Found:
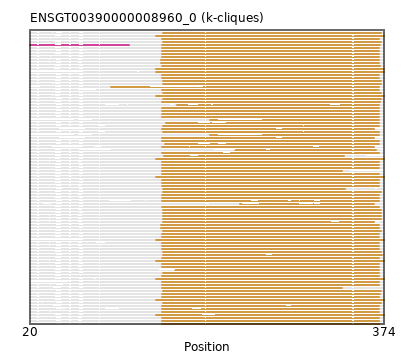
{"ENSTSYP00000032717": [["Domain_1", 152, 369, "PF01027"]], "ENSSARP00000008351": [["Domain_1", 146, 374, "PF01027"]], "ENSHGLP00000019053": [["Domain_1", 156, 369, "PF01027"]], "ENSHGLP00000006770": [["Domain_1", 152, 369, "PF01027"]], "ENSHGLP00100017239": [["Domain_1", 152, 364, "PF01027"]], "ENSDORP00000001042": [["Domain_1", 101, 369, "PF01027"]], "ENSCGRP00000006858": [["Domain_1", 151, 369, "PF01027"]], "ENSSBOP00000005278": [["Domain_1", 152, 369, "PF01027"]], "ENSMNEP00000038897": [["Domain_1", 152, 369, "PF01027"]], "ENSCHOP00000000218": [["Domain_1", 146, 374, "PF01027"]], "ENSHGLP00000023904": [["Domain_1", 153, 369, "PF01027"]], "ENSMODP00000002071": [["Domain_1", 153, 335, "PF01027"]], "ENSOARP00000011175": [["Domain_1", 153, 371, "PF01027"]], "ENSHGLP00100020931": [["Domain_1", 152, 369, "PF01027"]], "ENSHGLP00100016130": [["Domain_1", 200, 369, "PF01027"]], "ENSMFAP00000024438": [["Domain_1", 152, 369, "PF01027"]], "ENSMICP00000035209": [["Domain_1", 152, 332, "PF01027"]], "ENSNGAP00000008054": [["Domain_1", 152, 369, "PF01027"]], "ENSRROP00000004742": [["Domain_1", 152, 369, "PF01027"]], "ENSFDAP00000003163": [["Domain_1", 167, 369, "PF01027"]], "ENSODEP00000015373": [["Domain_1", 151, 369, "PF01027"]], "ENSHGLP00000026200": [["Domain_1", 152, 364, "PF01027"]], "ENSCCAP00000006787": [["Domain_1", 152, 369, "PF01027"]], "ENSSBOP00000028648": [["Domain_1", 152, 332, "PF01027"]], "MGP_PahariEiJ_P0084373": [["Domain_1", 152, 369, "PF01027"]], "ENSSBOP00000003581": [["Domain_1", 152, 369, "PF01027"]], "ENSOCUP00000005275": [["Domain_1", 153, 371, "PF01027"]], "ENSCLAP00000000511": [["Domain_1", 152, 369, "PF01027"]], "ENSACAP00000015200": [["Domain_1", 153, 371, "PF01027"]], "ENSHGLP00100016103": [["Domain_1", 153, 369, "PF01027"]], "ENSJJAP00000009430": [["Domain_1", 225, 366, "PF01027"]], "ENSNLEP00000026533": [["Domain_1", 152, 369, "PF01027"]], "ENSECAP00000016553": [["Domain_1", 153, 371, "PF01027"]], "MGP_CAROLIEiJ_P0036267": [["Domain_1", 152, 369, "PF01027"]], "ENSJJAP00000016801": [["Domain_1", 154, 334, "PF01027"]], "ENSRBIP00000020874": [["Domain_1", 152, 369, "PF01027"]], "ENSCAFP00000030996": [["Domain_0", 20, 119, "PF12811"], ["Linker_0_1", 119, 152], ["Domain_1", 152, 370, "PF01027"]], "ENSPCOP00000008523": [["Domain_1", 152, 369, "PF01027"]], "ENSGALP00000060967": [["Domain_1", 152, 369, "PF01027"]], "ENSCGRP00001025424": [["Domain_1", 151, 369, "PF01027"]], "ENSPSIP00000003711": [["Domain_1", 153, 372, "PF01027"]], "ENSOCUP00000025069": [["Domain_1", 153, 364, "PF01027"]], "ENSFALP00000007120": [["Domain_1", 153, 371, "PF01027"]], "ENSJJAP00000006339": [["Domain_1", 152, 364, "PF01027"]], "ENSP00000361207": [["Domain_1", 152, 369, "PF01027"]], "ENSDORP00000024219": [["Domain_1", 152, 369, "PF01027"]], "ENSPTRP00000004697": [["Domain_1", 152, 369, "PF01027"]], "ENSSHAP00000007710": [["Domain_1", 153, 