Found:
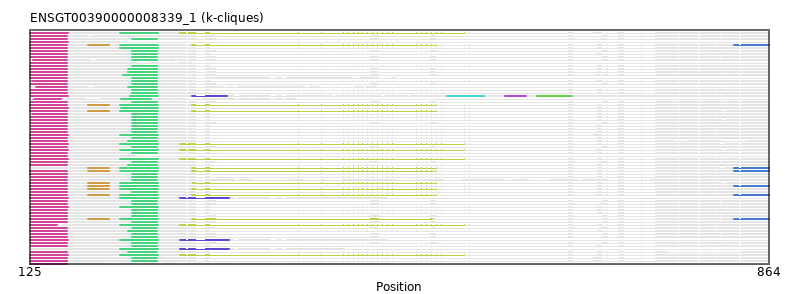
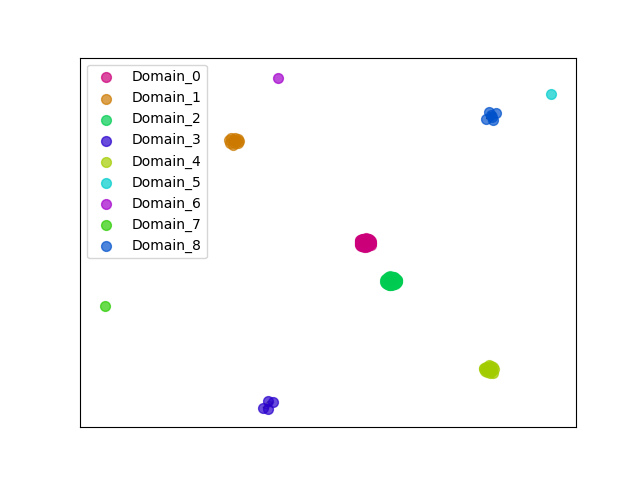
{"ENSHGLP00100022940": [["Domain_0", 126, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSCLAP00000010097": [["Domain_0", 126, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSAPLP00000011983": [["Domain_0", 125, 163, "PF00400"]], "ENSMAUP00000014306": [["Domain_0", 126, 162, "PF00400"], ["Domain_2", 223, 252, "PF00400"]], "ENSMICP00000015872": [["Domain_0", 126, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSCJAP00000059593": [["Domain_0", 131, 162, "PF00400"], ["Domain_2", 223, 252, "PF00400"]], "ENSPCOP00000003523": [["Domain_0", 126, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSJJAP00000005860": [["Domain_0", 126, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSCAPP00000017513": [["Domain_0", 126, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSANAP00000008839": [["Domain_0", 126, 162, "PF00400"]], "ENSTTRP00000002817": [["Domain_0", 125, 163, "PF00400"], ["Domain_2", 215, 253, "PF00400"], ["Linker_2_4", 253, 275], ["Domain_4", 275, 559, "PF00400"]], "ENSMFAP00000040304": [["Domain_0", 126, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSCHIP00000023533": [["Domain_0", 127, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSPVAP00000007100": [["Domain_0", 125, 152, "PF00400"], ["Domain_2", 215, 253, "PF00400"], ["Linker_2_4", 253, 275], ["Domain_4", 275, 559, "PF00400"]], "ENSCCAP00000008565": [["Domain_0", 128, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSOCUP00000021063": [["Domain_0", 125, 162, "PF00400"], ["Linker_0_1", 162, 183], ["Domain_1", 183, 204, "PF00400"], ["Linker_1_2", 204, 215], ["Domain_2", 215, 253, "PF00400"], ["Linker_2_4", 253, 287], ["Domain_4", 287, 531, "PF00400"]], "ENSMGAP00000003938": [["Domain_0", 125, 163, "PF00400"], ["Domain_2", 215, 253, "PF00400"], ["Linker_2_4", 253, 275], ["Domain_4", 275, 559, "PF00400"]], "ENSCGRP00001006954": [["Domain_0", 125, 162, "PF00400"], ["Domain_2", 218, 252, "PF00400"]], "ENSTSYP00000017661": [["Domain_0", 126, 162, "PF00400"], ["Domain_2", 223, 252, "PF00400"]], "ENSCANP00000037406": [["Domain_0", 126, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSOARP00000020114": [["Domain_0", 125, 162, "PF00400"], ["Linker_0_1", 162, 183], ["Domain_1", 183, 204, "PF00400"], ["Linker_1_2", 204, 215], ["Domain_2", 215, 253, "PF00400"], ["Domain_8", 829, 864, "PF00400"]], "ENSGGOP00000012864": [["Domain_0", 126, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSBTAP00000056302": [["Domain_0", 125, 162, "PF00400"], ["Linker_0_1", 162, 183], ["Domain_1", 183, 204, "PF00400"], ["Linker_1_2", 204, 215], ["Domain_2", 215, 253, "PF00400"], ["Linker_2_4", 253, 287], ["Domain_4", 287, 531, "PF00400"], ["Domain_8", 829, 864, "PF00400"]], "ENSCAFP00000022228": [["Domain_0", 125, 162, "PF00400"], ["Domain_2", 215, 253, "PF00400"]], "ENSMLEP00000034516": [["Domain_0", 126, 162, "PF00400"]], "ENSDORP00000000688": [["Domain_0", 126, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSFCAP00000013975": [["Domain_0", 127, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSRNOP00000001315": [["Domain_0", 125, 