Found:
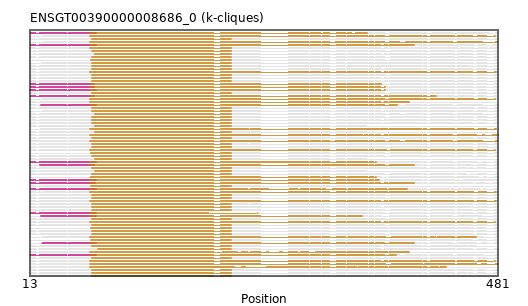
{"ENSETEP00000000219": [["Domain_1", 73, 392, "PF04495"]], "ENSMOCP00000021780": [["Domain_1", 78, 214, "PF04495"]], "ENSBTAP00000017590": [["Domain_0", 14, 79, "PF04495"], ["Domain_1", 73, 397, "PF04495"]], "ENSJJAP00000021213": [["Domain_1", 78, 214, "PF04495"]], "ENSPANP00000018493": [["Domain_1", 75, 214, "PF04495"]], "ENSTGUP00000008386": [["Domain_0", 14, 79, "PF04495"], ["Domain_1", 73, 379, "PF04495"]], "ENSSHAP00000014315": [["Domain_0", 25, 79, "PF04495"], ["Domain_1", 73, 397, "PF04495"]], "ENSPCOP00000010044": [["Domain_1", 75, 214, "PF04495"]], "ENSRROP00000021674": [["Domain_1", 75, 214, "PF04495"]], "ENSTSYP00000013323": [["Domain_1", 75, 214, "PF04495"]], "ENSMLUP00000001872": [["Domain_1", 73, 478, "PF04495"]], "ENSCPOP00000029003": [["Domain_1", 75, 214, "PF04495"]], "ENSRNOP00000036881": [["Domain_1", 74, 214, "PF04495"]], "ENSXETP00000040858": [["Domain_1", 73, 429, "PF04495"]], "ENSPTRP00000021582": [["Domain_1", 75, 214, "PF04495"]], "ENSCGRP00001013034": [["Domain_1", 75, 214, "PF04495"]], "ENSHGLP00100023930": [["Domain_1", 75, 214, "PF04495"]], "MGP_SPRETEiJ_P0054264": [["Domain_1", 75, 214, "PF04495"]], "ENSCANP00000011528": [["Domain_1", 75, 214, "PF04495"]], "ENSCSAP00000011097": [["Domain_0", 14, 77, "PF04495"], ["Domain_1", 73, 364, "PF04495"]], "ENSNLEP00000042726": [["Domain_1", 75, 214, "PF04495"]], "ENSLAFP00000023713": [["Domain_1", 73, 479, "PF04495"]], "ENSFALP00000002049": [["Domain_0", 24, 79, "PF04495"], ["Domain_1", 73, 380, "PF04495"]], "ENSPEMP00000028808": [["Domain_1", 75, 214, "PF04495"]], "ENSOPRP00000011382": [["Domain_1", 73, 479, "PF04495"]], "ENSACAP00000016239": [["Domain_0", 14, 79, "PF04495"], ["Domain_1", 73, 350, "PF04495"]], "ENSPPAP00000018993": [["Domain_1", 75, 214, "PF04495"]], "ENSCAFP00000018596": [["Domain_1", 73, 214, "PF04495"]], "ENSMPUP00000000808": [["Domain_0", 23, 77, "PF04495"], ["Domain_1", 73, 397, "PF04495"]], "ENSGALP00000060748": [["Domain_1", 78, 214, "PF04495"]], "ENSMODP00000040859": [["Domain_0", 14, 79, "PF04495"], ["Domain_1", 73, 359, "PF04495"]], "ENSOGAP00000014877": [["Domain_0", 14, 79, "PF04495"], ["Domain_1", 73, 390, "PF04495"]], "ENSSARP00000006976": [["Domain_1", 73, 459, "PF04495"]], "ENSOCUP00000010904": [["Domain_0", 14, 78, "PF04495"], ["Domain_1", 73, 397, "PF04495"]], "ENSNGAP00000024273": [["Domain_1", 78, 214, "PF04495"]], "ENSDNOP00000024513": [["Domain_0", 13, 79, "PF04495"], ["Domain_1", 73, 368, "PF04495"]], "ENSFDAP00000013227": [["Domain_1", 75, 214, "PF04495"]], "ENSANAP00000022516": [["Domain_1", 75, 214, "PF04495"]], "ENSPPYP00000014426": [["Domain_0", 14, 