Found:
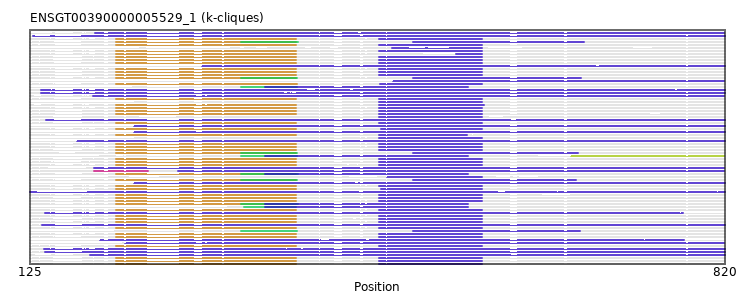
{"ENSMPUP00000016768": [["Domain_2", 336, 392, "PF08616"], ["Domain_3", 360, 563, "PF02141"], ["Linker_3_4", 563, 666], ["Domain_4", 666, 819, "PF07792"]], "ENSPCAP00000005493": [["Domain_3", 125, 820, "PF07792"]], "ENSODEP00000007928": [["Domain_3", 482, 577, "PF08616"]], "ENSANAP00000012687": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 577, "PF08616"]], "ENSCLAP00000007972": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 577, "PF08616"]], "ENSPSIP00000005179": [["Domain_2", 339, 391, "PF08616"], ["Domain_3", 360, 563, "PF02141"]], "ENSMOCP00000007712": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 577, "PF08616"]], "ENSLAFP00000003272": [["Domain_3", 141, 820, "PF07792"]], "ENSMEUP00000012202": [["Domain_3", 229, 820, "PF07792"]], "ENSSBOP00000035619": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 577, "PF08616"]], "ENSSTOP00000007071": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 577, "PF08616"]], "MGP_SPRETEiJ_P0037599": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 577, "PF08616"]], "ENSAMEP00000012461": [["Domain_3", 128, 820, "PF07792"]], "ENSP00000311401": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 577, "PF08616"]], "ENSPPYP00000015401": [["Domain_1", 211, 391, "PF09794"], ["Domain_2", 336, 393, "PF08616"], ["Domain_3", 360, 563, "PF02141"]], "ENSTGUP00000007555": [["Domain_3", 220, 819, "PF07792"]], "ENSBTAP00000024525": [["Domain_1", 211, 391, "PF09794"], ["Domain_2", 336, 393, "PF08616"], ["Linker_2_3", 393, 508], ["Domain_3", 508, 679, "PF08616"]], "ENSCGRP00001023168": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 577, "PF08616"]], "ENSDORP00000008279": [["Domain_1", 211, 392, "PF09794"], ["Linker_1_3", 392, 475], ["Domain_3", 475, 577, "PF08616"]], "ENSCGRP00000023546": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 577, "PF08616"]], "ENSMMUP00000024497": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 577, "PF08616"]], "ENSOGAP00000004762": [["Domain_1", 211, 391, "PF09794"], ["Domain_2", 336, 392, "PF08616"], ["Linker_2_3", 392, 508], ["Domain_3", 508, 671, "PF08616"]], "ENSRROP00000030647": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 577, "PF08616"]], "ENSNLEP00000008870": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 577, "PF08616"]], "ENSTSYP00000005937": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 577, "PF08616"]], "ENSFALP00000008729": [["Domain_3", 188, 819, "PF07792"]], "ENSMGAP00000006540": [["Domain_3", 229, 820, "PF07792"]], "ENSRBIP00000023565": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 577, "PF08616"]], "ENSMICP00000050405": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 562, "PF08616"]], "ENSJJAP00000010681": [["Domain_3", 474, 577, "PF08616"]], "ENSVPAP00000003616": [["Domain_3", 140, 820, "PF07792"]], "ENSCCAP00000038146": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 551], ["Domain_3", 551, 577, "PF08616"]], "ENSNGAP00000003700": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 577, "PF08616"]], "ENSOCUP00000000719": [["Domain_1", 211, 391, "PF09794"], ["Domain_2", 336, 393, "PF08616"], ["Domain_3", 360, 563, "PF02141"]], "ENSSSCP00000022987": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 577, "PF08616"]], "ENSCJAP00000037036": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 577, "PF08616"]], "ENSPPAP00000019159": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 577, "PF08616"]], "MGP_PahariEiJ_P0084007": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 