Found:
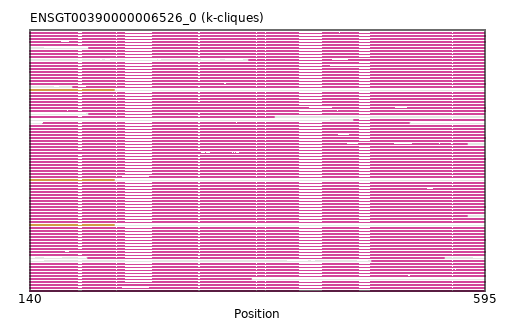
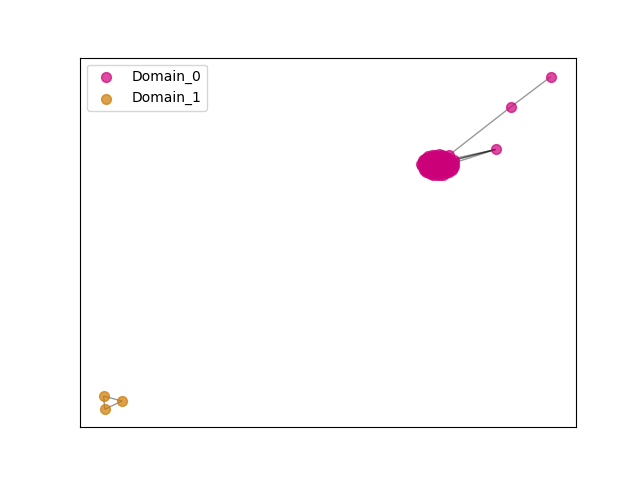
{"ENSCSAP00000002228": [["Domain_0", 140, 594, "PF01566"]], "ENSPANP00000045595": [["Domain_0", 140, 594, "PF01566"]], "ENSMLUP00000012648": [["Domain_0", 140, 595, "PF01566"]], "ENSCLAP00000001676": [["Domain_1", 140, 224, "PF01566"]], "ENSMOCP00000020805": [["Domain_0", 140, 594, "PF01566"]], "ENSTSYP00000033707": [["Domain_0", 140, 594, "PF01566"]], "ENSPCAP00000002647": [["Domain_0", 140, 577, "PF01566"]], "ENSLAFP00000013940": [["Domain_0", 140, 595, "PF01566"]], "ENSACAP00000012437": [["Domain_0", 140, 594, "PF01566"]], "MGP_CAROLIEiJ_P0039831": [["Domain_0", 140, 594, "PF01566"]], "ENSFDAP00000007555": [["Domain_0", 464, 594, "PF01566"]], "ENSBTAP00000052112": [["Domain_0", 140, 594, "PF01566"]], "ENSOARP00000018722": [["Domain_0", 140, 594, "PF01566"]], "ENSBTAP00000019132": [["Domain_0", 141, 594, "PF01566"]], "ENSCAFP00000011707": [["Domain_0", 140, 594, "PF01566"]], "ENSPVAP00000006126": [["Domain_0", 140, 595, "PF01566"]], "ENSNGAP00000002056": [["Domain_1", 140, 224, "PF01566"]], "ENSMICP00000033555": [["Domain_0", 140, 594, "PF01566"]], "ENSMICP00000018100": [["Domain_0", 140, 594, "PF01566"]], "ENSECAP00000018203": [["Domain_0", 140, 594, "PF01566"]], "ENSPCOP00000013109": [["Domain_0", 140, 594, "PF01566"]], "ENSSHAP00000001354": [["Domain_0", 198, 554, "PF01566"]], "ENSOARP00000018737": [["Domain_0", 141, 594, "PF01566"]], "ENSOANP00000004590": [["Domain_0", 140, 594, "PF01566"]], "ENSAPLP00000006969": [["Domain_0", 140, 595, "PF01566"]], "ENSPSIP00000010573": [["Domain_0", 140, 594, "PF01566"]], "ENSCHIP00000010506": [["Domain_0", 141, 594, "PF01566"]], "ENSOPRP00000005236": [["Domain_0", 140, 595, "PF01566"]], "ENSSSCP00000057157": [["Domain_0", 140, 594, "PF01566"]], "ENSSHAP00000004594": [["Domain_0", 482, 594, "PF01566"]], "MGP_PahariEiJ_P0043181": [["Domain_0", 140, 594, "PF01566"]], "ENSCPOP00000028217": [["Domain_0", 140, 594, "PF01566"]], "ENSFCAP00000004067": [["Domain_0", 140, 384, "PF01566"]], "ENSEEUP00000004799": [["Domain_0", 140, 595, "PF01566"]], "ENSSTOP00000011917": [["Domain_0", 140, 594, "PF01566"]], "ENSMPUP00000005310": [["Domain_0", 140, 594, "PF01566"]], "ENSFALP00000013426": [["Domain_0", 199, 594, "PF01566"]], "ENSP00000378364": [["Domain_0", 140, 594, "PF01566"]], "ENSMMUP00000039684": [["Domain_0", 140, 594, "PF01566"]], "ENSCATP00000014581": [["Domain_0", 140, 594, "PF01566"]], "ENSOARP00000018757": [["Domain_0", 141, 594, "PF01566"]], "ENSCGRP00000002597": [["Domain_0", 140, 594, "PF01566"]], "ENSCANP00000014172": [["Domain_0", 140, 