Found:
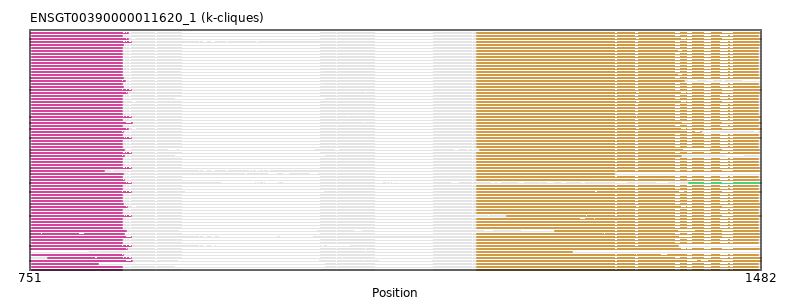
{"ENSPEMP00000015550": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSCANP00000021192": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSMMUP00000011401": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSMNEP00000024449": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSFALP00000011092": [["Domain_0", 752, 848, "PF07531"], ["Linker_0_1", 848, 1198], ["Domain_1", 1198, 1480, "PF05236"]], "ENSMICP00000003670": [["Domain_0", 753, 844, "PF07531"], ["Linker_0_1", 844, 1198], ["Domain_1", 1198, 1422, "PF05236"]], "ENSHGLP00000014479": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "MGP_CAROLIEiJ_P0056554": [["Domain_0", 753, 819, "PF07531"], ["Linker_0_1", 819, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "MGP_SPRETEiJ_P0058753": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSMEUP00000012473": [["Domain_0", 751, 853, "PF07531"], ["Linker_0_1", 853, 1198], ["Domain_1", 1198, 1482, "PF05236"]], "ENSODEP00000005479": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSNGAP00000014163": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSSBOP00000003724": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSEEUP00000007994": [["Domain_0", 751, 852, "PF07531"], ["Linker_0_1", 852, 1198], ["Domain_1", 1198, 1482, "PF05236"]], "ENSSHAP00000003029": [["Domain_0", 752, 847, "PF07531"], ["Linker_0_1", 847, 1276], ["Domain_1", 1276, 1480, "PF05236"]], "ENSTGUP00000015488": [["Domain_1", 1198, 1293, "PF05236"]], "ENSHGLP00100016105": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSAPLP00000013292": [["Domain_0", 751, 849, "PF07531"], ["Linker_0_1", 849, 1198], ["Domain_1", 1198, 1482, "PF05236"]], "ENSCAPP00000007345": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSCPOP00000027300": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSACAP00000015945": [["Domain_0", 752, 849, "PF07531"], ["Linker_0_1", 849, 1198], ["Domain_1", 1198, 1480, "PF05236"]], "ENSPVAP00000016903": [["Domain_0", 751, 852, "PF07531"], ["Linker_0_1", 852, 1228], ["Domain_1", 1228, 1482, "PF05236"]], "ENSRROP00000039901": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSPCAP00000004324": [["Domain_0", 751, 852, "PF07531"], ["Linker_0_1", 852, 1198], ["Domain_1", 1198, 1482, "PF05236"]], "ENSCCAP00000005244": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSCATP00000000530": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSSSCP00000027122": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSPCOP00000030566": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSCSAP00000011783": [["Domain_0", 752, 844, "PF07531"], ["Linker_0_1", 844, 1198], ["Domain_1", 1198, 1480, "PF05236"]], "ENSSHAP00000015895": [["Domain_0", 752, 845, "PF07531"], ["Linker_0_1", 845, 1198], ["Domain_1", 1198, 1480, "PF05236"]], "ENSPTRP00000023561": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSRNOP00000074471": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSDNOP00000033684": [["Domain_0", 752, 846, "PF07531"], ["Linker_0_1", 846, 1198], ["Domain_1", 1198, 1405, "PF05236"]], "ENSCAFP00000018423": [["Domain_0", 752, 844, "PF07531"], ["Linker_0_1", 844, 1198], ["Domain_1", 1198, 1480, "PF05236"]], "ENSPPYP00000024034": [["Domain_0", 752, 844, "PF07531"], ["Linker_0_1", 844, 1198], ["Domain_1", 1198, 1480, "PF05236"]], "ENSDORP00000006370": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSMLEP00000034843": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSMLUP00000009374": [["Domain_0", 751, 852, "PF07531"], ["Linker_0_1", 852, 1198], ["Domain_1", 1198, 1482, "PF05236"]], "ENSLAFP00000001512": [["Domain_0", 751, 852, "PF07531"], ["Linker_0_1", 852, 1198], ["Domain_1", 1198, 1481, "PF05236"]], "ENSNLEP00000019080": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSECAP00000020475": [["Domain_0", 