79, "PF04495"], ["Domain_1", 73, 240, "PF04495"]], "ENSCJAP00000052572": [["Domain_1", 75, 214, "PF04495"]], "ENSDNOP00000005632": [["Domain_0", 14, 77, "PF04495"], ["Domain_1", 73, 368, "PF04495"]], "ENSCCAP00000003235": [["Domain_1", 74, 214, "PF04495"]], "ENSTTRP00000004294": [["Domain_1", 73, 478, "PF04495"]], "ENSCHOP00000007685": [["Domain_1", 73, 479, "PF04495"]], "ENSECAP00000011471": [["Domain_0", 14, 77, "PF04495"], ["Domain_1", 73, 419, "PF04495"]], "ENSDORP00000009130": [["Domain_1", 75, 214, "PF04495"]], "ENSMICP00000022609": [["Domain_1", 75, 214, "PF04495"]], "ENSP00000234160": [["Domain_1", 75, 214, "PF04495"]], "MGP_CAROLIEiJ_P0052190": [["Domain_1", 75, 214, "PF04495"]], "ENSMAUP00000023190": [["Domain_1", 78, 214, "PF04495"]], "ENSFCAP00000016518": [["Domain_1", 75, 214, "PF04495"]], "ENSOARP00000002367": [["Domain_0", 14, 79, "PF04495"], ["Domain_1", 73, 362, "PF04495"]], "ENSHGLP00000015127": [["Domain_1", 75, 214, "PF04495"]], "ENSSBOP00000025126": [["Domain_1", 74, 214, "PF04495"]], "ENSMLEP00000037976": [["Domain_1", 75, 214, "PF04495"]], "ENSMGAP00000010551": [["Domain_1", 73, 480, "PF04495"]], "ENSODEP00000016102": [["Domain_1", 74, 214, "PF04495"]], "ENSCLAP00000008914": [["Domain_1", 75, 214, "PF04495"]], "ENSAMEP00000009210": [["Domain_1", 73, 478, "PF04495"]], "ENSMFAP00000028161": [["Domain_1", 75, 214, "PF04495"]], "ENSTBEP00000014190": [["Domain_1", 73, 392, "PF04495"]], "ENSCATP00000038238": [["Domain_1", 75, 214, "PF04495"]], "ENSCGRP00000008522": [["Domain_1", 75, 214, "PF04495"]], "ENSPCAP00000003144": [["Domain_1", 73, 479, "PF04495"]], "ENSEEUP00000010421": [["Domain_1", 73, 478, "PF04495"]], "ENSVPAP00000006692": [["Domain_1", 73, 480, "PF04495"]], "MGP_PahariEiJ_P0063186": [["Domain_1", 75, 214, "PF04495"]], "ENSCHIP00000012496": [["Domain_1", 75, 214, "PF04495"]], "ENSSSCP00000016896": [["Domain_1", 75, 214, "PF04495"]], "ENSMNEP00000014333": [["Domain_1", 75, 214, "PF04495"]], "ENSOANP00000013640": [["Domain_1", 73, 359, "PF04495"]], "ENSCAPP00000006921": [["Domain_1", 78, 214, "PF04495"]], "ENSSTOP00000003025": [["Domain_1", 81, 214, "PF04495"]], "ENSMMUP00000038168": [["Domain_1", 74, 214, "PF04495"]], "ENSPVAP00000014903": [["Domain_1", 73, 478, "PF04495"]], "ENSMUSP00000028509": [["Domain_1", 75, 214, "PF04495"]], "ENSRNOP00000072730": [["Domain_1", 75, 214, "PF04495"]], "ENSAPLP00000004299": [["Domain_1", 73, 480, "PF04495"]], "ENSGGOP00000021815": [["Domain_1", 75, 214, "PF04495"]], "ENSPSIP00000002377": [["Domain_0", 24, 79, "PF04495"], ["Domain_1", 73, 345, "PF04495"]], "ENSMEUP00000002600": [["Domain_1", 73, 481, "PF04495"]], "ENSRBIP00000037046": [["Domain_1", 75, 214, "PF04495"]]}