162, "PF00400"], ["Domain_2", 223, 252, "PF00400"]], "ENSETEP00000008061": [["Domain_0", 126, 163, "PF00400"], ["Domain_2", 216, 253, "PF00400"]], "ENSCCAP00000038967": [["Domain_0", 126, 162, "PF00400"]], "MGP_CAROLIEiJ_P0072120": [["Domain_0", 125, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSOANP00000012307": [["Domain_0", 125, 162, "PF00400"], ["Linker_0_1", 162, 183], ["Domain_1", 183, 204, "PF00400"], ["Linker_1_2", 204, 215], ["Domain_2", 215, 253, "PF00400"], ["Linker_2_4", 253, 288], ["Domain_4", 288, 531, "PF00400"]], "ENSANAP00000011309": [["Domain_0", 126, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSPPYP00000019467": [["Domain_0", 125, 162, "PF00400"], ["Linker_0_1", 162, 183], ["Domain_1", 183, 204, "PF00400"], ["Linker_1_2", 204, 215], ["Domain_2", 215, 253, "PF00400"], ["Linker_2_4", 253, 287], ["Domain_4", 287, 527, "PF00400"], ["Domain_8", 829, 864, "PF00400"]], "ENSECAP00000022584": [["Domain_0", 125, 162, "PF00400"], ["Linker_0_1", 162, 183], ["Domain_1", 183, 204, "PF00400"], ["Linker_1_2", 204, 215], ["Domain_2", 215, 253, "PF00400"], ["Linker_2_4", 253, 287], ["Domain_4", 287, 531, "PF00400"]], "ENSP00000389631": [["Domain_0", 126, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSLAFP00000011750": [["Domain_0", 125, 163, "PF00400"], ["Domain_2", 215, 253, "PF00400"]], "ENSNGAP00000013884": [["Domain_0", 126, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSPEMP00000018609": [["Domain_0", 125, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSSARP00000010568": [["Domain_0", 125, 163, "PF00400"], ["Domain_2", 215, 253, "PF00400"], ["Linker_2_3", 253, 275], ["Domain_3", 275, 324, "PF00400"]], "ENSSBOP00000003880": [["Domain_0", 126, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSFDAP00000018453": [["Domain_0", 126, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSCGRP00001022519": [["Domain_0", 125, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSPCAP00000012455": [["Domain_0", 125, 163, "PF00400"], ["Domain_2", 215, 253, "PF00400"]], "ENSMEUP00000007191": [["Domain_0", 125, 163, "PF00400"], ["Domain_2", 215, 253, "PF00400"], ["Linker_2_4", 253, 275], ["Domain_4", 275, 559, "PF00400"]], "ENSRROP00000006389": [["Domain_0", 126, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSODEP00000016113": [["Domain_0", 126, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "MGP_SPRETEiJ_P0074935": [["Domain_0", 125, 162, "PF00400"]], "ENSNLEP00000013372": [["Domain_0", 126, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSHGLP00000025000": [["Domain_0", 126, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSMODP00000001590": [["Domain_1", 183, 204, "PF00400"], ["Linker_1_2", 204, 215], ["Domain_2", 215, 253, "PF00400"], ["Linker_2_4", 253, 287], ["Domain_4", 287, 531, "PF00400"], ["Domain_8", 829, 864, "PF00400"]], "ENSDNOP00000034468": [["Domain_0", 128, 156, "PF00400"], ["Domain_2", 216, 246, "PF00400"]], "ENSTBEP00000008566": [["Domain_2", 215, 253, "PF00400"], ["Linker_2_3", 253, 275], ["Domain_3", 275, 324, "PF00400"]], "ENSPTRP00000033278": [["Domain_0", 126, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSMOCP00000010270": [["Domain_0", 