594, "PF01566"]], "ENSANAP00000031499": [["Domain_0", 140, 594, "PF01566"]], "ENSRNOP00000026531": [["Domain_0", 140, 594, "PF01566"]], "ENSVPAP00000007085": [["Domain_0", 140, 361, "PF01566"]], "MGP_SPRETEiJ_P0041665": [["Domain_0", 140, 594, "PF01566"]], "ENSPPAP00000031122": [["Domain_0", 140, 594, "PF01566"]], "ENSSBOP00000026234": [["Domain_0", 140, 594, "PF01566"]], "ENSHGLP00100023347": [["Domain_0", 140, 594, "PF01566"]], "ENSCAPP00000009241": [["Domain_0", 359, 594, "PF01566"]], "ENSPCOP00000019091": [["Domain_0", 140, 594, "PF01566"]], "ENSJJAP00000024729": [["Domain_0", 140, 594, "PF01566"]], "ENSMODP00000006316": [["Domain_0", 140, 594, "PF01566"]], "ENSDORP00000007986": [["Domain_0", 140, 594, "PF01566"]], "ENSDNOP00000000548": [["Domain_0", 140, 594, "PF01566"]], "ENSCHIP00000028319": [["Domain_0", 141, 594, "PF01566"]], "ENSMEUP00000014373": [["Domain_0", 140, 577, "PF01566"]], "ENSRROP00000037034": [["Domain_0", 140, 594, "PF01566"]], "ENSCCAP00000006748": [["Domain_0", 140, 594, "PF01566"]], "ENSHGLP00000022299": [["Domain_0", 140, 594, "PF01566"]], "ENSMFAP00000034894": [["Domain_0", 140, 594, "PF01566"]], "ENSSARP00000006841": [["Domain_0", 140, 595, "PF01566"]], "ENSGGOP00000008772": [["Domain_0", 140, 594, "PF01566"]], "ENSOCUP00000006709": [["Domain_0", 140, 594, "PF01566"]], "ENSCJAP00000023426": [["Domain_0", 183, 594, "PF01566"]], "ENSMNEP00000028789": [["Domain_0", 140, 594, "PF01566"]], "ENSMLEP00000002190": [["Domain_0", 140, 594, "PF01566"]], "ENSMUSP00000023774": [["Domain_0", 140, 594, "PF01566"]], "ENSOGAP00000000191": [["Domain_0", 140, 594, "PF01566"]], "ENSETEP00000001196": [["Domain_0", 140, 595, "PF01566"]], "ENSBTAP00000049571": [["Domain_0", 199, 594, "PF01566"]], "ENSPTRP00000071902": [["Domain_0", 140, 594, "PF01566"]], "ENSODEP00000015234": [["Domain_0", 140, 594, "PF01566"]], "ENSAMEP00000002360": [["Domain_0", 140, 595, "PF01566"]], "ENSTTRP00000013108": [["Domain_0", 140, 595, "PF01566"]], "ENSMAUP00000017029": [["Domain_0", 140, 594, "PF01566"]], "ENSCHIP00000031808": [["Domain_0", 140, 594, "PF01566"]], "ENSCHOP00000010205": [["Domain_0", 140, 595, "PF01566"]], "ENSPEMP00000014436": [["Domain_1", 140, 224, "PF01566"]], "ENSGALP00000047087": [["Domain_0", 153, 519, "PF01566"]], "ENSXETP00000021232": [["Domain_0", 140, 595, "PF01566"]], "ENSNLEP00000024992": [["Domain_0", 140, 594, "PF01566"]], "ENSTBEP00000001200": [["Domain_0", 140, 595, "PF01566"]], "ENSRBIP00000027485": [["Domain_0", 140, 594, "PF01566"]], "ENSPPYP00000024385": [["Domain_0", 140, 594, "PF01566"]], "ENSCGRP00001007617": [["Domain_0", 140, 594, "PF01566"]]}