Found:
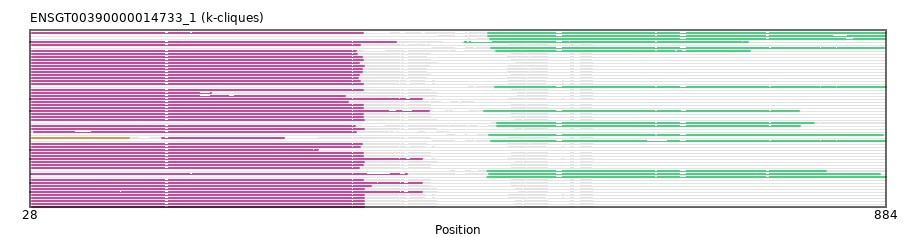
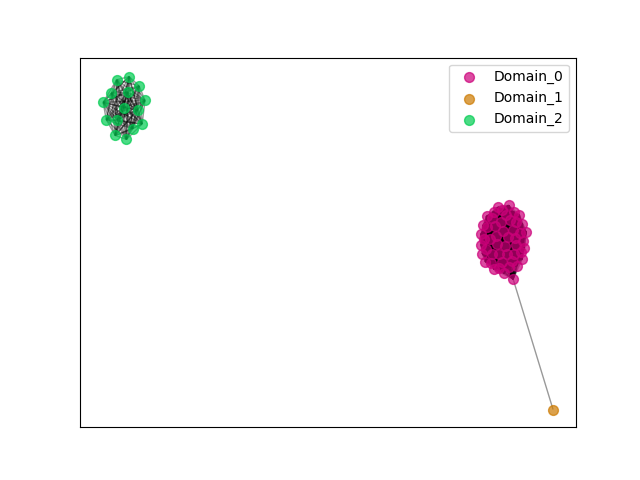
{"ENSBTAP00000041312": [["Domain_2", 489, 882, "PF05049"]], "ENSACAP00000004320": [["Domain_0", 30, 361, "PF05049"], ["Linker_0_2", 361, 486], ["Domain_2", 486, 884, "PF05049"]], "ENSPSIP00000011553": [["Domain_2", 485, 884, "PF05049"]], "ENSFCAP00000001023": [["Domain_0", 30, 361, "PF05049"]], "ENSSARP00000008430": [["Domain_0", 28, 420, "PF05049"]], "ENSCAFP00000032329": [["Domain_0", 30, 354, "PF05049"], ["Linker_0_2", 354, 494], ["Domain_2", 494, 748, "PF05049"]], "ENSCPOP00000000827": [["Domain_0", 30, 360, "PF05049"]], "ENSRBIP00000001198": [["Domain_0", 30, 369, "PF05049"]], "ENSCSAP00000019117": [["Domain_0", 30, 357, "PF05049"]], "ENSMMUP00000047913": [["Domain_0", 30, 361, "PF05049"]], "ENSP00000244314": [["Domain_0", 30, 361, "PF05049"]], "ENSPEMP00000025397": [["Domain_0", 30, 362, "PF05049"]], "ENSOANP00000013340": [["Domain_2", 487, 881, "PF05049"]], "ENSCATP00000004664": [["Domain_0", 30, 361, "PF05049"]], "ENSPANP00000008032": [["Domain_0", 30, 361, "PF05049"]], "ENSOPRP00000003909": [["Domain_0", 28, 316, "PF05049"]], "ENSCGRP00001012546": [["Domain_0", 30, 357, "PF05049"]], "ENSOARP00000009538": [["Domain_2", 489, 883, "PF05049"]], "MGP_PahariEiJ_P0046249": [["Domain_0", 30, 362, "PF05049"]], "ENSPPAP00000005438": [["Domain_0", 30, 361, "PF05049"]], "ENSMLUP00000008867": [["Domain_0", 28, 427, "PF05049"], ["Linker_0_2", 427, 482], ["Domain_2", 482, 797, "PF05049"]], "ENSMODP00000026055": [["Domain_2", 495, 812, "PF05049"]], "ENSMOCP00000012519": [["Domain_0", 30, 362, "PF05049"]], "ENSMUSP00000145975": [["Domain_0", 30, 362, "PF05049"]], "ENSCANP00000004511": [["Domain_0", 30, 355, "PF05049"]], "MGP_SPRETEiJ_P0081231": [["Domain_0", 30, 362, "PF05049"]], "ENSCHIP00000003612": [["Domain_0", 30, 362, "PF05049"]], "ENSPTRP00000019048": [["Domain_0", 30, 361, "PF05049"]], "ENSPVAP00000015937": [["Domain_0", 28, 420, "PF05049"]], "ENSHGLP00100012515": [["Domain_0", 30, 343, "PF05049"]], "ENSECAP00000004082": [["Domain_0", 30, 358, "PF05049"]], "ENSACAP00000005118": [["Domain_2", 486, 883, "PF05049"]], "ENSPCAP00000010017": [["Domain_0", 28, 420, "PF05049"]], "ENSMPUP00000006554": [["Domain_0", 30, 353, "PF05049"], ["Linker_0_2", 353, 495], ["Domain_2", 495, 798, "PF05049"]], "ENSMFAP00000012328": [["Domain_0", 30, 361, "PF05049"]], "ENSCLAP00000025547": [["Domain_0", 30, 360, "PF05049"]], "ENSGGOP00000023943": [["Domain_0", 30, 361, "PF05049"]], "MGP_CAROLIEiJ_P0077911": [["Domain_0", 30, 362, "PF05049"]], "ENSHGLP00000003858": [["Domain_0", 30, 209, "PF05049"]], "ENSMLEP00000002947": [["Domain_0", 30, 361, "PF05049"]], "ENSNGAP00000008160": [["Domain_0", 32, 354, "PF05049"]], "ENSLAFP00000002529": [["Domain_0", 28, 420, "PF05049"]], "ENSFCAP00000022926": [["Domain_2", 493, 884, "PF05049"]], "ENSOGAP00000018842": [["Domain_0", 30, 358, "PF05049"]], "ENSSTOP00000019772": [["Domain_0", 30, 362, "PF05049"]], "ENSSSCP00000003320": [["Domain_0", 30, 362, "PF05049"]], "ENSMNEP00000028547": [["Domain_0", 30, 361, "PF05049"]], "ENSOARP00000009529": [["Domain_1", 30, 127, "PF05049"], ["Linker_1_0", 127, 160], ["Domain_0", 160, 282, "PF05049"]], "ENSPPYP00000011270": [["Domain_0", 30, 357, "PF05049"]], "ENSDNOP00000019836": [["Domain_0", 30, 356, "PF05049"]], "ENSPSIP00000009348": [["Domain_0", 29, 405, "PF05049"], ["Linker_0_2", 405, 487], ["Domain_2", 487, 878, "PF05049"]], "ENSMAUP00000005849": [["Domain_0", 30, 346, "PF05049"]], "ENSBTAP00000018295": [["Domain_0", 30, 358, "PF05049"]], "ENSPSIP00000007503": [["Domain_2", 485, 824, "PF05049"]], "ENSAMEP00000004460": [["Domain_0", 28, 394, "PF05049"], ["Linker_0_2", 394, 462], ["Domain_2", 462, 746, "PF05049"]], "ENSACAP00000018673": [["Domain_2", 488, 883, "PF05049"]], "ENSCAPP00000011762": [["Domain_0", 30, 360, "PF05049"]], "ENSODEP00000013224": [["Domain_0", 30, 360, "PF05049"]], "ENSRNOP00000030295": [["Domain_0", 30, 362, "PF05049"]]}