125, 162, "PF00400"], ["Domain_2", 223, 252, "PF00400"]], "ENSFALP00000014414": [["Domain_0", 125, 162, "PF00400"], ["Linker_0_1", 162, 183], ["Domain_1", 183, 204, "PF00400"], ["Linker_1_2", 204, 215], ["Domain_2", 215, 253, "PF00400"], ["Linker_2_4", 253, 287], ["Domain_4", 287, 531, "PF00400"]], "ENSMPUP00000016419": [["Domain_0", 125, 162, "PF00400"], ["Linker_0_1", 162, 183], ["Domain_1", 183, 204, "PF00400"], ["Linker_1_2", 204, 215], ["Domain_2", 215, 253, "PF00400"], ["Linker_2_4", 253, 287], ["Domain_4", 287, 531, "PF00400"], ["Domain_8", 829, 864, "PF00400"]], "ENSMUSP00000082822": [["Domain_0", 125, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSPPAP00000012543": [["Domain_0", 126, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSMAUP00000003150": [["Domain_0", 125, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSSTOP00000001493": [["Domain_0", 126, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSRBIP00000005156": [["Domain_0", 126, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSCPOP00000020976": [["Domain_0", 126, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSRROP00000030917": [["Domain_0", 126, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSMLUP00000009970": [["Domain_0", 125, 163, "PF00400"], ["Domain_2", 215, 253, "PF00400"], ["Linker_2_4", 253, 275], ["Domain_4", 275, 559, "PF00400"]], "ENSPANP00000015119": [["Domain_0", 126, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSCGRP00000026865": [["Domain_0", 125, 162, "PF00400"], ["Domain_2", 223, 252, "PF00400"]], "ENSMNEP00000018063": [["Domain_0", 126, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSGALP00000050304": [["Domain_0", 126, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "MGP_PahariEiJ_P0060330": [["Domain_0", 125, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSAMEP00000006642": [["Domain_0", 125, 163, "PF00400"], ["Domain_2", 215, 253, "PF00400"], ["Linker_2_4", 253, 275], ["Domain_4", 275, 559, "PF00400"]], "ENSOGAP00000009730": [["Domain_0", 125, 162, "PF00400"], ["Linker_0_1", 162, 183], ["Domain_1", 183, 204, "PF00400"], ["Linker_1_2", 204, 215], ["Domain_2", 215, 253, "PF00400"], ["Linker_2_4", 253, 287], ["Domain_4", 287, 531, "PF00400"], ["Domain_8", 829, 864, "PF00400"]], "ENSCATP00000027862": [["Domain_0", 126, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSDNOP00000004638": [["Domain_0", 126, 163, "PF00400"], ["Domain_2", 225, 253, "PF00400"], ["Linker_2_3", 253, 287], ["Domain_3", 287, 322, "PF00400"], ["Linker_3_5", 322, 542], ["Domain_5", 542, 579, "PF00400"], ["Linker_5_6", 579, 600], ["Domain_6", 600, 621, "PF00400"], ["Linker_6_7", 621, 632], ["Domain_7", 632, 667, "PF00400"]], "ENSCJAP00000029194": [["Domain_0", 126, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]], "ENSCGRP00001006135": [["Domain_0", 125, 162, "PF00400"], ["Domain_2", 223, 252, "PF00400"]], "ENSOPRP00000012399": [["Domain_0", 125, 163, "PF00400"], ["Domain_2", 215, 253, "PF00400"], ["Linker_2_3", 253, 275], ["Domain_3", 275, 324, "PF00400"]], "ENSCGRP00000005693": [["Domain_0", 125, 162, "PF00400"], ["Domain_2", 227, 252, "PF00400"]]}