371, "PF01027"]], "ENSTBEP00000014211": [["Domain_1", 146, 374, "PF01027"]], "ENSOANP00000008592": [["Domain_1", 153, 371, "PF01027"]], "ENSCAPP00000009262": [["Domain_1", 152, 369, "PF01027"]], "ENSFCAP00000026156": [["Domain_1", 152, 370, "PF01027"]], "ENSRROP00000002511": [["Domain_1", 153, 369, "PF01027"]], "ENSPVAP00000012678": [["Domain_1", 146, 374, "PF01027"]], "ENSSSCP00000011032": [["Domain_1", 152, 370, "PF01027"]], "ENSPEMP00000010553": [["Domain_1", 152, 369, "PF01027"]], "ENSMOCP00000020474": [["Domain_1", 152, 369, "PF01027"]], "ENSMPUP00000008443": [["Domain_1", 153, 371, "PF01027"]], "ENSPCAP00000001586": [["Domain_1", 146, 374, "PF01027"]], "ENSLAFP00000008094": [["Domain_1", 146, 374, "PF01027"]], "ENSCHOP00000001396": [["Domain_1", 146, 374, "PF01027"]], "ENSTTRP00000014283": [["Domain_1", 146, 374, "PF01027"]], "ENSMMUP00000024428": [["Domain_1", 152, 369, "PF01027"]], "MGP_SPRETEiJ_P0037981": [["Domain_1", 152, 369, "PF01027"]], "ENSRROP00000002041": [["Domain_1", 165, 369, "PF01027"]], "ENSMLEP00000010429": [["Domain_1", 152, 369, "PF01027"]], "ENSOGAP00000010846": [["Domain_1", 153, 371, "PF01027"]], "ENSSTOP00000001993": [["Domain_1", 152, 369, "PF01027"]], "ENSBTAP00000043881": [["Domain_1", 153, 371, "PF01027"]], "ENSPPYP00000002657": [["Domain_1", 153, 371, "PF01027"]], "ENSTGUP00000006249": [["Domain_1", 153, 371, "PF01027"]], "ENSODEP00000026980": [["Domain_1", 151, 369, "PF01027"]], "ENSETEP00000001458": [["Domain_1", 146, 374, "PF01027"]], "ENSMUSP00000129712": [["Domain_1", 152, 369, "PF01027"]], "ENSDNOP00000027659": [["Domain_1", 153, 371, "PF01027"]], "ENSCATP00000004503": [["Domain_1", 152, 369, "PF01027"]], "ENSHGLP00100025936": [["Domain_1", 155, 369, "PF01027"]], "ENSCSAP00000005767": [["Domain_1", 153, 371, "PF01027"]], "ENSTSYP00000018034": [["Domain_1", 152, 369, "PF01027"]], "ENSMLUP00000004265": [["Domain_1", 146, 374, "PF01027"]], "ENSNGAP00000023005": [["Domain_1", 230, 364, "PF01027"]], "ENSPANP00000020226": [["Domain_1", 152, 369, "PF01027"]], "ENSANAP00000025676": [["Domain_1", 152, 369, "PF01027"]], "ENSGGOP00000010729": [["Domain_1", 152, 369, "PF01027"]], "ENSAMEP00000010275": [["Domain_1", 146, 374, "PF01027"]], "ENSMICP00000010573": [["Domain_1", 152, 369, "PF01027"]], "ENSMAUP00000000326": [["Domain_1", 152, 369, "PF01027"]], "ENSCANP00000014585": [["Domain_1", 152, 369, "PF01027"]], "ENSRNOP00000018723": [["Domain_1", 152, 369, "PF01027"]], "ENSFDAP00000010804": [["Domain_1", 152, 369, "PF01027"]], "ENSPPAP00000003432": [["Domain_1", 152, 369, "PF01027"]], "ENSCHIP00000000187": [["Domain_1", 152, 370, "PF01027"]], "ENSCPOP00000005795": [["Domain_1", 152, 369, "PF01027"]], "ENSNGAP00000021504": [["Domain_1", 152, 369, "PF01027"]], "ENSTSYP00000019571": [["Domain_1", 152, 369, "PF01027"]], "ENSHGLP00000009972": [["Domain_1", 200, 369, "PF01027"]], "ENSJJAP00000014555": [["Domain_1", 152, 369, "PF01027"]], "ENSOCUP00000001430": [["Domain_1", 153, 371, "PF01027"]]}