577, "PF08616"]], "ENSCAPP00000004977": [["Domain_3", 487, 577, "PF08616"]], "ENSCAFP00000011488": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 577, "PF08616"]], "ENSGGOP00000028719": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 577, "PF08616"]], "ENSMNEP00000000938": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 577, "PF08616"]], "ENSMFAP00000021858": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 476], ["Domain_3", 476, 577, "PF08616"]], "ENSOPRP00000013974": [["Domain_3", 229, 820, "PF07792"]], "ENSCATP00000018937": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 577, "PF08616"]], "MGP_CAROLIEiJ_P0035901": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 577, "PF08616"]], "ENSPANP00000024040": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 476], ["Domain_3", 476, 577, "PF08616"]], "ENSCPOP00000026862": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 577, "PF08616"]], "ENSEEUP00000006356": [["Domain_3", 136, 820, "PF07792"]], "ENSPTRP00000025998": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 577, "PF08616"]], "ENSHGLP00100010066": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 577, "PF08616"]], "ENSCSAP00000008450": [["Domain_1", 211, 391, "PF09794"], ["Domain_2", 336, 392, "PF08616"], ["Linker_2_3", 392, 508], ["Domain_3", 508, 676, "PF08616"]], "ENSFDAP00000014265": [["Domain_3", 474, 577, "PF08616"]], "ENSSARP00000004343": [["Domain_3", 137, 820, "PF07792"]], "ENSMAUP00000014650": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 577, "PF08616"]], "ENSOANP00000020382": [["Domain_3", 189, 819, "PF07792"]], "ENSPEMP00000028064": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 577, "PF08616"]], "ENSCHIP00000006201": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 577, "PF08616"]], "ENSMLEP00000040344": [["Domain_3", 474, 577, "PF08616"]], "ENSFCAP00000024596": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 577, "PF08616"]], "ENSDNOP00000024549": [["Domain_3", 488, 819, "PF07792"]], "ENSGALP00000009062": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 579, "PF08616"]], "ENSPVAP00000011010": [["Domain_3", 140, 778, "PF07792"]], "ENSTBEP00000006460": [["Domain_3", 195, 779, "PF07792"]], "ENSACAP00000002946": [["Domain_3", 190, 819, "PF07792"]], "ENSCHOP00000008749": [["Domain_3", 297, 820, "PF07792"]], "ENSETEP00000004850": [["Domain_3", 136, 820, "PF07792"]], "ENSECAP00000009940": [["Domain_2", 336, 392, "PF08616"], ["Domain_3", 360, 563, "PF02141"]], "ENSPCOP00000009955": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 577, "PF08616"]], "ENSSHAP00000020613": [["Domain_2", 336, 392, "PF08616"], ["Linker_2_3", 392, 508], ["Domain_3", 508, 675, "PF08616"]], "ENSMUSP00000039361": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 577, "PF08616"]], "ENSXETP00000018950": [["Domain_3", 185, 820, "PF07792"]], "ENSMODP00000000241": [["Domain_1", 211, 391, "PF09794"], ["Domain_2", 336, 392, "PF08616"], ["Linker_2_3", 392, 508], ["Domain_3", 508, 673, "PF08616"]], "ENSOARP00000014381": [["Domain_0", 189, 243, "PF07792"], ["Linker_0_3", 243, 273], ["Domain_3", 273, 819, "PF07792"]], "ENSTTRP00000014577": [["Domain_3", 139, 820, "PF07792"]], "ENSRNOP00000016233": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 577, "PF08616"]], "ENSHGLP00000014075": [["Domain_1", 211, 391, "PF09794"], ["Linker_1_3", 391, 474], ["Domain_3", 474, 577, "PF08616"]], "ENSMLUP00000007321": [["Domain_3", 172, 820, "PF07792"]]}