529, "PF01494"]], "ENSCAFP00000024844": [["Domain_0", 298, 410, "PF01494"], ["Linker_0_1", 410, 455], ["Domain_1", 455, 529, "PF01494"]], "ENSCGRP00000013373": [["Domain_1", 439, 529, "PF01494"]], "ENSGGOP00000014003": [["Domain_0", 298, 407, "PF01494"], ["Linker_0_1", 407, 453], ["Domain_1", 453, 525, "PF01494"]], "ENSMAUP00000010146": [["Domain_0", 298, 410, "PF01494"], ["Linker_0_1", 410, 439], ["Domain_1", 439, 529, "PF01494"]], "ENSCATP00000005411": [["Domain_0", 298, 407, "PF01494"], ["Linker_0_1", 407, 454], ["Domain_1", 454, 529, "PF01494"]], "ENSXETP00000006250": [["Domain_0", 139, 531, "PF01494"]], "ENSMMUP00000059909": [["Domain_0", 298, 407, "PF01494"], ["Linker_0_1", 407, 454], ["Domain_1", 454, 529, "PF01494"]], "ENSFDAP00000004661": [["Domain_0", 298, 409, "PF01494"], ["Linker_0_1", 409, 453], ["Domain_1", 453, 529, "PF01494"]], "ENSCPOP00000027731": [["Domain_0", 298, 410, "PF01494"], ["Linker_0_1", 410, 451], ["Domain_1", 451, 529, "PF01494"]], "ENSSARP00000003978": [["Domain_0", 139, 531, "PF01494"]], "ENSNGAP00000014749": [["Domain_0", 298, 412, "PF01494"], ["Linker_0_1", 412, 439], ["Domain_1", 439, 528, "PF01494"]], "ENSLAFP00000015188": [["Domain_0", 139, 531, "PF01494"]], "ENSPTRP00000011076": [["Domain_0", 298, 407, "PF01494"], ["Linker_0_1", 407, 453], ["Domain_1", 453, 529, "PF01494"]], "ENSMGAP00000019573": [["Domain_1", 440, 494, "PF01494"]], "MGP_PahariEiJ_P0082930": [["Domain_0", 298, 422, "PF01494"], ["Linker_0_1", 422, 439], ["Domain_1", 439, 529, "PF01494"]], "ENSPPAP00000030054": [["Domain_0", 298, 407, "PF01494"], ["Linker_0_1", 407, 453], ["Domain_1", 453, 529, "PF01494"]], "ENSMPUP00000009477": [["Domain_0", 298, 410, "PF01494"], ["Linker_0_1", 410, 452], ["Domain_1", 452, 529, "PF01494"]], "ENSCAPP00000003855": [["Domain_1", 450, 529, "PF01494"]], "ENSETEP00000015144": [["Domain_0", 139, 531, "PF01494"]], "ENSAMEP00000019286": [["Domain_0", 139, 531, "PF01494"]], "ENSPANP00000016688": [["Domain_0", 298, 407, "PF01494"], ["Linker_0_1", 407, 454], ["Domain_1", 454, 529, "PF01494"]], "ENSVPAP00000000738": [["Domain_0", 166, 531, "PF01494"]], "MGP_CAROLIEiJ_P0032814": [["Domain_0", 298, 415, "PF01494"], ["Linker_0_1", 415, 441], ["Domain_1", 441, 529, "PF01494"]], "ENSTBEP00000007218": [["Domain_1", 464, 494, "PF01494"]], "ENSRNOP00000014914": [["Domain_0", 298, 420, "PF01494"], ["Linker_0_1", 420, 441], ["Domain_1", 441, 529, "PF01494"]], "ENSMUSP00000105905": [["Domain_0", 298, 422, "PF01494"], ["Linker_0_1", 422, 439], ["Domain_1", 439, 529, "PF01494"]], "ENSAPLP00000013679": [["Domain_0", 156, 531, "PF01494"]], "ENSOARP00000000854": [["Domain_0", 298, 408, "PF01494"], ["Linker_0_1", 408, 453], ["Domain_1", 453, 529, "PF01494"]], "ENSPCOP00000022483": [["Domain_0", 298, 408, "PF01494"], ["Linker_0_1", 408, 450], ["Domain_1", 450, 529, "PF01494"]], "ENSNLEP00000019583": [["Domain_0", 298, 407, "PF01494"], ["Linker_0_1", 407, 454], ["Domain_1", 454, 529, "PF01494"]], "ENSECAP00000020826": [["Domain_0", 298, 404, "PF01494"], ["Linker_0_1", 404, 441], ["Domain_1", 441, 529, "PF01494"]], "ENSPVAP00000000605": [["Domain_0", 139, 531, "PF01494"]], "ENSPSIP00000016868": [["Domain_0", 301, 405, "PF01494"], ["Linker_0_1", 405, 437], ["Domain_1", 437, 522, "PF01494"]], "ENSMNEP00000026711": [["Domain_0", 298, 407, "PF01494"], ["Linker_0_1", 407, 454], ["Domain_1", 454, 529, "PF01494"]], "ENSCANP00000009166": [["Domain_1", 453, 529, "PF01494"]], "ENSOGAP00000005726": [["Domain_0", 298, 410, "PF01494"], ["Linker_0_1", 410, 453], ["Domain_1", 453, 529, "PF01494"]], "ENSCGRP00001026854": [["Domain_1", 439, 529, "PF01494"]], "ENSP00000333946": [["Domain_0", 298, 407, "PF01494"], ["Linker_0_1", 407, 453], ["Domain_1", 453, 529, "PF01494"]], "ENSMICP00000002738": [["Domain_0", 298, 409, "PF01494"], ["Linker_0_1", 409, 453], ["Domain_1", 453, 529, "PF01494"]], "ENSMEUP00000011604": [["Domain_0", 169, 531, "PF01494"]], "ENSCSAP00000008810": [["Domain_0", 298, 408, "PF01494"], ["Linker_0_1", 408, 454], ["Domain_1", 454, 529, "PF01494"]], "ENSRBIP00000030405": [["Domain_1", 454, 529, "PF01494"]]}