Found:
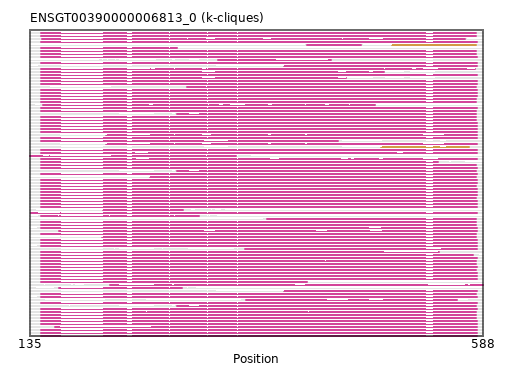
{"ENSMEUP00000005170": [["Domain_0", 282, 581, "PF00285"]], "ENSEEUP00000013109": [["Domain_0", 282, 581, "PF00285"]], "ENSCJAP00000020819": [["Domain_0", 146, 582, "PF00285"]], "ENSCSAP00000001504": [["Domain_0", 146, 581, "PF00285"]], "ENSHGLP00100010700": [["Domain_1", 486, 574, "PF00285"]], "ENSP00000342056": [["Domain_0", 146, 582, "PF00285"]], "ENSOANP00000031114": [["Domain_0", 372, 581, "PF00285"]], "ENSJJAP00000024952": [["Domain_0", 146, 543, "PF00285"]], "ENSNLEP00000026307": [["Domain_0", 208, 288, "PF00285"]], "ENSPPYP00000005303": [["Domain_0", 146, 581, "PF00285"]], "ENSCCAP00000030260": [["Domain_0", 389, 582, "PF00285"]], "ENSHGLP00000006570": [["Domain_0", 211, 582, "PF00285"]], "ENSMUSP00000005826": [["Domain_0", 146, 582, "PF00285"]], "ENSETEP00000009770": [["Domain_0", 146, 581, "PF00285"]], "ENSHGLP00100008907": [["Domain_0", 211, 582, "PF00285"]], "ENSPCOP00000016144": [["Domain_0", 146, 582, "PF00285"]], "ENSJJAP00000018666": [["Domain_0", 135, 341, "PF00285"]], "MGP_CAROLIEiJ_P0025139": [["Domain_0", 146, 582, "PF00285"]], "ENSDORP00000026016": [["Domain_0", 146, 582, "PF00285"]], "ENSHGLP00000009530": [["Domain_0", 146, 582, "PF00285"]], "ENSPVAP00000007162": [["Domain_0", 146, 581, "PF00285"]], "ENSMAUP00000023353": [["Domain_0", 146, 582, "PF00285"]], "ENSMPUP00000001312": [["Domain_0", 146, 581, "PF00285"]], "ENSCLAP00000021672": [["Domain_0", 146, 582, "PF00285"]], "ENSNGAP00000000856": [["Domain_0", 146, 582, "PF00285"]], "ENSCANP00000031835": [["Domain_0", 146, 582, "PF00285"]], "ENSFCAP00000011151": [["Domain_0", 146, 582, "PF00285"]], "ENSCAPP00000018536": [["Domain_0", 323, 436, "PF00285"]], "ENSMMUP00000054879": [["Domain_0", 146, 582, "PF00285"]], "ENSMICP00000026153": [["Domain_0", 146, 582, "PF00285"]], "ENSRNOP00000034921": [["Domain_0", 146, 582, "PF00285"]], "MGP_SPRETEiJ_P0026429": [["Domain_0", 146, 582, "PF00285"]], "ENSNLEP00000030811": [["Domain_0", 136, 582, "PF00285"]], "ENSPCOP00000021736": [["Domain_0", 350, 582, "PF00285"]], "ENSFALP00000000504": [["Domain_0", 146, 581, "PF00285"]], "ENSMFAP00000004989": [["Domain_0", 146, 582, "PF00285"]], "MGP_SPRETEiJ_P0025863": [["Domain_0", 146, 582, "PF00285"]], "ENSPSIP00000003402": [["Domain_0", 146, 581, "PF00285"]], "ENSPANP00000025221": [["Domain_0", 146, 582, "PF00285"]], "ENSSARP00000009135": [["Domain_0", 146, 412, "PF00285"]], "ENSAMEP00000004019": [["Domain_0", 146, 581, "PF00285"]], "ENSCGRP00000019179": [["Domain_0", 146, 582, "PF00285"]], "ENSSTOP00000014291": [["Domain_0", 146, 582, "PF00285"]], "MGP_PahariEiJ_P0088022": [["Domain_0", 146, 582, "PF00285"]], "ENSSBOP00000012270": [["Domain_0", 389, 582, "PF00285"]], "ENSJJAP00000002364": [["Domain_0", 146, 582, "PF00285"]], "ENSCHIP00000033452": [["Domain_0", 146, 582, "PF00285"]], "ENSHGLP00100022722": [["Domain_0", 146, 582, "PF00285"]], "ENSSARP00000012042": [["Domain_0", 215, 588, "PF00285"]], "ENSCGRP00001005320": [["Domain_0", 146, 451, "PF00285"]], "ENSTSYP00000004057": [["Domain_0", 146, 