Found:
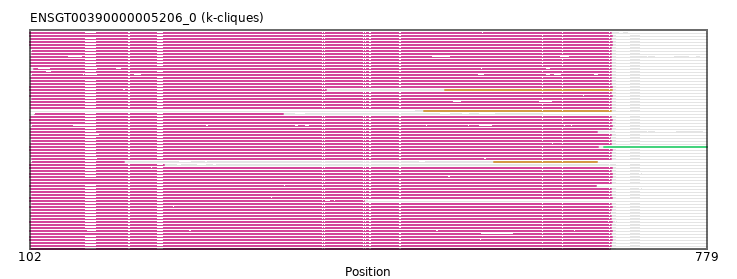
{"ENSFDAP00000018466": [["Domain_0", 103, 683, "PF00995"]], "ENSNLEP00000001339": [["Domain_0", 104, 683, "PF00995"]], "ENSMLEP00000029479": [["Domain_0", 103, 683, "PF00995"]], "ENSXETP00000059507": [["Domain_0", 102, 684, "PF00995"]], "ENSSHAP00000000095": [["Domain_0", 102, 684, "PF00995"]], "ENSOANP00000014958": [["Domain_0", 104, 196, "PF00995"], ["Linker_0_1", 196, 566], ["Domain_1", 566, 669, "PF00995"]], "ENSMPUP00000008165": [["Domain_0", 102, 684, "PF00995"]], "ENSGGOP00000002417": [["Domain_0", 103, 683, "PF00995"]], "ENSMICP00000007508": [["Domain_0", 103, 669, "PF00995"]], "ENSHGLP00100004455": [["Domain_1", 496, 683, "PF00995"]], "ENSMEUP00000007709": [["Domain_0", 104, 684, "PF00995"]], "ENSCHIP00000019682": [["Domain_0", 103, 683, "PF00995"]], "ENSPPAP00000029603": [["Domain_0", 104, 683, "PF00995"]], "ENSACAP00000016889": [["Domain_0", 102, 683, "PF00995"]], "ENSCHIP00000020058": [["Domain_0", 104, 683, "PF00995"]], "ENSCSAP00000004595": [["Domain_0", 102, 684, "PF00995"]], "ENSJJAP00000023722": [["Domain_0", 103, 683, "PF00995"]], "ENSMOCP00000013725": [["Domain_0", 103, 683, "PF00995"]], "MGP_PahariEiJ_P0046500": [["Domain_0", 103, 683, "PF00995"]], "ENSP00000413606": [["Domain_0", 103, 683, "PF00995"]], "ENSCCAP00000001598": [["Domain_0", 103, 683, "PF00995"]], "ENSOGAP00000020435": [["Domain_0", 102, 684, "PF00995"]], "ENSPCOP00000025540": [["Domain_0", 103, 683, "PF00995"]], "ENSCGRP00000018791": [["Domain_0", 103, 683, "PF00995"]], "ENSCHIP00000000028": [["Domain_0", 106, 683, "PF00995"]], "ENSCPOP00000016738": [["Domain_0", 103, 683, "PF00995"]], "ENSODEP00000016741": [["Domain_0", 103, 683, "PF00995"]], "ENSCAFP00000027039": [["Domain_0", 102, 684, "PF00995"]], "ENSRROP00000015178": [["Domain_0", 103, 683, "PF00995"]], "ENSCLAP00000003926": [["Domain_0", 103, 683, "PF00995"]], "MGP_CAROLIEiJ_P0083618": [["Domain_0", 103, 683, "PF00995"]], "ENSOCUP00000008722": [["Domain_0", 102, 684, "PF00995"]], "ENSMODP00000018743": [["Domain_0", 102, 670, "PF00995"], ["Linker_0_2", 670, 676], ["Domain_2", 676, 779, "PF06954"]], "ENSCCAP00000021071": [["Domain_0", 103, 683, "PF00995"]], "ENSHGLP00000008558": [["Domain_0", 103, 683, "PF00995"]], "ENSANAP00000000777": [["Domain_0", 103, 683, "PF00995"]], "ENSMUSP00000125405": [["Domain_0", 103, 683, "PF00995"]], "ENSPVAP00000006550": [["Domain_0", 102, 684, "PF00995"]], "ENSAMEP00000008985": [["Domain_0", 102, 684, "PF00995"]], "ENSSBOP00000022947": [["Domain_0", 104, 683, "PF00995"]], "ENSOARP00000002364": [["Domain_0", 102, 236, "PF00995"]], "ENSCATP00000045089": [["Domain_0", 104, 683, "PF00995"]], "ENSSSCP00000053276": [["Domain_0", 103, 683, "PF00995"]], "ENSOPRP00000013765": [["Domain_0", 102, 684, "PF00995"]], "ENSECAP00000014914": [["Domain_0", 102, 684, "PF00995"]], "ENSRBIP00000015728": [["Domain_0", 104, 683, "PF00995"]], "ENSDORP00000009057": [["Domain_0", 103, 683, "PF00995"]], "ENSTTRP00000003680": [["Domain_0", 102, 684, "PF00995"]], "ENSMMUP00000058147": [["Domain_0", 103, 682, "PF00995"]], "MGP_SPRETEiJ_P0087032": [["Domain_0", 103, 682, "PF00995"]], "ENSTBEP00000002226": [["Domain_0", 102, 684, "PF00995"]], "ENSCJAP00000042954": [["Domain_0", 103, 683, "PF00995"]], "ENSHGLP00100010365": [["Domain_0", 107, 355, "PF00995"]], "ENSFCAP00000001480": [["Domain_0", 104, 683, "PF00995"]], "ENSCGRP00001012094": [["Domain_0", 103, 683, "PF00995"]], "ENSTSYP00000020917": [["Domain_0", 103, 683, "PF00995"]], "ENSCANP00000016553": [["Domain_0", 103, 683, "PF00995"]], "ENSSTOP00000011571": [["Domain_0", 103, 683, "PF00995"]], "ENSMNEP00000022620": [["Domain_0", 104, 683, "PF00995"]], "ENSPPYP00000010630": [["Domain_0", 102, 684, "PF00995"]], "ENSPEMP00000027544": [["Domain_0", 103, 683, "PF00995"]], "ENSPCAP00000010268": [["Domain_0", 102, 668, "PF00995"]], "ENSNGAP00000005370": [["Domain_0", 103, 683, "PF00995"]], "ENSMAUP00000007720": [["Domain_0", 103, 683, "PF00995"]], "ENSCAPP00000018345": [["Domain_0", 104, 683, "PF00995"]], "ENSDNOP00000021830": [["Domain_0", 102, 398, "PF00995"], ["Linker_0_1", 398, 517], ["Domain_1", 517, 684, "PF00995"]], "ENSBTAP00000012094": [["Domain_0", 102, 684, "PF00995"]], "ENSPTRP00000064195": [["Domain_0", 104, 683, "PF00995"]], "ENSLAFP00000020259": [["Domain_0", 102, 684, "PF00995"]], "ENSPANP00000006970": [["Domain_0", 103, 683, "PF00995"]], "ENSPSIP00000007466": [["Domain_0", 102, 436, "PF00995"]], "ENSRNOP00000001322": [["Domain_0", 103, 682, "PF00995"]], "ENSMFAP00000025828": [["Domain_0", 103, 683, "PF00995"]]}