752, 844, "PF07531"], ["Linker_0_1", 844, 1198], ["Domain_1", 1198, 1480, "PF05236"]], "ENSTGUP00000008139": [["Domain_0", 752, 848, "PF07531"], ["Linker_0_1", 848, 1198], ["Domain_1", 1198, 1480, "PF05236"]], "ENSTTRP00000008707": [["Domain_0", 769, 852, "PF07531"], ["Linker_0_1", 852, 1198], ["Domain_1", 1198, 1482, "PF05236"]], "ENSCJAP00000026568": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSP00000252996": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSCGRP00001016803": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSCHIP00000009820": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSGALP00000008211": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSCGRP00000003439": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSOARP00000015964": [["Domain_0", 752, 825, "PF07531"], ["Linker_0_1", 825, 1198], ["Domain_1", 1198, 1480, "PF05236"]], "ENSMAUP00000015943": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSMODP00000020826": [["Domain_0", 752, 852, "PF07531"], ["Linker_0_1", 852, 1198], ["Domain_1", 1198, 1480, "PF05236"]], "ENSFCAP00000011127": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSJJAP00000007488": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSPTRP00000059890": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSPANP00000014497": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSBTAP00000039125": [["Domain_0", 752, 852, "PF07531"], ["Linker_0_1", 852, 1198], ["Domain_1", 1198, 1480, "PF05236"]], "ENSMOCP00000007701": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSOCUP00000019126": [["Domain_0", 752, 844, "PF07531"], ["Linker_0_1", 844, 1198], ["Domain_1", 1198, 1335, "PF05236"]], "ENSSTOP00000013236": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSCLAP00000005220": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSMFAP00000005325": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSMPUP00000007512": [["Domain_0", 752, 845, "PF07531"], ["Linker_0_1", 845, 1198], ["Domain_1", 1198, 1402, "PF05236"]], "ENSPPAP00000011422": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSGGOP00000002080": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSTBEP00000006763": [["Domain_0", 751, 852, "PF07531"], ["Linker_0_1", 852, 1198], ["Domain_1", 1198, 1399, "PF05236"]], "ENSMUSP00000038610": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSPSIP00000014648": [["Domain_0", 752, 848, "PF07531"], ["Linker_0_1", 848, 1198], ["Domain_1", 1198, 1480, "PF05236"]], "ENSSHAP00000018664": [["Domain_0", 752, 852, "PF07531"], ["Linker_0_1", 852, 1198], ["Domain_1", 1198, 1480, "PF05236"]], "ENSFDAP00000009034": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSOPRP00000004300": [["Domain_0", 751, 852, "PF07531"], ["Domain_2", 1410, 1482, "PF05236"]], "ENSANAP00000015950": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSMODP00000015054": [["Domain_0", 752, 845, "PF07531"], ["Linker_0_1", 845, 1201], ["Domain_1", 1201, 1480, "PF05236"]], "ENSRBIP00000030706": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSXETP00000012886": [["Domain_0", 751, 853, "PF07531"], ["Linker_0_1", 853, 1198], ["Domain_1", 1198, 1482, "PF05236"]], "ENSOANP00000005353": [["Domain_0", 752, 852, "PF07531"], ["Linker_0_1", 852, 1198], ["Domain_1", 1198, 1480, "PF05236"]], "MGP_PahariEiJ_P0067552": [["Domain_0", 753, 843, "PF07531"], ["Linker_0_1", 843, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSTSYP00000022441": [["Domain_0", 753, 844, "PF07531"], ["Linker_0_1", 844, 1198], ["Domain_1", 1198, 1479, "PF05236"]], "ENSOGAP00000021208": [["Domain_0", 752, 845, "PF07531"], ["Linker_0_1", 845, 1198], ["Domain_1", 1198, 1480, "PF05236"]], "ENSMGAP00000005176": [["Domain_0", 751, 849, "PF07531"], ["Linker_0_1", 849, 1198], ["Domain_1", 1198, 1482, "PF05236"]]}