582, "PF00285"]], "MGP_CAROLIEiJ_P0024641": [["Domain_0", 146, 582, "PF00285"]], "ENSDNOP00000033677": [["Domain_0", 148, 480, "PF00285"]], "ENSTTRP00000006754": [["Domain_0", 146, 581, "PF00285"]], "ENSCGRP00000006440": [["Domain_0", 146, 580, "PF00285"]], "ENSODEP00000007138": [["Domain_0", 146, 582, "PF00285"]], "MGP_PahariEiJ_P0087530": [["Domain_0", 160, 582, "PF00285"]], "ENSMOCP00000004247": [["Domain_0", 146, 582, "PF00285"]], "ENSSSCP00000046335": [["Domain_0", 351, 562, "PF00285"]], "ENSSHAP00000000753": [["Domain_0", 146, 581, "PF00285"]], "ENSECAP00000014421": [["Domain_0", 146, 581, "PF00285"]], "ENSMGAP00000000958": [["Domain_0", 255, 581, "PF00285"]], "ENSANAP00000002888": [["Domain_0", 147, 570, "PF00285"]], "ENSRROP00000010880": [["Domain_0", 146, 582, "PF00285"]], "ENSOANP00000008966": [["Domain_0", 146, 304, "PF00285"]], "ENSAPLP00000003725": [["Domain_0", 412, 466, "PF00285"], ["Linker_0_1", 466, 497], ["Domain_1", 497, 581, "PF00285"]], "ENSACAP00000007463": [["Domain_0", 146, 581, "PF00285"]], "ENSDNOP00000008458": [["Domain_0", 146, 581, "PF00285"]], "ENSFDAP00000009942": [["Domain_0", 146, 582, "PF00285"]], "ENSPPAP00000027449": [["Domain_0", 146, 582, "PF00285"]], "ENSCAFP00000000167": [["Domain_0", 146, 581, "PF00285"]], "ENSPEMP00000021454": [["Domain_0", 146, 578, "PF00285"]], "ENSTBEP00000005536": [["Domain_0", 282, 581, "PF00285"]], "ENSVPAP00000006795": [["Domain_0", 146, 581, "PF00285"]], "ENSCGRP00000008857": [["Domain_0", 146, 489, "PF00285"]], "ENSODEP00000007189": [["Domain_0", 164, 582, "PF00285"]], "ENSSBOP00000005225": [["Domain_0", 164, 287, "PF00285"]], "ENSBTAP00000005730": [["Domain_0", 146, 581, "PF00285"]], "ENSPEMP00000011281": [["Domain_0", 146, 544, "PF00285"]], "ENSOCUP00000006238": [["Domain_0", 146, 581, "PF00285"]], "ENSXETP00000020990": [["Domain_0", 146, 581, "PF00285"]], "ENSLAFP00000012446": [["Domain_0", 146, 581, "PF00285"]], "ENSOPRP00000001317": [["Domain_0", 207, 581, "PF00285"]], "ENSCPOP00000013435": [["Domain_0", 146, 582, "PF00285"]], "ENSRBIP00000016819": [["Domain_0", 146, 582, "PF00285"]], "ENSMLEP00000003571": [["Domain_0", 146, 582, "PF00285"]], "ENSJJAP00000024860": [["Domain_0", 146, 582, "PF00285"]], "ENSMUSP00000052373": [["Domain_0", 146, 582, "PF00285"]], "ENSMNEP00000024622": [["Domain_0", 146, 582, "PF00285"]], "ENSGALP00000053546": [["Domain_0", 146, 582, "PF00285"]], "ENSANAP00000037729": [["Domain_0", 146, 582, "PF00285"]], "ENSAPLP00000005268": [["Domain_0", 146, 282, "PF00285"]], "ENSOGAP00000001364": [["Domain_0", 146, 581, "PF00285"]], "ENSCGRP00001010200": [["Domain_0", 146, 582, "PF00285"]], "ENSCHOP00000000352": [["Domain_0", 292, 581, "PF00285"]], "ENSOARP00000010102": [["Domain_0", 146, 581, "PF00285"]], "ENSPTRP00000008664": [["Domain_0", 146, 582, "PF00285"]], "ENSHGLP00000020974": [["Domain_0", 146, 443, "PF00285"]], "ENSMLUP00000006129": [["Domain_0", 146, 581, "PF00285"]], "ENSCATP00000031996": [["Domain_0", 146, 582, "PF00285"]], "ENSSSCP00000038271": [["Domain_0", 146, 582, "PF00285"]], "ENSGGOP00000022736": [["Domain_0", 146, 582, "PF00285"]]}